Abstract
In vitro culture models have become an indispensable tool for assessing a vast variety of biological questions in many scientific fields. However, common in vitro cultures are maintained under static conditions, which do not reflect the in vivo situation and create a non-physiological environment. To assess whether the growth characteristics of cells cultured at pulsed-perfused versus static conditions differ, we observed the growth of differentially cultured cells in vitro by life-cell time-lapse imaging of recombinant HEK293YFPI152L cells, stably expressing yellow fluorescent protein. Cells were grown for ~ 30 h at 37 °C and ambient CO2 concentration in biochips mounted into a custom-designed 3D printed carrier and were imaged at a rate of ten images per hour using a fluorescence microscope with environment control infrastructure. Cells in one chip were maintained under static conditions whereas cells in another chip were recurrently perfused with fresh media. Generated image series were quantitatively analyzed using a custom-modified cell detection software. Imaging data averaged from four biological replicates per culturing condition demonstrate that cells cultured under conventional conditions exhibit an exponential growth rate. In contrast, cells cultured in periodic mode exhibited a non-exponential growth rate. Our data clearly indicate differential growth characteristics of cells cultured under periodic versus static conditions highlighting the impact of the culture conditions on the physiology of cells in vitro.
Keywords: Cell growth, Biochip, Microfluidics, HEK293, YFPI152L, Long-term time-lapse microscopy
Introduction
Common in vitro culture models in cell biology
In vitro culture models have become an indispensable tool for assessing a vast variety of biological questions in a broad spectrum of scientific fields. Common in vitro experiments applied for example in drug screening or chemical susceptibility testing, are based on the assessment of the function of cells, either grown in suspension, in a monolayer, or in three-dimensional configuration, e.g. in spheroids or in multi-cell-configuration such as organ-on-chip approaches (Alexander et al. 2018; Bhise et al. 2014; Cho and Yoon 2017; Gu et al. 2004; Walzik et al. 2015; Wiest et al. 2005, 2006). These cultures are typically being established by seeding cells in culture medium at defined density and volume into culture ware, such as flasks, dishes, multi-well plates or lab-on-a-chip devices, followed by incubation for several hours, days or even weeks according to the principles on ‘Good Cell Culture Practice’ (GCCP) (Pamies et al. 2017; Hartung et al. 2001). Typical in vitro culture models employed for the aforementioned applications include e.g. HEK293 (human embryonic kidney 293) cells. The cells are reported to represent an important manufacturing platform in bioengineering as this culture model allows production of recombinant proteins in large scale and at high efficiency (Hu et al. 2018).
Assay types for assessment of cellular growth characteristics
Cellular growth during the incubation period is typically measured by assessing the cell number using either label-free or label-based readout technologies in continuous mode (time-lapse evaluation) or by quantification of the cell number at experiment initiation and at the end of the incubation period (endpoint observation). Label-free technologies, such as optical or electrochemical methods measure the cell number by assessment of e.g. the cellular contrast in transmission light images, extrusion of H3O+, cellular consumption of dissolved oxygen, electrophysiological activity or changes in the dielectricity of cells (Fang 2007; Liu et al. 2014; Walzik et al. 2015; Weiss et al. 2013; Wiest et al. 2005). Label-based approaches analyse the cell number based on luminescence or fluorescence markers reflecting, e.g. intracellular ATP content (e.g. CellTiterGlo® Assay Kit), nuclear DNA content (e.g. Hoechst 33342 stain), activity of non-specific intracellular esterases (e.g. Calcein-AM) or the integrity and function of the cell membrane and membrane proteins such as ion channels (e.g. YFPI152L) (Kuenzel et al. 2016, 2017; Menzner et al. 2015; Gilbert and Boutros 2016; Gilbert et al. 2011). A generic approach for label-based evaluation of cellular growth in continuous mode uses fluorescent proteins such as variants of green fluorescent protein (GFP) for labeling. Fluorescent proteins are superior to label-based monitoring of cellular growth compared to e.g. loadable fluorescence indicators or luminescent markers, as these proteins are 1. typically fairly stable with respect to bleaching, 2. non-toxic, 3. cheap, i.e. resource effective, and 4. are not diluted by cell division, thus maintaining strong fluorescence intensity across several generations of cells. YFPI152L, a genetically engineered variant of yellow fluorescent protein (YFP), fulfills the requirements for fluorescence-based long-term life-cell analysis and has been successfully applied for a variety of biological questions including assessment of cellular growth in time-lapse experiments (Balansa et al. 2010, 2013a, b; Chung et al. 2010; Gebhardt et al. 2010; Gilbert et al. 2009a, b, d; Kruger et al. 2005; Talwar et al. 2013; Walzik et al. 2015). Using this assaying method, the growth or proliferation rate of cells is typically quantified by analyzing either the number of single cells or the confluence, i.e. the percentage of the overall area covered by cells, in images obtained from automated fluorescence microscopy using high-content fluorescence imaging infrastructure (Gilbert et al. 2009c; Schneidereit et al. 2017; Spitzer et al. 2016).
