Abstract
Background
One-third of depressed patients develop treatment-resistant depression with the related sequelae in terms of poor functionality and worse prognosis. Solid evidence suggests that genetic variants are potentially valid predictors of antidepressant efficacy and could be used to provide personalized treatments.
Methods
The present review summarizes genetic findings of treatment-resistant depression including results from candidate gene studies and genome-wide association studies. The limitations of these approaches are discussed, and suggestions to improve the design of future studies are provided.
Results
Most studies used the candidate gene approach, and few genes showed replicated associations with treatment-resistant depression and/or evidence obtained through complementary approaches (e.g., gene expression studies). These genes included GRIK4, BDNF, SLC6A4, and KCNK2, but confirmatory evidence in large cohorts was often lacking. Genome-wide association studies did not identify any genome-wide significant association at variant level, but pathways including genes modulating actin cytoskeleton, neural plasticity, and neurogenesis may be associated with treatment-resistant depression, in line with results obtained by genome-wide association studies of antidepressant response. The improvement of aggregated tests (e.g., polygenic risk scores), possibly using variant/gene prioritization criteria, the increase in the covering of genetic variants, and the incorporation of clinical-demographic predictors of treatment-resistant depression are proposed as possible strategies to improve future pharmacogenomic studies.
Conclusions
Genetic biomarkers to identify patients with higher risk of treatment-resistant depression or to guide treatment in these patients are not available yet. Methodological improvements of future studies could lead to the identification of genetic biomarkers with clinical validity.
Keywords: treatment-resistant depression, gene, pharmacogenomics, GWAS, antidepressant
Introduction
Major depressive disorder (MDD) is a primary health issue at both individual level and socio-economic level. In adolescents and young adults (aged between 15 and 39 years), depression is the third-leading cause of disability, while in middle-aged adults depression was reported to be the second cause of disability on a global scale (GBD 2015 Disease and Injury Incidence and Prevalence Collaborators, 2016). The heavy burden of the disease can be attributed both to the high lifetime prevalence (~13%) and to the insufficient response rates to antidepressant treatments. Complete symptom remission is achieved in approximately one-third of patients, while another approximately one-third develops treatment-resistant depression (TRD), but TRD estimates were up to 40% in other samples (Trivedi et al., 2006; Souery et al., 2011). The high percentage of treatment failure or incomplete remission is probably a consequence of intrinsic biological and environmental heterogeneity among MDD patients (Gratten et al., 2014), suggesting that biomarkers of antidepressant response would be useful to guide treatment at the individual level.
Antidepressant response was demonstrated to have a relevant genetic component by family studies and more recent approaches such as Genome-wide Complex Trait Analysis (Tansey et al., 2013). For this reason, genetic variants are considered theoretically optimal biomarkers to provide personalized antidepressant treatments and to reduce the proportion of patients that develops TRD.
TRD may have a different genetic make-up compared with milder nonresponse cases, but previous pharmacogenetic studies were mainly focused on a generic definition of response that did not consider the number of failed antidepressants. A significant genetic heterogeneity was reported among different MDD samples (Gratten et al., 2014), in line with the clinical observation that MDD is a heterogeneous entity. TRD patients were demonstrated to have some distinctive clinical features compared with non-TRD patients, such as higher symptom severity, more frequent suicidal risk, and comorbidity with anxiety (Souery et al., 2007; De Carlo et al., 2016; Kautzky et al., 2017). Despite the fact that these features may depend also on environmental factors or non-genetic biological factors, other clinical subtypes of MDD probably have a genetic basis, such as atypical vs typical MDD (Milaneschi et al., 2016). These findings together with the prognostic value of TRD suggest that TRD should be considered as a separate phenotype in genetic studies. On the ground of these observations, an increasing number of studies took into account not only antidepressant response/remission but also TRD or a measure of resistance stage (e.g., classes corresponding to the number/range of failed treatments). A relevant contribution to the field has been provided by The European Group for the Study of Resistant Depression, which has been working for over 15 years to study clinical and genetic variables associated with TRD, producing valuable data and anticipating a more recent spread of interest towards this phenotype (Schosser et al., 2012b).
TRD is usually defined as nonremission after at least 2 adequate antidepressant trials, but this standard definition may not reflect the underlying pathophysiological mechanisms or a reproducible set of genetic risk variants. The standard definition of TRD and the other available definitions have been formulated on the basis of clinical observations (Souery et al., 2007). For example, older definitions of TRD required nonremission to at least 2 adequate trials with antidepressants of different classes; then the different class criterium was abolished after several studies demonstrated no significant difference in switching to different types of treatments after the failure of the first treatment trial (Rush et al., 2008; Souery et al., 2011). Several more complex staging models have been developed, based on the duration of treatment, number, and type of treatments failed (McIntyre et al., 2014). Available genetic studies generally used the standard definition of TRD, but recent literature underlines that MDD and TRD are probably heterogeneous entities under the biological point of view, suggesting that classification systems should include information about the specific pathogenetic mechanisms involved (Akil et al., 2018), a concept that has been repeatedly underlined since the proposal of the Research Domain Criteria (Insel et al., 2010).
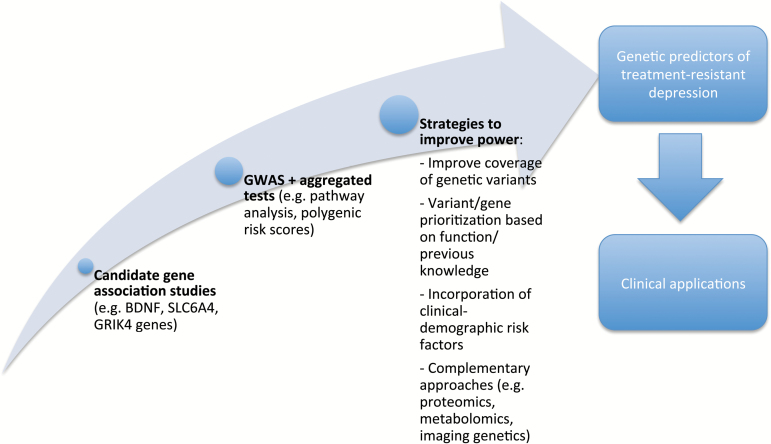
The present review provides an overview on the genetics of TRD, since the discussion on clinical relevance of this phenotype. Then, taking into account the pros and cons of previous studies, some possible strategies to improve future pharmacogenetic studies are discussed to contribute to the advances of this research field (Figure 1).
Figure 1.
Representation of previous approaches used in the study of the genetics of TRD (treatment-resistant depression) and possible strategies to implement in future studies to improve power of identifying and replicating significant associations.
Overview of Previous Findings
In the following 2 paragraphs, the results of genetic studies investigating TRD are summarized (Table 1). The most part of available studies used the candidate gene approach, that is, they genotyped relevant polymorphisms in genes having a known link with antidepressant mechanisms of action (pharmacodynamics) or metabolism (pharmacokinetics). More recently TRD was also investigated by some genome-wide association studies (GWAS). GWAS represent the technological answer to the hypothesis that antidepressant response has a polygenic nature, that is, thousands of polymorphisms across the genome are probably involved and many of them are supposedly outside genes or in genes with no known connection to antidepressant response. GWAS arrays polymorphisms throughout the genome, concentrated in relevant regions, and they can provide signals in previously unsuspected regions.
Table 1.
