 A novel cascade assembly reaction is applied to sensitively distinguish between low abundance miRNAs and pre-miRNAs in living cells.
A novel cascade assembly reaction is applied to sensitively distinguish between low abundance miRNAs and pre-miRNAs in living cells.
Abstract
Mature microRNAs (miRNAs) produced from precursor microRNAs (pre-miRNAs) by the RNase Dicer have showed significant potential for cancer diagnosis and prognosis due to their key regulatory roles in various pathological processes. However, discriminatory detection of low-abundance miRNAs and pre-miRNAs remains a key challenge since the mature sequence is also present in the pre-miRNA forms. Herein, we report a novel cascade reaction to sensitively distinguish miRNAs versus pre-miRNAs in living cells based on two pairs of programmable hairpin oligonucleotide probes with a simple sequence design. The programmable hairpin probes can metastably coexist until the introduction of miRNAs or pre-miRNAs, which can trigger a specific hybridization chain reaction (HCR), respectively, leading to the self-assembly of nicked DNA duplex structures and a remarkable specific fluorescence intensity increase. The system can readily and sensitively assess the miRNA or pre-miRNA abundance in a homogeneous solution. The intracellular miRNA and pre-miRNA expression level assessment in different living cells is realized. Thus, we provide a novel investigation tool for discriminatorily and accurately assessing miRNA and pre-miRNA abundance, which could be useful for the biomedical application of miRNAs.
Introduction
Mature microRNAs (miRNAs) are a class of short single-stranded noncoding RNAs with lengths of 19–23 nucleotides.1–3 They play important regulatory roles in gene expression of animals, plants, and viruses.4 The biogenesis of miRNAs is a multistep cellular process that begins in the nucleus where RNA polymerase II catalyzes the production of primary miRNAs (pri-miRNAs), which are cleaved by the RNase III Drosha to produce shorter stem-loop RNA precursor miRNAs (pre-miRNAs).5,6 The pre-miRNAs are then transported into the cytoplasm and processed into miRNA duplexes by the RNase Dicer.7–9 One strand of the duplex, termed mature miRNA, can form a RNA-induced silencing complex (RISC) with an auxiliary protein to partly and complementarily recognize the 3′-untranslated region of mRNA to suppress the translation or induce mRNA degradation.10
MiRNAs play pivotal roles in an array of physiological processes.11 Importantly, the aberrant expression of miRNAs is associated with various pathological developments such as cancer and cardiovascular disease.12–15 Thus, miRNAs have received considerable attention as valuable biomarkers and novel therapeutic targets.16,17 Owing to the imperfect matching between miRNAs and their target mRNAs, cellular miRNA-mediated gene suppression appears as a highly orchestrated mode in which one miRNA may regulate hundreds of mRNAs and many miRNAs may regulate one mRNA.18 Meanwhile, miRNAs exist as several distinct isoforms called isomiRs with different seed sequences and lengths, and the concentration of isomiRs and pre-miRNAs changes dynamically during various physiological and pathological processes.19 Thus, characterization of miRNAs and their pre-miRNA expression levels can provide significant clues on their physiological functions.
Nowadays, the most widely used strategy for assessing pre-miRNA and miRNA abundance simultaneously is northern analysis that separates them based on size, but it suffers from limited sensitivity and radioactive agents.20 An altered quantitative real-time PCR (qRT-PCR) protocol with unique primers has the potential to distinguish miRNA and pre-miRNA. However, it is time-consuming and cost-ineffective.21,22 Molecular beacon (MB) approaches offer a significant advantage to sense specific miRNA or DNA, and enable us to distinguish the pre-miRNA and miRNA since the MB elicits a signal only in response to the complementary sequence.23 Owing to the fact that the fluorescence intensity is directly correlated with the RNA hybridization event, the “one-to-one” signal manner cannot satisfy the low-abundance miRNA or pre-miRNA analysis. Therefore, the development of assays with high sensitivity and good discrimination ability remains a key challenge.
The hybridization chain reaction (HCR) in which a target triggers the cross-opening of two DNA hairpins to assemble DNA polymeric nanowires.24–27 It preserves the inherent high specific properties of the molecular hairpin, and notably possesses significant signal amplification functions due to the cascading displacement reaction and assembled nicked polymeric duplex structures, leading to great potential for bioanalysis.28–30 Weizmann et al. reported a real-time intracellular miRNA imaging method based on the HCR with programmable oligonucleotide probes generating fluorescence resonance energy transfer upon miRNA-triggered assembly.31 Hyper-branched HCR and nonlinear HCR systems with high signal amplification efficiency have also been explored.32,33 However, the HCR system could not be directly adapted to distinguish the miRNA and pre-miRNA due to the presence of the mature miRNA sequence in its pre-miRNA forms and false positive signals for miRNA detection. The HCR system with rationally programmable hairpin probes for pre-miRNA analysis and discrimination of pre-miRNA and miRNA has never been explored until now.
