Abstract
Cis-regulatory elements are critical for the precise spatiotemporal regulation of genes during development. However, identifying functional regulatory sites that drive cell differentiation in vivo has been complicated by the high numbers of cells required for whole-genome epigenetic assays. Here, we identified putative regulatory elements during sex determination by performing ATAC-seq and ChIP-seq for H3K27ac in purified XX and XY gonadal supporting cells before and after sex determination in mice. We show that XX and XY supporting cells initiate sex determination with similar chromatin landscapes and acquire sex-specific regulatory elements as they commit to the male or female fate. To validate our approach, we identified a functional gonad-specific enhancer downstream of Bmp2, an ovary-promoting gene. This work increases our understanding of the complex regulatory network underlying mammalian sex determination and provides a powerful resource for identifying non-coding regulatory elements that could harbor mutations that lead to Disorders of Sexual Development.
Keywords: sex determination, gonad development, regulatory elements, cell differentiation, enhancers, chromatin landscapes
INTRODUCTION
Sex determination is a powerful system to study cell differentiation. Although the chromosomal sex of a mammal is established at fertilization by inheritance of an X or Y chromosome from the father, XX and XY embryos are initially indistinguishable. Even the early gonad has no obvious sex-specific features, and initiates development as a bipotential organ with the ability to differentiate into either a testis (typically in XY individuals) or an ovary (typically in XX individuals) (Fig. 1A) (Burgoyne et al., 1995, Albrecht and Eicher, 2001). A subset of the somatic cells that compose the bipotential gonad, known as the supporting cell lineage, initiate gonadal sex determination by engaging either the male or the female pathway and promoting divergence from the progenitor state. Therefore, gonadal sex determination hinges on the ability of a population of bipotential cells to commit to one of two cell fates at a precise moment in development (Albrecht and Eicher, 2001).
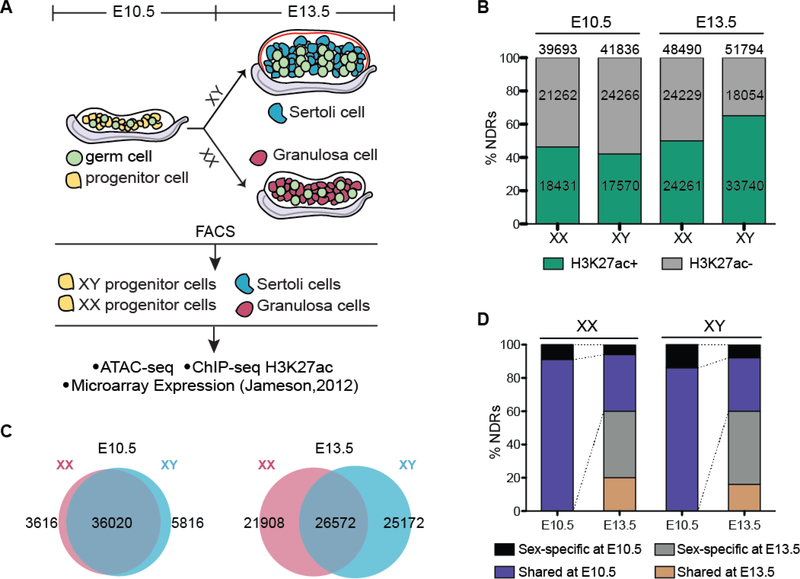
Figure 1. Chromatin architecture is remodeled during mammalian sex determination.
A) Overview of sex determination and work flow. Briefly, supporting progenitor cells (yellow) are bipotential and indistinguishable between XX and XY gonads at E10.5. Expression of the Y-linked Sry gene (at E11.5) directs Sertoli cell differentiation in the testis (XY, blue). Absence of Sry directs differentiation of granulosa cells in the ovary (XX, pink). XX and XY progenitor cells (E10.5), Sertoli cells (E13.5), and granulosa cells (E13.5) were FACS-purified and used for ATAC-seq and ChIP-seq for H3K27ac. Further analysis made use of microarray expression data from purified supporting cells (Jameson et al., 2012b). B) Percent (and number) of H3K27ac-negative (grey) and H3K27ac-postive (green) NDRs in XX and XY cells at E10.5 (left) and E13.5 (right). C) Venn diagrams of all NDRs in XX (pink) and XY (blue) supporting cells at E10.5 (left) and E13.5 (right). D) Percent of NDRs that are shared between XX and XY cells at E10.5 (purple) and at E13.5 (orange), or specific to either XX or XY cells at E10.5 (black) or at E13.5 (grey). The purple and black bars at E13.5 represent NDRs that were retained from E10.5, while the orange and grey represent newly acquired NDRs.
In eutharian and metatherian mammals, gonadal sex determination is triggered by expression of the Y-encoded gene Sry around mid-gestation (Gubbay et al., 1990, Sinclair et al., 1990, Koopman et al., 1991). Sry upregulates its downstream target Sox9, a transcription factor (TF) which then directs differentiation of Sertoli cells (Hacker et al., 1995, Bullejos and Koopman, 2001, Sekido et al., 2004). In XX gonads that lack Sry, the Wnt4/Rspo1/Ctnnb1 pathway is involved in directing the supporting progenitor cells to differentiate as granulosa cells (Fig. 1A) (Vainio et al., 1999, Parma et al., 2006, Maatouk et al., 2008). Importantly, canalization of the male or female pathway requires simultaneous repression of genes that promote the alternate fate (Kim et al., 2006, Barrionuevo et al., 2006, Jameson et al., 2012a, Bernard et al., 2012). This mutual antagonism is critical during sex determination, but also for maintaining Sertoli and granulosa cell identity even long after the initial fate commitment of the fetal gonad (Matson et al., 2011, Uhlenhaut et al., 2009).
Although it may appear that gonadal sex determination is simply defined by the presence or absence of Sry, a complex network of male- or female-promoting signaling pathways coexist at the bipotential stage that require tight regulation (Jameson et al., 2012b, Munger et al., 2013). Evidence that gene dosage must be tightly regulated comes from studies of humans with Disorders of Sex Developments (DSDs) that have duplications or deletions in the region upstream of the SOX9 locus, a region devoid of coding genes but enriched for regulatory elements. Duplications in XX individuals lead to female-to-male sex reversal, while deletions in XY individuals cause male-to-female sex reversal, potentially caused by increased or decreased SOX9 levels, respectively (Wagner et al., 1994, Benko et al., 2011, Lybaek et al., 2014, Kim et al., 2015). This highlights how a slight disruption to this network can be enough to send the system towards the opposite pathway. However, our inability to pinpoint the location of cis-regulatory elements limits our capacity to study the mechanisms that regulate the precise spatiotemporal expression of sex-determining genes. Additionally, only ~43% of individuals with DSDs will receive a genetic diagnosis (Eggers et al., 2016), partly due to mutations residing in non-coding regions that cannot be identified by standard diagnostic techniques such as karyotyping, sequencing of individual genes or even whole-exome sequencing.
To identify genomic elements that regulate sex determination, we developed a map of the chromatin accessibility landscape of the supporting cell lineage before and after commitment to the male or female fate. We purified XX and XY supporting cells before (E10.5) and after (E13.5) sex determination in mice, and performed Assay for Transposase-Accessible Chromatin (ATAC-seq) and Chromatin Immunoprecipitation followed by sequencing (ChIP-seq) for H3K27ac, a histone modification indicative of active enhancers (Creyghton et al., 2010). We show that XX and XY progenitor cells from E10.5 bipotential gonads have similar chromatin accessibility landscapes, and that these resolve into sex-specific patterns after differentiation into either granulosa (XX) or Sertoli (XY) cells. H3K27ac+ gonad-specific nucleosome-depleted regions (NDRs) are enriched around granulosa-promoting genes in granulosa cells, and Sertoli-promoting genes in Sertoli cells. Furthermore, these NDRs are enriched for binding motifs of transcription factors (TFs) linked to supporting cell differentiation, suggesting that gonad-specific NDRs are cis-regulatory elements that establish and/or maintain sex-specific transcriptional programs throughout sex determination. Finally, we demonstrate the power of our dataset to identify novel enhancers by validating the in vivo activity of an enhancer downstream of Bmp2, a female-specific gene (Nef et al., 2005, Kashimada et al., 2011). This work will increase our understanding of the complex regulatory network that drives mammalian sex determination and is an invaluable resource for identifying functional elements that could harbor non-coding mutations that cause DSDs and escape routine diagnostic techniques.
MATERIALS AND METHODS
Mice:
The Sf1-eGFP and TESCO-CFP, TESMS-CFP reporter mouse lines previously generated were maintained on a C57BL/6 (B6) background (Beverdam and Koopman, 2006, Sekido and Lovell-Badge, 2008, Gonen et al., 2018). Timed matings were established between reporter males and ICR females for their larger litter sizes. The morning of a vaginal plug was considered E0.5. Embryos were collected at E10.5/E13.5 and genetic sex was determined using PCR for the presence/absence of a region of the Y chromosome (pF: TGAAGCTTTTGGCTTTGAG, pR:CCGCTGCCAAATTCTTTGG).
Cell preparation:
Gonads were microdissected from E10.5 or E13.5 embryos and removed from the mesonephros. Isolated gonads were then incubated in 0.05% trypsin for 5–7 min at 37°C, rinsed, and mechanically dissociated in PBS/3% BSA. The cell suspension was strained through a 35μm filter top and the supporting cells were collected using fluorescence-activated cell sorting (FACS).
ATAC-seq:
ATAC-Seq libraries where prepared from approximately 60,000 FACS-purified cells as previously described with no modifications (Buenrostro et al., 2013). ATAC-seq was performed on two biological replicates per time point, with each replicate containing pooled FACS-sorted cells from gonads of multiple embryos. Single end ATAC-Seq reads were trimmed for quality and to remove adapters using Trimgalore with a stringency setting of 5. Trimmed reads were aligned to the mm9 genome using Bowtie with the parameters -m 1 and --best. MACS2 was used to call peaks using the --nomodel and --broad settings. Peaks were considered to be true positives if they were present in both replicates.