Summary of Genetic Studies Investigating TRD
| Gene | Polymorphisms | Sample Size | Main Findings | Reference |
|---|---|---|---|---|
| Glutamate ionotropic receptor NMDA type subunit 2B (GRIN2B) | rs1805502 | 178 TRD, 612 non-TRD, 779 HC | Increased risk of TRD in rs1805502 G allele carriers. | (Zhang et al., 2014) |
| Glutamate ionotropic receptor kainate type subunit 4 (GRIK4) | rs11218030, rs1954787 | 380 TRD, 247 non-TRD | Increased risk of TRD and psychotic symptoms during depressive episodes in rs11218030 G allele and rs1954787 GG genotype. | (Milanesi et al., 2015) |
| 100 TRD | Increased risk of non-response after ECT in rs11218030 G allele and rs1954787 GG genotype. | (Minelli et al., 2016) | ||
| Brain derived neurotrophic factor (BDNF) | rs6265 (Val66Met) | 62 TRD | Better response to ketamine in G (Val) allele. | (Laje et al., 2012) |
| 71 TRD | No association with ketamine response. | (Su et al., 2017) | ||
| 36 TRD | Better response to rTMS in G (Val) allele. | (Bocchio-Chiavetto et al., 2008) | ||
| BDNF, neurotrophic receptor tyrosine kinase 2 (NTRK2) | rs6265 (BDNF), rs1387923, rs2769605 and rs1565445 (NTRK2) | 644 non-TRD, 304 TRD | rs1565445 T allele, rs1565445 TT genotype, rs1565445 and rs1387923 T-T haplotype were associated with TRD. A genotypic combination at four loci in NTRK2 and BDNF (rs1387923-rs1565445- rs2769605-rs6265) was associated with TRD. | (Li et al., 2013) |
| CAMP responsive element binding protein 1 (CREB1) | rs2709376, rs2253206, rs7569963, rs7594560, rs4675690 | 119 non-TRD, 71 TRD | rs7569963 A allele, rs2253206-rs7569963 A-A and rs7569963-rs4675690 A-C haplotypes were associated with TRD. Negative results for the other SNPs. | (Serretti et al., 2011) |
| s889895, rs6740584, rs2551922, rs2254137 | 265 non-TRD, 102 TRD | No association was found for TRD. rs889895 GG was associated with remission. | (Calati et al., 2013) | |
| 147 non-TRD, 73 TRD | No association was found for TRD. rs2254137 AA was associated with remission. | (Fabbri et al., 2017) | ||
| Solute carrier family 6 member 4 (SLC6A4 or serotonin transporter) | 5-HTTLPR | 36 TRD | Better response to rTMS in LL genotype. | (Bocchio-Chiavetto et al., 2008) |
| 5-HTTLPR, rs25531 | 310 TRD, 284 HC | L(A)L(A) homozygote haplotype was more common in HC compared with TRD patients. | (Bonvicini et al., 2010) | |
| SLC6A4, solute carrier family 6 member 2 (SLC6A2 or norepinephrine transporter) |
5-HTTLPR (SLC6A4), rs2242446 (SLC6A2) | 119 TRD, 395 HC | 5-HTTLPR L/L in conjunction with SLC6A2 rs2242446 TT was less frequent in TRD patients compared with HC and in ECT non-responders compared with responders. | (Kautto et al., 2015) |
| Potassium two pore domain channel subfamily K member 2 (KCNK2) | rs12031300, rs10779646, rs17546779, rs12136349, rs2841616, rs7538655, rs2841608, rs7549184, rs10494996 | 264 non-TRD, 487 TRD | rs2841616, rs2841608, rs12136349, rs10494996 were associated with TRD in the whole cohort and in Caucasian patients. | (Perlis et al., 2008) |
| Protein phosphatase 3 catalytic subunit gamma (PPP3CC) | rs7430, rs10108011, rs11780915, rs2249098 | 276 non-TRD, 102 TRD | rs7430 and rs10108011 were associated with TRD. | (Fabbri et al., 2014) |
| PPP3CC | rs7430, rs10108011, rs11780915, rs2249098 | 147 non-TRD, 73 TRD | No association between genotypes and TRD. | (Fabbri et al., 2017) |
| PPP3CC, BDNF, 5-hydroxytryptamine receptor 2A (HTR2A or serotonin receptor 2A) | rs7430, rs10108011 (PPP3CC), rs6265, rs11030101, rs11030104, rs12273363 (BDNF), rs643627, rs6313 (HTR2A) | 76 non-TRD, 149 TRD | Using machine learning and clustering algorithms, a combination of 3 SNPs (rs7430 in PPP3CC, rs6265 in BDNF, rs6313 in HTR2A) and the clinical feature melancholia showed the best predictive performance of TRD. | (Kautzky et al., 2015) |
| HTR2A | rs643627, rs17288723, rs6313 | 276 non-TRD, 102 TRD | No association between these variants and TRD. | (Fabbri et al., 2014) |
| Catechol-O-methyltransferase (COMT) | rs4680 (Val108/158Met) | 100 TRD, 100 HC | The alternative allele A (Met) was more frequent in TRD than in HC and it was associated with worse ECT response. | (Lin et al., 2015) |
| 104 TRD | The A (Met) allele was associated with worse ECT response particularly regarding the core symptoms of depression and sleep-related symptoms. | (Domschke et al., 2010) | ||
| 90 TRD | No association between this variant and TMS response. | (Malaguti et al., 2011) | ||
| rs4680, rs174696 | 276 non-TRD, 102 TRD | No association between these variants and TRD. | (Fabbri et al., 2014) | |
| 5-hydroxytryptamine receptor 1A (HTR1A or serotonin receptor 1A) | rs6265 | 90 TRD | CC genotype was associated with higher symptom improvement after treatment with TMS. | (Malaguti et al., 2011) |
| HTR1A, BDNF | rs6295 (HTR1A), rs6265 (BDNF) | 119 TRD, 392 HC | The combination of rs6295 (HTR1A) GG and rs6265 (BDNF) GA + AA genotypes was more frequent in TRD compared with HC. | (Anttila et al., 2007) |
| Poly(A) binding protein cytoplasmic 4 like (PABPC4L) | GWAS (CNVs) with pathway analysis | 811 non-TRD, 452 TRD | A modest enrichment of duplications and a particular deletion spanning PABPC4L in TRD, but these findings were not significant after multiple- testing correction. Pathways regulating actin cytoskeleton were nominally associated with TRD. | (O’Dushlaine et al., 2014) |
| Calcium voltage-gated channel subunit alpha1 C (CACNA1C) (GO:0006942) | Pathway analysis in GWAS | 226 non-TRD, 394 TRD | The Gene Ontology term GO:0006942, including the CACNA1C gene, predicted the risk of TRD with a mean sensitivity of 0.83, specificity of 0.56, positive predictive value=0.77, negative predictive value=0.65 after cross-validation. | (Fabbri et al., 2018) |
| / | GWAS | 7795 non-TRD, 1311 TRD | No genome-wide significant finding. | (Li et al., 2016) |
Abbreviations: CNV,copy number variations; ECT,electroconvulsive therapy; HC,healthy controls; rTMS,repetitive transcranial magnetic stimulation.
Only candidate genes investigated by at least 2 independent studies and/or with complementary evidence of association with TRD (e.g., gene expression studies, in vitro or in vivo models) were reported. The results of GWAS were also reported. For each gene the nonabbreviated name is reported correspondence to the first occurrence.
Candidate Genes
Genes more convincingly involved in TRD are related to glutamatergic and monoaminergic neurotransmission as well as synaptic plasticity, as suggested by the antidepressant efficacy of the N-methyl-D-aspartate (NMDA) receptor antagonist ketamine and electroconvulsive therapy (ECT) in TRD (Kellner et al., 2012; de Sousa et al., 2017).
NMDA receptor (NMDAR) upregulation has been implicated in TRD pathogenesis (Franklin et al., 2015) and ketamine works by blocking this receptor. NMDAR is composed of a combination of individual protein subunits, including one called GluN2B, coded by the GRIN2B gene, which is predominant in the human cortex together with GluN2A. GluN2B-containing NMDARs directly suppress mammalian target of rapamycin signaling and repress protein synthesis involved in excitatory synaptic transmission in cortical neurons (Wang et al., 2011). Ketamine rapidly activates the mammalian target of rapamycin pathway, leading to increased synaptic signaling proteins and increased number and function of new spine synapses (Li et al., 2010). A key role of GluN2B in mediating ketamine effects was demonstrated by the observation that mice lacking GluN2B-containing receptors in their cortical neurons showed a reduced amount of depressive-like behavior, very similar to normal mice treated with ketamine (Miller et al., 2014). Consistent with these findings, the downstream genetic variant rs1805502 in the GRIN2B gene has been associated with TRD (Zhang et al., 2014).
The long-term changes in synaptic strength induced by ketamine are also dependent on NMDAR activation of AMPA/kainate glutamate receptors, since antagonists of these receptors block the antidepressant-like effects of ketamine (Maeng et al., 2008). The subunit 4 of the glutamate kainate receptor (coded by the GRIK4 gene) is expressed in the hippocampus (Darstein et al., 2003), where it exerts a modulatory effect on synaptic plasticity in conjunction with NMDARs (Lerma, 2006). Variants within GRIK4 (rs11218030 and rs1954787) were associated with TRD and the risk of developing psychotic symptoms during depressive episodes, a known clinical risk factor of TRD (Milanesi et al., 2015). The same polymorphisms were associated in a consistent direction with worse ECT response in TRD patients (Minelli et al., 2016). According to these studies, patients carrying the rs11218030 G allele or the rs1954787 GG genotype had increased risk of TRD and lack of response to ECT.
