ABSTRACT
Background
Lactase is an enzyme that hydrolyzes lactose into glucose and galactose in the small intestine, where they are absorbed. Hypolactasia is a common condition, primarily caused by genetic programming, that leads to lactose maldigestion and, in certain cases, lactose intolerance. Galactitol and galactonate are 2 products of hepatic galactose metabolism that are candidate markers for the intake of lactose-containing foods.
Objectives
The primary objective of the study was to explore the changes in serum and urine metabolomes during postprandial dairy product tests through the association between lactase persistence genotype and the postprandial dynamics of lactose-derived metabolites.
Methods
We characterized the 6-h postprandial serum kinetics and urinary excretion of lactose, galactose, galactitol, and galactonate in 14 healthy men who had consumed a single dose of acidified milk (800 g) which contained 38.8 g lactose. Genotyping of LCT-13910 C/T (rs4988235) was performed to assess primary lactase persistence.
Results
There were 2 distinct postprandial responses, classified as high and low metabolite responses, observed for galactose, and its metabolites galactitol and galactonate, in serum and urine. In all but 1 subject, there was a concordance between the high metabolite responses and genetic lactase persistence and between the low metabolite responses and genetic lactase nonpersistence (accuracy 0.92), galactitol and galactonate being more discriminative than galactose.
Conclusions
Postprandial galactitol and galactonate after lactose overload appear to be good proxies for genetically determined lactase activity. The development of a noninvasive lactose digestion test based on the measurement of these metabolites in urine could be clinically useful. This trial was registered at clinicaltrials.gov as NCT02230345.
Keywords: galactose metabolism, lactase activity, lactose, milk digestion, nutrigenetics; personalized nutrition
Introduction
Lactose is a disaccharide composed of glucose and galactose and is found mainly in milk and dairy products, but also in many manufactured food products (1). The digestion of lactose requires the presence of lactase, which hydrolyses the sugar in the small intestine brush border, after which the 2 monosaccharides are absorbed via passive diffusion or active transport. In most humans, a reduction in lactase activity (LA) occurs in late childhood through epigenetic programming (2). Consequently, undigested lactose reaches the colonic microbiota, resulting in the production of short-chain fatty acids, hydrogen, and methane (3). For between one-third and one-half of individuals with such lactose maldigestion (LM), these fermentation products can cause gastrointestinal symptoms such as diarrhea, abdominal pain, bloating, flatulence, and vomiting after the ingestion of milk products, thus defining the clinical manifestations of lactose intolerance (LI). On the other hand, ∼35% of the world's population retain intestinal LA similar to infantile levels throughout adulthood (4). This lactase-persistence phenotype is owing to polymorphisms in the promoter of the lactase gene (LCT) that eliminate the programmed reduction in activity of the promoter with age. Several prevalent mutations of the LCT promoter have been described in populations or ethnic groups that have a long history of herding animals, suggesting an evolutionary advantage of this phenotype in these groups.
Different methods are currently used to diagnose LM (5). Jejunal biopsy, by allowing the direct measurement of LA, is considered the gold standard method, but it is invasive, with patchy results. Conversely, genetic testing of primary lactase persistence is noninvasive but can lead to false results in the case of genetic heterogeneity (6) or where secondary causes of LM are present. The lactose tolerance test (LTT) and the recently developed gaxilose test (7) both allow indirect evaluation of LA through absorption testing, whereas the lactose hydrogen breath test (LHBT) is based on testing the colonic consequences of LM. These methods are widely used but can be unreliable and difficult to interpret in clinical practice (8–10). The development of a complementary method that would address some of the major limitations of the available tests could be clinically useful.
We have recently shown that both lactose and galactose are present in blood after the ingestion of milk and yogurt (11). Once absorbed, the majority of galactose enters the Leloir pathway, where it is metabolized to UDP-glucose. However, it can also enter 2 other hepatic pathways, to be either reduced to galactitol by aldehyde reductase or oxidized to galactonate by galactose dehydrogenase (12). Both galactitol and galactonate have been identified as candidate biomarkers of lactose-containing food intake in serum (13) and urine (14). We therefore hypothesized that these 2 metabolites might also be good proxies of LA .
Methods
Samples were obtained from a recently published crossover study that was designed to evaluate the metabolic impact of acute and prolonged intake of yogurt and acidified milk in 14 healthy men (11, 15–17). Supplemental Figure 1 provides a flowchart of the participants. Details on the study design have been published elsewhere (15).
Subjects
Subjects were healthy male volunteers (n = 14) who regularly consumed milk, with a mean ± SEM age of 25 ± 1.3 y (recruitment range of 18–40 y) and a mean ± SEM body mass index of 22 ± 0.5 kg/m2 (recruitment range of 18.5–25.0 kg/m2). Of the subjects, 13 were Caucasian and 1 was black African. A detailed tabulated description of these subjects has been published elsewhere (15). The overall study received ethical approval from the Commission cantonale d’éthique de la recherche sur l’être humain (CER-VD, approval number: 392/13, Vaud, Switzerland); the trial was registered at clinicaltrials.gov as NCT02230345.
