Abstract
How cellular contractile systems assemble has fascinated scientists for generations. The major molecule responsible for cellular force generation is the molecular motor, non-muscle myosin II (NMII). NMII molecules are organized into single myosin filaments and larger arrays of filaments called NMII I stacks, which are capable of generating increasing amounts of force. The textbook model of NMII stack assembly is the Network Contraction Model, where ensembles of distinct NMII filaments condense into a NMII stack by pulling on actin filaments. While this model has been widely accepted for ~20 years, it has been difficult to test inside cells due to the small size of NMII filaments. Recently, interest in how NMII stacks form has been reinvigorated by the advent of super-resolution microscopy techniques which have afforded unprecedented resolution of NMII filaments inside cells. A number of recent publications using these techniques have called into question key aspects of the Network Contraction Model, and our understanding of how NMII stacks assemble.
Keywords: Myosin II, Structured Illumination Microscopy, Expansion, Network Contraction, Myosin II Stacks
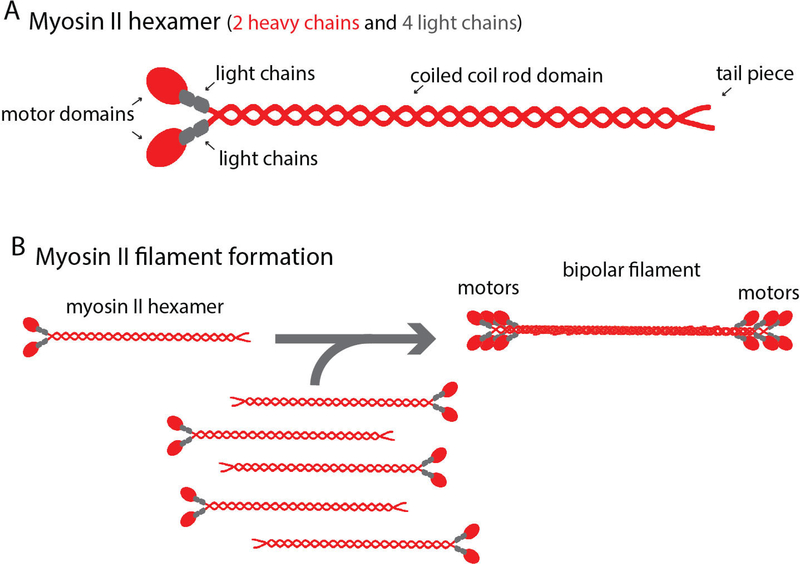
Force generation at the cellular level is critical for eukaryotic development, homeostasis, and the progression of force dependent diseases such as cancer (Babbin et al., 2009; Ma and Adelstein, 2014; Samuel et al., 2011; Tullio et al., 1997; Xia et al., 2012). Molecular motors coordinate with cytoskeletal elements, such as actin filaments, to drive force dependent processes such as cell migration, cell division, and muscle contraction (Even-Ram et al., 2007; Gupton and Waterman-Storer, 2006; Huxley, 1969; Mabuchi and Okuno, 1977; Sandquist et al., 2006; Spudich, 2014; Straight et al., 2003). Non-muscle myosin II (NMII) is the major molecular motor responsible for generating contractile forces within non-muscle cells. To carry out its wide variety of cellular functions, NMII assembles into filaments capable of generating force. A single NMII molecule is a hetero-hexamer composed of 2 heavy chains, 2 essential light chains, and 2 regulatory light chains (Fig. 1A) (Vicente-Manzanares et al., 2009). The N-terminal motor domains of the myosin II heavy chains contain both the actin binding and ATPase activity (Adelstein et al., 1971; Vicente-Manzanares et al., 2009). C-terminal electrostatic interactions of the rod domains from multiple NMII molecules creates a bipolar thick filament, positioning the N-terminal motor domains on opposite sides of the filament (Fig. 1B) (Vicente-Manzanares et al., 2009). The motor domains of each NMII molecule in a filament can bind and contract actin filaments, but the number of NMII molecules added to a NMII filament is limited (Billington et al., 2013; Pollard, 1975). Thus, the actin binding and force generating capabilities of a single NMII filament is limited. In order to increase the scale of force generation, cells organize NMII filaments into larger ensembles referred to as NMII stacks. Here we discuss the implications of recent work studying how cells organize NMII filaments into stacks.
