Abstract
Throughout the animal kingdom chemical senses are one of the primary means by which organisms make sense of their environment. To achieve perception of complex chemosensory stimuli large repertoires of olfactory and gustatory receptors are employed in bony vertebrates, which are characterized by high evolutionary dynamics in receptor repertoire size and composition. However, little is known about their evolution in earlier diverging vertebrates such as cartilaginous fish, which include sharks, skates, rays, and chimeras. Recently, the olfactory repertoire of a chimera, elephant shark, was found to be curiously reduced in odorant receptor number. Elephant sharks rely heavily on electroreception to localize prey; thus, it is unclear how representative their chemosensory receptor repertoire sizes would be for cartilaginous fishes in general. Here, we have mined the genome of a true shark, Scyliorhinus canicula (catshark) for olfactory and gustatory receptors, and have performed a thorough phylogenetic study to shed light on the evolution of chemosensory receptors in cartilaginous fish. We report the presence of several gustatory receptors of the TAS1R family in catshark and elephant shark, whereas TAS2R receptors are absent. The catshark olfactory repertoire is dominated by V2R receptors, with 5–8 receptors in the other three families (OR, ORA, TAAR). Species-specific expansions are mostly limited to the V2R family. Overall, the catshark chemosensory receptor repertoires are generally similar in size to those of elephant shark, if somewhat larger, showing similar evolutionary tendencies across over 400 Myr of separate evolution between catshark and elephant shark.
Keywords: catshark, Scyliorhinus canicula, chimera, phylogeny, evolution, taste receptor
Main Text
Catshark Possess Five of the Six Major Vertebrate Chemosensory Receptor Families
Bony vertebrates exhibit four major families of olfactory receptors (OR, TAAR, ORA, V2R) and two gustatory GPCR families, TAS1R and TAS2R (Bachmanov and Beauchamp 2007). We have performed a recursive search in the preliminary draft genome of catshark, Scyliorhinus canicula to delineate its complete chemosensory receptor repertoire, using representative protein sequences from all six families in several species as initial queries. Phylogenetic analysis was performed using a maximum likelihood approach, for details see Materials and Methods.
For all OR families catshark as well as elephant shark genes could be identified, with the exception of T2R receptors. Since the closely related ORA receptors were present, it is unlikely that t2r genes were not found for technical reasons. We conclude that T2R receptors are absent in both species, and possibly in all cartilaginous fish, for elephant shark consistent with earlier observations (Grus and Zhang 2009).
Our analysis identified between 5 and 40 receptors per chemosensory receptor family in catshark (table 1). Additionally, we found some new TAARs, ORs, V2Rs, and TAS1R gene sequences in the elephant shark genome beyond those previously published (Grus and Zhang 2009; Niimura 2009b; Venkatesh et al. 2014). In total, the catshark chemosensory receptor repertoire encompasses 65 genes, which is slightly larger than the elephant shark repertoire with 54 genes. Both are similar to the size of the chemosensory repertoire of the sea lamprey, which has been given as 59 genes (Libants et al. 2009), but several to many times smaller than the repertoires of bony vertebrates (Niimura and Nei 2006) suggesting that gene birth events are comparatively rare in jawless and cartilaginous fish chemosensory receptor families compared with the bony fish lineage, and in particular its tetrapod branch.
Table 1.
Chemosensory Receptor Repertoire Sizes
| Gene Family | No. of Genes in Catshark | No. of Genes in Elephant Shark | No. of Genes in Mouse | No. of Genes in Zebrafish |
|---|---|---|---|---|
| OR | 8 (1)b | 7a | 1037c | 154c |
| TAAR | 5b | 5a | 15d | 112d |
| ORA/V1R | 6b | 4a | 211c | 7e |
| V2R | 35b | 34a | 121c | 58f |
| V2RL | 5b | 3a | 0 | 2f |
| TAS1R | 6b | 4a | 3c | 4g |
| T2R | 0b | 0 | 33c | 4c |
Note.—Total gene numbers are given, in parentheses the number of pseudogenes.
Additional genes identified (elephant shark, three TAAR, four TAS1R, one OR, two V2R, two V2RL) compared with previously published numbers (Niimura 2009b; Venkatesh et al. 2014).
Refer to genes newly identified here (catshark). Superscripts refer to
Hussain et al. (2009).
Ahuja et al. (2018).
The Catshark Chemosensory Receptor Repertoire Is Dominated by V2Rs
In bony fish and tetrapods ORs constitute the dominant chemosensory family (Niimura and Nei 2006). It had therefore been surprising, when only six or genes were reported in the elephant shark genome (Venkatesh et al. 2014), but it had been unclear, how representative this reduced repertoire was for cartilaginous fish in general and true sharks in particular. Here, we report one additional or gene in elephant shark and a very similar size of eight or genes in catshark (table 1). This is considerably less than even the sea lamprey OR repertoire, reported as 27 genes (Libants et al. 2009) and suggests that the OR family has not undergone any major radiation in cartilaginous fish.
