Abstract
Genotype-specific contributions to the environmental tolerance and disease susceptibility of corals are widely accepted. Yet our understanding of how host genotype influences the composition and stability of the coral microbiome subjected to environmental fluctuations is limited. To gain insight into the community dynamics and environmental stability of microbiomes associated with distinct coral genotypes, we assessed the microbial community associated with Acropora tenuis under single and cumulative pressure experiments. Experimental treatments comprised either a single pulse of reduced salinity (minimum of 28 psu) or exposure to the cumulative pressures of reduced salinity (minimum of 28 psu), elevated seawater temperature (+2 °C), elevated pCO2 (900 ppm), and the presence of macroalgae. Analysis of 16S rRNA gene amplicon sequence data revealed that A. tenuis microbiomes were highly host-genotype specific and maintained high compositional stability irrespective of experimental treatment. On average, 48% of the A. tenuis microbiome was dominated by Endozoicomonas. Amplicon sequence variants (ASVs) belonging to this genus were significantly different between host individuals. Although no signs of stress were evident in the coral holobiont and the vast majority of ASVs remained stable across treatments, a microbial indicator approach identified 26 ASVs belonging to Vibrionaceae, Rhodobacteraceae, Hahellaceae, Planctomycetes, Phylobacteriaceae, Flavobacteriaceae, and Cryomorphaceae that were significantly enriched in corals exposed to single and cumulative stressors. While several recent studies have highlighted the efficacy of microbial indicators as sensitive markers for environmental disturbance, the high host-genotype specificity of coral microbiomes may limit their utility and we therefore recommend meticulous control of host-genotype effects in coral microbiome research.
Keywords: Coral microbiome, Host-genotype specificity, Endozoicomonas, Microbial indicators
Introduction
Corals contain abundant and diverse communities of microorganisms that together form a holobiont (Rohwer et al., 2002). The photoautotrophic dinoflagellate endosymbionts of the family Symbiodiniaceae are by far the best studied symbiotic partners of reef-building corals. Symbiodiniaceae lineages vary between coral species (Smith, Ketchum & Burt, 2017) and even between host genotypes of conspecific corals (Brener-Raffalli et al., 2018). Fine-scale adaptations of the Symbiodiniaceae lineages can influence the environmental sensitivity of their hosts (Baker, 2003), as some Symbiodiniaceae lineages are more thermo-tolerant and hence infer higher bleaching tolerance to corals (Rowan, 2004). Corals also harbor diverse communities of bacteria, archaea, and viruses (Bourne, Morrow & Webster, 2016; Hernandez-Agreda, Gates & Ainsworth, 2017; Thurber et al., 2017). Excessive environmental stress resulting in coral bleaching, tissue necrosis, and mortality, is often accompanied by a shift in the microbiome (Glasl, Herndl & Frade, 2016; Zaneveld, McMinds & Thurber, 2017). While the importance of the microbiome to coral fitness is well appreciated (Bourne, Morrow & Webster, 2016; Grottoli et al., 2018; Peixoto et al., 2017; Ziegler et al., 2017), the microbiome’s potential to expand the environmental tolerance of coral holobionts via microbial shuffling and switching is far less understood (Webster & Reusch, 2017). Endozoicomonas, a bacterial genus commonly associated with marine invertebrates, is considered a putative symbiont of corals as it can occur at high abundance in aggregates within the tissue (Neave et al., 2016b) and loss of Endozoicomonas is frequently seen in bleached or diseased corals (Bayer et al., 2013; Glasl, Herndl & Frade, 2016). Pangenome analysis of Endozoicomonas has revealed evidence for functional specificity between strains (Neave et al., 2017), hence fine-scale changes in the composition or relative abundance of different Endozoicomonas strains may contribute to variation in the environmental tolerance and disease susceptibility of conspecific corals.
A fundamental question in microbiome research is whether host intrinsic factors (e.g., genetics) or the environment are the main drivers of microbiome composition and stability (Spor, Koren & Ley, 2011; Wullaert, Lamkanfi & McCoy, 2018). The influence of host genetics and environmental factors on the community composition of a microbiome varies between host species and even between host compartments. For example, the rizhosphere microbiome of the perennial plant Boechera stricta are predominantly shaped by environmental factors, however, its leaf associated microbial community is largely controlled by host genetic factors (Wagner et al., 2016). Host-genotype specific factors also shape the gut microbiome of Drosophila melanogaster, a model system for animal-microbe interactions, and further mediate its nutritional phenotype (Chaston et al., 2016). While many coral microbiome studies have focused on the effect of environmental stress (e.g., elevated temperature, increased macroalgae abundance, anthropogenic pollution, and declining water quality (Garren et al., 2009; Vega Thurber et al., 2009; Zaneveld et al., 2016; Zhang et al., 2015)); the combined influence of host-genotype and environment on the microbial community composition remains largely unknown. This is a critical knowledge gap as microbiome-by-host genotype-by-environment interactions may have important implications for the resistance of corals to stress and disease. Considering the recent declines in coral reefs (De’ath et al., 2012; Hoegh-Guldberg et al., 2007; Hughes et al., 2017) and the key role microorganisms play in maintaining host health (Bourne, Morrow & Webster, 2016), disentangling the effect of environment and host-genotype on a coral’s microbiota is of utmost importance.
This study investigated the effect of host genotype-by-environment interactions on the microbiome of Acropora tenuis. The compositional variability of the A. tenuis microbiome associated with distinct host genotypes (individual coral colonies) was assessed with high taxonomic resolution based on amplicon sequence variants (ASV). The stability of the microbiome was further investigated by exposing corals to acute salinity fluctuations (ranging from 35 to 28 psu) under current (sea surface temperature of 27.5 °C and pCO2 of 400 ppm) and future (sea surface temperature of 29.5 °C, pCO2 of 900 ppm, and macroalgae) projected reef conditions. Stress treatments were designed to simulate environmental conditions that A. tenuis can experience in their natural environment. Both stress treatments (single and cumulative stress) consisted of a non-lethal low salinity pulse, mimicking freshwater influx into the reef as occurs after large rainfall events, often linked to cyclones that cross the Eastern Australian coastline and result in large riverine flows into the nearshore and mid-shelf reef areas of the Great Barrier Reef (Jones & Berkelmans, 2014; VanWoesik, DeVantier & Glazebrook, 1995).
Materials and Methods
Coral colony collection and experimental design
Nine A. tenuis colonies were collected from Davies Reef (Great Barrier Reef, Australia) in March 2017 and transported to the National Sea Simulator at the Australian Institute of Marine Science (Townsville, QLD, Australia). Corals were fragmented into coral nubbins, glued onto aragonite plugs and kept at control temperature (27.5 °C) and light (150 mol photons m−2 s−1) conditions in indoor flow-through aquaria for 3 weeks to allow healing. Corals were collected under the permit G12/35236.1 granted by the Great Barrier Reef Marine Park Authority to the Australian Institute of Marine Science.
