Abstract
The explosion in genome editing technologies that has occurred in the past decade has revolutionized cancer research and promises to improve cancer diagnosis and therapy. Ongoing efforts include engineering of CAR-T cells using CRISPR to generate a safer, more effective therapy with improved performance in immunologically “cold” tumors, as well as clever adaptations of CRISPR enzymes to allow fast, simple, and sensitive detection of specific nucleotide sequences. While still in their infancy, CRISPR-based cancer therapeutics and diagnostics are developing at an impressive speed and it is likely they will soon impact clinical practice. Here we summarize their history and the most recent developments.
At the core of the CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) system lie a series of RNA-guided programmable nucleases identified in a wide range of bacteria, where they provide adaptive immunity against viral infection1. In recent years, these systems have been co-opted by researchers across the globe to manipulate the mammalian genome in ways that would have sounded like science fiction until just a few years ago. Currently available CRISPR-based applications include methods for gene inactivation and site-specific mutagenesis2,3, chromosome engineering4–6, gene tagging7,8, regulation of gene expression (both gene activation and gene repression)9,10, site specific epigenetic modifications11,12, gene visualization13, molecular recording14, targeted RNA degradation15, and detection of specific DNA sequences for diagnostic purposes16.
Such versatility has not escaped the attention of cancer researchers, who quickly took advantage of the potential of genome editing to modify endogenous genes to generate improved models of human cancers. Some of the earliest applications took advantage of the ability of the CRISPR-Cas9 system to generate true gene knockouts as an effective alternative to RNAi for genetic screening purposes, and the use of CRISPR screens in cancer has been previously summarized17–19. The use of CRISPR for the creation of new and more accurate tumor models is also well established and has been reviewed elsewhere20–22. However, CRISPR has the potential to be much more than a research tool and CRISPR technologies are increasingly moving away from the bench and toward the clinic. Recent developments have highlighted the ability of CRISPR-based genome editing to improve cancer immunotherapies based on adoptive cell transfer, and have introduced novel, CRISPR-based, detection tools for DNA and RNA, with implications for tumor diagnosis and genotyping. Here, we summarize these emerging applications, starting with a brief overview of the basic molecular biology of CRISPR systems.
A (very) brief overview of CRISPR systems
All CRISPR-Cas systems share a general principle: to prevent phage infection, a nuclease effector complex (targeting either RNA or DNA) uses short RNA molecules with homology to the infecting viral genome to directly bind and cut invading nucleic acid sequences23,24. Based on effector protein organization, CRISPR-Cas systems are grouped into two major classes: class 1 and class 2, with each class further divided into various types and subtypes25–27. Class 1 systems are characterized by having multi-protein effector complexes, while class 2 CRISPR-Cas system have the appealing feature of utilizing a single protein effector that, coupled to the guide RNA, acts on the target nucleic acid.
Over the past few years, several class 2 systems have been adapted for use in eukaryotic cells and whole organisms. The most commonly used is the Cas9 enzyme from Streptococcus pyogenes (SpCas9), which utilizes a short crRNA in target recognition and a tracrRNA for structural assembly of the RNA-protein complex28. When delivered together with a hybrid synthetic guide RNA (gRNA) combining the crRNA and tracrRNA sequences, SpCas9 can be used to induce DNA double strand breaks (DSBs) in mammalian cells in vitro2,3,29 and in vivo30. SpCas9 recognizes a 20nt sequence with direct homology to the associated guide RNA (gRNA) that must be followed, in the genomic DNA, by a 3nt NGG protospacer adjacent motif (PAM) sequence. Repair of the DSB through the non-homologous end joining (NHEJ) pathway31,32 commonly results in the formation of small insertions and deletions (indels) that, when introduced into a protein coding sequence, can inactivate the gene by generating a frameshift33. Less frequently, the cell will repair the DSB via the homology directed repair (HDR) pathway, which uses the sister chromatid as a template to perfectly repair the break. HDR can be co-opted to introduce more specific mutations – including single nucleotide changes and the introduction of tags or loxP sites – by providing an exogenous ‘repair template’ (either single or double stranded) containing homology arms flanking the targeting site33.
Correcting cancer-driving mutations
The ability to directly edit the cellular genomes using CRISPR holds great promise for the field of gene therapy. Correcting oncogenic mutations in somatic tissues after they have arisen or editing the germline to correct cancer predisposition mutations in genetic diseases such as Nijmegen Breakage Syndrome34 or Ataxia–telangiectasia35 would represent breakthroughs in gene therapy for cancer. However, CRISPR technology in its current form is far from achieving these goals. Existing vehicles for the delivery of the CRISPR machinery have limited ability to reach most organs in the human body, and methods to efficiently transduce cancer cells in vivo are not yet available. Furthermore, the efficiency with which specific targeted mutations can be corrected using homology directed repair remains low, far below the efficiencies that would be needed to be effective in cancer treatment or prevention. Finally, the potential risk associated with off target activity by the SpCas9 nuclease and the generation of unwanted mutations, as well as the potential for SpCas9 activity to select for cells that have inactivated key tumor suppressor pathways36,37, need to be carefully considered when thinking about clinical applications.
Despite these caveats, in vivo editing of somatic cells has been proposed as a therapeutic approach in the context of cellular infection by human papillomavirus (HPV), an integrating DNA virus that is responsible for the vast majority of cervical cancers and a subset of other urogenital and head and neck cancers38. Expression of the viral proteins E6 and E7 by infected cells can promote cellular transformation by inactivation of the TP53 and Rb tumor suppressors, respectively39,40, and their repression using RNA interference can restore tumor suppressor activity and induce senescence or apoptosis of cervical cancer cells41–43. These viral oncogenes are attractive targets for CRISPR-based gene knockout, as they are tumor-specific with no endogenous mammalian homologue. Several groups have demonstrated that gRNAs targeting the viral E6 and E7 genes can induce apoptosis and growth inhibition in cancer cells infected with the relevant HPV subtype based on plasmid transfection with SpCas944,45. Based on these results, a clinical trial is currently planned in China using a topical gel containing plasmids expressing SpCas9 and the relevant E6/E7 gRNAs (NCT03057912). The gel will be applied directly to the cervix of HPV infected women twice weekly for 4 weeks, with patients evaluated for safety and efficacy based on changes in HPV DNA titers and cervical cytology. It remains to be seen whether this approach can edit a sufficient number of HPV-infected cells to effectively reduce the risk of cervical cancer progression.
Improving cell therapies with CRISPR
Another potential application of CRISPR-Cas genome editing is the editing of primary cells ex vivo for adoptive cell transfer applications. Modification of patient effector immune cells to target and eliminate cancer cells has recently emerged as an exciting therapeutic strategy, particularly in the context of hematologic cancers46,47. The vast majority of human tumors display a frustrating propensity to avoid detection and clearance by the adaptive immune system, which ideally would identify these mutant cells as “non-self” and eliminate them based on the expression of neo-antigens. By a variety of mechanisms, including downregulation of the major histocompatibility class I complex (MHC-I)48, growing tumors actively avoid immune detection and suppress immune cell activity. One proposed solution is the retraining of effector T-Cells using synthetic receptors engineered to recognize tumor cells. The generation of chimeric antigen receptor (CAR) T-Cells was first reported 25 years ago49. CAR-T cells utilize an artificial, specifically designed hybrid T-Cell receptor (TCR) recognizing a tumor-cell specific antigen50. In the current generation of CARs, a single-chain variable fragment (scFv) derived from a monoclonal antibody is linked through a transmembrane domain to an intracellular CD3-ζ domain from a conventional TCR and multiple co-stimulatory domains in a single polypeptide chain46,47. T-cells engineered to express these synthetic receptors are able to bind cell surface peptides on tumor cells without these peptides being presented in the context of the MHC-I, as demonstrated by the development of CAR-T cells targeting non-peptide glycosyl modifications51. Although the utility of CAR-T cells is dependent on the presence of a suitable, targetable antigen expressed on the surface of tumor cells, early clinical applications of CAR-T cells targeting the B-cell antigen CD19 have shown remarkable efficacy in treating a variety of hematological cancers52–54, culminating with the FDA approval of CAR-T therapies for diffuse large B-cell lymphoma (Yescarta™, Kite Pharma) and pediatric B-cell acute lymphoblastic leukemia (Kymriah™, Novartis). A variety of other CARs are currently being developed and evaluated for safety and efficacy in different tumor settings, although the majority of solid tumors have as yet proven refractory to CAR-T based therapy.
