Abstract
Steroid-resistant nephrotic syndrome (SRNS) represents the second most frequent cause of chronic kidney disease in the first three decades of life. It manifests histologically as focal segmental glomerulosclerosis (FSGS) and carries a 33% risk of relapse in a renal transplant. No efficient treatment exists. Identification of single-gene (monogenic) causes of SRNS has moved the glomerular epithelial cell (podocyte) to the center of its pathogenesis. Recently, mutations in >30 recessive or dominant genes were identified as causing monogenic forms of SRNS, thereby revealing the encoded proteins as essential for glomerular function. These findings helped define protein interaction complexes and functional pathways that could be targeted for treatment of SRNS. Very recently, it was discovered that in the surprisingly high fraction of ∼30% of all individuals who manifest with SRNS before 25 years of age, a causative mutation can be detected in one of the ∼30 different SRNS-causing genes. These findings revealed that SRNS and FSGS are not single disease entities but rather are part of a spectrum of distinct diseases with an identifiable genetic etiology. Mutation analysis should be offered to all individuals who manifest with SRNS before the age of 25 years, because (i) it will provide the patient and families with an unequivocal cause-based diagnosis, (ii) it may uncover a form of SRNS that is amenable to treatment (e.g. coenzyme Q10), (iii) it may allow avoidance of a renal biopsy procedure, (iv) it will further unravel the puzzle of pathogenic pathways of SRNS and (v) it will permit personalized treatment options for SRNS, based on genetic causation in way of ‘precision medicine’.
Keywords: clinical genetic testing, molecular genetics, monogenic disease, pathogenesis of nephrotic syndrome, steroid-resistant nephrotic syndrome (SRNS)
CLINICAL FEATURES OF : STEROID-RESISTANT NEPHROTIC SYNDROME
Nephrotic syndrome (NS) is a chronic kidney disease (CKD) that is defined by significant proteinuria (>40 mg/m2/hr) with resulting hypoalbuminemia, which in turn causes edema [1, 2]. The annual incidence of NS in children in the USA is 2–6 per 100 000 children, with a cumulative prevalence of 16 per 100 000 children [3, 4]. NS is classified by response or lack of response to a standardized corticosteroid therapy into ‘steroid-sensitive’ (SSNS) versus ‘steroid-resistant’ nephrotic syndrome (SRNS), respectively. SRNS accounts for ∼15% of childhood cases with NS and 40% of adult-onset cases with NS, and inevitably leads to CKD [3]. SRNS constitutes the second most frequent cause of CKD in children [5]. It carries a 33% risk of relapsing in a renal transplant, thereby causing recurrence of CKD [2]. No curative treatment is available. SRNS manifests histologically as focal segmental glomerulosclerosis (FSGS), a lesion characterized by sclerosis and podocyte foot process effacement in a few capillary segments of a certain fraction of glomeruli [6].
MONOGENIC CAUSES OF SRNS ELUCIDATE : THE PATHOGENESIS
The capillary tuft of the renal glomerular filtering apparatus consists of four major components: the fenestrated endothelial cell layer, the glomerular basement membrane (GBM), the epithelial podocyte layer, and the mesangial cells that help shape the glomerular tuft. In the last 15 years, >39 recessive or dominant genes have been discovered to cause SRNS in humans, if mutated (Table 1). Virtually all of the encoded proteins are localized in podocytes [43]. This discovery shifted the attention in the study of SRNS pathogenesis from mesangial cell dysfunction to podocyte dysfunction [1, 44]. It demonstrated that podocytes are essential for normal glomerular function. This notion is confirmed by rodent models of inducible podocyte depletion, which demonstrate that podocyte damage is sufficient to cause FSGS [45, 46]. The podocyte is a neuron-like cell. It branches off cellular processes to cover the outside of the glomerular capillary, with primary, secondary and tertiary cellular processes, the latter called ‘foot processes’. They interdigitate with foot processes from neighboring podocytes. The interdigitations form between them the glomerular slit membrane, which is critical for the filtering process and retention of protein in the blood stream (Figure 1). The integrity of the glomerular slit membrane is lost in NS. Thus, identification of single-gene causes of SRNS revealed dozens of proteins, each of which is an indispensable component of glomerular function, because loss of their function in a monogenic form of SRNS inescapably leads to proteinuria and FSGS. Thereby, the discovery of genes that if mutated cause monogenic forms of SRNS significantly increased our understanding of glomerular filtration barrier physiology and of pathogenic mechanisms of SRNS.
Table 1.
Recessive and dominant genes that cause monogenic SRNS, if mutated
| Gene | Protein | Accession number | Chromosome | Reference |
|---|---|---|---|---|
| Autosomal recessive | ||||
| ADCK4 | AarF domain containing kinase 4 | NM_024876.3 | 19 | [7] |
| ARHGDIA | Rho GDP dissociation inhibitor (GDI) alpha | NM_001185078.1 | 17 | [8] |
| CD2AP | CD2-associated protein | NM_012120.2 | 6 | [9] |
| CFH | Complement factor H | NM_000186.3 | 1 | [10] |
| COQ2 | Coenzyme Q2 4-hydroxybenzoate polyprenyltransferase | NM_015697.7 | 4 | [11] |
| COQ6 | Coenzyme Q6 mono-oxygenase | NM_182476.2 | 14 | [12] |
| CRB2 | Crumbs homolog 2 | NM_173689.5 | 9 | [13] |
| CUBN | Cubilin (intrinsic factor-cobalamin receptor) | NM_001081.3 | 10 | [14] |
| DGKE | Diacylglycerol kinase, epsilon | NM_003647.2 | 17 | [15] |
| EMP2 | Epithelial membrane protein 2 | NM_001424.4 | 16 | [16] |
| FAT1 | FAT tumor suppressor homolog 1 | NM_005245.3 | 4 | |
| ITGA3 | Integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) | NM_005501.2 | 17 | [17] |
| ITGB4 | Integrin, beta 4 | NM_000213.3 | 17 | [18] |
| KANK1 | KN motif and ankyrin repeat domain containing protein 1 | NM_001256876.1 | 9 | [19] |
| KANK2 | KN motif and ankyrin repeat domain containing protein 2 | NM_015493.6 | 19 | [19] |
| KANK4 | KN motif and ankyrin repeat domain containing protein 4 | NM_181712.4 | 1 | [19] |
| LAMB2 | Laminin, β2 | NM_002292.3 | 3 | [20] |
| MTTL1 | Mitochondrially encoded tRNA leucine 1 | NC_012920.1 | Mito | [21] |
| MYO1E | Homo sapiens myosin IE (MYO1E) | NM_004998.3 | 15 | [22] |
| NPHS1 | Nephrin | NM_004646.3 | 19 | [23] |
| NPHS2 | Podocin | NM_014625.2 | 1 | [24] |
| NUP93 | Nucleoporin 93 kDa | NM_014669.3 | 16 | |
| NUP107 | Nucleoporin 107 kDa | NM_020401.2 | 12 | [25] |
| NUP205 | Nucleoporin 205 kDa | NM_015135.2 | 7 | |
