Abstract
Since previous interspecies comparisons of Caulobacter genomes have revealed extensive genome rearrangements, we decided to compare the nucleotide sequences of four C. crescentus genomes, NA1000, CB1, CB2, and CB13. To accomplish this goal, we used PacBio sequencing technology to determine the nucleotide sequence of the CB1, CB2, and CB13 genomes, and obtained each genome sequence as a single contig. To correct for possible sequencing errors, each genome was sequenced twice. The only differences we observed between the two sets of independently determined sequences were random omissions of a single base in a small percentage of the homopolymer regions where a single base is repeated multiple times. Comparisons of these four genomes indicated that horizontal gene transfer events that included small numbers of genes occurred at frequencies in the range of 10−3 to 10−4 insertions per generation. Large insertions were about 100 times less frequent. Also, in contrast to previous interspecies comparisons, we found no genome rearrangements when the closely-related NA1000, CB1, and CB2 genomes were compared, and only eight inversions and one translocation when the more distantly related CB13 genome was compared to the other genomes. Thus, we estimate that inversions occur at a rate of one per 10 to 12 million generations in Caulobacter genomes. The inversions seem to be complex events that include the simultaneous creation of indels.
Introduction
Mutations occur in the DNA of all organisms both during DNA replication and in response to DNA damage. In bacteria, genomes also can change rapidly due to the deletion of large segments of the chromosome. Alternatively they can acquire large pieces of DNA due to horizontal gene transfer (HGT) [12, 18, 19]. HGT can result in the acquisition of plasmid DNA that confers new properties on its host, or it can involve the acquisition of a piece of chromosomal or environmental DNA that becomes incorporated into the bacterial chromosome. These acquired pieces of DNA can vary in size from a few hundred nucleotides to tens of thousands of nucleotides with the latter often mediated by bacteriophage or transposable elements [18].
It is clear that HGT impacts bacterial genome evolution but the magnitude of this impact has been debated [4]. Some have questioned whether a phylogenetic tree is even meaningful, citing the fact that gene trees can differ from genome trees [7]. Others have suggested that HGT occurs at a high frequency leading to few ubiquitous core genes [22]. However, Ochman et al. [18] estimated that E. coli acquires DNA by HGT at the rate of only 16 kb per million years, and recent data indicate that genomic segments obtained by HGT usually are obtained from close relatives [3]. Thus, it is important to compare the genomes of wild isolates of the same species to obtain accurate estimates about how HGT impacts genomes in natural situations. HGT has been well studied in laboratory experiments [26], but laboratory studies cannot incorporate either all of the variables that occur in the natural world, or the time scales of thousands or millions of years.
Recent developments in DNA sequencing technologies have greatly reduced the costs of sequencing bacterial genomes, and it is now practical to sequence genomes from large numbers of wild isolates of any type of bacteria or to sequence genomes of all members of any naturally occurring microbiome. Indeed, new bacterial genome sequences appear in the NCBI database on a daily basis. Unfortunately, most of these genomes are present in the database in draft form with the genome sequence present in hundreds of separate contigs (contiguous pieces of base pair sequence). This problem can be avoided by using high quality DNA and PacBio sequencing technologies to generate complete bacterial genome sequences. We and others have shown that the use of PacBio sequencing technologies to sequence a bacterial genome usually results in the entire genome being assembled into a single contig with any plasmids that are present assembled into additional contigs [21, 23, 25].
The Ely laboratory has had a longstanding interest in the genetics of Caulobacter [10], and recent studies have shown that the arrangement of the genes on the chromosome appears to be scrambled when the genomes of wild type strains of Caulobacter are compared [1, 24]. To better understand this phenomenon where gene order is maintained for only small blocks of genes, we decided to sequence and compare several genomes of a single Caulobacter species to minimize the genetic distances among the genomes being compared and hopefully minimize the extent of genome scrambling. Therefore, we determined the nucleotide sequence of three C. crescentus genomes and compared the resulting genomic sequences to that of the well-studied NA1000 genome version of the C. crescentus strain CB15 [16, 17]. Two of the C. crescentus wild type isolates, CB1 and CB2, were isolated by Dr. Jeanne S. Poindexter in 1960 from tap water [20]. CB13 and CB15 were isolated from the same pond near the UC Berkeley campus [20]. The available version of the original CB13 isolate is designated CB13B1a. When these four genome nucleotide sequences were compared, we found that genome rearrangements were observed only between distantly related members of the species, but HGT events were much more common.