The role of other genes coding for glutamate receptors was unfortunately not investigated, while other genes involved in synaptic plasticity were suggested as possible modulators of TRD. Brain derived neurotrophic factor (BDNF) has been extensively studied, since it plays a key role in promoting neuronal survival, differentiation, and growth. Peripheral BDNF expression levels were found to be decreased in TRD patients compared with antidepressant-responsive patients, in line with the hypothesis of a BDNF deficit in MDD and particularly in TRD (Hong et al., 2014). The Valine66Methionine, or rs6265 polymorphism, was the most investigated BDNF variant, since the Met protein is less efficiently secreted, resulting in lower interaction with BDNF targets (Chen et al., 2004). A preclinical study (Liu et al., 2012) and a small clinical pilot study in a sample of mainly European origin (Laje et al., 2012) suggested that the presence of the Met allele attenuates the antidepressant response to ketamine in TRD. A following study in a Chinese population did not confirm this hypothesis and found that TRD patients showed dose-related efficacy of ketamine independently from rs6265 genotype (Su et al., 2017). In this last study, the lower prevalence of the Val/Val genotype in Chinese subjects compared with Caucasians may have affected the power to detect a possible association with ketamine efficacy. Consistent with the lower functionality of the Met protein, a small pilot study suggested that patients with TRD may show better response to repetitive transcranial magnetic stimulation (rTMS) when they carry the Val/Val genotype compared with the Met allele (Bocchio-Chiavetto et al., 2008). Other studies investigated the interaction between rs6265 and polymorphisms in other genes in determining the risk of TRD. Interactions with polymorphisms in the NTRK2 gene (coding for BDNF receptor), PPP3CC gene, and serotonergic receptor genes (HTR1A and HTR2A) were reported to affect the risk of TRD, but these results were not replicated and they should be considered cautiously (Anttila et al., 2007; Li et al., 2013; Kautzky et al., 2015).
PPP3CC (protein phosphatase 3 catalytic subunit gamma) may have a role in the activation of a neuron-enriched phosphatase that regulates synaptic plasticity (Xia and Storm, 2005). It has been recently suggested as a candidate for TRD risk (Fabbri et al., 2014), possibly through an interaction with BDNF and HTR2A polymorphisms as reported above (Kautzky et al., 2015). CREB1 (CAMP responsive element binding protein 1) codes for a transcription factor that is a downstream effector of BDNF, and its function can affect BDNF signaling pathway. Some polymorphisms (rs2253206, rs7569963, rs4675690) in this gene were associated with TRD (Serretti et al., 2011), despite that subsequent studies did not genotype these variants and reported no association with TRD for other CREB1 variants but a possible effect on symptom remission (Calati et al., 2013; Fabbri et al., 2017).
Among other genes involved in the modulation of neurogenesis and neuroplasticity, it is worth mentioning preliminary results reported for growth-associated protein 43 and the Fas/FasL system (Fabbri et al., 2015; Santos et al., 2015). growth-associated protein 43 codes for a neuron-specific cytosolic protein involved in the development of neuronal growth cones and axonal regeneration (Leu et al., 2010). The Fas/FasL system is one of the best-known death-receptor mediated cell signaling systems, and it may be relevant to neurogenesis and neuroplasticity (Santos et al., 2015).
The monoaminergic theory, even though it only partially explains the pathogenesis of MDD, had a primary role in guiding genetic research in this field over the past few decades. Monoaminergic genes that were studied as possible predictors of TRD included the serotonin transporter (SLC6A4), the norepinephrine transporter (SLC6A2), serotonin receptors 2A and 1A (HTR2A and HTR1A), and catechol-O-methyltransferase (COMT). As previously reported, the effect of HTR2A and HTR1A on TRD risk was mainly studied in conjunction with variants in BDNF and PPP3CC genes, and results were unfortunately not replicated or negative. Thus, these genes are not further discussed (Anttila et al., 2007; Malaguti et al., 2011; Fabbri et al., 2014; Kautzky et al., 2015).
The insertion/deletion functional polymorphism (5-HTTLPR) of the SLC6A4 gene is a functional polymorphism which L (long) allele was associated with higher transcription of the gene and better antidepressant response compared with the S (short) allele in Caucasian samples (Porcelli et al., 2012). In line with these findings, TRD patients carrying the SS genotype were demonstrated to have smaller hippocampal volume compared with L carriers (Phillips et al., 2015), but the association between 5-HTTLRP and TRD is controversial since only a few studies considered this phenotype. Better response in TRD patients carrying the LL genotype or lower frequency of this genotype in TRD vs healthy controls was reported (Bocchio-Chiavetto et al., 2008; Bonvicini et al., 2010; Kautto et al., 2015), but confirmations in larger samples would provide higher confidence in these results. A reciprocal regulation with the serotonin transporter involving the 2-pore domain potassium channel TREK1 (KCNK2 gene) was recently found implicated in mood regulation. Genetic polymorphisms in KCNK2 were found to predict TRD in a quite large sample (Perlis et al., 2008) and TREK1 blockers have potential antidepressant effects (Ye et al., 2015).
Compared with the serotonin transporter, fewer data are available for the norepinephrine transporter (SLC6A2). TRD patients carrying the C variant at SLC6A2 rs2242446 were found to have smaller hippocampal volume compared with noncarriers (Phillips et al., 2015). The same allele of this polymorphism was observed more frequently in TRD patients vs healthy controls (Kautto et al., 2015), but replications are lacking.
COMT is responsible for one of the major catabolic pathways of monoamines. Variants in this gene were associated with increased risk of suicide in TRD (Schosser et al., 2012a), they were observed more frequently in TRD compared with healthy controls (Lin et al., 2015), and they may be associated with ECT response in TRD (Domschke et al., 2010). On the other hand, negative results were also reported (Malaguti et al., 2011; Fabbri et al., 2014).
Other putative genetic modulators of TRD are involved in antidepressant pharmacokinetics, and they include genes belonging to the cytochrome P450 (CYP450) superfamily and the P-glycoprotein (P-gp) coded by the ABCB1 gene.
Variants in genes coding for CYP450 enzymes may contribute to the risk of TRD despite there are no studies that investigated this association. Functional polymorphisms in CYP2D6 and CYP2C19 genes are the pharmacogenetic biomarkers with the highest support in current clinical guidelines (Clinical Pharmacogenetics Implementation Consortium, 2014) and in antidepressant labeling according to regulatory agencies (FDA, 2017). According to these recommendations, there is strong or moderate evidence supporting specific actions (choice of antidepressant and dose) based on the presence of functional variants in these 2 CYP450 genes when treatment is a tricyclic antidepressant, sertraline, citalopram, escitalopram, fluvoxamine, or paroxetine. For example, the Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines strongly recommend to avoid the prescription of paroxetine, amitriptyline, and nortriptyline in CYP2D6 ultrarapid metabolizers because of increased risk of treatment failure (Clinical Pharmacogenetics Implementation Consortium, 2014). However the complex nonlinear correlation between plasma level and clinical efficacy of selective serotonin reuptake inhibitors complicates the picture (Florio et al., 2017).
ABCB1 codes for an ATP-dependent efflux pump (P-gp), which limits the uptake and accumulation of some lipophilic drugs, including a number of antidepressants, into the brain. Some variants in this gene may impact on treatment response to antidepressants by affecting their transport across the brain-blood barrier. In line with this hypothesis, ABCB1 gene expression was associated with TRD (Breitfeld et al., 2017), and polymorphisms associated with P-gp increased activity may confer susceptibility to TRD in patients treated with normal doses of antidepressants targeted by P-gp (e.g., venlafaxine, paroxetine) (Rosenhagen and Uhr, 2010).
GWAS
Three GWAS have investigated the genetics of TRD, and they used different approaches that do not allow a straightforward comparison of findings. One study was focused on copy number variants (O’Dushlaine et al., 2014), another on common SNPs (single nucleotide polymorphisms) (Li et al., 2016), while the last considered pathways (Fabbri et al., 2018).