Test products
Both milk and yogurt were tested in a crossover design but, as the fermentation process led to large changes in lactose and galactose concentrations in yogurt (11), only data related to milk intake are presented in this report. The ingested milk was full fat, homogenized, and ultrahigh-temperature processed (provided by Emmi Mittelland Molkerei AG). The milk was acidified by the addition of 2% D-(+)-glucono-δ-lactone (≥99.0%; Jungbunzlauer AG) to mimic the texture of the test yogurt, as described previously (15).
Dietary intervention
The postprandial tests were performed at the Center of Clinical Research of CHUV in Lausanne. Briefly, each participant ingested a single 800 g dose of acidified milk, containing 38.8 g lactose. The test product was ingested within 15 min in the fasted state, and serum was taken postprandially at 10 time points (fasting, then at 15, 30, 60, 90, 120, 180, 240, 300, and 360 min). Urine was collected in the morning before the acidified milk ingestion, as well as during the 6-h postprandial phase (pooled sample). Samples were stored at −80°C prior to analysis. During the 3 wk preceding the test day, participants consumed 400 g of normal milk per day and followed a supervised diet (15).
Genetic test for lactase persistence
Genotyping of rs4988235, the most common single nucleotide polymorphism (SNP) associated with lactase persistence in Northern European population (LCT-13910 C>T polymorphism), was performed with the EliGene® Lactose Intolerance C-13910T LC Kit (Elisabeth® Pharmacon) according to manufacturer's instructions. Real-time polymerase chain reaction was carried out using the Lightcycler 96 (Roche), with internal standards for positive and negative controls. Homozygous carriers of the wild-type allele (CC) were considered to be lactase nonpersistent (n = 8), whereas heterozygous (CT) and homozygous carriers for the mutated allele (TT) were lactase persistent (n = 5). Subject F3_009, of African origin, was excluded from the statistical analyses because genotypes at rs4988235 were not relevant for diagnosis of lactase persistence in this population.
Serum and urine lactose, galactose, galactitol, and galactonate gas chromatography–mass spectrometry analysis
The primary endpoint of the original overall study reported by Pimentel et al. (16) was the untargeted metabolomic profile of serum after ingestion of yogurt and milk products. Secondary endpoints, including the blood cell transcriptome (17), as well as metabolic, inflammatory, and gut microbiota parameters, have been published separately (15). This report now investigates the postprandial behavior in serum and urine of 4 metabolites related to the metabolism of lactose (lactose, galactose, galactitol, and galactonate) after ingestion of the milk product as a function of the genetic profile for lactase persistence. Serum and urine samples were analyzed using an untargeted gas chromatography–mass spectrometry (GC–MS) metabolomics method. Supplemental Figure 2 provides examples of the chromatograms and spectra of lactose, galactose, galactitol, and galactonate. These molecules derive from lactose metabolism (Supplemental Figure 3) and were among the metabolites whose relative concentration changed the most following dairy intake. They were, therefore, evaluated for their capacity to indirectly reflect LA. Samples were prepared according to the Human Serum Metabolome Consortium (HUSERMET) procedure (18) with small modifications, as previously described (11, 14). Prior to analysis, each urine sample was diluted according to its specific gravity, as calculated from the refractive index by refractometry, with the use of an RE40 refractometer (Mettler-Toledo). GC-MS measurements were performed using an Agilent 7890B/5977A GC-MS system, 70 kV, equipped with a 60 m × 0.250 mm × 0.25 μm DB-5MS column (Agilent Technologies) (11, 13, 14). The selected metabolites were determined with level 1 identification (19) by the injection of pure standard solutions (14) of lactose and galactose from Merck and galactitol and galactonate from Sigma-Aldrich. The serum and urine samples were analyzed in batches, each batch containing the samples from one subject. The samples from each subject were randomized. The order of the analyses of the subjects was also randomized. The analytical laboratory was blinded to the sample identity.
Statistical analyses
All statistical analyses were performed using R (3.1.2), with 0.05 as the P value significance cutoff. Linear evaluation of the incremental area under the curve [iAUC; MESS package V.0.3.2 (20)] over 6 h was completed for each of the 4 metabolites in postprandial serum. The Wilcoxon's signed-rank test was used to compare the intensities of lactose, galactose, galactitol, and galactonate between subjects with and without lactase persistence genotypes, in serum (iAUC) and in urine (postprandial 6 h pool). A nonparametric longitudinal analysis (nparLD) was performed to confirm the presence of a time effect for the serum metabolites by lactase persistence genotype [package nparLD V.2.1 (21)]. A partial Spearman's correlation test, controlling for lactase persistence [ppcor package V.1.1 (22)], was used to assess the relationships between the metabolites in serum or urine and between serum and urine intensities of the same metabolite.