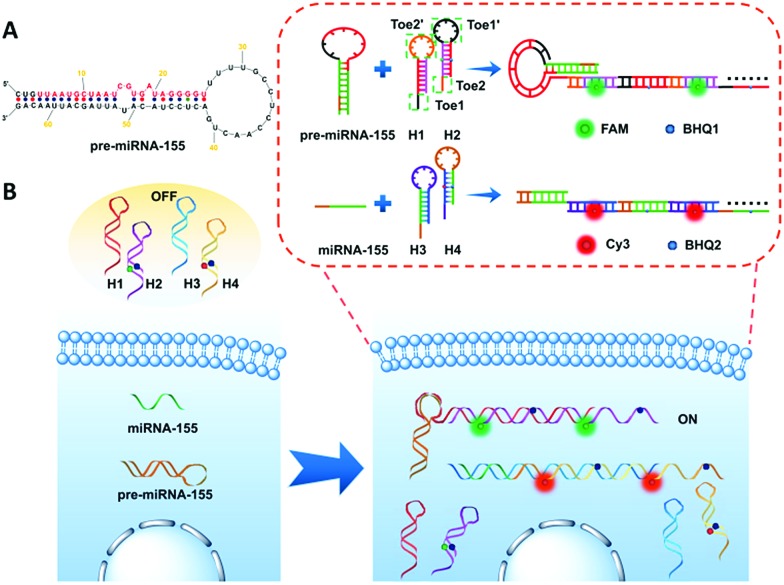
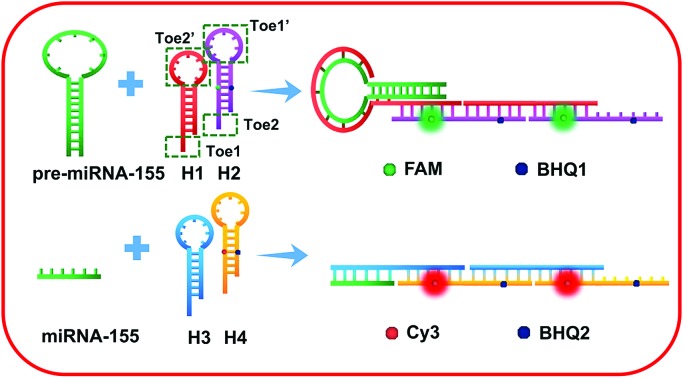
Herein, we design two pairs of programmable hairpin oligonucleotide probes that specifically self-assemble fluorescent nicked duplex structures responsive to miRNA or pre-miRNA to sensitively distinguish miRNA versus pre-miRNA. The red stem region of pre-miRNA-155 is the miRNA-155 sequence (Fig. 1A), and the large loop (∼15 base) of pre-miRNA-155 was designed as a trigger sequence of pre-miRNA-155 related to HCR detection, which avoided the false positive signal triggered by miRNA-155.34,35 As shown in Fig. 1B, the mutual reaction from the four hairpin probes was kinetically impeded due to the fact that the binding sites were sequestered in either small bulge loops or double-stranded forms. The trigger pre-miRNA-155 hybridized with H1 to expose an originally sequestered toehold (Toe2′) to H2, which induced the fluorescent reporter to be freed up from the quencher and expose Toe1′ to H1, leading to chain growth of assembled DNA and a strong increase in FAM fluorescence intensity. However, the miRNA-155 could only specifically trigger the assembly of H3 and H4 and produce a strong increase in Cy3 fluorescence intensity. Consequently, the specific fluorescence intensity increase can be employed to sensitively distinguish miRNA versus pre-miRNA abundance.
Fig. 1. Schematic illustration of (A) pre-miRNA-155; (B) imaging of pre-miRNA-155 and miRNA-155 in living cells based on the HCR.
Results and discussion
Feasibility of the pre-miRNA and miRNA triggered HCR
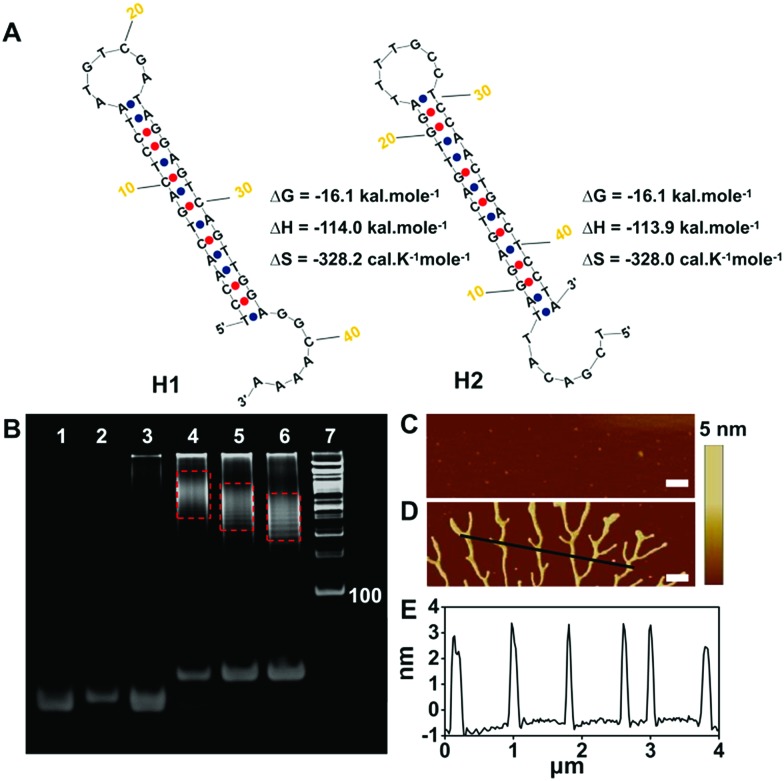
The in vitro feasibility of the pre-miRNA-155 and miRNA-155 triggered HCR was first characterized. The ideal structures and thermodynamic parameters of the H1, H2, H3 and H4 and relative duplex complexes were analyzed using an Oligo Analyzer 3.1 under the conditions of PBS buffer (pH 7.4, 10 mM) containing NaCl (137 mM) (Fig. 2A and S1A†). All the probes have lower enthalpy changes (ΔH), indicating that the probes designed are stable and have few other secondary structures. The lanes 1–3 in the native polyacrylamide gel electrophoresis (PAGE) image were H1, H2 and H1 plus H2, respectively, which indicated that the spontaneous reaction between H1 and H2 was negligible (Fig. 2B). On the contrary, the trigger pre-miRNA-155 could effectively initiate the hybridization of H1 and H2 to assemble nicked duplex structures (Fig. 2B, lanes 4–6). The average sizes of the HCR products were observed to change inversely to the trigger pre-miRNA-155 (lanes 4–6), in agreement with a previous report.36 Atomic force microscopy (AFM) analysis further confirmed the PAGE results (Fig. 2C and D). The hairpin probes H1 and H2 failed to spontaneously assemble and presented only tiny spots (Fig. 2C). In contrast, obvious assembled polymers appeared upon introduction of pre-miRNA-155 (Fig. 2D) with a thickness of 3.0 nm (Fig. 2E). Similar results were obtained for the miRNA-155 triggered HCR with H3 and H4 as monomers (Fig. S1†). Meanwhile, the stable coexistence of four programmable hairpin probes in a homogeneous solution was also realized (Fig. S2†). These results confirmed the good feasibility of the pre-miRNA and miRNA triggered HCR and its potential ability for distinguished sensitive detection.
Fig. 2. (A) Ideal structure and thermal kinetic parameters of H1, H2 and the HCR. (B) PAGE analysis of the HCR. Lanes 1–7 represent H1 (2 μM), H2 (2 μM), H1 + H2 (2 μM and 2 μM), H1 + H2 + pre-miRNA-155 (2 μM, 2 μM and 2 μM), H1 + H2 + 2 × pre-miRNA-155 (2 μM, 2 μM and 4 μM), H1 + H2 + 3 × pre-miRNA-155 (2 μM, 2 μM and 6 μM) and the DNA ladder marker, respectively. AFM phase images of the HCR (C) without and (D) with pre-miRNA-155. Scale bar: 500 nm. (E) Cross-sectional profile of the black line in (D).
Discrimination ability for pre-miRNA and miRNA analysis
The concentration ratio between H1 and H2 as well as H3 and H4 was first investigated in vitro to optimize the HCR performance. And the most appropriate concentration ratio between H1 (H3) and H2 (H4) was 1 : 1 (Fig. S3†). The catalytic amplification efficacy of our system was compared to a conventional molecular beacon (MB) without amplification (Fig. S4†), where H5 and H6 were the MBs recognizing pre-miRNA-155 and miRNA-155, respectively.
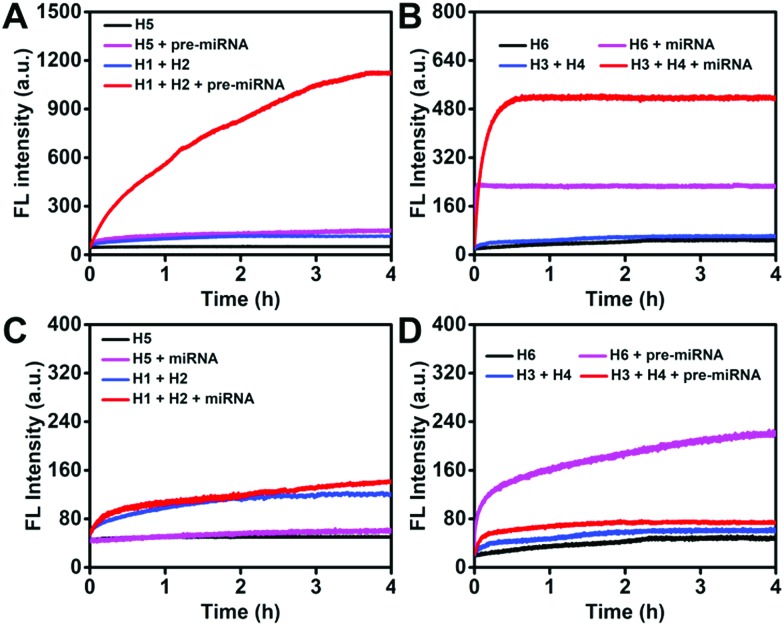
As indicated by time-dependent fluorescence analysis shown in Fig. 3A, both H5 and the mixture containing H1 and H2 exhibited a low fluorescence background signal. The addition of pre-miRNA-155 (20 nM) to the mixture containing H1 (100 nM) and H2 (100 nM) yielded a significant fluorescence increase (red curve), while a slight fluorescence intensity increase was observed when pre-miRNA-155 (20 nM) was added to the H5 aqueous solution (100 nM) (purple curve). The F/F0 value of the HCR system was 9.92 at 4 h, which was 3.29-fold higher than that of the MB strategy with a F/F0 value of 3.01. Both the HCR and MB systems for miRNA analysis exhibited a sharp and fast increase in fluorescence intensity after the addition of the miRNA (Fig. 3B). However, the plateau F/F0 value was 8.60 for the HCR which is 1.83-fold higher than that of MB detection. These results suggested good amplification efficacy of the HCR system for pre-miRNA and miRNA detection, especially for the pre-miRNA analysis.
