Abstract
Electroconvulsive therapy (ECT) is the treatment of choice for severe and treatment‐resistant depression; disorder severity and unfavorable treatment outcomes are shown to be influenced by an increased genetic burden for major depression (MD). Here, we tested whether ECT assignment and response/nonresponse are associated with an increased genetic burden for major depression (MD) using polygenic risk score (PRS), which summarize the contribution of disease‐related common risk variants. Fifty‐one psychiatric inpatients suffering from a major depressive episode underwent ECT. MD‐PRS were calculated for these inpatients and a separate population‐based sample (n = 3,547 healthy; n = 426 self‐reported depression) based on summary statistics from the Psychiatric Genomics Consortium MDD‐working group (Cases: n = 59,851; Controls: n = 113,154). MD‐PRS explained a significant proportion of disease status between ECT patients and healthy controls (p = .022, R2 = 1.173%); patients showed higher MD‐PRS. MD‐PRS in population‐based depression self‐reporters were intermediate between ECT patients and controls (n.s.). Significant associations between MD‐PRS and ECT response (50% reduction in Hamilton depression rating scale scores) were not observed. Our findings indicate that ECT cohorts show an increased genetic burden for MD and are consistent with the hypothesis that treatment‐resistant MD patients represent a subgroup with an increased genetic risk for MD. Larger samples are needed to better substantiate these findings.
Keywords: depression, electroconvulsive therapy, major depression, polygenic risk scores, treatment‐resistance
1. INTRODUCTION
Effective treatments for depression remain elusive because of poor understanding of the underlying etiology of this highly prevalent disorder. Electroconvulsive therapy (ECT) is the treatment of choice for severe and treatment‐resistant forms of depressive episodes (Fink & Taylor, 2007) and thus, patients assigned to ECT represent a specific subgroup selected for these factors. There is increasing evidence that severity of psychiatric disorder is associated with a higher genetic burden for the disorders, for example, (Amare et al., 2018; Frank et al., 2015). Recently, this has also been demonstrated in the largest genome‐wide association study (GWAS) for depression to date (Wray et al., 2018) which showed that major depression is a highly polygenic disorder, that is, a result of the contribution of many genetic variants. Polygenic risk score (PRS) profiling is an approach that uses the risk variants and corresponding effect sizes identified in large GWAS such as the above study as a “discovery sample” to generate risk scores in an independent “target sample,” reflecting the disease risk burden of each individual (Wray et al., 2014). Presently, the clinical utility of PRS remains limited at the level of the individual as they only explain a small share of variance in case–control status or symptom severity. However, they can be used as a research tool to dissect disease aetiology by investigating the association of genetic risk burden for a disorder with related subphenotypes. In Wray et al. 2018, higher PRS were associated with measures of increased severity such as early age at onset, symptom counts, and recurrent episodes (Appendix [Link]).
In the present study, we hypothesized that as ECT patients represent a severe and treatment‐resistant share of all MD patients, they should show an increased genetic burden for MD. We aimed to assess the feasibility of this approach to detect increased genetic risk of depression in a group of inpatients (n = 52) assigned to ECT as compared to population‐based controls. We generated PRS using results from the MD‐GWAS by Wray et al. (2018) (PGC‐MD2, Cases: n = 59,851; Controls: n = 113,154), testing whether these PRS were associated with MD ECT case–control status. In addition, we explored MD‐PRS in population based subjects with self‐reported MD, and MD‐PRS associations with clinical parameters in the ECT group.
2. MATERIALS AND METHODS
This study was approved by the ethics committee (II) at the Medical Faculty Mannheim, University of Heidelberg. All patients provided written consent. All procedures were performed in accordance with the Declaration of Helsinki.
2.1. ECT patients
Patients were recruited between 2014 and 2016 at the Department of Psychiatry and Psychotherapy of the Central Institute of Mental Health, Mannheim. Inclusion criteria were a present major depressive episode within the context of a diagnosis of either major depressive disorder or bipolar disorder according to DSM‐IV, age above 18 years and the clinical decision for an ECT treatment. Exclusion criteria were any substance‐related disorders, except tobacco and alcohol use. All participants were of Caucasian descent.
The criteria for assigning patients with a depressive episode to ECT were either treatment‐resistant depression defined as failure of two adequate dose‐duration antidepressants or psychotherapy from different classes in the current episode (Conway, George, & Sackeim, 2017) or positive experience to ECT from a previous episode, or severe depression with (a) psychotic symptoms, (b) severe suicidality, or (c) the refusal of food or fluid intake.
A total of 52 inpatients consented to participate in the present study. In 36 of the 52 included patients (69.2%) the indication for ECT was a current treatment‐resistant depressive episode. Six patients (11.5%) with a current depressive episode were assigned to ECT because of positive experience to ECT during a previous depressive episode, whereas five (9.6%) other patients received ECT because of depression with severe psychotic symptoms. In three patients (5.8%), the severe suicidality that was accompanied by the depressive episode was the main indication for ECT and in two patients (3.9%) the indication was refusal of food and fluid intake. In three cases, a legal guardian gave the formal consent to the study for the patient. All other patients gave their consent on their own.
A comorbid personality disorder (PD) was indicated when already diagnosed prior to the recent depressive episode. Generally, that diagnosis was either given after a Structured Clinical Interview for DSM‐IV Axis II Disorders (SCID‐II) interview, but in some patients based on a clinical judgment. Out of the fifteen patients with comorbid PDs, there were seven patients with Borderline PD (46.7%), three patients with a dependent PD (20.0%), two patients each with a histrionic (13.3%) and avoidant PD (13.3%), respectively and one patient with an obsessive–compulsive PD (6.7%).
Of 52 patients, 7 discontinued the treatment prematurely after one of the initial ECT sessions: four patients discontinued ECT after the first (n = 2), second (n = 1), or third (n = 1) session because of subjective intolerable side effects; one patient left the hospital against the medical advice after the fourth ECT session; in one patient ECT was stopped after suffering from a serotonergic syndrome because of ECT and concomitant medication; one patient dropped out due severe hyponatremia during the course of treatment and subsequent transfer to a hospital for internal medicine. Furthermore, we excluded one patient with diagnosis of schizophrenia from statistical analyses.
