Abstract
The histone acetyltransferase HBO1 (Histone acetyltransferase binding to origin recognition complex 1, Myst2/Kat7) participates in a range of life processes including DNA replication and tumorigenesis. Recent studies revealed that HBO1 is involved in gene transcriptional activation. However, the molecular behavior of HBO1 in inflammation is yet to be studied. Here we report that endotoxin lipopolysaccharide (LPS) elevates HBO1 protein level via up-regulating UPS25 (ubiquitin specific peptidase 25) and alters inflammatory gene transcription in THP-1 monocytes and in human primary macrophages. LPS protects HBO1 from ubiquitin proteasomal degradation without significantly altering its transcription. By immunoprecipitation, we identified that HBO1 associates with a deubiquitinating enzyme USP25 in THP-1 cells. LPS increases protein level of USP25 resulting in accumulation of HBO1 by suppression of HBO1 ubiquitination. Stabilized-HBO1 modulates inflammatory gene transcription in THP-1 cells. These findings indicate that USP25 promotes stability of HBO1 in bacterial infection thereby enhances HBO1-mediated inflammatory gene transcription.
Keywords: HBO1, USP25, ubiquitination, inflammation, gene transcription
1. Introduction
HBO1 (histone acetyltransferase binding to origin recognition complex 1, Myst2/Kat7) is a member of the histone acetyltransferase family. As a major histone acetylation enzyme, HBO1 is involved in a range of life processes. In the late M to early G1 phase of cell cycle, HBO1 incorporates with Cdt1, docking onto the chromatin to acetylate histone H3 at K14 and H4 at K12 [1, 2]. Histone H3/H4 acetylation triggers DNA replication licensing thereby initiating DNA replication [3, 4]. It has been reported that HBO1 overexpresses in many cancer cells and is related to aberrant progression of cell cycle and unlimited proliferation [5–9]. HBO1 plays a role in DNA repair after ultraviolet radiation [10]. The acetyltransferase activity of HBO1 is required for ubiquitin proteasomal degradation of the estrogen receptor, indicating HBO1 acetylates non-histone proteins as well [9, 11]. In HBO1-knockout mice, peripheral CD4+ and CD8+ T cells are substantially reduced which indicates HBO1 is critical in T cell development [12]. Recent studies indicate that HBO1 may modulate gene transcription. Knockout of HBO1 reduced more than 90% of the acetylation in H3K14, which is required for H3K14-dependent transcriptional activation of a panel of important genes for embryo development [13]. HBO1 directly interacts with transcription factor SIX1 and another histone acetyltransferase NCOA-3 to promote expression of corresponding glycolytic genes [14]. However, the role of HBO1 in inflammatory gene transcription in immune cells is not fully studied.
Post-translational modifications such as phosphorylation and ubiquitination have been reported in HBO1. A protein microarray analysis revealed that HBO1 is a substrate of the cyclin E/CDK2 complex. The cyclin E/CDK2 complex phosphorylates HBO1 at T88 which promotes the enrichment of cancer stem-like cells in breast cancer [15]. Phosphorylation of HBO1 at S57 by Polo-like kinase augments multi-drug resistance in pancreatic cancer cells [7]. Our previous work reported that HBO1 is an unstable protein with a half-life of approximate 3 h in lung epithelial cells. HBO1 is poly-ubiquitinated and regulated at the protein level by SCF-Fbxw15-mediated ubiquitin proteasomal degradation [16]. The molecular mechanisms of HBO1 regulation in distinct pathophysiological settings has yet to be understood.
Protein poly-ubiquitination is a reversible post-translational modification, a group of deubiquitinating enzymes remove the poly-ubiquitin chain. According to their distinct structures, deubiquitinating enzymes can be categorized into six sub-families: the ubiquitin-specific proteases (USPs), the ovarian tumor proteases (OTUs), the ubiquitin Carboxyl-terminal hydrolases (UCHs), the Josephin family, the motif interacting with ubiquitin (MIU)-containing novel DUB family (MINDYs), and the Zn-dependent JAB1/MPN/MOV34 metalloprotease DUBs (JAMMs) [17]. USP25, a member of ubiquitin-specific proteases sub-family, hydrolyzes both K48 and K63 linkages of polyubiquitin moieties where link to the substrate protein [18]. USP25 stabilizes tankyrases to enhance the Wnt/β-catenin signaling contributing to cancer development [19]. Aberrant expression of USP25 has been noted in breast cancer and non-small cell lung cancer patients as well [20, 21]. Interestingly, USP25 is a type 1 interferon responding gene; both viral infection and LPS treatment up-regulate USP25 expression [22–24]. Nevertheless, the signal transduction of USP25 under viral and bacterial infection requires further study.
Epigenetic mechanisms have exclusively and controversially emerged in every aspect of immunity including immune cell activation, lineage development and regulatory cell function [25, 26]. Among them, monocyte activation and proliferation are critical steps corresponding to bacterial infection [27]. Mounting evidence shows that epigenetics modulates host innate immunity against exogenous pathogens through multi-channel mechanisms involving in DNA methylation, histone post-translational modification and the activity of non-coding RNAs [28, 29]. For example, in mycobacterium tuberculosis infection, hyper-methylation located in promoter regions of IL17 family members has been observed in THP-1 monocytes [30]. Additionally, Histone H3K4me3 and H3K27me3 negatively regulate differentiation of monocytes in dendritic cells [31]. The molecular behavior of HBO1 in macrophage or monocyte in responding to bacterial infection has yet to be elucidated.
2. Materials and Methods
2.1. Cell line and reagents
Human monocyte THP-1 cells were cultured in RPMI1640 medium (500ml, ATCC 30-2001) supplemented with 10% (v/v) fetal bovine serum (FBS), 1% (v/v) Non-Essential Amino Acid (NEAA), 1% (v/v) sodium pyruvate, and 100 μg/mL streptomycin and 100 Units/mL penicillin. Human primary macrophages and their culture medium were from Celprogen (San Pedro, CA) and maintained in a 37°C incubator in the presence of 5% CO2.
The antibody against HBO1 (1:2000, cat#: ab70183) and recombinant human USP25 protein (ab188736) were purchased from Abcam. AcH3K14 antibody (1:1000, cat#: A-4023-050) were purchased from EpiGentek. Lamin A/C antibody was purchased from Cell signaling (1:1000, cat#:4C11). Antibodies purchased from Santa Cruz Biotechnology are listed in Table 1. β-actin antibody (1:10000, cat#: A5441, lot#: 116M4801V), LPS (cat#: L4391, lot#: 067M4036V) derived from Escherichia. coli (serum type: O111:B4), Actinomycin D (cat#: A9415, lot#: 026M4034V), and Leupeptin (cat#: L2884) were purchased from Sigma Aldrich. Protein A/G Pierce Protein A/G Agarose (cat#: 20422, lot#: SH254756) and Completed protease inhibitor cocktail (cat#: 88266, lot#: QE20062910) were purchased from Pierce. MG132 (cat#: F1101, lot#: F11042823) was purchased from UBPBio. Cycloheximide (cat#: ALX-380-269-G001) and ubiquitin aldehyde (cat#: BML-UW8450-0050, lot#: 07051723) were purchased from Enzo life Science. SYBR Select Master Mix (cat#: 4472920 ) was purchased from Applied Biosystem.
Table 1.
List of Antibodies used in the experiments.