While CAR-T cells show great promise, there are issues impinging on their efficacy and safety related in part to the methods by which they are generated. Typically, primary T-Cells are harvested from the patient’s peripheral blood and transduced with recombinant retrovirus carrying the CAR expression cassette55. One limitation of this strategy is that the random integration of the viral genome leads to the generation of non-uniform CAR-T cell populations with heterogeneous CAR expression and with the added risk of activating oncogenes at the integration site. Furthermore, to avoid rejection by the patient’s immune system and graft-versus-host disease (GVHD), CAR-T cells have only existed to date as an autologous therapy, with each patient generating their own cells for subsequent re-implantation. This increases the cost of this therapy, as well as the time required to develop a unique treatment for each patient. Autologous T-Cells also continue to express a variety of genes that can decrease their therapeutic efficacy in specific contexts. For patients diagnosed with acquired immune deficiency syndrome (AIDS), re-administered cells can be targeted and eliminated by the Human Immunodeficiency Virus (HIV) by virtue of continued expression of viral co-receptors, including CXCR4 and CCR556. This presents a significant issue for the use of CAR-T cells in the treatment of HIV-associated cancers such as Kaposi sarcoma, B-cell non-Hodgkin lymphoma, and cervical cancer57. As will be discussed in more detail below, many cancers actively inhibit T-Cell function by engaging immune-inhibitory receptors on the T-Cell surface. These receptors, including PD-1 and CTLA4, comprise an immune checkpoint that has been a major therapeutic focus in recent years. Continued expression of immune checkpoint genes in CAR-T cells may significantly reduce their efficacy in immunologically “cold” solid tumors. Finally, despite its efficacy, CAR-T cell therapy is not devoid of safety concerns. Anti-tumor activity is frequently correlated with a severe and potentially fatal systemic immune response known as cytokine release syndrome, which must be managed using anti-inflammatory agents52,58,59.
Improving CAR-T function
Generation of HIV-resistant T-Cells with homogenous CAR expression.
Gene editing approaches have been recently proposed, and in some cases proven effective, to address these major limitations of cellular therapies based on CAR T cells (Figure 1). The first efforts to apply genome editing technology to CAR-T cell generation utilized transcription activator-like effector nuclease (TALEN) technology to insert a CD19 CAR expression cassette into the CCR5 locus, with the goal of generating a uniform population of CAR-expressing T-cells that were negative for CCR5 and thus resistant to HIV infection. Work from David Rawlings and Andrew Scharnberg’s labs showed that directly inserting the chimeric receptor in the CCR5 locus using gene editing can address two problems at the same time: preventing killing of engineered cells by HIV and ensuring uniform expression of the chimeric receptor. They demonstrated this approach by using either a conventional TALEN, or a hybrid TALE DNA binding factor combined with a meganuclease with limited target site specificity, referred to as a megaTAL60. In order to introduce new information at the CCR locus, an HDR template was constructed as an adeno-associated virus (AAV) harboring the CAR expression cassette flanked by 1.3kb homology arms matching the endogenous CCR5 sequence immediately around the cut site. The CAR expression cassette was efficiently knocked-in the CCR5 locus with over 80% of edited cells displaying biallelic targeting. While this technology is powerful, the generation and qualification of specific zinc-finger nucleases can be challenging and time consuming. CRISPR technology, in contrast, can be used to rapidly test a variety of different genomic loci for potential CAR insertion. The initial challenge was developing a strategy for effectively delivering CRISPR machinery to primary T-Cells.
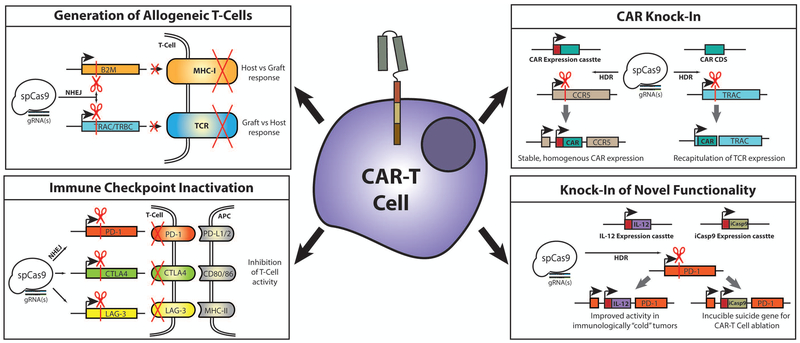
Figure 1 – Applications for CRISPR genome editing in CAR-T cells.
CRISPR can be used for the introduction of genetic knock-outs by disrupting the coding sequence of specific genes (by non-homologous end joining (NHEJ) repair following DSB induction), or for the generation of knock-ins by introducing novel genetic information at specific loci (by homology-directed repair (HDR) following DSB induction). Gene knock-out can be applied to CAR-T cells to generate allogeneic cells that would not need to be donor matched by targeting crucial components of the major histocompatibility type-I complex (MHC-I) or of the endogenous T-Cell receptor (TCR). CRISPR knock out can also to generate cells with reduced sensitivity to immune checkpoint pathways by targeting key receptors including PD-1, CTLA4, and LAG-3. Genetic knock-in can be used to introduce the CAR expression cassette into a defined locus, leading to more stable and homogenous expression of the receptor while avoiding potential mutational events associated with randomly integrating vectors. Alternately, the CAR coding sequence (CDS) can ben knocked into the TCR-alpha constant (TRAC) locus, disrupting the endogenous TCR while also producing a CAR with expression matched to endogenous receptor. Similarly, CRISPR knock-in can also be used to introduce novel effectors into CAR-T cells at defined loci. Potential effectors include those for IL-12 secretion, which can improve CAR-T function in immunologically suppressed or “cold” tumors, or for inducible Caspase-9 activity (iCasp9), allowing for selective ablation of CAR-T cells as an added safety measure.
Some of the earliest efforts to apply CRISPR genome editing in primary human T cells used plasmids expressing SpCas9 and gRNAs targeting specific genes that were delivered to primary T-Cells by nucleofection. The observed knockout efficiency was negligible with single gRNAs (1–5%) targeting each gene, and was only somewhat improved when specific gRNA pairs were employed to induce genomic deletions61. This work demonstrated that plasmid-based delivery of the SpCas9 nuclease and associated gRNAs was an inefficient system for editing of primary T-cells, and spurred exploration of alternate approaches. One issue with plasmid based delivery of CRISPR components is the prolonged presence of SpCas9 nuclease, which may lead to increased off-target activity, as well as the reported ability of primary T-cells to recognize cytosolic DNA and prompt anti-viral or apoptotic responses62. High levels of cellular toxicity have been associated with DNA plasmid delivery to primary human T-cells63, and are probably a major contributor to the low editing efficiency observed following plasmid nucleofection. This led to the investigation of non-DNA based systems for transient exposure of T-cells to SpCas9, similar to what had been previously demonstrated as effective using TALEN and megaTAL nucleases.
