SUMMARY
The ventricular-subventricular zone (V-SVZ) harbors adult neural stem cells. V-SVZ neural stem cells exhibit features of astrocytes, have a regional identity, and depending on their location in the lateral or septal wall of the lateral ventricle, generate different types of neuronal and glial progeny. We performed large-scale single-cell RNA sequencing to provide a molecular atlas of cells from the lateral and septal adult V-SVZ of male and female mice. This revealed regional and sex differences among adult V-SVZ cells. We uncovered lineage potency bias at the single-cell level among lateral and septal wall astrocytes toward neurogenesis and oligodendrogenesis, respectively. Finally, we identified transcription factor co-expression modules marking key temporal steps in neurogenic and oligodendrocyte lineage progression. Our data suggest functionally important spatial diversity in neurogenesis and oligodendrogenesis in the adult brain and reveal molecular correlates of adult NSC dormancy and lineage specialization.
Graphical Abstract
In Brief
Mizrak et al. performed large-scale, single-cell RNA sequencing of the adult ventricular-subventricular zone neural stem cell niche. They identify regional differences between the lateral wall and septal wall, as well as sex differences in cell types and signaling pathways.
INTRODUCTION
Somatic stem cells reside in complex niches and are important for tissue homeostasis and repair. Understanding the diversity of cell types in adult stem cell niches is essential for understanding stem cell dynamics and regulation. Neural stem cells reside in specialized niches in the adult mammalian brain, where they generate new neurons and glia throughout life. The ventricular-subventricular zone (V-SVZ), located adjacent to the lateral ventricles, is the largest germinal zone in the adult mouse brain and gives rise to olfactory bulb interneurons, as well as oligodendrocytes and astrocytes (Chaker et al., 2016). The V-SVZ is located along both the lateral wall, adjacent to the striatum, and the septal wall, next to the septum. Both walls harbor neural stem cells (NSCs) in the adult, although the septal wall has been far less studied. Adult V-SVZ NSCs co-exist in quiescent (qNSC) and actively dividing (aNSC) states (Chaker et al., 2016). Upon activation, they give rise to transit amplifying cells (TACs), which in turn generate neuroblasts (NBs) that migrate to the olfactory bulb (Doetsch et al., 1999). Although oligodendrocytes and astrocytes are also generated in the adult V-SVZ (Menn et al., 2006; Nait-Oumesmar et al., 1999; Ortega et al., 2013; Sohn et al., 2015; Xing et al., 2014), their lineage trajectories are less clear. Indeed, the extent to which individual NSCs in vivo are multipotent or exhibit lineage commitment to neuronal or glial fates is still unclear.
Adult V-SVZ NSCs are radial cells with many hallmarks of astrocytes (Doetsch et al., 1999). They contact the cerebrospinal fluid in the ventricles at the center of pinwheels formed by ependymal cells and extend a long process to the vasculature (Mirzadeh et al., 2008; Shen et al., 2008; Tavazoie et al., 2008). Adult V-SVZ NSCs are set aside during embryogenesis, remaining largely quiescent until adulthood (Fuentealba et al., 2015; Furutachi et al., 2015), and reside in a complex microenvironment comprising diverse cell types and local and long-range signals (Silva-Vargas et al., 2013). Importantly, V-SVZ NSCs have a regional identity and give rise to specific subtypes of olfactory bulb interneurons or glia depending on their spatial location (Chaker et al., 2016; Fiorelli et al., 2015). Moreover, spatially distinct pools of adult V-SVZ NSCs can be regulated by physiological states and distant neural circuits (Paul et al., 2017), highlighting the importance of regional differences between the lateral and septal walls regarding intrinsic NSC identity, as well as for niche signals in regulating stem cell behavior.
Single-cell RNA sequencing is a powerful approach to dissect cellular heterogeneity. Previous single-cell studies using low-throughput approaches have begun to illuminate the stages of qNSC activation in the adult V-SVZ (Basak et al., 2018; Dulken et al., 2017; Llorens-Bobadilla et al., 2015; Luo et al., 2015), focusing on NSCs purified by fluorescence-activated cell sorting (FACS) from the lateral wall and the neuronal lineage. However, the diversity of V-SVZ NSCs and niche compartments that support NSC self-renewal and differentiation require single-cell analysis of thousands of cells to dissect V-SVZ complexity. Importantly, no studies have examined the septal V-SVZ or sex differences in the V-SVZ. Here, we performed large-scale, single-cell profiling of all V-SVZ cell types in the lateral wall and in the septal wall of male and female adult mice without cell sorting and identify cell subtypes in V-SVZ stem cell lineages and niche cells. We uncover regional differences between the lateral and septal walls as well as some sex differences and identify markers and transcription factor modules underlying neurogenic and oligodendrogenic lineage potency and differentiation.
