Abstract
Tibetan sheep are the most common and widespread domesticated animals on the Qinghai–Tibetan Plateau (QTP) and have played an essential role in the permanent human occupation of this high-altitude region. However, the precise timing, route, and process of sheep pastoralism in the QTP region remain poorly established, and little is known about the underlying genomic changes that occurred during the process. Here, we investigate the genomic variation in Tibetan sheep using whole-genome sequences, single nucleotide polymorphism arrays, mitochondrial DNA, and Y-chromosomal variants in 986 samples throughout their distribution range. We detect strong signatures of selection in genes involved in the hypoxia and ultraviolet signaling pathways (e.g., HIF-1 pathway and HBB and MITF genes) and in genes associated with morphological traits such as horn size and shape (e.g., RXFP2). We identify clear signals of argali (Ovis ammon) introgression into sympatric Tibetan sheep, covering 5.23–5.79% of their genomes. The introgressed genomic regions are enriched in genes related to oxygen transportation system, sensory perception, and morphological phenotypes, in particular the genes HBB and RXFP2 with strong signs of adaptive introgression. The spatial distribution of genomic diversity and demographic reconstruction of the history of Tibetan sheep show a stepwise pattern of colonization with their initial spread onto the QTP from its northeastern part ∼3,100 years ago, followed by further southwest expansion to the central QTP ∼1,300 years ago. Together with archeological evidence, the date and route reveal the history of human expansions on the QTP by the Tang–Bo Ancient Road during the late Holocene. Our findings contribute to a depth understanding of early pastoralism and the local adaptation of Tibetan sheep as well as the late-Holocene human occupation of the QTP.
Keywords: Tibetan sheep; argali; introgression, HBB; RXFP2; adaptation; human expansion
Introduction
Together with human migration, particularly of nomads (Phillips 2001), domestic sheep (Ovis aries) have spread almost worldwide after their domestication in the Fertile Crescent ∼9,500–10,000 years before the present (BP) (Zeder 2008). During their domestication and subsequent dispersal, sheep and other major livestock species have precipitated the socioeconomic transformation from a hunter-gatherer lifestyle to seminomadic and nomadic pastoralism, and to sedentary agricultural settlements in human history (Chang and Koster 1986; Larson and Fuller 2014; Gaunitz et al. 2018). Given the comigration of humans and domesticated animals such as dog, sheep, cattle, and pig, tracing the expansions of domestic animals can inform the peopling patterns of humans (Larson et al. 2010; Wang et al. 2013; Decker et al. 2014; Zhao et al. 2017; Ní Leathlobhair et al. 2018). The Qinghai–Tibetan Plateau (QTP) forms the high-altitude core of Asia with an average elevation >4,000 m, encompassing an area of nearly 2.5 million km2. Prehistoric human colonization on the QTP can be traced back to at least 20,000 years BP. During the process, large-scale intermittent population expansions have occurred at various stages of the Holocene, facilitated by the advent of human technology (e.g., microliths) and agriculture (e.g., domesticated millet, barley, wheat, and sheep) (Chen et al. 2015; Qiu 2015; Zhang et al. 2016; Zhao et al. 2017). The novel agropastoral lifeway in the late Holocene enabled either permanent human occupation of the QTP (under the model of agriculture-facilitated occupation of the Tibetan Plateau after 3,600 years BP; Chen et al. 2015) or substantial population growth on the plateau (under the model of preagropastoral occupation of the central Tibetan Plateau at ∼7,400 years BP; Meyer et al. 2017), particularly the higher-elevation zones (e.g., above 2,000–3,000 m). However, the history of early human migrations, including the timing, route, and number of expansions, remains unclear and poorly understood because of the paucity of archeological data (Chen et al. 2015; Meyer et al. 2017).
Previous investigations have indicated that Tibetan sheep originated from northern China during the late Holocene, between 1,800–4,500 years BP (Lv et al. 2015; Yang et al. 2016; Zhao et al. 2017). Since then, Tibetan sheep have played an important role in the Tibetan pastoral economy, culture, and society (Chang and Koster 1986; Liu et al. 2016). Currently, Tibetan sheep are the most numerous livestock (>20 million) (Du 2011) on the QTP. They live and migrate closely with humans, providing a wide variety of raw sources for items such as food, clothing, fuel, transportation, and religious totems and ornaments for the indigenous population (i.e., Tibetan people) (Xian 2005; Chen et al. 2006; Peng 2010; Müller et al. 2012; Liu et al. 2016). Hence, the spread of Tibetan sheep onto the QTP represents an important episode in the late-Holocene human occupation of the plateau.
Genetic mechanism underlying high-altitude adaptation is a topic of great interest in recent years, and has important implications for understanding adaptation in extreme environments and hypoxia-related diseases (Qiu et al. 2012; Yang et al. 2016). Whole-genome selective scans have identified positive selection on a set of genes (e.g., EPAS1, EPO, EGLN1, EGLN2, and EGLN3) in the HIF-1 (hypoxia-induced factors) pathway associated with hypoxia response (Frede and Fandrey 2013) in humans (e.g., Tibetan, Simonson et al. 2010; Andean, Wolfson et al. 2017) and animals (e.g., Tibetan mastiff, Miao et al. 2017; Tibetan pig, Zhang et al. 2017; Tibetan schizothoracine fish, Xu et al. 2016; Tibetan sheep, Yang et al. 2016) living at the high elevations. Also, metabolic processes (e.g., energy metabolism) have been invoked to explain the high-altitude adaptation in animals such as yak (Qiu et al. 2012), Tibetan antelope (Ge et al. 2013), ground tit (Qu et al. 2013), and sheep (Yang et al. 2016). During their spread throughout the QTP, Tibetan sheep have adapted to various local environmental conditions, and three different ecotypes (oula, valley, and grassland sheep) have been developed (Du 2011; Liu et al. 2016). Additionally, particular morphological features (e.g., horn size and shape) have been shaped through strong artificial selection driven by local religious and cultural preferences. Only recently have the genetic origin and high-altitude adaptation of Tibetan sheep been studied (Liu et al. 2016; Wei et al. 2016; Yang et al. 2016; Zhao et al. 2017). However, previous investigations were heavily limited by a few sampling sites, small numbers of samples, and/or sparse genetic markers, and thus could not accurately characterize the whole-genome landscape or clearly reveal the local demographic history of Tibetan sheep.
Here, we generated a large genomic data set of domestic (1,083 samples from 45 Chinese sheep populations) and wild (four argali [O. ammon] and one European mouflon [O. musimon]) sheep. In particular, the data set consisted of whole-genome sequences, single nucleotide polymorphism (SNP) arrays, mitochondrial DNA (mtDNA), and Y-chromosomal variations from 986 Tibetan sheep throughout their distribution range across the QTP (fig. 1A and supplementary table S1, Supplementary Material online). We integrated these data with publicly available genomic data (Ovine SNP50K BeadChip or Ovine Infinium HD SNP BeadChip genotypes of 15 argali and 983 samples from 44 Chinese native sheep populations; 832 mtDNA fragments of Chinese native sheep; and Y-chromosomal variations in 34 argali and 587 Chinese native sheep samples; supplementary table S1, Supplementary Material online) (Zhang et al. 2014; Lv et al. 2015; Zhao et al. 2017). We also compiled previous ethnohistorical documentation and archeological records of domestic animals (e.g., sheep) and cultivated plants (e.g., barley and millet) on the QTP (supplementary table S2, Supplementary Material online). Based on the synthesis of genomic, archeological, and anthropological data, we aimed to 1) characterize the genomic landscape of different ecotypes of Tibetan sheep throughout their distribution range, 2) describe a refined picture of the genetic and demographic history of Tibetan sheep, 3) detect the genomic traces left by centuries of natural and artificial selection and adaptive introgression from argali in the genomes of Tibetan sheep, and 4) reveal the late-Holocene expansions of early sheep pastoralism and associated Tibetan people across the QTP.
Fig. 1.
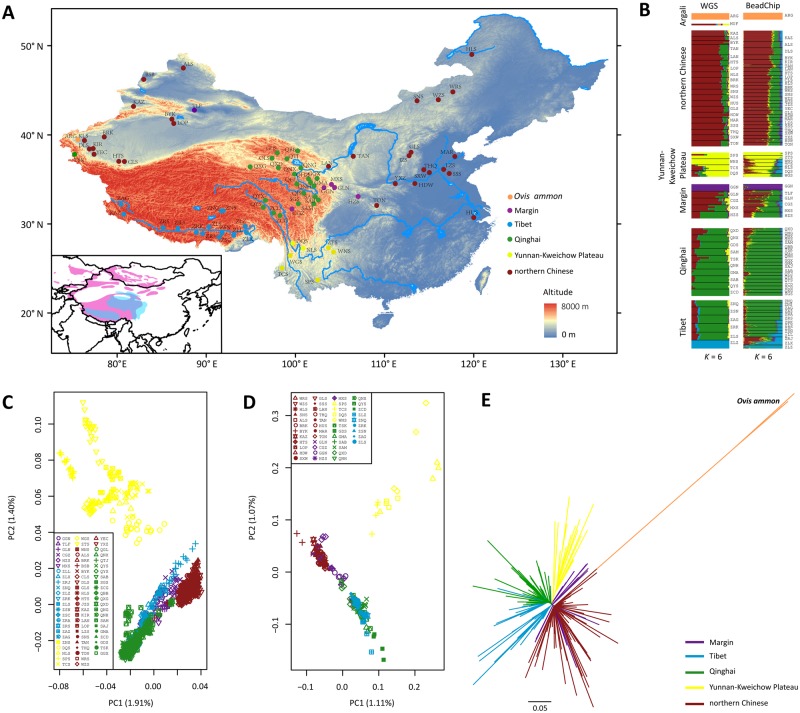
Geographic locations and population genetic structure of the sheep populations studied. (A) Sampling sites and geographic distribution of Chinese sheep and argali (Ovis ammon). The scaled graph in the lower left indicates the distribution region of Tibetan sheep (in skyblue) and argali (in pink). (B) Population genetic structure of Chinese sheep and particularly Tibetan sheep inferred from the ADMIXTURE analyses (K = 6) using whole-genome sequences (left) and Ovine SNP50K arrays (right). (C) Plots of principal components 1 and 2 from PCA analysis of 1,495 Chinese sheep individuals using the Ovine SNP50K arrays. (D) Plots of principal components 1 and 2 from PCA analysis of 181 Chinese sheep individuals using whole-genome sequences. (E) Neighbor-joining phylogenetic tree of 1,495 Chinese sheep individuals based on Reynolds’ distance estimated from the Ovine SNP50K array data, with O. ammon as the outgroup. See supplementary table S1, Supplementary Material online, for the abbreviations of the sheep populations.
Results and Discussion
Whole-Genome Sequencing and SNP Arrays
We generated whole-genome sequences of 181 domestic sheep and 5 wild sheep, totaling ∼21 billion raw reads and ∼2,078 Gb of aligned high-quality data with an average depth of 6.5× (supplementary table S3, Supplementary Material online). After SNP calling and subsequent stringent quality control (see Materials and Methods), we obtained 44,296,018 high-quality SNPs for all 186 individuals, with a range of 6,019,569–14,518,382 SNPs for each individual (supplementary table S4, Supplementary Material online). On average, 87.22% of the SNPs identified in the 186 individuals were confirmed in the sheep dbSNP database (supplementary table S5, Supplementary Material online). Of the 186 individuals, 109 have also been genotyped using the Illumina Ovine SNP50K BeadChip. Of the common SNP sites in the 109 individuals (13,090—24,493 SNPs in each individual) that had been analyzed with both methods, an average of 92.42% (85.49–96.78%) consistency was observed for the SNP genotypes per individual, demonstrating the general high reliability of our SNP calls (supplementary table S5, Supplementary Material online). Following the SnpEff annotation, we found that the largest number of SNPs were located in intergenic region (23,287,939, 52.57%), followed by those in the intronic (15,931,804, 35.97%), upstream gene (2,261,964, 5.11%) and downstream gene (1,829,762, 4.13%) regions (supplementary table S6, Supplementary Material online). For the integrated data set of SNP arrays, 48,383 SNPs and 1,618 individuals (from one argali population (15 individuals) and 75 domestic sheep populations) were retained in the population genetics analysis (supplementary table S1 and supplementary note, Supplementary Material online).
Genomic Variability and Linkage Disequilibrium
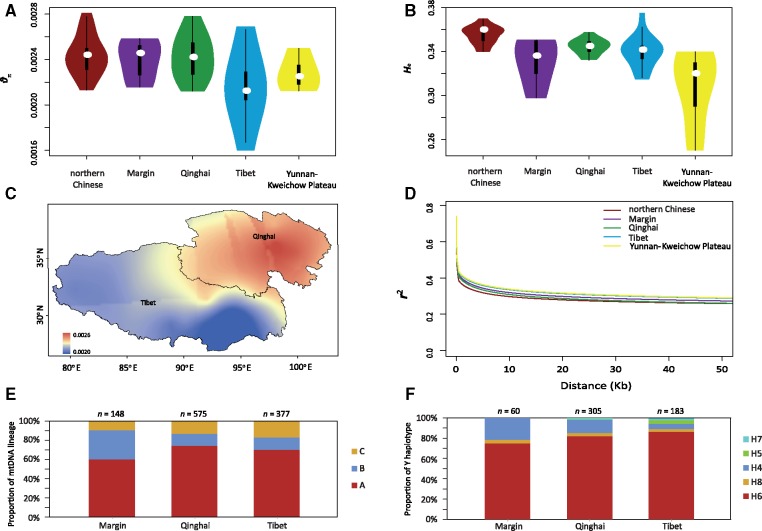
As represented by the genome-wide average θπ values from the sequences (fig. 2A and C) and the He values from the SNP arrays (fig. 2B and supplementary table S7, Supplementary Material online), the distribution of within-population genomic variability on the QTP and in the whole of China displayed a clear geographic pattern. Within the QTP, the margin and Qinghai subgroups of Tibetan sheep possessed clearly higher nucleotide diversity than the Tibet subgroup based on the θπ values (fig. 2A; see below the three subgroups of Tibetan sheep as inferred by the ADMIXTURE analysis). At the national scale, our results recapitulated previous findings (Yang et al. 2016; Zhao et al. 2017) that genomic variability was higher in the northern Chinese sheep compared with the Tibetan sheep and Yunnan–Kweichow sheep (fig. 2A and B). In contrast, we observed a lower level of genome-wide linkage disequilibrium (LD) in northern Chinese sheep and a higher level of LD in Yunnan–Kweichow sheep and the Tibet subgroup of Tibetan sheep (fig. 2D). Taken together, the observed clinal variations in LD and genomic variability from northern China to different areas on the QTP provided evidence for the fine-scale population structure of Tibetan sheep, suggesting the migration route of domestic sheep onto the QTP as inferred from the ADMIXTURE analysis below. Nevertheless, the differential LD and genomic variability among different sheep populations could be influenced by different husbandry practices. For example, Tibetan sheep populations have undergone long-term local breeding due to geographic isolation, whereas northern Chinese sheep populations have experienced extensive crossbreeding with the exotic fat-tailed sheep from Central Asia (Du 2011; Zhao et al. 2017). Also, the low level of genomic variability and high level of LD detected in the Tibet subgroup of Tibetan sheep could be explained by their lowest effective population size (see below the results of the population demographic history reconstruction) and possibly bottleneck and genetic drift during their expansions from Qinghai.