Substantially greater efficiency was obtained when purified SpCas9 ribonucleoparticles (RNPs – SpCas9 complexed with a specific gRNA7,64,65) were delivered to primary T cells66,67. Using a gRNA targeting the CXCR4 HIV co-receptor, a frequency of gene inactivation greater than 50% was observed. Furthermore, it was shown that SpCas9 RNPs could be combined with a single stranded oligonucleotide HDR donor template to introduce new DNA sequence information at a specific target site in CXCR4 with efficiency of approximately 25%. Equally encouraging results were obtained using an “all-RNA” approach, in which purified Cas9 mRNA and gRNA transcripts were electroporated into target T cells. This approach achieved high levels of gene knockout, particularly when the gRNAs were delivered sequentially (88.7–95.7% CD3 negative cells), and was amenable to multiplexed gRNA delivery for simultaneous knockout of multiple targets68. Editing using purified gRNA could be further improved by chemical modification of the gRNAs. 2’-O-methyl or 2’-O-methyl 3’ thioPACE modifications incorporated at both ends of the gRNA, which are predicted in increase intracellular stability, resulted in marked increases in indel frequency in primary T-cells when electroporated with purified Cas9 mRNA69. Finally, improved strategies to insert large DNA fragments into primary human T cells by electroporating SpCas9 RNPs and a long linear double or single stranded DNA repair templates have been recently developed and promise to further simplify and accelerate the development of patient specific CAR-T cells70.
The work discussed above demonstrated that CRISPR knock-in of the CD19-CAR into the CCR5 locus could generate a homogenous population of CAR-T cells with stable, uniform expression of the CAR. CRISPR technology also provides a way to achieve CAR expression levels much more closely matching those of endogenous TCRs, an approach pioneered by the group of Michel Sadelain. Using a gRNA targeting the first exon of the endogenous TCR-alpha TRAC locus, Eyquem and colleagues successfully knocked in the CD19-CAR to this genomic region, simultaneously achieving physiologic expression of the CAR and disruption of the endogenous TCR71. The resulting cells were directly compared to CD19-CAR-T cells generated via retroviral infection with or without concomitant loss of the TCR receptor, both in vitro and in vivo. CAR-T cells generated by CRISPR knock-in strategy clearly outperformed the traditionally generated CAR-T cells in an in vivo mouse model of pre-B cell acute lymphoblastic leukemia. A dramatic improvement in overall survival was accompanied by superior CAR-T retention and decreased signs of T-Cell exhaustion. These studies also demonstrated that the level of expression associated with CARs expressed from the endogenous TCR locus is associated with a receptor internalization and re-expression cycle that is much more closely matched to the normal receptor, ultimately leading to a more sustained and effective antitumor response. While these results are exciting, it will be important to confirm that they are broadly applicable to other CARs in different tumor contexts.
Generation of allogeneic CAR-T cells.
Generation of off-the-shelf CAR-T cells using CRISPR technology has the potential to generate truly allogeneic T-Cells for therapeutic applications. As mentioned earlier, CAR-T cells used to date are derived from T-cells that are purified from individual patients and modified to express the desired CAR ex vivo, expanded, and then re-infused55. However, in addition to cost and time considerations, many candidate patients with refractory cancers display compromised immune systems, and harvest of suitable T-Cells from these patients can be challenging. An appealing alternative would be the generation of CAR-T cells derived from healthy donors but engineered to avoid immune rejection or graft versus host disease72,73. Such an “off the shelf” CAR-T therapy has long been a goal for immuno-oncology, and CRISPR may provide the key for making it a reality. As mentioned earlier, however, the use of non-autologous T-Cells poses two major challenges: reactivity of the transplanted cells toward normal tissues in the recipient patient (GVHD74) and elimination of the donor-derived cells by the patients own immune system (host versus graft). The primary mediator of graft versus host reactivity is the endogenous T-cell receptor (TCR), which is still present on conventional CAR-T cells. The TCR complex consists of two polypeptide chains, TCR-alpha and TCR-beta, both of which feature variable and constant regions. Multiple studies have shown that complete loss of TCR surface expression can be achieved by zinc-finger nucleases75 or CRISPR68,76,77 targeting of TCR-alpha or beta subunits and T-Cells engineered to have lost TCR expression displayed reduced graft versus host toxicity when implanted into immunodeficient mice77.
On the other hand, the host versus graft response is dependent on the recognition, by the host immune system, of the MHC-I complex on the infused T cells. Here, CRISPR-Cas editing can be used to inactivate beta-2 microglobulin (B2M), a crucial subunit of MHC-I78,79. Generation of fully allogeneic T-Cells would require the simultaneous loss of both the TCR and MHC-I complexes, highlighting the need to develop systems for efficient multiplexed genome editing. Recombinant lentiviruses are a feasible means of delivering CRISPR machinery into primary CD4+ T-cells80. One approach for multiplexed gRNA expression in CAR-T cells involves generating a lentiviral construct expressing the CAR along with multiple tandem gRNAs driven by distinct U6 promoters to prevent recombination of tandem repeats. Infection of primary T cells with such a virus followed by Cas9 mRNA electroporation was used to generate functional CD19-CAR-T cells with >70% double knockout for the TRAC and B2M genes77. While B2M knockout should block elimination of transplanted CAR-T cells by endogenous CD8+ T-Cells, it is still possible that natural killer (NK) cells, which have been shown to target MHC-I negative cells81, could eliminate transplanted cells. Thus, further validation will be needed to confirm that TCR/MHC-I double negative CAR-T cells are truly allogeneic.
Inhibition of immune checkpoint signaling.
An additional challenge in the application of CAR-T cells has to do with the propensity of tumors to actively suppress T-Cell activity through the upregulation of inhibitory pathways. Immune suppression, T-cell exhaustion, and tolerance are thought to be a major reason for the limited clinical efficacy of CAR-T cells in solid tumors. In order to maintain immune system homeostasis and prevent autoimmunity, a variety of inhibitory signaling factors are expressed on the surface of T-Cells, the most well-characterized of which are CTLA-4, PD-1, and LAG-382. After binding of the TCR to antigen-loaded MHC-II, a variety of co-stimulatory signaling factors promote and reinforce downstream TCR signaling. Two of the most important are CD80 and CD86, which are expressed on the surface of antigen presenting cells and directly bind to CD28 on activated T-Cells. CTLA-4 directly competes with CD28 to bind CD80/86, blocking co-stimulation and inhibiting the T-Cell activation in lymphatic vessels83. PD-1 binds to the PD-L1 or PD-L2 cell surface receptors and negatively regulates TCR signaling through a less well understood mechanism. PD-L1 and PD-L2 have been shown to be upregulated in a variety of solid tumors as a means of avoiding immune surveillance84,85. LAG-3 is a homologue of CD4 that can directly bind to MHC-II and inhibit TCR signaling, although, as with PD-1, the mechanism is not entirely understood. These immune checkpoint factors have received a great deal of attention in recent years due to their role in blocking immune targeting of cancer cells. This has ultimately lead to the development of inhibitory antibody therapeutics targeting CTLA-4 (Ipilimumab, tremelimumab)86, PD-1 (nivolumab and pembrolizumab)87, and PD-L1 (atezolizumab). These immune checkpoint inhibitors have become one of most important new classes of cancer therapeutics, with FDA approval for CTLA-4 and PD-1 inhibitors covering an expanding variety of cancers including melanoma, renal cell carcinoma, non-small cell lung cancer, and urothelial cancer among others88. However, inhibition of CTLA-4 or PD-1/PD-L1 is associated with significant incidence of immune-related adverse events, and the combinatorial inhibition of both signaling pathways displayed significant toxicity89. An appealing possibility for reducing this toxicity while still preserving checkpoint inhibition is CRISPR-based genetic engineering of CAR-T cells to block major checkpoint pathways.
