Abstract
The five-membered PRS gene family of Saccharomyces cerevisiae is an example of gene duplication allowing the acquisition of novel functions. Each of the five Prs polypeptides is theoretically capable of synthesising PRPP but at least one of the following heterodimers is required for survival: Prs1/Prs3, Prs2/Prs5 and Prs4/Prs5. Prs3 contains a pentameric motif 284KKCPK288 found only in nuclear proteins. Deletion of 284KKCPK288 destabilises the Prs1/Prs3 complex resulting in a cascade of events, including reduction in PRPP synthetase activity and altered cell wall integrity (CWI) as measured by caffeine sensitivity and Rlm1 expression. Prs3 also interacts with the kinetochore-associated protein, Nuf2. Following the possibility of 284KKCPK288-mediated transport of the Prs1/Prs3 complex to the nucleus, it may interact with Nuf2 and phosphorylated Slt2 permitting activation of Rlm1. This scenario explains the breakdown of CWI encountered in mutants lacking PRS3 or deleted for 284KKCPK288. However, removal of NHR1–1 from Prs1 does not disrupt the Prs1/Prs3 interaction as shown by increased PRPP synthetase activity. This is evidence for the separation of the two metabolic functions of the PRPP-synthesising machinery: provision of PRPP and maintenance of CWI and is an example of evolutionary development when multiple copies of a gene were present in the ancestral organism.
Keywords: cell wall integrity, protein/protein interaction, human neuropathies, yeast genome duplication, PRPP synthetase
Manipulation of the Saccharomyces cerevisiae Prs1/Prs3 complex identifies three different functions: PRPP synthesis, CWI maintenance and intracellular transport.
ABBREVIATIONS
- aa
amino acid(s)
- BSA
bovine serum albumin
- 3-AT
3-amino-1,2,4-triazole
- CWI
cell wall integrity
- 5-FOA
5-fluoro-orotic acid
- GFP
green fluorescent protein
- o/n
overnight
- NHR
non-homologous region
- nt
nucleotide(s)
- ONPG
o-nitrophenyl-β-D-galactopyranoside
- PRPP
5-phosphoribosyl-1(α)-pyrophosphate
- Prs
5-phosphoribosyl-1(α)-pyrophosphate synthetase
- P-Prs5
phosphorylated Prs5
- P-Slt2
phosphorylated Slt2
- SC
synthetic complete
- WT
wild type
- YEPD
yeast extract peptone dextrose
- Y2H
yeast-two-hybrid analysis
INTRODUCTION
Yeast is an undisputed model organism for the investigation of the connection between genes and their functions in cell metabolism. As a result of the whole genome duplication that occurred approximately 100 million years ago, a tetraploid yeast was created (Wolfe and Shields 1997; Dujon et al.2004; Wolfe 2015). Over time this unstable tetraploid yeast lost more than 80% of the duplicated gene copies. However, at least 10% of the 6000 remaining genes that make up the genome of the current species of Saccharomyces cerevisiae are duplicated (Botstein and Fink 2011) and implies that these remaining gene duplications have made a positive contribution to the ancestral yeast's relative fitness.
Our research in S. cerevisiae has identified five copies of a gene each of which is theoretically capable of encoding the enzyme PRPP synthetase (EC 2.7.6.1; Prs: ATP:D-ribose-5-phosphate diphosphotransferase) responsible for the production of PRPP (5-phosphoribosyl-1(α)-pyrophosphate), a building block of nucleotides and aromatic amino acids (Hove-Jensen et al.2017). While bacterial genomes contain only one gene encoding PRPP synthetase plants, fungi and mammals contain between two and five PRS genes. For instance, in the genome of the filamentous fungus Eremothecium (Ashbya) gossypii four genes have been found whose products contribute to cell growth and the production of riboflavin, vitamin B2, an essential co-factor in human metabolism and often used as a food additive (Mateos Jiménez and Revuelta 2006; Jiménez, Santos and Revuelta 2008) demonstrating the ubiquity of the requirement of PRPP in cells.
Another filamentous fungus, Aspergillus nidulans, has three PRS genes, AnprsA, AnprsB and AnprsC, all of which are highly expressed during hyphal growth and sporulation. Genetic analysis revealed that AnprsB and AnprsC are essential or auxotrophic genes since the transformants were always heterokaryotic (Zhong et al.2012; Jiang et al.2017). Further analysis revealed that the auxotrophy caused by mutation in either AnprsB or AnprsC could be at least partially compensated by including histidine, tryptophan, pyrimidine and AMP in the media, thereby confirming the multiple auxotrophy associated with these mutants. The fact that mutation of AnprsA does not result in auxotrophy together with the relative transcription levels of AnprsB > AnprsC > AnprsA is commensurate with the hypothesis that the products of these genes may contribute unequally to Prs activity. Furthermore, the identity >80% of AnprsB with Prs2, Prs3 and Prs4 of S. cerevisiae together with the 70% identity of AnprsA and AnprsC to yeast Prs1 and Prs5, respectively, would suggest that PRPP synthesis in A. nidulans may, as in S. cerevisiae, be dependent on interactions between different Prs polypeptides.
An interesting discovery that a protein, HbPrs4, encoded in the rubber tree (Hevea brasiliensis Muell. Arg.) genome with 80% identity to Prs4 of Arabidopsis thaliania had a strong yeast-two-hybrid (Y2H) interaction with the anaphase promoting complex/cyclosome (APC/C), an E3 ubiquitin ligase regulating protein degradation through the ubiquitin/26 S proteasome pathway of the rubber tree, raising the possibility of an interaction between HbPrs4 and the cell cycle. A BlastN search of the rubber tree genome database revealed another sequence with 84% identity to the HbPRS4 gene. PRPP production is important for the nucleotide and protein biosynthesis required for latex generation. HbPRS4 is highly expressed in the bark and responded to ethylene treatment, used in natural rubber production, to increase latex production. Ethylene treatment was found to increase both HbPrs4 expression and the ATP/ADP content of latex cells (Amalou, Bangratz and Chrestin 1992; Yu et al.2017).
A recent publication has highlighted that increased expression of the five-membered PRS gene family of S. cerevisiae correlates with xylose utilisation in a genetically engineered industrial strain of S. cerevisiae, emphasising the central role of PRPP synthesis in the metabolic pathway for biofuel production from lignocellulose (Feng et al.2018).
Spinacia oleracea has four PRS genes. An aa sequence in the N-terminal region in spinach isozyme 2 is consistent with its localisation in the chloroplast, whereas isozyme 3 is postulated to be localised to the mitochondria. Isozyme 4 has no extension and is considered to be located in the cytoplasm (Krath and Hove-Jensen 1999, 2001).
The five paralogous genes, each of which is theoretically capable of encoding the enzyme PRPP synthetase that produces PRPP in S. cerevisiae, have, in accordance with S. cerevisiae nomenclature, been designated PRS1–5 (Carter et al.1997). Extensive genetic analysis indicates that in order to survive, the yeast genome must contain at least one of three minimal functional subunits, namely Prs1/Prs3, Prs2/Prs5 and Prs4/Prs5. The Prs1/Prs3 subunit is the most important since mutant strains relying on either of the other two subunits are severely impaired in their growth and PRPP-synthesising capacity (Hernando, Parr and Schweizer 1998; Hernando et al.1999). Furthermore, simultaneous deletion of PRS1 and PRS5 or PRS3 and PRS5 causes synthetic lethality (Hernando, Parr and Schweizer 1998). Both PRS1 and PRS5 genes contain non-identical in-frame insertions, not present in PRS2, PRS3 or PRS4 (Schneiter et al.2000; Wang et al.2004; Vavassori et al.2005a,b; Kleineidam et al.2009; Ugbogu et al.2013, 2016). The region corresponding to non-homologous region 1-1 (NHR1–1) of Prs1 is required for bringing the Prs1/Prs3 subunit into contact with an element of the cell wall integrity (CWI) pathway as shown by Y2H and co-immunoprecipitation (Wang et al.2004; Ugbogu et al.2016) and may explain why strains lacking PRS1 and PRS3 do not survive at elevated temperature (Ugbogu et al.2013). PRS5 contains two insertions, one of which is characterised by the presence of a cluster of three phosphorylatable serine residues (Ficarro et al.2002). Mutation of these residues, either singly or multiply, influences two endpoints of the CWI pathway at both ambient and elevated temperatures (Ugbogu et al.2016). Clearly, the insertion in PRS1 and at least one in PRS5 are not gratuitous but fulfil essential functions in the cell. Although not ubiquitous in PRS genes NHR sequences are also found in eukaryotic organisms which have multiple PRS genes. Aspergillus nidulans and E. (Ashbya) gossypii genomes (www.aspgd.org, https://www.agd.unibas.ch/) (Hermida et al.2005; Jiménez, Santos and Revuelta 2008) contain three and four PRS genes, respectively. Two of the three PRS genes in A. nidulans, AnPrsA and AnPrsC, contain NHR sequences at similar positions to those in S. cerevisiae Prs1 and Prs5. Two of the four genes in E. (Ashbya) gossypii also contain NHRs and are considered to be homologues of S. cerevisiae Prs1 and Prs5. Schizosaccharomyces pombe has three PRS genes (https://www.pombase.org), two of which, S. pombe Prs1 and Prs2, contain insertions. The insertion in S. pombe Prs1 is at a similar position to that of NHR5–1 of S. cerevisiae Prs5 and S. pombe Prs2 has an insertion at the same relative position as NHR5–2 of S. cerevisiae Prs5.