RESULTS
Comparison of Lateral and Septal Adult V-SVZ by Large-Scale Single-Cell Profiling
To elucidate regional differences and molecular heterogeneity among adult V-SVZ NSCs and niche cell types, we performed large-scale single-cell profiling of >41,000 bulk V-SVZ cells from separately dissected lateral and septal walls from adult male and female mice (Figures 1A and 1B). This approach allowed us to capture stem cells and their progeny at each stage of differentiation, as well as niche cells. Importantly, individual cells were randomly sampled without cell sorting to maximally preserve in vivo composition. An automated microwell-array-based platform was used to capture and simultaneously profile thousands of single cells from each sample (Bose et al., 2015; Yuan and Sims, 2016; Figure 1A). Following cell lysis and mRNA capture on commercially available barcoded mRNA capture beads (Macosko et al., 2015), on-chip reverse transcription labeled cDNA product from each cell with a unique barcode sequence. We obtained more than 41,000 raw single-cell RNA sequencing (RNA-seq) profiles from two biological replicates of each sample after removal of cell doublets and red blood cells. Importantly, cell clusters contained contributions from each sample and replicate with similar molecular capture rates in all samples (Figures S1C–S1E). We used Louvain community detection as implemented by phenograph (Levine et al., 2015) to identify molecularly distinct cell clusters in an unsupervised manner (Figures S1A and S1B), and t-distributed stochastic neighbor embedding (t-SNE) (van der Maaten and Hinton, 2008) and force directed graphs for data visualization. To avoid over-interpretation of small compositional differences potentially due to differential sampling, we focused on compositional differences in cell types with more than 800 cells in the follow-up orthogonal validation experiments.
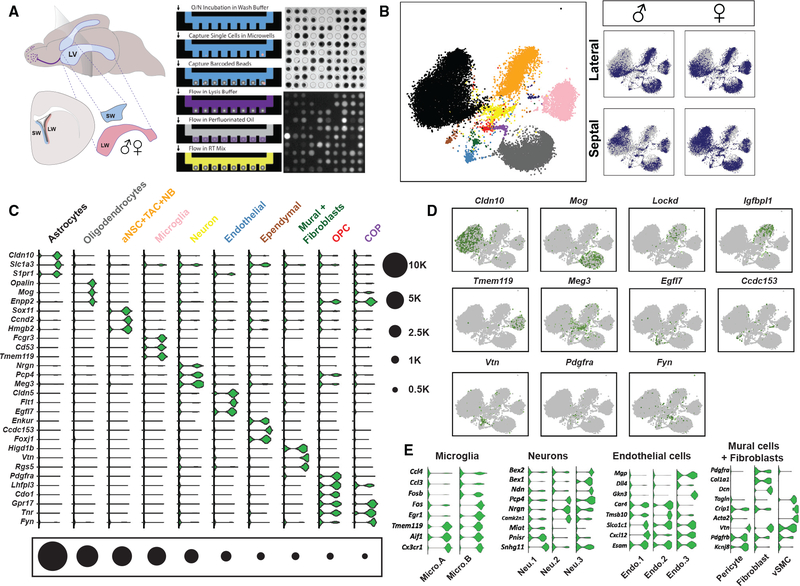
Figure 1. Large-Scale scRNA-Seq Reveals Major Cell Types in the V-SVZ.
(A) (Left) Schema of adult mouse brain showing location of the lateral ventricles (light blue, LV). Plane indicates level of coronal schema (below left) highlighting location of lateral wall V-SVZ (LW, red) adjacent to the striatum and septal wall V-SVZ (SW, blue) adjacent to the septum. Whole mounts of the entire LW (red) and SW (blue) were dissected from adult male and female mice, dissociated to single cells, and loaded onto a microwell-array-based platform. (Right) Workflow of single-cell capture and RT in microwell-array-based platform. Drop-seq beads and calcein-AM-stained V-SVZ cells were imaged to check for lysis efficiency during RNA capture step. (Top right) Bright-field image of cell- and bead-loaded microwells is shown. (Bottom right) Fluorescent image of lysed V-SVZ cells is shown.
(B) t-SNE projections of major cell types. Color-coding corresponds to cell types shown in (C). T cells are midnight blue. (Right) Individual samples are highlighted in blue on the combined dataset background (gray). Note that astrocytes comprise two large astrocyte clusters (Astrocyte A and Astrocyte B), which have been grouped in the t-SNE plot (black; see Figure S1). All t-SNE visualizations were prepared using the larger first replicate dataset (see STAR Methods for details).