Results
The postprandial serum kinetics of lactose in serum showed an increase between 2 and 3 h after ingestion of milk and a return to nearly baseline levels by 6 h for most subjects (Figure 1A). Although postprandial responses were more significant for the CC carriers (n = 8) (nparLD, P = 1.56 × 10−17) than for the CT and TT carriers (n = 2 and 3, respectively) (nparLD, P = 2.49 × 10−6), there was no significant difference between the 2 groups when comparing the iAUCs (P = 0.72, Figure 1E), as previously described (11).
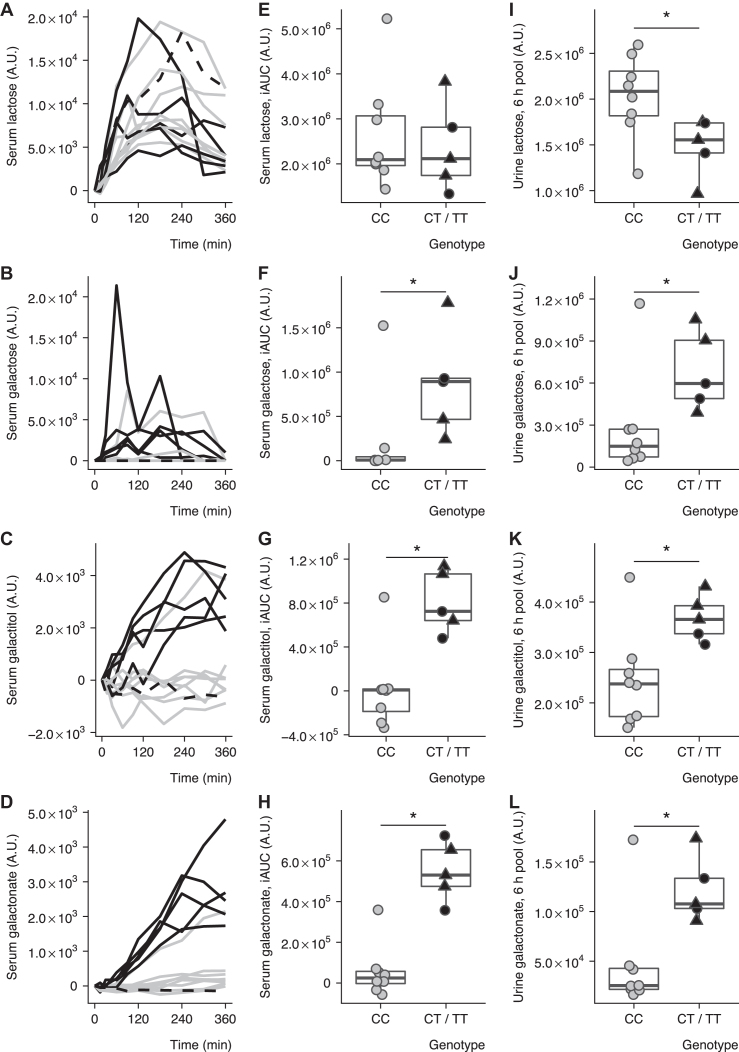
FIGURE 1.
Genetic dependency of the postprandial behavior of lactose-derived metabolites after intake of acidified milk. Serum lactose (A), galactose (B), galactitol (C), and galactonate (D) after milk intake in 14 healthy subjects. Participants with CC (solid grey, n = 8) and CT or TT (solid black, n = 5) or nondetermined (black dashed, n = 1) genotype at rs4988235. Comparison of postprandial serum (iAUC) lactose (E), galactose (F), galactitol (G), and galactonate (H) after milk intake between CC (grey circles, n = 8), CT (black circles, n = 2), and TT (black triangles, n = 3) carriers in 13 healthy subjects. Comparison of postprandial urine (6-h pool) lactose (I), galactose (J), galactitol (K), and galactonate (L) after milk intake between CC (grey circles, n = 8), CT (black circles, n = 2), and TT (black triangles, n = 3) carriers in 13 healthy subjects. Boxplots indicate medians and interquartile ranges. The Box-Whiskers plots indicate medians, interquartile ranges, and minimum and maximum values excluding outliers. *P < 0.05, Wilcoxon's signed-rank test between CC and CT or TT genotypes. Subject F3_009, who was of African origin, was excluded from the statistical analyses of panels E–L because genotypes at rs4988235 were not relevant for diagnosis of lactase persistence in this population. iAUC, incremental area under the curve.