| Name | Ratio of dilution | Catalog number | Lot number | epitope | Manufacturer |
|---|---|---|---|---|---|
| USP4 | 1:250 | sc-376000 | G1217 | 708–779 (h) | Santa Cruz |
| USP5 | 1:250 | sc-390943 | G1617 | 66–185 (h) | Santa Cruz |
| USP6 | 1:250 | sc-377306 | H1717 | 1117–1406 (h) | Santa Cruz |
| USP10 | 1:250 | sc-365828 | G0616 | 67–95 (h) | Santa Cruz |
| USP11 | 1:250 | sc-365528 | E3017 | 661–740 (h) | Santa Cruz |
| USP13 | 1:250 | sc-514416 | G1217 | 71–195 (h) | Santa Cruz |
| USP14 | 1:250 | sc-398009 | G1617 | 195–494 (h) | Santa Cruz |
| USP15 | 1:250 | sc-515688 | G1917 | 621–697 (h) | Santa Cruz |
| USP21 | 1:250 | sc-515911 | D2717 | 76–98 (h) | Santa Cruz |
| USP25 | 1:250 | sc-398414 | H0416 | 851–990 (h) | Santa Cruz |
| USP32 | 1:250 | sc-374465 | G1517 | 361–660 (h) | Santa Cruz |
| USP40 | 1:250 | sc-514248 | G1817 | 456–654 (h) | Santa Cruz |
| USP47 | 1:250 | sc-100633 | H2217 | 203–302 (h) | Santa Cruz |
| OTUB1 | 1:250 | sc-130458 | G0617 | recombinant | Santa Cruz |
| HAUSP | 1:250 | sc-137008 | G0616 | 11–210(h) | Santa Cruz |
| Ubp-M | 1:250 | sc-390683 | G2417 | 757–813 (h) | Santa Cruz |
| UBP-Y | 1:250 | sc-376130 | G1517 | 1–300(h) | Santa Cruz |
| UCH-L3 | 1:250 | sc-100340 | F3017 | 131–231 (h) | Santa Cruz |
| UCH-L5 | 1:250 | sc-271002 | G1817 | 220–329 (h) | Santa Cruz |
| Ubquitin | 1:500 | sc-166553 | A2710 | 1–76 (h) | Santa Cruz |
| NF-κB | 1:500 | sc-8008 | K2515 | 1–286 (h) | Santa Cruz |
| MyD88 | 1:500 | sc-74532 | K0917 | 1–296(h) | Santa Cruz |
| MyD88 | 1:500 | sc-74532 | K0917 | 1–296(h) | Santa Cruz |
| TLR4 | 1:500 | sc-293072 | D0518 | 198–395(m) | Santa Cruz |
| histoneH3 | 1:500 | sc-8654 | A0516 | C-terminus | Santa Cruz |
| histoneH4 | 1:500 | sc-10810 | B1010 | 7–103(h) | Santa Cruz |
2.2. Plasmid and DsiRNA transfection
DsiRNA or plasmids were transfected into THP-1 cells using electroporation executed with a nuclear transfection apparatus (Amaxa Biosystem, Gaithersburgh, MD, USA). Dicer-Substrate Short Interfering RNAs (DsiRNA) against HBO1 or USP25, including USP25-DsiRNA-1, USP25-DsiRNA-2, USP25-DsiRNA-3, HBO1-DsiRNA and Scrambled-DsiRNA (cat#: 51-01-14-03), were purchased from Integrated DNA Technology (IDT). pCMV6-XL5/HBO1 plasmid (catalog No: SC115703) and pCMV6-XL5/USP25 plasmid (catalog No: SC115248) were purchased from Origene. Briefly, one million cells in 100 μL of transfection buffer (20mM HEPES in PBS buffer) were mixed with 10 pM of DsiRNA or 1 μg of plasmid. RPMI 1640 medium (2 mL) was added to the six-well plates or flasks for each cuvette after the electroporation. Preset program U-001 was used for THP-1 cells. Transfected cells were cultured 72 h for further treatment or analysis.
2.3. Immunoblotting and co-immunoprecipitation
Immunoblotting and co-immunoprecipitation were conducted as previously described [32]. Briefly, cell lysates were prepared with lysis buffer (1X PBS, 1: 500 proteinase inhibitor cocktail, 0.3 % of SDS). Whole cell lysates were pre-cleared and applied to SDS-PAGE. The proteins were transferred to membranes, blocked with 5% (w/v) non-fat milk in Tris-buffered saline, and probed with primary antibodies as indicated. An enhanced chemiluminescence (ECL) was used to develop the image and the images were acquired with a Kodak in vivo professional 4000 system. For immunoprecipitation, 1 mg of cell lysates (in PBS with 0.5% Tween 20, and protease inhibitors) were incubated with specific primary antibodies for overnight at 4°C. The mixture was added to 40 μL of protein A/G-agaroses for an additional 2 h at 4°C. The precipitated complex was washed three times with 0.5% Tween 20 in PBS and analyzed by immunoblotting as described above. In HBO1-Ub co-immunoprecipitation experiment, cells were pretreated with MG132 for 30 min, and ubiquitin aldehyde (5 μM) was added to the cell lysis buffer to inhibit DUB activity [33].
2.4. Subcellular protein fractionation
THP-1 cells were collected by centrifugation at 1500 rpm for 5 min and washed with cold PBS buffer. The pelleted cells were resuspended in 1 mL of cytoplasmic membrane lysis buffer (10 mM Hepes, pH 8.0, 1.5 mM MgCl2, 10 mM KCl, 1 mM DTT, 0.1% Igepal CA-630, and protease inhibitor cocktails) and incubated on ice for 10 min. The cell suspensions were subjected to centrifuge at 2,000 rpm for 5 min, and the supernatant was collected as the cytoplasmic proteins and condensed by Amicon Ultra-4 Centrifugal filters (Merck Millipore Ltd). The pellets were lysed with 100 μL of cell lysis buffer (1X PBS, 1: 500 proteinase inhibitor cocktail, 0.3% of SDS). After centrifuging at 13,000 rpm for 10 min, the supernatants were collected as the nuclear proteins, The subcellular proteins were subjected to immunoblotting analysis.
2.5. in vitro de-ubiquitination assay
Endogenous HBO1 protein was obtained by HBO1 immunoprecipitation as described above. Briefly, THP-1 cells were treated with MG132 for 30min prior to harvest and cell lysates were prepared with lysis buffer (1X PBS, 1: 500 proteinase inhibitor cocktail, 0.3% of SDS). The cell lysates (contains 1mg total protein) were incubated with HBO1 antibody (2 μg) for 2 h followed by Protein A/G agarose beads (30 μL) incubation (1 h). The protein A/G agarose beads were precipitated by centrifugation and washed once with de-ubiquitination buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, pH 8.00, 10 mM DTT). The beads in a 25 μL of de-ubiquitination buffer were incubated for 4 h at 37°C in the presence or absence of active recombinant human USP25 protein (3 μg, as recommended by the manufacturer) (Abcam). The reactions were stopped by boiling for 5 min. Samples were subsequently prepared for immunoblotting analysis with indicated antibodies.
2.6. The quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA was extracted by using RNeasy Mini Kit (Cat: 74104) purchased from QIAGEN, the reverse transcription reaction was performed by using High-Capacity RNA-to-DNA KIT (Cat: 4387406) from Applied Biosystems. Total RNA (1 μg) was used for reverse transcription in a 20 μL system according to the protocol provided by manufacturer. Then, the cDNA templates were used for PCR in SYBR select Master Mix for CFX system (Cat: 4472942). qRT-PCR analysis was performed according to the manufacture's recommendations (Denaturation at 95°C for 2 min, followed by 46 cycles of 95°C for 15 s, 60°C for 1 min) in a CFX96 Real-Time PCR Detection System (Bio-Rad, USA). The primers used in qRT-PCR were listed as Table 2 (Table 2). The comparative Ct method, also referred to as the 2−ΔΔCt, was used to present relative gene expression (ΔCt = Ct gene of interest Ct GAPDH, ΔΔCt = ΔCt sample of treated ΔCt sample of control) [34].
Table 2.
List of PCR primers and DsiRNAs used in the studies.
| Primer | Sequence |
|---|---|
| GAPDH forward | 5′-GGCATGGACTGTGGTCATGA-3′ |
| GAPDH reverse | 5′-TTCACCATGGAGAAGGC -3′ |
| HBO1 forward | 5′-AAAGAGACAGGACCTGATTG-3′ |
| HBO1 reverse | 5′-GCAACAGAGAAATGACAGAC-3′ |
| USP25 forward | 5′-AATGTGATTGATCTCACTGG-3′ |
| USP25 reverse | 5′TCATCAGTTATTCCAGTCTCC-3′ |
| IL-1β forward | 5′-CTAAACAGATGAAGTGCTCC-3′ |
| IL-1β reverse | 5′-GGTCATTCTCCTGGAAGG-3′ |
| IL-6 forward | 5′-GCAGAAAAAGCAAAGAATC-3′ |
| IL-6 reverse | 5′-CTACATTTGCCGAAGAGC-3′ |
| IL- 10 forward | 5′-GCCTTTAATAAGCTCCAAGAG-3′ |
| IL- 10 reverse | 5′-ATCTTCATTGTCATGTAGGC-3′ |
| DsiRNA | |
| USP25-DsiRNA-1 | 5′-AAGAGAAUCCCAAAGUACUUGAATA-3′ 3′-AGUUCUCUUAGGGUUUCAUGAACUUAU-5′ |
| USP25-DsiRNA-2 | 5′-ACAUGAUGAAGAAUUGAUAUCACAT-3′ 3′-CCUGUACUACUUCUUAACUAUAGUGUA-5′ |
| USP25-DsiRNA-3 | 5′-CACUUCUUGUUGGUACCAAAAGGAA-3′ 3′-ACGUGAAGAACAACCAUGGUUUUCCUU-5′ |
| HBO1-DsiRNA | 5′-GCUUGAUACCUGGUAUCAUUCUCCA-3′ 3′-CUCGAACUAUGGACCAUAGUAAGAGGU-5′ |
2.7. Chromatin immunoprecipitation (ChIP) and ChIP-qPCR
ChIP experiments were carried out by using Epitect ChIP One Day Kit (lot:7600755, ref :1063182, QIAGEN, USA) following manufacture’s instruction. Briefly, cells were cross-linked with 1% (v/v) formaldehyde for 10 min at 37°C. The cross-linking reaction was terminated by addition of stop buffer. To obtain DNA fragments approximately 500 to 1500 bp in length, we sonicated the cells on ice for 5 rounds using an Vibra-Cell Ultrasonic Liquid Processors (40% Amplification, 4 seconds on/15 seconds off, 16 seconds total time per round). The lysates were pre-cleared using 50ul protein A beads (provided by kit) slurry for 1 h at 4°C. acH3K14 antibody (4 μg) was added to the lysates with rotating at 4°C for overnight. The samples were then incubated with 60 μL protein A beads for 1 h at 4°C. IP samples were reverse-cross linked and the DNA were isolated and purified according to manufacturer’s protocol. The purified DNA were subjected to qPCR analysis. The primers for ChIP-qPCR are following: IL-1β forward: 5′-CCTGGACTCTCATTCATTCTAC-3′; IL-1β reverse 5′-TCCATCTGAGACTCTATCTCTT-3′; IL-6 forward: 5′-CACCCTCACCCTCCAAC-3′; IL-6 reverse: 5′-AATGAGCCTCAGACATCTCC-3′; IL-10 forward: 5′-GCCTCAGTTTGCTCACTATAA-3′; IL-10 reverse: 5′-CACAGTGACGTGGACAAAT-3′.