The mammalian PRPP synthetase-associated proteins Pap-39 and Pap-41 have 76% sequence identity (Ishizuka et al.1996; Katashima et al.1998) with each other and are thought to play a negative regulatory role in PRPP synthetase activity since, following their removal by gel filtration in the presence of 1 M MgCl2, Prs activity is increased (Tatibana et al.1995; Becker 2001). Both Pap-39 and Pap-41 contain NHRs of 29 and 30 aa, respectively, occupying similar locations in the polypeptides to the locations of NHR1–1 and NHR5–2 of the corresponding S. cerevisiae genes. There is an interesting connection between human Prs (hPRPS1) and at least two human neuropathies, Arts syndrome and CMTX5 (Charcot–Marie–Tooth) and for which the corresponding associated mutations have been identified in hPRPS1 (de Brouwer et al.2010; Mittal et al.2015). We have exploited the high-sequence similarity of hPRPS1 and yeast Prs1 to create genocopies of Arts syndrome and CMTX5, and have examined their influence on Prs activity in our current investigations.
Prs3, which interacts with Prs1 (Wang et al.2004) to create the minimal functional unit Prs1/Prs3, harbours a sequence five aa in length that is unique to Prs3 and close to the C-terminus of the polypeptide. When deleted, there is a negative impact on yeast physiology, consistent with Prs1/Prs3 being the most important of the three genetically defined minimal functional subunits. Using a NADH-coupled enzyme test for PRPP synthetase in selected mutants and deletants, it has proved possible to separate the production of PRPP, the metabolic function, from the maintenance of CWI, thereby emphasising the scope for evolutionary development when multiple copies of a gene are present.
MATERIALS AND METHODS
Plasmids, in vitro mutagenesis and strains, growth conditions
Propagation of plasmids, Escherichia coli DH5α and standard DNA manipulations were performed as described previously (Ausubel et al.1995). The strains and plasmids used in this study are listed in Tables S1 and S2 (Supporting Information). Expression of fusion genes cloned in pGAD424-LEU2 was driven by the ADH1 promoter. The plasmid also contained the GAL4 DNA activation domain. In vitro mutagenesis with the plasmid pGAD-Prs3 as the template and the mutagenic primers was carried out using the Quick-ChangeR site-directed mutagenesis kit (Agilent, Stockport, UK) following the manufacturer's protocol. Mutagenic primer sequences are provided under Supplementary Data (Table S3, Supporting Information). Mutations were confirmed by double-stranded sequencing of the mutagenised plasmids. Yeast genetic manipulations were performed and media was prepared as described in Guthrie and Fink (1991) and Kaiser, Michaelis and Mitchell (1994). Yeast cultures were grown in YEPD (1% yeast extract, 2% peptone 2% glucose) or selective media, SC (0.67% yeast nitrogen base without aa and (NH4)2SO4, 2% glucose, 0.5% (NH4)2SO4) supplemented with the appropriate nutrients to select for plasmids. Yeast transformation was performed by the ‘plate’ method (Elble 1992). For Y2H analysis, the high efficiency transformation protocol described in Gietz and Woods (2002) was performed. YN96–77 was transformed with pGAD-Prs3, Tc4(pGAD-prs3Δ-KKCPK) or Tc10(pGAD-prs3-ΔKKCPK). Caffeine-mediated CWI pathway activation was determined as described previously (Ugbogu et al.2013).
Y2H analysis
The host strain PJ69–4A (James, Halladay and Craig 1996) was transformed with the interacting plasmids. At least three independent transformants for each combination were tested for their ability to grow on selective media lacking adenine or histidine plus 150 mM 3-AT and assayed for β-galactosidase activity using ONPG (o-nitrophenyl-β-D-galactopyranoside) as the substrate. The specific activity of β-galactosidase was calculated according to Wang et al. (2004).
Determination of Rlm1 expression
Rlm1 activation was measured in the transformed strains YN96–77 co-transformed with the reporter plasmid pHPS100-URA (Kirchrath et al.2000). The transformants were selected on appropriate media, and three independent colonies were inoculated individually into 10 ml of selective media. Following overnight (o/n) incubation, the cells were diluted to OD600 ≈ 0.5 in 2 × 50 ml of the same media in 250 ml Erlenmeyer flasks. One flask from each strain was incubated at 30°C and the other at 37°C until OD660 ≈ 1 was attained. Rlm1 expression was measured with the Thermo Scientific Yeast β-galactosidase assay kit according to the manufacturer's protocol (Fisher Thermo ScientificR, Renfrew, UK) and quantified according to Eq. 1:
Equation 1
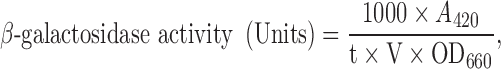 |
where V is the volume of cells (ml) used in the assay and t is the reaction time in min.
Rescue of synthetic lethality
YN97–18, YN97–19 and YN97–20, which contain the synthetically lethal deletion of PRS3 and PRS5 and are kept alive by the presence of pVT3 (Table S2, Supporting Information), were transformed with pGAD-Prs3, Tc4(pGAD-prs3-ΔKKCPK) or Tc10(pGAD-prs3-ΔKKCPK) and the transformants were selected for their ability to grow in the presence of 5-fluoro-orotic acid (5-FOA) for the counterselection of the URA3 plasmid pVT3.
Western blotting
Western blotting was performed essentially, as described in Ugbogu et al. (2013), on YN98–11 transformed with either pGAD-Prs3, Tc4(pGAD-prs3-ΔKKCPK) or Tc10(pGAD-prs3-ΔKKCPK) (Table S1, Supporting Information). For each transformant, crude extracts equivalent to 15 μg protein per lane were loaded and separated on 4-15% SDS-PAGE gel. Two gels were run simultaneously, one was stained with Coomassie Blue and the other, following transfer to a PVDF membrane, was probed with specific anti-GFP (green fluorescent protein) antibodies (Santa Cruz sc-57587, Biotechnology, Inc., Wembley, UK) and sc-2060 as primary and secondary antibodies, respectively. Successful separation and transfer to the membrane was checked by Ponceau S. Commercially available recombinant GFP (Roche, Welwyn Garden City, UK) was used as a positive control. GFP signals were detected using the ECL + Plus system by chemiluminescence (Bio-Rad, Perth, UK). After processing, the protein gel images were obtained with the ChemiDoc XRS+ imager using the Image Lab software, V4.1 (Bio-Rad), for the detection of bands corresponding to the separated polypeptides. The blots were reproduced at least once in independent experiments with a representative image shown.
Measurement of Prs activity
Single colonies of the strains to be tested for PRPP synthetase activity on SC-leu (YN96–77 transformants) or SC-trp (YN96–66 transformants) were grown o/n in 10 ml of the appropriate media. An aliquot of the o/n culture was added to 50 ml of the appropriate media to give an OD600 0.2 and incubated at 30°C until an OD600 ≈ 1.0–1.2 was attained. After harvesting, the cells were resuspended in sterile dist. H2O, washed once and the wet weight determined. Cell pellets were frozen at −80°C until cells were broken. Cell pellets were resuspended individually in 200 μl extraction buffer (50 mM KH2PO4/KH2 PO4 pH 7.5, 10% glycerol, 0.1% Triton X-100, 5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM PMSF, 5 mM DTT). The cell suspension was transferred into an Eppendorf tube containing 400 mg acid-washed glass beads (425–600 μm) (Sigma-Aldrich, Irvine, UK). The tubes were vortexed four times for 45 s with 1 min incubation on ice between vortexing. The disrupted cells were centrifuged for 10 000 g for 10 min at 4°C, and the supernatants were removed by pipetting to fresh Eppendorf tubes. Protein content of the crude extracts was determined by the Quick Start Bradford protein assay (Bradford 1976) using a standard curve prepared with BSA stock solutions of known concentrations.
The specific activity of PRPP synthetase was measured in a spectrophotometric coupled-enzyme test using myokinase, pyruvate kinase and lactate dehydrogenase (Sigma-Aldrich) (Donini et al.2017; Jiang et al.2017) as the decrease of NADH oxidised and expressed as μmol min−1 mg−1 protein using the molar extinction coefficient of 6220 M−1 cm−1 for NADH by monitoring absorption at 340 nm in a SpectraMax M series microplate reader. A conversion factor of 2 is required to calculate the specific activity since 2 moles of β-NADH are oxidised for each mole of PRPP produced. A 30-min preincubation of the reaction buffer consisting of 50 mM KH2PO4/KH2 PO4 pH 7.5, 5 mM MgCl2 × 6H2O, 2 mM ATP, 0.2 mM NADH, 3.75 mM phosphoenol-pyruvate, 1.5 U myokinase, 3 U pyruvate kinase and 1.5 U lactate dehydrogenase without 3.75 mM ribose-5-phosphate was carried out at 37°C. Following the addition of ribose-5-phosphate, the reaction was started by adding an appropriate volume of crude extract (0.2–0.3 μg/ml) to the reaction mixture and was monitored continuously over a period of 20 min with measurement being recorded at intervals of 26 s. Varying amounts of crude extract were used in independent experiments, thus compensating for the different specific activities of PRPP synthetase of the mutants tested.