Conversely, the postprandial kinetics of galactose (Figure 1B) and its metabolites galactitol (Figure 1C) and galactonate (Figure 1D) in serum showed 2 distinct patterns according to the LCT genotypes. For the CT and TT carriers, galactose increased significantly, although variably, from 30 min to 3 h after ingestion of acidified milk compared with fasting values, before returning to fasting levels (nparLD P = 9.50 × 10−5), whereas galactitol and galactonate both increased continuously from the first hour and during the 6 h (nparLD, P = 4.36 × 10−15 and nparLD, P = 7.76 × 10−22, respectively). For the CC carriers, the intensities of galactose and galactitol remained unchanged compared with fasting values (nparLD P > 0.05), whereas galactonate showed a small but significant postprandial response (nparLD P = 1.49 × 10−3). These postprandial responses appeared to be consistent between the subjects of each genotype group, with the exception of 1 subject (subject F3_015) who was homozygotic for the wild-type allele (CC) and who displayed postprandial responses similar to that of the CT and TT carriers.
In accordance with these observations, the iAUCs for galactose (Figure 1F), galactitol (Figure 1G), and galactonate (Figure 1H) were significantly higher in CT and TT carriers than in CC carriers (Wilcoxon's signed-rank test, P = 0.023, P = 0.011, and P = 0.0031, respectively). A cutoff value for each of the 3 metabolites could clearly separate the subjects from the 2 groups, with the exception of subject F3_015, as mentioned above. Statistics on serum data comparing CT and TT carriers and CC carriers at each time point are given in Supplemental Table 1.
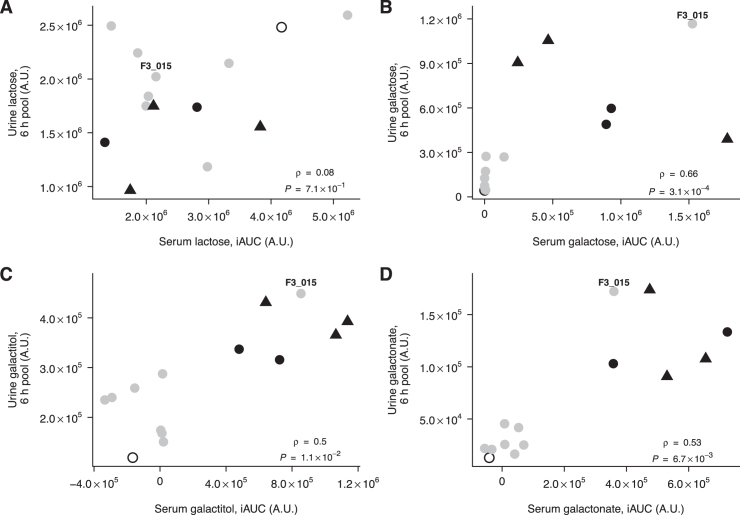
Interindividual variability in the postprandial responses of lactose (Figure 1I), galactose (Figure 1J), galactitol (Figure 1K), and galactonate (Figure 1L) was also observed in urine. In contrast to the observations in serum, CC carriers had significantly higher postprandial urinary levels of lactose than CT and TT carriers (P = 0.019) (see Figure 1I, Supplemental Table 1), although there was no clear cutoff level to separate the 2 groups. Conversely, the galactose, galactitol, and galactonate intensities in urine were significantly higher in CT and TT carriers than in CC carriers (Wilcoxon's signed-rank test, P = 0.030, P = 0.030, and P = 0.019, respectively). In addition, a cutoff could clearly separate the subjects from the 2 groups, with the exception of subject F3_015, who showed a discrepancy between genotype and urine metabolite responses similar to the response observed in serum. Significant positive correlations between serum and urine intensities were observed for galactose, galactitol, and galactonate (P = 3.10 × 10−4, P = 1.06 × 10−2, and P = 6.70 × 10−3, respectively), with a clear bimodal separation between lactase-persistent and -nonpersistent subjects (Figure 2), the best separation being observed for galactonate (see Figure 2D). In the comparisons made between galactose, galactonate, and galactitol, subject F3_009 (African origin) clearly clustered with the lactase-nonpersistent genotype group, whereas subject F3_015 (CC genotype) clustered with the lactase-persistent genotype group.
FIGURE 2.
Correlation between postprandial serum and urine lactose (A), galactose (B), galactitol (C), and galactonate (D) after milk intake in 14 subjects. Partial Spearman's correlation test controlling for lactase persistence. Participants with CC (grey circles, n = 8), CT (black circles, n = 2), TT (black triangles, n = 3), or nondetermined (unfilled circle, n = 1) genotype at rs4988235. Subject F3_015 is indicated on the figure. iAUC, incremental area under the curve.