2.8. Statistical analysis
Statistical comparisons were performed using One-way ANOVA with post-hoc Tukey HSD or Bonferroni and Holm multiple comparison provided by Online Web Statistical Calculators (http://astatsa.com) . P<0.05 indicates statistical significance.
3. Results
3.1. LPS increased HBO1 protein levels in THP-1 monocytes and human primary macrophages
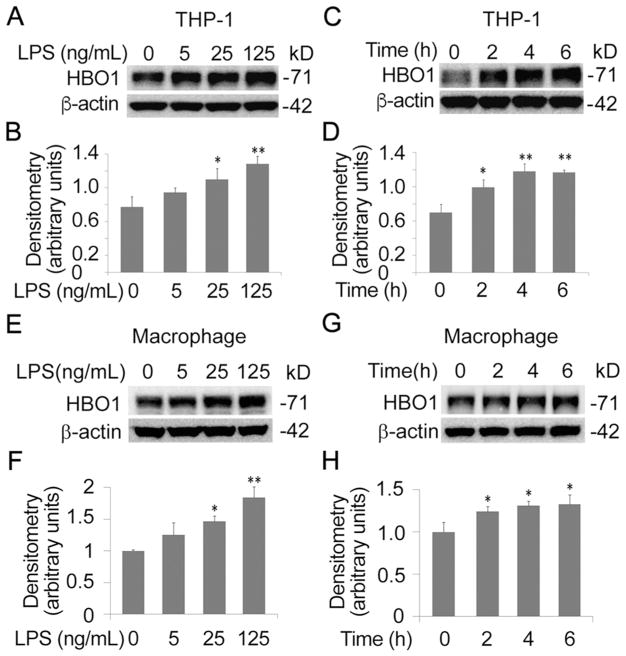
Previously we reported that HBO1 is unstable and LPS triggers HBO1 ubiquitin proteasomal degradation in mouse lung epithelial cells [16]. To test if LPS alters HBO1 protein levels in THP-1 monocytes, we treated THP-1 cells with LPS. Unexpectedly, the immunoblotting analysis indicated that LPS increased HBO1 protein levels in a concentration-dependent manner where 25 ng/mL of LPS was enough to significantly increase HBO1 over a 6 h treatment (Fig. 1A, B). To confirm this observation, we conducted a time course study. LPS with a concentration of 125 ng/mL markedly increased HBO1 protein levels at 2 h. LPS accumulated HBO1 in a time-dependent manner. In addition, LPS increased HBO1 protein levels approximately one-fold at the time point of 4 h (Fig. 1C, D). We repeated the experiments in human primary macrophages and obtained similar results (Fig. 1E–H). LPS increased HBO1 protein in human primary macrophages in a time and concentration dependent manner. These data demonstrated that LPS differentially elevated HBO1 protein levels in THP-1 monocytes and human primary macrophages, contrary to that in lung epithelial cells.
Fig. 1. LPS increased HBO1 protein levels in THP-1 cells and Human primary macrophages.
(A, B) THP-1 cells were treated with LPS in a range of concentrations as indicated for 6 h. Cell lysates were analyzed using immunoblotting for HBO1 or β-actin. The densitometry results of A were plotted in B. (C, D) THP-1 cells were treated with LPS (125 ng/mL) at varying time points. Cells lysates were analyzed with HBO1 or β-actin immunoblotting. The densitometry results of C were plotted in D. (E, F) Human primary macrophages were treated with LPS in a range of concentrations for 6 h. Cell lysates were analyzed using HBO1 or β-actin immunoblotting. The densitometry results of E were plotted in F. (G, H) Human primary macrophages were treated with LPS (125 ng/mL) for different time points. Cells lysates were analyzed with HBO1 or β-actin immunoblotting. The densitometry results of G were plotted in H. Data represents n=3 separate experiments. Graphs show mean ± SD, “*” denotes p<0.05 and “**” denotes p<0.01.
3.2. LPS did not alter HBO1 mRNA synthesis and stability
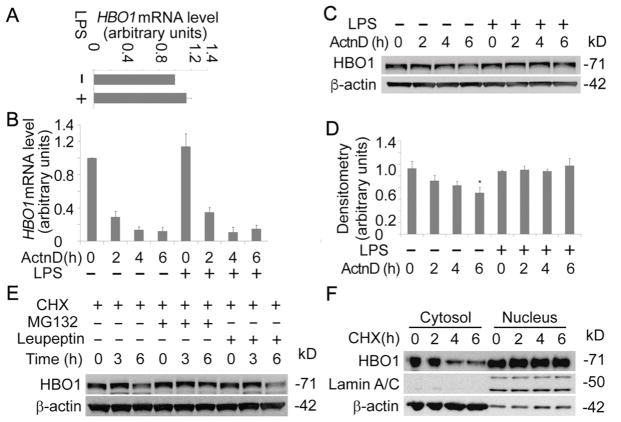
LPS may increase HBO1 protein levels in several major aspects, i.e., at transcriptional level/ translational processes and at the posttranslational level. We first accessed if LPS alters HBO1 transcription by determination of HBO1 mRNA levels in THP-1 cells. qRT-PCR results showed that LPS slightly increased HBO1 mRNA levels, but the changes were not significant compared to untreated controls (Fig. 2A). To study if LPS affects HBO1 mRNA stability, we treated cells with transcription inhibitor actinomycin D in the presence or absence of LPS. qRT-PCR results showed that LPS did not alter HBO1 mRNA turnover. HBO1 mRNA also decays at a similar rate in untreated control and LPS treated cells (Fig. 2B). To validate this observation, we conducted HBO1 immunoblotting analysis in actinomycin D and LPS treated cells above. Actinomycin D decreased HBO1 protein levels in a time-dependent manner. HBO1 proteins decreased approximate 50% in 4 h treatment of actinomycin D, similar to the turnover speed we observed in lung epithelial cells [16]. The actinomycin D-induced HBO1 decrease could be rescued via addition of LPS, suggesting that LPS probably elevates HBO1 protein levels after post-translation (Fig. 2C, D). To explore if in THP-1 cells HBO1 has a similar half-life as that in lung epithelial cells, we treated THP-1 cells with cycloheximide (CHX) to inhibit protein synthesis. Immunoblotting results showed that the half-life of HBO1 is ~ 4 h in THP-1 cells, longer than that in lung epithelial cells (~3 h) (Fig. 2E). HBO1 degradation could be rescued by MG132 but not leupeptin, indicating that HBO1 degradation is via proteasomal but not lysosomal pathway in THP-1 cells (Fig. 2E). To study the subcellular compartment(s) where HBO1 is degraded, we isolate the cytoplasmic protein and the nuclear proteins in THP-1 cells. Interestingly, the cytoplasmic HBO1 showed a shorter half-life (~2 h), albeit the nuclear HBO1 is stable without change in 6 h (Fig. 2F). Overall, since these results indicate that LPS did not significantly affect HBO1 mRNA transcription and stability, LPS probably impacts degradation of HBO1 protein in THP-1 cells.
Fig. 2. LPS did not change HBO1 mRNA production and stability.
(A) Total RNA was isolated from untreated and LPS-treated THP-1 cells (125 ng/mL for 6 h). HBO1 mRNA levels were determined with qRT-PCR. (B) THP-1 cells were treated with Actinomycin D (5 μg/mL) or co-treated with Actinomycin D and LPS (125 ng/mL) for varying time points as indicated. Total RNA was extracted, and the HBO1 mRNA levels were determined using qRT-PCR. (C, D) Above treated cells were lysed, and cell lysates were subjected to HBO1 or β-actin immunoblotting. The densitometry results of C were plotted in D. (E) THP-1 cells were treated with CHX, CHX + MG132 or CHX + Leupeptin for varying time points as indicated. Cell lysates were subjected to HBO1 or β-actin immunoblotting. (F) THP-1 cells were treated with CHX for different time points, the cytoplasmic proteins and the nuclear proteins were separated and subjected to HBO1, Lamin A/C or β-actin immunoblotting. Lamin A/C was used as a nuclear marker, and β-actin was as a cytoplasmic marker. Data represents n=3 separate experiments. Graphs show mean ± SD and “*” denotes p<0.05.
