Abstract
In this paper, we report the epidemic characteristics of the three co-circulating influenza viruses (i.e., A/H1N1, A/H3N2, and B) in two tropical African cities—Kampala and Entebbe, Uganda—over an eight-year period (2008–2015). Using wavelet methods, we show that influenza epidemics recurred annually during the study period. In most months, two or more influenza viruses co-circulated at the same time. However, the epidemic timing differed by influenza (sub)type. Influenza A/H3N2 caused epidemics approximately every 2 years in both cities and tended to alternate with A/H1N1 or B. Influenza A/H1N1 and B produced smaller but more frequent epidemics and biennial epidemics of these two viruses tended to be synchronous. In addition, epidemics of A/H3N2 were more synchronized in the two cities (located ca.37 km apart) than that of A/H1N1 or influenza B.
Keywords: Influenza, tropical regions, (sub)type specific, Uganda, epidemic frequency
1. Introduction
Influenza causes substantial morbidity and mortality worldwide, including in tropical African populations (McMorrow et al., 2015; Ortiz et al., 2012; Radin et al., 2012). In contrast to the annual wintertime epidemics in temperate regions, the transmission dynamics of influenza in the tropics are more diverse. Recent studies have reported an association of increased influenza circulation during rainy seasons in tropical climates (Cummings et al., 2016; Lutwama et al., 2012; Ng and Gordon, 2015; Soebiyanto et al., 2014; Tamerius et al., 2011; Tamerius et al., 2013); however, the long-term epidemiology of influenza in tropical Africa remain poorly characterized (Yazdanbakhsh and Kremsner, 2009).
Observations of influenza infection are typically sparse in tropical Africa for a number of reasons. First, resources are scarce in this region and thus prevention and intervention priorities are typically given to more severe diseases such as HIV/AIDS, tuberculosis, and malaria (Yazdanbakhsh and Kremsner, 2009). As a consequence, long-term surveillance datasets documenting respiratory infectious disease incidence, including influenza, are rare. Second, when established, surveillance systems of influenza are often syndromic and laboratory testing to confirm the etiologic agent has rarely been conducted. Third, individuals often do not seek clinical care for relatively mild infections such as influenza, due to a lack of access to healthcare systems in these regions. As a result, the numbers of influenza infection recorded by surveillance systems are usually low and scattered. These issues altogether create challenges for studying the transmission dynamics of influenza in tropical Africa.
To improve understanding of the long-term epidemiology of influenza in tropical Africa, here we analyze influenza surveillance data collected in two major cities in Uganda, Kampala and Entebbe, over a period of eight years (2008–2015). These data were collected by a nationwide influenza surveillance network in Uganda, with laboratory-confirmed influenza (sub)type information (Cummings et al., 2016; Lutwama et al., 2012). Using wavelet analysis, a signal processing technique, we examine the epidemiological characteristics for each influenza (sub)type as well as compare epidemiological differences among the three influenza viruses (i.e., A/H1N1, A/H3N2, and B).
2. Materials and Methods
2.1. Data
Influenza surveillance data were collected from a nationwide influenza surveillance network established by the Uganda Virus Research Institute during 2007 (Cummings et al., 2016; Lutwama et al., 2012). At each sentinel site, clinicians identified patients with influenza-like illness (ILI) and severe acute respiratory infection (SARI) using an established protocol (Cummings et al., 2016; Lutwama et al., 2012); specimens were taken from identified ILI/SARI patients and all specimens were tested for influenza virus by (sub)type specific reverse transcription polymerase chain reaction (RT-PCR). Verbal consent to participate in the study was obtained from patients older than 18 years of age. Verbal assent and consent was obtained from participant’s guardians in the case of patients younger than 18 years of age. Ethical review was performed by institutional review boards at UVRI, the Uganda National Council for Science and Technology, and Columbia University Medical Center.