Fig. 3. Monitoring the fluorescence increase of the HCR (each hairpin probe: 100 nM) and MB (100 nM) detection systems as a function of time in response to (A) pre-miRNA-155 (20 nM) and (B) miRNA-155 (20 nM). Monitoring the fluorescence increase of the HCR (each hairpin probe: 100 nM) and MB (100 nM) detection systems (C) miRNA-155 (250 nM) and pre-miRNA-155 (250 nM). The fluorescence intensities were normalized.
The capability of distinguishing miRNA and pre-miRNA of the HCR and MB system was then systematically investigated. As shown in Fig. 3C, both of the HCR and MB strategy for pre-miRNA-155 detection displayed a negligible increase (F/F0 value of 1.17 for the HCR and 1.18 for the MB at 4 h) in fluorescence intensity after the addition of miRNA-155, indicating the good discrimination ability of these systems for pre-miRNA detection. As for the miRNA analysis, a similar slight fluorescence increase was observed after the addition of pre-miRNA-155 into the HCR detection system (Fig. 3D, red curve). On the contrary, the pre-miRNA-155 induced a significant fluorescence increase (Fig. 3D, purple curve) (F/F0 value of 1.25 for the HCR and 4.59 for the MB at 4 h) for the MB-based miRNA detection system due to a similar sequence present in its pre-miRNA form. This demonstrated the good discrimination ability of the HCR system for miRNA and pre-miRNA detection as a result of the inherent specific ability of hairpin probes and the rational sequence design that avoided the influence of a similar sequence present in both miRNA and pre-miRNA on the HCR processes.
The HCR system in response to pre-miRNA-155 and miRNA-155 with different concentrations in vitro was also explored by measuring the FAM and Cy3 fluorescence intensity after 4 h incubation as shown in Fig. S5.† The fluorescence intensity increased with the concentration increase of pre-miRNA-155 (Fig. S5A†) and miRNA-155 (Fig. S5B†). It showed a good linear relationship between F/F0 and the logarithm of pre-miRNA-155 (Fig. S5A,† inset)/miRNA-155(Fig. S5B,† inset) concentration ranging from 1 nM to 100 nM and 1 nM to 100 nM, respectively. The limit of detection (LOD) was calculated to be 820 pM/680 pM by using three times the standard deviation of the control, while the LOD of MB-based detection was 19.75 nM and 8.16 nM, indicating the superior sensitivity of the proposed system (Fig. S5C and D†).
Furthermore, the base-mismatch discrimination ability of the proposed strategy was investigated. The single-base-mismatched strand (mis-1), three-base-mismatched strand (mis-3) and five-base-mismatched strand (mis-5) presented a slight fluorescence intensity increase compared to the control (Fig. S6†). In contrast, the complementary pre-miRNA-155 target showed a much higher fluorescence intensity increase in comparison with other groups (Fig. S6A†). Similar results were observed for the miRNA-155 detection (Fig. S6B†). The F/F0 value of CM pre-miRNA-155 was about 3.93-fold and 7.00-fold higher than those of mis-1 and mis-3 strands (Fig. S6C†), respectively. For the miRNA analysis, the CM miRNA-155 presented a 6.07-fold and 9.18-fold higher F/F0 value than mis-1 and mis-3 strands, respectively (Fig. S6D†), which revealed the good base discrimination ability and potential of the proposed HCR for isomiR detection.
Discrimination and detection of pre-miRNA and miRNA in living cells
The standard colorimetric MTT assay was performed on MCF-7 cells treated with HCR probes at different times. As shown in Fig. S7,† the HCR probes exhibited low cytotoxicity toward MCF-7 cells, and the cell viability was maintained above 85% even at 10 h. These results suggested that the HCR probes could potentially be used for live cell real-time pre-miRNA and miRNA detection. The incubation time for pre-miRNA/miRNA intracellular detection was investigated. H1, H2, H3 and H4 were transfected into A549 via lipofectamine 2000, and the real-time monitoring of intracellular fluorescence related to pre-miRNA and miRNA detection was carried out to optimize the incubation time. Both of FAM and Cy3 fluorescence associated with pre-miRNA and miRNA gradually increased along with the increase of the incubation time and reached saturation at 4 h, and then decreased due to the degradation of the hairpin oligonucleotide probes in the intracellular environment (Fig. S8A†). A more visual result of the mean fluorescence intensities could be observed in Fig. S8B and C†. They revealed that strong FAM (Fig. S8B†) and Cy3 (Fig. S8C†) fluorescence intensities were obtained after 4 h incubation; thus, 4 h was used for intracellular imaging.
