Abstract.
The goal of understanding the architecture of neural circuits at the synapse level with a brain-wide perspective has powered the interest in high-speed and large field-of-view volumetric imaging at subcellular resolution. Here, we developed a method combining tissue expansion and light-sheet fluorescence microscopy to allow extended volumetric super resolution high-speed imaging of large mouse brain samples. We demonstrate the capabilities of this method by performing two color fast volumetric super resolution imaging of mouse CA1 and dentate gyrus molecular-, granule cell-, and polymorphic layers. Our method enables an exact evaluation of granule cell and neurite morphology within the context of large cell ensembles spanning several orders of magnitude in resolution. We found that imaging a brain region of in super resolution using light-sheet fluorescence expansion microscopy is about 17-fold faster than imaging the same region by a current state-of-the-art high-resolution confocal laser scanning microscope.
Keywords: connectomics, super resolution, tissue expansion, dentate gyrus, light-sheet fluorescence microscopy
1. Introduction
One of the prime goals in today’s neuroscience is the volumetric architectural mapping of neural circuits spanning several magnitudes of resolution, i.e., imaging large fields of view up to several millimeters in super resolution, preferably using a single, straightforward light microscopic process. This challenge has drawn the attention of scientists beginning with Ramon y Cajal, who used the Golgi technique to draw detailed pictures of neurons containing most of their neurites1 and culminates in today’s electron microscopy (EM), which allows to decode the finest details of neuronal circuit structure.2 EM’s key advantage is the possibility to identify and distinguish presynaptic active zones containing synaptic vesicles from apposed postsynaptic structures and the visualization of even the finest axonal branches. However, EM connectivity mapping is extremely time consuming and requires expensive high-tech systems, both for image acquisition and image analysis. Most importantly, the sample contrast preparation is largely incompatible with rich molecular phenotyping, which can provide critical information on cell and synapse type. Thus, an ideal analysis method would generate super resolution data sets, allowing to reconstruct the critical details of synaptic connectivity from intact brain samples linked to the molecular information on the types of cells and synapses and even to dynamic information on natural activity pattern history, which is causally relevant to animal behavior.
To date light-based imaging approaches combined with specific genetic labeling methods are important tools to visualize the structural and functional architecture of nervous tissue in high resolution.3 Confocal microscopy allows high-resolution 3-D reconstruction of nervous tissue without the need for ultrathin physical sectioning.4 The development of two-photon microscopy further increased imaging depths to several hundred micrometers and the use of adaptive optics has improved imaging depth even further.5,6
However, due to the opaqueness of nervous system tissue and the size of a mouse brain—several millimeters in all spatial dimensions—light microscopy remains limited for volumetric imaging at nanometer resolution through intact brain samples. A possible solution to this problem is to slice the brain into thin sections followed by confocal-, or 2P imaging.7,8 However, detailed labeling and high-resolution connectivity reconstruction of thin sections is limited to small volumes and/or by the diffraction barrier of conventional light microscopes. Thus, an ideal integrative approach into this direction would be to selectively, fluorescently label individual structures within extended brain volumes and image these intact large brain samples at nanoscale resolution, that is, below the light microscope diffraction limit. A first promising step into this direction was the development of chemical tissue clearing approaches that largely eliminate light scattering (reviewed by Refs. 9–12). While some clearing methods rely on genetic labeling of cells, others also allow the accessibility of the native antigens to antibody and nucleic acid probes after the clearing process.13,14 Yet, none of these methods achieved resolutions in the nanoscale range.
Recently, expansion microscopy, a radically new technique that virtually enhances the resolution of light microscopy by rather increasing the size of the tissue sample than equipmentwise, was introduced.15–17 To bypass hardware limitations, the tissue is modified to utilize water adsorbent polymers to physically expand enzymatically treated tissue samples in an isotropic manner. As a result of this expansion, fluorescent moieties spaced closer than the optical diffraction limit () can be optically resolved resulting in effective super resolution images. Notably, the heavy adsorption of water during the expansion process renders the expanded tissue fully transparent. Thus, due to its transparency, expanded tissue represents an ideal object for light microscopic imaging at nanoscale resolution.18 Still one caveat is now the large size of the expanded tissue sample.
Therefore, to image large neuronal populations at high speed and at high-resolution volumetric imaging methods are required. The approach of light-sheet fluorescence microscopy (LSFM) has emerged as a useful platform for meeting these goals and has already been used to image entire fluorescently labeled mouse brains at cellular resolution.19–22 In LSFM, the sample is illuminated with a thin sheet of light and the emitted signal detected by an orthogonally arranged wide-field detection arm.23,24 This setup allows for ultrafast imaging by simultaneous sampling of an entire optical plane that is visualized by a sensitive camera. LSFM features a gentle fluorophore excitation with very low photobleaching compared to other fluorescence microscopy techniques, such as wide-field or confocal microscopy, since excitation is confined to the detection plane.25 This creates also an intrinsic optical sectioning. Image contrast is improved compared to techniques such as epi-illumination microscopy and can even further be amended by using a digitally scanned laser beam for illumination and a correspondingly moving line detection using rolling shutter readout by a scientific CMOS (sCMOS) camera, which produces a confocal line detection scheme.20,26
Volumetric data are acquired by moving the sample through a stationary light sheet. However, this light microscopic approach is fundamentally limited in resolution to about 250 nm laterally due to the diffraction barrier. Thus, only the combination of tissue expansion and light-sheet microscopy provides a methodological platform that allows high-speed light microscopic multicolor fluorescent imaging of large nervous tissue samples in super resolution. Here, we show the feasibility of this approach by imaging fluorescently labeled hippocampal neurons with neurites of the CA1 as well as the mouse dentate gyrus through all different layers at once and in super resolution. Moreover, we show data demonstrating two color labeling of pre- and postsynaptic proteins. We also identified GABAergic cells as postsynaptic targets of mossy fibers by identifying filopodial extensions of mossy terminals contacting GABAergic neurites.
Based on our results, we calculated that this approach would allow a nanoscale volumetric anatomical analysis of fluorescently and densely labeled neurons along the entire DG () in 110 h with our current LSFM setup. Using an optimally tailored instrument, this time can be reduced to assuming an imaging frame rate of 40 Hz.
2. Results
In this study, we focused on a super resolution analysis of large GFP-labeled granule cells ensembles in mouse dorsal DG. Coronal DG sections were prepared for tissue expansion and LSFM analysis from a PROX1-cre mouse injected with rAAV-DIO-EGFP-WPRE to achieve selective expression of EGFP in DG granule cells. Details of the sample preparation are given in Sec. 4.