Figure 1. Myosin II filament formation.
A) Schematic of a single NMII molecule and a NMII filament. Heavy and light chains are colored red and gray, respectively. Motor and rod domains are denoted by arrows. B) Bipolar filaments arise from non-covalent electrostatic rod domain binding.
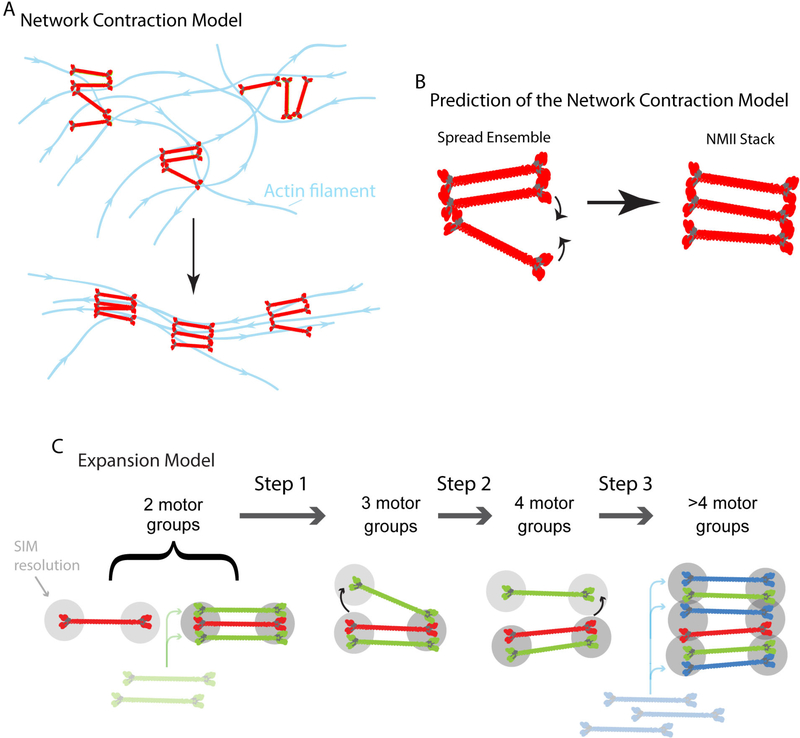
The organization of NMII filament stacks was reported in a series of elegant papers by Gary Borisy and colleagues in the 1990’s (Svitkina et al., 1997; Verkhovsky and Borisy, 1993; Verkhovsky et al., 1995). Borisy’s group adapted an electron microscopy (EM) technique termed platinum replica EM to facilitate the exquisite visualization of the cytoskeleton’s architecture within motile rat fibroblasts and fish keratocytes (Svitkina et al., 1997; Verkhovsky and Borisy, 1993; Verkhovsky et al., 1995). Key findings from these studies revealed a ‘non-sarcomeric” organization of NMII filaments at the protruding edge of cells in which NMII filaments were arranged in splayed out arrays apparently connected by their motor domains, and NMII filament stacks toward the cell center (Fig. 2A) (Verkhovsky and Borisy, 1993; Verkhovsky et al., 1995). These observations led to the “Network Contraction” Model (Fig. 2A) to explain how the actin and NMII architecture in cells was formed (Verkhovsky et al., 1999). The Network Contraction Model posits that tension generated by NMII organizes the unaligned actin architecture found at the leading edge, into the more organized and aligned actin filaments found further back in the lamella (Fig. 2A) (Verkhovsky et al., 1999). One of the key predictions of the Network Contraction model is that NMII filaments in the splayed configuration would come together as they pull on actin filaments (Fig. 2B). For ~20 years, the Network Contraction Model has become the textbook model for actin stress fiber assembly and myosin II stack assembly. Indeed, the Network Contraction Model is still being promoted as the major model of NMII stack formation (Svitkina, 2018). Work utilizing super-resolution microscopy has recently rekindled interest in how NMII filament ensembles assemble and has also called into question key aspects of the Network Contraction Model.
Figure 2. Conceptual framework of the “Network Contraction” and “Expansion” models.