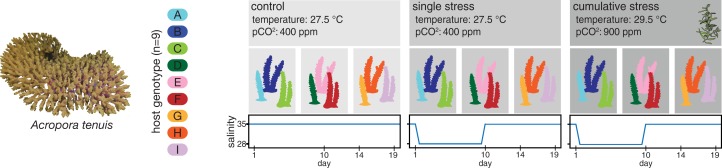
The experimental design consisted of three treatment conditions: (1) control, (2) single stress, and (3) cumulative stress treatment (Fig. 1). Nubbins of all nine A. tenuis genotypes (A–I) were exposed to all three treatment conditions to explore microbiome variation according to host genotype. Each experimental aquarium (three aquaria per treatment) held nubbins of three A. tenuis genotypes (four nubbins per genotype, total of 12 nubbins per aquarium). Coral nubbins were acclimated to experimental aquaria for 3 weeks during which corals in the cumulative stress treatment were gradually ramped to 29.5 °C and 900 ppm pCO2 over a period of 12 days. Corals in the control and single stressor treatments were kept at stable temperature (27.5 °C) and ambient (400 ppm) pCO2 conditions throughout the experiment.
Figure 1. Conceptual overview of the experimental design. Acropora tenuis colonies (n = 9) were fragmented and coral nubbins of each host genotype (A–I) were exposed to three different treatment conditions (control, single stress, and cumulative stress) and sampled on a regular basis throughout the experiment (day 1, 10, 14, and 19).
Image credit: Bettina Glasl.
Salinity was ramped down over 3 h to a minimum of 28 psu and oscillated between 28 and 30 psu in a 6 h rhythm to simulate natural fluctuations occurring on reefs (tidal influences). Temperature and pCO2 adjusted freshwater (0.2 μm filtered) was used to lower salinities prior to supplying the low saline seawater to the aquaria tanks. After 7 days of low salinity, the salinity was ramped up (3 h) to 35 psu. In the cumulative stress treatment, corals were additionally exposed to elevated temperature (29.5 °C), pCO2 (900 ppm), and macroalgae (Sargassum sp.), as predicted for the end of the 21st century (IPCC, 2014).
Samples were collected regularly throughout the experiment (see Fig. 1), including 24 h before the salinity pulse was induced (day 1) and at three time points (day 10, 14, and 19) after the low-salinity stress exposure. All nubbins were processed as follows: effective quantum yield was measured (pulse amplitude modulation (PAM) fluorometry), photographed, inspected for visual signs of stress (tissue lesions, bleaching, and necrosis), rinsed with 0.2 μm filter-sterilized seawater, snap frozen in liquid nitrogen and stored at −80 °C until further processing.
Coral nubbins were defrosted on ice before tissue was removed with an airgun in 1 × PBS (pH = 7.4), homogenized for 1 min at 12.5 rpm with a hand-held tissue homogenizer (Heidolph Silent Crusher M) and subsequently aliquoted for the quantification of Symbiodiniaceae cell density, chlorophyll a, protein concentration, and DNA extraction for amplicon-based sequencing of the 16S rRNA gene. Aliquots (500 μl) for Symbiodiniaceae cell counts were fixed with formaldehyde (final concentration 1.5%) and stored in the dark at room temperature. Aliquots for chlorophyll a, protein, and DNA extraction (1 ml each) were centrifuged for 10 min at 16,000×g, the supernatant was discarded and the remaining pellet was snap frozen in liquid nitrogen and stored at −80 °C until further processing. Coral nubbin surface area was assessed by a single paraffin wax dipping for 2 s followed by 5 min air-drying. The weight of each coral nubbin before and after dipping was quantified and the surface area was calculated against a standard curve.
Physiology of Symbiodiniaceae and the coral holobiont
The effective quantum yield of the Symbiodiniaceae was measured using PAM fluorometry. Corals were light adapted (5 h) before measuring the response of the photosystem II effective quantum yield (ΔF/Fm′) with a Heinz Walz™ Imaging PAM as previously described (Chakravarti, Beltran & Van Oppen, 2017). Coral nubbins were exposed to a saturation pulse and the minimum and maximum fluorescence was recorded and effective quantum yield was calculated (see Eq. S1).
Symbiodiniaceae cell densities were manually counted under a stereomicroscope using formaldehyde fixed tissue samples (final c = 1.5%). Samples were briefly vortexed and 9 μl of each sample was added to either side of two haemocytometers and the density of symbiont cells was quantitatively normalized to the tissue blastate and aliquot volume, and standardized to the nubbin’s surface area.
Chlorophyll a was extracted and concentrations were measured using a spectrophotometric assay. Tissue pellets were defrosted on ice, centrifuged at 16,000×g for 10 min at 4 °C, and remaining supernatant was discarded. Pellets were re-suspended in 1 ml of 100% acetone and incubated in the dark for 24 h at 4 °C after which they were centrifuged at 16,000×g for 10 min and supernatant (200 μl) was pipetted into a 96-well plate in triplicate. Absorbance at 630 and 663 nm was measured using a BioTek™ microplate reader and chlorophyll a concentration was calculated (see Eq. S1), quantitatively normalized to the tissue blastate and aliquot volume, and standardized to the nubbin’s surface area.
Total protein concentration was quantified using a Pierce™ BCA Protein Assay Kit (Thermo Scientific, Waltham, MA, USA) following the manufacturer’s instruction. Absorbance was measured in triplicate for each sample at 562 nm in a BioTek™ Plate reader. Standard curves were calculated using a bovine serum albumin (BSA) solution, creating a working range between 20 and 2,000 μg ml−1 and total protein was calculated against the BSA standard curve, quantitatively normalized to the tissue blastate and aliquot volume, and standardized to the surface area of each individual nubbin.
DNA extraction, 16S rRNA gene sequencing and analysis
DNA of all coral samples was extracted using the DNeasy PowerBiofilm Kit (QIAGEN, Venlo, Netherlands) following the manufacturer’s instructions. Blank extractions were included to control for kit contamination. Coral DNA extracts were stored at −80 °C until shipment on dry ice to Ramaciotti Centre (University of New South Wales, Australia) for sequencing. The V1–V3 region of the 16S rRNA gene was amplified using primers 27F (5′-AGAGTTTGATCMTGGCTCAG-3′; Lane, 1991) and 519R (5′-GWATTACCGCGGCKGCTG-3′; Turner et al., 1999) and libraries were prepared with the Illumina TruSeq protocol, followed by Illumina MiSeq 2 × 300 bp sequencing (see Table S1).
