Abstract
Mitochondrial ribosomes (mitoribosomes) are essential components of all mitochondria that synthesize proteins encoded by the mitochondrial genome. Unlike other ribosomes, mitoribosomes are highly variable across species. The basis for this diversity is not known. Here, we examine the composition and evolutionary history of mitoribosomes across the phylogenetic tree by combining three-dimensional structural information with a comparative analysis of the secondary structures of mitochondrial rRNAs (mt-rRNAs) and available proteomic data. We generate a map of the acquisition of structural variation and reconstruct the fundamental stages that shaped the evolution of the mitoribosomal large subunit and led to this diversity. Our analysis suggests a critical role for ablation and expansion of rapidly evolving mt-rRNA. These changes cause structural instabilities that are “patched” by the acquisition of pre-existing compensatory elements, thus providing opportunities for rapid evolution. This mechanism underlies the incorporation of mt-tRNA into the central protuberance of the mammalian mitoribosome, and the altered path of the polypeptide exit tunnel of the yeast mitoribosome. We propose that since the toolkits of elements utilized for structural patching differ between mitochondria of different species, it fosters the growing divergence of mitoribosomes.
Keywords: RNA evolution, RNA structure, ribosome, mitochondria, eukaryotes
Introduction
Mitochondria are organelles that perform multiple functions within eukaryotic cells including the production of chemical energy. They contain their own mitochondrial genome (mt-genome) and translational machinery. The mt-genome descended from an ancient translation-sufficient bacterium that was engulfed and farmed by a primordial eukaryote (Sagan 1967; Andersson et al. 1998; Lazcano and Pereto 2017; Zachar et al. 2018). Various genomic and phylogenetic studies are consistent with a monophyletic origin of the mt-genome from α-proteobacteria (Gray et al. 1998; Gray 2015) or its sister group (Martijn et al. 2018). Phylogenetic analyses suggest that the obligate intracellular bacterial parasite Rickettsia prowazekii (Andersson et al. 1998) or the obligate bacterial aerobic heterotroph HIMB59 (Thrash et al. 2011; Rodríguez-Ezpeleta and Embley 2012; Viklund et al. 2013) are phylogenetically the most similar to all mitochondria by their bacteria-derived components. Yet, mitochondria also contain a substantial fraction of a nonproteobacterial proteome (Björkholm et al. 2015) indicating a merger with other pre-existing mitochondrial “parents” of nonproteobacterial origin (Gray 2015; Husnik and McCutcheon 2016). Thus, mitochondria appear to have arisen from multiple endosymbiotic events (Poole and Gribaldo 2014; Husnik and McCutcheon 2016) that were followed by a comprehensive transfer of genes from the mt-genome to the eukaryotic host’s nuclear DNA and acquisition of antecedent proteins from the host (Gray 2014). The origin of nonproteobacterial components as well as the complete history of mitochondrial evolution remain a subject of ongoing studies (Gray 2014; Poole and Gribaldo 2014; Björkholm et al. 2015; Gray 2015; Harish and Kurland 2017).
Mitoribosomes synthesize proteins encoded by the mt-genome. The general consensus, based on phylogenetic and structural data on mitoribosomal RNA (mt-rRNA; Ferla et al. 2013), mitoribosomal proteins (Ban et al. 2014; Greber and Ban 2016; Martijn et al. 2018), mitoribosomal translation factors (Andersen et al. 2000; Atkinson and Baldauf 2011; Kuzmenko et al. 2014), and genomic organization (Lang et al. 1997) point to α-proteobacterial origin of the mitochondrial translation system (Gray 2015). Despite the proposed common ancestry, mitoribosomes are morphologically diverse and vary in protein and mt-rRNA content in different species (Amunts et al. 2015; Greber et al. 2015; Desai et al. 2017; Ramrath et al. 2018). The lengths of mt-RNAs tend to be shorter than those of extant bacteria. However, mt-rRNAs of some species contain additional insertions, which resemble eukaryotic expansion segments (Gerbi 1996), but have mitochondria-specific locations. Mitoribosomes are also enriched with mitochondria-specific proteins (Amunts et al. 2015; Greber et al. 2015; Desai et al. 2017). Even defining functional and structural features of mitoribosomes have been affected, as epitomized by differences in the polypeptide exit tunnels of the yeast (Amunts et al. 2014) and mammalian mitoribosomes (Brown et al. 2014; Greber et al. 2014; Amunts et al. 2015; Greber et al. 2015). The diversity of mitoribosomes is unprecedented in homologous systems, and the structures of cytosolic ribosomes from different species are highly similar (Ben-Shem et al. 2011; Klinge et al. 2011; Rabl et al. 2011; Wong et al. 2014).
It has recently become apparent that mitoribosomes have undergone substantial changes within a relatively short evolutionary period in response to a host environment and subsequent diversification of the eukaryotic species (van der Sluis et al. 2015). Here, we investigate the molecular pathways that led to species-specific mitoribosomes. Since fossilized records for intermediates in the evolutionary development of mitoribosomes do not exist, we combine the available structural information with genomic, proteomic and phylogenetic data to infer the steps that led to the architectural and functional diversity of mitoribosomes. The most pronounced changes took place in the exit tunnel and the central protuberance (CP) of the mitoribosomal large subunit (mt-LSU). Therefore, we use the high-resolution three-dimensional structures of the mt-LSU from Saccharomyces cerevisiae (Amunts et al. 2014), Homo sapiens (Brown et al. 2014), and Sus scrofa (Greber et al. 2015) to describe possible evolutionary scenarios that have resulted in the remodeling of the exit tunnel and CP in the metazoan and fungal branches. We dissect the relationship between mt-rRNA evolution and the acquisition of mitoribosomal proteins and investigate the roles of some of the most conserved of these proteins to identify possible evolutionary drivers. Our analysis suggests that mitoribosomes evolved, and gained new functions, through a “structural patching” of the archetypical ribosome driven by destabilizing changes in mt-rRNA.
Results
Phylogenetic Analysis of mt-rRNA
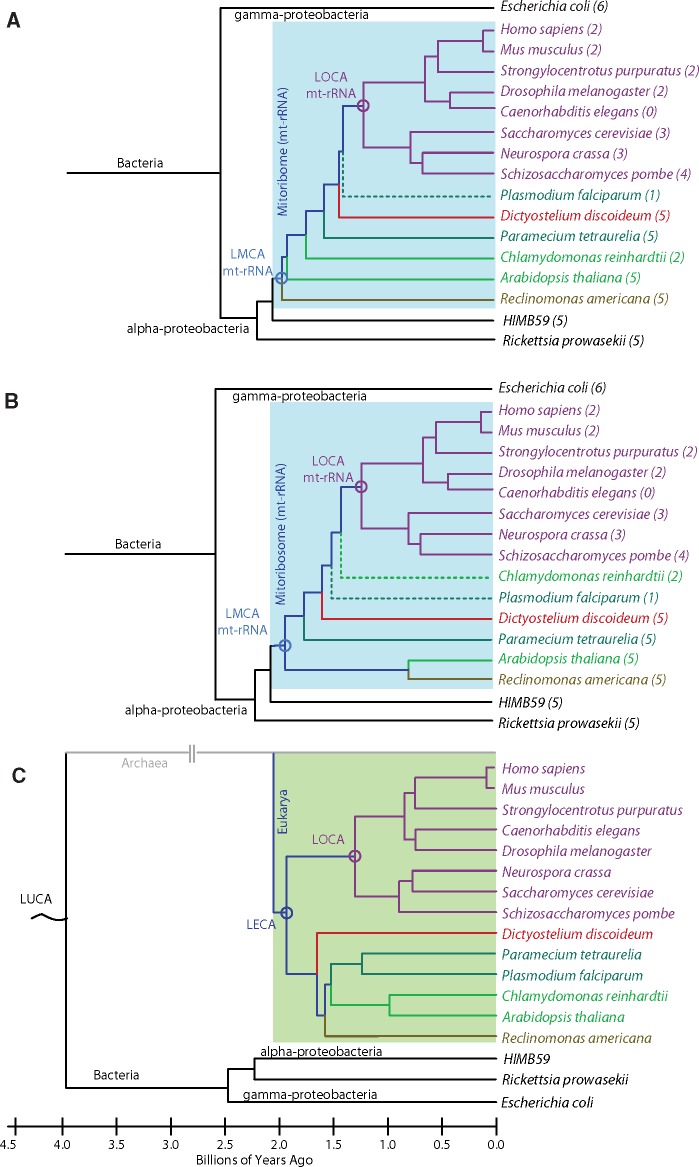
Here, we use phylogenetic reconstructions to trace the evolutionary history of mitoribosomes. To do this we first selected eukaryotic species with fully sequenced nuclear and mitochondrial genomes, and either a three-dimensional structure or a secondary structure prediction for their mt-LSU rRNA (Cannone et al. 2002). The 14 species selected represent five out of the eight major clades of eukaryotic organisms (Opisthokonta, Alveolata, Amoebozoa, Archaeplastida, and Excavata). For each species, statistical data for the mt-genome including DNA length, G/C content, and the number of proteins encoded are collated in supplementary table S1, Supplementary Material online together with data for the mt-LSU including rRNA length, designated LSU zone (see later), and accession codes for the structural database. The rRNA alignment for these species is provided as supplementary data set 1, Supplementary Material online. We also selected α-proteobacteria R. prowazekii and HIMB59, and the γ-proteobacterium Escherichia coli as references. The ribosome of E. coli is well characterized by detailed secondary and three-dimensional structures and is therefore used in the current study as a framework for understanding evolutionary changes of mitoribosomes.