| PDSS2 | Prenyl (decaprenyl) diphosphate synthase, subunit 2 | NM_020381.3 | 6 | [26] |
| PLCE1 | Phospholipase C, epsilon 1 | NM_016341.3 | 10 | [27] |
| PTPRO | Protein tyrosine phosphatase, receptor type, O | NM_030667.2 | 12 | [28] |
| SCARB2 | Scavenger receptor class B, member 2 | NM_005506.3 | 4 | [29] |
| SMARCAL1 | SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily a- like 1 | NM_014140.3 | 2 | [30] |
| WDR73 | WD repeat domain 73 | NM_032856.2 | 15 | [31–33] |
| XPO5 | Exportin 5 | NM_020750.2 | 6 | |
| Autosomal dominant | ||||
| ACTN4 | Actinin, alpha 4 | NM_004924.4 | 19 | [34] |
| ANLN | Anillin, actin binding protein | NM_018685.2 | 7 | [35] |
| ARHGAP24 | Rho GTPase-activating protein 24 | NM_001025616.2 | 4 | [36] |
| INF2 | Inverted formin, FH2 and WH2 domain containing | NM_022489.3 | 14 | [37] |
| LMX1B | LIM homeobox transcription factor 1, beta | NM_00117414.1 | 9 | [38, 39] |
| MYH9 | Myosin heavy chain 9 | NM_002473.4 | 22 | [40] |
| TRPC6 | Transient receptor potential cation channel, subfamily C, member 6 | NM_004621.5 | 11 | [41] |
| WT1 | Wilms tumor 1 | NM_024426.4 | 11 | [42] |
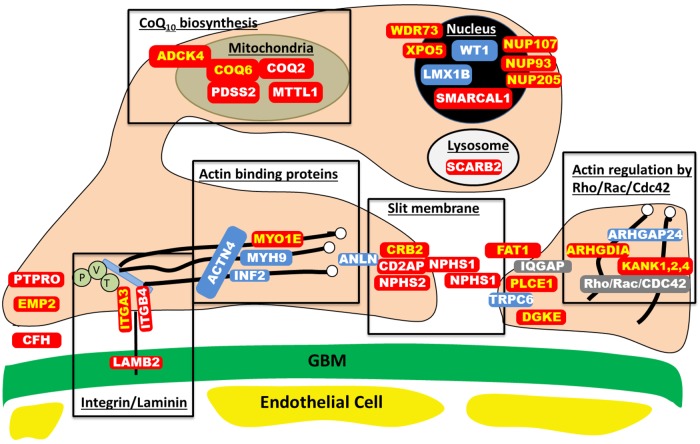
FIGURE 1:
Proteins involved in single-gene causes and pathogenic pathways of SRNS. Identification of single-gene (monogenic) causes of SRNS has revealed the renal glomerular epithelial cell, the podocyte, as the center of action in the pathogenesis of SRNS, because all of the related genes are highly expressed in podocytes. In this way, identification of genes that, if mutated, cause SRNS revealed certain proteins and functional pathways as essential for glomerular function, because a mutation in any single one of them is sufficient to cause SRNS. This figure depicts a simplified cross-section through two neighboring podocyte foot processes, which attach to the GBM via laminin/integrin receptors. Proteins that if mutated cause recessive monogenic forms of SRNS are in red, and proteins that if mutated cause dominant forms of SRNS are in blue. These SRNS-related proteins were found to be part of protein–protein interaction complexes that participate in defined structural components or signaling pathways of podocyte function (black frames). These proteins include: laminin/integrin receptors (focal adhesions), actin-binding proteins, glomerular slit membrane-associated components, actin-regulating small GTPases of the Rho/Rac/Cdc42 family, lyposomal proteins, nuclear transcription factors and proteins involved in coenzyme Q10(CoQ10) biosynthesis. Proteins that are encoded by recessive SRNS genes are marked in red: ADCK4, AarF domain containing kinase 4; ARHGDIA, Rho GDP dissociation inhibitor (GDI) alpha; CD2AP, CD2-associated protein; CFH, Complement factor H; COQ2, coenzyme Q2 4-hydroxybenzoate polyprenyltransferase; COQ6, coenzyme Q6 monooxygenase 6; CRB2, Crumbs family member 2; DGKE, Diacylglycerol kinase, epsilon EMP2, epithelial membrane protein 2; GBM, glomerular basement membrane; ITGA3, integrin, alpha 3; ITGB4, integrin, beta 4; KANK, KN motif And Ankyrin Repeat Domains 1/2/4; LAMB2, laminin, β2; MTTL1, mitochondrial tRNA leucine 1; MYO1E, Homo sapiens myosin 1e; NPHS1, nephrin; NPHS2, podocin; NUP93, Nucleoporin 93 kDa; NUP107, Nucleoporin 107 kDa; NUP205, Nucleoporin 205 kDA; PDSS2, prenyl (decaprenyl) diphosphate synthase, subunit 2; PLCE1, phospholipase C, epsilon 1; PTPRO, protein tyrosine phosphatase, receptor type, O; SCARB2, scavenger receptor class B, member 2; SMARCAL1, SWI/SNF related, matrix associated, actin-dependent regulator of chromatin, subfamily a-like 1; WDR73, WD repeat domain 73; XPO5, Exportin 5. Proteins that are encoded by dominant SRNS genes are marked in blue: ACTN4, actinin, alpha 4; ANLN, anillin; ARHGAP24, Rho GTPase-activating protein 24; INF2, inverted formin, FH2 and WH2 domain containing; LMX1B, LIM homeobox transcription factor 1-beta; MYH9, Myosin, heavy chain 9; TRPC6, transient receptor potential cation channel, subfamily C, member 6; WT1, Wilms tumor 1. IQGAP, IQ motif containing GTPase activating protein 1; P, Paxillin; V, Vinculin and T, Talin.
This discovery started with the genes encoding the slit membrane proteins nephrin (NPHS1) [23] and podocin (NPHS2) [24]. In the meantime, acceleration of next-generation sequencing has permitted identification of >27 monogenic causes of SRNS, and this number is rapidly increasing (see references in Table 1) (Figure 1). Fascinatingly, the encoded proteins begin to map back onto distinct structural protein complexes and signaling pathways that reveal what is essential for glomerular function (Figure 1). These functional complexes include besides glomerular slit membrane components, laminin/integrin signaling components, actin-binding proteins, actin-regulating small GTPases, lysosomal proteins, transcription factors and proteins of coenzyme Q10 biosynthesis.
A SURPRISINGLY HIGH FRACTION OF : EARLY-ONSET SRNS IS CAUSED BY MONOGENIC MUTATIONS
We have previously shown that 85% of SRNS cases with onset by 3 months of age and 66% of cases with onset by 1 year of age can be explained by recessive mutations in one of four genes only (NPHS1, NPHS2, LAMB2 or WT1) [47]. However, a significant proportion of cases with later-onset SRNS were still molecularly unsolved [47].
When examining 48 individuals with early-onset SRNS for monogenic mutations in 16 recessive and 5 dominant SRNS genes, we detected causative mutations in 16 of the 21 genes in 33% of the individuals [48]. More recently, we examined a large worldwide cohort of 1783 individuals from different families with SRNS manifesting before 25 years of age. We performed exon sequencing in all 27 genes known at the time to cause monogenic SRNS, if mutated [49]. We employed a strategy of high-throughput/low-cost multiplex PCR that we developed [50, 51]. It permits rapid sequencing of all ∼600 exons of the 27 genes, barcoding PCR products by patient using the Fluidigm™ platform with consecutive next-generation sequencing. Twenty-one of the genes were recessive and six were dominant. We detected a single-gene cause of SRNS in the surprisingly high fraction of 29.5% (526 of 1783) of the individuals from different families with SRNS worldwide, who manifested before 25 years of age (Table 2) [49]. We applied stringent criteria for calling mutations ‘disease causing’ (see Boxes 1 and 2). Causative mutations were found in 21 of the 27 genes examined (Table 2). Mutations were most frequently found in NPHS2, NPHS1 and WT1 (Table 2). This surprisingly high fraction of individuals with SRNS (manifesting before 25 years), in whom a causative mutation may be detected, was recently confirmed by three other groups [52–54].
Table 2.