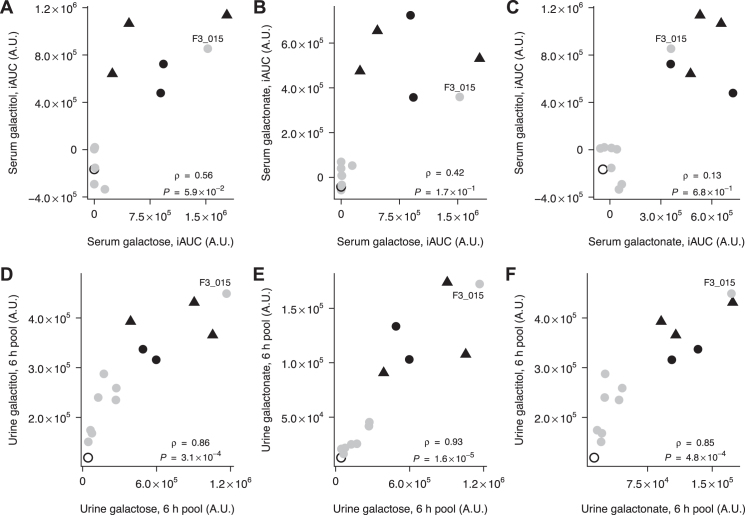
Combining pairs of metabolites either in serum (upper panels of Figure 3) or in urine (lower panels of Figure 3) even strengthened the capacity to separate the 2 profiles, although galactose was the least discriminative of the 3 metabolites in both fluids, and galactonate was the most discriminative in urine.
FIGURE 3.
Correlation between postprandial serum galactose and galactitol (A), galactose and galactonate (B), and galactonate and galactitol (C), and between postprandial urine galactose and galactitol (D), galactose and galactonate (E), and galactonate and galactitol (F), after milk intake in 14 subjects. Partial Spearman's correlation test controlling for lactase persistence. Participants with CC (grey circles, n = 8), CT (black circles, n = 2), TT (black triangles, n = 3), or nondetermined (unfilled circle, n = 1) genotype at rs4988235. Subject F3_015 is indicated on the figure. iAUC, incremental area under the curve.
Of note, the CT and TT carriers (n = 2 and n = 3, respectively) appeared to exhibit similar metabolite profiles for the 4 metabolites both in serum and in urine, although this could not be statistically confirmed owing to the small number of samples (see panels E–L of Figure 1, and also Figures 2 and 3).
The accuracy of galactose, galactitol, and galactonate serum or urine measurements after milk ingestion to predict lactase persistence determined genetically was 0.92 in this study (12 out of 13 subjects were concordant).
Discussion
We have shown that galactose and 2 of its metabolites, galactitol and galactonate, present a bimodal (high or low) postprandial response after milk ingestion both in serum and in urine. In 92% of our cases, these responses matched the genotype at rs4988235 for lactase persistence. We therefore propose that the measurement of these 3 metabolites after a lactose overload could be a good proxy for LA. We could not identify distinct metabolite profiles between heterozygous (CT) and homozygous (TT) carriers of the mutated T allele that confers lactase persistence. This is in line with a previous report of similar blood glucose response after lactose overload in both groups (23). However, it is not excluded that by studying a larger number of subjects it would be possible to identify an intermediate phenotype for the LCT C/T heterozygotes, in accordance with the gene dosage effect on jejunal LA that has been recently described (24).
For 1 subject (F3_015), the observed phenotype did not correspond with the phenotype expected for his lactase-nonpersistent genotype; instead, a high LA was suggested by the metabolite postprandial responses. However, we typed only the most common polymorphism associated with lactase persistence in Northern European populations, and the subjects we recruited were of diverse origins. It is therefore likely that we missed a relevant polymorphism in this subject owing to genetic heterogeneity. Indeed, one of the major limitations in lactase genetic testing is that, in genetically diverse populations (such as in Europe), targeted analysis of SNPs may not comprehensively evaluate lactase persistence (25). In this context, we excluded subject F3_009, who was of African origin, from the analyses because the SNP typed in this study was specific to Caucasians; however, we observed that this volunteer presented a postprandial metabolite response suggestive of a low LA, which is the most common phenotype in this population (26). For both subjects, the metabolic challenge would have been more specific or sensitive than the available genetic test to detect lactase persistence or nonpersistence.
Of the existing LM tests, several involve the administration of a lactose load; in the LTT, 50 g of lactose is usually administered, whereas for the LHBT, the recommended lactose dose has recently been reduced to 25 g (27). In the present challenge, an intake of 38.8 g lactose was sufficient to separate 2 metabolic profiles with clear cutoffs for galactose, galactitol, and galactonate, both in serum and in urine. Galactose is rapidly metabolized after absorption mainly via the Leloir pathway and does not accumulate in the organism in normal conditions (see Supplemental Figure 2) (28). The Leloir pathway is regulated by the rate-limiting enzyme galactokinase, but 2 alternative pathways can be activated that generate either galactitol or galactonate from galactose, 2 metabolites that are not further metabolized but are instead eliminated in urine. It is therefore not surprising that these downstream metabolites were more discriminant than galactose. Although the measurements in urine were completed on 6-h pooled samples, a body of evidence suggests that a measurement of galactitol and galactonate in a 3-h urine pool should be sufficient to discriminate high and low LA: 1) the kinetics of the metabolites in serum indicate discrimination of LA at 3 h (see panels C and D in Figure 1); 2) serum and urine measurements were highly correlated (see Figure 2); and 3) the kinetics of galactitol in urine in the previous study of Münger et al. (14) indicate that the excretion of galactitol peaks between 2 and 4 h after the ingestion of 600 ml of milk. The suitability of 3-h urine pools, however, needs to be confirmed in a larger study.