ChIP-seq:
ChIP-seq was performed with no modifications as in Van Galen et al. 2016 (van Galen et al., 2016) on ~100K FACS-purified cells. ChIP-seq was performed on two biological replicates, each replicate containing pooled FACS-sorted cells from gonads of multiple embryos. Unindexed chromatin from ~400K Drosophila S2 cells was added per IP as carrier chromatin. Chromatin was digested using 150 units of MNase (NEB #M0247S) and ChIP was performed using 3μl of H3 antibody (Active Motif #39763) or 2μl of H3K27ac antibody (Active Motif #39133). Alignment to the mm9 mouse genome was performed using Bowtie. For visualization on the UCSC genome browser, replicates were concatenated to maximize number of reads. Peaks were called using MACS2/2.1.0 with the “--broad” setting, and BigWig files were created using bedGraphToBigWig. H3 ChIP was used as input. To identify regions significantly enriched for H3K27ac compared to flanking regions, HOMER was used applying the findPeaks function and settings “--style histone”, with a size of 1000. The setting “-C 0” was used in MNase ChIP to disable fold enrichment limit of expected unique tag positions.
NDR analyses:
All NDR analyses were performed with HOMER. Categorization of ubiquitous NDRs was determined by identifying overlapping NDRs using the mergePeaks function between our datasets and previously published DNaseI-seq data performed in mouse fibroblasts, ESCs, heart, kidney, brain and liver (Maatouk et al., 2017). The remaining non-overlapping NDRs were considered gonad-specific NDRs. MergePeaks was then used between gonad-specific NDRs and H3K27ac ChIP-seq to identify gonad-specific H3K27ac+ NDRs. MergePeaks was also used to identify resolved and de novo NDRs within our datasets. The annotatePeaks.pl function was used to generate a list of genes associated to each NDR (each NDR is associated to its nearest TSS), as well as their genomic allocation and distance to the TSS. NDRs were categorized into intergenic, intronic, exonic (exon, 5’ UTR and 3’ UTR) or promoter (TSS and promoter). To determine the average number of NDRs associated to Sertoli-specific or granulosa-specific genes, we first identified Sertoli-specific genes (genes expressed >1.5 fold higher in E13.5 Sertoli cells compared to E13.5 granulosa cells), and granulosa-specific genes (genes expressed >1.5 fold higher in E13.5 granulosa cells compared to E13.5 Sertoli cells) from a previous microarray study (Jameson et al., 2012b), then counted the number of NDRs associated to each Sertoli- or granulosa-specific TSS. TF motif analyses were performed using HOMER with default settings and the -mask option.
Cloning the enhancer-reporter plasmids:
The putative enhancer was amplified by PCR and cloned into the NotI site of the pSfi-Hsp68–LacZ reporter vector (Addgene #33351). The primers used for the cloning were TgBmp2_DNG/F_inf: ACCGCGGTGGCGGCCGCATAGAAGATTGCCAGACTCC and TgBmp2_DNG/R_inf: TCCACTAGTTCTAGAACAGGGATTCTCTGTATAGC. Cloning was carried out using In-Fusion HD (Clonetech #639646). All plasmids were sequenced using Sanger sequencing to verify the correct sequence was inserted.
Microinjection of the enhancer-reporter plasmids:
To prepare DNA for zygote injection, 50 μg of the Enhancer-Hsp68-LacZ plasmid was linearized with NotI-HF and XhoI and gel purified by electroelution. The DNA was phenol-chloroform extracted, ethanol precipitated and resuspended in TE buffer (10 mM Tris-HCl, 1 mM disodium EDTA, pH 8.0). The DNA was further purified on a DNA-cleanup column (Qiagen PCR Purification Kit) and eluted again in TE Buffer. 5 ng/μl of the purified plasmid was injected into the pronucleus of each C57BL/6Jx CBA hybrid zygote. Embryos were transferred on the same day into oviducts pseudopregnant CD1 recipient mother. Injections were performed by the Crick Genetic Manipulation Service to generate transient transgenic lines. Embryos were harvested at E13.5.
X-Gal staining:
Embryos were dissected at E13.5 and genotyped for the LacZ gene via PCR (pF: GGCGTTAACTCGGCGTTTCAT, pR:CGGTTAACGCCTCGAATCAGC). The embryo bodies and dissected gonads were fixed in fresh cold fixative (0.2% Glutaraldehyde, 2% Formahdehyde) in buffer L0 (100 mM Phosphate buffer pH7.2, 2 mM MgCl2 and 5 mM EGTA) for 30 minutes at 4oC. Embryos were washed three times in L0 buffer, each wash for 5 minutes. Embryos were then moved to a freshly made and filtered X-Gal staining solution containing 100 mM Phosphate buffer pH7.2, 2 mM MgCl2, 5 mM EGTA, 0.02% NP40, 0.01% SodiumDeoxycholate, 50 mM K3Fe(CN)6, 50 mM K4Fe(CN)6.3H2O and 1 mg/ml X-Gal. Incubation in X-Gal staining solution was done in 37°C and varied from a few hours up to overnight, depending on the strength of the β-Gal expression. The next day, embryos were washed three times in PBS and images of the gonads and bodies were taken.
RESULTS
The supporting cell lineage acquires sex-specific chromatin landscapes during sex determination
Regulatory elements lie within regions of open chromatin that are typically devoid of nucleosomes, known as nucleosome-depleted regions (NDRs) (Lee et al., 2004). The open chromatin conformation allows DNA-binding motifs to be accessed by transcription factors, transcriptional repressors, DNA-binding proteins with insulator activity, or in the case of promoters, by RNA Polymerase II (Song et al., 2011). To investigate the chromatin accessibility landscape of gonadal supporting cells at the time of gonad formation and after their differentiation into either Sertoli cells or granulosa cells, we performed ATAC-seq, a highly sensitive protocol capable of identifying NDRs in as few as 50K cells (Buenrostro et al., 2013). As NDRs correlate with a number of different genomic elements, such as enhancers, silencers, insulators and promoters, we also performed low-cell ChIP-seq for H3K27ac, a histone modification indicative of active enhancers (Creyghton et al., 2010, Rada-Iglesias et al., 2011, van Galen et al., 2016). ATAC-seq and ChIP-seq were performed in FACS-purified XX and XY bipotential progenitor cells from E10.5 gonads of Sf1-GFP (Beverdam and Koopman, 2006), differentiated Sertoli cells from E13.5 gonads of TESCO-CFP (Sekido and Lovell-Badge, 2008), and differentiated granulosa cells from gonads of E13.5 TESMS-CFP transgenic mice (Gonen et al., 2018) (Fig. 1A). ATAC-seq and ChIP-seq were performed on 2 biological replicates (each replicate contained pooled cells from multiple embryos) and only NDRs present in both were used in our analyses (Fig. S1).
ATAC-seq identified ~40,000 NDRs in XX and XY E10.5 progenitor cells (Fig. 1B). After sex determination, the number of NDRs increased from 39,693 NDRs in XX progenitor cells to 48,490 in differentiated granulosa cells, and from 41,836 NDRs in XY progenitor cells to 52,794 NDRs in Sertoli cells (Fig. 1B). In addition, ChIP-seq revealed that ~40% of NDRs were also marked by H3K27ac in XX and XY progenitor cells, and after sex determination, the number of active enhancers increased to 50% in granulosa cells, and 65% in Sertoli cells (Fig. 1B).
Previous transcriptional profiling showed that E10.5-E11.0 XY and XX progenitor cells were mostly indistinguishable at this stage (Fig. S2A and S2C) (Nef et al., 2005, Jameson et al., 2012b, Munger et al., 2013). Accordingly, we found that the vast majority of NDRs were shared between XX and XY progenitor cells (90% of XX NDRs (36,020/39,693) and 86% of XY NDRs (36,020/41,836)) (Fig. 1C and 1D), suggesting that these cells are not only morphologically and transcriptionally indistinguishable, but also have similar chromatin landscapes. Differentiation into either Sertoli or granulosa cells is accompanied by the upregulation of a number of Sertoli- or granulosa-promoting genes (Fig. S2B and S2C) (Jameson et al., 2012b). Consistently, there was an increase in total numbers of NDRs, from 39,393 at E10.5 to 48,490 at E13.5 in XX cells, and from 41,836 at E10.5 to 51,794 at E13.5 in XY cells (Fig. 1B). However, the number of overlapping NDRs between XX and XY cells decreased from 36,020 at E10.5 to 26,572 at E13.5 (55% of XX and XY NDRs) (Fig. 1B and 1C). Only ~40% of NDRs that were present at E10.5 were retained at E13.5; therefore, the majority (~60%) of NDRs at E13.5 were newly acquired (Fig. 1D). Of these newly acquired NDRs, one third were shared between Sertoli and granulosa cells, and two thirds were specific to either Sertoli or granulosa cells (Fig. 1D). Together, these data suggest that differentiation of Sertoli and granulosa cells is accompanied by an increase in Sertoli- and granulosa-specific NDRs that may promote sex-specific transcriptional programs as the cells diverge from their progenitor state into distinct cell types.