Materials and Methods
Growth of Bacteria
C. crescentus wild type strains CB1 and CB13B1a were obtained from the Ely laboratory stock collection where they had been frozen at −72 C since 1975. C. crescentus strain CB2 was obtained from the American Type Culture Collection (ATCC 15252). Cultures of CB1, CB2, and CB13 were grown in 5 mL PYE Broth (0.2 % peptone, 0.1 % yeast extract, 0.5 mM CaCl2, and 0.8 mM MgSO4) overnight at 30°C on a rotator to allow aeration [14].
DNA Isolation
DNA was extracted from 2 ml of a pelleted and re-suspended overnight bacteria culture using the QIAamp DNA Mini Kit by Qiagen. The protocol provided by the manufacturer was utilized with the exception of adding a 1:1 ratio of 100% ethanol instead of 2:1 ratio of sample mixture to ethanol. The optional RNase treatment step was also completed.
Sequencing and Assembly
Extracted DNA was sent to the University of Washington, the Icahn School of Medicine, or the Delaware Bioinformatics Institute for genome sequencing and assembly using a Pacific Biosciences platform.
Confirmation of Correct Sequence via Sanger Sequencing
Representative homopolymer regions were resequenced utilizing Sanger sequencing. Primers for the regions of interest were created utilizing the Integrated DNA Technologies (IDT) Primer Quest Tool. PCR was carried out on an Eppendorf MasterCycler epgradient S. PCR amplification of the regions of interest were performed as follows: an initial denaturing time of 3 minutes at 94°C, the second denaturation at 94°C for 20 seconds, 20 seconds annealing time at 51°C, extension time of 30 seconds at 72°C, and final extension time of 5 minutes at 72°C after 35 cycles. The PCR products were sent to Eurofins Genomics for Sanger sequencing. Results were aligned against each copy of the available CB1 and CB2 sequences to identify which sequence was correct. The corrected genome sequences are available in the NCBI database as CP023313 (CB2), CP023314 (CB1), and CP023315 (CB13B1a). The raw reads SRA from the PacBio SMRT Cells are also uploaded under those same accession numbers.
Genome comparisons
Genome comparisons were performed by comparing the CB1, CB2, and CB13 genome sequences to the NA1000 reference sequence using Mauve [8]. The number of generations since a common ancestor was estimated from the number of SNPs that were present when compared to the NA1000 reference genome using an estimate of 0.003 mutations per generation [9]. NA1000 has a doubling time of about 3 hours in defined medium at 35 C [11], and Hentchel et al. [13] recently demonstrated that C. crescentus strain CB15 has a 5 hr doubling time in lake water when grown in the laboratory at 30 C. Lower temperatures would also greatly impact doubling times, and we have shown that CB15 has a one week doubling time when grown at 10 C in PYE broth (unpublished). Taken together, these results suggest that naturally occurring Caulobacter strains might have a minimum of a 3 to 4 day doubling time since water temperatures will vary seasonally and with latitude.
Results
Genome sequencing
Since, the genome nucleotide sequences of CB1, CB2, and CB13 were determined by PacBio sequencing, the long reads generated by this method enabled us to assemble each genome into a single contig that encompassed the entire genome. In contrast to some of the other Caulobacter genomes [1, 24], no plasmids were observed in any of the strains. The CB1 genome was the most closely related to the NA1000 genome. Since the NA1000 genome nucleotide sequence is thought to be accurate [16], we used this sequence as the reference to help us determine the accuracy of the CB1 sequence. Compared to the NA1000 reference sequence, the CB1 genome sequence contained small numbers of SNPs and single base indels that could be due to some combination of accumulated mutational differences between the two wild isolates and sequence errors generated by the PacBio sequencing technology. Since sequencing errors are not likely to be reproducible, we determined the genome sequence of the CB1 strain a second time. When the two independent CB1 genome sequences were compared, the sequences were identical except for 23 single base deletions that were present in only one of the two sequences. Interestingly, all 23 were in GC homopolymer regions consisting of 5 to 16 identical consecutive bases. This result indicates that the PacBio sequencing technology generates relatively accurate nucleotide sequences since no SNPs and only 23 single base indels in homopolymer regions were observed in two independent determinations of a 4 Mb genome sequence.
When the 23 CB1 indels were compared to the NA1000 genome sequence, some from each genome sequence matched the homopolymer sequence found in the NA1000 genome, while the matching homopolymer region in the other CB1 genome sequence was one base pair shorter. Therefore, we concluded that those 22 indels were due to sequencing errors where a single GC base pair was omitted from a homopolymer sequence in one or the other of the two sequence determinations. The 23rd indel was in a region that was not present in the NA1000 reference genome so we could not determine if it was the result of the loss or a gain of a base pair. Based on these results, we concluded that the only detectable errors in our CB1 genome sequences were occasional omissions of a single base in a homopolymer region and that these errors could be detected and corrected by comparison to an independent re-sequencing of the genome.
