Abstract
Background
The ZHX family has recently been in the spotlight as an integrator and an indispensable node in carcinogenesis, whose expression is frequently dysregulated in multiple cancers. The current study provides a novel investigation of the expression profiles of ZHX factors in breast cancer.
Materials and methods
The mRNA levels of ZHXs and follow-up periods in breast cancer patients were mined through the Oncomine, Cancer Cell Line Encyclopedia, bc-GenExMiner, cBioPortal and Kaplan–Meier plotter databases. In addition, ZHX3 protein expression was examined in 98 primary tumor samples by immunohistochemistry to investigate its association with clinicopathological parameters and patient outcomes.
Results
We found that the transcriptional levels of ZHX1, ZHX2 and ZHX3 were not significantly altered in tumor tissues compared with those in nontumor tissues. ZHX2 and ZHX3 mRNA levels were observed to be positively correlated with estrogen receptor and progesterone receptor expression, while ZHX2 mRNA levels were negatively associated with HER2 expression. Survival analyses revealed that high mRNA levels of ZHX2 and ZHX3 correlated with better overall survival in patients with breast cancer. Immunohistochemical analysis revealed that patients with decreased ZHX3 protein levels had poorer outcomes. Multivariate analysis exhibited that ZHX3 expression may serve as an independent high-risk prognostic predictor.
Conclusion
Dysregulated expression of ZHXs may be involved in the progression of breast cancer and could serve as a novel biomarker and potential target for breast cancer.
Keywords: ZHX, breast cancer, data mining, immunohistochemistry, prognosis
Introduction
As the top-ranking malignancy and the main reason for cancer death in women worldwide, breast cancer has been established to be a complicated disease of many biological subtypes with various clinical, pathological and molecular characteristics.1–3 According to the expression status of estrogen receptor/progesterone receptor (ER/PR) and EGF receptor 2 (HER2), the molecular subtyping of breast cancer provides distinct predictive/prognostic meanings and therapeutic clues.4 These studies suggest a more precise molecular classification of patients and allow us to more sufficiently know the clinical behaviors and therapeutic targets.5–7 However, irrespective of such advances in diagnosis and treatment for breast cancer, the confined success of the current therapeutic strategies needs new molecular biomarkers with more reliable prediction of patient survival and novel molecular targets which are more promising for cancer therapy.
ZHX protein is a set of transcription factors containing two zinc-finger motifs and five homeobox DNA-binding domains and is localized in the cell nucleus.8–13 From their identification, ZHX family members have been identified as transcriptional repressors through interacting with the α-subunit of the nuclear factor Y (NF-YA) and forming homodimers and heterodimers with each other.9–13 Increasing evidence has revealed that ZHX factors are major transcriptional mediators involved in the events including development and differentiation of hematopoietic cells, maintenance of neural progenitors and osteogenic differentiation of mesenchymal stem cells.8,14,15 Dysregulation of ZHX factors has been found to be correlated with initiation and progression of diverse diseases such as neurological, hematological and glomerular diseases.8,16,17 Data from the relevant studies also reveal that ZHXs may be involved in the progression of many cancer types.8 The crucial roles of the ZHX family in cancer initiation and progression derived from in vitro and in vivo assays give the reason for the ZHX members as biomarker candidates that may be utilized for cancer diagnosis, survival prediction and therapeutic surveillance. Yet, despite such great potential, ZHXs have been so far mostly unknown in breast cancer. In the current study, we employed multiple approaches to examine the expression patterns of ZHX family members in breast cancer and to examine their prognostic values and possible therapeutic implications, using both in silico data-mining approaches and immunohistochemical analysis.
Materials and methods
Bioinformatics analyses
The mRNA expression status of ZHXs in different cancer types was examined using the Oncomine cancer microarray online database (www.oncomine.org).18 When the mRNA levels of ZHXs in cancer tissues were compared with those in normal tissues, we defined the cutoffs as 0.01 and 2 for P-values and fold changes, respectively.
The mRNA levels of ZHXs in several cancer cell lines were examined using the Cancer Cell Line Encyclopedia (CCLE) database (http://portals.broadinstitute.org/ccle). The CCLE database is an online encyclopedia of data collection of gene expression, copy numbers and massively parallel sequences from more than 1,000 human cancer cell lines.
Breast Cancer Gene-Expression Miner v4.0 (bc-GenEx-Miner v4.0) contains 36 genomic datasets with annotation and three unique functions for statistical data mining.19 The expression and prognostic modules were utilized to compare the expression of target genes with clinical criteria and assess their prognostic values for breast cancer.
The prognostic impacts of mRNA levels of ZHXs were assayed using the Kaplan–Meier plotter online database (www.kmplot.com), which contains gene expression and patients’ outcome information from 5,143 clinical breast cancer patients.20,21 To investigate the overall survival (OS) and relapse-free survival (RFS) of breast cancer patients, clinical samples were divided into two groups on the basis of median gene expression (high vs low expression) and assessed by a Kaplan–Meier survival plot.
The impacts of genomic alterations of ZHXs containing gene mutations and copy number variance on the OS and disease-free survival (DFS) of breast cancer patients were calculated using cBioPortal online database (www.cbioportal.org).22,23 A breast invasive carcinoma dataset (The Cancer Genome Atlas [TCGA], provisional) containing pathological and prognostic data from 1,105 breast cancer patients was chosen for this analysis.
Patients and tissue specimens
Ninty-eight formalin-fixed paraffin-embedded specimens were collected from patients with breast cancer (median age, 53 years; range, 29–88 years) which were randomly collected from the Affiliated Cancer Hospital of Shantou University Medical College. All patients underwent curative surgery without any preoperative treatment including chemotherapy or radiotherapy between 2006 and 2007. The median follow-up period was 60 months (range, 43–111 months) from the date of surgery. During the follow-up period, 40 (40.8%) patients had died because of disease recurrence and distant metastasis. Tumor tissues (n=20) and their adjacent noncancerous tissues (n=20) were immediately snap frozen in liquid nitrogen and stored at −80°C until Western blot analyses. These specimens were harvested from another cohort of breast cancer patients who underwent surgery at the same hospital between May 2010 and July 2012.
Tumor grade and stage were identified according to the American Joint Committee on Cancer (AJCC) pathological TNM classification, sixth edition. The clinicopathological parameters are summarized in Table 1. No patients had received any preoperative treatment including chemotherapy or radiotherapy. Written informed consents were acquired prior to tissue sample collection according to the principles in the Declaration of Helsinki. The current study was approved by the institutional review board (# 04–070) of the Affiliated Cancer Hospital of Shantou University Medical College.
Table 1.