International cohort of 526 of the 1783 families, in whom a single-gene cause of SRNS was detected in 1 of 21 monogenic SRNS genes [49]
| Gene causing SRNS | Mode of inheritance | Total SRNS families with molecular diagnosis |
|---|---|---|
| NPHS2 | AR | 177 (9.93) |
| NPHS1 | AR | 131 (7.34) |
| WT1 | AD | 85 (4.77) |
| PLCE1 | AR | 37 (2.17) |
| LAMB2 | AR | 20 (1.12) |
| SMARCAL1 | AR | 16 (0.89) |
| INF2 | AD | 9 (0.5) |
| TRPC6 | AD | 9 (0.53) |
| COQ6 | AR | 8 (0.45) |
| ITGA3 | AR | 5 (0.28) |
| MYO1E | AR | 5 (0.28) |
| CUBN | AR | 5 (0.28) |
| COQ2 | AR | 4 (0.22) |
| LMX1B | AD | 4 (0.22) |
| ADCK4 | AR | 3 (0.17) |
| DGKE | AR | 2 (0.11) |
| PDSS2 | AR | 2 (0.11) |
| ARHGAP24 | AD | 1 (0.06) |
| ARHGDIA | AR | 1 (0.06) |
| CFH | AR | 1 (0.06) |
| ITGB4 | AR | 1 (0.06) |
| Total | 526 (29.5) |
AR, autosomal recessive; AD, autosomal dominant.
Box 1. Assignment of autosomal recessive mutations as being disease causing.
- Include allele as disease causing if:
- – Truncating mutation (stop, abrogation of start or stop, obligatory splice, frame shift) in an expressed gene (well-annotated mRNA, sequence conservation, protein expression) or
- – Missense mutation if:
- ° Continuously conserved at least up to Danio rerio (zebrafish) and
- ° Loss of function in human allele is supported by functional data.
- Exclude allele as disease causing if:
- – Heterozygous allele frequency >1% (in EVS server: 13 000 control chromosomes) or single homozygous reported
- – Non-segregation (e.g. ‘compound heterozygous’ in cis; affected family member is without the variant; unaffected parent is with homozygous variant).
Baseline assumptions: (i) Full penetrance (age related). (ii) Defined clinical phenotype. (iii) ‘Mutation’ implies that an allele changes the phenotype. (iv) Known genes with similar phenotype have been excluded.
Standard criteria for how genetic variants are filtered to exclude benign changes or single-nucleotide polymorphisms when applying mutation analysis to monogenic disease genes. Alleles included for recessive gene analysis are then confirmed via Sanger sequencing and by segregation via parental or other affected family member DNA.
Box 2. Assignment of autosomal dominant mutations as being disease causing.
- Include allele as disease causing if:
- – Truncating mutation (stop, abrogation of start or stop, obligatory splice, frame shift) in an expressed gene (well-annotated mRNA, sequence conservation, protein expression) and
- ° Continuously conserved to at least up to Danio rerio (zebrafish) or
- – Missense mutation if:
- ° Continuously conserved to Danio rerio, and
- ° Human allele is supported by functional data, and
- ° Full segregation exists (≥11 affected), and
- ° Known genes with similar phenotype have been excluded.
- Exclude allele as disease causing if:
- – Heterozygous allele frequency >0.1%
- – Non-segregation—i.e. affected family member is without the allele
- – Caveat regarding non-segregation: if an unaffected family member is with the allele, consider incomplete penetrance and variable expressivity.
Standard criteria for how genetic variants are filtered to exclude benign changes or single-nucleotide polymorphisms when applying mutation analysis to monogenic disease genes. Alleles included for dominant gene analysis are then confirmed via Sanger sequencing and by segregation via parental or other affected family member DNA.
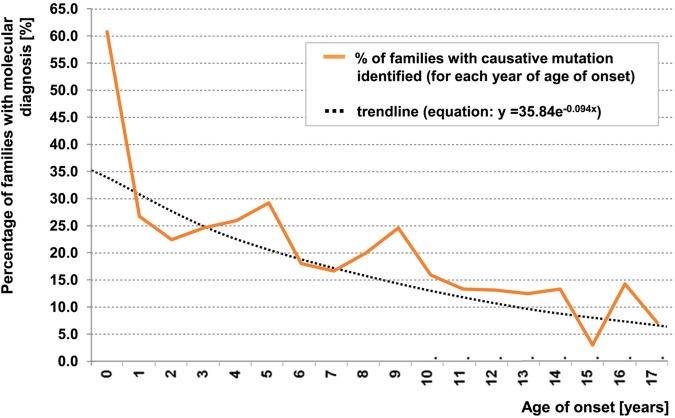
We found that the fraction of individuals in whom a single-gene cause was identified inversely correlated with age at manifestation (Figure 2). This fraction was related to age of onset as follows: onset in the first 3 months of life (69.4%), from 4 to 12 months (49.7%), from 1 to 6 years (25.3%), from 7 to 12 years (17.8%) and from 13 to 18 years (10.8%). For the PLCE1 gene, specific mutations correlated with age of onset [49]. The frequencies in which mutations of SRNS genes were found were also dependent on the fraction of consanguineous marriages in the cohorts examined, most likely due to the increased risk for recessive disease in offspring of consanguineous unions. Specifically, in non-consanguineous families, the detection rate was ∼25% of cases, whereas it was ∼50% in consanguineous families (Figure 3) [49]. We also detected founder mutations that had a higher occurrence rate in certain regions of the world [49]. For instance, the R138Q mutation of NPHS2 occurred frequently in Western Europe and the USA. These findings will be important for the establishment of genotype–phenotype correlations in clinical settings at distinct locations around the world. There was no gender difference for the likelihood of detecting the disease-causing mutation.
FIGURE 2:
Age of onset distribution (in years) for 1589 of 1783 examined families with SRNS [49]. A total of 1589 individuals from different families manifested with SRNS before 18 years of age. Graph indicates percentage of solved families per year of age of onset. Black dotted line represents a binomial fit of age-related percent of families with causative mutation (in families with more than one affected family member, the mean age of onset from all affected individuals was used).
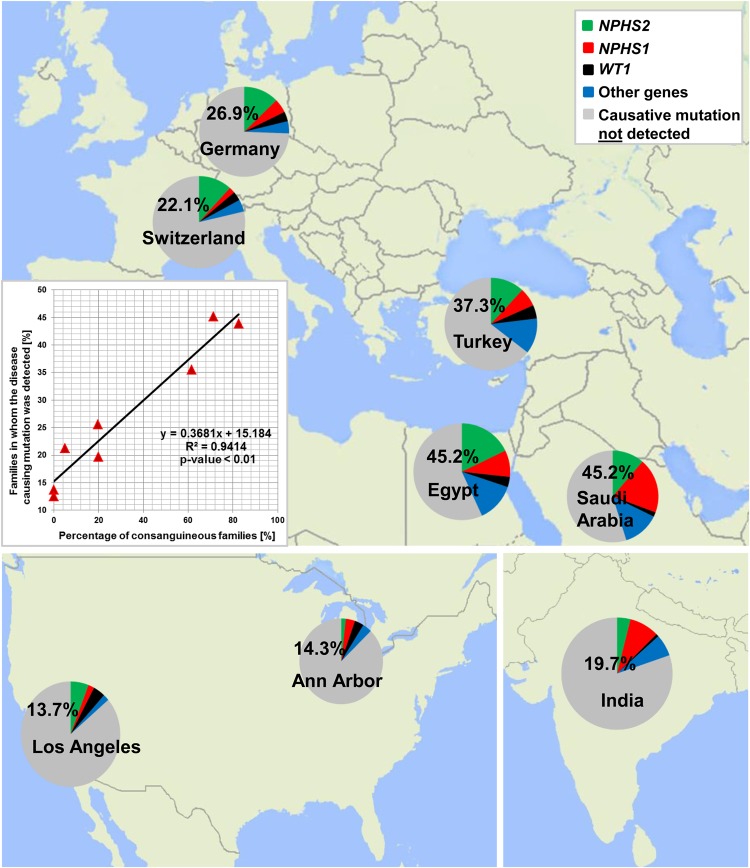
FIGURE 3:
Percentage of genetic findings in SRNS families from the eight largest contributing centers. We obtained samples from 1783 SRNS families worldwide and detected the disease-causing mutation in 506 families (28.4%). For eight centers, we detected the disease-causing mutations in the following fractions (families in whom we detected the causative mutation/total families examined from this center): Saudi-Arabia (45.2%, 28/62), Egypt (43.8%, 64/146), Turkey (35.5%, 60/169), Germany (25.6%, 117/457), Switzerland (21.3%, 20/94), India (19.7%, 25/127), Ann Arbor (12.5%, 7/56) and Los Angeles (13.7%, 7/51). Inset: the detection rate of the disease-causing mutations strongly correlates with the rate of consanguinity between the different centers (R2 = 0.9414) [49].
