Abstract
Crop rotation and intercropping with Allium plants suppresses Fusarium wilt in various crops. However, the mechanisms underlying this phenomenon have not been fully elucidated. This study was designed to assess the role of microorganisms inhabiting Allium rhizospheres and antifungal compounds produced by Allium roots in Fusarium wilt suppression by Allium cultivation. Suppression of cucumber Fusarium wilt and the pathogen multiplication by Allium (Welsh onion and/or onion)-cultivated soils were eliminated by heat treatment at 60 °C, whereas those by Welsh onion-root extract were lost at 40 °C. The addition of antibacterial antibiotics eliminated the suppressive effect of Welsh onion-cultivated soil on pathogen multiplication, suggesting the contribution of antagonistic gram-negative bacteria to the soil suppressiveness. The Illumina MiSeq sequencing of 16S rRNA gene amplicons revealed that genus Flavobacterium was the predominant group that preferentially accumulated in Allium rhizospheres. Flavobacterium species recovered from the rhizosphere soils of these Allium plants suppressed Fusarium wilt on cucumber seedlings. Furthermore, confocal laser scanning microscopy revealed that Flavobacterium isolates inhibited the multiplication of the pathogen in soil. Taken together, we infer that the accumulation of antagonistic Flavobacterium species plays a key role in Fusarium wilt suppression by Allium cultivation.
Introduction
Fusarium oxysporum has an extremely broad range of hosts and is one of the most devastating soil-borne pathogens, causing symptoms such as damping-off, root rot, and vascular wilt in crop plants1,2. It can saprophytically survive on soil and plant debris in the absence of a host3 and remain viable for a long time by producing chlamydospores, thereby making Fusarium wilt very difficult to control. Although effective control measures for Fusarium wilt include the use of resistant cultivars or rootstock4,5, this resistance is often overcome by new races of pathogens; moreover, the development of new resistant cultivars is time-consuming6. Another strategy for control includes the fumigation of soil using chemicals, such as chloropicrin7; however, this approach often negatively affects the environment as well as human health8.
The consecutive monocultures of agricultural crops lead to the accumulation of soil-borne fungal pathogens, including F. oxysporum9. Therefore, crop rotation and intercropping have received increasing attention in recent years because they have potential for managing soil-borne diseases10–13. In Japan and China, crop rotation and intercropping with Allium plants, such as Welsh onion (Allium fistulosum), onion (A. cepa), and Chinese chive (A. tuberosum), reportedly prevent the Fusarium wilt of bottle gourds (Lagenaria siceraria), spinach (Spinacia oleracea), tomato (Solanum lycopersicum), and banana (Musa spp.)10,11,14,15.
Two hypotheses that explain the mechanisms responsible for the suppression of Fusarium wilt by Allium cultivation have been proposed. The first hypothesis implicates the involvement of antimicrobial compounds released from roots of Allium plants16. However, there is limited evidence that the antimicrobial compounds released from the roots of Allium plants indeed reduce the incidence of Fusarium wilt. The second hypothesis, based on the known importance of the soil microbiome in the suppression of soil-borne diseases17,18, implicates microorganisms associated with Allium plants in the suppression of Fusarium wilt. However, although intercropping with Allium plants, such as onion and garlic (A. sativum), changes the bacterial diversity and structure of the soil19,20, no explicit evidence indicates that these changes play a role in the suppression of Fusarium wilt. Rhizosphere microbial communities are directly influenced by the root exudates of host plants and differ across plant species21–23. Therefore, we hypothesized that rhizospheres of Allium plants harbor unique microbial communities and that some of the predominant microorganisms are involved in the suppression of Fusarium wilt induced by Allium cultivation.
In this study, we first investigated whether microorganisms inhabiting Allium rhizospheres and antifungal compounds produced by Allium roots contribute to the suppression of cucumber Fusarium wilt caused by F. oxysporum f. sp. cucumerinum (Focu) isolate GUS77, which was used as a representative pathogenic isolate of F. oxysporum. We further identified the predominant rhizobacterial groups of Allium plants by the Illumina MiSeq sequencing of 16S rRNA gene amplicons. The identified bacteria were then isolated and assessed for their ability to suppress cucumber Fusarium wilt by culture-dependent measures to elucidate the importance of the predominant rhizobacteria of Allium plants in Fusarium wilt suppression.
Results
Fusarium wilt suppressiveness of plant-cultivated soils and of soil amended with Welsh onion root-extract
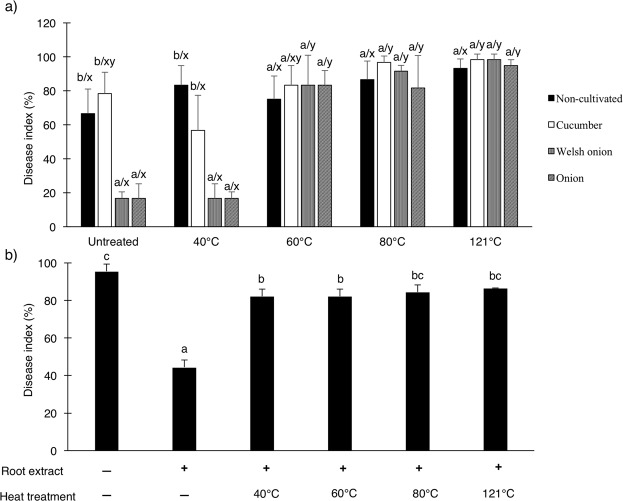
The severity of cucumber Fusarium wilt was significantly reduced in Allium (Welsh onion and onion)-cultivated soils compared with non-cultivated soil and cucumber-cultivated soil (P < 0.01) (Fig. 1a), indicating that Allium cultivation conferred cucumber Fusarium wilt suppressiveness to soil. Similarly, cucumber Fusarium wilt severity in soil amended with an aqueous extract of Welsh onion roots was also significantly lower than that in unamended soil (P < 0.01) (Fig. 1b). The suppressive effect of Allium-cultivated soils on cucumber Fusarium wilt was not diminished after heat treatment at 40 °C. However, when Allium-cultivated soils were treated at 60 °C, 80 °C, or 121 °C, they lost their ability to suppress cucumber Fusarium wilt (Fig. 1a). In contrast, the suppressiveness of soil supplemented with Welsh onion root extract was reduced almost completely after heat treatment at 40 °C (Fig. 1b).