2.1. Confocal Airyscan Imaging Is Not Practical for a Detailed Volumetric Reconstruction of Extended Fluorescently Labeled Connectivity Maps
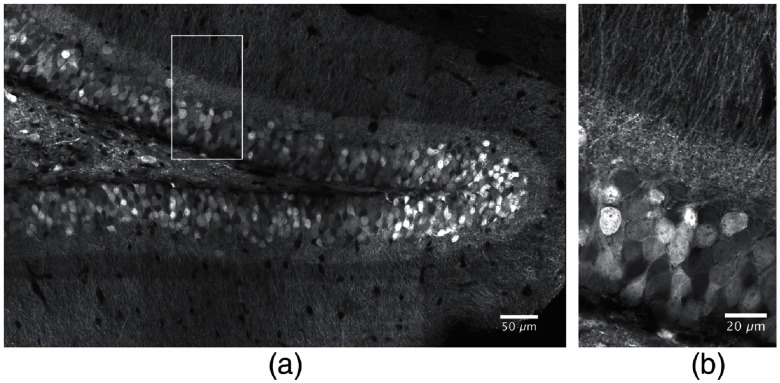
In a first approach, we explored the capabilities of state-of-the-art confocal laser scanning microscopy to obtain detailed high-resolution volumetric images of extended DG neuronal networks. For imaging EGFP-labeled DG granule cells (GC), we used a Zeiss LSM 880 with Airyscan detector, which is able to achieve a lateral full width at half maximum (FWHM) resolution of 120 nm under very optimal conditions.27 Figure 1(a) shows a single high-resolution confocal plane of a -thick coronal section through the dorsal DG, containing EGFP-labeled granule cells. The Airyscan-processed image was acquired using a water immersion (WI) objective lens with a numerical aperture (NA) of 1.2 and a pixel size of 50 nm. This image was generated from individual tiles comprising each. Tiles were acquired with 10% overlap and stitched using the ZEN software (ZEN Black 2.3 SP1, Carl Zeiss Microscopy GmbH). The magnification demonstrates the excellent resolution of the Airyscan processed image, that allows to separate densely packed dendrites [Fig. 1(b)]. However, we encountered two principal limitations for extended volumetric neural network reconstructions performing these imaging experiments. First and most importantly, it is well known that image acquisition of confocal microscopes is inherently slow due to the sequential, pixelwise data acquisition. Thus, the total imaging duration of a single confocal plane with a resolution of covering a field of required about 10.1 s, when no averaging was performed. Acquisition of a complete image field of , as shown in Fig. 1(a), required 400 s, since the tiles were acquired with 10% overlap for stitching. Thus, imaging of a -thick volume with this field size using the optimal axial step size of according to the Nyquist theorem required 1020 min or 17 h. Extrapolation of this data acquisition time to the total DG volume of yields a total duration of more than 1900 h. Moreover, post-hoc 3-D rendering of a large region of interest (ROI) demonstrated that the achieved resolution was not sufficient to allow the unambiguous evaluation of the 3-D structure of the densely labeled granule cells due to melding together of individual neurites, although the image acquisition was done as carefully as possible. An additional increase of the applied laser intensity from 0.65 to improved the signal-to-noise ratio but not the effective resolution. In addition, the higher laser intensity caused notable bleaching of the sample during confocal image acquisition (see Sec. 4).
Fig. 1.
Coronal section of a dorsal mouse dentate gyrus containing EGFP-expressing granule cells imaged by high-resolution confocal microscopy. The endogenous EGFP fluorescence was enhanced by antibody staining against EGFP (see Sec. 4). (a) A single confocal plane acquired using an Airyscan detector and a 1.2 NA WI imaging objective lens. The total field size in (a) was , achieved by combining single image stacks comprising in each frame. The individual tile stacks were acquired with 10% spatial overlap to enable efficient stitching using the algorithm described in Ref. 28. (b) Magnification of the ROI marked in Fig. 1(a).
In conclusion, we here used the latest high-resolution confocal laser scanning instrument with Airyscan detector and were able to laterally resolve neuronal structures as small as 160 nm (FWHM, see Sec. 4). The achieved axial resolution was 810 nm.
2.2. Light-Sheet Fluorescence Expansion Microscopy for Volumetric Imaging
LSFM is known for its high-speed imaging requiring only low irradiances.29 Unfortunately, volumetric imaging of mouse brain tissue by LSFM is complicated since it requires transparent specimens due to the necessity to illuminate the object from the side with a thin sheet of light to generate the optical sections. To overcome this problem and to increase the effective resolution, we decided to expand the mouse brain sections before light sheet imaging to achieve virtual optical super resolution in large tissue volumes at an imaging speed outperforming the currently fastest point scanning devices. Note that expansion of tissue has the great advantage that the heavy adsorption of water renders the samples completely transparent.
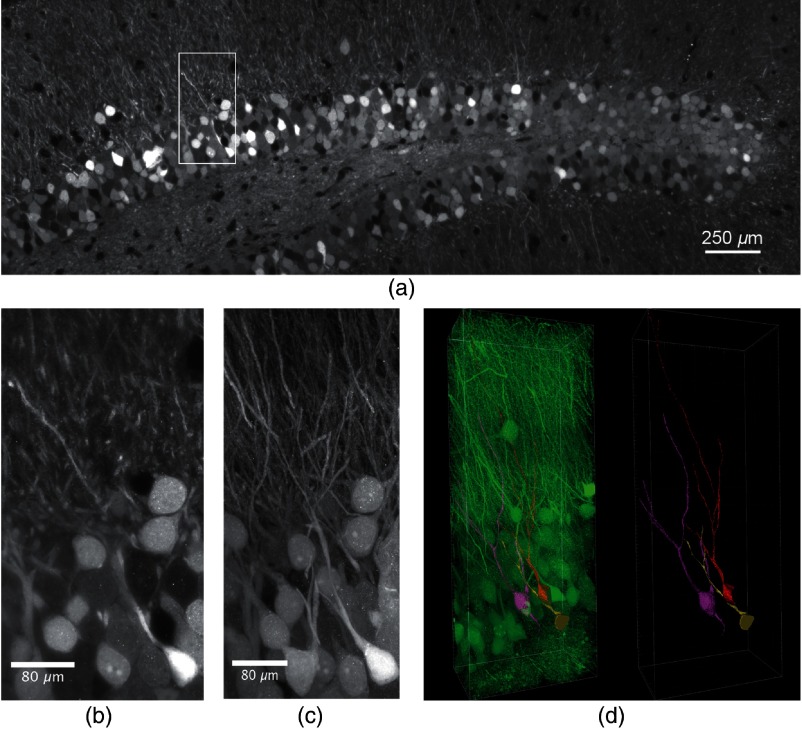
Expansion of a -thick anti-EGFP-labeled mouse brain slice from a PROX1-cre mouse, injected with rAAV-DIO-EGFP-WPRE to achieve selective expression of EGFP in DG granule cells was performed according to Ref. 16. The refractive index of the sample gel matched almost that of pure water and the expanded samples became completely transparent, excellently suited for LSFM using WI detection objectives.15,17 Sample expansion was isotropic15 and enlarged our DG sample by a factor of 3.9 (see Sec. 4). We employed a custom-built LSFM to image respective samples of expanded DG sections. A objective lens with a NA of 1.1 was used together with a magnification lens and yielded a field of view of . Thus, images of expanded DG samples had to be acquired in a mosaic fashion. For each mosaic tile, a -stack of 300 images with a was acquired with a frame rate of 1.8 Hz. After imaging, the tile stacks were contrast-adjusted and stitched yielding views of the dorsal DG sections of unprecedented resolution (Fig. 2). Figure 2(a) shows a single LSFM plane of the stitched volume at a depth of . The field size was after expansion corresponding to a size of about before expansion considering the magnification factor of (see Sec. 4). Figure 2(b) shows the magnification of the ROI marked in Fig. 2(a), and the possibility to detect individual spines demonstrates the remarkable resolution of the image. Note, that only small segments of the dendrites are visible in this single optical section. However, a maximum projection of the complete region that was imaged revealed the 3-D outline of entire dendritic fields [Fig. 2(c)]. The high virtual resolution of 100 nm laterally and 415 nm axially (see Sec. 4) and the superb contrast allowed to trace and segment the dendritic trees of three individual granule cells revealing their detailed morphoanatomical shape [Fig. 2(d)]. Thus, we achieved a lateral resolution about twofold lower than STED microscopy (25 to 80 nm),30 but clearly better than structured illumination microscopy (130 to 160 nm)30,31 or high-resolution confocal microscopy (,27) using an Airyscan detector. Our axial resolution was lower than achievable by STED and structured illumination microscopy (150 to 600 nm and 250 nm, respectively30), but higher than for high-resolution confocal microscopy (,27,30 and Sec. 4).
Fig. 2.