Data for Kampala were compiled from six sentinel sites located in the city (i.e., IHK/Surgery, Kibuli, Kisenyi, Kiswa, Kitebi, and Nsambya) and those for Entebbe were compiled from one sentinel site. The sentinel site in Entebbe reported regularly from 2007 to 2015. For Kampala, while at least one site was reporting at any time during 2008–2015, the number of sites reporting fluctuated. However, the time series of influenza positive rate (i.e. the proportion of ILI/SARI specimens testing positive for influenza virus) closely tracked the raw influenza positive counts (Figure S1), even though monthly counts of ILI/SARI and influenza positive patient visits varied over time. Therefore, to control for the fluctuation in reporting, we used the proportions of specimens testing positive for any influenza virus, A/H1N1, A/H3N2, or B, respectively, among all ILI/SARI specimens as proxies for the activity of influenza overall and the three individual influenza (sub)types. Due to the low numbers recorded, we aggregated the data to monthly intervals. To facilitate comparison between the two cities, here we focus on the time period from 2008 to 2015.
2.2. Wavelet transform and wavelet power spectrum: finding the dominant periodicity
As the timing of influenza epidemics in the tropical regions is highly irregular, we use wavelet methods to the decompose the influenza time series in both the time and frequency domains. Each of the eight aforementioned monthly time series [i.e., three influenza (sub)types + one for all of them combined, in Kampala and Entebbe, respectively] was first transformed using the following formula (Torrence and Compo, 1998; van Panhuis et al., 2015):
| [1] |
where Wn(s) is the wavelet transform at time point n (ranging from 1 to N, the total number of time points) and scale s. xk is the viral positive rate for a given influenza virus in month-k. Ψ is the wavelet function and (*) represents its complex conjugate; here we used the Morlet wavelet function (Torrence and Compo, 1998), with a frequency (ω0) of 6. δt is the length of the time step (i.e. 1 month here). The scale (s) was chosen per the formula sj = 2δt 2jδj, j = 0,1, …, δj−1(log2 N − 1), where δj, the spacing between successive scales, was set to 1/12. In doing so, the one-dimensional time series, xk, was transformed into a two-dimensional plane with time on the x-axis (indexed by n) and scale (or period, an inverse function of frequency) on the y-axis (indexed by s; Figures S2 and S3).
The local wavelet power spectrum, defined as |Wn(s)|2, represents the importance of a given oscillatory period [corresponding to the scale s; e.g., for the Morlet wavelet used here, the period is 1.03 times of the scale s (Torrence and Compo, 1998)] at time point n. For easier comparison of different wavelet power spectra, the wavelet power spectrum is commonly normalized by the variance of the time series (σ2) (Torrence and Compo, 1998). Therefore, we computed the normalized wavelet power spectrum as |Wn(s)|2/σ2. To obtain the dominant periodicity for each time series, we computed the average wavelet power spectrum for each scale s over all time points, i.e. the global wavelet spectrum (Grenfell et al., 2001; Torrence and Compo, 1998); the dominant periodicity was then the maximum of the global wavelet spectrum.
2.3. Wavelet phase angle difference: epidemic contribution of individual influenza (sub)type and inter-(sub)type interactions
Another useful feature of complex wavelet transform (e.g., the Morlet wavelet function used here) is the phase angle, defined as:
| [2] |
where is the imaginary, and the real part of Wn(s) (Torrence and Compo, 1998). As the notation indicates, the phase angle is specific for each time point n and scale s. The pattern of absolute phases or phase differences may reflect the relationships between time series (Grenfell et al., 2001; van Panhuis et al., 2015). For instance, a 0° phase difference between two epidemic curves of two circulating viruses indicates the two viruses circulate at the same time (i.e. in phase), while a 180° difference indicates the two viruses do not co-circulate (i.e. out of phase). Therefore, the phase angle difference provides a way to examine the lag relation between two epidemic time series.