Because of the established role in PD1/PD-L1 signaling in suppressing T-Cell tumor clearance, multiple efforts to deliver CRISPR machinery to primary T-Cells used gRNAs designed to induce PD1 knockout as readouts for CRISPR efficacy68,77,90. These results demonstrated the feasibility of generating PD1-null T-Cells using genome editing. Additionally, Carl J. June’s and Yangbing Zhao’s groups showed that CRISPR-mediated PD1 inactivation in T-Cells expressing a prostate stem cell antigen (PSCA) CAR resulted in increased ex vivo and in vivo activity toward human PC3 prostate cancer cells (which were engineered to express high levels of PD-L1)68. Similar results were also observed using a CD19 CAR in a Nalm6-leukemia model. Subsequently, work from Rupp and colleagues showed that knockout of PD1 in CD19-CAR-T cells results in increased therapeutic efficacy in a mouse xenograft model of myelogenous leukemia when tumor cells were induced to overexpress PD-L191. Similarly, Zhang and colleagues reported that CRISPR-induced LAG-3 knockout in human umbilical cord blood-derived primary T-Cells resulted in increased efficacy in a mouse xenograft model of lymphoma92. LAG-3 knockout CD19-CAR-T cells provided a modest increase in overall survival relative to LAG-3 wild type CD19-CAR-T in this model. While these qualification efforts are very preliminary and have only been performed in immune-compromised mouse models, they suggest that further engineering of CAR-T cells may improve their in vivo efficacy. Further information may soon be forthcoming, as multiple clinical trials have been initiated in the United States and China using CRISPR-edited CAR-T cells. A trial sponsored by the University of Pennsylvania and aimed at multiple myeloma plans to use CAR-T cells where the TCR and the PD-1 receptor have been deleted using CRISPR (NCT03399448)93.
Other applications of CRISPR to improve CAR-T therapies.
Currently, immuno-oncology therapeutic approaches involving engineered cell-based therapy are poised for a revolution using CRISPR based approaches. Effective protocols for delivering CRISPR machinery via electroporation of purified RNAs or pre-assembled Cas9-gRNA RNPs have been established and the advantage of engineered expression of the chimeric antigen receptor are clear. With these tools in hand, it seems likely that major breakthroughs in CAR-T technology will be forthcoming. Specific combinations of CAR knock-in and immune checkpoint factor knock-out may be the key for improving CAR-T efficacy toward solid tumors, which have to date proven resistant to this form of therapy. This may also be an excellent time to pursue more open-ended approaches. For example, a CRISPR knockout screen in CAR-T cells to identify novel negative regulators of T-cell activity, potentially using CAR-T retention in a solid tumor mouse model as a readout for positive hits in the screen could identify previously unknown suppressors of T-Cell infiltration. Along with knocking out inhibitory factors, there is also the possibility of knocking in additional functionality to the CAR-T cell. Engineering the cells to secrete pro-inflammatory mediators such as IL-12 can potentially improve therapeutic efficacy toward immunologically “cold” solid tumors that display substantial cellular heterogeneity94. Using retroviral approaches, these so-called “armored CARs” have been demonstrated to be less susceptible to a immune-suppressive tumor microenvironment95. Introduction of an inducible caspase-9 construct to act as a suicide gene could also act as a safety measure96,97, allowing the elimination of transplanted CAR-T cells in the event of severe adverse effects in the clinic. CRISPR-based knock-in of these expression constructs could help to generate a more homogeneous CAR-T population, in a manner similar to early efforts to knock-in the CAR expression construct itself. Additionally, these expression constructs could be introduced into coding loci to be knocked-out, such as B2M or PD-1, achieving two editing goals at once.
Reducing off-targets
The risk of introducing unwanted and potentially harmful mutations remains a major obstacle to the translation of CRISPR-based genome editing to the clinic. Three possible sources of off-targets need to be considered. First, a gRNA designed against a specific site could have additional unknown perfect matches elsewhere in the genome. In principle, this type of off target should be easy to avoid. After all, the sequence of the human genome is known and gRNAs design algorithms that guarantee a single perfect match have been developed98. In practice, however, the genetic variability between patients needs to be taken into account and relying exclusively on the reference genome when designing the gRNAs is dangerous.
A second source of off-target events comes from the ability of gRNAs to direct Cas9 to cleave at sites with partial complementarity. Studies with SpCas9 showed that off-target sites with as many as 5 mismatches can be cleaved, although cleavage is more likely when the off-target site has three mismatches or fewer99–101. Shortening the window during which cells are exposed to Cas9/gRNA activity is one approach that can reduce off-target activity102. This can be achieved using purified SpCas9 RNP complexes rather than plasmids to generate transient, “burst-like” kinetics64,65. Another approach involves using a modified SpCas9 (“nickase”) that induces DNA single strand breaks paired with two gRNAs designed to target nearby sites on the plus and minus DNA strands. This system greatly reduces off-target activity since concomitant cleavage by both gRNAs is required to induce a DSB103,104. The obvious limitation in this case is that two gRNAs need to be designed to generate a single mutation event, increasing the complexity of the experimental design and reducing the number of genomic sites can be successfully targeted.
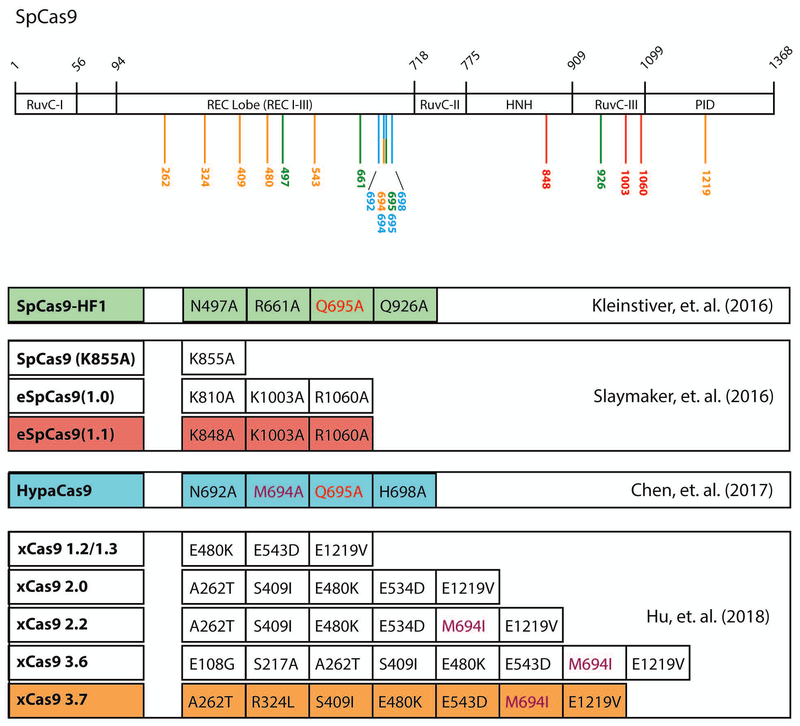
More promising approaches to reduce this type of off-target effect are aimed at improving the specificity of the Cas9 enzyme by rational protein engineering. Based on the crystal structure of SpCas9 in complex with gRNA and target DNA105,106, multiple attempts have been made to rationally design SpCas9 variants with higher on-target specificity. Under the presumption that the affinity of SpCas9 for target DNA could be reduced without significant reduction in on-target activity, Kleinstiver and colleagues selectively mutated a set of 4 residues that are predicted to directly interact with the target DNA backbone by hydrogen bonding107. Alanine substitution at all four positions resulted in an SpCas9 variant with a similar ability to induce inactivating mutations as the wild type, but greatly reduced off-target activity. Naming this variant SpCas9-HF1, the authors went on to show that its use could dramatically reduce the number of off-target events, even for gRNAs previously demonstrated to be highly promiscuous. Slaymaker and colleagues hypothesized that a positively charged non-target strand groove within spCas9 might mediate binding to the opposite DNA strand and maintain strand separation, which is crucial for DSB generation108. Mutation of key positively charged residues within this groove to alanine generated variants (eSpCas9, eSpCas9(1.0), eSpCas9(1.1)) with greatly reduced off-target activity but similar on-target activity relative to wild type SpCas9.