A modest enrichment of duplications was found in TRD, and a particular deletion spanning the PABPC4L (poly(A) binding protein cytoplasmic 4 like) gene was identified. PABPC4L is involved in the degradation of abnormal mRNA molecules, but this association did not survive multiple-testing correction (O’Dushlaine et al., 2014). The analysis of common SNPs did not provide more encouraging results, since no genome-wide significant or suggestive signal was identified (Li et al., 2016), suggesting that possible methodological limitations of these GWAS should be considered. The most known one is inadequate statistical power, given that a sample of ~2000 subjects was estimated to provide adequate power to identify individual variants associated with a binary trait with heritability ~40%, while ~5000 are required for a binary trait with heritability of 20% (Visscher et al., 2014). The heritability of TRD was estimated to be ~17% (Li et al., 2016), despite other GWAS of antidepressant response suggesting that the contribution of genes could be higher (Tansey et al., 2013). On the basis of these considerations, previous GWAS had inadequate power except one that may have been limited by an unbalance between the number of TRD and non-TRD patients (Li et al., 2016). Other relevant limitations of previous GWAS have been the small effect size of common risk loci associated with TRD and the relatively limited coverage of common human genetic variation, issues that were demonstrated to significantly affect power independently from heritability (Spencer et al., 2009). Possible alternative strategies to the recruitment of large samples include the use of aggregated tests and genotype imputation using large and diverse reference panels as discussed more in detail elsewhere. A previous GWAS applied pathway analysis and machine learning to investigate possible gene sets (pathways) implicated in the pathogenesis of TRD, with promising findings (Fabbri et al., 2018). A gene set (GO:0006942) including the CACNA1C gene showed a trend of association with TRD. Machine learning models showed that independent SNPs in this gene set predicted TRD with a mean sensitivity of 0.83 and specificity of 0.56 after 10-fold cross validation repeated 100 times. CACNA1C encodes for the α-1C subunit of the L-type voltage-dependent calcium channel; it transiently increases calcium-mediated membrane depolarization and modulates intracellular signaling, gene transcription, and synaptic plasticity. This gene has been associated with multiple psychiatric phenotypes, including schizophrenia, bipolar disorder, and MDD, suggesting it plays a pleiotropic role in psychiatric disorders (Cross-Disorder Group of the Psychiatric Genomics Consortium, 2013). A number of other genes in the identified pathway (33 genes out of a total of 72 genes) were linked to long-term potentiation, neural survival, neurogenesis, and neuroplasticity, but also to MDD and antidepressant efficacy (Fabbri et al., 2018). Among these genes, some regulate glutamate receptors involved in long-term potentiation, a form of persistent strengthening of synapses (e.g., TRPM4, PIK3CG, SUMO1), or calcium currents modulating the same process (e.g., CACNA1C, CAMK2D, FKBP1B, P2RX4, RYR2). Examples of genes previously associated with antidepressant action include adrenergic receptors alpha 1A and 1B and CTNNA3, coding for a protein involved in cell-cell adhesion that may be relevant in antidepressant-induced hippocampal cell proliferation (Mostany et al., 2008; Fabbri et al., 2018). It is interesting to note that a previous GWAS identified a gene set involved in inorganic cation transmembrane transporter activity (GO:0022890) as a possible modulator of antidepressant response in 2 samples (Cocchi et al., 2016). This gene set includes CACNA1C and other calcium-channel coding genes such as CACNB2; the latter gene was also identified as involved in the shared genetic susceptibility to several psychiatric traits including MDD (Cross-Disorder Group of the Psychiatric Genomics Consortium, 2013).
Strategies to Improve Future Studies
The previous paragraphs outline that pharmacogenetic research provided often nonreplicated results in the study of TRD and antidepressant response. Understanding the strengths and limitations of previous studies is a key issue to facilitate advances in the field, including clinical applications. Both candidate gene studies and GWAS show pros and cons that should be taken into account. The candidate gene approach allows the detailed study of variants in genes with higher pretest probability of association with TRD, but it is limited by previous knowledge (many signals may come from genes with unknown function or unknown link with antidepressant action) and by the number of polymorphisms that can be studied. GWAS are suitable to study polygenic traits such as TRD, but previous studies included only a relatively small proportion of known variants in the human genome and they had mostly inadequate power to detect signals with small effect size. For example, previous GWAS of antidepressant response included ~1 to 9 million common variants (GENDEP Investigators et al., 2013; Li et al., 2016), while ~40 million common variants were identified in the human genome using whole-genome sequence data from a number of cohorts (McCarthy et al., 2016). Previous GWAS included only a fraction of known genetic variants, but distinguishing genuine small effects from false positives was a relevant issue because of the strict multiple-testing correction needed to have acceptable false positive risk. This issue can be addressed in different ways: increase sample size or use analysis approaches that allow the increase of power (e.g., aggregated approaches). Consortia such as the Psychiatric Genomics Consortium are working on the former, but in the meantime analysis approaches aimed to maximize power should also be developed and implemented.
In the next 2 paragraphs, the advantages of improving the coverage of genetic variants and other methodological strategies to improve power are discussed (Figure 1).
Methods to Improve the Coverage of Genetic Variants: Imputation and Sequencing
In 2001 the first draft of the human genomic sequence was available thanks to the Human Genome Project (International Human Genome Sequencing Consortium, 2004). The Human Genome Project was the world’s largest collaborative biological project; it was a $3.8 billion investment and launched the genomic revolution. After 2001, 3 different generations of DNA sequencing technologies can be identified that provide an output orders of magnitude higher than the first sequencing technique and dramatically reduce cost per base (Gut, 2013). According to the US National Human Genome Research Institute, the cost for sequencing one human genome dropped from $100,000,000 in 2001 to $1000 in 2015, and cost dropping exceeded the Moore’s Law (which describes a long-term trend in the computer hardware industry that involves the doubling of “compute power” every 2 years) around 2008, meaning that more than excellent technological improvement was achieved (Human Genome Research Institute, 2017).
The use of sequencing is becoming widespread in genetic research thanks to the described technological improvements and drop of costs. The growth of publicly available genome sequencing datasets allowed the development of new and more comprehensive reference panels for genotype imputation, a method used to improve the coverage of common genetic variants in GWAS. In the last 2 to 3 years, the reference databases made available by the Haplotype Reference Consortium (HRC) and by the Trans-Omics for Precision Medicine (TOPMed) Program represent the largest and most diverse whole genome sequence datasets available (McCarthy et al., 2016; NIH, 2018). The use of these reference datasets for imputation is expected to provide the coverage of the most part of human common genetic variation in an accurate manner and to increase the percent of explained heritability in a given sample size, or in other words improve power (McCarthy et al., 2016).
Imputation can provide very good coverage of common genetic variants (at least in samples of European and Asian ancestry), but sequencing is needed to investigate the contribution of rare variants. Several projects are planned to perform sequencing at the population level to promote the development of precision medicine, that is, a new medical approach where each patient is considered a unique individual, with his/her own genetic signature and other unique biomarkers. Some examples are “deCODE genetics” in Iceland (deCODE, 2017) and “All of Us” in the United States (NIH, 2017).
The use of genome sequencing is becoming not only feasible on a relatively large scale but also supported by the recent omnigenic hypothesis. According to this theory, the genetic component of complex traits such as TRD is spread across most of the genome, including regions without an obvious connection to the trait of interest (Boyle et al., 2017). For example, 71%– to 100% of 1-MB windows in the genome were estimated to contribute to heritability for schizophrenia (Loh et al., 2015). The effect size of each associated locus was calculated to be approximately one-tenth the median effect size of genome-wide significant variants, suggesting that the most part of the relevant signals is not captured using the traditional GWAS approach (Boyle et al., 2017).