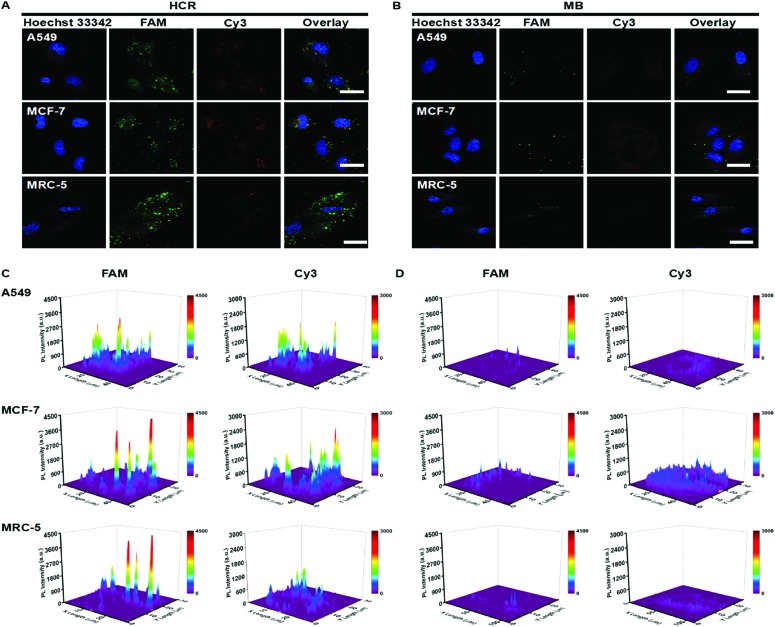
Two cancer cell lines (A549 and MCF-7) and a human embryonic lung fibroblast (MRC-5) line were employed to evaluate the intracellular pre-miRNA/miRNA detection of the HCR system. As a control, MB-based detection was also performed under the same conditions. All three cell lines showed a strong green fluorescence signal related to pre-miRNA-155, while strong red fluorescence derived from miRNA-155 recognition was only observed in the A549 and MCF-7 tumor cells and weakly appeared in the MRC-5 cell (Fig. 4A and C). This suggested that although all the cell lines possessed a similar abundance of pre-miRNA-155, the abundance of miRNA was different. Aberrant high expression levels of miRNA-155 were observed for both A549 and MCF-7 cancer cells, which could act as a potential biomarker. All the fluorescence intensities from the MB system were much weaker compared to the counterparts detected using HCR strategies, which further indicated the higher sensitivity of the HCR system (Fig. 4B and D). These results suggested that the HCR strategy could efficiently distinguish the pre-miRNA-155 and miRNA-155 abundance for all three cells, while the MB system could not satisfy the discrimination of intracellular miRNA-155 and pre-miRNA-155.
Fig. 4. Confocal laser scanning microscopy (CLSM) images of A549 cells, MCF-7 and MRC-5 cells incubated with the (A) HCR (100 nM) and (B) MB (100 nM). (C and D) Corresponding fluorescence intensities of these cells. The scale bar was 40 μm.
The abundance change of oncogenic miRNA-155 in MCF-7 cells was further investigated using miRNA-155 mimics and inhibitors as regulators. As shown in Fig. S9A and B,† the FAM fluorescence intensity related to pre-miRNA-155 showed a negligible change for the MCF-7 cells treated with miRNA-155 inhibitors or mimics compared to the control. However, the Cy3 fluorescence intensity associated with miRNA-155 showed a significant increase and decrease after treatment with miRNA-155 mimics and miRNA-155 inhibitors, respectively (Fig. S9A and C†). These results suggested the good feasibility of the HCR system to monitor the expression change of intracellular miRNA, even for low-abundance nucleic acids. To further validate its feasibility, the HCR system was also employed for analyzing miRNA let-7a and its pre-miRNA detection. It demonstrated good feasibility for sensitively discriminating miRNA let-7a from pre-miRNA let-7a in vitro (Fig. S10†). Furthermore, the CLSM imaging analysis showed that the strong green fluorescence signal related to pre-miRNA let-7a was observed in both MCF-7 and NHDF cells, while the strong red fluorescence signal associated with miRNA let-7a only was observed in NHDF cells (Fig S11A†). This suggested that the miRNA let-7a was at a high expression level in normal cells and at a low expression level in cancer cells (Fig. S11B†), which was different from the expression pattern of miRNA-155. These results suggested that the HCR system was readily extended to other miRNA detections with different expression levels.
