Abstract
CRISPR-Cas systems provide bacteria and archaea with adaptive immunity against invasion by bacteriophages and other mobile genetic elements. Short fragments of invader DNA are stored as immunological memories within CRISPR (clustered regularly interspaced short palindromic repeats) arrays in the host chromosome. These arrays provide a template for RNA molecules that can guide CRISPR-associated (Cas) proteins to specifically neutralize viruses upon subsequent infection. Over the past 10 years, our understanding of CRISPR-Cas systems has benefited greatly from a number of model organisms. In particular, the study of several members of the gram-negative Enterobacteriaceae family, especially Escherichia coli and Pectobacterium atrosepticum, have provided enormous insights into the mechanisms of CRISPR-Cas immunity. In this review, we provide an overview of CRISPR-Cas systems present in members of Enterobacteriaceae. We also detail the current mechanistic understanding of the type I-E and type I-F CRISPR-Cas systems that are commonly found in enterobacteria. Finally, we discuss how phages can escape or inactivate CRISPR-Cas systems, and the measures bacteria can enact to counter these types of events.
Introduction
In 1987, while analyzing the nucleotide sequence of the alkaline phosphatase isozyme conversion gene iap in the Escherichia coli K-12 genome, the group of Atsuo Nakata at Osaka University discovered an “unusual structure” just downstream of the iap gene, comprising five 29-bp repeating sequences with dyad symmetry, each separated by a 32-bp sequence (1). Subsequent analysis uncovered the same repeating sequences in two other enterobacteria, Shigella dysenteriae and Salmonella typhimurium (2). These early discoveries preceded the identification of similar loci in a wide range of bacteria and archaea throughout the 1990s (3, 4). However, the function of these loci remained a mystery for nearly 20 years. In the early 2000s, bioinformatic analysis of newly available bacterial and phage genomes facilitated the identification of the extrachromosomal origin of the repeat-interrupting sequences and the presence of a diverse but conserved set of genes associated with these loci (5–9). Together, these systems were termed CRISPR-Cas (clustered regularly interspaced short palindromic repeats-CRISPR associated) to designate the repeat-containing locus and the associated genes, respectively. CRISPR-Cas systems were hypothesized to comprise a bacterial defense mechanism against phage and other mobile genetic elements (10), which was first experimentally demonstrated in Streptococcus thermophilus in 2007 (11). The following year saw the publication of mechanistic CRISPR-Cas studies from several other organisms (12, 13), including E. coli (14), setting off a wave of research that has established the mechanistic underpinnings of these immune systems.
Throughout the ~30-year history of the CRISPR-Cas field, from the initial discovery of a CRISPR locus to the detailed elucidation of each step of immune response, E. coli has been an ideal and vital model organism for the study of CRISPR-Cas immunity. In this review, we highlight the central role E. coli and other members of the Enterobacteriaceae family have played in shaping our mechanistic understanding of adaptive immunity in bacteria.
Overview of CRISPR-Cas immune systems
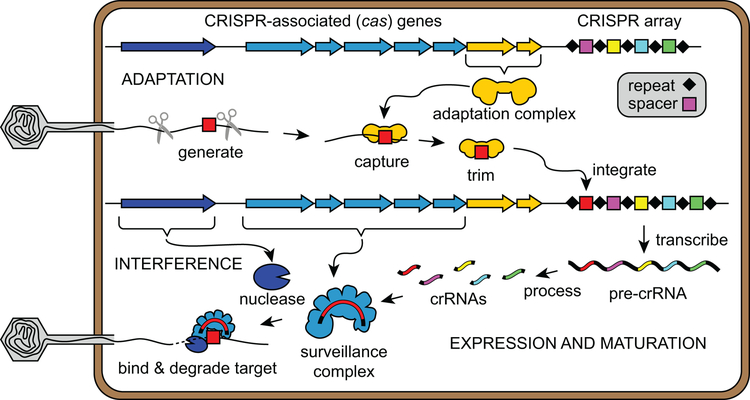
A functional CRISPR-Cas immune system consists of the CRISPR array, comprising a series of repeats interspaced by variable “spacer” sequences acquired from invasive nucleic acids (5–7), and a set of cas genes that serve as effectors for immune response (8–10) (Figures 1 and 2). The pathway to immunity can be divided into three stages: adaptation, expression and maturation, and interference (Figure 1). In the adaptation stage, Cas proteins capture short DNA fragments from “protospacer” regions of invasive nucleic acids and integrate these fragments as spacers into the host CRISPR array (11, 15, 16). The inserted fragment forms a genetic memory of the infection for subsequent immunity. During the expression and maturation stage, the CRISPR array is transcribed into a long pre-CRISPR RNA (pre-crRNA) that is further processed to generate CRISPR RNAs (crRNAs) (12, 14). Each crRNA assembles with Cas effector proteins to form a surveillance complex (17). During the interference stage, the surveillance complex searches the cell for the target protospacer based on complementary base-pairing with the crRNA spacer sequence (13, 14, 18). Target binding triggers nucleolytic cleavage and subsequent degradation of the target nucleic acid (13, 19, 20), thus neutralizing the infection.
Figure 1.
Overview of the three stages of type I CRISPR-Cas systems. CRISPR-Cas operons contain CRISPR arrays and CRISPR-associated (cas) genes. In the adaptation stage, short DNA fragments generated from phage DNA are captured by the adaptation complex. Following trimming of excess DNA, the adaptation complex integrates these fragments into the CRISPR array. During the expression and maturation stage, the CRISPR array is transcribed into long pre-CRISPR RNAs (pre-crRNAs). The pre-crRNAs are cleaved within the repeat regions to generate mature CRISPR RNAs (crRNAs). Each crRNA assembles with Cas proteins to form a surveillance complex. During the interference stage, the surveillance complex recognizes the target by complementary base-pairing with the crRNA sequence. Target binding triggers recruitment of a nuclease, which catalyzes degradation of the target nucleic acid.
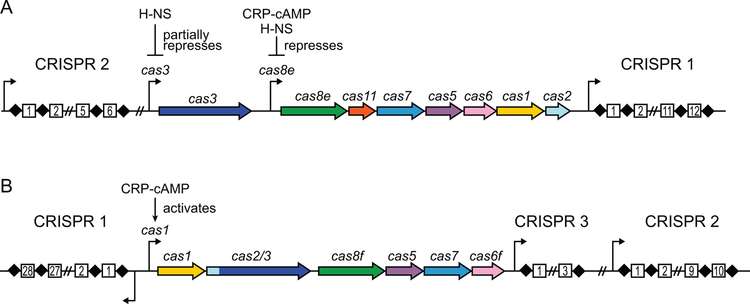
Figure 2.
CRISPR-Cas operons of E. coli K-12 and P. atrosepticum. A. Type I-E CRISPR-Cas operon of E. coli K-12. Promoters are shown as arrows. Repressors of the cas3 and cas8e promoters are indicated. B. Type I-F CRISPR-Cas operon of P. atrosepticum. CRISPR 1 is expressed on the minus strand. Activator of the cas1 promoter, which controls expression of all cas genes, is indicated.
CRISPR-Cas systems are extremely diverse due to constant co-evolution with phages and other mobile genetic elements (21). Despite this diversity, casl and cas2 are notably conserved across almost all known CRISPR-Cas systems. Cas1 and Cas2 are the only Cas proteins required for genetic recording of infections through spacer acquisition from invader DNA (15, 16), indicating a common memory generation mechanism across all CRISPR-Cas systems. In contrast, cas gene families involved in crRNA maturation and interference are diversified between different CRISPR-Cas systems (21), reflecting different mechanisms for these stages. Based on signature proteins and cas locus architecture, CRISPR-Cas systems are currently divided into two classes (class1 and class 2) that are subdivided into six types (types I-VI) and 33 subtypes (e.g. types I-A to I-F) (21, 22). Class 1 CRISPR-Cas systems (types I, III and IV) are characterized by multi-subunit surveillance complexes, while class 2 systems (types II, V and VI) utilize single-protein surveillance complexes. The relative simplicity of the class 2 single-protein effectors make them ideal candidates for repurposing as biotechnology tools for programmable cleavage of DNA or RNA targets (23). However, class 1 systems are far more prevalent in nature, and are present in ~90% of bacteria and archaea containing CRISPR-Cas systems (24).
Members of the Enterobacteriaceae family commonly contain type I-E and type I-F systems (Figure 2) (22, 25). These sub-types belong to class 1 and feature multiprotein surveillance complexes termed Cascade (CRISPR-associated complex for antiviral defense) in type I-E and Csy (CRISPR system Yersinia) complex in type I-F (Table 1) (14, 26, 27). Type I systems also feature a signature Cas3 nuclease/helicase protein that is required for target degradation during interference (19, 28–31). Cas3 is not a member of the surveillance complex and must be recruited to the complex following target recognition (19, 29, 32, 33). Thus, type I systems are unique amongst all CRISPR-Cas types in that the surveillance complex does not directly degrade the target. Here, we focus on the mechanisms of CRISPR-mediated immunity in types I-E and I-F, with emphasis on the lessons learned using enterobacteria as model organisms.
Table 1.
Cas proteins of type I-E and I-F CRISPR-Cas system.
| Name | Sub-type specifica | Function | |
|---|---|---|---|
| I-E | I-F | ||
| Cas1 | Cas1 | Cas1 | Integrase subunit of adaptation complex |
| Cas2 | Cas2 | Cas2/3 | Structural scaffold of adaptation complex |
| Cas3 | Cas3 | Nuclease/helicase for target dsDNA unwinding and degradation | |
| Cas5 | Cas5 (CasD) |
Cas5 (Csy2) |
Cascade/Csy subunit that recognizes 5’-handle of crRNA |
| Cas6 | Cas6 (Cse3) (CasE) |
Cas6f (Csy4) |
Cascade/Csy subunit required for crRNA maturation |
| Cas7 | Cas7 (Cse4) (CasC) |
Cas7 (Csy3) |
Cascade/Csy subunit forming backbone of complex, six copies present in complex |
| Cas8 | Cas8e (Cse1) (CasA) |
Cas8f (Csy1) |
Cascade/Csy complex subunit that recognizes PAM sequence and recruits Cas3 (or Cas2/3) |
| Cas11 | Cas11 (SS) (Cse2) (CasB) |
N/A | Cascade small subunit (SS), stabilizes non-target strand in R-loop, two copies present in complex |
Alternative names are shown in parentheses when applicable.
Overview of type I-E and I-F systems in enterobacteria
E. coli K-12 contains a type I-E system, and consequently this sub-type has served as a model system for understanding CRISPR-based immunity (14). The CRISPR-Cas locus in E. coli K-12 consists of a cas operon (8 genes) and two CRISPR arrays (CRISPR 1 and CRISPR 2) (Table 1, Figure 2A) (34, 35). Adaptation is mediated by the Cas1-Cas2 complex (15, 16, 36), while interference requires Cascade and Cas3 (14, 19). Cascade comprises five stoichiometrically unequal proteins that assemble with a 61-nt crRNA (Table 1) (14, 37). Surprisingly, under typical laboratory conditions, expression of the E. coli K-12 type I-E system is strongly repressed by the histone-like nucleoid-structuring (H-NS) protein, a global transcriptional repressor (34, 38–40). The two promoters that control cas gene expression (cas3 and cas8e promoters) are both at least partially under control of H-NS (Figure 2A) (34, 38, 40, 41). In addition, the cas8e promoter is repressed by the cAMP receptor protein (CRP)-cAMP complex (42). Production of crRNAs is also decreased in the presence of H-NS, either through the repression of the CRISPR promoters or through the lack of the Cas6e endonuclease that is required for crRN–A biogenesis (34, 38). Overall, native expression of the cas operon is insufficient to provide defense against phage infection or plasmid transformation (34, 40). Therefore, it is unclear how and under what conditions the E. coli K-12 CRISPR-Cas system can act as a defense mechanism. In vivo studies of E. coli K-12 CRISPR-Cas immunity have relied on heterologous expression in E. coli BL21-AI (14, 15, 43, 44), deletion of the hns gene (34, 40, 44–49), overexpression of the H-NS antagonist LeuO (40), or replacement of the cas3 and cas8e promoters (50–55). Nevertheless, these studies have provided remarkable insights into the mechanisms of immune response by the type I-E system.
The potato pathogen Pectobacterium atrosepticum, a member of Enterobacteriaceae, contains a type I-F system that has been studied extensively both in vivo and in vitro. The P. atrosepticum CRISPR-Cas locus contains three CRISPR arrays and six cas genes (Table 1, Figure 2B) (56). In type I-F systems, the cas2 and cas3 genes are fused into a single open reading frame (22), indicating a functional connection between the adaptation and interference stages of CRISPR-Cas immunity. Indeed, Cas1 and the Cas2/3 fusion protein assemble into a Cas1-Cas2/3 complex that has been implicated in adaptation (30, 57). Immunity in type I-F is mediated by the Csy complex, which contains four stoichiometrically unequal proteins that assemble with a 62-nt crRNA (26, 27, 58). Although the Csy complex is less well-studied than Cascade, recent structural investigations of the Csy complex from Pseudomonas aeruginosa have elucidated the mechanisms for target recognition and binding in type I-F (59, 60). Unlike E. coli, the CRISPR-Cas system in P. atrosepticum is an active immune system that is expressed in typical laboratory conditions and is activated by CRP-cAMP (Figure 2B) (56, 61). Thus, along with P. aeruginosa, P. atrosepticum has been the primary model organism for the study of type I-F systems.
Although CRISPR-Cas systems are widespread in other enterobacteria, it remains unclear whether these systems are active immune mechanisms as in P. atrosepticum, or repressed systems as in E. coli K-12. In general, expression and regulation of CRISPR-Cas systems varies substantially by species and sub-type (reviewed in (62)). For example, two recent studies revealed that CRISPR-Cas systems can be regulated through quorum sensing, including in the enterobacterium Serratia sp. ATCC39006 (63, 64). Although prediction of CRISPR-Cas regulation can be challenging, comparison of CRISPR loci can provide information on the history of immune activity. Differences in spacer sequences at the 5’-end of the CRISPR array indicate recent adaptation events during infection, while loss of spacers can also occur over time (65). The rapid evolution of CRISPR arrays enables discrimination between different strains of a single species, and can also provide insight into their recent immune activity (reviewed in (66)). CRISPR-based subtyping has been especially useful for discerning isolates of pathogenic enterobacteria, including plague-causing Yersiniapestis (6, 67), the plant pathogen Erwinia amylovora (68), and food-borne Salmonella and E. coli pathogens (69–71). Intriguingly, analysis of CRISPR sequences from diverse Salmonella and E. coli strains suggest that, similar to E. coli K-12, these systems are repressed by H-NS and are not currently active immune systems, although they may have been so at some point in evolutionary history (72–74). Indeed, spacer sequences from E. coli harvested from the remains of a 42,000-year old mammoth match spacers present in CRISPR arrays of modern E. coli (75). These observations suggest that the CRISPR-Cas systems of Salmonella and E. coli may function as alternative defense or regulatory mechanisms, as has been suggested by studies of E. coli K-12 and observed for other organisms (76–79).
