Abstract
Within the economically important plant family Solanaceae, Jaltomata is a rapidly evolving genus that has extensive diversity in flower size and shape, as well as fruit and nectar color, among its ∼80 species. Here, we report the whole-genome sequencing, assembly, and annotation, of one representative species (Jaltomata sinuosa) from this genus. Combining PacBio long reads (25×) and Illumina short reads (148×) achieved an assembly of ∼1.45 Gb, spanning ∼96% of the estimated genome. Ninety-six percent of curated single-copy orthologs in plants were detected in the assembly, supporting a high level of completeness of the genome. Similar to other Solanaceous species, repetitive elements made up a large fraction (∼80%) of the genome, with the most recently active element, Gypsy, expanding across the genome in the last 1–2 Myr. Computational gene prediction, in conjunction with a merged transcriptome data set from 11 tissues, identified 34,725 protein-coding genes. Comparative phylogenetic analyses with six other sequenced Solanaceae species determined that Jaltomata is most likely sister to Solanum, although a large fraction of gene trees supported a conflicting bipartition consistent with substantial introgression between Jaltomata and Capsicum after these species split. We also identified gene family dynamics specific to Jaltomata, including expansion of gene families potentially involved in novel reproductive trait development, and loss of gene families that accompanied the loss of self-incompatibility. This high-quality genome will facilitate studies of phenotypic diversification in this rapidly radiating group and provide a new point of comparison for broader analyses of genomic evolution across the Solanaceae.
Keywords: comparative genomics, de novo assembly, floral evolution, potato, tomato, transposable element
Introduction
Understanding the genetic substrate of trait diversification is a longstanding goal in evolutionary biology. Diversification can involve a range of genetic changes, including point mutations in coding or regulatory regions, or structural variation such as chromosomal inversions or gene duplications (Stapley et al. 2010; Berner and Salzburger 2015). Until recently, these data have been challenging to generate for all but the best developed model species. However, the emergence of next-generation and single molecule sequencing technologies now allows the rapid generation of diverse genomic resources, including whole-genome sequences, transcriptome sequences, and genome-wide marker panels for a much broader range of taxa (Stapley et al. 2010; Ellegren 2014; Bleidorn 2016). Comparative genomic analyses of related organisms provide opportunities to quantify species differences in genome size, complexity, noncoding features, and molecular evolution within genic regions, as well as structural differences in the number and identity of members of specific gene families, or in classes of transposable elements (TEs). In combination with data on specific phenotypic and functional trait variation, comparative genomic analyses can also evaluate the role of different genomic changes in both general patterns of lineage diversification and lineage-specific adaptive evolution. In addition to deciphering mechanisms of genome evolution, these data can be used to address the genetics of phenotypic diversity associated with adaptation and speciation, across groups of closely related, ecological diverse, species (Brawand et al. 2014; Zhang et al. 2014; Novikova et al. 2016; Yin et al. 2018).
The plant genus Jaltomata is one such rapidly radiating clade, consisting of 60–80 species estimated to have arisen within the last 5 Myr or less (Mione 1992; Miller et al. 2011; Wu et al. 2018). Jaltomata is closely related to both Solanum (which includes tomato, potato, and eggplant) and Capsicum (peppers), and together these three form a clade that is sister to the rest of the Solanaceae, a highly diverse plant family that also contains other economically important genera such as Nicotiana (tobacco) and Petunia (Bohs and Olmstead 1997; Olmstead et al. 1999; Walsh and Hoot 2001; Olmstead et al. 2008; Särkinen et al. 2013). Because multiple Solanaceous species already have whole-genome sequences with gene annotations, an additional high-quality genome in the key phylogenetic position held by Jaltomata provides a valuable resource for clarifying the evolutionary relationships among these important clades, and for comparative analyses of genomic, genetic, and phenotypic evolution across the family. For example, although most Solanaceae species have the same base number of chromosomes (X = 12; Petunia is the exception, with X = 7), estimated genome sizes vary drastically (e.g., 4-fold genome-size difference between tomato and hot pepper) (Kim et al. 2014; Xu et al. 2017) suggesting that repetitive element divergence might be a key contributor to genome-size variation. Further, comparative analyses of gene family evolution and sequence divergence could identify changes associated with important ecological trait variation (e.g., identification of tandemly duplicated genes involved in the capsaicin biosynthesis pathway specifically in hot pepper [Kim et al. 2014; Qin et al. 2014]). Comparative genomic analysis including Jaltomata therefore could reveal both large- and small-scale genetic changes responsible for genomic, phenotypic, and functional differentiation among these important clades.
Apart from its key position within the Solanaceae, the genus Jaltomata itself varies widely in ecological range, vegetative characters, physiology, and reproductive form and function (Haak et al. 2014), making it a valuable emerging model for studies of adaptive diversification and evolution of novel traits. Like its close relatives in Solanum and Capsicum, Jaltomata has its highest diversity in the Andean region of South America, although some species’ ranges extend into Central America and the southwestern United States. Its species occur in a broad range of habitats, including tropical forests, coastal lowlands, and lomas (discrete fog-misted communities surrounded by arid desert) (Mione 1992; Mione and Yacher 2005). Unlike its close relatives, Jaltomata lineages exhibit a striking and unique diversity of derived floral traits, especially in corolla (petal) shapes—that include rotate, campanulate (bell-shaped), and tubular forms (Miller et al. 2011; Kostyun and Moyle 2017)—and in the amount and color of nectar produced, which ranges from small amounts of nectar that is essentially colorless to copious amounts of deep red nectar (Hansen et al. 2007). In comparison, close relatives Solanum and Capsicum predominantly have flatter rotate corollas, and either colorless to pale yellow nectar (Capsicum) or no floral nectar at all (Solanum) (Knapp et al. 2004). Jaltomata species also vary in mature fruit color, including species that have either purple, red, orange, or green fruit at maturity; this fruit color variation appears to characterize three major subclades within the genus as separate dark purple-, red-, and orange-fruited clades (Miller et al. 2011; Särkinen et al. 2013; Wu et al. 2018). The orange-fruited clade (comprising ∼50 species) is the subgroup containing most novel derived floral trait variation and is estimated to have diverged within the last 1.5 Myr, consistent with a very rapid recent radiation of floral and reproductive diversity that likely drew upon multiple sources of genetic variation (Wu et al. 2018). Jaltomata is also distinctive among Solanaceae genera in that all examined species are self-compatible (SC) (Mione 1992) (Kostyun JL and Mione T, unpublished data), whereas most other genera—including Solanum and Capsicum—exhibit genetically determined self-incompatibility (SI) in some or all species (Goldberg et al. 2010). The availability of a high-quality genome in this genus could thus help to assess genome features specific to Jaltomata to identify genetic changes that might accompany or drive its unique and rapid trait evolution.
In this study, we generated a high-coverage and almost complete genome of one Jaltomata species, J. sinuosa, by adopting a hybrid assembly strategy using PacBio long reads and Illumina short reads. Using this newly assembly genome, we performed comparative genomic analyses with six additional high-quality genomes in the Solanaceae. We found that different topologies of Jaltomata, Solanum, and Capsicum were supported by a large number of individual gene trees, suggesting a complex history of rapid divergence and hybridization in the common ancestors of these three genera. Within Jaltomata, we identified a recent expansion of Gypsy elements, a superfamily of long terminal repeat-retrotransposons (LTR-RTs), around 1–2 Ma that likely contributed to the genome-size expansion of Jaltomata. In addition, assessing genome features specific to Jaltomata identified genetic changes that could have contributed to rapid trait evolution in the genus, including loci with lineage-specific patterns of adaptive evolution, the loss of gene families that accompanied the loss of self-incompatibility, and the expansion of gene families potentially involved in novel trait development in Jaltomata. We discuss the significance of this genome assembly in the light of phenotypic diversity in this rapidly radiating group and genomic evolution across the economically important plant family Solanaceae.
Materials and Methods
Species Selection and Tissue Sampling
Among the 60–80 species within the genus Jaltomata, J.sinuosa was chosen for whole-genome sequencing because estimated heterozygosity within this species is the lowest of all evaluated species (Wu et al. 2018 and this article), potentially facilitating genome assembly. Further, although J. sinuosa itself has rotate corollas with light-yellow nectar (i.e., similar to ancestral trait states in the genus; Miller et al. 2011; Kostyun et al. 2017), it belongs within the recently diverged (<1.5 Myr) and highly diverse (∼50 species) orange-fruited clade, which incorporates the majority of novel floral diversity within the genus (Miller et al. 2011; Wu et al. 2018). To obtain genomic DNA, young leaf tissue was collected from a single individual of J. sinuosa grown in the Indiana University greenhouse (voucher available at IND herbarium), and flash frozen with liquid nitrogen. DNA was extracted using Qiagen DNeasy Plant Kits, purified with ethanol precipitation, and quality checked using Nanodrop and gel electrophoresis. Approximately 60-μg genomic DNA was provided to the Duke University Sequencing and Genome Technologies facility for library preparation and sequencing: 4 large-insert (15–20 kb) libraries were constructed and sequenced in 67 SMRT cells on the Pacific Biosciences (PacBio) platform.