Demultiplexed paired end reads were analyzed in QIIME2 (Version 2017.9.0; https://qiime2.org) as previously described by Glasl et al. (2018). In brief, forward and reverse reads were truncated at their 3′ end at the 296 and 252 sequencing positions, respectively. Samples were checked for chimeras and grouped into features based on 100% sequence similarity, from here on referred to as ASV, using DADA2 (Callahan et al., 2016). Multiple de novo sequence alignments of the representative sequences were performed using MAFFT (Katoh et al., 2002). Non-conserved and highly gapped columns from the alignment were removed using default settings of the mask option in QIIME2. Unrooted and rooted trees were generated for phylogenetic diversity analysis using FastTree. For taxonomic assignment, a Naïve-Bayes classifier was trained on the SILVA v123 99% operational taxonomic units, where reference sequences only included the V1–V2 regions (27F/519R primer pair) of the 16S rRNA genes. The trained classifier was applied to the representative sequences to assign taxonomy. A total of 11,063,364 reads were retrieved from 100 sequenced samples and clustered into 4,624 ASVs (Table 1). Chloroplast and Mitochondria derived sequence reads and singletons were removed from the dataset and the feature table was rarefied to an even sequencing depth of 3,506 sequencing reads, leading to the exclusion of four samples. Demultiplexed sequences and metadata are available from the NCBI Sequence Read Archives under accession number PRJNA492377.
Table 1. Sequencing and sample overview.
| Host-genotype | Total no. of samples | No. of sequences | Richnessa | Evennessa | Shannon Indexa |
|---|---|---|---|---|---|
| A | 12 | 54,352 (±18,259) | 71 (±64) | 0.63 (±0.05) | 2.53 (±0.48) |
| B | 12 | 31,702 (±19,058) | 51 (±44) | 0.66 (±0.14) | 2.49 (±0.86) |
| C | 12 | 26,421 (±26,065) | 108 (±86) | 0.73 (±0.11) | 3.23 (±0.65) |
| D | 12 | 59,543 (±28,560) | 101 (±102) | 0.64 (±0.07) | 2.74 (±0.80) |
| E | 12 | 27,348 (±24,386) | 100 (±110) | 0.69 (±0.10) | 2.97 (±0.81) |
| F | 12 | 36,097 (±21,293) | 108 (±103) | 0.73 (±0.08) | 3.18 (±0.84) |
| G | 4 | 55,460 (±35,822) | 126 (±74) | 0.75 (±0.07) | 3.46 (±0.74) |
| H | 12 | 44,101 (±19,488) | 92 (±63) | 0.65 (±0.14) | 2.81 (±0.64) |
| I | 12 | 51,998 (±23,968) | 109 (±73) | 0.63 (±0.08) | 2.82 (±0.65) |
Note:
Diversity indices (average ± SD) for each host genotype are based on a non-rarefied ASV table from which chloroplast and mitochondria derived reads were removed.
Statistical analysis
Statistical analysis was performed in R (R Development Core Team, 2008). Holobiont health metadata were z-score standardized and variation between treatments and host genotypes was evaluated using Analysis of Variance (ANOVA) and if applicable, variations were further assessed with a Tukey post hoc test. Multivariate statistical approaches including Multivariate Homogeneity of Group Dispersion (“vegan package”; Oksanen et al., 2013), Permutation Multivariate Analysis of Variance (PERMANOVA, “vegan package”; Oksanen et al., 2013), Non-metric multidimensional scaling (“phyloseq package”; McMurdie & Holmes, 2013) and distance based Redundancy Analysis (db-RDA “phyloseq package”; McMurdie & Holmes, 2013) were based on Bray–Curtis dissimilarities. Mantel statistics based on Pearson’s product-moment correlation (mantel test, “vegan package” (Oksanen et al., 2013) were used to evaluate whether sample-to-sample dissimilarities in microbiome composition and physiological holobiont health parameters (protein concentration, chlorophyll a concentration, Symbiodiniaceae cell densities, and effective quantum yield) were correlated. Holobiont health parameters were z-score standardized and dissimilarity matrices were based on Bray–Curtis dissimilarities.
Alpha diversity measures including richness and Shannon diversity for the Endozoicomonas community were analyzed using the “phyloseq package” (McMurdie & Holmes, 2013). Variation in the total relative abundance of all Endozoicomonas ASVs per sample between treatments, over time and between host-genotypes was assessed using ANOVAs with arcsine-square-root transformed relative abundance data. The phylogenetic tree of the 11 most abundant Endozoicomonas ASVs was produced with phyloseq (McMurdie & Holmes, 2013) using the Newick rooted tree generated in QIIME2 (version 2017.9.0; https://qiime2.org).
Indicator value analysis (IndVal, “indispecies” package; De Cáceres & Legendre, 2009) was used to identify ASVs significantly associated with treatment groups (control, single stress, and cumulative stress) based on their occurrence and abundance distribution. Day 1 samples were excluded from the IndVal analysis to restrict the dataset to ASVs significantly associated with coral tissue after stress exposure (day 10, 14, and 19).
Graphs were created in R using ggplot2 (Wickham, 2009) and phyloseq packages (McMurdie & Holmes, 2013). Alluvial diagram was generated in RAWGraph (Mauri et al., 2017).
Results
Coral holobiont physiological response
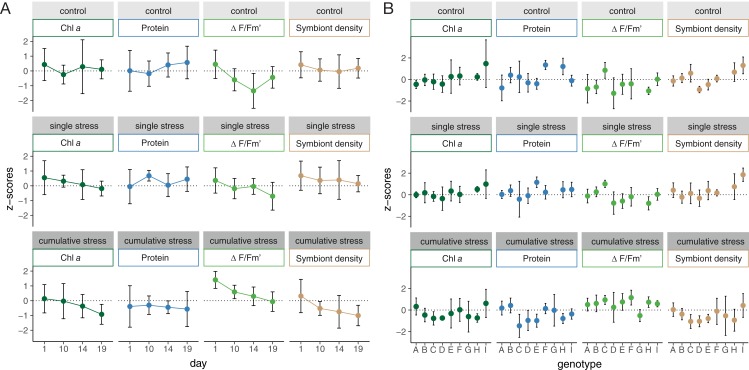
Corals showed no visual signs of stress (change in pigmentation, bleaching, tissue necrosis, and/or mortality) in any treatment. Chlorophyll a concentrations remained stable between treatments (one-way ANOVA with sampling time point as blocking factor, F(2/94) = 2.707, p = 0.072), however, effective quantum yield (ΔF/Fm′; one-way ANOVA with sampling time point as blocking factor, F(2/94) = 15.52, p = 1.49 × 10−6), symbiont cell densities (one-way ANOVA with sampling time point as blocking factor, F(2/94) = 8.83, p = 3.06 × 10−4) and protein concentration (one-way ANOVA with sampling time point as blocking factor, F(2/94) = 5.563, p = 5.21 × 10−3) varied significantly between treatments within sampling time points (Fig. 2A). Coral nubbins in the cumulative stress treatment contained significantly lower protein and symbiont cell densities, while displaying significantly higher effective quantum yield compared to nubbins in the control and single stressor treatments (Tukey post hoc test; Table S2). Furthermore, effective quantum yield (one-way ANOVA, F(8/91) = 2.688, p = 0.0106), symbiont cell densities (one-way ANOVA, F(8/91) = 4.334, p = 1.86 × 10−4) and chlorophyll a concentrations (one-way ANOVA, F(8/91) = 2.773, p = 8.64 × 10−3) varied significantly between host genotypes (Fig. 2B). Protein concentration, however, was unaffected by host genotype (one-way ANOVA, F(8/91) = 1.783, p = 0.0906) and hence was the only holobiont health parameter solely affected by treatment.
