Abstract
Background
An estimated 5%–10% of all cutaneous melanoma cases occur in families. This review describes susceptibility genes currently known to be involved in melanoma predisposition, genetic testing of familial melanoma patients, and management implications.
Results
CDKN2A is the major high-penetrance susceptibility gene with germline mutations identified in 20%–40% of melanoma families. A positive CDKN2A mutation status has been associated with a high number of affected family members, multiple primary melanomas, pancreatic cancer, and early age at melanoma onset. Mutations in the other melanoma predisposition genes—CDK4, BAP1, TERT, POT1, ACD, TERF2IP, and MITF—are rare, overall contributing to explain a further 10% of familial clustering of melanoma. The underlying genetic susceptibility remains indeed unexplained for half of melanoma families. Genetic testing for melanoma is currently recommended only for CDKN2A and CDK4, and, at this time, the role of multigene panel testing remains under debate. Individuals from melanoma families must receive genetic counseling to be informed about the inclusion criteria for genetic testing, the probability of an inconclusive result, the genetic risk for melanoma and other cancers, and the debatable role of medical management. They should be counseled focusing primarily on recommendations on appropriate lifestyle, encouraging skin self-examination, and regular dermatological screening.
Conclusions
Genetic testing for high-penetrance melanoma susceptibility genes is recommended in melanoma families after selection of the appropriate candidates and adequate counseling of the patient. All patients and relatives from melanoma kindreds, irrespective of their mutation status, should be encouraged to adhere to a correct ultraviolet exposure, skin self-examination, and surveillance by physicians.
Keywords: familial melanoma, CDKN2A, CDK4, genetic testing, genetic counseling, cancer screening
Introduction
Cutaneous melanoma is one of the most aggressive human cancers, with an increasing incidence worldwide [1]. When detected at an early stage, high survival rates are reported 5 years after diagnosis [2]. Although new therapies are currently available for metastatic disease, survival for patients with advanced disease is still poor.
Melanoma pathogenesis is complex and heterogeneous, with environmental, phenotypic, and genetic factors contributing to its development. The main risk factors involved in the etiopathogenesis of cutaneous melanoma are a large number of common acquired melanocytic nevi, atypical melanocytic nevi, light skin phenotype, exposure to ultraviolet (UV) radiation, and a family history of melanoma [3–5].
An estimated 5%–10% of all cutaneous melanoma cases occur in families. Familial melanoma is defined as a family in which either 2 first-degree relatives or 3 or more melanoma patients on the same side of the family (irrespective of degree of relationship) are diagnosed with melanoma [5]. In these families the pattern of heritability is consistent with an autosomal dominant inheritance with incomplete penetrance. Germline susceptibility has been associated with mutations in high-penetrance melanoma predisposition genes, CDKN2A (cyclin-dependent kinase 2A) and less frequently in CDK4 (cyclin-dependent kinase 4), BAP1 (breast cancer associated protein-1), TERT (telomerase reverse transcriptase), and POT1 (protection of telomeres 1), or with variants in intermediate-risk genes, MC1R (melanocortin 1 receptor) and MITF (microphthalmia-associated transcription factor) [6–9].
Currently, genetic testing is recommended in high-risk melanoma patients and families to improve early detection and reduce mortality. Individuals from high-risk melanoma families must receive genetic counseling so that they receive full information about the inclusion criteria for genetic testing, the probability of an inconclusive result, the genetic risk for melanoma and other cancers, and the debatable role of medical management.
This review describes susceptibility genes known to be involved in melanoma predisposition, genetic testing of familial melanoma patients, and management implications.
Melanoma Susceptibility Genes
Unlike other cancer predisposition syndromes, melanoma is not linked to a single gene, but several high- and intermediate-penetrance melanoma susceptibility genes have been identified to date (Table 1). Penetrance relates to the lifetime risk for a mutation carrier of developing melanoma and reflects the overall contribution of a specific gene alteration to the risk of melanoma.
Table 1.
Overview of High- and Intermediate-Penetrance Genes Involved in Melanoma Susceptibility
| Gene Penetrance | Gene | Encoded Protein | Role | Mutation Prevalence | References |
|---|---|---|---|---|---|
| High-penetrance | CDKN2A | p16INK4a | Cell cycle regulator | ~20%–40% of families | 10–13 |
| p14ARF | Cell cycle regulator | ~1% of families | 9,14,15 | ||
| CDK4 | CDK4 | Cell cycle regulator | 17 families | 16,17 | |
| TERT | Catalytic subunit of telomerase | Telomere elongation | 2 families | 18,19 | |
| POT1 | POT1 | Telomere maintenance | 14 families | 20–22 | |
| Intermediate-penetrance | MC1R | MC1R | Melanin synthesis and melanocyte proliferation | NA | 23,24 |
| MITF | MITF | Melanocyte development and differentiation | NA | 25,26 |
CDK4 = cyclin-dependent kinase 4; CDKN2A = cyclin-dependent kinase 2A; MC1R = melanocortin 1 receptor; MITF = microphthalmia- associated transcription factor; NA = not applicable; POT1 = protection of telomeres 1; TERT = telomerase reverse transcriptase.