Genome Assembly
We adopted a hybrid assembly approach (Koren et al. 2012) in which relatively high-accuracy Illumina paired-end reads (148×) were used to trim and correct low base-call-accuracy PacBio long reads (25×). Initially, we used two different genome-assembly strategies DBG2OLC (Ye et al. 2016) and MaSuRCA v3.2.2 (Zimin et al. 2017), and then evaluated the completeness of genome assembly for each using 1,515 plant near-universal single-copy ortholog within BUSCO v3 (Simão et al. 2015). We found the assembly output from MaSuRCA was much better than that from DBG2OLC, in terms of assembly coverage, contig N50, and genome completeness (supplementary table S1, Supplementary Material online). Thus, the initial assembly from MaSuRCA was used for all further analyses. Genome size was also estimated within the MaSuRCA pipeline based on the k-mer abundance distribution.
To remove potential contaminants in our assembly, all scaffolds were aligned against the NCBI nonredundant nucleotide sequence database using BlastN with a cutoff of E-5. Scaffolds were assigned to the closest reference based on the best combined hit score, and any scaffolds assigned to a nonplant species were removed (Bolger et al. 2014). In total, 22 small scaffolds were filtered out, consisting of 240,347 bases, leaving 7,667 scaffolds in the genome assembly. To estimate assembly accuracy at the nucleotide level, we estimated the discrepancy between the long-read sequences used in the initial genome assembly and the high-quality Illumina reads, the latter of which were used to error correct the long-read base calls (see supplementary text, Supplementary Material online).
Repeat Annotation
We followed the “Repeat Library Construction-Advanced” steps from the MAKER-P pipeline (Campbell et al. 2014) to generate a Jaltomata-specific repeat library. Briefly, miniature inverted TEs (MITEs) were detected by MITE-Hunter (Han and Wessler 2010); LTRs were constructed using LTRharvest (Ellinghaus et al. 2008) followed by LTR_retriever (Ou and Jiang 2018); and other repetitive sequences were identified using RepeatModeler (http://www.repeatmasker.org/RepeatModeler.html; last accessed January 9, 2019). RepeatMasker (http://www.repeatmasker.org) was then used to mask repeat elements in the assembled genome by searching for all homologous repeats in the species-specific library. To compare the recent activity of LTR-RTs, we also applied the same approach (i.e., LTRharvest followed by LTR_retriever) to annotate the full-length LTR-RTs in four other genomes from Solanum and Capsicum, including S. lycopersicum, S. tuberosum, S. pennellii, and C. annuum (see supplementary text, Supplementary Material online). The timing of LTR-RT bursts was based on the distribution of times inferred for each full-length element based on sequence divergence between its direct repeats, in LTR_retriever (Ou and Jiang 2018). Briefly, insertion time (T) of each full-length LTR-RT was estimated as T = K/2μ, where K is the divergence rate between the two terminal repeats and μ was set to be 1.3 × 10−8 mutations per site per year (Ma and Bennetzen 2004).
Gene Structure Annotation
We followed the MAKER-P pipeline (Campbell et al. 2014) to annotate gene models in the assembled genome using three classes of evidence: RNA-seq data, protein homology, and ab initio gene prediction (see supplementary text, Supplementary Material online). MAKER-P synthesized all information from these three different classes and produced final annotations with high evidence-based quality, requiring annotation evidence distance (which measures the goodness of fit of an annotation to the RNA/protein-alignment evidence supporting it) score <0.6. To assign gene functions, we followed the pipeline AHRD (https://github.com/groupschoof/AHRD) to automatically select the most concise, informative, and precise function annotation (see supplementary text, Supplementary Material online). For each gene, the associated gene ontology (GO) annotation(s) were assigned according to the predicted protein domain(s) (http://www.geneontology.org/external2go/interpro2go; last accessed January 9, 2019). To investigate the putative chromosomal locations of predicted genes in the genome, we performed whole-genome synteny alignment against the tomato genome using Satsuma v3.1.0 (Grabherr et al. 2010) and recorded the genes unambiguously associated with one identified syntenic region in tomato.
Inference of Homologous and Orthologous Gene Clusters
We downloaded the annotated gene/protein-coding sequences of six other diploid Solanaceae species (including S. lycopersicum, S. tuberosum, C.annuum, Nicotiana attenuata, Petunia axillaris, and P. inflata) from the SolGenomics database (http://solgenomics.net) and used Arabidopsis thaliana as the outgroup species. The six Solanaceae species were chosen because they each have one well-assembled and annotated genome (i.e., contig/scaffold size >10 kb and gene completeness >95%) with a comparable number of annotated genes (∼35,000 genes). Identification of homologous gene clusters was performed using orthoMCL v2.0.9 (Li et al. 2003) with the default options. We obtained 3,103 single-copy 1-to-1 orthologous clusters that each contained one sequence from all seven species for downstream phylogenetic analyses. The coding sequences (CDS) of those 1-to-1 orthologs were aligned using PRANK v.150803 (Löytynoja and Goldman 2005) with codons enforced. As a quality check on all multiple sequence alignments, we removed poorly aligned regions using a sliding window approach that masked any 15-bp window from alignment if it had more than six mismatches (not counting indels/gaps, which were masked to N) among all the investigated species. After this process, any alignment with more than 20% of its sequence masked was removed from the analysis.
Phylogenetic Analyses
We reconstructed evolutionary relationships between Jaltomata and the six other Solanaceous species for which we had whole-genome data. We used four different, but complementary, inference approaches to perform phylogenetic reconstruction: 1) maximum-likelihood applied to concatenated alignments (in RAxML v8.23; Stamatakis 2006), 2) consensus of gene trees (RAxML V8.23, with Majority Rule Extended [Salichos and Rokas 2013]), 3) quartet-based gene tree reconciliation (using ASTRAL v4.10.9) (Mirarab and Warnow 2015), and 4) Bayesian concordance of gene trees (using MrBayes v3.2 [Huelsenbeck and Ronquist 2001] followed by BUCKy v1.4.4 [Larget et al. 2010]) (see supplementary text, Supplementary Material online). Using these different tree reconstruction approaches allowed us to evaluate the extent to which they generated phylogenies that disagreed, as well as to identify the specific nodes and branches that were robust to all methods of phylogenetic reconstruction. In addition, we also reconstructed and annotated the chloroplast and mitochondrial genomes in Jaltomata and performed phylogenetic analyses of J. sinuosa with S. lycopersicum, C. annuum, and N. attenuata (the species for which chloroplast DNA and mitochondrial DNA sequences were readily available) using the concatenated sequences of mitochondrial or chloroplast genes in RAxML v8.23 (see supplementary text, Supplementary Material online).
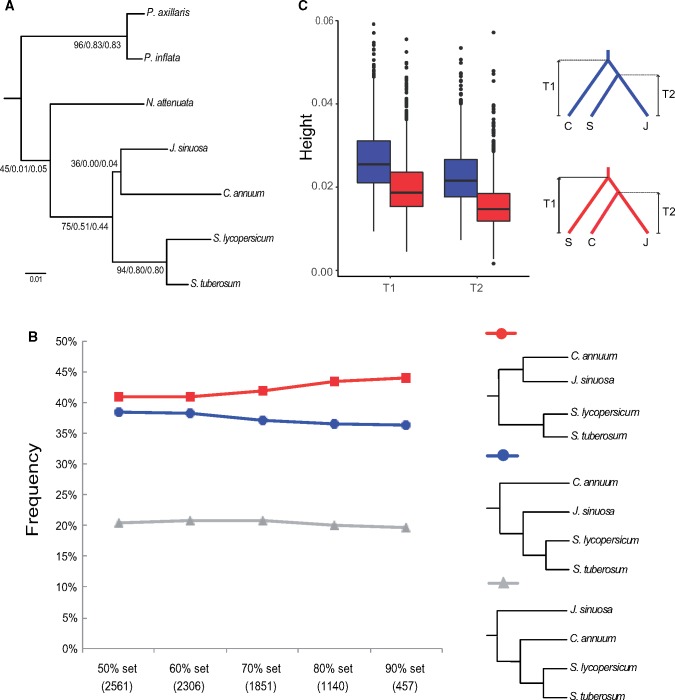
Our phylogenetic analyses indicated that gene trees supported two conflicting bipartitions among Jaltomata, Solanum, and Capsicum (i.e., “[Jaltomata, Solanum], Capsicum” or “[Jaltomata, Capsicum], Solanum”) at roughly equal frequencies (see the Results section). To differentiate which of these topologies most likely represented the initial pattern of lineage splitting (i.e., the “true” species tree) rather than relatedness due to subsequent introgression among species, we compared the relative divergence times (node depths) among species, using sets of gene trees that each supported one of the two most conflicting bipartitions, with N.attenuata as the outgroup. Gene trees constructed from nonintrogressed (initial branching order) sequences are expected to have deeper mean divergence times at the two internal nodes (T1 and T2; fig. 3) than those constructed from introgressed sequences, because introgression will reduce sequence divergence between the two lineages that have exchanged genes (Fontaine et al. 2015). Internal node depths (divergence times) for each gene tree that supported one of these two alternative bipartitions were calculated from biallelic informative sites; per gene estimates were used to calculate the genome-wide means of divergence times T1 and T2, to determine which of the two topologies had higher average divergence times (see further, supplementary text, Supplementary Material online).
