Abstract
Background
Angiogenesis plays a critical role in the progression of glioma. Previous studies have indicated that RNA-binding proteins (RBPs) interact with RNAs and participate in the regulation of the malignant behaviors of tumors. As a type of endogenous non-coding RNAs, circular RNAs (circRNAs) are abnormally expressed in various cancers and are involved in diverse tumorigeneses including angiogenesis.
Methods
The expression levels of FUS, circ_002136, miR-138-5p, SOX13, and SPON2 were determined using quantitative real-time PCR (qRT-PCR) and western blot. Transient cell transfection was performed using the Lipofectamine 3000 reagent. The RNA-binding protein immunoprecipitation (RNA-IP) and the RNA pull-down assays were used to detect the interaction between FUS and circ_002136. The dual-luciferase reporter assay system was performed to detect the binding sites of circ_002136 and miR-138-5p, miR-138-5p and SOX13. The chromatin immunoprecipitation (ChIP) assays were used to examine the interactions between transcription factor SOX13 and its target proteins .
Results
We demonstrated that down-regulation of FUS or circ_002136 dramatically inhibited the viability, migration and tube formation of U87 glioma-exposed endothelial cells (GECs). MiR-138-5p was down-regulated in GECs and circ_002136 functionally targeted miR-138-5p in an RNA-induced silencing complex (RISC). Inhibition of circ_002136, combined with the restoration of miR-138-5p, robustly reduced the angiogenesis of GECs. As a target gene of miR-138-5p, SOX13 was overexpressed in GECs and was proved to be involved in circ_002136 and miR-138-5p-mediated angiogenesis in gliomas. In addition, we found that SOX13 was directly associated with and activated the SPON2 promoter, thereby up-regulating the expression of SPON2 at the transcriptional level. Knockdown of SPON2 suppressed the angiogenesis in GECs. More important, SOX13 activated the FUS promoter and increased its expression, forming a feedback loop.
Conclusion
Our data suggests that the feedback loop of FUS/circ_002136/miR-138-5p/SOX13 played a crucial role in the regulation of angiogenesis in glioma. This also provides a potential target and an alternative strategy for combined glioma therapy.
Electronic supplementary material
The online version of this article (10.1186/s13046-019-1065-7) contains supplementary material, which is available to authorized users.
Keywords: Angiogenesis, FUS; circ_002136, miR-138-5p, SOX13, SPON2
Background
Malignant glioma is the most common primary tumor in the human central nervous system. Despite many advances in surgical techniques, chemotherapeutics and radiation therapy, the prognosis of patients with malignant gliomas remains obstinately poor. Glioma progression depends on tumor blood vessel growth. Glioma cells secrete vascular endothelial growth factor and other pro-angiogenic factors to promote the growth of vascular endothelial cells. Moreover, glial vascular endothelial cells also secrete a variety of factors that promote tumor growth. The interactions of these secreted factors can promote glioma growth [1]. Therefore, anti-angiogenesis therapeutics are considered as important treatments for malignant glioma, and an in-depth study of angiogenesis in glioma is in need.
The FUS (Fused in sarcoma) gene is located at chromosome 16p11.2 and consists of 15 exons encoding a protein of 526 amino acids belonging to the FET (FUS/EWS/TAF15) protein family. As a DNA/RNA-binding protein with a gene regulation function, it is involved in regulating intracellular RNA transport, mRNA synthesis, alternative splicing, and polyadenylation site selection [2]. It has been found that FUS mRNA or protein expression is up-regulated in liposarcoma [3], breast cancer [4], cervical cancer [5], and other cells. FUS can promote the malignant progression of non-small cell lung cancer [6]. Silencing of the FUS expression inhibits the proliferation and migration of neuroblastoma cells and increased their chemosensitivity to cisplatin [7]. A recent study has confirmed that FUS regulates the expression of 19 circRNAs, including circ_3279 and circ_5306, via binding to introns flanking the splicing junction [8]. But the function of FUS in vascular endothelial cells has not yet been reported.
CircRNA is a non-coding RNA with a covalent loop structure, which can perform biological functions via various modes of regulation. For example, circRNAs can affect gene expression or transcription by regulating transcription and alternative splicing [9]. CircRNAs may also act as molecular sponges of microRNAs (miRNAs) or competitive endogenous RNAs to regulate translation of the target genes [10]. Previous studies have shown that circRNAs play regulatory roles in the malignant biological behavior of glioma cells. For example, circ-TTBK2 and circ-HIPK3 promote malignant progression of glioma cells [11, 12]. Hsa_circ_0000177 is significantly up-regulated in human glioma cells, promoting cell proliferation, invasion in vitro, and growth of glioma in vivo [13]. Human circ_002136 (hsa_circ_0000005, hsa_circCDK11A_001) formed through the looping of linear CDK11A-VT1 (Homo sapiens cyclin-dependent kinase 11A transcript variant 1; GenBank: NM_024011) is located at chromosome 1 and is 49,639 bps in length. To date, the function and mechanism of circ_002136 have not been clarified.
MiRNAs regulate the expression of target genes at the post-transcriptional level via binding to the 3′-untranslated region (3’-UTR) of the target genes. Studies have shown that over-expression of miR-138-5p suppresses tumor cell proliferation, invasiveness and induces apoptosis in pancreatic cancer [14] and bladder cancer [15]. The SOX13 gene is located at chromosome 1q31.3–32.1 and consists of 14 exons belonging to the SOX gene family (Sex-related region Y, Sry-related high-mobility group box). It has been found that SOX13 is highly expressed in oligodendroglioma [16] and can regulate angiogenesis [17]. However, the roles of miR-138-5p and SOX13 in glial vascular endothelial cells and their potential molecular mechanisms remain poorly defined.
SPON2 (Spondin 2, Mindin) is a member of the Mindin F-Spondin superfamily, which encodes secreted proteins and extracellular matrix proteins. SPON2 is highly expressed in ovarian cancer [18] and breast cancer [19]. In patients with hepatocellular carcinoma, the expression of SPON2 is positively correlated with prognosis [20]. These suggest that SPON2 plays an essential role in the development of various tumors. However, the function of SPON2 in vascular endothelial cells of glioma is still unknown.
In our study, we first identified the endogenous expressions of FUS, circ_002136, miR-138-5p, SOX13, and SPON2 in GECs, and explored possible regulatory interactions among the factors mentioned above and their impacts on the GEC angiogenesis. Our results provide a new insight into the mechanism of angiogenesis in glioma as well as new strategies for anti-angiogenesis treatment of glioma.
Methods
Cell culture and the preparation of glioblastoma (GBM) cell-conditioned medium
The immortalized human cerebral microvascular endothelial cell (EC) line hCMEC/D3 was provided by Dr. Couraud from the Institut Cochin, Paris, France. Cells were cultured in endothelial basal medium (EBM-2) (Lonza, Walkersville, MD, USA), supplemented with 5% fetal bovine serum (FBS) “Gold” (PAA Laboratories, Pasching, Austria), 1% penicillin-streptomycin (Life Technologies, Paisley, UK), 1% chemically defined lipid concentrate (Life Technologies), 1 ng/mL human basic fibroblast growth factor (bFGF) (Sigma-Aldrich, Beijing, China), 1.4 μM hydrocortisone, 5 μg/mL ascorbic acid (Sigma-Aldrich), and 10 mM N-2-hydroxyethylpiperazine-N-ethane-sulphonic acid (HEPES) (PAA Laboratories). ECs were limited with the passage below 30. The human glioblastoma cell line (U87MG) and human embryonic kidney 293 T (HEK293T) cell line were purchased from the Shanghai Institutes for Biological Sciences Cell Resource Center (Shanghai, China) and were cultured in high-glucose Dulbecco’s modified Eagle medium (DMEM, GIBCO, Carlsbad, CA, USA), supplemented with 10% FBS. All cells were maintained in a humidified incubator at 37 °C with 5% CO2.
GBM cell-conditioned medium was collected from the human glioblastoma cell line U87 plated in 100-mm-diameter Petri dishes. Cells that had grown to near confluency were washed twice with serum-free medium and incubated in serum-free EBM-2 medium for 24 h. The supernatant was harvested, centrifuged at 2000×g at 4 °C for 10 min and supplemented with 5% FBS, 1% penicillin-streptomycin, 1% chemically defined lipid concentrate, 1 ng/ml bFGF, 1.4 μM hydrocortisone, 5 μg/ml ascorbic acid, 10 mM HEPES, and stored at 4 °C. The GBM cell-conditioned medium was used to culture human cerebral microvascular endothelial cells (hCMEC/D3) for 24 h to produce GECs.
Quantitative real-time PCR
Total RNAs were extracted from ECs and GECs with Trizol reagent (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s description. RNA concentration and quality were determined for each sample with a Nanodrop Spectrophotometer (ND-100; Thermo Fisher Scientific, Waltham, MA, USA) using the 260/280 nm ratio. One Step SYBR PrimeScript RT-PCR Kit (Takara Biomedical Technology, Dalian, China) was used to qualify the expression levels of FUS (NM_004960.3), circ_002136(NM_024011), CDK11A(NM_024011.3), SOX13(NM_005686.2), and SPON2(NM_012445.3). In addition, RNase R was used to confirm the existence of circ_002136 and eliminate the influence of linear RNAs. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an endogenous control. The expression levels of miR-138-5p (NR_029700.1) were detected using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) and TaqMan Universal Master Mix II (Life Technologies). The U6 housekeeping gene was included as an endogenous control. All qRT-PCR reactions were performed using the 7500 Fast RT-PCR System (Applied Biosystems). The relative quantification 2-ΔΔCt method was applied to calculate the gene expression values. Primers and probes used in this study are shown in Table 1.
Table 1.
Primers and probes used for RT-qPCR
| Primer or Probe | Gene | Sequence (5′- > 3′) or Assay ID |
|---|---|---|
| Primer | FUS | F: GCCAAGATCAATCCTCCATGAGTAGTG |
| R: TCCACGGTCCTGCTGTCCATAG | ||
| circ_002136 | F: CTTTCCGAGACATTTGCTGG | |
| R: CATGGAGATCACAATAAGGAACTC | ||
| P:FAM + TCTTCTTCTCCTCTGTCTTCC+MGB | ||
| GAPDH | F: GGACCTGACCTGCCGTCTAG | |
| R: TAGCCCAGGATGCCCTTGAG | ||
| P:FAM + CCTCCGACGCCTGCTTCACCACCT+Eclipse | ||
| CDK11A | F: AGAGGAAGAGGAGGAGGAGGAGAC | |
| R: CGAACCGTGACTCTGGAACAACC | ||
| SOX13 | F: CTGGACTTCAACCGAAATTTGA | |
| R: GTTCCTTCCTAGAAACCTCTCC | ||
| SPON2 | F: GATTGTAGACAGCGCCTCAGTTCC | |
| R: GACGCACTCAGCCTCTTCTTCG | ||
| Probe | MiR-138-5p U6 | 002284(Applied Biosystems) |
| 001973(Applied Biosystems) |
Western blot assay
Total protein was extracted from cells using ice-cold radioimmunoprecipitation assay (RIPA) buffer (Beyotime Institute of Biotechnology, Jiangsu, China) supplemented with protease inhibitors (10 mg/mL aprotinin, 10 mg/mL phenyl-methylsulfonyl fluoride [PMSF], and 50 mM sodium orthovanadate). The BCA protein assay kit (Beyotime Institute of Biotechnology) was used to determine the protein concentration of the supernatant. Equal amounts of protein samples (50 μg) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and electrically transferred onto polyvinylidene difluoride (PVDF) membrane (Millipore, Shanghai, China). Non-specific binding was blocked by incubation with 5% fat-free milk in Tris-buffered saline containing 0.1% Tween-20 (TBST) at room temperature for two hours.