Overview of Adaptation Mechanisms
In order to mount a specific immune response against infection, CRISPR immunity must begin with the acquisition of spacers from the invading genome, a process known as adaptation (Figure 1). In type I CRISPR-Cas systems, adaptation can occur through two different routes termed naïve and primed spacer acquisition (or priming) (15, 45, 50). Naïve adaptation occurs during infection by an invader that has not been previously encountered. In the E. coli type I-E system, Cas1 and Cas2 are the only Cas proteins required for naïve spacer acquisition (15, 16); however, studies of the P. aeruginosa type I-F system suggest that Cas1, Cas2/3 and the Csy complex are all required for non-primed adaptation (80). Primed adaptation occurs when a preexisting spacer is present in the CRISPR array, which can strongly promote spacer acquisition from the same invader (45, 50, 81). In this type of adaptation, the surveillance complex recognizes the target and directs spacer acquisition against the invader, a process that also requires Cas1, Cas2 and Cas3 (or Cas2/3) (50, 80). Studies in the native P. atrosepticum system indicate that primed adaptation occurs >500-times more efficiently than naïve adaptation (82), suggesting that most spacers are acquired through priming. Despite the difference between naïve and primed adaptation, both mechanisms can be divided into three steps: the generation of “prespacer” DNA fragments (Figure 3), the capture of prespacers by the adaptation complex (Figure 4), and the integration of the prespacer into the CRISPR locus (Figure 5). Although the mechanisms of prespacer generation and capture varies between naïve and primed adaptation, the mechanism for prespacer integration is thought to be the same. In this section, we summarize the current understanding of these three stages of adaptation in the type I-E and type I-F systems.
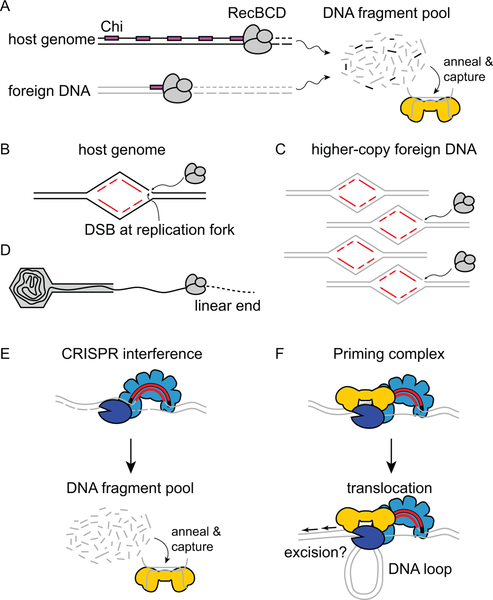
Figure 3.
Prespacer generation during CRISPR-Cas adaptation. A. During naïve adaptation, RecBCD products serve as substrates for the Cas1-Cas2 adaptation complex (gold). The abundance of Chi sites (magenta) prevents accumulation of DNA fragments from the host genome, while lack of Chi sites in foreign DNA leads to a larger pool from this source. B-C. Replication forks are common sites for double-strand breaks (DSB), which are initiation sites for RecBCD degradation sites. Higher-copy foreign DNA contains more replication forks, and therefore leads to more DSBs and RecBCD recruitment. D. Phage DNA is often injected in a linear form, and these ends are subject to degradation by RecBCD. E-F. Two potential mechanisms for primed adaptation. E. During interference-dependent primed adaptation, the surveillance complex (light blue) directs Cas3 (dark blue) degradation of the foreign DNA. Cas3 products create a DNA fragment pool that provides prespacer substrates for Cas1-Cas2. F. During interference-independent primed adaptation, both Cas3 and Cas1-Cas2 (or Cas1-Cas2/3) are recruited to the target-bound surveillance complex. The Cas3 helicase domain extrudes a DNA loop as the priming complex searches for prespacer substrates and excises them, either through the nuclease activity of Cas1, Cas3 or both subunits.
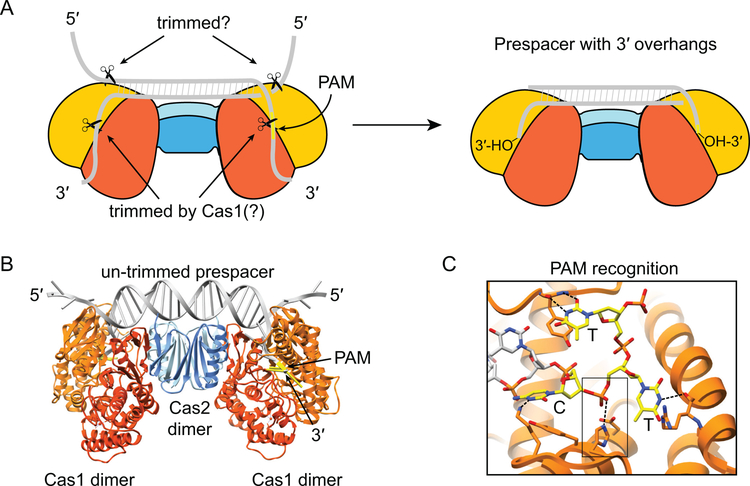
Figure 4.
Prespacer trimming during adaptation. (A) Schematic illustration of E. coli Cas1-Cas2 bound to 23-bp duplex with splayed ends. Individual subunits of Cas1 are showed in orange or gold, individual subunits of Cas2 are shown in light or darker blue, DNA substrate is shown in gray, PAM is shown in yellow. The 3’-overhangs, one of which contains the PAM, are located in the Cas1 active sites, suggesting trimming of these ends may be carried out by Cas1. The 5’-overhangs are exposed, and may be trimmed by another factor. The trimmed product is a prespacer containing a 23-bp duplex with 5-nt 3’-overhangs containing exposed 3’-hydroxyl groups. B. Structure of the Cas1-Cas2 complex bound to an untrimmed substrate (PDB ID: 5DQZ) (93). Colored as in A. C. Close-up of Cas1 active site recognition of 5’-CTT-3’ PAM (yellow) region of untrimmed prespacer. Hydrogen-bonding interactions are shown with dashed lines. The boxed region shows the scissile phosphate, which is coordinated by the catalytic histidine and aspartate residues of the HD domain. Following cleavage, only the cytosine is retained as the last nucleotide of the trimmed substrate.
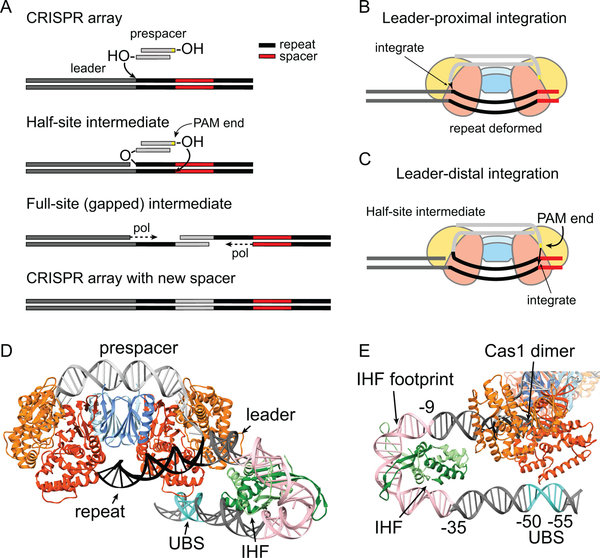
Figure 5.
Integration of prespacers during adaptation. A. Schematic overview of integration process in E. coli. A 3’-OH group of the prespacer attacks the backbone at the leader-proximal end of the first repeat within the CRISPR array, forming a half-site intermediate. The end of the prespacer containing the cytosine from the PAM attacks at the leader-distal end of the opposite strand of the repeat, forming a full-site intermediate. The single-stranded repeats on either end of the integrated spacer are filled by DNA polymerase I, and the nicks are ligated to create and intact CRISPR array. B-C. Schematic of leader-proximal and leader-distal integration by E. coli Cas1-Cas2 (colored as in Figure 4). Cas1-Cas2 senses the sequence-dependent deformation of the repeat to facilitate integration at the correct sites within the repeat. D-E. Cryo-EM structure of Cas1-Cas2 bound to half-site intermediate containing IHF-bound leader (PDB 5WFE) (109). IHF (dark and light green subunits) binds positions −9 to −35 of the leader (pink) and bends the DNA by ~180°. This bending brings the upstream binding element (UBS, turquoise) into proximity of the leader-proximal Cas1 dimer, an interaction that ensures integration at the leader-proximal repeat.
(a). Prespacer generation
During the first stage of adaptation, short fragments of DNA must be generated to serve as substrates for capture and integration by the adaptation complex. Importantly, these prespacers must be preferentially generated from the foreign DNA to limit toxicity of the immune system. Spacers acquired from the host chromosome cause autoimmunity and cell death (44). An early study of naïve acquisition in E. coli revealed preferential acquisition of spacers from foreign DNA, even though the system used in the study lacked the CRISPR interference machinery and therefore had no selective pressure against self-acquisition (15). This study suggested the presence of an intrinsic mechanism for biasing spacer acquisition against foreign DNA. A later study in E. coli showed that this bias is at least partially based on prespacer generation through RecBCD-catalyzed DNA degradation (Figure 3A) (83). When double-stranded breaks (DSBs) form, RecBCD binds the free DNA ends and processes it to the nearest Chi site (84). Chi sites are more abundant in the host genome, resulting in a limited pool of potential chromosomal spacers. Compared to the host DNA, extrachromosomal DNA from phage or plasmids are replicated more frequently, resulting in a higher likelihood for stalled replication forks, which are common sites for DSBs (Figure 3B–C) (85–87). In addition, Chi sites are relatively rare in extrachromosomal DNA, resulting in continuous degradation by RecBCD (Figure 3A). As a result, more potential substrates for spacer acquisition are generated from extrachromosomal DNA. Indeed, spacer acquisition from foreign DNA can be reduced by adding Chi sites into the foreign DNA (83). Some extrachromosomal DNA, such as phage genomes, enter the host cell as linear dsDNA (Figure 3D). The free end can be recognized by RecBCD directly, resulting in continuous degradation and the formation of potential substrates for spacer acquisition.
Although RecBCD has been implicated in spacer generation, naïve adaptation can occur in strains where components of the RecBCD complex are deleted, indicating that other events can also promote naïve adaptation (83). However, spacer acquisition in these deletion strains is no longer biased toward extrachromosomal DNA, indicating that other spacer generation mechanisms are equally effective against both host and foreign DNA. The reduced bias of spacer acquisition from extrachromosomal DNA in RecBCD depleted cells further indicates that self vs non-self spacer acquisition is directly related to the mechanism by which substrates are generated during naïve adaptation.
During primed adaptation, prespacer generation is coupled with the interference machinery (Figure 3E–F) (50, 80). Currently, there are two proposed spacer generation pathways during primed adaptation. In the interference-dependent pathway, target degradation by Cas3 supplies substrates for primed adaptation (Figure 3E) (51, 54, 82, 88), similar to RecBCD degradation providing substrates for naïve adaptation. Following target binding by the surveillance complex, Cas3 (or Cas2/3) is recruited by the Cas8 subunit and degrades the target DNA by unwinding and nicking one strand at various intervals (28, 32, 89, 90). The products form a pool of potential substrates that can be captured by Cas1-Cas2 (52, 88). In the interference-independent pathway, a priming complex between Cas3 and Cas1-Cas2 is recruited by the surveillance complex following target recognition (Figure 3F) (30, 32, 57, 90). This complex translocates along the DNA and directly excises double-stranded prespacers from the foreign DNA (32, 90). For both pathways, primed adaptation is triggered only upon Cascade-target recognition, ensuring that prespacers are derived only from the foreign DNA. Thus, primed adaptation is inherently biased toward non-self DNA, and can serve as a mechanism to boost self vs. non-self discrimination during adaptation.
Although the putative Cas1-Cas2-Cas3 priming complex has not been directly observed in the E. coli type I-E system, several lines of evidence support the interference-independent pathway. Some target mutations completely block degradation by Cas3, but these targets can still promote priming in E. coli (32, 46, 47, 91) (discussed further in section “Effects of target mutations“). Consistently, within the type I-F Cas1-Cas2/3 complex, Cas1 inhibits target degradation by Cas2/3 (30), while in the E. coli type I-E system, Cas3 nuclease activity is blocked when it is recruited to target-bound Cascade in the presence of Cas1-Cas2 (32). These studies suggest that recruitment of Cas3 in conjunction with the Cas1-Cas2 complex prevents processive degradation of the target and instead elicits more precise prespacer excision (Figure 3F). Primed spacer acquisition studies in type I-F have also indicated that the helicase activity of Cas2/3 may enable translocation of the Cas1-Cas2/3 complex following recruitment to the surveillance complex, based on the proximity of acquired spacers to the original target site (80, 82). Recent single-molecule studies of the Thermodifida fusca type I-E system confirm this model (90). In this system, target-bound Cascade directly recruits Cas1-Cas2 and Cas3, and this overall complex translocates processively along the DNA. Cascade remains bound to the target, so translocation occurs through extrusion of a DNA loop, similar to DNA looping that has been observed for the E. coli type I-E system (Figure 3F) (32). This mechanism is thought to allow the priming complex to search for and excise double-stranded substrates for spacer integration, although these steps in the process have not yet been demonstrated.
(b). Prespacer capture and trimming by Cas1-Cas2
Following prespacer generation by RecBCD, Cas3 or other nucleases, the potential prespacers must be captured by the Cas1-Cas2 complex for integration into the CRISPR array. Cas1-Cas2 is a heterohexamer containing a Cas2 dimer sandwiched by two Cas1 dimers (Figure 4A–B) (36). The optimal substrate for E. coli Cas1-Cas2 is a 33-bp prespacer, which contains a central 23-bp double-stranded duplex with 5 nt overhangs on each side splayed into ssDNA ends by tyrosine wedges in Cas1 (92, 93). The length of the central double-stranded duplex is governed by the fixed distances between the two Cas1 wedges. The length of the prespacer is determined by the distance between two Cas1 integrase active sites. The 23-bp duplex sits on the flat surface of Cas1-Cas2, which positions the 3’ ends into the catalytic site of each Cas1 (Figure 4B–C).