Fig. 3.
—Phylogenetic relationships among the seven investigated Solanaceae species. (A) Concatenated RAxML tree rooted by A. thaliana. The values above each internal branch indicate the proportion of gene trees supported (%), internode certainty, and tree certainty all supporting each node in the majority-rule consensus tree, from BUCKy (Larget et al. 2010). (B) The percentage of gene trees supporting the three alternative phylogenetic positions of Jaltomata relative to S. lycopersicum, S. tuberosum, and C. annuum (assuming Solanum is monophyletic). Five different sets of gene trees were used, which were selected based on the average bootstrap cutoff across the gene tree. (C) Evaluation of the relative node depths (ages) in the two alternative bipartitions among Jaltomata (J), Solanum (S), and Capsicum (C). The higher average height (depth) of T1 and T2 indicate that Jaltomata and Solanum are sister clades, whereas shallower gene trees supporting Jaltomata and Capsicum as sister are likely to be influenced by introgression.
Gene Family Analyses
To investigate changes in gene family sizes, we investigated the families that are significantly rapidly evolving among the six Solanaceae species including Jaltomata. (In this analysis, C. annuum was excluded because of the complex relationships among Jaltomata, Capsicum, and Solanum; see the Results section.) We determined the significantly expanded or contracted gene families along each branch of phylogeny using the program CAFE v3.0 (Han et al. 2013) with P value cutoff of 0.01. The input gene families were generated from the OrthoMCL program (Li et al. 2003). Phylogenetic relationships were based on the output from the RAxML analysis of the concatenated data set of 3,103 coding-sequence alignments. Divergence times among different species were directly retrieved from previous estimates among Solanaceae species (Särkinen et al. 2013). In addition, we further investigated the expansion of a specific candidate gene within Jaltomata, SEUSS, by examining gene trees, primary mapped Illumina reads (i.e., using only the best alignment of multimapped reads), PacBio reads spanning more than one gene copy, and expression patterns supported by RNA-seq from 14 Jaltomata species examined in our previous study (Wu et al. 2018) (see supplementary text, Supplementary Material online).
Positive Selection Analyses
To infer positively selected genes in the Jaltomata genome, we tested 3,103 single-copy 1-to-1 orthologous genes (excluding C. annuum) whose gene tree topologies were the same as the inferred species tree. For each investigated gene, we inferred putative adaptive evolution (i.e., dN/dS > 1) using the branch-site (BS) model (model = 2 and NS sites = 2) in PAML v4.4 (Yang 2007) on the terminal branch leading to J. sinuosa. This inference uses a likelihood ratio test to determine whether the alternative test model (fixed_omega = 0) is significantly better than the null model (fixed_omega = 1), and identifies putative positively selected genes as those with a likelihood ratio test P value <0.01 and a false discovery rate <0.2 (Benjamini and Hochberg 1995). Because multinucleotide mutations (MNMs) can cause false inferences of positive selection in the PAML BS test (Venkat et al. 2017), for all significant genes we further applied a more conservative BS model (BS + MNM) (Venkat et al. 2017) in which another parameter δ is incorporated to represent the relative instantaneous rate of double mutations to that of single mutations (Venkat et al. 2017). We then assessed how many and which BS-significant genes remained significant (P < 0.01) in the BS + MNM test. A GO-enrichment analysis was performed on these remaining putative selected genes using ONTOLOGIZER v2.0 with the parent–child analysis and a cutoff P value of 0.01 (Bauer et al. 2008).
Results
Genome Assembly and Annotation
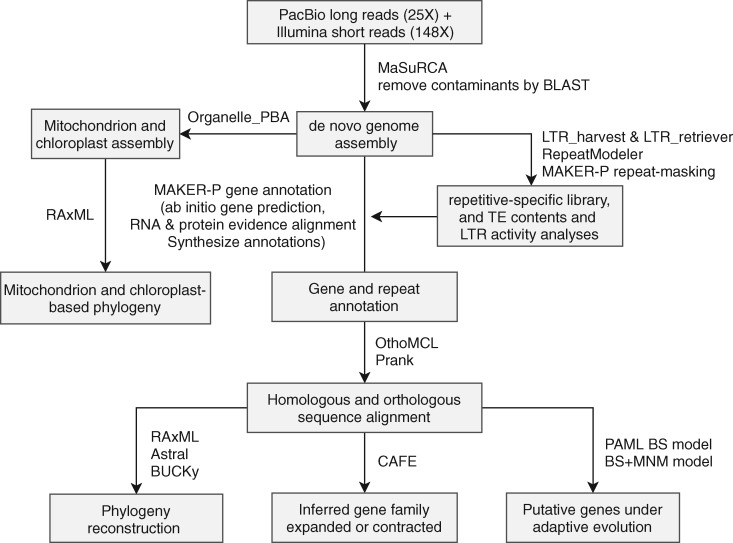
Following the workflow (fig. 1), we generated the first genome assembly of a Jaltomata species. The genome of J. sinuosa is estimated to be ∼1,512 Mb based on k-mer frequencies, which is consistent with the estimated size from flow cytometry (∼1,650 Mb; Haak DC, unpublished data). Using the MaSuRCA genome-assembly pipeline, we generated an assembly of ∼1,456 Mb, in which 96.6% of 1,515 BUSCO universal single-copy orthologous genes could be found (table 1). The assembly comprises 7,667 scaffolds, with a contig and scaffold N50 of 364.9 and 397.6 kb, respectively (table 1). In the ∼1,389 Mb (91.9%) of assembled reference genome that was also covered by at least five Illumina reads, only 68,203 sites had a base-call different from the base-call that was consistently indicated by the aligned Illumina reads (i.e., cases in which all Illumina reads support a site to be, for example, “A” rather than the base-call “T” in the assembly, indicating a potential error in the assembly; this criterion was applied to both mismatches and indels and excluded any sites where the Illumina reads indicated two different alleles at a site). This examination of base-call error rate across 91.9% of assembly, exclusively targeted for homozygous sites, suggested a lower bound of base-call error rate to be ∼0.005%. The upper bound of error rates was estimated to be 0.28% by counting the total discrepancies between all the primarily aligned reads and the assembled reference (including mismatches due to heterozygosity), although the error rate might be higher in genomic regions that are not well covered by Illumina data (Schmidt et al. 2017). Overall, the high coverage of the plant conserved single-copy genes, low base-calling error rate, and the high mapping rate of RNA-seq reads (96.2%, see below), indicate a high-quality assembly for the J. sinuosa genome. We also assembled the chloroplast genome into a single contig with a total length of 156 kb (Guanine–Cytosine [GC] content of 37.91%) with 82 annotated genes; the mitochondrial genome was assembled into a single contig with a total length of 317 kb (GC content of 42.05%) and 21 annotated genes.
Fig. 1.
—Workflow of assembly of J. sinuosa genome and downstream comparative genomic analyses.
Table 1.
Summary of the J. sinuosa Genome Assembly
| Assembly features | |
| Number of scaffolds | 7,667 |
| Genome size (Mb) | 1,512 |
| Assembly size (Mb) | 1,454 |
| Plant_CEGs (BUSCO) (%)a | 87.0 (+8.7 + 0.9) |
| Contig N50 length (kb) | 364.9 |
| Scaffold N50 length (kb) | 397.6 |
| GC content % | 37.94 |
| RNA-seq mapped (%)b | 90.6 (+5.6) |
| Structure annotation | |
| Numbers of protein-coding genes | 34,726 |
| Mean CDS length (bp) | 1,101 |
| Number of exons per gene | 5.3 |
Plant_CEGs (Clusters of Essential Genes) shows the percentage of complete single-copy orthologs plus the percentage of duplicated orthologs and fragmented orthologs.
RNA-seq mapped indicates the percentage of uniquely mapped reads plus the percentage of multimapped reads.