Datasets of ZHX family members in breast cancer (Oncomine database)
| Gene | Dataset | Normal (cases) | Tumor (cases) | Fold change | t-test | P-value |
|---|---|---|---|---|---|---|
| ZHX1 | Ma 4 | Breast (14) | Ductal breast carcinoma in situ (9) | 1.347 | 2.150 | 0.027 |
| Curtis | Breast (144) | Invasive breast carcinoma (21) | 1.064 | 2.209 | 0.018 | |
| Breast (144) | Invasive breast carcinoma (1,556) | 1.026 | 2.247 | 0.013 | ||
| Breast (144) | Tubular breast carcinoma (67) | 1.062 | 3.372 | 4.73E-4 | ||
| ZHX2 | TCGA | Breast (61) | Invasive lobular breast carcinoma (36) | 1.202 | 3.114 | 0.001 |
| Breast (61) | Invasive ductal breast carcinoma (389) | 1.117 | 2.820 | 0.003 | ||
| Breast (61) | Invasive breast carcinoma (76) | 1.136 | 2.351 | 0.010 | ||
| Breast (61) | Mixed lobular and ductal breast carcinoma | 1.510 | 2.613 | 0.019 | ||
| ZHX3 | Ma 4 | Breast (14) | Invasive breast carcinoma in situ (9) | 1.337 | 2.000 | 0.035 |
| TCGA | Breast (61) | Invasive ductal breast carcinoma (389) | 1.317 | 4.649 | 6.83E-6 | |
| Breast (61) | Invasive breast carcinoma (76) | 1.256 | 3.392 | 4.83E-4 | ||
| Breast (61) | Invasive ductal and lobular carcinoma (3) | 1.389 | 6.499 | 0.009 |
Abbreviation: TCGA, The Cancer Genome Atlas.
Western blot analysis
This procedure has been described previously.24,25 Sixty micrograms of proteins from each sample was subjected to SDS-PAGE and then transferred to a polyvinylidene difluoride (PVDF) membrane (EMD Millipore, Billerica, MA, USA). After 1-hour incubation in blocking buffer (Tris-buffered saline with 0.1% Tween and 5% nonfat dry milk), the membranes were incubated with a rabbit ZHX3 antibody (Abcam, Cambridge, MA, USA; dilution, 1:500) and a horseradish peroxidase-conjugated secondary antibody against rabbit IgG (Abcam; dilution, 1:1,000). Signals were captured with the enhanced chemiluminescence system following the manufacturer’s instructions (Amersham Pharmacia, Piscataway, NJ, USA). The membranes were reprobed with an anti-actin monoclonal antibody (Abcam; dilution, 1:1,000) to assure the equal loading of each sample. The intensity of ZHX3 was quantified using a Bio-Rad Quantity One quantitation software, with the ratio between the tumor and the paired noncancerous tissues of less than twofolds, suggesting downregulated ZHX3 expression.
Immunohistochemistry and evaluation
Immunohistochemical analysis of ZHX3 protein expression was performed using a standard EnVision complex method as described previously.24–27 Following deparaffinization, rehydration and antigen retrieval, 4 µm sections cut from formalin-fixed paraffin-embedded specimens were incubated with a rabbit polyclonal anti-ZHX3 antibody (Abcam; dilution, 1:200). Immunohistochemical staining was conducted by an EnVision antibody complex (anti-mouse/rabbit) method in conjunction with an Envision™ Detection kit (ZSGB-BIO, Beijing, China) with 3,3′-diaminobenzidine as the chromogen substrate. Nuclei were counterstained with hematoxylin. Sections immunostained with rabbit IgG as the primary antibody were used as negative controls.
Ten random microscopic fields per slide at a magnification of ×400 were evaluated by two independent observers who were blinded to the clinical information. ZHX3 staining was assessed using the semi-quantitative approach, which combines the staining intensity and percentage of positive cells. The mean percentage of positively stained cells was scored as follows: 0%–5% (0); 5%–25% (1); 26%–50% (2); 51%–75% (3) and 76%–100% (4). Staining intensity was categorized as follows: absent (0); weak (1); moderate (2) and strong (3). The multiplication of staining intensity and percentage of positive cells was used as the final staining score. For statistical evaluation, the tumor samples with a final staining score of <3 were classed as negative ZHX3 expression and those with scores ≥3 as positive ZHX3 expression.
Statistical analyses
Statistical analysis was performed using SPSS 17.0 statistical software package (SPSS Inc., Chicago, IL, USA). The statistical significance of differences in numerical data was calculated using Student’s t-test. The differences in the expression levels of ZHXs were correlated with different clinical variables through Fisher’s exact test or Pearson chi-squared test, whichever was appropriate. OS was defined as the time from diagnosis to the date of last contact or of death from any cause. Survival curves were generated using the Kaplan–Meier method with a log-rank test. The prognostic impacts of clinicopathological variables were analyzed by univariate and multivariate regression analyses with a Cox hazard model. P<0.05 (two-tailed) was considered to indicate a statistically significant difference.
Ethics approval and consent to participate
Signed informed consent was obtained from the patients prior to tissue sample collection. The study protocol conformed to the ethical guidelines outlined in the Declaration of Helsinki and was approved by the Institutional Review Board (no. 07-170) of Ningxia Hui Autonomous Region People’s Hospital.
Results
Transcriptional levels of ZHX family members in breast cancer
To overview the transcriptional expression differences of ZHXs between cancer tissues and normal tissues in several cancer types, we set out to perform an online examination using the Oncomine database. As shown in Figure 1, a total of 308, 434 and 416 unique analyses were included for ZHX1, ZHX2 and ZHX3, respectively. ZHX1 mRNA levels were downregulated in cancer tissues compared to that in normal tissues in three studies, whereas two analyses indicated an upregulated ZHX1 expression. Downregulation and overexpression of ZHX2 were observed to be equal in amounts based on 22 studies. Compared with normal tissues, ZHX3 expression was reduced in cancer tissues as demonstrated in seven unique analyses and was increased in three datasets. Regarding breast cancer, unfortunately, only two studies revealed lower expression of ZHX3. We next examined their expression profiles in breast cancer tissues compared with those in normal tissues from 12 datasets. However, no obvious difference was observed according to the defined criteria for the fold changes, despite their significant P-values (Table 1). Moreover, CCLE database analysis revealed that the mRNA expression levels of ZHX1, ZHX2 and ZHX3, in breast cancer cells, listed in the 9th, 7th and 17th position among all cancer types, respectively (Figure 2).
Figure 1.