We propose that the measurement of galactitol and galactonate in urine could be used as a new noninvasive test for the accurate evaluation of LA and subsequent diagnosis of normal lactasia or primary or secondary hypolactasia. The test showed high sensitivity (87.5%) and high specificity (100%) in our study, in which only genetic testing was used as the reference for LA. It is likely that the sensitivity would have been higher if the LA had been accurately assessed by other methods, such as LTT, LHBT, or jejunal biopsy, for all the subjects in this study. The advantage of this new test is that its performance would not be altered in clinical conditions that limit or invalidate the interpretation of other available LM diagnostic tests. For example, the precision of the test is not reduced owing to the presence of impaired glucose tolerance as for the LTT, the test is not overly influenced by microbiota modifications like the LHBT, and the test is not affected by genetic heterogeneity, which limits the use of the SNP-based genetic testing. In addition, given that the ingestion of lactose is required for the test, the evaluation of gastrointestinal symptoms could be used to concomitantly diagnose LI. This would be an advantage compared with the gaxilose test, which uses a synthetic analog of lactose not found in food matrices as a substrate for lactase and which does not allow the assessment of LI symptoms.
There are nevertheless some potential limitations to our proposed test. First, urinary concentrations of galactitol and galactonate will be influenced by subjects’ renal excretion rates and urine dilution. Adjusting the concentrations based on urine creatinine content or specific gravity, as done in the present study, might therefore be necessary to reduce interindividual variability. Second, the validity of the test relies on the correct functionality of enzymes that act in 2 distinct metabolic processes, namely, gastrointestinal hydrolysis of lactose and hepatic metabolism of galactose. As such, the test is likely to be affected by any gastrointestinal, hepatic, or renal dysfunctions. Indeed, the course of galactose elimination capacity is used to monitor survival in patients with cirrhosis (29). Conversely, in patients with galactosemia, who present a defect in enzymes of the Leloir pathway, alternative pathways are activated, resulting in accumulation of galactitol and galactonate (30). In such cases, the test would not be sensitive to LM, and hence would not detect it. Third, although a test based on metabolites has an advantage over the genetic test in being phenotypic, and hence clinically more relevant, GC-MS-based tests are costly, and such technologies are not available in many centers. However, enzymatic tests based on bacterial enzymes specifically hydrolyzing galactose metabolites, in particular galactonate (31), may be validated as alternatives to the GC-MS-based test.
The identification of 2 different galactose metabolite excretion profiles that depend on LA raises the question of whether these metabolites are appropriate food intake biomarkers of lactose-containing foods in subjects with lactase nonpersistence (13, 14). In a pilot study, we were able to identify galactitol and galactonate in urine after the ingestion of only 10 g of lactose, with a dose response (ingestion of 20 and 40 g of lactose) in subjects with lactase persistence but not in subjects with lactase nonpersistence (G Vergères, unpublished data). This suggests that absorption of dietary portions of lactose can activate pathways of galactose metabolism that are alternatives to the Leloir pathway in all subjects regardless of their lactase status, but that the measurement of these metabolites in urine would be only qualitative and not quantitative in subjects with lactase nonpersistence. Thus, our proposed test would likely not be discriminative of LA if the dose of lactose was much lower than 40 g.
In conclusion, hypolactasia is a highly prevalent disorder that is often difficult to diagnose. An accurate assessment of LA in subjects experiencing gastrointestinal discomfort could help to avoid inappropriate food restriction. The evaluation of galactitol and galactonate in urine could be used as a noninvasive test to determine LA. Additional, larger studies are needed to confirm these results in different populations, including populations with different genotypes, age groups (children, adults, elderly), and genders. These studies should also confirm the accuracy of the test in 3-h pooled urine and help to refine the discriminative cutoff. Once validated, this indirect assessment of LA could be highly relevant for clinical assessment of lactose metabolism.
Supplementary Material
Acknowledgements
The authors’ contributions were as follows—NV, KJB, GP, FPP, and GV: designed the research; LHM, CF, KJB, and RB: conducted the research; GP: analyzed the data; NV, KJB, GP, and GV: wrote the manuscript; NV and GV: had primary responsibility for the final content; and all authors: read and approved the final manuscript. None of the authors has a conflict of interest.
Notes
The milk used in this study was provided by Emmi Mittelland Molkerei AG.