High-Penetrance Genes
CDKN2A was the first familial melanoma predisposition gene to be identified and is mutated in approximately 20%–40% of high-risk families, depending on selection criteria and on geographic region of the families [12,13,27–32].
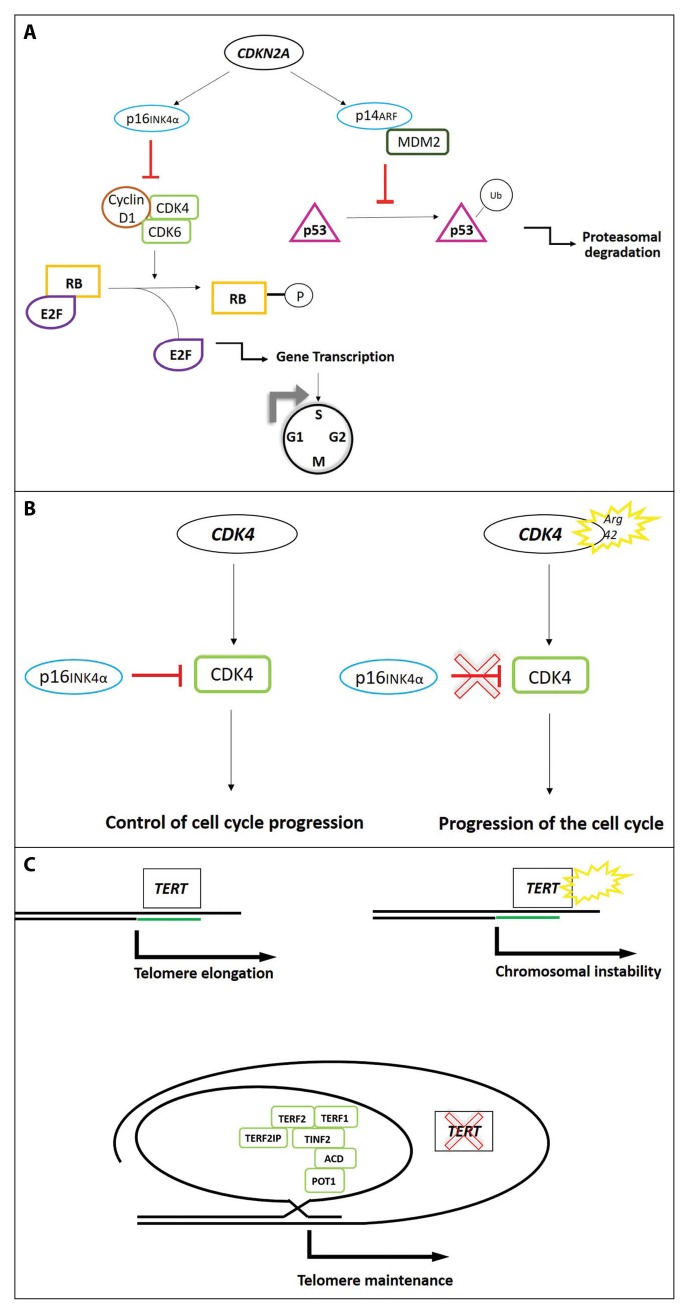
The CDKN2A tumor suppressor gene is located at the 9p21 locus and encodes 2 different proteins, p16INK4A (p16) and p14ARF (p14), both regulating cell cycle (Figure 1A). The p16 promotes cell cycle arrest in the G1 phase by inhibiting retinoblastoma (RB) protein phosphorylation through cyclin-dependent kinase 4 (CDK4). p14 is also a tumor suppressor and acts through the p53 pathway inducing cell cycle arrest or favoring apoptosis [33].
Figure 1.
Pathways of high-risk genes involved in melanoma susceptibility. (A) CDKN2A encodes 2 proteins: p16INK4a and p14ARF. Mutations in CDKN2A gene allow the cells to escape from cell cycle arrest. In detail, p16INK4a inhibits cyclin D1/CDK4/6 complex to release E2F through RB phosphorylation. p14ARF interacts with MDM2 to block p53 ubiquitination, thus promoting apoptosis. When mutated, CDKN2A produces 2 dysfunctional proteins inducing cell cycle progression and avoiding p53 degradation. (B) Mutations in CDK4 promote the G1 to S phase transition, escaping the p16INK4a inhibition. (C) TERT encodes the telomerase reverse transcriptase, involved in the maintenance of telomere length. Mutations in the promoter region of TERT increase telomerase activity resulting in chromosomal instability. POT1 interacts with the shelterin complex acting as protective structure which prevents access of TERT to telomeres. The S270N mutation in the POT1 gene has been associated with familial melanoma. CDK = cyclin-dependent kinase; CDKN2A = cyclin-dependent kinase inhibitor 2A; MDM2 = mouse double minute 2; POT1 = protection of telomeres 1; RB = retinoblastoma. [Copyright: ©2019 Rossi et al.]