The membranes were subsequently incubated with primary antibodies as follows: FUS (1:2000; Proteintech, Chicago, IL, USA), SOX13 (1:500; Proteintech), SPON2 (1:1000; Affinity, Cincinnati, OH, USA), and GAPDH (1:10,000; Proteintech) at 4 °C overnight. The membranes were washed and incubated with HRP-conjugated secondary antibodies (Santa Cruz Biotechnology, Santa Cruz, CA, USA), diluted at 1:5000 at room temperature for two hours. Immunoblots were visualized using an enhanced chemiluminescence kit (ECL; Santa Cruz Biotechnology) and detected by ECL Detection Systems (Thermo Scientific, Beijing, China) and then scanned using Chemi Imager 5500 V2.03 software. The relative integrated density values (IDVs) were calculated by FluorChem 2.0 software and then normalized to that of GAPDH.
Plasmid construction and cell transfection
Short-hairpin RNAs (shRNAs) directed against human FUS, circ_002136, CDK11A, SOX13, and SPON2 gene were constructed in pGPU6/GFP/Neo vector (GenePharma, Shanghai, China) to generate silencing plasmids. The human SOX13 gene coding sequence was ligated into the pIRES2-EGFP vector (GenScript, Piscataway, NJ, USA) to construct the SOX13-over-expression plasmid. pGPU6/GFP/Neo and pIRES2-EGFP empty vectors without targeting sequences were used as negative controls (NCs). GECs were seeded into 24-well plates and transfected with the plasmids via Opti-MEM I and Lipofectamine LTX and Plus Reagents (Life Technologies) when they reached approximately 80% confluence. The stable transfected cell lines were created by selection on culture medium containing 0.4 mg/mL geneticin (G418) (Sigma-Aldrich, St. Louis, MO, USA). G418-resistant cell clones were established after approximately four weeks of use. For co-transfection of FUS (−) and circ_002136 (−), cells with stably knocked down FUS were transfected with pGPU6/circ_002136 (−)/Blasticidin. G418 and Blasticidin dual-resistant clone were selected. qRT-PCR was then performed to measure the transfected efficiencies.
Furthermore, miR-138-5p agomir [miR-138-5p (+); GenePharma], miR-138-5p antagomir [miR-138-5p (−)], and their respective NCs were transiently transfected into GECs using Lipofectamine 3000 Reagents (Life Technologies). For co-transfection of circ_002136 (−) and miR-138-5p agomir/ antagomir or SOX13 (+) and miR-138-5p agomir, cells with stably knocked down circ_002136 or overexpressed SOX13, or their respective NCs, were transiently transfected with miR-138-5p agomir/antagomir or their NCs. All transient-transfected cells were harvested after 48 h. The sequences of all shRNA templates are shown in Table 2. The transfection efficiencies are shown in Additional file 1: Figure S1.
Table 2.
Sequences of shRNA template
| Gene | Sequence(5′- > 3′) | |
|---|---|---|
| FUS | Sense | CACCGCCCTACGGACAGCAGAGTTTCAAGAGAACTCTGCTGTCCGTAGGGTTTTTTG |
| Antisense | CGGGATGCCTGTCGTCTCAAAGTTCTCTTGAGACGACAGGCATCCCAAAAAACCTAG | |
| Circ_002136 | Sense | CACCGCTATGGAAGACAGAGGAGAAGTTCAAGAGACTTCTCCTCTGTCTTCCATAGTTTTTTG |
| Antisense | CGATACCTTCTGTCTCCTCTTCAAGTTCTCTGAAGAGGAGACAGAAGGTATCAAAAAACCTAG | |
| CDK11A | Sense | CACCAGAUCUACAUCGUGAUGAATTTTCAAGAGAUUCAUCACGAUGUAGAUCUTGTTTTTTG |
| Antisense | GATCCAAAAAAAGAUCUACAUCGUGAUGAATTTCTCTTGAAUUCAUCACGAUGUAGAUCUTG | |
| SOX13 | Sense | CACCGGAAGATCCTGCAAGCCTTCCTTCAAGAGAGGAAGGCTTGCAGGATCTTCCTTTTTTG |
| Antisense | GATCCAAAAAAGGAAGATCCTGCAAGCCTTCCTCTCTTGAAGGAAGGCTTGCAGGATCTTCC | |
| SPON2 | Sense | CACCGGGCGCTGATGAAGGAGATCGTTCAAGAGACGATCTCCTTCATCAGCGCCCTTTTTTG |
| Antisense | CCCGCGACTACTTCCTCTAGCAAGTTCTCTGCTAGAGGAAGTAGTCGCGGAAAAAACCTAG | |
| NC | Sense | CACCGTTCTCCGAACGTGTCACGTCAAGAGATTACGTGACACGTTCGGAGAATTTTTTG |
| Antisense | GATCCAAAAAATTCTCCGAACGTGTCACGTAATCTCTTGACGTGACACGTTCGGAGAAC | |
Cell viability assay
Cell Counting Kit-8 (CCK-8, Beyotime Institute of Biotechnology) assay was conducted to determine the viability of GECs. Cells were plated in 96-well plates at a density of 2000 cells/well and incubated in GBM cell-conditioned medium for 24 h. Each well was incubated with 10 μL CCK-8 solution at 37 °C for 2 h. Optical density values were evaluated at 450 nm using the SpectraMax M5 microplate reader (Molecular Devices, San Jose, CA, USA).
Cell migration assay
The migration ability of GECs was assessed using a 6.5 mm Transwell with 8.0 μm Pore Polycarbonate Membrane Insert (#3422, Corning, NY, USA). The upper chamber was used to incubate cells resuspended in 200 μL serum-free medium at a density of 5 × 105 cells/mL. The lower chamber was filled with 600 μL GBM cell-conditioned medium.
After incubation at 37 °C for 48 h, non-migrated cells on the top surface of the membrane were carefully removed. Migrated cells on the lower surface of the membrane were fixed with methanol and glacial acetic acid at the ratio of 3:1 and stained with 10% Giemsa solution (Dinguo, Beijing, China). Then, images of the stained cells were taken with an inverted microscope, and the cell numbers in five randomly selected fields were counted for statistical analysis in each well.
Tube formation assay
Matrigel assay was performed to evaluate the tube formation of GECs. Pre-chilled 96-well plates were coated with 100 μL Matrigel (BD Biosciences, Bedford, MA, USA) per well and incubated to polymerize at 37 °C for 30 min. ECs were resuspended in 100 μL GBM cell-conditioned medium and seeded onto the surface of the polymerized Matrigel at a density of 4 × 105 cells/mL, followed by incubation at 37 °C for 6 h. Olympus DP71 microscopy (Olympus, Tokyo, Japan) was used to acquire three or more images at random from each culture, and ImageJ software was used to measure the total tubule length and the number of branches.
Reporter vector construction and luciferase reporter assays
The putative binding sequences and mutant sequences of miR-138-5p in circ_002136 and SOX13 3′-UTR were amplified by PCR and cloned downstream of the pmirGLO Dual-luciferase miRNA Target Expression Vector (Promega, Madison, WI, USA) to construct a dual-luciferase reporter vector (circ_002136Wt/circ_002136Mut and SOX13–3’-UTR-Wt/SOX13–3’-UTR-Mut, GenePharma). Circ_002136Wt/circ_002136Mut or SOX13–3’-UTR-Wt/SOX13–3’-UTR-Mut dual-luciferase vectors and miR-138-5p agomir (or agomir NC) plasmids were co-transfected into HEK293T cells using Lipofectamine 3000. The dual-luciferase activity was measured 48 h after transfection. The Dual-Luciferase Reporter Assay System (Promega) was used to analyse luciferase activity. Relative luciferase activity was expressed as the ratio of firefly luciferase activity to Renilla luciferase activity.
For the SOX13-SPON2 reporter constructs, the SPON2 promoter region (− 1, 000 to + 200 bp) was amplified from human genomic DNA by PCR and then subcloned into pGL3-Basic-Luciferase vector (Promega) containing a firefly luciferase reporter gene, yielding the wide-type plasmid (SPON2-Wt). In addition, to test the binding specificity, corresponding mutants of putative SOX13 binding sites were created to form the reporter vector SPON2-mutated-types (SPON2-Mut1, SPON2-Mut2, SPON2-Mut3 and SPON2-Mut4) (GenePharma). The human full-length SOX13 gene was constructed into pEX3 (pGCMV/MCS/Neo) plasmid vector (GenePharma). HEK293T cells were co-transfected with pGL3 vector (either with wide-type promoter regions or mutated promoter regions) and pEX3-SOX13 (or pEX3 empty vector) using Lipofectamine 3000. The promoter activity of constructed plasmid was normalized with the co-transfected reference vector (pRL-TK) and expressed as relative to the activity of pEX3 empty vector, which the activity set to 1.
RNA pull-down assay
The interaction between RBP FUS and circ_002136 introns was detected using Pierce Magnetic RNA-Protein Pull-Down Kit (ThermoFisher) according to the manufacturer’s protocols. In brief, circ_002136 transcripts were transcribed using T7 RNA polymerase (Ambion Life, Taoyuan, Taiwan). Biotin RNA Labeling Mix (Ambion Life) was used to biotin-label the purified RNAs, and then the positive control (Input), negative control (Antisense RNA), and biotinylated RNAs were mixed and co-incubated with proteins extracted from ECs at room temperature. Magnetic beads were added to prepare a probe–magnetic bead complex. Then the bead complex was washed with Handee spin columns and boiled in SDS buffer. Finally, retrieved proteins were detected by western blot, including GAPDH as a control.
RNA-binding protein immunoprecipitation assay
GECs lysates from different groups were incubated with RIP buffer containing magnetic beads conjugated with anti-human argonaute 2 (Ago2) antibodies (Millipore, Billerica, MA, USA). Normal mouse IgG (Millipore) was used as NCs. The samples were incubated with Proteinase K, and then immunoprecipitated RNAs were isolated. The RNA concentration was measured using a spectrophotometer (NanoDrop, ThermoScientific), and the RNA quality was assessed using a bioanalyzer (Agilent, Santa Clara, CA, USA). Purified RNAs were extracted and applied in qPCR for reverse transcription analysis.
ChIP assay
ChIP assays were performed using the Simple ChIP Enzymatic Chromatin IP Kit (Cell Signaling Technology, Danvers, MA, USA) according to the manufacturer’s protocol. Briefly, GECs were crosslinked with EBM-2 containing 1% formaldehyde and collected in a lysis buffer containing 1% PMSF. Chromatin was digested by micrococcal nuclease, and 2% aliquots of lysate were used as an input control. Lysates were incubated with 3 μg anti- SOX13 antibody (Proteintech) or normal rabbit IgG, followed by immunoprecipitation with protein G agarose beads and incubation at 4 °C overnight with gentle shaking. DNA crosslink was reversed by the addition of 5 mol/L NaCl and Proteinase K at 65 °C for 2 h, and finally, DNA was purified. Immunoprecipitated DNA was amplified by PCR using specific primers. Primers used for ChIP PCR are shown in Table 3.
Table 3.