A) Network contraction model redrawn from Figure 5 of Verkhovsky et al. 1999. Myosin II filaments and actin filaments are red and blue, respectively. Arrow indicates the transition between splayed myosin and actin filament arrays and aligned arrays. B) A key prediction of the Network Contraction model is that spread myosin II filaments in small clusters move together (curved arrows). C) Expansion Model. Gray circles denote SIM limited resolution and schematics of myosin filaments were drawn from EM studies by the Borisy group and 3D PALM from Fenix et al. 2016. During expansion, a tight bundle of NMIIFs splits asymmetrically with one side spreading out in space (step 1, arrow) to form a “3 motor group” before the other side spreads (step 2, arrow) forming a “4 motor group” NMII-F stack. Finally, additional rounds of splitting and filament addition lead to the larger stacks we refer to as 4+ motor groups (step 3, arrow) (Fenix et al., 2016). Of note, (Beach et al., 2017) also noted that they occasionally saw filaments Expanding directly from two-motor groups to four-motor groups. While expansion is the dominant mechanism, a small percentage of NMIIA filament ensembles came together in a process called “Concatenation”, where long distance movements (up to microns) of distinct NMIIA filaments or NMIIA filaments stacks align to form a larger NMIIA filament stack. Interestingly, this process was not mutually exclusive with Expansion), and different cell types may display a difference on whether they rely more heavily on Expansion or Concatenation (Fenix et al., 2016).
There are three paralogs of NMII filaments in mammals: A, B, and C (Vicente-Manzanares et al., 2009). For the purposes of discussing the Network Contraction Model, we will focus on NMIIA, based on several reasons. First, NMIIA has been shown to be the major NMII paralog which drives force production in non-muscle cells (Cai et al., 2006). NMIIA is also the first paralog to create filaments and is present in every filament at the leading edge of motile cells. In addition, NMIIA can be found in the same filaments as the other paralog at the edge of motile cells, NMIIB (Beach et al., 2017; Shutova et al., 2014), and knocking down NMIIA with siRNA resulted in the absence of NMIIB filament ensembles at the edge (Fenix et al., 2016). Of note, knocking down NMIIB does not result in the absence of NMIIA (Shutova et al., 2017). This data suggests that NMIIA filaments may act as a template for the creation of NMIIB filaments. However, a template-mechanism is likely to be complicated, as NMIIA is not formally required for NMIIB filament assembly in all cell types. For example, some non-motile cells, like Cos 7, lack NMIIA and still form NMIIB filaments. Though taken together, it is clear that NMIIA is a good marker for NMII filaments in motile cells, and may be the dominant paralog at the edge where myosin II filaments are found in the spread conformation relevant to the Network Contraction Model.
The dimensions of single NMII filaments have been thoroughly characterized in vitro using electron microscopy. The length of a NMII filament is ~280 nm (Figure 1A) (Billington et al., 2013; Pollard, 1975), which puts the total length of a NMII filament right at the diffraction limit. Therefore, it is difficult to impossible to reveal the required structurally dynamic information needed to test the Network Contraction Model using diffraction limited light microscopy. With the advent of new, super-resolution microscopy techniques amenable to live-cell imaging however, questions on how NMII filament ensembles and the greater cytoskeletal architecture are dynamically formed can now be explored (Gustafsson, 2005).
In 2014, the Lippincott-Schwartz group demonstrated that structured illumination microscopy (SIM), which yields ~110–130 nm resolution, could resolve motor groups on opposite sides of a NMIIA filament (Burnette et al., 2014). This work also showed organized bundles of actin containing the actin bundling protein α-actinin-1 are present before NMII accumulation at the leading edge of cells, indicating that the actin bundle precedes the appearance of NMII, questioning whether NMII filament aggregation is forming the actin bundle as the network contraction model predicts. Though this work did not directly address how NMIIA stacks were forming, the data showed NMIIA filament stacks appeared to form by the enlargement of a NMIIA filament rather than by aggregation with separate NMIIA filaments (Burnette et al., 2014). This observation also seemed to contradict the Network Contraction model, and led to more thorough investigations of NMIIA stack formation.