GENOTYPE–PHENOTYPE CORRELATIONS : IN MONOGENIC SRNS
Steroid-sensitive nephrotic syndrome
It was shown early on that individuals with monogenic mutations will hardly ever exhibit steroid sensitivity of their NS [55, 56]. Very recently, however, we discovered mutations in the EMP2 gene as a rare cause of SSNS [16, 57].
Recessive versus dominant disease
One of the most important genotype–phenotype correlations in SRNS is the distinction between recessive versus dominant SRNS genes (Table 1). In recessive mutations, family history is most likely negative, because parents of individuals with recessive mutations will be healthy heterozygous carriers, and no one in the ancestry will have had disease (because, if there is any inherited mutation, it will be heterozygous only). In contrast, in dominant disease, one of the parents of an affected individual will most likely be affected, and the disease may have been handed down through multiple generations (except for situations of de novo mutations, or incomplete penetrance, which can occur in autosomal dominant genes). Thereby, the detection of dominant mutations has important clinical implications, e.g. in situations of a planned living-related donor kidney transplantation. Here, it will be important to exclude the presence of the disease-causing mutation in the related donor, in whom SRNS may not yet have manifested.
Gene-specific phenotypes
Specific SRNS genes or specific mutations (alleles) in the same SRNS gene may cause characteristic phenotypes. This may pertain to age of onset of disease [49, 58]. For instance, mutations in the recessive genes NPHS1, LAMB2 or PLCE1 lead to onset of SRNS in early childhood, whereas mutations in other recessive genes, such as NPHS2, lead to onset in later childhood. As a rule, mutations in dominant SRNS genes (ACTN4, TRPC6, INF2, ANLN and ARHGAP24) cause adult-onset SRNS, with the exception of WT1 [49].
Allele-specific phenotypes
Within the same gene, specific mutations may determine a range for age of onset of SRNS that is dependent on the specific mutation. The phenomenon is known as ‘multiple allelism’. For instance, in NPHS2 mutations, the mutation R138Q causes onset in early childhood [56, 59], whereas the mutation R229Q in compound heterozygosity with specific second mutations causes adult-onset SRNS [60]. Whereas currently only ‘strong’ mutations are called disease causing (Boxes 1 and 2), it is very likely that a high percentage of adult-onset SRNS is caused by ‘weak’ recessive alleles (such as R229Q of NPHS2), which have not yet been revealed as deleterious. One of the most important tasks in the future of renal genetics is to define deleteriousness of ‘weak’ recessive mutations that are present in the population using cell-based and animal model systems.
Allele-specific clinical and syndromic features
Specific SRNS genes, if mutated, may cause distinct clinical phenotypes in a gene-specific and/or allele-specific way [58]. This is especially apparent for WT1, where mutations in the KTS domain cause Frasier syndrome, whereas missense mutations can cause Denys–Drash syndrome or isolated NS [61]. LAMB2 mutations are usually associated with neuronal or retinal involvement [20, 62]. INF2 mutations can lead either to isolated NS or can be present in individuals with Charcot–Marie–Tooth disease [63]. Individuals with LMX1B mutations usually present with Nail-Patella syndrome. However, specific LMX1B mutations have been found in individuals with isolated SRNS [64]. It is important that individuals with SRNS and certain clinical phenotypes obtain early clinical genetic testing, because this may have important consequences for clinical management, as in WT1 where there is a risk for developing gonadoblastoma [61].
Correlation between mutated gene and renal biopsy pattern
For specific genes, and for specific mutations within the same mutated gene, there can be a correlation between genotype and renal histologic pattern. Individuals with congenital onset NS (i.e. onset within the first 90 days of life) and with the renal histology of diffuse mesangial sclerosis (DMS) usually have mutation in one out of the four following genes: LAMB2, WT1, NPHS1 or PLCE1. In contrast, we did not detect any specific genotypes for the histologic pattern of FSGS in SRNS [49].
An allelic spectrum for DMS versus FSGS
Data from human genetics and from mouse models of SRNS/FSGS show that the renal histologic patterns of DMS and FSGS lie at different ends of a spectrum of a shared pathogenesis. This means that ‘severe’ recessive mutations (protein truncating mutations) of NPHS2 cause a fetal-onset renal ‘developmental’ phenotype of immature glomeruli (i.e. DMS), whereas ‘mild’ mutations (missense mutations) in the same gene cause the renal ‘degenerative’ phenotype of FSGS [58].
CLINICAL CONSEQUENCES FROM : MUTATION ANALYSIS
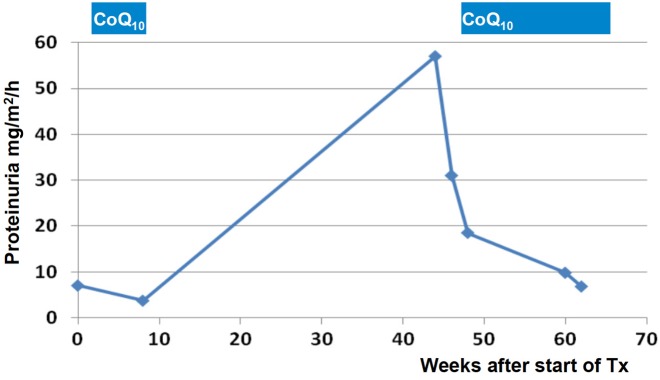
Besides the genotype–phenotype correlations discussed above, single-gene mutations in SRNS genes may have therapeutic consequences in some cases. For instance, most individuals with a single-gene cause of SRNS will not respond to steroid treatment [55, 56]. Furthermore, identification of the causative mutation may reveal that a potential therapy is available for some rare single-gene causes of SRNS. For example, if a mutation in a gene encoding enzymes of the coenzyme Q10 biosynthesis is detected (COQ2, COQ6, ADCK4 or PDSS2), experimental treatment with coenzyme Q10 may be warranted [12, 65], because a partial response to treatment with coenzyme Q10 has been described in individuals with SRNS and mutations in COQ2 [65], COQ6 [12] and ADCK4 [7] (Figure 4). Likewise, a patient with recessive mutations in PLCE1 responded fully to treatment with steroids or cyclosporine A [27]. Finally, individuals with mutations of CUBN may be amenable to treatment with vitamin B12, and individuals with ARHGDIA may theoretically be responsive to the eplerenone treatment [66]. The detection of WT1 mutations often has clinical consequences as, for instance, KTS+ mutations depending on karyotype may confer a risk for gonodoblastoma [61]. TRPC6 mutations may potentially be amenable to treatment with calcineurin inhibitors [67].
FIGURE 4:
Response of SRNS to oral coenzyme Q10(CoQ10) in monogenic SRNS due to a mutation in an enzyme of the coenzyme Q10 biosynthesis pathway. In a 5-year-old girl with SRNS and a causative homozygous mutation in the COQ6 gene, treatment with coenzyme Q10 was commenced during remission. Following inadvertent interruption of coenzyme Q10 administration, proteinuria rose into the nephrotic range. Following reinstitution of therapy, proteinuria normalized [12].