Aggregated Tests and Other Strategies to Improve Power
High throughput data (GWAS and sequencing data) provide the valuable opportunity to implement aggregated approaches to study polygenic/omnigenic traits such as antidepressant response. Polymorphisms do no act as single units, but they interact among each other, within the same gene, and also across different genes. In other words, the effect of a single variant could be nullified or modified by the concomitant presence of other variants, and the cumulative effect of a number of polymorphisms is expected to possibly alter the function of a gene or gene set (pathway). For this reason, in pathway analysis the unit of analysis is a set of genes functionally connected among each other (e.g., part of the same chemical or cellular process or involved in protein-protein interactions; Li et al., 2017). The identification of pathways associated with TRD can be helpful not only to develop polygenic biomarkers but also to contribute to the clarification of the pathogenetic mechanisms involved in TRD. Several GWAS applied pathway analysis to the study of antidepressant response and remission. They reported that pathways involved in neuroplasticity, neurogenesis, and inflammation/immune response probably contribute to antidepressant response/remission (Fabbri et al., 2016). To the best of our knowledge, only 2 studies implemented pathway analysis in the study of TRD, using rare variants (O’Dushlaine et al., 2014) or common variants (Fabbri et al., 2018). The former study found a possible role of pathways regulating actin cytoskeleton that did not survive multiple-testing correction (O’Dushlaine et al., 2014). Several studies demonstrated the importance of actin cytoskeleton in dendritic spine morphology, synaptic plasticity, and psychiatric disorders such as depression, providing a possible biological rationale (Piubelli et al., 2011; Grintsevich, 2017). The second study was already mentioned; it found that a gene ontology gene set (GO:0006942) including the CACNA1C gene predicted TRD risk with a mean sensitivity of 0.83, specificity of 0.56, positive predictive value=0.77, nd predictive value=0.65 after 10-fold cross validation repeated 100 times (Fabbri et al., 2018). Thirty-three genes of a total 72 genes in this gene set were previously associated with long-term potentiation, neural survival, neurogenesis, and neuroplasticity, but also MDD and antidepressant efficacy. One of the main limitations of this study was the lack of result validation in an independent sample.
Pathways associated with TRD can also be used to prioritize polygenic risk scores, which are calculated as the sum of alleles associated with the trait of interest, weighted by effect sizes, for polymorphisms with P values less than predefined thresholds. Previous GWAS applied this approach to the prediction of antidepressant response with unsatisfying results (GENDEP Investigators et al., 2013; García-González et al., 2017). A possible explanation is the lack of statistical power and insufficient coverage of variants, which could be partly addressed by prioritizing variants with higher pretest probability of exerting an effect on TRD, such as variants in pathways previous associated with this trait or antidepressant response. Prioritization can be performed by assigning incremental weights to variants based on the results of previous GWAS but also functional considerations. The incorporation of variant functional annotation including enrichment for expression quantitative trait loci, methylation quantitative trait, cis-regulatory elements (CREs), and pleiotropy across different traits was reported to improve the prediction of complex traits (Shi et al., 2016).
The integration of different types of -omics data (e.g., genomics, transcriptomics, and proteomics) with molecular, behavioral, imaging, environmental, and clinical data is also a possible approach to increase power and replication of findings. This approach is the key feature of the TOPMed program, which answers to many of the objectives of the 2016 strategic vision released by the US NIH (NIH, 2018). For example, the incorporation of clinical information in genetic studies should not be overlooked, and clinical risk factors for TRD should not be considered pertinent to clinicians only. A number of clinical and socio-demographic factors were consistently associated with TRD by several studies, for example older age, chronic depression, moderate-severe suicidal ideation, high level of anxiety symptoms or comorbidity with anxiety disorder, lower education, being single, or divorced (Perlis, 2013; De Carlo et al., 2016; Kautzky et al., 2017). As discussed in the Introduction, some of these risk factors (e.g., severity, suicidal ideation, anxiety comorbidity) may have a genetic base that overlaps with the genetics of TRD, but others are independent (socio-demographic factors) or probably independent (e.g., duration of the depressive episode) from the effect of genetic variants. The lack of consideration of the latter group’s influence on TRD may bias the results of pharmacogenetic studies and be responsible for false negative or false positive findings.
Discussion
Few studies have investigated the genetics of TRD compared with overall antidepressant efficacy and results were often obtained by candidate gene studies in relatively small samples. The most robust results in terms of replication, evidence using complementary approaches (e.g., gene expression, neuroimaging), and biological rationale are variants in GRIK4, BDNF, SLC6A4, and KCNK2 genes. Functional variants in CYP450 genes may hypothetically play a role, but no study specifically investigated this question. Only 3 GWAS studied the genetics of TRD (O’Dushlaine et al., 2014; Li et al., 2016; Fabbri et al., 2018), and no genome-wide significant signal was identified at single variant level. The lack of genome-wide significant polymorphisms should be interpreted in light of similar results obtained by GWAS of antidepressant response. The few significant hits identified by these studies were inconsistent across independent samples (Fabbri et al., 2016), supporting the hypothesis that some major limitations affected GWAS. As discussed previously, lack of power was among these limitations and this was both due to relative small sample size but also to the poor implementation of analysis approaches able to maximize power. For example, results obtained by gene set (pathway) analysis showed higher similarity across different GWAS and higher biological rationale than signals at variant level, since they pointed towards the involvement of pathways mediating neural plasticity, neurogenesis, and inflammation (Fabbri et al., 2016). Pathways possibly associated with TRD are comparable, because of the importance of actin cytoskeleton in dendritic spine morphology/synaptic plasticity and the role of GO:0006942 genes in neural plasticity/neurogenesis (O’Dushlaine et al., 2014; Fabbri et al., 2018). These findings support the hypothesis that antidepressant response and TRD are polygenic traits, and the methodological improvement of aggregated tests should be pursued to disentangle the whole contribution of genetic variants to these traits. On the other hand, these results should not be interpreted as the proof that antidepressant action depends on a few pathways and these could be sufficient to explain the genetics of this trait, but these pathways probably have a higher weight in determining TRD. This idea leads to the hypothesis that not all genes should be considered equally in GWAS, but information about gene and polymorphism function should be incorporated in genetic studies to assign specific weights and prioritize variants/genes. The inclusion of this information in aggregated tests such as polygenic risk scores could allow an increase in prediction and ability to replicate findings.
Another limitation of previous GWAS was the covering of a limited proportion of human common genetic variants (not more than ~7–9 millions vs the ~40 millions of common variants so far identified; McCarthy et al., 2016). Genotype imputation using large and diverse reference panels (e.g., Haplotype Reference Consortium and TOPMed) is expected to allow the inclusion of more and more common variants as improvements in the method and growth of reference panels proceed, but rare variants are largely not considered in genome-wide arrays. For this reason and for the evidence suggesting that complex traits may have a genetic component spread in small signals across the most part of the genome (Boyle et al., 2017), the collection of sequencing data would be helpful to perform polygenic analysis able to capture the complexity of TRD. No study has applied whole genome sequencing to the study of TRD so far, leaving this area unexplored, but national projects such as “All of Us” in the United States may allow this kind of study on large population samples (NIH, 2017).
Finally, clinical-demographic factors associated with TRD should not be overlooked in genetic studies. Some clinical features that are commonly observed in TRD, such as high suicidal risk and anxiety comorbidities, may have a genetic basis that overlaps with TRD genetics and in this case they would represent the features of an endophenotype of MDD. Other known clinical and socio-demographic predictors of TRD risk have no genetic basis (e.g., poor education, old age), and the noninclusion of these modulators in pharmacogenetic studies may lead to biases in results.
Currently, there are no recommended genetic biomarkers to predict the risk of TRD or to guide treatment choice in TRD patients. Clinical guidelines such as CPIC guidelines recommend that CYP2D6 and CYP2C19 functional genotypes are taken into account for some antidepressant treatments (Clinical Pharmacogenetics Implementation Consortium, 2014). We suggest that genetic testing of polymorphisms in these 2 genes may be helpful in some patients with TRD treated with drugs metabolized by these cytochromes to exclude a major metabolic alteration. Monitoring drug plasma levels may also be helpful. On the other hand, a number of variants in genes involved in antidepressant pharmacodynamics are also involved in determining antidepressant action; thus, focusing on CYP450 genes is not expected to be sufficient in most cases. The implementation of future studies that include the improvements suggested by this review (Figure 1) may provide more valid genetic biomarkers of TRD, probably sets of hundreds or thousands of polymorphisms selected to maximize the sensitivity and specificity of the prediction test. The availability of this kind of clinical application would help in guiding treatment choice and dramatically reduce the individual and socio-economic burden resulting from poor antidepressant response in MDD.