To follow the evolutionary history of the mt-LSU rRNA, we generated a phylogenetic tree using mt-LSU rRNA sequences using maximum-parsimony (fig. 1A) and maximum-likelihood (fig. 1B) methods based on rRNA sequences from supplementary data set S1, Supplementary Material online. More complete trees derived from supplementary data set S2, Supplementary Material online (containing a subset of archaeal, bacterial and mitochondrial sequences) using the maximum-parsimony and maximum-likelihood methods are presented in supplementary figure S1A and B, Supplementary Material online. The trees shown in figure 1A and B are similar to one another and share a similar topology to the tree reconstructed from other universal mitochondria-encoded genes (cox1 and cob; Gray et al. 1999; Kurland and Andersson 2000). The trees recapitulate major features of the eukaryotic branch of the canonical tree (Katz 2012; Letunic and Bork 2016) shown in figure 1C. The main difference among these trees is the topology of species representing Bikonta clades. All LSU rRNA-derived trees suggest that mt-LSU rRNA is clustered within α-proteobacteria, and reveal the closest resemblance between mt-LSU rRNA of Reclinomonas americana (Lang et al. 1997; Ferla et al. 2013) and LSU rRNA of HIMB59. We note that the inspection of the bootstrap replicate values in the extended trees (supplementary fig. S1A and B, Supplementary Material online) suggests that some other members of the α-proteobacterial clade (e.g., R. prowazekii) may also share common ancestry with mt-LSU rRNA.
Fig. 1.
Phylogenies of mitoribosomal and eukaryotic evolution. Phylogenetic trees generated from reconstruction of mt-LSU rRNA sequences by (A) the maximum parsimony and (B) maximum likelihood methods using MEGA7 (Kumar et al. 2016). The numbers in parentheses indicate mt-rRNA zone number as depicted in figure 2. (C) Canonical phylogenetic tree of a subset of eukaryotic species obtained from the interactive tree of life (iTOL; (Letunic and Bork 2016)) mapped onto an evolutionary timeline and colored by clade: Opisthokonta (purple), Alveolata (teal), Amoebozoa (red), Archaeplastida (green), and Excavata (brown). The Plasmodium falciparum and Chlamydomonas reinhardtii branches are dashed in the mt-LSU rRNA reconstructions, as they do not follow the canonical topology. Bacterial ancestors and extant bacterial species are shown in black. Approximate ages of nodes were obtained from the Time Tree project (Hedges et al. 2015; Kumar et al. 2017). Last universal common ancestor (LUCA), last eukaryotic common ancestor (LECA), last mitochondrial common ancestor of rRNA (LMCA mt-rRNA), last opisthokontal common ancestor of mt-rRNA (LOCA mt-rRNA).
As the three-dimensional fold of mt-rRNA governs the function of the mitoribosome, we next examined the structural similarity of the mt-LSU rRNA across the eukaryotic branch of the phylogenetic tree to identify regions that changed during evolution. For this analysis, the three-dimensional structures of E. coli ribosomes and the yeast and mammalian mitoribosomes served as templates to map the available secondary structure information (Cannone et al. 2002) for the species representing major eukaryotic clades and the α-proteobacteria R. prowazekii and HIMB59. The comparison shows that these α-proteobacterial ribosomes contain a reduced LSU rRNA content compared with that of E. coli (supplementary fig. S2A and B, Supplementary Material online). In particular, helices H58, H59, and H98 are deleted and H54, H16-18, and H63 are shortened. Inspection of a multiple sequence alignment created for 13 α-proteobacterial species sampled from all major clades (supplementary data set 3, Supplementary Material online, obtained from the SEREB database, Bernier et al. 2018) confirms that these reductions exist in all α-proteobacterial LSU rRNAs. Furthermore, a similar reduction pattern is also observed in the LSU mt-rRNA of R. americana (supplementary fig. S2C, Supplementary Material online), which is evolutionarily the closest to all bacterial ribosomes. Irrespective of the uncertainty surrounding the origin of mitochondrial genome (Kurland and Andersson 2000; Thrash et al. 2011; Martijn et al. 2018), the structural similarity of all α-proteobacterial rRNAs with that of R. americana prompt a conclusion that the LSU mt-rRNA of the last mitochondrial common ancestor (LMCA) likely contained a reduced rRNA compared with that of the majority of bacteria (including E. coli). Therefore, we use two-dimensional rRNA projections of HIMB59 or R. americana (supplementary fig. S2B and C, Supplementary Material online) together with the available three-dimensional structure of the E. coli ribosome as a starting point to trace the evolutionary history of mitoribosomes.
Evolution of mt-rRNA
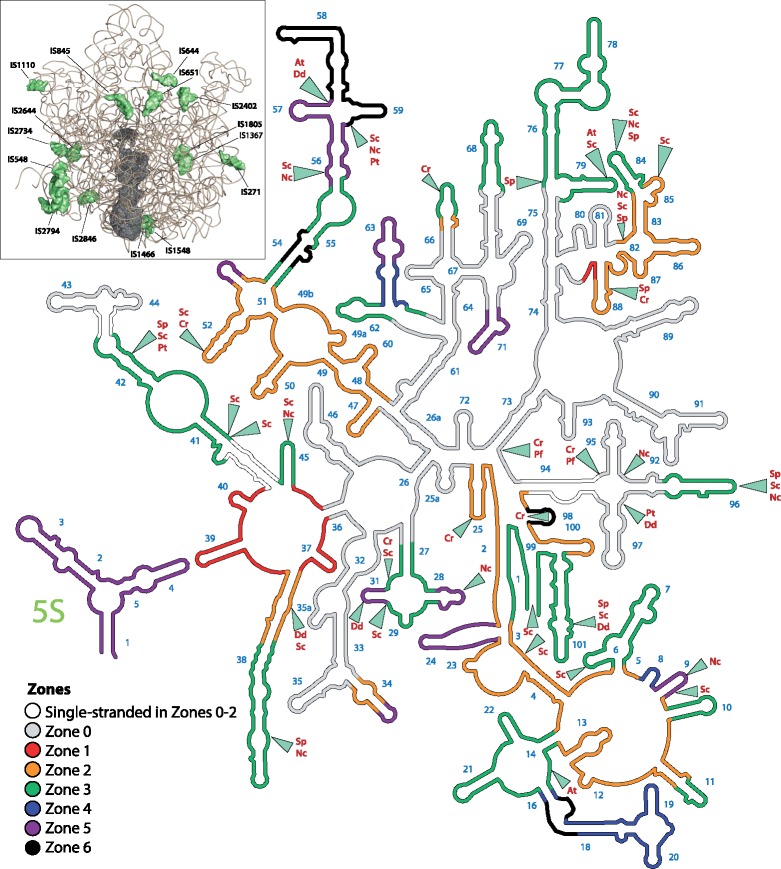
To visualize species-dependent changes, we analyzed deletions and extensions of mt-rRNA (compiled in the supplementary data set S4, Supplementary Material online). We further superimposed mt-rRNAs and grouped mt-rRNA alterations that are related to each other using E. coli LSU rRNA as a template (supplementary table S1, Supplementary Material online). This mapping led to the identification of seven zones, where zone 0 represents minimal mt-rRNA and zone 6 represents E. coli LSU rRNA (fig. 2).