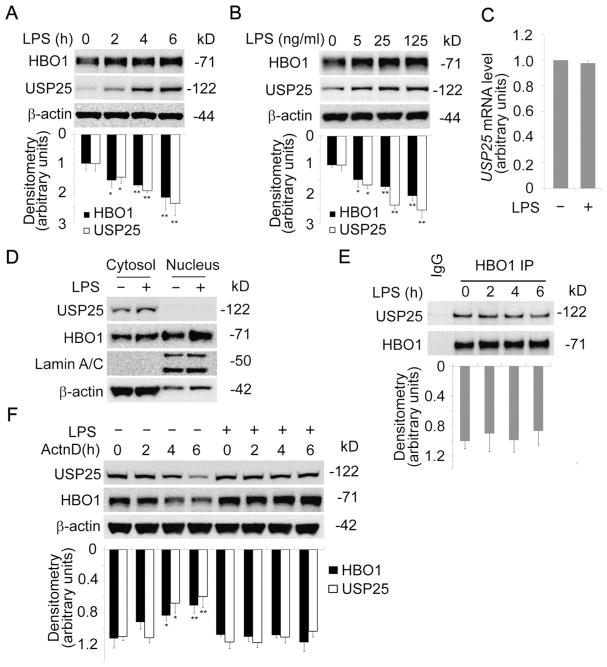
3.3. USP25 associated with and deubiquitinated HBO1
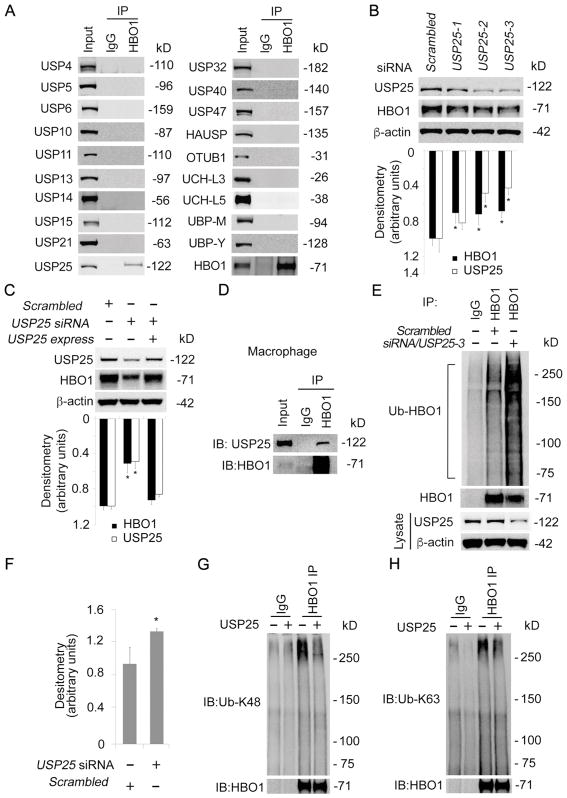
Protein destined to turnover is in general poly-ubiquitinated. However, protein poly-ubiquitination is reversible, the poly-ubiquitin moieties could be removed by the action of a family of enzymes referred as deubiquitinating enzymes [17]. To test if a deubiquitinating enzyme contributes to HBO1 stability, we performed HBO1 co-immunoprecipitation using THP-1 cell lysates and analyzed the precipitates with immunoblotting of deubiquitinating enzymes. Results from HBO1 immunoblotting analysis demonstrated that HBO1 was successfully immunoprecipitated (Fig. 3A). Among the 19 deubiquitinating enzymes screened, we identified that USP25 associated with HBO1. To further elucidate the assumption that USP25 deubiquitination stabilizes HBO1, we knocked down USP25 with DsiRNA. Among the tested DsiRNA, both USP25-2 and USP25-3 silenced USP25 efficiently (Fig. 3B, upper panel). As predicted, depletion of USP25 remarkably reduced the protein level of HBO1 (Fig. 3B). Over-expression of USP25 successfully rescued HBO1 from degradation in USP25 silenced cells, indicating that USP25-3 siRNA is specific (Fig. 3C). In addition, we performed HBO1 co-immunoprecipitation and confirmed HBO1 association with USP25 in human primary macrophages (Fig. 3D). To observe if USP25 affects ubiquitination level of HBO1, we conducted HBO1 co-immunoprecipitation and analyzed the precipitates against a ubiquitin antibody. HBO1 protein was slightly ubiquitinated in the scrambled control group, but the ubiquitination was substantially enhanced in UPS25-silenced group (Fig. 3E, F). Results from in vitro deubiquitination assay showed that USP25 hydrolyzed both K48-linked and K63-linked polyubiquitin chains within HBO1 (Fig. 3G, H). These findings demonstrated that USP25 associates with and deubiquitinates HBO1.
Fig. 3. USP25 interacted with and deubiquitinated HBO1.
(A) THP-1 cell lysates were co-immunoprecipitated with HBO1 antibody. The immunoprecipitates were analyzed with DUBs immunoblotting (see table 1). (B) THP-1 cells were transfected with USP25-DsiRNA sets and scrambled-DsiRNA for 72 h. The cell lysates were analyzed with USP25, HBO1 and β-actin immunoblotting. The densitometry results were plotted in lower panel. (C) THP-1 cells were transfected with scrambled-DsiRNA, USP25-DsiRNA or USP25-DsiRNA plus pCMV6-XL5/USP25 plasmid for 72 h. Cell lysates were subjected to HBO1, USP25, or β-actin immunoblotting. The densitometry results were plotted in lower panel. (D) Human primary macrophages were harvested, cell lysates (1mg of total protein) were subjected to HBO1 immunoprecipitation followed by USP25 and HBO1 immunoblotting as indicated. (E–F) THP-1 cells were transfected with USP25-DsiRNA-3 or scrambled-DsiRNA for 72 h. Equal amount of cell lysates (1mg of total protein) were subjected to HBO1 immunoprecipitation followed by immunoblotting as indicated. The efficacy of USP25 knockdown in each group was determined by immunoblotting as indicated. The densitometry results of E were plotted in F. (G, H) THP-1 were treated with MG132 for 30 min, cell lysates were immunoprecipitated with a HBO1 antibody and incubated for 4 h in a deubiquitination buffer in the presence or absence of the recombinant USP25 proteins. Immunoblotting analyses were probed with the indicated antibodies. Data represents n=3 separate experiments. Graphs show mean ± SD and “*” denotes p<0.05.
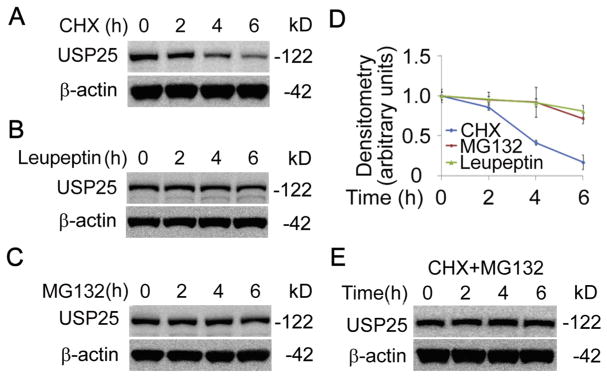
3.4. LPS stabilized HBO1 via elevation of a labile protein USP25
As a deubiquitinating enzyme, USP25 itself may be regulated at protein level. USP25 immunoblotting analysis of CHX-treated cells showed that USP25 is unstable with a half-life of approximate 3 h (Fig. 4A, D). Interestingly, USP25 proteins tended to decrease with either MG132 or leupeptin treated cells (Fig. 4B–D). MG132 could rescue CHX-induced degradation of USP25, suggesting that USP25 may be mainly degraded via proteasome (Fig. 4E). We assumed that LPS may increase USP25 protein level to stabilize HBO1 via enhancing HBO1 deubiquitination. As predicted, LPS increased USP25 protein level in a time-dependent manner in THP-1 cells, consistent with other reports [23, 35] (Fig. 5A). A low concentration of LPS (5 ng/mL) was sufficient to increase USP25 protein level (Fig. 5B). We extracted the total RNA from THP-1 cells in the presence or absence of LPS for qRT-PCR analysis. Results showed that USP25 mRNA levels were not remarkably changed by LPS, suggesting that LPS elevates USP25 protein levels post-translationally (Fig. 5C). USP25 has been reported as a cytoplasmic protein [36], but HBO1 locates mainly in the nucleus. We isolated the subcellular proteins and identified that LPS increased cytoplasmic USP25, LPS did not relocate USP25 to the nucleus (Fig. 5D). LPS increased HBO1 protein levels both in the cytoplasmic and the nuclear compartments (Fig. 5D). LPS elevated USP25 may stabilize the cytoplasmic HBO1 thereby more HBO1 molecules migrate into the nucleus. Results from co-immunoprecipitation studies showed that LPS does not dynamically alter HBO1-USP25 interaction (Fig. 5E). Actinomycin D treatment showed similar results as we observed in Fig. 2C whereby LPS stabilized both HBO1 and USP25 proteins (Fig. 5F).