We first computed the circular mean phase angle at each time point at a given periodicity (e.g. 1-yr) spanning an arbitrary ±6 month window (Grinsted et al., 2004). For instance, the mean phase angle for the 1-yr period at n=1 (i.e. Jan 2008) is the circular mean of all with periods between 6 and 18 months [again, note that for the Morlet wavelet function with a frequency ω0=6, the period is 1.03 times of the scale s (Torrence and Compo, 1998)]. Based on our analysis in subsection 2.2, the dominant periodicity was either 1 or 2 years for the three influenza (sub)types. Therefore, we computed the mean phase angles at the 1-yr and 2-yr periodicity, respectively. We then performed two analyses using the mean phase angles for each epidemic curve. In the first, we computed the phase angle differences between the epidemic curve of all influenza viruses combined and that of individual influenza (sub)type to examine the contribution of each influenza (sub)type to the overall influenza epidemic. The epidemic curve of the predominant influenza (sub)type would be expected to have the same phase angles as the overall influenza epidemic curve and hence near zero phase differences. In the second, we computed the phase angle differences between pairs of influenza viruses to examine the interactions between co-circulating influenza (sub)types. As in (Grenfell et al., 2001), we constrained the raw phase difference (θ) with ±180° to remove spurious jumps in phase difference, using the formula: (θ+540 mod 360) – 180, where “mod” gives the remainder following division.
2.4. Software
All statistical analyses were done in R language (R Foundation for Statistical Computing, Vienna, Austria). The wavelet analyses were performed using the “biwavelet” package in R (Gouhier et al., 2016).
3. Results
Our study included two cities in Uganda, Kampala and Entebbe. Kampala is the capital of Uganda with a population of approximately 1.5 million (2014 census), and Entebbe is approximately 37 km southwest of Kampala and has a population of ca. 70 thousand (2014 census) (Uganda Bureau of Statistics). During the study period (2008–2015), the surveillance network detected 420 influenza cases among 2807 ILI/SARI visits in Kampala, with 130 (31.0%) A/H1N1, 181 (43.1%) A/H3N2, and 108 (25.7%) B cases (the remaining influenza case was an unsubtypeable A infection). The viral composition in Entebbe was similar to Kampala, with 141 (30.9%) A/H1N1, 215 (47.0%) A/H3N2, and 99 (21.7%) B cases detected among 4051 ILI/SARI visits (the remaining 0.4% consisted of one A/H3N2 and A/H1N1 coinfection and one unsubtypeable A infection). Figure 1 shows the viral positive rates among all ILI/SARI visits for the two cities. The epidemic curves of A/H3N2 in the two cities closely tracked each other, with a Pearson correlation coefficient (r) of 0.70 [0.59, 0.79] (mean and 95% confidence interval). In comparison, A/H1N1 and B epidemics in the two cities were less correlated (r=0.41 [0.23, 0.57] for A/H1N1 and 0.45 [0.27, 0.59] for B).
Figure 1.
Monthly time series for influenza overall (A), A/H1N1 (B), A/H3N2 (C), and B (D) from Jan 2008 to Dec 2015 in Kampala and Entebbe.
Wavelet transforms on the monthly time series showed that epidemic patterns of influenza in the two cities during 2008–2015 were in general similar, but had noticeable differences (Kampala in Figure S2 and Entebbe in Figure S3). For both cities, epidemics of influenza recurred annually during 2008–2015 (Figure 2A); the dominant epidemic period was 12 months in Kampala and 11 months in Entebbe. In addition, both cities experienced A/H3N2 epidemics every 22 months (Figure 2C). However, the epidemic patterns of A/H1N1 and B were more diverse in the two cities. Kampala experienced a much larger outbreak of A/H1N1 during the 2009 pandemic than Entebbe (Figure 1B; Figure S2B vs Figure S3B). As a result, the dominant A/H1N1 epidemic period was 12 months, compared to 22 months in Entebbe. Note, however, that the global power spectrum for A/H1N1 in Entebbe at 12-month was only slightly lower than that at 22-month, suggesting similar annual recurrence. For influenza B, the dominant epidemic period was 12 months in Kampala and 25 months in Entebbe; in addition, the wavelet transform also showed larger power spectra at shorter time periods (2 to 12 months) in Kampala than Entebbe (Figure S2D vs S3D). Findings above are based on viral positive rates and in general consistent with those based on raw counts of positive specimens (Figure S4).
Figure 2.