Primers used for ChIP experiments
| Gene | Binding site or Control | Sequence (5′- > 3′) | Product size (bp) | Annealing temperature (°C) |
|---|---|---|---|---|
| SPON2 | PCR1 | F:TTTACCGAGTGCTAGAGCCG | 165 | 59.6 |
| R: AGGCTGCTGTGGCTGTTT | ||||
| PCR2 | F: CTTACGACGCAGGGTCTGG | 237 | 59.8 | |
| R:CGGCTCTAGCACTCGGTAAA | ||||
| PCR3 | F:ACCCAAGAAAATCAGCCAAAGC | 211 | 60.2 | |
| R:TCACTGTGGAATCGCGTGAG | ||||
| PCR4 | F:GTGCCCAGCATCTATTCTGGT | 207 | 59.9 | |
| R: CTACAGCGTCCCACAGACC | ||||
| FUS | PCR1 | F:AGTGTTTTGCAGTTACAAGACCTG | 97 | 59.8 |
| R:GGAAAGTGAGACTCAGAGACCC | ||||
| PCR2 | F: CGTCTTGGCTCACTGCAACT | 167 | 60.5 | |
| R:GTCAGGAGTTCGAGACCAGC |
In vivo Matrigel plug assay
Four-week-old male BALB/C athymic nude mice were purchased from the Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). The animals were fed with autoclaved food and water during the experiment. All animal procedures were performed in strict accordance with the protocol approved by the Administrative Panel on Laboratory Animal Care of China Medical University (Shenyang, China). In brief, 3 × 106 GECs resuspended in 400 μL solution containing 80% Matrigel were subcutaneously injected. Plugs were harvested after 4 days and then weighed, photographed, and dispersed in 400 μL PBS (with overnight incubation at 4 °C) to collect the hemoglobin. Hemoglobin content was measured using Drabkin’s reagent solution (Sigma-Aldrich) according to the manufacturer’s instructions.
Statistical analysis
Quantitative data were presented as mean ± standard deviation (SD). GraphPad Prism v5.01 (GraphPad, La Jolla, CA, USA) software was used for statistical analysis. Student’s t-test (two-tailed) or one-way ANOVA, followed by Bonferroni’s post-hoc test, was employed to evaluate all statistical analyses. Differences were considered statistically significant when P < 0.05.
Results
FUS is up-regulated in GECs; FUS silencing suppresses angiogenesis of glioma; FUS binds to and promotes the production of circ_002136 to regulate glioma angiogenesis
This study first examined the expression of FUS in ECs and GECs. The results showed that the mRNA and protein expression levels of FUS in GECs were significantly higher than those in ECs (Fig. 1A, B). The shRNA of FUS was then transfected into GECs to silence FUS, and the transfection efficiency was evaluated by western blot (Additional file 1: Figure S1A). The cell viability of GECs was detected by the CCK8 assay. The results showed that, compared to the FUS(˗)NC group, cell viability in the FUS(−) group was significantly reduced (Fig. 1C). The GEC cell migration assay showed that compared to the FUS(−)NC group, the migration capability of the FUS(−) group was significantly lower (Fig. 1D). The results of the Matrigel three-dimensional angiogenesis assay showed that compared to the FUS(−)NC group, relative tubule length and relative numbers of branches in the FUS(−) group were significantly lower (Fig. 1E).
Fig. 1.
Knockdown of FUS suppressed GEC angiogenesis in U87 glioma. (a-b) The mRNA and protein expression levels of FUS in ECs and GECs were evaluated by qRT-PCR and western blot. GAPDH was used as an endogenous control. IDVs represents integrated density values. Data are presented as the means ± SD (n = 5, each group). **P < 0.01 vs. ECs group. (c) The effects of FUS knockdown on GEC viability were detected by the CCK-8 assay. Data are presented as the means ± SD (n = 5, each group). **P < 0.01 vs. FUS (−) NC group. (d) The effects of FUS knockdown on GECs were detected by the transwell assay. Data are presented as the means ± SD (n = 5, each group). **P < 0.01 vs. FUS (−) NC group. The scale bar represents 100 μm. (e) The effects of FUS knockdown on GECs tube formation were measured by the Matrigel tube formation assay (black arrow, tube structures; grey arrow, tube branches). Data are presented as the means ± SD (n = 5, each group). **P < 0.01 vs. FUS (−) NC group. The scale bar represents 100 μm. (f) The relative enrichment of circ_002136 in anti-IgG and anti-FUS was detected by an RNA-IP assay. Data are presented as the means ± SD (n = 3, each group). **P < 0.01 vs. anti-IgG group. (g) FUS and GAPDH protein levels immunoprecipitation with circ_002136 were evaluated by western blot. The expression levels of FUS and GAPDH proteins are shown. (h) The effects of FUS knockdown on circ_002136 expression levels were detected by qRT-PCR. Data are presented as the means ± SD (n = 5, each group). **P < 0.01 vs. FUS (−) NC group. (i) The effects of FUS knockdown on CDK11A expression levels were detected by qRT-PCR. Data are presented as the means ± SD (n = 5, each group)
RNA-IP assay was performed to investigate whether binding occurred between FUS and circ_002136. The relative enrichment of circ_002136 in the FUS co-precipitation group was significantly increased compared to that in the IgG immunoprecipitation group (Fig. 1F). The results of RNA pull-down assays demonstrated that FUS combined with circ_002136 and promoted the generation of circ_002136 (Fig. 1G). The expression of circ_002136 in GECs with stable knockdown of FUS was then further examined. The results showed that compared to the FUS(−)NC group, the expression of circ_002136 in the FUS(−) group was significantly reduced (Fig. 1H). The expression of linear CDK11A, the circ_002136 parental gene, was also detected in GECs with stable knockdown of FUS. The results showed no significant difference in the expression of CDK11A in the FUS(−) group compared to the FUS(−)NC group (Fig. 1I).
Furthermore, circ_002136 was silenced in GECs stably transfected with the FUS silencing vector. Rescue experiments showed that the expression of miR-138-5p was significantly increased both in the FUS(−) + circ_002136(−)NC and the FUS(−)NC + circ_002136(−) groups compared to the FUS(−)NC + circ_002136(−)NC group. Compared to the FUS(−) + circ_002136(−)NC group, the expression of miR-138-5p was further significantly increased in the FUS(−) + circ_002136(−) group (Additional file 2: Figure S2A). However, the mRNA and protein expression levels of SOX13 and SPON2 were significantly decreased both in the FUS(−) + circ_002136(−)NC and FUS(−)NC + circ_002136(−) groups compared to the FUS(−)NC + circ_002136(−)NC group. In comparison with the FUS (−) + circ_002136(−)NC group, the mRNA and protein expression levels of SOX13 and SPON2 were further significantly decreased in the FUS(−) + circ_002136(−) group (Additional file 2: Figure S2B-D). In addition, the effects of dual silencing of FUS and circ_002136 on viability, migration and tube formation of GECs were consistent with the changes in SOX13 and SPON2 expression (Additional file 2: Figure S2E-G).
Circ_002136, but not linear CDK11A, is up-regulated in GECs, while silencing of circ_002136 inhibits glioma angiogenesis
The expression of circ_002136 in ECs and GECs was examined. The results showed significantly up-regulated expression of circ_002136 in the GECs group compared to the ECs group (Fig. 2A). However, the difference in expression of CDK11A was not statistically significant between the ECs and GECs groups (Fig. 2B).
Fig. 2.
Knockdown of circ_002136 inhibited GEC angiogenesis in U87 glioma. (a) The relative expression of circ_002136 was detected in ECs and GECs by qRT-PCR. Data are presented as the means ± SD (n = 5, each group). **P < 0.01 vs. ECs group. (b) The relative expression of CDK11A was detected in ECs and GECs by qRT-PCR. Data are presented as the means ± SD (n = 5, each group). (c) The effects of circ_002136 knockdown on the viability of GECs were detected by the CCK-8 assay. Values represent the means ± SD (n = 5, each group). **P < 0.01 vs. circ_002136(−)NC group. (d) The effects of circ_002136 knockdown on the migration of GECs were determined by the transwell assay. Values represent the means ± SD (n = 5, each group). **P < 0.01 vs. circ_002136(−)NC group. The scale bar represents 100 μm. (E) The effects of circ_002136 knockdown on the tube formation of GECs were evaluated by the Matrigel tube formation assay. Values represent the means ± SD (n = 5, each group). **P < 0.01 vs. circ_002136(−)NC group. The scale represents 100 μm
RNase R is an RNA exonuclease that degrades linear RNAs with short 3′ tails regardless of their secondary structure, but it does not degrade circular forms. In this study, RNase R was used to confirm the circular form of RNAs. Circ_002136 was resistant to RNase R treatment, whereas linear CDK11A was significantly reduced in ECs and GECs treated with RNase R (Figs. S1B, C). Silencing of circ_002136 was performed to further investigate the potential effect of circ_002136 in GECs and transfection efficiency was evaluated by qRT-PCR (Fig. S1D). In addition, the expression of CDK11A was detected after silencing of circ_002136 to confirm that the circular form, rather than the linear form, of CDK11A was inhibited. As shown in Fig. S1E, there was no significant difference in CDK11A expression between the circ_002136(−) and circ_002136(−)NC groups. Next, CDK11A was silenced to detect whether sh-CDK11A influenced circ_002136 expression. The transfection efficiency of CDK11A was verified (Additional file 1: Figure S1F) and there was no significant change in circ_002136 expression between the sh-CDK11A and sh-NC groups (Additional file 1: Figure S1G). Furthermore, cell viability, migration and tube formation abilities were significantly reduced in the circ_002136(−) group compared to the circ_002136(−)NC group (Fig. 2C–E).
MiR-138-5p is down-regulated in GECs and over-expression of miR-138-5p inhibits gliomas angiogenesis
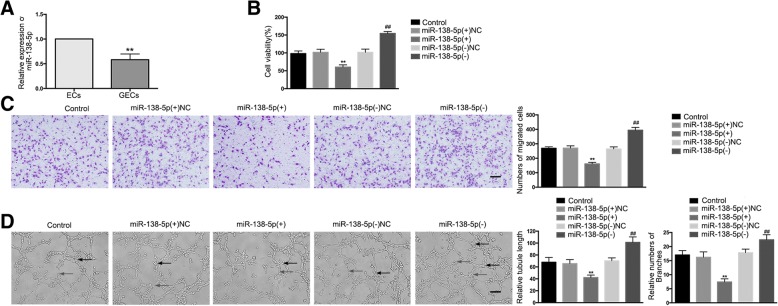
The endogenous expression of miR-138-5p in ECs and GECs was detected by qRT-PCR. The expression of miR-138-5p was significantly down-regulated in the GECs group compared to the ECs group (Fig. 3A). In this study, over-expression or silencing of miR-138-5p was performed to further understand its role in the angiogenesis of GECs, with the transfection efficiency shown in Additional file 1: Figure S1H. The results demonstrated that, compared to the miR-138-5p(+)NC group, cell viability, migration, and tube formation abilities were significantly decreased in the miR-138-5p(+) group, whereas compared to the miR-138-5p(−)NC group, these abilities were significantly increased in the miR-138-5p(−) group (Fig. 3B–D).
Fig. 3.