Figure 1.
Effect of heat treatment on the suppressive ability of Allium-cultivated soils and soil supplemented with root extract of Welsh onion against cucumber Fusarium wilt. (a) Disease index in each cultivated soil and non-cultivated soil. Bars represent mean ± SD (n = 4), and the different letters above the bars indicate statistically significant differences between plant treatments (a–b) and between heat treatments (x–y) (P < 0.01, Tukey’s test). (b) Disease index in soil supplemented with root extract of Welsh onion. Bars represent mean ± SD (n = 3), and the different letters above the bars indicate statistically significant differences (P < 0.01, Tukey’s test).
Impact of heat treatment on culturable bacterial and fungal populations in Allium-cultivated soils
The population densities of culturable gram-negative bacteria in non-heat-treated Welsh onion-cultivated soil and non-heat-treated onion-cultivated soil were 0.7–1.3 × 108 and 3.4–6.0 × 107 cfu/g dry soil, respectively (Table 1). Heat treatment at 40 °C did not influence the population densities of culturable gram-negative bacteria. However, the population densities of culturable gram-negative bacteria in Allium-cultivated soils treated at a temperature of 60 °C or higher were reduced more than 10-fold compared to the non-heat-treated cultivated soils. Similarly, the population densities of culturable fungi in both cultivated soils decreased sharply when the soil was treated at a temperature ≥ 60 °C. In contrast, the population densities of culturable gram-positive bacteria in both cultivated soils did not change after heat treatment at a temperature between 40 °C and 80 °C.
Table 1.
Effect of heat treatment on population densities of culturable bacteria and fungi in Allium plant-cultivated soils (cfu gram−1 dry soil).
| Rhizosphere soil | Microorganism | Heat treatment | ||||
|---|---|---|---|---|---|---|
| No treatment | 40 °C | 60 °C | 80 °C | 121 °C | ||
| Welsh onion | Bacteria | |||||
| Gram-negative | 0.7–1.3 × 108 | 0.6–1.1 × 108 | <2.8 × 106 | ND | ND | |
| Gram-positive | 0.7–1.0 × 108 | 5.0–9.7 × 107 | 2.0–3.9 × 107 | 1.9–3.5 × 107 | ND | |
| Fungi | 0.8–3.1 × 106 | 0.5–2.4 × 106 | 0.3–1.8 × 104 | <1.5 × 102 | ND | |
| Onion | Bacteria | |||||
| Gram-negative | 3.4–6.0 × 107 | 2.6–4.7 × 107 | <1.9 × 106 | ND | ND | |
| Gram-positive | 4.0–6.7 × 107 | 2.1–3.0 × 107 | 2.0–3.0 × 107 | 2.3–2.7 × 107 | ND | |
| Fungi | 2.4–2.7 × 105 | 1.5–1.8 × 105 | 2.3–3.7 × 103 | <2.3 × 102 | ND | |
The culturable bacterial densities were estimated using 1/10 strength tryptic soy agar, and Gram reaction was determined using the KOH method. The culturable fungal densities were estimated using rose bengal-streptomycin agar. The experiment was repeated three times. ND: not detected.
Inhibitory effect of plant-cultivated soils and root extracts on multiplication of Fusarium oxysporum
In Experiment 1, multiplication of FocuGFP-10, a green fluorescent protein (GFP)-tagged isolate of GUS77, in liquid medium was significantly inhibited by the supplementation of soil suspensions, regardless of soil type, when compared with the control (P < 0.01) (Table 2). When comparing Welsh onion-cultivated soil with the other two soils (i.e., non-cultivated soil and cucumber-cultivated soil), the inhibitory effect of the former was significantly higher than that of the latter two types of soil (P < 0.01). This inhibitory effect of Welsh onion-cultivated soil was not affected by heat treatment at 40 °C. However, when the suspension of Welsh onion-cultivated soil was treated at 60 °C, the inhibitory effect on FocuGFP-10 multiplication was diminished. Additionally, in Experiment 2, treatment with antibacterial antibiotics completely abolished the inhibitory effect of Welsh onion-cultivated soil. In Experiment 3, amendment with Welsh onion root extract also significantly inhibited FocuGFP-10 multiplication in liquid medium compared with the control (P < 0.01). In contrast, amendment with cucumber root extract did not affect FocuGFP-10 multiplication. The inhibitory effect of Welsh onion-root extract was abolished after heat treatment at both 40 °C and 60 °C.
Table 2.
Inhibitory effect of Welsh onion-cultivated soil and root extracts on multiplication of Fusarium oxysporum f. sp. cucumerinum in liquid medium.
| Experiment | Type of amendment in the liquid mediuma | Heat/Antibiotics treatment | Focu density (log spores ml−1)e |
|---|---|---|---|
| Experiment 1 | Welsh onion-cultivated soilb | No treatment | 5.76 ± 0.10 a |
| 40 °C | 5.82 ± 0.09 a | ||
| 60 °C | 6.13 ± 0.02 b | ||
| Cucumber-cultivated soilb | No treatment | 6.11 ± 0.04 b | |
| Non-cultivated soilb | No treatment | 6.21 ± 0.05 b | |
| Control (SDW) | No treatment | 6.58 ± 0.03 c | |
| Experiment 2 | Welsh onion-cultivated soilb | No treatment | 5.78 ± 0.23 a |
| Antibioticsd | 6.30 ± 0.08 b | ||
| Control (SDW) | No treatment | 6.77 ± 0.09 b | |
| Antibiotics | 6.63 ± 0.12 b | ||
| Experiment 3 | Welsh onion-root extractc | No treatment | 6.01 ± 0.37 a |
| 40 °C | 6.78 ± 0.16 b | ||
| 60 °C | 6.75 ± 0.16 b | ||
| Cucumber-root extractc | No treatment | 6.86 ± 0.09 b | |
| Control (SDW) | No treatment | 6.81 ± 0.03 b |
aLiquid medium: potato sucrose broth including the spores of Fusarium oxysporum f. sp. cucumerinum.
bWelsh onion-cultivated soil, cucumber-cultivated soil, or non-cultivated soil was 1000-fold diluted with sterile distilled water, and then a 0.3 ml of each 1000-fold dilution of soils was added into the liquid medium.
cThe concentrations of Welsh onion- and cucumber-root extract were 50 mg root material per ml. A 1.5 ml of each root extract was added into the liquid medium.
dAntibiotics means a mixture of antibacterial antibiotics comprising ampicillin (300 µg ml−1), imipenem (300 µg ml−1), and chloramphenicol (300 µg ml−1).
eMean ± SD shown (n = 3). For each experiment, figures followed by different letters indicate significant differences (P < 0.01, Tukey’s test).