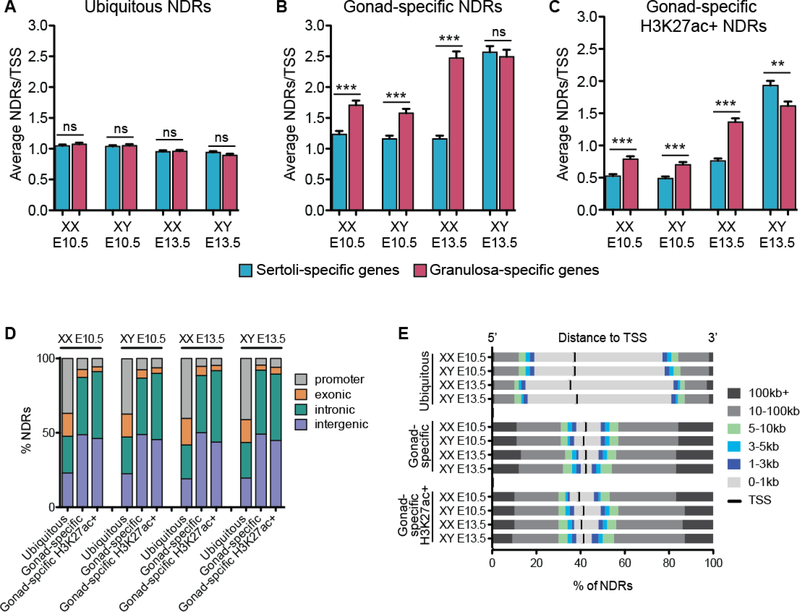
Gonad-specific NDRs are distal regulatory elements that neighbor Sertoli- and granulosa-promoting genes
To identify which NDRs are specific to the supporting cell lineage and more likely to regulate sex-determining genes, we removed NDRs that were also present in six other mouse tissues (primary skin fibroblasts, embryonic stem cells and adult kidney, liver, heart and brain) (Maatouk et al., 2017) from all NDRs identified in both sexes and timepoints. NDRs shared between all cell types were termed “ubiquitous NDRs,” whereas those found only in the supporting cell lineage were termed “gonad-specific NDRs.” We predicted that gonad-specific NDRs would be enriched around Sertoli-promoting genes in Sertoli cells and around granulosa-promoting genes in granulosa cells. To test this hypothesis, we utilized the set of genes that becomes specific in either Sertoli or granulosa cells at E13.5 (Jameson et al., 2012b) and determined the number of NDRs associated with each gene by annotating them to the closest TSS using Hypergeometric Optimization of Motif Enrichment (HOMER). Although regulatory elements can regulate TSSs at a distance, to date the only way of properly assigning a regulatory element to its target TSS is by chromatin-conformation capture analyses. However, these same assays have shown that looping largely occurs between genes and proximal enhancers within Topologically Associated Domains (TADs) (Dekker et al., 2002). On average, the TSSs of both Sertoli- or granulosa-promoting genes neighbored one ubiquitous NDR (possibly corresponding to the promoter) in both time points and sexes, suggesting that ubiquitous NDRs are not preferentially enriched around Sertoli- or granulosa-promoting genes (Fig. 2A). Interestingly, gonad-specific and H3K27ac+ gonad-specific NDRs were enriched around granulosa-promoting genes as compared to Sertoli-promoting genes in both XX and XY cells at E10.5, suggesting that progenitor cells are predisposed towards the granulosa cells fate (Fig. 2B and 2C).
Figure 2. H3K27ac+ gonad-specific NDRs are distal enhancers that neighbor granulosa-and Sertoli-promoting genes.
A-C) Graphs showing the average number of NDRs associated to the nearest TSS as determined by HOMER. Based on their cell-type specificity, NDRs were classified as ubiquitous (A), gonad-specific (B) or H3K27ac+ gonad-specific (C). NDRs were associated to the nearest TSS of genes that become granulosa cell-specific (pink, 823 total genes) and Sertoli cell-specific (blue, 1026 total genes). Granulosa-specific and Sertoli-specific genes were obtained from (Jameson et al., 2012b). Error bars represent the standard error of the mean (SEM) and ** represents p<0.001 as determined by a student’s t test. D) Ubiquitous, gonad-specific and gonad-specific H3K27ac-positive NDRs were subcategorized based on their genomic location (promoter, grey; exonic, orange; intronic, green; intergenic, purple). E) Ubiquitous, gonad-specific and gonad-specific H3K27ac-positive NDRs were subcategorized based on their 5’ (left) and 3’ (right) distance to the associated TSS (TSS, black middle line; 0–1kb, light grey; 1–3kb, purple; 3–5kb, blue; 5–10kb, green; 10–100kb, medium grey; >100kb, dark grey).
In accordance with our hypothesis, differentiation of granulosa cells was accompanied by an increase in gonad-specific NDRs neighboring granulosa-promoting genes (~2.5 NDRs/TSS), but not Sertoli-promoting genes (~1 NDR/TSS) (Fig. 2B). H3K27ac+ NDRs were similarly enriched around granulosa- (~1.5 NDRs/TSS) but not Sertoli-promoting genes (<1 NDR/TSS) (Fig. 2C). In contrast, differentiation of Sertoli cells was accompanied by an increase in gonad-specific NDRs neighboring both granulosa- and Sertoli-promoting genes (~2.5 NDRs/TSS) (Fig. 2B). However, H3K27ac+ NDRs, indicative of active enhancers, were significantly enriched mostly around Sertoli-promoting genes in Sertoli cells (~2 NDRs/TSS vs 1.6 NDRs/TSS around granulosa-promoting genes) (Fig. 2C). Therefore, H3K27ac-negative NDRs neighboring granulosa-promoting genes in Sertoli cells may be bound by repressors that block upregulation of the female-determining pathway (Li et al., 2014).
Our ATAC-seq data revealed that the genomic distribution of ubiquitous NDRs differed from gonad-specific NDRs in both time points and sexes. Around 40% of ubiquitous NDRs overlapped promoters, ~25% overlapped an intron and ~25% fell within an intergenic region (Fig. 2D). Accordingly, the majority (~60%) of ubiquitous NDRs fell within 1kb of the TSS (Fig. 2E). In contrast, only ~10% of all gonad-specific and H3K27ac+ gonad-specific NDRs overlapped a promoter, whereas ~40% fell within an intron and ~50% were intergenic (Fig. 2D). Accordingly, ~10% of all gonad-specific and H3K27ac+ gonad-specific NDRs fell within 1kb of the TSS, while the majority of gonad-specific NDRs were >10kb away (Fig. 2E). Our data suggests that NDRs that are common amongst different cell types largely represent promoters, whereas NDRs that are specific to the supporting cell lineage are mostly intergenic, representative of distal regulatory elements, and primarily neighbor granulosa- or Sertoli-promoting genes.
Gonad-specific NDRs are enriched for transcription factor binding motifs that promote supporting cell development
To identify transcription factor binding sites that potentially bind putative granulosa and Sertoli regulatory elements, we conducted a TF binding motif analysis using HOMER. The TF binding motifs identified within ubiquitous NDRs were very similar between both time points and sexes, with CTCF being the top represented motifs in all categories (Table 1 and Table S1). The prevalence of CTCF binding sites in ubiquitous NDRs is consistent with previous literature that shows high conservation of CTCF binding sites amongst different cell types (Kim et al., 2007, Xi et al., 2007). Other motifs identified in ubiquitous NDRs were for several members of the E26 transformation-specific (ETS) family of transcription factors (Table 1 and Table S1). These ubiquitous transcription factors have a wide range of functions during development including cell differentiation, cell cycle control, proliferation and apoptosis (Sharrocks, 2001). In addition to ETS factors, binding motifs for SP1, YY1, and GABPA identified in ubiquitous NDRs are commonly found in promoters, further supporting our finding that ubiquitous NDRs largely represent promoters (Fig. 2D, E) (Lin et al., 2007).
Table 1. Transcription factor motif analysis.
TF motif analysis performed in HOMER for ubiquitous, all gonad-specific, and H3K27ac+ gonad-specific NDRs in XX (white) and XY (grey) supporting cells at E10.5 (top two rows) and E13.5 (bottom two rows). The top five transcription factor motif families (sorted by p-value; p<e10) are represented, along with the members of that family that are expressed in the gonadal supporting cell lineage in the corresponding sex and stage (Nef et al., 2005, Jameson et al., 2012b). Underlined genes have a critical role in gonadal development and/or sex determination (see text for citations). Additional TF motifs and information are listed in Table S1.
| Ubiquitous NDRs | Gonad-specific NDRs | H3K27ac+ gonad-specific NDRs | ||||
|---|---|---|---|---|---|---|
| Motif Family | Family members with gonadal expression | Motif Family | Family members with gonadal expression | Motif Family | Family members with gonadal expression | |
| E10.5 XX | CTCF | Ctcf | CTCF | Ctcf | HOX | Hoxa5, Hoxa9, Hoxb3,Hoxc4, Hoxc5, Hoxc6,Hoxc8, Hoxc9, Hoxd8,Hoxd9 |
| ETS | Elf1, Elf2, Elk3, Elk4,Ets1, Ets2, Etv3, Etv6,Ewsr1, Gabpa | HOX | Hoxa5, Hoxa9, Hoxb3,Hoxc4, Hoxc5, Hoxc6,Hoxc8, Hoxc9, Hoxd8,Hoxd9 | SOX | Sox4, Sox11, Sox12,Sox6, Sox9 | |
| Sp/KLF | Sp1, Klf11, Klf12, Klf6,Klf7 | REST | Rest | TEAD | Tead1, Tead2 | |
| NFY | Nfya, Nfyb, Nfyc | LHX | Lhx9 | LHX | Lhx9 | |
| YY | Yy1 | TEAD | Tead1, Tead2 | GATA | Gata2, Gata4, Gata6 | |
| E10.5 XY | CTCF | Ctcf | CTCF | Ctcf | HOX | Hoxa5, Hoxa9, Hoxb3, |
| ETS | Elf1, Elf2, Elk3, Elk4,Ets1, Ets2, Etv3, Etv6,Ewsr1, Gabpa | HOX | Hoxa5, Hoxa9, Hoxb3,Hoxc4, Hoxc5, Hoxc6,Hoxc8, Hoxc9, Hoxd8,Hoxd9 | TEAD | Tead1, Tead2 | |
| NFY | Nfya, Nfyb, Nfyc | REST | Rest | SOX | Sox4, Sox11, Sox12,Sox6, Sox9 | |
| Sp/KLF | Sp1, Klf11, Klf12, Klf13, Klf6, Klf7 | TEAD | Tead1, Tead2 | GATA | Gata2, Gata4, Gata6 | |
| YY | Yy1 | LHX | Lhx9 | LHX | Lhx9 | |
| E13.5 XX | CTCF | Ctcf | NR5A | Nr5a1 | NR5A | Nr5a1 |
| ETS | Elf1, Elf2, Elk4, Gabpa,Etv3, Etv6, Ets1, Ets2,Elf4, Elk1, Etv1 | NF1 | Nf1, Nf2 | NF1 | Nf1, Nf2 | |
| NFY | Nfya, Nfyb, Nfyc | CTCF | CTCF | GATA | Gata4, Gata6 | |
| Sp/KLF | Sp1, Klf11, Klf12, Klf4,Klf6, Klf7, Klf13, Klf9,Klf16, Klf2, Klf3, Klf8 | GATA | Gata4, Gata6 | LHX | Lhx9 | |
| YY | Yy1 | LHX | Lhx9 | TEAD | Tead1, Tead2 | |
| E13.5 XY | CTCF | Ctcf | NR5A | Nr5a1 | NR5A | Nr5a1 |
| ETS | Elf1, Elf2, Elk3, Elk4,Gabpa, Etv3, Etv5,Etv6, Ets2, Elf4, Elk1,Etv1 | CTCF | Ctcf | GATA | Gata4, Gata6 | |
| NFY | Nfya, Nfyb, Nfyc | NF1 | Nf1, Nf2 | SOX | Sox4, Sox11, Sox12,Sox8, Sox9 | |
| Sp/KLF | Sp1, Klf11, Klf4, Klf6,Klf7, Klf13, Klf9, Klf16, Klf2, Klf3, Klf8 | GATA | Gata4, Gata6 | NF1 | Nf1, Nf2 | |
| YY | Yy1 | DMRT | Dmrt1, Dmrt3 | DMRT | Dmrt1, Dmrt3 | |
CTCF binding sites were also enriched amongst gonad-specific NDRs, but not amongst H3K27ac+ gonad-specific NDRs (Table 1 and Table S1), reinforcing the idea that H3K27ac+ NDRs function as enhancers and not as other regulatory elements such as insulators (Creyghton et al., 2010, Rada-Iglesias et al., 2011).