Dominant periodicity. The lines show the global power spectra for influenza overall (A), A/H1N1 (B), A/H3N2 (C), and influenza B (D) in Kampala (red line) and Entebbe (blue line). Vertical dashed lines indicate the dominant periodicity, i.e. the period with the maximum global power spectrum (also labeled for each epidemic time series).
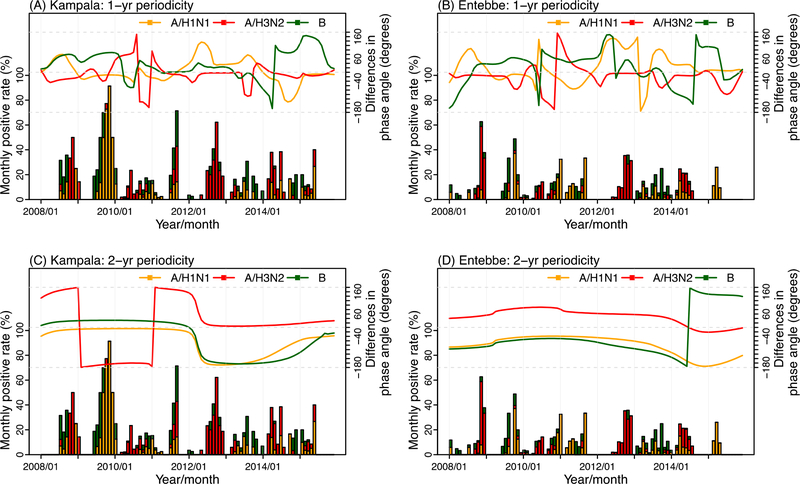
Figure 3 shows the wavelet phase differences between the epidemic curve of all influenza viruses combined and that of individual influenza (sub)type. For the annual cycles (i.e. 1-yr periodicity), in both cities, among the three influenza viruses, one tended to predominate with concurrent circulation of another one or two viruses; however, the combination of co-circulating influenza viruses varied over time without a clear pattern. For instance, in Kampala (Figure 3A), during 2008–2010 the phase angle differences for A/H1N1 were close to zero, indicating A/H1N1 was the predominant subtype; during the same period of time, either A/H3N2 or B co-circulated with A/H1N1. In 2011, all three viruses co-circulated such that all three had nearly zero phase angle differences. Staring 2012, A/H3N2 became the predominant subtype while B or A/H1N1 the co-circulating virus.
Figure 3.
Wavelet phase angle differences between the overall influenza epidemic and each of the three influenza (sub)types. The lines show the wavelet phase angle differences at 12±6 month periods (upper panel: A for Kampala and B for Entebbe) or 24±6 month periods (lower panel: C for Kampala and D for Entebbe) between the overall influenza epidemic and A/H1N1 (in orange), A/H3N2 (in red), or B (in green). The bars show monthly influenza viral positive rates, with each colored segment representing viral positive rate for each of the three influenza (sub)types: A/H1N1 (in orange), A/H3N2 (in red), and B (in green).
For the biennial cycles (i.e. 2-yr periodicity), two clear patterns are evident from Figure 3 (C for Kampala and D for Entebbe). First, for both cities, A/H1N1 and B tended to co-circulate at the same time, such that the phase angle difference time series for the two viruses closely tracked each other. Second, for both cities, A/H3N2 tended to alternate with A/H1N1 and B as the predominant strain(s) such that the phase angle for the A/H3N2 time series was approximately 180° different from that for A/H1N1 or B for most months of the study period. For instance, in Kampala (Figure 3C), A/H1N1 and B were the predominant strains during 2008–2011 (0° phase angle difference compared to the overall influenza epidemic) while A/H3N2 was the predominant during 2012–2015. A similar pattern of predominant strain alternation was seen for Entebbe, although the timing of dominancy was different (Figure 3D).