Over-expression of miR-138-5p suppressed GEC angiogenesis in U87 glioma. (a) The relative expression of miR-138-5p in ECs and GECs was detected by qRT-PCR. U6 was used as an internal control. Data are presented as the means ± SD (n = 5, each group). **P < 0.01 vs. ECs group. (b) The effects of miR-138-5p on the viability of GECs were determined by the CCK-8 assay. Values represent the means ± SD (n = 5, each group). **P < 0.01 vs. miR-138-5p(+)NC group; ##P < 0.01 vs. miR-138-5p(−)NC group. (c) The effects of miR-138-5p on the migration of GECs were determined by the transwell assay. Values represent the means ± SD (n = 5, each group). **P < 0.01 vs. miR-138-5p(+)NC group; ##P < 0.01 vs. miR-138-5p(−)NC group. The scale bar represents 100 μm. (d) The effects of miR-138-5p on tube formation of GECs were evaluated by the Matrigel tube formation assay. Values represent the means ± SD (n = 5, each group). **P < 0.01 vs. miR-138-5p(+)NC group; ##P < 0.01 vs. miR-138-5p(−)NC group. The scale bar represents 100 μm
MiR-138-5p targets circ_002136 but not CDK11A, and reverses circ_002136-mediated gliomas angiogenesis
The sequence of circ_002136 (chr1: 1586822–1,650,894) was characterized by alignment with the CDK11A genomic DNA sequence (Fig. 4A). The expression of miR-138-5p was significantly enhanced in the circ_002136(−) group compared to the circ_002136(−)NC group (Fig. 4B). Similarly, it was found that over-expression or silencing of miR-138-5p resulted in significantly decreased or increased expression of circ_002136 in GECs (Fig. 4C). However, over-expression or silencing of miR-138-5p did not affect the expression of linear CDK11A (Additional file 1: Figure S1I).
Fig. 4.
MiR-138-5p functionally targeted circ_002136. (a) A schematic representation of how circ_002136 arose from the CDK11A gene as determined by scanning CDK11A genomic DNA and circBase. (b) The expression of miR-138-5p was measured after knockdown of circ_002136 by qRT-PCR. Values represent the means ± SD (n = 5, each group). **P < 0.01 vs. circ_002136(−)NC group. (c) MiR-138-5p regulated the expression of circ_002136 in GECs. Values represent the means ± SD (n = 5, each group). **P < 0.01 vs. miR-138-5p(+)NC group; ##P < 0.01 vs. miR-138-5p(−)NC group. (d) The putative binding site between circ_002136 and miR-138-5p was predicted, and the relative luciferase activity was expressed as firefly/renilla luciferase activity. Values represent the means ± SD (n = 5, each group). **P < 0.01 vs. circ_002136 Wt + miR-138-5p(+)NC group. (e-f) MiR-138-5p was identified in the circ_002136-RISC complex. The relative expression of circ_002136 and miR-138-5p was measured using qRT-PCR. Data are presented as the means ± SD (n = 5, each group). **P < 0.01 vs. anti-IgG group, ##P < 0.01 vs. anti-Ago2 in control group. (g) The co-effects of circ_002136 and miR-138-5p on the viability of GECs were evaluated by the CCK-8 assay. Data are presented as the means ± SD (n = 5, each group). **P < 0.01 vs. circ_002136(−)NC + miR-138-5p(+)NC group. (h) The co-effects of circ_002136 and miR-138-5p on the migration of GECs were evaluated by the transwell assay. Data are presented as the means ± SD (n = 5, each group). **P < 0.01 vs. circ_002136(−)NC + miR-138-5p(+)NC group. The scale bar represents 100 μm. (I) The co-effects of circ_002136 and miR-138-5p on the tube formation of GECs were evaluated by the Matrigel tube formation assay. Data are presented as the means ± SD (n = 5, each group). **P < 0.01 vs. circ_002136(−)NC + miR-138-5p(+)NC group. The scale bar represents 100 μm
Using the bioinformatics software Starbase v2.0 (http://starbase.sysu.edu.cn/), it was predicted that circ_002136 might possess a binding site for miR-138-5p. This prediction was verified by a dual-luciferase reporter assay system. The results showed that compared to the circ_002136Wt + miR-138-5p(+)NC group, the relative luciferase activity of the circ_002136Wt + miR-138-5p(+) group was significantly suppressed. Nevertheless, there were no significant differences in relative luciferase activity between the circ_002136Mut + miR-138-5p(+) and circ_002136Mut + miR-138-5p(+)NC groups (Fig. 4D). Furthermore, RNA-IP results showed that compared to the anti-IgG group, the relative abundance of circ_002136 and miR-138-5p in the anti-Ago2 group significantly increased, whereas in the miR-138-5p (−) group, the relative abundance of circ_002136 and miR-138-5p immunoprecipitated with Ago2 was lower than that in the control group. This suggests that circ_002136 and miR-138-5p were co-enriched in the expected RISC (Fig. 4E, F).
To further investigate whether circ_002136 regulates glioma angiogenesis by modulating miR-138-5p, GECs with stable knockdown of circ_002136 were transiently transfected with miR-138-5p agomir or antagomir. Rescue experiments showed that cell viability, migration, and tube formation abilities were significantly reduced in the circ_002136(−) + miR-138-5p(+) group compared to the circ_002136(−)NC + miR-138-5p(+)NC group, whereas there was no significant difference between the circ_002136(−) + miR-138-5p(−) and circ_002136(−)NC + miR-138-5p(−)NC groups (Fig. 4G–I).
SOX13 is a target gene of miR-138-5p and is involved in circ_002136 and miR-138-5p-mediated gliomas angiogenesis
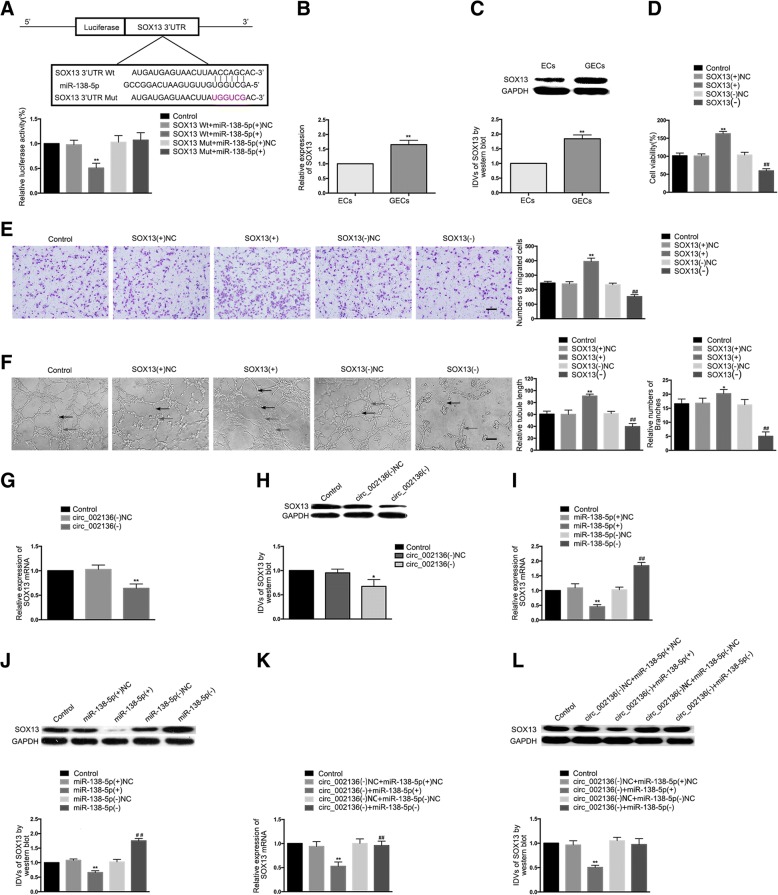
Using the bioinformatics software microRNA.org-target program (http://www.microrna.org/microrna/home.do), it was predicted that a putative binding site for miR-138-5p exists in the 3’-UTR of SOX13 mRNA, which was a complementary sequence of the miR-138-5p seed region. The results of the dual-luciferase reporter assay showed that compared to the SOX13–3’-UTR-Wt + miR-138-5p(+)NC group, the relative luciferase activity in the SOX13–3’-UTR-Wt + miR-138-5p(+) group was significantly reduced, while compared to the SOX13–3’-UTR-Mut + miR-138-5p(+)NC group, the relative luciferase activity in the SOX13–3’-UTR-Mut + miR-138-5p(+) group showed no significant change (Fig. 5A).
Fig. 5.
SOX13, a target of miR-138-5p, regulated the GEC angiogenesis in U87 glioma. (a) A putative binding site between SOX13 and miR-138-5p was predicted and the relative luciferase activity was expressed as the firefly/renilla luciferase activity. Values represent the means ± SD (n = 5, each group). **P < 0.01 vs. SOX13 Wt + miR-138-5p(+)NC group. (b-c) The mRNA and protein expression levels of SOX13 were measured in ECs and GECs by qRT-PCR and western blot. Values represent the means ± SD (n = 5, each group). **P < 0.01 vs. ECs group. (d) SOX13 regulated the viability of GECs. Values represent the means ± SD (n = 5, each group). **P < 0.01 vs. SOX13(+)NC group; ##P < 0.01 vs. SOX13(−)NC group. (e) SOX13 regulated the migration of GECs. Values represent the means ± SD (n = 5, each group). **P < 0.01 vs. SOX13(+)NC group; ##P < 0.01 vs. SOX13(−)NC group. The scale bar represents 100 μm. (f) SOX13 regulated the tube formation of GECs. Values represent the means ± SD (n = 5, each group). *P < 0.05, **P < 0.01 vs. SOX13(+)NC group; ##P < 0.01 vs. SOX13(−)NC group. The scale bar represents 100 μm. (g-h) The mRNA and protein expression levels of SOX13 were inhibited after knockdown of circ_002136. Values represent the means ± SD (n = 3, each group). *P < 0.05, **P < 0.01 vs. circ_002136(−)NC group. (i-j) The mRNA and protein expression levels of SOX13 were regulated by miR-138-5p. Values represent the means ± SD (n = 3, each group). **P < 0.01 vs. miR-138-5p(+)NC group. ##P < 0.01 vs. miR-138-5p(−)NC group. (K-L) The mRNA and protein expression levels of SOX13 were co-regulated by both circ_002136 and miR-138-5p. Values represent the means ± SD (n = 3, each group). **P < 0.01 vs. circ_002136(−)NC + miR-138-5p(+)NC group
The mRNA and protein expression levels of SOX13 in the GECs group were significantly up-regulated compared to the ECs group (Fig. 5B, C). In this study, both over-expression and silencing of SOX13 were performed in GECs to further explore the effect of SOX13 on the angiogenesis of GECs. The stable transfection efficiency of SOX13 was also verified (Additional file 1: Figure S1J). As shown in Fig. 5D-F, compared to the SOX13(+)NC group, the cell viability, migration, and tube formation abilities of the SOX13(+) group were significantly improved, while compared to the SOX13(−)NC group, these abilities in the SOX13(−) group were significantly reduced.
This study further investigated whether SOX13 is involved in the regulatory network of circ_002136 and miR-138-5p in glioma angiogenesis. The results showed that the mRNA and protein expression levels of SOX13 were significantly reduced in the circ_002136(−) group compared to the circ_002136(−)NC group (Fig. 5G, H). Compared to the miR-138-5p(+)NC group, however, the mRNA and protein expression levels of SOX13 in the miR-138-5p(+) group were significantly reduced. Compared to the miR-138-5p(−)NC group, the mRNA and protein expression levels of SOX13 in the miR-138-5p(−) group were significantly increased (Fig. 5I, J). Furthermore, the rescue experiments showed that the mRNA and protein expression levels of SOX13 were significantly reduced in the circ_002136(−) + miR-138-5p(+) group compared to the circ_002136(−)NC + miR-138-5p(+)NC group, but showed no significant differences in the circ_002136(−) + miR-138-5p(−) group compared to the circ_002136(−)NC + miR-138-5p(−)NC group (Fig. 5K, L).