In contrast, 40 v2r genes were observed in the catshark genome, slightly larger than the 37 genes we detected in the elephant shark genome (table 1). This is roughly comparable to mammalian and fish repertoire sizes (Young and Trask 2007; Ahuja et al. 2018) and more than all other chemosensory families combined. v2r genes have not been found in jawless fish (Libants et al. 2009), thus the origin of the family appears to be in the most recent common ancestor (MRCA) of jawed fish.
Phylogenetic analysis shows a small subgroup of catshark and elephant shark v2r genes orthologous to zebrafish V2R-like OlfCa1 and OlfCb1 (fig. 1). We therefore suggest to name these genes as v2rl, V2R-like. The v2rl subgroup is most closely related to type 1 taste receptors, TAS1Rs, from which they segregate with maximal branch support (fig. 1a). We report five such genes for catshark and three for elephant shark (fig. 1b). The maximal branch support within the V2RL clade allows the deduction of two ancestral v2rl genes already in the MRCA of cartilaginous and bony fish. Subsequently, small gene expansions specific to the cartilaginous lineage generated the extant v2rl gene numbers, which are considerably larger than present in zebrafish (two genes, olfCa1, olfCb1) and mammals (one gene, gprc6).
Fig. 1.
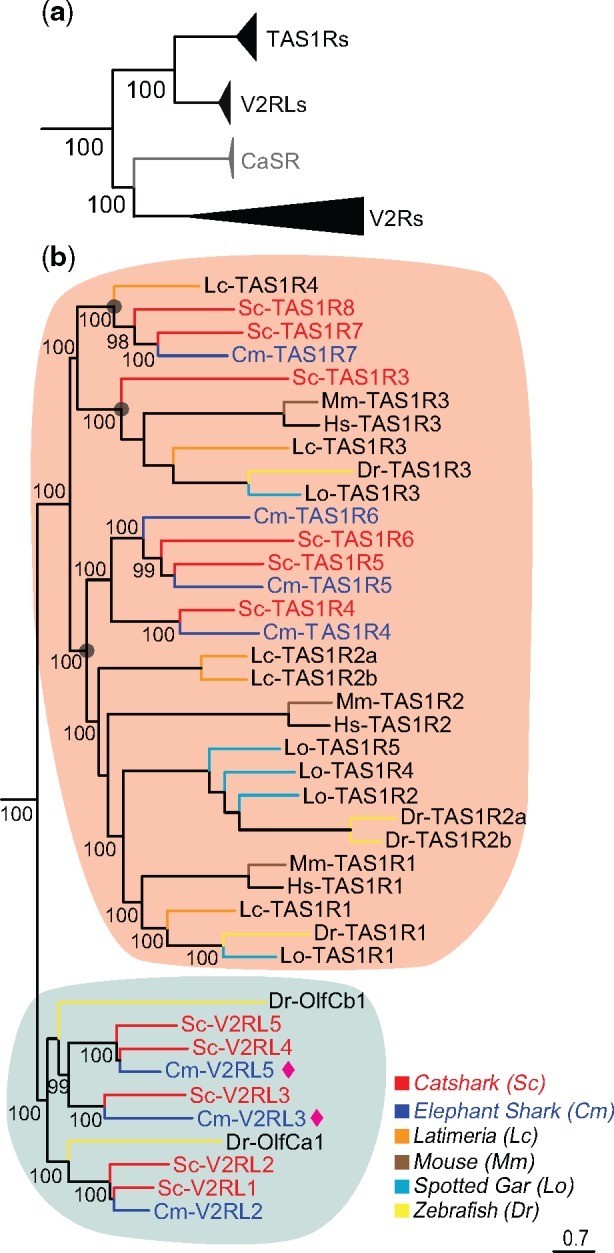
—Two to three ancestral tas1r genes already present in the MRCA of cartilaginous and bony vertebrates. Phylogenetic tree of TAS1Rs (orange), V2RLs (grey) and V2Rs of catshark and elephant shark. CaSR and TAS1Rs as outgroups for V2Rs and V2RLs respectively. (a) All gene groups are shown in collapsed representation to emphasize the basal nodes. Note that v2rl genes are the sister group to TAS1Rs and CaSR represents the sister group for the main group of V2Rs. The phylogenetic tree was generated using a maximum likelihood method (PhyML-aLRT) with SPR setting for tree optimization and chi square-based aLRT for branch support (given as percentage). Note the maximal branch support for all nodes. (b) The TAS1R and V2RL node of (a) shown in detail. Branch support shown as percentage. Branches are color-coded for catshark (red) and elephant shark (blue) along with zebrafish (yellow), mouse (brown), spotted gar (cyan), and Latimeria (orange). Transparent grey circles denote the clades corresponding to the three predicted ancestral tas1r genes in the MRCA of cartilaginous and bony vertebrates. Two new v2rl genes were found in elephant shark (purple spades).
The main group of V2Rs is most closely related to the calcium-sensing receptor (CaSR), from which it segregates with maximal branch support (fig. 1a). There are 35 catshark genes in this group and nearly the same number (34) of elephant shark genes (table 1). Interestingly these numbers are reached by several species-specific gene duplications generating subclades of up to 7 catshark and 12 elephant shark genes, which just happen to result in a very similar total number. In several cases, direct orthologs of catshark and elephant shark V2Rs are observed, for example, V2R2 (fig. 2).