To be recognized by Cas1-Cas2, it has been proposed that ssDNA substrates generated from naïve acquisition (RecBCD degradation) or interference-dependent primed acquisition (Cas3 degradation) must anneal to form double stranded Cas1-Cas2 substrates (Figure 3A, E) (83, 93, 94). Both RecBCD and Cas3 degrade DNA asymmetrically, yielding various length ssDNA fragments for both strands. These DNA fragments putatively re-anneal to provide a dsDNA substrate pool for Cas1-Cas2, although it remains unclear whether annealing occurs spontaneously or is assisted by Cas1-Cas2 or another host factor. Another possibility is that the Cas1-Cas2 complex initially binds to ssDNA, and then the second strand may be generated by a DNA polymerase to form a dsDNA (94). Although it has been shown that the E. coli Cas1-Cas2 complex cannot bind or integrate ssDNA in vitro (16, 95), it is possible that other host enzymes may assist with the use of these types of substrates. Consistently, DNA fragments bound by E.coli Cas1-Cas2 in vivo are resistant to digestion by restriction endonucleases, suggesting that Cas1-Cas2 can associate with non-dsDNA in vivo (52). In addition, the Cas1-Cas2/3 complex in the type I-F system can bind short ssDNA substrates directly with similar affinities as for dsDNA, suggesting that Cas3 can assist with the capturing of ssDNA by Cas1-Cas2 (57).
Early analysis of protospacers targeted by CRISPR-Cas systems revealed a conserved 3-bp protospacer adjacent motif (PAM), present in the foreign DNA but not in the spacer within the host CRISPR array (96). Subsequent studies have shown that the PAM is required for optimal target binding by Cascade or the Csy complex and cleavage by Cas3 (discussed further below) (28, 29, 32, 49, 91, 97). The requirement for a PAM during CRISPR interference necessitates that prespacers be selected from sites with correct PAM sequences during adaptation. In the E. coli type I-E system this specificity is achieved through intrinsic PAM recognition by the Cas1-Cas2 complex (Figure 4C), which has high binding affinity for prespacer substrates with a canonical PAM (5’-CTT-3’) (93). However, Cas1-Cas2 can also capture prespacers with non-canonical PAMs, albeit with lower affinity (83, 98, 99). Depending on Cas1 and Cas2 expression level, a substantial fraction (>50%) of acquired spacers may have non-canonical PAMs (83, 99), resulting in reduced immunity. In contrast, primed spacer acquisition is highly accurate for selecting protospacers with canonical PAM sequences, even under high cas gene expression (50, 100). Cas3 preferentially cleaves T-rich sequences, generating 30–100 nt ssDNA fragments that are enriched for 5’-NTT-3’ PAMs at the 3’-end (88). Cas1-Cas2 selection of prespacer substrates from this 5’-NTT-3’ PAM enriched pool may account for the higher PAM specificity of primed adaptation.
In P. atrosepticum, both naïve and primed adaptation are comparably accurate for selecting protospacers with correct PAM sequences (5’-GG-3’) (82). This may be due to the native expression of Cas1-Cas2/3, in comparison to Cas1-Cas2 overexpression experiments performed in E. coli. It may also reflect a higher intrinsic specificity for PAM sequences by the type I-F Cas1-Cas2/3 complex in comparison to the type I-E Cas1-Cas2 complex. Notably, Cascade is highly promiscuous for PAM sequences, and can recognize targets with >20 different PAMs depending on the crRNA sequence (46, 47, 101). Although the PAM specificity of the Csy complex is less well studied, it is possible that both the adaptation and interference machinery of type I-F have evolved more stringent PAM specificity than their type I-E counterparts.
Because RecBCD and Cas3 products are generally longer than the optimal prespacer substrate, it is likely that they must be trimmed to the correct size prior to integration. Both type I-E and I-F Cas1 can cleave prespacers site-specifically, albeit with low activity (57, 93). In other type I sub-types, an additional protein, Cas4, is required for efficient prespacer trimming, which ensures that only processed substrates are integrated (102–105). The lack of Cas4 in type I-E and I-F suggests that Cas1 alone may be responsible for prespacer trimming, or that another non-CRISPR-associated factor may be required for optimal trimming activity. In the E. coli substrate-bound Cas1-Cas2 structure, the 3’-ssDNA ends are located in the symmetric Cas1 active sites on either end of the heterohexameric complex, consistent with cleavage activity of these ends by Cas1 (Figure 4C) (92, 93). The 5’-ssDNA ends do not interact with Cas1-Cas2 and are exposed to solvent, suggesting they could potentially be degraded by other endo-or exonucleases (Figure 4A–B). It is also possible that re-annealed dsDNA substrates can be processed to suitable protospacer substrates before binding by Cas1-Cas2 through an unknown mechanism (94).
Prespacer generation and capture are likely more intimately linked during interference-independent primed adaptation, as the Cas1-Cas2 complex may directly excise and capture prespacers as part of the priming complex (Figure 3F). It remains unknown whether Cas1 or Cas3 nuclease activities is involved in the excision of prespacers through this mechanism, and whether further trimming of the prespacer is necessary following this excision. It also remains unclear whether Cas1-Cas2 alone recognizes PAM sequences during this process, or whether Cas3 and/or Cascade also contribute to PAM selection. In the type II CRISPR-Cas system, the Cas9 surveillance complex is an essential member of the adaptation complex and is required for selection of prespacers with correct PAM sequences (106). Future studies are required to determine whether Cascade and the Csy complex play similar roles during primed adaptation in type I-E and I-F systems.
(c). Spacer Integration
Once loaded with the prespacer, Cas1-Cas2 recognizes the CRISPR array and catalyzes spacer integration (Figure 5A). New spacers are integrated at the leader-proximal end of the CRISPR array (15, 45, 50, 83, 98, 107). This polarization of the CRISPR array can result in higher abundance of crRNAs containing newly incorporated spacers, ensuring that the most recent immunization events provide optimal anti-viral response (108). The Cas1-Cas2 complex positions the two 3’-OH groups of the prespacer to catalyze two nucleophilic attacks on each strand at either end of the leader-proximal repeat (Figure 5A–C) (16, 92, 93, 109, 110). The two transesterification reactions result in two single-stranded repeats flanking the newly integrated spacer (Figure 5A) (16, 110). This gapped intermediate is putatively repaired by DNA polymerase I and DNA ligase to yield an intact CRISPR (55).
The Cas1-Cas2 complex must specifically recognize the CRISPR locus to ensure correct genetic recording for optimal immunity. Early studies of adaptation in E. coli revealed that the last 60-bp of the leader are essential for spacer acquisition (15), suggesting that the adaptation complex recognizes sites within the leader. Subsequent biochemical and structural studies have shown that Cas1 directly reads the sequence just upstream of the leader-repeat junction (positions −1 to −4) to ensure newly incoming spacers are inserted at the leader-proximal repeat instead of other repeat sites (16, 109, 111). An additional protein, integration host factor (IHF), is also required for efficient and site-specific spacer integration in both type I-E and I-F systems (57, 107, 112). IHF is a histone-like protein that binds at positions −9 to −35 of the leader in E. coli (107). The cryo-EM structure of E. coli Cas1-Cas2 and IHF bound to the leader shows that IHF bends the leader DNA by ~180° (Figure 5D–E) (109). This bending directs an upstream recognition motif located at positions −50 to −55 of the leader close to Cas1 for a second specificity check prior to integration. Like IHF, the upstream recognition motif is essential for efficient integration at the leader-repeat junction (109).
Aside from integrating spacers at the correct site, Cas1-Cas2 must also ensure that spacers are integrated in the correct orientation. The PAM-proximal end of the prespacer must be integrated at the leader-distal end of the repeat to produce a crRNA targeting the correct protospacer strand (Figure 5A–C). Although it has been shown that the presence of a PAM in the incoming spacer is sufficient for correct orientation in vivo (113), the mechanism guiding spacer orientation remains unclear. In E. coli, the first nucleotide of the 5’-CTT-3’ PAM is retained at the 3’-end of the prespacer following trimming (Figure 5B–C). This cytosine is integrated as part of the new spacer, but functionally serves as the last nucleotide of the repeat (45, 50, 114). Interestingly, it has been shown that Cas1-Cas2 preferentially positions prespacer ends containing 3’-cytosines to the repeat-spacer site for nucleophilic attack, resulting in correct orientation of the incoming spacer (111). However, in the P. atrosepticum type I-F system, prespacer trimming occurs adjacent to the 5’-GG-3’ PAM, and no portion of the PAM is retained in the substrate (57). This suggests that PAM-end processing of the prespacer may not occur until after Cas1-Cas2 orients itself at the CRISPR (115).
Another factor that may dictate spacer orientation is the order in which the two transesterification reactions are catalyzed. E. coli Cas1-Cas2 catalyzes both integration and disintegration at the leader-repeat junction significantly faster compared to the repeat distal end in vitro, indicating that the first attack occurs at the leader-repeat junction to form a half-site intermediate (Figure 5A, C) (107, 111). The initiation of integration from the leader-repeat junction may result from stabilizing contacts between Cas1 and the leader (109). Consistently, Cas1-Cas2 complexes in the type I-C and II systems preferentially catalyze spacer integration at the leader-repeat junction, even in the absence of other host factors (102, 116, 117). However, P. atrosepticum Cas1-Cas2/3 shows no bias toward integration at either end of the repeat, suggesting that other host factors are required to dictate the orientation of spacer integration in the type I-F system (57).
After the first nucleophilic attack, Cas1-Cas2 must orient the other 3’ end of the substrate for the second attack to generate the full-site integration product. Early studies suggested that the palindromic sequences within the repeat may form a cruciform structure to accommodate two nucleophilic attacks at either end of the repeat (118). However, Cas1-Cas2 cannot bind a repeat in which the palindromic sequences are swapped (95). The swapped repeat also abolishes spacer integration in vivo (110). These results suggest that Cas1-Cas2 recognizes the repeat sequence itself, instead of the putative cruciform structure. This was further supported by recent structures of E. coli Cas1-Cas2 bound to half-site and pseudo-full-site integration products (Figure 5D) (109). In these structures, Cas1-Cas2 contacts the duplex repeat instead of a cruciform structure. Interestingly, Cas1-Cas2 interacts with the repeat in a non-sequence-dependent manner and Cas1-Cas2 binding causes severe bending and underwinding of the repeat DNA. These structures suggest that Cas1-Cas2 recognizes the repeat through indirect readout based on the sequence-dependent deformability of the repeat.
Overview of crRNA Maturation and Interference
Following integration of a spacer into the CRISPR array, the CRISPR must be transcribed and processed to form mature crRNAs (Figure 1, 6) (14, 119). These guide molecules assemble with Cas proteins to form the Cascade or Csy surveillance complex (Figure 6) (14, 26, 60, 120–122). The complex must search the milieu of the cellular DNA to locate the rare target sequences, a daunting task that is simplified by first searching for PAM sequences (Figure 7) (32, 43, 49, 90, 123). Upon location of the correct site, the surveillance complex unwinds the DNA, facilitating hybridization of the crRNA and target strand of the DNA, and displacing the non-target strand to form an R-loop (37, 59, 124–128). Formation of the R-loop locks the complex at the target site and enables recruitment of Cas3, which processively degrades the DNA (Figure 8) (19, 28, 29, 32, 89, 90, 129, 130). In this section, we discuss the steps that lead to CRISPR interference.
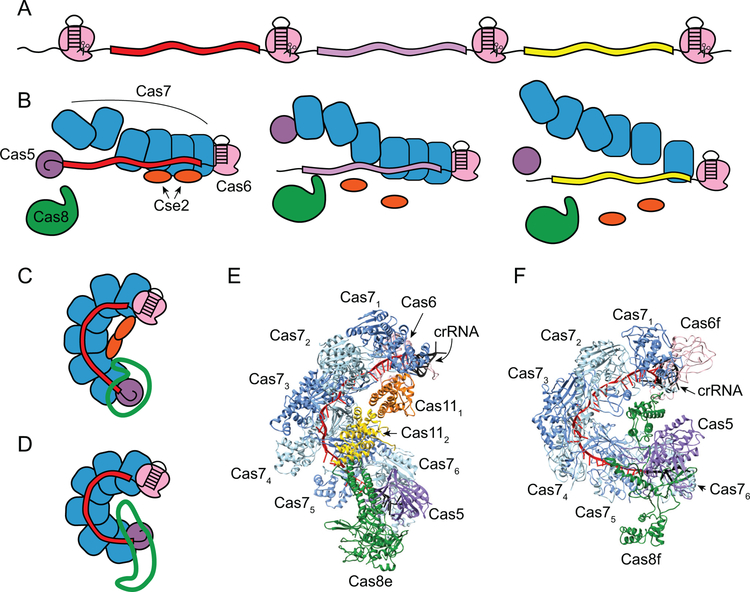
Figure 6.
Expression and maturation of crRNAs and assembly of Cascade and Csy surveillance complexes. A. The product of transcription of the CRISPR is a pre-crRNA containing stem-loops within the repeats. Cas6 endoribonucleases (pink) recognize the stem-loops and cleave at the base to form mature crRNAs. B. Cas6 subunits remain bound to the stem-loop at the 3’-end of the crRNA following cleavage. Six Cas7 subunits (blue) assemble along the length of the crRNA spacer (red), and the Cas5 subunit (purple) caps the Cas7 backbone by specifically recognizing the 5’-repeat-derived handle. In type I-E Cascade, two subtype-specific subunits, Cse2 (orange), assemble on the belly of the complex. The Cas8 subunit (green) caps the complex on the Cas5 end. C. Schematic of assembled Cascade (C) or Csy (D) complex. Subunits are colored as in B, except for Cas8e (C) or Cas8f (D), which are shown in green outline. E-F. Structures of fully assembled E. coli Cascade (PDB ID 4TVX) and P. aeruginosa Csy (PDB ID 6B45) complex (59, 120).