Fig. 2.
Diversity of LSU mt-rRNAs. The secondary structure diagram of Escherichia coli 23S rRNA is divided into seven zones (0–6) defined in supplementary table S1, Supplementary Material online based on the presence of a particular helix in a given subset of species. Zone 0 represents a minimal mt-rRNA. Unshaded segments represent helices that are single stranded or separated due to fragmentation in the species of Zones 0–2. Segments highlighted in black (Zone 6) do not exist in α-proteobacterial species (including Rickettsia prowazekii and HIMB59) or in the mt-LSU rRNA of Reclinomonas americana. Sites of expansion (insertion sites) are marked with triangles and labeled with two-letter abbreviations for the species in which that expansion occurs. Abbreviations are as follows: Sp, Schizosaccharomyces pombe; Sc, Saccharomyces cerevisiae; Nc, Neurospora crassa; At, Arabidopsis thaliana; Pt, Paramecium tetraurelia; Dd, Dictyostelium discoideum, and Cr, Chlamydomonas reinhardtii. Inset: locations of insertion sites across the phylogeny mapped onto the tertiary structure of E. coli LSU rRNA. Insertion sites are prefixed with “IS” followed by the position of the insertion in the rRNA according to E. coli numbering.
Reduction of rRNA has occurred in the majority of species but is most noticeable in the mitoribosomes of metazoans (Zones 0–2) as exemplified here by mt-rRNA of H. sapiens (supplementary fig. S3, Supplementary Material online). Moderate mt-rRNA deletion occurred in species representing nonopisthokonts (Zones 3–5). The majority of deletions is in domains I and III of the LSU mt-rRNA at regions proximal to the ribosomal surface. The deleted sections are characterized by either complete removal of folded rRNA helices or their partial unwinding resulting in formation of single-stranded regions. As described below, newly added elements (primarily mitoribosomal proteins) typically adapt to the remaining mt-rRNA, serving as “structural patches” for functionally important regions. The mt-rRNA deletions are not a simple rewinding of ribosomal evolution before LUCA (Petrov et al. 2014), suggesting that subsequent events rendered reversion impossible. Sections that are never deleted or reduced are mostly functionally essential elements that include: 1) the peptidyl transfer center and the adjacent initial part of the polypeptide exit tunnel (H26, 26a, 32, 33, 36, and 61); 2) the intersubunit interface (H64-H68); 3) tRNA “rail” (H69); 4) the L7/L12 stalk base (H43-44); and 5) the sarcin-ricin loop (H95). In Plasmodium falciparum and Chlamydomonas reinhardtii, the mt-rRNA has not only reduced, but become fragmented.
Interestingly, mt-rRNAs of some species also contain expansions, which are similar to those observed for eukaryotic ribosomes in the cytosol (Gerbi 1996), and located at the mitoribosomal surface. These expansions are most pronounced in fungal species including S. cerevisiae (supplementary fig. S4, Supplementary Material online), Schizosaccharomyces pombe, and Neurospora crassa, but also occur in some nonopisthokonts (fig. 2). The locations of the expansion segments (ESs) were mapped onto the three-dimensional structure of the LSU rRNA of E. colifig. 2, inset; (supplementary table S2, Supplementary Material online). ESs located near the exit tunnels of mitoribosomes from Arabidopsis thaliana, Dictyostelium discoideum, Paramecium tetraurelia, and C. reinhardtii may have functional consequences, potentially mediating interactions with the inner mitochondrial membrane (Pfeffer et al. 2015) or mitoribosomal proteins. In contrast, mammalian mt-rRNAs generally lack ESs.
Evolution of the Mitoribosomal Proteome
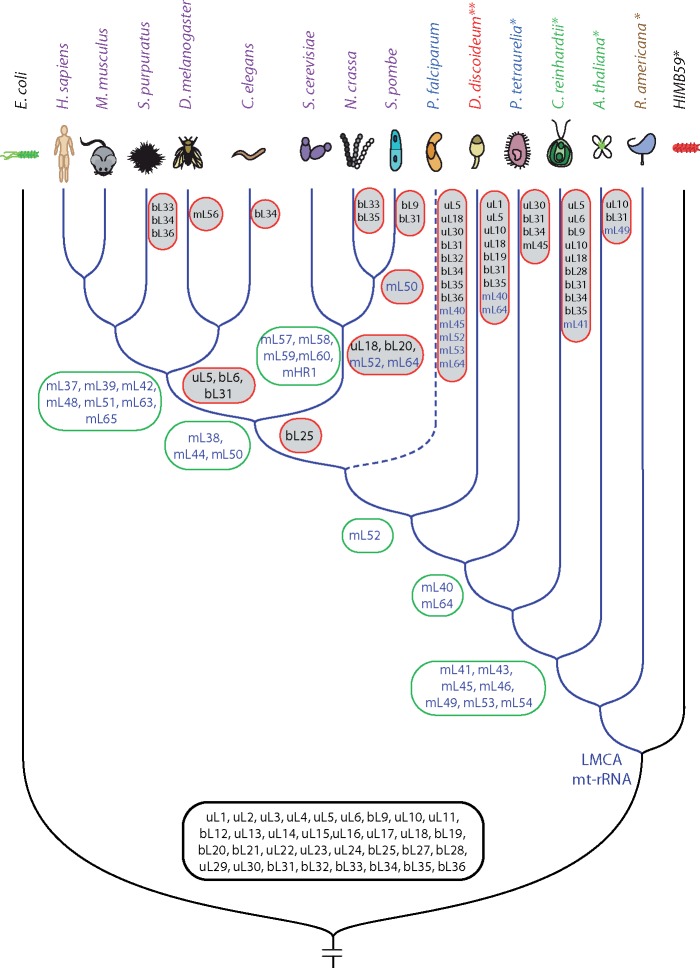
Unlike bacterial ribosomes, for which the LSU contains 36 highly conserved proteins (uL1-bL36; nomenclature from (Ban et al. 2014), mitoribosomes of different species exhibit high variability in protein content. Along with having a subset of homologs of bacterial proteins, mitoribosomes have additional proteins that vary in number and identity. Given a high similarity between the mt-rRNA-derived phylogenetic trees (fig. 1A and B), we used the tree obtained by the maximum parsimony algorithm (fig. 1A) as an internal clock for mitoribosomal evolution. To explore how the mitoribosomal proteome changed during the evolution, we then mapped acquired and lost mitoribosomal proteins onto the timeline of mt-rRNA (fig. 3), expanding upon previous analyses (Smits et al. 2007; Desmond et al. 2011). Although all known mitoribosomal proteins were included, it is likely that the more diverse taxa have lineage-specific proteins that are yet to be identified.
Fig. 3.
Changes to the mitoribosomal proteome from the analysis of (Desmond et al. 2011) manually mapped to the mt-rRNA-derived phylogenetic tree (as in fig. 1A). Proteins of bacterial origin (prefixed with a “u” or a “b” depending on whether they are universal or specific to bacterial ribosomes) are shown in black and proteins exclusive to mitoribosomes (prefixed with a “m”) are shown in blue. Proteins inherited from a bacterial ancestor are outlined with a black box. Proteins acquired by mitoribosomes are outlined with green boxes. Proteins lost by mitoribosomes are outlined with red boxes and shaded in gray. Species names are colored as in figure 1. Branches of mt-LSU rRNAs are in blue and those of the bacterial outgroup are in black. The tree depicts only a few bacterial species relevant for the current study. Species that contain 5S mt-rRNA are marked with an asterisk (*). Dictyostelium discoideum (marked with **) contains a rRNA gene with structural characteristics similar to those of 5S rRNA (Bullerwell et al. 2010), but which does not appear to be associated with the mitoribosome (Pi et al. 1998). Suffix “m” of mitoribosomal proteins of bacterial or universal origin is omitted for clarity in all labels of all figures (e.g., uL5m is labeled as uL5).