Repetitive Element Annotation and LTR Insertion Age Distribution
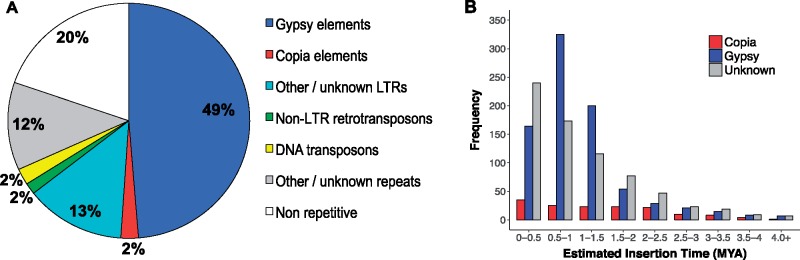
The assembled J. sinuosa genome contains a total of ∼1,158 Mb (80.29% of the assembly) of repetitive sequences (supplementary table S2, Supplementary Material online). LTR-RTs are the major source of repetitive sequences, accounting for 64.43% of the genome assembly (fig. 2A); of those LTR-RTs that could be unambiguously classified by LTR_retriever (∼190 Mb of genomic regions were annotated as unknown LTR-RTs; see supplementary text, Supplementary Material online), Gypsy elements are much more abundant (∼703 Mb) than Copia elements (∼34 Mb) (fig. 2A and supplementary table S2, Supplementary Material online). To evaluate the recent activity of LTR-RTs, we identified 1,682 full-length LTR-RTs within our data set (including 823 Gypsy and 151 Copia; supplementary table S3, Supplementary Material online) and, using the distribution of sequence divergence between the two terminal repeats within each LTR element, we infer a recent burst of Gypsy element activity around 1–2 Ma (fig. 2B). In comparison, the C. annuum and three Solanum genomes (S. lycopersicum, S. tuberosum, and S. pennellii) show many fewer recent insertions of Gypsy elements (supplementary fig. S1B–D, Supplementary Material online). Instead, Gypsy elements in C. annuum are inferred to have been most active around 3 Ma (supplementary fig. S1A, Supplementary Material online), whereas a higher abundance of recently active Copia elements was detected in S. pennellii (supplementary fig. S1D, Supplementary Material online), consistent with a previous study (Bolger et al. 2014).
Fig. 2.
—Landscape of repetitive sequences in the J. sinuosa genome. (A) The repeat contents in the assembly. (B) The estimated insertion age distribution of full-length LTR-RTs in the assembly.
Gene Annotation and Transcription
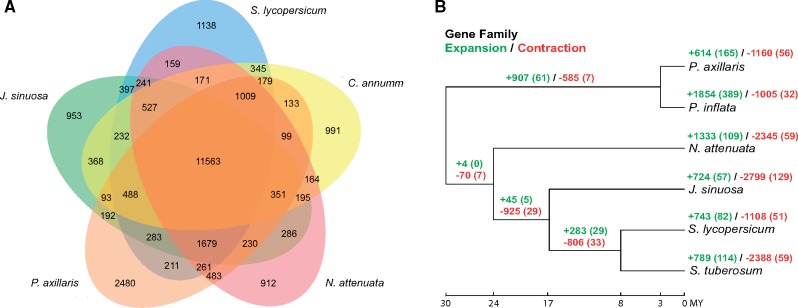
A total of 34,726 high-confidence (annotation evidence distance <0.6) protein-coding genes with 37,106 transcripts were predicted (supplementary table S4, Supplementary Material online), which is similar to the 35,768 predicted genes in the domesticated tomato, S. lycopersicum (Tomato Genome Consortium 2012). The protein-coding sequences (CDS) in the annotated genes have an average length of 1,101 bp and the predicted genes have an average of 5.3 exons (supplementary table S5, Supplementary Material online); both are similar to the average CDS length (∼1,027 bps) and exon number (∼4.9) in the tomato annotation ITAG3.2 (Tomato Genome Consortium 2012). Through whole-genome synteny alignment, 23,013 (66.3%) of these predicted Jaltomata genes were associated with unambiguous syntenic regions within the tomato genome (supplementary table S6, Supplementary Material online). Among all annotated genes, nearly all of them (99.90%) were functionally annotated through the AHRD pipeline (supplementary table S7, Supplementary Material online). The transcriptome-wide RNA-seq reads (Wu et al. 2018) from J. sinuosa were mapped against the assembly with an alignment rate of 96.2%, with 67.81% of annotated genes having more than 0.5 transcripts per million. Because RNA-seq data from one species might only sample a subset of expressed genes, we also mapped the RNA-seq reads from 13 other Jaltomata lineages (Wu et al. 2018) back to the assembly and identified 82.83% of annotated genes that have transcripts per million >0.5 in at least one species. We identified 11,563 gene families that were shared among J. sinuosa, S. lycopersicum, C. annuum, N.attenuate, and P. axillaris, whereas a total of 953 gene families were specific to J. sinuosa (fig. 4A). GO term enrichment analysis indicated these J. sinuosa-specific genes are significantly over-represented in negative regulation of metabolic process or catalytic activity (supplementary table S9, Supplementary Material online).
Fig. 4.
—Comparative gene family analyses. (A) Unique and homologous gene families. The numbers of unique or shared gene families are shown in the corresponding diagram components. (B) Gene family expansion and contraction patterns across the Solanaceae. The numbers of expanded (green) and contracted (red) gene families are shown in each branch, and numbers in the brackets are the number of significantly expanded or contracted gene families (P < 0.01).
Complex Phylogenetic Relationships between Jaltomata, Solanum, and Capsicum
The four phylogeny-reconstruction methods all supported the same topology among the seven Solanaceae species that included J. sinuosa as more closely related to C. annuum than to S. lycopersicum (fig. 3A and supplementary fig. S2, Supplementary Material online). However, there was also substantial gene tree discordance observed at this node: only 42% of gene trees supported the branch that groups J. sinuosa together with C. annuum. An internode certainty value of zero on this branch indicates that the number of gene trees supporting this bipartition is almost equal to the number of gene trees supporting the most common conflicting bipartition (fig. 3A). A similar pattern was also observed at the internode that splits the two Petunia species from the other investigated Solanaceae species (fig. 3A).
In order to further investigate the phylogenetic placement of Jaltomata relative to Solanum and Capsicum, we examined the proportion of gene trees supporting the each of the three different possible topologies, using five groups of genes which had progressively higher bootstrap support (i.e., average % bootstrap support across the RAxML gene trees of greater than 50%, 60%, 70%, 80%, or 90%). For the largest (>50% bootstrap) group, we found that ∼42% of gene trees supported the most common bipartition (i.e., [Jaltomata, Capsicum], Solanum), but that ∼38% gene trees supported the conflicting topology that places Jaltomata as sister to Solanum; this general pattern is consistent across sets of genes with increasingly higher power (fig. 3B). Our phylogenies constructed from the concatenated data of 10 mitochondrial genes supported Jaltomata as the sister clade of Capsicum, whereas the concatenated data from 72 chloroplast genes supported Jaltomata as closer to Solanum (supplementary fig. S3 and table S10, Supplementary Material online). However, there were very few (only four) informative sites in the mitochondrial data set that can unambiguously resolve the relationship (supplementary fig. S3B, Supplementary Material online), thus we interpret this apparent conflict between plastid genomes is due to a lack of power (too few informative variable sites) specifically in the mitochondria data.
To differentiate which of the two majority conflicting topologies is due to the initial lineage splitting events versus subsequent introgression, we compared the estimated divergence times among the three species from those genes supporting either of the two conflicting topologies. The gene trees supporting “Solanum, (Capsicum, Jaltomata)” have lower mean divergence times T1 and T2 relative to those from the gene trees supporting “Capsicum, (Jaltomata, Solanum)” (fig. 3C). Because introgression reduces sequence divergence between species exchanging genes (see the Materials and Methods section), our data suggest that the initial species branching order was “Capsicum, (Jaltomata, Solanum),” whereas the excess of gene trees supporting “Solanum, (Capsicum, Jaltomata)” is due to subsequent introgression between Capsicum and Jaltomata.
Dynamic Evolution of Gene Families in Jaltomata
We identified 129 rapidly evolving gene families that contracted specifically on the branch leading to J. sinuosa (fig. 4B). Interestingly, of these we found a gene family (Cluster 7; supplementary table S11, Supplementary Material online) that is functionally involved in pollen–pistil interactions (GO: 0048544) for which there are only 21 genes in the J. sinuosa genome, whereas the other five investigated Solanaceae genomes have 38–46 genes (supplementary table S12, Supplementary Material online). This contracted gene cluster included receptorlike kinase family proteins with an S-locus glycoprotein domain, which are involved in plant reproduction and signaling in pollen–pistil interactions (supplementary table S11, Supplementary Material online). Based on our analysis of syntenic blocks with the tomato genome, the genes in this cluster are distributed across multiple chromosomes in the genome (supplementary table S11, Supplementary Material online). The loss of multiple putative S-locus receptor kinase family proteins is consistent with the ancestral loss of self-incompatibility in all Jaltomata lineages (see the Discussion section). Compared with Jaltomata, contraction of different gene families was detected in the S. lycopersicum and N. attenuata genomes (supplementary table S13, Supplementary Material online).