Microbial community analysis based on 16S rRNA gene amplicons obtained by Illumina MiSeq sequencing
The sequencing resulted in a total of 1,466,567 raw reads detected from 12 soil DNA samples (See Supplementary Table S1). After merging forward and reverse reads using dada2, the number of merged reads for each of the samples ranged from 28,691 to 12,061. The number of OTUs varied among the different samples from 810 to 294. The rarefaction curve suggested that the number of reads was sufficient to assess the diversity of bacteria in the rhizosphere communities (see Supplementary Fig. S1).
The α-diversity (Shannon index) of bacteria of Allium rhizosphere (Welsh onion and onion) and cucumber rhizosphere revealed no significant differences, while those of onion and cucumber were significantly lower than that of non-cultivated soil (P < 0.05) (see Supplementary Table S1). Clustering analysis based on the OTUs according to the Bray–Curtis index (β-diversity) indicated that bacterial community structures in rhizosphere soils of Allium plants were closely related and clearly separated from those in rhizosphere of cucumber and those in non-cultivated soil (Fig. 2). At genus level, the predominant rhizobacterial groups of both Allium plants (relative abundance of more than 1.0%) were Devosia, Flavobacterium, and Kaistobacter (Table 3). Among these bacterial genera, only Flavobacterium species were significantly abundant in the rhizosphere soils of Allium plants compared with cucumber rhizosphere soil and non-cultivated soil (P < 0.05).
Figure 2.
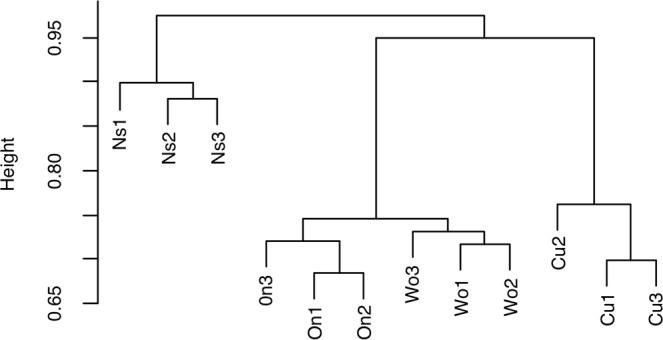
Contrasted clustering of each rhizosphere soil and non-cultivated soil samples based on OTUs composition according to Bray–Curtis similarity. Wo: Welsh onion rhizosphere soil, On: onion rhizosphere soil, Cu: cucumber rhizosphere soil, and Ns: non-cultivated soil. There were three replicates for each type of soils.
Table 3.
Relative abundances of genera comprising more than 1.0% of genera either in each rhizosphere soil (Welsh onion, onion, or cucumber), or non-cultivated soil.
| Genus | Welsh onion | Onion | Cucumber | Non-cultivated |
|---|---|---|---|---|
| Agrobacterium | 0.3 ± 0.1 b | 0.1 ± 0.2 b | 1.0 ± 0.3 a | 0.0 ± 0.0 b |
| Bacillus | 1.7 ± 0.1 a | 0.7 ± 0.3 b | 2.3 ± 0.4 a | 1.9 ± 0.4 a |
| Chitinophaga | 0.2 ± 0.0 b | 0.2 ± 0.2 b | 1.1 ± 0.1 a | 0.4 ± 0.3 b |
| Devosia | 1.9 ± 0.1 ab | 2.4 ± 0.6 a | 2.4 ± 0.9 a | 0.8 ± 0.2 b |
| Flavisolibacter | 0.6 ± 0.3 b | 0.5 ± 0.2 b | 1.4 ± 0.0 a | 0.4 ± 0.1 b |
| Flavobacterium | 3.4 ± 0.3 a | 1.7 ± 0.4 b | 0.2 ± 0.2 c | 0.4 ± 0.3 c |
| Kaistobacter | 1.3 ± 0.3 a | 1.5 ± 0.3 a | 2.1 ± 0.9 a | 2.1 ± 0.8 a |
| Methylibium | 0.8 ± 0.1 b | 1.8 ± 0.2 a | 0.3 ± 0.3 bc | 0.1 ± 0.1 c |
| Nitrospira | 0.3 ± 0.1 b | 0.1 ± 0.1 b | 0.5 ± 0.0 b | 1.2 ± 0.3 a |
| Novosphingobium | 0.1 ± 0.1 b | 0.0 ± 0.0 b | 2.0 ± 0.6 a | 0.3 ± 0.1 b |
| Opitutus | 0.4 ± 0.1 b | 0.1 ± 0.1 b | 1.6 ± 0.5 a | 0.2 ± 0.1 b |
| Pseudoxanthomonas | 1.0 ± 0.3 a | 0.8 ± 0.1 a | 0.9 ± 0.3 a | 0.0 ± 0.0 b |
| Rhodoplanes | 0.6 ± 0.3 a | 0.6 ± 0.1 a | 0.6 ± 0.3 a | 1.1 ± 0.3 a |
| Steroidobacter | 0.5 ± 0.1 b | 0.5 ± 0.1 b | 1.4 ± 0.3 a | 0.6 ± 0.2 b |
| Streptomyces | 0.7 ± 0.5 b | 1.2 ± 0.2 ab | 1.1 ± 0.1 ab | 1.8 ± 0.4 a |
| Thermomonas | 1.1 ± 0.5 a | 0.4 ± 0.2 ab | 0.7 ± 0.2 ab | 0.2 ± 0.0 b |
Mean ± SD shown (n = 3). Different lowercase letters within a row indicate statistically significant differences (P < 0.05, Tukey’s test).