Static versus periodic growth conditions
While many aspects of experimental conditions such as cell culture media composition, cell culture ware, composition of gaseous phase, mechanical stimulation and shear stress have been investigated in detail, the difference of static culture conditions versus periodic exchange of cell culture medium has only raised marginal interest (Yao and Asayama 2017; McGillicuddy et al. 2018; van der Valk et al. 2018; van Midwoud et al. 2012; Abolpour Mofrad et al. 2016; Huh et al. 2010; Inamdar and Borenstein 2011; Khademhosseini and Langer 2016; Liu et al. 2006, 2014; Pfister et al. 2015; Demmel et al. 2015; Mahto et al. 2010). Static culture types are probably the most frequently employed in vitro model, but the culturing conditions obviously may create a non-physiological environment. Due to the fact, that the medium remains unchanged throughout several days, the cells presumably suffer from decreasing availability of nutrients, build-up of cellular metabolites, and accumulation of indicators of cell stress as well as altered physiology and viability, compared to cells cultured under in vivo-like conditions that mimic the native environment of tissue. It is well known that the extracellular micro-environment acidifies in static cultures within minutes (McConnell et al. 1992), creating a non-physiological situation. Cells in their native environment in vivo are continuously perfused with extracellular fluid, creating a dynamic extracellular microenvironment of physiological abundance of nutrients and metabolites. To address the issue encountered with conventional in vitro cultures described above, (micro-) fluidics devices, allowing for continuous or periodical perfusion with fresh media, are increasingly being developed and proposed for in vitro models used e.g. in the context of drug susceptibility testing (Eklund et al. 2004; Marx et al. 2016; McConnell et al. 1992; Weltin et al. 2014; Wolf et al. 1998).
To assess whether the growth characteristics of cells cultured at periodic i.e. dynamically changing, conditions differ from conventional, i.e. static, conditions, we aimed to observe the growth of differentially cultured cells in vitro by life-cell time-lapse fluorescence imaging. For assessing the growth characteristics, we aimed to employ recombinant HEK293YFPI152L cells, stably expressing a variant of yellow fluorescent protein, and to quantify the growth area or confluence in image series. In order to evaluate whether the growth rates differ between the employed approaches, we further aimed to calculate the fold-change in growth area after 30 h culturing duration.
Materials and methods
Reagents
Poly-d-lysine (PDL) was obtained from Sigma (Taufkirchen, Germany). PDL was prepared as 10× stock in water and stored at 4 °C.
Cell line
HEK293 cells (CRL-1573™) were purchased from The American Type Culture Collection (ATCC, Manassas, VA, USA). Generation of the recombinant HEK293YFPI152L cell line is described in Walzik et al. (2015).
Cell culture
Recombinant HEK293YFPI152L were maintained in DMEM (Invitrogen, Darmstadt, Germany) supplemented with 10% fetal bovine serum (FBS, Biochrom, Berlin, Germany) and penicillin (100 U/ml)/streptomycin (100 mg/ml) (Invitrogen) and were cultured in T75 flasks (TPP, Trasadingen, Switzerland) at 37 °C, 5% CO2 according to standard procedures. Cells were passaged every 2–3 days and used in long term imaging experiments when approx. 80–90% confluent.
Preparation of cell chips
As a preparatory step prior to long-term imaging experiments and in order to promote adherence of cells onto the growth surface of the employed cell chips (Cellasys GmbH, Kronburg, Germany), cleaned and sterilized chips were filled with 200 µl 1× poly-d-lysine (PDL, Sigma) and were incubated for 10 min at room temperature in a laminar flow hood. Upon aspiration of the PDL solution the chips were left in the laminar flow hood for approx. 10 min for drying. In a next step, PDL-coated chips were filled with 300 µl standard cell culture medium supplemented with a total of 6 × 104 cells and were placed into an incubator for 12–24 h.
Preparation of long-term imaging experiments
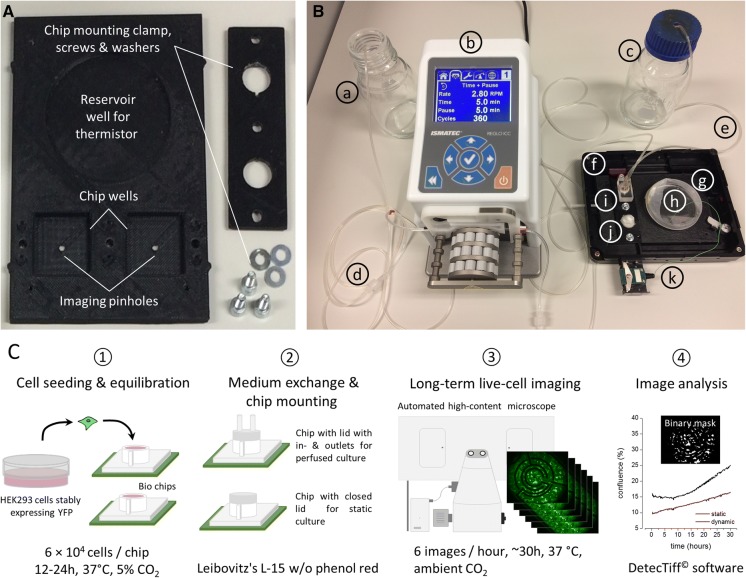
Approx. 30 min prior to long-term imaging experiments, the culture medium within the chips was replaced by 300 µl Leibovitz’s L-15 medium (Merck, Darmstadt, Germany) without phenol red, supplemented with 10% fetal bovine serum and penicillin (100 U/ml)/streptomycin (100 mg/ml). To prevent evaporation of the culture medium during long-term experimentation, the chip to be used in static culture mode was closed with a plastic conical cap and was sealed with parafilm. The second chip to be used in periodic or pulsed-perfused culture mode was closed with a different lid, equipped with in- and outlets to allow for media exchange during experimentation. Both chips were mounted into a purpose-designed chip carrier and the assembled chip carrier was transferred to the motorized stage of a of a high-content long-term imaging system (Nikon Eclipse Ti, Nikon, Tokyo, Japan) equipped with a cell culture incubator (Okolab, Pozzuoli, Italy) capable of maintaining a constant temperature during the time-lapse experiment. An image of the set-up including the assembled chip carrier is shown in Fig. 1a.