The CDKN2A gene is the major melanoma susceptibility gene with more than 60 germline mutations identified to date, the majority of which are missense mutations in the p16 transcript [6,34]. CDKN2A mutation penetrance varies between geographical areas, according to the population incidence rate of melanoma, ranging from 58% in Europe to 76% in the United States and to 91% in Australia by age 80 years [35]. The likelihood of detecting a CDKN2A mutation in melanoma families increases with the number of affected members (approximately 10% for 2-case melanoma families and 30%–40% for families with 3 or more cases of melanoma), with the presence within the family of relatives with multiple primary melanoma (MPM), pancreatic cancer, or early age at melanoma onset [36]. In addition, CDKN2A mutations are also detected in individuals with MPM in the absence of a family history of melanoma in 8.3%, 15%, and 57% in United States, North America, and Greece, respectively [37,38]. The association between pancreatic cancer and melanoma is often observed in CDKN2A-mutated melanoma families and has been proposed as a hereditary cancer syndrome [39–45]. Annual surveillance for pancreatic cancer is therefore warranted in high-risk melanoma families with CDKN2A mutations [46]. Besides pancreatic cancer, families with CDKN2A mutations have been reported to also have an increased risk of developing breast, lung, and other tobacco-related cancers [42].
Few melanoma kindreds worldwide have been found to carry mutations of the CDK4 oncogene, the second identified high-risk melanoma susceptibility gene, encoding one of the binding partners of p16.
The CDK4 oncogene plays an important role at the G1/S phase cell cycle checkpoint (1B). When CDK4 is mutated, p16 cannot inhibit the CDK4 kinase activity resulting in increased phosphorylation of RB bound to E2F transcription factors with higher E2F release. E2F activates the transcription of pro-S phase cell cycle genes, promoting G1/S phase transition.
All CDK4 pathogenetic mutations cluster in codon 24 of exon 2, a critical site for p16 binding [16,47]. The phenotype of CDK4-mutated families is indistinguishable from the CDKN2A phenotype, with early-onset cutaneous melanoma, development of MPMs, and presence of atypical nevi [48].
In families negative for mutations in known high-risk genes, the introduction of next-generation sequencing methodologies led to the identification of germline mutations in a small number of novel high-penetrance melanoma susceptibility genes involved in pathways other than those mediated by known melanoma risk factors.
The BAP1 gene regulates differentiation of melanocytes and is part of the DNA damage response. Cutaneous melanoma is considered as part of the phenotype associated with the BAP1 cancer susceptibility syndrome, characterized by multiple skin-colored spitzoid melanocytic tumors, uveal melanoma, and cutaneous melanoma, recently expanded to include mesothelioma, renal cell carcinoma, and basal cell carcinoma [49–53]. The elevated number of cancers in BAP1-mutated families suggests that this gene is a critical regulator of oncogenesis.
Telomere maintenance has been recently discovered as a key pathway in melanoma predisposition (Figure 1C). The TERT gene encodes the catalytic subunit of telomerase, which is the ribonucleoprotein complex that maintains telomere length. TERT mutations induce increased expression of telomerase, thus promoting telomere stabilization and acting on cell aging, turnover, and senescence. A novel mutation occurring in the promoter region of the TERT gene, encoding the catalytic subunit of telomerase, has been recently identified in 2 melanoma families [18,19].
POT1 is a crucial member of the shelterin complex proteins, important for telomere maintenance. Mutations in the POT1 gene have been recently identified in a total of 12 CDKN2A-negative melanoma families [20,21]. Additional shelterin complex genes, such as ACD and TERF2IP, were later found to be mutated in familial melanoma patients [54]. Overall, germline mutations in POT1, ACD, and TERF2IP are detected in approximately 9% of high-density families without mutations in known high-penetrance genes. Families carrying POT1, ACD, and TERF2IP mutations often present with MPM and early-onset melanoma.
Intermediate-Penetrance Genes
In almost half of highly dense melanoma families, the underlying genetic basis is still unexplained. Besides the possibility of rare mutations in a few additional unknown high-penetrance genes, a polygenic susceptibility as result of coinheritance of multiple intermediate- and/or low-risk alleles or an interplay between susceptibility genes and genetic modifiers (other genes, phenotypic characteristics, and/or environmental risk factors) has been suggested. Two intermediate-penetrance genes, MC1R and MITF, predisposing to melanoma have been identified to date (Table 1).