Expanded, antibody-stained mouse brain slice imaged by LSFM. The sample was expanded and imaged with a custom-built light-sheet microscope. In total 70 -stacks with a step size of , covering a depth of with 30% overlap were stitched to generate the image. (a) Single plane of the stitched volume at a depth of . The total volume shown after expansion was (Video 1, MP4, 32 MB [URL: https://doi.org/10.1117/1.NPh.6.1.015005.1]). (b) Magnification of the region marked in (a), lateral field size . (c) Maximum projection of the selected region marked in (a) comprising 76 slices of the stack, lateral field size . Single granule cells and dendrites can well be distinguished and separated. The stack was median-filtered to remove staining artifacts before the maximum projection. (d) Segmentation and tracing of the neurites of three selected granule cells (Video 2, MP4, 19 MB [URL: https://doi.org/10.1117/1.NPh.6.1.015005.2]). Left panel: segmented GCs in the neuronal network, right panel: segmented GCs with traced neurites.
Note that a sample volume of would be transformed by expansion to a volume of (). The step size of imaging should be in order to fulfill the Nyquist criterion, since the FWHM of our axial resolution was (see Sec. 4). This yields a total number of 5000 sections. The lateral extension of such a sample requires the use of tiles each having a field of view of . Thus, a total number of 720.000 images will cover the complete volume, what would require 111 h for imaging at a rate of 1.8 Hz. Thus, light-sheet fluorescence expansion microscopy (LSFEM) imaging of a complete dentate gyrus is faster by a factor of 17 compared to Airyscan confocal microscopy.
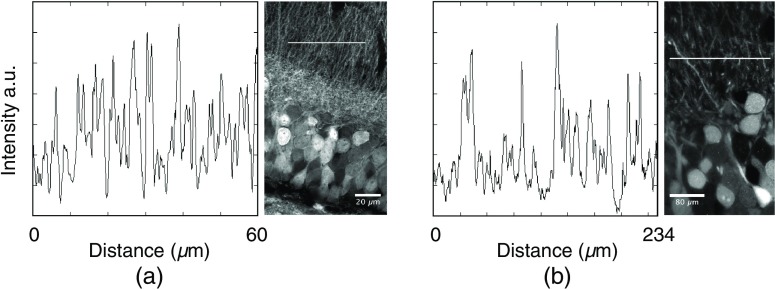
Figure 1, which was acquired by Airyscan confocal laser scanning microscopy, showed a brain region of similar spatial extension before tissue expansion. Visual comparison between Figs. 1 and 2 readily reveals the gain in contrast and axial resolution achieved by light-sheet expansion microscopy. This was demonstrated by plotting line profiles of the dense neurite region of Figs. 1(b) and 2(b) (Fig. 3). The figure demonstrates the lower number of dendrites containing in an optical slice of the LSFEM with its effective thickness of 415 nm compared to the optical slice of the Airyscan image with its axial extension of 810 nm.
Fig. 3.
Contrast in Airyscan and LSFM microscopy images. (a) Line profile determined in a -wide line with a length of . The right panel shows the position of the line in Fig. 1(b). (b) Line profile determined in a wide line with a length of . The right panel shows the position of the line in Fig. 2(b).
2.3. Sparsely Labeled Expanded Mouse Brain Slices
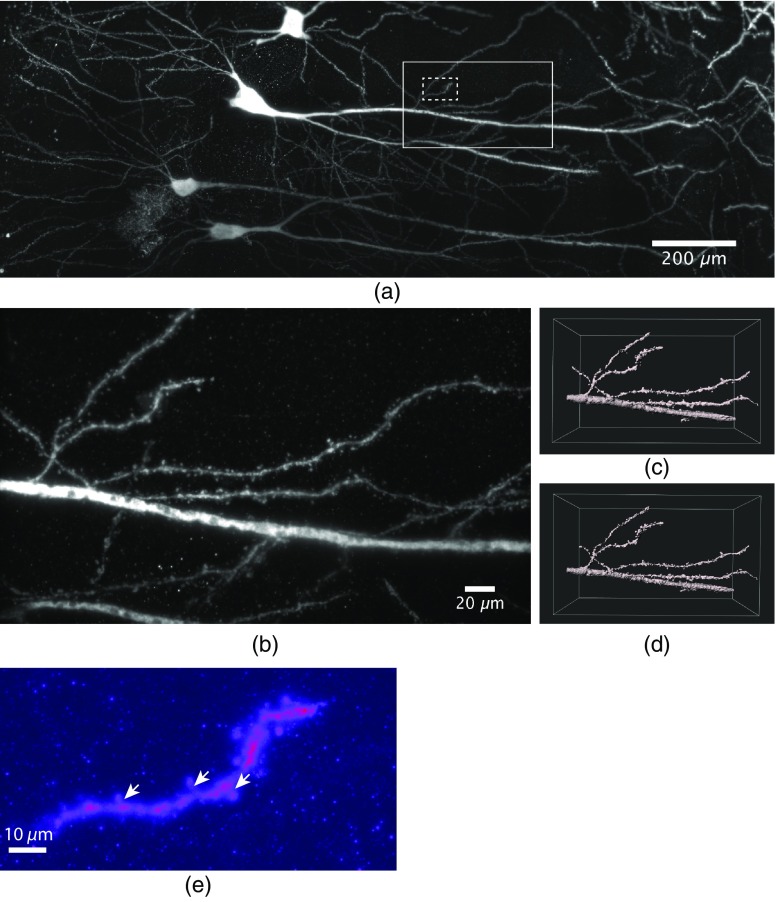
GC in brain slices of dorsal DG display an extremely narrow arrangement of cell bodies and neurites, as can be seen in Figs. 1 and 2. Individual neurites may optically be resolved, yet, tracing them over their entire spatial extension is possible only in rare cases. This is due to the fact that entire neurons are unlikely to be entirely contained within a single tissue section and, although we achieve effective super resolution, melding together of closely together lying individual structures can virtually not be avoided in densely packed structures. One solution to this problem is using a more “sparse labeling” approach. To this end, we infected the hippocampal CA1 region of wt mice with a rAAV-expressing EGFP under control of the human synapsin1 promoter by stereotaxic virus injection and stained afterward with an antibody against EGFP (for details, see Sec. 4). The use of low virus titers (10E8/ml) resulted in a stochastic and relatively sparse labeling of CA1 pyramidal neurons. This experimental approach allowed us to unequivocally identify individual dendritic networks and follow them over 1.3 mm in this experiment [Fig. 4(a) and Video 3]. Note that the image stack shown in Fig. 4 was obtained using an axial step size of only revealing even the tiniest dendritic structures in nanoscale resolution. E.g., individual dendritic spines can be visualized in the high-resolution images [Figs. 4(b)–4(d) and Video 4]. Figure 4(e) demonstrates that even spine necks can be recognized.
Fig. 4.
Dendritic segments of sparsely labeled pyramidal neurons in CA1. (a) Maximum projection of a total of 24 stacks with an axial step size of covering a depth of were acquired and stitched together (Video 3, MP4, 39 MB [URL: https://doi.org/10.1117/1.NPh.6.1.015005.3]). The sparse labeling and small axial imaging step size allowed to reconstruct the labeled granule cells and dendrites over distances of 1.3 mm after expansion. (b) Magnification of the large ROI marked in (a). The high virtual optical resolution allowed to identify finest detail, e.g., the morphology of individual synaptic spines. (c) 3-D surface rendering of a region of the data shown in (b), the dimensions were . (d) Deconvolution of the image data using the experimental PSF yielded a significant increase in data quality (Video 4, MP4, 4 MB [URL: https://doi.org/10.1117/1.NPh.6.1.015005.4]). (e) A maximum projection of 250 frames of a magnified region of the image stack shown in (a) (see dashed ROI) containing a single dendrite demonstrates that even dendritic spine necks (arrows) can be recognized.