Fig. 1.
Setup and workflow for long-term culture and parallel time-lapse imaging. a 3D printed chip carrier. The carrier was printed using the biocompatible thermoplast ABS and has the dimensions of a standard multi-titer plate. It provides wells for two biochips and a standard 6 cm cell culture dish (see ‘h’ in b). During life-cell imaging the culture dish is filled with 4 ml water, serving as a reference for the thermistor (see ‘k’ in b) and the environment control infrastructure of the microscope. Cells are imaged through a pinhole at the bottom of the carrier. b Setup for establishing static and periodic culturing conditions during time-lapse long-term imaging. (a) medium waste bottle, (b) peristaltic pump, (c) medium reservoir bottle, (d) waste tubing, (e) perfusion tubing, (f) imaging chamber, (g) 3D printed chip carrier, (h) water reservoir for thermistor, (i) chip for perfusion culture, (j) chip for static culture, (k) thermistor cable and connector. c Experimental workflow for comparative cell growth analysis. Details see text
Long-term imaging experiments
Cells were imaged with a 10× objective (CFI Plan Fluor DL 10X Phase, N.A. 0.30, Nikon). Illumination from a xenon lamp (Lambda LS, Sutter Instruments, Novato, CA, USA), passing through a filter block (C-FL Epi-FL FITC, EX 465–495, DM 505, BA 515–555, Olympus, Tokyo, Japan) was used to excite and detect the YFPI152L fluorescence signal. Fluorescence was imaged by a sCMOS camera (NEO, Andor, Belfast, Northern Ireland, UK) and digitized to disk onto a computer (Dell Precision T3500, Dell, Round Rock, TX, USA) with Windows 7 operating System (Microsoft Corporation, USA). The primary resolution of the camera was 2560 × 2160 pixel. The experimental protocol involved imaging each chip every 6 min for a total of 30 h. Cells cultured in periodic culture mode were perfused repeatedly every 10 min for a period of 5 min and at a rate of 60 µl per minute with a total volume of 300 µl Leibovitz’s L-15 medium without phenol red, supplemented with 10% fetal bovine serum and penicillin (100 U/ml)/streptomycin (100 mg/ml). Liquid-handling was performed with a peristaltic pump (Ismatec Reglo ICC 4CH, Cole-Parmer, Wertheim, Germany).
Image analysis
Image series of fluorescent HEK293YFPI152L cells were automatically analyzed using a modified version of DetecTIFF® software (Gilbert et al. 2009c) written in LabView (National Instruments, Dublin, Ireland). In brief, images were segmented using an iterative size and intensity-based thresholding algorithm and the image area covered by cells, i.e. the confluence, was calculated as percentage of the overall image area.
Data analysis and visualization
Image data were annotated in Microsoft Excel as well as analyzed and plotted using Origin 7G (OriginLab Corporation, Northampton, MA, USA). For comparison of cell growth, the fold-change, i.e. the increase of the growth area covered by cells within a period of 30 h, was calculated using the following equation:
where Areainit is the growth area observed at experiment initiation and Areafinal is the growth area calculated after 30 h culture duration. Statistical analysis was done based on one-way ANOVA tests, checking for data normality and performing post hoc tests (Dunn or Bonferroni method). ‘n.s.’ in the graph indicates that the difference between the analyzed populations is not statistically significant.
Linear and exponential fits were obtained using Origin 7G (OriginLab Corporation). The fit accuracy is expressed as coefficient of determination (R2).
Results
Custom infrastructure for comparative culture analysis
To assess whether the growth characteristics of cells cultured at periodically changing, conditions differ from conventional, i.e. static, conditions, we aimed to observe the growth of differentially cultured cells in vitro by life-cell time-lapse imaging. To support direct comparison of generated imaging data, we intended to observe the different cultures in the same experiment. As the dimensions of the employed biochips differ from those of standard culture ware, we purposely designed a chip carrier using computer assisted design (CAD, see Methods for details) and printed it with a custom-grade 3D printer. An image of the chip carrier without mounted chips is shown in Fig. 1a, mounted chips as prepared for automated imaging are shown in Fig. 1b.