2.2. Controls
Data from Heinz Nixdorf Recall (HNR) study, a population‐based study of individuals with homogeneous German ethnicity, comprised the control sample (n = 4,814, M:2395; F:2419). The HNR controls had been assessed for depression status using a computer‐assisted personal interview with the question: “Do you have or have you ever had depression? (Y/N)”. A total of n = 3,547 answered “no” and n = 426 answered “yes”, whereas answers for n = 841 were unknown.
2.3. Assessments
ECT patients were assessed for demographics, including: Age, sex (male/female) and body mass index.
Baseline clinical factors were also assessed: age at first disease onset, length of current episode (months), multiple drug therapy resistance (yes/no), presence of PD (yes/no), positive family history in first degree relatives for affective disorders (yes/no), type of depression (unipolar or bipolar depression), alcohol dependence or abuse (yes/no), and nicotine dependence (yes/no).
The 21‐item version of the Hamilton depression rating scale (HDRS) was administered pre‐ and post‐ECT treatment.
2.4. ECT
Right unilateral brief pulse ECT was performed with a Thymatron IV device (Somatics, LLC. Lake Bluff, IL). S‐ketamine (~1.0 mg/kg) or thiopental (~5 mg/kg) were used as anesthetic agents and succinylcholine (~1.0 mg/kg) for muscle relaxation. Seizure threshold was titrated at the initial session and stimulation dose at subsequent treatments was given at above 2.5 times the seizure threshold (Bumb et al., 2015; Hoyer et al., 2017). Charge was subsequently adjusted if seizures were considered as potentially insufficient during the ECT course (e.g., motor response time <15 s or electroencephalogram (EEG) seizure activity <25 s; Kranaster, Hoyer, Janke, & Sartorius, 2013).
The psychiatrist, who was responsible for the whole in‐patient treatment of the respective patient, made the clinical decision of when to terminate the ECT course. ECT was continued until the subject showed either a remission or a stable response or did not show a significant response after at least 12 ECT sessions. In the case of no further and relevant clinical improvement for 2 weeks (4–6 ECT sessions), ECT was terminated.
No specifications on the concomitant psychotropic medication were made.
2.5. Blood sampling, control data, genotyping and quality control
A venous blood sample was collected from participants for genome‐wide genotyping. Genotyping was performed using the Global Screening Array (Illumina, Inc., San Diego, CA). The HNR sample had also been genotyped using the Global Screening Array. The merged data set contained n = 642,553 overlapping SNPs. The data were subjected to a stringent quality control (QC) procedure, which included following parameters for retainment in data set: SNP missing rates <0.05 (prior to filtering individuals), individual missingness <0.02, autosomal heterozygosity deviation |F het| < 0.2, SNP missing rate < 0.02 (after filtering individuals), minor allele frequency > 0.01, Hardy–Weinberg equilibrium (Case: p > 1e−10, Control: p > 1e−6, Overall: p > 1e−6) and difference in missing rate between cases and controls <0.02. Ten principal components (PCs) were computed using principal component analysis (PCA) on a filtered subset of frequent (MAF > 0.05) autosomal SNPs in approximate linkage equilibrium (pairwise R 2 < 0.1 within a window of 250 SNPs) to find informative ancestry information and detect and remove genetic outliers (defined as those exceeding six standard deviations). A relatedness cutoff of Pi Hat = 0.125 was used to exclude related individuals. Filtering was performed using PLINK 1.90 (Chang et al., 2015). After QC, the data set comprised 44 ECT cases and 4,290 individuals from the HNR sample, with 485,607 variants remaining. Of the HNR individuals passing QC, n = 376 had self‐reported depression (HNR‐DEP) and n = 3,172 were healthy controls. Those with unknown depression status (n = 742) were removed from the analysis.
2.6. Data analysis
Statistical analyses were performed using IBM SPSS Statistics for Windows version 24. Descriptive statistics were calculated for all participants.
Given the sample size and uneven proportion of responders/nonresponders, we calculated nonparametric Spearman's rank correlations to examine factors related to response. Response was examined categorically (yes/no), defined as a 50% reduction in HDRS scores, and also a continuous variable (ΔHDRS score, the pre‐post difference between HDRS). Correlations with remission, defined as post‐treatment HDRS score > 10, were also examined.
2.7. Polygenic risk score calculation
PRS were calculated using genome‐wide association data from the Psychiatric Genomics Consortium (PGC‐MD2, Cases: n = 59,851; Controls: n = 113,154)(Wray et al., 2018) using PRsice v 1.25 (Euesden, Lewis, & O'Reilly, 2015). Clumping was carried out to retain only one representative variant per region of linkage disequilibrium (LD) using thresholds of p1 1, p2 1 an LD threshold of r 2 ≥ 0.1 and a distance threshold of 500 kb. The multi histocompatibility complex of chromosome 6 was excluded. Scores were calculated for a range of p‐value thresholds (5 × 10−8, 1 × 10−6, 1 × 10−4, 0.001, 0.01, 0.05, 0.1, 0.2, 0.5, 1.0). PRS were standardized to the mean and standard deviation of controls, that is, . (PRS − meancontrols)/standard deviationcontrols (Lewis & Vassos, 2017).
A binomial logistic regression analysis was carried out to determine the contribution of MD‐PRS to disease status. Case–control status was specified as the dependent variable. Proportion of variance explained by PRS was tested by comparing Nagelkerke's R 2 in an initial model including PCs informative of case–control status to a full model which additionally included PRS. Data from HNR‐DEP were not included in the case–control analysis.
In a next step, we included HNR‐DEP individuals passing QC (n = 376) and calculated PRS. Using the above method, binomial regression analyses were used to compare both ECT vs. HNR‐DEP and HNR‐DEP vs. controls.
Using partial correlations (accounting for PCs informative of case–control status), we tested whether MD‐PRS were correlated with ECT response and demographic/clinical factors in the ECT sample.
3. RESULTS
3.1. Descriptive statistics
Descriptive statistics are shown on Table 1.
Table 1.