Statistical analysis
All data are calculated as the mean ± SD. Statistical analysis was conducted using SPSS StatisticsS software, version 22. The statistics for the β-galactosidase assays were performed by two-factor ANOVA analysis in tandem with the Bonferroni test to determine the significance within the dataset. This approach was taken to test for significance within each transformant group by comparing Rlm1 expression in response to heat stress and between transformants relative to the respective culture conditions.
For the measurement of the specific PRPP synthetase activity, an independent samples t-test was conducted between the crude extracts of the WT and individual mutants. Robust tests for equality of variance between samples were assessed by the Levene test (Levene 1960). In the case of extracts with unequal variance, a correction was applied to the P-values. In all instances, a P-value of ≤ 0.05 was considered statistically significant.
RESULTS
Deletion of 284KKCPK288 in Prs3
There is a high degree of sequence similarity between all five Prs gene products. We have discovered that Prs3 contains the sequence (aa 284KKCPK288), i.e. two positively charged aa, lysine (K) flanking three residues, one of which is proline (P) located close to the C-terminus of the polypeptide and not present in the Prs1, Prs2, Prs4 or Prs5 polypeptides. Such a sequence is present in several yeast nuclear proteins but not in yeast cytoplasmic proteins (Herrero, Martinez-Campa and Moreno 1998). In a genome-wide Y2H assay, it was found that Prs3 interacts with the nuclear import protein, karyopherin α homologue, importin α encoded by the SRP1 gene (Ito et al.2001) (http://dbarchive.biosciencedbc.jp/en/yeast-y2h/download.html), implying that Prs3 has the potential to be at least temporarily a yeast nuclear protein. An obvious question is: What effect(s) does deletion of this site have on yeast physiology?
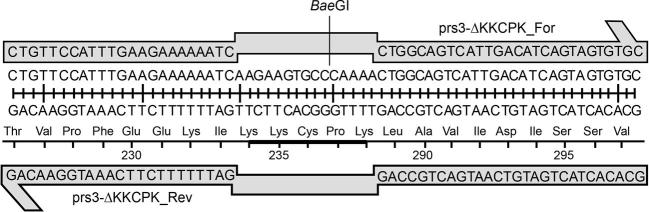
By means of specifically designed primers, the 15 nts corresponding to the motif 284KKCPK288 were deleted from the PRS3 sequence. Fortuitously, this sequence contained a BaeGI restriction endonuclease site, therefore allowing a pre-screening by restriction digest of the mutagenised plasmids prior to sequencing. Sequencing of the entire length of the selected plasmids confirmed the loss of 15 nt containing a unique BaeGI site. This region is illustrated in Fig. 1. Two plasmids Tc4(pGAD-prs3-ΔKKCPK) and Tc10(pGAD-prs3-ΔKKCPK) were selected for further experimentation.
Figure 1.
Schematic representation of the C-terminal region of Prs3 containing the 284KKCPK288 motif and indicating the position of the BaeGI site used for the initial screening of mutagenised plasmids. The mutagenic primers prs3_ΔKKCPK_For and Prs3-ΔKKCPK_Rev (Table S3, Supporting Information) responsible for the deletion of the 15 nt corresponding to 284KKCPK288 (highlighted as a black bar) are indicated above and below the nucleotide and protein sequences of 284KKCPK288 and its flanking regions.
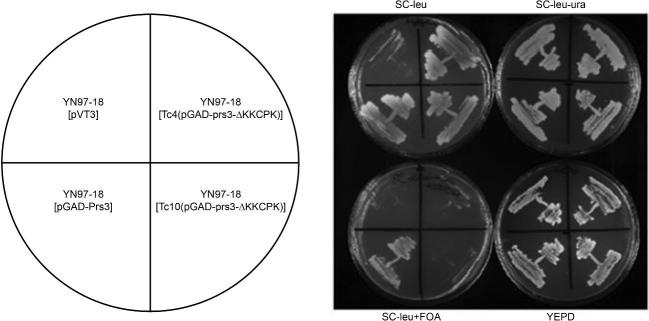
To determine whether or not deletion of 284KKCPK288 in Prs3 has an influence on yeast physiology, we carried out plasmid shuffling using a synthetically lethal prs3Δ prs5Δ double deletant (YN97–18) which is kept alive by the inclusion of a URA3-based plasmid, pVT3, carrying PRS3 (Hernando et al.1999). Transformation of this strain with the WT Prs3 plasmid, Tc4(pGAD-prs3-ΔKKCPK) or Tc10(pGAD-prs3-ΔKKCPK) followed by elimination of pVT3 on 5-FOA-containing media provided evidence that neither of the two plasmids lacking the sequence were capable of sustaining growth on 5-FOA-containing media (Fig. 2). Repeating this experiment using YN97–19 and YN97–20 yielded identical results, thus providing evidence that the 284KKCPK288 is essential for viability (data not shown).
Figure 2.
Rescue of synthetic lethality of YN97–18 (prs3Δ prs5Δ) [pVT3]. Plasmid shuffling was performed using FOA counterselection. The strains were streaked onto appropriate media as indicated above and below the photographed plates and incubated at 30°C for 5 days. The cartoon on the left indicates the plasmids used for testing the rescue of synthetic lethality.
Elimination of 284KKCPK288 disrupts the Prs1/Prs3 complex
We have previously shown that in a strain containing an integrated GFP-labelled version of PRS1 in combination with a deletion of PRS3 the GFP signal is no longer visible in a western blot. This is not the case if GFP-labelled PRS1 is combined with deletions of PRS2, PRS4 or PRS5 (Ugbogu et al.2013). Western blotting of transformants of YN98–11 with WT Prs3, Tc4(pGAD-prs3-ΔKKCPK) or Tc10(pGAD-prs3-ΔKKCPK) revealed that the GFP signal was visible only when WT Prs3 was present (Fig. 3). However, removal of the 284KKCPK288 results in the loss of the strength of the GFP signal to the level observed in a strain lacking Prs3 implying that the loss of the five aa alters the Prs3 polypeptide to such an extent that it can no longer combine with Prs1 to form a stable Prs1/Prs3 heterodimer. The lower GFP-responsive band has been observed in previous western blots and has been explained as an unknown protein which cross-hybridises with anti-GFP antibodies since it appeared in western blots of a WT strain containing no GFP-labelled proteins (Ugbogu et al.2013).
Figure 3.
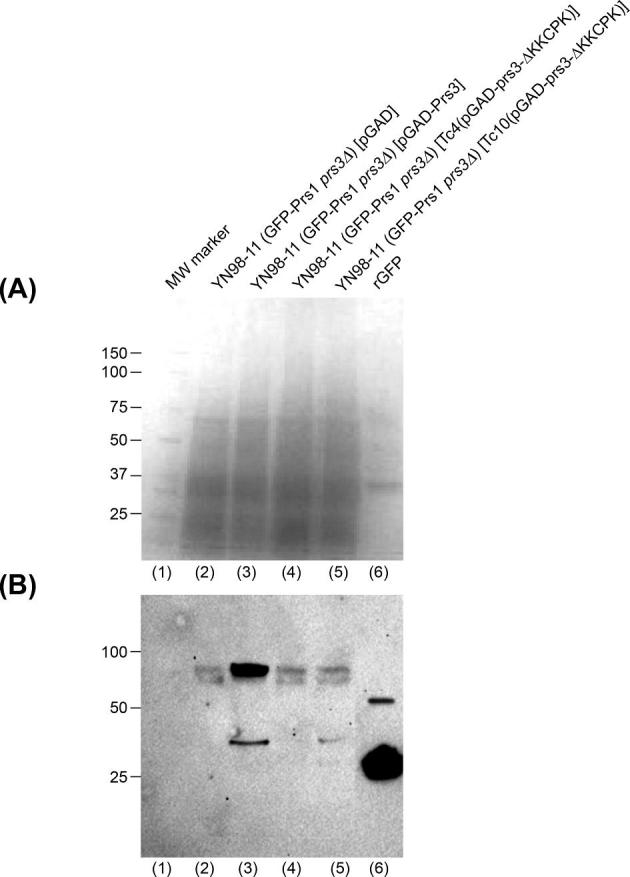
Western blotting reveals that the deletion of 284KKCPK288 from Prs3 results in the loss of the GFP-signal associated with Prs1 in YN98–11. Crude extracts equivalent to 15 μg protein per lane were prepared from YN98–11 transformed with either pGAD (lane 2), pGAD-Prs3 (lane 3), Tc4(pGAD-prs3-ΔKKCPK) (lane 4) or Tc10(pGAD-prs3-ΔKKCPK) (lane 5) and separated on two simultaneously run 4–15% SDS-PAGE gels. (A) Coomassie Blue stained gel as loading control and (B) following transfer of the duplicated gel to a PVDF membrane western blotting was performed with specific anti-GFP antibodies (Santa Cruz sc-57587) and sc-2060 as primary and secondary antibodies, respectively. Successful electrophoresis and transfer to a PVDF membrane were checked by Ponceau S staining. The GFP signal was detected by chemiluminescence with anti-GFP antibodies. (A) Lane (1) molecular weight marker from Coomassie-stained gel. Lane (6) 10 μg rGFP standard.