Fig. 2.
Geographic pattern of genetic diversity for Chinese sheep. (A) Nucleotide diversity (θπ) distributions in Chinese sheep groups and Tibetan sheep subgroups from various regions of China. The white dot indicates the median value of each group/subgroup. (B) Heterozygosity (He) distributions in Chinese sheep groups and Tibetan sheep subgroups from various regions of China. The white dot indicates the median value of each group/subgroup. (C) Synthetic maps illustrating the geographic variation in nucleotide diversity (θπ) in the Tibet and Qinghai subgroups of Tibetan sheep. (D) Linkage disequilibrium (LD) decay for five geographic groups/subgroups of populations measured by r2. (E) The proportions of mtDNA lineages in the three subgroups of Tibetan sheep (Tibet, Qinghai, and margin subgroups). (F) The proportions of Y-chromosome haplotypes in three subgroups of Tibetan sheep (Tibet, Qinghai, and margin subgroups).
Population Genetic Structure
We explored the population genetic structure of Tibetan sheep in the context of all Chinese sheep populations. In accordance with our earlier studies (Yang et al. 2016; Zhao et al. 2017), the principal component analysis (PCA), neighbor-joining (NJ) phylogeny, and ADMIXTURE analyses based on the SNP array and sequence data sets capitulated the major genetic division among the Chinese sheep populations from three large geographic regions: northern China, the Yunnan–Kweichow Plateau, and the QTP (fig. 1B–E, supplementary figs. S1–S3 and supplementary note, Supplementary Material online). Interestingly, our NJ phylogenetic analysis based on the SNP arrays (fig. 1E) supports a closer genetic affinity between Tibetan sheep and the Yunnan–Kweichow populations than that between Tibetan sheep and the northern Chinese populations (i.e., the ancestral population of Tibetan sheep), because the Yunnan–Kweichow sheep were clustered into the Tibetan sheep clade. This could be explained by the fact that the sheep populations on the QTP and the Yunnan–Kweichow Plateau represent the original thin-tailed sheep in China, while northern Chinese sheep have been greatly influenced by later introgressions of fat-tailed sheep from Central Asia and Mongolia (Zhao et al. 2017).
Within Tibetan sheep, the NJ tree based on the SNP arrays (fig. 1E) discerned a visible genetic differentiation among the three subgroups of Tibetan sheep from Qinghai, Tibet, and the marginal areas of the QTP. We noted that the Qinghai and Tibet subgroups were not fully separated clades, and a few individuals were roughly located in different branches on the NJ tree. In the analyses of sequences, FST estimates showed that the Tibet subgroup sheep had a closer relationship with the Qinghai subgroup than with the subgroup from the marginal areas of the QTP (supplementary table S8, Supplementary Material online). Further ADMIXTURE analyses showed no apparent difference in genetic makeup among most of the Tibetan sheep populations, but a gradual northeast-to-southwest decrease was observed in the genetic components represented by northern Chinese sheep in Tibetan sheep populations, with the margin subgroup showing the highest average proportions (45.84%), followed by a slightly higher level in the Qinghai subgroup (13.83%) than in the Tibet subgroup (12.12%). The observation supported a migration route of ancient sheep from northern China to the marginal areas of the QTP, then to Qinghai, and finally to Tibet during their colonization of the plateau. This migration route was also supported by the declining trend in the ancestral mtDNA lineages (lineages A and B) (Cai et al. 2007, 2011; Zhao et al. 2017) and Y-chromosomal haplotypes (haplotypes H4 and H6) (Zhang et al. 2014) in the three subgroups of the margin, Qinghai, and Tibet populations (fig. 2E and F, supplementary figs. S4–S8, supplementary tables S9–S13, and supplementary note, Supplementary Material online).
Population Demographic History Reconstruction
We used the approximate Bayesian computation (ABC) approach and whole-genome sequence data to reconstruct the population history of Tibetan sheep. We compared seven alternative demographic models (supplementary fig. S9, Supplementary Material online) to rigorously infer the most probable divergence pattern and migration route of the three subgroups of Tibetan sheep. Model-1 showed the best fit to the data because its Bayes factor was three times larger than those of the other six models (supplementary table S14, Supplementary Material online). Additionally, this model had the largest marginal density and a high capability of generating simulated data closest to the empirical data (supplementary fig. S10, Supplementary Material online), which provided strong statistical support for its superiority (supplementary note, Supplementary Material online).
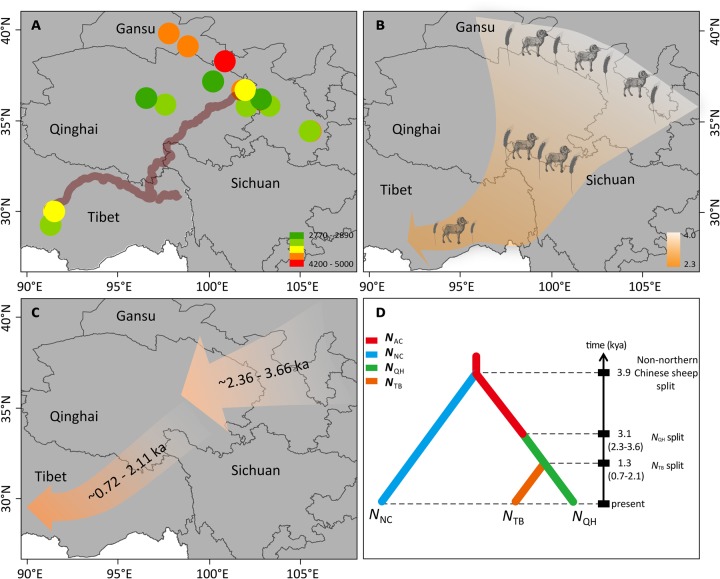
The best-supported Model-1 indicated a stepwise divergence pattern and a northeast-to-southwest migration route for Tibetan sheep. That is, the ancestral populations (NAC) first arrived in the northeastern area of the QTP and generated the Qinghai subgroup (NQH) ∼3,114 years ago (T1, 50% Highest Posterior Density [HPD]: 2,363‒3,666; supplementary table S15, Supplementary Material online). Subsequently, the NQH migrated southwest and generated the Tibet subgroup (NTB) ∼1,316 years ago (TTB, 50% HPD: 721‒2,110; supplementary table S15, Supplementary Material online). The estimates of effective population sizes were 77,306 (50% HPD: 55,306‒92,270), 46,189 (50% HPD: 31,820‒62,739), and 45,099 (50% HPD: 27,261‒65,712) for the NAC, NQH, and NTB populations, respectively (supplementary table S15, Supplementary Material online). Notably, we found that the estimated posterior distributions of all the parameters recaptured the empirical data in Model-1 (supplementary fig. S10, Supplementary Material online). Additionally, the Kolmogorov–Smirnov test showed an unbiased estimate for parameters GAMMA, NAC,NNC, and NTB, whereas estimates of the other parameters showed slight deviations from the uniform distribution (supplementary fig. S14, Supplementary Material online). Overall, these estimates implied the high credibility of our inferences on the demographic parameter in Model-1 (supplementary figs. S9–S14, supplementary tables S14–S18, and supplementary note, Supplementary Material online).
The Late-Holocene Human Occupation of the QTP
We compiled the evidence from a total of 62 archeological sites (supplementary table S2, Supplementary Material online) in previous literature and classified them into two main chronological stages within the late Holocene based on the delimitation of cultures, that is, 5,200–3,600 years BP in the late Neolithic culture (e.g., late Yangshao, Majiayao, and Qijia cultures) and 3,600–2,300 years BP in the Bronze Age culture (e.g., Kayue, Xindian, and Nuomuhong cultures) (Chen et al. 2015; Zhang et al. 2016). This synthesized evidence implied that during the late Neolithic (5,200–3,600 years BP), early human societies, mostly farming communities, inhabited the low-altitude northeastern marginal areas of the QTP and lived on a primary crop of millet (87.50% of all archeological sites) domesticated from the neighboring Loess Plateau (<2,500 m) (Guedes et al. 2014) and several domestic animals (e.g., sheep, cattle, pig, and dog; 28.13% of all archeological sites; supplementary table S2, Supplementary Material online). At the Bronze stage (3,600–2,300 years BP), humans colonized much higher elevations (>3,000 m) on the QTP and settled permanently, with the subsistence strategy replaced by growing cold-tolerant cereals such as barley and wheat (90.63% of all archeological sites) (Guedes and Butler 2014; Guedes et al. 2015) and more frequently raising domestic animals (e.g., sheep, cattle, pig, and dog; 34.38% of all archeological sites; supplementary table S2, Supplementary Material online). During this stage, the farming system was shifted from millet agriculture to a novel agropastoral system (Li 2012; Chen et al. 2015). In particular, sheep were the primary domestic animals accompanying humans during both the late Neolithic and Bronze stages (88.89% and 100% of all archeological sites containing animals). Therefore, the expansions of crops (e.g., millet, barley, and wheat) and domestic animals especially sheep occurred along with the permanent human occupation of the QTP, and accordingly the colonization history of Tibetan sheep could help to understand the early human expansions on the QTP.
Notably, our genomic inference regarding the demographic history of Tibetan sheep using whole-genome sequencing data and the ABC modeling framework was in good agreement with the archeological evidence presented above and the recorded human history in ancient China (fig. 3). More specifically, the inferred route for the spread of Tibetan sheep through the northeastern marginal area to the hinterland of the QTP (fig. 3) closely mirrored the routeway known as the Tang–Bo Ancient Road (Zhang et al. 2016), which extended from Shaanxi to Tibet via Gansu and Qinghai and was historically the easiest pathway for human occupation of the QTP (Chen et al. 2015; Zhang et al. 2016). Additionally, the inferred timing supported the hypothesis that sheep farming has facilitated permanent human residence on the QTP in the late Holocene era (fig. 3) (Chen et al. 2015). Interestingly, the inferred demographic history of Tibetan sheep revealed a two-step pattern for their colonization onto the QTP through an initial movement from northern China to the northeastern QTP (∼3,100 years BP), followed by a later expansion from the northeastern to the southwestern QTP (∼1,300 years BP) (fig. 3). Climate change could be the major driving force that prompted the colonization of sheep and associated settlement of humans on the QTP because it is renowned for its great impacts on biotic communities and human colonization worldwide, usually associated with latitudinal or altitudinal shifts in species distributions (Parmesan and Yohe 2003; deMenocal and Stringer 2016; Han et al. 2016). In particular, the QTP is a region highly sensitive to climate change (Yao et al. 2000), and the increasingly cold and dry climatic conditions after 3,600 years BP favored the expansions of alpine meadow vegetation (Marcott et al. 2013; Chen et al. 2015; Madsen 2016), which could enable the spread of sheep farming and human settlement on the QTP. Moreover, the establishment of cold-adapted agropastoral economy, including sheep, barley, and wheat, enabled the prehistoric humans to colonize the higher-elevation QTP during the cold and dry climate period after 3,600 years BP (Chen et al. 2015; Zhang et al. 2016). Due to a lack of archeological information, little is known about the settlement process on the QTP after 2,000 years BP. Here, our modeling inference for the second stage of colonization by Tibetan sheep ∼1,300 years BP provided evidence of human expansion and settlement during this period. In addition to climate change, demographic pressure was also a potential driver of the human expansion after 2,000 years BP because the cold-adapted agropastoral system yielded high productivity and led to a significant population increase in the northeastern margin of the QTP (Han et al. 2016). Thus, our findings contribute to a better understanding of the timing and phases of early human settlement on the QTP.
Fig. 3.
Integrated diagram of the inferred demographic history of the late-Holocene human and sheep populations on the Qinghai-Tibetan Plateau (QTP). (A) Distribution and approximate dates of prehistoric archeological sites of sheep in the QTP and surrounding areas. The thick brown line indicates the Tang–Bo Ancient Road (Chen et al. 2015; Zhang et al. 2017). (B) A proposed demographic scenario for the development of agriculture on the QTP based on archeological records of domestic animals (e.g., sheep, cattle, pig, and dog) and crops (e.g., millet, barley, and wheat) (supplementary table S2, Supplementary Material online), with sheep and wheat as representative species shown on the figure. (C) A proposed demographic scenario of sheep on the QTP based on genetic data in this study. (D) The best-supported demographic model of Chinese sheep populations inferred from the approximate Bayesian computation (ABC) approach.