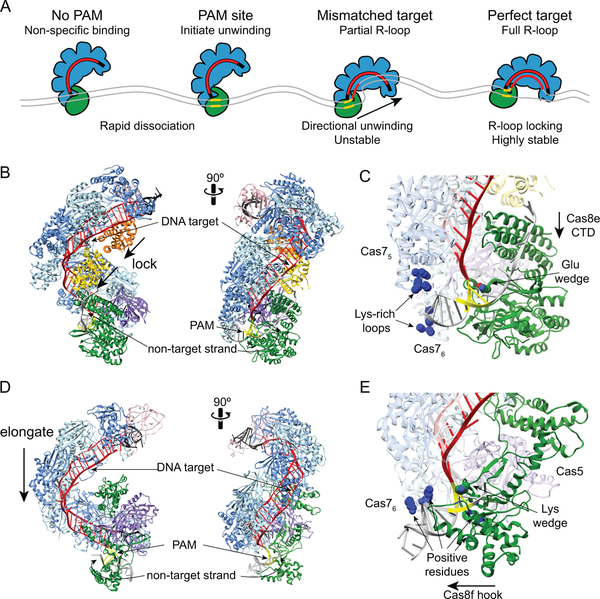
Figure 7.
Target search and R-loop formation by Cascade and Csy complexes. A. Overview of target search by E. coli Cascade. Non-specific interactions at sites with no PAM or PAMs adjacent to non-matching sequences result in rapid dissociation. At PAM sites, Cascade can initiate unwinding to enable crRNA strand invasion. At partially matching target, R-loops can form directionally away from the PAM until a mismatch is encountered. Depending on the location and thermodynamic barrier of the mismatch, the R-loop can stall, continue to form or Cascade can dissociate. At a perfect target, complete R-loop formation leads to Cascade locking and a highly stable interaction. B. Structure of E. coli Cascade bound to dsDNA (PDB ID 5H9F, colored as in Figure 6) (127). The Cas11 subunits move down along the belly during R-loop locking. The non-target strand is stabilized through interactions with Cas8e. C. Close-up of Cas8e-PAM and Cascade-dsDNA interactions. A glutamate residue acts as a wedge to facilitate DNA unwinding. The PAM is read through minor-groove interactions with Cas8e. Lys-rich loops in Cas75 and Cas76 subunits act as an electrostatic vise to hold the dsDNA during unwinding. The Cas8e CTD four-helix bundle moves toward the N-terminal domain during R-loop locking. D. Structure of P. aeruginosa Csy complex bound to dsDNA (PDB ID 6B44, colored as in Figure 6) (59). The Cas7 backbone elongates by ~20 Å upon DNA binding. E. Close-up of Cas8f-PAM and Csy-dsDNA interactions. A lysine residue acts as a wedge to facilitate DNA unwinding. The PAM is read through minor-groove interactions with Cas8f. Several positive residues in Cas76, Cas8f and Cas5 stabilize interactions with the dsDNA. The Cas8f hook elongates to accommodate the dsDNA.
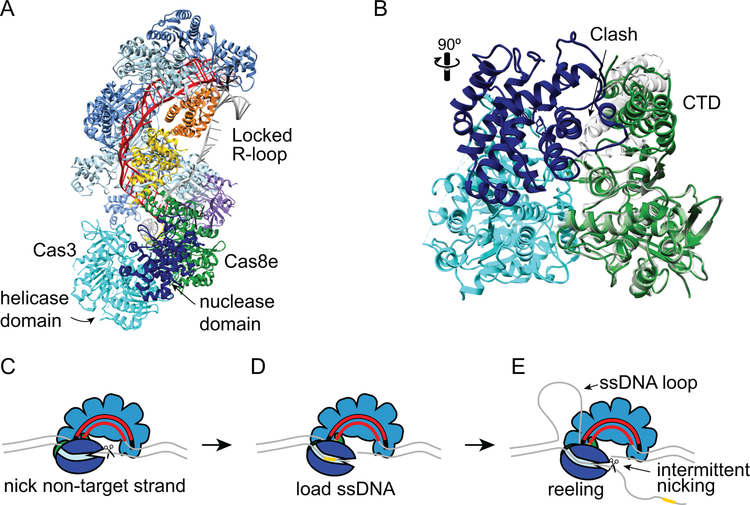
Figure 8.
Target degradation by Cas3 during CRISPR interference. A. Structure of type I-E T. fusca Cascade-dsDNA following Cas3 recruitment (PDB ID 6C66) (33). Cascade and DNA are colored as in Figures 6 and 7, Cas3 is shown in cyan (helicase and C-terminal domains) and dark blue (HD nuclease domain). Cas3 senses the conformation of Cas8e following R-loop locking. Comparison of Cas8e structure in the locked (PDB ID 6C66, green) and unlocked (PDB ID 5U07, gray) conformations (33, 128). Prior to R-loop locking and movement of the CTD, a loop and helix of the Cas8e CTD would cause steric clashes with the Cas3 nuclease domain. C-E. Schematic of E. coli Cas3 DNA reeling and degradation. C. Following its recruitment to the R-loop, Cas3 nicks the non-target strand. D. The nicked strand is loaded into the Cas3 helicase domain, stabilizing the Cas3-Cascade interaction. E. The Cas3 helicase domain reels the non-target strand, while the target strand is extruded and forms a ssDNA loop. The HD nuclease domain can nick the non-target strand intermittently, creating products that could serve as prespacer substrates for Cas1-Cas2 (Figure 3E), or it can release the DNA and repeat the reeling motion. In type I-F systems, two Cas2/3 molecules may be recruited to the Csy-complex, allowing for bidirectional degradation of both strands of the DNA.
(a). CRISPR RNA biogenesis and surveillance complex formation
The core component of Cas surveillance complexes is the mature crRNA, which guides the complex to foreign nucleic acid, triggering immunity (12, 14). CRISPR arrays are first transcribed into long precursor CRISPR RNA (pre-crRNA), that often contains stem-loops in the palindromic repeats (Figure 6A). In type I-E and I-F, the pre-crRNA is recognized and cleaved by a Cas6 endoribonuclease to yield mature crRNA (14, 56, 119, 131–133). Cas6 specifically recognizes the stem-loop within the repeat and cleave at the base of the stem, ensuring that cleavage only occurs within the repeat sequences of the corresponding pre-crRNA (119, 131, 132). Cas6 proteins are metal-independent endoribonucleases that use general acid/base catalytic mechanisms, in which the nucleophilic attack is initiated by the deprotonated hydroxyl at the 2’ position of the ribose adjacent to the scissile phosphate (134).
Following cleavage, the mature crRNA is composed of a single spacer sequence, flanked on either end by segments of the repeat (Figure 6B). Both the type I-E and I-F Cas6 endoribonucleases have been shown to remain tightly bound to the stem-loop at the 3’-end of the cleaved crRNA, allowing these subunits to nucleate formation of their respective surveillance complexes (131, 135). The remaining subunits of the complexes assemble along the length of the mature crRNA (Figure 6B–F) (26, 37, 59, 60, 120–122, 127, 136). Six copies of Cas7 subunits polymerize non-specifically along the crRNA to form the backbone of the complex. Cas5 specifically recognizes the repeat-derived 5’ handle of the mature crRNA, capping the Cas7 backbone of the complex. Notably, for the E. coli Cascade complex, the stoichiometry of Cas7 subunits can be altered by lengthening or shortening the spacer region within the crRNA (137, 138), indicating that the distance between the Cas5 and Cas6 repeat-derived binding sites determines the stoichiometry of the complex. In Cascade, two copies of Cas11 small subunits interact with Cas7 subunits through salt bridges to form the belly that helps stabilize the crRNA and target DNA (Figure 6C, E) (120, 122, 126–128, 139, 140). While Cas11 subunits are found in surveillance complexes from sub-types within both type I and III, type I-F lacks a cas11 gene. Lastly, the Cas8 large subunit caps the Cas5 end of the complex (Figure 6C–F).
(b). PAM searching and recognition
The first step during interference is target recognition by the surveillance complex (Figure 7A). To initiate timely response against phage, the surveillance complex must be able to efficiently locate targets among the vast amount of DNA in the cell. At the same time, the surveillance complex has to avoid targeting complementary sequences in the bacterial genome. The PAM sequence allows Cascade and the Csy complex to distinguish foreign targets from the complementary sequence in the CRISPR array (43, 49, 97, 141). Because there is no PAM present in the CRISPR array, the surveillance complex cannot bind the CRISPR template strand that is complementary to the crRNA spacer. In E. coli, the last three base pairs of the repeat protect the CRISPR array from self-targeting. E. coli Cascade has broad specificity for PAM sequences (46, 47, 101), but it cannot recognize the 5’-CCG-3’ sequence found at the end of the repeat. In particular, the cytosines at the −2 and −3 positions are deleterious to Cascade binding (47, 101). Thus, the end of the repeat sequence prevents both interference and primed adaptation against the host chromosome (47).
Aside from their importance for self-versus-non-self discrimination, PAM sequences are also essential for efficient location of the target sequence. An early study of CRISPR immunity in E. coli revealed that phages could escape interference by developing spontaneous mutations in the PAM region, as well as the first several positions of the target proximal to the PAM (49). In addition, PAM sequences were shown to be required for high-affinity Cascade binding to dsDNA, but not ssDNA, targets (43). These observations suggested a model in which Cascade first searches for PAM sequences and initiates target unwinding and binding from that site (Figure 7A). Recognition of sequences that are sequestered within dsDNA requires at least partial unwinding of the duplex to probe for complementarity with the crRNA. The “PAM first” model predicts that Cas surveillance complexes only initiate unwinding at PAM sites that may or may not be located next to the target (43). This strategy substantially decreases the number of sites at which the DNA must be unwound, minimizing the time and energy required for target location.
In subsequent years, considerable evidence has accumulated in support of the PAM first model in types I, II and V, all of which target dsDNA (32, 142–144). Early bulk biochemical assays of E. coli Cascade revealed that the complex binds “nonspecific” dsDNA that lacks a target site, interactions that were hypothesized to be part of a PAM scanning mechanism (Figure 7A) (37, 43). Consistently, the binding efficiency of both Cascade and Csy complex decreases in the presence of PAM-rich competitor DNA sequences (97, 123). Single molecule approaches have confirmed that Cascade and other dsDNA-targeting surveillance complexes sample DNA through both nonspecific and PAM-specific interactions (32, 123, 142–144). While these studies suggest that E. coli Cascade searches for PAM sequences via three-dimensional collisions, single-molecule DNA curtains studies of T. fusca Cascade revealed one-dimensional diffusion for that complex based on a conserved positively charged patch in the Cas8 subunit (90). Although E. coli Cascade may also diffuse along short stretches of DNA, bulk biochemical and single molecule studies suggest the target searching process by both Cascade and Csy complexes is accelerated by rapid dissociation and association rates at nonspecific PAM sites along the dsDNA (32, 97, 123).
The Cas8 subunit is responsible for PAM recognition in both Cascade (Figure 7B,C) and the Csy complex (Figure 7D,E) (43, 59, 127). Multiple peptide motifs within Cas8 read out the minor groove of the PAM (59, 127). Specific interactions between Cas8 and PAM sequences stall the surveillance complex at PAM sites and initiate DNA unwinding (Figure 7A) (32, 90, 123). Two additional motifs in E. coli Cascade have been implicated in nonspecific and PAM-specific interactions. A lysine-rich P-hairpin in Cas8e helps to position the dsDNA for PAM readout (123). In addition, two lysine-rich loops in adjacent Cas75 and Cas76 subunits of the Cascade backbone form a positively-charged vise that contacts the DNA backbone through nonspecific electrostatic interactions (Figure 7C) (126). This motif is required for nonspecific DNA sampling and for specific target binding and is thought to hold the DNA in place during PAM recognition and DNA unwinding (123, 126). Similarly, a positively-charged cleft comprising regions of Cas8f, Cas5 and Cas76 creates a vise for positioning dsDNA during PAM recognition by the Csy complex (Figure 7E) (59).
(c). DNA unwinding and R-loop formation
During PAM recognition, the surveillance complex initiates DNA unwinding, allowing for interrogation of the adjacent DNA for complementarity with the crRNA. When Cascade locates a PAM site, favorable interactions between Cas8e and the PAM are thought to enable dsDNA bending, destabilizing the dsDNA at the PAM-proximal region (43, 123, 126–128). The subsequent unwinding of the bent dsDNA is helped by the insertion of wedge-like motifs in both Cas8e and Cas8f (Figure 7C, E) (59, 127). This destabilization may drive local dsDNA melting, enabling strand invasion and the formation of a crRNA-DNA heteroduplex to initiate R-loop formation (Figure 7A). Thus, the energy loss during DNA bending and unwinding is compensated by base pair formation between the crRNA and target DNA. Notably, every sixth nucleotide of the crRNA-DNA duplex remains unpaired to accommodate the underwound hybrid duplex in the nucleic acid binding channel (Figure 7B, D) (46, 59, 121, 136). A recent cryo-EM structure of T. fusca Cascade captured an intermediate partial R-loop containing two 5-bp crRNA-DNA stretches (128). Based on this structure, it was hypothesized that the dsDNA may melt in 6-bp steps, forming the shorter 5-bp duplexes en route to full R-loop formation.
Single molecule magnetic tweezers studies have confirmed that DNA melting and R-loop formation occur in a directional manner (Figure 7A) (124, 125). R-loop formation originates within the PAM-proximal “seed” region, which acts as the next checkpoint for target binding following PAM recognition. As a result, the seed sequence is especially important for high-affinity target binding by both Cascade and Csy complexes (26, 49). The R-loop expands from the PAM-proximal to the PAM-distal end by zipping the crRNA and target strand (124, 125). R-loop formation stalls when it meets non-complementary sequence (Figure 7A). In some cases, the surveillance complex can overcome the non-complementary energy barrier and continue R-loop formation, triggering further directional unwinding of the target DNA. Alternatively, the complex may be unable to overcome the energy barrier leading to R-loop collapse and dissociation from the target. Targets with mismatches at the PAM-proximal end, especially within the seed sequence, are more likely to dissociate, while mismatches at the PAM-distal end are tolerated, although they may form unstable partial R-loops.
Complete base pairing between the crRNA and target sequence triggers conformational changes in both Cascade and the Csy complex, resulting in a highly stable R-loop structure (59, 127, 128). In Cascade, the Cas7 backbone of the complex remains rigid, while the Cas11 and Cas8e subunits undergo changes to widen the DNA-binding channel (127). The Cas11 dimer slides toward the tail of the complex (Figure 7B), pushing down on the alpha-helical C-terminal domain (CTD) of Cas8e, which moves toward the PAM-interacting N-terminal domain (NTD) (Figure 7C) (126–128). The non-target DNA strand is stabilized by the backside of the Cas11 dimer (126, 128, 140). Cascade does not undergo a conformational change to lock the R-loop until full target binding, as observed in the Cascade partial R-loop structure and single molecule magnetic tweezers studies (124, 125, 128). The Csy complex also undergoes significant changes upon DNA binding. However, unlike Cascade, the entire Cas7 backbone of the Csy complex is elongated by ~20Å, based on displacement of each subunit to accommodate the RNA-DNA hybrid (Figure 7D) (59, 60). The Cas8f subunit also rearranges its hook domain, which is involved in dsDNA binding (Figure 7E) (59). The non-target strand is stabilized by a domain within the Cas7 subunits, compensating for the lack of Cas11 subunits in the Csy complex.