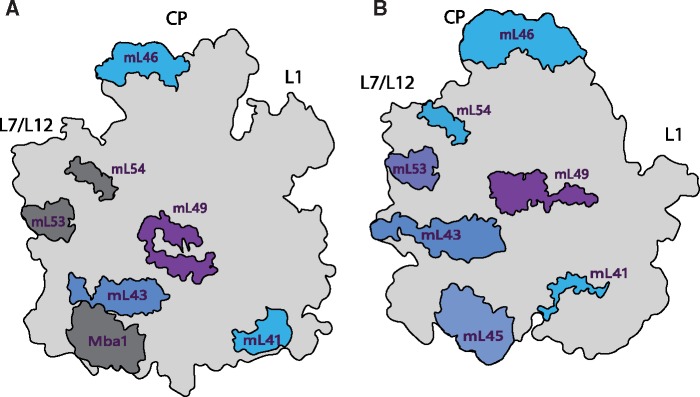
Mitoribosomes contain variations of a bacterial subset of proteins (uL1m-bL36m). While the majority of genomic and phylogenetic studies (Lang et al. 1997; Gray et al. 1998; Desmond et al. 2011; Melnikov et al. 2018) suggests that these proteins were inherited from a bacterial genome, alternative scenarios of mitochondrial origin (Björkholm et al. 2015; Harish and Kurland 2017), point to their inheritance from LUCA. In addition to these proteins, mitoribosomes of all but R. americana species contain a near-universal subset of seven mitochondria-specific proteins: mL41, mL43, mL45, mL46, mL49, mL53, and mL54. This implies that these seven proteins were incorporated into mitoribosomes during the early stages of mitochondrial evolution. Inspection of the positions of these proteins in the available structures (fig. 4) reveals that they are localized to functionally important regions of the mitoribosome: the polypeptide exit tunnel (mL41 and mL45), a central H16-H20 binding region (mL43 and mL49), the CP (mL46), and the L7/L12 stalk (mL53 and mL54). To determine the possible origins of this subset of mitoribosomal proteins, we queried the sequence and structure of each member against the ECOD (Cheng et al. 2014) and NCBI’s nonredundant protein sequence (Benson et al. 2000) databases. The results, summarized in supplementary tables S3 and S4, Supplementary Material online, show that these mitoribosomal proteins (except mL41 and mL54) share folds and sequence similarity with proteins found elsewhere. Specifically, homologous proteins have been found within bacteria (mL44, mL46, and mL49), the eukaryotic translational machinery (mL49), and the inner mitochondrial membrane (mL43, mL45, mL53), suggesting that the near-universal subset of mitoribosomal proteins may have been recruited from a pool of pre-existing proteins as a result of gene transfer or gene duplication (Smits et al. 2007; supplementary table S4, Supplementary Material online). Mitochondria-specific ribosomal proteins have evolved further in a species-specific manner, particularly due to the emergence of extensions that interact with neighboring proteins or mt-rRNA.
Fig. 4.
Positions of the near-universal set of mitochondria-specific mitoribosomal proteins in the (A) Saccharomyces cerevisiae mt-LSU and (B) Homo sapiens mt-LSU. Mitoribosomal proteins Mba1 (mL45), mL53, and mL54 that exist in the S. cerevisiae mitoribosome but are not resolved in the cryo-EM map EMD-2566 (Amunts et al. 2014), are placed at locations inferred from the mt-LSU of H. sapiens and are highlighted in dark gray.
As unikonts evolved, their mitoribosomes became enriched with mL40, mL52, and mL64, which are present in both the Amoebozoa and Opisthokonta phyla (fig. 3). Opisthokonta then further acquired mitoribosomal proteins mL38, mL44, and mL50, whose precursors were also available within mitochondria (supplementary table S4, Supplementary Material online). These proteins differ between the metazoan and fungal branches, and these differences correlate with variations in the structures of their mt-rRNAs (fig. 2) suggesting that mt-rRNA and proteins co-evolved. Similar evolutionary mechanisms have triggered the growth of idiosyncratic extensions in the mitoribosomal proteins of bacterial origin (supplementary tables S5, S6, Supplementary Material online). While metazoan-specific proteins substitute for reductions in mt-rRNA, the proteins specific to the fungal species tend to interact with ESs.
Resurrecting the History of Mitoribosomal Evolution
To explore the relationship between changes in mt-rRNA and mitoribosomal proteome at the structural level, we focus on the polypeptide exit tunnel and the CP, the two regions of the mt-LSU that have undergone the most substantial remodeling. For each region, we first combine the data from the previously presented phylogenetic and proteomic analysis with all available secondary structure and three-dimensional information to describe a plausible chronological sequence of events from the putative bacterial precursor (using the E. coli ribosome as a reference) to a hypothetical Last Opisthokontal Common Ancestor (LOCA). We then follow the independent evolutionary paths of mitoribosomes along the metazoan and fungal branches, using the mammalian and yeast mitoribosomes as examples.
Early Evolution of the Exit Tunnel
The polypeptide exit tunnel supports the emergence of newly synthesized proteins from the ribosome and in doing so facilitates their initial folding and interaction with chaperones. Given its fundamental role in protein synthesis, the exit tunnel is among the most conserved regions of ribosomes, except in mitoribosomes. Adaptations in the regions neighboring the polypeptide exit tunnel likely started during the early stages of mitoribosomal evolution (supplementary fig. S5, Supplementary Material online) and included the reduction of mt-rRNA (revealed by shortening of H7, H16-20, and H54-55) and incorporation of new proteins (mL41, mL43, mL45, and mL49).
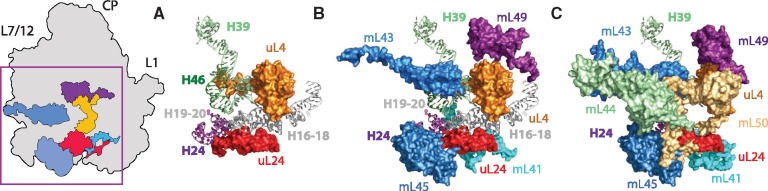
The early addition of mL43 and mL49 appears to be related to the reduction of helices H16-20 of mt-rRNA (fig. 5). In the E. coli ribosome, H16-20 are located at the ribosomal surface and bind proteins uL4 and uL24 (fig. 5A) and rRNA helices H24, H39, and H46. In comparison to E. coli, H16-18 in the α-proteobacterial LSU and the R. americana mt-LSU are shorter (supplementary fig. S2, Supplementary Material online). This reduction introduced instability during early mitoribosomal evolution, which may have led to the incorporation of mL43 and mL49, potentially to provide rigidity to a structurally weakened region (fig. 5B). mL49 provides additional interactions for uL4m allowing it to maintain the same position as in the E. coli ribosome, whereas mL43 performs a similar function for helices H39 and H46 (supplementary fig. S4, Supplementary Material online).
Fig. 5.
Structural changes related to deletion of rRNA helices H16-20 in the Opisthokontan mitoribosomes. Three-dimensional models of the central region around H16-20 in the LSU of (A) the Escherichia coli ribosome (PDB ID: 4V9D), (B) early mitoribosome (Bikonts), and (C) Opisthokontan mitoribosome modeled by combining structures of the bacterial ribosome and the human mitoribosome (PDB ID: 3J9M). rRNA is shown in surface representation and proteins are drawn as cartoons.
The incorporation of mL45 to the perimeter of the exit tunnel (supplementary fig. S5, Supplementary Material online) is likely to have been driven by its ability to confer membrane-binding capabilities to the mitoribosome (Greber et al. 2014) as well as by its role in cementing the uL24m/H19 interactions affected by reduction of H16-18. As mitoribosomes synthesize mostly transmembrane proteins, being tethered to the inner mitochondrial membrane would potentially provide an evolutionary advantage for their cotranslational insertion into the membrane (Pfeffer et al. 2015; Greber and Ban 2016; Ott et al. 2016). The early recruitment of mL45 to the mitoribosome suggests that specialization for membrane-protein synthesis was already apparent at the early stages of the mitoribosomal evolution. This statement is further supported by the early acquisition of mL41, whose incorporation may have also been simultaneously driven by two factors: to stabilize the interactions between the mitoribosome and the inner mitochondrial membrane and to compensate for the reduction of H54-55 that had occurred in all mitoribosomes and the α-protobacterial species. Despite the changes described above, the exit tunnel path and the architecture of the exit tunnel of the nonopisthokontan species resemble the bacterial precursor.