Adaptive Introgression from Argali in Tibetan Sheep
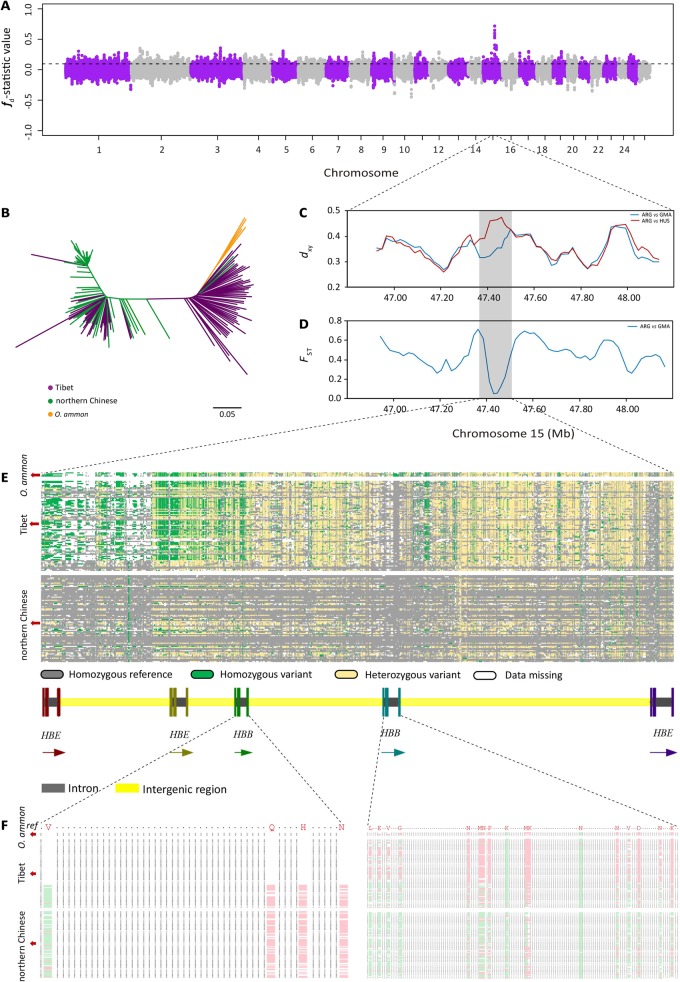
The TreeMix and f3-statistic analyses based on the SNP arrays identified eight Tibetan sheep populations (i.e., QNG in TreeMix, fig. 4D; QXD, ZLS, QNX, ZRJ, GGX, QTJ, and QXG in f3-statistics, fig. 4E) as being admixed with argali. To more comprehensively and accurately test for genomic evidence of potential argali introgression in Tibetan sheep, we employed whole-genome sequences to calculate the efficient D statistics (Green et al. 2010) for each combination of Tibetan sheep and argali using the form [[[HUS, TIB], ARG], Bighorn]. In the form, Bighorn (i.e., Bighorn sheep) represents the outgroup, ARG (i.e., argali) represents the candidate introgressor, and TIB (i.e., Tibetan sheep) and HUS (i.e., Hu sheep) refer to the tested and reference domestic sheep populations from high and low altitudes, respectively. The results showed that genomic segments of argali have introgressed into the genomes of Tibetan sheep, with a significant signal observed in 36 out of 1,472 groups (D = 0.0101–0.0146; Z > 3; supplementary table S19, Supplementary Material online). To further locate the introgressed genomic regions in the genomes of Tibetan sheep, we computed the modified f-statistic (fd) value (Martin et al. 2015) for each 100-kb window (with a 20-kb step) across the genomes. Based on the cutoff P < 0.05 and the combination of overlapped introgressed regions, we detected 972–1,098 nonoverlapping significantly introgressed genomic tracts from argali for each Tibetan sheep population (supplementary table S20, Supplementary Material online), which have a total length of 136,910,467–151,541,991 bp and cover ∼5.23–5.79% of the whole genomes (fig. 5A, supplementary fig. S15 and supplementary table S21, Supplementary Material online). As a putatively introgressed tract could be a product of either genetic introgression or incomplete lineage sorting (Huerta-Sánchez et al. 2014), we estimated the probability of incomplete lineage sorting for the introgressed regions identified above (supplementary table S20, Supplementary Material online). Using a generation time of 3 years (Zhao et al. 2017) and a recombination rate of 1.0 × 10−8 (Kijas et al. 2012) for sheep and a divergence time of 2.36 Ma between argali and sheep (Yang et al. 2017), the expected length of a shared ancestral tract was L = 1/(1.0 × 10−8 × [2.36 × 106/3]) = 127.12 bp and the probability of a length of at least 80,377 bp (i.e., the observed introgressed regions were 80,377–479,930 bp) was 1 − GammaCDF(80,377, shape = 2, rate = 1/L) = 0. Thus, the identified introgressed regions in Tibetan sheep were unlikely to be due to incomplete lineage sorting. The proportion of 5.23–5.79% introgression is higher than that observed in Eurasian people (1–4% from Neanderthals) (Green et al. 2010), but lower than the upper bounds reported for North American brown bears (3–8% from polar bear) (Liu et al. 2014; Cahill et al. 2015) and canid (1.4–27.3% from Taimyr wolf to dog; up to 25% from dog to Eurasian wolf) (Skoglund et al. 2015; Fan et al. 2016).
Fig. 4.
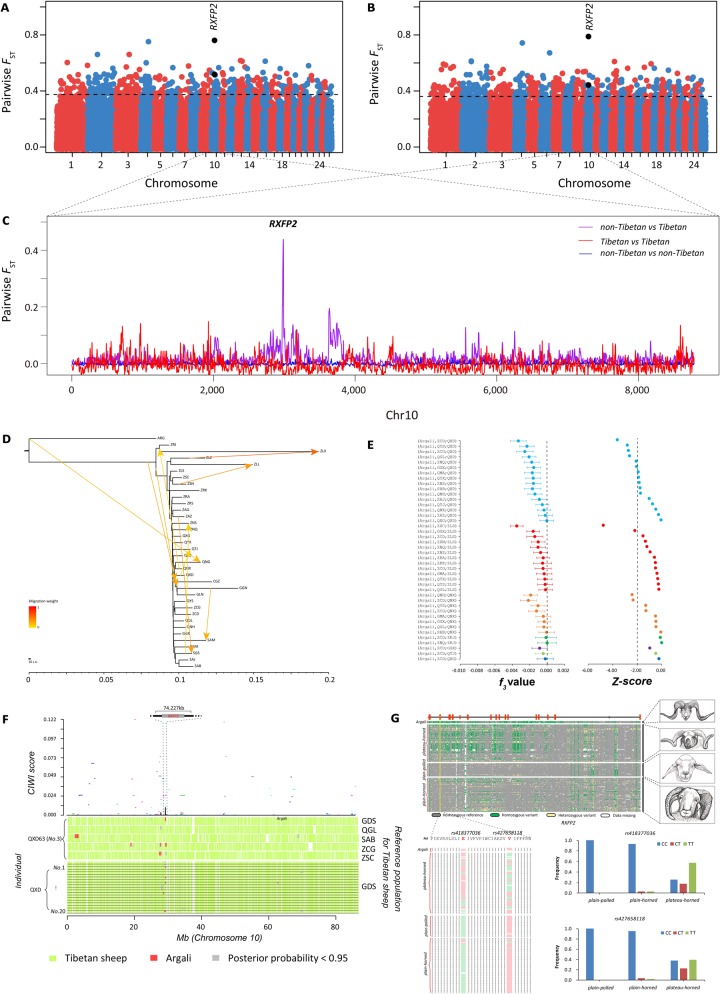
Identification of genomic regions at the RXFP2 locus introgressed into Tibetan sheep from argali. (A) Genome-wide distribution of pairwise FST values between plain-polled and plateau-horned sheep computed by the Ovine SNP50K arrays (see detailed information in supplementary table S36, Supplementary Material online). (B) Genome-wide distribution of pairwise FST values between plain-horned and plateau-horned sheep computed by the Ovine SNP50K arrays (see detailed information in supplementary table S37, Supplementary Material online). (C) Smoothed FST values between the Tibetan and non-Tibetan sheep populations across chromosome 10. (D) Inferred neighbor-joining phylogenetic tree showing the genetic relationships among Chinese sheep populations with 10 migration edges. ARG (Ovis ammon) was used as the outgroup to root the tree. (E) Outgroup f3 statistics between ARG and all Tibetan sheep populations using the Ovine SNP50K arrays, with significant results (f3 value < 0) shown on the figure. (F) Evidence of introgression at the RXFP2 locus identified by PCAdmix and CIWI approaches. The distribution of the concordance score (A-scores) and CIWI score across chromosome 10 is plotted at the top of the panel and indicates the concordance of ancestry assignment among 20 individuals from a Tibetan sheep population (Dulan population, QXD). The A-scores estimated by five alternative reference populations (Guide [GDS], Maqin [QGL], Hongyuan [SAB], Gonjo [ZCG], and Comai population [ZSC]) are displayed by five colored segments, and the CIWI score inferred from the A-score is shown by the black line. Distribution of PCAdmix results for QXD63 (No. 3) across chromosome 10 is shown on the middle of the panel. PCAdmix results of QXD63 (No. 3) were estimated by five alternative reference populations (GDS, QGL, SAB, ZCG, and ZSC). The distribution of PCAdmix results of 20 individuals from QXD across chromosome 10 is plotted at the bottom of the panel. GDS was used as a reference population to perform the PCAdmix analysis. (G) The patterns of genotypes of the introgressed genomic region (chr10: 29,435,112–29,509,145) among 165 domestic sheep and 4 argali as well as the horn type phenotype (also see supplementary fig. S21, Supplementary Material online) are shown on the top. The alignment of protein sequences from the RXFP2 genes from 165 domestic sheep and 4 argali is plotted at the lower left. The amino acids of two nonsynonymous substitutions (rs418377036 and rs427658118) in the sheep reference genome Oar v.4.0 are marked in red. Amino acids identical to or different from those in the reference genome are indicated by green and pink, respectively. The genotype frequency distributions of rs418377036 and rs427658118 among sheep groups with different horn phenotypes are plotted at the lower right.
Fig. 5.
Identification of genomic regions at the HBB locus introgressed into Tibetan sheep from argali. (A) Genome-wide distribution of fd values calculated for 100-kb windows with a 20-kb step across the genome for a Tibetan sheep population (Gansu Maqu population, GMA). Each dot represents a 100-kb window and the dashed line indicates the significance threshold (P < 0.05). (B) Neighbor joining (NJ) tree of 165 domestic sheep individuals from northern China and Tibet and 4 argali individuals based on pairwise p-distances using the SNPs in the introgressed genomic region (chr15: 47,400,001–47,500,000). (C) Mean pairwise sequence divergence (dxy) of the introgressed genomic region on chromosome 15 between argali and two Chinese sheep populations. The Hu sheep (HUS) and Gansu Maqu (GMA) populations represent northern Chinese sheep from the low-altitude region and Tibetan sheep with signatures of argali introgression, respectively. (D) Population differentiation (FST) around the introgressed genomic region on chromosome 15 between argali and Gansu Maqu (GMA) populations. (E) The pattern of genotypes among 165 domestic sheep and 4 argali of the introgressed genomic region (chr15: 47,431,866–47,485,446). (F) Alignment of protein sequences in the HBB genes from 165 domestic sheep and 4 argali. The amino acids of 21 nonsynonymous substitutions in the sheep reference genome Oar v.4.0 are marked in red. Amino acids identical to or different from those in the reference genome are indicated by green and pink, respectively.
To identify the most significantly introgressed genomic regions, we extracted the blocks showing the top 10 fd values for each of the 19 Tibetan sheep populations tested, and then pinpointed the compilation of the top 10 blocks across all 19 populations (supplementary table S22, Supplementary Material online). Functional annotation of the 43 genes located within the most significant genomic regions using DAVID v.6.7 (Huang et al. 2009a, 2009b) revealed major Gene Ontology (GO) enrichment associated with blood oxygen metabolism (GO:0005833, hemoglobin complex, P = 1.26E-06; GO:0005344, oxygen transporter activity, P = 3.77E-06; GO:0019825, oxygen binding, P = 5.78E-06; GO:0020037, heme binding, P = 1.74E-03) and olfactory transduction (oas04740, olfactory transduction, P = 8.28E-04; GO:0004984, olfactory receptor activity, P = 1.62E-03) (supplementary table S23, Supplementary Material online). These GO categories are biologically relevant to the high-altitude adaptation of Tibetan sheep because they imply potential beneficial changes in either the oxygen transportation system or olfactory sensation. Oxygen transportation system is relevant to the rates of blood flow for oxygen delivery and the subsequent supply of oxygen to cells (Beall 2007), thus could provide an adaptive response to the high-altitude hypoxia environment for Tibetan sheep. Olfactory sensation is involved in the sensing of external environments for scent-mediated chemical communication (Qiu et al. 2012), which is critical for Tibetan sheep to get adaptation to an increased number of predators (e.g., wolf) in the high-altitude alpine meadow. These adaptive mechanisms have been recognized in many species such as human (Simonson et al. 2010), yak (Qiu et al. 2012), Tibetan antelope (Ge et al. 2013), Tibetan wild boar (Li et al. 2013), and Tibetan mastiff (Miao et al. 2017) on the QTP.
In particular, we focused on the top introgressed genomic region encompassing chr15: 47,400,001–47,500,000 because it showed the highest and significant (P < 0.05) fd values in 14 of the 19 Tibetan sheep populations tested (supplementary table S22, Supplementary Material online). We inspected the region to confirm that it is in fact a product of genetic introgression as opposed to shared ancestral polymorphism. We computed the mean pairwise sequence divergence (dxy) between the argali and either the Tibetan sheep or the northern Chinese sheep (e.g., Hu sheep in East China) from low-altitude region. The results showed that the dxy value in the focused region was significantly reduced in Tibetan sheep but highly elevated in the northern Chinese sheep (fig. 5C and supplementary fig. S16A, Supplementary Material online), which is expected under a scenario of genetic introgression. We also estimated the genetic differentiation (FST) between argali and Tibetan sheep, and found that the FST value was lower in the top introgressed genomic region than in the rest of the genome (fig. 5D and supplementary fig. S16B, Supplementary Material online). Additionally, the NJ phylogenetic tree based on the pairwise genetic distance (i.e., p-distance) of the top introgressed genomic region displayed a clear divergence pattern in which most Tibetan sheep were much closer to argali (O. ammon) than to the northern Chinese sheep (fig. 5B). Hence, these results collectively supported that the shared pattern of the focused region was due to argali introgression into Tibetan sheep.