Suppressive effect of Flavobacterium isolates on cucumber Fusarium wilt
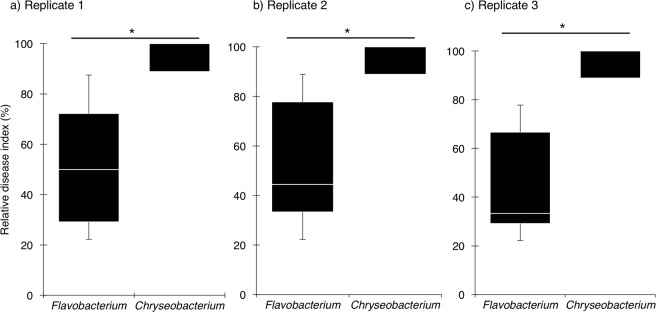
In replicate 1, the median relative disease index (RDI; %) in the soil treated with Chryseobacterium isolates, which served as bacterized controls, was 88.9% (Fig. 3a). Flavobacterium isolates obtained from Allium rhizospheres exhibited significant suppressive effects on cucumber Fusarium wilt than Chryseobacterium isolates (P < 0.01). The median RDI of Flavobacterium treatments was 50.0%. The similar results were obserbed in each replicate (Fig. 3b,c).
Figure 3.
Box-plot showing the suppressive effect of Flavobacterium isolates and Chryseobacterium isolates against cucumber Fusarium wilt. Nineteen isolates of Flavobacterium species and 15 isolates of Chryseobacterium species were assessed. The box-plot shows the minimum, maximum, 25–75%, and median values of the relative disease index. Vertical bars extending beyond the boxes represent the 5th and 95th percentiles. Asterisks indicate statistically significant difference (P < 0.01, Mann–Whitney U-test).
Inhibitory effect of Flavobacterium isolates on hyphal growth of Focu in soil
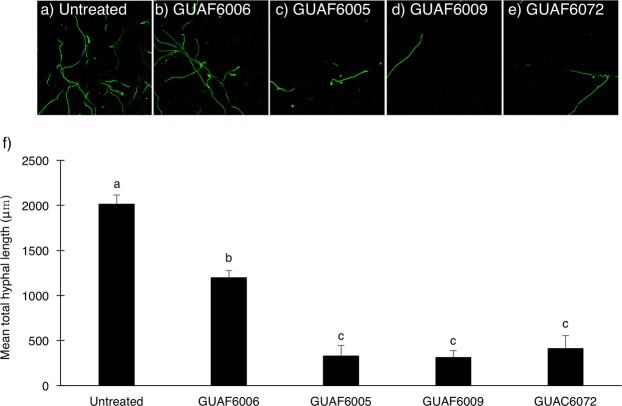
In autoclaved non-bacterized soils, FocuGFP-10 grew hyphae vigorously and the average total hyphal length/camera field reached 2015 μm (Fig. 4). In contrast, hyphal growth was significantly suppressed in soils treated with bacterial isolates, regardless of the bacterial genus (P < 0.01) (Fig. 4). However, when comparing Flavobacterium and Chryseobacterium treatments, the former displayed a significantly stronger inhibitory effect (with the average total hyphal length/camera field ranging from 312 to 411 μm) than the latter (mean = 1195 μm) (P < 0.01).
Figure 4.
Confocal laser scanning microscopy of FocuGFP-10 in bacterized soil. (a) Non-bacterized, (b) Chryseobacterium isolate GUAF6006, (c) Flavobacterium isolate GUAF6006, (d) Flavobacterium isolate GUAF6009, and (e) Flavobacterium isolate GUAC6072. Data are representative of nine images. (f) Mean total length of FocuGFP-10 hyphae in each camera field of view. Bars represent mean of three replications, and the different letters above the bars indicate statistically significant differences (P < 0.01, Tukey’s test).
Discussion
Our study demonstrated that the cultivation of Welsh onion and onion conferred suppressiveness to soil with respect to Fusarium wilt in cucumber plants (Fig. 1a). This result indicates that Allium cultivation alters the microbial or chemical properties of soil in a manner that suppresses pathogenic F. oxysporum. Generally, the role of microorganisms in soil suppressiveness to soil-borne diseases has been made apparent by the fact that the disease suppressive effects of soils are lost upon pasteurization1,24. We found that the Fusarium wilt suppressiveness of Allium-cultivated soils disappeared after heat treatment at ≥60 °C (Fig. 1a). Similarly, the inhibitory effect of Welsh onion-cultivated soil on the multiplication of Focu in liquid medium was lost after heat treatment at 60 °C but not at 40 °C (Table 2). Heat treatment at 60 °C reduced the population density of culturable gram-negative bacteria and fungi in Allium-cultivated soils but not that of culturable gram-positive bacteria, whereas treatment at 40 °C did not affect the populations of those microorganisms (Table 1). In addition, supplementation with antibacterial antibiotics abolished the inhibitory effect of Welsh onion-cultivated soil on Focu multiplication. Taken together, these results suggest that gram-negative antagonistic bacteria accumulated in Allium-cultivated soils may be a major factor in cucumber Fusarium wilt suppressiveness.
Cluster analysis based on the data from the Illumina MiSeq sequencing of 16S rRNA gene amplicons demonstrated that the bacterial community structures of Allium rhizosphere soils were similar to each other and different from those of cucumber rhizosphere soil and non-cultivated soil (Fig. 2). Interestingly, the gram-negative genus Flavobacterium was predominant in (relative abundance >1.0%) and characteristic of the bacterial communities of Welsh onion and onion rhizosphere soil (Table 3). Shen, et al.25 recently reported that Flavobacterium was one of the most abundant bacterial genera present in the soil of banana fields in which Fusarium wilt decline had been occurring. Therefore, we hypothesized that the accumulation of Flavobacterium species is a key component of Fusarium wilt suppressiveness of Allium-cultivated soils. Indeed, Flavobacterium isolates recovered from rhizospheres of Welsh onion and onion exhibited significant suppressive effects against Fusarium wilt on cucumber seedlings (Fig. 3). In addition, isolates having a strong disease-suppressing effect significantly inhibited the growth of Focu hyphae in soil (Fig. 4), suggesting that the accumulation of antagonistic Flavobacterium species in soil is an important mechanism of Fusarium wilt suppression by Allium cultivation. Although little is known about the suppressive ability of Flavobacterium species against soil-borne fungal pathogens, some species of Flavobacterium isolated from rhizosphere and soil have been reported to produce antimicrobial substances such as hydrogen cyanide, chitinase, and siderophore26–28. We are currently investigating the mode of action of our Flavobacterium isolates against Fusarium wilt pathogen.