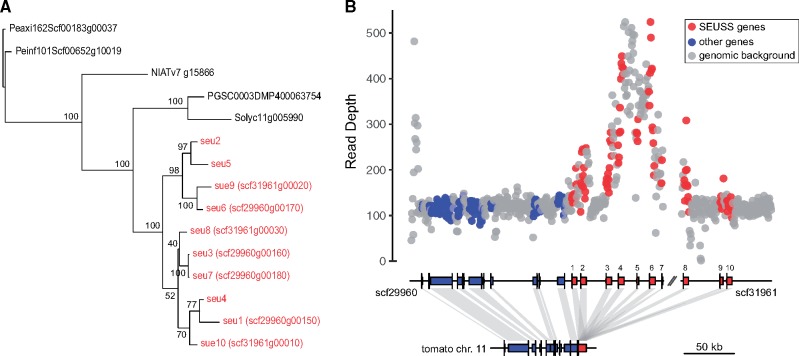
We also identified 57 rapidly evolving gene families that expanded specifically on the branch leading to J. sinuosa (fig. 4B). These gene families were involved in a broad range of functions, including genes responsible for plant stress-related response, such as disease resistance, response to wounding, and regulation of nitrogen compound metabolic process (supplementary table S14, Supplementary Material online). Similar to J. sinuosa, significant lineage-specific expansion of various stress-related gene families was also found in other Solanaceae species, including genes involved in heat shock, oxidative stress, and resistance to fungi in S. lycopersicum and N. attenuata genomes (supplementary table S15, Supplementary Material online). Of the expanded families specific to the Jaltomata genome, one particularly interesting example involved the transcription factor SEUSS, which is known to play an important role in floral organ development in model systems (see the Discussion section). Although only one copy of SEUSS was identified in each other Solanaceae genome, an estimated ten copies (seven of them were annotated by the MAKER pipeline) were detected in the J. sinuosa genome (fig. 5A), and the gene tree of these copies indicates the expansion of SEUSS happened recently and specifically in the Jaltomata lineage (fig. 5A). The inferred ten copies of Jaltomata SEUSS gene were found on two scaffolds, suggesting the expansion of SEUSS gene copy number is due to recent tandem duplication (fig. 5B).
Fig. 5.
—Evolutionary dynamics of copy number of the gene SEUSS in J. sinuosa. (A) Lineage-specific expansion of SEUSS in J. sinuosa. The ten duplicated copies were labeled according to their relative position order along the located scaffolds. Among them, seven copies were annotated by the MAKER pipeline. (B) Validation that there are multiple copies of SEUSS using read depth of Illumina DNA-seq reads which is at least equal or above the genomic background and adjacent single-copy genes. Each dot represents read depth from a 1 kb sliding window. The red dots indicate the windows that overlapped with SEUSS genes, whereas the blue dots indicate the windows that overlapped with other single-copy genes. The gray dots are the windows that contain other (intergenic) regions.
Several methods validated the presence of >1 SEUSS copies in Jaltomata. We identified five PacBio long reads spanning two adjacent SEUSS copies. We also found that read depth around each SEUSS copy was equal to or higher than read depth in the adjacent single-copy loci or background genomic regions (fig. 5B). The higher depth of primary mapped reads at some of these inferred SEUSS copies suggests that there might be additional paralogous copies of these loci that we were unable to differentiate because their sequences are too similar (i.e., due to very recent duplication events). Using RNA-seq data from Wu et al. (2018), we found that four structurally intact copies (SEUSS 4, 6, 8, and 10) have >25 reads that uniquely map to them in at least one of 13 Jaltomata species, suggesting there are at least four putatively functional copies in the genus (supplementary table S16, Supplementary Material online). Across all 13 species, only RNA-seq reads from the sampled reproductive, but not vegetative, tissues mapped to SEUSS loci (supplementary table S16, Supplementary Material online), and expression of SUESS copies varied among species: three copies (SEUSS 4, 6, and 8) were expressed in all purple-fruited lineages (i.e., J. repandidentata, J. procumbens, and J. darcyana), whereas almost all of the ten inferred SEUSS copies had no reads mapped (i.e., very low expression) in most of the orange/green-fruited lineages (including J. sinuosa) (supplementary table S16, Supplementary Material online). This difference in number of reads mapped in different Jaltomata lineages cannot be explained by sequence divergence, because the purple-fruited lineages are more distant from J. sinuosa relative to other investigated Jaltomata lineages.
Detection of Genes Potentially under Positive Selection in Jaltomata
Using the BS model in PAML, we identified 89 genes out of 6,582 testable genes as putatively under positive selection (i.e., the lineage-specific selection model fit significantly better than the null model, P value <0.01 and false discovery rate <0.2; supplementary table S17, Supplementary Material online). After implementing the BS + MNM test (Venkat et al. 2017) on these 89 putative selected genes, 58 of them remained significant (P value <0.01; supplementary table S16, Supplementary Material online). Some of these 58 loci are involved in stress responses, including resistance to osmotic and oxidative stress, response to heat shock, and heavy metal transportation (supplementary table S17, Supplementary Material online); however, these putatively selected genes were not enriched for any specific GO terms (supplementary table S18, Supplementary Material online). Nonetheless, among these loci are several candidates for elements of Jaltomata-specific trait evolution, including loss of self-incompatibility, growth during (floral) development, and regulation of pigment biosynthesis (see the Discussion section).
Discussion
Here, we generated a high-quality genome sequence of a representative species (J. sinuosa) from within Jaltomata, a rapidly evolving, florally and reproductively diverse genus in the Solanaceae. We used these data to clarify a complex history of origin that Jaltomata shares with its two most closely related genera—Solanum and Capsicum—and to infer recent TE dynamics that might be responsible for genome-size evolution within the Solanaceae. We also identified overall patterns of significant gene family gain and loss, as well as adaptive molecular evolution, which could be implicated in the rapid reproductive trait evolution that is distinctive to Jaltomata among its Solanaceous relatives.
Comparative Phylogenomic Analysis Reveals Complex History of Divergence among Jaltomata and Its Closest Relatives
The Solanaceae is a highly speciose plant family, with an estimated 100 genera and 2,500 species that have all evolved within the last ∼30 Myr. Previous phylogenetic studies have confirmed that many lineages within the Solanaceae arose within a highly compressed time frame (Olmstead et al. 1999; Särkinen et al. 2013), making resolution of some evolutionary relationships challenging. In particular, previous molecular phylogenetic studies using chloroplast and nuclear loci indicated that Jaltomata is close to both Solanum and Capsicum, however the relationship among these three genera has varied depending upon the specific loci used in phylogenetic reconstruction (Bohs and Olmstead 1997; Olmstead et al. 1999; Walsh and Hoot 2001; Olmstead et al. 2008; Särkinen et al. 2013). The J.sinuosa genome therefore provided an opportunity to evaluate and clarify the historical evolutionary relationships among key genera within Solanaceae.
Just as with other recent phylogenomic studies of contemporary (Brawand et al. 2014; Lamichhaney et al. 2015; Novikova et al. 2016; Pease et al. 2016) or more ancient rapid radiations (Jarvis et al. 2014; Wickett et al. 2014; Suh et al. 2015; Yang et al. 2015), we detected evidence for substantial gene tree discordance in relationships among the seven Solanaceous species analyzed here. This included high discordance (internode uncertainty) at the internode that split the Petunia lineages from the remaining species (fig. 3), as well as the internodes separating Jaltomata, Solanum, and Capsicum. In particular, our concatenation-maximum-likelihood phylogeny supported a closer relationship between Jaltomata and Capsicum, but we also detected a similar number of individual gene trees supporting the alternative topology of Jaltomata and Solanum as more closely related (fig. 3). Genome-wide, the observed pattern of minority gene tree discordance was not consistent with the action of incomplete lineage sorting (ILS) alone (under ILS the two alternative minority trees are expected to be approximately equally represented) (Degnan and Rosenberg 2009), indicating that a substantial component of discordance was likely also due to introgression between lineages (Huson et al. 2005).
Following the logical framework in Fontaine et al. 2015, in which the younger (shallower) tree topology is inferred to be due to introgression, we used the relative depth (age) of the two alternative tree topologies to infer that Jaltomata is likely sister to Solanum (a relationship supported by the tree with the older/deeper mean node depths). In contrast, an excess of gene trees supporting a sister relationship between Jaltomata and Capsicum (the tree with on average shorter/younger node depths) is likely to be due to introgression since the split of the three species; this is despite the observation that the latter tree is marginally more frequent across all gene trees compared with the next most common bipartition (fig. 3). Our use of whole-genome sequence data from Jaltomata therefore enabled us to disentangle the likely complex history of origin at the base of the clade that unites these three lineages.