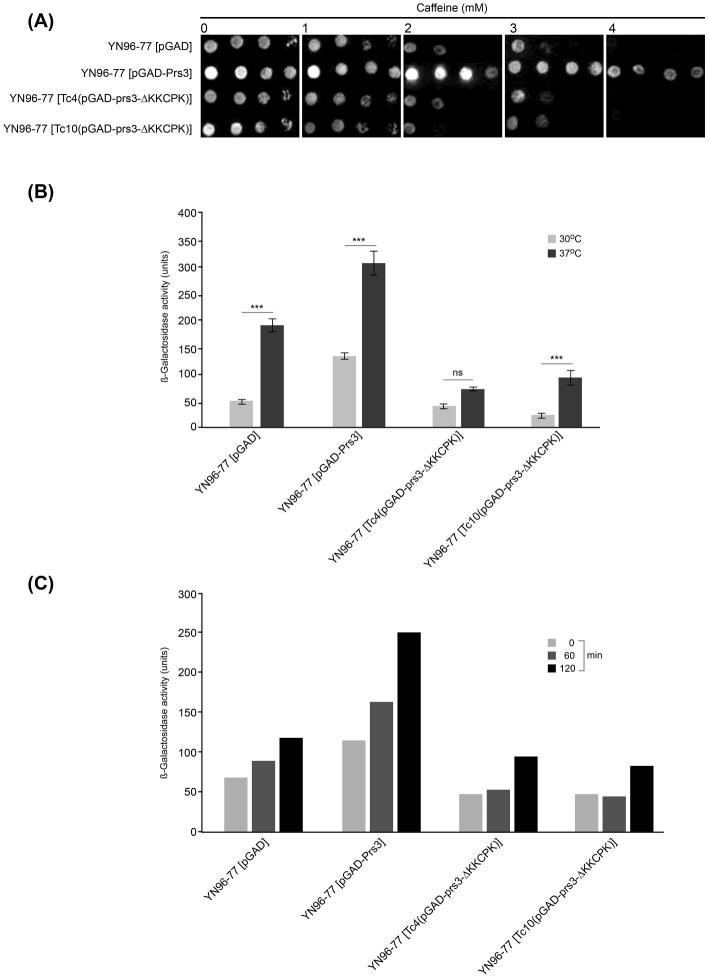
Further investigation of the transformants containing one or other of the plasmids lacking the 284KKCPK288 sequence yielded results consistent with a breakdown of CWI maintenance. Transformants of the strain YN96–77 lacking PRS3 with the deleted plasmids do not reverse the caffeine-sensitive phenotype of this strain. As shown in Fig. 4A, the two transformants containing a prs3-ΔKKCPK plasmid show a similar degree of caffeine sensitivity, i.e. cessation of growth at 2–3 mM caffeine comparable with that of the negative control YN96–77 [pGAD]. However, when the WT Prs3 is present good growth is still visible at 4 mM caffeine. Growth at 37°C is severely impaired in the presence or absence of 284KKCPK288 but in contrast to the transformant containing WT Prs3 the addition of 1 M sorbitol to the media does not fully restore growth in YN96–77 containing either Tc4(pGAD-prs3-ΔKKCPK) or Tc10(pGAD-prs3-ΔKKCPK) (Tatsiana Chyker, unpublished data).
Figure 4.
(A) The sequence 284KKCPK288 is required for growth on caffeine. Cultures of YN96–77 (prs3Δ) transformed with pGAD, pGAD-Prs3, Tc4(pGAD-prs3-ΔKKCPK) or Tc10(pGAD-prs3-ΔKKCPK) were adjusted to OD600 0.5 and 3 μl of 10-fold serial dilutions thereof were spotted onto SC-leu caffeine-containing media at the caffeine concentrations indicated. Plates were incubated at 30°C for 3 days prior to documentation in a ChemiDoc XRS + imager. The experiment was repeated four times, and a representative image is shown. (B) The effect of deletion of 284KKCPK288 on Rlm1 expression. The four strains described in (A) were transformed with the reporter plasmid pHPS100-URA that expresses β-galactosidase as a measure of Rlm1 expression. The cultures were grown at either 30°C or 37°C to an OD600 1.0, and Rlm1 expression was measured as β-galactosidase activity using the Thermo Scientific yeast β-galactosidase kit. Nine independent repeat experiments were performed. The data are presented as the mean ± SEM. Asterisks represent significant temperature-dependent Rlm1 expression within transformants (P-value < 0.01 = ***), ns = not significant. (C) Kinetics of Rlm1 expression in response to heat stress. The strains described in the legend of Figure 4(B) were grown at 30°C to an OD600 1.0 and an aliquot taken for β-galactosidase measurement prior to shifting the cultures to 37°C. Aliquots for β-galactosidase measurements were removed after 60 and 120 min at 37°C.
Effect of deletion of 284KKCPK288 on Rlm1 expression and PRPP synthetase activity
Using a reporter plasmid for Rlm1 expression following Slt2 activation, there is a significant reduction in Rlm1 expression at 30°C of over 70% in transformants of YN96–77 (prs3Δ) [pHPS100-URA] with Tc4(pGAD-prs3-ΔKKCPK) or Tc10(pGAD-prs3-ΔKKCPK) in comparison to a transformant of the same strain carrying a WT version of Prs3. Incubation of the same strains at 37°C resulted in a 1.5- to 4.0-fold increase in Rlm1 expression whether or not the 284KKCPK288 sequence is present (Fig. 4B).
In the absence of 284KKCPK288 the response of Rlm1 expression with respect to time is also affected (Fig. 4C). Transformants of YN96–77 (prs3Δ) [pHPS100-URA] were grown at 30°C before shifting to 37°C. Aliquots of each culture were removed at 60 and 120 min for measurement of Rlm1 expression at 37°C. Under these culture conditions, both the vector control and the WT Prs3 showed a steady increase in Rlm1 expression whereas removal of 284KKCPK288 disrupts this pattern. Removal of 284KKCPK288 has apparently a 2-fold effect: a reduction in the β-galactosidase activity at time zero and no steady increase over time (Fig. 4C).
Prs activity in YN96–77 (Fig. 5) was examined with the transformants harbouring WT Prs3, the vector and Tc4(pGAD-prs3-ΔKKCPK) or Tc10(pGAD-prs3-ΔKKCPK). When Prs3 was present, Prs activity is equivalent to the WT. However, in the absence of 284KKCPK288 the enzyme activity was significantly reduced in comparison to that of the WT and YN96–77 [pGAD-Prs3]. Specifically, with Tc4(pGAD-prs3-ΔKKCPK) or Tc10(pGAD-prs3-ΔKKCPK) the enzyme activity was only 25% higher than that of the vector control which measures Prs activity produced by the complexes Prs2/Prs5 and Prs4/Prs5. This is further evidence that the deletion of 284KKCPK288 has a deleterious effect on the Prs1/Prs3 complex, a result that is not unexpected since it has been shown in western blotting that neither of these two plasmids are capable of restoring the GFP signal as evidence of a Prs1/Prs3 heterodimer (Fig. 3).
Figure 5.