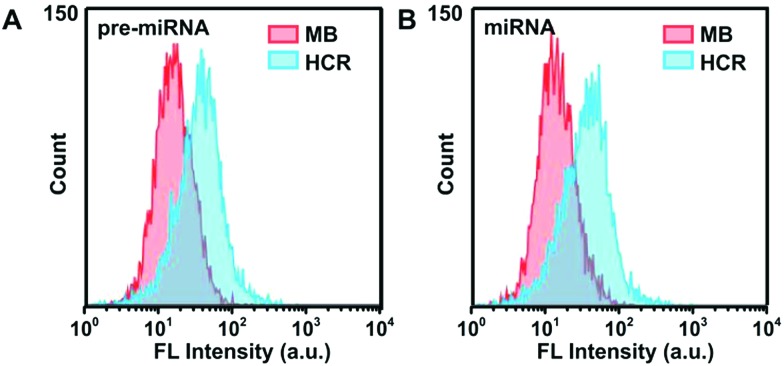
Furthermore, flow cytometry analysis was employed to characterize the FAM and Cy3 fluorescence intensity in A549 cells (Fig. 5), which was consistent with the results of CLSM measurements. The HCR strategy presented much stronger fluorescence signals than the MB-based system, indicating the high sensitivity of the HCR system. The concentrations of pre-miRNA-155 and miRNA-155 in these different cell lysate samples were validated using the qRT-PCR with unique primers, which demonstrated a similar pre-miRNA-155 abundance among all three cells, and a lower abundance of miRNA in MRC-5 than in A549 and MCF-7 cells (Fig S12†), in agreement with the HCR measurements. These results further confirmed the high sensitivity and good discrimination ability of the proposed HCR system for pre-miRNA/miRNA detection.
Fig. 5. Flow cytometric assay of A549 cells incubated with HCR probes or MB probes in response to (A) pre-miRNA-155 and (B) miRNA-155, respectively.
Conclusions
In summary, we have engineered a programmable hairpin oligonucleotide probe based HCR to sensitively distinguish low-abundant miRNA versus pre-miRNAs in living cells. The HCR strategy inherited the inherent specificity of hairpin probes and possessed significant signal amplification functions due to the cascade assembly reaction. It avoided complicated cell lysis and RNA isolation processes involved in northern analysis and qRT-PCR, and realized analysis of miRNA and its pre-miRNA in situ with cell heterogeneity. It exhibited superior sensitivity and specificity to conventional MB for synthetic pre-miRNA and miRNA discrimination. Notably, the miRNA-155 and pre-miRNA-155 expression levels among different cells difficultly assessed by the MB system were efficiently and readily detected by the proposed HCR strategy, which was validated by qRT-PCR. To the best of our knowledge, this is the first time that the HCR system has been explored with a simple sequence design for intracellular pre-miRNA detection and pre-miRNA/miRNA abundance discrimination assessment, which hold great promise for the biomedical application of miRNAs.
Experimental procedures
Materials and reagents
All the DNA oligonucleotides were synthesized by Sangon Biological Engineering Technology & Co., Ltd (Shanghai, China) and purified using high-performance liquid chromatography. All the miRNA sequences and primers were synthesized by Gene-Pharma Co., Ltd. (Shanghai, China). These sequences are listed in Table S1.† The miRcute miRNA Isolation Kit, Fastking gDNA Dispelling RT SuperMix, Talent qPCR PreMix (SYBR Green), miRcute Plus miRNA First-Strand cDNA Synthesis Kit and miRcute Plus miRNA qPCR Detection Kit (SYBR Green) were purchased from Tiangen (China). Phosphate buffer saline (PBS, pH 7.4) was purchased from Life Technologies Corporation (Los Angeles, CA, USA). Lipofectamine 2000 was purchased from Invitrogen (MA, USA). All other reagents were of analytical grade. Ultrapure water (≥18 MΩ) was used in all the experiments and it was obtained from a Millipore water purification system.
Characterization
The morphologies of assembled DNA duplexes were examined with AFM (Nanoscope IIIa, USA). The gel electrophoresis was imaged on an Alliance Ld2 (Uvitec, Cambridge, U.K.). Fluorescence measurements were carried out on a Hitachi F-4500 fluorescence spectrofluorometer (Tokyo, Japan). Confocal fluorescence imaging measurements were performed on a confocal laser scanning fluorescence microscope (CLSM, FV1200, Olympus, Japan). Flow cytometric analysis was performed on a BD FACSAria (Becton, Dickinson and Company, USA). The qRT-PCR was performed on a CFX96 touch RT-PCR detection system (Bio-Rad, USA).
Native polyacrylamide gel electrophoresis
Oligonucleotide assembled products were analyzed by 8% PAGE. To prepare the hydrogel, 2.7 mL 30% acrylamide/bis-acrylamide gel solution (29 : 1), 2 mL 5× TAE buffer (pH 8.3), 90 μL 10% ammonium persulfate (APS), 10 μL N,N,N′,N′-tetramethylethylenediamine (TEMED) and 6.2 mL deionized water were mixed. The gel was polymerized for 5 h at room temperature. Then, 5 μL of each sample was mixed with 1 μL of 6× loading buffer and added to the native polyacrylamide gel. The PAGE was performed in baths of ice at 100 V for about 90 min. After staining in 2× Gel-Red™ Nucleic Acid Gel Stain solution (Biotium, USA) for 30 min, the gel was imaged using an Alliance Ld2 (Uvitec, Cambridge, U.K.).
AFM characterization
First, 10 μL sample solution was dropped onto a mica surface and washed off with 30 μL of 2 mM Mg2+ solution after 10 min. Then the sample was dried with nitrogen and was imaged on an atomic force microscope (Nanoscope IIIa, USA).