Fig. 2.
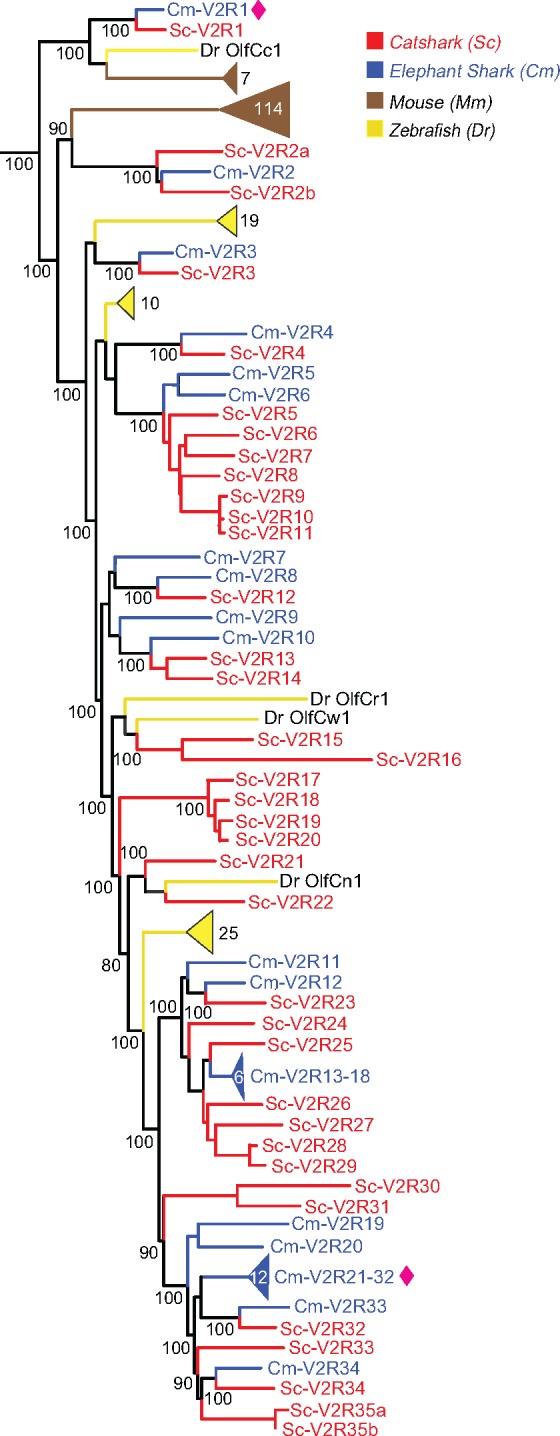
—The catshark chemosensory receptor repertoire is dominated by V2Rs. Largest family of V2Rs in catshark comprising of thirty-five genes in catshark (red) were compared with thirty-four elephant shark V2Rs (blue) along with zebrafish (yellow), and mouse (brown). The most basal gene, Sc-V2R1, Cm-V2R1, is orthologous to zebrafish OlfCc1 and the mammalian V2R2 subfamily. The phylogenetic tree was generated as described in figure 1 and branch support is given as percentage. Sequences were named according to named orthologs or closest paralogs from other species otherwise according to phylogenetic relationship. Sequences are named a, b where exon 3 and exon 6 might be derived from the same gene. New genes in elephant shark are marked with purple spade.
The most basal gene, Sc-V2R1, Cm-V2R1, is orthologous to zebrafish OlfCc1 and the mammalian V2R2 subfamily (fig. 2). It may serve as coreceptor in zebrafish and mouse (Martini et al. 2001; DeMaria et al. 2013) and it will be interesting to investigate, whether such a function might also be conserved in cartilaginous fish. The second most basal gene, Sc-V2R2, Cm-V2R2, is orthologous to all remaining mouse v2r genes (a single clade), but appears to have been lost in zebrafish (fig. 2). The remaining catshark/elephant gene expansion is intermingled with six zebrafish clades comprising 1–19 olfC genes, suggesting a similar number of ancestral v2r genes in the MRCA of cartilaginous and bony fish, all of which appear to have been lost in tetrapods (fig. 2).
Two to Three Gustatory tas1r Genes Present in the MRCA of Cartilaginous and Bony Vertebrates
The gustatory tas1r genes are close relatives of the olfactory v2r genes and, like these, belong to class C GPCRs, which are characterized by a large, extracellular N-terminus and a characteristic six exon structure (Sainz et al. 2001). The mammalian taste receptor 1 (TAS1R) family is best understood. It comprises three members TAS1R1, TAS1R2, and TAS1R3 (Voigt et al. 2012), which hetero-oligomerize to TAS1R1/TAS1R3 and TAS1R2/TAS1R3, functioning as umami and sweet taste receptor, respectively (Zhao et al. 2003). Teleost fish possess the direct orthologs of TAS1R1 and TAS1R3, but have expanded TAS1R2 to 2–3 genes (Ishimaru et al. 2005). Interestingly, the TAS1R2/TAS1R3 hetero-oligomers of teleosts also react to amino acids, not to sugars like their mammalian counterparts (Oike et al. 2007). In Latimeria, one TAS1R1, two TAS1R2, and two TAS1R3 have been described (Picone et al. 2014). The evolutionary relationships of these genes are not clear so far, because the TAS1R repertoire of earlier-diverging species has not been available so far.