Although CB2 was more distantly related to NA1000, we decided to use a similar approach to correct the CB2 sequence. When a second independent genome nucleotide sequence was determined and compared to the first, again the two sequences were identical except for the presence of one base indels in homopolymer regions. This time 150 homopolymer differences were observed between the two genome sequence determinations. When we compared the 150 homopolymer sequences to the NA1000 genome sequence, we found only 67 instances where the corresponding region was present in NA1000. As we had observed with CB1, in each of the 67 instances, the homopolymer sequence in one of the two CB2 genome sequences matched that of NA1000 while the other was one base pair shorter each time. To verify this conclusion, we designed primers and amplified seven of the 67 regions of the CB2 genome. The amplified DNA was then subjected to Sanger sequencing, and in each case, the Sanger sequence matched the larger of the two homopolymer regions. Together, these results confirmed our conclusion that the PacBio sequencing system produces accurate bacterial genome sequences except for a small number of single base pair omissions in homopolymer regions. Since these single base pair omissions occur randomly in homopolymer regions during each sequencing run, two independent sequencing determinations could be used to generate an accurate genome nucleotide sequence by assuming that wherever there was a difference, the longer homopolymer contained the correct number of bases. This approach was used to obtain an accurate CB13 genome sequence as well [2].
Genome comparisons: CB1
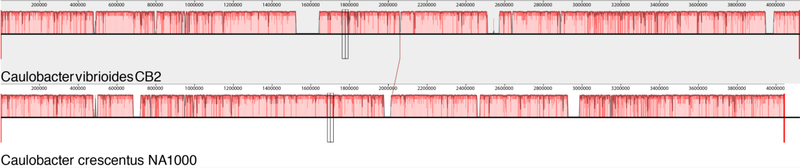
After correcting the homopolymer sequencing errors, the CB1, CB2, and CB13 genomes were compared to the NA1000 reference genome. All four genomes were similar except that the CB2 genome was approximately 0.5 Mb larger (Table 1). The corrected CB1 genome was most similar to that of NA1000 with only 26 SNPs, 12 one-base indels, three small indels, and four larger indels (Table 2, Fig. S1). If C. crescentus strains accumulate mutations at the rate of 0.003 mutations per generation as proposed by Drake [9], then CB1 would be separated from NA1000 only by about 10,000 generations or about 100 years if these strains had been doubling at an average rate of once per 3 to 4 days. The CB1 genome contains all of the genes present in NA1000 (Table 3), but it also includes 92 additional genes that are present in a 94 kb indel (Fig. 1, Supplementary Table 1). At one end, the indel includes a UGA translation stop codon of the preceding glutamine-hydrolyzing GMP synthase gene while NA1000 has a UAG stop codon at this position. Further inspection revealed a P4 type integrase gene 320 bp downstream from the changed stop codon. A BLASTp comparison of the amino acid sequence of the integrase revealed that it has 99% amino acid identity to a P4 integrase gene in Caulobacter sp. K31. The integrase gene was followed by 34 additional genes that are also present after the K31 integrase gene, but they are not present in other Caulobacter genomes. A second region that includes a set of nine genes that are present in K31 and involved in conjugative transfer was found towards the middle of the CB1 indel, but the final 37 kb of the CB1 indel was not present in the K31 genome. In contrast, the genes in the region preceding the integrase gene are the same in the K31 genome and the four C. crescentus genomes included in this study, but the integrase gene and the rest of the indel are not present in the NA1000, CB2, or CB13 genomes. Therefore, we thought that the CB1 indel probably resulted from an insertion event. However, the P4 integrase gene is also present, usually 319 bp from the GMP synthase gene stop codon in five of the other Caulobacter genomes that have been sequenced including that of C. segnis. The integrase is also present in three Sphingobium genomes. In each case, it is adjacent to the glutamine-hydrolyzing GMP synthase gene as it is in the CB1 and K31 genomes. This position conservation in 10 genomes from two related genera suggests that the glutamine-hydrolyzing GMP synthase – P4 integrase gene pair seems to be an ancient combination that has been deleted in NA1000 and most other Caulobacter genomes, but was still present in some Caulobacter genomes. This conclusion is consistent with the observation that deletions of inserted phage genes and other genes that do not provide useful functions are commonly observed in bacterial genomes [18].
Table 1.