The idea that these variants display increased specificity through reduced overall target affinity has however been challenged by Jennifer Doudna’s group. Using single-molecule Förster resonance energy transfer (FRET) experiments, they demonstrated SpCas9-HF1 and eSpCas9(1.1) had similar on-target and off-target affinity as wild type SpCas9109. Instead, they proposed that in these mutants the catalytic nuclease domain shifts from an inactive to active conformation with much slower kinetics. The previously uncharacterized REC3 domain was proposed to act as the gatekeeper, facilitating the conformational shift of the nuclease domain in response to on-target binding. Rational mutation of key residues within REC3 produced a new variant, HypaCas9, with higher fidelity that either SpCas9-HF1 or eSpCas9(1.1).
While the studies detailed above used a rational approach, mutating specific residues within key SpCas9 functional domains, directed evolution approaches to change Cas9 specificity have also proven successful. One such approach, phage-assisted continuous evolution (PACE)110 was employed to generate SpCas9 variants with broadened PAM specificity in order to expand the universe of targetable sites in the mammalian genome111. By linking viral propagation to SpCas9 PAM promiscuity, the authors generated a series of mutants, dubbed “xCas9”, which could effectively recognize non-canonical PAM sites. Surprisingly, these mutants also displayed far higher on-target specificity than wild type SpCas9, indicating that target fidelity was selected along with broadened PAM recognition during viral evolution. While the increased PAM usage for the xCas9 variants may increase the difficulty of identifying truly unique 20nt target sequences, it should be possible to generate new variants retaining increased on-target specificity, but having more restricted PAM usage.
It should be noted that in the four approaches listed above there is very little overlap in the specific SpCas9 amino acids mutated to generate more accurate variants (Figure 2). Only residues Q695 and M694 are common to more than one of the various SpCas9 high-fidelity mutants reported so far, suggesting that there may be room for further improvements.
Figure 2 – Relative positions of SpCas9 mutations generating improved specificity.
SpCas9 contains two domains essential for nuclease activity, HNH and RuvC. Additionally, the REC lobe mediates binding to gRNA-target DNA heteroduplex and the PAM interacting domain (PID) makes contact with the NGG PAM sequence. Directed and unbiased efforts to generate SpCas9 mutants with lower off-target activity have generated a series of mutant proteins with surprisingly little overlap in target amino acid mutation. Representative mutants from the four major published studies are highlighted, with the position of the individual mutations relative to the SpCas9 domain structure indicated by color. While the majority of targeted amino acids are localized to the REC lobe, only residues Q695 and M694 are common between multiple mutants.
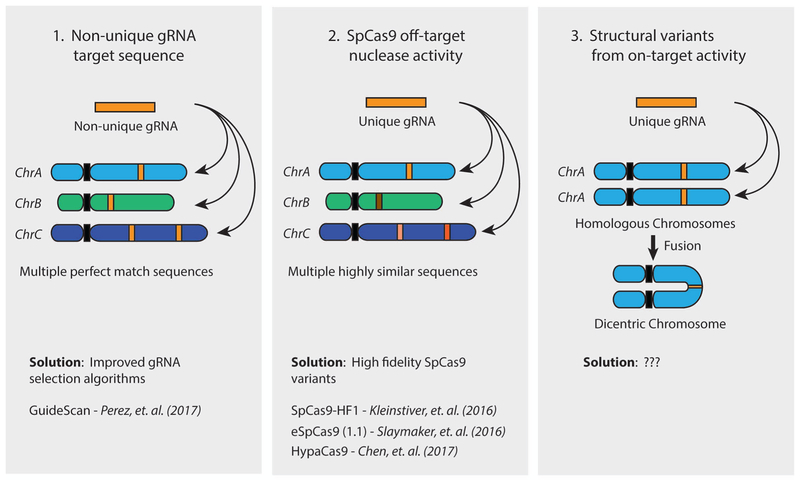
If the two sources of off-target activity described so far can be, at least in principle, fully addressed by improving gRNA design and by increasing the specificity of Cas enzymes, there is a third, often overlooked, source that is unfortunately much harder if not impossible to avoid. The ease with which the simultaneous induction of multiple DSBs by CRISPR can lead to structural rearrangements within or between chromosomes raises a disturbing possibility: since an active on-target gRNA is expected to induce breaks in both alleles of a target gene, there is a significant potential for two homologous chromosomes or, if the cell has replicated its DNA, two sister chromatids to subsequently form a fused dicentric chromosome112. The well-characterized breakage-fusion-bridge cycle that results from the formation of such dicentric chromosomes can lead to a variety of unpredictable consequences, including either apoptosis or cellular transformation (Figure 3).
Figure 3 – Sources of SpCas9 off-target activity.
The activity of SpCas9 in human cells can produce off-target consequences through three mechanisms, based on the gRNA being used. (1) If the 20nt gRNA target sequence is not unique in the human genome, SpCas9 will induce a DNA double strand break (DSB) at each locus containing a perfect match, resulting in the formation of non-targeted indels or structural rearrangements. This can be avoided through the use of gRNA selection algorithms specifically designed to avoid non-unique sequences, such as GuideScan. (2) SpCas9 can also induce DSBs at highly similar sequences containing 3 or fewer mismatches to the gRNA. These off-target breaks can result in non-targeted indels and structural rearrangements either alone or in combination with on-target SpCas9 activity. The propensity for SpCas9 to cut at mismatched target sites can be reduced through the use of reported high-fidelity SpCas9 variants, including SpCas9-HF1, eSpCas9, and HypaCas9. (3) Even completely on-target activity from a single gRNA has the potential to introduce off-target structural rearrangements through simultaneous cutting of the same locus on homologous chromosomes or sister chromatids. Chromosome head-to-head fusion is one potential outcome of this event, resulting in the formation of dicentric chromosome. No solution for this type of off-target event has yet been proposed.
Novel CRISPR-based tools for cancer diagnostics
A multitude of different CRISPR systems are being identified in prokaryotes, and their study is suggesting exciting new applications for this technology25,113. Among class 2 CRISPR systems, three major types have been identified: Type II, Type V, and most recently Type VI. Type VI systems are particularly interesting, as they are characterized by an effector molecule (Cas13) that targets and single stranded RNA (ssRNA) rather than dsDNA as Type II and Type V15,27. But Cas13 has another surprising feature: once activated by binding to the target sequence recognized by the associated crRNA, Cas13 not only cleaves the target strand, but also begins to degrade non-targeted ssRNAs it finds in its proximity, a phenomenon aptly named “collateral RNA cleavage”15,114. How this activity is regulated in bacteria and what role it plays in adaptive immunity is not fully clear, but the potential for collateral RNA cleavage to be adapted as a highly sensitive nucleic acid detection technology was quickly recognized and implemented by several groups16,114–116.
SHERLOCK (Specific High-Sensitivity Enzymatic Reporter UnLOCKing) was the first nucleic acid detection system taking advantage of collateral RNA cleavage16. The central idea underlying SHERLOCK is the use of an RNA reporter consisting of ssRNA containing a fluorophore and a quencher. In the first step of the detection process, the sample nucleic acid is amplified isothermally and transcribed into RNA. The RNA pool is then incubated with the reporter RNA, Cas13, and a crRNA designed to recognize the DNA species one wants to detect. Binding of the Cas13a complex to its target sequence activates collateral RNA degradation, cleaving the reporter and freeing the fluorophore from the quencher. Using this technique ssRNA target sequences could be detected at concentrations as low as ~2aM (~2×10−18 mol/L)16.