Descriptive and clinical statistics of ECT patients
| Descriptives | Total (n) | Mean (SD) | |
|---|---|---|---|
| Age, years | 45 | 58.38 (18.722) | |
| Body mass index | 38 | 25.71 (4.165) | |
| Age at initial disease onset | 38 | 41.29 (19.324) | |
| Current episode length, months | 37 | 11.38 (12.722) | |
| Yes | No | ||
| Sex (male/female) | 45 | 22 | 23 |
| Alcohol use disorder | 42 | 6 | 36 |
| Tobacco | 42 | 12 | 30 |
| Positive family history | 38 | 19 | 19 |
| Personality disorder | 38 | 15 | 23 |
| Response | 37 | 30 | 7 |
| Remission | 37 | 14 | 23 |
| HDRS baseline | 42 | 27.26 (6.356) | |
| HDRS final | 38 | 10.58 (6.832) | |
| Diagnosis | 52 | MDD: 32 (7 excluded), BD: 12, SCZ: 1 | |
| Bilateral ECT | 45 | 8 | 37 |
The correlation analysis revealed that categorical response (50% reduction in HDRS) was statistically significantly correlated with being male (rho = 0.332, p = .045, df = 35), having a positive family history for affective disorders (rho = 0.358, p = .029, df = 35), and negatively correlated with diagnosis of PD (rho = −0.335, p = .043, df = 35). Tobacco use was negatively correlated with response (rho = −0.290, p = .082, df = 35) at the trend level. No other variables showed statistically significant correlation with response.
Examining response as a continuous variable (ΔHDRS score) yielded similar findings with respect to male sex (rho = 0.373, p = .021, df = 36) and PD (rho = −0.335, p = .043, df = 35). Additionally, ΔHDRS score was associated with increased age (rho = 0.363, p = .001, df = 36), negatively associated with length of current episode (rho = −0.348, p = .035, df = 35 and positively correlated with increased age at first disease onset (rho = 0.370, p = .022, df = 36). No other variables showed statistically significant correlation with ΔHDRS score.
Remission was positively correlated with age (rho = 0.426, p = .009, df = 35), age at first disease onset (rho = 0.494, p = .002, df = 36), and at the trend level with having bipolar disorder (rho = 0.328, p = .051, df = 34).
3.2. PRS
Although removed from the response analysis above, the genotype data from the dropouts were used in the PRS analysis as they still represent cases assigned to ECT.
For case–control status, a p‐value threshold of 1.0 was found to be the most informative threshold (see Figure 1a). Statistically significantly higher PRS were found in ECT cases than controls (p = .022) (see Figure 1b, left and right bar), explaining ΔNagelkerke R 2 = 1.173% of variance, using information from n = 83,066 SNPs.
Figure 1.
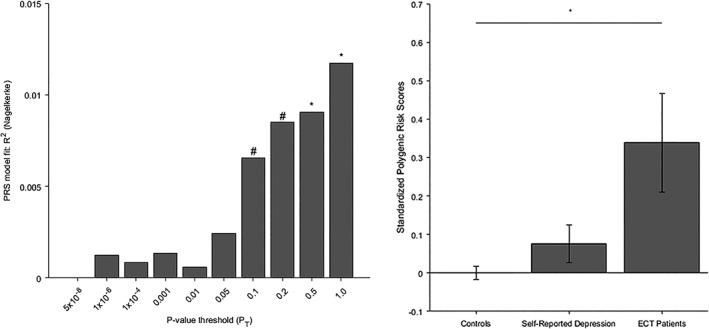
(a) Model fit for case–control status of MD‐PRS calculated at different p‐value thresholds. *p < .05, #p < .10. (b) Standardized polygenic risk scores in: healthy controls (left, n = 172); individuals with self‐reported depression, (middle, n = 376); ECT patients (right, n = 44). Error bars denote standard error of mean
Descriptively, PRS scores in HNR‐DEP were intermediate to ECT patients and controls (see Figure 1(b), middle bar). No statistically significant differences were observed between ECT patients and HNR‐DEP (p = .237), or HNR‐DEP and controls (p = .150).
In a partial correlation analysis we examined whether MD‐PRS differed in responders (coded 1) and nonresponders (coded 0) to treatment. A statistically significant correlation was not observed (rho = −0.189, p = .300, df = 30) but descriptively, the direction was for nonresponders to have higher PRS for MD than responders. The correlation between MD‐PRS and response coded as Δ HDRS score was also not statistically significant (rho = −0.016, p = .930, df = 31). A statistically significant correlation was observed between MD‐PRS and alcohol dependence/abuse (rho = 0.372, p = .023, df = 35), but no other demographic or clinical variables showed statistically significant correlations with MD‐PRS.
4. DISCUSSION
The present feasibility study represents the first usage of a whole‐genome (PRS) approach in an ECT sample. Our findings using a multi‐marker technique to characterize an important subgroup of depressed patients show that patients assigned to ECT hold potential for further exploration using a molecular genetics approach. These patients are usually suffering from severe or therapy‐resistant forms of depressive episodes, which appears to be consistent with having an increased genetic burden of disease. Individuals from the HNR cohort self‐reporting depression had scores intermediate to ECT patients and controls, suggesting that although they indicated that they had depression, these individuals had less genetic burden of MD.
The ability of the PRS to predict case–control status, while small (p = .022, ΔNagelkerke R 2 = 1.173%), is similar to that of other studies using similar approaches in psychiatric genetics (on the order of 10−2 to 10−3, see also Wray et al., 2018). Although not clinically informative at this stage, these results are consistent with depression being a polygenic trait and suggest the potential utility of the PRS approach to characterize patient subgroups in samples of larger size.
We did not observe statistically significant correlations between MD‐PRS and response. Descriptively, the direction was for nonresponders to have higher PRS, but conclusions cannot yet be drawn as our analysis was underpowered: because of the efficacy of ECT, the proportion of nonresponders is necessarily small, rendering statistical comparison a challenge, especially in a sample of the present size. Interestingly, we observed increased MD‐PRS in patients with a history of alcohol dependence/abuse, which is consistent with a large body of research describing comorbidity between depression and alcohol dependence at the clinical and genetic levels and supports recent reports suggesting that genetic pleiotropy may be responsible for this disease comorbidity (Andersen et al., 2017; Foo et al., 2018). In a recent study, we observed that alcohol use disorder is a positive predictor of ECT response (Aksay et al., 2017). We did not find any such evidence in the current study, most likely because of the limited number of nonresponders and small proportion of patients with alcohol dependence/abuse. Caution is needed when generalizing these findings and confirmation in a larger sample awaits.
It is also worth mentioning that our finding that presence of comorbid PDs was negatively correlated with the antidepressant response to ECT corroborates previous data (de Vreede, Burger, & van Vliet, 2005; Kaster, Goldbloom, Daskalakis, Mulsant, & Blumberger, 2018; Rasmussen, 2015).