Late Evolution of the Exit Tunnel in Opisthokonta
Upon incorporation of mL43 and mL49 into the mitoribosomes, the structural integrity of the mt-LSU was no longer reliant on H16-20, allowing further shortening of these helices. Furthermore, as Opisthokonta species branched off the rest of unikonts, mitoribosomal proteins mL44 and mL50 were incorporated into the mt-LSU to provide additional stability to mL43 and mL49, respectively. As a result, H16-20 disappeared from the majority of the Opisthokontan ribosomes (fig. 5C).
As Opisthokonta bifurcates into the Metazoa and Fungi branches, mitoribosomal evolution takes two separate paths. In metazoan species (represented here by the mammalian mitoribosome), new protein moieties (mL39, mL51, mL63, and extension of uL23m and uL24m) accumulate and partially compensate for mt-rRNA in the regions that are either lost (H1, H2, H9, H10, H28, H29, H39, H46, H52, H53, H96, and H101) or partially shortened and unwound (H41, H42, H94; (fig. 6B, supplementary fig. S4, Supplementary Material online). These changes cause remodeling of the mitoribosomal periphery, but do not affect the path of the polypeptide exit tunnel, which remains unaltered from that in bacterial ribosomes and the hypothetical mitoribosome of the LOCA.
Fig. 6.
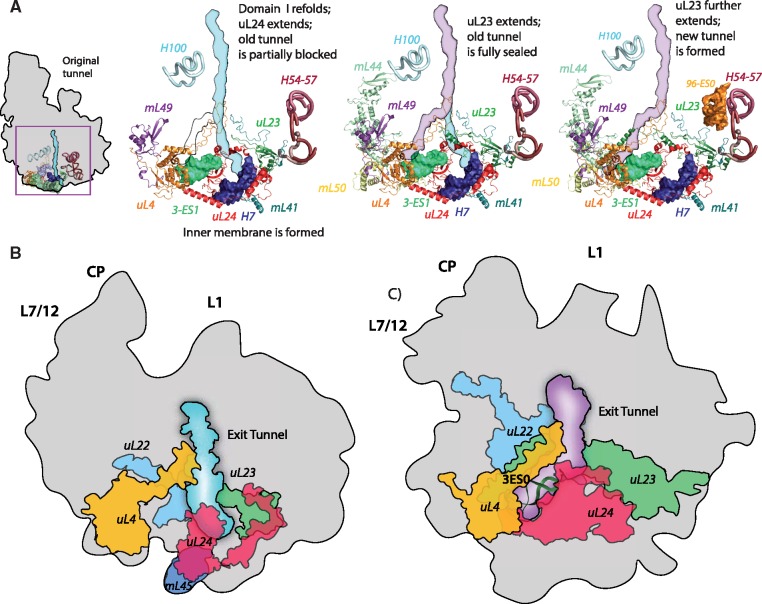
Evolution of the polypeptide exit tunnel in mitoribosomes. (A) Schematic model for formation of the new exit tunnel in the Saccharomyces cerevisiae mitoribosome starting from a bacterial precursor. Representation of the mitoribosomal components that constitute the exit tunnels of (B) Homo sapiens (PDB ID: 3J9M) and (C) S. cerevisiae (PDB ID: 3J6B). Plausible paths and detailed evolutionary events that led to the remodeling of the exit tunnel are described in supplementary figure S4, Supplementary Material online.
In contrast, structural changes around the mitoribosomal exit tunnel in fungal species (represented here by the yeast mitoribosome) led to a diverted path fig. 6C; (Amunts et al. 2014). Remarkably, considering the extent of mt-rRNA expansion in S. cerevisiae (∼200 kDa), the single evolutionary event that appears to have ignited the remodeling of the exit tunnel was the loss of a short stretch of H24, just eight nucleotides long. This loss formed an alternative exit for a nascent polypeptide from the mitoribosome. The fact that this exit developed into a functional tunnel suggests that it was used by a nascent polypeptide shortly after its formation. This raises the questions: 1) to what extent were both tunnels co-active; 2) what structural re-arrangements allowed the alternative tunnel to gain a selective advantage; and 3) at what stage did the original tunnel stop functioning?
To address these questions, we analyzed the structural elements that shape both tunnels. H24 deletion forms a shortcut to the mitoribosomal surface located further away from the membrane than the original exit (supplementary fig. S6A and B, Supplementary Material online). This deletion may have caused (or was caused by) a small positional shift of uL24m, which resulted in a partial blockage of the original exit tunnel. Since this alone is unlikely to be immediately adaptive, it implies that the downstream remodeling of the exit tunnel that provided extra shielding and direction for a diverted nascent polypeptide, took place shortly after the H24 deletion. This remodeling may have happened cooperatively as a partial refolding of domain I of the LSU that included H3-H10 (supplementary fig. S6B and D, Supplementary Material online). As a result of this refolding H5, H6 and H10 were shortened, whereas H9 was elongated by mt-rRNA expansion 9-ES1. Additionally, a new helical expansion, 3-ES1 was formed as an intrusion in H3 (supplementary fig. S6C and D, Supplementary Material online). This element became a new rRNA binding site of proteins uL4m and uL24m, which had been originally (in the bacterial ancestor) bound to H19 and H24, respectively.
A new (partially constructed) exit tunnel was formed because of this remodeling event (fig. 6C). The average diameter of this tunnel (>20 Å) is wider than in bacterial ribosomes (∼14 Å), potentially providing more space for cotranslational folding while also contributing to the accessibility of a nascent polypeptide. As the new tunnel evolved, the original one was blocked by a helical extension of uL23m, diverting the nascent peptides to the new exit route. Further extensions of uL23m and the previously incorporated mL50 assisted in the completion of the new tunnel architecture (fig. 6A). Therefore, the original tunnel was inactivated before the new tunnel fully matured.
Taken together, the data suggest: 1) protein additions adapted the tunnel exit for binding to the membrane in LOCA; 2) the alternative exit in the yeast mitoribosome was formed by several rapid mt-rRNA rearrangements, primarily as a result of deletion in H24; 3) the new tunnel had a functional advantage from its onset, and the original exit was blocked before the full formation of the new tunnel; 4) subsequent structural additions, primarily of uL23m and mL50, have refined the shape of the yeast mitoribosomal exit tunnel.
Early Evolution of the CP
The CP is a structural element of the ribosomal LSU that interacts with the head of the small subunit. In all known cytosolic ribosomes, helices H38 and H84 within the CP are embraced by 5S rRNA and stabilized by proteins. The remodeling of the CP likely began with the incorporation of mL46, which triggered the loss of bL31m in the majority of species (fig. 3, supplementary fig. S7, Supplementary Material online). Since both bL31m and mL46 exist in the CP of the yeast mitoribosome, the deletion of bL31m occurred independently within each lineage and was also facilitated by shortening of mt-rDNA in some species. Incorporation of mL40 into the CP has also been discovered in P. tetraurelia (fig. 3), suggesting its relatively early origin.
As a result of further mt-rDNA shortening, the 5S rRNA gene has vanished from some mt-genomes (fig. 3), For example, 5S rRNA is lost from most Unikonts and some Bikonts but retained in some plants and some protists (Gray et al. 1998). The deletion of 5S rRNA likely coincided with the loss of bL25m, followed by an independent species-specific deletion of uL18m (fig. 3), as both proteins are tightly associated with 5S rRNA. Given the central role of 5S rRNA in the CP and that bacterial ribosomes reconstituted without 5S rRNA have severe protein-synthesis defects (Erdmann et al. 1971), it is difficult to rationalize an evolutionary scenario in which 5S rRNA was absent without a pre-existing element that could be incorporated as a compensatory mechanism. Our analysis shows that mL46 (and in some instances mL40) were acquired before 5S rRNA loss figs. 3 and 7, (supplementary fig. S7, Supplementary Material online), suggesting that it has been partially compensating for the loss of 5S rRNA from the early proto-mitoribosome.