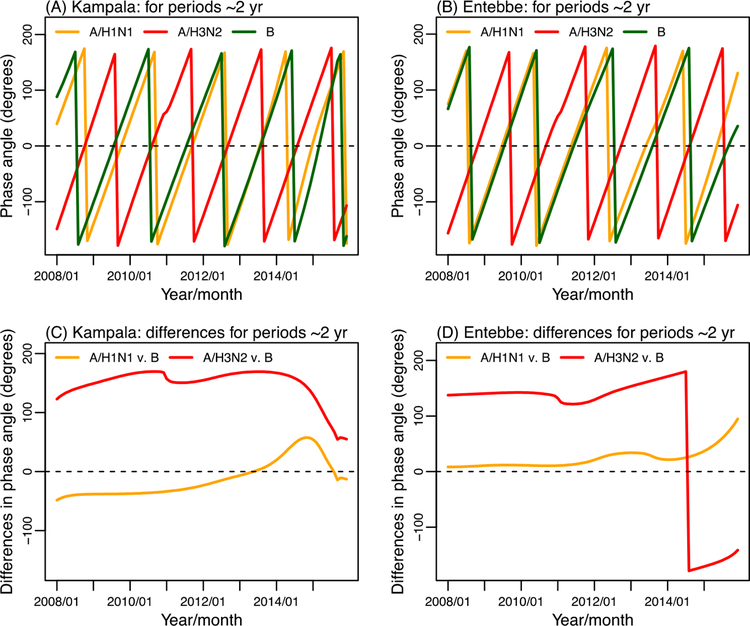
Figure 4 further shows the wavelet phase angles at the 2-yr periodicity for the three influenza (sub)types, respectively. Consistent with Figure 3 C and D, the phase angle time series of A/H1N1 and B closely tracked each other for both cities, suggesting these two influenza viruses tended to circulate during the same months. In contrast, the phase angles of A/H3N2 were approximately 180° different from the other two (sub)types, indicating A/H3N2 incidence when the other two (sub)types were not in circulation.
Figure 4.
Wavelet phase angles for the three influenza (sub)types and phase angle differences. Upper panel: wavelet phase angles at 24±6 month periods for A/H1N1, A/H3N2, or B in Kampala (A) and Entebbe (B). Lower panel: phase angle differences between either of the A subtypes and type B in Kampala (C) and Entebbe (D).
4. Discussion
Unlike the regular wintertime epidemics experienced in temperate regions, influenza epidemics typically exhibit much more diverse patterns in tropical regions. Here we have identified a number of outstanding epidemic characteristics in two Ugandan tropical cities. Using wavelet methods, we computed the dominant periodicity of overall influenza incidence, as well as for each of the three currently circulating influenza (sub)types. We show that during 2008–2015 influenza epidemic recurred annually in both Uganda cities with the three influenza (sub)types circulating in alternative years. In addition, we show that, over a shorter-term (roughly annual), one influenza virus tended to predominant with another one or two co-circulating at the same time; over a longer-term (roughly biennial), A/H1N1 tended to co-circulate with influenza B and that these two influenza viruses were out of phase with A/H3N2.
The two Ugandan cities included in our study, Kampala and Entebbe, are only approximately 37 km apart. However, highly similar epidemic activity was only observed for A/H3N2, in both short and long term (Figures 1C and 2C). In comparison, epidemic activity for A/H1N1 or B was less in concert between the two cities. This finding is consistent with previous studies in temperate regions (e.g. the US) reporting higher synchrony of A/H3N2 across geolocations (Viboud et al., 2006). It suggests that, similar to populations in temperate regions, A/H3N2 is likely more transmissible than the other two influenza (sub)types in tropical populations. Indeed, during our study period (2008–2015), A/H3N2 was the predominant strain in both studied cities.