At E10.5, other TF motifs enriched in gonad-specific NDRs in both sexes were for the Homeobox family of genes (HOX) and its subfamily Lim homeobox genes (LHX), GATA-binding transcription factors, and the TEA domain family (TEAD) (Table 1 and Table S1). While the HOX family of transcription factors are important for morphogenesis in all tissues, the LHX and GATA motifs are likely more specific to gonad morphogenesis, as Lhx9 and Gata4 are required for early gonad development in both sexes (Birk et al., 2000, Hu et al., 2013). Although TEAD motifs were highly represented, their role during gonad development is still unknown. Interestingly, at E10.5, SOX motifs, which likely represent the binding sites for the key Sertoli-promoting TFs SRY, SOX9 and SOX8 (Chaboissier et al., 2004, Barrionuevo et al., 2006), were only represented in the H3K27ac+ gonad-specific category in both XX and XY cells (Table 1 and Table S1).
At E13.5, LHX, GATA, and TEAD motifs were still present in gonad-specific and H3K27ac+ gonad-specific NDRs in both sexes, with the addition of binding motifs for Nuclear Factor 1 (NF1) and Nuclear receptor subfamily 5 group A member 2 (NR5A2), the latter as the top represented motif (Table 1 and Table S1). The Nr5a2 motif likely represents its sibling nuclear receptor Steroidogenic Factor 1 (SF1 or NR5A1), which is required for early gonad formation in both sexes and is important for the upregulation of Sox9 in Sertoli cells (Sekido and Lovell-Badge, 2008).
TCF, LHX and FOX motifs were prevalent in gonad-specific NDRs in granulosa cells (Table 1 and Table S1). Wnt signaling, which regulates target genes through downstream interactions between β-catenin and TCF/LEF factors, is essential for ovarian development (Behrens et al., 1996, Vainio et al., 1999). Likewise, the forkhead box member Foxl2 encodes a transcription factor that is required for the long-term maintenance of granulosa cell identity, as well as the repression of the male fate (Uhlenhaut et al., 2009). Finally, although Lhx9 is required for proper gonad formation of both sexes, Lhx9 becomes highly expressed in granulosa cells during sex determination (Birk et al., 2000). In contrast to granulosa cells, gonad-specific NDRs in Sertoli cells were highly enriched for SOX and DMRT motifs (Table 1 and Table S1). As mentioned previously, Sox9 and Sox8 play a crucial role in driving differentiation of progenitor cells towards the Sertoli cell lineage (Chaboissier et al., 2004, Barrionuevo et al., 2006), and Dmrt1 is required to prevent postnatal transdifferentiation of Sertoli cells into granulosa cells by repressing the female pathway (Matson et al., 2011). Taken together, our data shows that NDRs that are specific to gonads are initially enriched for binding motifs of TFs required for early gonad formation (likely LHX9, GATA4 and SF1), and they acquire NDRs enriched for binding motifs of TFs that drive either granulosa (likely TCF1, LHX9 and FOXL2) or Sertoli (likely SRY, SOX9, SOX8 and DMRT1).
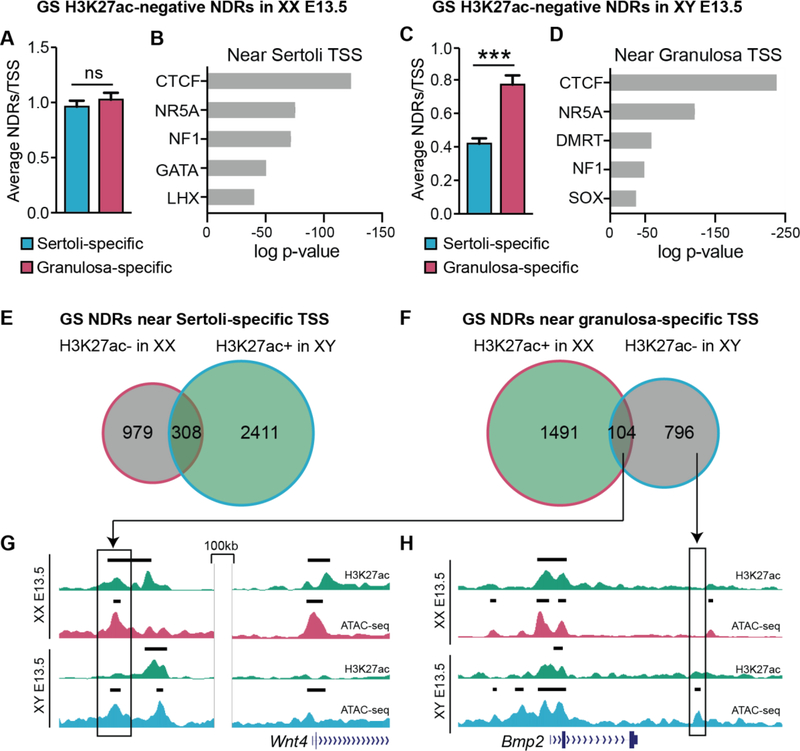
Gonad-specific H3K27ac-negative NDRs neighbor granulosa-promoting genes in Sertoli cells
As canalization of the male or female pathway requires both the upregulation of male- or female-promoting genes and simultaneous repression of the alternate fate, we sought to determine whether gonad-specific H3K27ac-negative NDRs are enriched around genes that promote the alternate pathway in Sertoli and granulosa cells, potentially acting as silencers. In granulosa cells, H3K27ac-negative NDRs were equally distributed around granulosa- and Sertoli-promoting genes, with an average of ~1 NDR per TSS (Fig. 3A). TF motif analysis of H3K27ac-negative NDRs around Sertoli-promoting genes in granulosa cells showed enrichment of motifs for the boundary proteins CTCF and NF1, and for the NR5A, GATA and LHX families (Fig. 3B). In contrast, H3K27ac-negative NDRs were significantly enriched around granulosa-promoting genes in Sertoli cells (~0.9 vs ~0.4 NDRs/TSS), suggesting that these NDRs could function as silencers of the granulosa cell fate (Fig. 3C). TF motif analysis of these potential silencers found that the DMRT and SOX motifs were highly represented (Fig. 3D). As DMRT1 and SOX9 are TFs that have previously been shown to block the upregulation of granulosa-promoting genes (Barrionuevo et al., 2006, Matson et al., 2011), this finding further supports a silencing role for H3K27ac-negative NDRs near granulosa-promoting genes in Sertoli cells.
Figure 3. Gonad-specific H3K27ac-negative NDRs are potential silencers of the alternate fate.
Analysis of gonad-specific (GS) H3K27ac-negative NDRs in granulosa cells (A&B), and GS H3K27ac-negative NDRs in Sertoli cells (C&D). A&C) Graphs showing the average number of NDRs associated to the nearest TSS as determined by HOMER. NDRs were associated to the nearest TSS of genes that become granulosa cell-specific (pink) and Sertoli cell-specific (blue). Error bars represent the standard error of the mean (SEM) and *** represents p<0.0001 as determined by a student’s t test. B&D) Top five motifs identified by TF binding motif analysis of protein families with members expressed in the gonad ranked in order of significance (p<e10, additional motifs in Table S1). E&F) Venn diagrams of H3K27ac-negative (grey) and H3K27ac-positive (green) NDRs in granulosa cells (pink outline) and Sertoli cells (blue outline), near Sertoli-specific TSS (E) and granulosa-specific TSS (F). G&H) Genome Browser tracks of ATAC-seq in granulosa (dark pink) and Sertoli (dark blue), and corresponding ChIP-seq for H3K27ac (green). Bold black lines above tracks represent significant enrichment compared to flanking regions as determined by HOMER. An NDR near the granulosa-promoting gene Wnt4 that is H3K27ac-positive in granulosa cells and H3K27ac-negative in Sertoli cells (G) and an NDR near the granulosa-promoting gene Bmp2 that is H3K27ac-negative in Sertoli cells and is not shared in granulosa cells (H) are shown.
Interestingly, we found that there is little overlap between gonad-specific NDRs that are H3K27ac-positive in one sex and H3K27ac-negative in the opposite sex (Fig. 3E and 3F). This suggests that while some regulatory elements can function as either enhancers or silencers depending on the sex (example Fig. 3G), most enhancers and silencers occupy distinct genomic regions (for example Fig. 3H). Taken together, our results suggest that gonad-specific NDRs that neighbor sex-determining genes and lack H3K27ac enrichment might function as silencers by binding repressors of the alternate fate.