Non-specific collateral nuclease activity is also a key feature of the CRISPR Type V effector Cas12a/Cpf1, with the notable difference that the Cas12a/crRNA complex recognizes and degrades DNA, not RNA. Cas12a is at the core of DETECTR (DNA Endonuclease Targeted CRISPR Trans Reporter), an alternative nucleic acid detection system developed by Jennifer Doudna and colleagues in 2018115. Rather than targeting ssRNA, Cas12a’s collateral activity is targeted toward ssDNA, abrogating the need for RNA transcription as part of the detection system. Otherwise the protocols are similar, with amplified DNA being incubated with Cas12a/gRNA complex in combination with a fluorescently labeled ssDNA reporter.
That this is a fertile area of investigation is reflected by the number of variations and improvements that were quickly incorporated in the SHERLOCK-2 system116. Cas13 enzymes from different bacterial species display divergent dinucleotide preferences for collateral nuclease activity, and these differing preferences were used to develop a multiplexed assay, mixing four different effector nucleases (PsmCas13b, LwaCas13a, CcaCas13b, AsCas12a) with four different fluorescent reporter strands. Four different target sequences could be simultaneously detected in a single reaction at sensitivity of comparable to non-multiplexed SHERLOCK.
To increase the sensitivity of the SHERLOCK system, a two-step signal amplification protocol was developed using two different CRISPR effectors. Collateral ssRNA degradation by the Type III RNAse effector Csm6 is activated by the production of key second messengers117,118 that can be generated by the activity of Cas13. Using two different fluorescent reporters in the same channel with specificity to Csm6 or Cas13a, the sensitivity of SHERLOCK 2 could be further improved reaching nearly single molecule detection116.
One of the most appealing aspects of the SHERLOCK detection system is its versatility and simplicity. The components of the detection reaction can be freeze-dried and paper-spotted to generate lateral-flow test strips (analogous to those used in pregnancy tests sold in drug stores across the country) that can be produced cheaply for use outside of a laboratory setting with only a slight reduction in sensitivity (20aM versus 2aM)16,116.
Given their sensitivity and ease of use, the SHERLOCK and DETECTR systems could prove useful in multiple settings in the context of cancer diagnosis and treatment. For example, DETECTR was demonstrated to effectively detect specific strains Human Papillomavirus associated with cervical cancer initiation (HPV16, HPV18) in human anal swab DNA extracts. The systems could also be used to rapidly test for cancer-predisposing germline mutations. The extreme sensitivity of these systems makes them amenable to cancer genotyping approaches using cell-free DNA (cfDNA) fragments isolated from blood samples. SHERLOCK was demonstrated to operate within the detection range necessary for these types of approaches and was also used to detect specific mutations in liquid biopsy samples from non-small cell lung cancer patients, effectively identifying specific tumor-associated EGFR mutations. The extreme ease of use associated with lateral flow test strips, along with the relatively low cost of production, could offer accurate cancer genotyping to communities and markets that have been previously underserved the by the cancer genomics revolution.
As CRISPR-based technologies continue to mature, it is becoming increasingly apparent that the applications of this system in cancer research and therapy have by no means been exhausted. Instead, this versatile and surprising tool is being modified, improved, and applied in an ever-increasing number of novel and exciting ways that promise to reshape the way that cancer is researched, treated, and diagnosed.
Acknowledgments
We apologize to the many colleagues whose work we couldn’t cite due to space constraints. Genome editing work in the Ventura lab is supported by grants from the MSKCC Brain Tumor Consortium, the Geoffrey Beene Cancer Research Foundation, the Pershing Square Sohn Cancer Research Alliance, the STARR consortium, and the NIH/NCI (Grant P30-CA008748).
References
- 1.Mojica FJ, Rodriguez-Valera F. The discovery of CRISPR in archaea and bacteria. FEBS J. September 2016;283(17):3162–3169. [DOI] [PubMed] [Google Scholar]
- 2.Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. February 15 2013;339(6121):819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science. February 15 2013;339(6121):823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blasco RB, Karaca E, Ambrogio C, et al. Simple and rapid in vivo generation of chromosomal rearrangements using CRISPR/Cas9 technology. Cell Rep. November 20 2014;9(4):1219–1227. [DOI] [PubMed] [Google Scholar]
- 5.Choi PS, Meyerson M. Targeted genomic rearrangements using CRISPR/Cas technology. Nat Commun. April 24 2014;5:3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maddalo D, Manchado E, Concepcion CP, et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature. December 18 2014;516(7531):423–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho SW, Lee J, Carroll D, Kim JS, Lee J. Heritable gene knockout in Caenorhabditis elegans by direct injection of Cas9-sgRNA ribonucleoproteins. Genetics. November 2013;195(3):1177–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ratz M, Testa I, Hell SW, Jakobs S. CRISPR/Cas9-mediated endogenous protein tagging for RESOLFT super-resolution microscopy of living human cells. Sci Rep. April 20 2015;5:9592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilbert LA, Larson MH, Morsut L, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. July 18 2013;154(2):442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. CRISPR RNA-guided activation of endogenous human genes. Nat Methods. October 2013;10(10):977–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thakore PI, D’Ippolito AM, Song L, et al. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat Methods. December 2015;12(12):1143–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu X, Tao Y, Gao X, et al. A CRISPR-based approach for targeted DNA demethylation. Cell Discov. 2016;2:16009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen B, Gilbert LA, Cimini BA, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. December 19 2013;155(7):1479–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheth RU, Yim SS, Wu FL, Wang HH. Multiplex recording of cellular events over time on CRISPR biological tape. Science. December 15 2017;358(6369):1457–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abudayyeh OO, Gootenberg JS, Konermann S, et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. August 5 2016;353(6299):aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gootenberg JS, Abudayyeh OO, Lee JW, et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. April 28 2017;356(6336):438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chow RD, Chen S. Cancer CRISPR Screens In Vivo. Trends Cancer. May 2018;4(5):349–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fellmann C, Gowen BG, Lin PC, Doudna JA, Corn JE. Cornerstones of CRISPR-Cas in drug discovery and therapy. Nat Rev Drug Discov. February 2017;16(2):89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurata M, Yamamoto K, Moriarity BS, Kitagawa M, Largaespada DA. CRISPR/Cas9 library screening for drug target discovery. J Hum Genet. February 2018;63(2):179–186. [DOI] [PubMed] [Google Scholar]
- 20.Kannan R, Ventura A. The CRISPR revolution and its impact on cancer research. Swiss Med Wkly. 2015;145:w14230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mou H, Kennedy Z, Anderson DG, Yin H, Xue W. Precision cancer mouse models through genome editing with CRISPR-Cas9. Genome Med. 2015;7(1):53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tschaharganeh DF, Lowe SW, Garippa RJ, Livshits G. Using CRISPR/Cas to study gene function and model disease in vivo. FEBS J. September 2016;283(17):3194–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrangou R, Fremaux C, Deveau H, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. March 23 2007;315(5819):1709–1712. [DOI] [PubMed] [Google Scholar]
- 24.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. September 25 2012;109(39):E2579–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hille F, Richter H, Wong SP, Bratovic M, Ressel S, Charpentier E. The Biology of CRISPR-Cas: Backward and Forward. Cell. March 8 2018;172(6):1239–1259. [DOI] [PubMed] [Google Scholar]