In summary, the benefit of identifying single-gene causes of SRNS lies in the fact that it provides an unequivocal molecular genetic diagnosis for the patients and families involved; it helps define important genotype–phenotype correlations. Furthermore, discovery of specific mutations can reveal rare monogenic causes of SRNS that may be amenable to treatment such as mutations in the coenzyme Q10 biosynthesis pathway.
WHEN TO INITIATE MUTATION ANALYSIS?
Because of the high likelihood of finding a causative monogenic mutation in SRNS with onset before 25 years of age, and because of the many important implications for disease management, it is advisable to suggest clinical genetic testing to all individuals with FSGS or with persistent proteinuria that manifests before age 25 years. For practicability and cost containment, the entire panel of ∼39 monogenic genes that are known to cause SRNS if mutated can now be examined (see below) [51]. Because the likelihood of detecting a causative mutation in SRNS is inversely related to age of onset (Figure 2) [49, 59], initiation of genetic testing should be considered especially in childhood onset SRNS. However, even in young adults, the likelihood of detecting a monogenic cause of SRNS is still substantial, being >10% [49]. Mutations in recessive disease genes are found more frequently in early-onset disease, whereas mutations in dominant genes more frequently cause adult-onset disease [49]. In addition, the likelihood of finding a causative (recessive) mutation is very high in individuals with SRNS from consanguineous marriages (Figure 3).
HOW TO INITIATE MUTATION ANALYSIS?
Clinical testing versus research-based testing
‘Clinical genetic testing’ is the term for mutation analysis directed at finding the causative mutation(s) in a monogenic disease. It is usually performed by a certified clinical genetic testing laboratory (e.g. Clinical Laboratory Improvement Amendments, CLIA certification). Due to the rarity of some disease-causing genes, and due to the dynamic process by which thousands of novel monogenic disease genes are still discovered daily by research laboratories around the world, many diseases still require mutation analysis that is run by a research lab (‘research-based testing’). The research lab makes a decision on whether the genetic variant identified explains the disease phenotype, i.e. whether the variant can be viewed as ‘disease causing’. However, no clinical decision-making can be based on research-based findings. Therefore, the findings from research-based testing, if related ‘informed consent’ from the patient is in place, can then be utilized by the patient with the help of their physician to obtain a CLIA certified report through ‘specific mutation confirmation’ for the mutation. In this process, the physician will refer the patient to genetic counseling, during which the patient may confirm that he/she requests clinical genetic testing. A certified clinical genetic testing lab can then contact the research lab on behalf of the patient with the question, which specific gene and exon to examine for independent ‘specific mutation confirmation (SMC)’. The certified clinical testing lab can then generate a mutation analysis report for the patient. This report can be a basis for clinical decision-making.
METHODS OF CLINICAL GENETIC TESTING : IN SRNS
Recent advances in high-throughput sequencing
Before considering screening for a causative mutation in NS, the clinician may want to be aware of the different methods of sequencing patient DNA. Recent advances in high-throughput sequencing, and the continuous reduction in cost for whole-exome sequencing (WES), have made mutation analysis less time and cost intensive. Sanger sequencing for detection of single-gene cause of SRNS has been replaced by indication-driven gene panel analysis, i.e. instead of testing for one gene at a time, >30 genes can be tested simultaneously by using high-throughput PCR amplification and sequencing approaches [49–51, 68].
Multiple commercial laboratories offer clinical genetic testing in SRNS. An internet search for ‘clinical genetic testing for nephrotic syndrome’ will reveal some of these sources. Cost ranges widely, e.g. from $600 to $3500 for examination of 26–30 SRNS genes. Cost may also vary depending on the institution from which the sample is sent, as individual rates can sometimes be negotiated. As more genes are being discovered mutation analysis will be performed using whole exome sequencing, therefore the cost for mutation analysis will further decrease.
Gene panels
Gene panel analysis is currently considered the most cost-effective approach to indication-driven mutation analysis [48–51, 68]. In this approach, all protein-coding exons of a panel of ∼30 genes that are assumed to bear causative mutations in patients with a certain phenotype (e.g. proteinuria or SRNS) are sequenced. The analysis is usually based on classic Sanger sequencing within a multiplex-PCR format (i.e. amplifying multiple exons in the same PCR). For this purpose, microfluidic systems can be employed that allow thousands of reactions to occur simultaneously in nanoliter volumes [48–51, 68]. Once amplified, the samples are individually given a unique barcode for identification and then pooled for sequencing using a next-generation sequencer. This technique allows targeted sequencing at ∼5% of what a traditional Sanger sequence would cost [51]. However, the number of genes that can be examined using this technique is limited to ∼30 genes.
Whole exome sequencing
In contrast to gene panel sequencing, WES allows sequencing of all ∼330 000 exons in the human genome (i.e. the ‘exome’). Exon-containing fragments of DNA are first enriched from the patient's DNA sample, using solid-phase/array-based hybridization of the patient's DNA fragments with bait probes that represent the sequences of all exons to capture the entire exome [69]. It is currently assumed that WES offers a theoretical likelihood of 86% of detecting the disease-causing mutation in a recessive disease [70, 71]. Besides its use to detect mutations in an established list of known disease-causing genes, WES was also very successfully applied to detecting novel disease-causing genes (ADCK4, KANK2, EMP2, CRB2 and CEP164).
Following exome capture, all exon fragments are sequenced using a high-throughput next-generation sequencing platform [72]. The sequences are then compared with the human reference genome (www.genome.ucsc.edu) for genetic sequence variants that differ between the patient and a normal reference sequence. Any given two individuals differ by ∼2000 genetic variants [51, 73]. However, in a monogenic disease, only one or two variants in a single gene represent the causative mutation. Finding the relevant causative genetic mutation requires an elaborate a priori reduction process, either by genetic mapping [74], by application of stringent genetic criteria (see Boxes 1 and 2) or by applying algorithms on ‘deleteriousness’ of genetic variants (see below ‘Calling genetic variants mutations’).
Whole genome sequencing
Whole genome sequencing (WGS) uses massively parallel DNA enrichment technology to sequence the entire genome of an individual, including the 99% of the genome that is non-coding and highly variable. WGS is the most inclusive method of sequencing a DNA sample of an individual for genetic variants, but it is still the most costly method. Currently, there is no significant advantage of its use over WES in monogenic diseases, as all mutations that lie outside exons or exon splice sites are difficult to implicate as causative for disease, due to their very indirect role for protein function. This situation may change within the next 10–15 years, as more informed algorithms for allele calling may evolve, together with deeper sources of WGS sequence data from worldwide human populations.
Sanger sequencing
The classic chain-termination method of DNA sequencing (‘Sanger sequencing’) still plays a very important role for confirmation of genetic variants that result from high-throughput sequencing techniques such as WES. It also plays a role in SMC following research-based genetic testing (see above).
CALLING GENETIC VARIANTS MUTATIONS
The term ‘genetic variant’ is used for any difference in the DNA sequence between two individuals, e.g. a patient and a ‘normal reference individual’. In contrast, the term ‘mutation’ is used only if there is a strong level of certainty that a genetic variant alters the phenotype of an individual, i.e. causes disease. In a potentially monogenic disease such as SRNS, very stringent genetic and biological criteria should be followed before a mutation is called disease causing. The criteria that we follow for mutation calling in recessive and dominant renal diseases are summarized in Boxes 1 and 2.