Experimental workflow for comparative cell growth analysis
In a first step prior to comparative imaging, recombinant and fluorescent HEK293YFPI152L cells were seeded into biochips at defined density in DMEM, supplemented with fetal calf serum and antibiotics (see Methods for details) and were incubated at standard culture conditions over night (see step 1 in Fig. 1c). We decided to use recombinant HEK293YFPI152L cells as the HEK293 (human embryonic kidney-derived) host cell line is robust and modest with respect to culturing conditions. Furthermore, HEK293 cells are a commonly employed culture model for answering a vast variety of biological questions in a broad range of laboratories worldwide. In addition, the cells stably express a variant of yellow fluorescent protein (YFP), exhibiting a high signal-to-noise ratio in fluorescence microscopy, and thus enable quantitative and automated analysis of cellular growth based on fluomicrographs obtained from life-cell time-lapse imaging. The next day, the standard culture medium was replaced by 300 µl Leibowitz’s L-15 medium without phenol red. We used Leibowitz’s L-15 medium without phenol red as it allows culturing cells at ambient CO2 concentration and minimizes background contamination through scattered light in fluorescence microscopy as compared to phenol red-containing medium. Prepared chips were mounted into the chip carrier (see image in Fig. 1b and step 2 in Fig. 1c) and were transferred to the motorized stage of an automated Nikon Eclipse Ti microscope with cell culture incubator for maintaining a constant temperature during time-lapse experimentation. In this configuration, the cells were recurrently imaged every 6 min for a total of ~ 30 h at 37 °C and ambient CO2 concentration (see step 3 in Fig. 1c). Cell chips used in static mode were imaged in 300 µl Leibowitz’s L-15 medium added prior imaging initiation as described above. Chips used in periodic mode were recurrently perfused with new medium during automated imaging using the setup shown in Fig. 1b as detailed in the Methods section. Generated images sequences were subsequently quantitatively analyzed using a modified version of DetecTiff© software (Gilbert et al. 2009c) (see step 4 in Fig. 1c).
Growth of cells cultured under periodic versus static conditions
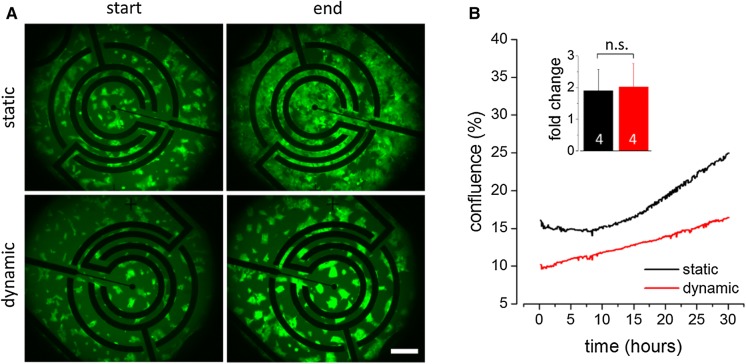
Figure 2a shows representative images of recombinant HEK293YFPI152L cells at experiment initiation (start) and after 30 h imaging duration (end) captured from cells cultured in cell chips in static (upper row) and periodic culture mode (bottom row). For comparison of the growth characteristics of differentially cultured HEK293YFPI152L cells, the growth area or cellular confluence was quantified in single images of the generated image series using image analysis software. Figure 2b shows time courses of the mean confluence (in %, ± SD) of HEK293YFPI152L cells, cultured in static (black) and periodic (red) culture mode, respectively, averaged from a total of four biological replicates each. These data demonstrate that cells cultured under conventional, i.e. static conditions exhibit an exponential growth rate. Exponential growth of cells in vitro has previously been reported for different cell lines, including HEK293 cells and HEK293YFPI152L cells and thus, was somewhat expected (Walzik et al. 2015). In contrast, cells cultured in periodic mode exhibited a non-exponential, linear growth rate within the observed time period. In order to evaluate whether the differences observed for the cellular growth rate also affected the overall increase in confluence, we calculated the average fold change in growth area for each culturing mode based on the first and last image taken during long-term experimentation. The histogram inset in Fig. 1b clearly indicates that the fold change values (mean ± SD, N = 4) calculated for cells cultured in periodic (1.8 ± 0.6) and static mode (2.0 ± 0.7), respectively, are not significantly different. This result is expected as normalization of the depicted time courses to the initial confluence value reveals intersecting curves at approx. 30 h culture duration.
Fig. 2.
Comparative analysis of cellular growth characteristics of HEK293YFPI152L cells at static versus periodic culturing conditions. a Representative fluomicrographs of HEK293YFPI152L cells cultured in multi-parametric cell chips in static (upper row) and periodic (bottom row) culture mode at experiment initiation (start) and after 30 h imaging duration (end). Scale bar: 200 µm. b Time courses of the average confluence (in %, mean ± SD, N = 4) of cells cultured in static (black) and periodic (red) culture mode, respectively, calculated from images as shown in (a). The time courses indicate exponential and linear growth for cells cultured in static and periodic mode, respectively. The inset histogram displays the average fold-change in growth area (mean ± SD, N = 4) and indicates that the final growth area is comparable for both culture conditions after 30 h culture duration. ‘n.s.’: not significant. (Color figure online)
In order to provide an additional and more comprehensive indicator, also reflecting the time-course of the growth rate, we fitted the generated time-course data using linear and exponential functions for assessing time-resolved growth characteristics of cells grown under dynamic and static conditions, respectively (see Methods for details). The fit accuracy is expressed as coefficient of determination (R2) and was 0.97 ± 0.27 and 0.99 ± 0.17 for cells cultured in periodic mode and under static conditions, respectively.