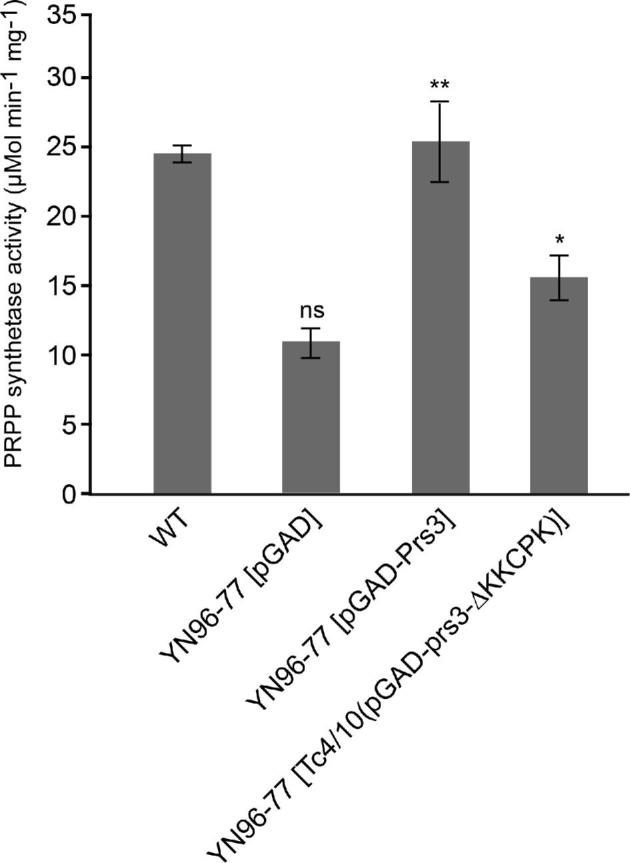
Deletion of 284KKCPK288 from Prs3 reduces PRPP synthetase activity. YN94–1 (WT) and YN96–77 (prs3Δ) transformed with pGAD, pGAD-Prs3, Tc4(pGAD-prs3-ΔKKCPK) or Tc10(pGAD-prs3-ΔKKCPK) were grown o/n from a single colony at 30°C in 10 ml SC-leu or YEPD for WT. Cell extracts were prepared as described in the Materials and Methods section. Following protein determination and pre-incubation of the reaction buffer at 37°C for 30 min, the reaction was started by the addition of ribose-5-phosphate and appropriate amounts of extracts dispensed into a 96-well plate. The results are expressed as μmol min−1 mg−1. The significance levels of YN96–77 [pGAD-Prs3] and YN96–77 [Tc4/10(pGAD-prs3-ΔKKCPK)] are calculated with respect to the specific activity of YN96–77 [pGAD]. Data are shown as mean ± SD. Number of measurements for each transformant: WT, pGAD-Prs3, Tc4(pGAD-prs3-ΔKKCPK) (n = 3) and for pGAD, Tc10(pGAD-prs3-ΔKKCPK) (n = 5). For illustration purposes, the data for Tc4(pGAD-prs3-ΔKKCPK) and Tc10(pGAD-prs3-ΔKKCPK) have been combined (Tc4/Tc10(pGAD-prs3-ΔKKCPK). P-values: * = P ≤ 0.05, ** = P ≤ 0.01, ns = not significant.
NHR1-1 is not required for binding of Prs1 with Prs2 and Prs3
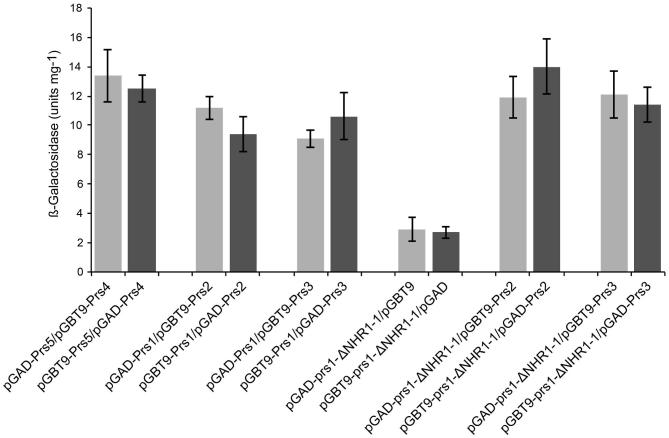
In contrast to the impact of the deletion of 284KKCPK288 on the integrity of the Prs1/Prs3 complex, removal of NHR1–1 of Prs1 does not alter this complex. Y2H analysis of prs1-ΔNHR1–1 with Prs3 or Prs2 was carried out, and the results are shown in Fig. 6. The transformants with both pairwise combinations were tested for β-galactosidase activity (Fig. 6) and their ability to grow on media lacking either adenine or histidine (data not shown). It is clear that NHR1–1 does not play a part in the interactions between Prs1 and Prs2 or Prs3 emphasising that the interaction of Prs1 with Slt2 is solely dependent on the presence of NHR1–1 (Wang et al.2004).
Figure 6.
β-Galactosidase activity of strains testing for interactions between NHR1–1 less Prs1 with Prs2 and Prs3. Cell extracts from strains assessing the indicated interactions were analysed using the ONPG β-galactosidase assay. The columns represent the mean of values obtained in three independent experiments, and the error bars are representative of the SD between them.
Interaction of Prs3 with the kinetochore-associated protein Nuf2
A connection between PRPP-synthesising machinery and the cell cycle is suggested by the altered morphology of prs3Δ and prs5Δ strains, the former appears as highly vacuolated, large spherical cells (Schneiter et al.2000) that suggested a possible defect in polarised growth or cell division. It was observed that a prs3Δ strain failed to arrest the cell cycle in G1 upon nutrient deprivation (Binley et al.1999). These observations gained more importance in light of the fact that the kinetochore-associated protein Nuf2 interacted with Prs2 in a genome-wide Y2H assay (Uetz et al.2000). On the basis of this result, we undertook a Y2H analysis of Nuf2 with Prs1-Prs5 which was carried out in both orientations. The data for the combination Nuf2Gal4BD and Prs1–5Gal4AD are summarised in Table 1. Only the interaction between Nuf2 and Prs4 could not be confirmed by histidine or adenine prototrophy and had a lower β-galactosidase activity than the negative control with the empty vectors. Interestingly, the interaction between Nuf2 and each of the five Prs polypeptides as measured by β-galactosidase activity can be summarised as Prs3/Nuf2 > Prs2/Nuf2 > Prs5/Nuf2 > Prs1/Nuf2. These results are compatible with each of the Prs polypeptides being at least temporarily in the nucleus by virtue of interaction with the nuclear resident protein Nuf2 (Suzuki et al.2016).
Table 1.
Y2H interactions of Prs polypeptides with Nuf2 in PJ69–4A.
| Gal4BD | Gal4AD | β-Galactosidase filter assay | Histidine prototrophy | Adenine prototrophy | β-Galactosidase activity |
|---|---|---|---|---|---|
| – | – | – | – | – | 2.8 ± 0.3 |
| Snf4 | Snf1 | + | + | + | 11.4 ± 0.7 |
| Prs5 | Prs4 | + | + | + | 11.1 ± 0.1 |
| Nuf2 | Prs1 | + | + | + | 25.7 ± 1.3 |
| Nuf2 | Prs2 | + | + | + | 39.9 ± 0.5 |
| Nuf2 | Prs3 | + | + | + | 56.3 ± 0.6 |
| Nuf2 | Prs4 | + | – | – | 1.7 ± 0.1 |
| Nuf2 | Prs5 | + | + | + | 26 ± 0.3 |
The β-galactosidase activity (nmole ONPG min−1 mg−1) represented the mean ± SD from at least three independent experiments.
NHR1-1 of Prs1 is essential for maintenance of CWI but its loss does not impair PRPP synthetase activity
We have shown that NHR1–1 of Prs1 is essential for rescuing the synthetic lethality of a prs1Δ pr5Δ strain, most likely by virtue of its interaction with Slt2 since when NHR1–1 is removed from Prs1 there is no longer any interaction with Slt2 (Wang et al.2004). As shown above (Fig. 6), loss of NHR1–1 does not interfere with the interaction of Prs1 with Prs3. However, in contrast removal of NHR1–1 caused a significant increase in PRPP synthetase activity in comparison to the Prs1 plasmid in YN96–66 (Fig. 7B). Furthermore, deletion of the NHR1–1 results in bringing the divalent-cation-binding site and the PRPP-binding site (ribose-5-phosphate loop) close together, resembling the situation in Prs3, Prs2 and Prs4 which could in fact give rise to increased enzyme activity. Nevertheless, this result is particularly interesting since for the first time it has been shown that the removal of NHR1–1 disrupts the maintenance of CWI (Ugbogu et al.2013) but does not have a negative effect on PRPP synthetase activity.
Figure 7.
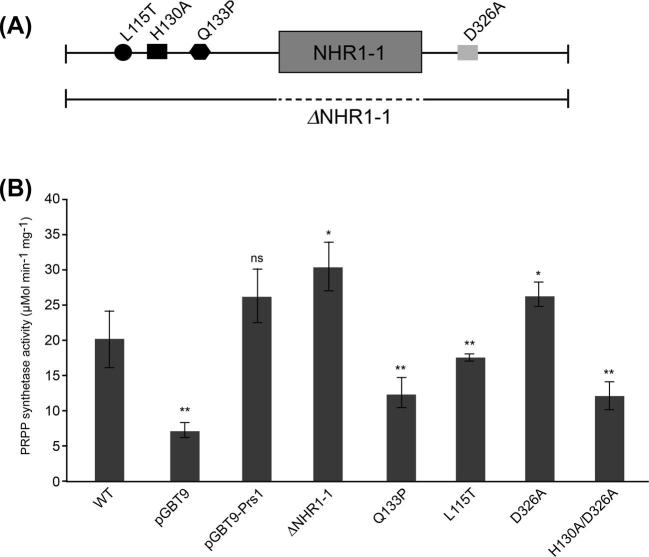
(A) Schematic representation of Prs1 showing the positions of the genocopies corresponding to CMTX5 (L115T), Arts syndrome (Q133P), the divalent-cation-binding site (H130A), the PRPP-binding site (ribose-5-phosphate) (D326A) of PRPP synthetase, the location of NHR1–1 (grey box) and the extent of its deletion as a broken line. (B) PRPP synthetase of the constructs illustrated in (A) was measured as described in Figure 5. Transformants of YN96–66 with indicated plasmids were grown in SC-trp or YEPD for WT. Data are shown as mean ± SD. Number of measurements for each transformant: WT and pGBT9 (n = 5); pGBT9-Prs1 (n = 6); ΔNHR1–1 and Q133P (n = 4); L115T (n = 3); D326A (n = 8) and H130A/D326A (n = 10). P-values: * = P ≤ 0.05; ** = P ≤ 0.01, ns = not significant with respect to pGBT9-Prs1.