Notably, these images were acquired using a NA1.1 objective lens combined with a 1.5× magnification, which achieved a theoretical FWHM lateral optical resolution of 310 nm and a nominal FWHM axial resolution of 1010 nm at the emission maximum of the used dye, ATTO647N (664 nm). The expansion process magnified all labeled object structures by a factor of 3.9, thus transforming these values into an expected virtual lateral and axial resolution of and 260 nm, respectively. Experimentally, we did not reach the theoretical expectation, but rather determined values of 135 and 590 nm for the FWHM values of the lateral and axial virtual optical resolution, respectively (see Sec. 4). Presumably, this discrepancy was partly due to the fact that a dipping objective was used for imaging because it featured a high NA and a long working distance. It contained a correction collar for adjusting it also for the use with a cover slip and was employed in this way. We suspect, however, that in this configuration the objective did not deliver its full performance. Hence, we assume that it is optimal for imaging expanded samples using long working distance dipping objective lenses in an upright optical detection path.
2.4. Multicolor Imaging of Sparsely Labeled DG Neurons After Tissue Expansion
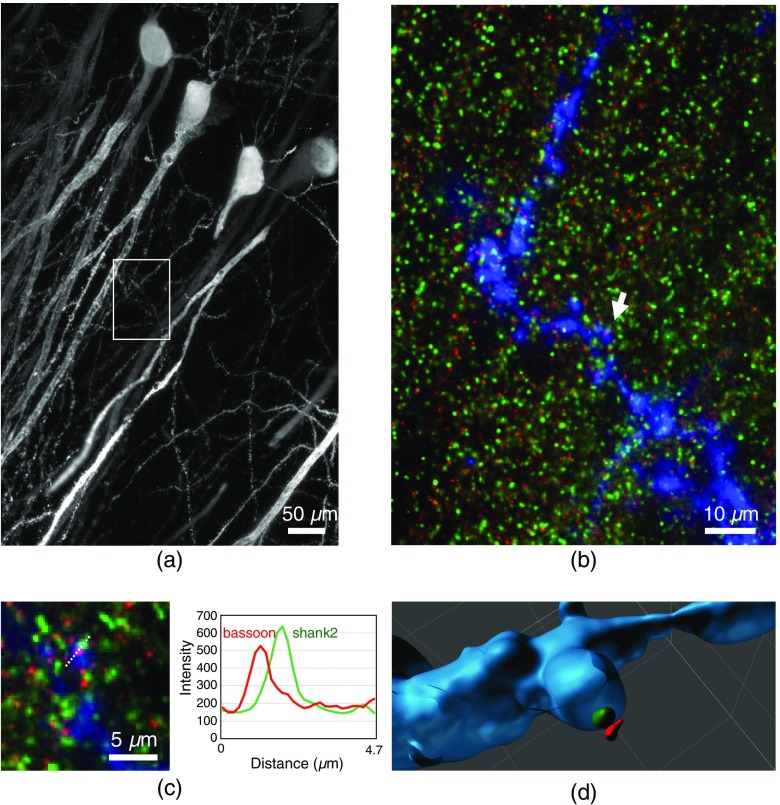
In order to show the possibility of neural connectivity mapping, we performed experiments demonstrating multicolor labeling of pre- and postsynaptic proteins. Briefly, we labeled virus-infected EGFP-positive pyramidal neurons with antibodies against EGFP to outline neurite morphology and subsequently stained for the postsynaptic protein shank2 (Alexa568, green) and the presynaptic protein Bassoon (Alexa 647, red). Figure 5(a) shows a region with an axial extension of , which was imaged using 1300 sections. Figure 5(b) shows a magnification of the ROI marked in (a) as maximum projection of 13 images extending over a distance of . Figure 5(c) shows a contrast-enhanced magnification of a region marked in (b) by a white arrow with a lateral field size of , indicating synaptic connectivity. Figure 5(d) shows a 3-D surface rendering of the synapse region shown in (c) to demonstrate the spatial arrangement of the fluorescent labels within the context of the synapse. Using the image data shown in Fig. 5, we measured the distances between 10 shank2-bassoon pairs [see Fig. 5(d)]. The maximum intensity projection of those regions was used from 10 to 15 planes. The intensity profiles—obtained by drawing a line perpendicular to the synapse surface—were fitted by Gaussian functions to determine the respective maximum positions of the label distribution. This yielded the expected distance of between pre- and postsynaptic proteins. Altogether, Fig. 5 demonstrates the capability of our approach to visualize details of synaptic connectivity, which is a prerequisite for systematic connectivity analysis.
Fig. 5.
Multicolor imaging of sparsely labeled pyramidal neurons in CA1 imaged by LSFEM. (a) Maximum intensity projection of an expanded mouse brain slice expressing EGFP. The endogenous EGFP fluorescence was enhanced by antibody staining against EGFP, the secondary antibody was conjugated to Alexa 488. The total axial extension was . (b) Magnification of the ROI marked in (a). Dendrites were stained with Alexa488 (blue), postsynaptic proteins (shank2) stained with Alexa568 (green), presynaptic proteins (bassoon) stained with Alexa647 (red). The shown images were deconvolved. (c) Magnification a region indicated in (b) (white arrow), lateral field size , indicating synaptic connectivity. The right panel in (c) shows the intensity profiles along the dotted line. (d) 3-D surface rendering of the synapse region shown in (c).
In addition, we generated DG samples containing sparsely expressing EGFP-positive GCs in order to visualize the mossy fibers within hilus of the DG. We then performed antibody stainings against parvalbumin on these EGFP-positive, anti-EGFP stained sections to selectively label hilar GABAergic interneurons in red (Alexa 568). We then performed two-color LSFEM volumetric imaging and used differential color surface rendering to identify GABAergic cells as postsynaptic targets of mossy fibers boutons. Figures 6(a) and 6(b) show an overview of the DG hilus with EGFP-positive mossy fibers in green and parvalbumin-positive GABAergic interneurons in red. Figure 6(c) shows a zoom identifying filopodia on mossy fiber boutons that contact dendrites of GABAergic hilar interneurons. Collectively, we present evidence that the spatial resolution using LSFEM is sufficient to study neuronal connectivity within the context of large neuronal ensembles.
Fig. 6.
Two-color imaging of mossy fibers and GABAergic interneurons in DG. (a) Mossy fibers in the hilus area expressing EGFP. The endogenous EGFP fluorescence was enhanced by antibody staining against EGFP, the secondary antibody was conjugated to Alexa 488 (green). Parvalbumin staining identified GABAergic interneurons shown in red (Video 5, MP4, 65 MB [URL: https://doi.org/10.1117/1.NPh.6.1.015005.5]). 1500 optical slices were acquired with a step size of , the shown data were deconvolved. Volume size . (b) Side view of the data shown in (a). (c) Segmented parvalbumin cells and mossy fibers reconstructed in 3-D. Magnification of the ROI marked in (b), showing connection between the cells.
For performing the differential color surface rendering, the volume was deconvolved using Huygens with the corresponding measured PSF for each channel. The deconvolved image data were loaded into Imaris to perform a 3-D rendering of the volume of interest, for the GABAeric cells and for the mossy fibers boutons. The connectivity area shown in Fig. 6 was obtained using the surface tool over both channels choosing a surface grain size of and a manually defined intensity threshold.
3. Discussion
Wide scale volumetric analysis of the topology of neuronal circuits requires a fast and high-resolution nanoscale imaging approach. Here, we developed and explored LSFEM by combining expansion microscopy and LSFM to achieve this goal.
The idea of tissue expansion combined with conventional light microscopy was initially introduced by Chen et al. in 201515 and termed expansion microscopy. Using this technique, nanoscale imaging of microtubuli in cultured cells as well as of Bassoon and Homer1 in a Thy1-YFP mouse brain using conventional confocal microscopy was demonstrated.