Genome statistics for four C. crescentus genomes
| Genome Features | NA1000 | CB1 | CB2 | CB13 |
|---|---|---|---|---|
| Base Pairs (Mb) | 4.02 | 4.14 | 4.67 | 4.14 |
| G/C content (%) | 67.2 | 67.2 | 67.2 | 67.1 |
| tRNA genes | 51 | 51 | 52 | 51 |
| Protein-coding Genes | ~3900 | ~4000 | ~3900 | ~3900 |
Table 2.
Indels and SNPs compared to the NA1000 reference genome
| Genome | SNPs | 1 bp indels | 2–20 bp indels | >20 bp indels | Generations apart |
|---|---|---|---|---|---|
| CB1 | 26 | 12 | 3 | 4 | 10,000 |
| CB2 | 73,859 | 644 | 2 | 348 | 25 × 106 |
| CB13 | 301,289 | 1885 | 2727 | 1492 | 100 × 106 |
Table 3.
Gene acquisition and loss compared to the NA1000 reference genome
| Genome | Shared genes | Genes not in NA1000 | NA1000 Genes absent |
|---|---|---|---|
| CB1 | 3886 | 93 | 0 |
| CB2 | 3575 | 360 | 317 |
| CB13 | 3345 | 583 | 541 |
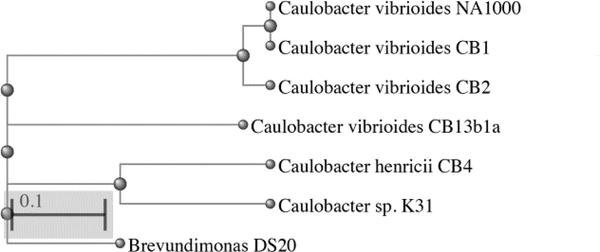
Fig. 1.
A phylogenetic tree of the amino acid sequences of the conserved 20 gene ribosomal protein operon depicting the relationships among the genomes described in this study relative to other Caulobacter genomes. Brevundimonas sp. DS20 is included as an outgroup.
The other three intermediate sized indels in the comparison of CB1 and NA1000 genomes (196 bp, 235 bp and 325 bp) appear to be deletions between short repeated sequences in the NA1000 genome. The 325 bp deletion resulted in a shortened form of the CB1 cspA cold shock protein gene converting it into a cspC gene in the NA1000 genome (Fig. S2). The 196 bp and the 235 bp deletions occurred in intergenic regions. Two of the small (12 bp) indels also appear to have resulted from recombination between nearby repeated regions, and the remaining 7 bp indel causes a frameshift in the CB1 surF gene so that two smaller proteins would be produced instead of a larger protein that was twice the size.
Genome comparisons: CB2
The CB2 genome was more distantly related to the NA1000 genome with SNPs comprising approximately 2% of the genome and 994 indels of various lengths (Table 2, Fig. 2). Thus, it appears to be separated from NA1000 by approximately 25 million generations or about 250,000 years. Interestingly, the number of one bp indels did not increase in proportion to the number of SNPs and nearly all of the observed one bp indels were in intergenic regions. This result is probably due to selection since one bp indels will change the reading frame so that defective proteins are produced if the indel occurs in a protein coding region. In contrast SNPs in coding regions often result in the substitution of a compatible amino acid or in no amino acid change so a functional protein would still be produced.
Fig. 2.
The region of the CB1 genome that contains the 90 kb insertion is represented by the white region of the CB1 genome. The regions of homology between the two genomes are aligned on the left side of the figure and the alignment ends with the GMP synthase gene. The CB1 GMP synthase and the adjacent Integrase genes are shaded. The region that is homologous to the corresponding region in the Caulobacter sp. K31 genome is labeled “present in K31”. A region that contains genes that are homologous to genes involved in plasmid transfer is marked “conjugative transfer”.
In addition to the single base changes, we identified more than 300 larger indels which were located primarily in intergenic regions as observed in other bacteria [19]. These indels resulted in 317 genes that were present in NA1000 but not in CB2, and 360 genes that were present in CB2 but not in NA1000 (Table 3). The largest indel was a 120 kb insertion into the CB2 genome that codes for 111 genes including a large number of heavy metal resistance genes (Fig. 2, Supplementary Table 2). The CB2 genome also contains two sets of phage genes that are embedded in two insertions (42 kb and a 45 kb) that are not present in NA1000 (Supplementary Tables 3 and 4). Both sets are insertions are adjacent to a tRNA gene. Similarly, a 12 kb set of genes containing an integrase gene and three modification/restriction genes also was inserted into a tRNA gene. In one additional case, a 6 kb segment containing a recombinase gene was inserted into a tRNA gene in the CB2 genome and resulted in a duplication of the tRNA gene so that identical tRNA genes flank the insertion. As a result, the CB2 genome contains an extra tRNA gene compared to the other three genomes (Table 1). Consistent with the observations of Williams [27], these results indicate that tRNA genes are often targets of insertion events that are mediated by integrase or recombinase genes.