Statement of Interest
Dr Souery has received grant/research support from GlaxoSmithKline and Lundbeck and has served as a consultant or on advisory boards for AstraZeneca, Bristol-Myers Squibb, Eli Lilly, Janssen, and Lundbeck. Dr Montgomery has been a consultant or served on advisory boards for AstraZeneca, Bionevia, Bristol Myers Squibb, Forest, GlaxoSmithKline, Grunenthal, Intellect Pharma, Johnson & Johnson, Lilly, Lundbeck, Merck, Merz, M’s Science, Neurim, Otsuka, Pierre Fabre, Pfizer, Pharmaneuroboost, Richter, Roche, Sanofi, Sepracor, Servier, Shire, Synosis, Takeda, Theracos, Targacept, Transcept, UBC, Xytis, and Wyeth. Dr Kasper received grants/research support, consulting fees, and/or honoraria within the last 3 years from Angelini, AOP Orphan Pharmaceuticals AG, AstraZeneca, Eli Lilly, Janssen, KRKA-Pharma, Lundbeck, Neuraxpharm, Pfizer, Pierre Fabre, Schwabe, and Servier. Dr Zohar has received grant/research support from Lundbeck, Servier, Brainsway, and Pfizer; has served as a consultant or on advisory boards for Servier, Pfizer, Abbott, Lilly, Actelion, AstraZeneca, and Roche; and has served on speakers’ bureaus for Lundbeck, Roch, Lilly, Servier, Pfizer, and Abbott. Dr Mendlewicz is a member of the Board of the Lundbeck International Neuroscience Foundation and of the advisory board of Servier. Dr Serretti is or has been consultant/speaker for Abbott, Abbvie, Angelini, Astra Zeneca, Clinical Data, Boheringer, Bristol Myers Squibb, Eli Lilly, GlaxoSmithKline, Innovapharma, Italfarmaco, Janssen, Lundbeck, Naurex, Pfizer, Polifarma, Sanofi, and Servier. The other authors declare no potential conflict of interest.
Acknowledgments
None.
References
- Akil H, Gordon J, Hen R, Javitch J, Mayberg H, McEwen B, Meaney MJ, Nestler EJ(2018)Treatment resistant depression: a multi-scale, systems biology approach. Neurosci Biobehav Rev 84:272–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anttila S, Huuhka K, Huuhka M, Rontu R, Hurme M, Leinonen E, Lehtimäki T(2007)Interaction between 5-HT1A and BDNF genotypes increases the risk of treatment-resistant depression. J Neural Transm (Vienna) 114:1065–1068. [DOI] [PubMed] [Google Scholar]
- Bocchio-Chiavetto L, Miniussi C, Zanardini R, Gazzoli A, Bignotti S, Specchia C, Gennarelli M(2008)5-HTTLPR and BDNF val66met polymorphisms and response to rtms treatment in drug resistant depression. Neurosci Lett 437:130–134. [DOI] [PubMed] [Google Scholar]
- Bonvicini C, Minelli A, Scassellati C, Bortolomasi M, Segala M, Sartori R, Giacopuzzi M, Gennarelli M(2010)Serotonin transporter gene polymorphisms and treatment-resistant depression. Prog Neuropsychopharmacol Biol Psychiatry 34:934–939. [DOI] [PubMed] [Google Scholar]
- Boyle EA, Li YI, Pritchard JK(2017)An expanded view of complex traits: from polygenic to omnigenic. Cell 169:1177–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitfeld J, Scholl C, Steffens M, Laje G, Stingl JC(2017)Gene expression and proliferation biomarkers for antidepressant treatment resistance. Transl Psychiatry 7:e1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calati R, Crisafulli C, Balestri M, Serretti A, Spina E, Calabrò M, Sidoti A, Albani D, Massat I, Höfer P, Amital D, Juven-Wetzler A, Kasper S, Zohar J, Souery D, Montgomery S, Mendlewicz J(2013)Evaluation of the role of MAPK1 and CREB1 polymorphisms on treatment resistance, response and remission in mood disorder patients. Prog Neuropsychopharmacol Biol Psychiatry 44:271–278. [DOI] [PubMed] [Google Scholar]
- Chen ZY, Patel PD, Sant G, Meng CX, Teng KK, Hempstead BL, Lee FS(2004)Variant brain-derived neurotrophic factor (BDNF) (met66) alters the intracellular trafficking and activity-dependent secretion of wild-type BDNF in neurosecretory cells and cortical neurons. J Neurosci 24:4401–4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical Pharmacogenetics Implementation Consortium (2014)CPIC guidelines Available at: https://cpicpgx.org/guidelines/. Retrieved 27 Nov 2017.
- Cocchi E, Fabbri C, Han C, Lee SJ, Patkar AA, Masand PS, Pae CU, Serretti A(2016)Genome-wide association study of antidepressant response: involvement of the inorganic cation transmembrane transporter activity pathway. BMC Psychiatry 16:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics Consortium (2013)Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 381:1371–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darstein M, Petralia RS, Swanson GT, Wenthold RJ, Heinemann SF(2003)Distribution of kainate receptor subunits at hippocampal mossy fiber synapses. J Neurosci 23:8013–8019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Carlo V, Calati R, Serretti A(2016)Socio-demographic and clinical predictors of non-response/non-remission in treatment resistant depressed patients: a systematic review. Psychiatry Res 240:421–430. [DOI] [PubMed] [Google Scholar]
- deCODE (2017)DeCODE genetics Available at: https://www.decode.com/research. Retrieved 27 Nov 2017.
- de Sousa RT, Loch AA, Carvalho AF, Brunoni AR, Haddad MR, Henter ID, Zarate CA, Machado-Vieira R(2017)Genetic studies on the tripartite glutamate synapse in the pathophysiology and therapeutics of mood disorders. Neuropsychopharmacology 42:787–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domschke K, Zavorotnyy M, Diemer J, Nitsche S, Hohoff C, Baune BT, Deckert J, Arolt V, Zwanzger P(2010)COMT val158met influence on electroconvulsive therapy response in major depression. Am J Med Genet B Neuropsychiatr Genet 153B:286–290. [DOI] [PubMed] [Google Scholar]
- Fabbri C, Marsano A, Albani D, Chierchia A, Calati R, Drago A, Crisafulli C, Calabrò M, Kasper S, Lanzenberger R, Zohar J, Juven-Wetzler A, Souery D, Montgomery S, Mendlewicz J, Serretti A(2014)PPP3CC gene: a putative modulator of antidepressant response through the B-cell receptor signaling pathway. Pharmacogenomics J 14:463–472. [DOI] [PubMed] [Google Scholar]
- Fabbri C, Crisafulli C, Gurwitz D, Stingl J, Calati R, Albani D, Forloni G, Calabrò M, Martines R, Kasper S, Zohar J, Juven-Wetzler A, Souery D, Montgomery S, Mendlewicz J, Girolamo GD, Serretti A(2015)Neuronal cell adhesion genes and antidepressant response in three independent samples. Pharmacogenomics J 15:538–548. [DOI] [PubMed] [Google Scholar]
- Fabbri C, Crisafulli C, Calabrò M, Spina E, Serretti A(2016)Progress and prospects in pharmacogenetics of antidepressant drugs. Expert Opin Drug Metab Toxicol 12:1157–1168. [DOI] [PubMed] [Google Scholar]
- Fabbri C, Crisafulli C, Calati R, Albani D, Forloni G, Calabrò M, Martines R, Kasper S, Zohar J, Juven-Wetzler A, Souery D, Montgomery S, Mendlewicz J, Serretti A(2017)Neuroplasticity and second messenger pathways in antidepressant efficacy: pharmacogenetic results from a prospective trial investigating treatment resistance. Eur Arch Psychiatry Clin Neurosci 267:723–735. [DOI] [PubMed] [Google Scholar]
- Fabbri C, Corponi F, Albani D, Raimondi I, Forloni G, Schruers K, Kasper S, Kautzky A, Zohar J, Souery D, Montgomery S, Cristalli CP, Mantovani V, Mendlewicz J, Serretti A(2018)Pleiotropic genes in psychiatry: calcium channels and the stress-related FKBP5 gene in antidepressant resistance. Prog Neuropsychopharmacol Biol Psychiatry 81:203–210. [DOI] [PubMed] [Google Scholar]
- FDA (2017)Table of pharmacogenomic biomarkers in drug labeling Available at: https://www.fda.gov/Drugs/ScienceResearch/ucm572698.htm. Retrived 27 Nov 2017.