(d). Cas3 recruitment and target DNA degradation
The conformational changes in Cascade and Csy complex lock the R-loop and serve as recruitment signals for the Cas3 nuclease/helicase (Figure 8). In particular, the conformational changes that occur within the Cas8e subunit upon R-loop locking are required for Cas3 recruitment by Cascade (Figure 7C) (33, 91, 127, 140). In some type I systems, Cas3 and Cas8 are fused into a single protein, similar to the Cas2/3 fusion in type I-F, indicating the importance of Cas8 in Cas3 recruitment (19). A negative stain electron microscopy structure of the E. coli Cascade-Cas3 structure confirmed the direct interaction between Cas8e and Cas3 (29). A recent high-resolution cryo-EM structure of the T. fusca Cascade-Cas3 structure revealed that the rotation of the CTD of Cas8e during R-loop locking is responsible for Cas3 recruitment, through the exposure of a Cas8e region that directly interacts with Cas3 (Figure 8A–B) (33). Comparison of Cas8e conformations from the partial and full R-loop structures shows significant clashes between the CTD and Cas3 when Cas8e is in the unlocked conformation (Figure 8B). Although recruitment of Cas2/3 by the Csy complex is less well understood, it is possible that Cas8f hook domain rearrangement or the elongation of the Cas7 backbone may act as recruitment signals (59).
Once recruited to the target site, Cas3 is responsible for target degradation (Figure 1, 8C–E). The N-terminal HD nuclease domain exhibits metal-dependent endonuclease activity against ssDNA and the C-terminal helicase domain has ATP- and magnesium-dependent 3’−5’ unwinding activity (31, 129, 130, 145). Both activities are required for target degradation (28). During this process, Cas3 processively unwinds the target DNA using its helicase activity, cleaving the resulting ssDNA through the activity of the HD nuclease.
Recent structural, biochemical and single molecule studies of type I-E and I-F systems have elucidated the detailed mechanism of target degradation by Cas3 (Figure 8C–E). Following recruitment, Cas3 (or Cas2/3) initiates degradation by nicking the non-target strand (Figure 8C) (28, 30, 89). The cryo-EM structure of the full T. fusca Cascade R-loop revealed that positions 8–16 of the non-target strand are bulged out and exposed for Cas3 nicking (128). This nicking activity is ATP-independent and resistant to EDTA for the type I-E Cas3 and type I-F Cas2/3, respectively (28, 30). The nicked DNA provides a substrate for dsDNA unwinding by Cas3. A recent single-molecule study of Cas3 unwinding and cleavage suggested that the initial nick is required to stabilize Cas3 interactions with Cascade, potentially based on loading of the single-stranded non-target strand into the Cas3 helicase domain (Figure 8D) (89).
As Cas3 unwinds the DNA, it remains associated with Cascade (89, 90). Thus, although Cas3 translocates along the DNA away from the target site, it does so by reeling the non-target strand of the DNA into its helicase domain and looping out the unwound target-strand DNA (Figure 8E). Single-molecule studies of DNA reeling revealed that Cas3 unwinds the DNA in 1-bp steps, but translocates 3-bp at a time (89). During this process, Cas3 repeatedly loops the DNA, allowing the nickase activity of the HD domain to cleave the non-target strand at various intervals. Based on the thymidine preference for Cas3 cleavage (88), it is possible that nicking occurs primarily in T-rich stretches. It has also been proposed that repetitive reeling may allow Cas3 to generate spacer-length fragments used by Cas1-Cas2 during interference-dependent primed adaptation (89). Once a nick is generated, additional Cas3 can be recruited to the ssDNA gap and processively degrade upstream of the target in the 3’−5’ direction, resulting in unidirectional degradation of the target (28, 32).
Although type I-F Cas2/3 is less well understood, a recent biochemical study revealed some mechanistic differences in comparison to type I-E. Like E. coli Cas3, Cas2/3 nicks the non-target strand to initiate degradation (30). However, rather than unidirectional degradation of the non-target strand, Cas2/3 degrades both strands of DNA. As part of the Cas14:Cas2/32 complex, two copies of Cas2/3 are initially recruited to the Csy complex. Although it is not clear whether the Cas1-Cas2/3 complex remains partially intact during interference, this initial stoichiometry suggests that two copies of Cas2/3 may be responsible for reeling and degradation of both strands of DNA in opposite directions away from the protospacer. Consistently, while primed spacer acquisition in type I-E is strand biased (45, 50, 100), with spacers mainly acquired from the same strand as the original protospacer, type I-F priming occurs from both strands (80–82). Spacers are also acquired preferentially from sites close to the original protospacer (80, 82), strongly suggesting that prespacers are formed from Cas3 products during bidirectional degradation originating at the target site.
Cas3 is a non-specific endonuclease, and its expression and activity must be tightly controlled to ensure that it is activated only upon binding at the correct target site. In E. coli, Cas3 nuclease activity is apparently attenuated, as nicking events occur rarely in single-molecule experiments performed at low nM concentrations (89). Complete degradation of a target requires higher concentrations in vitro (28, 29), and similarly, rapid interference in vivo requires Cas3 overexpression (41). In type I-F, Cas1 inhibits Cas2/3 nuclease activity within the Cas1-Cas2/3 complex, and Cas2/3 is only activated upon association with the Csy complex (30). It is unclear whether a similar mechanism exists in type I-E, although single molecule studies have suggested that Cas3 cleavage activity is inhibited in the presence of Cas1-Cas2 (32).
Molecular warfare during CRISPR immunity
CRISPR-Cas systems can provide robust defense against viral infection; however, phages have multiple mechanisms for avoiding these immune mechanisms. Rapid evolution of phages can render a spacer sequence obsolete over time, especially as viral particles harboring escape mutations propagate in the population (49, 146, 147). Indeed, CRISPR immunity has been shown to be a key factor driving phage evolution (148, 149). Primed adaptation allows type I CRISPR-Cas systems to counteract escape events (50), through rapid acquisition of new spacers directed against the invader. In addition, diversity of spacer sequences within a population of bacteria can limit the ability of phages to escape CRISPR immunity through the development of single point mutations (147). Therefore, phages have evolved more sophisticated methods for overcoming CRISPR immunity. Phage-encoded anti-CRISPR proteins can inactivate the CRISPR-Cas machinery through a variety of mechanisms, neutralizing the host response and enabling continued infection (150–152). In this section, we discuss experimental in vivo approaches that have been developed to study CRISPR immune response, the various mechanisms by which phages can escape immunity, and how CRISPR-Cas systems have evolved to keep up in this molecular arms race.
(a). Experimental approaches toward understanding CRISPR immunity
Our understanding of type I-E and I-F systems has been greatly enhanced by the ability to study these systems in model Enterobacteria. Most in vivo studies have relied on one of two approaches to examine CRISPR immune response: phage-challenge or plasmid-based assays. Each type of assay has several benefits and potential limitations. During phage challenge, the positive selection of cells that have been immunized makes it easier to detect rare naïve acquisition events than in plasmid-based assays (45, 50). Although methods have been developed to positively select cells upon spacer acquisition from plasmids (98, 153), only a few studies have investigated naïve acquisition from plasmids at native Cas1-Cas2 expression levels (82). Nevertheless, plasmid-based adaptation studies are beneficial for gaining a comprehensive view of spacer acquisition. Because some acquired spacers may provide more effective immunity than others, the full range of spacer acquisition events may not be detected during phage challenge as cells with the most effective spacers will dominate the population. Instead, several studies have relied on adaptation against plasmids coupled with high-throughput sequencing of the acquired spacers (51, 53, 82, 99, 100, 154). These analyses have enabled quantification of features from 100,000s of newly acquired spacers, including spacer sequence motifs, spacer length, PAM sequences, protospacer location and source DNA.
An additional benefit of plasmid-based acquisition is that these assays can be performed in the absence of the interference machinery, which is required for providing the immune response necessary for cell survival in a phage challenge assay. These types of experiments are especially useful for determining spacer source, either from the plasmid or the genomic DNA, without worry of autoimmunity upon acquisition from the genome (99). Because the interference machinery is required for primed acquisition, these types of experiments can only be used to study naïve acquisition events and often rely on the overexpression of Cas1-Cas2. When the interference machinery is present, plasmids bearing “priming targets” can enable high levels of spacer acquisition even at native expression levels (82). Intriguingly, analyses of primed acquisition from a plasmid target have revealed biased acquisition of spacers from “hot” protospacer regions (51, 154). It remains unclear whether protospacer sequence, location or other factors may contribute to preferential acquisition of hot protospacers.
Plasmid-based assays have also been useful for studying CRISPR interference. During phage challenge, robust CRISPR interference response is required for cell survival. As discussed in the next section, phages harboring mutations in the target often escape immunity through reduction of CRISPR interference (49, 147). Plasmid-based CRISPR interference assays can detect intermediate levels of interference that could allow for phage escape (46, 47, 91). These types of experiments have revealed that not all mutations cause the same level of defect in CRISPR interference. In addition, the use of plasmid-based target libraries has enabled high-throughput analysis of PAM specificity and mutational tolerance within the target region (46, 47, 101). These studies highlight the importance of plasmid-based assays for elucidating the molecular details of CRISPR immunity.
(b). Effects of target mutations
Single mutations in the seed or PAM region strongly inhibit CRISPR interference, and these locations are the primary sites for spontaneous escape mutations (51). However, these mutations do not fully abolish the ability of the surveillance complex to recognize the target, enabling priming to occur from mutated targets for which interference has been reduced or fully blocked (50). Numerous high-throughput studies have revealed the sequence requirements for E. coli Cascade-target binding (101, 155), as well as the degree to which mutations can be tolerated for interference and/or priming (46, 47, 156, 157). These studies have revealed the promiscuity of PAM recognition by Cascade. Although only a small number of “canonical” PAM sequences (5–7 depending on spacer sequence) can promote rapid interference (19, 157), numerous other sequences (>20) can enable primed acquisition from the target site (46, 47). The Cascade-dsDNA structure explains this promiscuity, as the PAM is mainly read out through minor-groove contacts that do not provide high sequence specificity (127, 128). Similarly, seed mutations can also be tolerated to some degree, although the type of mismatch, location of the mismatch, and sequence of the crRNA all contribute to the level of tolerance (46, 47). In some situations, up to two seed mutations can be tolerated (47), while targets with as many as 13 mutations can still trigger priming (46). A recent genome-wide investigation in E. coli revealed that Cascade can associate with >100 sites with very limited homology within the host chromosome; however, at least 18 bp are required to elicit an immune response, ensuring that the chromosome remains protected against autoimmunity (156).
Priming was initially discovered as a mechanism for overcoming escape mutations (50). However, subsequent studies in both E. coli and P. atrosepticum have shown that priming is far more efficient from perfect targets in comparison to targets containing mutations (51, 82). Nevertheless, overall spacer acquisition is relatively rare from unmutated targets in comparison to targets with a PAM or seed mutation (47). Reduced priming from perfect targets likely occurs because the source for prespacer substrates rapidly dwindles due to target degradation by Cas3 (51, 88). Indeed, while low levels of priming can be observed against perfect targets in E. coli K-12 hns deletion strains where Cas3 expression is limiting (41, 45, 47), priming against perfect targets cannot be observed through standard PCR-based methods when Cas3 is overexpressed (50, 51). These results are consistent with a model in which rapid interference against a perfect target inhibits priming due to consumption of the prespacer pool. In contrast, mutations in the target slow interference, creating a continuous prespacer pool and a relative increase in the amount of priming.
The decreased rate of interference for mutant targets can be caused by multiple factors. Target mutations can elicit alternative binding modes by Cascade that may favor priming over interference (91, 158). For mutations in the PAM and the first few nucleotides of the seed, the Cas8e subunit adopts an alternative conformation that prevents Cas8e locking and inhibits direct recruitment of Cas3, leading to decreased rate of interference (54, 91). Recruitment of Cas3 may also be impaired due to dissociation of Cas8e, as has been observed for T. fusca Cascade binding to PAM and seed mutants (159). Consistently, for some PAM mutants, interference is completely blocked both in vivo and in vitro (29, 32, 47, 91, 158), suggesting that Cas8e is fully non-functional in Cas3 recruitment. These types of targets can still promote priming, and likely do so through the interference-independent pathway (32, 47, 91). It has been proposed that the alternative conformation of Cas8e when bound to PAM mutant targets prevents direct Cas3 recruitment in the absence of Cas1-Cas2, but favors indirect loading of Cas3 in the presence of Cas1-Cas2 (32, 91). Thus, the unlocked Cas8e conformation may promote the formation of a priming complex that can translocate and search for prespacers, as has been observed for the E. coli and T. fusca type I-E systems (32, 90).
However, it is likely that the alternative conformation of Cas8e does not account for all priming. Cascade can form fully locked R-loops at targets containing seed mutations that are further from the PAM (54). In these cases, the kinetics of target binding and R-loop locking are reduced, which contributes to slower target degradation and an increase in primed adaptation. It was recently proposed that priming at fully locked R-loops occurs mainly through the interference-dependent pathway, based on the observation that priming rates are similar regardless of Cascade conformation (54). It is also likely that Cas1-Cas2 can be recruited at fully locked R-loops, as has been observed for both the type I-E and I-F system (30, 32, 90). These observations support a previously described model in which Cas3 can be recruited alone or in conjunction with Cas1-Cas2 at fully locked R-loops, but can only be recruited in the presence of Cas1-Cas2 at R-loops with unlocked Cas8e (91).
(c). Population-based avoidance of escape mutations
Primed adaptation can counteract phage evolution and provide broader specificity against closely related phages. Another benefit of priming is the ability to generate a population with a diverse set of spacer sequences (82, 100). Naïve adaptation is rare, and stringent selective pressure for cells that have acquired or inherited spacers from an infectious virus likely results in a limited number of spacers present in the population. Primed adaptation is more rapid, and can occur from either perfect or mutated targets, allowing various cells in the population to acquire different spacer sequences.