The initial approach was based on the use of trifunctional labels comprising a linker to the hygroscopic gel, a fluorophore for visualization, and an oligonucleotide that was hybridized to a complementary sequence attached to a secondary antibody. This rather complicated approach was simplified by Chozinski et al. who developed a strategy to perform expansion microscopy using conventional immunostaining,16 thus circumventing the complicated labeling procedure using custom synthetized trifunctional dyes. They demonstrated nanoscale fluorescence imaging of a THY1- YFP-H mouse brain slice immunostained for YFP, the presynaptic marker protein Bassoon, and the postsynaptic marker Homer1. Further methodological developments demonstrated that expansion could also be performed on samples containing genetically encoded fluorescent proteins, that partly retained their fluorescence after the expansion procedure.32 This was an important step toward large-volume imaging, since it overcomes the necessity of antibody stainings, that are usually limited by penetration constraints of brain tissue.
Finally, Chang et al. introduced iterative expansion microscopy, which was principally a sequential creation of two different expandable polymer gels, one within the mesh of the other.17 This approach yielded linear expansion factors of about 20, allowing to image the structural details of dendritic spines.
In recent years, LSFM became a standard method for the analysis of large and extended biological specimen, e.g., fluorescently labeled mouse brains.33 For fixed specimen, one of the requirements was that the tissue was optically cleared before imaging. Meanwhile, over 10 different clearing agents are in use (for review, see Refs. 9, 11, and 34). While a suitable and effective clearing method was essential for LSFM in the case of nonexpanded samples to achieve the required transparency, this aspect became irrelevant when combining it with expansion to LSFEM, because expanded samples were intrinsically transparent due to their high water content.
The LSFEM pipeline introduced here allows to rapidly obtain super-resolved neuronal connectivity maps using a diffraction-limited light-sheet microscope. As a proof of principle, we imaged and reconstructed large and densely fluorescently labeled subregions of the mouse hippocampal DG and CA1 regions. Notably, in the densely packed granule cell layer (gcl) of the dorsal hippocampus, we could image and segment individual granule cells together with their neurites extending deep into the molecular layer in super-resolution revealing their fine structural details.
Note that the imaging resolution was high enough to visualize individual dendritic spines. Moreover, we could also show data demonstrating two color labeling of pre- and postsynaptic proteins and identified GABAergic cells as postsynaptic targets of mossy fibers by visualizing filopodial extensions of mossy terminals contacting GABAergic neurites. The implementation of multicolor staining is especially important in the case of neural network reconstructions that partly rely on the possibility to image the tight association between pre- and postsynaptic specializations. As shown synaptic contacts can be identified by proximity measurements of the pre- to the postsynapse, which affords differential labeling of both structures for reliable connectivity predictions.
A comparison between high-resolution confocal laser scanning microscopy using an Airyscan detector (HR-CLSM) and LSFEM suggests that HR-CLSM is not as practical for volumetric connectivity analysis as our LSFEM approach with regard to the achievable optical resolution and the imaging speed (see Figs. 1 and 2). While high-resolution (high NA) CLSM objectives are superior to the objective used in our LSFM device, these objectives are not optimal to acquire large 3-D scans of the type we are performing to study neuronal connectivity maps. This is because of the relatively short working distance as well as the relatively low imaging speed, and also the relatively high bleaching rates of classical point scanning devices. The high imaging speed is especially beneficial for expanded tissue samples, since it minimizes bleaching of the fluorophores. We could show that imaging of a mosaic tile of by Airyscan imaging using a state-of-the-art CLSM required about 480 s, when optimal lateral and axial resolution should be achieved, whereas imaging the same object field (multiplied by the expansion factor 3.9) by LSFEM required only about 30 s.
Extended samples must be imaged in a mosaic-tile fashion and subsequently stitched together. The stitching of the stacks is performed using published algorithms and reconstructed the complete dorsal dentate gyrus of a mouse contained on a -thick coronal section out of 70 stacks. Mosaic imaging allowed the use of high NA detection objectives to achieve an optimal optical resolution. The magnification of the objective and the size of the camera chip led to a resolution that well satisfied the Nyquist theorem. Hence, biological details in the order of 100 nm before expansion could well be resolved, which correlates with the length scales necessary for neuronal connectivity mapping.
We would also like to point out that a fair comparison of scanning speed/resolution by CLSM on expanded samples over a large volume is not practical, since the limited working distance and the high rate of bleaching (of the point scanning device) does not allow elaborate volumetric scans to be made.
It is important to mention that sample preparation needs to be done carefully. Especially mounting of the expanded sample requires care since the expanded sample is quite unstable. To avoid destruction of the sample during mounting, we developed a dedicated sample chamber, into which the cover slip with the sample could directly be inserted. Since imaging large volumes at super resolution takes a considerable amount of time, even when using LSFEM important parameters for accurate circuit reconstruction are sample drift and residual probe swelling. Concerning sample drift our instrument was designed for optimum stability and displayed a drift of about only. However, the positional precision was limited by the motors, which were used to move the sample during image acquisition. The motors displayed a unidirectional repeatability of 100 nm. Yet, any lateral sample drift or movement was of no concern for circuit reconstruction, since it was a requirement to stitch the various image stacks (see Sec. 4). The stitching process corrected for any possible sample movement. Concerning a potential residual swelling of the sample after the expansion process, we did not observe any further swelling when it was prepared according to our given protocol. A size change of the samples was in the order of magnitude of the measurement precision as determined by repeated imaging of fluorescent beads embedded within the expanded gel.
Another important aspect of LSFEM analysis of large volumes is the amount of raw data that is generated. Although the amount of data produced is relatively large (raw data for the image shown in Fig. 2 was 100 GB), it still can be handled with desktop computers equipped with fast graphic cards and sufficient memory (see Sec. 4). The amount of data generated is depending on the volume and the density of labeled neurons. The produced data are in the terabyte range. An issue still remaining is, however, how to compare and analyze connectivity matrices of large brain regions imaged in super resolution. This will likely require new mathematical algorithms for graph analysis as well as statistical tests to compare different neural circuits with each other and to identify anatomical connectivity motifs.
Finally, we would like to stress here that a decisive factor in imaging extended neural networks by expansion light-sheet microscopy is imaging duration. Thus, we are currently optimizing our setup and expect to increase the imaging rate by a factor of 10 to 20. Such a device would allow nanoscale imaging of our specimen () in about 10 h. Also, we believe that the use of an appropriately designed lattice light-sheet microscope would allow to further increase the achievable resolution, especially when used in its structured illumination mode.35
4. Material and Methods
4.1. Mice
Mice were maintained on a 12-h light/12-h dark cycle with food and water always available. All the experiments were carried out in accordance with the German animal protection law (TierSCHG), FELASA and were approved by the animal welfare committee of the University of Bonn.
C57BL/6 mice were purchased from Charles River (Sulzfeld, Germany). The Prox-1 cre line36 is maintained in the local animal facility (Haus für Experimentelle Therapie).
4.2. Virus Injection
Viral injections were performed under aseptic conditions in Prox1::cre or C57BL/6 mice up to 10 months old. The mice were anesthetized with a mixture of Fentanyl (Rotexmedica, Germany), Midazolam (Rotexmedica, Germany), and Domitor (Orion Pharma, Finland) via intraperitoneal injection (). Analgesia ( of buprenorphine; Buprenovet, Bayer, Germany) was administered intraperitoneally prior to the injection, and Xylocain (AstraZeneca, Germany) was used for local anesthesia. Stereotaxic injections were performed using an injection frame (WPI Benchmark/Kopf) and a microprocessor-controlled minipump (World Precision Instruments, Sarasota, Florida), 1000 nl of the viral solution was injected bilaterally into the hippocampus (rAAV-DIO-eGFP; rAAV-syn-GFP; coordinates: rostrocaudal: from the Bregma; mediolateral: from the midline; dorsoventral: ). After the injection, the scalp was sutured with PERMA-HAND Silk Suture (Ethicon), and an antibacterial ointment (Refobacin®, Almirall, Germany) was applied, followed by the intraperitoneal injection of a mixture of Naloxon (B. Braun, Germany), Flumazenil (B. Braun, Germany), and Antisedan (Orion Pharma, Finland) (). To prevent the wound pain, analgesia was administered on the 3 following days.