To ensure identification of disease-causing variants, there is a process of quality control that minimizes the technical errors in sequencing and gene annotation that have been introduced throughout the process of DNA sequencing. In addition, there is an elaborate process for reduction of variants from the many genetic variants that result when comparing exonic sequences from a patient with a normal reference sequence. Clinical exome sequencing studies have been published that feature a tiered pipeline to sort allelic variants into different categories of disease based on an a priori gene list generated from the clinical phenotype [75]. Variants are then assigned different designations to reflect likelihood of pathogenicity ranging from ‘probable’ to ‘unknown significance’. Between clinical genetic diagnostic programs and research laboratories, there is little difference in the actual variant reduction process, once the variant list is generated [51, 69, 70, 73, 75–77].
Genetic criteria that we use for reduction of genetic variants and identification of the unique disease-causing mutation(s) are listed separately for recessive versus dominant genes in Boxes 1 and 2. In general, the decision that a genetic variant can be called a disease-causing mutation is made if a genetic variant truncates the encoded protein or represents an obligatory splice site altering variant. In missense mutations (non-synonymous variants), the degree of evolutionary conservation of the related amino acid residue is evaluated. The frequency of the variant is also compared with population databases of healthy control individuals (‘1000 genomes’, www.1000genomes.org; ‘Exome Variant Server’, evs.gs.washington.edu; and the ExAC Browser, exac.broadinsitute.org). Variants in dominant disease genes should be absent from these databases even in the heterozygous state, except when incomplete penetrance is expected for a given gene. In recessive disease genes, heterozygous variants may be present in healthy control populations. A minor allele frequency of <1% is usually expected when judging a recessive variant as potentially disease causing. It is also reviewed if a certain variant has been published as disease causing before (e.g. HGMD Database, www.hgdm.cf.ac.uk). Finally, it is judged if loss of function for a variant has been tested in a cell-based or animal model system, which usually requires a sizable investment of work by a research laboratory. Electronic prediction programs that integrate the above criteria are also consulted (e.g. PolyPhen-2 database; genetics.bwh.harvard.edu/pph2).
Variants called as potentially disease causing will then have to be confirmed by Sanger sequencing. Segregation analysis in parental DNA samples will show that the two compound heterozygous mutations in a recessive gene come from one parent each and are not inherited from the same parent. If the latter was the case, this, which would exclude at least one of the variants as not disease causing if both parents are healthy, is usually the case in a recessive disease.
HOW TO INTERPRET RESULTS FROM : MUTATION ANALYSIS
Variants of unknown significance
It is very important to understand that a finding of ‘variants of unknown significance (VUS)’ resulting from clinical genetic testing has absolutely no heuristic bearing on the diagnostic test and should be considered inconclusive. Often these findings are misinterpreted in the way that there is ‘a little bit of an abnormal finding’, where the correct interpretation would be that those VUS are meaningless biologically and clinically, because the hypothesis that these variants could explain disease had to be rejected according to a priori interpretation criteria.
Recessive versus dominant
Similarly, when interpreting the meaning of results from clinical genetic testing, it is very important to have a clear understanding of the action of recessive versus dominant disease genes. In dominant diseases, a heterozygous mutation alone is sufficient to cause disease (with the exception that there can be incomplete disease penetrance in a relative or ancestor of the patient). In contrast, in a recessive disease gene, both gene copies need to be mutated, either in a homozygous way (i.e. the same mutation is inherited from father and mother) or in a compound heterozygous way (i.e. two different mutations in the same gene are inherited from each parent).
Challenges to clinical genetic testing
Because gene panel sequencing and WES generate a problem of ‘multiple testing’, it is very important to assure, when interpreting clinical genetic testing results, that the interpreting lab follows strict genetic interpretation criteria (see Boxes 1 and 2) in order to avoid false-positive results, which may lead to unwarranted actions in the clinical management of the patient. Even if the sequencing process correctly annotates the variant, the reference sequence in itself contains built-in errors, including reference errors such as pseudogenes annotated as coding regions.
FUTURE DIRECTIONS
The recent identification of single-gene causes of SRNS in a surprisingly high fraction of individuals worldwide has shown that SRNS and FSGS are not single disease entities but rather represent a spectrum of distinct diseases with clearly defined etiology. Gene identification will offer many advantages for future management of SRNS. With the available sequencing technology and the continuous reduction in sequencing cost, panel genetic testing should be offered to every patient with persistent proteinuria occurring before 25 years of age if the patient consents for clinical genetic testing for the following reasons:
Gene panel sequencing became technically feasible only very recently and therefore represents a modern approach to the diagnostics of SRNS.
It was discovered very recently that in SRNS with onset before 25 years of age, there is a very high likelihood to successfully reveal a causative mutation [49].
Mutation analysis will provide the patient and families with an unequivocal cause-based (etiologic) diagnosis, potentially enabling etiology-based ‘personalized’ clinical management of SRNS.
Mutation analysis may allow avoidance of a renal biopsy procedure.
Mutation analysis may reveal a form of SRNS that is amenable to treatment, as exemplified by rare genetic forms of SRNS that can be treated with coenzyme Q10 (Figure 4).
Clinical, epidemiologic, and treatment studies should always attempt to reveal the etiologic type of SRNS by identifying any causative mutation in a monogenic gene, in order to stratify those cohorts according to ‘etiologic’ criteria. Study data should not be obscured by generating an unspecific ‘genetic burden analysis’ that does not take the action of Mendelian (monogenic) genes and mutations into account.
Clear definition of the ‘monogenic landscape’ in large cohorts of patients with SRNS will help further unravel the puzzle of pathogenic pathways of SRNS.
Specific disease-causing mutations from patients with SRNS may be studied in cell-based and animal model systems to develop ‘personalized’ treatment approaches.
In the near future we should expect personalized treatment options for SRNS (‘precision medicine’) based on genetic causation, as exemplified by mutation in genes of the coenzyme Q10 biosynthesis pathway (COQ2, COQ6, ADCK4 or PDSS2) where treatment with coenzyme Q10 can be attempted [7, 12].
Recently, a single-gene cause of SSNS has been discovered by revealing mutation in EMP2. It is expected that more SSNS genes will be discovered and that the discovery of these genes may offer inroads into understanding the therapeutic actions of glucocorticoids in SSNS.