Figure 2. Physiological response of Acropora tenuis under control, single stress, and cumulative stress treatments.
Variations in the chlorophyll a (Chl a) concentration, protein concentration, effective quantum yield (ΔF/Fm′) and symbiont cell density (Symbiont density) of A. tenuis (A) over time (day 1, 10, 14, and 19) and (B) between individual host-genotypes (A–I). Physiological parameters are z-score standardized and error bars represent standard deviations.
Microbial community response
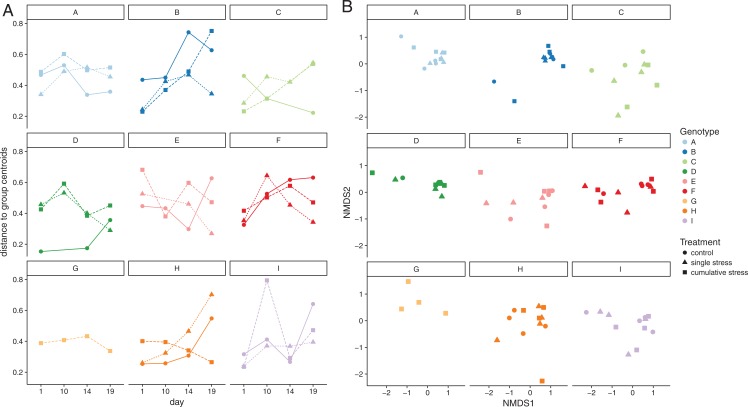
The microbiome of A. tenuis remained highly stable across treatments, with no significant changes in the heterogeneity, also referred to as multivariate dispersion (one-way ANOVA, F(2/93) = 1.2107, p = 0.3026; Fig. 3A), or in community composition (PERMANOVA, p = 0.5156, 10,000 permutations; Fig. 3B). However, the microbiome composition varied significantly between individual host genotypes (PERMANOVA, p = 9.99 × 10−5, 10,000 permutations), but was unaffected by treatment, sampling time point or tank effects when tested for each genotype individually (PERMANOVA with host-genotype as blocking factor, 10,000 permutations; Table S3). Similar results were obtained using presence/absence data (Fig. S1). Host genotype was the only significant factor, explaining 32.4% of the observed community variation (permutational ANOVA for db-RDA based on 1,000 permutations, p = 9.99 × 10−4; Fig. S2). Treatment and holobiont health parameters did not significantly contribute to the microbiome variation (Table S4). Furthermore, no significant correlation between similarity matrices based on microbiome composition and physiological holobiont health parameters was observed (chlorophyll a, protein, effective quantum yield, and symbiont cell density; Mantel statistic based on Pearson’s product-moment correlation r = −0.0238, p = 0.6243, 10,000 permutations).
Figure 3. Configurational and compositional stability of Acropora tenuis microbiome.
(A) Multivariate dispersion (heterogeneity) measured by the distance to the group centroid for each host-genotype (A–I) within each treatment (control, acute stress, and cumulative stress) over time (day 1, 10, 14, and 19). (B) Non-metric multidimensional scaling (NMDS) illustrating compositional similarity of sample replicates of each host-genotype (A–I) under different treatment conditions (control, single stress, and cumulative stress).
Endozoicomonas assemblage
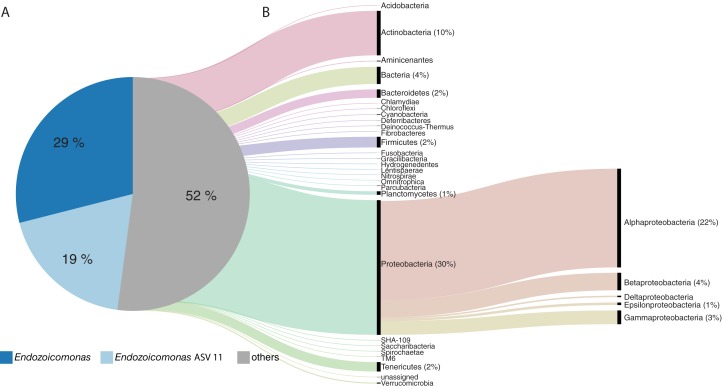
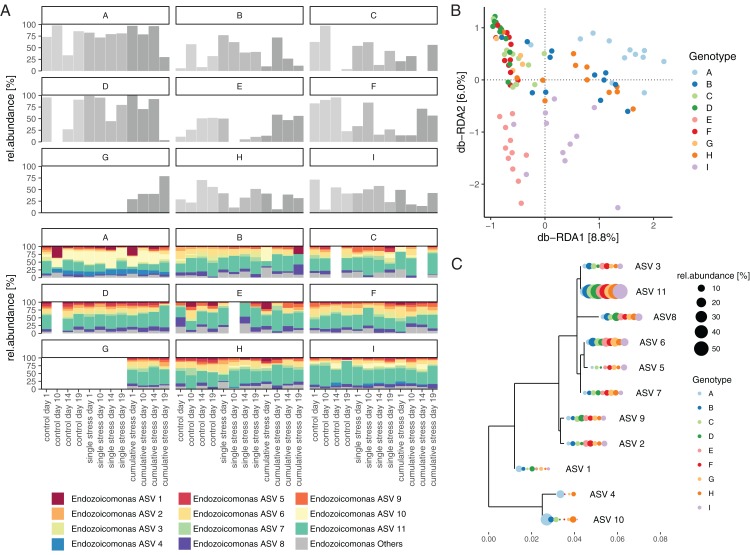
Endozoicomonas affiliated sequences comprised the majority of the A. tenuis microbiome, representing 48% (±29%) of the community (based on proportion of reads) and comprising 133 unique ASVs. One Endozoicomonas strain (ASV 11) was consistently present (100% of all samples) and highly abundant (19% ± 12%) throughout the experiment (Fig. 4). The A. tenuis microbiome also contained diverse bacteria affiliated with phyla including Proteobacteria (30%), Actinobacteria (10%), Firmicutes (2.4%), and Bacteroidetes (1.9%; Fig. 4).
Figure 4. The taxonomic composition of the Acropora tenuis microbiome.
(A) The A. tenuis microbiome was dominated by the bacterial genus Endozoicomonas (average relative abundance of 48%), with one Endozoicomonas ASV (ASV 11) present in all samples (average relative abundance of 19%). (B) The average contribution of the remaining microbiome (others) is displayed as an alluvial diagram, depicting the proportional contribution of bacterial phyla (classes for Proteobacteria). Mean relative abundances (%) are provided for bacterial taxa >1%.