Present addresses: LHM: ETH Zürich, Institute of Food, Nutrition and Health, Laboratory of Food Biochemistry, Schmelzbergstrasse 9, CH-8092 Zurich, Switzerland.
KJB: Agroscope, Federal Department of Economic Affairs, Education and Research, Schwarzenburgstrasse 161, 3003 Bern, Switzerland.
FPP: La Tour Hospital, Av J.-D Maillard 3, 1217 Meyrin, Switzerland.
Supplemental Figures 1–3 and Supplemental Table 1 are available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at https://academic.oup.com/ajcn/.
Abbreviations used: GC-MS, gas chromatography–mass spectrometry; iAUC, incremental area under the curve; LA, lactase activity; LHBT, lactose hydrogen breath test; LI, lactose intolerance; LM, lactose maldigestion; LTT, lactose tolerance test; nparLD, nonparametric longitudinal analysis; SNP, single nucleotide polymorphism.
References
- 1. Johnson JM, Conforti FD. Lactose A2. In: Caballero B.editor. Encyclopedia of Food Sciences and Nutrition. 2nd ed Oxford: Academic Press; 2003:3472–6. [Google Scholar]
- 2. Labrie V, Buske OJ, Oh E, Jeremian R, Ptak C, Gasiunas G, Maleckas A, Petereit R, Zvirbliene A, Adamonis K et al.. Lactase nonpersistence is directed by DNA-variation-dependent epigenetic aging. Nat Struct Mol Biol. 2016;23(6):566–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. He T, Priebe MG, Harmsen HJ, Stellaard F, Sun X, Welling GW, Vonk RJ. Colonic fermentation may play a role in lactose intolerance in humans. J Nutr. 2006;136(1):58–63. [DOI] [PubMed] [Google Scholar]
- 4. Gerbault P, Liebert A, Itan Y, Powell A, Currat M, Burger J, Swallow DM, Thomas MG. Evolution of lactase persistence: an example of human niche construction. Philos Trans R Soc Lond B Biol Sci. 2011;366(1566):863–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Misselwitz B, Pohl D, Fruhauf H, Fried M, Vavricka SR, Fox M. Lactose malabsorption and intolerance: pathogenesis, diagnosis and treatment. United European Gastroenterol J. 2013;1(3):151–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ingram CJ, Mulcare CA, Itan Y, Thomas MG, Swallow DM. Lactose digestion and the evolutionary genetics of lactase persistence. Hum Genet. 2009;124(6):579–91. [DOI] [PubMed] [Google Scholar]
- 7. Aragon JJ, Hermida C, Martinez-Costa OH, Sanchez V, Martin I, Sanchez JJ, Codoceo R, Cano JM, Cano A, Crespo L et al.. Noninvasive diagnosis of hypolactasia with 4-galactosylxylose (gaxilose): a multicentre, open-label, phase IIB–III nonrandomized trial. J Clin Gastroenterol. 2014;48(1):29–36. [DOI] [PubMed] [Google Scholar]
- 8. Simren M, Stotzer PO. Use and abuse of hydrogen breath tests. Gut. 2006;55(3):297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gasbarrini A, Corazza GR, Gasbarrini G, Montalto M, Di Stefano M, Basilisco G, Parodi A, Usai-Satta P, Vernia P, Anania C et al.. Methodology and indications of H2-breath testing in gastrointestinal diseases: the Rome Consensus Conference. Aliment Pharmacol Ther. 2009;29 Suppl 1:1–49. [DOI] [PubMed] [Google Scholar]
- 10. Perets TT, Hamouda D, Layfer O, Ashorov O, Boltin D, Levy S, Niv Y, Dickman R. Small intestinal bacterial overgrowth may increase the likelihood of lactose and sorbitol but not fructose intolerance false positive diagnosis. Ann Clin Lab Sci. 2017;47(4):447–51. [PubMed] [Google Scholar]
- 11. Pimentel G, Burton KJ, Rosikiewicz M, Freiburghaus C, von Ah U, Munger LH, Pralong FP, Vionnet N, Greub G, Badertscher R et al.. Blood lactose after dairy product intake in healthy men. Br J Nutr. 2017;118(12):1070–7. [DOI] [PubMed] [Google Scholar]
- 12. Coelho AI, Berry GT, Rubio-Gozalbo ME. Galactose metabolism and health. Curr Opin Clin Nutr Metab Care. 2015;18(4):422–7. [DOI] [PubMed] [Google Scholar]
- 13. Trimigno A, Münger L, Picone G, Freiburghaus C, Pimentel G, Vionnet N, Pralong FP, Capozzi F, Badertscher R, Vergères G. GC-MS based metabolomics and NMR spectroscopy investigation of food intake biomarkers for milk and cheese in serum of healthy humans. Metabolites. 2018;8(2):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Münger LH, Trimigno A, Picone G, Freiburghaus C, Pimentel G, Burton KJ, Pralong FP, Vionnet N, Capozzi F, Badertscher R et al.. Identification of urinary food intake biomarkers for milk, cheese, and soy-based drink by untargeted GC-MS and NMR in healthy humans. J Proteome Res. 2017;16(9):3321–35. [DOI] [PubMed] [Google Scholar]