REFERENCES
- 1. Wiggins RC. The spectrum of podocytopathies: a unifying view of glomerular diseases. Kidney Int 2007; 71: 1205–1214 [DOI] [PubMed] [Google Scholar]
- 2. Hildebrandt F. Genetic kidney diseases. Lancet 2010; 375: 1287–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Benoit G, Machuca E, Antignac C. Hereditary nephrotic syndrome: a systematic approach for genetic testing and a review of associated podocyte gene mutations. Pediatr Nephrol 2010; 25: 1621–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Caridi G, Trivelli A, Sanna-Cherchi S et al. . Familial forms of nephrotic syndrome. Pediatr Nephrol 2010; 25: 241–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Smith JM, Stablein DM, Munoz R et al. . Contributions of the transplant registry: the 2006 Annual Report of the North American Pediatric Renal Trials and Collaborative Studies (NAPRTCS). Pediatr Transplant 2007; 11: 366–373 [DOI] [PubMed] [Google Scholar]
- 6. Benoit G, Machuca E, Heidet L et al. . Hereditary kidney diseases: highlighting the importance of classical Mendelian phenotypes. Ann N Y Acad Sci 2010; 1214: 83–98 [DOI] [PubMed] [Google Scholar]
- 7. Ashraf S, Gee HY, Woerner S et al. . ADCK4 mutations promote steroid-resistant nephrotic syndrome through CoQ10 biosynthesis disruption. J Clin Invest 2013; 123: 5179–5189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gupta IR, Baldwin C, Auguste D et al. . ARHGDIA: a novel gene implicated in nephrotic syndrome. J Med Genet 2013; 50: 330–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lowik MM, Groenen PJ, Pronk I et al. . Focal segmental glomerulosclerosis in a patient homozygous for a CD2AP mutation. Kidney Int 2007; 72: 1198–1203 [DOI] [PubMed] [Google Scholar]
- 10. Sethi S, Fervenza FC, Zhang Y et al. . Secondary focal and segmental glomerulosclerosis associated with single-nucleotide polymorphisms in the genes encoding complement factor H and C3. Am J Kidney Dis 2012; 60: 316–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Diomedi-Camassei F, Di Giandomenico S, Santorelli FM et al. . COQ2 nephropathy: a newly described inherited mitochondriopathy with primary renal involvement. J Am Soc Nephrol 2007; 18: 2773–2780 [DOI] [PubMed] [Google Scholar]
- 12. Heeringa SF, Chernin G, Chaki M et al. . COQ6 mutations in human patients produce nephrotic syndrome with sensorineural deafness. J Clin Invest 2011; 121: 2013–2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ebarasi L, Ashraf S, Bierzynska A et al. . Defects of CRB2 cause steroid-resistant nephrotic syndrome. Am J Hum Genet 2015; 96: 153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ovunc B, Otto EA, Vega-Warner V et al. . Exome sequencing reveals cubilin mutation as a single-gene cause of proteinuria. J Am Soc Nephrol 2011; 22: 1815–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ozaltin F, Li B, Rauhauser A et al. . DGKE variants cause a glomerular microangiopathy that mimics membranoproliferative GN. J Am Soc Nephrol 2013; 24: 377–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gee HY, Ashraf S, Wan X et al. . Mutations in EMP2 cause childhood-onset nephrotic syndrome. Am J Hum Genet 2014; 94: 884–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Has C, Sparta G, Kiritsi D et al. . Integrin alpha3 mutations with kidney, lung, and skin disease. N Engl J Med 2012; 366: 1508–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kambham N, Tanji N, Seigle RL et al. . Congenital focal segmental glomerulosclerosis associated with beta4 integrin mutation and epidermolysis bullosa. Am J Kidney Dis 2000; 36: 190–196 [DOI] [PubMed] [Google Scholar]
- 19. Gee HY, Zhang F, Ashraf S et al. . KANK deficiency leads to podocyte dysfunction and nephrotic syndrome. J Clin Invest 2015; 125: 2375–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zenker M, Aigner T, Wendler O et al. . Human laminin beta2 deficiency causes congenital nephrosis with mesangial sclerosis and distinct eye abnormalities. Hum Mol Genet 2004; 13: 2625–2632 [DOI] [PubMed] [Google Scholar]
- 21. Yasukawa T, Suzuki T, Ueda T et al. . Modification defect at anticodon wobble nucleotide of mitochondrial tRNAs(Leu)(UUR) with pathogenic mutations of mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes. J Biol Chem 2000; 275: 4251–4257 [DOI] [PubMed] [Google Scholar]
- 22. Mele C, Iatropoulos P, Donadelli R et al. . MYO1E mutations and childhood familial focal segmental glomerulosclerosis. N Engl J Med 2011; 365: 295–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kestila M, Lenkkeri U, Mannikko M et al. . Positionally cloned gene for a novel glomerular protein–nephrin–is mutated in congenital nephrotic syndrome. Mol Cell 1998; 1: 575–582 [DOI] [PubMed] [Google Scholar]
- 24. Boute N, Gribouval O, Roselli S et al. . NPHS2, encoding the glomerular protein podocin, is mutated in autosomal recessive steroid-resistant nephrotic syndrome. Nat Genet 2000; 24: 349–354 [DOI] [PubMed] [Google Scholar]
- 25. Miyake N, Tsukaguchi H, Koshimizu E et al. . Biallelic mutations in nuclear pore complex subunit NUP107 cause early-childhood-onset steroid-resistant nephrotic syndrome. Am J Hum Genet. 2015; 97: 555–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lopez LC, Schuelke M, Quinzii CM et al. . Leigh syndrome with nephropathy and CoQ10 deficiency due to decaprenyl diphosphate synthase subunit 2 (PDSS2) mutations. Am J Hum Genet 2006; 79: 1125–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hinkes B, Wiggins RC, Gbadegesin R et al. . Positional cloning uncovers mutations in PLCE1 responsible for a nephrotic syndrome variant that may be reversible. Nat Genet 2006; 38: 1397–1405 [DOI] [PubMed] [Google Scholar]
- 28. Ozaltin F, Ibsirlioglu T, Taskiran EZ et al. . Disruption of PTPRO causes childhood-onset nephrotic syndrome. Am J Hum Genet 2011; 89: 139–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Berkovic SF, Dibbens LM, Oshlack A et al. . Array-based gene discovery with three unrelated subjects shows SCARB2/LIMP-2 deficiency causes myoclonus epilepsy and glomerulosclerosis. Am J Hum Genet 2008; 82: 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Boerkoel CF, Takashima H, John J et al. . Mutant chromatin remodeling protein SMARCAL1 causes Schimke immuno-osseous dysplasia. Nat Genet 2002; 30: 215–220 [DOI] [PubMed] [Google Scholar]
- 31. Colin E, Huynh Cong E, Mollet G et al. . Loss-of-function mutations in WDR73 are responsible for microcephaly and steroid-resistant nephrotic syndrome: Galloway-Mowat syndrome. Am J Hum Genet 2014; 95: 637–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vodopiutz J, Seidl R, Prayer D et al. . WDR73 mutations cause infantile neurodegeneration and variable glomerular kidney disease. Hum Mutat 2015; 36: 1021–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jinks RN, Puffenberger EG, Baple E et al. . Recessive nephrocerebellar syndrome on the Galloway-Mowat syndrome spectrum is caused by homozygous protein-truncating mutations of WDR73. Brain : a journal of neurology 2015; 138: 2173–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kaplan JM, Kim SH, North KN et al. . Mutations in ACTN4, encoding alpha-actinin-4, cause familial focal segmental glomerulosclerosis. Nat Genet 2000; 24: 251–256 [DOI] [PubMed] [Google Scholar]
- 35. Gbadegesin RA, Hall G, Adeyemo A et al. . Mutations in the gene that encodes the F-actin binding protein anillin cause FSGS. J Am Soc Nephrol 2014; 25: 1991–2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Akilesh S, Suleiman H, Yu H et al. . Arhgap24 inactivates Rac1 in mouse podocytes, and a mutant form is associated with familial focal segmental glomerulosclerosis. J Clin Invest 2011; 121: 4127–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brown EJ, Schlondorff JS, Becker DJ et al. . Mutations in the formin gene INF2 cause focal segmental glomerulosclerosis. Nat Genet 2010; 42: 72–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vollrath D, Jaramillo-Babb VL, Clough MV et al. . Loss-of-function mutations in the LIM-homeodomain gene, LMX1B, in nail-patella syndrome. Hum Mol Genet 1998; 7: 1091–1098 [DOI] [PubMed] [Google Scholar]