Fig. 4. USP25 is a labile protein in THP-1 cells.
(A–C) THP-1 cells were treated with protein synthesis inhibitor cycloheximide (CHX) (A), proteasome inhibitor MG132 (B), or lysosome inhibitor Leupeptin (C) in various time points as indicated. Cell lysates were analyzed using immunoblotting for USP25 or β-actin. (D) The densitometry results of A–C were plotted in D. (E) THP-1 cells were treated with CHX and MG132 as indicated, cell lysates were analyzed using USP25 and β-actin immunoblotting. Data represents n=3 separate experiments. Graphs show mean ± SD.
Fig. 5. LPS stabilized USP25 protein in THP-1 cells.
(A) THP-1 cells were treated with LPS at different time points as indicated. Cells lysates were analyzed with immunoblotting for HBO1, USP25, or β-actin. The densitometry results were plotted in lower panel. (B) THP-1 cells were treated with LPS at varying concentrations as indicated. Cells lysates were analyzed by immunoblotting for HBO1, USP25 or β-actin. The densitometry results were plotted in lower panel. (C) Total RNA was isolated from untreated and LPS-treated THP-1 cells (125 ng/mL for 6 h). USP25 mRNA levels were determined with qRT-PCR. (D) THP-1 cells were treat with LPS (125 ng/mL for 6 h), the cytoplasmic and the nuclear proteins were separated and subjected to HBO1, USP25, Lamin A/C or β-actin immunoblotting. (E) THP-1 cells were treated with LPS (125 ng/mL) in a range of time points. Equal amount of cell lysates (1mg of total protein) were subjected to HBO1 immunoprecipitation followed by immunoblotting as indicated. The densitometry results were plotted in lower panel. (F) THP-1 cells were treated with Actinomycin D (5 μg/mL) or co-treated with Actinomycin D and LPS (125 ng/mL) for varying time points. Cell lysates were subjected to immunoblotting as indicated. The densitometry results were plotted in lower panel. Data represents n=3 separate experiments. Graphs show mean ± SD, “*” denotes p<0.05 and “**” denotes p<0.01.
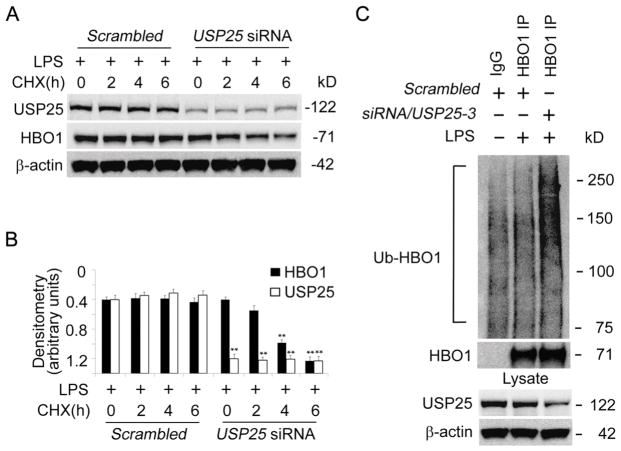
To understand if LPS-induced accumulation of HBO1 is mediated by an elevation of USP25, we tested if LPS blocked HBO1 degradation in USP25 silenced THP-1 cells. LPS treatment significantly reduced CHX-mediated HBO1 degradation in THP-1 cells (Fig. 6A, B). LPS did not halt HBO1 degradation in USP25 silenced cells (Fig. 6A, B). LPS did not suppress HBO1 ubiquitination in USP25 silenced cells (Fig. 6C). In all, these data indicate that LPS upregulates USP25 to keep HBO1 in a deubiquitinated status, thus elevates HBO1 protein level in THP-1 cells.
Fig. 6. LPS stabilized HBO1 via USP25-mediated deubiquitination.
(A, B) Scrambled-DsiRNA or USP25-DsiRNA were introduced into THP-1 cells for 72 h. The cells were treated with CHX at different time points in the presence of LPS (125 ng/mL). Cell lysates were analyzed with USP25, HBO1 or β-actin immunoblotting. The densitometry results of A were plotted in B. (C) Scrambled-DsiRNA or USP25-DsiRNA were introduced into THP-1 cells for 72 h. The cells were treated with LPS (125 ng/mL for 6 h). Equal amount of cell lysates (1 mg) were subjected to HBO1 immunoprecipitation followed by Ubiquitin and HBO1 immunoblotting. The corresponding cell lysates were subjected to immunoblotting to analyze the efficacy of USP25 knockdown. Data in each panel represents n=3 separate experiments. Graphs show mean ± SD, “*” denotes p<0.05 and “**” denotes p<0.01.
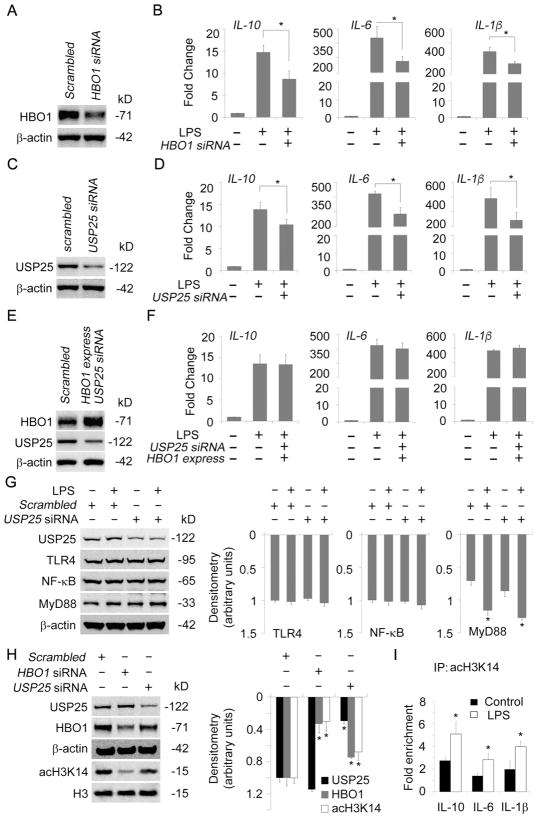
3.5. HBO1 stabilized by LPS regulated inflammatory gene transcription
Recent studies reported that HBO1 may affect gene transcription [37–39]. We theorized that LPS may regulate inflammatory gene transcription via the USP25/HBO1 axis. First, we tested if silence of HBO1 may affect LPS-induced gene transcription. HBO1-silenced cells and scrambled control cells were used for the study (Fig. 7A). qRT-PCR results showed the transcription levels of a panel of LPS-induced cytokine genes including IL-1β IL-6, and IL-10 were significantly lower in HBO1-silenced THP-1 cells as compared to that in control cells (Fig. 7B). Secondly, we depleted USP25 to determine if HBO1-mediated inflammation gene transcription would be affected. Depletion of USP25 led to reduced transcription of IL-1β IL-6, and IL-10 under LPS stimulation, compared to scrambled control (Fig. 7C, D). Finally, we tested whether ectopic expression of HBO1 could rescue the repressed cytokine transcription by depletion of USP25. Ectopic expression of HBO1 did reverse the cytokine gene transcription to a similar level of LPS only group in USP25-silenced THP-1 cells (Fig. 7E, F). To evaluate the impact of USP25 depletion on LPS-TLR4 pathway, we checked the protein levels of TLR4, MyD88, and NF-κB in USP25 silenced THP-1 cells. Immunoblotting results showed that LPS-activated MyD88 was not dramatically altered by depletion of USP25 (Fig. 7G). We did not observe obvious changes of TLR4 and total NF-κB proteins in LPS treated THP-1 cells, probably due to the low concentration of LPS we used and early time window we observed (Fig. 7G). Depletion of USP25 or HBO1 by siRNA resulted in a decrease of H3K14 acetylation (Fig. 7H). CHIP-qPCR results showed that LPS did enhance the association of acH3K14 with the genes of IL-1β IL-6, and IL-10 (Fig. 7I). These data indicate that the USP25/HBO1 axis is crucial in LPS-induced transcription of IL-1β IL-6, and IL-10.
Fig. 7. HBO1 stabilized by LPS regulated inflammatory gene transcription.