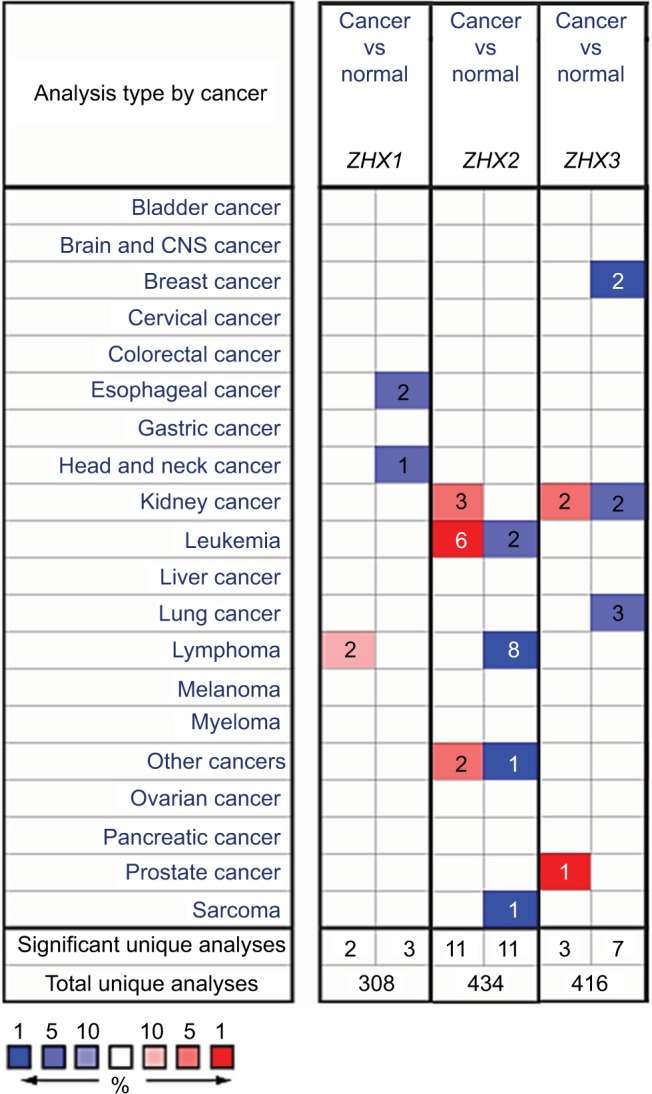
Transcriptional levels of ZHX family members in different cancer types and cancer cell lines.
Notes: This graphic was obtained from Oncomine (www.oncomine.org) indicates the numbers of datasets with significant overexpression (red) or downexpression (blue) of ZHXs at transcriptional levels in cancer tissues compared with those in corresponding normal tissues. Cell color was determined by the best gene rank percentile for the analyses within the cell, and the gene rank was analyzed by the percentile of target genes in the top of all genes measured in each research.
Abbreviation: CNS, central nervous system.
Figure 2.
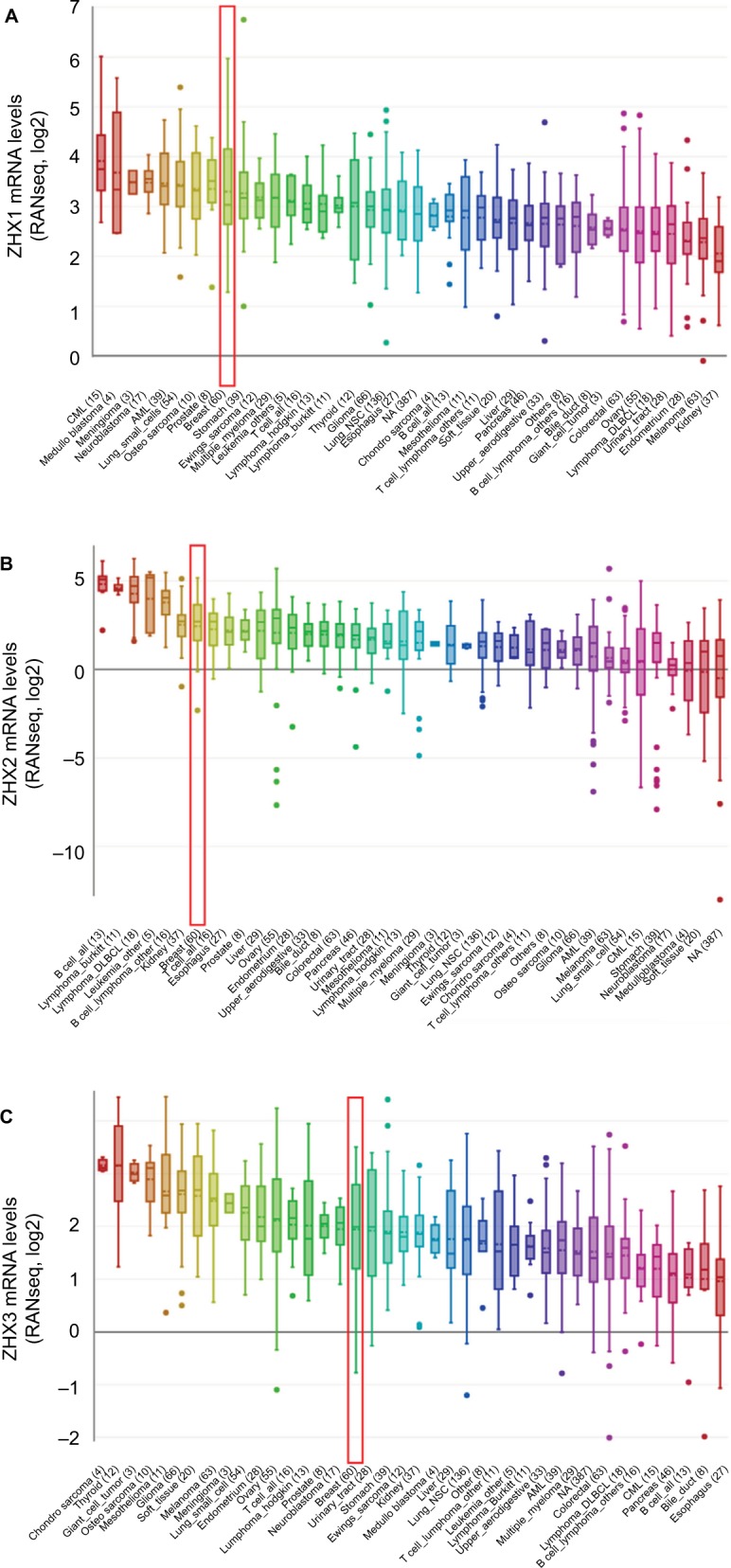
ZHXs were distinctively highly expressed in breast cancer cell lines from Cancer Cell Line Encyclopedia analysis.