SOX13 binds to the SPON2 promoter region to promote expression of SPON2
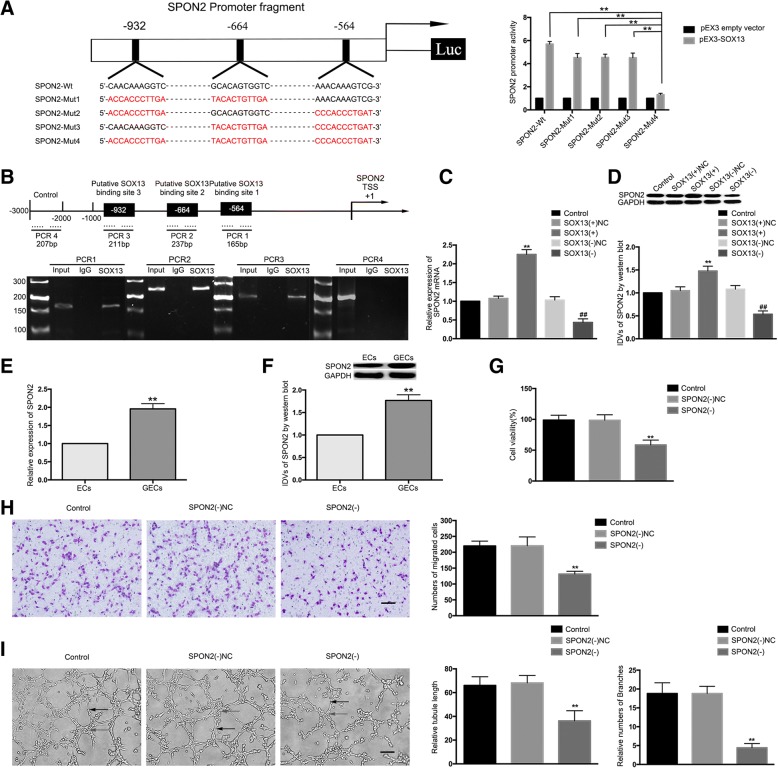
A search of the bioinformatics software JASPAR (http://jaspar.genereg.net) database predicted three putative SOX13-binding sites at loci 564, 664, and 932 in the 1000-bp region upstream and 200-bp region downstream of the SPON2 transcription start site (TSS). To determine the regions responsible for the SOX13 activated promoter of SPON2 in GECs, a series of mutation constructs were generated and luciferase reporter assays were performed. Wild-type and mutation constructs are indicated in Fig. 6A. As shown in Fig. 6A, SPON2 promoter activities were up-regulated after co-transfecting with pEX3-SOX13. Mutation of all the putative SOX13 binding sites induced a significant change in SPON2 promoter activity, but mutation of any two did not. These results established that SOX13 stimulated the promoter activity of SPON2 and indicated that the SPON2 responsive elements, which were necessary for high promoter activity, were likely to reside within the 564, 664, and 932 site regions. To further determine whether SOX13 was directly associated with the SPON2 promoter, ChIP assays were performed. ChIP assays revealed that SOX13 binds to the SPON2 promoter region, with no relationship observed in the control region (Fig. 6B). Subsequently, the mRNA and protein expression levels of SPON2 were examined in the GECs with stable over-expression or silencing of SOX13. Compared to the SOX13(+)NC group, the mRNA and protein expression levels of SPON2 in the SOX13(+) group were significantly increased, while the mRNA and protein expression levels of SPON2 in the SOX13(−) group were significantly decreased compared to those in the SOX13(−)NC group (Fig. 6C, D).
Fig. 6.
SPON2 was a target of SOX13; SPON2 knockdown suppressed the GEC angiogenesis in U87 glioma. (a) A schematic depiction of the predicted binding sites for SOX13 on the promoter fragment of SPON2 and the site mutagenesis design for the reporter assay. Promoter activities were measured to determine responsive SOX13-binding sites in the promoter of SPON2 using luciferase reporter assays. Data are presented as the means ± SD (n = 3, each group). **P < 0.01 vs. SPON2-Mut4 group. (b) A schematic representation of the SPON2 promoter region 3000 bp upstream or 200 bp downstream of the TSS, designated as + 1. ChIP PCR products for the putative SOX13 binding sites and an upstream region not expected to associate with SOX13 are depicted with bold lines. (c-d) The qRT-PCR and western blot analysis of the SOX13 regulation of SPON2 mRNA and protein expression levels. Values represent the means ± SD (n = 3, each group). **P < 0.01 vs. SOX13(+)NC group; ##P < 0.01 vs. SOX13(−)NC group. (e-f) The mRNA and protein expression levels of SPON2 in ECs and GECs were evaluated by qRT-PCR and western blot. Data are presented as the means ± SD (n = 5, each group). **P < 0.01 vs. ECs group. (g) The effects of SPON2 knockdown on the viability of GECs were detected by the CCK-8 assay. Values represent the means ± SD (n = 5, each group). **P < 0.01 vs. SPON2(−)NC group. (h) The effects of SPON2 knockdown on the migration of GECs were determined by the transwell assay. Values represent the means ± SD (n = 5, each group). **P < 0.01 vs. SPON2(−)NC group. The scale bar represents 100 μm. (i) The effects of SPON2 knockdown on the tube formation of GECs were evaluated by the Matrigel tube formation assay. Values represent the means ± SD (n = 5, each group). **P < 0.01 vs. SPON2(−)NC group. The scale bar represents 100 μm
In this study, the expression of SPON2 in ECs and GECs was further examined. The results showed that the mRNA and protein expression levels of SPON2 in the GECs group were significantly up-regulated compared to the ECs group (Fig. 6E, F). SPON2 was then silenced to further investigate its function in GECs. The stable transfection efficiency of SPON2 is shown in Additional file 1: Figure S1K. Cell viability, migration, and tube formation in the SPON2(−) group were significantly reduced compared to the SPON2(−)NC group (Fig. 6G–I).
MiR-138-5p suppresses SOX13-mediated angiogenesis and the expression of SPON2; SOX13 promotes FUS transcription to form a feedback loop regulating glioma angiogenesis
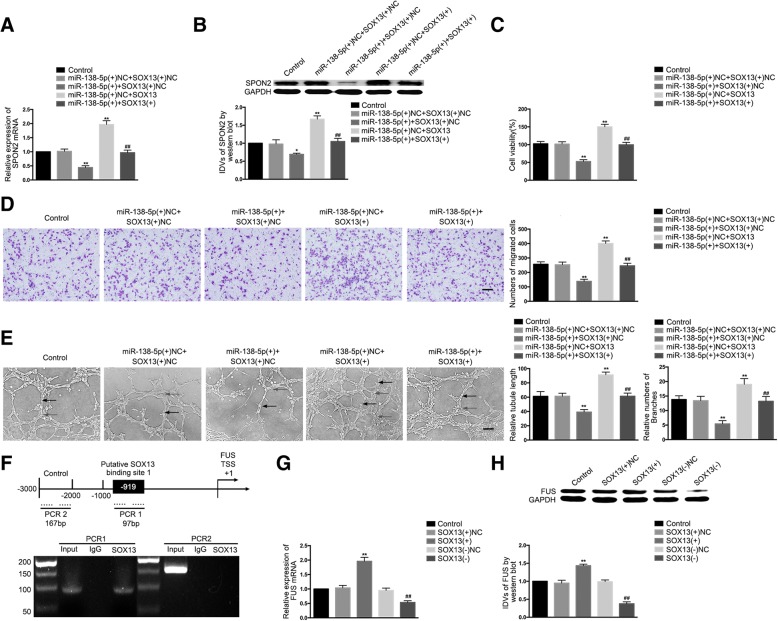
MiR-138-5p was overexpressed in GECs stably transfected with the SOX13 over-expression vector, and the effect of double over-expression of miR-138-5p and SOX13 on mRNA and protein expression of SPON2 was examined. Compared to the miR-138-5p(+)NC + SOX13(+)NC group, the mRNA and protein expression levels of SPON2 were significantly decreased in the miR-138-5p(+) + SOX13(+)NC group and significantly increased in the miR-138-5p(+)NC + SOX13(+) group, whereas SPON2 expression in the miR-138-5p(+) + SOX13(+) group was significantly increased compared to the miR-138-5p(+) + SOX13(+)NC group (Fig. 7A, B). In addition, the changes in viability, migration, and tube formation were consistent with the changes in SPON2 expression (Fig. 7C–E).
Fig. 7.
Co-effects of miR-138-5p and SOX13; SOX13 promoted transcription of FUS forming a feedback loop. (a-b) The co-effects of miR-138-5p and SOX13 on the expression levels of SPON2 were evaluated by qRT-PCR and western blot. Data are presented as the means ± SD (n = 3, each group). *P < 0.05, **P < 0.01 vs. miR-138-5p(+)NC + SOX13(+)NC group. ##P < 0.01 vs. miR-138-5p(+) + SOX13(+)NC group. (c) The co-effects of miR-138-5p and SOX13 on the viability of GECs were evaluated by the CCK8 assay. Data are presented as the means ± SD (n = 5, each group). **P < 0.01 vs. miR-138-5p(+)NC + SOX13(+)NC group; ##P < 0.01 vs. miR-138-5p(+) + SOX13(+)NC group. (d) The co-effects of miR-138-5p and SOX13 on the migration of GECs were evaluated by the transwell assay. Data are presented as the means ± SD (n = 5, each group). **P < 0.01 vs. miR-138-5p(+)NC + SOX13(+)NC group; ##P < 0.01 vs. miR-138-5p(+) + SOX13(+)NC group. The scale bar represents 100 μm. (e) The co-effects of miR-138-5p and SOX13 on the tube formation of GECs were evaluated by the Matrigel tube formation assay. Data are presented as the means ± SD (n = 5, each group). *P < 0.05, **P < 0.01 vs. miR-138-5p(+)NC + SOX13(+)NC group; ##P < 0.01 vs. miR-138-5p(+) + SOX13(+)NC group. The scale bar represents 100 μm. (f) Schematic representation of the human FUS promoter region 3000 bp upstream or 200 bp downstream of the TSS, designated as + 1. ChIP PCR products for the putative SOX13 binding site and an upstream region not expected to associate with SOX13 are depicted with bold lines. (g-h) The qRT-PCR and western blot analysis of SOX13 regulation of the expression levels of FUS. Values are presented as the means ± SD (n = 3, each group). **P < 0.01 vs. SOX13(+)NC group; ##P < 0.01 vs. SOX13(−)NC group
One putative SOX13 binding site was predicted at loci 919 in the 1000-bp region upstream and 200-bp region downstream of the FUS TSS using the JASPAR database. ChIP assays revealed that SOX13 and FUS bound at this putative binding site, but no relationship was observed in the control region (Fig. 7F). Similarly, the mRNA and protein expression levels of FUS were examined in GECs with stable over-expression or silencing of SOX13. Compared to the SOX13(+)NC group, the mRNA and protein expression levels of FUS in the SOX13(+) group were significantly increased, whereas compared to the SOX13(−)NC group, the mRNA and protein expression levels of FUS in the SOX13(−) group were significantly decreased (Fig. 7G, H).