With its short time course and striking therapeutic effects, ECT offers a good model to explore fundamental biological mechanisms (i.e., immunological, neurotrophic, epigenetic) underlying changes in depressive symptomatology observed as a result of treatment. Clinical findings about the role of genetic factors suggest a possible role in gene variation in the mediation of response to ECT (Kellner, Popeo, Pasculli, Briggs, & Gamss, 2012); while supporting this idea, existing data remains preliminary, highlighting the need for large‐scale confirmatory studies (Benson‐Martin, Stein, Baldwin, & Domschke, 2016). Investigations so far have only explored the candidate gene level and to go beyond “tentative knowledge,” systematic genome‐wide studies which can identify unequivocally contributing genes are needed (Sullivan, 2017).
Our study has several limitations. First, while ECT cohorts have the advantage of being well‐phenotyped and characterized, only severe cases are assigned, leading to necessarily limited sample sizes. The sample used in the current study, while large for an ECT sample, is limited when considered in the perspective of GWAS. On the other hand, GWAS studies often suffer from limited phenotyping at the expense of larger numbers to gain statistical power. Further investigations which tackle both of these issues and investigate well‐characterized, larger samples are expected to give the power needed to clarify underlying mechanisms. For example, even samples not deeply phenotyped but including health record information indicating that ECT was performed can be included.
Next, descriptively we found that population‐based individuals who had self‐reported depression had lower PRS for MD than patients assigned to ECT. It should be noted that the self‐report depression status is not equivalent to a clinical diagnosis, and this group is potentially heterogeneous. While it has been shown that self‐reports of depression carry enough signal to be reflected in genetics (e.g., Wray et al., 2018), comparison to a sample of expert‐diagnosed patients with MDD/BD not undergoing ECT would offer more refined insight.
It should also be noted that our ECT cohort comprised both patients with MDD and BD. In a post‐hoc test, we examined whether or not this affected the results of the comparison of ECT patients and controls. After repeating the calculation with bipolar patients excluded, we found that results did not change substantially (R 2 = 1.228%, p = .037).
Here, we have shown the potential utility of a PRS approach to examine genetic risk for MD in patients assigned to ECT. It is important to move in the direction of taking advantage of ECT as a model to examine the etiology of antidepressant response as it provides a clear pre‐post treatment longitudinal design which can be investigated using time‐sensitive gene expression and epigenetic/epigenomic methods. Further research taking advantage of such a longitudinal design is expected to allow more in‐depth exploration into both phenotypic changes observed and the underlying biology and eventually will inform treatment strategies.
CONFLICT OF INTEREST
The authors have no conflicts of interest to report.
ACKNOWLEDGMENTS
MR and MMN were supported by the German Federal Ministry of Education and Research (BMBF) through grants BMBF 01ZX1311A (to MR and MMN), through grants 01ZX1314A (to MMN) and 01ZX1314G (to MR) within the e:Med research program, and by the German Research Foundation via the Excellence Cluster ImmunoSensation, NO246/10‐1 (to MMN) and RI 908/11‐1 (to MR). The PGC has received major funding from the US National Institute of Mental Health and the US National Institute on Drug Abuse (U01 MH109528 and U01 MH1095320).
Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium
Naomi R. Wray* 1, 2
Stephan Ripke* 3, 4, 5
Manuel Mattheisen* 6, 7, 8, 9
Maciej Trzaskowski* 1
Enda M. Byrne 1
Abdel Abdellaoui 10
Mark J. Adams 11
Esben Agerbo 9, 12, 13
Tracy M. Air 14
Till F. M. Andlauer 15, 16
Silviu‐Alin Bacanu 17
Marie Bækvad‐Hansen 9, 18
Aartjan T. F. Beekman 19
Tim B. Bigdeli 17, 20
Elisabeth B. Binder 15, 21
Douglas H. R. Blackwood 11
Julien Bryois 22
Henriette N. Buttenschøn 8, 9, 23
Jonas Bybjerg‐Grauholm 9, 18
Na Cai 24, 25
Enrique Castelao 26
Jane Hvarregaard Christensen 7, 8, 9
Toni‐Kim Clarke 11
Jonathan R. I. Coleman 27
Lucía Colodro‐Conde 28
Baptiste Couvy‐Duchesne 2, 29
Nick Craddock 30
Gregory E. Crawford 31, 32
Gail Davies 33
Ian J. Deary 33
Franziska Degenhardt 34, 35
Eske M. Derks 28
Nese Direk 36, 37
Conor V. Dolan 10
Erin C. Dunn 38, 39, 40
Thalia C. Eley 27
Valentina Escott‐Price 41
Farnush Farhadi Hassan Kiadeh 42
Hilary K. Finucane 43, 44
Jerome C. Foo 45
Andreas J. Forstner 34, 35, 46, 47
Josef Frank 45
Héléna A. Gaspar 27
Michael Gill 48
Fernando S. Goes 49
Scott D. Gordon 28
Jakob Grove 7, 8, 9, 50
Lynsey S. Hall 11, 51
Christine Søholm Hansen 9, 18
Thomas F. Hansen 52, 53, 54
Stefan Herms 34, 35, 47
Ian B. Hickie 55
Per Hoffmann 34, 35, 47
Georg Homuth 56
Carsten Horn 57
Jouke‐Jan Hottenga 10
David M. Hougaard 9, 18
Marcus Ising 58
Rick Jansen 19
Ian Jones 59
Lisa A. Jones 60
Eric Jorgenson 61
James A. Knowles 62
Isaac S. Kohane 63, 64, 65
Julia Kraft 4
Warren W. Kretzschmar 66
Jesper Krogh 67
Zoltán Kutalik 68, 69
Yihan Li 66
Penelope A. Lind 28
Donald J. MacIntyre 70, 71
Dean F. MacKinnon 49
Robert M. Maier 2
Wolfgang Maier 72
Jonathan Marchini 73
Hamdi Mbarek 10
Patrick McGrath 74
Peter McGuffin 27
Sarah E. Medland 28
Divya Mehta 2, 75