Fluorescence assays in a homogeneous solution
All the hairpin probes were preprocessed to ensure the formation of a stable hairpin structure. For the detection of pre-miRNA-155, a mixture containing 100 nM H1 and 100 nM H2 was prepared, and different concentrations of pre-miRNA-155 were then added to the solution at 37 °C for 4 h. The fluorescence intensities were measured on a Hitachi F-4500 fluorescence spectrometer (Tokyo, Japan). MiRNA-155 was detected using a mixture of 100 nM H3 and 100 nM H4 with a similar procedure. For MB based pre-miRNA-155 and miRNA-155 analysis, different concentrations of pre-miRNA or miRNA-155 solution were added to the corresponding MB (100 nM) solution and reacted for a desired time for fluorescence detection.
Cytotoxicity
The cytotoxicity of the HCR probes was assessed using a standard MTT assay. First, MCF-7 cells were cultured in 96-well plates at 37 °C at a density of 104 cells per well for 24 h. Then H1, H2, H3 and H4 (each hairpin probe: 100 nM) were transfected with lipofectamine 2000 and incubated for 2 h, 4 h, 6 h, 8 h and 10 h. Subsequently, the cells were washed with PBS buffer (pH 7.4, 10 mM) twice, and 100 μL of 0.5 mg mL–1 MTT was added into each well and incubated at 37 °C for 4 h. Finally, 100 μL DMSO was added, and the absorbance was recorded at 492 nm using an Anthos 2010 microplate reader (manufactured by Biochrom Ltd., Cambridge CB4 OFJ, England).
CLSM imaging and flow cytometric assay
A549 cells, MCF-7 cells and MRC-5 cells were cultured according to our previous report.37 Transfection assays were performed according to the manufacturer's protocol. Briefly, transfection was carried out using 12 μL lipofectamine 2000 in 488 μL Opti-MEM at room temperature for 5 min. Then, 100 nM H1, 100 nM H2, 100 nM H3 and 100 nM H4 were mixed in 496 μL Opti-MEM. Afterwards, the two solutions were mixed at room temperature for another 20 min and added to the cell culture dish to cultivate for 4 h. The resulting cells were washed twice using PBS solution (10 mM, pH7.4) and fresh DMEM medium (1 mL) was added. The cells were imaged using a CLSM system (FV1200, Olympus, Japan). The flow cytometric assays were performed in PBS (10 mM, pH 7.4) with BD Calibur (Becton, Dickinson and Company, USA).
qRT-PCR analysis
Three cell lines (A549, MCF-7 and MRC-5) were used for the extraction of total RNAs. The total RNA extracted procedure from cell lysates was operated according to our previous report.38 The extracted RNA was re-dispersed in 30 mL of diethylpyrocarbonate (DEPC)-treated water. For pre-miRNA-155 analysis, the cDNA was prepared using Fastking gDNA Dispelling RT SuperMix, which was detected with Talent qPCR PreMix (SYBR Green). The miRcute Plus miRNA First-Strand cDNA Synthesis Kit (Tiangen, China) and miRcute Plus miRNA qPCR Detection Kit (SYBR Green) were used for miRNA-155 analysis.
Conflicts of interest
The authors declare no competing financial interest.
Supplementary Material
Acknowledgments
The work was supported by National Natural Science Foundation of China (21874008, 21475008), Special Foundation for State Major Research Program of China (Grant No. 2016YFC0106602 and 2016YFC0106601); the Open Research Fund Program of Beijing Key Lab of Plant Resource Research and Development, Beijing Technology and Business University (PRRD-2016-YB2); the Fundamental Research Funds for the Central Universities (Grant No. FRF-BD-17-016A) and Beijing Municipal Science and Technology Commission (Grant No. z131102002813058).