- Florio V, Porcelli S, Saria A, Serretti A, Conca A(2017)Escitalopram plasma levels and antidepressant response. Eur Neuropsychopharmacol 27:940–944. [DOI] [PubMed] [Google Scholar]
- Franklin M, Hlavacova N, Babic S, Pokusa M, Bermudez I, Jezova D(2015)Aldosterone signals the onset of depressive behaviour in a female rat model of depression along with SSRI treatment resistance. Neuroendocrinology 102:274–287. [DOI] [PubMed] [Google Scholar]
- García-González J, et al. (2017)Pharmacogenetics of antidepressant response: a polygenic approach. Prog Neuropsychopharmacol Biol Psychiatry 75:128–134. [DOI] [PubMed] [Google Scholar]
- GBD 2015. Disease and Injury Incidence and Prevalence Collaborators (2016) Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990–2015: a systematic analysis for the global burden of disease study 2015. Lancet 388:1545–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GENDEP Investigators, MARS Investigators, STAR*D Investigators (2013)Common genetic variation and antidepressant efficacy in major depressive disorder: a meta-analysis of three genome-wide pharmacogenetic studies. Am J Psychiatry 170:207–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratten J, Wray NR, Keller MC, Visscher PM(2014)Large-scale genomics unveils the genetic architecture of psychiatric disorders. Nat Neurosci 17:782–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grintsevich EE.(2017)Remodeling of actin filaments by drebrin A and its implications. Adv Exp Med Biol 1006:61–82. [DOI] [PubMed] [Google Scholar]
- Gut IG.(2013)New sequencing technologies. Clin Transl Oncol 15:879–881. [DOI] [PubMed] [Google Scholar]
- Hong W, Fan J, Yuan C, Zhang C, Hu Y, Peng D, Wang Y, Huang J, Li Z, Yu S, Liu X, Wu Z, Chen J, Yi Z, Xu L, Fang Y(2014)Significantly decreased mrna levels of BDNF and MEK1 genes in treatment-resistant depression. Neuroreport 25:753–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Genome Research Institute (2017)Sequencing cost data Available at: https://www.genome.gov/sequencingcostsdata/. Retrieved 27 Nov 2017.
- Insel T, Cuthbert B, Garvey M, Heinssen R, Pine DS, Quinn K, Sanislow C, Wang P(2010)Research domain criteria (rdoc): toward a new classification framework for research on mental disorders. Am J Psychiatry 167:748–751. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium (2004)Finishing the euchromatic sequence of the human genome. Nature 431:931–945. [DOI] [PubMed] [Google Scholar]
- Kautto M, Kampman O, Mononen N, Lehtimäki T, Haraldsson S, Koivisto PA, Leinonen E(2015)Serotonin transporter (5-HTTLPR) and norepinephrine transporter (NET) gene polymorphisms: susceptibility and treatment response of electroconvulsive therapy in treatment resistant depression. Neurosci Lett 590:116–120. [DOI] [PubMed] [Google Scholar]
- Kautzky A, Baldinger P, Souery D, Montgomery S, Mendlewicz J, Zohar J, Serretti A, Lanzenberger R, Kasper S(2015) The combined effect of genetic polymorphisms and clinical parameters on treatment outcome in treatment-resistant depression. Eur Neuropsychopharmacol 25:441–453. [DOI] [PubMed] [Google Scholar]
- Kautzky A, Baldinger-Melich P, Kranz GS, Vanicek T, Souery D, Montgomery S, Mendlewicz J, Zohar J, Serretti A, Lanzenberger R, Kasper S(2017)A new prediction model for evaluating treatment-resistant depression. J Clin Psychiatry 78:215–222. [DOI] [PubMed] [Google Scholar]
- Kellner CH, Greenberg RM, Murrough JW, Bryson EO, Briggs MC, Pasculli RM(2012)ECT in treatment-resistant depression. Am J Psychiatry 169:1238–1244. [DOI] [PubMed] [Google Scholar]
- Laje G, Lally N, Mathews D, Brutsche N, Chemerinski A, Akula N, Kelmendi B, Simen A, McMahon FJ, Sanacora G, Zarate C Jr(2012)Brain-derived neurotrophic factor val66met polymorphism and antidepressant efficacy of ketamine in depressed patients. Biol Psychiatry 72:e27–e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerma J.(2006)Kainate receptor physiology. Curr Opin Pharmacol 6:89–97. [DOI] [PubMed] [Google Scholar]
- Leu B, Koch E, Schmidt JT(2010)GAP43 phosphorylation is critical for growth and branching of retinotectal arbors in zebrafish. Dev Neurobiol 70:897–911. [DOI] [PubMed] [Google Scholar]
- Li N, Lee B, Liu RJ, Banasr M, Dwyer JM, Iwata M, Li XY, Aghajanian G, Duman RS(2010)Mtor-dependent synapse formation underlies the rapid antidepressant effects of NMDA antagonists. Science 329:959–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QS, Tian C, Seabrook GR, Drevets WC, Narayan VA(2016)Analysis of 23andme antidepressant efficacy survey data: implication of circadian rhythm and neuroplasticity in bupropion response. Transl Psychiatry 6:e889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Wernersson R, Hansen RB, Horn H, Mercer J, Slodkowicz G, Workman CT, Rigina O, Rapacki K, Stærfeldt HH, Brunak S, Jensen TS, Lage K(2017)A scored human protein-protein interaction network to catalyze genomic interpretation. Nat Methods 14:61–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhang Y, Wang Z, Chen J, Fan J, Guan Y, Zhang C, Yuan C, Hong W, Wang Y, Wu Z, Huang J, Hu Y, Cao L, Yi Z, Cui D, Yu S, Fang Y(2013)The role of BDNF, NTRK2 gene and their interaction in development of treatment-resistant depression: data from multicenter, prospective, longitudinal clinic practice. J Psychiatr Res 47:8–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, He H, Zhang C, Wang Z, Jiang M, Li Q, Lan X, Zhang M, Huang X(2015)Influence of val108/158met COMT gene polymorphism on the efficacy of modified electroconvulsive therapy in patients with treatment resistant depression. Cell Biochem Biophys 71:1387–1393. [DOI] [PubMed] [Google Scholar]
- Liu RJ, Lee FS, Li XY, Bambico F, Duman RS, Aghajanian GK(2012)Brain-derived neurotrophic factor val66met allele impairs basal and ketamine-stimulated synaptogenesis in prefrontal cortex. Biol Psychiatry 71:996–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh PR, Bhatia G, Gusev A, Finucane HK, Bulik-Sullivan BK, Pollack SJ, de Candia TR, Lee SH, Wray NR, Kendler KS, O’Donovan MC, Neale BM, Patterson N, Price AL, Schizophrenia Working Group of Psychiatric Genomics Consortium (2015)Contrasting genetic architectures of schizophrenia and other complex diseases using fast variance-components analysis. Nat Genet 47:1385–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeng S, Zarate CA Jr, Du J, Schloesser RJ, McCammon J, Chen G, Manji HK(2008)Cellular mechanisms underlying the antidepressant effects of ketamine: role of alpha-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptors. Biol Psychiatry 63:349–352. [DOI] [PubMed] [Google Scholar]
- Malaguti A, Rossini D, Lucca A, Magri L, Lorenzi C, Pirovano A, Colombo C, Smeraldi E, Zanardi R(2011)Role of COMT, 5-HT(1A), and SERT genetic polymorphisms on antidepressant response to transcranial magnetic stimulation. Depress Anxiety 28:568–573. [DOI] [PubMed] [Google Scholar]
- McCarthy S, et al. , Haplotype Reference Consortium (2016)A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet 48:1279–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre RS, Filteau MJ, Martin L, Patry S, Carvalho A, Cha DS, Barakat M, Miguelez M(2014)Treatment-resistant depression: definitions, review of the evidence, and algorithmic approach. J Affect Disord 156:1–7. [DOI] [PubMed] [Google Scholar]
- Milaneschi Y, Lamers F, Peyrot WJ, Abdellaoui A, Willemsen G, Hottenga JJ, Jansen R, Mbarek H, Dehghan A, Lu C, Boomsma DI, Penninx BW, CHARGE inflammation working group (2016)Polygenic dissection of major depression clinical heterogeneity. Mol Psychiatry 21:516–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milanesi E, Bonvicini C, Congiu C, Bortolomasi M, Gainelli G, Gennarelli M, Minelli A(2015)The role of GRIK4 gene in treatment-resistant depression. Genet Res (Camb) 97:e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller OH, Yang L, Wang CC, Hargroder EA, Zhang Y, Delpire E, Hall BJ(2014)Glun2b-containing NMDA receptors regulate depression-like behavior and are critical for the rapid antidepressant actions of ketamine. Elife 3:e03581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minelli A, Congiu C, Ventriglia M, Bortolomasi M, Bonvicini C, Abate M, Sartori R, Gainelli G, Gennarelli M(2016)Influence of GRIK4 genetic variants on the electroconvulsive therapy response. Neurosci Lett 626:94–98. [DOI] [PubMed] [Google Scholar]
- Mostany R, Valdizán EM, Pazos A(2008)A role for nuclear beta-catenin in SNRI antidepressant-induced hippocampal cell proliferation. Neuropharmacology 55:18–26. [DOI] [PubMed] [Google Scholar]
- National Institutes of Health (2017)All of us Available at: https://allofus.nih.gov/.