SIM has subsequently been used to resolve the spread organization of NMIIA filaments as shown previously by Borisy’s group via EM (Fenix et al., 2016). Each ensemble of NMIIA filaments were characterized based on the number of SIM limited-spots that were present when imaging an expressed construct in which the motor domain of NMIIA was fused to a fluorescent protein (Fenix et al., 2016). We refer to each of these spots as a “motor-group” because each SIM limited region could — and probably does — contain multiple bundled NMII filaments, as argued in (Beach et al., 2017) (Figure 2C). SIM revealed four major categories of NMIIA filament ensembles, which were called 2-motor-groups, 3-motor-groups, 4-motor-groups and >4-motor-groups (Fig. 2C) (Fenix et al., 2016). The 2-motor-groups represented a single NMIIA filament or tight bundle of NMIIA filaments and the 3, 4, and >4 motor-groups represented spread organizations and stacks. Based on the Network Contraction Model, the expectation was that the more spread conformations would condense into a stack-like ensemble (Fig. 2B). Surprisingly, live-cell imaging revealed that larger motor-groups only rarely, if ever, condensed into smaller motor-groups. Instead the majority (85%) of smaller ensembles (e.g., 2-motor-groups) somehow obtained new motor-groups, thus transforming them into 3, 4, and >4 motor-groups (Figure 2C) (Fenix et al., 2016). This phenomenon was termed “Expansion” (Fenix et al., 2016).
The asymmetry in the way the 2-motor-groups transitioned into larger ensembles hinted at the mechanisms underlying Expansion. Time-lapse imaging using SIM, single particle tracking photo-activated microscopy and photo-activation experiments supported the hypothesis that NMIIA filaments were moving apart from one another in a step wise manner (Fenix et al., 2016). That is, the motor-groups on one side of the ensemble would move apart first followed by the motor-groups moving apart on the other side (Fig. 2C). Indeed, higher temporal sampling using time-lapse SIM imaging further supported the notion that Expansion involved filaments in NMIIA ensembles moving away from each other (Beach et al., 2017). Unfortunately, the resolution limit of SIM precludes a detailed discussion of the initial organization of filament(s) in the 2-motor-groups. It is clear that multiple filaments must exist as they eventually move apart from one another. How these filaments assemble so close to one another remains an interesting and open question, which will require the development and application of new imaging modalities to fully explore. However, the subsequent steps of the Expansion Model can easily be resolved with SIM and can be utilized immediately to elucidate the mechanisms controlling NMIIA filament stack formation. Indeed, experiments have already revealed some major mechanisms of Expansion. NMIIA motor activity, actin filament density around NMIIA filaments, and RhoA associated kinase are involved (Beach et al., 2017; Fenix et al., 2016). In addition, there also appears to be competition between RhoA associated kinase and myosin light chain kinase regulating Expansion (Beach et al., 2017). Excitingly, these studies have just scratched the surface of how Expansion occurs.
Many questions about Expansion are yet to be addressed. For example, it is clear that more NMIIA molecules are being added to the growing NMIIA filament ensembles during Expansion (Fig. 2C) (Beach et al., 2017; Fenix et al., 2016). How this occurs and is regulated remains unknown. There are phosphorylation sites along both the NMIIA heavy chain and regulatory light chain (Vicente-Manzanares et al., 2009), and some of these sites could be regulating the addition of new NMIIA molecules and/or possibly the physical movement of NMIIA filaments away from one another. In addition, it is also clear that actin filaments are an important player during myosin II stack formation as reducing the density of the actin filament network reduces stack size (Fenix et al., 2016). In addition, combined inhibition of the Arp2/3 complex and formin-mediated actin polymerization, decreased frequency of Expansion events (Beach et al., 2017). Knocking down a of a number of actin binding proteins, including actin bundling, polymerizing factors, and disassembly factors also appeared to perturb the ability of cells to organize their NMIIA stacks (Hu et al., 2017). However, a caveat to the knockdown experiments was that no quantification of organization or stack size was provided (Hu et al., 2017). Taken together, the results above suggest that future studies focusing on the interplay between actin filaments, actin binding proteins, myosin regulatory proteins, and Expansion are likely to yield interesting results.
An additionally intriguing line of research will be to explore how conserved Expansion is across cellular processes. So far, work on Expansion has primarily focused on the formation of NMIIA filament ensembles in crawling cells during interphase. Similar NMIIA ensembles (i.e., arranged as stacks) can be present in the cleavage furrow during cytokinesis, where they also appear to Expand (Fenix et al., 2016). Given that the leading edge of crawling cells and the cleavage furrow are just two of several cellular structures that contain myosin II stacks (e.g., tight junctions of epithelia cells and the sarcomeres of striated muscle cells), there is potential for Expansion to play a role in the ability of cells to generate contractile forces in many cellular contexts.