Here, we identified in total six tas1r genes in the catshark genome, and four in the elephant shark. We assume these numbers to be final for both catshark and elephant shark, because the current genomic coverage is 200× and 19.25×, respectively (Wyffels et al. 2014). All elephant shark tas1r genes possess orthologs in catshark. Interestingly, one of the catshark tas1r genes, Sc-TAS1R3, appears to have been lost in elephant shark. Furthermore, catshark TAS1R7 and TAS1R8 appear to result from a gene duplication within the true shark lineage, because elephant shark has a single gene, TAS1R7, in this subnode. All confirmed TAS1R candidates show the characteristic exon structure (data not shown), although due to the preliminary nature of the genomic assembly not all six exons could be identified in each case.
The phylogeny shown here allows some conclusions concerning the origin and relationship of mammalian and teleost TAS1R receptors. Mouse and zebrafish TAS1R3 possess a direct ortholog in catshark, Sc-TAS1R3 (fig. 1b), suggesting this gene to be already present in the MRCA of cartilaginous and bony vertebrates, whereas TAS1R1 and TAS1R2 appear to have originated in a duplication event within the bony lineage (fig. 1b). In the 420 Myr since divergence of chimeras and true sharks (Heinicke et al. 2009) the evolutionary dynamic has been very small (three gene birth event in catshark, two in elephant shark, all except one in the MRCA of true sharks and chimeras), which parallels the slow evolution of this family in bony fish and tetrapods. This is very different from the evolutionary history of the closely related V2Rs, which often exhibit species-specific repertoires (Hashiguchi and Nishida 2006). The intermingling of cartilaginous fish TAS1Rs with bony fish TAS1Rs in the phylogenetic tree (fig. 1b) allows to estimate the number of ancestral TAS1Rs in the MRCA of cartilaginous and bony vertebrates. The most parsimonious explanation of the observed tree assumes a gene loss event for bony fish in the Sc-TAS1R7, eight subclade, which results in a prediction of three ancestral tas1r genes in the MRCA of cartilaginous and bony vertebrates (fig. 1). The origin of the TAS1R family cannot be exactly deduced, but should have happened within the jawed lineage, since TAS1Rs were not found in lamprey (Grus and Zhang 2009).
Small Repertoires for OR, TAAR, and ORA Receptor Families in Catshark and Elephant Shark
OR genes are the largest gene family in bony vertebrates (Niimura and Nei 2006), but have only undergone very limited gene expansion in cartilaginous fish (table 1; fig. 3). In mammals, class I and class II ORs have been distinguished, with class I orthologous to a zebrafish subfamily of five genes, and class II possessing a single zebrafish ortholog, Dr3OR5.4. Both classes exhibit a single catshark ortholog gene, Sc-OR1 and Sc-OR2, respectively (fig. 3) suggesting the origin of these two genes in the MRCA of cartilaginous and bony fish. Three more zebrafish genes or subclades are orthologous to a catshark and/or elephant shark gene, suggesting in total the presence of at least five or genes in the MRCA of cartilaginous and bony fish, of which elephant shark appears to have lost two genes and catshark one. Thirty-two putatively functional OR genes were identified from the sea lamprey genome (Niimura 2009b), whereas in elephant shark we identified seven ORs (Cm-OR1 and Cm-OR3-8). Previously in elephant shark or8 and or1 gene have been reported as real ORs but others, or3-7 as nonORs (Niimura and Nei 2006; Venkatesh et al. 2014). Four elephant shark or genes have a direct ortholog in catshark, that is, for these four gene pairs not a single gene birth or death event happened in the last 420 Myr (Heinicke et al. 2009) another gene is a singleton in elephant shark (Cm-OR7), but has undergone a single duplication in catshark, resulting in Sc-OR7, Sc-OR8 (fig. 3). Overall the evolutionary dynamics of the OR family appear to be extremely limited in cartilaginous fish, in stark contrast to the very dynamic evolution in bony vertebrates.
Fig. 3.