Christel M. Middeldorp 10, 76, 77
Evelin Mihailov 78
Yuri Milaneschi 19
Lili Milani 78
Francis M. Mondimore 49
Grant W. Montgomery 1
Sara Mostafavi 79, 80
Niamh Mullins 27
Matthias Nauck 81, 82
Bernard Ng 80
Michel G. Nivard 10
Dale R. Nyholt 83
Paul F. O'Reilly 27
Hogni Oskarsson 84
Michael J. Owen 59
Jodie N. Painter 28
Carsten Bøcker Pedersen 9, 12, 13
Marianne Giørtz Pedersen 9, 12, 13
Roseann E. Peterson 17, 85
Erik Pettersson 22
Wouter J. Peyrot 19
Giorgio Pistis 26
Danielle Posthuma 86, 87
Jorge A. Quiroz 88
Per Qvist 7, 8, 9
John P. Rice 89
Brien P. Riley 17
Margarita Rivera 27, 90
Saira Saeed Mirza 36
Robert Schoevers 91
Eva C. Schulte 92, 93
Ling Shen 61
Jianxin Shi 94
Stanley I. Shyn 95
Engilbert Sigurdsson 96
Grant C. B. Sinnamon 97
Johannes H. Smit 19
Daniel J. Smith 98
Hreinn Stefansson 99
Stacy Steinberg 99
Fabian Streit 45
Jana Strohmaier 45
Katherine E. Tansey 100
Henning Teismann 101
Alexander Teumer 102
Wesley Thompson 9, 53, 103, 104
Pippa A. Thomson 105
Thorgeir E. Thorgeirsson 99
Matthew Traylor 106
Jens Treutlein 45
Vassily Trubetskoy 4
André G. Uitterlinden 107
Daniel Umbricht 108
Sandra Van der Auwera 109
Albert M. van Hemert 110
Alexander Viktorin 22
Peter M. Visscher 1, 2
Yunpeng Wang 9, 53, 104
Bradley T. Webb 111
Shantel Marie Weinsheimer 9, 53
Jürgen Wellmann 101
Gonneke Willemsen 10
Stephanie H. Witt 45
Yang Wu 1
Hualin S. Xi 112
Jian Yang 2, 113
Futao Zhang 1
Volker Arolt 114
Bernhard T. Baune 115
Klaus Berger 101
Dorret I. Boomsma 10
Sven Cichon 34, 47, 116, 117
Udo Dannlowski 114
EJC de Geus 10, 118
J. Raymond DePaulo 49
Enrico Domenici 119
Katharina Domschke 120
Tõnu Esko 5, 78
Hans J. Grabe 109
Steven P. Hamilton 121
Caroline Hayward 122
Andrew C. Heath 89
Kenneth S. Kendler 17
Stefan Kloiber 58, 123, 124
Glyn Lewis 125
Qingqin S. Li 126
Susanne Lucae 58
Pamela A. F. Madden 89
Patrik K. Magnusson 22
Nicholas G. Martin 28
Andrew M. McIntosh 11, 33
Andres Metspalu 78, 127
Ole Mors 9, 128
Preben Bo Mortensen 8, 9, 12, 13
Bertram Müller‐Myhsok 15, 16, 129
Merete Nordentoft 9, 130
Markus M. Nöthen 34, 35
Michael C. O'Donovan 59
Sara A. Paciga 131
Nancy L. Pedersen 22
Brenda W. J. H. Penninx 19
Roy H. Perlis 38, 132
David J. Porteous 105
James B. Potash 133
Martin Preisig 26
Marcella Rietschel 45
Catherine Schaefer 61
Thomas G. Schulze 45, 93, 134, 135, 136
Jordan W. Smoller 38, 39, 40
Kari Stefansson 99, 137
Henning Tiemeier 36, 138, 139
Rudolf Uher 140
Henry Völzke 102
Myrna M. Weissman 74, 141
Thomas Werge 9, 53, 142
Cathryn M. Lewis 27, 143
Douglas F. Levinson 144
Gerome Breen 27, 145
Anders D. Børglum 7, 8, 9
Patrick F. Sullivan 22, 146, 147
1, Institute for Molecular Bioscience, The University of Queensland, Brisbane, QLD, AU
2, Queensland Brain Institute, The University of Queensland, Brisbane, QLD, AU
3, Analytic and Translational Genetics Unit, Massachusetts General Hospital, Boston, MA, US
4, Department of Psychiatry and Psychotherapy, Universitätsmedizin Berlin Campus Charité Mitte, Berlin, DE
5, Medical and Population Genetics, Broad Institute, Cambridge, MA, US
6, Centre for Psychiatry Research, Department of Clinical Neuroscience, Karolinska Institutet, Stockholm, SE
7, Department of Biomedicine, Aarhus University, Aarhus, DK
8, iSEQ, Centre for Integrative Sequencing, Aarhus University, Aarhus, DK
9, iPSYCH, The Lundbeck Foundation Initiative for Integrative Psychiatric Research, DK
10, Dept of Biological Psychology & EMGO+ Institute for Health and Care Research, Vrije Universiteit Amsterdam, Amsterdam, NL
11, Division of Psychiatry, University of Edinburgh, Edinburgh, GB
12, Centre for Integrated Register‐based Research, Aarhus University, Aarhus, DK
13, National Centre for Register‐Based Research, Aarhus University, Aarhus, DK
14, Discipline of Psychiatry, University of Adelaide, Adelaide, SA, AU
15, Department of Translational Research in Psychiatry, Max Planck Institute of Psychiatry, Munich, DE
16, Munich Cluster for Systems Neurology (SyNergy), Munich, DE
17, Department of Psychiatry, Virginia Commonwealth University, Richmond, VA, US
18, Center for Neonatal Screening, Department for Congenital Disorders, Statens Serum Institut, Copenhagen, DK
19, Department of Psychiatry, Vrije Universiteit Medical Center and GGZ inGeest, Amsterdam, NL
20, Virginia Institute for Psychiatric and Behavior Genetics, Richmond, VA, US
21, Department of Psychiatry and Behavioral Sciences, Emory University School of Medicine, Atlanta, GA, US
22, Department of Medical Epidemiology and Biostatistics, Karolinska Institutet, Stockholm, SE
23, Department of Clinical Medicine, Translational Neuropsychiatry Unit, Aarhus University, Aarhus, DK
24, Human Genetics, Wellcome Trust Sanger Institute, Cambridge, GB
25, Statistical Genomics and systems Genetics, European Bioinformatics Institute (EMBL‐EBI), Cambridge, GB
26, Department of Psychiatry, University Hospital of Lausanne, Prilly, Vaud, CH
27, Social Genetic and Developmental Psychiatry Centre, King's College London, London, GB
28, Genetics and Computational Biology, QIMR Berghofer Medical Research Institute, Brisbane, QLD, AU
29, Centre for Advanced Imaging, The University of Queensland, Brisbane, QLD, AU
30, Psychological Medicine, Cardiff University, Cardiff, GB
31, Center for Genomic and Computational Biology, Duke University, Durham, NC, US
32, Department of Pediatrics, Division of Medical Genetics, Duke University, Durham, NC, US
33, Centre for Cognitive Ageing and Cognitive Epidemiology, University of Edinburgh, Edinburgh, GB