Notes: The mRNA expression levels of ZHX1 (A), ZHX2 (B) and ZHX3 (C) in breast cancer cells, ranks in the 9th, 7th and 17th among all cancer cell types (shown in red frame).
Abbreviations: CML, chronic myelocytic leukemia; NSC, non-small cell; DLBCL, diffuse large B cell lymphoma; AML, acute myelocytic leukemia; NA, not applicable.
Relationship between mRNA levels of ZHXs and clinicopathological parameters in breast cancer
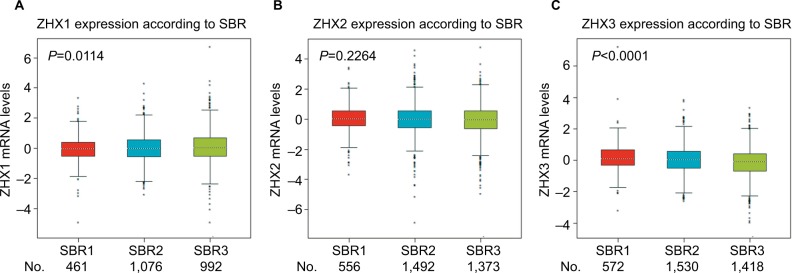
We next examined the correlation between the mRNA expression levels of ZHX factors and clinicopathological variables in patients with breast cancer using the bc-GenExMiner online database. As summarized in Table 2, no significant difference was observed between patients aged ≤51 years and >51 years. Patients showing positive nodal status had lower ZHX3 mRNA levels than those showing negative nodal status. ER expression was observed to be correlated with elevated ZHX2 and ZHX3 expression, while HER2 expression was found to be correlated with lower ZHX1 mRNA expression levels. In addition, we found that ZHX2 and ZHX3 mRNA levels were significantly decreased in triple-negative breast cancer (TNBC) patients. Of note, according to Scarff, Bloom and Richardson (SBR) grade status classification, there was a significant association between more advanced SBR grade and higher mRNA level of ZHX1 and lower mRNA level of ZHX3, respectively (Figure 3).
Table 2.
Datasets of ZHX family in breast cancer from bc-GenExMiner v4.1
| Variables | ZHX1 | ZHX2 | ZHX3 | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| n | mRNA | P-value | n | mRNA | P-value | n | mRNA | P-value | |
|
| |||||||||
| Age (years) | |||||||||
| ≤51 | 868 | – | 0.3985 | 1,361 | – | 0.5445 | 1,392 | – | 0.2618 |
| >51 | 1,764 | – | 2,142 | 2,210 | – | ||||
| Nodal status | |||||||||
| Negative | 1,322 | – | 0.6801 | 2,447 | – | 0.5825 | 2,493 | – | 0.0145 |
| Positive | 1,141 | – | 1,509 | – | 1,562 | ↓ | |||
| ER (IHC) | |||||||||
| Negative | 985 | – | 0.7029 | 1,525 | – | 0.0012 | 1,559 | – | <0.0001 |
| Positive | 2,487 | – | 3,923 | ↑ | 3,988 | ↑ | |||
| PR (IHC) | |||||||||
| Negative | 635 | – | 0.8158 | 946 | – | 0.0014 | 946 | – | <0.0001 |
| Positive | 1,009 | – | 1,439 | ↑ | 1,439 | ↑ | |||
| HER2 (IHC) | |||||||||
| Negative | 671 | – | 0.0185 | 1,409 | – | 0.1462 | 1,409 | – | 0.4722 |
| Positive | 160 | ↓ | 201 | – | 201 | – | |||
| Triple-negative status (IHC) | |||||||||
| Not | 2,625 | – | 0.3568 | 4,099 | – | 0.0008 | 4,164 | – | <0.0001 |
| TNBC | 197 | – | 374 | ↓ | 374 | ↓ | |||
Abbreviations: ER, estrogen receptor; IHC, immunohistochemistry; PR, progesterone receptor; TNBC, triple-negative breast cancer.
Figure 3.
Correlation of mRNA expression of ZHX factors with SBR grade status.
Notes: Global significant differences of ZHX1 (A), ZHX2 (B) and ZHX3 (C) between groups were assessed by Welch’s test to generate P-values, along with Dunnett– Tukey–Kramer’s tests for pairwise comparison when a global significant difference exists.
Abbreviation: SBR, Scarff, Bloom and Richardson.
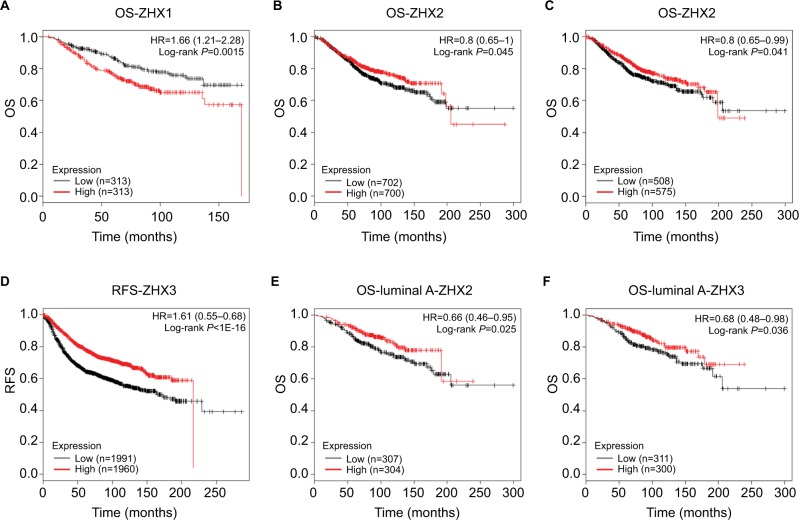
Increased ZHX2/3 mRNA levels associate with better OS in breast cancer
The prognostic impacts of ZHXs on patient survival were characterized using the Kaplan–Meier plotter survival analysis. We observed that high ZHX1 mRNA levels predicted a shorter OS for breast cancer patients (Figure 4A). By contrast, high expression of ZHX2 and ZHX3 was significantly correlated with better OS of breast cancer patients (Figure 4B and C). Patients with high ZHX3 mRNA expression also exhibited a favorable RFS (Figure 4D). In particular, sub-analysis revealed that high expression of ZHX2 and ZHX3 was correlated with better OS in luminal A subtype breast cancer separately (Figure 4E and F).
Figure 4.
Prognostic value of mRNA levels of ZHX factors in breast cancer patients (OS and RFS in Kaplan–Meier plot).
Notes: The impact of ZHX1 (A), ZHX2 (B) and ZHX3 (C) on OS of breast cancer patients. (D) The impact of ZHX3 on RFS of breast cancer patients. (E) The impact of ZHX2 on OS in luminal A subtype. (F) The impact of ZHX3 on OS in luminal A subtype.