The MC1R gene, encoding a G-protein coupled receptor with a high affinity for the α-melanocyte-stimulating hormone (αMSH), has a key role in cutaneous pigmentation. Binding of αMSH to MC1R stimulates cAMP-induced tyrosinase activity resulting in eumelanin synthesis. MC1R is a highly polymorphic gene in the Caucasian population, with more than 100 variants identified [55,56]. Specific MC1R variants (R142H, R151C, R160W, and D294H) resulting in a reduced receptor function with a switch from eumelanin to pheomelanin synthesis are classified as red hair color (RHC) or “R” variants and have been strongly associated with fair skin, freckling, UV radiation sensitivity, and inability to tan [24,57].
MC1R variants, mainly R alleles, have been associated with an increased risk of melanoma independently of phenotypic features [56]. Notably, a stronger risk has been reported for patients with darkly pigmented skin [23]. It is indeed recognized that MC1R influences melanoma risk not only through its effect on pigmentation and UV sensitivity but also through additional biological pathways, including induction of antioxidant defenses, DNA repair mechanisms, and melanocyte proliferation, regulation, and differentiation [58]. In addition, inheritance of MC1R variants with CDKN2A mutations has been shown to increase penetrance of melanoma in families carrying CDKN2A mutations [23,24]. Finally, carrying R variants has been associated with specific clinico-dermoscopic features of melanoma such as hypopigmentation, structureless areas, atypia, and vessels [59–61].
The MITF gene is a master regulator of melanocyte homeostasis encoding a lineage-specific transcription factor, involved in cell survival, differentiation, and proliferation [62]. A rare functional variant p.E318K in the MITF gene has been implicated in familial melanoma and in melanoma/renal cell carcinoma susceptibility [25,26]. The p.E318K mutation alters MITF sumoylation, increasing the MITF transcriptional activity with upregulation of downstream genes. Clinically, a high nevus count, development of MPM, onset of melanoma before the age of 40, and nonblue eye color are phenotypic characteristics that have been associated with the p.E318K mutation [25,63,64].
Genetic Counseling and Testing
Genetic counseling should be offered to melanoma patients with hereditary predisposition so that they may better understand the meaning of the disease, pattern of inheritance, option of genetic testing, possible results and implications for other family members, and recommendations for primary and secondary prevention of melanoma and for psychological assessment [34,65].
To date, CDKN2A and CDK4 are the only genes recommended to be tested as single genes for genetic screening of melanoma predisposition [8,31,45]. Genetic testing of the CDKN2A gene has been available for a long time and can be offered to patients with familial melanoma and/or MPM who are likely to carry a CDKN2A mutation. However, its use in clinical practice has been controversial because of the reported variation in the estimates of CDKN2A mutation penetrance, depending on selection criteria of patients, ethnic background, environmental exposure, and coinheritance of low-intermediate predisposing genes such as MC1R variants. Leachman et al [31] proposed a useful rule to select appropriate candidates eligible for genetic testing in melanoma with regard to the specific population or geographic area (Table 2): the rule of 2 for countries with a low incidence of melanoma (Southern Europe) and the rule of 3 for countries with a moderate–high incidence of melanoma (United States and Northern Europe). A rule of 4 for countries with a very high incidence of melanoma (Australia) has been suggested [65].
Table 2.
Selection Criteria for CDKN2A Genetic Testing According to Melanoma Incidence [31]
| Geographic area/population with low melanoma incidence | Geographic area/population with moderate–high melanoma incidence |
|---|---|
|
|
To reduce the risk of uninformative negative results, it is important to identify the best candidate in the family for genetic testing, usually an individual with a personal presentation most suggestive of a CDKN2A mutation, such as young-onset or MPM patients [34]. If a CDKN2A mutation is detected in a family, screening of other family members is recommended. If no CDKN2A mutation is identified within a melanoma family, it should be stressed that the family is still at increased risk of melanoma on the basis of the family history. Patients should be aware of the difficulty of interpreting the results and the potential limited impact on clinical management.
In patients with a strong family history but negative for CDKN2A and CDK4 mutations, testing for other melanoma-associated genes can be performed: BAP1 in the presence of typical cutaneous melanocytic lesions, ocular melanoma, or other associated cancers described in the BAP1-cancer susceptibility syndrome or MITF in the presence of renal cell carcinoma.
Panel testing of melanoma predisposition genes is an attractive option for hereditary melanoma, especially for high-penetrance genes, and by now numerous panel tests are available at the same cost as a single gene test. However, routine screening of intermediate- or low-penetrance genes is questionable because of the uncertainty of predicting clinical outcome of disease development. However, a tailored genetic testing approach with multigene panels based on the cancer profile observed in the family could be performed if in the families there are cases of breast cancer, prostate cancer, ovarian cancer, and/or colon cancer [45].