The total relative abundance of Endozoicomonas was not affected by treatment (two-way ANOVA, F(2/84) = 0.473, p = 0.625), sampling time point (two-way ANOVA, F(3/84) = 0.588, p = 0.625) or the interaction of treatment-by-sampling time point (two-way ANOVA, F(6/84) = 0.696, p = 0.654). However, total relative Endozoicomonas abundance varied significantly between host genotypes (one-way ANOVA, F(8/87) = 3.741, p = 2.04 × 10−4) and remained stable between treatments when tested for each genotype individually (within subject ANOVA, F(2/85) = 0.756, p = 0.473); Fig. 5A).
Figure 5. Composition and distribution of Endozoicomonas assemblages.
(A) Total relative abundance of Endozoicomonas and the relative abundance distribution of the 11 most abundant Endozoicomonas ASVs associated with individual coral nubbins of each host genotype (A–I) under control, single stress, and cumulative stress conditions over time (day 1, 10, 14, and 19). (B) Distance-based Redundancy Analysis (db-RDA) quantifying the contribution of host-genotype to significantly explaining the observed compositional variation of the Endozoicomonas microbiome. (C) Phylogenetic tree of the 11 most abundant Endozoicomonas ASVs (including the ubiquitously present ASV 11) and their average relative abundance within a host genotype.
The Endozoicomonas community composition also varied significantly between host genotypes (PERMANOVA, p = 9.99 × 10−5, 10,000 permutations; Fig. 5), however, was unaffected by treatment, sampling time point or tank (PERMANOVA with host-genotype as blocking factor, 10,000 permutations; Table S5). Furthermore, host-genotype significantly explained 26.4% of the observed compositional variability of the Endozoicomonas community (permutational ANOVA for db-RDA based on 1,000 permutations, p = 9.99 × 10−5; Fig. 5B).
Microbial indicators for environmental stress
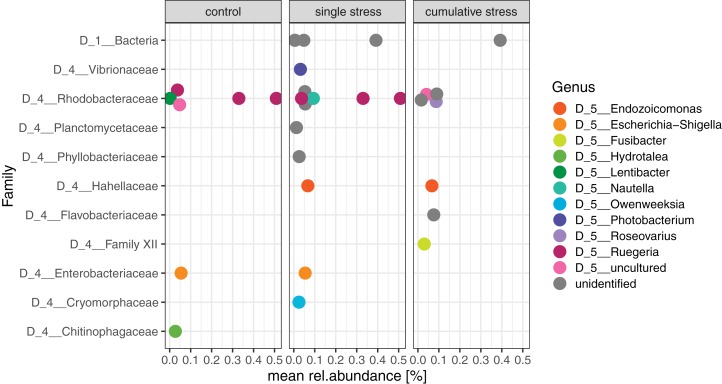
Indicator value analysis was performed to assess if specific ASVs could be identified as indicators for environmental stress treatments. Despite the vast majority of ASVs (i.e., 4,598 ASVs) showing no response to experimental treatment, 26 ASVs were significantly associated (p < 0.05) with one and/or two treatment groups (Fig. 6; Table S6). The identified indicator ASVs belonged to the bacterial families Vibrionaceae, Rhodobacteraceae, Hahellaceae (genus Endozoicomonas), Planctomycetes, Phylobacteriaceae, Flavobacteriaceae, and Cryomorphaceae (Fig. 6).
Figure 6. Microbial indicators significantly associated with one and/or two treatments.
Indicators were identified based on their occurrence and abundance in coral tissue post stress exposure (excluding samples collected at day 1) using Indicator Value analysis. Each dot represents a single amplicon sequence variant (ASV), labeled with the taxonomic affiliation and their average relative abundance in the treatment group.
Discussion
Elucidating the effect of host genotype on microbiome composition and understanding consequences of environmental change for holobiont stability is central to predicting the influence of host genetics on the stress tolerance of corals. Here, we followed the compositional stability of microbiomes associated with nine distinct A. tenuis genotypes when exposed to control, single and cumulative stress treatments over time. The A. tenuis microbiome varied significantly between coral genotypes, with host genotype being a much stronger driver of microbiome variation than environment. Similar host-genotype specificities have recently been described for sponge microbiomes (Glasl et al., 2018) and are also frequently reported for plant, crustacean, and human microbiomes (Balint et al., 2013; Macke et al., 2017; Spor, Koren & Ley, 2011). Traditional coral health parameters targeting the coral algal symbiont (i.e., chlorophyll a concentrations, symbiont cell densities, effective quantum yield) were also significantly affected by host-genotype, although no correlation between these parameters and the microbiome was observed. This suggests that the A. tenuis microbiome composition remains largely unaffected by the performance and density of the algal symbiont, and that other host intrinsic factors (e.g., genetics) and/or the environmental life-history of individual genotypes fine-tune the microbiome composition.
Endozoicomonas form symbiotic partnerships with diverse marine invertebrates (Neave et al., 2016a). In corals, Endozoicomonas occur as dense clusters within the coral tissue and in some bacterial 16S rRNA gene profiling studies they can reach relative abundances as high as 95% of retrieved sequences (Bayer et al., 2013; Neave et al., 2016a; Pogoreutz et al., 2018). Loss of Endozoicomonas from the coral microbiome has been correlated with negative health outcomes for the coral host, though their direct effects on host fitness are unknown (Bourne et al., 2008; Glasl, Herndl & Frade, 2016; Ziegler et al., 2016). In A. tenuis, we detected no significant change in the relative frequency, alpha diversity, richness, and community composition of Endozoicomonas following exposure to non-lethal environmental stress. These results are consistent with findings for Pocillopora verrucosa, where Endozoicomonas remains the dominant symbiont even under bleaching conditions (Pogoreutz et al., 2018). In our study, the Endozoicomonas community generally exhibited high host-genotype specificity at the ASV level, though a single Endozoicomonas strain (ASV 11) was consistently shared among all coral nubbins and genotypes (including field control samples—data not shown). This ubiquitous strain likely represents a stable and consistent member of the resident Endozoicomonas community. A stable core is often described as a key feature of a symbiotic coral microbiome (Ainsworth et al., 2015; Hernandez-Agreda, Gates & Ainsworth, 2017), and despite being ubiquitously persistent between conspecific corals, the core characteristically only comprises a few members of the whole microbiome (Hernandez-Agreda et al., 2018).
While the Endozoicomonas community as a whole was not significantly affected by environmental treatment, one Endozoicomonas ASV was identified as a significant indicator for environmental stress. Similar environmental sensitivity has been reported for two prevalent Endozoicomonas species following exposure to elevated dissolved organic carbon (Pogoreutz et al., 2018). Although these Endozoicomonas affiliated ASVs show high sequence identity, small variations in the rRNA gene sequence can impact the biology and pathogenicity of bacteria (Cilia, Lafay & Christen, 1996; Fukushima, Kakinuma & Kawaguchi, 2002), hence single nucleotide variations (ASV level) may affect the functional role of microbes with flow on consequences for the coral holobiont. Shuffling and switching of Endozoicomonas strains may therefore provide the coral holobiont with an enhanced capacity to cope with shifting environmental conditions (Neave et al., 2017), although characterization of the symbiotic contribution made by Endozoicomonas to the coral host is required to better understand the ecological significance of these findings.