Late Evolution of the CP in Opisthokonta
Once Opisthokonta branched off from the other Unikonts, mL38 was recruited to the CP (fig. 3) to further compensate for the reduction of 5S rRNA. The precursors of these proteins were pre-existing (supplementary tables S3 and S4, Supplementary Material online) and readily available in the mitochondrial matrix. Thus, the loss of 5S rRNA has been compensated for quickly, allowing a consequent reaction chain of structural rearrangements to happen. By LOCA, the H38 and H84 of the CP are stabilized by bacterial proteins uL5m, uL16m, bL18m, bL27m, uL30m, and mitoribosomal proteins mL38, mL40, and mL46 (supplementary fig. S6, Supplementary Material online).
Starting from LOCA, evolutionary events have taken different approaches to compensate for the loss of 5S rRNA. In yeast, whose mt-genome switched to the expansion mode, the gap left by 5S rRNA was filled by expansions of H84 (84-ES1-3) cemented in place by pre-existing protein uL5 and recently acquired mL38 and mL40 (fig. 7 and supplementary fig. S6, Supplementary Material online). The emergence of these expansions resulted in the loss of uL18m from the CP. Thus, this cluster was further stabilized by expansion of protein uL16m and cemented by rRNA expansion 38-ES1. Finally, the 82-ES cluster embraced proteins mL38, mL40, and mL46 from the outside, providing structural integrity to the remodeled CP. The extension of bL27m further cemented the entire rRNA/protein cluster. Overall, these expansions successfully maintained the contacts between the CP and the small subunit and therefore retained the functionality of the CP.
Fig. 7.
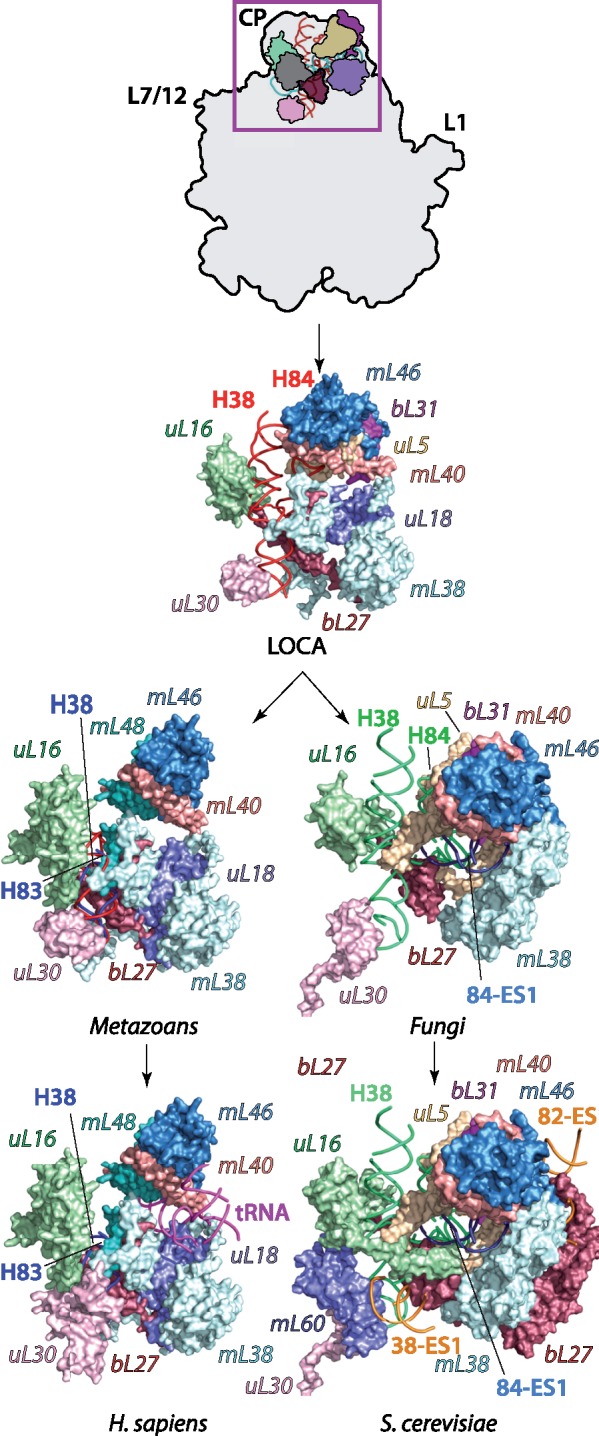
Evolution of the mitoribosomal central protuberance (CP). By LOCA, mL40, and mL46 are incorporated into the CP, resulting in the loss of 5S rRNA (cyan, inset) and bL25m (dark gray, inset). The deletion of 5S rRNA is compensated by two different mechanisms. In Homo sapiens (Mammals), the CP is patched with protein mL48 that compensates for deletion of bL31, and a mt-tRNA molecule that cements the mL40, mL46, mL48 cluster. In Saccharomyces cerevisiae, the expansion 84-ES1 partially fills a region of the CP previously occupied by 5S rRNA. The CP is further stabilized by expansions 38-ES1 and 82-ES1-5 and incorporation of mL60 and expansions of uL16m and bL27m. Plausible paths and detailed evolutionary events that led to the remodeling of the CP are described in supplementary figure S6, Supplementary Material online.
The likely evolutionary scenario for the remodeling of the CP appears to be more complex in metazoans. Upon loss of 5S rRNA, the evolutionary constraints continued to progressively deplete the mt-genome, which led to shortening of H38 and H82-84 of mt-rRNA in the CP region. Protein mL48, highly homologous to uS10m, was incorporated to reinforce the CP, impaired due to initial shortening of H84. However, as the reduction of mt-rRNA in these helices further progressed, the structural integrity and mechanistic function of the CP could not be maintained by the newly incorporated mitoribosomal proteins. Therefore, the stability of the CP in mammalian mitoribosomes was reestablished though the acquisition of a mt-tRNA (fig. 7). Modern mammalian mitoribosomes feature different mt-tRNAs (Rorbach et al. 2016), suggesting that the exact identity does not affect their structural role. The mt-tRNA became incorporated into the CP via interactions with uL18m and mL38, enhancing the binding of mL40 and mL46 (supplementary fig. S6, Supplementary Material online). The analysis of the late evolution of the CP in mammalian mitoribosomes suggests that the structural elements required for remodeling were readily available in advance.
Discussion
Our combined phylogenetic and structural analysis indicates that the primary driving force in the evolution of mitoribosomes is the erosion of mt-genomes. This erosion is caused by: 1) loss of dispensable genes from the mt-genome (Gray et al. 1999; Gray 2015); 2) transferal of essential genes from the mt-genome to the nucleus including those that encode mitoribosomal proteins (Berg and Kurland 2000; Kurland and Andersson 2000); and 3) pressure to minimize the overall length of mt-genomes resulting in the reduction of mt-rRNA. The extent and rates of these interconnected events have been affected by independent selective pressures in different eukaryotic species.
The genetic erosion has considerable effects on mitoribosomal structure and function. mt-rRNA loss destabilizes mitoribosomes and the structural vulnerability of the metastable intermediates appears to make them more receptive to accommodating new elements that stabilize the structure (fig. 8). The patching essentially occurs by accretion in an onion-like fashion (Hsiao et al. 2009), where new elements are accreted onto a pre-existing core. These structural patches are the addition of species-specific and pre-existing nonribosomal macromolecules. The examples of structural patching occurring as a result of the loss of 5S rRNA from the CP and loss of H16-20 in the evolution of the exit tunnel illustrate the principle by which rapid genetic changes that could have destabilized macromolecular complexes actually promoted functional fitness in mitoribosomes. Thus, the mitoribosome is a molecular symbiont with a complex evolutionary history.
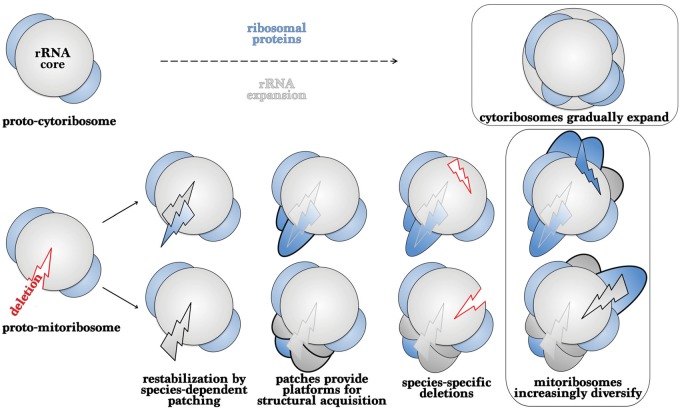
Fig. 8.