Interestingly, despite the lack of synchrony for A/H1N1 or B between the two cities, the biennial epidemic cycles of these two viruses within the same city were highly synchronized. We show that not only did the wavelet phase angles of the two viruses at the 2-yr periodicity closely track each other, the dominant epidemic periodicities were also highly similar (Figure 2 B and D). In contrast, for both cities, biennial epidemics of A/H3N2 tended to occur out-of-phase from the other two influenza (sub)types (Figure 4 C and D). These findings suggest that there are likely strong epidemic interactions among the three influenza (sub)types at the population level. These viral interactions are likely a result of cross-immunity conferred by prior infections of antigenically similar strains (Bodewes et al., 2011; Ekiert et al., 2009; Ekiert et al., 2011; Epstein, 2006; Katz et al., 2009; McMichael et al., 1983; Miller et al., 2012; Pica and Palese, 2013; Sandbulte et al., 2007) and types (Dreyfus et al., 2012; Laurie et al., 2016; Stanekova and Vareckova, 2010).
We recognize a number of limitations in this study. First, due to a lack of information on the number of all-cause patient visits to each sentinel site or the catchment population size (denominators commonly used in previous studies), we used influenza viral positive rate as a proxy for influenza epidemic activity. The number of ILI/SARI visits (i.e. the denominator here) could fluctuate due to respiratory infections other than influenza, exhibit differing seasonal pattern, and as such confound the seasonality of influenza. However, this effect is likely minimal for our study cities, as here the influenza viral positive rates indeed closely reflected changes in actual counts of influenza positive specimens (Figure S1). In addition, results based on the actual counts were consistent with those based on viral positive rates (Figure S4). Second, due to sparse observations, we aggregated the data into monthly intervals. As influenza is an acute infection with a relatively short epidemic duration (e.g., 8 to 20 weeks in temperate regions), studies using data with finer time resolution (e.g. weekly data) will likely provide a more accurate characterization of influenza epidemiology among tropical populations. Third, our study period only lasted for eight years (i.e. 2008–2015). Given the highly diverse epidemic activity observed for influenza, in particular, in tropical regions, a longer study period would allow stronger inference of epidemic features and more reliable wavelet extraction of key signals across longer time scales (e.g., multiannual cycles). Lastly, due to a lack of long-term surveillance, we only examined influenza epidemics in two cities. Nevertheless, our study also has a number of strengths, including the (sub)type specific analysis and fine spatial resolution (i.e. at the city level), both of which have rarely been assessed previously.
In summary, we have characterized the epidemic features of influenza overall and by (sub)type over an eight-year period (2008–2015) in two tropical populations, and have identified key interactions among the three co-circulating influenza (sub)types in those populations. To achieve better understanding of influenza epidemics in tropical regions, in particular, in tropical Africa, improved, long-term surveillance in those regions is needed to support more detailed epidemiological studies.
Supplementary Material
Acknowledgement
This study was supported by US NIH grants GM100467, GM110748, and ES009089, as well as the Irving Institute for Clinical and Translational Research at Columbia University through the National Center for Advancing Translational Sciences, National Institutes of Health (UL1TR000040). Surveillance activities carried out by UVRI were funded through Ministry of Health, Uganda and the World Health Organization Country Office and Regional Office for Africa and a cooperative agreement with the U.S. Centers for Disease Control and Prevention.
Footnotes
Conflict of Interest: Jeffrey Shaman disclosed partial ownership of SK Analytics. No other disclosures were reported.