Abbreviations: OS, overall survival; RFS, relapse-free survival.
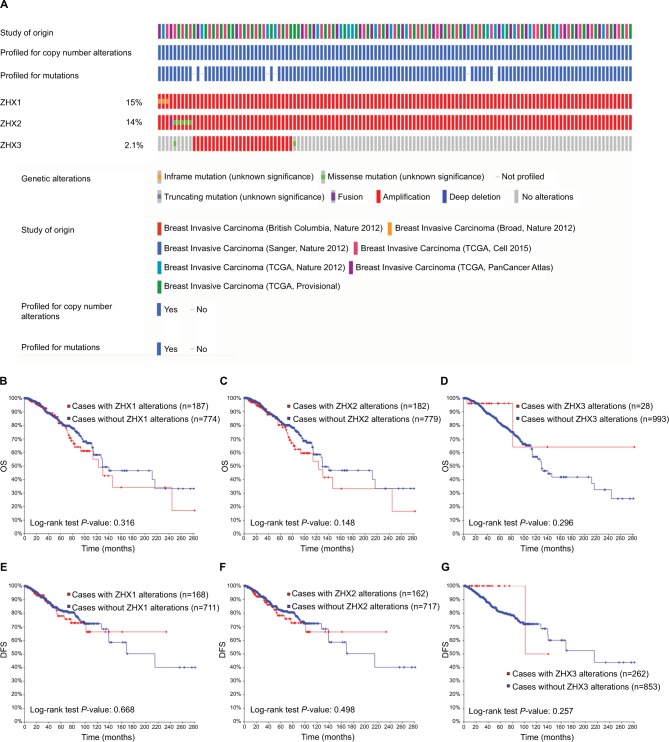
Association between genetic alterations of ZHX factors and patient survival
Genetic alterations of ZHXs occurred in 678 (17%) of 4,077 patients with invasive breast carcinoma (Figure 5A). After analyses by Kaplan–Meier plot and log-rank test, no significant relationship was found between OS and DFS in the breast cancer patients with or without the alterations in ZHXs (Figure 5B–G).
Figure 5.
Genetic alterations of ZHX gene expression and their association with patient survival in breast invasive carcinoma (cBioPortal).
Notes: (A) Oncoprint in cBioPortal represented the proportion and distribution of samples with alterations in ZHX factors. The figure was cropped on the right to exclude samples without alterations. (B–D) Kaplan–Meier plots comparing OS in cases with/without ZHX1 (B), ZHX2 (C) and ZHX3 (D) alterations. (E–G) Kaplan–Meier plots comparing DFS in cases with/without ZHX1 (E), ZHX2 (F) and ZHX3 (G) alterations.
Abbreviations: DFS, disease-free survival; OS, overall survival; TCGA, The Cancer Genome Atlas.
ZHX3 expression is an independent prognostic factor in breast cancer
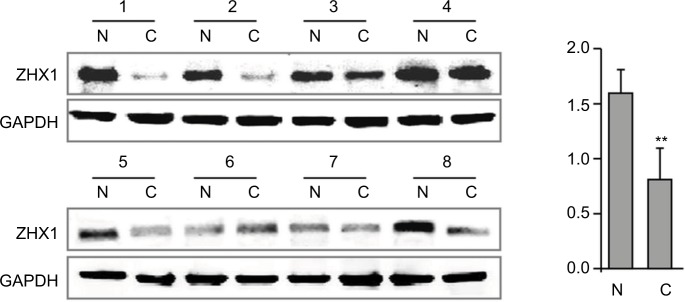
In support of the abovementioned findings, we further determined whether ZHX3 protein expression is downregulated in tumors vs noncancerous tissues. Figure 6 shows the representative results in a cohort specimen (n=20) by Western blot analysis. Seventy percent (14/20) of tumor specimens had significantly lower ZHX3 protein levels than their matching adjacent noncancerous tissues. We then examined the expression profile of ZHX3 in 98 formalin-fixed paraffin-embedded specimens by using immunohistochemistry. We observed positive ZHX3 immunostaining in the nucleus of tumor cells in 45.9% (45/98) of breast cancer samples tested (Figure 7). We found that negative ZHX3 expression was associated with lymph node metastasis, advanced tumor stage, poor differentiation and positive ER expression (Table 3). Kaplan–Meier survival analyses showed that patients with positive ZHX3 expression exhibited a better OS than those without ZHX3 expression (Figure 8). On univariate analysis, lymph node metastasis, advanced tumor stages and ZHX3 expression were associated with an unfavorable OS (Table 4). After adjustment for the prognostic variables obtained in univariate analyses, only lymph node metastasis and ZHX3 expression maintained the independent significance in multivariate analysis (Table 4). Altogether, our results suggest that ZHX3 expression is an independent prognostic indicator for breast cancer patients.
Figure 6.
ZHX3 protein levels were determined by Western blot analysis in primary breast cancer tissues (C) vs matching adjacent noncancerous tissues (N).
Notes: A quantitative analysis of ZHX3 expression normalized by β-actin is shown in the right panel (n=20). **P<0.01.
Figure 7.
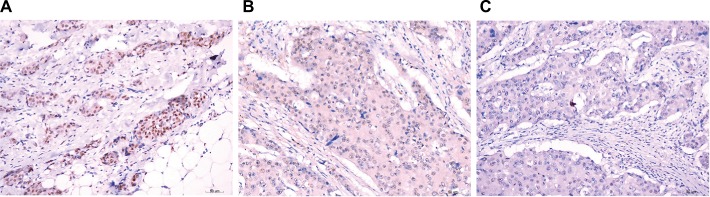
Representative immunohistochemical staining for ZHX3 in breast cancer.
Notes: (A) Strong staining of ZHX3 in breast cancer tissue. (B) Moderate staining of ZHX3 in breast cancer tissue. (C) Weak staining of ZHX3 in breast cancer tissue. Original magnification, ×400.
Table 3.