(A, B) Scrambled-DsiRNA or HBO1-DsiRNA were introduced into THP-1 cells for 72 h. The cells were treated with LPS (125 ng/mL) for 6 h. Efficacy of HBO1 knockdown was determined using immunoblotting (A). Total RNA was extracted and the mRNA levels of IL-1β, IL-6 and IL-10 were determined (B). (C, D) Scrambled-DsiRNA or USP25-DsiRNA were introduced into THP-1 cells for 72 h. The cells were treated with LPS (125 ng/mL) for 6 h. Efficacy of USP25 knockdown was determined using immunoblotting (C). Total RNA was extracted and the mRNA levels of IL-1β, IL-6 and IL-10 were determined (D). (E, F) Scrambled-DsiRNA, pCMV6-XL5/HBO1 plasmid or USP25-DsiRNA were delivered into THP-1 cells using electroporation as indicated. After 72 h, the cells were treated with LPS (125 ng/mL) for 6 h. Efficacy of ectopic HBO1 expression and USP25 knockdown were analyzed using immunoblotting (E). Total RNA was extracted and the mRNA levels of IL-1β, IL-6 and IL-10 were determined (F). (G) Scrambled-DsiRNA or USP25-DsiRNA were introduced into THP-1 cells for 72 h. The cells were treated with LPS (125 ng/mL) for 6 h. Cell lysates were subjected to USP25, TLR4, NF-κB, MyD88, and β-actin immunoblotting. The densitometry results were plotted in right panels. (H) THP-1 cells were introduced with scrambled-DsiRNA, HBO1-DsiRNA or USP25-DsiRNA for 72h. Cell lysates were subjected to USP25, HBO1, β-actin, acH3K14, histone H3 immunoblotting. The densitometry results were plotted in right panel. (I) THP-1 cells were treated with LPS (125 ng/mL) for 6 h. The cells were subjected to ChIP-qPCR, qPCR signals were normalized to that of IgG. Data are representative of three independent experiments. Graphs show mean ± SD, and “*” denotes p<0.05.
4. Discussion
The original findings in this study are: (i) LPS differentially elevated HBO1 protein in THP-1 cells; (ii) USP25 interacted with and deubiquitinated HBO1; (iii) LPS stabilized USP25 to accumulate HBO1; and (iv) USP25/HBO1 signaling is important for LPS-induced inflammatory gene transcription in THP-1 cells (Fig. 8). Ample evidence demonstrates that epigenetic changes are required for transcription of inflammatory genes. Histone acetylation decondenses the structure of chromatin that facilitates transcriptional factors to access the loci of target genes for the subsequent transcription [40, 41]. HBO1-initiated DNA replication occurs in late G1 and early S phases of cell cycle. However, HBO1-mediated acetylation is responsible for 90% of H3K14 acetylation [42]. Considering the protein abundance of HBO1 is relatively stable in different phases of the cell cycle, it is conceivable that HBO1 may be involved in distinct life processes such as gene transcription. Several studies reported that HBO1 mediated histone H3 and H4 acetylation may be involved in gene transcription [13, 14, 43]. Pathological stimuli may impel transcriptional factors into decondensed chromatin to process transcription of corresponding genes. In this study, we demonstrate that HBO1 regulates inflammatory gene transcription in THP-1 monocytes. HBO1 may be an important player in epigenetically regulating inflammatory gene transcription in immune cells.
Fig. 8. USP25/HBO1 axis is crucial in LPS-induced inflammatory gene transcription.
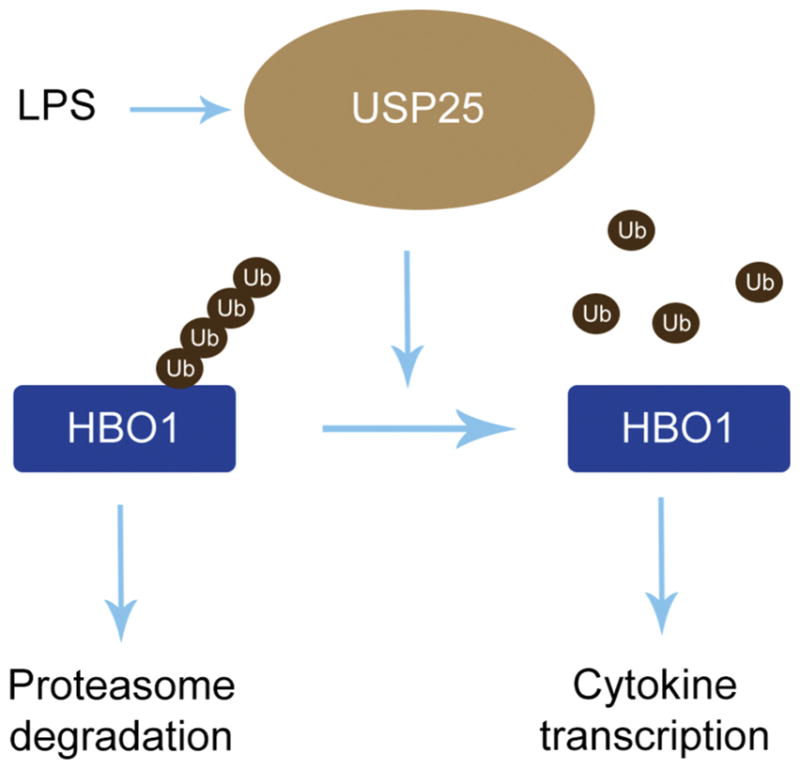
HBO1 is dynamic at protein level that is stringently monitored by ubiquitin proteasomal degradation machinery. Endotoxin LPS influences on the turnover of USP25 that leads to elevation of USP25 at protein levels. Elevated USP25 interacts with and deubiquitinates HBO1 that stabilizes HBO1. Accumulated HBO1 in turn promotes inflammatory gene transcription.
The ubiquitin system regulates a plethora of cellular processes through modulating cellular availability of functional proteins [44]. The availability of a protein could be modulated to enhance or abate specific pathways under specific stress condition. Protein polyubiquitination is a reversible posttranslational modification, and deubiquitinating enzymes play an indispensable role in this system [17]. Among the deubiquitinating enzymes, USP25 plays an important role in immune response under inflammatory stress. USP25 is recruited to deubiquitinate tumor necrosis factor receptor-associated factor 3 (TRAF3) and tumor necrosis factor receptor-associated factor 6 (TRAF6) after TLR4 is activated by LPS [23]. USP25 is also a negative regulator in the IL-17-triggered signaling pathway but a positive regulator in RNA and DNA virus-triggered innate immune responses [45]. How USP25 is regulated in these pathophysiological settings is not fully understood. Here, we identified that USP25 is a labile protein with a half-life of approximate 3 h in THP-1 cells. LPS elevated USP25, in part, through impacting its turnover. Elevation of USP25 boosts deubiquitination of HBO1 leading to HBO1 stabilization and accumulation. Up-regulated HBO1 promotes inflammatory gene transcription in THP-1 cells. Previously, we reported that HBO1 is unstable in mouse lung epithelial cells when treated with LPS, and SCF-Fbxw15, an ubiquitin E3 ligase, catalyzes HBO1 ubiquitination and subsequent proteasomal degradation [16]. Distinct type of cells may differentially react to the same stimulus. Notably, THP-1 cells and lung epithelial cells respond to LPS at different concentration ranges. THP-1 cells are more sensitive to LPS at nanogram/mL, while lung epithelial cells respond to LPS at microgram/mL levels. Pulmonary alveoli, where lung epithelial cells are located, are directly exposed to exogenous pathogens, therefore, lung epithelial cells may be more tolerant to the LPS. This may partially explain the distinct molecular behaviors between THP-1 cells and lung epithelial cells.
Endotoxin LPS, an out-wall component of Gram-negative bacteria, is a causative factor for the inflammatory gene expression in immune cells. Numerous regulatory networks has been reported to elucidate the complexity of the subsequent cytokine gene transcription induced by LPS. LPS interacts with its legitimated toll-like receptor 4 to culminate downstream signaling transduction through MyD88 or TRIF. As TLR4 signaling is activated, a range of transcriptional factors including NF-κB or Stat3 migrate into the nucleus to initiate inflammatory gene transcription [46, 47]. At this stage, concomitant epigenetic actions of chromatin modulators are required. Histone acetyltransferases p300 and CBP have been reported to be crucial in gene transcription mediated by NF-κB or Stat [48–51]. In this study, we observed that another histone acetyltransferase HBO1, originally believed as a DNA replication initiator, functions as a regulator of inflammatory gene transcription. HBO1 participates in regulation of IL-1β IL-6, and IL-10 gene transcription responding to LPS in THP-1 monocytes. Knockdown of HBO1 partially abates LPS-mediated cytokine transcription, suggesting that HBO1 may be one of critical players in regulating LPS-mediated cytokine transcription at epigenetic level. Inflammatory gene expression is cell type specific, it is known that LPS stimulates gene transcription of IL-1, IL-6, IL-10, and TNFα in THP-1 cells. Similarly, we identified that expression of IL-1β IL-6, and IL-10 is regulated by USP25/HBO1 axis. A RNA-seq analysis may be helpful to elicit the profile of HBO-1 dependent inflammatory genes. Results from ChIP-qPCR suggest that HBO1-catalyzed H3K14ac associates with the inflammatory genes, supporting that HBO1 may epigenetically regulate transcription of inflammatory genes. Our limited evidence suggests that NF-κB signaling may not be essential in HBO1 mediated inflammatory gene transcription in THP-1 cells. HBO1 is reported to suppress NF-κB signaling via its non-acetyltransferase enzymatic function [52]. In line with this observation, we did not observe a significant change of TNFα at transcriptional level in HBO1 silenced THP-1 cells, which is believed to be a NF-κB dependent inflammatory gene. LPS may activate other nuclear factors such as Stat3 to regulate inflammatory gene transcription via USP25/HBO1 axis. The acetyltransferase activity of HBO1 may directly modify acetylation status of the nuclear factors to synergize their function in inflammatory gene transcription as well. Overall, these findings may shed light on pharmaceutical targeting on inflammatory cytokine transcription in infectious diseases, particularly in systemically bacterial infectious illness.