References:
- Bodewes R, Kreijtz JH, Geelhoed-Mieras MM, van Amerongen G, Verburgh RJ, van Trierum SE, Kuiken T, Fouchier RA, Osterhaus AD, Rimmelzwaan GF, 2011. Vaccination against seasonal influenza A/H3N2 virus reduces the induction of heterosubtypic immunity against influenza A/H5N1 virus infection in ferrets. J. Virol 85, 2695–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings MJ, Bakamutumaho B, Kayiwa J, Byaruhanga T, Owor N, Namagambo B, Wolf A, Wamala JF, Morse SS, Lutwama JJ, O’Donnell MR, 2016. Epidemiologic and Spatiotemporal Characterization of Influenza and Severe Acute Respiratory Infection in Uganda, 2010–2015. Ann Am Thorac Soc 13, 2159–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, Lee JH, Metlagel Z, Bujny MV, Jongeneelen M, van der Vlugt R, Lamrani M, Korse HJWM, Geelen E, Sahin O, Sieuwerts M, Brakenhoff JPJ, Vogels R, Li OTW, Poon LLM, Peiris M, Koudstaal W, Ward AB, Wilson IA, Goudsmit J, Friesen RHE, 2012. Highly Conserved Protective Epitopes on Influenza B Viruses. Science 337, 1343–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert DC, Bhabha G, Elsliger MA, Friesen RH, Jongeneelen M, Throsby M, Goudsmit J, Wilson IA, 2009. Antibody recognition of a highly conserved influenza virus epitope. Science 324, 246–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert DC, Friesen RH, Bhabha G, Kwaks T, Jongeneelen M, Yu W, Ophorst C, Cox F, Korse HJ, Brandenburg B, Vogels R, Brakenhoff JP, Kompier R, Koldijk MH, Cornelissen LA, Poon LL, Peiris M, Koudstaal W, Wilson IA, Goudsmit J, 2011. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science 333, 843–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein SL, 2006. Prior H1N1 Influenza Infection and Susceptibility of Cleveland Family Study Participants during the H2N2 Pandemic of 1957: An Experiment of Nature. J. Infect. Dis 193, 49–53. [DOI] [PubMed] [Google Scholar]
- Gouhier TC, Grinsted A, Gouhier MTC, 2016. Package ‘biwavelet’. [Google Scholar]
- Grenfell BT, Bjornstad ON, Kappey J, 2001. Travelling waves and spatial hierarchies in measles epidemics. Nature 414, 716–723. [DOI] [PubMed] [Google Scholar]
- Grinsted A, Moore JC, Jevrejeva S, 2004. Application of the cross wavelet transform and wavelet coherence to geophysical time series. Nonlinear processes in geophysics 11, 561–566. [Google Scholar]
- Katz J, Hancock K, Veguilla V, Zhong W, Lu XH, Sun H, Butler E, Dong L, Liu F, Li ZN, DeVos J, Gargiullo P, Cox N, 2009. Serum cross-reactive antibody response to a novel influenza A (H1N1) virus after vaccination with seasonal influenza vaccine. MMWR. Morb. Mortal. Wkly. Rep 58, 521–524. [PubMed] [Google Scholar]
- Laurie K, Guarnaccia T, Carolan L, Horman W, Yan A, Aban M, Petrie S, Cao P, Heffernan J, McVernon J, Mosse J, Kelso A, McCaw J, Barr I, 2016. The time-interval between infections and viral hierarchies are determinants of viral interference following influenza virus infection in a ferret model. Eur J Immunol 46, 685–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutwama JJ, Bakamutumaho B, Kayiwa JT, Chiiza R, Namagambo B, Katz MA, Geissler AL, 2012. Clinic- and hospital-based sentinel influenza surveillance, Uganda 2007–2010. The Journal of infectious diseases 206 Suppl 1, S87–93. [DOI] [PubMed] [Google Scholar]
- McMichael AJ, Gotch FM, Noble GR, Beare PA, 1983. Cytotoxic T-cell immunity to influenza. N. Engl. J. Med 309, 13–17. [DOI] [PubMed] [Google Scholar]