Management of Familial Melanoma Patients
Carriers of a CDKN2A mutation are at high risk of developing multiple melanomas and, in some families, pancreatic cancer [12,13]. The identification of a deleterious CDKN2A mutation suggests that carriers should be included in intensive skin surveillance programs with skin examination, also including scalp, oral and genital mucosa, performed every 6 months. However, the frequency of dermatological visit (3-month, 6-month, or 1-year intervals) should be planned on the basis of the patient’s risk factors; digital dermoscopy and clinical photography would be helpful for monitoring these patients. With regard to pancreatic cancer, patients should be aware of the current lack of effective screening guidelines [46,66]. Overall, CDKN2A carriers are candidates for annual pancreatic cancer screening via endoscopic ultrasonography or magnetic resonance cholangiopancreatography.
The recommendation of avoiding smoking in cancer prevention programs has been recently suggested for CDKN2A mutation carriers after the description of an increased prevalence of tobacco-associated cancers in CDKN2A-mutated families [67].
First-degree relatives (parents, siblings, children) will have a 50% chance of harboring the same mutation and risk. It is also prudent for children from familial melanoma kindreds to undergo routine skin examinations beginning at puberty.
Increased skin cancer screening, patients’ skin self-examination education, and surveillance by physicians should be encouraged in all patients and relatives, irrespective of mutation status, for early detection of melanoma.
Footnotes
Funding: None.
Competing interests: The authors have no conflicts of interest to disclose.
Authorship: All authors have contributed significantly to this publication.
References
- 1.Nikolaou V, Stratigos AJ. Emerging trends in the epidemiology of melanoma. Br J Dermatol. 2014;170(1):11–19. doi: 10.1111/bjd.12492. [DOI] [PubMed] [Google Scholar]
- 2.Balch CM, Buzaid AC, Soong SJ, et al. Final version of the American Joint Committee on Cancer staging system for cutaneous melanoma. J Clin Oncol. 2001;19(16):3635–3648. doi: 10.1200/JCO.2001.19.16.3635. [DOI] [PubMed] [Google Scholar]
- 3.Gandini S, Sera F, Cattaruzza MS, et al. Meta-analysis of risk factors for cutaneous melanoma, I: common and atypical naevi. Eur J Cancer. 2005;41(1):28–44. doi: 10.1016/j.ejca.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 4.Gandini S, Sera F, Cattaruzza MS, et al. Meta-analysis of risk factors for cutaneous melanoma, II: sun exposure. Eur J Cancer. 2005;41(1):45–60. doi: 10.1016/j.ejca.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 5.Gandini S, Sera F, Cattaruzza MS, et al. Meta-analysis of risk factors for cutaneous melanoma, III: family history, actinic damage and phenotypic factors. Eur J Cancer. 2005;41(14):2040–2059. doi: 10.1016/j.ejca.2005.03.034. [DOI] [PubMed] [Google Scholar]
- 6.Goldstein AM. Familial melanoma, pancreatic cancer and germline CDKN2A mutations. Hum Mutat. 2004;23(6):630. doi: 10.1002/humu.9247. [DOI] [PubMed] [Google Scholar]
- 7.Psaty EL, Scope A, Halpern AC, Marghoob A. Defining the patient at high risk for melanoma. Int J Dermatol. 2010;49(4):362–376. doi: 10.1111/j.1365-4632.2010.04381.x. [DOI] [PubMed] [Google Scholar]
- 8.Potrony M, Badenas C, Aguilera P, et al. Update in genetic susceptibility in melanoma. Ann Transl Med. 2015;3(15):210. doi: 10.3978/j.issn.2305-5839.2015.08.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aoude LG, Wadt KA, Pritchard AL, Hayward NK. Genetics of familial melanoma: 20 years after CDKN2A. Pigment Cell Melanoma Res. 2015;28(2):148–160. doi: 10.1111/pcmr.12333. [DOI] [PubMed] [Google Scholar]
- 10.Hussussian CJ, Struewing JP, Goldstein AM, et al. Germline p16 mutations in familial melanoma. Nat Genet. 1994;8(1):15–21. doi: 10.1038/ng0994-15. [DOI] [PubMed] [Google Scholar]
- 11.Kamb A, Shattuck-Eidens D, Eeles R, et al. Analysis of the p16 gene (CDKN2) as a candidate for the chromosome 9p melanoma susceptibility locus. Nat Genet. 1994;8(1):23–26. doi: 10.1038/ng0994-22. [DOI] [PubMed] [Google Scholar]
- 12.Goldstein AM, Chan M, Harland M, et al. High-risk melanoma susceptibility genes and pancreatic cancer, neural system tumors, and uveal melanoma across GenoMEL. Cancer Res. 2006;66(20):9818–9828. doi: 10.1158/0008-5472.CAN-06-0494. [DOI] [PubMed] [Google Scholar]