Highlights.
LPS differentially upregulates HBO1 protein in THP-1 monocytes.
USP25 interacts with and deubiquitinates HBO1.
LPS induces elevation of USP25 protein that leads to accumulation of HBO1 at protein level.
LPS induces inflammatory gene transcription via HBO1/USP25 axis.
Acknowledgments
This work was supported, in part, by a National Institutes of Health R01 grant HL125435 (to C.Z.).
Abbreviation
- HBO1
Histone acetyltransferase binding to origin recognition complex 1
- UPS25
Ubiquitin specific peptidase 25
- LPS
Lipopolysaccharide
- Cdt1
Chromatin Licensing And DNA Replication Factor 1
- HDAC11
Histone deacetylase 11
- MCM
Minichromosome Maintenance Complex
- SIX1
Sine Oculis Homeobox Homolog1
- NCOA-3
Nuclear Receptor Coactivator 3
- CDK2
Cyclin Dependent Kinase 2
- SCF
Skp, Cullin, F-box containing complex
- DUB
Deubiquitinating enzyme
- H3K4me3
Histone H3 lysine 4 trimethylation
- H3K27me3
Histone H3 lysine 27 trimethylation
- acH3K14
Histone H3 lysine 14 acetylation
- DsiRNA
Dicer-Substrate Short Interfering RNAs
- CHX
Cycloheximide
- TLR4
Toll Like Receptor 4
- MyD88
Myeloid Differentiation Primary Response 88
- TRIF
TIR-domain-containing adapter-inducing interferon-β
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Iizuka M, Matsui T, Takisawa H, Smith MM. Regulation of replication licensing by acetyltransferase Hbo1. Molecular and cellular biology. 2006;26:1098–1108. doi: 10.1128/MCB.26.3.1098-1108.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miotto B, Struhl K. HBO1 histone acetylase is a coactivator of the replication licensing factor Cdt1. Genes & development. 2008;22:2633–2638. doi: 10.1101/gad.1674108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miotto B, Struhl K. HBO1 histone acetylase activity is essential for DNA replication licensing and inhibited by Geminin. Molecular cell. 2010;37:57–66. doi: 10.1016/j.molcel.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iizuka M, Stillman B. Histone acetyltransferase HBO1 interacts with the ORC1 subunit of the human initiator protein. The Journal of biological chemistry. 1999;274:23027–23034. doi: 10.1074/jbc.274.33.23027. [DOI] [PubMed] [Google Scholar]
- 5.Chen Z, Zhou L, Wang L, Kazobinka G, Zhang X, Han X, Li B, Hou T. HBO1 promotes cell proliferation in bladder cancer via activation of Wnt/beta-catenin signaling. Molecular carcinogenesis. 2018;57:12–21. doi: 10.1002/mc.22715. [DOI] [PubMed] [Google Scholar]
- 6.Iizuka M, Takahashi Y, Mizzen CA, Cook RG, Fujita M, Allis CD, Frierson HF, Jr, Fukusato T, Smith MM. Histone acetyltransferase Hbo1: catalytic activity, cellular abundance, and links to primary cancers. Gene. 2009;436:108–114. doi: 10.1016/j.gene.2009.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song B, Liu XS, Rice SJ, Kuang S, Elzey BD, Konieczny SF, Ratliff TL, Hazbun T, Chiorean EG, Liu X. Plk1 phosphorylation of orc2 and hbo1 contributes to gemcitabine resistance in pancreatic cancer. Molecular cancer therapeutics. 2013;12:58–68. doi: 10.1158/1535-7163.MCT-12-0632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo LL, Yu SY, Li M. Functional analysis of HBO1 in tumor development and inhibitor screening. International journal of molecular medicine. 2016;38:300–304. doi: 10.3892/ijmm.2016.2617. [DOI] [PubMed] [Google Scholar]
- 9.Iizuka M, Susa T, Takahashi Y, Tamamori-Adachi M, Kajitani T, Okinaga H, Fukusato T, Okazaki T. Histone acetyltransferase Hbo1 destabilizes estrogen receptor alpha by ubiquitination and modulates proliferation of breast cancers. Cancer science. 2013;104:1647–1655. doi: 10.1111/cas.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niida H, Matsunuma R, Horiguchi R, Uchida C, Nakazawa Y, Motegi A, Nishimoto K, Sakai S, Ohhata T, Kitagawa K, Moriwaki S, Nishitani H, Ui A, Ogi T, Kitagawa M. Phosphorylated HBO1 at UV irradiated sites is essential for nucleotide excision repair. Nature communications. 2017;8:16102. doi: 10.1038/ncomms16102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iizuka M, Susa T, Tamamori-Adachi M, Okinaga H, Okazaki T. Intrinsic ubiquitin E3 ligase activity of histone acetyltransferase Hbo1 for estrogen receptor alpha. Proceedings of the Japan Academy. Series B Physical and biological sciences. 2017;93:498–510. doi: 10.2183/pjab.93.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Newman DM, Voss AK, Thomas T, Allan RS. Essential role for the histone acetyltransferase KAT7 in T cell development, fitness, and survival. Journal of leukocyte biology. 2017;101:887–892. doi: 10.1189/jlb.1MA0816-338R. [DOI] [PubMed] [Google Scholar]
- 13.Kueh AJ, Dixon MP, Voss AK, Thomas T. HBO1 is required for H3K14 acetylation and normal transcriptional activity during embryonic development. Molecular and cellular biology. 2011;31:845–860. doi: 10.1128/MCB.00159-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li L, Liang Y, Kang L, Liu Y, Gao S, Chen S, Li Y, You W, Dong Q, Hong T, Yan Z, Jin S, Wang T, Zhao W, Mai H, Huang J, Han X, Ji Q, Song Q, Yang C, Zhao S, Xu X, Ye Q. Transcriptional Regulation of the Warburg Effect in Cancer by SIX1. Cancer cell. 2018;33:368–385e367. doi: 10.1016/j.ccell.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 15.Duong MT, Akli S, Macalou S, Biernacka A, Debeb BG, Yi M, Hunt KK, Keyomarsi K. Hbo1 is a cyclin E/CDK2 substrate that enriches breast cancer stem-like cells. Cancer research. 2013;73:5556–5568. doi: 10.1158/0008-5472.CAN-13-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zou C, Chen Y, Smith RM, Snavely C, Li J, Coon TA, Chen BB, Zhao Y, Mallampalli RK. SCF(Fbxw15) mediates histone acetyltransferase binding to origin recognition complex (HBO1) ubiquitin-proteasomal degradation to regulate cell proliferation. The Journal of biological chemistry. 2013;288:6306–6316. doi: 10.1074/jbc.M112.426882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mevissen TET, Komander D. Mechanisms of Deubiquitinase Specificity and Regulation. Annual review of biochemistry. 2017;86:159–192. doi: 10.1146/annurev-biochem-061516-044916. [DOI] [PubMed] [Google Scholar]
- 18.Shi L, Wen Y, Zhang N. (1)H, (1)(3)C and (1)(5)N backbone and side-chain resonance assignments of the N-terminal ubiquitin-binding domains of USP25. Biomolecular NMR assignments. 2014;8:255–258. doi: 10.1007/s12104-013-9495-1. [DOI] [PubMed] [Google Scholar]
- 19.Xu D, Liu J, Fu T, Shan B, Qian L, Pan L, Yuan J. USP25 regulates Wnt signaling by controlling the stability of tankyrases. Genes & development. 2017;31:1024–1035. doi: 10.1101/gad.300889.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng S, Zhou H, Xiong R, Lu Y, Yan D, Xing T, Dong L, Tang E, Yang H. Over-expression of genes and proteins of ubiquitin specific peptidases (USPs) and proteasome subunits (PSs) in breast cancer tissue observed by the methods of RFDD-PCR and proteomics. Breast cancer research and treatment. 2007;104:21–30. doi: 10.1007/s10549-006-9393-7. [DOI] [PubMed] [Google Scholar]
- 21.Li J, Tan Q, Yan M, Liu L, Lin H, Zhao F, Bao G, Kong H, Ge C, Zhang F, Yu T, Li J, He X, Yao M. miRNA-200c inhibits invasion and metastasis of human non-small cell lung cancer by directly targeting ubiquitin specific peptidase 25. Molecular cancer. 2014;13:166. doi: 10.1186/1476-4598-13-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin D, Zhang M, Zhang MX, Ren Y, Jin J, Zhao Q, Pan Z, Wu M, Shu HB, Dong C, Zhong B. Induction of USP25 by viral infection promotes innate antiviral responses by mediating the stabilization of TRAF3 and TRAF6. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:11324–11329. doi: 10.1073/pnas.1509968112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhong B, Liu X, Wang X, Liu X, Li H, Darnay BG, Lin X, Sun SC, Dong C. Ubiquitin-specific protease 25 regulates TLR4-dependent innate immune responses through deubiquitination of the adaptor protein TRAF3. Science signaling. 2013;6:ra35. doi: 10.1126/scisignal.2003708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ren Y, Zhao Y, Lin D, Xu X, Zhu Q, Yao J, Shu HB, Zhong B. The Type I Interferon-IRF7 Axis Mediates Transcriptional Expression of Usp25 Gene. The Journal of biological chemistry. 2016;291:13206–13215. doi: 10.1074/jbc.M116.718080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alvarez-Errico D, Vento-Tormo R, Sieweke M, Ballestar E. Epigenetic control of myeloid cell differentiation, identity and function. Nature reviews Immunology. 2015;15:7–17. doi: 10.1038/nri3777. [DOI] [PubMed] [Google Scholar]