To further dissect the genetic and functional characteristics of the top introgressed genomic region, we compared the genotypes of the SNPs in the region for argali, Tibetan sheep, and northern Chinese sheep. Notably, we found that the genotype pattern of Tibetan sheep was highly similar to that of argali but strikingly different from that of the northern Chinese sheep (fig. 5E). Functional annotation analysis further revealed that this region encompassed the genes HBE (hemoglobin subunit epsilon) (Jiang et al. 2015) and HBB (hemoglobin subunit beta) (Manca et al. 2006). Within the HBB gene, 21 SNPs showed changes in the amino acid sequence (e.g., Val2Met, Gln40Arg), codon (e.g., GTG > ATG, CAG > CGG), and the nucleotide sequence (e.g., 47448175 G>A, 47448419 A>G) as well as significant differences in the frequencies of specific alleles between the high-altitude sheep (Tibetan sheep, n = 84) and the low-altitude sheep (northern Chinese sheep breeds in the low-altitude region, n = 81) (fig. 5F and supplementary table S24, Supplementary Material online), suggesting the importance of HBB gene for sheep living in the high-altitude, low-oxygen environment. In fact, the beta-globin genes are known to play an important role in hypoxia adaptation because of their crucial functions in oxidative metabolism (Aguileta et al. 2004; Opazo et al. 2008). In particular, previous works have widely reported that variants in the HBB gene could alter the oxygen affinity of hemoglobin in Tibetans (Yi et al. 2010) and high-altitude animals (Natarajan et al. 2013; Projecto-Garcia et al. 2013; Miao et al. 2017), and thus facilitate the adaptive response to hypoxia. Additionally, the introgression of the HBB gene from wild to domestic animals has been observed in several species such as cattle (Bos taurus) (Tanaka et al. 2011) and Tibetan mastiff (Canis lupus familiaris) (Miao et al. 2017). These pieces of evidence suggested that this introgression from argali could have improved the oxygen transport properties of Tibetan sheep and enabled them to rapidly adapt to the hypoxic plateau environment.
We also tested for genetic introgression using the “Consistently Introgressed Windows of Interest (CIWI)” approach (Barbato et al. 2017) based on the SNP arrays and identified 1,956 genes that exhibited strong introgression signals in five or more Tibetan sheep populations (supplementary tables S25–S28 and supplementary note, Supplementary Material online). To explore the introgression signals common to the SNP array and sequencing data, we extracted the genomic regions that showed the top 10% fd values in at least five Tibetan sheep populations (i.e., the same criterion as the SNP arrays) from the genetic introgression test based on sequences (supplementary note, Supplementary Material online). This extraction yielded 7,209 argali-introgressed genomic regions harboring 1,590 genes from the sequencing data. Interestingly, a large number (i.e., 37–362; supplementary table S29, Supplementary Material online) of these genes were common to the introgressed genes reported for other domesticated animals from the phylogenetically related species (Chen et al. 2018; Wu et al. 2018), including genes related to nervous system (e.g., CNTN5, PTPRZ1, and UNC5C) and body shape (e.g., GNPDA2, CPE, and SGCB) and hypoxia-associated genes (e.g., NOS2, IL1A, and ANGPT1) in the HIF-1 pathway (supplementary table S29, Supplementary Material online). For the 263 genes shared between the sequencing (263/1,590 = 16.54%) and SNP array (263/1,956 = 13.45%) data (supplementary table S30, Supplementary Material online), we performed GO annotation analysis and found that the main functional enrichments of these genes were associated with nervous system, brain and muscle development, obesity phenotype and diabetic retinopathy (supplementary table S31, Supplementary Material online), which could be explained by the importance of a quick response to external environment, muscle function, body shape, and visual perception for Tibetan sheep to cope with the harsh environment on the QTP, as indicated in a recent study (Pan et al. 2018). There is a discrepancy between the two sets of introgression genes, which could be resulted from the differences in methodologies, density of genetic markers, and sample sizes.
Among the common genes detected in the two different data sets, RXFP2, a major gene for sheep horn status (e.g., presence or absence, morphology in domestic and wild sheep) (Kijas et al. 2012; Johnston et al. 2013; Kardos et al. 2015; Wiedemar and Drögemüller 2015; Pan et al. 2018), is prominent in both introgression tests (fig. 4F). Additionally, the SNP genotypes in a specific genomic region (chr10: 29,435,112–29,481,215) of the RXFP2 gene exhibited patterns that were clearly similar between argali and Tibetan sheep (e.g., plateau-horned), but different from those in the low-altitude sheep (e.g., plain-horned and plain-polled; fig. 4G). Moreover, two missense variants, rs418377036 (Glu667Lys/1999 C>T) and rs427658118 (Val653Met/1957 C>T), were found to have significantly differentiated genotype frequencies among the plain-polled, plain-horned, and plateau-horned populations (fig. 4G). Further characterization of the genotypes of the two missense SNPs in the wild and domestic sheep populations (argali, n = 4; plain-polled sheep, n = 24; plain-horned sheep, n = 62; and plateau-horned sheep, n = 68) showed a more frequent T allele in the high-altitude sheep (e.g., plateau-horned Tibetan sheep), which may serve as an advantage allele for sheep living in the high-altitude environment.
Hybridization between Tibetan sheep and argali has been a common breeding practice adopted by local nomad communities (Zhu 2010). Both argali and Tibetan sheep show the phenotypes of large and spiral horns compared with the relatively small horns in the low-altitude sheep populations (fig. 4G) (Subbotin et al. 2007; Du 2011; Yang et al. 2017). Our finding indicated that the large and spiral horn phenotype and the associated major gene RXFP2 in Tibetan sheep are most likely derived from the argali introgression. Interestingly, a recent study on Chinese sheep detected signals of rapid evolution of the horn-related RXFP2 gene in semiferal Tibetan sheep populations and proposed an alternative explanation that semiferalization is the major factor responsible for the large and spiral horn phenotype and the unique RXFP2 haplotype observed (Pan et al. 2018). Regardless of the potential different origins of the large and spiral horns, this phenotype is favored by the local Tibetan people because of its important religious and cultural roles (e.g., totem and trophy) throughout history (Fan 1131; Huang 1989; He 2000). Additionally, the large horn size is an advantageous phenotype for sheep to obtain more food resources through intraspecific or interspecific competitions in general and for rams to outcompete for more courtships in particular (Preston et al. 2003; Bro-Jorgensen 2007).
Selective Signatures Associated with High-Altitude Adaptation in Sheep
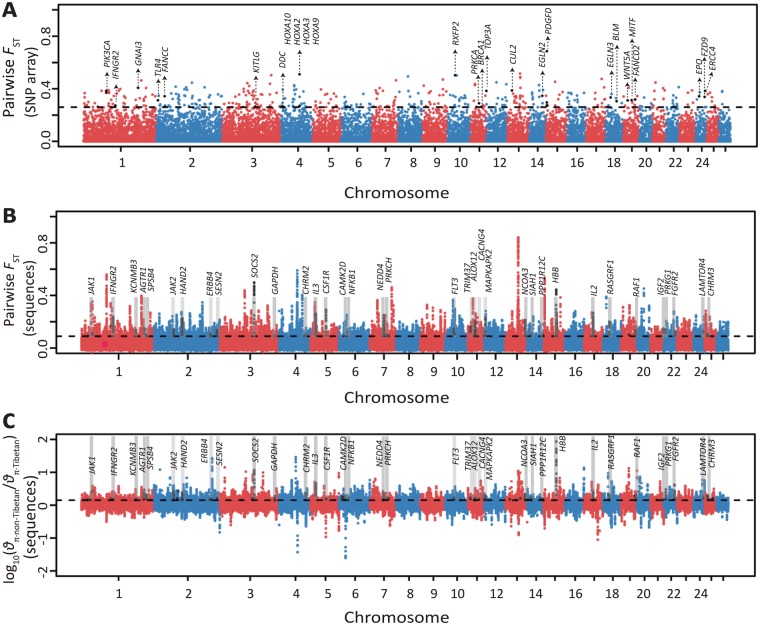
To capture potential genes under divergence selection associated with high-altitude adaptation, we estimated the genome-wide FST values (Weir and Cockerham 1984) between Tibetan sheep on the QTP and northern Chinese sheep from low-altitude region (i.e., defined as non-Tibetan sheep) based on the SNP array data of 1,323 samples from 61 sheep populations (supplementary table S32, Supplementary Material online). With a top 1% FST cutoff (FST ≥ 0.26), we detected a total of three hundred and two 300-kb genomic regions comprising 921 candidate genes (fig. 6A and supplementary table S33, Supplementary Material online). Strong selective signals (FST ≥ 0.2722) were highlighted in eight genes (EPO, TLR4, PIK3CA, PRKCA, EGLN3, EGLN2, IFNGR2, and CUL2; fig. 6A) known to be important components of the HIF-1 (hypoxia-induced factors) pathway (Frede and Fandrey 2013), indicating potential roles for these genes in high-altitude hypoxia adaptation in Tibetan sheep. Specifically, one prominent candidate gene EPO encodes a secreted glycosylated cytokine and binds to the erythropoietin receptor to control erythrocyte production. This gene was reported to play a role in successful vascular adaptation in pregnant Andean women (Wolfson et al. 2017) and in the hypoxia adaptation of a Tibetan schizothoracine fish (Xu et al. 2016) at high altitudes. Furthermore, the genes EGLN2 and EGLN3, which belong to the EGLN family, are cellular oxygen sensors that catalyze and hydroxylate HIF alpha proteins. The EGLN family was demonstrated to be involved in EPO production and erythropoiesis in murine zygotes (Fisher et al. 2009), and the transcriptomic and proteomic changes of EGLN3 were linked to high-altitude hypoxic adaptation in Tibetan pig (Zhang et al. 2017).
Fig. 6.
Manhattan plot of selective sweeps in the Tibetan sheep. (A) Genome-wide distribution of pairwise FST values between Tibetan sheep and non-Tibetan sheep computed using the Ovine SNP50K arrays (see detailed information in supplementary table S33, Supplementary Material online). The threshold value corresponding to the top 1% of the FST value (FST = 0.26) is shown as the horizontal dashed line. The black dots indicate the significant sites that harbor candidate positively selected genes. (B) Genome-wide distribution of pairwise FST values between Tibetan sheep and non-Tibetan sheep computed using the sequence data set (see detailed information in supplementary table S34, Supplementary Material online). (C) Distribution of log10(θπ·ratios) between non-Tibetan sheep and Tibetan sheep estimated by the sequence data set (see detailed information in supplementary table S34, Supplementary Material online). The top 5% genome-wide significant threshold values of pairwise FST (FST = 0.0846) and log10(θπ·non-Tibetan/θπ·Tibetan) [log10(θπ ratio) = 0.1502] for the sequence data set are shown separately by black dashed lines in (B) and (C), respectively.
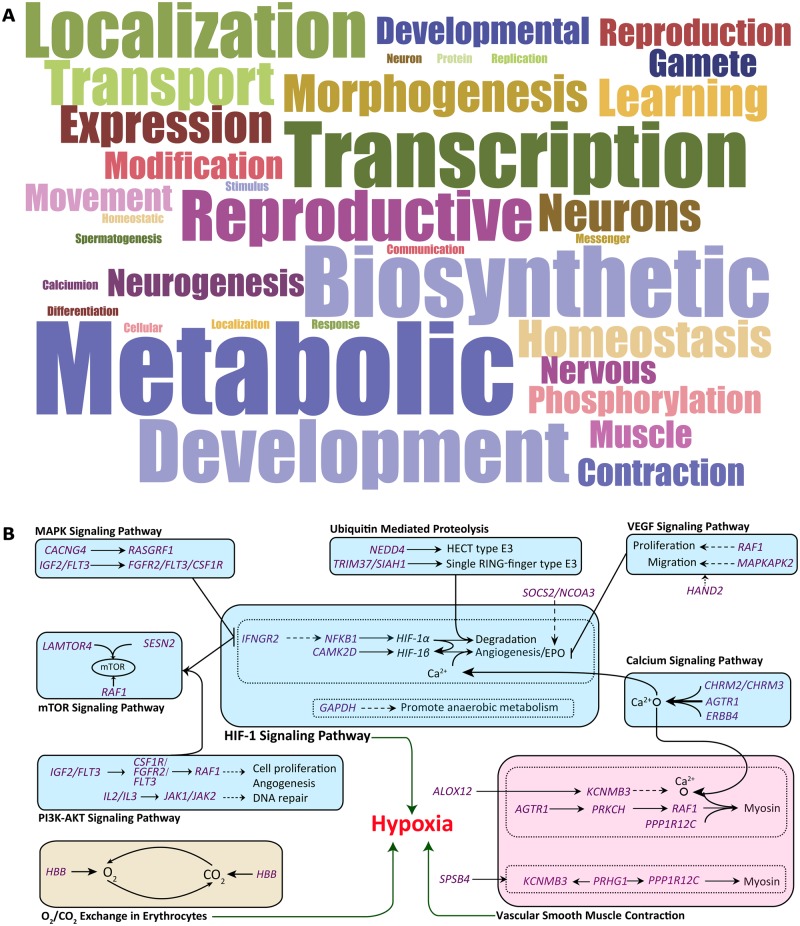
Based on the sequencing data, a whole-genome selection scan in 12 representative sheep populations from the QTP (i.e., Tibetan sheep) and the low-altitude regions (i.e., non-Tibetan sheep) identified a total of 3,194 genomic regions (FST ≥ 0.0846, log10(θπ·non-Tibetan/θπ·Tibetan) ≥ 0.1502; fig. 6B and C), encompassing 747 candidate genes (supplementary table S34, Supplementary Material online) under positive selection in Tibetan sheep. Notably, 114 of the argali-introgressed genes (e.g., HBB, HBE, and RXFP2) detected above were also identified to be positively selected (supplementary table S35, Supplementary Material online), providing strong evidence for the adaptive role of such introgressed genes in high-altitude adaptation. From the word cloud analysis, major enrichments of the biological process GO terms of the candidate genes were observed to be associated with metabolic process, biosynthetic process, transcription regulation, and nervous system development and function (fig. 7A), which have been reported to affect high-altitude environment adaptation in humans and animals (Storz et al. 2009, 2010; Frede and Fandrey 2013; Natarajan et al. 2013; Projecto-Garcia et al. 2013; Wang et al. 2014; Yang et al. 2016; Miao et al. 2017). Among the candidate genes obtained from the sequencing data, we highlighted a list of genes (e.g., HBB, IFNGR2, GAPDH, CAMK2D, NFKB1, SOCS2, NCOA3, and MITF; supplementary table S34 and supplementary note, Supplementary Material online) that were most likely associated with the adaptation of sheep to high altitude. These genes are functionally related to the hypoxia-induced HIF-1 pathway (Frede and Fandrey 2013; Yang et al. 2016), O2/CO2 exchange in erythrocytes (Carlson et al. 2008), and resistance to ultraviolet radiation (Hornyak et al. 2009). For instance, the gene SOCS2 was recently shown to interact closely with the hypoxic EPO response in Tibetan sheep (Yang et al. 2016), and its potential role in high-altitude adaptation was implicated in Tibetan goat (Wang et al. 2016) as well. Additionally, we found that 35 candidate genes are located in the central positions of the HIF-1 signaling and its surrounding pathways (including the ubiquitin mediated proteolysis pathway and the VEGF, MAPK, mTOR, P13K-AKT, and calcium signaling pathways) and the pathways of vascular smooth muscle contraction and O2/CO2 exchange in erythrocytes (fig. 7B). The complex pattern of pathway interactions provided further in-depth insight into the molecular mechanisms underlying the high-altitude hypoxia adaptation of Tibetan sheep (supplementary note, Supplementary Material online).