Shen, et al.18 reported that bacterial diversity might be an important factor in soil suppressiveness because bacterial diversities were significantly higher in suppressive soils than in conducive soils. However, our data revealed that there were no significant differences in the α-diversity of bacterial communities (Shannon indices) from rhizosphere soils of Allium (i.e., Welsh onion and onion) and cucumber (See Supplementary Table S1), indicating that bacterial diversity is not a crucial factor in the suppression of Fusarium wilt by Allium cultivation.
Antifungal compounds released from Allium roots are recognized to play a role in Fusarium wilt suppression conferred by intercropping or rotation with Allium plants16,29. In accordance with these reports, the aqueous extract of Welsh onion roots significantly suppressed Focu multiplication in liquid medium and Fusarium wilt on cucumber seedlings (Fig. 1b and Table 2). These suppressive effects of Welsh onion root extract were almost completely lost after heat treatment at 40 °C. These results demonstrated that the suppression of cucumber Fusarium wilt in sterilized soil by supplementation with Welsh onion root extract may be attributed to the direct inhibition of Focu multiplication by heat-sensitive compounds, possibly volatiles, such as 2-methyl-2-pentenal and organosulfur compounds (dimethyl trisulfide, dimethyl disulfide, dipropyl disulfide, and dipropyl trisukfide)16,30–32. Zhang, et al.16 suggested that these antifungal volatiles potentially play a role in the suppression of banana Fusarium wilt when Chinese chive is used as a rotating or intercropping plant. Similarly, Li, et al.12 found that suppression of Fusarium root rot of peanut by intercropping with a medicinal herb (Atractylodes lancea) was mainly due to antifungal volatiles released by the below-ground parts of A. lancea. However, given that the suppressive effects of Welsh onion- and onion-cultivated soils were retained even after heat treatment at 40 °C (Fig. 1a), antifungal volatiles in root exudate of Allium plants may be partially involved but are not a major factor responsible for the suppression of cucumber Fusarium wilt by Allium cultivation.
In conclusion, the findings of this study clearly demonstrate that the suppression of Fusarium wilt by Allium cultivation is mainly due to the accumulation of antagonistic Flavobacterium species. We believe that our results provide important insights into multitrophic interactions among plants, soil-borne pathogens, and natural antagonistic bacteria in crop rotation/intercropping systems. Moreover, we also believe that the elucidation of mechanisms underlying the recruitment and accumulation of antagonistic bacteria by Allium plants may lead to the development of novel eco-friendly Fusarium wilt management strategies.
Materials and Methods
Preparation of pathogen inoculum
Fusarium oxysporum f. sp. cucumerinum (Focu) isolate GUS77 used in this study as the challenging pathogen was previously recovered from cucumber fields and was chosen based on its performance on aggressiveness tests. FocuGFP-10, a green fluorescent protein (GFP)-tagged isolate of GUS77, was used for fluorescent microscopy and confocal laser scanning microscopy. Full description is reported in the supplementary information.
Preparation of soils cultivated with Allium and cucumber plants
In order to confirm whether Allium cultivation induces soil suppressiveness against pathogenic F. oxysporum, we evaluated soils cultivated with Allium plants or cucumber and a non-cultivated soil. For this, we prepared soils cultivated with Welsh onion, onion, and cucumber (Cucumis sativus). Full description is reported in the supplementary information.
Preparation of root extracts from Welsh onion and cucumber
Root extracts of Welsh onion and cucumber were prepared. Full description is reported in the supplementary information.
Impact of heat treatment on Fusarium wilt suppressiveness by plant-cultivated soils and soil amended with Welsh onion root-extract
Each type of plant-cultivated soil and the non-cultivated soil, prepared as described above, was mixed with double-autoclaved potting soil (Ikubyou-baido: Takii seed and seedling, Kyoto, Japan) and with double-autoclaved river sand in a ratio of 4:1:1 (w/w/w). Six grams of each type of soil mixture were then placed in flat-bottom glass tubes (3 cm in diameter and 12 cm high, Iwaki Glass, Chiba, Japan) and heat-treated in one of the following ways: 1) no treatment, 2) incubation in a water bath at 40 °C for 30 min, 3) incubation in a water bath at 60 °C for 30 min, 4) incubation in a water bath at 80 °C for 30 min, and 5) autoclaving at 121 °C for 60 min.
For the evaluation of soil amended with Welsh onion-root extract, a 1-ml aliquot of the root extract of Welsh onion was added to a flat-bottom glass tube (3 cm in diameter and 12 cm high) containing 6 g (dry weight) of a double-autoclaved soil mixture, prepared by blending field soil sieved through a 2-mm mesh sieve, commercial potting soil (Ikubyou-baido), and river sand in a ratio of 1: 1: 1 (w/w/w). The soil mixture amended with the root extract was then subjected to five types of heat treatments as described above. All soils were then inoculated with Focu by pouring a 4 ml aliquot of a spore suspension (1.5 × 104 spores ml−1) into each glass tube and the soil surface was covered with 2 g of sterile vermiculite. Surface-sterilized and pre-germinated cucumber seeds were then planted in this vermiculite layer (1 seed per tube), covered with a small amount of sterile vermiculite, and fertilized with 2 ml of 500-fold-diluted Hyponex solution (Type: 6–10–5, Hyponex Japan, Osaka, Japan). The tubes were then placed in a chamber with controlled conditions (25 °C, 12 h of day light) for 20 days. The severity of the disease developing on the seedlings was assessed using a scale of 0 to 3, where 0 = healthy plant, 1 = cotyledon leaf yellowing and/or vascular browning, 2 = hypocotyl browning, and 3 = seedling dead, and the results were expressed as a disease index (%), calculated using the following formula: {Σ (disease severity of cucumber seedlings)/(the number of replicated tubes × 3)} × 100. There were five replicated tubes (one seedling per tube) per treatment and the experiment was repeated at least three times.