Spacer diversity is critical for ensuring bacterial populations can limit phage propagation. A study of the type I-F P. aeruginosa system revealed that populations with diverse CRISPR arrays are able to drive phage to extinction, while phages persist when the bacterial population contains a limited number of spacers (147). Viral persistence can be attributed to development of escape mutants, which are more likely to occur if only a limited number of mutations must be present to escape the few spacers present in the bacterial population. However, as the number of spacers increases, the likelihood of a virus developing mutations to escape all spacers decreases. While a single mutation may allow a phage to escape from individual cells, the population as a whole will provide continued immunity through the presence of other spacers against unmutated regions of the phage genome. These insights explain the diversity of CRISPR arrays in environmental samples and highlight the importance of multiple adaptation mechanisms for providing robust immunity.
(d). Anti-CRISPR proteins
While escape mutation provides one potential line of defense against CRISPR-Cas systems, phages have also evolved anti-CRISPR proteins that directly inactivate the bacterial immune response (reviewed in (152)). Anti-CRISPR genes were first discovered in prophages of P. aeruginosa PA14 (150). These prophages were found to render the PA14 type I-F system incapable of providing defense against normally CRISPR-sensitive phages. Specifically, genes located within a variable region of the prophage genomes proved to be the components necessary to inactivate CRISPR-Cas immunity. Since this initial discovery, anti-CRISPR genes have been found against several sub-types within type I and II systems from diverse bacteria and archaea, including several members of Enterobacteriaceae (160–165). These genes generally encode small proteins (50–150 amino acids) with little sequence similarity to other proteins or to each other, highlighting the selective pressure that has driven their evolution.
Biochemical and structural studies have revealed that immune inactivation by anti-CRISPR proteins commonly occurs by blocking the interference stage (59, 60, 151, 166–171). Various anti-CRISPR proteins have been shown to inhibit target binding by the surveillance complex, often through DNA mimicry (59, 60, 151, 167, 169, 171). In addition, anti-CRISPR proteins that bind directly to Cas3 can prevent recruitment to the target-bound surveillance complex (151, 166). Despite these mechanistic insights, it remained unclear how phages could express enough anti-CRISPR proteins to inactivate the CRISPR system, which would likely already be expressed and ready to launch an interference response during phage infection. Two recent studies confirm that phages must build tolerance to CRISPR interference over time, through the expression of anti-CRISPR genes from multiple phage genomes (172, 173). Thus, early infection events are likely to result in phage neutralization through CRISPR interference, but continued exposure to the phages will eventually enable expression of sufficient levels of anti-CRISPR proteins for inactivation. Similar to CRISPR-Cas systems, which provide benefit to a community of bacteria while often sacrificing individual cells, anti-CRISPR mechanisms require population-level response to mount a counterattack against CRISPR immunity.
Conclusions
The type I-E and I-F systems of enterobacteria and other proteobacteria have served as important model systems to understand the molecular mechanisms of CRISPR-Cas systems. Studies in E. coli and P. atrosepticum have provided the foundation of our knowledge of type I CRISPR-Cas systems, and have revealed several key similarities and differences between subtypes. In the future, these model organisms will continue to be indispensable for answering mechanistic questions that remain unresolved. They will also be critical for solving a longstanding mystery in the field: What is the endogenous role of the CRISPR-Cas systems of the most well-studied enterobacteria, E. coli and Salmonella? Beyond mechanistic studies, understanding how, and under what circumstances, these systems function in E. coli may uncover new roles for CRISPR-Cas systems beyond host defense.
Acknowledgements
We thank members of the Sashital lab for helpful discussions and Ailong Ke for providing coordinates for the Cascade-Cas3 structure.
References
- 1.Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. 1987. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol 169:5429–5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakata A, Amemura M, Makino K. 1989. Unusual nucleotide arrangement with repeated sequences in the Escherichia coli K-12 chromosome. J Bacteriol 171:3553–3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mojica FJM, Díez-Villaseñor C, Soria E, Juez G. 2002. Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria. Mol Microbiol 36:244–246. [DOI] [PubMed] [Google Scholar]
- 4.Hermans PWM, Van Soolingen D, Bik EM, De Haas PEW, Dale JW, Van Embden JDA. 1991. Insertion element IS987 from Mycobacterium bovis BCG is located in a hot-spot integration region for insertion elements in Mycobacterium tuberculosis complex strains. Infect Immun 59:2695–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. 2005. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151:2551–2561. [DOI] [PubMed] [Google Scholar]
- 6.Pourcel C, Salvignol G, Vergnaud G. 2005. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 151:653–663. [DOI] [PubMed] [Google Scholar]
- 7.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E. 2005. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol 60:174–182. [DOI] [PubMed] [Google Scholar]
- 8.Jansen R, van Embden JD, Gaastra W, Schouls LM. 2002. Identification of a novel family of sequence repeats among prokaryotes. OMICS 6:23–33. [DOI] [PubMed] [Google Scholar]
- 9.Haft DH, Selengut J, Mongodin EF, Nelson KE. 2005. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLOS Comput Biol 1:e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. 2006. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct 1:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712. [DOI] [PubMed] [Google Scholar]
- 12.Carte J, Wang R, Li H, Terns RM, Terns MP. 2008. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev 22:3489–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marraffini LA, Sontheimer EJ. 2008. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322:1843–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brouns SJJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJH, Snijders APL, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321:960–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yosef I, Goren MG, Qimron U. 2012. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res 40:5569–5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nuñez JK, Lee ASY, Engelman A, Doudna JA. 2015. Integrase-mediated spacer acquisition during CRISPR-Cas adaptive immunity. Nature 519:193–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson RN, Wiedenheft B. 2015. A Conserved Structural Chassis for Mounting Versatile CRISPR RNA-Guided Immune Responses. Mol Cell 58:722–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP. 2009. RNA-Guided RNA Cleavage by a CRISPR RNA-Cas Protein Complex. Cell 139:945–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Westra ER, van Erp PBG, Kunne T, Wong SP, Staals RHJ, Seegers CLC, Bollen S, Jore MM, Semenova E, Severinov K, de Vos WM, Dame RT, de Vries R, Brouns SJJ, van der Oost J. 2012. CRISPR Immunity Relies on the Consecutive Binding and Degradation of Negatively Supercoiled Invader DNA by Cascade and Cas3. Mol Cell 46:595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadan AH, Moineau S. 2010. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468:67–71. [DOI] [PubMed] [Google Scholar]
- 21.Koonin EV, Makarova KS, Zhang F. 2017. Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol 37:67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJJ, Charpentier E, Haft DH, Horvath P, Moineau S, Mojica FJM, Terns RM, Terns MP, White MF, Yakunin AF, Garrett RA, van der Oost J, Backofen R, Koonin EV. 2015. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol 13:722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murugan K, Babu K, Sundaresan R, Rajan R, Sashital DG. 2017. The Revolution Continues: Newly Discovered Systems Expand the CRISPR-Cas Toolkit. Mol Cell 68:15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makarova KS, Zhang F, Koonin EV. 2017. SnapShot: Class 1 CRISPR-Cas Systems. Cell 168:946–946.e1. [DOI] [PubMed] [Google Scholar]
- 25.Medina-Aparicio L, Dávila S, Rebollar-Flores JE, Calva E, Hernández-Lucas I. 2018. The CRISPR-Cas system in Enterobacteriaceae. Pathog Dis 76:fty002. [DOI] [PubMed] [Google Scholar]
- 26.Wiedenheft B, van Duijn E, Bultema J, Waghmare S, Zhou K, Barendregt A, Westphal W, Heck A, Boekema E, Dickman M, Doudna JA. 2011. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci 108:10092–10097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richter C, Gristwood T, Clulow JS, Fineran PC. 2012. In Vivo Protein Interactions and Complex Formation in the Pectobacterium atrosepticum Subtype I-F CRISPR/Cas System. PLoS One 7:e49549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mulepati S, Bailey S. 2013. In vitro reconstitution of an Escherichia coli RNA-guided immune system reveals unidirectional, ATP-dependent degradation of DNA Target. J Biol Chem 288:22184–22192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hochstrasser ML, Taylor DW, Bhat P, Guegler CK, Sternberg SH, Nogales E, Doudna J a. 2014. CasA mediates Cas3-catalyzed target degradation during CRISPR RNA-guided interference. Proc Natl Acad Sci 111:6618–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rollins MF, Chowdhury S, Carter J, Golden SM, Wilkinson RA, Bondy-Denomy J, Lander GC, Wiedenheft B. 2017. Cas1 and the Csy complex are opposing regulators of Cas2/3 nuclease activity. Proc Natl Acad Sci 114:E5113–E5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sinkunas T, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. 2011. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J 30:1335–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Redding S, Sternberg SH, Marshall M, Gibb B, Bhat P, Guegler CK, Wiedenheft B, Doudna JA, Greene EC. 2015. Surveillance and Processing of Foreign DNA by the Escherichia coli CRISPR-Cas System. Cell 163:854–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao Y, Luo M, Dolan AE, Liao M, Ke A. 2018. Structure basis for RNA-guided DNA degradation by Cascade and Cas3. Science 361:eaat0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pougach K, Semenova E, Bogdanova E, Datsenko KA, Djordjevic M, Wanner BL, Severinov K. 2010. Transcription, processing and function of CRISPR cassettes in Escherichia coli. Mol Microbiol 77:1367–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Díez-Villaseñor C, Almendros C, García-Martínez J, Mojica FJM. 2010. Diversity of CRISPR loci in Escherichia coli. Microbiology 156:1351–1361. [DOI] [PubMed] [Google Scholar]
- 36.Nuñez JK, Kranzusch PJ, Noeske J, Wright AV, Davies CW, Doudna JA. 2014. Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity. Nat Struct Mol Biol 21:528–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jore MM, Lundgren M, van Duijn E, Bultema JB, Westra ER, Waghmare SP, Wiedenheft B, Pul U, Wurm R, Wagner R, Beijer MR, Barendregt A, Zhou K, Snijders APL, Dickman MJ, Doudna JA, Boekema EJ, Heck AJR, van der Oost J, Brouns SJJ. 2011. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol 18:529–536. [DOI] [PubMed] [Google Scholar]
- 38.Pul Ü, Wurm R, Arslan Z, GeiBen R, Hofmann N, Wagner R. 2010. Identification and characterization of E. coli CRISPR-cas promoters and their silencing by H-NS. Mol Microbiol 75:1495–1512. [DOI] [PubMed] [Google Scholar]
- 39.Oshima T, Ishikawa S, Kurokawa K, Aiba H, Ogasawara N. 2006. Escherichia coli Histone-Like Protein H-NS Preferentially Binds to Horizontally Acquired DNA in Association with RNA Polymerase. DNA Res 13:141–153. [DOI] [PubMed] [Google Scholar]
- 40.Westra ER, Pul Ü, Heidrich N, Jore MM, Lundgren M, Stratmann T, Wurm R, Raine A, Mescher M, Van Heereveld L, Mastop M, Wagner EGH, Schnetz K, Van Der Oost J, Wagner R, Brouns SJJ. 2010. H-NS-mediated repression of CRISPR-based immunity in Escherichia coli K12 can be relieved by the transcription activator LeuO. Mol Microbiol 77:1380–1393. [DOI] [PubMed] [Google Scholar]
- 41.Majsec K, Bolt EL, Ivancic-Bace I. 2016. Cas3 is a limiting factor for CRISPR-Cas immunity in Escherichia coli cells lacking H-NS. BMC Microbiol 16:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang CD, Chen YH, Huang HY, Huang H Da, Tseng CP. 2014. CRP represses the CRISPR/Cas system in Escherichia coli: Evidence that endogenous CRISPR spacers impede phage P1 replication. Mol Microbiol 92:1072–91. [DOI] [PubMed] [Google Scholar]
- 43.Sashital DG, Wiedenheft B, Doudna JA. 2012. Mechanism of foreign DNA selection in a bacterial adaptive immune system. Mol Cell 46:606–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yosef I, Goren MG, Kiro R, Edgar R, Qimron U. 2011. High-temperature protein G is essential for activity of the Escherichia coli clustered regularly interspaced short palindromic repeats (CRISPR)/Cas system. Proc Natl Acad Sci 108:20136–20141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swarts DC, Mosterd C, van Passel MWJ, Brouns SJJ. 2012. CRISPR interference directs strand specific spacer acquisition. PLoS One 7:e35888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fineran PC, Gerritzen MJH, Suarez-Diez M, Kunne T, Boekhorst J, van Hijum SaFT, Staals RHJ, Brouns SJJ. 2014. Degenerate target sites mediate rapid primed CRISPR adaptation. Proc Natl Acad Sci 111:E1629–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xue C, Seetharam AS, Musharova O, Severinov K, J Brouns SJ, Severin AJ, Sashital DG. 2015. CRISPR interference and priming varies with individual spacer sequences. Nucleic Acids Res 43:10831–10847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edgar R, Qimron U. 2010. The Escherichia coli CRISPR system protects from lambda lysogenization, lysogens, and prophage induction. J Bacteriol 192:6291–6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Semenova E, Jore MM, Datsenko KA, Semenova A, Westra ER, Wanner B, van der Oost J, Brouns SJJ, Severinov K. 2011. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci 108:10098–10103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Datsenko KA, Pougach K, Tikhonov A, Wanner BL, Severinov K, Semenova E. 2012. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat Commun 3:945. [DOI] [PubMed] [Google Scholar]