- 26.Koonin EV. Evolution of RNA- and DNA-guided antivirus defense systems in prokaryotes and eukaryotes: common ancestry vs convergence. Biol Direct. February 10 2017;12(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shmakov S, Abudayyeh OO, Makarova KS, et al. Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Mol Cell. November 5 2015;60(3):385–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deltcheva E, Chylinski K, Sharma CM, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. March 31 2011;471(7340):602–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. Elife. January 29 2013;2:e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yin H, Xue W, Chen S, et al. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol. June 2014;32(6):551–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burma S, Chen BP, Chen DJ. Role of non-homologous end joining (NHEJ) in maintaining genomic integrity. DNA Repair (Amst). September 8 2006;5(9–10):1042–1048. [DOI] [PubMed] [Google Scholar]
- 32.Valerie K, Povirk LF. Regulation and mechanisms of mammalian double-strand break repair. Oncogene. September 1 2003;22(37):5792–5812. [DOI] [PubMed] [Google Scholar]
- 33.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. November 2013;8(11):2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Demuth I, Digweed M. The clinical manifestation of a defective response to DNA double-strand breaks as exemplified by Nijmegen breakage syndrome. Oncogene. December 10 2007;26(56):7792–7798. [DOI] [PubMed] [Google Scholar]
- 35.Lavin MF. Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nat Rev Mol Cell Biol. October 2008;9(10):759–769. [DOI] [PubMed] [Google Scholar]
- 36.Haapaniemi E, Botla S, Persson J, Schmierer B, Taipale J. CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat Med. June 11 2018. [DOI] [PubMed] [Google Scholar]
- 37.Ihry RJ, Worringer KA, Salick MR, et al. p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat Med. June 11 2018. [DOI] [PubMed] [Google Scholar]
- 38.Schiffman M, Doorbar J, Wentzensen N, et al. Carcinogenic human papillomavirus infection. Nat Rev Dis Primers. December 1 2016;2:16086. [DOI] [PubMed] [Google Scholar]
- 39.Howley PM. Role of the human papillomaviruses in human cancer. Cancer Res. September 15 1991;51(18 Suppl):5019s–5022s. [PubMed] [Google Scholar]
- 40.Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. December 21 1990;63(6):1129–1136. [DOI] [PubMed] [Google Scholar]
- 41.Butz K, Ristriani T, Hengstermann A, Denk C, Scheffner M, Hoppe-Seyler F. siRNA targeting of the viral E6 oncogene efficiently kills human papillomavirus-positive cancer cells. Oncogene. September 4 2003;22(38):5938–5945. [DOI] [PubMed] [Google Scholar]
- 42.Hall AH, Alexander KA. RNA interference of human papillomavirus type 18 E6 and E7 induces senescence in HeLa cells. J Virol. May 2003;77(10):6066–6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horner SM, DeFilippis RA, Manuelidis L, DiMaio D. Repression of the human papillomavirus E6 gene initiates p53-dependent, telomerase-independent senescence and apoptosis in HeLa cervical carcinoma cells. J Virol. April 2004;78(8):4063–4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu Z, Yu L, Zhu D, et al. Disruption of HPV16-E7 by CRISPR/Cas system induces apoptosis and growth inhibition in HPV16 positive human cervical cancer cells. Biomed Res Int. 2014;2014:612823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kennedy EM, Kornepati AV, Goldstein M, et al. Inactivation of the human papillomavirus E6 or E7 gene in cervical carcinoma cells by using a bacterial CRISPR/Cas RNA-guided endonuclease. J Virol. October 2014;88(20):11965–11972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fesnak AD, June CH, Levine BL. Engineered T cells: the promise and challenges of cancer immunotherapy. Nat Rev Cancer. August 23 2016;16(9):566–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sadelain M, Riviere I, Riddell S. Therapeutic T cell engineering. Nature. May 24 2017;545(7655):423–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Algarra I, Cabrera T, Garrido F. The HLA crossroad in tumor immunology. Hum Immunol. January 2000;61(1):65–73. [DOI] [PubMed] [Google Scholar]
- 49.Eshhar Z, Waks T, Gross G, Schindler DG. Specific activation and targeting of cytotoxic lymphocytes through chimeric single chains consisting of antibody-binding domains and the gamma or zeta subunits of the immunoglobulin and T-cell receptors. Proc Natl Acad Sci U S A. January 15 1993;90(2):720–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lim WA, June CH. The Principles of Engineering Immune Cells to Treat Cancer. Cell. February 9 2017;168(4):724–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Posey AD Jr., Schwab RD, Boesteanu AC, et al. Engineered CAR T Cells Targeting the Cancer-Associated Tn-Glycoform of the Membrane Mucin MUC1 Control Adenocarcinoma. Immunity. June 21 2016;44(6):1444–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brentjens RJ, Davila ML, Riviere I, et al. CD19-targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia. Sci Transl Med. March 20 2013;5(177):177ra138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kalos M, Levine BL, Porter DL, et al. T cells with chimeric antigen receptors have potent antitumor effects and can establish memory in patients with advanced leukemia. Sci Transl Med. August 10 2011;3(95):95ra73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Porter DL, Levine BL, Kalos M, Bagg A, June CH. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N Engl J Med. August 25 2011;365(8):725–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang X, Riviere I. Clinical manufacturing of CAR T cells: foundation of a promising therapy. Mol Ther Oncolytics. 2016;3:16015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Berger EA, Murphy PM, Farber JM. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. [DOI] [PubMed] [Google Scholar]
- 57.Yarchoan R, Uldrick TS. HIV-Associated Cancers and Related Diseases. N Engl J Med. March 15 2018;378(11):1029–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Davila ML, Riviere I, Wang X, et al. Efficacy and toxicity management of 19–28z CAR T cell therapy in B cell acute lymphoblastic leukemia. Sci Transl Med. February 19 2014;6(224):224ra225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kochenderfer JN, Dudley ME, Feldman SA, et al. B-cell depletion and remissions of malignancy along with cytokine-associated toxicity in a clinical trial of anti-CD19 chimeric-antigen-receptor-transduced T cells. Blood. March 22 2012;119(12):2709–2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sather BD, Romano Ibarra GS, Sommer K, et al. Efficient modification of CCR5 in primary human hematopoietic cells using a megaTAL nuclease and AAV donor template. Sci Transl Med. September 30 2015;7(307):307ra156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mandal PK, Ferreira LM, Collins R, et al. Efficient ablation of genes in human hematopoietic stem and effector cells using CRISPR/Cas9. Cell Stem Cell. November 6 2014;15(5):643–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Monroe KM, Yang Z, Johnson JR, et al. IFI16 DNA sensor is required for death of lymphoid CD4 T cells abortively infected with HIV. Science. January 24 2014;343(6169):428–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu L, Johnson C, Fujimura S, Teque F, Levy JA. Transfection optimization for primary human CD8+ cells. J Immunol Methods. September 30 2011;372(1–2):22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim S, Kim D, Cho SW, Kim J, Kim JS. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. June 2014;24(6):1012–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zuris JA, Thompson DB, Shu Y, et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat Biotechnol. January 2015;33(1):73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schumann K, Lin S, Boyer E, et al. Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc Natl Acad Sci U S A. August 18 2015;112(33):10437–10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Seki A, Rutz S. Optimized RNP transfection for highly efficient CRISPR/Cas9-mediated gene knockout in primary T cells. J Exp Med. March 5 2018;215(3):985–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ren J, Liu X, Fang C, Jiang S, June CH, Zhao Y. Multiplex Genome Editing to Generate Universal CAR T Cells Resistant to PD1 Inhibition. Clin Cancer Res. May 1 2017;23(9):2255–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hendel A, Bak RO, Clark JT, et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat Biotechnol. September 2015;33(9):985–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Roth TL, Puig-Saus C, Yu R, et al. Reprogramming human T cell function and specificity with non-viral genome targeting. Nature. July 2018;559(7714):405–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Eyquem J, Mansilla-Soto J, Giavridis T, et al. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature. March 2 2017;543(7643):113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marcus A, Eshhar Z. Allogeneic chimeric antigen receptor-modified cells for adoptive cell therapy of cancer. Expert Opin Biol Ther. July 2014;14(7):947–954. [DOI] [PubMed] [Google Scholar]