Impact of heat treatment on culturable bacterial and fungal populations in Allium-cultivated soils
In parallel with the assessment of an effect of heat treatment on soil suppressiveness, we investigated possible changes in the bacterial and fungal populations in Allium-cultivated soil. Full description is reported in the supplementary information.
Inhibitory effect of plant-cultivated soils and of the root extract on multiplication of Fusarium oxysporum
To clarify whether the accumulation of antagonistic bacteria and/or antifungal compounds produced by Allium plants play a role in soil suppressiveness against cucumber Fusarium wilt, the following three experiments (Experiment 1–3) were performed. In Experiment 1, the inhibitory effect of Welsh onion-cultivated soil against Focu multiplication was compared with those of cucumber-cultivated soil and non-cultivated soil. Additionally, the impact of heat treatment on the inhibitory effect of Welsh onion-cultivated soil was also investigated. For heat treatment, 10 g of Welsh onion-cultivated soil, prepared as described before, were suspended in 90 ml of SDW in a 200-ml Erlenmeyer flask and treated at 40 °C and 60 °C, as described above. To evaluate the inhibitory effect of soils against Focu multiplication, a 0.3 ml of each 1000-fold dilution of soils was added into test tubes containing 0.2 ml of potato sucrose broth and 1.5 ml of SDW. Each tube was then inoculated with 1 ml of spore suspension of FocuGFP-10 (3 × 105 spores ml−1) and shaken for 16 h at 25 °C on a rotary shaker at 150 rpm. After incubation, the concentration of spores was determined using a haemocytometer under a fluorescence microscope. The spores were counted in five fields of view per sample. There was one tube per treatment and the experiment was repeated three times.
In Experiment 2, the impact of an antibacterial treatment on the inhibitory effect of Welsh onion-cultivated soil was investigated. For the antibacterial treatment, a 100 µl of 100-fold diluted suspension of Welsh onion-cultivated soil was mixed with 900 µl of a mixture of antibacterial antibiotics comprising ampicillin (300 µg ml−1), imipenem (300 µg ml−1), and chloramphenicol (300 µg ml−1), and then incubated for 1 h on a rotatory shaker at 25 °C. The inhibitory effect of the soils against Focu multiplication was tested by the same procedures in Experiment 1. There was one tube per treatment and the experiment was repeated three times.
In Experiment 3, the inhibitory effect of Welsh onion-root extract against Focu multiplication was compared with that of cucumber-root extract. Simultaneously, the impact of heat treatment on the inhibitory effect of Welsh onion-root extract was also investigated. For heat treatment, Welsh onion-root extract was treated at 40 °C and 60 °C as described above. Either 1.5 ml of each root extract (Welsh onion or cucumber) or SDW was added to a test tube containing 1 ml of potato sucrose broth, and this was then inoculated with 500 µl of Focu suspension (6 × 106 spores ml−1) and shaken at 150 rpm for 30 h at 25 °C. After the incubation, the spores were counted using a hemocytometer. There was one tube per treatment and the experiment was repeated three times.
Microbial community analysis based on 16S rRNA gene amplicons obtained by Illumina MiSeq sequencing
This analysis was carried out through the Illumina MiSeq sequencing of 16S rRNA gene amplicons, using soil DNA extracted from non-cultivated soil and from rhizosphere soils of Welsh onion, onion, and cucumber plants. Non-cultivated soil was prepared as described before. Rhizosphere soils were collected from each of three plants of Welsh onion, onion, and cucumber, which were grown in vinyl pots containing field soil for 70 days as described before. The rhizosphere soils and non-cultivated soil were stored at −80 °C until DNA extraction. The DNA extraction was conducted using a FastDNA SPIN Kit (MP Biomedicals, CA, USA) following the manufacturer’s protocol with little modifications for the first step. Briefly, 0.4 g of either each rhizosphere soil or non-cultivated soil was added to lysing matrix tubes containing 878 µl of sodium phosphate buffer, 122 µl of MT buffer, and 100 µl of 20% skim milk solution. The following processes were conducted according to the manufacture’s protocol. The DNA extraction was repeated three times. The V3–V4 region of each DNA sample was amplified with specific primers33 and paired-end sequenced following the manufacturer’s protocol (https://support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/16 s/16s-metagenomic-library-prep-guide-15044223-b.pdf) on an Illumina MiSeq (Illumina, CA, USA).
Sequence processing was conducted using Qiime2 (version 2018.2) with demux-summarize, dada234, and feature-table. A pre-trained Naive Bayes classifier based on the Greengenes 13_8 99% operational taxonomic units (OTUs) database (http://greengenes.secondgenome.com), which had been trimmed to include V3–V4 regions of 16S rRNA gene, bound by the 465 F/805 R primer pair, was applied to paired-end sequence reads to make taxonomy tables.
Community dissimilarity according to the Bray–Curtis index was calculated based on the OTUs data processed by coverage-based rarefaction35 using the pvclust package in R 3.3.1 software. Predominant bacterial groups of rhizosphere soils of Allium plants and cucumber, and non-cultivated soil, with relative abundance of more than 1.0% at the genus level were selected. Furthermore, rhizobacterial groups predominant only in Allium plants were selected as potential antagonistic bacteria. Rhizobacterial sequence data were deposited in the Sequence Read Archive database under accession numbers DRX121065– DRX121076 (BioProject: PRJDB6419).
Isolation of Flavobacterium species from Allium rhizosphere
It was postulated that Flavobacetrium species were involved in Fusarium wilt suppression by Allium-cultivation. To test this hypothesis, Flavobacterium species were isolated from the rhizosphere soils of Welsh onion and onion. Both plants were grown in vinyl pots as described above. Rhizosphere soils of these plants were collected, diluted with SDW, and then spread on the surface of a semi-selective medium (namely PSR2A-C/T agar)36 for species of Flavobacterium and Chryseobacterium. Simultaneously, Chryseobacterium species were also recovered from Welsh onion rhizosphere soil. Flavobacterium and Chryseobacterium species belong to the same family, i.e., Flavobacteriaceae.
Suppressive effect of Flavobacterium isolates on cucumber Fusarium wilt
Flavobacterium isolates obtained from Allium rhizospheres were evaluated for their ability to suppress cucumber Fusarium wilt in the cucumber seedling assay. For comparison, suppressive effect of Chryseobacterium isolates obtained from Welsh onion were also tested (designated as “bacterized control”). Full description is reported in the supplementary information.