Footnotes
†Electronic supplementary information (ESI) available. See DOI: 10.1039/c8sc03305f
References
- Dong H., Lei J., Ding L., Wen Y., Ju H., Zhang X. Chem. Rev. 2013;113:6207–6233. doi: 10.1021/cr300362f. [DOI] [PubMed] [Google Scholar]
- Wark A. W., Lee H. J., Corn R. M. Angew. Chem., Int. Ed. 2008;47:644–652. doi: 10.1002/anie.200702450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W., Fu P., Sun M., Xu L., Kuang H., Xu C. J. Am. Chem. Soc. 2017;139:11752–11759. doi: 10.1021/jacs.7b03617. [DOI] [PubMed] [Google Scholar]
- Barte D. P. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Radmark O., Kim S., Kim V. N. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Ha M., Kim V. N. Nat. Rev. Mol. Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- Kim Y. K., Kim B., Kim V. N. PNAS. 2016;113:E1881–E1889. doi: 10.1073/pnas.1602532113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Kandasamy S. K., Fukunaga R. Nucleic Acids Res. 2018;46:3726–3741. doi: 10.1093/nar/gky043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starega-Roslan J., Galka-Marciniak P., Krzyzosiak W. J. Nucleic Acids Res. 2015;43:10939–10951. doi: 10.1093/nar/gkv968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J., Loedige I., Filipowicz W. Nat. Rev. Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- He L., Hannon G. J. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Kasinski A. L., Slack F. J. Nat. Rev. Cancer. 2011;11:849–864. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Getz G., Miska E. A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B. L., Mak R. H., Ferrando A. A., Downing J. R., Jacks T., Horvitz H. R., Golub T. R. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Lujambio A., Lowe S. W. Nature. 2012;482:347–355. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M., Toli J., Abdellatif M. Curr. Opin. Cardiol. 2011;26:181–189. doi: 10.1097/HCO.0b013e328345983d. [DOI] [PubMed] [Google Scholar]
- Cheng C. J., Bahal R., Babar I. A., Pincus Z., Barrera F., Liu C., Svoronos A., Braddock D. T., Glazer P. M., Engelman D. M., Saltzman W. M., Slack F. J. Nature. 2015;518:107–110. doi: 10.1038/nature13905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Geissler D., Qiu X., Wegner K. D., Hildebrandt N. Angew. Chem., Int. Ed. 2015;54:10024–10029. doi: 10.1002/anie.201504887. [DOI] [PubMed] [Google Scholar]
- Jonas S., Izaurralde E. Nat. Rev. Genet. 2015;16:421–433. doi: 10.1038/nrg3965. [DOI] [PubMed] [Google Scholar]
- Telonis A. G., Loher P., Jing Y., Londin E., Rigoutsos I. Nucleic Acids Res. 2015;43:9158–9175. doi: 10.1093/nar/gkv922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varallyay E., Burgyan J., Havelda Z. Nat. Protoc. 2008;3:190–196. doi: 10.1038/nprot.2007.528. [DOI] [PubMed] [Google Scholar]
- Schmittgen T. D., Jiang J., Liu Q., Yang L. Nucleic Acids Res. 2004;32:3–110. doi: 10.1093/nar/gnh040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T. D., Lee E. J., Jiang J., Sarkar A., Yang L., Elton T. S., Chen C. Methods. 2008;44:31–38. doi: 10.1016/j.ymeth.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James A. M., Baker M. B., Bao G., Searles C. D. Theranostics. 2017;7:634–646. doi: 10.7150/thno.16840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Liu J., Xu J. J., Zhang S. S., Chen H. Y. Chem. Soc. Rev. 2018;47:1996–2019. doi: 10.1039/c7cs00573c. [DOI] [PubMed] [Google Scholar]
- Ren K., Xu Y., Liu Y., Yang M., Ju H. ACS Nano. 2018;12:263–271. doi: 10.1021/acsnano.7b06200. [DOI] [PubMed] [Google Scholar]
- Bi S., Yue S., Zhang S. Chem. Soc. Rev. 2017;46:4281–4298. doi: 10.1039/c7cs00055c. [DOI] [PubMed] [Google Scholar]
- Wang J., Chao J., Liu H., Su S., Wang L., Huang W., Willner I., Fan C. Angew. Chem., Int. Ed. 2017;56:2171–2175. doi: 10.1002/anie.201610125. [DOI] [PubMed] [Google Scholar]
- Li L., Feng J., Liu H., Li Q., Tong L., Tang B. Chem. Sci. 2016;7:1940–1945. doi: 10.1039/c5sc03909f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Liu G. Q., Yang X. L., Jiang J. H. J. Am. Chem. Soc. 2015;137:6829–6836. doi: 10.1021/jacs.5b01778. [DOI] [PubMed] [Google Scholar]
- Li B., Jiang Y., Chen X., Ellington A. D. J. Am. Chem. Soc. 2012;134:13918–13921. doi: 10.1021/ja300984b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheglakov Z., Cronin T. M., He C., Weizmann Y. J. Am. Chem. Soc. 2015;137:6116–6119. doi: 10.1021/jacs.5b01451. [DOI] [PubMed] [Google Scholar]
- Bi S., Chen M., Jia X., Dong Y., Wang Z. Angew. Chem., Int. Ed. 2015;54:8144–8148. doi: 10.1002/anie.201501457. [DOI] [PubMed] [Google Scholar]
- Xuan F., Hsing I. M. J. Am. Chem. Soc. 2014;136:9810–9813. doi: 10.1021/ja502904s. [DOI] [PubMed] [Google Scholar]
- Duan R., Zuo X., Wang S., Quan X., Chen D., Chen Z., Jiang L., Fan C., Xia F. Nat. Protoc. 2014;9:597–607. doi: 10.1038/nprot.2014.036. [DOI] [PubMed] [Google Scholar]
- Jin C., Fu T., Wang R., Liu H., Zou J., Zhao Z., Ye M., Zhang X., Tan W. Chem. Sci. 2017;8:7082–7086. doi: 10.1039/c7sc02819a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirks R. M., Pierce N. A. PNAS. 2004;101:15275–15278. doi: 10.1073/pnas.0407024101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Dong H., Yang Z., Zhong X., Chen Y., Dai W., Zhang X. ACS Appl. Mater. Interfaces. 2017;9:159–166. doi: 10.1021/acsami.6b13150. [DOI] [PubMed] [Google Scholar]
- Zhang K., Dong H., Dai W., Meng X., Lu H., Wu T., Zhang X. Anal. Chem. 2017;89:648–655. doi: 10.1021/acs.analchem.6b02858. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.