4.3. Perfusion and Slicing
Three weeks after the injection, the mice were anesthetized with a mixture of xylazine (; Bayer Vital, Germany) and ketamine (; bela-pharm GmbH & Co. KG, Germany). Using a peristaltic pump (Laborschlauchpumpe PLP33, Mercateo, Germany), the mice were perfused transcardially with PBS followed by 4% paraformaldehyde (PFA) in 1 × PBS. All solutions were stored on ice prior to the perfusion. Brains were removed from the skull and post-fixed in 4% paraformaldehyde overnight (ON) at . After fixation, the brains were moved into PBS containing 0.01% sodium azide and stored at .
4.4. Immunochemistry
Immunohistochemistry was performed according to standard protocols. Briefly, the fixed brains were sectioned coronally (70 or ) using a vibratome (Leica VT1000 S). To prevent unspecific binding of the primary antibody, the sections were incubated in blocking buffer ( PBS containing 0.1% TritonX-100 and 5% normal goat serum) on a shaker for 6 h at room temperature. After blocking, the sections were incubated ON in primary antibody (chicken anti-GFP; 1:5000 in blocking buffer; Abcam, ab13970; rabbit anti-parvalbumin diluted 1:1000 in blocking buffer, Swant, PV27) at . The following day, slices were washed at room temperature in blocking buffer three times for 20 min and incubated ON in Alexa Fluor® 488-conjugated goat antibody against chicken IgY, or Alexa Fluor® 568 goat anti-rabbit (1:400 in blocking buffer; Life Technologies, A-11039, A-11011) at .
4.5. Labeling of Pre- and Postsynaptic Proteins
For pre- and postsynaptic labeling sections were stained against presynaptic active zone protein bassoon (primary antibody mouse anti-bassoon diluted 1:200 in blocking buffer, Enzo Life Sciences, SAP7F407; secondary antibody goat anti-mouse conjugated with biotin diluted 1:200 in blocking buffer, Jackson ImmunoResearch, 115-067-003 followed by Alexa Fluor® 647 Streptavidin, Jackson ImmunoResearch, 016-600-084) and postsynaptic scaffold protein Shank2 (primary antibody guinea pig anti-Shank2 diluted 1:200 in blocking buffer, Synaptic Systems, 162 204; secondary antibody goat anti-guinea pig conjugated with Alexa Fluor® 568 diluted 1:200 in blocking buffer, Life Technologies, A-11075).
4.6. Gelation and Expansion
The expansion microscopy protocol was adopted from Refs. 15 and 16. The immunostained sections were incubated with 1 mM methylacrylic acid-NHS (Sigma Aldrich) linker for 1 h. After washing three times in PBS, the sections were incubated for 45 min in the monomer solution (8.6% sodium acrylate, 2.5% acrylamide, 0.15% N,N’- methylenebisacrylamide, and 11.7% NaCl in PBS), followed by 2 h incubation at 37°C in a gelling solution. The gelling solution was prepared by adding 4-hydroxy-TEMPO (0.01%), TEMED (0.2%) and ammonium persulfate (0.2%) to the monomer solution. After the gel formation, the samples were incubated at 37°C ON in the digestion buffer (50 mM Tris, 1 mM EDTA, 0.5% Triton-X100, 0.8M guanidine HCl, and of proteinase K; pH 8.0). The next day, the digestion buffer was removed and the sections were washed with deionized water for 2.5 h. Subsequently, the samples were stored in deionized water at until imaging. For microscopic examination, the expanded gel sample was fixed on the bottom coverslip of the imaging chamber with poly-l-lysine to avoid movements during the measurement and the chamber was filled with deionized water.
4.7. Light Microscopy
For light-sheet microscopy, we used a custom-built setup based on a Nikon Eclipse Ti-U inverted microscope (Nikon, Düsseldorf, Germany). A previous version of this instrument was described by Ref. 26. Scanned illumination and sample stage were custom-designed, whereas the Eclipse Ti-U provided the detection path. For fluorescence excitation, three fiber-coupled lasers emitting at 488, 638 (Obis LS, Coherent, Santa Clara), and 532 nm (LasNova Green 50 Series, Lasos, Jena, Germany) were employed. Laser light was regulated by an acousto-optical tunable filter (AOTF, TF-525-250-6-3-GH18A, Gooch&Housego, Ilminster). The horizontally scanned light sheet was generated by a galvanometer system with silver-coated mirrors. The adjustment of the beam waist position within the sample chamber was realized by a relay optics mounted on a linear precision stage. The beam waist in the object plane was adjusted to a diameter of for the 488 nm, for the 532 nm, and for the 638-nm laser lines. The irradiance at the sample plane amounted to 240, 340, and at 488, 532, and 638 nm, respectively. Our custom-designed sample chamber featured an illumination window with coverslip thickness (0.17 mm) and a bottom glass coverslip for light detection. The sample chamber could be moved in three spatial directions by motorized microtranslation stages. For illumination we used a Mitutoyo NA 0.28 air objective. The detection objective was a NA 1.1 WI objective with cover slip correction (Nikon). The Eclipse Ti-U had a built-in optional magnification, which was employed as indicated. We used a sCMOS camera (, pixel size , Orca Flash 4.0 V2, Hamamatsu Photonics K.K., Hamamatsu City, Japan) for data acquisition in rolling shutter mode. The object field pixel size was corresponding to a field of view of . All electronic components were controlled by a custom-written LabView program.
The sample was mounted in a custom built sample chamber. The excitation laser beam entered the chamber through the window at the left-hand side, which was formed by a conventional coverslip with 0.17 mm thickness glued to the outer chamber wall. The sample was mounted on a coverslip, which was slid into the interstice in the chamber walls from the front. Eventually, the chamber was closed by a third coverslip at the front side and filled with deionized water. For the waterproofing of the chamber, the slits between the coverslips and the chamber walls were sealed with 2% agarose. The chamber was mounted on a three-axis motor.
The image acquisition speed of our instrument was not optimized for high speed, since it was an experimental system. Image acquisition time depended on the line exposure time and the slit width of the rolling shutter, respectively, the waiting time before line activation. For a typical exposure time of 20 ms per line and a confocal slit width of 256 pixels, we reached a total exposure time of 180 ms for one frame. Due to various signal processing tasks of the instrument, the total time required to acquire one frame amounted in summary to about 500 ms. Altogether, the acquisition time for a stack of 400 frames amounted to 222.7 s corresponding to a frame rate of 1.8 Hz. Acquisition of a complete stack comprising 1500 images using a step size of at the irradiance of resulted in a bleaching of 2% in a central image.
The sample size greatly exceeded the lateral object field size of . Therefore, the data were acquired in a mosaic-like fashion. To this end, we acquired image tiles in the direction perpendicular to the laser beam ( direction). The propagation direction of the laser beam corresponded to the direction. This ensured that the focus of the beam remained always in the center of the field of view. To enable a subsequent stitching of the tiles, two neighboring tiles had 30% spatial overlap. When the border of the sample was reached, the sample chamber was shifted along the optical axis to start the acquisition of the next stacks, but displaced along the optical axis. Due to the air/water interface in the illumination beam path, the laser focus shifted along the optical axis when the sample chamber was moved in direction, and it had to be readjusted prior to image acquisition. So far, this readjustment was done manually. To obtain a mosaic of stacks, the laser focus had to be adjusted times. The total acquisition time for a mosaic comprising stacks with 300 images per stack was 5 h. This mosaic covered a volume of .
High-resolution confocal microscopy was performed using a Zeiss LSM 880 equipped with Airyscan detector. Images were acquired using a WI objective lens with NA 1.2 according to standard protocols. For Airyscan imaging, a laser power of was used routinely (0.3% of the available power). After imaging of a complete stack comprising 160 images, the topmost frame was bleached by 25%.