Discussion
For the sake of feasibility and cost reasons, in vitro cultures are conventionally maintained under static rather than under periodic culture condition, because periodic cultures require a more complex infrastructure that is also much more resource-intensive compared to the conventional culturing approach. Static culture conditions, however, create a non-physiological environment presumably altering physiology and viability compared to in vivo-like culture conditions. The initial benefit of static cultures may thus be compromised by subsequent time-consuming and cost-intensive validation of experimental results using individual assay types and different culture approaches. Using long-term time-lapse fluorescence microscopy we have analyzed the growth of cells in vitro cultured under static versus periodic condition. Our data demonstrate that HEK293 cells cultured under conventional conditions exhibit an exponential growth rate, whereas HEK293 cells cultured in periodic mode exhibited a non-exponential, rather linear, growth rate. Exponential growth of cells in vitro has previously been reported for different cell lines, including stem cells and culture models such as HEK293YFPI152L, RKO, HCT116, Lim1215 or HT29 cells (Shekar and Ranganathan 2012; Witzel et al. 2015; Walzik et al. 2015; Yates et al. 2017)_ENREF_46. Furthermore, exponential growth is initially expected as a single parent cell divides into two daughter cells during cell division.
There are numerous possible explanations for the phenomenon of the observed non-exponential and rather linear growth behavior of periodically cultured cells. For example, mechanical stress or stimulation of the cells may explain the observed differences. During cell division, adherent cells in vitro change their morphotype from flat and outspread to round and loosely attached. When the cells are perfused during cell division, the loosely attached cells may be mechanically dislodged from the bottom of the biochip and removed from the quantified cell population. Visual observation would be the gold standard and generally suitable for testing this hypothesis, but the acquired image series are not suitable as the employed sampling rate (ten images per hour) is too small for identification of cells being washed away during parallel cell division and perfusion. Also, a phenomenon called contact inhibition of proliferation, describing the inhibition of cell proliferation in a density dependent manner, may be involved in the observed linear proliferation characteristics in perfused cultures (Stoker and Rubin 1967). Mechanosensitive ion channels such as piezo proteins, have been associated with cellular development, volume regulation, cellular migration, proliferation, and elongation and may be activated through the liquid flow and hence, interact with cellular growth (Bagriantsev et al. 2014). Besides mechanical stress or stimulation of the cells, another mechanism may explain the observed linear growth characteristics. Signaling molecules, secreted by the cells and involved in cell cycle progression, division and proliferation such as ligands of intracellular signaling pathways and growth factors may be accumulated at lower concentration in perfused cultures compared to static cultures and thus may cause slower cell cycle progression. Another consequence of the aforementioned accumulation of signaling molecules and factors may also affect synchronization of the cell cycle of the HEK293 cells in culture. However, none of the hypothesized mechanisms has been evaluated in detail as this was not within the scope of this study. We aimed at comparing the growth characteristics of HEK293 cells cultured under static versus periodic conditions and providing the results to the community for several reasons. First, to bring the relevance of perfusion cultures in the focus of up-to-date cell biology emphasizing the importance of the nutritional status of cells in vitro and in the context of GCCP (Coecke et al. 2005; Hartung et al. 2001; Pamies et al. 2017) and second, to spark the interest for further experimentation towards gaining a deeper understanding of the phenomenon observed in the course of this study.
Despite the fact that our observation of a non-exponential and rather linear growth behavior of periodically cultured cells has been reproduced in a total of four independently conducted biological replicate experiments with recombinant HEK293 cells, further experimentation with additional in vitro culture models would be required for validation of our observations as a general phenomenon occurring in cultures of periodically cultured cells.
While this work focuses on in vitro cultures of HEK293 cells, the methodological approach could also be adapted for other cell lines and strategies, such as drug or toxicity screening, e.g. in the context of the 3Rs (Russell and Burch 1959; Alexander et al. 2018).
Acknowledgements
The authors gratefully acknowledge funding of the Staedtler Stiftung and ongoing support from the Erlangen Graduate School in Advanced Optical Technologies (SAOT) by the German Research Foundation (DFG) in the framework of the German Excellence Initiative. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Authors’ contribution
J.W. and D.F.G. conceived the study. S.A.M. and D.F.G. conducted imaging experiments. D.F.G. analyzed and displayed imaging data. D.F.G. and J.W. and wrote the paper. All authors commented and agreed on the manuscript.
Compliance with ethical standards
Conflict of interest
JW is CEO and shareholder of Cellasys GmbH.