Available data following the advent of amenable live-cell super-resolution microscopy points to Expansion as a major mechanism of NMIIA filament stack formation at the leading edge of migrating cells and in the cleavage furrow of dividing cells (Beach et al., 2017; Fenix et al., 2016). This does not mean that the Network Contraction Model is not a potential mechanism of NMII stack assembly in other cellular contexts. For example, the Network Contraction Model may form stacks where NMII filaments are denser (e.g., ventral stress fibers) and not as amenable to being resolved by SIM. Indeed, there is currently no reason to suggest that Expansion and Network Contraction should be considered mutually exclusive. However, future evidence will be required to show when and where Network Contraction occurs.
Finally, other potential mechanisms of NMII stack assembly will also need to be more thoroughly studied in the context of Expansion. For example, separate NMII filament ensembles can run into each other to form a NMII stack in a process called Concatenation (Fenix et al., 2016), which was later confirmed by (Hu et al., 2017). Of interest, Concatenation was not mutually exclusive with Expansion (i.e., Expansion could occur in two NMII ensembles as they Concatenated) (Fenix et al., 2016). While Concatenation was rare at the leading edge, it also could be more dominant within denser contractile arrays. Future investigations of NMII assemblies in diverse cellular systems will be required to reveal the extent of the Expansion model and how it relates to longer-range interactions driving stack formation such as Concatenation.
References
- Adelstein RS, Pollard TD, and Kuehl WM. 1971. Isolation and characterization of myosin and two myosin fragments from human blood platelets. Proceedings of the National Academy of Sciences of the United States of America. 68:2703–2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babbin BA, Koch S, Bachar M, Conti MA, Parkos CA, Adelstein RS, Nusrat A, and Ivanov AI. 2009. Non-muscle myosin IIA differentially regulates intestinal epithelial cell restitution and matrix invasion. Am J Pathol. 174:436–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach JR, Bruun KS, Shao L, Li D, Swider Z, Remmert K, Zhang Y, Conti MA, Adelstein RS, Rusan NM, Betzig E, and Hammer JA. 2017. Actin dynamics and competition for myosin monomer govern the sequential amplification of myosin filaments. Nature cell biology. 19:85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billington N, Wang A, Mao J, Adelstein RS, and Sellers JR. 2013. Characterization of three full-length human nonmuscle myosin II paralogs. The Journal of biological chemistry. 288:33398–33410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette DT, Shao L, Ott C, Pasapera AM, Fischer RS, Baird MA, Der Loughian C, Delanoe-Ayari H, Paszek MJ, Davidson MW, Betzig E, and Lippincott-Schwartz J. 2014. A contractile and counterbalancing adhesion system controls the 3D shape of crawling cells. J Cell Biol. 205:83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Biais N, Giannone G, Tanase M, Jiang G, Hofman JM, Wiggins CH, Silberzan P, Buguin A, Ladoux B, and Sheetz MP. 2006. Nonmuscle myosin IIA-dependent force inhibits cell spreading and drives F-actin flow. Biophysical journal. 91:3907–3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Even-Ram S, Doyle AD, Conti MA, Matsumoto K, Adelstein RS, and Yamada KM. 2007. Myosin IIA regulates cell motility and actomyosin-microtubule crosstalk. Nature cell biology. 9:299–309. [DOI] [PubMed] [Google Scholar]
- Fenix AM, Taneja N, Buttler CA, Lewis J, Van Engelenburg SB, Ohi R, and Burnette DT. 2016. Expansion and concatenation of non-muscle myosin IIA filaments drive cellular contractile system formation during interphase and mitosis. Mol Biol Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupton SL, and Waterman-Storer CM. 2006. Spatiotemporal feedback between actomyosin and focal-adhesion systems optimizes rapid cell migration. Cell. 125:1361–1374. [DOI] [PubMed] [Google Scholar]