4.8. Characterization of Optical Resolution
The lateral resolution theoretically achievable with an objective lens is given by the Rayleigh criterion, , which quantifies the distance between the maximum and first minimum of the point spread function (PSF):
Here, denotes the wavelength of the detected light and NA is the numerical aperture of the objective. Experimentally, this value is difficult to determine. Commonly the full width at half maximum (,) is used instead, which is related to as
Similarly, the axial detection resolution is given as
where denotes the refractive index of the medium. The value can be related to the FWHM along axial direction, , as
The axial width of the LSFM PSF is given by the product of the excitation and detection probability distributions.
In order to measure the resolution realizable with our LSFEM setup, we used fluorescent beads with subresolution diameters and the PSF extraction feature of the deconvolution software Huygens (Scientific Volume Imaging, Hilversum, The Netherlands).
We prepared samples of green and red fluorescent beads with diameters of 100 and 176 nm in 1% agarose gels, respectively. We acquired -stacks of these samples with both the Airyscan confocal microscope and the light-sheet microscope and determined the PSF using Huygens (Table 1). From the lateral and axial intensity profiles of the PSFs, we derived the respective FWHM values as follows:
Table 1.
Optical resolutions of the utilized microscopes.
| LSM/ excitation | Theoretical FWHM resolution (nm)a | Experimental FWHM resolution (nm)b | Experimental virtual FWHM resolution (nm)c | |||
|---|---|---|---|---|---|---|
| Lateral | Axial | Lateral | Axial | Lateral | Axial | |
| Airyscan 488 nm | — | — | 160 | 810 | — | — |
| LSFM 488 nm | 242 | 790 | 380 | 1625 | 100 | 415 |
| LSFM 640 nm | 310 | 1010 | 520 | 2300 | 135 | 590 |
The theoretical FWHM values were determined using the procedures given above and emission wavelengths of 520 and 665 nm for green and red excitation, respectively.
All values have errors of maximally 5%.
The virtual resolution was calculated taking the average expansion factor of 3.9 into account.
Note that the PSF was measured using a lateral object field pixel size of 173 nm. We are restricted to this value, because it is determined by the objective magnification (), the supplemental magnification lens () and the camera pixel size (). Simulating the imaging of subresolution objects with this pixel size shows that this slight violation of the Nyquist theorem leads to an underestimation of the resolution by 15% to 20%.
For comparison, we also determined the PSF of the Airyscan LSM 880 using a oil immersion objective with NA 1.4. Here, we obtained FWHM values for the PSF of 135 and 515 nm laterally and axially, respectively.
4.9. Image Processing
3-D stacks of raw 16-bit images were processed using custom-written MATLAB scripts, which allowed parallel data processing. In a first step, the intensity histograms were adjusted to homogenize brightness and contrast throughout the complete data set. Every 3-D stack was first scanned to find its minimum and maximum intensity values. With the respective values, a linear intensity adjustment was performed to cover the full dynamic range.
To achieve complete representations of the mouse DG, several 3-D data sets were stitched together. For this purpose, the stitching plugin of Fiji was used.28 Stitching was performed in two steps to optimize the processing speed. First, substacks of the 3-D data sets were created using a MATLAB script. Each substack contained 15% of the full stack. In a second step, each substack was stitched to its respective neighboring substack yielding the best overlap in terms of the cross correlation measure. Based on this information, the full 3-D stacks were stitched.
A final step to improve the contrast throughout the 3-D data was performed after stitching. This was done to compensate possible intensity variations of the sample in axial direction. To this end, a histogram equalization was performed in every image plane of the stitched data set. For calculation of -projections, the maximum intensity projection algorithm of Fiji was used.
Selected image stacks as outlined in the results section were spatially deconvolved using Huygens software (Professional version 17.04, Scientific Volume Imaging, The Netherlands) using theoretical PSFs and the classical maximum likelihood estimation algorithm. The deconvolution software provides as output the PSF, which was determined by analysis of fluorescent microbeads embedded in 1% agarose gel.
The 3-D representation of the data was achieved using the Surpass view in Imaris (Version 9.10 Bitplane Inc., Zurich, Switzerland). Data processing was performed on a workstation equipped with two Intel Xeon Platinum 8160 CPU (2.1 GHz, 24 cores), 512 GB memory, and an Nvidia Quadro P5000 GPU (16 GB GDDR5X) running under Windows 10 Pro.
4.10. Determination of the Expansion Factor
The magnification factor of expansion microscopy was experimentally determined by comparing the short width of the cell soma of labeled granule cells in images obtained from samples before expansion (Fig. 1) with those after expansion (Fig. 2). From Fig. 1, we obtained a soma width of (). For three different expansion experiments, we determined a soma width of , , and . Thus, the average expansion factor was .
Supplementary Material
Acknowledgments
This work was supported by the German Research Foundation (Grant Nos. KU 2474/13-1, INST 217/886-1, SCHW 1578/2-1 and SFB 1089/P03).
Biographies
Jana Bürgers received her bachelor’s and master’s degrees in physics from the University of Bonn in 2014 and 2016, respectively. Now, she is PhD student in the Biophysical Chemistry Group of the University Bonn. Her research interests include light sheet, lattice light sheet, and expansion microscopy.
Irina Pavlova received her master’s degree in neurosciences from the University of Bonn in 2017. Currently, she is a PhD student in the Functional Neuroconnectomics Group at the Institute of Experimental Epileptology and Cognition Research, University of Bonn Medical School. In the present time, her research interests are focused on the odor-driven centrifugal modulation of the olfactory perception.
Juan E. Rodriguez-Gatica studied electronic engineering and received her master’s degree from the University of Concepcion, Chile, in 2014, doing internships at Columbia University, USA, and University of Waterloo, Canada. Until 2016, he worked at the SCIAN-Lab at the Universidad de Chile. Currently, he is making his PhD in the Biophysical Chemistry Group at the University of Bonn. His research interests include light sheet and expansion microscopy and analysis of 2-D/3-D microscopic images.
Christian Henneberger studied medicine at the Humboldt and Free University Berlin and received his degree in 2003. After completing his thesis in developmental neurobiology and postdoctoral research at the Charité (Berlin, Germany) and the University College London (United Kingdom), he became professor at the University of Bonn, Germany, in 2011. His research interests include learning processes, the underlying neuronal and non-neuronal processes and their investigation using novel optical tools.
Marc Oeller received his bachelor’s and master’s degrees from the University of Bonn, Germany, in 2016 and 2018, respectively. Currently, he is a PhD candidate in Prof. Vendruscolo’s Group in the Centre for Misfolding Diseases at the University of Cambridge, UK. His main research focus is protein solubility and biophysical analysis of protein folding.
Jan A. Ruland received his bachelor’s and master’s degrees from the University Bonn in 2014 and 2016, respectively. Now, he is a PhD student at University Bonn, since 2016, in the Biophysical Chemistry Group. His research interests include single molecule tracking, confocal microscopy, and ribosome biogenesis.
Jan P. Siebrasse received his diploma from the Institute of Immunology of the University of Münster, Germany, and his doctoral degree from the Institute of Medical Physics and Biophysics. He was a postdoc in the Nano Channel Optronics Group at the Center for Nanotechnology in Münster and at the Research & Development Department of Teutopharma GmbH, Glandorf, Germany, before joining the Biophysical Chemistry Group of the University Bonn.
Ulrich Kubitscheck leads the Department of Biophysical Chemistry at the University of Bonn, Germany. He obtained his academic degrees from the University of Bremen, spent his career working at The Weizmann Institute of Science, Israel, and the University of Münster, Germany, before taking up his position in Bonn. He develops light sheet fluorescence microscopic and single molecule imaging techniques for studying neuroconnectomics as well as mRNA processing and nucleocytoplasmic transport within living cells.