- 13.Goldstein A, Chan M, Harland M, et al. Features associated with germline CDKN2A mutations: a GenoMEL study of melanoma-prone families from three continents. J Med Genet. 2007;44(2):99–106. doi: 10.1136/jmg.2006.043802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bahuau M, Vidaud D, Jenkins RB, et al. Germ-line deletion involving the INK4 locus in familial proneness to melanoma and nervous system tumors. Cancer Res. 1998;58(11):2298–2303. [PubMed] [Google Scholar]
- 15.Pellegrini C, Maturo MG, Martorelli C, et al. Characterization of melanoma susceptibility genes in high-risk patients from Central Italy. Melanoma Res. 2017;27(3):258–267. doi: 10.1097/CMR.0000000000000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuo L, Weger J, Yang Q, et al. Germline mutations in the p16INK4A binding domain of CDK4 in familial melanoma. Nat Genet. 1996;12(1):97–99. doi: 10.1038/ng0196-97. [DOI] [PubMed] [Google Scholar]
- 17.Puntervoll HE, Yang XR, Vetti HH, et al. Melanoma prone families with CDK4 germline mutation: phenotypic profile and associations with MC1R variants. J Med Genet. 2013;50(4):264–270. doi: 10.1136/jmedgenet-2012-101455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horn S, Figl A, Rachakonda PS, et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339(6122):959–961. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- 19.Harland M, Petljak M, Robles-Espinoza CD, et al. Germline TERT promoter mutations are rare in familial melanoma. Fam Cancer. 2016;15(1):139–144. doi: 10.1007/s10689-015-9841-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robles-Espinoza CD, Harland M, Ramsay AJ, et al. POT1 loss-of-function variants predispose to familial melanoma. Nat Genet. 2014;46(5):478–481. doi: 10.1038/ng.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi J, Yang XR, Ballew B, et al. Rare missense variants in POT1 predispose to familial cutaneous malignant melanoma. Nat Genet. 2014;46(5):482–486. doi: 10.1038/ng.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Müller C, Krunic M, Wendt J, von Haeseler A, Okamoto I. Germline variants in the POT1-gene in high-risk melanoma patients in Austria. G3 (Bethesda) 2018;8(5):1475–1480. doi: 10.1534/g3.117.300394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pasquali E, García-Borrón JC, Fargnoli MC, et al. MC1R variants increased the risk of sporadic cutaneous melanoma in darker-pigmented Caucasians: a pooled-analysis from the M-SKIP project. Int J Cancer. 2015;136(3):618–631. doi: 10.1002/ijc.29018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fargnoli MC, Gandini S, Peris K, et al. MC1R variants increase melanoma risk in families with CDKN2A mutations: a meta-analysis. Eur J Cancer. 2010;46(8):1413–1420. doi: 10.1016/j.ejca.2010.01.027. [DOI] [PubMed] [Google Scholar]
- 25.Yokoyama S, Woods SL, Boyle GM, et al. A novel recurrent mutation in MITF predisposes to familial and sporadic melanoma. Nature. 2011;480(7375):99–103. doi: 10.1038/nature10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bertolotto C, Lesueur F, Giuliano S, et al. A SUMOylation-defective MITF germline mutation predisposes to melanoma and renal carcinoma. Nature. 2011;480(7375):94–98. doi: 10.1038/nature10539. [DOI] [PubMed] [Google Scholar]
- 27.Begg C, Orlow I, Hummer A, et al. Lifetime risk of melanoma in CDKN2A mutation carriers in a population-based sample. J Natl Cancer Inst. 2005;97(20):1507–1515. doi: 10.1093/jnci/dji312. [DOI] [PubMed] [Google Scholar]
- 28.Bruno W, Ghiorzo P, Battistuzzi L, et al. Clinical genetic testing for familial melanoma in Italy: a cooperative study. J Am Acad Dermatol. 2009;61(5):775–782. doi: 10.1016/j.jaad.2009.03.039. [DOI] [PubMed] [Google Scholar]
- 29.Maubec E, Chaudru V, Mohamdi H, et al. Familial melanoma: clinical factors associated with germline CDKN2A mutations according to the number of patients affected by melanoma in a family. J Am Acad Dermatol. 2012;67(6):1257–1264. doi: 10.1016/j.jaad.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 30.Harland M, Cust AE, Badenas C, et al. Prevalence and predictors of germline CDKN2A mutations for melanoma cases from Australia, Spain and the United Kingdom. Hered Cancer Clin Pract. 2014;12(1):20. doi: 10.1186/1897-4287-12-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leachman S, Carucci J, Kohlmann L, et al. Selection criteria for genetic assessment of patients with familial melanoma. J Am Acad Dermatol. 2009;61(4):677.e1–14. doi: 10.1016/j.jaad.2009.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lang J, Boxer M, MacKie RM. CDKN2A mutations in Scottish families with cutaneous melanoma: results from 32 newly identified families. Br J Dermatol. 2005;153(6):1121. doi: 10.1111/j.1365-2133.2005.06846.x. [DOI] [PubMed] [Google Scholar]