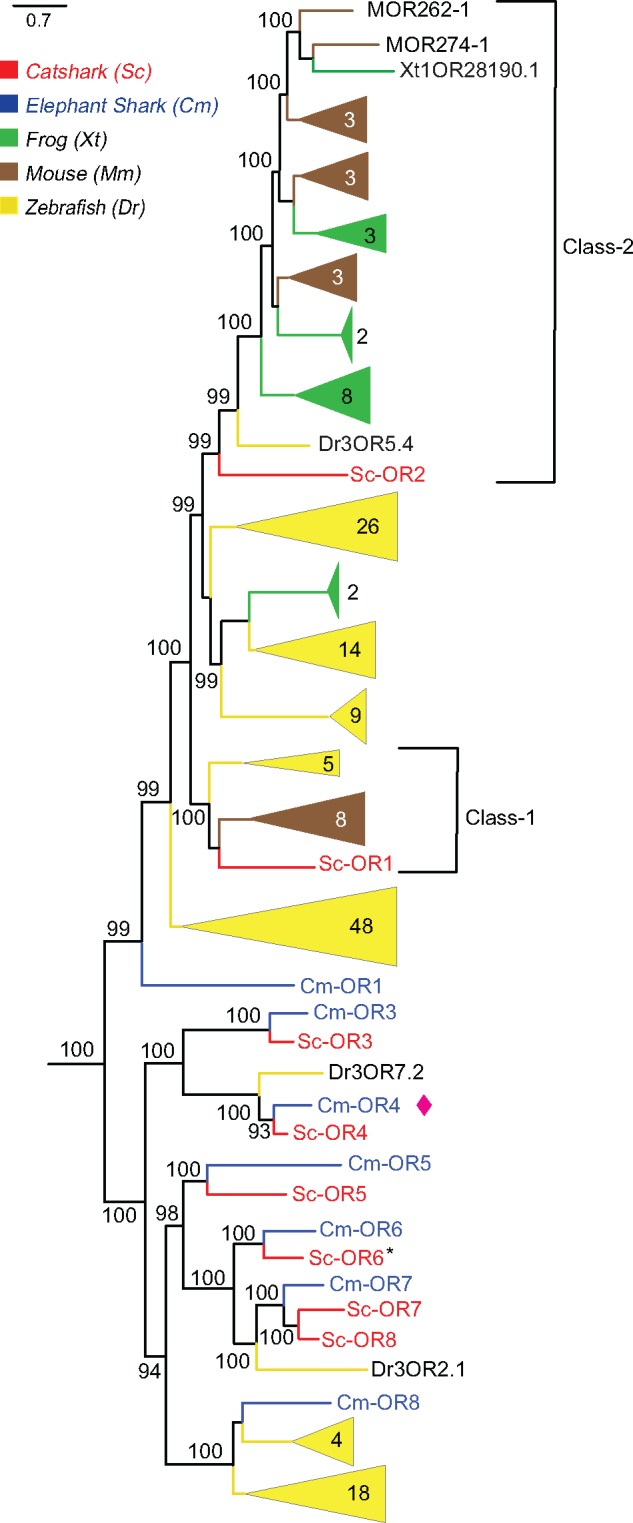
—Sharks possess a small odorant receptor repertoire. Eight or genes of catshark (red), were compared with elephant shark (blue), frog (green), zebrafish (yellow), and mouse (brown). Phylogenetic tree was generated as described in figure 1 and branch support is given as percentage. Potential pseudogenes indicated by asterisk or genes are named by class to which they belong, eight genes are labelled one to eight. One new or gene found in elephant shark marked with purple spade.
The TAAR family is large in teleost fish, of medium size in tetrapods, and was reported as just two genes in elephant shark, based on analysis of an initial assembly (Hussain et al. 2009). We found five taar genes for elephant shark (Cm-Taar1a-Cm-Taar4) and report a similar size of five genes for catshark (Sc-Taar1a-Sc-Taar4), see table 1. Figure 4 shows the phylogeny of representative TAARs from zebrafish, mouse, frog, catshark, and elephant shark. This phylogeny clusters TAAR into three monophyletic groups. The most basal group tarl3 and tarl4 genes is clearly clustered separately from others. However, two of these genes in catshark (tarl 3 and tarl 4) and one in elephant shark (tarl 4), do not exhibit the characteristic TAAR motif present in TM7 (Hussain et al. 2009). Since they are a sister group to the validated taar genes (taar1a-1b), which do possess the motif, we refer to them as taar-like genes (tarl). There is a clear ortholog relationship between cartilaginous taar1 and teleost taar1 genes. taar 3-4 genes of sharks are more similar to vertebrate taar 2-4. This could point to the retention of ancestral characteristics by taar2-4.
Fig. 4.
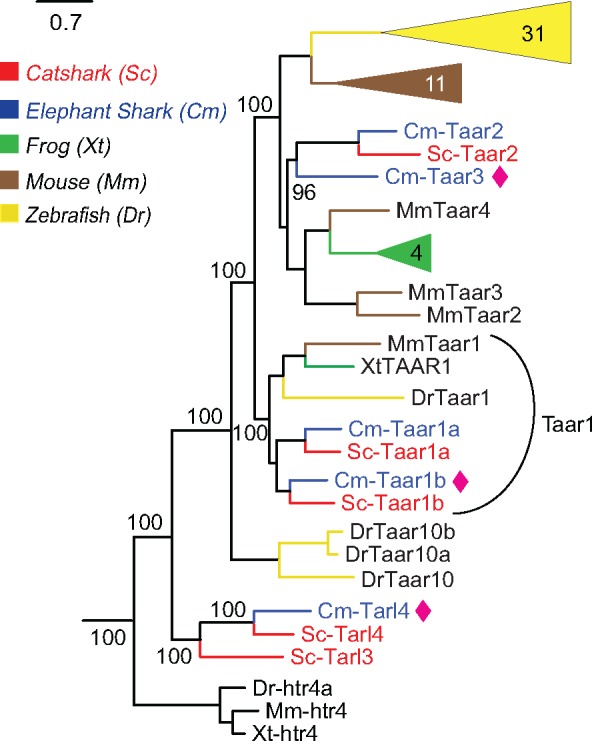
—The TAAR repertoire of catshark and elephant shark consists of five genes each. Phylogenetic tree of five taar genes of both catshark (red) and elephant shark (blue), frog, zebrafish, and mouse (species and color code as given in fig. 2). The phylogenetic tree was generated as described in figure 1 and branch support is given as percentage. TAARs are named according to class and orthologs they are located with. Aminergic receptors are used as outgroup, only the closest outgroup (htr4) is shown.