Inhibitory effect of Flavobacterium isolates on hyphal growth of Focu in soil
To determine whether the Flavobacterium isolates could antagonize Focu in soil, hyphal growth in soil bacterized with Flavobacterium isolates was examined by confocal laser scanning microscopy (CLSM) using FocuGFP-10 as the challenging pathogen. The soil was bacterized with Flavobacterium isolates GUAF6005 (from Welsh onion), GUAF6009 (from Welsh onion), or GUAC6072 (from onion), or with Chryseobacterium isolate GUAF6006 (from Welsh onion). The Flavobacerium isolates used here exhibited a strong suppressive effect against cucumber Fusarium wilt (RDI < 50%) in the cucumber seedling assay, whereas the GUAF6006 Chryseobacterium isolate slightly reduced cucumber Fusarium wilt severity (RDI = 88.9%). As illustrated in Supplementary Fig. S2, a chamber was filled with seven grams of the double-autoclaved soil mixture (field soil: river sand: potting soil = 1:1:1 w/w/w) on a coverglass (Cell Imaging Coverglass 1 Chamber, Eppendorf North America Inc., NY, USA). A small hole was drilled into one of the shorter sides of the chamber. Four milliliters of FocuGFP-10 spore suspension (1.8 × 105 spores ml−1) was inoculated into the soil mixture. Subsequently, a 3-ml aliquot of each bacterial isolate (ca. 0.5–1.0 × 109 cells ml−1) or SDW was separately applied to the FocuGFP-10-inoculated soil mixtures. Surface-sterilized and germinated cucumber seeds were then planted into the holes (one seed per chamber), and soil surfaces were covered with a small amount of sterile vermiculite. The sides of the coverglasses were covered with aluminum foil and the chambers were incubated in a controlled environmental chamber (25 °C, 12 h of daylight) for 1 week. After incubation, images of FocuGFP-10 hyphae in the soil mixtures were captured at three different locations by CLSM (LSM710, Carl Zeiss, Jena, Germany). The mean of total length of FocuGFP-10 hyphae in each camera field-of-view was then measured using LIA32 software (https://www.agr.nagoya-u.ac.jp/~shinkan/LIA32/). There were three replicates for each treatment.
Statistical analyses
Statistical analyses were performed using BellCurve for Excel (version 2.13) (Social Survey Research Information, Tokyo, Japan). Full description is reported in the supplementary information.
Supplementary information
Acknowledgements
The authors are grateful to Dr. Keiko Inaba-Hasegawa (Genome Sequencing Facility of Gifu University, Japan) and Ms. Izumi Nomura (Faculty of Applied Biological Sciences, Gifu University, Japan) for the amplicon sequencings with MiSeq. This work was mainly supported by a Grant-in-Aid for Young Scientists (B) (Grant Number. 24780317) and for JSPS Fellows (Grant Number. 953381) from the Ministry of Education, Culture, Sports, Science and Technology of Japan. This work was also partly supported by JST ERATO Grant Number JPMJER1502, Japan.
Author Contributions
T.N., H.S. and M.S. designed the study. T.N. and M.M. contributed to experiments. T.N., I.K., Y.K., K.Y. and H.T. contributed to data analysis. T.N. and M.S. wrote the manuscript and all authors reviewed the manuscript.
Data Availability
The datasets supporting the conclusions of this study are included within this manuscript and its supplementary files.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-37559-7.
References
- 1.Cha J-Y, et al. Microbial and biochemical basis of a Fusarium wilt-suppressive soil. ISME J. 2016;10:119–129. doi: 10.1038/ismej.2015.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fravel D, Olivain C, Alabouvette C. Fusarium oxysporum and its biocontrol. New Phytol. 2003;157:493–502. doi: 10.1046/j.1469-8137.2003.00700.x. [DOI] [PubMed] [Google Scholar]
- 3.Akhter A, Hage-Ahmed K, Soja G, Steinkellner S. Potential of Fusarium wilt-inducing chlamydospores, in vitro behaviour in root exudates and physiology of tomato in biochar and compost amended soil. Plant Soil. 2016;406:425–440. doi: 10.1007/s11104-016-2948-4. [DOI] [Google Scholar]
- 4.Pavlou GC, Vakalounakis DJ. Biological control of root and stem rot of greenhouse cucumber, caused by Fusarium oxysporum f. sp. radicis-cucumerinum, by lettuce soil amendment. Crop Prot. 2005;24:135–140. doi: 10.1016/j.cropro.2004.07.003. [DOI] [Google Scholar]
- 5.Pavlou GC, Vakalounakis DJ, Ligoxigakis EK. Control of root and stem rot of cucumber, caused by Fusarium oxysporum f. sp. radicis-cucumerinum, by grafting onto resistant rootstocks. Plant Dis. 2002;86:379–382. doi: 10.1094/PDIS.2002.86.4.379. [DOI] [PubMed] [Google Scholar]
- 6.Yetisir H, Kurt S, Sari N, Tok FM. Rootstock potential of Turkish Lagenaria siceraria germplasm for watermelon: plant growth, graft compatibility, and resistance to Fusarium. Turkish J. Agric. For. 1994;31:381–388. [Google Scholar]
- 7.Takehara T, Kuniyasu K, Mori M, Hagiwara H. Use of a nitrate-nonutilizing mutant and selective media to examine population dynamics of Fusarium oxysporum f. sp. spinaciae in soil. Phytopathology. 2003;93:1173–1181. doi: 10.1094/PHYTO.2003.93.9.1173. [DOI] [PubMed] [Google Scholar]
- 8.Ayed F, Daami-Remadi M, Jabnoun-Khiareddine H, Mahjoub M. El. Potato vascular Fusarium wilt in Tunisia: incidence and biocontrol by Trichoderma spp. Plant Pathol. J. 2006;5:92–98. doi: 10.3923/ppj.2006.92.98. [DOI] [Google Scholar]
- 9.Li Xgang, Ding Cfeng, Zhang Tlin, Wang Xxiang. Fungal pathogen accumulation at the expense of plant-beneficial fungi as a consequence of consecutive peanut monoculturing. Soil Biol. Biochem. 2014;72:11–18. doi: 10.1016/j.soilbio.2014.01.019. [DOI] [Google Scholar]