Recent studies have highlighted the potential for coral microbiomes to act as sensitive markers for environmental disturbance (Glasl, Webster & Bourne, 2017; Roitman, Joseph Pollock & Medina, 2018). Here, we showed that a small number of ASVs, including taxa commonly reported to increase under host stress (i.e., Vibrionaceae, Rhodobacteraceae (Ben-Haim et al., 2003; Bourne, Morrow & Webster, 2016; Sunagawa, Woodley & Medina, 2010)), were significantly associated with the tissue of A. tenuis exposed to single and cumulative stress treatments. However, despite the potential diagnostic value of these ASVs, host genotype overwhelmed any overarching effect of environment on the coral microbiome. This high divergence in the microbiome between conspecific corals is likely to hinder our ability to detect fine-scale variation of sensitive microbial indicator taxa. Therefore, unless host-genotype independent microbial indicators can be identified and validated, the efficacy of integrating microbial community data into coral health monitoring initiatives appears unfeasible due to high compositional variability between microbiomes of conspecific corals.
Conclusions
This study shows that the A. tenuis microbiome varies significantly between host individuals (genotypes) and that these genotype-specific communities persist during exposure to non-lethal environmental disturbances. Consideration of microbiome-by-host genotype-by-environment effects is therefore needed to elucidate how intraspecific variations of the microbiome affect the susceptibility of corals to environmental stress and disease. Furthermore, microbial variability between individual coral genotypes may cloud our ability to identify universal microbial changes during periods of adverse environmental conditions. This is particularly relevant if establishing sensitive microbial indicators for sub-lethal environmental disturbances (tested in this study), since the observed stability of the coral microbiome combined with the host genotype specificity likely precludes the robust assignment of microbial indicators across broad scales.
Supplemental Information
Acknowledgments
We thank Victoria Lydick and Andrew Ball for their help during the experiment. We also acknowledge the technical support provided by the National SeaSimulator, and are particularly grateful to Craig Humphrey and Andrea Severati for their support with the experimental system. We thank Pedro R. Frade for inspiring scientific discussions on the coral microbiome and photobiology of corals and Jonathan Barton for his insights into coral husbandry.
Funding Statement
The study was funded by the Advance Queensland PhD scholarship, AIMS@JCU PhD scholarship and the GBRMPA Science for Management Award awarded to Bettina Glasl. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare that they have no competing interests.
Author Contributions
Bettina Glasl conceived and designed the experiments, performed the experiments, analyzed the data, contributed reagents/materials/analysis tools, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Caitlin E. Smith conceived and designed the experiments, performed the experiments, approved the final draft.
David G. Bourne conceived and designed the experiments, authored or reviewed drafts of the paper, approved the final draft.
Nicole S. Webster conceived and designed the experiments, authored or reviewed drafts of the paper, approved the final draft.
Field Study Permissions
The following information was supplied relating to field study approvals (i.e., approving body and any reference numbers):
Corals were collected under the permit G12/35236.1 granted by the Great Barrier Reef Marine Park Authority to the Australian Institute of Marine Science.
Data Availability
The following information was supplied regarding data availability:
Demultiplexed sequences and metadata are available from the Sequence Read Archives (SRA) under accession number PRJNA492377.
Furthermore, ASV abundance table, metadata and ASV taxonomies are available as Supplemental Files.
R code is available from the GitHub repository under https://github.com/bglasl/Git_4_Coral.
References
- Ainsworth et al. (2015).Ainsworth TD, Krause L, Bridge T, Torda G, Raina J-B, Zakrzewski M, Gates RD, Padilla-Gamino JL, Spalding HL, Smith C, Woolsey ES, Bourne DG, Bongaerts P, Hoegh-Guldberg O, Leggat W. The coral core microbiome identifies rare bacterial taxa as ubiquitous endosymbionts. ISME Journal. 2015;9(10):2261–2274. doi: 10.1038/ismej.2015.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker (2003).Baker AC. Flexibility and specificity in coral-algal symbiosis: diversity, ecology, and biogeography of Symbiodinium. Annual Review of Ecology, Evolution, and Systematics. 2003;34(1):661–689. doi: 10.1146/annurev.ecolsys.34.011802.132417. [DOI] [Google Scholar]
- Balint et al. (2013).Balint M, Tiffin P, Hallstrom B, O’Hara RB, Olson MS, Fankhauser JD, Piepenbring M, Schmitt I. Host genotype shapes the foliar fungal microbiome of balsam poplar (Populus balsamifera) PLOS ONE. 2013;8(1):e53987. doi: 10.1371/journal.pone.0053987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer et al. (2013).Bayer T, Neave MJ, Alsheikh-Hussain A, Aranda M, Yum LK, Mincer T, Hughen K, Apprill A, Voolstra CR. The microbiome of the Red Sea coral Stylophora pistillata is dominated by tissue-associated Endozoicomonas bacteria. Applied and Environmental Microbiology. 2013;79(15):4759–4762. doi: 10.1128/aem.00695-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Haim et al. (2003).Ben-Haim Y, Thompson F, Thompson C, Cnockaert M, Hoste B, Swings J, Rosenberg E. Vibrio coralliilyticus sp. nov., a temperature-dependent pathogen of the coral Pocillopora damicornis. International Journal of Systematic and Evolutionary Microbiology. 2003;53(1):309–315. doi: 10.1099/ijs.0.02402-0. [DOI] [PubMed] [Google Scholar]
- Bourne et al. (2008).Bourne DG, Iida Y, Uthicke S, Smith-Keune C. Changes in coral-associated microbial communities during a bleaching event. ISME Journal. 2008;2(4):350–363. doi: 10.1038/ismej.2007.112. [DOI] [PubMed] [Google Scholar]
- Bourne, Morrow & Webster (2016).Bourne DG, Morrow KM, Webster NS. Coral holobionts: insights into the coral microbiome: underpinning the health and resilience of reef ecosystems. Annual Reviews of Microbiology. 2016;70:317–340. doi: 10.1146/annurev-micro-102215-095440. [DOI] [PubMed] [Google Scholar]