- National Institutes of Health (2018)Trans-Omics for Precision Medicine (TOPMed) Program Available at: https://www.nhlbi.nih.gov/science/trans-omics-precision-medicine-topmed-program. Accessed 27 Feb 2018.
- O’Dushlaine C, Ripke S, Ruderfer DM, Hamilton SP, Fava M, Iosifescu DV, Kohane IS, Churchill SE, Castro VM, Clements CC, Blumenthal SR, Murphy SN, Smoller JW, Perlis RH(2014)Rare copy number variation in treatment-resistant major depressive disorder. Biol Psychiatry 76:536–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlis RH.(2013)A clinical risk stratification tool for predicting treatment resistance in major depressive disorder. Biol Psychiatry 74:7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlis RH, Moorjani P, Fagerness J, Purcell S, Trivedi MH, Fava M, Rush AJ, Smoller JW(2008)Pharmacogenetic analysis of genes implicated in rodent models of antidepressant response: association of TREK1 and treatment resistance in the STAR(*)D study. Neuropsychopharmacology 33:2810–2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JL, Batten LA, Tremblay P, Aldosary F, Du L, Blier P(2015)Impact of monoamine-related gene polymorphisms on hippocampal volume in treatment-resistant depression. Acta Neuropsychiatr 27:353–361. [DOI] [PubMed] [Google Scholar]
- Piubelli C, Gruber S, El Khoury A, Mathé AA, Domenici E, Carboni L(2011)Nortriptyline influences protein pathways involved in carbohydrate metabolism and actin-related processes in a rat gene-environment model of depression. Eur Neuropsychopharmacol 21:545–562. [DOI] [PubMed] [Google Scholar]
- Porcelli S, Fabbri C, Serretti A(2012)Meta-analysis of serotonin transporter gene promoter polymorphism (5-HTTLPR) association with antidepressant efficacy. Eur Neuropsychopharmacol 22:239–258. [DOI] [PubMed] [Google Scholar]
- Rosenhagen MC, Uhr M(2010)Single nucleotide polymorphism in the drug transporter gene ABCB1 in treatment-resistant depression: clinical practice. J Clin Psychopharmacol 30:209–211. [DOI] [PubMed] [Google Scholar]
- Rush AJ, Wisniewski SR, Warden D, Luther JF, Davis LL, Fava M, Nierenberg AA, Trivedi MH(2008)Selecting among second-step antidepressant medication monotherapies: predictive value of clinical, demographic, or first-step treatment features. Arch Gen Psychiatry 65:870–880. [DOI] [PubMed] [Google Scholar]
- Santos M, Carvalho S, Lima L, Mota-Pereira J, Pimentel P, Maia D, Correia D, Gomes S, Cruz A, Medeiros R(2015)FAS -670A>G genetic polymorphism is associated with treatment resistant depression. J Affect Disord 185:164–169. [DOI] [PubMed] [Google Scholar]
- Schosser A, Calati R, Serretti A, Massat I, Kocabas NA, Papageorgiou K, Linotte S, Mendlewicz J, Souery D, Zohar J, Juven-Wetzler A, Montgomery S, Kasper S (2012a) The impact of COMT gene polymorphisms on suicidality in treatment resistant major depressive disorder–a European multicenter study. Eur Neuropsychopharmacol 22:259–266. [DOI] [PubMed] [Google Scholar]
- Schosser A, Serretti A, Souery D, Mendlewicz J, Zohar J, Montgomery S, Kasper S (2012b) European group for the study of resistant depression (GSRD)–where have we gone so far: review of clinical and genetic findings. Eur Neuropsychopharmacol 22:453–468. [DOI] [PubMed] [Google Scholar]
- Serretti A, Chiesa A, Calati R, Massat I, Linotte S, Kasper S, Lecrubier Y, Antonijevic I, Forray C, Snyder L, Bollen J, Zohar J, De Ronchi D, Souery D, Mendlewicz J(2011)A preliminary investigation of the influence of CREB1 gene on treatment resistance in major depression. J Affect Disord 128:56–63. [DOI] [PubMed] [Google Scholar]
- Shi J, et al. (2016)Winner’s curse correction and variable thresholding improve performance of polygenic risk modeling based on genome-wide association study summary-level data. Plos Genet 12:e1006493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souery D, Oswald P, Massat I, Bailer U, Bollen J, Demyttenaere K, Kasper S, Lecrubier Y, Montgomery S, Serretti A, Zohar J, Mendlewicz J, Group for the Study of Resistant Depression (2007)Clinical factors associated with treatment resistance in major depressive disorder: results from a European multicenter study. J Clin Psychiatry 68:1062–1070. [DOI] [PubMed] [Google Scholar]
- Souery D, Serretti A, Calati R, Oswald P, Massat I, Konstantinidis A, Linotte S, Bollen J, Demyttenaere K, Kasper S, Lecrubier Y, Montgomery S, Zohar J, Mendlewicz J(2011)Switching antidepressant class does not improve response or remission in treatment-resistant depression. J Clin Psychopharmacol 31:512–516. [DOI] [PubMed] [Google Scholar]
- Spencer CC, Su Z, Donnelly P, Marchini J(2009)Designing genome-wide association studies: sample size, power, imputation, and the choice of genotyping chip. Plos Genet 5:e1000477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su TP, Chen MH, Li CT, Lin WC, Hong CJ, Gueorguieva R, Tu PC, Bai YM, Cheng CM, Krystal JH(2017)Dose-related effects of adjunctive ketamine in Taiwanese patients with treatment-resistant depression. Neuropsychopharmacology 42:2482–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tansey KE, Guipponi M, Hu X, Domenici E, Lewis G, Malafosse A, Wendland JR, Lewis CM, McGuffin P, Uher R(2013)Contribution of common genetic variants to antidepressant response. Biol Psychiatry 73:679–682. [DOI] [PubMed] [Google Scholar]
- Trivedi MH, Rush AJ, Wisniewski SR, Nierenberg AA, Warden D, Ritz L, Norquist G, Howland RH, Lebowitz B, McGrath PJ, Shores-Wilson K, Biggs MM, Balasubramani GK, Fava M, STAR*D Study Team (2006)Evaluation of outcomes with citalopram for depression using measurement-based care in STAR*D: implications for clinical practice. Am J Psychiatry 163:28–40. [DOI] [PubMed] [Google Scholar]
- Visscher PM, Hemani G, Vinkhuyzen AA, Chen GB, Lee SH, Wray NR, Goddard ME, Yang J(2014)Statistical power to detect genetic (co)variance of complex traits using SNP data in unrelated samples. Plos Genet 10:e1004269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CC, Held RG, Chang SC, Yang L, Delpire E, Ghosh A, Hall BJ(2011)A critical role for glun2b-containing NMDA receptors in cortical development and function. Neuron 72:789–805. [DOI] [PubMed] [Google Scholar]
- Xia Z, Storm DR(2005)The role of calmodulin as a signal integrator for synaptic plasticity. Nat Rev Neurosci 6:267–276. [DOI] [PubMed] [Google Scholar]
- Ye D, Li Y, Zhang X, Guo F, Geng L, Zhang Q, Zhang Z(2015)TREK1 channel blockade induces an antidepressant-like response synergizing with 5-HT1A receptor signaling. Eur Neuropsychopharmacol 25:2426–2436. [DOI] [PubMed] [Google Scholar]
- Zhang C, Li Z, Wu Z, Chen J, Wang Z, Peng D, Hong W, Yuan C, Wang Z, Yu S, Xu Y, Xu L, Xiao Z, Fang Y(2014)A study of N-methyl-D-aspartate receptor gene (GRIN2B) variants as predictors of treatment-resistant major depression. Psychopharmacology (Berl) 231:685–693. [DOI] [PubMed] [Google Scholar]