Most NDRs arise de novo in differentiated Sertoli or granulosa cells
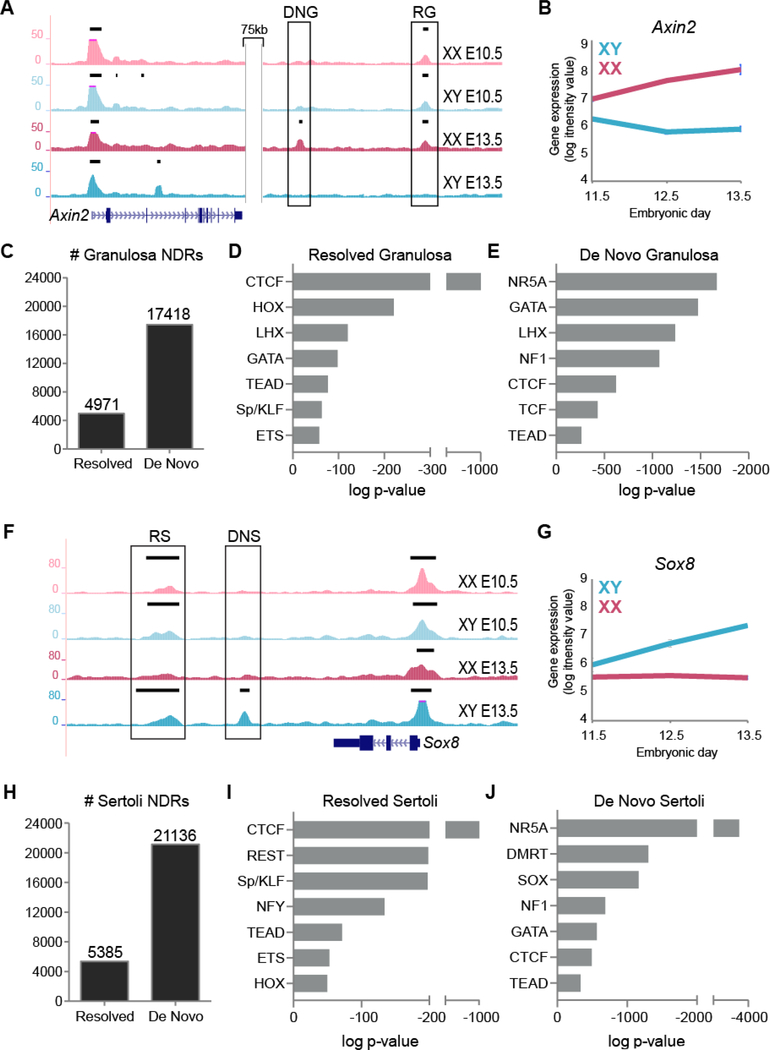
We next asked whether NDRs that are sex-specific at E13.5 are primarily inherited from the progenitor state or if chromatin is opening at regulatory elements during differentiation. To do this, we identified four classes of sex-specific NDRs: 1) NDRs that were present in both XX and XY E10.5 progenitor cells and remained open in granulosa cells but closed in Sertoli cells (resolved granulosa), 2) NDRs that were present in both XX and XY E10.5 progenitor cells and remained open in Sertoli cells but closed in granulosa cells (resolved Sertoli), 3) NDRs that were not present at E10.5 in either sex but appeared de novo in granulosa cells (de novo granulosa), and 4) NDRs that were not present at E10.5 in either sex but appeared de novo in Sertoli cells (de novo Sertoli). For example, both a granulosa-specific resolved and a granulosa-specific de novo NDR lie downstream of Axin2, a gene that becomes upregulated in granulosa cells (Fig. 4A and 4B). Similarly, a Sertoli-specific resolved and a Sertoli-specific de novo NDR lie downstream of Sox8, a Sertoli-promoting gene (Fig. 4F and 4G). Our data shows that de novo NDRs were ~4 times more prevalent than resolved NDRs in both granulosa and Sertoli cells (Fig. 4C and 4H).
Figure 4. Gonad-specific NDRs are either retained from the progenitor state or arise de novo.
A) Genome Browser tracks of ATAC-seq in XX E10.5 (light pink), XY E10.5 (light blue), granulosa (dark pink) and Sertoli (dark blue) purified cells of the genomic location surrounding Axin2. Bold black lines above tracks represent significant enrichment compared to flanking regions as determined by HOMER. A resolved granulosa (RG) and a de novo granulosa (DNG) NDR (boxed) lie downstream of Axin2. B) Axin2 gene expression profile in purified XX (pink) and XY (blue) supporting cells throughout sex determination (E11.5-E13.5) (Jameson et al., 2012b). C) Number of resolved and de novo NDRs in granulosa cells. D and E) Top motifs of families with members expressed in the gonad identified by TF binding motif analysis in resolved and de novo NDRs in granulosa cells ranked in order of significance (additional motifs in Table S1). F) Genome Browser tracks of ATAC-seq in XX E10.5 (light pink), XY E10.5 (light blue), granulosa (dark pink) and Sertoli (dark blue) purified cells of the genomic location surrounding Sox8. Bold black lines above tracks represent significant enrichment compared to flanking regions as determined by HOMER. A resolved Sertoli (RS) and a de novo Sertoli (DNS) NDR (boxed) lie downstream of Sox8. G) Sox8 gene expression profile in purified XX (pink) and XY (blue) supporting cells throughout sex determination (E11.5-E13.5) (Jameson et al., 2012b). H) Number of resolved and de novo NDRs in Sertoli cells. I and J) Top motif families (with members expressed in the gonad) identified by TF binding motif analysis in resolved and de novo NDRs in Sertoli cells ranked in order of significance (additional motifs in Table S1).
To understand whether resolved NDRs and de novo NDRs could have different biological functions, we performed TF motif analyses for each category using HOMER. Interestingly, we found that resolved NDRs were enriched for different TF motifs than de novo NDRs in both male and females. Furthermore, NDRs which resolved as either Sertoli- or granulosa-specific also differed from each other (with the exception of CTCF) despite being present in both sexes at E10.5 (Fig. 4D-E, and 4I-J, and Table S1).
In granulosa cells, resolved NDRs were enriched for HOX, LHX and GATA motifs, similar to motifs identified in gonad-specific NDRs at E10.5, that play an important role in early gonad development (Fig. 4D, Table 1 and Table S1). Interestingly, resolved Sertoli NDRs were enriched for different TFs, such as RE1 Silencing Transcription Factor (REST), Specificity Protein 1 (Sp1), Nuclear Transcription Factor Y (Nfy), TEAD factors, and several Kruppel-like Factors (Klf), which likely represent KLF4, a transcription factor involved in Sertoli cell differentiation (Godmann et al., 2008) (Fig. 4I and Table S1).
In de novo granulosa NDRs, HOX motifs were no longer enriched, but GATA and LHX binding sites remained highly represented along with motifs for NR5A and NF1 (Fig. 4E). TCF motifs were also identified amongst the top 25 represented motifs, but surprisingly, no FOX motifs were present in either resolved nor de novo granulosa NDRs (Table S1). In de novo Sertoli NDRs, binding sites for NR5A, SOX and DMRT genes dominated (Fig. 4J). Amongst the top 25 motifs were also members of the ETS family of proteins (Table S1), which can function downstream of the Fgf signaling cascade, a pathway important for repressing the female fate and promoting Sertoli cell development (Kim et al., 2006).
Our data show that the majority of granulosa- and Sertoli-specific NDRs arise de novo during sex determination and are distinct from each other as they contain binding sites of TFs that promote either granulosa or Sertoli cell differentiation. A smaller proportion (around 25%) of NDRs were present in both sexes prior to sex determination and retained only in one sex after sex determination. These NDRs were also enriched for different TF binding motifs between granulosa and Sertoli cells. Together, our data suggests that NDRs that become sex-specific by E13.5 promote the divergence of granulosa and Sertoli cells from their common progenitor, whether or not they were present prior to differentiation.
ATAC-seq identifies a novel enhancer downstream of Bmp2
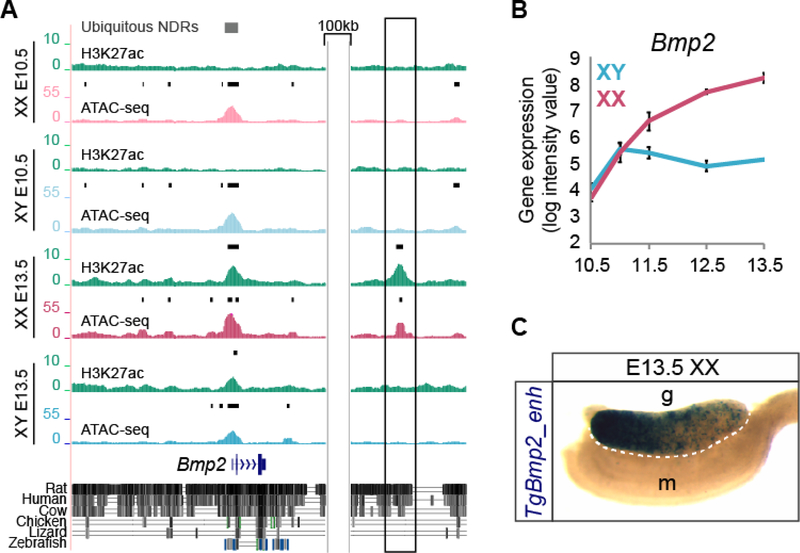
Until now, only a few distal regulatory elements have been identified for sex-determining genes (Sekido and Lovell-Badge, 2008, Maatouk et al., 2017, Gonen et al., 2018). Our data set provides an excellent resource to identify distal regulatory elements, including functional enhancers that regulate Sertoli or granulosa cell development. To demonstrate this, we performed a transient transgenic assay with a putative enhancer downstream of Bmp2. We identified a 922bp-long, H3K27ac+ gonad-specific NDR that arises de novo in granulosa cells and lies 139kb downstream of the Bmp2 TSS (its closest TSS) (Fig. 5A). Previous studies have shown that Bmp2 functions downstream of Wnt4 during ovarian development and synergizes with FOXL2 to upregulate Fst, a female-specific gene which causes sex reversal when disrupted (Kashimada et al., 2011, Yao et al., 2004). As Bmp2 becomes upregulated in granulosa cells during sex determination (Fig. 5B) (Jameson et al., 2012b), we speculated that this putative enhancer might be regulating female-specific expression of Bmp2. To test the activity of this putative enhancer in vivo, we cloned it upstream of an Hsp68-LacZ reporter plasmid and used this to generate transient transgenic mice. Despite some variation in the patterns of LacZ expression in our reporter embryos, which might be due to differences in the copy number or the integration site of our reporter plasmid, our reporter assay showed that this putative Bmp2 enhancer was active in XX E13.5 gonads (7/10 (70%) of transgenic embryos examined), but not in the mesonephros, consistent with its cell-type specificity (Fig. 5C and Fig. S3). Despite this NDR being present only in granulosa cells, our transgenic reporter was also active in 2/12 (16%) of XY gonads of transgenic embryos (Fig. S4). We identified 6 SOX, 3 TCF and 8 LHX consensus binding motifs within the putative Bmp2 enhancer (Fig. S5), possibly explaining how this enhancer-reporter construct can function in both granulosa and Sertoli cells depending on which TF it interacts with during early gonadal development. The ability to drive LacZ expression in gonads of both sexes despite the putative enhancer being present in only one sex is consistent with previous findings (Gonen et al., 2018) and further highlights the bipotential nature of early supporting progenitors. Finally, the putative Bmp2 enhancer shows sequence conservation with humans, suggesting that it may have an important role during human sex determination (Fig. 5A). These results demonstrate that our dataset can be used to identify functional gonad-specific enhancers that may be playing a critical role during gonadal sex determination by establishing granulosa- and Sertoli-specific transcriptional programs.