Representation of mitoribosomal evolution. Top panel, cytosolic ribosomes (cytoribosomes) expand through the gradual acquisition of rRNA and proteins. Bottom panel, mitoribosomes adapt to erratic deletions of mt-rRNA through the rapid acquisition of locally abundant elements or by mt-rRNA expansion. Those patches provide structural platforms for further acquisitions that might promote functional fitness and become embedded in the structure. Species-specific deletions and acquisitions foster the diversity of mitoribosomes.
Condensed mt-genomes also result in a reduced set of mt-mRNAs that need to be translated. This changes the evolutionary pressures on the mitoribosomes leading to further specialization. For example, the incorporation of mL45 to mammalian mitoribosomes facilitates the interactions between the exit tunnel and the inner mitochondrial membrane in an apparent response to synthesizing exclusively membrane proteins. Further complexity likely comes from constructive neutral evolution, in which structural features become fixed into the mitoribosome through subsequent codependent mutations without the apparent evolution of novel functions (Stoltzfus 1999; Gray et al. 2010; Finnigan et al. 2012). Proteins incorporated into mitoribosomes in this way may subsequently gain function. Additional structural and phylogenetic analyses will be required to track the specific details of early evolutionary changes within mitoribosomes of bikont species.
Conclusions
Our analyses suggest that ablation and expansion of mt-rRNA generates metastable regions of mitoribosomes that require patching by pre-existing elements that may confer new functions. The extent and type of modifications that can be made in different species are determined by the structural toolkits available. Fungal mitoribosomes have a structural toolkit that includes mt-rRNA expansion and protein acquisition, whereas metazoans appear to be restricted to only adding existing RNA elements and proteins. The variable extent and rate of changes to the mt-genome and the different toolkits available in different species are responsible for promoting mitoribosomal diversity.
Materials and Methods
Sequences of rRNAs were obtained from the CRW website (Cannone et al. 2002). The sequences were aligned by MAFFT (Kuraku et al. 2013) using a subset of a previously published bacterial LSU rRNA alignments (Petrov et al. 2014) as a seed and manually adjusted and verified using available 2D and 3D data (Bernier et al. 2018). The phylogenetic trees from rRNA genes were constructed using the MEGA7 program (Kumar et al. 2016). The phylogenetic distribution of mitoribosomal proteins was obtained from the study by Desmond et al. (2011). Superimpositions of the ribosomal 3D structures of mt-S. cerevisiae (Amunts et al. 2014), mt-H. sapiens (Amunts et al. 2015), and E. coli (Selmer et al. 2006), as well as the comparative 3D analysis of mitoribosomal evolution were performed using PyMOL 1.9 (Schrodenger, LLC 2016). Mapping of the data onto 2D and 3D structures was performed using RiboVision (Bernier et al. 2014). Additional supporting information is available in Materials and Methods, Supplementary Material online.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This study was supported by the Swedish Research Council (NT_2015: 04107), the Swedish Foundation for Strategic Research (Future Leaders Grant FFL15: 0325), the Ragnar Söderberg Foundation (Fellowship in Medicine M44/16). The authors would also like to thank Prof. Venki Ramakrishnan for his support and Prof. Loren Williams for helpful comments and discussion.
Author Contributions
A.S.P., A.B., and A.A. designed research; A.S.P., E.C.W., C.R.B., and A.M.N. performed research; A.S.P., C.R.B., A.M.N., and A.A. analyzed data; A.S.P. and E.C.W. prepared the figures; and A.S.P., E.C.W., A.B., and A.A. wrote the manuscript.
References
- Amunts A, Brown A, Bai X-C, Llácer JL, Hussain T, Emsley P, Long F, Murshudov G, Scheres SH, Ramakrishnan V.. 2014. Structure of the yeast mitochondrial large ribosomal subunit. Science 3436178:1485–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amunts A, Brown A, Toots J, Scheres SH, Ramakrishnan V.. 2015. The structure of the human mitochondrial ribosome. Science 3486230:95–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen GR, Thirup S, Spremulli LL, Nyborg J.. 2000. High resolution crystal structure of bovine mitochondrial EF-Tu in complex with GDP. J Mol Biol. 2972:421–436. [DOI] [PubMed] [Google Scholar]
- Andersson SGE, Zomorodipour A, Andersson JO, Sicheritz-Ponten T, Alsmark UCM, Podowski RM, Naslund AK, Eriksson A-S, Winkler HH, Kurland CG.. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 3966707:133–140. [DOI] [PubMed] [Google Scholar]
- Atkinson GC, Baldauf SL.. 2011. Evolution of elongation factor G and the origins of mitochondrial and chloroplast forms. Mol Biol Evol. 283:1281–1292. [DOI] [PubMed] [Google Scholar]
- Ban N, Beckmann R, Cate JHD, Dinman JD, Dragon F, Ellis SR, Lafontaine DLJ, Lindahl L, Liljas A, Lipton JM, et al. 2014. A new system for naming ribosomal proteins. Curr Opin Struct Biol. 24:165–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M.. 2011. The structure of the eukaryotic ribosome at 3.0 A resolution. Science 3346062:1524–1529. [DOI] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Rapp BA, Wheeler DL. (Benson2000 co-authors). 2000. GenBank. Nucl Acids Res. 28(1):15–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg OG, Kurland C.. 2000. Why mitochondrial genes are most often found in nuclei. Mol Biol Evol. 176:951–961. [DOI] [PubMed] [Google Scholar]
- Bernier CR, Petrov AS, Waterbury CC, Jett J, Li F, Freil LE, Xiong X, Wang L, Migliozzi BL, Hershkovits E, et al. 2014. RiboVision suite for visualization and analysis of ribosomes. Faraday Discuss. 169:195–207. [DOI] [PubMed] [Google Scholar]
- Bernier CR, Petrov AS, Kovacs NA, Penev P, Williams LD.. 2018. Translation: the universal structural core of life. Mol Biol Evol. 35(8):2065–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björkholm P, Harish A, Hagström E, Ernst AM, Andersson SGE.. 2015. Mitochondrial genomes are retained by selective constraints on protein targeting. Proc Natl Acad Sci USA. 11233:10154–10161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A, Amunts A, Bai X-C, Sugimoto Y, Edwards PC, Murshudov G, Scheres SH, Ramakrishnan V.. 2014. Structure of the large ribosomal subunit from human mitochondria. Science 3466210:718–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullerwell CE, Burger G, Gott JM, Kourennaia O, Schnare MN, Gray MW.. 2010. Abundant 5S rRNA-like transcripts encoded by the mitochondrial genome in amoebozoa. Eukaryot Cell. 95:762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, et al. 2002. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 31:2.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Schaeffer RD, Liao Y, Kinch LN, Pei J, Shi S, Kim B-H, Grishin NV.. 2014. ECOD: an evolutionary classification of protein domains. PLoS Comput Biol. 1012:e1003926.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai N, Brown A, Amunts A, Ramakrishnan V.. 2017. The structure of the yeast mitochondrial ribosome. Science 3556324:528–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmond E, Brochier-Armanet C, Forterre P, Gribaldo S.. 2011. On the last common ancestor and early evolution of eukaryotes: reconstructing the history of mitochondrial ribosomes. Res Microbiol. 1621:53–70. [DOI] [PubMed] [Google Scholar]
- Erdmann VA, Fahnestock S, Higo K, Nomura M.. 1971. Role of 5S RNA in the functions of 50S ribosomal subunits. Proc Natl Acad Sci USA. 6812:2932–2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferla MP, Thrash JC, Giovannoni SJ, Patrick WM.. 2013. New rRNA gene-based phylogenies of the Alphaproteobacteria provide perspective on major groups, mitochondrial ancestry and phylogenetic instability. PLoS One 812:e83383.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnigan GC, Hanson-Smith V, Stevens TH, Thornton JW.. 2012. Evolution of increased complexity in a molecular machine. Nature 4817381:360–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbi SA. 1996. Expansion segments: regions of variable size that interrupt the universal core secondary structure of ribosomal RNA In: Zimmermann RA, Dahlberg AE, editors. Ribosomal RNA—structure, evolution, processing, and function in protein synthesis. Boca Raton (FL: ): CRC Press; p. 71–87. [Google Scholar]