Fig. 7.
Functional enrichments of the candidate positively selected genes detected in the Tibetan sheep. (A) Word cloud summarizing 117 biological process terms with significant (P < 0.05) enrichments from the GO analysis of 747 candidate genes under positive selection in the Tibetan sheep. These GO terms are mainly related to metabolic process, biosynthetic process, transcription regulation, and nervous system development and function. (B) Proposed mechanisms of signaling pathways for hypoxia adaptation in sheep. Positively selected genes in the Tibetan sheep are indicated in purple. Pathways related to hypoxia-induced factors (HIF-1) signaling and its surrounding pathway, O2/CO2 exchange, and vascular smooth muscle contraction are indicated in light blue, light yellow, and light pink, respectively.
In the selection test between Tibetan sheep and northern Chinese sheep at the low altitudes (i.e., non-Tibetan sheep) based on the SNP arrays, we also detected strong selective signals in a few genes (e.g., RXFP2, BMP2, PDGFD, VRTN, and HOXA gene family; fig. 4A–C, supplementary fig. S17, supplementary table S33 and supplementary note, Supplementary Material online) associated with two iconic morphological features, horn shape and tail configuration, which could be due to long-term artificial and natural selection and the adaptive introgression from argali as described above. Apart from these genes, several novel genes (e.g., EVX1, APOA1BP, and APOA2) and GO categories and pathways (e.g., the GO terms appendage morphogenesis, limb morphogenesis, embryonic skeletal system development and cellular response to oxygen-containing compound, and the regulation of lipolysis in adipocytes and regulation of actin cytoskeleton pathways) were also detected in the pairwise selection tests between populations with different horn and tail phenotypes (supplementary fig. S17, supplementary tables S36–S42 and supplementary note, Supplementary Material online). Interestingly, the homologous genes of APOA1BP and APOA2 (e.g., APOB and APOE) have recently been reported to affect lipid levels in humans (Lu et al. 2017), which are functionally similar to the roles of APOA1BP and APOA2 in the fat deposition in sheep tail.
Selective Signatures Associated with Ecotypes
In the analyses comparing Tibetan sheep populations of the three ecotypes (supplementary fig. S18 and supplementary tables S1 and S32, Supplementary Material online) based on the SNP array data set, we observed much higher FST values between the grassland and valley sheep than between the oula and valley sheep (supplementary figs. S19A and S20A and supplementary tables S43 and S44, Supplementary Material online). Interestingly, we detected body weight-associated candidate genes in both oula (e.g., FGF21 and SLC37A3) (Benton et al. 2015; Talukdar et al. 2016) and grassland (e.g., GHR, CPN1, and LEP) (Saeed et al. 2015; Johns et al. 2017; Lin et al. 2018) sheep. Further genomic characterization of the ecotype-associated selective signals based on the sequence data revealed 371 and 360 candidate selected genomic regions harboring 167 and 154 genes in the grassland and oula populations, respectively (supplementary figs. S19B, S19C, S20B, and S20C and supplementary tables S45 and S46, Supplementary Material online). Notably, we found several highlighted genes that are functionally related to body growth and energy metabolism in the genomes of both grassland (body growth: PRKAA2, GHR, NR3C1, and SOCS7; energy metabolism: GBE1 and NCOA2) (Martens et al. 2005; Tang et al. 2011; Lu et al. 2015; Said et al. 2016; Lin et al. 2018) and oula sheep (body growth: PRKAA2, ESR1, and SOX9; energy metabolism: ADIPOQ) (Zhou et al. 2006; Razquin et al. 2010; te Winkel et al. 2010; Tang et al. 2011).
In the functional enrichments of the candidate genes, we found that the significantly enriched GO term regulation of anatomical structure morphogenesis (GO:0022603, P-value = 0.0035) and growth (GO:0040007, P-value = 0.0364; supplementary tables S47 and S48, Supplementary Material online) in the oula genome appeared to be functionally relevant to body growth. Thus, these results suggest a breeding practice of local communities targeted at traits such as the body size and weight of the grassland and oula sheep, which have long been serving as the main meat-use animals for Tibetans (Guo et al. 2017). This could further imply that the phenotypic differences among the different ecotypes of Tibetan sheep were most likely the result of the artificial selective forces rather than environmental adaptive pressures as previously thought (Du 2011).
Conclusions
We performed the first comprehensive and in-depth genomic investigation of Tibetan sheep. Population genomics analyses have unveiled their genomic variability, population structure, demographic history (e.g., timing and route of population colonization of Tibetan sheep), adaptive introgression from argali, and phenotypic variation. Coupled with the archaeological or genetic evidence of humans, plants, and animals on the QTP, our results revealed that permanent human occupation of the QTP occurred in the late Holocene through the Tang–Bo Ancient Road (∼3,100 years BP, from northern China to the northeastern areas of the QTP; and ∼1,300 years BP, from the northeastern areas to southwestern areas of the QTP), likely triggered by climate change, the advent of the agropastoral economy, and the increasing human population. Additionally, our results add the evolutionary mechanisms of adaptive gene introgression from argali into Tibetan sheep (e.g., HBB, HBE, and RXFP2) accounting for their rapid local adaptation. The newly generated genome-wide data are valuable resources for the future genetic improvement of Tibetan sheep and comparative genomic analyses to elucidate the common genetic mechanisms underlying high-altitude genetic adaptation across species.
Materials and Methods
Sample Collection and DNA Extraction
We sampled blood or ear tissue from a total of 1,083 unrelated individuals representing 65 populations of Chinese native sheep (fig. 1A and supplementary table S1, Supplementary Material online), including 81 samples from 20 populations in northern China, 16 from 4 populations from the Yunnan–Kweichow Plateau, and 986 from 41 populations of Tibetan sheep from the QTP. These samples of Tibetan sheep represent the most comprehensive sampling throughout their distribution range and the largest sample size for this local breed in any investigation to date. In addition, four argali (O. ammon) individuals from the Pamirs in Xinjiang were collected following the Wildlife Protection Law of China (supplementary table S1, Supplementary Material online). The unrelatedness of the domestic sheep samples was ensured by knowledge from local herdsmen and pedigrees. The sample information, such as species/population names and codes, sampling sites and sample sizes, is summarized in figure 1A and supplementary table S1, Supplementary Material online. All the sampled tissues were preserved in 95% ethanol and stored at −80 °C until DNA extraction. Genomic DNA was extracted from the blood or ear samples using a standard phenol–chloroform method (Sambrook and Russell 2001).
mtDNA Sequencing and Y-Chromosomal Genotyping
A 749-bp fragment of the mtDNA control region was amplified by polymerase chain reaction (PCR). Detailed information on the primers, PCR protocols, sequencing, and sequence quality trimming is provided by Lv et al. (2015). After aligning these newly generated sequences of Tibetan sheep (n = 914, MH261383–MH262296) and publicly available sequences of Chinese sheep (n = 832), we obtained a 422-bp partial sequence of the control region in 1,746 individuals for use in subsequent analyses, including the estimation of sequence variation, the detection of population expansion signatures, and haplotype network construction (supplementary figs. S4–S7, supplementary tables S9–S11, and supplementary note, Supplementary Material online).
We genotyped 458 rams from 33 Tibetan sheep populations (supplementary table S1, Supplementary Material online) using three Y-chromosomal genetic markers (two SNPs oY1 and oY2 and one microsatellite SRYM18) (Meadows et al. 2006; Zhang et al. 2014). The protocols for SNP and microsatellite genotyping are detailed by Zhang et al. (2014). By combining the genotypes obtained here and those available publicly (Zhang et al. 2014), we constructed the haplotypes and calculated the haplotype diversity (supplementary fig. S8, supplementary tables S12 and S13, and supplementary note, Supplementary Material online).
SNP BeadChip Genotyping and Statistical Analyses
A total of 620 samples representing 31 populations of Tibetan sheep were genotyped with the Illumina Ovine SNP50K (54,241 SNPs) BeadChip. Details of the genotyping procedures for the SNP BeadChip can be found by Kijas et al. (2012). We applied the same procedures and criteria for quality control as used by Kijas et al. (2012) and Zhao et al. (2017). We combined this newly generated data set with publicly available Ovine SNP50K data (Zhao et al. 2017), and the integrated genotypes of 998 individuals (from 1 argali population and 75 Chinese native sheep populations) were included in the analyses, which included the estimation of within-population genetic variability, characterization of population structure (e.g., model-based clustering, PCA, and phylogenetic relationship reconstruction), detection of selection signatures, and identification of gene flow between species or populations (supplementary note, Supplementary Material online).
Whole-Genome Sequencing
For whole-genome resequencing, we selected 186 representative individuals from the whole set of samples based on the Ovine SNP50K BeadChip data, comprising 81 animals of 20 populations from northern China, 16 animals of 4 populations from the Yunnan–Kweichow Plateau, 84 animals from Tibetan sheep, 4 argali (O. ammon), and 1 European mouflon (O. musimon) (supplementary table S1, Supplementary Material online). We used >18 µg (i.e., >20 ng/µl and >900 µl) of genomic DNA per sample to construct the libraries. DNA was sheared into fragments of 200–800 bp with the Covaris system (Covaris, Woburn, MA). The fragments were then treated following the Illumina DNA sample preparation protocol, with a process of end repaired, A-tailed, ligated to paired-end adaptors and PCR amplification with 350-bp inserts. The constructed libraries were sequenced on the Illumina HiSeq X Ten platform (Illumina, San Diego, CA) for 150-bp paired-end reads. All samples were sequenced at ∼6.5× coverage.
Sequence Alignment and Variant Calling
Raw sequence reads were discarded when they contained ≥10% unidentified nucleotides, ≥ 50% low-quality read bases (Q-score < 5), ≥10 nucleotides overlapped with the adapter sequences, or duplicate reads (Yang et al. 2016). The program Burrows-Wheeler Aligner v.0.7.4 (BWA-MEM algorithm) was then used to map the clean reads to the sheep reference genome Oar v.4.0 with the default parameters. The mapped reads were further filtered by removing multiply aligned reads, and the remaining high-quality reads were kept for variant calling. We used the software Genome Analysis Toolkit (GATK) v.3.3.0 (McKenna et al. 2010) to call SNPs according to previous recommendations (Li and Durbin 2010; Nielsen et al. 2011; Van der Auwera et al. 2013). In more detail, RealignerTargetCreator and BaseRecalibrator tools were employed to detect misalignments and recalibrate base quality scores, and then HaplotypeCaller and GenotypeGVCFs algorithms were jointly used to call variants. Moreover, to minimize the influence of sequencing and mapping bias, variant sites with QD < 2.0; FS > 60.0; MQ < 40.0; HaplotypeScore > 13.0; MappingQualityRankSum < −12.5; ReadPosRankSum < −8.0 were discarded. Sites showing an extremely low (<2×) or high average coverage (>15×) were also filtered out. Following the SNP calling, the toolbox SnpEff v.4.3 (Cingolani et al. 2012) was used for SNP annotations.
To assess the accuracy of our SNP calling, the SNPs identified here were compared with those from Build 147 of the sheep dbSNP database of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/SNP, last accessed September 11, 2018). Moreover, the genotypes of the SNPs were compared with those on the Illumina Ovine SNP50K BeadChip array (Illumina) for 109 individuals with array data available. The SNP called using the GATK approach was used in subsequent analyses of LD, demographic reconstruction, selection signature detection, and genomic introgression.
ANGSD Analyses
To avoid potential errors caused by the relatively low sequencing depth in the analyses of genomic diversity and population genetic structure such as PCA, population structuring, and NJ trees, SNP discovery was also performed based on the genotype likelihoods (GLs) calculated from the program ANGSD v.0.915 (Korneliussen et al. 2014), which can take genotype uncertainty into account. In summary, the SAMTools model (-GL 1) (Li et al. 2009) implemented in ANGSD was used to compute the GLs after mapping the reads, with the parameters “-uniqueOnly 1 -remove_bads 1 -only_proper_pairs 1 -trim 0 -C 50 -baq 1 -minMapQ 30 -minQ 20.” A maximum likelihood method was then employed to infer major/minor states based on the GLs, which were further used to estimate allele frequency based on Kim’s method (Kim et al. 2011). SNP discovery was subsequently implemented with a likelihood ratio test statistic for the χ2 distribution of the allele frequencies and a P-value threshold of 1 × 10−3.