FUS or circ_002136 knockdown and miR-138-5p over-expression significantly suppress glioma angiogenesis in vivo both individually and combined
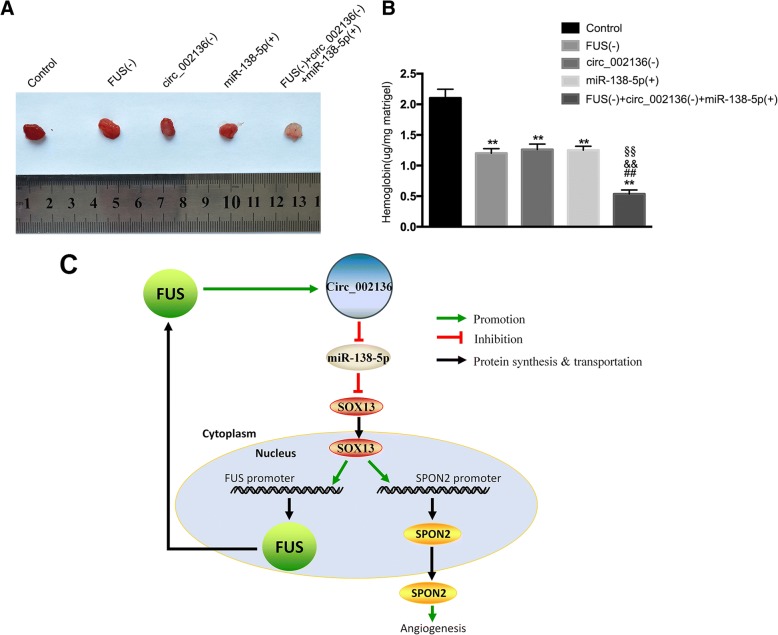
A matrigel plug assay was applied to further verify the effects of FUS or circ_002136 knockdown and miR-138-5p over-expression in vivo. Co-transfection was conducted prior to the assessment of angiogenesis in vivo. The results showed that the amount of hemoglobin in the FUS(−), circ_002136(−), and miR-138-5p(+) groups was significantly decreased, compared to the control group. The amount of hemoglobin was lowest in the FUS(−) + circ_002136(−) + miR-138-5p(+) group (Fig. 8A, B). These findings suggested that silencing of FUS and circ_002136, combined with miR-138-5p over-expression, suppressed glioma angiogenesis in vivo. A schematic representation of the mechanism by which the FUS/circ_002136/miR-138-5p/SOX13/SPON2 axis affects the angiogenesis of GECs is presented in Fig. 8C.
Fig. 8.
FUS and circ_002136 knockdown combined with miR-138-5p over-expression suppressed angiogenesis in vivo (a) The co-effect of FUS, circ_002136, and miR-138-5p on angiogenesis in vivo was evaluated by the Matrigel plug assay. (a) The amount of hemoglobin was measured after the combined treatment of FUS, circ_002136, and miR-138-5p. Data are presented as the means ± SD (n = 3, each group). **P < 0.01 vs. control group; ##P < 0.01 vs. the FUS (−) group; &&P < 0.01 vs. the circ_002136 (−) group. §§P < 0.01 vs. the miR-138-5p (+) group. (c) A schematic representation of the FUS/circ_002136/miR-138-5p/SOX13/SPON2 axis involvement in the mechanism mediating the GEC angiogenesis
Discussion
This study demonstrated for the first time that FUS, circ_002136, SOX13 and SPON2 were highly expressed, while miR-138-5p was under-expressed in GECs. FUS bound to circ_002136, while circ_002136 acted as a molecular sponge for miR-138-5p, which had a negative regulatory effect on SOX13 and regulated glioma angiogenesis. SOX13 promoted the expression of SPON2 and increased the angiogenic capacity of GECs. SOX13 bound to the FUS promoter region to up-regulate the expression of FUS, forming a positive feedback loop that promoted glioma angiogenesis.
This study found that FUS was highly expressed in GECs and that FUS silencing inhibited the viability, migration, and tube formation of GECs. It has been reported that FUS plays an important role in regulating various biological processes. For example, FUS and DNA damage-inducible transcript 3 are able to form a fusion oncogene FUS-CHOP, which increases the invasive capability of human mucus-like and round cell liposarcoma cells [21]. FUS protein is also a pathological hallmark of neurodegenerative diseases such as amyotrophic lateral sclerosis and frontotemporal lobe degeneration [22, 23]. This study further found that circ_002136 was highly expressed in GECs and that circ_002136 silencing was reduced in GEC angiogenesis, suggesting that circ_002136 functioned as an oncogene in GECs.
The regulation of glioma development by circRNAs has become a major research focus. Circ_0001649 regulates glioma cell growth, colony formation, and apoptosis [24]. Circ-FBXW7 encodes FBXW7-185aa, which can shorten the half-life of c-Myc and slow the proliferation of glioma cells [25]. CircRNAs also play an important role in regulating EC function. Circ_0003575 is up-regulated in oxLDL-induced human umbilical vein endothelial cells (HUVECs) and promotes HUVEC proliferation and angiogenic capacity [26]. CircHECTD1 accelerates silica-induced transformation from pulmonic ECs to mesenchymal cells [27]. As previously reported, CDK11A is abnormally expressed in penile squamous cell carcinoma [28] and hepatocellular carcinoma [29]. In this study, we observed that the expression of linear CDK11A was not up-regulated in GECs and that the degradation of CDK11A by RNase R did not affect the expression of circ_002136, suggesting that circ_002136 and linear CDK11A are two mutually independent RNAs that might perform different functions, a finding consistent with the function of circ-SHKBP1 [30].
The present study revealed that FUS contained a binding site for circ_002136, which was proven through RNA-IP and RNA pull-down assays. It was further confirmed that FUS silencing significantly reduced the expression of circ_002136, while the expression of CDK11A was not affected. Previous studies have shown that RBPs can interact with circRNAs to perform diverse biological functions. In doxorubicin-induced heart failure, the RBP Qki5 interacts with circRNAs Ttn, Fhod3, and Strn3 to improve cardiac function [31]. In lung cancer cells, the RBP TNRC6A binds to and regulates the formation of circ_0006916 accelerating lung cancer cell growth [32].
Our findings demonstrated that miR-138-5p was down-regulated in GECs and that miR-138-5p over-expression significantly reduced the angiogenic capacity of GECs, and vice versa, suggesting that miR-138-5p acted as a tumor suppressor in GECs. These findings followed those of other research groups. MiR-138-5p inhibits proliferation and enhances radiation-induced DNA damage and autophagy in nasopharyngeal carcinoma [33]. MiR-138-5p contributed to the TNF-α-induced insulin resistance through inducing autophagy in HepG2 cells by regulating SIRT1 [34]. Our study also verified that a binding site existed between circ_002136 and miR-138-5p, indicating that circ_002136 might act as a miR-138-5p sponge to modulate its functions in GECs. Further studies manifested that knockdown of circ_002136 significantly up-regulated the expression of miR-138-5p. MiR-138-5p silencing increased the expression of circ_002136 and vice versa. These findings, as well as those from the RNA-IP assays, suggested reciprocal repression between circ_002136 and miR-138-5p, which might operate in a RISC manner. Furthermore, the co-effect of circ_002136 and miR-138-5p on the angiogenesis of GECs was verified. The results showed that miR-138-5p reversed circ_002136 knockdown-mediated inhibition of viability, migration, and tube formation of GECs. Accumulated evidence confirms that circRNAs serve as miRNA sponges by targeting miRNAs. For example, circRNA ZNF609 functions as a competitive endogenous RNA to regulate FOXP4 expression by sponging miR-138-5p in renal carcinoma [35].CircRNA MYLK binds to miR-29a and promotes epithelial-mesenchymal transition and xenograft growth, angiogenesis, and metastasis of bladder cancer [36].
This study confirmed high expression of SOX13 in GECs. SOX13-silencing attenuated the angiogenic capacity of GECs and vice versa, indicating that SOX13 might act as an oncogene in GECs. SOX13 acts as an effective regulator of embryonic development, stem cell maintenance, tissue homeostasis, and multiple cancer development [37]. Previous studies have confirmed that SOX13 is up-regulated in tumors such as renal clear cell carcinoma [38] and colorectal cancer [39]. SOX13 regulates T lymphocyte differentiation by promoting the development of γδ T cells and inhibiting the differentiation of αβ T cells [40].
One of the most common modes of miRNAs action is the inhibition of gene expression at the transcriptional and post-transcriptional levels by binding to the 3′-UTR of the target mRNA [41]. Our results revealed that SOX13 was a target gene for miR-138-5p and that miR-138-5p binds to the ACCAGC sequence in the 3’-UTR of SOX13. In GECs, circ_002136 silencing significantly reduced the expression of SOX13, while over-expression and silencing of miR-138-5p significantly reduced or increased, respectively, the expression of SOX13. Further studies revealed that the reduction in SOX13 expression mediated by circ_002136 knockdown was reversed by miR-138-5p silencing. MiR-138-5p performs many biological functions by regulating target genes. For example, in non-small cell lung cancer, miR-138-5p reduces the expression of GPR124 and suppresses Gefitinib resistance [42].
Our present study demonstrated that SPON2 performed oncogene functions in GECs as a result of SPON2 up-regulating angiogenesis. This was confirmed by the inhibition of angiogenesis in SPON2-knockdown GECs. SPON2 is an innate immune modulator of host cells that recruits inflammatory cells and regulates neuronal development. Previous studies have shown that SPON2 plays an important role in Egr-1-mediated inhibition of proliferation, migration, and tube formation of vascular ECs in colorectal cancer [43]. In addition, luciferase reporter assays and ChIP assays certified that SOX13 was directly associated with and activated the SPON2 promoter, thereby up-regulating SPON2 expression. Further findings demonstrated that over-expression of SOX13 significantly increased the expression of SPON2 and that SOX13 promoted the angiogenesis of GECs by transcriptionally up-regulating SPON2. Moreover, our data indicated that miR-138-5p down-regulated the expression of SPON2 and inhibited the viability, migration, and tube formation of GECs by negatively regulating SOX13. Based on the above results, knockdown of circ_002136 negatively regulated the expression of SOX13 by targeting miR-138-5p and further down-regulated SPON2 expression to inhibit the glioma angiogenesis.
Based on our prediction, ChIP assays were performed to confirm that SOX13 bound to the FUS promoter and promoted FUS transcription. Over-expression of SOX13 up-regulated FUS expression, whereas inhibition of SOX13 diminished FUS expression, validating our hypothesis that a positive feedback loop was formed between FUS/circ_002136/miR-138-5p/SOX13 to regulate the angiogenesis of GECs. Similar positive feedback loops have been reported in previous studies. In the TDP43/SNHG12/miR-195/SOX5 pathway of glioma cells, SOX5 promotes the transcription of SNHG12 and forms a positive feedback loop to regulate the biological behavior of glioma cells [44]. In the GAS5/miR-196a-5p/FOXO1/PID1 (MIIP) pathway of glioma stem cells, FOXO1 promotes GAS5 transcription and forms a positive feedback loop that regulates the biological behavior of glioma stem cells [45]. Regulation of the positive feedback loop and its effect on the function of glioma cells and angiogenesis is now an important research focal point in biomedical science.
Conclusions
In summary, the present study revealed that knockdown of FUS and circ_002136 inhibited the viability, migration, and tube formation of GECs. MiR-138-5p exerted angiogenesis-suppressive function by decreasing SOX13 expression. The results of this study for the first time provided insights into the multi-level molecular regulatory networks formed by FUS (RBP), circ_002136 (circRNAs), miR-138-5p (miRNAs), SOX13 (transcription factor), and SPON2 (target gene). In conclusion, our findings provide a better understanding of RBP-circRNA-miRNA- transcription factor feedback loop function in glioma angiogenesis. FUS/circ_002136/miR-138-5p/SOX13/SPON2 may aid in slowing the gliomagenesis in patients and provide an alternative strategy for glioma treatment.