References
- Abolpour Mofrad S, Kuenzel K, Friedrich O, Gilbert DF. Optimizing neuronal differentiation of human pluripotent NT2 stem cells in monolayer cultures. Dev Growth Differ. 2016;58:664–676. doi: 10.1111/dgd.12323. [DOI] [PubMed] [Google Scholar]
- Alexander F, Jr, Eggert S, Wiest J. A novel lab-on-a-chip platform for spheroid metabolism monitoring. Cytotechnology. 2018;70:375–386. doi: 10.1007/s10616-017-0152-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagriantsev SN, Gracheva EO, Gallagher PG. Piezo proteins: regulators of mechanosensation and other cellular processes. J Biol Chem. 2014;289:31673–31681. doi: 10.1074/jbc.R114.612697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balansa W, Islam R, Fontaine F, Piggott AM, Zhang H, Webb TI, Gilbert DF, Lynch JW, Capon RJ. Ircinialactams: subunit-selective glycine receptor modulators from Australian sponges of the family Irciniidae. Bioorg Med Chem. 2010;18:2912–2919. doi: 10.1016/j.bmc.2010.03.002. [DOI] [PubMed] [Google Scholar]
- Balansa W, Islam R, Fontaine F, Piggott AM, Zhang H, Xiao X, Webb TI, Gilbert DF, Lynch JW, Capon RJ. Sesterterpene glycinyl-lactams: a new class of glycine receptor modulator from Australian marine sponges of the genus Psammocinia. Org Biomol Chem. 2013;11:4695–4701. doi: 10.1039/c3ob40861b. [DOI] [PubMed] [Google Scholar]
- Balansa W, Islam R, Gilbert DF, Fontaine F, Xiao X, Zhang H, Piggott AM, Lynch JW, Capon RJ. Australian marine sponge alkaloids as a new class of glycine-gated chloride channel receptor modulator. Bioorg Med Chem. 2013;21:4420–4425. doi: 10.1016/j.bmc.2013.04.061. [DOI] [PubMed] [Google Scholar]
- Bhise NS, Ribas J, Manoharan V, Zhang YS, Polini A, Massa S, Dokmeci MR, Khademhosseini A. Organ-on-a-chip platforms for studying drug delivery systems. J Control Release. 2014;190:82–93. doi: 10.1016/j.jconrel.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S, Yoon JY. Organ-on-a-chip for assessing environmental toxicants. Curr Opin Biotechnol. 2017;45:34–42. doi: 10.1016/j.copbio.2016.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung SK, Vanbellinghen JF, Mullins JG, Robinson A, Hantke J, Hammond CL, Gilbert DF, Freilinger M, Ryan M, Kruer MC, Masri A, Gurses C, Ferrie C, Harvey K, Shiang R, Christodoulou J, Andermann F, Andermann E, Thomas RH, Harvey RJ, Lynch JW, Rees MI. Pathophysiological mechanisms of dominant and recessive GLRA1 mutations in hyperekplexia. J Neurosci. 2010;30:9612–9620. doi: 10.1523/JNEUROSCI.1763-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coecke S, Balls M, Bowe G, Davis J, Gstraunthaler G, Hartung T, Hay R, Merten OW, Price A, Schechtman L, Stacey G, Stokes W. Guidance on good cell culture practice. a report of the second ECVAM task force on good cell culture practice. Altern Lab Anim. 2005;33:261–287. doi: 10.1177/026119290503300313. [DOI] [PubMed] [Google Scholar]
- Demmel F, Brischwein M, Wolf P, Huber F, Pfister C, Wolf B. Nutrient depletion and metabolic profiles in breast carcinoma cell lines measured with a label-free platform. Physiol Meas. 2015;36:1367–1381. doi: 10.1088/0967-3334/36/7/1367. [DOI] [PubMed] [Google Scholar]
- Eklund SE, Taylor D, Kozlov E, Prokop A, Cliffel DE. A microphysiometer for simultaneous measurement of changes in extracellular glucose, lactate, oxygen, and acidification rate. Anal Chem. 2004;76:519–527. doi: 10.1021/ac034641z. [DOI] [PubMed] [Google Scholar]
- Fang Y. Non-invasive optical biosensor for probing cell signaling. Sensors (Basel, Switzerland) 2007;7:2316–2329. doi: 10.3390/s7102316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt FM, Mitrovic AD, Gilbert DF, Vandenberg RJ, Lynch JW, Dodd PR. Exon-skipping splice variants of excitatory amino acid transporter-2 (EAAT2) form heteromeric complexes with full-length EAAT2. J Biol Chem. 2010;285:31313–31324. doi: 10.1074/jbc.M110.153494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DF, Boutros M. A protocol for a high-throughput multiplex cell viability assay. Methods Mol Biol. 2016;1470:75–84. doi: 10.1007/978-1-4939-6337-9_6. [DOI] [PubMed] [Google Scholar]
- Gilbert D, Esmaeili A, Lynch JW. Optimizing the expression of recombinant alphabetagamma GABAA receptors in HEK293 cells for high-throughput screening. J Biomol Screen. 2009;14:86–91. doi: 10.1177/1087057108328017. [DOI] [PubMed] [Google Scholar]
- Gilbert DF, Islam R, Lynagh T, Lynch JW, Webb TI. High throughput techniques for discovering new glycine receptor modulators and their binding sites. Front Mol Neurosci. 2009;2:17. doi: 10.3389/neuro.02.017.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DF, Meinhof T, Pepperkok R, Runz H. DetecTiff: a novel image analysis routine for high-content screening microscopy. J Biomol Screen. 2009;14:944–955. doi: 10.1177/1087057109339523. [DOI] [PubMed] [Google Scholar]