The V1R/ORA gene family also shows opposing evolutionary characteristics in tetrapods versus teleosts. Here, the tetrapod families can be very large, but the teleost family is highly conserved, with 6–7 genes in many species (Zapilko and Korsching 2016). We identified six ora genes in catshark, and confirmed four ORAs for elephant shark (table 1). This conforms to the general tendency for catshark receptor repertoires to be somewhat larger than those of elephant shark. Nevertheless, catshark has lost one of the genes present in elephant shark, ORA1, consistent with both gene birth and gene death events sculpting the ORA repertoire in catshark. Interestingly, this is the gene giving rise to all of tetrapod ORAs (fig. 5). The remaining three elephant shark genes possess direct orthologs in catshark, whose different terminal branch lengths suggest individually different evolutionary rates for these three genes (fig. 5). All of these genes are lost in tetrapods, with the exception of a single Xenopus gene, ORA15. Furthermore, we identified three additional ora genes in catshark that cluster with teleost ora5 and ora6. The absence of such genes in lamprey (Grus and Zhang 2009) and elephant shark (Venkatesh et al. 2014), confirmed here, had raised doubts as to the evolutionary origin of ORA5-6 compared with ORA1-4, whose orthologs are present in elephant shark. Now it can be concluded that the ancestral gene of the ORA5/6 clade was already present in the MRCA of cartilaginous fish and bony fish.
Fig. 5.
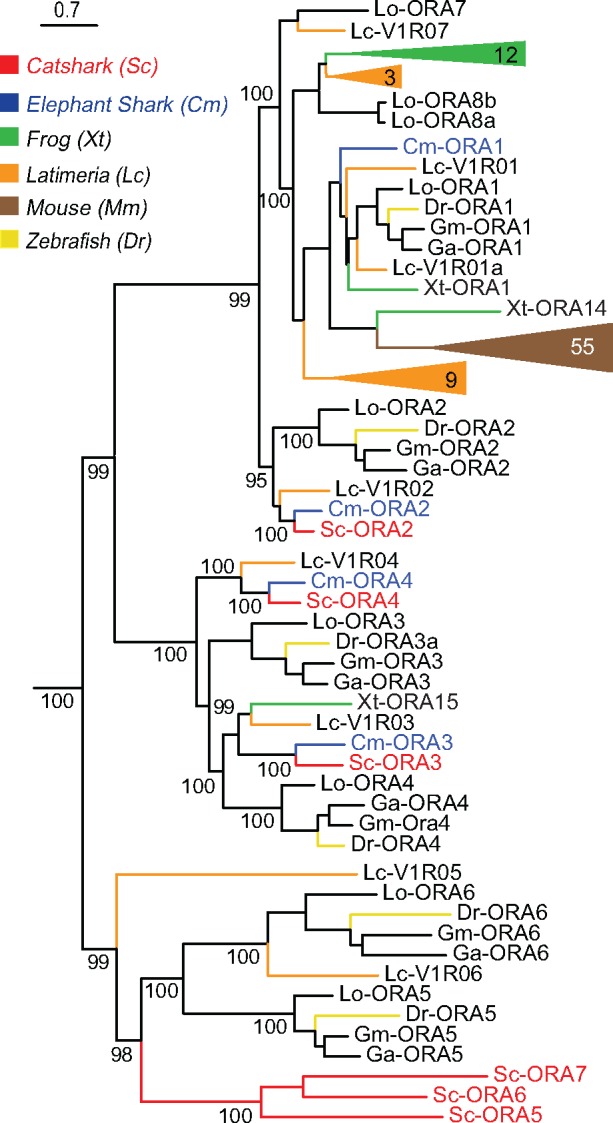
—The catshark ORA repertoire shows an ancient origin of the ORA5/6 subclade. Six ora genes from catshark (red) and four from elephant shark (blue) were used along with the followings: frog, zebrafish, Latimeria, and mouse (species and color code as given in fig. 2). The phylogenetic tree was generated as described in figure 1 and branch support is given as percentage. Catshark ORAs were named according to the orthologs from two to seven. TAS2Rs were used as outgroup (not shown here).
Taken together, all three families (OR, TAAR, ORA) show only minor gene birth and death events in a shark and a chimera species, in stark contrast to the evolutionary dynamics of these families in bony vertebrates. Thus, in sharks these three receptor families seem to play a much reduced role in olfaction as compared with bony vertebrates. In contrast, the shark V2R family exhibits extensive gene birth events very similar to the evolutionary characteristics of V2Rs in bony vertebrates, consistent with the hypothesis that odor detection in both true sharks and chimeras depends heavily on the V2R family of ORs. Although no shark V2Rs have been deorphanized so far, they may well comprise amino acid receptors like their teleost counterparts (Speca et al. 1999; Oike et al. 2007). Thus, one may expect odor detection via V2Rs to help in food localization. The large evolutionary divergence of 420 Myr notwithstanding, both catshark and elephant shark are benthic predators of small invertebrates (Cox and Francis 1997; Valls et al. 2011), consistent with an important role of V2Rs in prey detection.