Associations between ZHX3 expression and clinicopathological features in breast cancer
| Variables | No. of patients | ZHX3 expression
|
P-value | |
|---|---|---|---|---|
| Negative, n (%) | Positive, n (%) | |||
|
| ||||
| Age (years) | ||||
| <45 | 45 | 22 (48.9) | 23 (51.1) | 0.417 |
| ≥45 | 53 | 31 (58.5) | 22 (41.5) | |
| T (primary tumor) | ||||
| T1/T2 | 47 | 26 (55.3) | 34 (44.7) | 0.842 |
| T3/T4 | 51 | 27 (52.9) | 11 (47.1) | |
| N (regional lymph nodes) | ||||
| N0 | 61 | 27 (44.3) | 34 (55.7) | 0.014 |
| N1–N3 | 37 | 26 (70.3) | 11 (29.7) | |
| Stage of tumors | ||||
| I/II | 79 | 37 (46.8) | 42 (53.2) | 0.004 |
| III/IV | 19 | 16 (84.2) | 3 (15.8) | |
| Histological type | ||||
| Non-ductal | 23 | 12 (53.2) | 11 (47.8) | 0.510 |
| Ductal | 75 | 41 (54.7) | 34 (45.3) | |
| Differentiation | ||||
| Well/moderate | 60 | 27 (45.0) | 33 (55.0) | 0.037 |
| Poor | 38 | 26 (68.4) | 12 (31.6) | |
| ER status | ||||
| Negative | 47 | 34 (72.3) | 13 (27.7) | 0.001 |
| Positive | 51 | 19 (37.3) | 32 (62.7) | |
| PR status | ||||
| Negative | 29 | 20 (69.0) | 9 (31.0) | 0.076 |
| Positive | 69 | 33 (47.8) | 36 (52.2) | |
| HER2 status | ||||
| Negative | 59 | 30 (50.8) | 29 (49.2) | 0.535 |
| Positive | 39 | 23 (59.0) | 16 (41.0) | |
Abbreviations: ER, estrogen receptor; PR, progesterone receptor.
Figure 8.
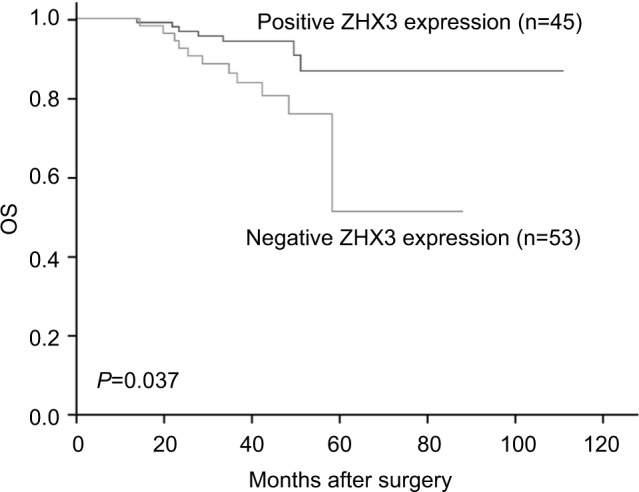
Kaplan–Meier survival curves with univariate analyses (log-rank) according to the expression status of ZHX3 in patients with breast cancer.
Note: The OS of patients with ZHX3-positive tumors was significantly higher than that of patients with ZHX3-negative tumors (P=0.037).
Abbreviation: OS, overall survival.
Table 4.
Univariate and multivariate Cox proportional hazard model for the survival of breast cancer patients
| Variables | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
|
| ||||
| HR (95% CI) | P-value | HR (95% CI) | P-value | |
|
| ||||
| Age (years) | ||||
| ≥45 vs <45 | 1.886 (0.626–5.684) | 0.260 | ||
| Tumor size | ||||
| T3/T4 vs T1/T2 | 2.840 (0.892–9.040) | 0.447 | ||
| Lymph node metastasis | ||||
| N1–N3 vs N0 | 4.343 (1.263–14.933) | 0.020 | 4.40 (1.238–15.673) | 0.022 |
| Stage of tumors | ||||
| III/IV vs I/II | 2.572 (1.238–5.342) | 0.011 | ||
| Histological type | ||||
| Ductal vs non-ductal | 1.985 (0.878–4.489) | 0.100 | ||
| ER status | ||||
| Positive vs negative | 0.737 (0.285–1.906) | 0.530 | ||
| PR status | ||||
| Positive vs negative | 0.559 (0.214–1.458) | 0.235 | ||
| HER2 status | ||||
| Positive vs negative | 1.765 (0.661–4.716) | 0.257 | ||
| ZHX3 expression | ||||
| Negative vs positive | 2.749 (1.065–7.097) | 0.037 | 3.663 (1.371–9.784) | 0.010 |
Abbreviations: ER, estrogen receptor; PR, progesterone receptor.
Discussion
The development of microarray techniques promotes the research levels relating to studies of RNA and DNA and it has also become a key tool for cancer biological and biomedical research.28 In the current study, we analyzed the expression pattern and prognostic impacts of different ZHX factors in breast cancer using in silico analysis and immunohistochemical studies. Our results indicate that dysregulation of ZHXs is involved in tumor progression and that the expression of ZHXs may be associated with the outcomes of breast cancer patients.
ZHX1, the first identified factor of this family, has been proposed as a tumor suppressor in many cancer types.29–33 However, two recent studies suggested that ZHX1 may serve as an oncogene in glioblastoma and cholangiocarci- noma.34,35 Upregulated ZHX1 expression in tumor specimens compared with that in normal tissues has been found to be correlated with reduced survival of cancer patients.34,35 The observations in glioblastoma and cholangiocarcinoma are consistent with our findings, ie, patients with high ZHX1 expression showed significantly prolonged OS than those with low ZHX1 expression. In addition, we found that ZHX1 mRNA level was significantly upregulated in patients with higher SBR grade, which predicted fast-growing and spreading tumors. It has been reported that ZHX1 is a downstream molecule of cytokines and enables cells to respond to the changing environment.36 We inferred that this disparity, at least in part, may be due to the complexity of tumor microenvironment and the intrinsic differences in each type of tumors.
ZHX2 and ZHX3, forming heteromeric complexes with ZHX1, have been noted in the transcriptional suppression of cancer markers in normal hepatocytes.37 It has been also found that silencing ZHX2 expression by gene promoter methylation is an epigenetic event in hepatocellular carcinoma and the overexpression of ZHX2 inhibits proliferation and enhances the chemosensitivity of hepatocellular carcinoma cells.38–40 HBV has been reported to repress ZHX2 expression and facilitate the proliferation of hepatocellular carcinoma via activating miR-155 and conversely, ZHX2 restricts HBV replication via epigenetic and non-epigenetic manners.41,42 These results suggest the tumor suppressor functions of ZHX2. However, another report revealed that ZHX2 protein expression was observed only in cancer tissues and associated with advanced clinical stage and metastasis of hepatocellular carcinoma.43 Another recent study showed downregulated ZHX3 expression and upregulated ZHX2 expression in renal cell carcinoma, and the expression levels of ZHX1 and ZHX3 were significantly associated with pathological stage.32 Decreased ZHX2 expression has also been found to be correlated with an unfavorable outcome in multiple myeloma.44 In the current study, despite no significant change found in the mRNA levels of ZHX2/3 in cancer tissues compared with normal tissues, we observed that high ZHX2/3 mRNA expression was associated with better OS in patients with breast cancer. Further, our analyses revealed that high mRNA levels of ZHX2/3 were also correlated with a favorable OS in patients with luminal A subtype breast cancer. We do not have any clues for this disparity, but the correlation between ZHX2/3 and positive ER expression in breast cancer might be a possible explanation. It has been established that loss of ER-alpha in breast cancer patient is associated with poor outcome, promoted recurrence after treatment and increased metastasis.45 Moreover, in support of the results by data mining approaches, our analysis of immunohistochemistry identified strongly correlated tendency between decreased ZHX3 protein expression and an unfavorable OS. We conclude that ZHX3 protein expression may serve as a prognostic predictor for breast cancer.