- Gilbert DF, Wilson JC, Nink V, Lynch JW, Osborne GW. Multiplexed labeling of viable cells for high-throughput analysis of glycine receptor function using flow cytometry. Cytom A. 2009;75:440–449. doi: 10.1002/cyto.a.20703. [DOI] [PubMed] [Google Scholar]
- Gilbert DF, Erdmann G, Zhang X, Fritzsche A, Demir K, Jaedicke A, Muehlenberg K, Wanker EE, Boutros M. A novel multiplex cell viability assay for high-throughput RNAi screening. PLoS ONE. 2011;6:e28338. doi: 10.1371/journal.pone.0028338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu MB, Mitchell RJ, Kim BC. Whole-cell-based biosensors for environmental biomonitoring and application. Adv Biochem Eng Biotechnol. 2004;87:269–305. doi: 10.1007/b13533. [DOI] [PubMed] [Google Scholar]
- Hartung T, Gstraunthaler G, Coecke S, Lewis D, Blanck O, Balls M. Good cell culture practice (GCCP)–an initiative for standardization and quality control of in vitro studies. The establishment of an ECVAM task force on GCCP. Altex. 2001;18:75–78. [PubMed] [Google Scholar]
- Hu J, Han J, Li H, Zhang X, Liu L, Chen F, Zeng B. Human embryonic kidney 293 cells: a vehicle for biopharmaceutical manufacturing, structural biology, and electrophysiology. Cells Tissues Organs. 2018;205:1–8. doi: 10.1159/000485501. [DOI] [PubMed] [Google Scholar]
- Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE. Reconstituting organ-level lung functions on a chip. Science. 2010;328:1662–1668. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inamdar NK, Borenstein JT. Microfluidic cell culture models for tissue engineering. Curr Opin Biotechnol. 2011;22:681–689. doi: 10.1016/j.copbio.2011.05.512. [DOI] [PubMed] [Google Scholar]
- Khademhosseini A, Langer R. A decade of progress in tissue engineering. Nat Protoc. 2016;11:1775–1781. doi: 10.1038/nprot.2016.123. [DOI] [PubMed] [Google Scholar]
- Kruger W, Gilbert D, Hawthorne R, Hryciw DH, Frings S, Poronnik P, Lynch JW. A yellow fluorescent protein-based assay for high-throughput screening of glycine and GABAA receptor chloride channels. Neurosci Lett. 2005;380:340–345. doi: 10.1016/j.neulet.2005.01.065. [DOI] [PubMed] [Google Scholar]
- Kuenzel K, Friedrich O, Gilbert DF. A recombinant human pluripotent stem cell line stably expressing halide-sensitive YFP-I152L for GABAAR and GlyR-targeted high-throughput drug screening and toxicity testing. Front Mol Neurosci. 2016;9:51. doi: 10.3389/fnmol.2016.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuenzel K, Mofrad SA, Gilbert DF (2017) Phenotyping cellular viability by functional analysis of ion channels: GlyR-targeted screening in NT2-N cells. In: Gilbert DF, Friedrich O (eds) Cell viability assays. Methods in Molecular Biology 1601:205–214. Humana Press, New York, NY [DOI] [PubMed]
- Liu L, Cash TP, Jones RG, Keith B, Thompson CB, Simon MC. Hypoxia-induced energy stress regulates mRNA translation and cell growth. Mol Cell. 2006;21:521–531. doi: 10.1016/j.molcel.2006.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Wu C, Cai H, Hu N, Zhou J, Wang P. Cell-based biosensors and their application in biomedicine. Chem Rev. 2014;114:6423–6461. doi: 10.1021/cr2003129. [DOI] [PubMed] [Google Scholar]
- Mahto SK, Yoon TH, Rhee SW (2010) A new perspective on in vitro assessment method for evaluating quantum dot toxicity by using microfluidics technology. Biomicrofluidics 4:pii:034111. 10.1063/1.3486610 [DOI] [PMC free article] [PubMed]
- Marx U, Andersson TB, Bahinski A, Beilmann M, Beken S, Cassee FR, Cirit M, Daneshian M, Fitzpatrick S, Frey O, Gaertner C, Giese C, Griffith L, Hartung T, Heringa MB, Hoeng J, De Jong WH, Kojima H, Kuehnl J, Leist M, Luch A, Maschmeyer I, Sakharov D, Sips AJ, Steger-Hartmann T, Tagle DA, Tonevitsky A, Tralau T, Tsyb S, Van De Stolpe A, Vandebriel R, Vulto P, Wang J, Wiest J, Rodenburg M, Roth A. Biology-inspired microphysiological system approaches to solve the prediction dilemma of substance testing. Altex. 2016;33:272–321. doi: 10.14573/altex.1603161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcconnell HM, Owicki JC, Parce JW, Miller DL, Baxter GT, Wada HG, Pitchford S. The cytosensor microphysiometer: biological applications of silicon technology. Science. 1992;257:1906–1912. doi: 10.1126/science.1329199. [DOI] [PubMed] [Google Scholar]
- McGillicuddy N, Floris P, Albrecht S, Bones J. Examining the sources of variability in cell culture media used for biopharmaceutical production. Biotechnol Lett. 2018;40:5–21. doi: 10.1007/s10529-017-2437-8. [DOI] [PubMed] [Google Scholar]
- Menzner AK, Abolpour Mofrad S, Friedrich O, Gilbert DF. Towards in vitro DT/DNT testing: assaying chemical susceptibility in early differentiating NT2 cells. Toxicology. 2015;338:69–76. doi: 10.1016/j.tox.2015.10.007. [DOI] [PubMed] [Google Scholar]