- 73.Yang Y, Jacoby E, Fry TJ. Challenges and opportunities of allogeneic donor-derived CAR T cells. Curr Opin Hematol. November 2015;22(6):509–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shlomchik WD. Graft-versus-host disease. Nat Rev Immunol. May 2007;7(5):340–352. [DOI] [PubMed] [Google Scholar]
- 75.Torikai H, Reik A, Liu PQ, et al. A foundation for universal T-cell based immunotherapy: T cells engineered to express a CD19-specific chimeric-antigen-receptor and eliminate expression of endogenous TCR. Blood. June 14 2012;119(24):5697–5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Osborn MJ, Webber BR, Knipping F, et al. Evaluation of TCR Gene Editing Achieved by TALENs, CRISPR/Cas9, and megaTAL Nucleases. Mol Ther. March 2016;24(3):570–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ren J, Zhang X, Liu X, et al. A versatile system for rapid multiplex genome-edited CAR T cell generation. Oncotarget. March 7 2017;8(10):17002–17011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Grey HM, Kubo RT, Colon SM, et al. The small subunit of HL-A antigens is beta 2-microglobulin. J Exp Med. December 1 1973;138(6):1608–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Peterson PA, Rask L, Lindblom JB. Highly purified papain-solubilized HL-A antigens contain beta2-microglobulin. Proc Natl Acad Sci U S A. January 1974;71(1):35–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang W, Ye C, Liu J, Zhang D, Kimata JT, Zhou P. CCR5 gene disruption via lentiviral vectors expressing Cas9 and single guided RNA renders cells resistant to HIV-1 infection. PLoS One. 2014;9(12):e115987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Orr MT, Lanier LL. Natural killer cell education and tolerance. Cell. September 17 2010;142(6):847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. March 22 2012;12(4):252–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rudd CE, Taylor A, Schneider H. CD28 and CTLA-4 coreceptor expression and signal transduction. Immunol Rev. May 2009;229(1):12–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dong H, Strome SE, Salomao DR, et al. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med. August 2002;8(8):793–800. [DOI] [PubMed] [Google Scholar]
- 85.Sharpe AH, Wherry EJ, Ahmed R, Freeman GJ. The function of programmed cell death 1 and its ligands in regulating autoimmunity and infection. Nat Immunol. March 2007;8(3):239–245. [DOI] [PubMed] [Google Scholar]
- 86.Ascierto PA, Marincola FM, Ribas A. Anti-CTLA4 monoclonal antibodies: the past and the future in clinical application. J Transl Med. November 13 2011;9:196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Swaika A, Hammond WA, Joseph RW. Current state of anti-PD-L1 and anti-PD-1 agents in cancer therapy. Mol Immunol. October 2015;67(2 Pt A):4–17. [DOI] [PubMed] [Google Scholar]
- 88.Hodi FS, O’Day SJ, McDermott DF, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. August 19 2010;363(8):711–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Larkin J, Chiarion-Sileni V, Gonzalez R, et al. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N Engl J Med. July 2 2015;373(1):23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gwiazda KS, Grier AE, Sahni J, et al. High Efficiency CRISPR/Cas9-mediated Gene Editing in Primary Human T-cells Using Mutant Adenoviral E4orf6/E1b55k “Helper” Proteins. Mol Ther. September 29 2016;24(9):1570–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rupp LJ, Schumann K, Roybal KT, et al. CRISPR/Cas9-mediated PD-1 disruption enhances anti-tumor efficacy of human chimeric antigen receptor T cells. Sci Rep. April 7 2017;7(1):737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang Y, Zhang X, Cheng C, et al. CRISPR-Cas9 mediated LAG-3 disruption in CAR-T cells. Front Med. December 2017;11(4):554–562. [DOI] [PubMed] [Google Scholar]
- 93.Baylis F, McLeod M. First-in-human Phase 1 CRISPR Gene Editing Cancer Trials: Are We Ready? Curr Gene Ther. 2017;17(4):309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chmielewski M, Hombach AA, Abken H. Of CARs and TRUCKs: chimeric antigen receptor (CAR) T cells engineered with an inducible cytokine to modulate the tumor stroma. Immunol Rev. January 2014;257(1):83–90. [DOI] [PubMed] [Google Scholar]
- 95.Yeku OO, Purdon TJ, Koneru M, Spriggs D, Brentjens RJ. Armored CAR T cells enhance antitumor efficacy and overcome the tumor microenvironment. Sci Rep. September 5 2017;7(1):10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Di Stasi A, Tey SK, Dotti G, et al. Inducible apoptosis as a safety switch for adoptive cell therapy. N Engl J Med. November 3 2011;365(18):1673–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhou X, Di Stasi A, Tey SK, et al. Long-term outcome after haploidentical stem cell transplant and infusion of T cells expressing the inducible caspase 9 safety transgene. Blood. June 19 2014;123(25):3895–3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Perez AR, Pritykin Y, Vidigal JA, et al. GuideScan software for improved single and paired CRISPR guide RNA design. Nat Biotechnol. April 2017;35(4):347–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. August 17 2012;337(6096):816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kim D, Kim S, Kim S, Park J, Kim JS. Genome-wide target specificities of CRISPR-Cas9 nucleases revealed by multiplex Digenome-seq. Genome Res. March 2016;26(3):406–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tsai SQ, Zheng Z, Nguyen NT, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. February 2015;33(2):187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cao J, Wu L, Zhang SM, et al. An easy and efficient inducible CRISPR/Cas9 platform with improved specificity for multiple gene targeting. Nucleic Acids Res. November 2 2016;44(19):e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mali P, Aach J, Stranges PB, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. September 2013;31(9):833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ran FA, Hsu PD, Lin CY, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. September 12 2013;154(6):1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. September 25 2014;513(7519):569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nishimasu H, Ran FA, Hsu PD, et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. February 27 2014;156(5):935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kleinstiver BP, Pattanayak V, Prew MS, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. January 28 2016;529(7587):490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. January 1 2016;351(6268):84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chen JS, Dagdas YS, Kleinstiver BP, et al. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature. October 19 2017;550(7676):407–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Esvelt KM, Carlson JC, Liu DR. A system for the continuous directed evolution of biomolecules. Nature. April 28 2011;472(7344):499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hu JH, Miller SM, Geurts MH, et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. April 5 2018;556(7699):57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Frock RL, Hu J, Meyers RM, Ho YJ, Kii E, Alt FW. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat Biotechnol. February 2015;33(2):179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Murugan K, Babu K, Sundaresan R, Rajan R, Sashital DG. The Revolution Continues: Newly Discovered Systems Expand the CRISPR-Cas Toolkit. Mol Cell. October 5 2017;68(1):15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.East-Seletsky A, O’Connell MR, Knight SC, et al. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature. October 13 2016;538(7624):270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chen JS, Ma E, Harrington LB, et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. April 27 2018;360(6387):436–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gootenberg JS, Abudayyeh OO, Kellner MJ, Joung J, Collins JJ, Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. April 27 2018;360(6387):439–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kazlauskiene M, Kostiuk G, Venclovas C, Tamulaitis G, Siksnys V. A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science. August 11 2017;357(6351):605–609. [DOI] [PubMed] [Google Scholar]
- 118.Niewoehner O, Garcia-Doval C, Rostol JT, et al. Type III CRISPR-Cas systems produce cyclic oligoadenylate second messengers. Nature. August 31 2017;548(7669):543–548. [DOI] [PubMed] [Google Scholar]