- Gray MW. 2014. The pre-endosymbiont hypothesis: a new perspective on the origin and evolution of mitochondria. Cold Spring Harbor Perspect Biol. 63: a016097.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW. 2015. Mosaic nature of the mitochondrial proteome: implications for the origin and evolution of mitochondria. Proc Natl Acad Sci USA. 11233:10133–10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW, Burger G, Lang BF.. 1999. Mitochondrial evolution. Science 2835407:1476–1481. [DOI] [PubMed] [Google Scholar]
- Gray MW, Lukeš J, Archibald JM, Keeling PJ, Doolittle WF.. 2010. Irremediable complexity? Science 3306006:920–921. [DOI] [PubMed] [Google Scholar]
- Gray MW, Lang BF, Cedergren R, Golding GB, Lemieux C, Sankoff D, Turmel M, Brossard N, Delage E, Littlejohn TG, et al. 1998. Genome structure and gene content in protist mitochondrial DNAs. Nucleic Acids Res. 264:865–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber BJ, Bieri P, Leibundgut M, Leitner A, Aebersold R, Boehringer D, Ban N.. 2015. Ribosome. The complete structure of the 55S mammalian mitochondrial ribosome. Science 3486232:303–308. [DOI] [PubMed] [Google Scholar]
- Greber BJ, Boehringer D, Leitner A, Bieri P, Voigts-Hoffmann F, Erzberger JP, Leibundgut M, Aebersold R, Ban N.. 2014. Architecture of the large subunit of the mammalian mitochondrial ribosome. Nature 5057484:515–519. [DOI] [PubMed] [Google Scholar]
- Greber BJ, Ban N.. 2016. Structure and function of the mitochondrial ribosome. Annu Rev Biochem. 85:103–132. [DOI] [PubMed] [Google Scholar]
- Harish A, Kurland CG.. 2017. Mitochondria are not captive bacteria. J Theor Biol. 434:88–98. [DOI] [PubMed] [Google Scholar]
- Hedges SB, Marin J, Suleski M, Paymer M, Kumar S.. 2015. Tree of life reveals clock-like speciation and diversification. Mol Biol Evol. 324:835–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao C, Mohan S, Kalahar BK, Williams LD.. 2009. Peeling the onion: ribosomes are ancient molecular fossils. Mol Biol Evol. 2611:2415–2425. [DOI] [PubMed] [Google Scholar]
- Husnik F, McCutcheon JP.. 2016. Repeated replacement of an intrabacterial symbiont in the tripartite nested mealybug symbiosis. Proc Natl Acad Sci U S A. 11337:E5416–E5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz LA. 2012. Origin and diversification of eukaryotes. Annu Rev Microbiol. 66:411–427. [DOI] [PubMed] [Google Scholar]
- Klinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N.. 2011. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science 3346058:941–948. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Suleski M, Hedges SB.. 2017. TimeTree: a resource for timelines, timetrees, and divergence times. Mol Biol Evol. 347:1812–1819. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K.. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 337:1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraku S, Zmasek CM, Nishimura O, Katoh K.. 2013. aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res. 41(W1): W22–W28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland C, Andersson S.. 2000. Origin and evolution of the mitochondrial proteome. Microbiol Mol Biol Rev. 644:786–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmenko A, Atkinson GC, Levitskii S, Zenkin N, Tenson T, Hauryliuk V, Kamenski P.. 2014. Mitochondrial translation initiation machinery: conservation and diversification. Biochimie 100:132–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang BF, Burger G, O'Kelly CJ, Cedergren R, Golding GB, Lemieux C, Sankoff D, Turmel M, Gray MW.. 1997. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature 3876632:493–497. [DOI] [PubMed] [Google Scholar]
- Lazcano A, Pereto J.. 2017. On the origin of mitosing cells: a historical appraisal of Lynn Margulis endosymbiotic theory. J Theor Biol. 434:80–87. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P.. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44(W1): W242–W245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martijn J, Vosseberg J, Guy L, Offre P, Ettema TJG.. 2018. Deep mitochondrial origin outside the sampled alphaproteobacteria. Nature 5577703:101–105. [DOI] [PubMed] [Google Scholar]
- Melnikov S, Manakongtreecheep K, Söll D.. 2018. Revising the structural diversity of ribosomal proteins across the three domains of life. Mol Biol Evol. 357:1588–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott M, Amunts A, Brown A.. 2016. Organization and regulation of mitochondrial protein synthesis. Annu Rev Biochem. 85:77–101. [DOI] [PubMed] [Google Scholar]
- Petrov AS, Bernier CR, Hsiao C, Norris AM, Kovacs NA, Waterbury CC, Stepanov VG, Harvey SC, Fox GE, Wartell RM, et al. 2014. Evolution of the ribosome at atomic resolution. Proc Natl Acad Sci USA. 11128:10251–10256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S, Woellhaf MW, Herrmann JM, Förster F.. 2015. Organization of the mitochondrial translation machinery studied in situ by cryoelectron tomography. Nat Commun. 6: 6019. [DOI] [PubMed] [Google Scholar]
- Pi M, Morio T, Urushihara H, Tanaka Y.. 1998. Characterization of a novel small RNA encoded by Dictyostelium discoideum mitochondrial DNA. Mol Gener Genet MGG. 2572:124–131. [DOI] [PubMed] [Google Scholar]
- Poole AM, Gribaldo S.. 2014. Eukaryotic origins: how and when was the mitochondrion acquired? Cold Spring Harb Perspect Biol. 612: a015990.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N.. 2011. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science 3316018:730–736. [DOI] [PubMed] [Google Scholar]
- Ramrath DJF, Niemann M, Leibundgut M, Bieri P, Prange C, Horn EK, Leitner A, Boehringer D, Schneider A, Ban N.. 2018. Evolutionary shift toward protein-based architecture in trypanosomal mitochondrial ribosomes. Science 362(6413):eaau7735. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Ezpeleta N, Embley TM.. 2012. The SAR11 group of alpha-proteobacteria is not related to the origin of mitochondria. PLoS One 71:e30520.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorbach J, Gao F, Powell CA, D’Souza A, Lightowlers RN, Minczuk M, Chrzanowska-Lightowlers ZM.. 2016. Human mitochondrial ribosomes can switch their structural RNA composition. Proc Natl Acad Sci USA. 11343:12198–12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagan L. 1967. On the origin of mitosing cells. J Theor Biol. 143:225–IN226. [DOI] [PubMed] [Google Scholar]
- Schrodinger LLC. 2016. The PyMOL Molecular Graphics System, Version 1.9.
- Selmer M, Dunham CM, Murphy FV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V.. 2006. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 3135795:1935–1942. [DOI] [PubMed] [Google Scholar]
- Smits P, Smeitink JA, van den Heuvel LP, Huynen MA, Ettema TJ.. 2007. Reconstructing the evolution of the mitochondrial ribosomal proteome. Nucleic Acids Res. 3514:4686–4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltzfus A. 1999. On the possibility of constructive neutral evolution. J Mol Evol. 492:169–181. [DOI] [PubMed] [Google Scholar]
- Thrash JC, Boyd A, Huggett MJ, Grote J, Carini P, Yoder RJ, Robbertse B, Spatafora JW, Rappé MS, Giovannoni SJ.. 2011. Phylogenomic evidence for a common ancestor of mitochondria and the SAR11 clade. Sci Rep. 1:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Sluis EO, Bauerschmitt H, Becker T, Mielke T, Frauenfeld J, Berninghausen O, Neupert W, Herrmann JM, Beckmann R.. 2015. Parallel structural evolution of mitochondrial ribosomes and OXPHOS complexes. Genome Biol Evol. 75:1235–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viklund J, Martijn J, Ettema TJG, Andersson SGE.. 2013. Comparative and phylogenomic evidence that the alphaproteobacterium HIMB59 is not a member of the oceanic SAR11 clade. PLoS One 811:e78858.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong W, Bai X-C, Brown A, Fernandez IS, Hanssen E, Condron M, Tan YH, Baum J, Scheres SHW.. 2014. Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine. eLife 3:e03080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachar I, Szilágyi A, Számadó S, Szathmáry E.. 2018. Farming the mitochondrial ancestor as a model of endosymbiotic establishment by natural selection. Proc Natl Acad Sci USA. 115(7):E1504–E1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.