(C) Violin plots showing log-transformed CPM (count per million) values of pervasive and known cell type markers (fold change [FC] > 2; false discovery rate [FDR] < 0.05). Black circles indicate numbers of cells in each major cell type cluster.
(D) Scaled expression of key markers on t-SNEs (one marker per cell type except Lockd and Igfbpl1 for aNSC+TAC+NB).
(E) Violin plots showing pervasive markers for subtypes of microglia, neurons, endothelial cells, mural cells, and fibroblasts. (FDR < 0.05). vSMC, vascular smooth muscle cell.
Major Cell Types in the Adult V-SVZ
To identify the major cell types in the adult V-SVZ, we performed unsupervised clustering. The major tissue-resident cell types in the combined dataset comprised astrocytes, neuronal progenitors (aNSCs, TACs, and NBs), oligodendrocyte progenitor cells (OPCs), committed oligodendrocyte precursors (COPs), oligodendrocytes, microglia, neurons, ependymal cells, endothelial cells, mural cells (pericytes and vascular smooth muscle cells), and fibroblasts (Figures 1B and 1C). We also detected a small cluster of cells, which are either circulating or resident T lymphocytes. Binomial specificity analyses identified pervasive and specific markers for each cell type (Figures 1C and S1A). We used this large-scale dataset to explore regional differences in neurogenesis and gliogenesis between lateral and septal walls in males and females and to define molecularly distinct subclusters of V-SVZ cell types, including niche cells, as outlined in the next sections.
Niche cells are important regulators of V-SVZ stem cells and their progeny. To resolve molecularly distinct subpopulations, we performed clustering and expression specificity analysis on major niche cell types (microglia, neurons, endothelial cells, and mural cells). Among the microglial cell cluster (5,525 cells total from two biological replicates), we identified two microglial subtypes with distinct transcriptional signatures (microglia A and microglia B; Figure 1E), likely representing surveying (ramified) and activated states of microglia (Friedman et al., 2018; Nomaru et al., 2014). Both Micro.A and Micro.B expressed microglia markers Tmem119, Aif1, and Cx3cr1, whereas Micro.B was also enriched in immediate early transcription factors as well as pro-inflammatory chemokines (e.g., Ccl3 and Ccl4; Friedman et al., 2018; Matcovitch-Natan et al., 2016; Mathys et al., 2017; Figure 1E; Table S1). Microglial branching morphology decreases during activation (Kettenmann et al., 2011). To visualize V-SVZ microglia in vivo, we performed immunostaining for Iba1 and quantified ramified and unramified microglia in the lateral and septal walls of the V-SVZ in male and female mice. Overall, microglia were more enriched in the lateral wall (Figure S1H). Moreover, the V-SVZ contained predominantly unramified microglia (Figures S1F and S1H), as previously reported (Ribeiro Xavier et al., 2015), although ramified microglia were also present (Figures S1G and S1H). As V-SVZ microglia are important for the migration and the survival of neural progenitors (Ribeiro Xavier et al., 2015), our data showing microglia enrichment in the lateral wall are consistent with higher levels of neuronal line-age progenitors in the lateral wall over the septal wall (Figures 1B, 3A, and 3B).
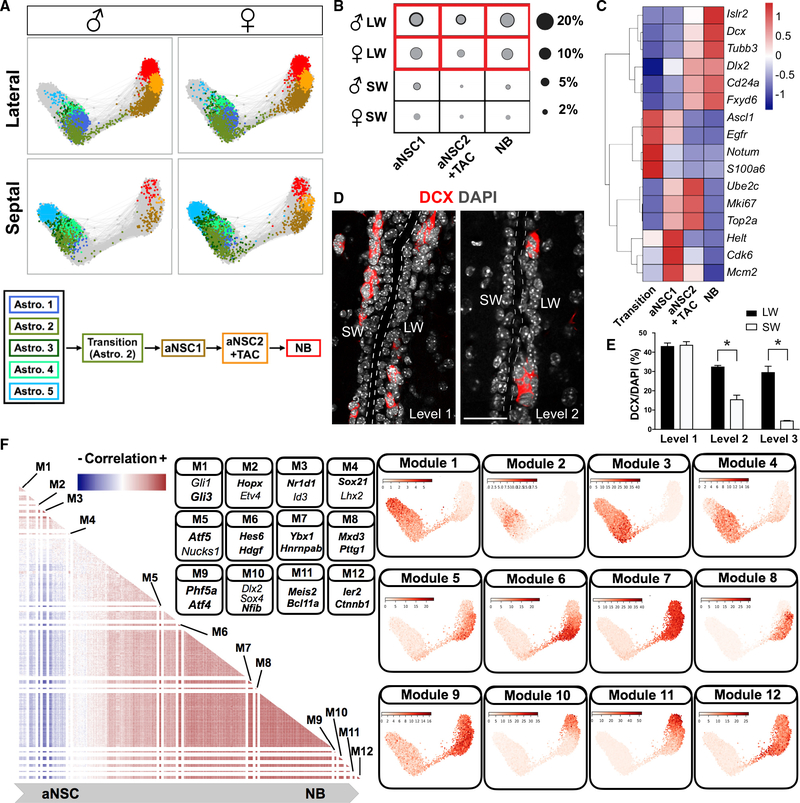
Figure 3. Adult V-SVZ Neuronal Lineage and Temporal Regulatory Steps.