- 51.Semenova E, Savitskaya E, Musharova O, Strotskaya A, Vorontsova D, Datsenko KA, Logacheva MD, Severinov K. 2016. Highly efficient primed spacer acquisition from targets destroyed by the Escherichia coli type I-E CRISPR-Cas interfering complex. Proc Natl Acad Sci U S A 113:7626–7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Musharova O, Klimuk E, Datsenko KA, Metlitskaya A, Logacheva M, Semenova E, Severinov K, Savitskaya E. 2017. Spacer-length DNA intermediates are associated with Cas1 in cells undergoing primed CRISPR adaptation. Nucleic Acids Res 45:3297–3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shmakov S, Savitskaya E, Semenova E, Logacheva MD, Datsenko KA, Severinov K. 2014. Pervasive generation of oppositely oriented spacers during CRISPR adaptation. Nucleic Acids Res 42:5907–5916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krivoy A, Rutkauskas M, Kuznedelov K, Musharova O, Rouillon C, Severinov K, Seidel R. 2018. Primed CRISPR adaptation in Escherichia coli cells does not depend on conformational changes in the Cascade effector complex detected in Vitro. Nucleic Acids Res 46:4087–4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ivančić-Bace I, Cass SD, Wearne SJ, Bolt EL. 2015. Different genome stability proteins underpin primed and Naïve adaptation in E. Coli CRISPR-Cas immunity. Nucleic Acids Res 43:10821–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Przybilski R, Richter C, Gristwood T, Clulow JS, Vercoe RB, Fineran PC. 2011. Csy4 is responsible for CRISPR RNA processing in Pectobacterium atrosepticum. RNA Biol 8:517–28. [DOI] [PubMed] [Google Scholar]
- 57.Fagerlund RD, Wilkinson ME, Klykov O, Barendregt A, Pearce FG, Kieper SN, Maxwell HWR, Capolupo A, Heck AJR, Krause KL, Bostina M, Scheltema RA, Staals RHJ, Fineran PC. 2017. Spacer capture and integration by a type I-F Cas1-Cas2–3 CRISPR adaptation complex. Proc Natl Acad Sci 114:E5122–E5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Duijn E, Barbu IM, Barendregt A, Jore MM, Wiedenheft B, Lundgren M, Westra ER, Brouns SJ, Doudna JA, van der Oost J, Heck AJ. 2012. Native tandem and ion mobility mass spectrometry highlight structural and modular similarities in clustered-regularly-interspaced shot-palindromic-repeats (CRISPR)-associated protein complexes from Escherichia coli and Pseudomonas aeruginosa. Mol Cell Proteomics 11:1430–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guo TW, Bartesaghi A, Yang H, Falconieri V, Rao P, Merk A, Eng ET, Raczkowski AM, Fox T, Earl LA, Patel DJ, Subramaniam S. 2017. Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex. Cell 171:414–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chowdhury S, Carter J, Rollins MCF, Golden SM, Jackson RN, Hoffmann C, Nosaka L, Bondy-Denomy J, Maxwell KL, Davidson AR, Fischer ER, Lander GC, Wiedenheft B. 2017. Structure Reveals Mechanisms of Viral Suppressors that Intercept a CRISPR RNA-Guided Surveillance Complex. Cell 169:47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Patterson AG, Chang JT, Taylor C, Fineran PC. 2015. Regulation of the type I-F CRISPR-Cas system by CRP-cAMP and GalM controls spacer acquisition and interference. Nucleic Acids Res 43:6038–6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Patterson AG, Yevstigneyeva MS, Fineran PC. 2017. Regulation of CRISPR-Cas adaptive immune systems. Curr Opin Microbiol 37:1–7. [DOI] [PubMed] [Google Scholar]
- 63.Høyland-Kroghsbo NM, Paczkowski J, Mukherjee S, Broniewski J, Westra E, Bondy-Denomy J, Bassler BL. 2016. Quorum sensing controls the Pseudomonas aeruginosa CRISPR-Cas adaptive immune system. Proc Natl Acad Sci USA 114:131–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Patterson AG, Jackson SA, Taylor C, Evans GB, Salmond GPC, Przybilski R, Staals RHJ, Fineran PC. 2016. Quorum Sensing Controls Adaptive Immunity through the Regulation of Multiple CRISPR-Cas Systems. Mol Cell 64:1102–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tyson GW, Banfield JF. 2008. Rapidly evolving CRISPRs implicated in acquired resistance of microorganisms to viruses. Environ Microbiol 10:200–207. [DOI] [PubMed] [Google Scholar]
- 66.Shariat N, Dudley EG. 2014. CRISPRs: Molecular Signatures Used for Pathogen Subtyping. Appl Environ Microbiol 80:430–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cui Y, Li Y, Gorge O, Platonov ME, Yan Y, Guo Z, Pourcel C, Dentovskaya SV., Balakhonov SV, Wang X, Song Y, Anisimov AP, Vergnaud G, Yang R. 2008. Insight into microevolution of Yersinia pestis by clustered regularly interspaced short palindromic repeats. PLoS One 3:e2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McGhee GC, Sundin GW. 2012. Erwinia amylovora CRISPR elements provide new tools for evaluating strain diversity and for microbial source tracking. PLoS One 7:e41706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu F, Kariyawasam S, Jayarao BM, Barrangou R, Gerner-Smidt P, Ribot EM, Knabel SJ, Dudley EG. 2011. Subtyping Salmonella enterica serovar enteritidis isolates from different sources by using sequence typing based on virulence genes and clustered regularly interspaced short palindromic repeats (CRISPRs). Appl Environ Microbiol 77:4520–4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu F, Barrangou R, Gerner-Smidt P, Ribot EM, Knabel SJ, Dudley EG. 2011. Novel virulence gene and clustered regularly interspaced short palindromic repeat (CRISPR) multilocus sequence typing scheme for subtyping of the major serovars of Salmonella enterica subsp. enterica. Appl Environ Microbiol 77:1946–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Delannoy S, Beutin L, Fach P. 2012. Use of clustered regularly interspaced short palindromic repeat sequence polymorphisms for specific detection of enterohemorrhagic Escherichia coli strains of serotypes O26:H11, O45:H2, O103:H2, O111:H8, O121:H19, O145:H28, and O157:H7 by real-time PCR. J Clin Microbiol 50:4035–4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shariat N, Timme RE, Pettengill JB, Barrangou R, Dudley EG. 2015. Characterization and evolution of Salmonella CRISPR-Cas systems. Microbiology 161:374–386. [DOI] [PubMed] [Google Scholar]
- 73.Touchon M, Charpentier S, Clermont O, Rocha EPC, Denamur E, Branger C. 2011. CRISPR distribution within the Escherichia coli species is not suggestive of immunity-associated diversifying selection. J Bacteriol 193:2460–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Medina-Aparicio L, Rebollar-Flores JE, Gallego-Hernández AL, Vázquez A, Olvera L, Gutiérrez-Ríos RM, Calva E, Hernández-Lucas I. 2011. The CRISPR/Cas immune system is an operon regulated by LeuO, H-NS, and leucine-responsive regulatory protein in Salmonella enterica serovar Typhi. J Bacteriol 193:2396–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Savitskaya E, Lopatina A, Medvedeva S, Kapustin M, Shmakov S, Tikhonov A, Artamonova II, Logacheva M, Severinov K. 2017. Dynamics of Escherichia coli type I-E CRISPR spacers over 42 000 years. Mol Ecol 26:2019–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sampson TR, Weiss DS. 2013. Alternative roles for CRISPR/Cas systems in bacterial pathogenesis. PLOS Pathog 9:e1003621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sampson TR, Napier BA, Schroeder MR, Louwen R, Zhao J, Chin CY, Ratner HK, Llewellyn AC, Jones CL, Laroui H, Merlin D, Zhou P, Endtz HP, Weiss DS. 2014. A CRISPR-Cas system enhances envelope integrity mediating antibiotic resistance and inflammasome evasion. Proc Natl Acad Sci 111:11163–11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Perez-Rodriguez R, Haitjema C, Huang Q, Nam KH, Bernardis S, Ke A, Delisa MP. 2011. Envelope stress is a trigger of CRISPR RNA-mediated DNA silencing in Escherichia coli. Mol Microbiol 79:584–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sampson TR, Saroj SD, Llewellyn AC, Tzeng YL, Weiss DS. 2013. A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature 497:254–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vorontsova D, Datsenko KA, Medvedeva S, Bondy-Denomy J, Savitskaya EE, Pougach K, Logacheva M, Wiedenheft B, Davidson AR, Severinov K, Semenova E. 2015. Foreign DNA acquisition by the I-F CRISPR-Cas system requires all components of the interference machinery. Nucleic Acids Res 43:10848–10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Richter C, Dy RL, McKenzie RE, Watson BNJ, Taylor C, Chang JT, McNeil MB, Staals RHJ, Fineran PC. 2014. Priming in the Type I-F CRISPR-Cas system triggers strand-independent spacer acquisition, bi-directionally from the primed protospacer. Nucleic Acids Res 42:8516–8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Staals RHJ, Jackson SA, Biswas A, Brouns SJJ, Brown CM, Fineran PC. 2016. Interference-driven spacer acquisition is dominant over naive and primed adaptation in a native CRISPR-Cas system. Nat Commun 7:12853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Levy A, Goren MG, Yosef I, Auster O, Manor M, Amitai G, Edgar R, Qimron U, Sorek R. 2015. CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature 520:505–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dillingham MS, Kowalczykowski SC. 2008. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol Mol Biol Rev 72:642–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.George JW, Stohr BA, Tomso DJ, Kreuzer KN. 2001. The tight linkage between DNA replication and double-strand break repair in bacteriophage T4. Proc Natl Acad Sci U S A 98:8290–8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kuzminov A 2001. Single-strand interruptions in replicating chromosomes cause double-strand breaks. Proc Natl Acad Sci 98:8241–8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Michel B, Flores M-J, Viguera E, Grompone G, Seigneur M, Bidnenko V. 2001. Rescue of arrested replication forks by homologous recombination. Proc Natl Acad Sci 98:8181–8188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kunne T, Kieper SN, Bannenberg JW, Vogel AIM, Miellet WR, Klein M, Depken M, Suarez-Diez M, Brouns SJJ. 2016. Cas3-Derived Target DNA Degradation Fragments Fuel Primed CRISPR Adaptation. Mol Cell 63:852–864. [DOI] [PubMed] [Google Scholar]
- 89.Loeff L, Brouns SJJ, Joo C. 2018. Repetitive DNA Reeling by the Cascade-Cas3 Complex in Nucleotide Unwinding Steps. Mol Cell 70:385–394.e3. [DOI] [PubMed] [Google Scholar]
- 90.Dillard KE, Brown MW, Johnson NV, Xiao Y, Dolan A, Hernandez E, Dahlhauser SD, Kim Y, Myler LR, Anslyn EV, Ke A, Finkelstein IJ. 2018. Assembly and Translocation of a CRISPR-Cas Primed Acquisition Complex. Cell 175:934–946.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xue C, Whitis NR, Sashital DG. 2016. Conformational Control of Cascade Interference and Priming Activities in CRISPR Immunity. Mol Cell 64:826–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nuñez JK, Harrington LB, Kranzusch PJ, Engelman AN, Doudna JA. 2015. Foreign DNA capture during CRISPR-Cas adaptive immunity. Nature 527:535–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang J, Li J, Zhao H, Sheng G, Wang M, Yin M, Wang Y, Wang J, Li J, Zhao H, Sheng G, Wang M, Yin M, Wang Y. 2015. Structural and Mechanistic Basis of PAM-Dependent Spacer Acquisition in CRISPR-Cas Systems. Cell 163:840–853. [DOI] [PubMed] [Google Scholar]
- 94.Amitai G, Sorek R. 2016. CRISPR-Cas adaptation: Insights into the mechanism of action. Nat Rev Microbiol 14:67–76. [DOI] [PubMed] [Google Scholar]
- 95.Moch C, Fromant M, Blanquet S, Plateau P. 2017. DNA binding specificities of Escherichia coli Cas1-Cas2 integrase drive its recruitment at the CRISPR locus. Nucleic Acids Res 45:2714–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mojica FJM, Diez-Villasenor C, Garcia-Martinez J, Almendros C. 2009. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155:733–740. [DOI] [PubMed] [Google Scholar]
- 97.Rollins MF, Schuman JT, Paulus K, Bukhari HST, Wiedenheft B. 2015. Mechanism of foreign DNA recognition by a CRISPR RNA-guided surveillance complex from Pseudomonas aeruginosa. Nucleic Acids Res 43:2216–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Diez-Villasenor C, Guzman NM, Almendros C, Garcia-Martinez J, Mojica FJM. 2013. CRISPR-spacer integration reporter plasmids reveal distinct genuine acquisition specificities among CRISPR-Cas I-E variants of Escherichia coli. RNA Biol 10:792–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yosef I, Shitrit D, Goren MG, Burstein D, Pupko T, Qimron U. 2013. DNA motifs determining the efficiency of adaptation into the Escherichia coli CRISPR array. Proc Natl Acad Sci 110:14396–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Savitskaya E, Semenova E, Dedkov V, Metlitskaya A, Severinov K. 2013. High-throughput analysis of type I-E CRISPR/Cas spacer acquisition in E. coli. RNA Biol 10:716–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Leenay RT, Maksimchuk KR, Slotkowski RA, Agrawal RN, Gomaa AA, Briner AE, Barrangou R, Beisel CL. 2016. Identifying and Visualizing Functional PAM Diversity across CRISPR-Cas Systems. Mol Cell 62:137–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee H, Zhou Y, Taylor DWDW, Sashital DGDG. 2018. Cas4-Dependent Prespacer Processing Ensures High-Fidelity Programming of CRISPR Arrays. Mol Cell 70:48–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kieper SN, Almendros C, Behler J, McKenzie RE, Nobrega FL, Haagsma AC, Vink JNA, Hess WR, Brouns SJJ. 2018. Cas4 Facilitates PAM-Compatible Spacer Selection during CRISPR Adaptation. Cell Rep 22:3377–3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rollie C, Graham S, Rouillon C, White MF. 2018. Prespacer processing and specific integration in a Type I-A CRISPR system. Nucleic Acids Res 46:1007–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shiimori M, Garrett SC, Graveley BR, Terns MP. 2018. Cas4 Nucleases Define the PAM, Length, and Orientation of DNA Fragments Integrated at CRISPR Loci. Mol Cell 70:814–824.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Heler R, Samai P, Modell JW, Weiner C, Goldberg GW, Bikard D, Marraffini LA. 2015. Cas9 specifies functional viral targets during CRISPR-Cas adaptation. Nature 519:199–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nuñez JK, Bai L, Harrington LB, Hinder TL, Doudna JA. 2016. CRISPR Immunological Memory Requires a Host Factor for Specificity. Mol Cell 62:824–833. [DOI] [PubMed] [Google Scholar]