- McMorrow ML, Wemakoy EO, Tshilobo JK, Emukule GO, Mott JA, Njuguna H, Waiboci L, Heraud JM, Rajatonirina S, Razanajatovo NH, Chilombe M, Everett D, Heyderman RS, Barakat A, Nyatanyi T, Rukelibuga J, Cohen AL, Cohen C, Tempia S, Thomas J, Venter M, Mwakapeje E, Mponela M, Lutwama J, Duque J, Lafond K, Nzussouo NT, Williams T, Widdowson MA, 2015. Severe Acute Respiratory Illness Deaths in Sub-Saharan Africa and the Role of Influenza: A Case Series From 8 Countries. Journal of Infectious Diseases 212, 853–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MS, Tsibane T, Krammer F, Hai R, Rahmat S, Basler CF, Palese P, 2012. 1976 and 2009 H1N1 Influenza Virus Vaccines Boost Anti-Hemagglutinin Stalk Antibodies in Humans. J. Infect. Dis 207, 98–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S, Gordon A, 2015. Influenza Burden and Transmission in the Tropics. Curr Epidemiol Rep 2, 89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz JR, Lafond KE, Wong TA, Uyeki TM, 2012. Pandemic influenza in Africa, lessons learned from 1968: a systematic review of the literature. Influenza and other respiratory viruses 6, 11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pica N, Palese P, 2013. Toward a universal influenza virus vaccine: prospects and challenges. Annu Rev Med 64, 189–202. [DOI] [PubMed] [Google Scholar]
- Radin JM, Katz MA, Tempia S, Talla Nzussouo N, Davis R, Duque J, Adedeji A, Adjabeng MJ, Ampofo WK, Ayele W, Bakamutumaho B, Barakat A, Cohen AL, Cohen C, Dalhatu IT, Daouda C, Dueger E, Francisco M, Heraud JM, Jima D, Kabanda A, Kadjo H, Kandeel A, Bi Shamamba SK, Kasolo F, Kronmann KC, Mazaba Liwewe ML, Lutwama JJ, Matonya M, Mmbaga V, Mott JA, Muhimpundu MA, Muthoka P, Njuguna H, Randrianasolo L, Refaey S, Sanders C, Talaat M, Theo A, Valente F, Venter M, Woodfill C, Bresee J, Moen A, Widdowson MA, 2012. Influenza surveillance in 15 countries in Africa, 2006–2010. The Journal of infectious diseases 206 Suppl 1, S14–21. [DOI] [PubMed] [Google Scholar]
- Sandbulte MR, Jimenez GS, Boon AC, Smith LR, Treanor JJ, Webby RJ, 2007. Cross-reactive neuraminidase antibodies afford partial protection against H5N1 in mice and are present in unexposed humans. PLoS Med 4, e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soebiyanto RP, Clara W, Jara J, Castillo L, Sorto OR, Marinero S, de Antinori ME, McCracken JP, Widdowson MA, Azziz-Baumgartner E, Kiang RK, 2014. The role of temperature and humidity on seasonal influenza in tropical areas: Guatemala, El Salvador and Panama, 2008–2013. Plos One 9, e100659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanekova Z, Vareckova E, 2010. Conserved epitopes of influenza A virus inducing protective immunity and their prospects for universal vaccine development. Virol J 7, 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamerius J, Nelson M, Zhou S, Viboud C, Miller M, Alonso W, 2011. Global Influenza Seasonality: Reconciling Patterns Across Temperate and Tropical Regions. Environ. Health Perspect 119, 439–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamerius JD, Shaman J, Alonso WJ, Bloom-Feshbach K, Uejio CK, Comrie A, Viboud C, 2013. Environmental predictors of seasonal influenza epidemics across temperate and tropical climates. PLoS Pathog 9, e1003194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrence C, Compo GP, 1998. A practical guide to wavelet analysis. Bulletin of the American Meteorological Society 79, 61–78. [Google Scholar]
- Uganda Bureau of Statistics, Census 2014
- van Panhuis WG, Choisy M, Xiong X, Chok NS, Akarasewi P, Iamsirithaworn S, Lam SK, Chong CK, Lam FC, Phommasak B, Vongphrachanh P, Bouaphanh K, Rekol H, Hien NT, Thai PQ, Duong TN, Chuang JH, Liu YL, Ng LC, Shi Y, Tayag EA, Roque VG Jr., Lee Suy LL, Jarman RG, Gibbons RV, Velasco JM, Yoon IK, Burke DS, Cummings DA, 2015. Region-wide synchrony and traveling waves of dengue across eight countries in Southeast Asia. Proc Natl Acad Sci U S A 112, 13069–13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viboud C, Bjornstad ON, Smith DL, Simonsen L, Miller MA, Grenfell BT, 2006. Synchrony, waves, and spatial hierarchies in the spread of influenza. Science 312, 447–451. [DOI] [PubMed] [Google Scholar]
- Yazdanbakhsh M, Kremsner PG, 2009. Influenza in Africa. PLoS Med 6, e1000182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.