34, Institute of Human Genetics, University of Bonn, Bonn, DE
35, Life&Brain Center, Department of Genomics, University of Bonn, Bonn, DE
36, Epidemiology, Erasmus MC, Rotterdam, Zuid‐Holland, NL
37, Psychiatry, Dokuz Eylul University School Of Medicine, Izmir, TR
38, Department of Psychiatry, Massachusetts General Hospital, Boston, MA, US
39, Psychiatric and Neurodevelopmental Genetics Unit (PNGU), Massachusetts General Hospital, Boston, MA, US
40, Stanley Center for Psychiatric Research, Broad Institute, Cambridge, MA, US
41, Neuroscience and Mental Health, Cardiff University, Cardiff, GB
42, Bioinformatics, University of British Columbia, Vancouver, BC, CA
43, Department of Epidemiology, Harvard T.H. Chan School of Public Health, Boston, MA, US
44, Department of Mathematics, Massachusetts Institute of Technology, Cambridge, MA, US
45, Department of Genetic Epidemiology in Psychiatry, Central Institute of Mental Health, Medical Faculty Mannheim, Heidelberg University, Mannheim, Baden‐Württemberg, DE
46, Department of Psychiatry (UPK), University of Basel, Basel, CH
47, Human Genomics Research Group, Department of Biomedicine, University of Basel, Basel, CH
48, Department of Psychiatry, Trinity College Dublin, Dublin, IE
49, Psychiatry & Behavioral Sciences, Johns Hopkins University, Baltimore, MD, US
50, Bioinformatics Research Centre, Aarhus University, Aarhus, DK
51, Institute of Genetic Medicine, Newcastle University, Newcastle upon Tyne, GB
52, Danish Headache Centre, Department of Neurology, Rigshospitalet, Glostrup, DK
53, Institute of Biological Psychiatry, Mental Health Center Sct. Hans, Mental Health Services Capital The Region of Denmark, Copenhagen, DK
54, iPSYCH, The Lundbeck Foundation Initiative for Psychiatric Research, Copenhagen, DK
55, Brain and Mind Centre, University of Sydney, Sydney, NSW, AU
56, Interfaculty Institute for Genetics and Functional Genomics, Department of Functional Genomics, University Medicine and Ernst Moritz Arndt University Greifswald, Greifswald, Mecklenburg‐Vorpommern, DE
57, Roche Pharmaceutical Research and Early Development, Pharmaceutical Sciences, Roche Innovation Center Basel, F. Hoffmann‐La Roche Ltd, Basel, CH
58, Max Planck Institute of Psychiatry, Munich, DE
59, MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, GB
60, Department of Psychological Medicine, University of Worcester, Worcester, GB
61, Division of Research, Kaiser Permanente Northern California, Oakland, CA, US
62, Psychiatry & The Behavioral Sciences, University of Southern California, Los Angeles, CA, US
63, Department of Biomedical Informatics, Harvard Medical School, Boston, MA, US
64, Department of Medicine, Brigham and Women's Hospital, Boston, MA, US
65, Informatics Program, Boston Children's Hospital, Boston, MA, US
66, Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford, GB
67, Department of Endocrinology at Herlev University Hospital, University of Copenhagen, Copenhagen, DK
68, Institute of Social and Preventive Medicine (IUMSP), University Hospital of Lausanne, Lausanne, VD, CH
69, Swiss Institute of Bioinformatics, Lausanne, VD, CH
70, Division of Psychiatry, Centre for Clinical Brain Sciences, University of Edinburgh, Edinburgh, GB
71, Mental Health, NHS 24, Glasgow, GB
72, Department of Psychiatry and Psychotherapy, University of Bonn, Bonn, DE
73, Statistics, University of Oxford, Oxford, GB
74, Psychiatry, Columbia University College of Physicians and Surgeons, New York, NY, US
75, School of Psychology and Counseling, Queensland University of Technology, Brisbane, QLD, AU
76, Child and Youth Mental Health Service, Children's Health Queensland Hospital and Health Service, South Brisbane, QLD, AU
77, Child Health Research Centre, University of Queensland, Brisbane, QLD, AU
78, Estonian Genome Center, University of Tartu, Tartu, EE
79, Medical Genetics, University of British Columbia, Vancouver, BC, CA
80, Statistics, University of British Columbia, Vancouver, BC, CA
81, DZHK (German Centre for Cardiovascular Research), Partner Site Greifswald, University Medicine, University Medicine Greifswald, Greifswald, Mecklenburg‐Vorpommern, DE
82, Institute of Clinical Chemistry and Laboratory Medicine, University Medicine Greifswald, Greifswald, Mecklenburg‐Vorpommern, DE
83, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, AU
84, Humus, Reykjavik, IS
85, Virginia Institute for Psychiatric & Behavioral Genetics, Virginia Commonwealth University, Richmond, VA, US
86, Clinical Genetics, Vrije Universiteit Medical Center, Amsterdam, NL
87, Complex Trait Genetics, Vrije Universiteit Amsterdam, Amsterdam, NL
88, Solid Biosciences, Boston, MA, US
89, Department of Psychiatry, Washington University in Saint Louis School of Medicine, Saint Louis, MO, US
90, Department of Biochemistry and Molecular Biology II, Institute of Neurosciences, Center for Biomedical Research, University of Granada, Granada, ES
91, Department of Psychiatry, University of Groningen, University Medical Center Groningen, Groningen, NL
92, Department of Psychiatry and Psychotherapy, Medical Center of the University of Munich, Campus Innenstadt, Munich, DE
93, Institute of Psychiatric Phenomics and Genomics (IPPG), Medical Center of the University of Munich, Campus Innenstadt, Munich, DE
94, Division of Cancer Epidemiology and Genetics, National Cancer Institute, Bethesda, MD, US
95, Behavioral Health Services, Kaiser Permanente Washington, Seattle, WA, US