Genes missing from CB2 include a 60 kb region flanked by an integrase gene and a set of transposase genes in the NA1000 genome, another 39 kb set of transposase genes and nitrogen metabolism genes that were inserted into an NA1000 tRNA gene, a 36 kb region that contains several transport genes flanked by an integrase and a transposase gene, and an 11 kb region containing an integrase and a set of conjugal transfer genes that were inserted into a tRNA gene (Fig. 2).
In addition, 65 other indels contain two to ten genes that are present in the NA1000 genome but not in the CB2 genome or vice versa. These indels total 642,741 bp which is equivalent to about 16% of the total base pairs in one of their genomes. This difference is reflected in the fact that they their share 92% of their genes with the other 8% of the genes being unique to each genome (Table 3). Genes present in the smaller indels that differ in these two wild type strains include 45 transposase genes and 10 toxin/antitoxin pairs indicating that these genes tend to be more mobile than other genes.
Genome comparisons: CB13
Relative to the other three genomes, the CB13 genome is the most distantly related (Fig. 3). It has more than 300,000 SNPs relative to NA1000 indicating that the two isolates are separated by about 100 million generations or approximately one million years (Table 2). However CB13 shares 3345 genes with NA1000 which is equivalent to 86% of the NA1000 genome (Table 3). Despite this degree of gene sharing, none of the confirmed mobile elements found in the NA1000 genome, was present in the CB13 genome. The 1492 indels (Table 3) that are greater than 20 bp, total more than 1.45 million base pairs indicating that about one sixth of each of the two genomes has been deleted or replaced by HGT). Since the loss of shared regions of the genomes would result in a reduction in the apparent number of SNPs between the two genomes, the number of generations separating CB13 from NA1000 may be an underestimate.
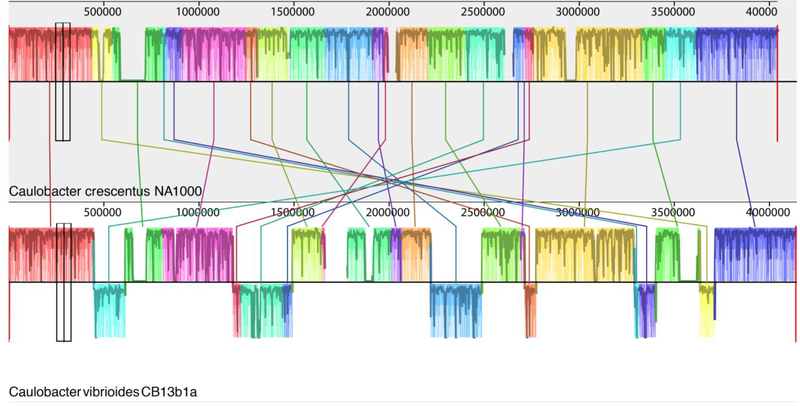
Fig. 3.
A comparison of the entire CB2 (top) and NA1000 (bottom) genomes. White spaces within the shaded bars represent the location and size of the indels described in the text.
Relative to NA1000, CB13 contains eight inversions and one translocation without an inversion (Fig. 3). Thus, these events appear to have occurred at a rate of about one per 10 to 12 million generations. No inversions or translocations were observed when the CB1 and CB2 genomes were compared to the NA1000 genome. It is not clear what causes these inversions and translocations, but previous comparisons of more distantly related Caulobacter and Brevundimonas genomes indicated that most inversions included the chromosomal origin of replication [1, 24]. In addition, most inversions are bordered by genes that are present in only one of the two genomes. For example, one inversion breakpoint occurs at nucleotide position 437325 in NA1000 and 440705 in CB13 (Fig. 3). At this position, the NA1000 genome contains two genes that are not found in the CB13 genome, and the CB13 genome contains one gene that is not found in the NA1000 genome. Thus, the inversion process may be a complex process that simultaneously creates an insertion or deletion at each end of the inversion.
In contrast, some regions of the CB13 genome are very highly conserved among all four genomes indicating that selection plays an important role in the conservation of the genomes. For example, the entire nucleotide sequence of the 21 gene, 9087 bp ribosomal protein operons of NA1000 and CB1 are identical, and the CB2 operon contains only 15 SNPs relative to the NA1000 operon. The more distantly related CB13 ribosomal protein operon contains an 18 bp insert in the early part of the first gene in the operon and 152 SNPs relative to NA1000. Thus, despite all of the indels and rearrangements throughout the genome, the CB13 ribosomal operon has 98% nucleotide identity in this highly conserved region of the genome.