- 15. Burton KJ, Rosikiewicz M, Pimentel G, Butikofer U, von Ah U, Voirol MJ, Croxatto A, Aeby S, Drai J, McTernan PG et al.. Probiotic yogurt and acidified milk similarly reduce postprandial inflammation and both alter the gut microbiota of healthy, young men. Br J Nutr. 2017;117(9):1312–22. [DOI] [PubMed] [Google Scholar]
- 16. Pimentel G, Burton KJ, von Ah U, Bütikofer U, Pralong FP, Vionnet N, Portmann R, Vergères G. Metabolic footprinting of fermented milk consumption in serum of healthy men. J Nutr. 2018;148(6):851–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Burton KJ, Pimentel G, Zangger N, Vionnet N, Drai J, McTernan PG, Pralong FP, Delorenzi M, Vergeres G. Modulation of the peripheral blood transcriptome by the ingestion of probiotic yoghurt and acidified milk in healthy, young men. PLoS One. 2018;13(2):e0192947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dunn WB, Broadhurst D, Begley P, Zelena E, Francis-McIntyre S, Anderson N, Brown M, Knowles JD, Halsall A, Haselden JN et al.. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat Protoc. 2011;6(7):1060–83. [DOI] [PubMed] [Google Scholar]
- 19. Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, Fan TW, Fiehn O, Goodacre R, Griffin JL et al.. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics. 2007;3(3):211–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ekstrom C. MESS: Miscellaneous Esoteric Statistical Scripts. R package version 0.3-2[Internet] 2014. Available from: https://github.com/cran/MESS. [Google Scholar]
- 21. Noguchi K, Gel YR, Brunner E, Konietschke F. nparLD: an R software package for the nonparametric analysis of longitudinal data in factorial experiments. J Stat Softw. 2012;50:1–12.25317082 [Google Scholar]
- 22. Kim S. ppcor: an R package for a fast calculation to semi-partial correlation coefficients. Commun Stat Appl Methods. 2015;22(6):665–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dzialanski Z, Barany M, Engfeldt P, Magnuson A, Olsson LA, Nilsson TK. Lactase persistence versus lactose intolerance: is there an intermediate phenotype?. Clin Biochem. 2016;49(3):248–52. [DOI] [PubMed] [Google Scholar]
- 24. Enattah NS, Kuokkanen M, Forsblom C, Natah S, Oksanen A, Jarvela I, Peltonen L, Savilahti E. Correlation of intestinal disaccharidase activities with the C/T-13910 variant and age. World J Gastroenterol. 2007;13(25):3508–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ingram CJ, Raga TO, Tarekegn A, Browning SL, Elamin MF, Bekele E, Thomas MG, Weale ME, Bradman N, Swallow DM. Multiple rare variants as a cause of a common phenotype: several different lactase persistence associated alleles in a single ethnic group. J Mol Evol. 2009;69(6):579–88. [DOI] [PubMed] [Google Scholar]
- 26. Itan Y, Jones BL, Ingram CJ, Swallow DM, Thomas MG. A worldwide correlation of lactase persistence phenotype and genotypes. BMC Evol Biol. 2010;10:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rezaie A, Buresi M, Lembo A, Lin H, McCallum R, Rao S, Schmulson M, Valdovinos M, Zakko S, Pimentel M. Hydrogen and methane-based breath testing in gastrointestinal disorders: the North American Consensus. Am J Gastroenterol. 2017;112(5):775–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Keiding S, Johansen S, Winkler K, Tonnesen K, Tygstrup N. Michaelis-Menten kinetics of galactose elimination by the isolated perfused pig liver. Am J Physiol. 1976;230(5):1302–13. [DOI] [PubMed] [Google Scholar]
- 29. Merkel C, Marchesini G, Fabbri A, Bianco S, Bianchi G, Enzo E, Sacerdoti D, Zoli M, Gatta A. The course of galactose elimination capacity in patients with alcoholic cirrhosis: possible use as a surrogate marker for death. Hepatology. 1996;24(4):820–3. [DOI] [PubMed] [Google Scholar]
- 30. Ficicioglu C, Hussa C, Gallagher PR, Thomas N, Yager C. Monitoring of biochemical status in children with Duarte galactosemia: utility of galactose, galactitol, galactonate, and galactose 1-phosphate. Clin Chem. 2010;56(7):1177–82. [DOI] [PubMed] [Google Scholar]
- 31. Szumilo T. Purification and properties of D-galactonate dehydratase from Mycobacterium butyricum. Biochim Biophys Acta. 1981;661(2):240–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.