- Pamies D, Bal-Price A, Simeonov A, Tagle D, Allen D, Gerhold D, Yin D, Pistollato F, Inutsuka T, Sullivan K, Stacey G, Salem H, Leist M, Daneshian M, Vemuri MC, Mcfarland R, Coecke S, Fitzpatrick SC, Lakshmipathy U, Mack A, Wang WB, Yamazaki D, Sekino Y, Kanda Y, Smirnova L, Hartung T. Good cell culture practice for stem cells and stem-cell-derived models. Altex. 2017;34:95–132. doi: 10.14573/altex.1607121. [DOI] [PubMed] [Google Scholar]
- Pfister C, Bozsak C, Wolf P, Demmel F, Brischwein M. Cell shape-dependent shear stress on adherent cells in a micro-physiologic system as revealed by FEM. Physiol Meas. 2015;36:955–966. doi: 10.1088/0967-3334/36/5/955. [DOI] [PubMed] [Google Scholar]
- Russell WMS, Burch RL. The principles of humane experimental technique. London: Methuen; 1959. [Google Scholar]
- Schneidereit D, Kraus L, Meier JC, Friedrich O, Gilbert DF. Step-by-step guide to building an inexpensive 3D printed motorized positioning stage for automated high-content screening microscopy. Biosens Bioelectron. 2017;92:472–481. doi: 10.1016/j.bios.2016.10.078. [DOI] [PubMed] [Google Scholar]
- Shekar R, Ranganathan K. Phenotypic and growth characterization of human mesenchymal stem cells cultured from permanent and deciduous teeth. Indian J Dent Res. 2012;23:838–839. doi: 10.4103/0970-9290.111281. [DOI] [PubMed] [Google Scholar]
- Spitzer P, Condic M, Herrmann M, Oberstein TJ, Scharin-Mehlmann M, Gilbert DF, Friedrich O, Grömer T, Kornhuber J, Lang R, Maler JM. Amyloidogenic amyloid-β-peptide variants induce microbial agglutination and exert antimicrobial activity. Sci Rep. 2016;6:32228. doi: 10.1038/srep32228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoker MGP, Rubin H. Density dependent inhibition of cell growth in culture. Nature. 1967;215:171–172. doi: 10.1038/215171a0. [DOI] [PubMed] [Google Scholar]
- Talwar S, Lynch JW, Gilbert DF. Fluorescence-based high-throughput functional profiling of ligand-gated ion channels at the level of single cells. PLoS ONE. 2013;8:e58479. doi: 10.1371/journal.pone.0058479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Valk J, Bieback K, Buta C, Cochrane B, Dirks WG, Fu J, Hickman JJ, Hohensee C, Kolar R, LiebschM Pistollato F, Schulz M, Thieme D, Weber T, Wiest J, Winkler S, Gstraunthaler G. Fetal Bovine Serum (FBS): Past - Present - Future. ALTEX. 2018;35:99–118. doi: 10.14573/altex.1705101. [DOI] [PubMed] [Google Scholar]
- Van Midwoud PM, Janse A, Merema MT, Groothuis GM, Verpoorte E. Comparison of biocompatibility and adsorption properties of different plastics for advanced microfluidic cell and tissue culture models. Anal Chem. 2012;84:3938–3944. doi: 10.1021/ac300771z. [DOI] [PubMed] [Google Scholar]
- Walzik MP, Vollmar V, Lachnit T, Dietz H, Haug S, Bachmann H, Fath M, Aschenbrenner D, Abolpour Mofrad S, Friedrich O, Gilbert DF. A portable low-cost long-term live-cell imaging platform for biomedical research and education. Biosens Bioelectron. 2015;64:639–649. doi: 10.1016/j.bios.2014.09.061. [DOI] [PubMed] [Google Scholar]
- Weiss D, Brischwein M, Grothe H, Wolf B, Wiest J. Label-free monitoring of whole cell vitality. Conf Proc IEEE Eng Med Biol Soc. 2013;2013:1607–1610. doi: 10.1109/EMBC.2013.6609823. [DOI] [PubMed] [Google Scholar]
- Weltin A, Slotwinski K, Kieninger J, Moser I, Jobst G, Wego M, Ehret R, Urban GA. Cell culture monitoring for drug screening and cancer research: a transparent, microfluidic, multi-sensor microsystem. Lab Chip. 2014;14:138–146. doi: 10.1039/C3LC50759A. [DOI] [PubMed] [Google Scholar]
- Wiest J, Brischwein M, Ressler J, Otto AM, Grothe H, Wolf B. Cellular assays with multiparametric bioelectronic sensor chips. CHIMIA Int J Chem. 2005;59:243–246. doi: 10.2533/000942905777676623. [DOI] [Google Scholar]
- Wiest J, Stadthagen T, Schmidhuber M, Brischwein M, Ressler J, Raeder U, Grothe H, Melzer A, Wolf B. Intelligent mobile lab for metabolics in environmental monitoring. Anal Lett. 2006;39:1759–1771. doi: 10.1080/00032710600714089. [DOI] [Google Scholar]
- Witzel F, Fritsche-Guenther R, Lehmann N, Sieber A, Bluthgen N. Analysis of impedance-based cellular growth assays. Bioinformatics. 2015;31:2705–2712. doi: 10.1093/bioinformatics/btv216. [DOI] [PubMed] [Google Scholar]
- Wolf B, Brischwein M, Baumann W, Ehret R, Kraus M. Monitoring of cellular signalling and metabolism with modular sensor-technique: the physiocontrol-microsystem (PCM) Biosens Bioelectron. 1998;13:501–509. doi: 10.1016/S0956-5663(97)00136-X. [DOI] [PubMed] [Google Scholar]
- Yao T, Asayama Y. Animal-cell culture media: history, characteristics, and current issues. Reprod Med Biol. 2017;16:99–117. doi: 10.1002/rmb2.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates CA, Ford MJ, Mort RL. A multi-stage representation of cell proliferation as a markov process. Bull Math Biol. 2017;79:2905–2928. doi: 10.1007/s11538-017-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]