Figure 7B also displays the values for Prs activity in strains representing genocopies of human neuropathies and sites essential for enzyme activity. The mutation Q133P is the yeast genocopy of the mutation associated with Arts syndrome (de Brouwer et al.2007, 2010). This mutation Q133P could only relieve the synthetic lethality of a prs1Δ pr5Δ strain to a limited extent (Ugbogu et al.2013) and reduces PRPP synthetase activity by over 50% of that measured in YN96–66 [pGBT9-Prs1]. CMTX5, the human neuropathy, is associated with the mutation M115T in hPRPS1 (Kim et al.2007; Synofzik et al.2014), and the yeast genocopy (L115T) still retains 80% Prs activity in comparison to YN96–66 [pGBT9-Prs1]. The Prs activity was also measured in two further mutations, one affecting the ribose-5-phosphate binding site at position 326 (D326A) and the double mutation (H130A/D326A) affecting both the divalent-cation and the PRPP-binding (ribose-5-phosphate) sites. There was no measurable effect on PRPP synthetase activity when the ribose-5-phosphate site was mutated, whereas the double mutation H130A/D326A had an effect on enzyme activity similar to that of the Arts syndrome (Q133P) genocopy. The importance of the H130A divalent-cation-binding site in contrast to the D326A PRPP-binding site (ribose-5-phosphate) has been observed previously in another scenario since H130A was incapable of rescuing a prs1Δ pr5Δ strain whereas D326A was more efficient than the WT (Ugbogu et al.2013).
DISCUSSION
PRPP is an extremely important metabolite due to its crucial role in the synthesis of purine and pyrimidine nucleotides and the fact that it links carbon and nitrogen metabolism. Our collection of mutants and deletions in the five PRS-encoding genes existing in the S. cerevisiae genome, which affect both PRPP synthesis and CWI, has provided the opportunity to investigate their relative contributions to these two vital aspects of yeast physiology. We have shown that the integrity of the minimal functional unit Prs1/Prs3 is dependent on the presence of Prs3 (Ugbogu et al.2013). The results presented here demonstrate that the 284KKCPK288 sequence close to the C-terminus of Prs3 and not found in any of the other four Prs polypeptides of S. cerevisiae is essential for the maintenance of the Prs1/Prs3 subcomplex. Removal of these five aa, as is the case with a PRS3 deletion, also results in the loss of the GFP-Prs1 signal (Fig. 3). Furthermore, the failure of a Prs3 plasmid lacking the 284KKCPK288 sequence to rescue the synthetic lethality of prs3Δ prs5Δ strain (Fig. 2) is further evidence for the sequence 284KKCPK288 being necessary for the integrity of the heterodimer, Prs1/Prs3, the most important of the three minimal functional units. Furthermore, it supports the hypothesis that a prs3Δ prs5Δ strain is, in fact, a triple deletant, prs1Δ prs3Δ prs5Δ, as postulated previously (Ugbogu et al.2013). The existence of such a triple deletant is incompatible with the requirement for the presence of at least one of the minimal functional units, Prs1/Prs3, Prs2/Prs5 or Prs4/Prs5 since any one of these subunits must be capable of synthesising PRPP at a level necessary for viability and to maintain CWI. The most important connection between PRPP synthesis and CWI is dependent on the NHR1–1 of Prs1 that directly interacts with the MAPK Slt2 when this is dually phosphorylated (Ugbogu et al.2016). However, the two minimal functional units, Prs2/Prs5 and Prs4/Prs5, can, by virtue of NHR5–2 of Prs5, play at least a supporting role in the maintenance of CWI. In addition, Y2H analysis has revealed that each of the four remaining polypeptides interacts with Slt2. Although it has been shown that Prs1 is co-immunoprecipitated with phosphorylated Slt2 (Ugbogu et al.2016), the Y2H interaction between Slt2 and either Prs3 or Prs1 was stronger than between Slt2 and Prs2 or Prs4 or Prs5 (Stefano Vavassori, unpublished data) emphasising the importance of the Prs1/Prs3 complex and the necessity for cooperation of the five Prs polypeptides in the maintenance of cell viability. We have evidence that the phosphorylation status of Slt2 is different from the pattern found in the WT when one or more of the three phosphorylation sites in NHR5–2 are mutated. In a premature truncation of Prs5 that removed most of NHR5–2, impaired phosphorylation of Slt2 following mild heat shock was shown by western blotting (Ugbogu et al.2016). Mutation of three specific serine residues, S364A, S367A and S369A, within NHR5–2 results in hyperphosphorylation of Slt2, and this was reflected in the expression of Rlm1, an endpoint of the CWI pathway and Fks2, a component of the 1,3-β-glucan synthase, which is expressed only under conditions of stress (Levin 2011). A connection between Fks2 and Prs polypeptides is feasible via the Paf1 transcription complex. Elimination of PAF1 results in a 50% reduction of the transcription of Prs1 (Chang et al.1999), and there is an interaction at the FKS2 promoter between Paf1 and Slt2/Swi4/Swi6 when FKS2 is transcribed (Kim and Levin 2011).
Alignment of the sequence of PRPP synthetase of Bacillus subtilis (Eriksen et al.2000; Hove-Jensen et al.2017) and the sequences of the five Prs polypeptides from yeast shows that NHR1–1 and NHR5–2 have locations external to the hexameric complex of B. subtilis Prs, implying that the interactions of these two NHRs with other proteins are likely to be favoured. In a strain in which the sequence of NHR1–1 has been deleted from Prs1, a 50% increase in Prs activity in comparison to the WT grown in YEPD is achieved (Fig. 7B). A lower, albeit possibly a more accurate factor, 15%, for the increase in Prs activity in the absence of NHR1–1 is obtained by comparing the activities of YN96–66 [pGBT9-Prs1] and YN96–66 [pGBT9-prs1-ΔNHR1–1] since in both instances the constructs are plasmid borne. Taking into consideration the alignment of Prs3 with B. subtilis Prs, it is feasible that the presence of NHR1–1 could interfere with binding of Prs1 with Prs3. However, the presence of NHR1–1 and its ability to interact with phosphorylated Slt2 ensure two important properties of cellular homeostasis, PRPP synthesis and maintenance of CWI, are upheld, even though NHR1–1 is located C-terminally within the catalytic flexible loop of Prs1 (Fig. 8). We know that the presence of NHR1–1 is essential for rescuing synthetic lethality of a prs1Δ prs5Δ strain and have hypothesised that the cause of synthetic lethality for such a strain when PRS1 and PRS5 are simultaneously deleted is a breakdown of CWI (Ugbogu et al.2013). In addition, we have shown that removal of NHR1–1 does not compromise the Prs1/Prs3 interaction as demonstrated by Y2H (Fig. 6) and strengthens the argument that the synthetic lethality associated with simultaneous deletion of PRS1 and PRS5 is due to a breakdown of the CWI pathway. This is the first time it has been demonstrated that it is possible to separate Prs activity and maintenance of CWI as shown by the increase of Prs activity when NHR1–1 is no longer present. This is further substantiated by the inability of a strain lacking NHR1–1 to increase Rlm1 expression in response to elevated temperature (Ugbogu et al.2013).
Figure 8.
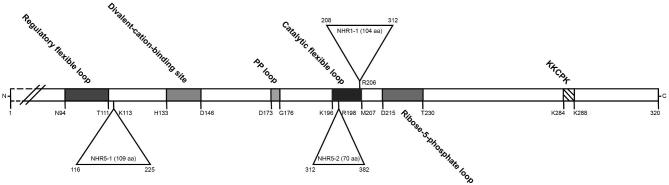
Schematic distribution of characteristic motifs of Prs polypeptides. The Prs3 polypeptide consists of 320 aa, and the position of 284KKCPK288 is indicated by a hatched box. The positions of the diagnostic features of a Prs polypeptide are indicated on the open bar, and their coordinates below the open bar were obtained by comparing the sequence of B. subtilis Prs lacking its initial methionine with that of S. cerevisiae Prs3. The insertion of Prs1, NHR1–1, is indicated by a triangle above the open bar. The two insertions in Prs5, NHR5–1 and NHR5–2, are indicated by triangles below the open bar. The coordinates of the insertions refer to their positions in the sequences of Prs1 and Prs5, respectively. The numbering of their insertion points corresponds to the sequence of the Prs3 polypeptide. All sequence comparisons were performed using the CLUSTALW multiple alignment tool (weight matrix = Gonnet) available in Expasy (NPS@: Network Protein Sequence Analysis (Combet et al.2000).