The olfactory organ of elephant shark has not been described so far, and together with the known specialization in electroception of this species (Didier 1995; Lisney 2010) this raised doubts how representative the OR repertoire of this species might be. Catshark, on the other hand, exhibit a complex olfactory organ (Theisen et al. 1986) and do not appear as specialized for electroception as elephant shark. The overall similarity of the chemosensory repertoires of catshark and elephant shark we describe here suggests now that the elephant shark repertoire is no outlier. The slightly larger chemosensory receptor repertoire of catshark is consistent with a somewhat larger dependence on olfaction for catshark.
Materials and Methods
In order to delineate the olfactory and gustatory genes, scaffolds (sf., see supplementary data set S1, Supplementary Material online) from the draft of the catshark genome (to be published elsewhere) and recent elephant shark genome (Venkatesh et al. 2014) were obtained by genome-wide searches using TBlastN with the representative TAS1R, TAS2R, OR, TAAR, ORA, and V2R sequences from mouse, frog, elephant shark, Latimeria and zebrafish as queries, and recursively in follow-up searches. For TAS1R phylogeny we additionally searched for spotted gar sequences. Homology regions above 200 amino acid length were considered further. Splicing predictions were made by comparing related protein sequences to genomic DNA sequences with the online-tool GeneWise (Birney et al. 2004). Sequence data used in this article are included in supplementary file (data set S2), Supplementary Material online. Sequences were aligned with MAFFT 7 (Katoh and Standley 2013) an online version of the multiple alignment tool MAFFT (Katoh et al. 2002) using the E-INS-I strategy with the default parameters. Clustal Omega (Sievers et al. 2011) was also used for alignment. The multiple sequence alignment was edited using Gap Strip Squeeze to remove regions with gaps in over 90% of sequences (https://www.hiv.lanl.gov/content/sequence/GAPSTREEZE/gap.html, last accessed January 22, 2019).
The phylogenetic trees were calculated using a Maximum likelihood algorithm, PhyML-aLRT with SPR setting for tree optimization and chi square-based aLRT (Guindon et al. 2010) for branch support on Phylemon server available online (Sanchez et al. 2011). Branch support above 80% was considered significant. TAS1R, CasR, nonOR rhodopsin-like GPCR genes, htr, and T2Rs of zebrafish, mouse, xenopus, human and latimeria served as outgroups for V2RL, V2R, OR, TAAR, and ORA, respectively. Treefiles for figure 1a, figure 2 (Treefile 1); figure 1b (Treefile 2); figure 3 (Treefile 3); figure 4 (Treefile 4); figure 5 (Treefile 5) are given in supplementary file (data set S3), Supplementary Material online. Trees were drawn using FigTree (http://tree.bio.ed.ac.uk/software/figtree/, last accessed January 22, 2019). Newly predicted genes were named according to previously named orthologs or closest paralogs from other species, starting with more basal genes. Gene with one or more stop codons was labelled as pseudogene. One or gene may either represent pseudogenes or databank inaccuracies due to the preliminary assembly (fig. 2, supplementary data set 1, Supplementary Material online). Fifteen genes are full or nearly full length (above 700 aa), three are partial (between 550 and 700 aa), fifteen and nine sequences were restricted to one of the large exons (exon 3 and exon 6, respectively). In those cases, where exon 3 and exon 6 might be derived from the same gene, we distinguished with a letter, for example, Sc-V2R2a and Sc-V2R2b.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank the Genoscope-Centre National de Séquençage (Evry, France) for sequencing the catshark draft genome used in this work, Hélène Mayeur (UMR 7232, Observatoire Océanologique, Banyuls sur Mer, France) and Jean-Marc Aury (Genoscope) for assembling these data. This work was supported by the German Science foundation (grant KO 1046/7-1 to S.I.K.).
Author Contributions
The experiments were conceived by S.F., S.M., and S.I.K., designed and performed by K.S. and A.S.S. Illustrations were drafted by K.S. and S.I.K. Data analysis was done by K.S., A.S.S., and K.S., S.F. and S.I.K. wrote the paper.