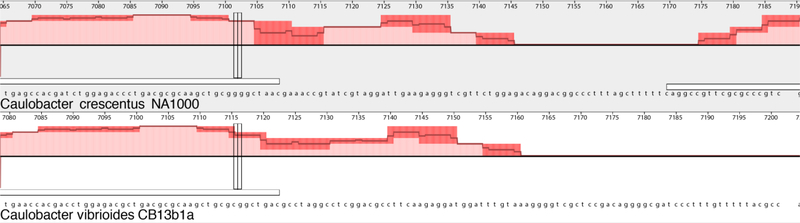
The dnaA gene nucleotide sequence is also identical in NA1000 and CB1 as well. Relative to these two dnaA genes, the CB2 dnaA gene contains 16 SNPS scattered throughout the gene and has a cluster of additional changes in the last 125 bp of the gene that may be due to an adjacent recombination event associated with an indel (Fig. 4). Similarly, the nucleotide sequence of the CB13 dnaA gene is 91% identical to the NA1000 gene with a different cluster of changes at the end associated with a different indel. Thus, insertions in these genomes sometimes change the coding sequence in terminal regions of a gene by replacing the terminus of the gene, in this case without changing the predicted amino acid sequence of the three versions of the dnaA gene.
Fig. 4.
A comparison of the entire CB13 (bottom) and NA1000 (top) genomes. Shaded blocks representing corresponding segments of each genome are connected by a line. Blocks below the horizontal black line correspond to regions that are inverted relative to the NA1000 genome.
The CB13 contains two unrelated sets of phage genes, while CB2 contains three additional clusters of phage genes that are not closely related to any of the others or to any other sequences in the GenBank database. The best matches are usually to genes present in other Caulobacter genomes with amino acid identities that range from 40% to 70% amino acid identity. All five of these phage gene clusters are included in insertions that contain other genes, and they tend to have different combinations of phage genes that are generally in the same order. None of the phage gene clusters appear to be an intact prophage. However, these results suggest that many of these large clusters of genes that are observed to be inserted into Caulobacter genomes may have originally been derived from chromosomal gene segments that contained prophage as suggested by Chen et al. [5].
Essential genes
To determine if all of the genes that were determined to be essential for growth of NA1000 in PYE [6] were present in the CB1, CB2, and CB13 genomes, we compared the aligned genomes using the progressiveMauve alignment tool. The CB1 genome contained all of the essential genes since it contains all of the genes found in the NA1000 genome. Six essential genes were missing from both the CB2 and CB13 genomes (Table 4). Four of these genes CCNA_00465–467 and CCNA_00469 are part of an insertion in the NA1000 genome [24] and are also missing from three other Caulobacter genomes. Since it is present only in the NA1000 and CB1 genomes, the expression of one of the other genes in this region probably causes a requirement for these four genes. One other missing essential gene codes for an antitoxin that is only essential if the adjacent toxin gene is expressed. This pair of genes also appears to be present only in CB1 and NA1000. A second antitoxin gene is missing from the CB13 genome, but this toxin/antitoxin pair of genes is present in the CB2 (Table 4) and strain K31 [24] genomes in addition to the NA1000 and CB1 genomes. The fourth essential gene that is missing from both the CB2 and CB13 genomes codes for a hypothetical protein, and two additional essential genes that are missing only from CB13 code for hypothetical proteins as well (Table 4). Thus, it is likely that all of the missing essential genes are required only when some other gene is present.
Table 4.
NA1000 essential genes that are not present in CB2 or CB13.
| NA1000 gene | Annotation | CB2 | CB13 |
|---|---|---|---|
| CCNA_00465 | UDP-galactopyranose mutase | absent | absent |
| CCNA_00466 | Glycosyltransferase | absent | absent |
| CCNA_00467 | Oligosaccharide translocase/flippase | absent | absent |
| CCNA_00469 | Glycosyltransferase | absent | absent |
| CCNA_00761 | Hypothetical protein | present | absent |
| CCNA_02841 | Hypothetical protein | absent | absent |
| CCNA_02844 | parD3 antitoxin | present | absent |
| CCNA_03274 | Hypothetical protein | present | absent |
| CCNA_03630 | socA antitoxin protein | absent | absent |
Discussion
Each strain used in this study had been stored lyophilized or frozen for more than 40 years, but it is possible that some changes to these genomes occurred during laboratory culture. For example, the CB13 isolate that we used was designated CB13B1a indicating that it had been cultured in Dr. Poindexter’s laboratory and that on three occasions a sub-clone was isolated from the then current culture. Nevertheless, a thorough study of CB15/NA1000 strains that had been sub-cultured for various lengths of time in six different laboratories indicated that only eight SNPs and two single base insertion/deletions (indels) were observed among the nine genomes that were sequenced [16]. In addition, strains designated CB15 had lost a 26 kb mobile element. Thus, we believe that all or nearly all of the differences that we have observed among these four wild type strains were present in the original isolates.