There are still a few limitations regarding the results of the current study. Further validation including physiological and molecular mechanism will enhance our understanding of the clinical behavior of ZHXs. In addition, only in silico analysis was performed to detect the mRNA levels and the prognostic impacts of this family. Their protein expression profile may be required for more description. It also should be understood in the context of the gene expression as prognostic biomarkers in breast cancer included in MammaPrint, the integrative clusters, and others. Therefore, the exact function of ZHXs in tumors should be further examined in cancer initiation and progression.
Conclusion
Collectively, in the current study, we comprehensively examined the expression status and prognostic impact of ZHX factors in breast cancer. Our findings contribute to the systematic understanding of the biological functions of ZHXs in breast cancer as well as provide the evidence that the members of this family can be utilized as new prognostic biomarkers and promising molecular targets for breast cancer treatment.
Data sharing statement
The dataset used and/or analyzed in the current study is available from the corresponding authors on reasonable request.
Acknowledgments
We thank Mr Zhen Zhang (Department of Biochemistry and Molecular Biology, University of Kansas Medical Center) for his careful reading of this manuscript and kind suggestions. This study was supported in part by the National Natural Science Foundation of China (grant nos. 81860426 and 81760440) and the Natural Science Foundation of Ningxia, China (grant no. 2018AAC02016).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Perou CM, Sørlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 3.Di Cosimo S, Baselga J. Management of breast cancer with targeted agents: importance of heterogeneity [corrected] Nat Rev Clin Oncol. 2010;7(3):139–147. doi: 10.1038/nrclinonc.2009.234. [DOI] [PubMed] [Google Scholar]
- 4.Onitilo AA, Engel JM, Greenlee RT, Mukesh BN. Breast cancer subtypes based on ER/PR and HER2 expression: comparison of clinicopathologic features and survival. Clin Med Res. 2009;7(1–2):4–13. doi: 10.3121/cmr.2009.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peppercorn J, Perou CM, Carey LA. Molecular subtypes in breast cancer evaluation and management: divide and conquer. Cancer Invest. 2008;26(1):1–10. doi: 10.1080/07357900701784238. [DOI] [PubMed] [Google Scholar]
- 6.van ‘t Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 7.Hong CQ, Zhang F, You YJ, et al. Elevated C1orf63 expression is correlated with cdk10 and predicts better outcome for advanced breast cancers: a retrospective study. BMC Cancer. 2015;15(1):548. doi: 10.1186/s12885-015-1569-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y, Ma D, Ji C. Zinc fingers and homeoboxes family in human diseases. Cancer Gene Ther. 2015;22(5):223–226. doi: 10.1038/cgt.2015.16. [DOI] [PubMed] [Google Scholar]
- 9.Barthelemy I, Carramolino L, Gutiérrez J, Barbero JL, Márquez G, Zaballos A. zhx-1: a novel mouse homeodomain protein containing two zinc-fingers and five homeodomains. Biochem Biophys Res Commun. 1996;224(3):870–876. doi: 10.1006/bbrc.1996.1114. [DOI] [PubMed] [Google Scholar]
- 10.Hirano S, Yamada K, Kawata H, et al. Rat zinc-fingers and homeoboxes 1 (ZHX1), a nuclear factor-YA-interacting nuclear protein, forms a homodimer. Gene. 2002;290(1–2):107–114. doi: 10.1016/s0378-1119(02)00553-x. [DOI] [PubMed] [Google Scholar]
- 11.Kawata H, Yamada K, Shou Z, et al. Zinc-fingers and homeoboxes (ZHX) 2, a novel member of the ZHX family, functions as a transcriptional repressor. Biochem J. 2003;373(Pt 3):747–757. doi: 10.1042/BJ20030171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamada K, Kawata H, Shou Z, et al. Analysis of zinc-fingers and homeoboxes (ZHX)-1-interacting proteins: molecular cloning and characterization of a member of the ZHX family, ZHX3. Biochem J. 2003;373(Pt 1):167–178. doi: 10.1042/BJ20021866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawata H, Yamada K, Shou Z, Mizutani T, Miyamoto K. The mouse zinc-fingers and homeoboxes (ZHX) family; ZHX2 forms a heterodimer with ZHX3. Gene. 2003;323:133–140. doi: 10.1016/j.gene.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 14.Suehiro F, Nishimura M, Kawamoto T, et al. Impact of zinc fingers and homeoboxes 3 on the regulation of mesenchymal stem cell osteogenic differentiation. Stem Cells Dev. 2011;20(9):1539–1547. doi: 10.1089/scd.2010.0279. [DOI] [PubMed] [Google Scholar]
- 15.Liu G, Clement LC, Kanwar YS, Avila-Casado C, Chugh SS. ZHX proteins regulate podocyte gene expression during the development of nephrotic syndrome. J Biol Chem. 2006;281(51):39681–39692. doi: 10.1074/jbc.M606664200. [DOI] [PubMed] [Google Scholar]
- 16.Clement LC, Liu G, Perez-Torres I, Kanwar YS, Avila-Casado C, Chugh SS. Early changes in gene expression that influence the course of primary glomerular disease. Kidney Int. 2007;72(3):337–347. doi: 10.1038/sj.ki.5002302. [DOI] [PubMed] [Google Scholar]
- 17.Nagel S, Ehrentraut S, Meyer C, Kaufmann M, Drexler HG, Macleod RA. Aberrantly expressed OTX homeobox genes deregulate B-cell differentiation in Hodgkin lymphoma. PLoS One. 2015;10(9):e0138416. doi: 10.1371/journal.pone.0138416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jézéquel P, Campone M, Gouraud W, et al. bc-GenExMiner: an easy-to-use online platform for gene prognostic analyses in breast cancer. Breast Cancer Res Treat. 2012;131(3):765–775. doi: 10.1007/s10549-011-1457-7. [DOI] [PubMed] [Google Scholar]