Literature Cited
- Ahuja G, et al. 2018. Overlapping but distinct topology for zebrafish V2R-like olfactory receptors reminiscent of odorant receptor spatial expression zones. BMC Genomics 19:383.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alioto TS, Ngai J.. 2006. The repertoire of olfactory C family G protein-coupled receptors in zebrafish: candidate chemosensory receptors for amino acids. BMC Genomics 7:309.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmanov AA, Beauchamp GK.. 2007. Taste receptor genes. Annu Rev Nutr. 27:389–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Clamp M, Durbin R.. 2004. GeneWise and genomewise. Genome Res. 14(5):988–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox GJ, Francis M.. 1997. Sharks and rays of New Zealand. New Zealand: Canterbury University Press. [Google Scholar]
- DeMaria S, et al. 2013. Role of a ubiquitously expressed receptor in the vertebrate olfactory system. J Neurosci. 33(38):15235–15247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didier DA. 1995. Phylogenetic systematics of extant chimaeroid fishes (Holocephali, Chimaeroidei). New York, NY: American Museum Novitates; no. 3119. [Google Scholar]
- Grus WE, Zhang J.. 2009. Origin of the genetic components of the vomeronasal system in the common ancestor of all extant vertebrates. Mol Biol Evol. 26(2):407–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321. [DOI] [PubMed] [Google Scholar]
- Hashiguchi Y, Nishida M.. 2006. Evolution and origin of vomeronasal-type odorant receptor gene repertoire in fishes. BMC Evol Biol. 6:76.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinicke M, Naylor G, Hedges S.. 2009. Cartilaginous fishes (Chondrichthyes). Timetree Life. 9:320–327. [Google Scholar]
- Hussain A, Saraiva LR, Korsching SI.. 2009. Positive Darwinian selection and the birth of an olfactory receptor clade in teleosts. Proc Natl Acad Sci U S A. 106(11):4313–4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru Y, et al. 2005. Two families of candidate taste receptors in fishes. Mech Dev. 122(12):1310–1321. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T.. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libants S, et al. 2009. The sea lamprey Petromyzon marinus genome reveals the early origin of several chemosensory receptor families in the vertebrate lineage. BMC Evol Biol. 9:180.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisney TJ. (Lisney2010 co-authors). 2010. A review of the sensory biology of chimaeroid fishes (Chondrichthyes; Holocephali). Rev Fish Biol Fish. 20(4):571–590. [Google Scholar]
- Martini S, Silvotti L, Shirazi A, Ryba NJ, Tirindelli R.. 2001. Co-expression of putative pheromone receptors in the sensory neurons of the vomeronasal organ. J Neurosci. 21(3):843–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y. 2009a. Evolutionary dynamics of olfactory receptor genes in chordates: interaction between environments and genomic contents. Hum Genomics. 4(2):107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y. 2009b. On the origin and evolution of vertebrate olfactory receptor genes: comparative genome analysis among 23 chordate species. Genome Biol Evol. 1:34–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y, Nei M.. 2006. Evolutionary dynamics of olfactory and other chemosensory receptor genes in vertebrates. J Hum Genet. 51(6):505–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oike H, et al. 2007. Characterization of ligands for fish taste receptors. J Neurosci. 27(21):5584–5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picone B, et al. 2014. Taste and odorant receptors of the coelacanth – a gene repertoire in transition. J Exp Zool B Mol Dev Evol. 322(6):403–414. [DOI] [PubMed] [Google Scholar]
- Sainz E, Korley JN, Battey JF, Sullivan SL.. 2001. Identification of a novel member of the T1R family of putative taste receptors. J Neurochem. 77(3):896–903. [DOI] [PubMed] [Google Scholar]
- Sanchez R, et al. 2011. Phylemon 2.0: a suite of web-tools for molecular evolution, phylogenetics, phylogenomics and hypotheses testing. Nucleic Acids Res. 39(Web Server issue):W470–W474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraiva LR, Korsching SI.. 2007. A novel olfactory receptor gene family in teleost fish. Genome Res. 17(10):1448–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 7:539.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speca DJ, et al. 1999. Functional identification of a goldfish odorant receptor. Neuron 23(3):487–498. [DOI] [PubMed] [Google Scholar]
- Theisen B, Zeiske E, Breucker H.. 1986. Functional morphology of the olfactory organs in the spiny dogfish (Squalus acanthias L.) and the small-spotted catshark (Scyliorhinus canicula (L.)). Acta Zool. 67(2):73–86. [Google Scholar]
- Valls M, Quetglas A, Ordines F, Moranta J.. 2011. Feeding ecology of demersal elasmobranchs from the shelf and slope off the Balearic Sea (Western Mediterranean). Sci Mar. 75:7. [Google Scholar]
- Venkatesh B, et al. 2014. Elephant shark genome provides unique insights into gnathostome evolution. Nature 505(7482):174–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt A, et al. 2012. Genetic labeling of Tas1r1 and Tas2r131 taste receptor cells in mice. Chem Senses. 37(9):897–911. [DOI] [PubMed] [Google Scholar]
- Wyffels J, et al. 2014. SkateBase, an elasmobranch genome project and collection of molecular resources for chondrichthyan fishes. F1000Res. 3:191.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JM, Trask BJ.. 2007. V2R gene families degenerated in primates, dog and cow, but expanded in opossum. Trends Genet. 23(5):212–215. [DOI] [PubMed] [Google Scholar]
- Zapilko V, Korsching SI.. 2016. Tetrapod V1R-like ora genes in an early-diverging ray-finned fish species: the canonical six ora gene repertoire of teleost fish resulted from gene loss in a larger ancestral repertoire. BMC Genomics 17:83.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao GQ, et al. 2003. The receptors for mammalian sweet and umami taste. Cell 115(3):255–266. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