- Brener-Raffalli et al. (2018).Brener-Raffalli K, Clerissi C, Vidal-Dupiol J, Adjeroud M, Bonhomme F, Pratlong M, Aurelle D, Mitta G, Toulza E. Thermal regime and host clade, rather than geography, drive Symbiodinium and bacterial assemblages in the scleractinian coral Pocillopora damicornis sensu lato. Microbiome. 2018;6(1):39. doi: 10.1186/s40168-018-0423-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan et al. (2016).Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: high-resolution sample inference from illumina amplicon data. Nature Methods. 2016;13(7):581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarti, Beltran & Van Oppen (2017).Chakravarti LJ, Beltran VH, Van Oppen MJH. Rapid thermal adaptation in photosymbionts of reef-building corals. Global Change Biology. 2017;23(11):4675–4688. doi: 10.1111/gcb.13702. [DOI] [PubMed] [Google Scholar]
- Chaston et al. (2016).Chaston JM, Dobson AJ, Newell PD, Douglas AE. Host genetic control of the microbiota mediates the drosophila nutritional phenotype. Applied and Environmental Microbiology. 2016;82(2):671–679. doi: 10.1128/aem.03301-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cilia, Lafay & Christen (1996).Cilia V, Lafay B, Christen R. Sequence heterogeneities among 16S ribosomal RNA sequences, and their effect on phylogenetic analyses at the species level. Molecular Biology and Evolution. 1996;13(3):451–461. doi: 10.1093/oxfordjournals.molbev.a025606. [DOI] [PubMed] [Google Scholar]
- De Cáceres & Legendre (2009).De Cáceres M, Legendre P. Associations between species and groups of sites: indices and statistical inference. Ecology. 2009;90(12):3566–3574. doi: 10.1890/08-1823.1. [DOI] [PubMed] [Google Scholar]
- De’ath et al. (2012).De’ath G, Fabricius KE, Sweatman H, Puotinen M. The 27-year decline of coral cover on the Great Barrier Reef and its causes. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(44):17995–17999. doi: 10.1073/pnas.1208909109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima, Kakinuma & Kawaguchi (2002).Fukushima M, Kakinuma K, Kawaguchi R. Phylogenetic analysis of Salmonella, Shigella, and Escherichia coli strains on the basis of the gyrB gene sequence. Journal of Clinical Microbiology. 2002;40(8):2779–2785. doi: 10.1128/jcm.40.8.2779-2785.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garren et al. (2009).Garren M, Raymundo L, Guest J, Harvell CD, Azam F. Resilience of coral-associated bacterial communities exposed to fish farm effluent. PLOS ONE. 2009;4(10):e7319. doi: 10.1371/journal.pone.0007319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasl, Herndl & Frade (2016).Glasl B, Herndl GJ, Frade PR. The microbiome of coral surface mucus has a key role in mediating holobiont health and survival upon disturbance. ISME Journal. 2016;10(9):2280–2292. doi: 10.1038/ismej.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasl et al. (2018).Glasl B, Smith CE, Bourne DG, Webster NS. Exploring the diversity-stability paradigm using sponge microbial communities. Scientific Reports. 2018;8(1):8425. doi: 10.1038/s41598-018-26641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasl, Webster & Bourne (2017).Glasl B, Webster NS, Bourne DG. Microbial indicators as a diagnostic tool for assessing water quality and climate stress in coral reef ecosystems. Marine Biology. 2017;164(4):91. doi: 10.1007/s00227-017-3097-x. [DOI] [Google Scholar]
- Grottoli et al. (2018).Grottoli AG, Martins PD, Wilkins MJ, Johnston MD, Warner ME, Cai WJ, Melman TF, Hoadley KD, Pettay DT, Levas S, Schoepf V. Coral physiology and microbiome dynamics under combined warming and ocean acidification. PLOS ONE. 2018;13(1):e0191156. doi: 10.1371/journal.pone.0191156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Agreda, Gates & Ainsworth (2017).Hernandez-Agreda A, Gates RD, Ainsworth TD. Defining the core microbiome in corals’ microbial soup. Trends in Microbiology. 2017;25(2):125–140. doi: 10.1016/j.tim.2016.11.003. [DOI] [PubMed] [Google Scholar]
- Hernandez-Agreda et al. (2018).Hernandez-Agreda A, Leggat W, Bongaerts P, Herrera C, Ainsworth TD. Rethinking the coral microbiome: simplicity exists within a diverse microbial biosphere. MBio. 2018;9(5):e00812-18. doi: 10.1128/mBio.00812-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoegh-Guldberg et al. (2007).Hoegh-Guldberg O, Mumby PJ, Hooten AJ, Steneck RS, Greenfield P, Gomez E, Harvell CD, Sale PF, Edwards AJ, Caldeira K, Knowlton N, Eakin CM, Iglesias-Prieto R, Muthiga N, Bradbury RH, Dubi A, Hatziolos ME. Coral reefs under rapid climate change and ocean acidification. Science. 2007;318:1737–1742. doi: 10.1126/science.1152509. [DOI] [PubMed] [Google Scholar]
- Hughes et al. (2017).Hughes TP, Barnes ML, Bellwood DR, Cinner JE, Cumming GS, Jackson JBC, Kleypas J, Van De Leemput IA, Lough JM, Morrison TH, Palumbi SR, Van Nes EH, Scheffer M. Coral reefs in the Anthropocene. Nature. 2017;546:82–90. doi: 10.1038/nature22901. [DOI] [PubMed] [Google Scholar]
- IPCC (2014).IPCC . In: Climate change 2014: Synthesis report. Contribution of working groups I, II and III to the fifth assessment report of the intergovernmental panel on climate change [core writing team, R.K. Pachauri and L.A. Meyer (eds.)] Pachauri RK, Meyer LA, editors. Geneva: IPCC; 2014. [Google Scholar]
- Jones & Berkelmans (2014).Jones AM, Berkelmans R. Flood impacts in Keppel Bay, southern Great Barrier Reef in the aftermath of cyclonic rainfall. PLOS ONE. 2014;9(1):e84739. doi: 10.1371/journal.pone.0084739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh et al. (2002).Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Research. 2002;30(14):3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane (1991).Lane DJ. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic Acid Techniques in Bacterial Systematics. New York: John Wiley and Sons; 1991. pp. 115–175. [Google Scholar]
- Macke et al. (2017).Macke E, Callens M, De Meester L, Decaestecker E. Host-genotype dependent gut microbiota drives zooplankton tolerance to toxic cyanobacteria. Nature Communications. 2017;8(1):1608. doi: 10.1038/s41467-017-01714-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauri et al. (2017).Mauri M, Elli T, Caviglia G, Uboldi G, Azzi M. RAWGraphs: a visualisation platform to create open outputs. Proceedings of the 12th Biannual Conference on Italian SIGCHI Chapter (CHItaly ’17); New York: ACM; 2017. [Google Scholar]