While it cannot be determined if the reduction of Rlm1 expression in prs3Δ strains containing either Tc4(pGAD-prs3-ΔKKCPK) or Tc10(pGAD-prs3-ΔKKCP) is due to the lack of the five aa or the concomitant loss of Prs1, there is evidence that Prs1, Prs3 and Prs5 all interact with Rlm1 (Ugbogu et al.2016), a transcription factor always resident in the nucleus (Levin 2005, 2011; Engelberg, Perlman and Levitzki 2014). It is tempting to hypothesise that Prs3 on account of its 284KKCPK288 sequence is responsible for the transport of itself, Prs1 and Prs5 into the nucleus, thereby activating expression of Rlm1 and Fks2 and ensuring the maintenance of CWI. The interaction of Prs3 with Nuf2, a component of the kinetochore Ndc80 complex that is localised in the nucleus, provides further support to our hypothesis that Prs1, Prs3 and Prs5 must be transported to the nucleus. In our Y2H-based analysis of Nuf2 with each of the five Prs polypeptides, the strongest interaction was between Nuf2 and Prs3 followed by Prs2 > Prs5 > Prs1 (Table 1). The interaction between Prs4 and Nuf2 was below the level of the negative control. These results support our working hypothesis that Prs3 can function as a transport protein in times of stress bringing a Prs1/Prs3/Prs5 complex into the nucleus, thereby ensuring Prs1 via NHR1–1 comes into contact with activated Slt2. In an earlier publication (Schneiter et al.2000), western blotting with Prs1-specific antibodies revealed the presence of Prs1 in the nuclear fraction.
Furthermore, it would appear that the loss of 284KKCPK288 causes such a conformational change in Prs3 that it prevents stabilisation of the Prs1/Prs3 dimer. The Prs3 polypeptide, although incapable per se, as are the four other Prs polypeptides, of PRPP synthesis nevertheless makes a major contribution to yeast survival by stabilising the most important minimal functional subunit, Prs1/Prs3, and ensuring its transport, with or without Prs5, to the nucleus in order to fulfil the requirement for the maintenance of CWI. Some support for this interpretation is provided by the measurement of Prs activity in transformants of YN96–77. Introduction of Prs3 into the prs3Δ strain YN96–77 restores Prs activity to that of the WT but introduction of either Tc4(pGAD-prs3-ΔKKCPK) or Tc10(pGAD-prs3-ΔKKCPK) into YN96–77 does not bring Prs activity up to the level measured in YN96–77 [pGAD-Prs3] (Fig. 5). Taken together, the average Prs activity of the transformants carrying the deleted plasmids displays a reduced Prs activity. In comparison to the activity of YN96–77 [pGAD] in which Prs activity is supplied by the Prs2/Prs5 and/or Prs4/Prs5 minimal subunits, a mean activity of 70% is achieved with Tc4(pGAD-prs3-ΔKKCPK) or Tc10(pGAD-prs3-ΔKKCPK) relative to YN96–77 [pGAD] (Fig. 5). This is in agreement with the inability of either of these two constructs to support the existence of a Prs1/Prs3 complex.
Alterations in the specific activity of Prs have been shown to be associated with the human neuropathies, Arts syndrome and CMTX5 (de Brouwer et al.2010; Mittal et al.2015). Genocopies in yeast of the human mutations associated with both of these neuropathies had a negative influence on Prs activity; the mutation Q133P associated with Arts syndrome resulted in halving the Prs activity measured (Fig. 7B). It is known that proline is a helix breaker and the substitution of glutamine (Q) by proline (P) prevents helix stabilisation through intermolecular H-bonding (de Brouwer et al.2007, 2010). The CMTX5 genocopy in yeast, L115T, has a less severe effect on Prs activity, reducing it by approximately 35%. In keeping with its influence on Prs activity in humans, the L115T mutation is found to be more adept than Q133P in rescuing the synthetic lethality of a prs1Δ prs5Δ strain. The double mutation H130A/D326A affecting the divalent-cation- and PRPP-binding (ribose-5-phosphate) sites was unable to rescue the synthetic lethality (Ugbogu et al.2013) and, like Q133P, caused >50% reduction in Prs activity as measured in YN96–66 [pGBT9-Prs1] and would suggest that the integrity of the divalent-cation-binding site is vital for Prs activity whereas D326A alone does not affect Prs activity.
CONCLUSIONS
The NHRs of Prs1 and Prs5 are responsible for interaction with components of the CWI pathway (Ugbogu et al.2013; Ugbogu 2016), and the 284KKCPK288 sequence of Prs3, which should perhaps be renamed NHR3–1, is essential for the stability of Prs1 and possibly necessary for transport of Prs1 and Prs5 to the nucleus. These findings provide a further explanation for the requirement of at least one heterodimeric complex for the survival of the yeast cell, since in addition to synthesising PRPP, there has to be a link to the maintenance of CWI. Support for such a link is provided by interaction of Prs polypeptides with Nuf2, a component of the kinetochore complex and Srp1 (importin α). Slt2 is found both in the nucleus and cytoplasm (Kamada et al.1995; Hahn and Thiele 2002). Given the interaction of Prs1 with Slt2, Prs3 and Rlm1, it could well be that Prs3 facilitates the entry of Slt2 into the nucleus under conditions of stress. Therefore, Prs3 may be considered to both stabilise the Prs1/Prs3 minimal functional unit and ensure, under conditions of stress, together with Prs5 entry into the nucleus where the transient four-component complex, Prs1/Prs3/P-Prs5/P-Slt2, comes into contact with Rlm1 to initiate the stress response. It is also likely that there is an interaction with the Paf1 transcription complex that has responsibility for the transcriptional elongation of Fks2 under conditions of stress. The interaction of Prs1 and Prs3 is not compromised upon removal of NHR1–1. Our finding that removal of NHR1–1 from Prs1 results in a significant increase in Prs activity is a first time demonstration that the functions, CWI maintenance and PRPP production, associated with the Prs gene family can be separated.
Supplementary Material
Acknowledgements
We are grateful to Craig Nicol for help with figure preparation and Nilesh Kanese for help with the plate reader.
AUTHORS’ CONTRIBUTION
Maëlle S, AR, RC, FH, SV, KW, GB, CGN, TC and EE conducted the experiments, LMS and MS conceived the idea of the project. LMS and MS interpreted the data and wrote the paper.
FUNDING
This work was supported by a donation account B16D1018 (MS), the Eda Lady Jardine Trust (SV), the Henry Lester Trust Ltd., London (KW), an internship funded by the University of La Rochelle (Maëlle S), an Erasmus exchange internship with the University of Marburg (GB) and a Wellcome Trust vacation scholarship (EE).
Conflict of interest. None declared.