Figure 5. Transient transgenic analysis of a granulosa-specific NDR downstream of Bmp2.
A) Genome Browser tracks of ATAC-seq in XX E10.5 (light pink), XY E10.5 (light blue), granulosa (dark pink) and Sertoli (dark blue) purified cells and corresponding H3K27ac ChIP-seq profiles (green) at the genomic location surrounding Bmp2. Bold black lines above tracks represent significant enrichment compared to flanking regions as determined by HOMER. Grey boxes represent ubiquitous NDRs (top). Sequence conservation between rat, human, cow, chicken, lizard, and zebrafish are shown below. A de novo gonad-specific H3K27ac+ granulosa NDR downstream of Bmp2 (boxed) was selected for transient transgenic analysis and cloned into an hsp68-LacZ reporter cassette. B) Gene expression profile in purified XX (pink) and XY (blue) supporting cells throughout sex determination (E10.5-E13.5) showing that Bmp2 becomes granulosa-specific (Nef et al., 2005). C) An E13.5 XX gonad from a transgenic embryo shows β-galactosidase expression (TgBmp2_enh, dark blue) in the gonad (g), but not the mesonephros (m). TgBmp2_enh expression is stronger in the anterior pole of the gonad (left).
DISCUSSION AND CONCLUSIONS
Cis-regulatory elements, such as enhancers and silencers, play a critical role during development by coordinating the precise spatiotemporal expression of each gene within a transcriptional network. Consequently, disruption of these elements has the potential to cause congenital disorders. To increase our understanding of the complex and antagonistic developmental networks underlying mammalian sex determination, we identified in vivo cis-regulatory elements by employing whole-genome approaches.
We used ATAC-seq to assay for regions of open chromatin coupled with ChIP-seq for H3K27ac, a marker of active enhancers and promoters (Creyghton et al., 2010), in purified XX and XY supporting cells before (E10.5) and after (E13.5) gonadal sex determination. We show that prior to sex determination, XX and XY progenitor cells have similar chromatin accessibility patterns that resolve into sex-specific patterns as they differentiate into either granulosa or Sertoli cells. To distinguish NDRs common to many cell types (ubiquitous) from those found only in the supporting cell lineage (gonad-specific), we made use of previously published DNaseI-seq data from mouse ESCs, fibroblasts, heart, liver, kidney, and brain (Maatouk et al., 2017). Although these NDRs were identified by DNaseI-seq, ATAC-seq was previously shown to have a high correlation with DNaseI-seq, with the advantage of requiring very few numbers of cells (Buenrostro et al., 2013). We found that, in contrast to ubiquitous NDRs, gonad-specific NDRs as well as H3K27ac+ gonad-specific NDRs were enriched around granulosa-promoting genes in granulosa cells and Sertoli-promoting genes in Sertoli cells. Furthermore, we show that binding motifs for TFs that promote granulosa- or Sertoli-development are enriched in gonadspecific NDRs that neighbor granulosa- or Sertoli-promoting genes, respectively. Interestingly, we found that gonad-specific H3K27ac-negative NDRs were more likely to neighbor granulosa-promoting genes than Sertoli-promoting genes in Sertoli cells, and were enriched for DMRT and SOX TF binding motifs, known repressors of the female pathway. Finally, we demonstrate the power of our dataset to identify enhancers by characterizing a novel enhancer downstream of Bmp2 that functions in gonads of transgenic mice as predicted.
The progenitor cells of the gonad exist in a bipotential state with the ability to differentiate into either Sertoli (XY) or granulosa (XX) cells depending on the developmental signal they receive (Albrecht and Eicher, 2001). Extensive transcriptional profiling of these cells at E10.5 has shown that less than 1% of genes are transcribed in a sexually dimorphic way and are likely Y-linked genes or X-linked genes that escape X inactivation (Nef et al., 2005, Jameson et al., 2012b, Munger et al., 2013). Therefore, autosomal genes that promote Sertoli or granulosa cell development (such as SOX and Wnt signaling genes) are initially transcribed at similar levels in both XX and XY progenitor cells. However, despite increasing evidence that cell differentiation is epigenetically regulated, it remains unclear whether the chromatin state of progenitor cells plays a role in maintaining their bipotential nature and establishing their male or female fate. We predicted that the chromatin accessibility landscape of XX and XY progenitor cells would also be similar. Accordingly, we found that the vast majority (~90%) of NDRs are shared between males and females prior to gonadal sex determination, and the enrichment patterns of TF binding motifs within these NDRs are nearly identical in XX and XY supporting cells at this stage. Interestingly, gonad-specific NDRs were enriched around granulosa-promoting genes in both sexes at E10.5, suggesting that both XX and XY progenitor cells are predisposed towards the granulosa cell lineage. This is consistent with previous findings that show that there are more granulosa-promoting genes expressed at the bipotential stage than there are Sertoli-promoting genes (Jameson et al., 2012b).
An intriguing finding was an enrichment of SOX binding motifs in H3K27ac+ gonad-specific NDRs at E10.5 in both sexes. This suggests that regions for SOX binding sites are not only accessible but active at E10.5. As SRY and SOX genes share at least 50% homology in their DNA binding domains, the presence of SOX motifs at E10.5 could also represent binding sites for SRY itself (Denny et al., 1992). This finding indicates that the bipotential supporting lineage is poised to rapidly engage the male pathway upon Sry’s transient activation and downstream Sox9 upregulation, regardless of its chromosomal complement. In support of this, transgenic expression of Sry in early XX gonads is sufficient to drive Sertoli cell development (Koopman et al., 1991, Hiramatsu et al., 2009). In absence of Sry or if Sry expression is delayed outside of its functional window (~12 to 15 tail somites), the pro-ovarian Wnt signaling pathway is not repressed and the female pathway becomes engaged (Hiramatsu et al., 2009). Therefore, the absence of SOX motifs in granulosa cell NDRs likely represents a closure of early SOX motif-containing NDRs that were not bound by Sry nor Sox9 (Vainio et al., 1999, Chassot et al., 2008). The similarity of the enhancer distribution and binding sites within these enhancers between XX and XY cells prior to sex determination is consistent with, and possibly contributes to, their similar transcriptional profiles and ability to engage either pathway.
As enhancers play a critical role in establishing tissue-specific gene expression patterns during development, we predicted that differentiation of Sertoli and granulosa cells would require enhancers that upregulate Sertoli- and granulosa-promoting genes, and that these enhancers would be gonad-specific. In accordance with our hypothesis, we found only 55% of NDRs were shared between Sertoli and granulosa cells at E13.5 in contrast to ~90% at E10.5. As expected, H3K27ac+ gonad-specific NDRs were more likely to neighbor granulosa-promoting genes in granulosa cells, and Sertoli-promoting genes in Sertoli cells, suggesting that these enhancers establish granulosa- and Sertoli-specific transcriptional profiles. In support of this idea, our data shows that granulosa-specific NDRs are enriched with binding sites of TFs important for granulosa cell differentiation such as TCF and LHX factors, whereas Sertoli-specific NDRs are enriched for SOX and DMRT motifs, TFs required for Sertoli cell development and maintenance. The sibling nuclear receptors SF1 (NR5A1) and NR5A2 share a binding motif, identified in the HOMER database as NR5A2, which was the most significantly represented motif in both XX and XY gonad-specific and H3K27ac+ gonad-specific NDRs. Sf1 is crucial for early gonadogenesis in both sexes as well as for the initiation of the male pathway through Sox9 signaling (Sekido and Lovell-Badge, 2008). Importantly, FOXL2, which is required for ovarian development and maintenance, has also been shown to bind this motif (Benayoun et al., 2008). Since the precise binding motif for FOXL2 is not included in HOMER’s TF motif database, we speculate that the NR5A2 motifs identified in Sertoli and granulosa cell NDRs in our analyses represent both SF1 and FOXL2; however, further analyses using ChIP is needed to determine which TFs are binding these sites.
Supporting cells can transdifferentiate into their alternate fate long after sex determination has occurred, emphasizing the need for repressive mechanisms even during adulthood (Matson et al., 2011, Uhlenhaut et al., 2009). We found that gonad-specific NDRs which lack H3K27ac and are therefore unlikely to function as enhancers, were more likely to neighbor granulosa-promoting genes than Sertoli-promoting genes in Sertoli cells. Enrichment of binding motifs for DMRT1 and SOX9 suggest that these NDRs might function as silencers by binding repressors of the granulosa cell fate. Interestingly, H3K27ac-negative NDRs were similarly enriched around granulosa- and Sertoli-promoting genes in granulosa cells. This data support previous findings that show that initiation of the male pathway requires upregulation of the testis pathway and simultaneous repression of the ovarian pathway, while commitment to the female fate mostly depends on the continuous activation of female-promoting genes (Munger et al., 2013). This suggests that repression of female-promoting genes is a more critical component of male fate commitment. Future studies on repressive histone modifications such as H3K27me3 will provide more evidence for the silencing function of these H3K27ac-negative NDRs.