- Gustafsson MG 2005. Nonlinear structured-illumination microscopy: wide-field fluorescence imaging with theoretically unlimited resolution. Proceedings of the National Academy of Sciences of the United States of America. 102:13081–13086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Dasbiswas K, Guo Z, Tee YH, Thiagarajan V, Hersen P, Chew TL, Safran SA, Zaidel-Bar R, and Bershadsky AD. 2017. Long-range self-organization of cytoskeletal myosin II filament stacks. Nature cell biology. 19:133–141. [DOI] [PubMed] [Google Scholar]
- Huxley HE 1969. The mechanism of muscular contraction. Science. 164:1356–1365. [PubMed] [Google Scholar]
- Ma X, and Adelstein RS. 2014. The role of vertebrate nonmuscle Myosin II in development and human disease. Bioarchitecture. 4:88–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabuchi I, and Okuno M. 1977. The effect of myosin antibody on the division of starfish blastomeres. J Cell Biol. 74:251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD 1975. Electron microscopy of synthetic myosin filaments. Evidence for cross-bridge. Flexibility and copolymer formation. J Cell Biol. 67:93–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel MS, Lopez JI, McGhee EJ, Croft DR, Strachan D, Timpson P, Munro J, Schroder E, Zhou J, Brunton VG, Barker N, Clevers H, Sansom OJ, Anderson KI, Weaver VM, and Olson MF. 2011. Actomyosin-mediated cellular tension drives increased tissue stiffness and beta-catenin activation to induce epidermal hyperplasia and tumor growth. Cancer Cell. 19:776–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandquist JC, Swenson KI, Demali KA, Burridge K, and Means AR. 2006. Rho kinase differentially regulates phosphorylation of nonmuscle myosin II isoforms A and B during cell rounding and migration. The Journal of biological chemistry. 281:35873–35883. [DOI] [PubMed] [Google Scholar]
- Shutova MS, Asokan SB, Talwar S, Assoian RK, Bear JE, and Svitkina TM. 2017. Self-sorting of nonmuscle myosins IIA and IIB polarizes the cytoskeleton and modulates cell motility. J Cell Biol. 216:2877–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shutova MS, Spessott WA, Giraudo CG, and Svitkina T. 2014. Endogenous species of mammalian nonmuscle myosin IIA and IIB include activated monomers and heteropolymers. Current biology : CB. 24:1958–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich JA 2014. Hypertrophic and dilated cardiomyopathy: four decades of basic research on muscle lead to potential therapeutic approaches to these devastating genetic diseases. Biophysical journal. 106:1236–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Cheung A, Limouze J, Chen I, Westwood NJ, Sellers JR, and Mitchison TJ. 2003. Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science. 299:1743–1747. [DOI] [PubMed] [Google Scholar]
- Svitkina TM 2018. Ultrastructure of the actin cytoskeleton. Curr Opin Cell Biol. 54:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina TM, Verkhovsky AB, McQuade KM, and Borisy GG. 1997. Analysis of the actin-myosin II system in fish epidermal keratocytes: mechanism of cell body translocation. J Cell Biol. 139:397–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullio AN, Accili D, Ferrans VJ, Yu ZX, Takeda K, Grinberg A, Westphal H, Preston YA, and Adelstein RS. 1997. Nonmuscle myosin II-B is required for normal development of the mouse heart. Proceedings of the National Academy of Sciences of the United States of America. 94:12407–12412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkhovsky AB, and Borisy GG. 1993. Non-sarcomeric mode of myosin II organization in the fibroblast lamellum. J Cell Biol. 123:637–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkhovsky AB, Svitkina TM, and Borisy GG. 1995. Myosin II filament assemblies in the active lamella of fibroblasts: their morphogenesis and role in the formation of actin filament bundles. J Cell Biol. 131:989–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkhovsky AB, Svitkina TM, and Borisy GG. 1999. Network contraction model for cell translocation and retrograde flow. Biochem Soc Symp. 65:207–222. [PubMed] [Google Scholar]
- Vicente-Manzanares M, Ma X, Adelstein RS, and Horwitz AR. 2009. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nature reviews. Molecular cell biology. 10:778–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia ZK, Yuan YC, Yin N, Yin BL, Tan ZP, and Hu YR. 2012. Nonmuscle myosin IIA is associated with poor prognosis of esophageal squamous cancer. Dis Esophagus. 25:427–436. [DOI] [PubMed] [Google Scholar]