- 26.Lim PS, Li J, Holloway AF, Rao S. Epigenetic regulation of inducible gene expression in the immune system. Immunology. 2013;139:285–293. doi: 10.1111/imm.12100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoeksema MA, de Winther MP. Epigenetic Regulation of Monocyte and Macrophage Function. Antioxidants & redox signaling. 2016;25:758–774. doi: 10.1089/ars.2016.6695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mehta S, Jeffrey KL. Beyond receptors and signaling: epigenetic factors in the regulation of innate immunity. Immunology and cell biology. 2015;93:233–244. doi: 10.1038/icb.2014.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hennessy C, McKernan DP. Epigenetics and innate immunity: the 'unTolld' story. Immunology and cell biology. 2016;94:631–639. doi: 10.1038/icb.2016.24. [DOI] [PubMed] [Google Scholar]
- 30.Zheng L, Leung ET, Wong HK, Lui G, Lee N, To KF, Choy KW, Chan RC, Ip M. Unraveling methylation changes of host macrophages in Mycobacterium tuberculosis infection. Tuberculosis. 2016;98:139–148. doi: 10.1016/j.tube.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 31.Huang Y, Min S, Lui Y, Sun J, Su X, Liu Y, Zhang Y, Han D, Che Y, Zhao C, Ma B, Yang R. Global mapping of H3K4me3 and H3K27me3 reveals chromatin state-based regulation of human monocyte-derived dendritic cells in different environments. Genes and immunity. 2012;13:311–320. doi: 10.1038/gene.2011.87. [DOI] [PubMed] [Google Scholar]
- 32.Zou C, Synan MJ, Li J, Xiong S, Manni ML, Liu Y, Chen BB, Zhao Y, Shiva S, Tyurina YY, Jiang J, Lee JS, Das S, Ray A, Ray P, Kagan VE, Mallampalli RK. LPS impairs oxygen utilization in epithelia by triggering degradation of the mitochondrial enzyme Alcat1. Journal of cell science. 2016;129:51–64. doi: 10.1242/jcs.176701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y, Lear T, Zhao Y, Zhao J, Zou C, Chen BB, Mallampalli RK. F-box protein Fbxl18 mediates polyubiquitylation and proteasomal degradation of the pro-apoptotic SCF subunit Fbxl7. Cell death & disease. 2015;6:e1630. doi: 10.1038/cddis.2014.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nature protocols. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 35.Ding C, Li F, Long Y, Zheng J. Chloroquine attenuates lipopolysaccharide-induced inflammatory responses through upregulation of USP25. Canadian journal of physiology and pharmacology. 2017;95:481–491. doi: 10.1139/cjpp-2016-0303. [DOI] [PubMed] [Google Scholar]
- 36.Bosch-Comas A, Lindsten K, Gonzalez-Duarte R, Masucci MG, Marfany G. The ubiquitin-specific protease USP25 interacts with three sarcomeric proteins. Cellular and molecular life sciences : CMLS. 2006;63:723–734. doi: 10.1007/s00018-005-5533-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiology and molecular biology reviews : MMBR. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ochiai N, Nishizuka M, Osada S, Imagawa M. Fad24, a Positive Regulator of Adipogenesis, Is Required for S Phase Re-entry of C2C12 Myoblasts Arrested in G0 Phase and Involved in p27(Kip1) Expression at the Protein Level. Biological & pharmaceutical bulletin. 2016;39:807–814. doi: 10.1248/bpb.b15-00954. [DOI] [PubMed] [Google Scholar]
- 39.Wright DG, Marchal C, Hoang K, Ankney JA, Nguyen ST, Rushing AW, Polakowski N, Miotto B, Lemasson I. Human T-cell leukemia virus type-1-encoded protein HBZ represses p53 function by inhibiting the acetyltransferase activity of p300/CBP and HBO1. Oncotarget. 2016;7:1687–1706. doi: 10.18632/oncotarget.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tessarz P, Kouzarides T. Histone core modifications regulating nucleosome structure and dynamics. Nature reviews Molecular cell biology. 2014;15:703–708. doi: 10.1038/nrm3890. [DOI] [PubMed] [Google Scholar]
- 41.Stasevich TJ, Hayashi-Takanaka Y, Sato Y, Maehara K, Ohkawa Y, Sakata-Sogawa K, Tokunaga M, Nagase T, Nozaki N, McNally JG, Kimura H. Regulation of RNA polymerase II activation by histone acetylation in single living cells. Nature. 2014;516:272–275. doi: 10.1038/nature13714. [DOI] [PubMed] [Google Scholar]
- 42.Hung T, Binda O, Champagne KS, Kuo AJ, Johnson K, Chang HY, Simon MD, Kutateladze TG, Gozani O. ING4 mediates crosstalk between histone H3 K4 trimethylation and H3 acetylation to attenuate cellular transformation. Molecular cell. 2009;33:248–256. doi: 10.1016/j.molcel.2008.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao S, Qi X, Li J, Sang L. Upregulated KAT7 in synovial fibroblasts promotes Th17 cell differentiation and infiltration in rheumatoid arthritis. Biochemical and biophysical research communications. 2017;489:235–241. doi: 10.1016/j.bbrc.2017.05.143. [DOI] [PubMed] [Google Scholar]
- 44.Ciechanover A. The unravelling of the ubiquitin system. Nature reviews Molecular cell biology. 2015;16:322–324. doi: 10.1038/nrm3982. [DOI] [PubMed] [Google Scholar]
- 45.Zhong B, Liu X, Wang X, Chang SH, Liu X, Wang A, Reynolds JM, Dong C. Negative regulation of IL-17-mediated signaling and inflammation by the ubiquitin-specific protease USP25. Nature immunology. 2012;13:1110–1117. doi: 10.1038/ni.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Plociennikowska A, Hromada-Judycka A, Borzecka K, Kwiatkowska K. Co-operation of TLR4 and raft proteins in LPS-induced pro-inflammatory signaling. Cellular and molecular life sciences : CMLS. 2015;72:557–581. doi: 10.1007/s00018-014-1762-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu YC, Yeh WC, Ohashi PS. LPS/TLR4 signal transduction pathway. Cytokine. 2008;42:145–151. doi: 10.1016/j.cyto.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 48.Horvai AE, Xu L, Korzus E, Brard G, Kalafus D, Mullen TM, Rose DW, Rosenfeld MG, Glass CK. Nuclear integration of JAK/STAT and Ras/AP-1 signaling by CBP and p300. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:1074–1079. doi: 10.1073/pnas.94.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim RH, Flanders KC, Birkey Reffey S, Anderson LA, Duckett CS, Perkins ND, Roberts AB. SNIP1 inhibits NF-kappa B signaling by competing for its binding to the C/H1 domain of CBP/p300 transcriptional co-activators. The Journal of biological chemistry. 2001;276:46297–46304. doi: 10.1074/jbc.M103819200. [DOI] [PubMed] [Google Scholar]
- 50.Zhuang S. Regulation of STAT signaling by acetylation. Cellular signalling. 2013;25:1924–1931. doi: 10.1016/j.cellsig.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mukherjee SP, Behar M, Birnbaum HA, Hoffmann A, Wright PE, Ghosh G. Analysis of the RelA:CBP/p300 interaction reveals its involvement in NF-kappaB-driven transcription. PLoS biology. 2013;11:e1001647. doi: 10.1371/journal.pbio.1001647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Contzler R, Regamey A, Favre B, Roger T, Hohl D, Huber M. Histone acetyltransferase HBO1 inhibits NF-kappaB activity by coactivator sequestration. Biochemical and biophysical research communications. 2006;350:208–213. doi: 10.1016/j.bbrc.2006.09.030. [DOI] [PubMed] [Google Scholar]