- McMurdie & Holmes (2013).McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLOS ONE. 2013;8(4):e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neave et al. (2016a).Neave MJ, Apprill A, Ferrier-Pages C, Voolstra CR. Diversity and function of prevalent symbiotic marine bacteria in the genus Endozoicomonas. Applied Microbiology and Biotechnology. 2016a;100(19):8315–8324. doi: 10.1007/s00253-016-7777-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neave et al. (2017).Neave MJ, Michell CT, Apprill A, Voolstra CR. Endozoicomonas genomes reveal functional adaptation and plasticity in bacterial strains symbiotically associated with diverse marine hosts. Scientific Reports. 2017;7(1):40579. doi: 10.1038/srep40579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neave et al. (2016b).Neave MJ, Rachmawati R, Xun L, Michell CT, Bourne DG, Apprill A, Voolstra CR. Differential specificity between closely related corals and abundant Endozoicomonas endosymbionts across global scales. ISME Journal. 2016b;11(1):186–200. doi: 10.1038/ismej.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen et al. (2013).Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H. vegan: community ecology package. R package version 2.0-9http://CRAN.R-project.org/package=vegan 2013
- Peixoto et al. (2017).Peixoto RS, Rosado PM, Leite DCD, Rosado AS, Bourne DG. Beneficial microorganisms for corals (BMC): proposed mechanisms for coral health and resilience. Frontiers in Microbiology. 2017;8:341. doi: 10.3389/fmicb.2017.00341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogoreutz et al. (2018).Pogoreutz C, Radecker N, Cardenas A, Gardes A, Wild C, Voolstra CR. Dominance of Endozoicomonas bacteria throughout coral bleaching and mortality suggests structural inflexibility of the Pocillopora verrucosa microbiome. Ecology and Evolution. 2018;8:2240–2252. doi: 10.1002/ece3.3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2008).R Development Core Team . R: a language and environment for statistical computing. Vienna: The R Foundation for Statistical Computing; 2008. [Google Scholar]
- Rohwer et al. (2002).Rohwer F, Seguritan V, Azam F, Knowlton N. Diversity and distribution of coral-associated bacteria. Marine Ecology Progress Series. 2002;243:1–10. doi: 10.3354/meps243001. [DOI] [Google Scholar]
- Roitman, Joseph Pollock & Medina (2018).Roitman S, Joseph Pollock F, Medina M. Coral microbiomes as bioindicators of reef health. In: Rajora OP, editor. Population Genomics. Cham: Springer; 2018. pp. 1–19. [Google Scholar]
- Rowan (2004).Rowan R. Coral bleaching: thermal adaptation in reef coral symbionts. Nature. 2004;430:742–742. doi: 10.1038/430742a. [DOI] [PubMed] [Google Scholar]
- Smith, Ketchum & Burt (2017).Smith EG, Ketchum RN, Burt JA. Host specificity of Symbiodinium variants revealed by an ITS2 metahaplotype approach. ISME Journal. 2017;11(6):1500–1503. doi: 10.1038/ismej.2016.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spor, Koren & Ley (2011).Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nature Reviews Microbiology. 2011;9(4):279–290. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- Sunagawa, Woodley & Medina (2010).Sunagawa S, Woodley CM, Medina M. Threatened corals provide underexplored microbial habitats. PLOS ONE. 2010;5(3):e9554. doi: 10.1371/journal.pone.0009554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurber et al. (2017).Thurber RV, Payet JP, Thurber AR, Correa AMS. Virus–host interactions and their roles in coral reef health and disease. Nature Reviews Microbiology. 2017;15(4):205–216. doi: 10.1038/nrmicro.2016.176. [DOI] [PubMed] [Google Scholar]
- Turner et al. (1999).Turner S, Pryer KM, Miao VP, Palmer JD. Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. Journal of Eukaryotic Microbiology. 1999;46(4):327–338. doi: 10.1111/j.1550-7408.1999.tb04612.x. [DOI] [PubMed] [Google Scholar]
- VanWoesik, DeVantier & Glazebrook (1995).VanWoesik R, DeVantier LM, Glazebrook JS. Effects of cyclone ‘Joy’ on nearshore coral communities of the Great Barrier Reef. Marine Ecology Progress Series. 1995;128:261–270. doi: 10.3354/meps128261. [DOI] [Google Scholar]
- Vega Thurber et al. (2009).Vega Thurber RL, Willner-Hall D, Rodriguez-Mueller B, Desnues C, Edwards RA, Angly F, Dinsdale EA, Kelly L, Rohwer F. Metagenomic analysis of stressed coral holobionts. Environmental Microbiology. 2009;11(8):2148–2163. doi: 10.1111/j.1462-2920.2009.01935.x. [DOI] [PubMed] [Google Scholar]
- Wagner et al. (2016).Wagner MR, Lundberg DS, Del Rio TG, Tringe SG, Dangl JL, Mitchell-Olds T. Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nature Communications. 2016;7(1):12151. doi: 10.1038/ncomms12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster & Reusch (2017).Webster NS, Reusch TBH. Microbial contributions to the persistence of coral reefs. ISME Journal. 2017;11(10):2167–2174. doi: 10.1038/ismej.2017.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham (2009).Wickham H. ggplot2: elegant graphics for data analysis. New York: Springer; 2009. [Google Scholar]
- Wullaert, Lamkanfi & McCoy (2018).Wullaert A, Lamkanfi M, McCoy KD. Defining the impact of host genotypes on microbiota composition requires meticulous control of experimental variables. Immunity. 2018;48(4):605–607. doi: 10.1016/j.immuni.2018.04.001. [DOI] [PubMed] [Google Scholar]
- Zaneveld et al. (2016).Zaneveld JR, Burkepile DE, Shantz AA, Pritchard CE, McMinds R, Payet JP, Welsh R, Correa AMS, Lemoine NP, Rosales S, Fuchs C, Maynard JA, Thurber RV. Overfishing and nutrient pollution interact with temperature to disrupt coral reefs down to microbial scales. Nature Communications. 2016;7(1):11833. doi: 10.1038/ncomms11833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaneveld, McMinds & Thurber (2017).Zaneveld JR, McMinds R, Thurber RV. Stress and stability: applying the Anna Karenina principle to animal microbiomes. Nature Microbiology. 2017;2(9):17121. doi: 10.1038/nmicrobiol.2017.121. [DOI] [PubMed] [Google Scholar]
- Zhang et al. (2015).Zhang YY, Ling J, Yang QS, Wang YS, Sun CC, Sun HY, Feng JB, Jiang YF, Zhang YZ, Wu ML, Dong JD. The diversity of coral associated bacteria and the environmental factors affect their community variation. Ecotoxicology. 2015;24(7–8):1467–1477. doi: 10.1007/s10646-015-1454-4. [DOI] [PubMed] [Google Scholar]
- Ziegler et al. (2016).Ziegler M, Roik A, Porter A, Zubier K, Mudarris MS, Ormond R, Voolstra CR. Coral microbial community dynamics in response to anthropogenic impacts near a major city in the central Red Sea. Marine Pollution Bulletin. 2016;105(2):629–640. doi: 10.1016/j.marpolbul.2015.12.045. [DOI] [PubMed] [Google Scholar]
- Ziegler et al. (2017).Ziegler M, Seneca FO, Yum LK, Palumbi SR, Voolstra CR. Bacterial community dynamics are linked to patterns of coral heat tolerance. Nature Communications. 2017;8:14213. doi: 10.1038/ncomms14213. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following information was supplied regarding data availability:
Demultiplexed sequences and metadata are available from the Sequence Read Archives (SRA) under accession number PRJNA492377.
Furthermore, ASV abundance table, metadata and ASV taxonomies are available as Supplemental Files.
R code is available from the GitHub repository under https://github.com/bglasl/Git_4_Coral.