Our analyses also highlighted the extensive phylogenetic incongruence among these and other Solanaceae genera more generally, a history that should be accounted for in comparative studies. In particular, assessing the level and distribution of incongruence is critical when making inferences about trait evolution in radiating lineages, as substantial gene tree discordance can contribute to incorrect inferences of convergence (“hemiplasy”) at both the phenotypic and molecular level (Hahn and Nakhleh 2016; Wu et al. 2018).
TEs Contribute to Genome-Size Evolution across the Solanaceae
We also used the J. sinuosa genome assembly to examine possible causes of genome-size evolution, specifically variation in TE history. The estimated genome size of J. sinuosa (∼1.5 Gb) is >50% larger than the tomato genome (∼0.9 Gb), but less than half of the hot pepper genome (∼3.5 Gb), despite no accompanying change in ploidy level, suggesting that TEs might be an important contributor to genome-size variation in the Solanaceae. Indeed, we determined that nearly 80% of the assembly consists of repetitive elements, with the vast majority belonging to the Gypsy superfamily of LTR-RTs (fig. 2). We also inferred a recent proliferation of Gypsy elements that occurred around 1–2 Ma, which might have contributed to the larger genome size in Jaltomata lineages relative to Solanum. Previous studies have revealed a similar pattern of relatively recent Gypsy proliferation in other Solanaceae species, including a substantial excess of Gypsy elements (12-fold more frequent than Copia elements) within the hot pepper genome compared with domestic tomato (Kim et al. 2014). Similarly, genome-size variation scales with variation in the number of Gypsy repeats among four Nicotiana species (Xu et al. 2017). In contrast, within Solanum, the larger genome size of wild species S. pennellii compared with domesticated tomato is associated with a recent proliferation of Copia-like elements (Bolger et al. 2014). Overall, this and other recent analyses indicate that differential activity of LTR-RTs contributed substantially to differences in genome size across the Solanaceae and suggest that future studies examining these and broader TE activity and DNA loss (Kapusta et al. 2017) could reveal important dynamics of genome-size and content variation in this group.
Both Gene Family Evolution and Specific Molecular Changes Contribute to Unique Reproductive Trait Variation within Jaltomata
Unlike close relatives Solanum and Capsicum, species in Jaltomata exhibit extensive floral diversity in corolla shape and nectar volume and color (Miller et al. 2011) in addition to an apparently ancestral transition to self-compatibility (Mione 1992) (Kostyun and Mione, unpublished data). Another goal in developing a representative genome sequence for Jaltomata was therefore to identify genetic mechanisms that might have contributed to the evolutionary origin of these derived floral and reproductive states. In our previous phylogenomic study using transcriptome data, we investigated patterns of molecular evolution in several thousand loci across 14 species within Jaltomata and found that relatively few of the genes that showed a signal of positive selection (i.e., with dN/dS > 1) had functional associations with floral development. However, extensive gene tree discordance and very low sequence divergence (<1% among the species in the radiating group that displays the derived floral traits) only allowed a small subset of genes to be tested at some internal branches (Wu et al. 2018). Moreover, this transcriptome-based study only focused on molecular variation in coding sequences, although regulatory changes and structural variation, such as gene duplications, are also known contributors to the evolution of novel traits (Hoekstra and Coyne 2007). The generation of a reference genome in this group therefore allowed us to address the possible role of additional genetic variation that could not be investigated previously.
Interestingly, a major inference from our whole-genome analysis is that each of the seven Solanaceae lineages examined has experienced substantial expansion and contraction of gene families (fig. 4B), and several of these might have contributed to the distinctive reproductive trait evolution in Jaltomata. First, we detected significant contractions in gene families functionally associated with pollen–pistil interactions, including the putative S-locus receptor kinase family proteins, which could be a genomic signature of an ancestral transition to self-compatibility in this genus. Within the Solanaceae, self-incompatibility is mediated by the “S-locus” which encodes a single female/stylar S-determinant (i.e., S-RNase) and one or more pollen-expressed F-box proteins (Iwano and Takayama 2012) that interact to cause pollen rejection when the male and female parent share the same allele at the S-locus (McClure et al. 1989). Multiple studies have shown that loss of the pistil-side S-RNase mediates the transition from SI to SC in Solanaceous species, often followed by the subsequent loss of one or more pollen-side F-box proteins and other components of the SI-machinery (Charlesworth and Charlesworth 1979; Stone 2002; Takayama and Isogai 2005; Covey et al. 2010). Similarly in Brassicaceae, breakdown of SI appears to occur via loss of pistil-side factors, although the specific genetic mechanisms controlling SI are distinct between these two families (Kusaba et al. 2001; Sherman-Broyles et al. 2007; Tang et al. 2007; Shimizu et al. 2008). Several analyses in both these families have documented genome-wide effects of loss of SI (Hu et al. 2011; Slotte et al. 2013), however in most cases the associated transition to selfing has been recent. In contrast to these studies, all examined Jaltomata species are self-compatible (Mione 1992) (Kostyun and Mione, unpublished data), indicating that the loss of self-incompatibility occurred early within Jaltomata, prior to the origin of the major subgroups within this clade (i.e., >3 Ma). Consistent with an early loss of SI in this genus, we found that heterozygosity in each of 14 Jaltomata species (inferred by remapping RNA-seq reads from Wu et al. [2018] to the genome assembly) is comparable to that found in self-compatible species of Solanum, and much lower than in self-incompatible species of Solanum (Pease et al. 2016) (see supplementary table S8, Supplementary Material online). This early loss of SI might explain why we identified significant contractions of gene families related to pollen–pistil interactions specifically in Jaltomata. A similar pattern was observed in the genome of Caenorhabditis nigoni—a self-fertile nematode species that split from its outcrossing sibling species C. briggsae ∼3.5 Ma (Thomas et al. 2015)—in which several hundred genes mediating protein–protein interactions, including the genes important for sperm–egg interactions, appear to have been lost (Yin et al. 2018).
Unlike the contraction of gene families potentially associated with relaxed selection, we also identified expansion of gene families that might be associated with the evolution of novel floral traits. In particular, we detected recent tandem duplications of one interesting candidate gene SEUSS (fig. 5). SEUSS is a transcription factor, which in A. thaliana interacts with LEUNIG within a transcriptional corepressor complex to negatively regulate the expression of homeotic MADS-box gene AGAMOUS (Franks et al. 2002, 2006; Sridhar et al. 2006). Mutations in SEUSS cause ectopic and precocious expression of AGAMOUS mRNA, leading to partial homeotic transformation of floral organs in the outer two whorls (i.e., sepal and petal) (Franks et al. 2002). LEUNIG and SEUSS also have a more general role in lateral organ patterning and boundary formation in Arabidopsis, possibly through interactions with transcriptional factors in the YABBY and KNOX gene families (Stahle et al. 2009; Lee et al. 2014).
These functional roles are especially interesting with respect to the origin of floral diversity in Jaltomata, as the two derived corolla shapes (i.e., campanulate and tubular) are formed due to differential accelerated growth rates and variation in petal fusion (i.e., boundary formation) (Kostyun et al. 2017). Thus, the function of SEUSS is closely related to the mechanism generating the corolla shape diversity in Jaltomata. In general, duplication and divergence of floral identity genes appear to play an important role in the evolution of floral morphology in plants, such as the expansion of MIKCc-type MADS-box genes (Hileman et al. 2006; Panchy et al. 2016). Although we do not yet have expression data for individual tissues (such as individual floral organs), we found that SEUSS copies were expressed only in the reproductive tissues of some lineages, and that species differ in the expression of these SEUSS copies. Interestingly, the SEUSS genes showed more limited expression in the orange-fruited lineages (supplementary table S15, Supplementary Material online) with the derived floral traits (i.e., higher extent of corolla fusion), suggesting that downregulation of some or all copies of this negative regulator of floral development might have contributed to the evolution of novel floral (corolla) morphs in this group. Future work with tissue-specific expression and function across different Jaltomata species with distinct corolla shapes will allow us to further assess whether SEUSS variants are strongly implicated in floral trait divergence across Jaltomata.
Apart from gene family contractions and expansions, we also detected loci with patterns of adaptive molecular evolution that could be functionally implicated in reproductive trait changes. Although many of the 58 genes that are inferred to be under positive selection specifically within the Jaltomata lineage (supplementary table S16, Supplementary Material online) are involved in general plant processes, some are from functional classes associated with the unique reproductive trait variation observed within this clade. For example, several candidates are associated with pollen–pistil interactions, including an S-ribonuclease binding protein that mediates the degradation of self-pollen (Sims and Ordanic 2001; Kao and Tsukamoto 2004). Novel changes in these proteins might have followed the early loss of self-incompatibility in this genus, perhaps as a result of reduced constraint on previous functions and the adoption of new roles under the altered reproductive environment of self-compatibility. Other potentially interesting candidates include an auxin response factor, a WRKY transcription factor, and a NAC transcription factor, all of which function in multiple plant developmental processes (Johnson and Lenhard 2011). Although the plant NAC gene family is quite large and its members play key roles in numerous developmental and stress response pathways, this putative candidate is especially interesting as NAM (a NAC protein) has been shown to function in organ boundary specification, including in flowers of closely related Petunia (Souer et al. 1996; Zhong et al. 2016), and development of the novel floral forms in Jaltomata appears to be specifically associated with altered floral organ boundaries (Kostyun et al. 2017).