96, Faculty of Medicine, Department of Psychiatry, University of Iceland, Reykjavik, IS
97, School of Medicine and Dentistry, James Cook University, Townsville, QLD, AU
98, Institute of Health and Wellbeing, University of Glasgow, Glasgow, GB
99, deCODE Genetics / Amgen, Reykjavik, IS
100, College of Biomedical and Life Sciences, Cardiff University, Cardiff, GB
101, Institute of Epidemiology and Social Medicine, University of Münster, Münster, Nordrhein‐Westfalen, DE
102, Institute for Community Medicine, University Medicine Greifswald, Greifswald, Mecklenburg‐Vorpommern, DE
103, Department of Psychiatry, University of California, San Diego, San Diego, CA, US
104, KG Jebsen Centre for Psychosis Research, Norway Division of Mental Health and Addiction, Oslo University Hospital, Oslo, NO
105, Medical Genetics Section, CGEM, IGMM, University of Edinburgh, Edinburgh, GB
106, Clinical Neurosciences, University of Cambridge, Cambridge, GB
107, Internal Medicine, Erasmus MC, Rotterdam, Zuid‐Holland, NL
108, Roche Pharmaceutical Research and Early Development, Neuroscience, Ophthalmology and Rare Diseases Discovery & Translational Medicine Area, Roche Innovation Center Basel, F. Hoffmann‐La Roche Ltd, Basel, CH
109, Department of Psychiatry and Psychotherapy, University Medicine Greifswald, Greifswald, Mecklenburg‐Vorpommern, DE
110, Department of Psychiatry, Leiden University Medical Center, Leiden, NL
111, Virginia Institute for Psychiatric & Behavioral Genetics, Virginia Commonwealth University, Richmond, VA, US
112, Computational Sciences Center of Emphasis, Pfizer Global Research and Development, Cambridge, MA, US
113, Institute for Molecular Bioscience; Queensland Brain Institute, The University of Queensland, Brisbane, QLD, AU
114, Department of Psychiatry, University of Münster, Münster, Nordrhein‐Westfalen, DE
115, Department of Psychiatry, Melbourne Medical School, University of Melbourne, Melbourne, AU
116, Institute of Medical Genetics and Pathology, University Hospital Basel, University of Basel, Basel, CH
117, Institute of Neuroscience and Medicine (INM‐1), Research Center Juelich, Juelich, DE
118, Amsterdam Public Health Institute, Vrije Universiteit Medical Center, Amsterdam, NL
119, Centre for Integrative Biology, Università degli Studi di Trento, Trento, Trentino‐Alto Adige, IT
120, Department of Psychiatry and Psychotherapy, Medical Center, University of Freiburg, Faculty of Medicine, University of Freiburg, Freiburg, DE
121, Psychiatry, Kaiser Permanente Northern California, San Francisco, CA, US
122, Medical Research Council Human Genetics Unit, Institute of Genetics and Molecular Medicine, University of Edinburgh, Edinburgh, GB
123, Department of Psychiatry, University of Toronto, Toronto, ON, CA
124, Centre for Addiction and Mental Health, Toronto, ON, CA
125, Division of Psychiatry, University College London, London, GB
126, Neuroscience Therapeutic Area, Janssen Research and Development, LLC, Titusville, NJ, US
127, Institute of Molecular and Cell Biology, University of Tartu, Tartu, EE
128, Psychosis Research Unit, Aarhus University Hospital, Risskov, Aarhus, DK
129, University of Liverpool, Liverpool, GB
130, Mental Health Center Copenhagen, Copenhagen Universtity Hospital, Copenhagen, DK
131, Human Genetics and Computational Biomedicine, Pfizer Global Research and Development, Groton, CT, US
132, Psychiatry, Harvard Medical School, Boston, MA, US
133, Psychiatry, University of Iowa, Iowa City, IA, US
134, Department of Psychiatry and Behavioral Sciences, Johns Hopkins University, Baltimore, MD, US
135, Department of Psychiatry and Psychotherapy, University Medical Center Göttingen, Goettingen, Niedersachsen, DE
136, Human Genetics Branch, NIMH Division of Intramural Research Programs, Bethesda, MD, US
137, Faculty of Medicine, University of Iceland, Reykjavik, IS
138, Child and Adolescent Psychiatry, Erasmus MC, Rotterdam, Zuid‐Holland, NL
139, Psychiatry, Erasmus MC, Rotterdam, Zuid‐Holland, NL
140, Psychiatry, Dalhousie University, Halifax, NS, CA
141, Division of Epidemiology, New York State Psychiatric Institute, New York, NY, US
142, Department of Clinical Medicine, University of Copenhagen, Copenhagen, DK
143, Department of Medical & Molecular Genetics, King's College London, London, GB
144, Psychiatry & Behavioral Sciences, Stanford University, Stanford, CA, US
145, NIHR Maudsley Biomedical Research Centre, King's College London, London, GB
146, Genetics, University of North Carolina at Chapel Hill, Chapel Hill, NC, US
147, Psychiatry, University of North Carolina at Chapel Hill, Chapel Hill, NC, US
Foo JC, Streit F, Frank J, et al. Evidence for increased genetic risk load for major depression in patients assigned to electroconvulsive therapy. Am J Med Genet Part B. 2019;180B:35–45. 10.1002/ajmg.b.32700
Funding information National Institute of Mental Health, Grant/ Award Number: U01 MH109528; National Institute on Drug Abuse, Grant/Award Number: U01 MH1095320; Deutsche Forschungsgemeinschaft, Grant/Award Numbers: RI 908/11‐1, NO246/10‐1; Bundesministerium für Bildung und Forschung, Grant/Award Numbers: 01ZX1314G, 01ZX1314A, 01ZX1311A
REFERENCES
- Aksay, S. S. , Hambsch, M. , Janke, C. , Bumb, J. M. , Kranaster, L. , & Sartorius, A. (2017). Alcohol use disorder as a possible predictor of electroconvulsive therapy response. The Journal of ECT, 33(2), 117–121. 10.1097/YCT.0000000000000366 [DOI] [PubMed] [Google Scholar]