- 33.Chudnovsky Y, Khavari PA, Adams AE. Melanoma genetics and the development of rational therapeutics. J Clin Invest. 2005;115(4):813–824. doi: 10.1172/JCI24808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gabree M, Patel D, Rodgers L. Clinical applications of melanoma genetics. Curr Treat Options Oncol. 2014;15(2):336–350. doi: 10.1007/s11864-014-0282-8. [DOI] [PubMed] [Google Scholar]
- 35.Bishop DT, Demenais F, Goldstein AM, et al. Geographical variation in the penetrance of CDKN2A mutations for melanoma. J Natl Cancer Inst. 2002;94(12):894–903. doi: 10.1093/jnci/94.12.894. [DOI] [PubMed] [Google Scholar]
- 36.Kefford RF, Newton-Bishop JA, Bergman W, Ticker MA. Counseling and DNA testing for individuals perceived to be genetically predisposed to melanoma: a consensus statement of the Melanoma Genetics Consortium. J Clin Oncol. 1999;17(10):3245–3251. doi: 10.1200/JCO.1999.17.10.3245. [DOI] [PubMed] [Google Scholar]
- 37.Blackwood MA, Holmes R, Synnestvedt M, et al. Multiple primary melanoma revisited. Cancer. 2002;94(8):2248–2255. doi: 10.1002/cncr.10454. [DOI] [PubMed] [Google Scholar]
- 38.Nikolaou V, Kang X, Stratigos A, et al. Comprehensive mutational analysis of CDKN2A and CDK4 in Greek patients with cutaneous melanoma. Br J Dermatol. 2011;165(6):1219–1222. doi: 10.1111/j.1365-2133.2011.10551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bergman W, Gruis N. Familial melanoma and pancreatic cancer. N Engl J Med. 1996;334(7):471–472. [PubMed] [Google Scholar]
- 40.Goldstein AM, Fraser MC, Struewing JP, et al. Increased risk of pancreatic cancer in melanoma-prone kindreds with p16INK4A mutations. N Engl J Med. 1995;333(15):970–974. doi: 10.1056/NEJM199510123331504. [DOI] [PubMed] [Google Scholar]
- 41.Ghiorzo P, Ciotti P, Mantelli M, et al. Characterization of Ligurian melanoma families and risk of occurrence of other neoplasia. Int J Cancer. 1999;83(4):441–448. doi: 10.1002/(sici)1097-0215(19991112)83:4<441::aid-ijc2>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 42.Borg A, Sandberg T, Nilsson K, et al. High frequency of multiple melanomas and breast and pancreas carcinomas in CDKN2A mutation-positive melanoma families. J Natl Cancer Inst. 2000;92(15):1260–1266. doi: 10.1093/jnci/92.15.1260. [DOI] [PubMed] [Google Scholar]
- 43.Vasen HF, Gruis NA, Frants NN, et al. Risk of developing pancreatic cancer in families with familial atypical multiple mole melanoma associated with a specific 19 deletion of p16 (p16-Leiden) Int J Cancer. 2000;87(6):809–811. [PubMed] [Google Scholar]
- 44.Rulyak SJ, Brentnall TA, Lynch HT, Austin MA. Characterization of the neoplastic phenotype in the familial atypical multiple-mole melanoma-pancreatic carcinoma syndrome. Cancer. 2003;98(4):798–804. doi: 10.1002/cncr.11562. [DOI] [PubMed] [Google Scholar]
- 45.Lynch HT, Brand RE, Hogg D, et al. Phenotypic variation in eight extended CDKN2A germline mutation familial atypical multiple mole melanoma-pancreatic carcinoma syndrome. Cancer. 2002;94(1):84–96. doi: 10.1002/cncr.10159. [DOI] [PubMed] [Google Scholar]
- 46.Parker JF, Florell SR, Alexander A, et al. Pancreatic carcinoma surveillance in patients with familial melanoma. Arch Dermatol. 2003;139(8):1019–1025. doi: 10.1001/archderm.139.8.1019. [DOI] [PubMed] [Google Scholar]
- 47.Molven A, Grimstvedt MB, Steine SJ, et al. A large Norwegian family with inherited malignant melanoma, multiple atypical nevi, and CDK4 mutation. Genes Chromosomes Cancer. 2005;44(1):10–18. doi: 10.1002/gcc.20202. [DOI] [PubMed] [Google Scholar]
- 48.Puntervoll HE, Yang XR, Vetti HH, et al. Melanoma prone families with CDK4 germline mutation: phenotypic profile and associations with MC1R variants. Med Genet. 2013;50(4):264–270. doi: 10.1136/jmedgenet-2012-101455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aoude LG, Wadt K, Bojesen A, et al. A BAP1 mutation in a Danish family predisposes to uveal melanoma and other cancers. PLoS One. 2013;8(8):e72144. doi: 10.1371/journal.pone.0072144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abdel-Rahman MH, Pilarski R, Cebulla CM, et al. Germline BAP1 mutation predisposes to uveal melanoma, lung adenocarcinoma, meningioma, and other cancers. J Med Genet. 