- 10.Igarashi C, et al. Suppression of spinach Fusarium wilt by intercropping with Allium plants. Japanese J. Phytopathol. 2017;83:87–94. doi: 10.3186/jjphytopath.83.87. [DOI] [Google Scholar]
- 11.Huang YH, et al. Control of Fusarium wilt in banana with Chinese leek. Eur. J. Plant Pathol. 2012;134:87–95. doi: 10.1007/s10658-012-0024-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li X, et al. Suppression of soil-borne Fusarium pathogens of peanut by intercropping with the medicinal herb Atractylodes lancea. Soil Biol. Biochem. 2018;116:120–130. doi: 10.1016/j.soilbio.2017.09.029. [DOI] [Google Scholar]
- 13.Yang M, et al. Plant-plant-microbe mechanisms involved in soil-borne disease suppression on a maize and pepper intercropping system. PLoS One. 2014;9:1–22. doi: 10.1371/journal.pone.0115052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kijima T, et al. Study about use of antibiotic microorganisms (in Japanese) Bull Tochigi Agr Exp Stn. 1988;35:95–128. [Google Scholar]
- 15.Liu S, Wu F, Wen X. Allelopathic effects of root exudates of Chinese onion on tomato growth and the pathogen Fusarium oxysporum (Sch1) f. sp. lycopersici. Allelopathy. 2013;31:387–404. [Google Scholar]
- 16.Zhang H, Mallik A, Zeng R. Sen. Control of panama disease of banana by rotating and intercropping with Chinese chive (Allium tuberosum Rottler): role of plant volatiles. J. Chem. Ecol. 2013;39:243–252. doi: 10.1007/s10886-013-0243-x. [DOI] [PubMed] [Google Scholar]
- 17.Rosenzweig N, Tiedje JM, Quensen JF, Meng Q, Hao JJ. Microbial communities associated with potato common scab-suppressive soil determined by pyrosequencing analyses. Plant Dis. 2012;96:718–725. doi: 10.1094/PDIS-07-11-0571. [DOI] [PubMed] [Google Scholar]
- 18.Shen Z, et al. Soils naturally suppressive to banana Fusarium wilt disease harbor unique bacterial communities. Plant Soil. 2015;393:21–33. doi: 10.1007/s11104-015-2474-9. [DOI] [Google Scholar]
- 19.Ahmad I, et al. Effect of pepper-garlic intercropping system on soil microbial and bio-chemical properties. Pakistan J. Bot. 2013;45:695–702. [Google Scholar]
- 20.Zhou X, Yu G, Wu F. Effects of intercropping cucumber with onion or garlic on soil enzyme activities, microbial communities and cucumber yield. Eur. J. Soil Biol. 2011;47:279–287. doi: 10.1016/j.ejsobi.2011.07.001. [DOI] [Google Scholar]
- 21.Gardner T, et al. Soil rhizosphere microbial communities and enzyme activities under organic farming in Alabama. Diversity. 2011;3:308–328. doi: 10.3390/d3030308. [DOI] [Google Scholar]
- 22.Mendes R, et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science. 2011;332:1097–100. doi: 10.1126/science.1203980. [DOI] [PubMed] [Google Scholar]
- 23.Li XG, et al. Soil sickness of peanuts is attributable to modifications in soil microbes induced by peanut root exudates rather than to direct allelopathy. Soil Biol. Biochem. 2014;78:149–159. doi: 10.1016/j.soilbio.2014.07.019. [DOI] [Google Scholar]
- 24.Scher FM, Baker R. Mechanism of biological control in a Fusarium-suppressive soil. Phytopathology. 1980;70:412. doi: 10.1094/Phyto-70-412. [DOI] [Google Scholar]
- 25.Shen Z, et al. Banana Fusarium wilt disease incidence is Influenced by shifts of soil microbial communities under different monoculture spans. Microb. Ecol. 2018;75:739–750. doi: 10.1007/s00248-017-1052-5. [DOI] [PubMed] [Google Scholar]
- 26.Belimov AA, et al. Cadmium-tolerant plant growth-promoting bacteria associated with the roots of Indian mustard (Brassica juncea L. Czern.) Soil Biol. Biochem. 2005;37:241–250. doi: 10.1016/j.soilbio.2004.07.033. [DOI] [Google Scholar]
- 27.Kharade SS, McBride MJ. Flavobacterium johnsoniae chitinase ChiA is required for chitin utilization and is secreted by the type IX secretion system. J. Bacteriol. 2014;196:961–70. doi: 10.1128/JB.01170-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soltani A-A, et al. Plant growth promoting characteristics in some Flavobacterium spp. isolated from soils of Iran. J. Agric. Sci. 2010;2:106–115. [Google Scholar]
- 29.Xu N, et al. Composition of Welsh onion (Allium fistulosum L.) root exudates and their allelopathy on cucumber sprouts and Fusarium oxysporum f. sp. cucumerinum. Allelopathy. 2013;32:243–256. [Google Scholar]
- 30.Benkeblia N, Lanzotti V. Allium thiosulfinates: chemistry, biological properties and their potential utilization in food preservation. Food j. 2007;2:193–201. [Google Scholar]
- 31.Pyun M-S, Shin S. Antifungal effects of the volatile oils from Allium plants against Trichophyton species and synergism of the oils with ketoconazole. Phytomedicine. 2006;13:394–400. doi: 10.1016/j.phymed.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 32.Tapiero H, Townsend DM, Tew KD. Organosulfur compounds from alliaceae in the prevention of human pathologies. Biomed. Pharmacother. 2004;58:183–193. doi: 10.1016/j.biopha.2004.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klindworth A, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1–e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Callahan BJ, et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chao A, Jost L. Coverage-based rarefaction and extrapolation: standardizing samples by completeness rather than size. Ecology. 2012;93:2533–2547. doi: 10.1890/11-1952.1. [DOI] [PubMed] [Google Scholar]
- 36.Nishioka T, et al. Development of culture medium for the isolation of Flavobacterium and Chryseobacterium from rhizosphere soil. Microbes Environ. 2016;31:104–110. doi: 10.1264/jsme2.ME15144. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting the conclusions of this study are included within this manuscript and its supplementary files.