Interestingly, our data also found that the majority (~75%) of sex-specific NDRs were not inherited from a progenitor state, but rather appeared de novo following sex determination. When we analyzed the TF motifs enriched in NDRs that resolved from a progenitor state into sex-specific NDRs versus those that arise de novo, we found that each category was enriched for different TFs. De novo NDRs were enriched for TFs that drive supporting cell development, similar to the gonad-specific NDRs discussed above. Of interest, binding sites for CTCF were present in both resolved granulosa and resolved Sertoli NDRs, suggesting that different CTCF sites are maintained during sex determination depending on whether bipotential cells engage the male or female pathway. CTCF factors are important for the formation of TADs, higher-order chromatin structures that regulate promoter-enhancer interactions by confining them into specific regions (Ghirlando and Felsenfeld, 2016). Taken together, our data suggests that chromatin undergoes extensive remodeling within the supporting cell lineage throughout sex determination, including loss and gain of sex-specific NDRs that interact with different TFs and promoters.
In addition to increasing our understanding of the role that chromatin remodeling plays in vivo during a key developmental event, this dataset will provide a useful tool for the identification of functional gonad-specific enhancers and associated TFs that drive mammalian sex determination. As proof of concept, we identified a de novo granulosa enhancer 139kb downstream of the granulosa-promoting gene Bmp2 and showed that it was functional in ovaries of transgenic mice by performing an in vivo reporter assay. Our finding that the majority of gonad-specific NDRs fall >10kb away from a TSS highlights the importance of using unbiased, genome-wide techniques for the identification of critical regulatory elements. In addition to this putative Bmp2 enhancer, our dataset led to the identification of an enhancer 565kb upstream of Sox9 (named Enhancer 13) that, when deleted, caused complete male-to-female sex reversal (Gonen et al., 2018). Although identified in mice, Enh13 maps to a region of the human genome known as XY SR, deletions and duplications of which have resulted in human DSDs (Kim et al., 2015). Thus, our dataset is not only predictive of enhancers driving expression of sex-determining genes in mice but has the potential to increase the number of diagnoses in human DSDs with unknown etiologies.
Supplementary Material
Highlights.
Supporting gonadal cells initiate sex determination with similar chromatin landscapes
Differentiated Sertoli and granulosa cells acquire sex-specific chromatin landscapes
Enhancers are enriched for motifs of TFs that drive Sertoli or granulosa cell fate
Silencers neighbor granulosa genes in Sertoli cells
ATAC-seq and ChIP-seq identified a novel enhancer downstream of Bmp2
ACKNOWLEDGEMENTS
We thank the NU Sequencing core for their help with ChIP-seq and ATAC-seq, and the R. H. Lurie Comprehensive Cancer Center Flow Cytometry Facility for their help with FACS-sorting. We thank the Genetic Modification Service of the Francis Crick Institute for their help with the transient transgenics. We’d like to thank Peter van Galen for his help with the ChIP-seq protocol, and Lindsey Barske and Blanche Capel for their comments on the manuscript. Research was partially supported by the NIH to SAGM (NIH T32 GM008061), and partly by Francis Crick core funding (Cancer Research UK, the U.K. Medical Research Council, and the Wellcome Trust: FC001107).
Footnotes
COMPETING INTERESTS
The authors have no competing interests to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- ALBRECHT KH & EICHER EM 2001. Evidence that Sry is expressed in pre-Sertoli cells and Sertoli and granulosa cells have a common precursor. Dev Biol, 240, 92–107. [DOI] [PubMed] [Google Scholar]
- BARRIONUEVO F, BAGHERI-FAM S, KLATTIG J, KIST R, TAKETO MM, ENGLERT C & SCHERER G 2006. Homozygous inactivation of Sox9 causes complete XY sex reversal in mice. Biol Reprod, 74, 195–201. [DOI] [PubMed] [Google Scholar]
- BEHRENS J, VON KRIES JP, KUHL M, BRUHN L, WEDLICH D, GROSSCHEDL R & BIRCHMEIER W 1996. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature, 382, 638–42. [DOI] [PubMed] [Google Scholar]
- BENAYOUN BA, CABURET S, DIPIETROMARIA A, BAILLY-BECHET M, BATISTA F, FELLOUS M, VAIMAN D & VEITIA RA 2008. The identification and characterization of a FOXL2 response element provides insights into the pathogenesis of mutant alleles. Hum Mol Genet, 17, 3118–27. [DOI] [PubMed] [Google Scholar]
- BENKO S, GORDON CT, MALLET D, SREENIVASAN R, THAUVIN-ROBINET C, BRENDEHAUG A, THOMAS S, BRULAND O, DAVID M, NICOLINO M, LABALME A, SANLAVILLE D, CALLIER P, MALAN V, HUET F, MOLVEN A, DIJOUD F, MUNNICH A, FAIVRE L, AMIEL J, HARLEY V, HOUGE G, MOREL Y & LYONNET S 2011. Disruption of a long distance regulatory region upstream of SOX9 in isolated disorders of sex development. J Med Genet, 48, 825–30. [DOI] [PubMed] [Google Scholar]
- BERNARD P, RYAN J, SIM H, CZECH DP, SINCLAIR AH, KOOPMAN P & HARLEY VR 2012. Wnt signaling in ovarian development inhibits Sf1 activation of Sox9 via the Tesco enhancer. Endocrinology, 153, 901–12. [DOI] [PubMed] [Google Scholar]
- BEVERDAM A & KOOPMAN P 2006. Expression profiling of purified mouse gonadal somatic cells during the critical time window of sex determination reveals novel candidate genes for human sexual dysgenesis syndromes. Hum Mol Genet, 15, 417–31. [DOI] [PubMed] [Google Scholar]
- BIRK OS, CASIANO DE, WASSIF CA, COGLIATI T, ZHAO L, ZHAO Y, GRINBERG A, HUANG S, KREIDBERG JA, PARKER KL, PORTER FD & WESTPHAL H 2000. The LIM homeobox gene Lhx9 is essential for mouse gonad formation. Nature, 403, 909–13. [DOI] [PubMed] [Google Scholar]
- BUENROSTRO JD, GIRESI PG, ZABA LC, CHANG HY & GREENLEAF WJ 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods, 10, 1213–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BULLEJOS M & KOOPMAN P 2001. Spatially dynamic expression of Sry in mouse genital ridges. Dev Dyn, 221, 201–5. [DOI] [PubMed] [Google Scholar]
- BURGOYNE PS, THORNHILL AR, BOUDREAN SK, DARLING SM, BISHOP CE & EVANS EP 1995. The genetic basis of XX-XY differences present before gonadal sex differentiation in the mouse. Philos Trans R Soc Lond B Biol Sci, 350, 253–60 discussion 260–1. [DOI] [PubMed] [Google Scholar]
- CHABOISSIER MC, KOBAYASHI A, VIDAL VI, LUTZKENDORF S, VAN DE KANT HJ, WEGNER M, DE ROOIJ DG, BEHRINGER RR & SCHEDL A 2004. Functional analysis of Sox8 and Sox9 during sex determination in the mouse. Development, 131, 1891–901. [DOI] [PubMed] [Google Scholar]
- CHASSOT AA, RANC F, GREGOIRE EP, ROEPERS-GAJADIEN HL, TAKETO MM, CAMERINO G, DE ROOIJ DG, SCHEDL A & CHABOISSIER MC 2008. Activation of beta-catenin signaling by Rspo1 controls differentiation of the mammalian ovary. Hum Mol Genet, 17, 1264–77. [DOI] [PubMed] [Google Scholar]
- CREYGHTON MP, CHENG AW, WELSTEAD GG, KOOISTRA T, CAREY BW, STEINE EJ, HANNA J, LODATO MA, FRAMPTON GM, SHARP PA, BOYER LA, YOUNG RA & JAENISCH R 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A, 107, 21931–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DEKKER J, RIPPE K, DEKKER M & KLECKNER N 2002. Capturing chromosome conformation. Science, 295, 1306–11. [DOI] [PubMed] [Google Scholar]
- DENNY P, SWIFT S, BRAND N, DABHADE N, BARTON P & ASHWORTH A 1992. A conserved family of genes related to the testis determining gene, SRY. Nucleic Acids Res, 20, 2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EGGERS S, SADEDIN S, VAN DEN BERGEN JA, ROBEVSKA G, OHNESORG T, HEWITT J, LAMBETH L, BOUTY A, KNARSTON IM, TAN TY, CAMERON F, WERTHER G, HUTSON J, O'CONNELL M, GROVER SR, HELOURY Y, ZACHARIN M, BERGMAN P, KIMBER C, BROWN J, WEBB N, HUNTER MF, SRINIVASAN S, TITMUSS A, VERGE CF, MOWAT D, SMITH G, SMITH J, EWANS L, SHALHOUB C, CROCK P, COWELL C, LEONG GM, ONO M, LAFFERTY AR, HUYNH T, VISSER U, CHOONG CS, MCKENZIE F, PACHTER N, THOMPSON EM, COUPER J, BAXENDALE A, GECZ J, WHEELER BJ, JEFFERIES C, MACKENZIE K, HOFMAN P, CARTER P, KING RI, KRAUSZ C, VAN RAVENSWAAIJ-ARTS CM, LOOIJENGA L, DROP S, RIEDL S, COOLS M, DAWSON A, JUNIARTO AZ, KHADILKAR V, KHADILKAR A, BHATIA V, DUNG VC, ATTA I, RAZA J, THI DIEM CHI N, HAO TK, HARLEY V, KOOPMAN P, WARNE G, FARADZ S, OSHLACK A, AYERS KL & SINCLAIR AH 2016. Disorders of sex development: insights from targeted gene sequencing of a large international patient cohort. Genome Biol, 17, 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GHIRLANDO R & FELSENFELD G 2016. CTCF: making the right connections. Genes Dev, 30, 881–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GODMANN M, KATZ JP, GUILLOU F, SIMONI M, KAESTNER KH & BEHR R 2008. Kruppel-like factor 4 is involved in functional differentiation of testicular Sertoli cells. Dev Biol, 315, 552–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GONEN N, FUTTNER CR, WOOD S, GARCIA-MORENO SA, SALAMONE IM, SAMSON SC, SEKIDO R, POULAT F, MAATOUK DM & LOVELL-BADGE R 2018. Sex reversal following deletion of a single distal enhancer of Sox9. Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUBBAY J, COLLIGNON J, KOOPMAN P, CAPEL B, ECONOMOU A, MUNSTERBERG A, VIVIAN N, GOODFELLOW P & LOVELL-BADGE R 1990. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature, 346, 245–50. [DOI] [PubMed] [Google Scholar]