Genomic Diversity, Linkage Disequilibrium, and Population Genetic Structure
The nucleotide diversity (θπ) was calculated based on a maximum likelihood estimate of the folded Site Frequency Spectrum (SFS) (Kim et al. 2011) using a sliding window approach (50-kb windows with 10-kb steps). The SFS was computed through a two-step procedure implemented in the program ANGSD v.0.915 (Korneliussen et al. 2014): 1) The site allele frequency likelihood was calculated and the associated files (.saf) were generated by the option “-dosaf 1”; and 2) the allele frequency likelihood files (.saf) were optimized using the realSFS program to compute the SFS. To assess the genome-wide LD pattern, the program PopLDdecay v.3.29 (https://github.com/BGI-shenzhen/PopLDdecay, last accessed September 11, 2018) (Li et al. 2018) was used to calculate the average r2 value with the default parameters.
For the estimation of population structure, PCA was performed using the ngsTools suite implemented in the program ANGSD v.0.915, which accounted for the genotype uncertainties of relatively low-coverage sequencing data. The covariance matrix between individuals was approximated by weighting the posterior probability of each genotype, and eigenvalues for the covariance matrix were generated with the function “eigen” in the R platform. Additionally, phylogenetic NJ tree was constructed based on the pairwise genetic distances estimated using NgsDist (Vieira et al. 2016), and population structuring was inferred using NGSadmix (Skotte et al. 2013), which was run with a GL-based maximum likelihood method and the number of assumed ancestral populations K ranging from 2 to 10. In addition, the FST values between the subgroups of Tibetan sheep were estimated using Weir and Cockerham’s method (Weir and Cockerham 1984), with nonoverlapping 50-kb windows sliding in 10-kb increments across the genome.
Demographic History Reconstruction
Fine-scale population genetic structure analysis indicated that Tibetan sheep could be divided into three genetic subgroups, which generally corresponded to their geographic origins (i.e., Qinghai, Tibet, and the marginal areas of the QTP) (fig. 1). Intriguingly, the subgroups showed apparent differences in whole-genome genetic variability (fig. 2A–C), mtDNA haplotype frequency (fig. 2E), and Y-chromosomal constitution (fig. 2F). To elucidate the demographic history of Tibetan sheep, we employed the ABC approach and whole-genome sequences to test seven competing divergence pattern scenarios. According to the evidence from ancient DNA (Cai et al. 2007, 2011) and the previously inferred demographic history of Chinese sheep (Yang et al. 2016; Zhao et al. 2017), in our model, we set the ancestral sheep (NAC) as an unsampled “ghost” population that first arrived in China ∼4,800 years BP and subsequently evolved into northern Chinese (NNC) and nonnorthern Chinese groups ∼3,900 years ago. In Model-1 and Model-2, we assumed a stepwise divergence pattern, in which the nonnorthern Chinese group first gave rise to one of the main subgroups of Tibetan sheep (i.e., Qinghai [NQH] or Tibet [NTB]), and the other main subgroup was later derived from the earlier established subgroup (supplementary fig. S9, Supplementary Material online). In Model-3, Model-4, and Model-5, both main subgroups of Tibetan sheep (NQH and NTB) were supposed to be derived from the nonnorthern Chinese group, but the timing of the divergences in each model was set to be different (supplementary fig. S9, Supplementary Material online). In Model-6 and Model-7, the nonnorthern Chinese group first gave rise to one of the main subgroups of Tibetan sheep (NQH or NTB), and later, the other main subgroup was formed by an admixture between the earlier established subgroup and the northern Chinese group (NNC) (supplementary fig. S9, Supplementary Material online). The samples and genetic data set included in the modeling, detailed description of the scenarios, and specific steps of the modeling procedures are presented in the supplementary information (supplementary figs. S9–S14, supplementary tables S14–S18, and supplementary note, Supplementary Material online).
Genome-Wide Scan of Selective Signatures
Given the low density of SNP markers in the Ovine SNP50K BeadChip data (supplementary note, Supplementary Material online), we performed an in-depth scan of selection signals associated with high-altitude adaptation in the whole-genome sequences. Twelve representative populations from Tibetan sheep and northern Chinese breeds in the low-altitude region (i.e., defined as non-Tibetan sheep; supplementary table S32, Supplementary Material online) were selected for the analysis. We screened the sheep genomes with nonoverlapping 50-kb windows sliding in 10-kb increments, and estimated the FST and θπ ratio (θπ·non-Tibetan/θπ·Tibetan) values for each window using vcftools v.0.1.13 (Danecek et al. 2011). The overlapped windows within the top 5% of the FST and θπ ratio (log10-transformed) (Yang et al. 2016) empirical distributions were considered as the candidate selective regions and were subsequently examined for the candidate genes. The candidate genes were then assigned to known KEGG pathways (http://www.kegg.jp, last accessed September 11, 2018) involved in hypoxia adaptation to reveal potential molecular mechanisms. To glean the main information from the functional enrichment results, an online Wordcloud generator (https://www.jasondavies.com/wordcloud/, last accessed September 11, 2018) was applied to create a word cloud for the significantly (P-value < 0.05) enriched GO terms.
We also employed the whole-genome sequencing data to explore potential selective signals between populations from different ecotypes (e.g., grassland sheep vs. valley sheep and oula sheep vs. valley sheep; supplementary table S32, Supplementary Material online) within the Tibetan sheep. We implemented the same analyses as above by calculating the FST and θπ ratio values per window for the tested pairwise populations. The threshold for selective signatures was set as the top 1% of the FST and θπ ratio empirical distributions. GO annotation and KEGG pathway analyses of the selective genes were implemented using DAVID v.6.7 (Huang et al. 2009a, 2009b).
Whole-Genome Analysis of Genomic Introgression
We used Patterson’s D statistic (Green et al. 2010; Durand et al. 2011) to detect high-altitude sheep that shared more derived alleles with argali than with low-altitude sheep. In a typical tree topology [[[P1, P2], P3], O] with “O” as the outgroup, the null hypothesis is that P1 and P2 shared the same ancestor with P3, and that there was no gene flow between P3 and either P2 or P1 after their ancestors split from each other. If the results deviated from the null hypothesis (e.g., gene flow occurred), a positive D value would be expected when P2 shared more derived alleles with P3 and a negative D value when P1 and P3 had more common alleles.
In this study, we used the D statistic [[[HUS, TIB], ARG], Bighorn sheep], in which Bighorn sheep (O. canadensis) is the outgroup (SRA accession number: SRR501858), ARG (i.e., argali) represents the candidate introgressor, and HUS (i.e., Hu sheep) and TIB (i.e., Tibetan sheep) represent the domestic populations at low and high altitudes, respectively. We tested for the signatures of argali introgression in the genomes of 19 Tibetan sheep populations and used a 5-Mb block size to calculate the D value, and the standard error was measured using the weighted block jackknife approach (Martin et al. 2015). The statistical significance of the D value was evaluated using a two-tailed Z-test, with an absolute Z-score larger than 3 considered to be significant (Durand et al. 2011). Furthermore, we examined the locations of introgressed argali genomes in Tibetan sheep with a modified fd value (Martin et al. 2015; Teng et al. 2017), using a 100-kb sliding window with 20-kb increments across the genome. The P-value was evaluated from the Z-transformed fd value, and a region with P < 0.05 was defined as a significantly introgressed genomic region (Teng et al. 2017).
Since the shared genetic components between species could result from either introgression or common ancestry, we further verified the top introgressed genomic region (see Results) by the statistics of mean pairwise sequence divergence (dxy). According to the theoretical expectation, introgression would cause reduced dxy in the target regions, whereas shared ancestry would not lead to lower dxy values (Martin et al. 2015). Also, the FST value of the top introgressed genomic region was calculated between Tibetan sheep and argali, and lower FST value would be expected in the introgressed genomic regions as compared with the adjacent nonintrogressed genomic regions. In addition, we constructed NJ trees for the domestic sheep and argali based on the pairwise genetic distances among the top introgressed genomic region estimated from the program NgsDist (Vieira et al. 2016).
To dissect the genomic architecture of the top introgressed genomic region (see Results), we extracted the SNP genotypes for the region from the whole-genome sequences and compared the pattern of SNP genotypes between argali, Tibetan sheep, and northern Chinese sheep breeds in the low-altitude region. Furthermore, we searched for nonsynonymous mutations representing putative functional variants within the candidate gene (HBB) in the top introgressed genomic region. In more detail, the genotypes of 2,471 SNPs within HBB (chr15: 47,431,866–47,485,446) were extracted from 165 domestic sheep (including 84 Tibetan sheep and 81 sheep from northern Chinese sheep breeds in the low-altitude region) and 4 argali. Twenty-one nonsynonymous SNP mutations were then identified, and the genotype frequency distribution and the amino acid and nucleotide changes of these SNPs were compared among the argali and different groups of domestic sheep. We also summarized the allele frequencies of missense mutations in the introgressed genomic regions for the different groups.
For the another prominent introgressed gene RXFP2 (e.g., a major gene for horn status; see Results), we extracted the genotypes of 896 SNPs for this gene (chr10: 29,435,112–29,509,145) from the resequencing data and compared the pattern of the genotypes between the domestic sheep (i.e., plateau-horned sheep, 68 individuals; plain-polled sheep, 24 individuals; plain-horned sheep, 62 individuals) and wild sheep (argali, four individuals) groups. Of these SNPs, two nonsynonymous SNP mutations within RXFP2 gene were identified and the genotypic distribution of the two SNPs was compared among different sheep groups. In addition, the allele frequencies of missense mutations in the introgressed genomic region of RXFP2 were summarized for different groups.
Probability of Incomplete Lineage Sorting
Following the methodology described by Huerta-Sánchez et al. (2014), we calculated the probability of the argali-introgressed tracts in Tibetan sheep due to incomplete lineage sorting. We let r be the recombination rate per generation per base pair in Tibetan sheep, m be the length of the introgressed tracts, and t be the length of argali and Tibetan sheep branches since divergence. The expected length of a shared ancestral sequence is L = 1/(r × t). The probability of a length of at least m is 1 − GammaCDF (m, shape = 2, r = 1/L), in which GammaCDF is the Gamma distribution function.
We calculated the length of the introgressed tracts (m) and filter for those that are too short (i.e., < L) to be confidently introgressed. Firstly, we combined the overlapped introgressed genomic regions to obtain nonoverlapping introgressed tracts. We measured the length for each introgressed tract via the distance between the starting SNP and ending SNP in the tract (supplementary table S20, Supplementary Material online). We then excluded the introgressed tracts shorter than L, and calculated the total length of the remaining introgressed tracts for each of the 19 Tibetan sheep populations tested (supplementary table S20, Supplementary Material online). We estimated the proportion of Tibetan sheep genome that is introgressed from argali using the total introgressed length divided by the sheep reference genome Oar v.4.0 (2,615,516,299 bp; supplementary table S21, Supplementary Material online).
Archeological and Genetic Evidence
To investigate the process and pattern of permanent human occupation on the QTP in the late Holocene (5,200–2,300 years BP) (Chen et al. 2015; Zhang et al. 2016), we compiled information from the published literature about prehistoric archeological sites on the QTP and its surrounding areas (Gansu and Sichuan provinces, China) (Aldenderfer 2011; Chen et al. 2015). Specifically, we performed an extensive literature review by searching archeological publications in “China National Knowledge Infrastructure” and “Web of Science” and original briefings that included the keywords millet, wheat, barley, and sheep (supplementary table S2, Supplementary Material online). Due to the paucity of archeological evidence for humans on the QTP in the late Holocene, we also collected reported genetic evidence of human population history on the QTP (supplementary table S49, Supplementary Material online). Then, we synthesized all the collected archeological and genetic evidence for humans and the domesticated species in terms of time, location, and the direction of movement, which revealed the temporal change in subsistence strategy, economic mode, and culture type during the late-Holocene human permanent occupation on the QTP. By integrating the previous evidence and the sheep demographic history inferred in this study, we expect to provide a full picture of prehistoric permanent human settlement on the QTP.
Data Access
The whole-genome sequencing, SNPs, mtDNA, and Y-chromosomal data reported in this study are available upon request for research purpose.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This study was funded by grants from the National Natural Science Foundation of China (Nos. 91731309 and 31272413), the Taishan Scholars Program of Shandong Province (No. ts201511085), the External Cooperation Program of Chinese Academy of Sciences (152111KYSB20150010), and Chinese Government contribution to CAAS-ILRI Joint Laboratory on Livestock and Forage Genetic Resources in Beijing. We thank Juha Kantanen from Nature Resources Institute Finland (Luke) for providing the European mouflon sample. We are grateful to the late Zhi-Fa Wang and a number of other persons for sampling. We thank Min Zhang for her help in DNA extraction, Ze-hui Chen for drawing the sketches of sheep horns, and Xue Ren, Rasmus Nielsen, Huajing Teng, Xin Li and Mario Barbato for their technical help with the statistical analyses. The paper contributes to the CGIAR Research Program on Livestock and Fish.
Author Contributions
M.-H.L. conceived and supervised the study. X.-J.H., X.-L.X., F.-H.L., and Y.-H.C. analyzed the data. J.Y., X.-J.H., X.-L.X., and M.-H.L. wrote the paper with contribution from J.-L.H., and F.-H.L. W.-R.L., M.-J.L., Y.-T.W., J.-Q.L., Y.-G.L., Y.-L.R., Z.-Q.S., F.W., E.-E.H., and J.-L.H. contributed samples or provided help during the sample collection. X.-J.H. and X.-L.X. conducted the laboratory work. All the authors reviewed and approved the final manuscript.