PacBio sequencing generates accurate sequences of bacterial genomes that are readily assembled into single contigs that encompass the entire genome. The only sequencing errors we observed were that a small percentage of the homopolymer regions were missing a single base. This comparison of four accurately-sequenced, closely-related genomes provided snapshots of the evolutionary processes that occur in natural bacterial communities and showed that missense and single base indels accumulate continually. Indels containing a few genes included genes horizontally transferred from closely related species and seemed to occur at a frequency of 10−3 to 10−4 insertions per generation. They may involve homologous recombination since they occurred primarily in intergenic regions, and they often replaced the ends of adjacent genes. Larger insertions were even less frequent and often involved a tRNA gene. The four insertions that are present in NA1000 and CB1, but not in CB2 or CB13 include a total of 150 genes that account for about half of the genes that are present in the NA1000 genome but not in the CB2 genome. Similarly, the four insertions that are present in the CB2 genome but are not present in the other genomes include a total of 249 genes that account for 69% of the genes that are only found in the CB2 genome. Thus, these acquisitions of new genes are more frequent than the previous estimate of 16 kb per million years for E. coli [18], but much less common than other authors have implied [22].
In contrast to the Caulobacter genome scrambling described in our previous studies [1, 24], we found no inversions when the NA1000, CB1, and CB2 genomes were compared. However, we did find eight inversions when the CB13 genome was compared to the other genomes. Therefore, inversions are rare events, occurring at a rate of about one per 10 to 12 million generations, and only more distantly related Caulobacter genomes appear to have their genomes scrambled.
None of the insertions that contain phage genes appear to contain intact prophage genomes, and none of the phage genes match any of the Caulobacter phage genomes that have been sequenced to date. Thus it appears that the large insertions present in these C. crescentus genomes often include fragments of phage genomes that appear to be unrelated to each other and may have originated with phage that have yet to be discovered or that no longer exist. These large insertions also included most of the strain-specific genes that were annotated as hypothetical (61% in CB2). Some of the insertions contained heavy metal resistance genes, and we have shown previously that changes in the number of heavy metal resistance genes can contribute to measurable differences in the concentrations of heavy metals that are tolerated by individual strains [1]. We did not observe any other genes whose annotation indicated that they might confer a benefit to the host genome.
Supplementary Material
Fig. 5.
The aligned nucleotide sequence of the NA10000 and CB13b1a dnaA genes showing the stop codon TAA (UAA for NA1000 mRNA) and TGA (UGA for CB13 mRNA) at the end of the white bar that represents the 3’-end of each gene. Note that the nucleotide sequence is completely after the T that corresponds to the beginning of the stop codon.
Table 5.
Genes present in the NA1000 or CB2 genomes, but not in both.
| Gene category | NA1000 | CB2 |
|---|---|---|
| Hypothetical | 121 | 198 |
| Transposase | 40 | 9 |
| Toxin/antitoxin | 14 | 5 |
Acknowledgements:
This work was funded in part by NIH grant GM076277 and by the NIH Institutional Development Award (IDeA) grant number P20GM103446 to DCS and by NIH grant GM076277 to BE.
Footnotes
Conflict of Interest: The authors declare that they have no conflict of interest.