Martin K. Schwarz received his diploma from the Institute of Molecular Pathology (IMP) in Vienna in 1995 and his doctoral degree from the University of Vienna for his thesis carried out at the Max-Planck Institute for Biophysical Chemistry in Goettingen in 1998. Now, he is leading his Independent Research Group at the Institute of Experimental Epileptology and Cognition Research at the University of Bonn. His research interests include neuroconnectomics, biomedical optics, and their applications.
Disclosures
No conflicts of interest, financial or otherwise, are declared by the authors.
References
- 1.Ramón S., Cajal Y., Textura del sistema nervioso del hombre y de los vertebrados, Imprenta y Librería de Nicolás Moya, Madrid: (1899–1904). [Google Scholar]
- 2.Mikula S., Denk W., “High-resolution whole-brain staining for electron microscopic circuit reconstruction,” Nat. Methods 12(6), 541–546 (2015). 10.1038/nmeth.3361 [DOI] [PubMed] [Google Scholar]
- 3.Osten P., Margrie T. W., “Mapping brain circuitry with a light microscope,” Nat. Methods 10(6), 515–523 (2013). 10.1038/nmeth.2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conchello J.-A., Lichtman J. W., “Optical sectioning microscopy,” Nat. Methods 2(12), 920–931 (2005). 10.1038/nmeth815 [DOI] [PubMed] [Google Scholar]
- 5.Helmchen F., Denk W., “Deep tissue two-photon microscopy,” Nat. Methods 2(12), 932–940 (2005). 10.1038/nmeth818 [DOI] [PubMed] [Google Scholar]
- 6.Tang J., Germain R. N., Cui M., “Superpenetration optical microscopy by iterative multiphoton adaptive compensation technique,” Proc. Natl. Acad. Sci. U. S. A. 109(22), 8434–8439 (2012). 10.1073/pnas.1119590109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Micheva K. D., et al. , “Single-synapse analysis of a diverse synapse population: proteomic imaging methods and markers,” Neuron 68(4), 639–653 (2010). 10.1016/j.neuron.2010.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ragan T., et al. , “Serial two-photon tomography for automated ex vivo mouse brain imaging,” Nat. Methods. 9(3), 255–258 (2012). 10.1038/nmeth.1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu D., et al. , “Recent progress in tissue optical clearing,” Laser Photonics Rev. 7(5), 732–757 (2013). 10.1002/lpor.2013.7.issue-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richardson D. S., Cell J. L., “Clarifying tissue clearing,” Cell 162(2), 246–257 (2015). 10.1016/j.cell.2015.06.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tainaka K., et al. , “Chemical principles in tissue clearing and staining protocols for whole-body cell profiling,” Annu. Rev. Cell Dev. Biol. 32(1), 713–741 (2016). 10.1146/annurev-cellbio-111315-125001 [DOI] [PubMed] [Google Scholar]
- 12.Vigouroux R. J., Belle M., Chédotal A., “Neuroscience in the third dimension: shedding new light on the brain with tissue clearing,” Mol. Brain 10(1), 33 (2017). 10.1186/s13041-017-0314-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim S.-Y., Chung K., Deisseroth K., “Light microscopy mapping of connections in the intact brain,” Trends Cogn. Sci. (Regul. Ed.) 17(12), 596–599 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Chung K., et al. , “Structural and molecular interrogation of intact biological systems,” Nature 497(7449), 332–337 (2013). 10.1038/nature12107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen F., Tillberg P. W., Boyden E. S., “Optical imaging. Expansion microscopy,” Science 347(6221), 543–548 (2015). 10.1126/science.1260088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chozinski T. J., et al. , “Expansion microscopy with conventional antibodies and fluorescent proteins,” Nat. Methods 13(6), 485–488 (2016). 10.1038/nmeth.3833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang J.-B., et al. , “Iterative expansion microscopy,” Nat. Methods 14(6), 593–599 (2017). 10.1038/nmeth.4261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dodt H.-U., “Microscopy. The superresolved brain,” Science 347(6221), 474–475 (2015). 10.1126/science.aaa5084 [DOI] [PubMed] [Google Scholar]
- 19.Dodt H.-U., et al. , “Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain,” Nat. Methods 4(4), 331–336 (2007). 10.1038/nmeth1036 [DOI] [PubMed] [Google Scholar]
- 20.Silvestri L., et al. , “Confocal light sheet microscopy: micron-scale neuroanatomy of the entire mouse brain,” Opt. Express 20(18), 20582–20598 (2012). 10.1364/OE.20.020582 [DOI] [PubMed] [Google Scholar]
- 21.Schwarz M. K., et al. , “Fluorescent-protein stabilization and high-resolution imaging of cleared, intact mouse brains,” PLoS One 10(5), e0124650 (2015). 10.1371/journal.pone.0124650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doerr J., et al. , “Whole-brain 3D mapping of human neural transplant innervation,” Nat. Commun. 8, 14162 (2017). 10.1038/ncomms14162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Voie A. H., Burns D. H., Spelman F. A., “Orthogonal-plane fluorescence optical sectioning: three-dimensional imaging of macroscopic biological specimens,” J. Microsc. 170(Pt 3), 229–236 (1993). 10.1111/jmi.1993.170.issue-3 [DOI] [PubMed] [Google Scholar]
- 24.Huisken J., et al. , “Optical sectioning deep inside live embryos by selective plane illumination microscopy,” Science 305(5686), 1007–1009 (2004). 10.1126/science.1100035 [DOI] [PubMed] [Google Scholar]
- 25.Stelzer E. H. K., “Light-sheet fluorescence microscopy for quantitative biology,” Nat. Methods 12(1), 23–26 (2015). 10.1038/nmeth.3219 [DOI] [PubMed] [Google Scholar]
- 26.Baumgart E., Kubitscheck U., “Scanned light sheet microscopy with confocal slit detection,” Opt. Express 20(19), 21805–21814 (2012). 10.1364/OE.20.021805 [DOI] [PubMed] [Google Scholar]
- 27.Korobchevskaya K., et al. , “Exploring the potential of airyscan microscopy for live cell imaging,” Photonics 4, 41 (2017). 10.3390/photonics4030041 [DOI] [Google Scholar]
- 28.Preibisch S., Saalfeld S., Tomancak P., “Globally optimal stitching of tiled 3D microscopic image acquisitions,” Bioinformatics 25(11), 1463–1465 (2009). 10.1093/bioinformatics/btp184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Power R. M., Huisken J., “A guide to light-sheet fluorescence microscopy for multiscale imaging,” Nat. Methods 14(4), 360–373 (2017). 10.1038/nmeth.4224 [DOI] [PubMed] [Google Scholar]
- 30.Toomre D., Bewersdorf J., “A new wave of cellular imaging,” Annu. Rev. Cell Dev. Biol. 26(1), 285–314 (2010). 10.1146/annurev-cellbio-100109-104048 [DOI] [PubMed] [Google Scholar]
- 31.Sawada K., et al. , “Super-resolution structural analysis of dendritic spines using three-dimensional structured illumination microscopy in cleared mouse brain slices,” Eur. J. Neurosci. 47(9), 1033–1042 (2018). 10.1111/ejn.2018.47.issue-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tillberg P. W., et al. , “Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies,” Nat. Biotechnol. 34(9), 987–992 (2016). 10.1038/nbt.3625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keller P. J., Dodt H.-U., “Light sheet microscopy of living or cleared specimens,” Curr. Opin. Neurobiol. 22(1), 138–143 (2012). 10.1016/j.conb.2011.08.003 [DOI] [PubMed] [Google Scholar]
- 34.Zhu X., et al. , “Optical brain imaging: a powerful tool for neuroscience,” Neurosci. Bull. 33, 95–102 (2017). 10.1007/s12264-016-0053-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen B.-C., et al. , “Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution,” Science 346, 439–453 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gong S., et al. , “Targeting Cre recombinase to specific neuron populations with bacterial artificial chromosome constructs,” J. Neurosci. 27(37), 9817–9823 (2007). 10.1523/JNEUROSCI.2707-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.