Additional files
Figure S1. Correlation between circ_002136, miR-138-5p and linear CDK11A and evaluation of the transfection efficiency. (A) The transfection efficiency of FUS. Data are presented as the means ± SD (n = 3, each group). **P < 0.01 vs. FUS (−) NC group. (B) The expression of circ_002136 with RNase R treatment. Data are presented as the means ± SD (n = 5, each group). **P < 0.01 vs. control group in ECs; ##P < 0.01 vs. RNase R group in ECs. (C) The expression of CDK11A with RNase R treatment. Data are presented as the means ± SD (n = 5, each group). **P < 0.01 vs. control group in ECs; ##P < 0.01 vs. control group in ECs. (D) The transfection efficiency of circ_002136 knockdown. Data are presented as the means ± SD (n = 5, each group). **P < 0.01 vs. circ_002136 (−) NC group. (E) The expression of CDK11A after circ_002136 knockdown. Data are presented as the means ± SD (n = 5, each group). (F) The transfection efficiency of CDK11A knockdown. Data are presented as the means ± SD (n = 5, each group). **P < 0.01 vs. sh-NC group. (G) The expression of circ_002136 after CDK11A knockdown. Data are presented as the means ± SD (n = 5, each group). (H) The transfection efficiency of miR-138-5p agomir or antagomir. Data are presented as the means ± SD (n = 5, each group). **P < 0.01 vs. miR-138-5p (+) NC group, ##P < 0.01 vs. miR-138-5p (−) NC group. (I) The expression of CDK11A after miR-138-5p over-expression or silencing. Data are presented as the means ± SD (n = 5, each group). (J) The transfection efficiency of SOX13. Data are presented as the means ± SD (n = 3, each group). **P < 0.01 vs. SOX13 (+) NC group, ##P < 0.01 vs. SOX13 (−) NC group. (K) The transfection efficiency of SPON2. Data are presented as the means ± SD (n = 3, each group). **P < 0.01 vs. SPON2 (−) NC group. (TIF 471 kb)
Figure S2. Co-effects of FUS and circ_002136 on the expression of downstream molecules and angiogenesis. (A) The expression of miR-138-5p was co-regulated by both FUS and circ_002136. Values represent the means ± SD (n = 5, each group). **P < 0.01 vs. FUS(−)NC + circ_002136(−)NC group. ##P < 0.01 vs. FUS (−) + circ_002136(−)NC group. (B-D) The co-effects of FUS and circ_002136 on the mRNA and protein expression levels of SOX13 and SPON2 in GECs were evaluated by qRT-PCR and western blot. Data are presented as the means ± SD (n = 3, each group). **P < 0.01 vs. FUS(−)NC + circ_002136(−)NC group. ##P < 0.01 vs. FUS(−) + circ_002136(−)NC group. (E) The co-effects of FUS and circ_002136 on the viability of GECs were evaluated by the CCK-8 assay. Data are presented as the means ± SD (n = 5, each group). **P < 0.01 vs. FUS(−)NC + circ_002136(−)NC group. ##P < 0.01 vs. FUS(−) + circ_002136(−)NC group. (F) The co-effects of FUS and circ_002136 on the migration of GECs were evaluated by the transwell assay. Data are presented as the means ± SD (n = 5, each group). **P < 0.01 vs. FUS(−)NC + circ_002136(−)NC group. ##P < 0.01 vs. FUS(−) + circ_002136(−)NC group. The scale bar represents 100 μm. (G) The co-effects of FUS and circ_002136 on the tube formation of GECs were evaluated by the Matrigel tube formation assay. Data are presented as the means ± SD (n = 5, each group). **P < 0.01 vs. FUS(−)NC + circ_002136(−)NC group. ##P < 0.01 vs. FUS(−) + circ_002136(−)NC group. The scale bar represents 100 μm. (TIF 1639 kb)
Acknowledgements
Not applicable.
Availability of data and material
The datasets used and analysed during the current study are available from the corresponding author on reasonable request.
Funding
This work was supported by grants from the Natural Science Foundation of China (81573010, 81872503, 81672511 and 81502179), the Liaoning Science and Technology Plan Project (No. 2017225020, 2015225007), the Project of Key Laboratory of Neuro-oncology in Liaoning Province (112–2400017005), and special developmental project guided by the central government of Liaoning Province (No. 2017011553–301).
Abbreviations
- 3’-UTR
3′-untranslated region
- CCK-8
Cell counting kit-8
- CDK11A
Cyclin-dependent kinase 11A
- ChIP
Chromatin immunoprecipitation
- circRNA
Circular RNA
- EC
Endothelial cell
- FUS
Fused in sarcoma
- GBM
Glioblastoma
- GEC
Glioma-exposed endothelial cells
- HEK293T
Human embryonic kidney 293 T
- miRNA
MicroRNA
- NC
Negative control
- qRT-PCR
Quantitative real-time PCR
- RBP
RNA-binding protein
- RISC
RNA-induced silencing complex
- RNA-IP
RNA-binding protein immunoprecipitation
- TSS
Transcription start site
Authors’ contributions
Study concept and design: ZH, YX, and YL. Acquisition of data: ZH, XR, XL, JZ, and YL. Analysis and interpretation of data: ZH, LL, and JM. Drafting of the manuscript: ZH, LS, and DW. Critical revision of the manuscript for important intellectual content: YX and YL. Final approval of the version to be published: YX. Administrative, technical, and material support: YX, YL, SS, and CY. All contributors agree to be accountable for all aspects of the work.
Ethics approval and consent to participate
The animal study was approved by the Administrative Panel on Laboratory Animal Care of China Medical University. IACUC Issue No.2018045.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zhenwei He, Email: donald19820315@hotmail.com.
Xuelei Ruan, Email: 18328583948@163.com.
Xiaobai Liu, Email: 12353258@qq.com.
Jian Zheng, Email: illusion25@163.com.
Yunhui Liu, Email: liuyh_cmuns@163.com.
Libo Liu, Email: 672270151@qq.com.
Jun Ma, Email: 525762390@qq.com.
Lianqi Shao, Email: lianqi_shao1990@sina.com.
Di Wang, Email: 89411940@qq.com.
Shuyuan Shen, Email: 1360909257@qq.com.
Chunqing Yang, Email: CQyang1990@163.com.
Yixue Xue, Email: xueyixue888@163.com.
References
- 1.Zhu C, Kros JM, Cheng C, Mustafa D. The contribution of tumor-associated macrophages in glioma neo-angiogenesis and implications for anti-angiogenic strategies. Neuro-Oncology. 2017;19(11):1435–1446. doi: 10.1093/neuonc/nox081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cassiday LA, Maher LJ., 3rd Having it both ways: transcription factors that bind DNA and RNA. Nucleic Acids Res. 2002;30(19):4118–4126. doi: 10.1093/nar/gkf512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spitzer JI, Ugras S, Runge S, Decarolis P, Antonescu C, Tuschl T, Singer S. mRNA and protein levels of FUS, EWSR1, and TAF15 are upregulated in liposarcoma. Genes Chromosomes Cancer. 2011;50(5):338–347. doi: 10.1002/gcc.20858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ke H, Zhao L, Feng X, Xu H, Zou L, Yang Q, Su X, Peng L, Jiao B. NEAT1 is required for survival of breast Cancer cells through FUS and miR-548. Gene Regul Syst Bio. 2016;10(Suppl 1):11–17. doi: 10.4137/GRSB.S29414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu H, Zheng T, Yu J, Zhou L, Wang L. LncRNA XIST accelerates cervical cancer progression via upregulating Fus through competitively binding with miR-200a. Biomed Pharmacother. 2018;105:789–797. doi: 10.1016/j.biopha.2018.05.053. [DOI] [PubMed] [Google Scholar]
- 6.Xiong D, Wu YB, Jin C, Li JJ, Gu J, Liao YF, Long X, Zhu SQ, Wu HB, Xu JJ, et al. Elevated FUS/TLS expression is negatively associated with E-cadherin expression and prognosis of patients with non-small cell lung cancer. Oncol Lett. 2018;16(2):1791–1800. doi: 10.3892/ol.2018.8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z, Lei H, Sun Q. MicroRNA-141 and its associated gene FUS modulate proliferation, migration and cisplatin chemosensitivity in neuroblastoma cell lines. Oncol Rep. 2016;35(5):2943–2951. doi: 10.3892/or.2016.4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Errichelli L, Dini Modigliani S, Laneve P, Colantoni A, Legnini I, Capauto D, Rosa A, De Santis R, Scarfo R, Peruzzi G, et al. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat Commun. 2017;8:14741. doi: 10.1038/ncomms14741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 10.Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, Zhou LY, Sun T, Wang M, Yu T, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. 2016;37(33):2602–2611. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- 11.Zheng J, Liu X, Xue Y, Gong W, Ma J, Xi Z, Que Z, Liu Y. TTBK2 circular RNA promotes glioma malignancy by regulating miR-217/HNF1beta/Derlin-1 pathway. J Hematol Oncol. 2017;10(1):52. doi: 10.1186/s13045-017-0422-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin P, Huang Y, Zhu P, Zou Y, Shao T, Wang O. CircRNA circHIPK3 serves as a prognostic marker to promote glioma progression by regulating miR-654/IGF2BP3 signaling. Biochem Biophys Res Commun. 2018. [DOI] [PubMed]
- 13.Chen Z, Duan X. hsa_circ_0000177-miR-638-FZD7-Wnt signaling Cascade contributes to the malignant behaviors in glioma. DNA Cell Biol. 2018. [DOI] [PubMed]
- 14.Yu C, Wang M, Li Z, Xiao J, Peng F, Guo X, Deng Y, Jiang J, Sun C. MicroRNA-138-5p regulates pancreatic cancer cell growth through targeting FOXC1. Cell Oncol (Dordr) 2015;38(3):173–181. doi: 10.1007/s13402-014-0200-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang R, Liu M, Liang H, Guo S, Guo X, Yuan M, Lian H, Yan X, Zhang S, Chen X, et al. miR-138-5p contributes to cell proliferation and invasion by targeting Survivin in bladder cancer cells. Mol Cancer. 2016;15(1):82. doi: 10.1186/s12943-016-0569-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schlierf B, Friedrich RP, Roerig P, Felsberg J, Reifenberger G, Wegner M. Expression of SoxE and SoxD genes in human gliomas. Neuropathol Appl Neurobiol. 2007;33(6):621–630. doi: 10.1111/j.1365-2990.2007.00881.x. [DOI] [PubMed] [Google Scholar]
- 17.McGary KL, Park TJ, Woods JO, Cha HJ, Wallingford JB, Marcotte EM. Systematic discovery of nonobvious human disease models through orthologous phenotypes. Proc Natl Acad Sci U S A. 2010;107(14):6544–6549. doi: 10.1073/pnas.0910200107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson GL, McIntosh M, Wu L, Barnett M, Goodman G, Thorpe JD, Bergan L, Thornquist MD, Scholler N, Kim N, et al. Assessing lead time of selected ovarian cancer biomarkers: a nested case-control study. J Natl Cancer Inst. 2010;102(1):26–38. doi: 10.1093/jnci/djp438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellsworth RE, Seebach J, Field LA, Heckman C, Kane J, Hooke JA, Love B, Shriver CD. A gene expression signature that defines breast cancer metastases. Clin Exp Metastasis. 2009;26(3):205–213. doi: 10.1007/s10585-008-9232-9. [DOI] [PubMed] [Google Scholar]