The Jaltomata Genome as a Tool for Future Comparative Analysis
Overall, the generation of a high-quality Jaltomata genome enabled us to evaluate genetic changes specific to this diverse lineage and across the Solanaceae more generally, as well as to clarify the historical evolutionary relationships among these important clades. In doing so, we inferred that Jaltomata has a complex history of divergence from its most closely related genera, which involves substantial introgression following initial lineage divergence. Based on quantification of recent TE activity, we infer that TE dynamics are likely an important contributor to genome content and genome-size differences among groups in the Solanaceae. Using comparisons of gene family expansion and contraction, and more conventional analyses of adaptive protein evolution, we identified several genetic changes that might have either facilitated or accompanied the unique reproductive trait evolution observed in this clade.
In the future, the interesting evolutionary features and central phylogenetic placement of Jaltomata within the Solanaceae offer a unique opportunity to examine patterns of coding, structural, and TE evolution across this diverse and speciose plant family, as well as to further evaluate mechanisms that could contribute to the rapid reproductive diversification observed specifically within Jaltomata. These future comparative genomic analysis could reveal additional large- and small-scale genetic changes responsible for genomic differentiation and divergent form and function among a set of economically important clades that are well-established models for developmental biology (Petunia), reproductive interactions (Nicotiana), secondary metabolite production (e.g., Capsicum), and ecophysiological responses (e.g., Solanum), as well as within this emerging model for analyzing the evolution of novel floral evolution.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
The authors thank David Haak for the flow cytometry estimate of J. sinuosa genome size, Shujun Ou for advice on transposable element analyses, Matthew Hahn for advice on analyses and inference, and two anonymous reviewers for helpful comments. This work was supported by the National Science Foundation (DEB 1136707 to L.C.M.).
Data deposition: The project has been deposited at NCBI BioProject database under the accession number PRJNA474403
Literature Cited
- Bauer S, Grossmann S, Vingron M, Robinson PN.. 2008. Ontologizer 2.0—a multifunctional tool for GO term enrichment analysis and data exploration. Bioinformatics 24(14):1650–1651. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y.. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 57(1): 289–300. [Google Scholar]
- Berner D, Salzburger W.. 2015. The genomics of organismal diversification illuminated by adaptive radiations. Trends Genet. 31(9):491–499. [DOI] [PubMed] [Google Scholar]
- Bleidorn C. 2016. Third generation sequencing: technology and its potential impact on evolutionary biodiversity research. Syst Biodivers. 14(1):1–8. [Google Scholar]
- Bohs L, Olmstead RG.. 1997. Phylogenetic relationships in Solanum (Solanaceae) based on ndhF sequences. Syst Bot. 22(1):5–17. [Google Scholar]
- Bolger A, et al. 2014. The genome of the stress-tolerant wild tomato species Solanum pennellii. Nat Genet. 46(9):1034–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawand D, et al. 2014. The genomic substrate for adaptive radiation in African cichlid fish. Nature 513(7518):375–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MS, et al. 2014. MAKER-P: a tool kit for the rapid creation, management, and quality control of plant genome annotations. Plant Physiol. 164(2):513–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B.. 1979. The evolution and breakdown of S-allele systems. Heredity 43(1):41. [Google Scholar]
- Covey PA, et al. 2010. Multiple features that distinguish unilateral incongruity and self‐incompatibility in the tomato clade. Plant J. 64(3):367–378. [DOI] [PubMed] [Google Scholar]
- Degnan JH, Rosenberg NA.. 2009. Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol Evol. 24(6):332–340. [DOI] [PubMed] [Google Scholar]
- Ellegren H. 2014. Genome sequencing and population genomics in non-model organisms. Trends Ecol Evol. 29(1):51–63. [DOI] [PubMed] [Google Scholar]
- Ellinghaus D, Kurtz S, Willhoeft U.. 2008. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics 9:18.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine MC, et al. 2015. Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science 347(6217):1258524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks RG, Liu Z, Fischer RL.. 2006. SEUSS and LEUNIG regulate cell proliferation, vascular development and organ polarity in Arabidopsis petals. Planta 224(4):801–811. [DOI] [PubMed] [Google Scholar]
- Franks RG, Wang C, Levin JZ, Liu Z.. 2002. SEUSS, a member of a novel family of plant regulatory proteins, represses floral homeotic gene expression with LEUNIG. Development 129(1):253–263. [DOI] [PubMed] [Google Scholar]
- Goldberg EE, et al. 2010. Species selection maintains self-incompatibility. Science 330(6003):493–495. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, et al. 2010. Genome-wide synteny through highly sensitive sequence alignment: satsuma. Bioinformatics 26(9):1145–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haak DC, Kostyun JL, Moyle LC.. 2014. Merging Ecology and Genomics to Dissect Diversity in Wild Tomatoes and Their Relatives In: Landry C., Aubin-Horth N. (eds) Ecological Genomics. Advances in Experimental Medicine and Biology, vol 781 Dordrecht: Springer. [DOI] [PubMed] [Google Scholar]
- Hahn MW, Nakhleh L.. 2016. Irrational exuberance for resolved species trees. Evolution 70(1):7–17. [DOI] [PubMed] [Google Scholar]
- Han MV, Thomas GW, Lugo-Martinez J, Hahn MW.. 2013. Estimating gene gain and loss rates in the presence of error in genome assembly and annotation using CAFE 3. Mol Biol Evol. 30(8):1987–1997. [DOI] [PubMed] [Google Scholar]
- Han Y, Wessler SR.. 2010. MITE-Hunter: a program for discovering miniature inverted-repeat transposable elements from genomic sequences. Nucleic Acids Res. 38:e199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen DM, Olesen JM, Mione T, Johnson SD, Müller CB.. 2007. Coloured nectar: distribution, ecology, and evolution of an enigmatic floral trait. Biol Rev. 82(1):83–111. [DOI] [PubMed] [Google Scholar]
- Hileman LC, et al. 2006. Molecular and phylogenetic analyses of the MADS-box gene family in tomato. Mol Biol Evol. 23(11):2245–2258. [DOI] [PubMed] [Google Scholar]
- Hoekstra HE, Coyne JA.. 2007. The locus of evolution: evo devo and the genetics of adaptation. Evolution 61(5):995–1016. [DOI] [PubMed] [Google Scholar]
- Hu TT, et al. 2011. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet. 43(5):476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F.. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17(8):754–755. [DOI] [PubMed] [Google Scholar]
- Huson, DH, Klöpper T, Lockhart PJ, Steel MA. 2005. Reconstruction of reticulate networks from gene trees. Annual International Conference on Research in Computational Molecular Biology. Berlin, Heidelberg: Springer. pp. 233–249. [Google Scholar]
- Iwano M, Takayama S.. 2012. Self/non-self discrimination in angiosperm self-incompatibility. Curr Opin Plant Biol. 15(1):78–83. [DOI] [PubMed] [Google Scholar]
- Jarvis ED, et al. 2014. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 346(6215):1320–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K, Lenhard M.. 2011. Genetic control of plant organ growth. New Phytol. 191(2):319–333. [DOI] [PubMed] [Google Scholar]
- Kao T-H, Tsukamoto T.. 2004. The molecular and genetic bases of S-RNase-based self-incompatibility. Plant Cell 16(Suppl 1):S72–S83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapusta A, Suh A, Feschotte C.. 2017. Dynamics of genome size evolution in birds and mammals. Proc Natl Acad Sci U S A. 114:E1460–E1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, et al. 2014. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat Genet. 46(3):270–278. [DOI] [PubMed] [Google Scholar]
- Knapp S, Bohs L, Nee M, Spooner DM.. 2004. Solanaceae—a model for linking genomics with biodiversity. Comp Funct Genomics. 5(3):285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S, et al. 2012. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat Biotechnol. 30(7):693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostyun JL, Moyle LC.. 2017. Multiple strong postmating and intrinsic postzygotic reproductive barriers isolate florally diverse species of Jaltomata (Solanaceae). Evolution 71:1556–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostyun JL, Preston JC, Moyle LC.. 2017. Heterochronic developmental shifts underlie floral diversity within Jaltomata (Solanaceae). Evodevo 8:17.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaba M, et al. 2001. Self-incompatibility in the genus Arabidopsis: characterization of the S locus in the outcrossing A. lyrata and its autogamous relative A. thaliana. Plant Cell 13(3):627–643. [PMC free article] [PubMed] [Google Scholar]