Literature cited
- 1).Ash K, Brown T, Watford T, et al. (2014) A comparison of the Caulobacter NA1000 and K31 genomes reveals extensive genome rearrangements and differences in metabolic potential. Open Biology 4:140128 DOI: 10.1098/rsob.140128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2).Berrios L, Ely B (2018) Achieving accurate sequence and annotation data for Caulobacter vibrioides CB13. Current Microbiology 75(12):1642–1648. DOI: 10.1007/s00284-018-1572-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3).Bolotin E, Hershberg R (2017) Horizontally acquired genes are often shared between closely related species. Front Microbiol 8:1536 DOI: 10.3389/fmicb.2017.01536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4).Boto L (2015) Evolutionary change and phylogenetic relationships in light of horizontal gene transfer. J Biosci 40:465–472. DOI: 10.1007/s12038-015-9514-8 [DOI] [PubMed] [Google Scholar]
- 5).Chen J, Quiles-Puchalt N, Chiang YN et al. (2018) Genome hypermobility by lateral transduction. Science 362:207–212 DOI: 10.1126/science.aat5867 [DOI] [PubMed] [Google Scholar]
- 6).Christen B, Abeliuk E, Collier J M et al. (2011) The essential genome of a bacterium. Mol Syst Biol 7:528 DOI : 10.1038/msb.2011.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7).Doolittle WF, Bapteste E (2007) Pattern pluralism and the tree of life hypothesis. Proc. Nat Aad Sci USA 104:2043–2049. DOI: 10.1073/pnas.0610699104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8).Darling AE, Mau B, Perna NT, (2010) progressiveMAUVE: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147 DOI: 10.1371/journal.pone.0011147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9).Drake JW, Charlesworth B, Charlesworth D, Crow JF (1998) Rates of spontaneous mutation. Genetics 148:1667–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10).Ely B (1991) Genetics of Caulobacter crescentus. Methods Enzymol. 204:372–384. [DOI] [PubMed] [Google Scholar]
- 11).Ferber DM, Ely B (1982) Resistance to amino acid inhibition in Caulobacter crescentus. Mol Gen Genet 187:446–452 [Google Scholar]
- 12).Friedman R, Ely B (2012) Codon usage methods for horizontal gene transfer detection generate an abundance of false positive and false negative results. Current Microbiology 65:639–642. doi: 10.1007/s00284-012-0205-5 [DOI] [PubMed] [Google Scholar]
- 13).Hentchel KL, Reyes Ruiz LM, Curtis PD, et al. (2018) Genome-scale fitness profile of Caulobacter crescentus grown in natural freshwater. The ISME Journal DOI: 10.1038/s41396-018-0295-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14).Johnson RC, Ely B (1977) Isolation of spontaneously derived mutants of Caulobacter crescentus. Genetics 86:25–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15).Koonin EV, Puigbo P, Wolf YI (2011) Comparison of Phylogenetic trees and search for a central trend in the “Forest of Life”. J Comput Biol 18:917–924. DOI: 10.1089/cmb.2010.0185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16).Marks ME, Castro-Rojas CM, Telling C, et al. (2010) The genetic basis of laboratory adaptation in Caulobacter crescentus. J. Bacteriol 192:3678–3688. DOI: 10.1128/JB.00255-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17).Nierman WC, Feldblyum TV, Laub MT, et al. (2001) Complete genome sequence of Caulobacter crescentus. Proc Natl Acad Sci USA 98:4136–4141. DOI: 10.1073/pnas.061029298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18).Ochman H, Lawrence JG, Groisman EA (2000) Lateral gene transfer and the nature of bacterial innovation. Nature 405:299–304 [DOI] [PubMed] [Google Scholar]
- 19).Oliveira PH, Touchon M, Cury J, Rocha EPC (2017) The chromosomal organization of horizontal gene transfer in bacteria. Nature communications 8:841 DOI: 10.1038/s41467-017-00808-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20).Poindexter JS (1964) Biological properties and classification of the Caulobacter group. Bacteriol Rev. 28:231–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21).Quail MA, Smith M, Coupland P (2012) A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics 13:341 DOI: 10.1186/1471-2164-13-341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22).Rocha EPC (2016) Using sex to cure the genome. PLoS Biol 14(3):e1002417 DOI: 10.1371/journal.pbio.1002417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23).Scott D, Ely B (2015). Comparison of genome sequencing technology and assembly methods for the analysis of a GC-rich bacterial genome. Current Microbiol 70: 338–344. DOI: 10.1007/s00284-014-0721-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24).Scott D, Ely B (2016) Conservation of the essential genome among Caulobacter and Brevundimonas species. Current Microbiol 72:503–510 DOI: 10.1007/s00284-014-0721-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25).Shin SC, Ahndo H, Kim SJ, et al. (2013) Advantages of single-molecule real-time sequencing in high-GC content genomes. PLoS One 8: e68824 DOI: 10.1371/journal.pone.0068824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26).Souza V, Turner P, Lenski RL (1997) Long term experimental evolution in Escherichia coli. V. Effects of recombination with immigrant genotypes on the rate of bacterial evolution. J Evol. Biol. 10 (5);7453–769 [Google Scholar]
- 27).Williams KP (2002) Integration sites for genetic elements in prokaryotic tRNA and tmRNA genes: sublocation preference of integrase subfamilies. Nucleic Acids Res. 30:866–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.