- 20.Zhang YL, Li Q, Yang XM, Fang F, Li J, Wang YH, Yang Q, Zhu L, Nie HZ, Zhang XL, et al. SPON2 promotes M1-like macrophage recruitment and inhibits hepatocellular carcinoma metastasis by distinct integrin-rho GTPase-hippo pathways. Cancer Res. 2018;78(9):2305–2317. doi: 10.1158/0008-5472.CAN-17-2867. [DOI] [PubMed] [Google Scholar]
- 21.Tornin J, Hermida-Prado F, Padda RS, Gonzalez MV, Alvarez-Fernandez C, Rey V, Martinez-Cruzado L, Estupinan O, Menendez ST, Fernandez-Nevado L, et al. FUS-CHOP promotes invasion in Myxoid Liposarcoma through a SRC/FAK/RHO/ROCK-dependent pathway. Neoplasia. 2018;20(1):44–56. doi: 10.1016/j.neo.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daigle JG, Lanson NA, Jr, Smith RB, Casci I, Maltare A, Monaghan J, Nichols CD, Kryndushkin D, Shewmaker F, Pandey UB. RNA-binding ability of FUS regulates neurodegeneration, cytoplasmic mislocalization and incorporation into stress granules associated with FUS carrying ALS-linked mutations. Hum Mol Genet. 2013;22(6):1193–1205. doi: 10.1093/hmg/dds526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsumoto A, Suzuki H, Fukatsu R, Shimizu H, Suzuki Y, Hisanaga K. An autopsy case of frontotemporal lobar degeneration with the appearance of fused in sarcoma inclusions (basophilic inclusion body disease) clinically presenting corticobasal syndrome. Neuropathology. 2016;36(1):77–87. doi: 10.1111/neup.12232. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Sui X, Zhao H, Cong L, Li Y, Xin T, Guo M, Hao W. Decreased circular RNA hsa_circ_0001649 predicts unfavorable prognosis in glioma and exerts oncogenic properties in vitro and in vivo. Gene. 2018. [DOI] [PubMed]
- 25.Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, Huang N, Yang X, Zhao K, Zhou H, et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst. 2018;110(3). [DOI] [PMC free article] [PubMed]
- 26.Li CY, Ma L, Yu B. Circular RNA hsa_circ_0003575 regulates oxLDL induced vascular endothelial cells proliferation and angiogenesis. Biomed Pharmacother. 2017;95:1514–1519. doi: 10.1016/j.biopha.2017.09.064. [DOI] [PubMed] [Google Scholar]
- 27.Fang S, Guo H, Cheng Y, Zhou Z, Zhang W, Han B, Luo W, Wang J, Xie W, Chao J. circHECTD1 promotes the silica-induced pulmonary endothelial-mesenchymal transition via HECTD1. Cell Death Dis. 2018;9(3):396. doi: 10.1038/s41419-018-0432-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou QH, Deng CZ, Li ZS, Chen JP, Yao K, Huang KB, Liu TY, Liu ZW, Qin ZK, Zhou FJ, et al. Molecular characterization and integrative genomic analysis of a panel of newly established penile cancer cell lines. Cell Death Dis. 2018;9(6):684. doi: 10.1038/s41419-018-0736-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Selli C, Pearce DA, Sims AH, Tosun M. Differential expression of store-operated calcium- and proliferation-related genes in hepatocellular carcinoma cells following TRPC1 ion channel silencing. Mol Cell Biochem. 2016;420(1–2):129–140. doi: 10.1007/s11010-016-2776-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He Q, Zhao L, Liu Y, Liu X, Zheng J, Yu H, Cai H, Ma J, Liu L, Wang P, et al. Circ-SHKBP1 regulates the angiogenesis of U87 glioma-exposed endothelial cells through miR-544a/FOXP1 and miR-379/FOXP2 pathways. Mol Ther Nucleic Acids. 2018;10:331–348. doi: 10.1016/j.omtn.2017.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gupta SK, Garg A, Bar C, Chatterjee S, Foinquinos A, Milting H, Streckfuss-Bomeke K, Fiedler J, Thum T. Quaking inhibits doxorubicin-mediated cardiotoxicity through regulation of cardiac circular RNA expression. Circ Res. 2018;122(2):246–254. doi: 10.1161/CIRCRESAHA.117.311335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dai X, Zhang N, Cheng Y, Yang T, Chen Y, Liu Z, Wang Z, Yang C, Jiang Y. RNA-binding protein trinucleotide repeat-containing 6A regulates the formation of circular RNA 0006916, with important functions in lung Cancer cells. Carcinogenesis. 2018. [DOI] [PubMed]
- 33.Gao W, Lam JW, Li JZ, Chen SQ, Tsang RK, Chan JY, Wong TS. MicroRNA-138-5p controls sensitivity of nasopharyngeal carcinoma to radiation by targeting EIF4EBP1. Oncol Rep. 2017;37(2):913–920. doi: 10.3892/or.2017.5354. [DOI] [PubMed] [Google Scholar]
- 34.Luan B, Sun C. MiR-138-5p affects insulin resistance to regulate type 2 diabetes progression through inducing autophagy in HepG2 cells by regulating SIRT1. Nutr Res. 2018;59:90–98. doi: 10.1016/j.nutres.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 35.Xiong Y, Zhang J, Song C. CircRNA ZNF609 functions as a competitive endogenous RNA to regulate FOXP4 expression by sponging miR-138-5p in renal carcinoma. J Cell Physiol. 2018. [DOI] [PubMed]
- 36.Zhong Z, Huang M, Lv M, He Y, Duan C, Zhang L, Chen J. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305–317. doi: 10.1016/j.canlet.2017.06.027. [DOI] [PubMed] [Google Scholar]
- 37.Roose J, Korver W, de Boer R, Kuipers J, Hurenkamp J, Clevers H. The sox-13 gene: structure, promoter characterization, and chromosomal localization. Genomics. 1999;57(2):301–305. doi: 10.1006/geno.1999.5779. [DOI] [PubMed] [Google Scholar]
- 38.Gu W, Wang B, Wan F, Wu J, Lu X, Wang H, Zhu Y, Zhang H, Shi G, Dai B, et al. SOX2 and SOX12 are predictive of prognosis in patients with clear cell renal cell carcinoma. Oncol Lett. 2018;15(4):4564–4570. doi: 10.3892/ol.2018.7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang P, Ma Y, Wang F, Yang J, Liu Z, Peng J, Qin H. Comprehensive gene and microRNA expression profiling reveals the crucial role of hsa-let-7i and its target genes in colorectal cancer metastasis. Mol Biol Rep. 2012;39(2):1471–1478. doi: 10.1007/s11033-011-0884-1. [DOI] [PubMed] [Google Scholar]
- 40.Melichar HJ, Narayan K, Der SD, Hiraoka Y, Gardiol N, Jeannet G, Held W, Chambers CA, Kang J. Regulation of gammadelta versus alphabeta T lymphocyte differentiation by the transcription factor SOX13. Science. 2007;315(5809):230–233. doi: 10.1126/science.1135344. [DOI] [PubMed] [Google Scholar]
- 41.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao Y, Fan X, Li W, Ping W, Deng Y, Fu X. miR-138-5p reverses gefitinib resistance in non-small cell lung cancer cells via negatively regulating G protein-coupled receptor 124. Biochem Biophys Res Commun. 2014;446(1):179–186. doi: 10.1016/j.bbrc.2014.02.073. [DOI] [PubMed] [Google Scholar]
- 43.Wang LF, Liu YS, Yang B, Li P, Cheng XS, Xiao CX, Liu JJ, Li S, Ren JL, Guleng B. The extracellular matrix protein mindin attenuates colon cancer progression by blocking angiogenesis via Egr-1-mediated regulation. Oncogene. 2018;37(5):601–615. doi: 10.1038/onc.2017.359. [DOI] [PubMed] [Google Scholar]
- 44.Liu X, Zheng J, Xue Y, Qu C, Chen J, Wang Z, Li Z, Zhang L, Liu Y. Inhibition of TDP43-mediated SNHG12-miR-195-SOX5 feedback loop impeded malignant biological behaviors of glioma cells. Mol Ther Nucleic Acids. 2018;10:142–158. doi: 10.1016/j.omtn.2017.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao X, Liu Y, Zheng J, Liu X, Chen J, Liu L, Wang P, Xue Y. GAS5 suppresses malignancy of human glioma stem cells via a miR-196a-5p/FOXO1 feedback loop. Biochim Biophys Acta Mol Cell Res. 2017;1864(10):1605–1617. doi: 10.1016/j.bbamcr.2017.06.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Correlation between circ_002136, miR-138-5p and linear CDK11A and evaluation of the transfection efficiency. (A) The transfection efficiency of FUS. Data are presented as the means ± SD (n = 3, each group). **P < 0.01 vs. FUS (−) NC group. (B) The expression of circ_002136 with RNase R treatment. Data are presented as the means ± SD (n = 5, each group). **P < 0.01 vs. control group in ECs; ##P < 0.01 vs. RNase R group in ECs. (C) The expression of CDK11A with RNase R treatment. Data are presented as the means ± SD (n = 5, each group). **P < 0.01 vs. control group in ECs; ##P < 0.01 vs. control group in ECs. (D) The transfection efficiency of circ_002136 knockdown. Data are presented as the means ± SD (n = 5, each group). **P < 0.01 vs. circ_002136 (−) NC group. (E) The expression of CDK11A after circ_002136 knockdown. Data are presented as the means ± SD (n = 5, each group). (F) The transfection efficiency of CDK11A knockdown. Data are presented as the means ± SD (n = 5, each group). **P < 0.01 vs. sh-NC group. (G) The expression of circ_002136 after CDK11A knockdown. Data are presented as the means ± SD (n = 5, each group). (H) The transfection efficiency of miR-138-5p agomir or antagomir. Data are presented as the means ± SD (n = 5, each group). **P < 0.01 vs. miR-138-5p (+) NC group, ##P < 0.01 vs. miR-138-5p (−) NC group. (I) The expression of CDK11A after miR-138-5p over-expression or silencing. Data are presented as the means ± SD (n = 5, each group). (J) The transfection efficiency of SOX13. Data are presented as the means ± SD (n = 3, each group). **P < 0.01 vs. SOX13 (+) NC group, ##P < 0.01 vs. SOX13 (−) NC group. (K) The transfection efficiency of SPON2. Data are presented as the means ± SD (n = 3, each group). **P < 0.01 vs. SPON2 (−) NC group. (TIF 471 kb)
Figure S2. Co-effects of FUS and circ_002136 on the expression of downstream molecules and angiogenesis. (A) The expression of miR-138-5p was co-regulated by both FUS and circ_002136. Values represent the means ± SD (n = 5, each group). **P < 0.01 vs. FUS(−)NC + circ_002136(−)NC group. ##P < 0.01 vs. FUS (−) + circ_002136(−)NC group. (B-D) The co-effects of FUS and circ_002136 on the mRNA and protein expression levels of SOX13 and SPON2 in GECs were evaluated by qRT-PCR and western blot. Data are presented as the means ± SD (n = 3, each group). **P < 0.01 vs. FUS(−)NC + circ_002136(−)NC group. ##P < 0.01 vs. FUS(−) + circ_002136(−)NC group. (E) The co-effects of FUS and circ_002136 on the viability of GECs were evaluated by the CCK-8 assay. Data are presented as the means ± SD (n = 5, each group). **P < 0.01 vs. FUS(−)NC + circ_002136(−)NC group. ##P < 0.01 vs. FUS(−) + circ_002136(−)NC group. (F) The co-effects of FUS and circ_002136 on the migration of GECs were evaluated by the transwell assay. Data are presented as the means ± SD (n = 5, each group). **P < 0.01 vs. FUS(−)NC + circ_002136(−)NC group. ##P < 0.01 vs. FUS(−) + circ_002136(−)NC group. The scale bar represents 100 μm. (G) The co-effects of FUS and circ_002136 on the tube formation of GECs were evaluated by the Matrigel tube formation assay. Data are presented as the means ± SD (n = 5, each group). **P < 0.01 vs. FUS(−)NC + circ_002136(−)NC group. ##P < 0.01 vs. FUS(−) + circ_002136(−)NC group. The scale bar represents 100 μm. (TIF 1639 kb)
Data Availability Statement
The datasets used and analysed during the current study are available from the corresponding author on reasonable request.