References
- Aguileta G, Bielawski JP, Yang Z.. 2004. Gene conversion and functional divergence in the β-globin gene family. J Mol Evol. 592: 177–189. [DOI] [PubMed] [Google Scholar]
- Aldenderfer M. 2011. Peopling the Tibetan Plateau: insights from archaeology. High Alt Med Biol. 122: 141–147. [DOI] [PubMed] [Google Scholar]
- Barbato M, Hailer F, Orozco-terWengel P, Kijas J, Mereu P, Cabras P, Mazza R, Pirastru M, Bruford MW.. 2017. Genomic signatures of adaptive introgression from European mouflon into domestic sheep. Sci Rep. 71: 7623.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall CM. 2007. Two routes to functional adaptation: Tibetan and Andean high-altitude natives. Proc Natl Acad Sci U S A. 104(Suppl 1): 8655–8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton MC, Johnstone A, Eccles D, Harmon B, Hayes MT, Lea RA, Griffiths L, Hoffman EP, Stubbs RS, Macartney-Coxson D.. 2015. An analysis of DNA methylation in human adipose tissue reveals differential modification of obesity genes before and after gastric bypass and weight loss. Genome Biol. 16:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bro-Jørgensen J. 2007. The intensity of sexual selection predicts weapon size in male bovids. Evolution 616:1316–1326. [DOI] [PubMed] [Google Scholar]
- Cahill JA, Stirling I, Kistler L, Salamzade R, Ersmark E, Fulton TL, Stiller M, Green RE, Shapiro B.. 2015. Genomic evidence of geographically widespread effect of gene flow from polar bears into brown bears. Mol Ecol. 246: 1205–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai DW, Han L, Zhang XL, Zhou H, Zhu H.. 2007. DNA analysis of archaeological sheep remains from China. J Archaeol Sci. 349: 1347–1355. [Google Scholar]
- Cai DW, Tang ZW, Yu HX, Han L, Ren XY, Zhao XB, Zhu H, Zhou H.. 2011. Early history of Chinese domestic sheep indicated by ancient DNA analysis of Bronze Age individuals. J Archaeol Sci. 384: 896–902. [Google Scholar]
- Carlson BE, Anderson JC, Raymond GM, Dash RK, Bassingthwaighte JB.. 2008. Modeling oxygen and carbon dioxide transport and exchange using a closed loop circulatory system. Adv Exp Med Biol. 614:353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Koster HA.. 1986. Beyond bones: toward an archaeology of pastoralism. Adv Archaeol Method Theory. 9:97–148. [Google Scholar]
- Chen FH, Dong GH, Zhang DJ, Liu XY, Jia X, An CB, Ma MM, Xie YW, Barton L, Ren XY, et al. 2015. Agriculture facilitated permanent human occupation of the Tibetan Plateau after 3600 B.P. Science 3476219: 248–250. [DOI] [PubMed] [Google Scholar]
- Chen N, Cai Y, Chen Q, Li R, Wang K, Huang Y, Hu S, Huang S, Zhang H, Zheng Z, et al. 2018. Whole-genome resequencing reveals world-wide ancestry and adaptive introgression events of domesticated cattle in East Asia. Nat Commun. 91: 2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SY, Duan ZY, Sha T, Xiangyu J, Wu SF, Zhang YP.. 2006. Origin, genetic diversity, and population structure of Chinese domestic sheep. Gene 3762: 216–223. [DOI] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM.. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 62: 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. 2011. The variant call format and VCFtools. Bioinformatics 2715: 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker JE, McKay SD, Rolf MM, Kim J, Molina Alcalá A, Sonstegard TS, Hanotte O, Götherström A, Seabury CM, Praharani L, et al. 2014. Worldwide patterns of ancestry, divergence, and admixture in domesticated cattle. PLoS Genet. 103: e1004254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deMenocal PB, Stringer C.. 2016. Climate and the peopling of the world. Nature 5387623: 49–50. [DOI] [PubMed] [Google Scholar]
- Du LX. 2011. Animal genetic resources in China (in Chinese). Beijing (China: ): China Agriculture Press. [Google Scholar]
- Durand EY, Patterson N, Reich D, Slatkin M.. 2011. Testing for ancient admixture between closely related populations. Mol Biol Evol. 288: 2239–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y. 1131. Hou Han Shu, Section on Xi Qiang Zhuan (in Chinese). Nanjing (China: ): Jiangnan East Road Transport Department. [Google Scholar]
- Fan ZX, Silva P, Gronau I, Wang SG, Armero AS, Schweizer RM, Ramirez O, Pollinger J, Galaverni M, Del-Vecchyo DO, et al. 2016. Worldwide patterns of genomic variation and admixture in gray wolves. Genome Res. 262: 163–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher TS, Lira PD, Stock JL, Perregaux DG, Brissette WH, Ozolins TRS, Li BY.. 2009. Analysis of the role of the HIF hydroxylase family members in erythropoiesis. Biochem Biophys Res Commun. 3884: 683–688. [DOI] [PubMed] [Google Scholar]
- Frede S, Fandrey J.. 2013. Cellular and molecular defenses against hypoxia In: Swenson ER, Bärtsch P, editors. High altitude: human adaptation to hypoxia. New York: Springer Press; p. 26. [Google Scholar]
- Gaunitz C, Fages A, Hanghøj K, Albrechtsen A, Khan N, Schubert M, Seguin-Orlando A, Owens IJ, Felkel S, Bignon-Lau O, et al. 2018. Ancient genomes revisit the ancestry of domestic and Przewalski’s horses. Science 3606384: 111–114. [DOI] [PubMed] [Google Scholar]
- Ge RL, Cai Q, Shen YY, San A, Ma L, Zhang Y, Yi X, Chen Y, Yang L, Huang Y, et al. 2013. Draft genome sequence of the Tibetan antelope. Nat Commun. 4:1858.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH-Y, et al. 2010. A draft sequence of the Neandertal genome. Science 3285979: 710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedes JAD, Lu HL, Hein AM, Schmidt AH.. 2015. Early evidence for the use of wheat and barley as staple crops on the margins of the Tibetan Plateau. Proc Natl Acad Sci U S A. 11218: 5625–5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedes JD, Butler EE.. 2014. Modeling constraints on the spread of agriculture to Southwest China with thermal niche models. Quat Int. 349:29–41. [Google Scholar]
- Guedes JD, Lu HL, Li YX, Spengler RN, Wu XH, Aldenderfer MS.. 2014. Moving agriculture onto the Tibetan plateau: the archaeobotanical evidence. Archaeol Anthrop Sci. 6:255–269. [Google Scholar]
- Guo X, Liu J, Zeng Y, Ding X, Bao P, Yan P, Pei J.. 2017. Study on complete mitochondrial genome of oula sheep (Ovis aries). Agric Sci Technol. 18:1365–1366. [Google Scholar]
- Han M, Brierley GJ, Cullum C, Li X.. 2016. Climate, vegetation and human land use interactions on the Qinghai–Tibet Plateau through the Holocene In: Brierley GJ, Li X, Cullum C, Gao J, editors. Landscape and ecosystem diversity, dynamics and management in the Yellow River Source Zone. Berlin (Germany: ): Springer Press; p. 253–274. [Google Scholar]
- He GY. 2000. Di Qiang Yuan Liu Shi (in Chinese). Nanchang (China: ): Jiangxi Education Press. [Google Scholar]
- Hornyak TJ, Jiang SL, Guzman EA, Scissors BN, Tuchinda C, He HB, Neville JD, Strickland FM.. 2009. Mitf dosage as a primary determinant of melanocyte survival after ultraviolet irradiation. Pigment Cell Melanoma Res. 223: 307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA.. 2009a. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 371: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA.. 2009b. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 41: 44–57. [DOI] [PubMed] [Google Scholar]
- Huang FS. 1989. Zang Zu Shi Lue (in Chinese). Beijing (China: ): Nationalities Publishing Press. [Google Scholar]
- Huerta-Sánchez E, Jin X, Asan, Bianba Z, Peter BM, Vinckenbosch N, Liang Y, Yi X, He M, Somel M, et al. 2014. Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature 5127513: 194–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Wang X, Kijas JW, Dalrymple BP.. 2015. Beta-globin gene evolution in the ruminants: evidence for an ancient origin of sheep haplotype B. Anim Genet. 465: 506–514. [DOI] [PubMed] [Google Scholar]
- Johns N, Stretch C, Tan BHL, Solheim TS, Sørhaug S, Stephens NA, Gioulbasanis I, Skipworth RJE, Deans DAC, Vigano A, et al. 2017. New genetic signatures associated with cancer cachexia as defined by low skeletal muscle index and weight loss. J Cachexia Sarcopenia Muscle. 81: 122–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston SE, Gratten J, Berenos C, Pilkington JG, Clutton-Brock TH, Pemberton JM, Slate J.. 2013. Life history trade-offs at a single locus maintain sexually selected genetic variation. Nature 5027469: 93–95. [DOI] [PubMed] [Google Scholar]
- Kardos M, Luikart G, Bunch R, Dewey S, Edwards W, McWilliam S, Stephenson J, Allendorf FW, Hogg JT, Kijas J.. 2015. Whole-genome resequencing uncovers molecular signatures of natural and sexual selection in wild bighorn sheep. Mol Ecol. 2422: 5616–5632. [DOI] [PubMed] [Google Scholar]
- Kijas JW, Lenstra JA, Hayes B, Boitard S, Neto LRP, San Cristobal M, Servin B, McCulloch R, Whan V, Gietzen K, et al. 2012. Genome-wide analysis of the world's sheep breeds reveals high levels of historic mixture and strong recent selection. PLoS Biol. 102: e1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Lohmueller KE, Albrechtsen A, Li Y, Korneliussen T, Tian G, Grarup N, Jiang T, Andersen G, Witte D, et al. 2011. Estimation of allele frequency and association mapping using next-generation sequencing data. BMC Bioinformatics 12:231.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneliussen TS, Albrechtsen A, Nielsen R.. 2014. ANGSD: analysis of next generation sequencing data. BMC Bioinformatics 15:356.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson G, Fuller DQ.. 2014. The evolution of animal domestication. Annu Rev Ecol Evol Syst. 451: 115–136. [Google Scholar]
- Larson G, Liu R, Zhao X, Yuan J, Fuller D, Barton L, Dobney K, Fan Q, Gu Z, Liu XH, et al. 2010. Patterns of East Asian pig domestication, migration, and turnover revealed by modern and ancient DNA. Proc Natl Acad Sci U S A. 10717: 7686–7691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CS, Huang YC, Huang RD, Wu YR, Wang WQ.. 2018. The genetic architecture of amylose biosynthesis in maize kernel. Plant Biotechnol J. 162: 688–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2010. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 265: 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 2516: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JS. 2012. The development of the original agriculture and animal husbandry on the Tibetan plateau from the archaeological materials. Agric Archaeol. 4:1–7. [Google Scholar]
- Li M, Tian S, Jin L, Zhou G, Li Y, Zhang Y, Wang T, Yeung CKL, Chen L, Ma J, et al. 2013. Genomic analyses identify distinct patterns of selection in domesticated pigs and Tibetan wild boars. Nat Genet. 4512: 1431–1438. [DOI] [PubMed] [Google Scholar]
- Lin S, Li C, Li C, Zhang X.. 2018. Growth hormone receptor mutations related to Individual Dwarfism. Int J Mol Sci. 195: 1433.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Ding X, Zeng Y, Yue Y, Guo X, Guo T, Chu M, Wang F, Han J, Feng R, et al. 2016. Genetic diversity and phylogenetic evolution of Tibetan Sheep based on mtDNA D-Loop sequences. PLoS One 117: e0159308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Lorenzen ED, Fumagalli M, Li B, Harris K, Xiong Z, Zhou L, Korneliussen TS, Somel M, Babbitt C, et al. 2014. Population genomics reveal recent speciation and rapid evolutionary adaptation in polar bears. Cell 1574: 785–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Peloso GM, Liu DJ, Wu Y, Zhang H, Zhou W, Li J, Tang CS, Dorajoo R, Li H, et al. 2017. Exome chip meta-analysis identifies novel loci and East Asian-specific coding variants that contribute to lipid levels and coronary artery disease. Nat Genet. 4912: 1722–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Habtetsion TG, Li Y, Zhang H, Qiao Y, Yu MD, Tang Y, Zhen Q, Cheng Y, Liu Y.. 2015. Association of NCOA2 gene polymorphisms with obesity and dyslipidemia in the Chinese Han population. Int J Clin Exp Pathol. 86: 7341–7349. [PMC free article] [PubMed] [Google Scholar]
- Lv FH, Peng WF, Yang J, Zhao YX, Li WR, Liu MJ, Ma YH, Zhao QJ, Yang GL, Wang F, et al. 2015. Mitogenomic meta-analysis identifies two phases of migration in the history of eastern Eurasian sheep. Mol Biol Evol. 3210: 2515–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen DB. 2016. Conceptualizing the Tibetan Plateau: environmental constraints on the peopling of the “Third Pole.” Archaeol Res Asia. 5:24–32. [Google Scholar]
- Manca L, Pirastru M, Mereu P, Multineddu C, Olianas A, el Sherbini el S, Franceschi P, Pellegrini M, Masala B.. 2006. Barbary sheep (Ammotragus lervia): the structure of the adult beta-globin gene and the functional properties of its hemoglobin. Comp Biochem Physiol B. 1452: 214–219. [DOI] [PubMed] [Google Scholar]
- Marcott SA, Shakun JD, Clark PU, Mix AC.. 2013. A reconstruction of regional and global temperature for the past 11,300 years. Science 3396124: 1198–1201. [DOI] [PubMed] [Google Scholar]
- Martens N, Uzan G, Wery M, Hooghe R, Hooghe-Peters EL, Gertler A.. 2005. Suppressor of cytokine signaling 7 inhibits prolactin, growth hormone, and leptin signaling by interacting with STAT5 or STAT3 and attenuating their nuclear translocation. J Biol Chem. 28014: 13817–13823. [DOI] [PubMed] [Google Scholar]