- 108.McGinn J, Marraffini LA. 2016. CRISPR-Cas Systems Optimize Their Immune Response by Specifying the Site of Spacer Integration. Mol Cell 64:616–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wright AV., Liu JJ, Knott GJ, Doxzen KW, Nogales E, Doudna JA. 2017. Structures of the CRISPR genome integration complex. Science 357:1113–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Arslan Z, Hermanns V, Wurm R, Wagner R, Pul U. 2014. Detection and characterization of spacer integration intermediates in type I-E CRISPR-Cas system. Nucleic Acids Res 42:7884–7893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rollie C, Schneider S, Brinkmann AS, Bolt EL, White MF. 2015. Intrinsic sequence specificity of the Cas1 integrase directs new spacer acquisition. Elife 4:e08716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yoganand KNR, Sivathanu R, Nimkar S, Anand B. 2017. Asymmetric positioning of Cas1–2 complex and Integration Host Factor induced DNA bending guide the unidirectional homing of protospacer in CRISPR-Cas type I-E system. Nucleic Acids Res 45:367–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shipman SL, Nivala J, Macklis JD, Church GM. 2016. Molecular recordings by directed CRISPR spacer acquisition. Science 353:aaf1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Goren MG, Yosef I, Auster O, Qimron U. 2012. Experimental definition of a clustered regularly interspaced short palindromic duplicon in escherichia coli. J Mol Biol 423:14–16. [DOI] [PubMed] [Google Scholar]
- 115.Jackson SA, McKenzie RE, Fagerlund RD, Kieper SN, Fineran PC, Brouns SJJ. 2017. CRISPR-Cas: Adapting to change. Science 356:eaal5056. [DOI] [PubMed] [Google Scholar]
- 116.Wright AV., Doudna JA. 2016. Protecting genome integrity during CRISPR immune adaptation. Nat Struct Mol Biol 23:876–883. [DOI] [PubMed] [Google Scholar]
- 117.Xiao Y, Ng S, Nam KH, Ke A. 2017. How type II CRISPR-Cas establish immunity through Cas1-Cas2-mediated spacer integration. Nature 550:137–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Babu M, Beloglazova N, Flick R, Graham C, Skarina T, Nocek B, Gagarinova A, Pogoutse O, Brown G, Binkowski A, Phanse S, Joachimiak A, Koonin EV., Savchenko A, Emili A, Greenblatt J, Edwards AM, Yakunin AF. 2011. A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair. Mol Microbiol 79:484–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. 2010. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science 329:1355–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jackson RN, Golden SM, van Erp PBG, Carter J, Westra ER, Brouns SJJ, van der Oost J, Terwilliger TC, Read RJ, Wiedenheft B. 2014. Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli. Science 345:1473–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mulepati S, Orr A, Bailey S. 2012. Crystal structure of the largest subunit of a bacterial RNA-guided immune complex and its role in DNA target binding. J Biol Chem 287:22445–22449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhao H, Sheng G, Wang J, Wang M, Bunkoczi G, Gong W, Wei Z, Wang Y. 2014. Crystal structure of the RNA-guided immune surveillance Cascade complex in Escherichia coli. Nature 515:147–150. [DOI] [PubMed] [Google Scholar]
- 123.Xue C, Zhu Y, Zhang X, Shin Y-K, Sashital DG. 2017. Real-Time Observation of Target Search by the CRISPR Surveillance Complex Cascade. Cell Rep 21:3717–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Szczelkun MD, Tikhomirova MS, Sinkunas T, Gasiunas G, Karvelis T, Pschera P, Siksnys V, Seidel R. 2014. Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc Natl Acad Sci 111:9798–9803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rutkauskas M, Sinkunas T, Songailiene I, Tikhomirova MS, Siksnys V, Seidel R. 2015. Directional R-Loop Formation by the CRISPR-Cas Surveillance Complex Cascade Provides Efficient Off-Target Site Rejection. Cell Rep 10:1534–1543. [DOI] [PubMed] [Google Scholar]
- 126.van Erp PBG, Jackson RN, Carter J, Golden SM, Bailey S, Wiedenheft B. 2015. Mechanism of CRISPR-RNA guided recognition of DNA targets in Escherichia coli. Nucleic Acids Res 43:8381–8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hayes RP, Xiao Y, Ding F, van Erp PBG, Rajashankar K, Bailey S, Wiedenheft B, Ke A. 2016. Structural basis for promiscuous PAM recognition in type I-E Cascade from E. coli. Nature 530:499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Xiao Y, Luo M, Hayes RP, Kim J, Ng S, Ding F, Liao M, Ke A. 2017. Structure Basis for Directional R-loop Formation and Substrate Handover Mechanisms in Type I CRISPR-Cas System. Cell 170:48–60.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Huo Y, Nam KH, Ding F, Lee H, Wu L, Xiao Y, Farchione MD, Zhou S, Rajashankar K, Kurinov I, Zhang R, Ke A. 2014. Structures of CRISPR Cas3 offer mechanistic insights into Cascade-activated DNA unwinding and degradation. Nat Struct Mol Biol 21:771–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gong B, Shin M, Sun J, Jung C-H, Bolt EL, van der Oost J, Kim J-S. 2014. Molecular insights into DNA interference by CRISPR-associated nuclease-helicase Cas3. Proc Natl Acad Sci 111:16359–16364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sashital DG, Jinek M, Doudna JA. 2011. An RNA-induced conformational change required for CRISPR RNA cleavage by the endoribonuclease Cse3. Nat Struct Mol Biol 18:680–687. [DOI] [PubMed] [Google Scholar]
- 132.Gesner EM, Schellenberg MJ, Garside EL, George MM, Macmillan AM. 2011. Recognition and maturation of effector RNAs in a CRISPR interference pathway. Nat Struct Mol Biol 18:688–692. [DOI] [PubMed] [Google Scholar]
- 133.Cady KC, O’Toole G a. 2011. Non-identity Targeting of Yersinia-Subtype CRISPR-Prophage Interaction Requires the Csy and Cas3 Proteins. J Bacteriol 193:2433–2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Haurwitz RE, Sternberg SH, Doudna JA. 2012. Csy4 relies on an unusual catalytic dyad to position and cleave CRISPR RNA. EMBO J 31:2824–2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sternberg SH, Haurwitz RE, Doudna JA. 2012. Mechanism of substrate selection by a highly specific CRISPR endoribonuclease. RNA 18:661–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wiedenheft B, Lander GC, Zhou K, Jore MM, Brouns SJJ, van der Oost J, Doudna JA, Nogales E. 2011. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature 477:486–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kuznedelov K, Mekler V, Lemak S, Tokmina-Lukaszewska M, Datsenko KA, Jain I, Savitskaya E, Mallon J, Shmakov S, Bothner B, Bailey S, Yakunin AF, Severinov K, Semenova E. 2016. Altered stoichiometry Escherichia coli Cascade complexes with shortened CRISPR RNA spacers are capable of interference and primed adaptation. Nucleic Acids Res 44:10849–10861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Luo ML, Jackson RN, Denny SR, Tokmina-Lukaszewska M, Maksimchuk KR, Lin W, Bothner B, Wiedenheft B, Beisel CL. 2016. The CRISPR RNA-guided surveillance complex in Escherichia coli accommodates extended RNA spacers. Nucleic Acids Res 44:7385–7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Mulepati S, Heroux A, Bailey S. 2014. Crystal structure of a CRISPR RNA-guided surveillance complex bound to a ssDNA target. Science 345:1479–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.van Erp PBG, Patterson A, KANT R, Berry L, Golden SM, Forsman BL, Carter J, Jackson RN, Bothner B, Wiedenheft B. 2017. Conformational Dynamics of DNA Binding and Cas3 Recruitment by the CRISPR RNA-guide Cascade Complex. ACS Chem Biol 13:481–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Westra ER, Semenova E, Datsenko KA, Jackson RN, Wiedenheft B, Severinov K, Brouns SJJ. 2013. Type I-E CRISPR-Cas Systems Discriminate Target from Non-Target DNA through Base Pairing-Independent PAM Recognition. PLOS Genet 9:e1003742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. 2014. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507:62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Singh D, Sternberg SH, Fei J, Ha T, Doudna JA. 2016. Real-time observation of DNA recognition and rejection by the RNA-guided endonuclease Cas9. Nat Commun 7:1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Singh D, Mallon J, Poddar A, Wang Y, Tippana R, Yang O, Bailey S, Ha T. 2018. Realtime observation of DNA target interrogation and product release by the RNA-guided endonuclease CRISPR Cpf1 (Cas12a). Proc Natl Acad Sci 115:5444–5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jackson RN, Lavin M, Carter J, Wiedenheft B. 2014. Fitting CRISPR-associated Cas3 into the Helicase Family Tree. Curr Opin Struct Biol 24:106–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Deveau H, Barrangou R, Garneau JE, Labonte J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. 2008. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol 190:1390–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.van Houte S, Ekroth AKE, Broniewski JM, Chabas H, Ashby B, Bondy-Denomy J, Gandon S, Boots M, Paterson S, Buckling A, Westra ER. 2016. The diversity-generating benefits of a prokaryotic adaptive immune system. Nature 532:385–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Paez-Espino D, Sharon I, Morovic W, Stahl B, Thomas BC, Barrangou R, Banfield JF. 2015. CRISPR immunity drives rapid phage genome evolution in Streptococcus thermophilus. MBio 6:e00262–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Tao P, Wu X, Rao V. 2018. Unexpected evolutionary benefit to phages imparted by bacterial CRISPR-Cas9. Sci Adv 4:eaar4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bondy-Denomy J, Pawluk A, Maxwell KL, Davidson AR. 2013. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 493:429–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bondy-Denomy J, Garcia B, Strum S, Du M, Rollins MF, Hidalgo-Reyes Y, Wiedenheft B, Maxwell KL, Davidson AR. 2015. Multiple mechanisms for CRISPR-Cas inhibition by anti-CRISPR proteins. Nature 526:136–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Borges AL, Davidson AR, Bondy-Denomy J. 2017. The Discovery, Mechanisms, and Evolutionary Impact of Anti-CRISPRs. Annu Rev Virol 4:37–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Amlinger L, Hoekzema M, Wagner EGH, Koskiniemi S, Lundgren M. 2017. Fluorescent CRISPR Adaptation Reporter for rapid quantification of spacer acquisition. Sci Rep 7:10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Semenova E, Kuznedelov K, Datsenko KA, Boudry PM, Savitskaya EE, Medvedeva S, Beloglazova N, Logacheva M, Yakunin AF, Severinov K. 2015. The Cas6e ribonuclease is not required for interference and adaptation by the E. coli type I-E CRISPR-Cas system. Nucleic Acids Res 43:6049–6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Xu Hua Fu B, Wainberg M, Kundaje A, Fire AZ. 2017. High-throughput characterization of cascade type i-e crispr guide efficacy reveals unexpected pam diversity and target sequence preferences. Genetics 206:1727–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Cooper LA, Stringer AM, Wade JT. 2018. Determining the Specificity of Cascade Binding, Interference, and Primed Adaptation In Vivo in the Escherichia coli Type I-E CRISPR-Cas System. MBio 9:e02100–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Caliando BJ, Voigt CA. 2015. Targeted DNA degradation using a CRISPR device stably carried in the host genome. Nat Commun 6:6989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Blosser TR, Loeff L, Westra ER, Vlot M, Kunne T, Sobota M, Dekker C, Brouns SJJ, Joo C. 2015. Two distinct DNA binding modes guide dual roles of a CRISPR-Cas protein complex. Mol Cell 58:60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Jung C, Hawkins JA, Jones SK, Xiao Y, Rybarski JR, Dillard KE, Hussmann J, Saifuddin FA, Savran CA, Ellington AD, Ke A, Press WH, Finkelstein IJ. 2017. Massively Parallel Biophysical Analysis of CRISPR-Cas Complexes on Next Generation Sequencing Chips. Cell 170:35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Pawluk A, Bondy-Denomy J, Cheung VHW, Maxwell KL, Davidson AR. 2014. A new group of phage anti-CRISPR genes inhibits the type I-E CRISPR-Cas system of Pseudomonas aeruginosa. MBio 5:e00896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Pawluk A, Staals RHJJ, Taylor C, Watson BNJJ, Saha S, Fineran PC, Maxwell KL, Davidson AR. 2016. Inactivation of CRISPR-Cas systems by anti-CRISPR proteins in diverse bacterial species. Nat Microbiol 1:16085. [DOI] [PubMed] [Google Scholar]
- 162.Pawluk A, Amrani N, Zhang Y, Garcia B, Hidalgo-Reyes Y, Lee J, Edraki A, Shah M, Sontheimer EJ, Maxwell KL, Davidson AR. 2016. Naturally Occurring Off-Switches for CRISPR-Cas9. Cell 167:1829–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Rauch BJ, Silvis MR, Hultquist JF, Waters CS, McGregor MJ, Krogan NJ, Bondy-Denomy J. 2016. Inhibition of CRISPR-Cas9 with Bacteriophage Proteins. Cell 168:P150–158.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.He F, Bhoobalan-Chitty Y, Van LB, Kjeldsen AL, Dedola M, Makarova KS, Koonin EV., Brodersen DE, Peng X. 2018. Anti-CRISPR proteins encoded by archaeal lytic viruses inhibit subtype I-D immunity. Nat Microbiol 3:461–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Guo T, Han W, She Q. 2018. Tolerance of Sulfolobus SMV1 virus to the immunity of I-A and III-B CRISPR-Cas systems in Sulfolobus islandicus. RNA Biol 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wang X, Yao D, Xu JG, Li AR, Xu J, Fu P, Zhou Y, Zhu Y. 2016. Structural basis of Cas3 inhibition by the bacteriophage protein AcrF3. Nat Struct Mol Biol 23:868–870. [DOI] [PubMed] [Google Scholar]
- 167.Shin J, Jiang F, Liu JJ, Bray NL, Rauch BJ, Baik SH, Nogales E, Bondy-Denomy J, Corn JE, Doudna JA. 2017. Disabling Cas9 by an anti-CRISPR DNA mimic. Sci Adv 3:e1701620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Harrington LB, Doxzen KW, Ma E, Liu JJ, Knott GJ, Edraki A, Garcia B, Amrani N, Chen JS, Cofsky JC, Kranzusch PJ, Sontheimer EJ, Davidson AR, Maxwell KL, Doudna JA. 2017. A Broad-Spectrum Inhibitor of CRISPR-Cas9. Cell 170:1224–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kim I, Jeong M, Ka D, Han M, Kim NK, Bae E, Suh JY. 2018. Solution structure and dynamics of anti-CRISPR AcrIIA4, the Cas9 inhibitor. Sci Rep 8:3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Yang H, Patel DJ. 2017. Inhibition Mechanism of an Anti-CRISPR Suppressor AcrIIA4 Targeting SpyCas9. Mol Cell 67:117–127.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Dong D, Guo M, Wang S, Zhu Y, Wang S, Xiong Z, Yang J, Xu Z, Huang Z. 2017. Structural basis of CRISPR-SpyCas9 inhibition by an anti-CRISPR protein. Nature 546:436–439. [DOI] [PubMed] [Google Scholar]
- 172.Borges AL, Zhang JY, Rollins MF, Osuna BA, Wiedenheft B, Bondy-Denomy J. 2018. Bacteriophage Cooperation Suppresses CRISPR-Cas3 and Cas9 Immunity. Cell 174:917–925.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Landsberger M, Gandon S, Meaden S, Rollie C, Chevallereau A, Chabas H, Buckling A, Westra ER, van Houte S. 2018. Anti-CRISPR Phages Cooperate to Overcome CRISPR-Cas Immunity. Cell 174:908–916.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]