2011;48(12):856–859. doi: 10.1136/jmedgenet-2011-100156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carbone M, Ferris LK, Baumann F, et al. BAP1 cancer syndrome: malignant mesothelioma, uveal and cutaneous melanoma, and MBAITs. J Transl Med. 2012;10:179. doi: 10.1186/1479-5876-10-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carbone M, Yang H, Pass HI, et al. BAP1 and cancer. Nat Rev Cancer. 2013;13(3):153–159. doi: 10.1038/nrc3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Battaglia A. The importance of multidisciplinary approach in early detection of BAP1 tumor predisposition syndrome: clinical management and risk assessment. Clin Med Insights Oncol. 2014;8:37–47. doi: 10.4137/CMO.S15239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aoude LG, Pritchard AL, Robles-Espinoza CD, et al. Nonsense mutations in the shelterin complex genes ACD and TERF2IP in familial melanoma. J Natl Cancer Inst. 2014;107(2) doi: 10.1093/jnci/dju408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gerstenblith MR, Goldstein AM, Fargnoli MC, Peris K, Landi MT. Comprehensive evaluation of allele frequency differences of MC1R variants across populations. Hum Mutat. 2007;28(5):495–505. doi: 10.1002/humu.20476. [DOI] [PubMed] [Google Scholar]
- 56.Kanetsky PA, Rebbeck TR, Hummer AJ, et al. Population-based study of natural variation in the melanocortin-1 receptor gene and melanoma. Cancer Res. 2006;66(18):9330–9337. doi: 10.1158/0008-5472.CAN-06-1634. [DOI] [PubMed] [Google Scholar]
- 57.Beaumont KA, Shekar SN, Newton RA, et al. Receptor function, dominant negative activity and phenotype correlations for MC1R variant alleles. Hum Mol Genet. 2007;16(18):2249–2260. doi: 10.1093/hmg/ddm177. [DOI] [PubMed] [Google Scholar]
- 58.Newton RA, Roberts DW, Leonard JH, Sturm RA. Human melanocytes expressing MC1R variant alleles show impaired activation of multiple signaling pathways. Peptides. 2007;28(12):2387–2396. doi: 10.1016/j.peptides.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 59.Cuéllar F, Puig S, Kolm I, et al. Dermoscopic features of melanomas associated with MC1R variants in Spanish CDKN2A mutation carriers. Br J Dermatol. 2009;160(1):48–53. doi: 10.1111/j.1365-2133.2008.08826.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fargnoli MC, Sera F, Suppa M, et al. Dermoscopic features of cutaneous melanoma are associated with clinical characteristics of patients and tumours and with MC1R genotype. J Eur Acad Dermatol Venereol. 2014;28(12):1768–1775. doi: 10.1111/jdv.12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Quint KD, van der Rhee JI, Gruis NA, et al. Melanocortin 1 receptor (MC1R) variants in high melanoma risk patients are associated with specific dermoscopic ABCD features. Acta Derm Venereol. 2012;92(6):587–592. doi: 10.2340/00015555-1457. [DOI] [PubMed] [Google Scholar]
- 62.Koludrovic D, Davidson I. MITF, the Janus transcription factor of melanoma. Future Oncol. 2013;9(2):235–244. doi: 10.2217/fon.12.177. [DOI] [PubMed] [Google Scholar]
- 63.Potrony M, Puig-Butille JA, Aguilera P, et al. Prevalence of MITF p.E318K in patients with melanoma independent of the presence of CDKN2A causative mutations. JAMA Dermatol. 2016;152(4):405–412. doi: 10.1001/jamadermatol.2015.4356. [DOI] [PubMed] [Google Scholar]
- 64.Bassoli S, Pellegrini C, Longo C, et al. Clinical, dermoscopic, and confocal features of nevi and melanomas in a multiple primary melanoma patient with the MITF p.E318K homozygous mutation. Melanoma Res. 2018;28(2):166–169. doi: 10.1097/CMR.0000000000000427. [DOI] [PubMed] [Google Scholar]
- 65.Badenas C, Aguilera P, Puig-Butillé JA, et al. Genetic counseling in melanoma. Dermatol Ther. 2012;25(5):397–402. doi: 10.1111/j.1529-8019.2012.01499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Canto MI, Harinck F, Hruban RH, et al. International Cancer of Pancreas Screening (CAPS) Consortium. International Cancer of the Pancreas Screening (CAPS) Consortium summit on the management of patients with increased risk for familial pancreatic cancer. Gut. 2013;62(3):339–347. doi: 10.1136/gutjnl-2012-303108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Potrony M, Puig-Butillé JA, Aguilera P, et al. Increased prevalence of lung, breast, and pancreatic cancers in addition to melanoma risk in families bearing the cyclin-dependent kinase inhibitor 2° mutation: implications for genetic counseling. J Am Acad Dermatol. 2014;71(5):888–895. doi: 10.1016/j.jaad.2014.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]