- Martin SH, Davey JW, Jiggins CD.. 2015. Evaluating the use of ABBA–BABA statistics to locate introgressed loci. Mol Biol Evol. 321: 244–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 209: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows JRS, Hanotte O, Drögemüller C, Calvo J, Godfrey R, Coltman D, Maddox JF, Marzanov N, Kantanen J, Kijas JW.. 2006. Globally dispersed Y chromosomal haplotypes in wild and domestic sheep. Anim Genet. 375: 444–453. [DOI] [PubMed] [Google Scholar]
- Meyer MC, Aldenderfer MS, Wang Z, Hoffmann DL, Dahl JA, Degering D, Haas WR, Schlütz F.. 2017. Permanent human occupation of the central Tibetan Plateau in the early Holocene. Science 3556320: 64–67. [DOI] [PubMed] [Google Scholar]
- Miao B, Wang Z, Li Y.. 2017. Genomic analysis reveals hypoxia adaptation in the Tibetan Mastiff by introgression of the gray wolf from the Tibetan Plateau. Mol Biol Evol. 343: 734–743. [DOI] [PubMed] [Google Scholar]
- Müller K, Lin L, Wang C, Glindemann T, Schiborra A, Schönbach P, Wan H, Dickhoefer U, Susenbeth A.. 2012. Effect of continuous v. daytime grazing on feed intake and growth of sheep grazing in a semi-arid grassland steppe. Animal 63: 526–534. [DOI] [PubMed] [Google Scholar]
- Natarajan C, Inoguchi N, Weber RE, Fago A, Moriyama H, Storz JF.. 2013. Epistasis among adaptive mutations in deer mouse hemoglobin. Science 3406138: 1324–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ní Leathlobhair M, Perri AR, Irving-Pease EK, Witt KE, Linderholm A, Haile J, Lebrasseur O, Ameen C, Blick J, Boyko AR, et al. 2018. The evolutionary history of dogs in the Americas. Science 3616397: 81–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Paul JS, Albrechtsen A, Song YS.. 2011. Genotype and SNP calling from next-generation sequencing data. Nat Rev Genet. 126: 443–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opazo JC, Hoffmann FG, Storz JF.. 2008. Differential loss of embryonic globin genes during the radiation of placental mammals. Proc Natl Acad Sci U S A. 10535: 12950–12955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Z, Li S, Liu Q, Wang Z, Zhou Z, Di R, Miao B, Hu W, Wang X, Hu X, et al. 2018. Whole-genome sequences of 89 Chinese sheep suggest role of RXFP2 in the development of unique horn phenotype as response to semi-feralization. GigaScience 7:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmesan C, Yohe G.. 2003. A globally coherent fingerprint of climate change impacts across natural systems. Nature 4216918: 37–42. [DOI] [PubMed] [Google Scholar]
- Peng W. 2010. A textual research on “sheep cart.” Cult Relics. 10:71–75. [Google Scholar]
- Phillips DJ. 2001. Peoples on the move: introducing the nomads of the world. Carlisle (United Kingdom: ): Pigquant. [Google Scholar]
- Preston BT, Stevenson IR, Pemberton JM, Coltman DW, Wilson K.. 2003. Overt and covert competition in a promiscuous mammal: the importance of weaponry and testes size to male reproductive success. Proc R Soc B Biol Sci. 2701515: 633–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Projecto-Garcia J, Natarajan C, Moriyama H, Weber RE, Fago A, Cheviron ZA, Dudley R, McGuire JA, Witt CC, Storz JF.. 2013. Repeated elevational transitions in hemoglobin function during the evolution of Andean hummingbirds. Proc Natl Acad Sci U S A. 11051: 20669–20674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J. 2015. Who are the Tibetans? Science 3476223: 708–711. [DOI] [PubMed] [Google Scholar]
- Qiu Q, Zhang G, Ma T, Qian W, Wang J, Ye Z, Cao C, Hu Q, Kim J, Larkin DM, et al. 2012. The yak genome and adaptation to life at high altitude. Nat Genet. 448: 946–949. [DOI] [PubMed] [Google Scholar]
- Qu Y, Zhao H, Han N, Zhou G, Song G, Gao B, Tian S, Zhang J, Zhang R, Meng X, et al. 2013. Ground tit genome reveals avian adaptation to living at high altitudes in the Tibetan plateau. Nat Commun. 4:2071.. [DOI] [PubMed] [Google Scholar]
- Razquin C, Martínez JA, Martínez-González MA, Salas-Salvadó J, Estruch R, Marti A.. 2010. A 3-year Mediterranean-style dietary intervention may modulate the association between adiponectin gene variants and body weight change. Eur J Nutr. 495: 311–319. [DOI] [PubMed] [Google Scholar]
- Saeed S, Bonnefond A, Manzoor J, Shabir F, Ayesha H, Philippe J, Durand E, Crouch H, Sand O, Ali M, et al. 2015. Genetic variants in LEP, LEPR, and MC4R explain 30% of severe obesity in children from a consanguineous population. Obesity 238: 1687–1695. [DOI] [PubMed] [Google Scholar]
- Said SM, Murphree MI, Mounajjed T, El-Youssef M, Zhang L.. 2016. A novel GBE1 gene variant in a child with glycogen storage disease type IV. Hum Pathol. 54:152–156. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW.. 2001. Molecular cloning: a laboratory manual. 3rd ed New York: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Simonson TS, Yang Y, Huff CD, Yun H, Qin G, Witherspoon DJ, Bai Z, Lorenzo FR, Xing J, Jorde LB, et al. 2010. Genetic evidence for high-altitude adaptation in Tibet. Science 3295987: 72–75. [DOI] [PubMed] [Google Scholar]
- Skoglund P, Ersmark E, Palkopoulou E, Dalén L.. 2015. Ancient wolf genome reveals an early divergence of domestic dog ancestors and admixture into high-latitude breeds. Curr Biol. 2511: 1515–1519. [DOI] [PubMed] [Google Scholar]
- Skotte L, Korneliussen TS, Albrechtsen A.. 2013. Estimating individual admixture proportions from next generation sequencing data. Genetics 1953: 693–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF, Runck AM, Moriyama H, Weber RE, Fago A.. 2010. Genetic differences in hemoglobin function between highland and lowland deer mice. J Exp Biol. 213(Pt 15): 2565–2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF, Runck AM, Sabatino SJ, Kelly JK, Ferrand N, Moriyama H, Weber RE, Fago A.. 2009. Evolutionary and functional insights into the mechanism underlying high-altitude adaptation of deer mouse hemoglobin. Proc Natl Acad Sci U S A. 10634: 14450–14455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbotin AE, Kapitanova DV, Lopatin AV.. 2007. Factors of craniometric variability in argali, using an example of Ovis ammon polii (Bovidae, Artiodactyla). Dokl Biol Sci. 416:400–402. [DOI] [PubMed] [Google Scholar]
- Talukdar S, Zhou Y, Li D, Rossulek M, Dong J, Somayaji V, Weng Y, Clark R, Lanba A, Owen BM, et al. 2016. A long-acting FGF21 molecule, PF-05231023, decreases body weight and improves lipid profile in non-human primates and type 2 diabetic subjects. Cell Metab. 233: 427–440. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Takizawa T, Dorji T, Amano T, Mannen H, Maeda Y, Yamamoto Y, Namikawa T.. 2011. Polymorphisms in the bovine hemoglobin-beta gene provide evidence for gene-flow between wild species of Bos (Bibos) and domestic cattle in Southeast Asia. Anim Sci J. 821: 36–45. [DOI] [PubMed] [Google Scholar]
- Tang H, Dong X, Hassan MM, Abbruzzese JL, Li D.. 2011. Body mass index and obesity- and diabetes-associated genes and risk for pancreatic cancer. Cancer Epidemiol. Biomarkers Prev. 205: 779–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Winkel ML, van Beek RD, de Muinck Keizer-Schrama SMPF, Uitterlinden AG, Hop WCJ, Pieters R, van den Heuvel-Eibrink MM.. 2010. Pharmacogenetic risk factors for altered bone mineral density and body composition in pediatric acute lymphoblastic leukemia. Haematologica 955: 752–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng HJ, Zhang YH, Shi CM, Mao FB, Cai WS, Lu L, Zhao FQ, Sun ZS, Zhang JX.. 2017. Population genomics reveals speciation and introgression between brown Norway rats and their sibling species. Mol Biol Evol. 349: 2214–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, et al. 2013. From FastQ data to high-confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 43:11.10.1–11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira FG, Lassalle F, Korneliussen TS, Fumagalli M.. 2016. Improving the estimation of genetic distances from Next-Generation Sequencing data. Biol J Linn Soc. 1171: 139–149. [Google Scholar]
- Wang GD, Fan RX, Zhai WW, Liu F, Wang L, Zhong L, Wu H, Yang HC, Wu SF, Zhu CL, et al. 2014. Genetic convergence in the adaptation of dogs and humans to the high-altitude environment of the Tibetan Plateau. Genome Biol Evol. 68: 2122–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GD, Zhai W, Yang HC, Fan RX, Cao X, Zhong L, Wang L, Liu F, Wu H, Cheng LG, et al. 2013. The genomics of selection in dogs and the parallel evolution between dogs and humans. Nat Commun. 4:1860.. [DOI] [PubMed] [Google Scholar]
- Wang XL, Liu J, Zhou GX, Guo JZ, Yan HL, Niu YY, Li Y, Yuan C, Geng RQ, Lan XY, et al. 2016. Whole-genome sequencing of eight goat populations for the detection of selection signatures underlying production and adaptive traits. Sci Rep. 6:38932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C, Wang H, Liu G, Zhao F, Kijas JW, Ma Y, Lu J, Zhang L, Cao J, Wu M, et al. 2016. Genome-wide analysis reveals adaptation to high altitudes in Tibetan sheep. Sci Rep. 6:26770.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC.. 1984. Estimating F-statistics for the analysis of population structure. Evolution 386: 1358–1370. [DOI] [PubMed] [Google Scholar]
- Wiedemar N, Drögemüller C.. 2015. A 1.8-kb insertion in the 3-UTR of RXFP2 is associated with polledness in sheep. Anim Genet. 464: 457–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfson GH, Vargas E, Browne VA, Moore LG, Julian CG.. 2017. Erythropoietin and soluble erythropoietin receptor: a role for maternal vascular adaptation to high-altitude pregnancy. J Clin Endocrinol Metab. 1021: 242–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu DD, Ding XD, Wang S, Wójcik JM, Zhang Y, Tokarska M, Li Y, Wang M-S, Faruque O, Nielsen R, et al. 2018. Pervasive introgression facilitated domestication and adaptation in the Bos species complex. Nat Ecol Evol. 27: 1139–1145. [DOI] [PubMed] [Google Scholar]
- Xian B. 2005. A rustic opinion on the natural-harmoniously traditional energy culture of the Zang (Tibetan) nationality (in Chinese). Hohhot (China): The annual meeting of the academic society of the history of philosophical and social ideas in Chinese minorities, Researches on the harmonious thought of Chinese minority nationalities. p. 169–179.
- Xu Q, Zhang C, Zhang D, Jiang H, Peng S, Liu Y, Zhao K, Wang C, Chen L.. 2016. Analysis of the erythropoietin of a Tibetan Plateau schizothoracine fish (Gymnocypris dobula) reveals enhanced cytoprotection function in hypoxic environments. BMC Evol Biol. 16:11.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Li WR, Lv FH, He SG, Tian SL, Peng WF, Sun YW, Zhao YX, Tu XL, Zhang M, et al. 2016. Whole-genome sequencing of native sheep provides insights into rapid adaptations to extreme environments. Mol Biol Evol. 3310: 2576–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Wang Y, Zhao Y, Zhang X, Li R, Chen L, Zhang G, Jiang Y, Qiu Q, Wang W, et al. 2017. Draft genome of the Marco Polo Sheep (Ovis ammon polii). GigaScience 612: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T, Liu X, Wang N, Shi Y.. 2000. Amplitude of climatic changes in Qinghai-Tibetan Plateau. Chin Sci Bull. 4513: 1236–1243. [Google Scholar]
- Yi X, Liang Y, Huerta-Sanchez E, Jin X, Cuo ZXP, Pool JE, Xu X, Jiang H, Vinckenbosch N, Korneliussen TS, et al. 2010. Sequencing of 50 human exomes reveals adaptation to high altitude. Science 3295987: 75–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeder MA. 2008. Domestication and early agriculture in the Mediterranean Basin: origins, diffusion, and impact. Proc Natl Acad Sci U S A. 10533: 11597–11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Chamba Y, Shang P, Wang ZX, Ma J, Wang LY, Zhang H.. 2017. Comparative transcriptomic and proteomic analyses provide insights into the key genes involved in high-altitude adaptation in the Tibetan pig. Sci Rep. 7:3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DJ, Dong GH, Wang H, Ren XY, Ha PP, Qiang MR, Chen FH.. 2016. History and possible mechanisms of prehistoric human migration to the Tibetan Plateau. Sci China Earth Sci. 599: 1765–1778. [Google Scholar]
- Zhang M, Peng WF, Yang GL, Lv FH, Liu MJ, Li WR, Liu YG, Li JQ, Wang F, Shen ZQ, et al. 2014. Y chromosome haplotype diversity of domestic sheep (Ovis aries) in northern Eurasia. Anim Genet. 456: 903–907. [DOI] [PubMed] [Google Scholar]
- Zhao YX, Yang J, Lv FH, Hu XJ, Xie XL, Zhang M, Li WR, Liu MJ, Wang YT, Li JQ, et al. 2017. Genomic reconstruction of the history of native sheep reveals the peopling patterns of nomads and the expansion of early pastoralism in East Asia. Mol Biol Evol. 349: 2380–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Zheng Q, Engin F, Munivez E, Chen Y, Sebald E, Krakow D, Lee B.. 2006. Dominance of SOX9 function over RUNX2 during skeletogenesis. Proc Natl Acad Sci U S A. 10350: 19004–19009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XG. 2010. Study on hybridization breeding of Xinjiang wild argali (in Chinese) [doctoral dissertation]. Shihezi University, Shihezi (China).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.