- 20.Győrffy B, Surowiak P, Budczies J, Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8(12):e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Györffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123(3):725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 22.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.You Y, Li H, Qin X, et al. Decreased CDK10 expression correlates with lymph node metastasis and predicts poor outcome in breast cancer patients - a short report. Cell Oncol. 2015;38(6):485–491. doi: 10.1007/s13402-015-0246-4. [DOI] [PubMed] [Google Scholar]
- 25.You Y, Li H, Qin X, Ran Y, Wang F. Down-regulated ECRG4 expression in breast cancer and its correlation with tumor progression and poor prognosis – A short Report. Cell Oncol. 2016;39(1):89–95. doi: 10.1007/s13402-015-0260-6. [DOI] [PubMed] [Google Scholar]
- 26.You Y, Yang W, Wang Z, et al. Promoter hypermethylation contributes to the frequent suppression of the cdk10 gene in human nasopharyngeal carcinomas. Cell Oncol. 2013;36(4):323–331. doi: 10.1007/s13402-013-0137-5. [DOI] [PubMed] [Google Scholar]
- 27.You Y, Yang W, Qin X, et al. ECRG4 acts as a tumor suppressor and as a determinant of chemotherapy resistance in human nasopharyngeal carcinoma. Cell Oncol. 2015;38(3):205–214. doi: 10.1007/s13402-015-0223-y. [DOI] [PubMed] [Google Scholar]
- 28.Sealfon SC, Chu TT. RNA and DNA microarrays. Methods Mol Biol. 2011;671:3–34. doi: 10.1007/978-1-59745-551-0_1. [DOI] [PubMed] [Google Scholar]
- 29.Wang J, Liu D, Liang X, et al. Construction of a recombinant eukaryotic human ZHX1 gene expression plasmid and the role of ZHX1 in hepatocellular carcinoma. Mol Med Rep. 2013;8(5):1531–1536. doi: 10.3892/mmr.2013.1700. [DOI] [PubMed] [Google Scholar]
- 30.Wang Z, Ma X, Cai Q, et al. MiR-199a-3p promotes gastric cancer progression by targeting ZHX1. FEBS Lett. 2014;588(23):4504–4512. doi: 10.1016/j.febslet.2014.09.047. [DOI] [PubMed] [Google Scholar]
- 31.Ma X, Huang M, Wang Z, Liu B, Zhu Z, Li C. ZHX1 inhibits gastric cancer cell growth through inducing cell-cycle arrest and apoptosis. J Cancer. 2016;7(1):60–68. doi: 10.7150/jca.12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwon RJ, Kim YH, Jeong DC, et al. Expression and prognostic significance of zinc fingers and homeoboxes family members in renal cell carcinoma. PLoS One. 2017;12(2):e0171036. doi: 10.1371/journal.pone.0171036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guan J, Liu Z, Xiao M, et al. MicroRNA-199a-3p inhibits tumorigenesis of hepatocellular carcinoma cells by targeting ZHX1/PUMA signal. Am J Transl Res. 2017;9(5):2457–2465. [PMC free article] [PubMed] [Google Scholar]
- 34.Kwon RJ, Han ME, Kim JY, et al. ZHX1 promotes the proliferation, migration and invasion of cholangiocarcinoma cells. PLoS One. 2016;11(11):e0165516. doi: 10.1371/journal.pone.0165516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwon RJ, Han ME, Kim YJ, et al. Roles of zinc-fingers and homeoboxes 1 during the proliferation, migration, and invasion of glioblastoma cells. Tumour Biol. 2017;39(3):101042831769457. doi: 10.1177/1010428317694575. [DOI] [PubMed] [Google Scholar]
- 36.Shou Z, Yamada K, Kawata H, Yokoyama O, Miyamoto K. A mechanism of induction of the mouse zinc-fingers and homeoboxes 1 (ZHX1) gene expression by interleukin-2. Biochem Biophys Res Commun. 2004;314(3):885–890. doi: 10.1016/j.bbrc.2003.12.162. [DOI] [PubMed] [Google Scholar]
- 37.Yamada K. ZHX2 and ZHX3 repress cancer markers in normal hepatocytes. Front Biosci. 2009:143724–3732. doi: 10.2741/3483. [DOI] [PubMed] [Google Scholar]
- 38.Lv Z, Zhang M, Bi J, Xu F, Hu S, Wen J. Promoter hypermethylation of a novel gene, ZHX2, in hepatocellular carcinoma. Am J Clin Pathol. 2006;125(5):740–746. doi: 10.1309/09B4-52V7-R76K-7D6K. [DOI] [PubMed] [Google Scholar]
- 39.Yue X, Zhang Z, Liang X, et al. Zinc fingers and homeoboxes 2 inhibits hepatocellular carcinoma cell proliferation and represses expression of cyclins A and E. Gastroenterology. 2012;142(7):1559–1570. doi: 10.1053/j.gastro.2012.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luan F, Liu P, et al. Reduced nucleic ZHX2 involves in oncogenic activation of glypican 3 in human hepatocellular carcinoma. Int J Biochem Cell Biol. 2014;55:129–135. doi: 10.1016/j.biocel.2014.08.021. [DOI] [PubMed] [Google Scholar]
- 41.Song X, Tan S, Wu Z, et al. HBV suppresses ZHX2 expression to promote proliferation of HCC through miR-155 activation. Int J Cancer. 2018;143(12):3120–3130. doi: 10.1002/ijc.31595. [DOI] [PubMed] [Google Scholar]
- 42.Xu L, Wu Z, Tan S, et al. Tumor suppressor ZHX2 restricts hepatitis B virus replication via epigenetic and non-epigenetic manners. Antiviral Res. 2018;153:114–123. doi: 10.1016/j.antiviral.2018.03.008. [DOI] [PubMed] [Google Scholar]
- 43.Hu S, Zhang M, Lv Z, Bi J, Dong Y, Wen J. Expression of zinc-fingers and homeoboxes 2 in hepatocellular carcinogenesis: a tissue microarray and clinicopathological analysis. Neoplasma. 2007;54(3):207–211. [PubMed] [Google Scholar]
- 44.Armellini A, Sarasquete ME, García-Sanz R, et al. Low expression of ZHX2, but not RCBTB2 or Ran, is associated with poor outcome in multiple myeloma. Br J Haematol. 2008;141(2):212–215. doi: 10.1111/j.1365-2141.2007.06956.x. [DOI] [PubMed] [Google Scholar]
- 45.Dhasarathy A, Kajita M, Wade PA. The transcription factor Snail mediates epithelial to mesenchymal transitions by repression of estrogen receptor-alpha. Mol Endocrinol. 2007;21(12):2907–2918. doi: 10.1210/me.2007-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]