- Amare, A. T. , Schubert, K. O. , Hou, L. , Clark, S. R. , Papiol, S. , Heilbronner, U. , … Baune, B. T. (2018). A polygenic score for schizophrenia and HLA and inflammation genes predict response to lithium in bipolar affective disorder. JAMA Psychiatry, 75(1), 65–74. https://jamanetwork.com/journals/jamapsychiatry/article-abstract/2663274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen, A. M. , Pietrzak, R. H. , Kranzler, H. R. , Ma, L. , Zhou, H. , Liu, X. , … Han, S. (2017). Polygenic scores for major depressive disorder and risk of alcohol dependence. JAMA Psychiatry, 74(11), 1153–1160. 10.1001/jamapsychiatry.2017.2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson‐Martin, J. J. , Stein, D. J. , Baldwin, D. S. , & Domschke, K. (2016). Genetic mechanisms of electroconvulsive therapy response in depression. Human Psychopharmacology, 31(3), 247–251. 10.1002/hup.2531 [DOI] [PubMed] [Google Scholar]
- Bumb, J. M. , Aksay, S. S. , Janke, C. , Kranaster, L. , Geisel, O. , Gass, P. , … Sartorius, A. (2015). Focus on ECT seizure quality: Serum BDNF as a peripheral biomarker in depressed patients. European Archives of Psychiatry and Clinical Neuroscience, 265(3), 227–232. 10.1007/s00406-014-0543-3 [DOI] [PubMed] [Google Scholar]
- Chang, C. C. , Chow, C. C. , Tellier, L. C. , Vattikuti, S. , Purcell, S. M. , & Lee, J. J. (2015). Second‐generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience, 4, 7. 10.1186/s13742-015-0047-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway, C. R. , George, M. S. , & Sackeim, H. A. (2017). Toward an evidence‐based, operational definition of treatment‐resistant depression: When enough is enough. JAMA Psychiatry, 74(1), 9–10. 10.1001/jamapsychiatry.2016.2586 [DOI] [PubMed] [Google Scholar]
- de Vreede, I. M. , Burger, H. , & van Vliet, I. M. (2005). Prediction of response to ECT with routinely collected data in major depression. Journal of Affective Disorders, 86(2–3), 323–327. 10.1016/j.jad.2005.03.008 [DOI] [PubMed] [Google Scholar]
- Euesden, J. , Lewis, C. M. , & O'Reilly, P. F. (2015). PRSice: Polygenic risk score software. Bioinformatics, 31(9), 1466–1468. 10.1093/bioinformatics/btu848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink, M. , & Taylor, M. A. (2007). Electroconvulsive therapy: Evidence and challenges. JAMA, 298(3), 330–332. 10.1001/jama.298.3.330 [DOI] [PubMed] [Google Scholar]
- Foo, J. C. , Streit, F. , Treutlein, J. , Ripke, S. , Witt, S. H. , Strohmaier, J. , … Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium . (2018). Shared genetic etiology between alcohol dependence and major depressive disorder. Psychiatric Genetics, 28(4), 66–70. 10.1097/YPG.0000000000000201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, J. , Lang, M. , Witt, S. H. , Strohmaier, J. , Rujescu, D. , Cichon, S. , … Rietschel, M. (2015). Identification of increased genetic risk scores for schizophrenia in treatment‐resistant patients. Molecular Psychiatry, 20(7), 913 10.1038/mp.2015.52 [DOI] [PubMed] [Google Scholar]
- Hoyer, C. , Sartorius, A. , Aksay, S. S. , Bumb, J. M. , Janke, C. , Thiel, M. , … Kranaster, L. (2017). Electroconvulsive therapy enhances the anti‐ageing hormone Klotho in the cerebrospinal fluid of geriatric patients with major depression. European Neuropsychopharmacology, 28(3), 428–435. 10.1016/j.euroneuro.2017.12.012 [DOI] [PubMed] [Google Scholar]
- Kaster, T. S. , Goldbloom, D. S. , Daskalakis, Z. J. , Mulsant, B. H. , & Blumberger, D. M. (2018). Electroconvulsive therapy for depression with comorbid borderline personality disorder or post‐traumatic stress disorder: A matched retrospective cohort study. Brain Stimulation, 11(1), 204–212. 10.1016/j.brs.2017.10.009 [DOI] [PubMed] [Google Scholar]
- Kellner, C. H. , Popeo, D. M. , Pasculli, R. M. , Briggs, M. C. , & Gamss, S. (2012). Appropriateness for electroconvulsive therapy (ECT) can be assessed on a three‐item scale. Medical Hypotheses, 79(2), 204–206. 10.1016/j.mehy.2012.04.036 [DOI] [PubMed] [Google Scholar]
- Kranaster, L. , Hoyer, C. , Janke, C. , & Sartorius, A. (2013). Bispectral index monitoring and seizure quality optimization in electroconvulsive therapy. Pharmacopsychiatry, 46(4), 147–150. 10.1055/s-0032-1331748 [DOI] [PubMed] [Google Scholar]
- Lewis, C. M. , & Vassos, E. (2017). Prospects for using risk scores in polygenic medicine. Genome Medicine, 9(1), 96 10.1186/s13073-017-0489-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen, K. G. (2015). Do patients with personality disorders respond differentially to electroconvulsive therapy? A review of the literature and consideration of conceptual issues. Journal of Electroconvulsive Therapy, 31(1), 6–12. 10.1097/YCT.0000000000000165 [DOI] [PubMed] [Google Scholar]
- Sullivan, P. F. (2017). How good were candidate gene guesses in schizophrenia genetics? Biological Psychiatry, 82(10), 696–697. 10.1016/j.biopsych.2017.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray, N. R. , Lee, S. H. , Mehta, D. , Vinkhuyzen, A. A. , Dudbridge, F. , & Middeldorp, C. M. (2014). Research review: Polygenic methods and their application to psychiatric traits. Journal of Child Psychology and Psychiatry, 55(10), 1068–1087. 10.1111/jcpp.12295 [DOI] [PubMed] [Google Scholar]
- Wray, N. R. , Ripke, S. , Mattheisen, M. , Trzaskowski, M. , Byrne, E. M. , Abdellaoui, A. , … Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium . (2018). Genome‐wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nature Genetics. 10.1038/s41588-018-0090-3 [DOI] [PMC free article] [PubMed] [Google Scholar]