- HACKER A, CAPEL B, GOODFELLOW P & LOVELL-BADGE R 1995. Expression of Sry, the mouse sex determining gene. Development, 121, 1603–14. [DOI] [PubMed] [Google Scholar]
- HIRAMATSU R, MATOBA S, KANAI-AZUMA M, TSUNEKAWA N, KATOH-FUKUI Y, KUROHMARU M, MOROHASHI K, WILHELM D, KOOPMAN P & KANAI Y 2009. A critical time window of Sry action in gonadal sex determination in mice. Development, 136, 129–38. [DOI] [PubMed] [Google Scholar]
- HU YC, OKUMURA LM & PAGE DC 2013. Gata4 is required for formation of the genital ridge in mice. PLoS Genet, 9, e1003629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JAMESON SA, LIN YT & CAPEL B 2012a. Testis development requires the repression of Wnt4 by Fgf signaling. Dev Biol, 370, 24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JAMESON SA, NATARAJAN A, COOL J, DEFALCO T, MAATOUK DM, MORK L, MUNGER SC & CAPEL B 2012b. Temporal transcriptional profiling of somatic and germ cells reveals biased lineage priming of sexual fate in the fetal mouse gonad. PLoS Genet, 8, e1002575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KASHIMADA K, PELOSI E, CHEN H, SCHLESSINGER D, WILHELM D & KOOPMAN P 2011. FOXL2 and BMP2 act cooperatively to regulate follistatin gene expression during ovarian development. Endocrinology, 152, 272–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KIM GJ, SOCK E, BUCHBERGER A, JUST W, DENZER F, HOEPFFNER W, GERMAN J, COLE T, MANN J, SEGUIN JH, ZIPF W, COSTIGAN C, SCHMIADY H, ROSTASY M, KRAMER M, KALTENBACH S, ROSLER B, GEORG I, TROPPMANN E, TEICHMANN AC, SALFELDER A, WIDHOLZ SA, WIEACKER P, HIORT O, CAMERINO G, RADI O, WEGNER M, ARNOLD HH & SCHERER G 2015. Copy number variation of two separate regulatory regions upstream of SOX9 causes isolated 46,XY or 46,XX disorder of sex development. J Med Genet, 52, 240–7. [DOI] [PubMed] [Google Scholar]
- KIM TH, ABDULLAEV ZK, SMITH AD, CHING KA, LOUKINOV DI, GREEN RD, ZHANG MQ, LOBANENKOV VV & REN B 2007. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell, 128, 1231–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KIM Y, KOBAYASHI A, SEKIDO R, DINAPOLI L, BRENNAN J, CHABOISSIER MC, POULAT F, BEHRINGER RR, LOVELL-BADGE R & CAPEL B 2006. Fgf9 and Wnt4 act as antagonistic signals to regulate mammalian sex determination. PLoS Biol, 4, e187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KOOPMAN P, GUBBAY J, VIVIAN N, GOODFELLOW P & LOVELL-BADGE R 1991. Male development of chromosomally female mice transgenic for Sry. Nature, 351, 117–21. [DOI] [PubMed] [Google Scholar]
- LEE CK, SHIBATA Y, RAO B, STRAHL BD & LIEB JD 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet, 36, 900–5. [DOI] [PubMed] [Google Scholar]
- LI Y, ZHENG M & LAU YF 2014. The sex-determining factors SRY and SOX9 regulate similar target genes and promote testis cord formation during testicular differentiation. Cell Rep, 8, 723–33. [DOI] [PubMed] [Google Scholar]
- LIN JM, COLLINS PJ, TRINKLEIN ND, FU Y, XI H, MYERS RM & WENG Z 2007. Transcription factor binding and modified histones in human bidirectional promoters. Genome Res, 17, 818–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LYBAEK H, DE BRUIJN D, DEN ENGELSMAN-VAN DIJK AH, VANICHKINA D, NEPAL C, BRENDEHAUG A & HOUGE G 2014. RevSex duplication-induced and sex-related differences in the SOX9 regulatory region chromatin landscape in human fibroblasts. Epigenetics, 9, 416–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAATOUK DM, DINAPOLI L, ALVERS A, PARKER KL, TAKETO MM & CAPEL B 2008. Stabilization of beta-catenin in XY gonads causes male-to-female sex-reversal. Hum Mol Genet, 17, 2949–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAATOUK DM, NATARAJAN A, SHIBATA Y, SONG L, CRAWFORD GE, OHLER U & CAPEL B 2017. Genome-wide identification of regulatory elements in Sertoli cells. Development, 144, 720–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MATSON CK, MURPHY MW, SARVER AL, GRISWOLD MD, BARDWELL VJ & ZARKOWER D 2011. DMRT1 prevents female reprogramming in the postnatal mammalian testis. Nature, 476, 101–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MUNGER SC, NATARAJAN A, LOOGER LL, OHLER U & CAPEL B 2013. Fine time course expression analysis identifies cascades of activation and repression and maps a putative regulator of mammalian sex determination. PLoS Genet, 9, e1003630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NEF S, SCHAAD O, STALLINGS NR, CEDERROTH CR, PITETTI JL, SCHAER G, MALKI S, DUBOIS-DAUPHIN M, BOIZET-BONHOURE B, DESCOMBES P, PARKER KL & VASSALLI JD 2005. Gene expression during sex determination reveals a robust female genetic program at the onset of ovarian development. Dev Biol, 287, 361–77. [DOI] [PubMed] [Google Scholar]
- PARMA P, RADI O, VIDAL V, CHABOISSIER MC, DELLAMBRA E, VALENTINI S, GUERRA L, SCHEDL A & CAMERINO G 2006. R-spondin1 is essential in sex determination, skin differentiation and malignancy. Nat Genet, 38, 1304–9. [DOI] [PubMed] [Google Scholar]
- RADA-IGLESIAS A, BAJPAI R, SWIGUT T, BRUGMANN SA, FLYNN RA & WYSOCKA J 2011. A unique chromatin signature uncovers early developmental enhancers in humans. Nature, 470, 279–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SEKIDO R, BAR I, NARVAEZ V, PENNY G & LOVELL-BADGE R 2004. SOX9 is up-regulated by the transient expression of SRY specifically in Sertoli cell precursors. Dev Biol, 274, 271–9. [DOI] [PubMed] [Google Scholar]
- SEKIDO R & LOVELL-BADGE R 2008. Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature, 453, 930–4. [DOI] [PubMed] [Google Scholar]
- SHARROCKS AD 2001. The ETS-domain transcription factor family. Nat Rev Mol Cell Biol, 2, 827–37. [DOI] [PubMed] [Google Scholar]
- SINCLAIR AH, BERTA P, PALMER MS, HAWKINS JR, GRIFFITHS BL, SMITH MJ, FOSTER JW, FRISCHAUF AM, LOVELL-BADGE R & GOODFELLOW PN 1990. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature, 346, 240–4. [DOI] [PubMed] [Google Scholar]
- SONG L, ZHANG Z, GRASFEDER LL, BOYLE AP, GIRESI PG, LEE BK, SHEFFIELD NC, GRAF S, HUSS M, KEEFE D, LIU Z, LONDON D, MCDANIELL RM, SHIBATA Y, SHOWERS KA, SIMON JM, VALES T, WANG T, WINTER D, ZHANG Z, CLARKE ND, BIRNEY E, IYER VR, CRAWFORD GE, LIEB JD & FUREY TS 2011. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res, 21, 1757–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UHLENHAUT NH, JAKOB S, ANLAG K, EISENBERGER T, SEKIDO R, KRESS J, TREIER AC, KLUGMANN C, KLASEN C, HOLTER NI, RIETHMACHER D, SCHUTZ G, COONEY AJ, LOVELL-BADGE R & TREIER M 2009. Somatic sex reprogramming of adult ovaries to testes by FOXL2 ablation. Cell, 139, 1130–42. [DOI] [PubMed] [Google Scholar]
- VAINIO S, HEIKKILA M, KISPERT A, CHIN N & MCMAHON AP 1999. Female development in mammals is regulated by Wnt-4 signalling. Nature, 397, 405–9. [DOI] [PubMed] [Google Scholar]
- VAN GALEN P, VINY AD, RAM O, RYAN RJ, COTTON MJ, DONOHUE L, SIEVERS C, DRIER Y, LIAU BB, GILLESPIE SM, CARROLL KM, CROSS MB, LEVINE RL & BERNSTEIN BE 2016. A Multiplexed System for Quantitative Comparisons of Chromatin Landscapes. Mol Cell, 61, 170–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WAGNER T, WIRTH J, MEYER J, ZABEL B, HELD M, ZIMMER J, PASANTES J, BRICARELLI FD, KEUTEL J, HUSTERT E, WOLF U, TOMMERUP N, SCHEMPP W & SCHERER G 1994. Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell, 79, 1111–20. [DOI] [PubMed] [Google Scholar]
- XI H, SHULHA HP, LIN JM, VALES TR, FU Y, BODINE DM, MCKAY RD, CHENOWETH JG, TESAR PJ, FUREY TS, REN B, WENG Z & CRAWFORD GE 2007. Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome. PLoS Genet, 3, e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YAO HH, MATZUK MM, JORGEZ CJ, MENKE DB, PAGE DC, SWAIN A & CAPEL B 2004. Follistatin operates downstream of Wnt4 in mammalian ovary organogenesis. Dev Dyn, 230, 210–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.