- 39. McIntosh I, Dreyer SD, Clough MV et al. . Mutation analysis of LMX1B gene in nail-patella syndrome patients. Am J Hum Genet 1998; 63: 1651–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Heath KE, Campos-Barros A, Toren A et al. . Nonmuscle myosin heavy chain IIA mutations define a spectrum of autosomal dominant macrothrombocytopenias: May-Hegglin anomaly and Fechtner, Sebastian, Epstein, and Alport-like syndromes. Am J Hum Genet 2001; 69: 1033–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Winn MP, Conlon PJ, Lynn KL et al. . A mutation in the TRPC6 cation channel causes familial focal segmental glomerulosclerosis. Science 2005; 308: 1801–1804 [DOI] [PubMed] [Google Scholar]
- 42. Jeanpierre C, Denamur E, Henry I et al. . Identification of constitutional WT1 mutations, in patients with isolated diffuse mesangial sclerosis, and analysis of genotype/phenotype correlations by use of a computerized mutation database. Am J Hum Genet 1998; 62: 824–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zenker M, Machuca E, Antignac C. Genetics of nephrotic syndrome: new insights into molecules acting at the glomerular filtration barrier. J Mol Med 2009; 87: 849–857 [DOI] [PubMed] [Google Scholar]
- 44. Somlo S, Mundel P. Getting a foothold in nephrotic syndrome. Nat Genet 2000; 24: 333–335 [DOI] [PubMed] [Google Scholar]
- 45. Kim YH, Goyal M, Kurnit D et al. . Podocyte depletion and glomerulosclerosis have a direct relationship in the PAN-treated rat. Kidney Int 2001; 60: 957–968 [DOI] [PubMed] [Google Scholar]
- 46. Wharram BL, Goyal M, Wiggins JE et al. . Podocyte depletion causes glomerulosclerosis: diphtheria toxin-induced podocyte depletion in rats expressing human diphtheria toxin receptor transgene. J Am Soc Nephrol 2005; 16: 2941–2952 [DOI] [PubMed] [Google Scholar]
- 47. Hinkes BG, Mucha B, Vlangos CN et al. . Nephrotic syndrome in the first year of life: two thirds of cases are caused by mutations in 4 genes (NPHS1, NPHS2, WT1, and LAMB2). Pediatrics 2007; 119: e907–e919 [DOI] [PubMed] [Google Scholar]
- 48. Lovric S, Fang H, Vega-Warner V et al. . Rapid detection of monogenic causes of childhood-onset steroid-resistant nephrotic syndrome. Clin J Am Soc Nephrol 2014; 9: 1109–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sadowski CE, Lovric S, Ashraf S et al. . A single-gene cause in 29.5% of cases of steroid-resistant nephrotic syndrome. J Am Soc Nephrol 2014; 26: 1279–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Halbritter J, Baum M, Hynes AM et al. . Fourteen monogenic genes account for 15% of nephrolithiasis/nephrocalcinosis. J Am Soc Nephrol 2015; 26: 543–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Halbritter J, Diaz K, Chaki M et al. . High-throughput mutation analysis in patients with a nephronophthisis-associated ciliopathy applying multiplexed barcoded array-based PCR amplification and next-generation sequencing. J Med Genet 2012; 49: 756–767 [DOI] [PubMed] [Google Scholar]
- 52. Giglio S, Provenzano A, Mazzinghi B et al. . Heterogeneous genetic alterations in sporadic nephrotic syndrome associate with resistance to immunosuppression. J Am Soc Nephrol 2015; 26: 230–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Trautmann A, Bodria M, Ozaltin F et al. . Spectrum of steroid-resistant and congenital nephrotic syndrome in children: The Podonet Registry Cohort. Clin J Am Soc Nephrol 2015; 10: 592–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lipska BS, Iatropoulos P, Maranta R et al. . Genetic screening in adolescents with steroid-resistant nephrotic syndrome. Kidney Int 2013; 84: 206–213 [DOI] [PubMed] [Google Scholar]
- 55. Weber S, Gribouval O, Esquivel EL et al. . NPHS2 mutation analysis shows genetic heterogeneity of steroid-resistant nephrotic syndrome and low post-transplant recurrence. Kidney Int 2004; 66: 571–579 [DOI] [PubMed] [Google Scholar]
- 56. Ruf RG, Lichtenberger A, Karle SM et al. . Patients with mutations in NPHS2 (podocin) do not respond to standard steroid treatment of nephrotic syndrome. J Am Soc Nephrol 2004; 15: 722–732 [DOI] [PubMed] [Google Scholar]
- 57. Wan X, Chen Z, Choi WI et al. . Loss of epithelial membrane protein 2 aggravates podocyte injury via upregulation of caveolin-1. J Am Soc Nephrol 2015; 27: 10.1681/ASN.2014121197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hildebrandt F, Heeringa SF. Specific podocin mutations determine age of onset of nephrotic syndrome all the way into adult life. Kidney Int 2009; 75: 669–671 [DOI] [PubMed] [Google Scholar]
- 59. Hinkes B, Vlangos C, Heeringa S et al. . Specific podocin mutations correlate with age of onset in steroid-resistant nephrotic syndrome. J Am Soc Nephrol 2008; 19: 365–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tory K, Menyhard DK, Woerner S et al. . Mutation-dependent recessive inheritance of NPHS2-associated steroid-resistant nephrotic syndrome. Nat Genet 2014; 46: 299–304 [DOI] [PubMed] [Google Scholar]
- 61. Chernin G, Vega-Warner V, Schoeb DS et al. . Genotype/phenotype correlation in nephrotic syndrome caused by WT1 mutations. Clin J Am Soc Nephrol 2010; 5: 1655–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hasselbacher K, Wiggins RC, Matejas V et al. . Recessive missense mutations in LAMB2 expand the clinical spectrum of LAMB2-associated disorders. Kidney Int 2006; 70: 1008–1012 [DOI] [PubMed] [Google Scholar]
- 63. Boyer O, Nevo F, Plaisier E et al. . INF2 mutations in Charcot-Marie-Tooth disease with glomerulopathy. N Engl J Med 2011; 365: 2377–2388 [DOI] [PubMed] [Google Scholar]
- 64. Boyer O, Woerner S, Yang F et al. . LMX1B mutations cause hereditary FSGS without extrarenal involvement. J Am Soc Nephrol 2013; 24: 1216–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Montini G, Malaventura C, Salviati L. Early coenzyme Q10 supplementation in primary coenzyme Q10 deficiency. N Engl J Med 2008; 358: 2849–2850 [DOI] [PubMed] [Google Scholar]
- 66. Gee HY, Saisawat P, Ashraf S et al. . ARHGDIA mutations cause nephrotic syndrome via defective RHO GTPase signaling. J Clin Invest 2013; 123: 3243–3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Schlondorff J, Del Camino D, Carrasquillo R et al. . TRPC6 mutations associated with focal segmental glomerulosclerosis cause constitutive activation of NFAT-dependent transcription. Am J Physiol Cell Physiol 2009; 296: C558–C569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Halbritter J, Porath JD, Diaz KA et al. . Identification of 99 novel mutations in a worldwide cohort of 1,056 patients with a nephronophthisis-related ciliopathy. Hum Genet 2013; 132: 865–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Teer JK, Mullikin JC. Exome sequencing: the sweet spot before whole genomes. Hum Mol Genet 2010; 19: R145–R151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ng SB, Turner EH, Robertson PD et al. . Targeted capture and massively parallel sequencing of 12 human exomes. Nature 2009; 461: 272–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lifton RP. Individual genomes on the horizon. N Engl J Med 2010; 362: 1235–1236 [DOI] [PubMed] [Google Scholar]
- 72. Otto EA, Hurd TW, Airik R et al. . Candidate exome capture identifies mutation of SDCCAG8 as the cause of a retinal-renal ciliopathy. Nat Genet 2010; 42: 840–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Yang Y, Muzny DM, Reid JG et al. . Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med 2013; 369: 1502–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Hildebrandt F, Heeringa SF, Ruschendorf F et al. . A systematic approach to mapping recessive disease genes in individuals from outbred populations. PLoS Genet 2009; 5: e1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lee H, Deignan JL, Dorrani N et al. . Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA 2014; 312: 1880–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. MacArthur DG, Manolio TA, Dimmock DP et al. . Guidelines for investigating causality of sequence variants in human disease. Nature 2014; 508: 469–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. MacArthur DG, Balasubramanian S, Frankish A et al. . A systematic survey of loss-of-function variants in human protein-coding genes. Science 2012; 335: 823–828 [DOI] [PMC free article] [PubMed] [Google Scholar]