REFERENCES
- Amalou Z, Bangratz J, Chrestin H. Ethrel (ethylene releaser)-induced increases in the adenylate pool and transtonoplast DeltapH within Hevea latex cells. Plant Physiol 1992;98:1270–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE et al. Current Protocols in Molecular Biology. New York: Greene Publishing Associates & Wiley-Interscience, 1995. [Google Scholar]
- Becker MA. Phosphoribosylpyrophosphate synthetase and the regulation of phosphoribosylpyrophosphate production in human cells. Prog Nucleic Acid Re 2001;69:115–48. [DOI] [PubMed] [Google Scholar]
- Binley KM, Radcliffe PA, Trevethick J et al. The yeastPRS3 gene is required for cell integrity, cell cycle arrest upon nutrient deprivation, ion homeostasis and the proper organization of the actin cytoskeleton. Yeast 1999;15:1459–69. [DOI] [PubMed] [Google Scholar]
- Botstein D, Fink GR. Yeast: an experimental organism for 21st century biology. Genetics 2011;189:695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976;72:248–54. [DOI] [PubMed] [Google Scholar]
- Carter AT, Beiche F, Hove-Jensen B et al. PRS1 is a key member of the gene family encoding phosphoribosylpyrophosphate synthetase in Saccharomyces cerevisiae. Mol Gen Genet 1997;254:148–56. [DOI] [PubMed] [Google Scholar]
- Chang M, French-Cornay D, Fan HY et al. A complex containing RNA polymerase II, Paf1p, Cdc73p, Hpr1p, and Ccr4p plays a role in protein kinase C signaling. Mol Cell Biol 1999;19:1056–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combet C, Blanchet C, Geourjon C et al. NPS@: Network protein sequence analysis. Trends Biochem Sci 2000;25:147–50. [DOI] [PubMed] [Google Scholar]
- de Brouwer AP, van Bokhoven H, Nabuurs SB et al. PRPS1 mutations: four distinct syndromes and potential treatment. Am J Hum Genet 2010;86:506–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Brouwer AP, Williams KL, Duley JA et al. Arts syndrome is caused by loss-of-function mutations in PRPS1. Am J Hum Genet 2007;81:507–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donini S, Garavaglia S, Ferraris DM et al. Biochemical and structural investigations on phosphoribosylpyrophosphate synthetase from Mycobacterium smegmatis. PLoS One 2017;12:e0175815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon B, Sherman D, Fischer G et al. Genome evolution in yeasts. Nature 2004;430:35–44. [DOI] [PubMed] [Google Scholar]
- Elble R. A simple and efficient procedure for transformation of yeasts. Biotechniques 1992;13:18–20. [PubMed] [Google Scholar]
- Engelberg D, Perlman R, Levitzki A. Transmembrane signaling in Saccharomyces cerevisiae as a model for signaling in metazoans: state of the art after 25 years. Cell Signal 2014;26:2865–78. [DOI] [PubMed] [Google Scholar]
- Eriksen TA, Kadziola A, Bentsen AK et al. Structural basis for the function of Bacillus subtilis phosphoribosyl-pyrophosphate synthetase. Nat Struct Biol 2000;7:303–8. [DOI] [PubMed] [Google Scholar]
- Feng Q, Liu ZL, Weber SA et al. Signature pathway expression of xylose utilization in the genetically engineered industrial yeast Saccharomyces cerevisiae. PLoS One 2018;13:e0195633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficarro SB, McCleland ML, Stukenberg PT et al. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat Biotechnol 2002;20:301–5. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol 2002;350:87–9. [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to Yeast Genetics and Molecular Biology. (Methods in Enzymology), vol. 194 San Diego: Academic Press Inc, 1991. [Google Scholar]
- Hahn JS, Thiele DJ. Regulation of the Saccharomyces cerevisiae Slt2 kinase pathway by the stress-inducible Sdp1 dual specificity phosphatase. J Biol Chem 2002;277:21278–84. [DOI] [PubMed] [Google Scholar]
- Hermida L, Brachat S, Voegeli S et al. The Ashbya Genome Database (AGD)-a tool for the yeast community and genome biologists. Nucleic Acids Res 2004;33:D348–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernando Y, Carter AT, Parr A et al. Genetic analysis and enzyme activity suggest the existence of more than one minimal functional unit capable of synthesizing phosphoribosyl pyrophosphate in Saccharomyces cerevisiae. J Biol Chem 1999;274:12480–7. [DOI] [PubMed] [Google Scholar]
- Hernando Y, Parr A, Schweizer M. PRS5, the fifth member of the phosphoribosyl pyrophosphate synthetase gene family in Saccharomyces cerevisiae, is essential for cell viability in the absence of either PRS1 or PRS3. J Bacteriol 1998;180:6404–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero P, Martinez-Campa C, Moreno F. The hexokinase 2 protein participates in regulatory DNA-protein complexes necessary for glucose repression of the SUC2 gene in Saccharomyces cerevisiae. FEBS Lett 1998;434:71–76. [DOI] [PubMed] [Google Scholar]
- Hove-Jensen B, Andersen KR, Kilstrup M et al. Phosphoribosyl diphosphate (PRPP): biosynthesis, enzymology, utilization, and metabolic significance. Microbiol Mol Biol R 2017;81: e00040–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka T, Kita K, Sonoda T et al. Cloning and sequencing of human complementary DNA for the phosphoribosylpyrophosphate synthetase-associated protein 39. BBA- Gene Struct Expr 1996;1306:27–30. [DOI] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R et al. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA 2001;98:4569–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 1996;144:1425–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P, Wei WF, Zhong GW et al. The function of the three phosphoribosyl pyrophosphate synthetase (Prs) genes in hyphal growth and conidiation in Aspergillus nidulans. Microbiology 2017;163:218–32. [DOI] [PubMed] [Google Scholar]
- Jiménez A, Santos MA, Revuelta JL. Phosphoribosyl pyrophosphate synthetase activity affects growth and riboflavin production in Ashbya gossypii. BMC Biotechnol 2008;8:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C, Michaelis S, Mitchell A. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Laboratory Press, 1994. [Google Scholar]
- Kamada Y, Jung US, Piotrowski J et al. The protein kinase C-activated MAP kinase pathway of Saccharomyces cerevisiae mediates a novel aspect of the heat shock response. Gene Dev 1995;9:1559–71. [DOI] [PubMed] [Google Scholar]
- Katashima R, Iwahana H, Fujimura M et al. Molecular cloning of a human cDNA for the 41-kDa phosphoribosylpyrophosphate synthetase-associated protein. BBA- Gene Struct Expr 1998;1396:245–50. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Sohn KM, Shy ME et al. Mutations in PRPS1, which encodes the phosphoribosyl pyrophosphate synthetase enzyme critical for nucleotide biosynthesis, cause hereditary peripheral neuropathy with hearing loss and optic neuropathy (CMTX5). Am J Hum Genet 2007;81:552–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KY, Levin DE. Mpk1 MAPK association with the Paf1 complex blocks Sen1-mediated premature transcription termination. Cell 2011;144:745–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchrath L, Lorberg A, Schmitz HP et al. Comparative genetic and physiological studies of the MAP kinase Mpk1p from Kluyveromyces lactis and Saccharomyces cerevisiae. J Mol Biol 2000;300:743–58. [DOI] [PubMed] [Google Scholar]
- Kleineidam A, Vavassori S, Wang K et al. Valproic acid- and lithium-sensitivity in prs mutants of Saccharomyces cerevisiae. Biochem Soc Trans 2009;37:1115–20. [DOI] [PubMed] [Google Scholar]
- Krath BN, Hove-Jensen B. Organellar and cytosolic localization of four phosphoribosyl diphosphate synthase isozymes in spinach. Plant Physiol 1999;119:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krath BN, Hove-Jensen B. Class II recombinant phosphoribosyl diphosphate synthase from spinach. Phosphate independence and diphosphoryl donor specificity. J Biol Chem 2001;276:17851–6. [DOI] [PubMed] [Google Scholar]
- Levene H. Robust tests for equality of variances. In: Contributions to Probability and Statistics: Essays in Honor of Harold Hotelling. Olkin I, Hotelling H (eds.) Redwood City, CA, USA: Stanford University Press, 1960, 278–92. [Google Scholar]
- Levin DE. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol Mol Biol R 2005;69:262–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin DE. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: the cell wall integrity signaling pathway. Genetics 2011;189:1145–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateos L, Jiménez A, Revuelta JL et al. Purine biosynthesis, riboflavin production, and trophic-phase span are controlled by a Myb-related transcription factor in the fungus Ashbya gossypii. Appl Environ Micro 2006;72:5052–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal R, Patel K, Mittal J et al. Association of PRPS1 mutations with disease phenotypes. Dis Markers 2015;2015:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiter R, Carter AT, Hernando Y et al. The importance of the five phosphoribosyl-pyrophosphate synthetase (Prs) gene products of Saccharomyces cerevisiae in the maintenance of cell integrity and the subcellular localization of Prs1p. Microbiology 2000;146:3269–78. [DOI] [PubMed] [Google Scholar]
- Synofzik M, Müller vom Hagen J, Haack TB et al. X-linked Charcot-Marie-Tooth disease, Arts syndrome, and prelingual non-syndromic deafness form a disease continuum: evidence from a family with a novel PRPS1 mutation. Orphanet J Rare Dis 2014;9:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Badger BL, Haase J et al. How the kinetochore couples microtubule force and centromere stretch to move chromosomes. Nat Cell Biol 2016;18:382–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatibana M, Kita K, Taira M et al. Mammalian phosphoribosyl-pyrophosphate synthetase. Adv Enzyme Regul 1995;35:229–49. [DOI] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 2000;403:623–7. [DOI] [PubMed] [Google Scholar]
- Ugbogu AE, Wang K, Schweizer LM et al. Metabolic gene products have evolved to interact with the cell wall integrity pathway in Saccharomyces cerevisiae. FEMS Yeast Res 2016;16:1–8. [DOI] [PubMed] [Google Scholar]
- Ugbogu EA, Wippler S, Euston M et al. The contribution of the nonhomologous region of Prs1 to the maintenance of cell wall integrity and cell viability. FEMS Yeast Res 2013;13:291–301. [DOI] [PubMed] [Google Scholar]
- Vavassori S, Wang K, Schweizer LM et al. In Saccharomyces cerevisiae, impaired PRPP synthesis is accompanied by valproate and Li+ sensitivity. Biochm Soc Trans 2005a;33:1154–7. [DOI] [PubMed] [Google Scholar]
- Vavassori S, Wang K, Schweizer LM et al. Ramifications of impaired PRPP synthesis in Saccharomyces cerevisiae. Biochem Soc Trans 2005b;33:1418–20. [DOI] [PubMed] [Google Scholar]
- Wang K, Vavassori S, Schweizer LM et al. Impaired PRPP-synthesizing capacity compromises cell integrity signalling in Saccharomyces cerevisiae. Microbiology 2004;150:3327–39. [DOI] [PubMed] [Google Scholar]
- Wolfe KH. Origin of the yeast whole-genome duplication. PLoS Biol 2015;13:e1002221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 1997;387:708–13. [DOI] [PubMed] [Google Scholar]
- Yu H, Zhang Y, Zhang D et al. Identification of a ribose-phosphate pyrophosphokinase that can interact in vivo with the anaphase promoting complex/cyclosome (APC/C). IJMS 2017;18:pii: E617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong G, Wei W, Guan Q et al. Phosphoribosyl pyrophosphate synthetase, as a suppressor of the sepH mutation in Aspergillus nidulans, is required for the proper timing of septation. Mol Microbiol 2012;86:894–907. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.