- Lamichhaney S, et al. 2015. Evolution of Darwin’s finches and their beaks revealed by genome sequencing. Nature 518(7539):371–375. [DOI] [PubMed] [Google Scholar]
- Larget BR, Kotha SK, Dewey CN, Ané C.. 2010. BUCKy: gene tree/species tree reconciliation with Bayesian concordance analysis. Bioinformatics 26(22):2910–2911. [DOI] [PubMed] [Google Scholar]
- Lee JE, Lampugnani ER, Bacic A, Golz JF.. 2014. SEUSS and SEUSS‐LIKE 2 coordinate auxin distribution and KNOXI activity during embryogenesis. Plant J. 80(1):122–135. [DOI] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS.. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13(9):2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löytynoja A, Goldman N.. 2005. An algorithm for progressive multiple alignment of sequences with insertions. Proc Natl Acad Sci U S A. 102(30):10557–10562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Bennetzen JL.. 2004. Rapid recent growth and divergence of rice nuclear genomes. Proc Natl Acad Sci U S A. 101(34):12404–12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure BA, et al. 1989. Style self-incompatibility gene products of Nicotlana alata are ribonucleases. Nature 342(6252):955. [DOI] [PubMed] [Google Scholar]
- Miller RJ, Mione T, Phan H-L, Olmstead RG.. 2011. Color by numbers: nuclear gene phylogeny of Jaltomata (Solanaceae), sister genus to Solanum, supports three clades differing in fruit color. Syst Bot. 36(1):153–162. [Google Scholar]
- Mione T. 1992. Systematics and evolution of Jaltomata (Solanaceae) [PhD dissertation]. [ Storrs (CT: ): University of Connecticut. [Google Scholar]
- Mione T, Yacher L.. 2005. Jaltomata (Solanaceae) of Costa Rica. A Festschrift for William G. D’Arcy: the legacy of a taxonomist. (Monographs in systematic botany from the Missouri Botanical Garden 104). St. Louis (MO: ): Missouri Botanical Garden; P. 117–130. [Google Scholar]
- Mione T, et al. 2017. Pollination decreases longevity of the protogynous flowers of Jaltomata sinuosa (Solanaceae). J Biol Nat. 7(3): 123–129. [Google Scholar]
- Mirarab S, Warnow T.. 2015. ASTRAL-II: coalescent-based species tree estimation with many hundreds of taxa and thousands of genes. Bioinformatics 31(12):i44–i52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikova PY, et al. 2016. Sequencing of the genus Arabidopsis identifies a complex history of nonbifurcating speciation and abundant trans-specific polymorphism. Nat Genet. 48(9):1077–1082. [DOI] [PubMed] [Google Scholar]
- Olmstead RG, Sweere JA, Spangler RE, Bohs L, Palmer JD.. 1999. Phylogeny and provisional classification of the Solanaceae based on chloroplast DNA. Solanaceae IV 1:1–137. [Google Scholar]
- Olmstead RG, et al. 2008. A molecular phylogeny of the Solanaceae. Taxon 57:1159–1181. [Google Scholar]
- Ou S, Jiang N.. 2018. LTR_retriever: a highly accurate and sensitive program for identification of LTR retrotransposons. Plant Physiol. 176(2):1410–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchy N, Lehti-Shiu M, Shiu S-H.. 2016. Evolution of gene duplication in plants. Plant Physiol. 171(4):2294–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pease JB, Haak DC, Hahn MW, Moyle LC.. 2016. Phylogenomics reveals three sources of adaptive variation during a rapid radiation. PLoS Biol. 14(2):e1002379.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C, et al. 2014. Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc Natl Acad Sci U S A. 111(14):5135–5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salichos L, Rokas A.. 2013. Inferring ancient divergences requires genes with strong phylogenetic signals. Nature 497(7449):327–331. [DOI] [PubMed] [Google Scholar]
- Särkinen T, Bohs L, Olmstead RG, Knapp S.. 2013. A phylogenetic framework for evolutionary study of the nightshades (Solanaceae): a dated 1000-tip tree. BMC Evol Biol. 13:214.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt MH-W, et al. 2017. De novo assembly of a new Solanum pennellii accession using nanopore sequencing. Plant Cell 29(10):2336–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman-Broyles S, et al. 2007. S locus genes and the evolution of self-fertility in Arabidopsis thaliana. Plant Cell 19(1):94–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu KK, Shimizu‐Inatsugi R, Tsuchimatsu T, Purugganan MD.. 2008. Independent origins of self‐compatibility in Arabidopsis thaliana. Mol Ecol. 17(2):704–714. [DOI] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics: 31:3210–3212. [DOI] [PubMed] [Google Scholar]
- Sims TL, Ordanic M.. 2001. Identification of a S-ribonuclease-binding protein in Petunia hybrida. Plant Mol Biol. 47(6):771–783. [DOI] [PubMed] [Google Scholar]
- Slotte T, et al. 2013. The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nat Genet. 45(7):831. [DOI] [PubMed] [Google Scholar]
- Souer E, van Houwelingen A, Kloos D, Mol J, Koes R.. 1996. The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85(2):159–170. [DOI] [PubMed] [Google Scholar]
- Sridhar VV, Surendrarao A, Liu Z.. 2006. APETALA1 and SEPALLATA3 interact with SEUSS to mediate transcription repression during flower development. Development 133(16):3159–3166. [DOI] [PubMed] [Google Scholar]
- Stahle MI, Kuehlich J, Staron L, von Arnim AG, Golz JF.. 2009. YABBYs and the transcriptional corepressors LEUNIG and LEUNIG_HOMOLOG maintain leaf polarity and meristem activity in Arabidopsis. Plant Cell 21(10):3105–3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22(21):2688–2690. [DOI] [PubMed] [Google Scholar]
- Stapley J, et al. 2010. Adaptation genomics: the next generation. Trends Ecol Evol. 25(12):705–712. [DOI] [PubMed] [Google Scholar]
- Stone J. 2002. Molecular mechanisms underlying the breakdown of gametophytic self-incompatibility. Q Rev Biol. 77(1):17–32. [DOI] [PubMed] [Google Scholar]
- Suh A, Smeds L, Ellegren H.. 2015. The dynamics of incomplete lineage sorting across the ancient adaptive radiation of neoavian birds. PLoS Biol. 13(8):e1002224.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama S, Isogai A.. 2005. Self-incompatibility in plants. Annu Rev Plant Biol. 56:467–489. [DOI] [PubMed] [Google Scholar]
- Tang C, et al. 2007. The evolution of selfing in Arabidopsis thaliana. Science 317(5841):1070–1072. [DOI] [PubMed] [Google Scholar]
- Thomas CG, et al. 2015. Full-genome evolutionary histories of selfing, splitting, and selection in Caenorhabditis. Genome Res. 25(5):667–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato Genome Consortium. 2012. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485:635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkat A, Hahn MW, Thornton JW.. 2017. Multinucleotide mutations cause false inferences of lineage-specific positive selection. Nat Ecol Evol. 2:1280–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh BM, Hoot SB.. 2001. Phylogenetic relationships of Capsicum (Solanaceae) using DNA sequences from two noncoding regions: the chloroplast atpB-rbcL spacer region and nuclear waxy introns. Int J Plant Sci. 162(6):1409–1418. [Google Scholar]
- Wickett NJ, et al. 2014. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc Natl Acad Sci U S A. 111(45):E4859–E4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Kostyun JL, Hahn MW, Moyle L.. 2018. Dissecting the basis of novel trait evolution in a radiation with widespread phylogenetic discordance. Mol Ecol. 27(16):3301–3316. [DOI] [PubMed] [Google Scholar]
- Xu S, et al. 2017. Wild tobacco genomes reveal the evolution of nicotine biosynthesis. Proc Natl Acad Sci U S A. 114(23):6133–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang Y, et al. 2015. Dissecting molecular evolution in the highly diverse plant clade Caryophyllales using transcriptome sequencing. Mol Biol Evol. 32(8):2001–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C, Hill CM, Wu S, Ruan J, Ma ZS.. 2016. DBG2OLC: efficient assembly of large genomes using long erroneous reads of the third generation sequencing technologies. Sci Rep 6:31900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin D, et al. 2018. Rapid genome shrinkage in a self-fertile nematode reveals sperm competition proteins. Science 359(6371):55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, et al. 2014. Comparative genomics reveals insights into avian genome evolution and adaptation. Science 346(6215):1311–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, Powell S, Preston J.. 2016. Organ boundary NAC‐domain transcription factors are implicated in the evolution of petal fusion. Plant Biol. 18(6):893–902. [DOI] [PubMed] [Google Scholar]
- Zimin AV, et al. 2017. Hybrid assembly of the large and highly repetitive genome of Aegilops tauschii, a progenitor of bread wheat, with the mega-reads algorithm. Genome Res. 29:2669–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.