(A) Force-directed graphs showing the lineage progression from dormant astrocytes to NBs and a schematic summary.
(B) Fraction of each cell type represented by circles (gray, mean; black, mean + SD; two biological replicates). The compositions of aNSC1, aNSC2+TAC, and NB populations were extracted from combined clustering of neuronal progenitors from two biological replicates.
(C) Heatmap (scaled by row) showing average expression of functional markers (FDR < 0.05) in different neuronal progenitor subtypes.
(D) Representative images of doublecortin (DCX) immunostaining at different rostro-caudal levels of the V-SVZ (level 1, bregma +1.41 mm; level 2, bregma +0.73 mm). Scale bar, 50 μm.
(E) Quantification of DCX+ cell proportions at different rostro-caudal levels of the V-SVZ (n = 3; mean + SEM) (p < 0.05). Level 3, bregma +0.1 mm.
(F) (Left) Heatmap showing correlation matrix for significant TF modules. Both axes show TFs with the same dimensionality. The correlation between the same TF was set to 0 for visualization purposes. (Right) Significant TF clusters (Figure S5A; Table S4) were ordered based on their dispersion on force-directed graphs. The average expressions of TF modules in each cell were plotted on force-directed graphs. Modules with distinct expression patterns were separated with white lines in the heatmap. Representative TFs from different modules are shown in the panel, and TFs associated with previously undescribed co-expression patterns in each module are highlighted in bold.
Resident neurons and long-range innervation provide neuro-transmitters that regulate adult NSCs and their progeny (Káradóttir and Kuo, 2018; Young et al., 2011). We detected three major neuronal subclusters in our datasets (3,202 cells; Figure 1E; Table S1), including one enriched in Chat (Neuron 3), which may include ChAT+ local neurons that regulate V-SVZ cells (Paez-Gonzalez et al., 2014). In addition, we detected multiple vascular cell types, which are key niche cells in the V-SVZ (Otsuki and Brand, 2017). Subclustering of the endothelial cluster (1,774 cells) revealed three distinct endothelial subtypes (Figure 1E; Table S1). Based on marker gene enrichment (Table S1), Endo.1 may correspond to endothelial capillary cells, Endo.2 to venous endothelial cells, and Endo.3 to arterial endothelial cells (Vanlandewijck et al., 2018). Finally, in the mural cell and fibro-blast cluster (958 cells), we detected three major cell types—pericytes, vascular smooth muscle cells, and fibroblasts expressing Dcn and Col1a1 (Vanlandewijck et al., 2018; Figure 1E; Table S1).
Astrocyte Subclusters Have Regional Distribution in the Adult V-SVZ
V-SVZ NSCs exhibit hallmark features of astrocytes. To dissect V-SVZ astrocyte heterogeneity, we performed clustering analysis on all astrocyte and ependymal cell clusters (14,762 cells). We identified five major subclusters of astrocytes, which were clearly distinct from ependymal cells (Figure 2A; Table S2). Using gene set enrichment analysis (GSEA), we further validated their classification as astrocytes and ependymal cells by comparing them to third ventricle astrocyte and ependymal gene sets from a large single-cell dataset from the murine third ventricle (Campbell et al., 2017; Subramanian et al., 2005; Figure S2A). All astrocyte subclusters, except Astro.2, were enriched in third ventricle astrocyte gene sets. In contrast, Astro.2 was depleted for astrocyte gene sets and enriched for “tanycyte 2” markers (Campbell et al., 2017; Figure S2A). In addition, genes upregulated in different injury paradigms were differentially enriched in distinct astrocyte subclusters with the highest enrichment in ependymal cells (Figure S2D; Llorens-Bobadilla et al., 2015; Zamanian et al., 2012).
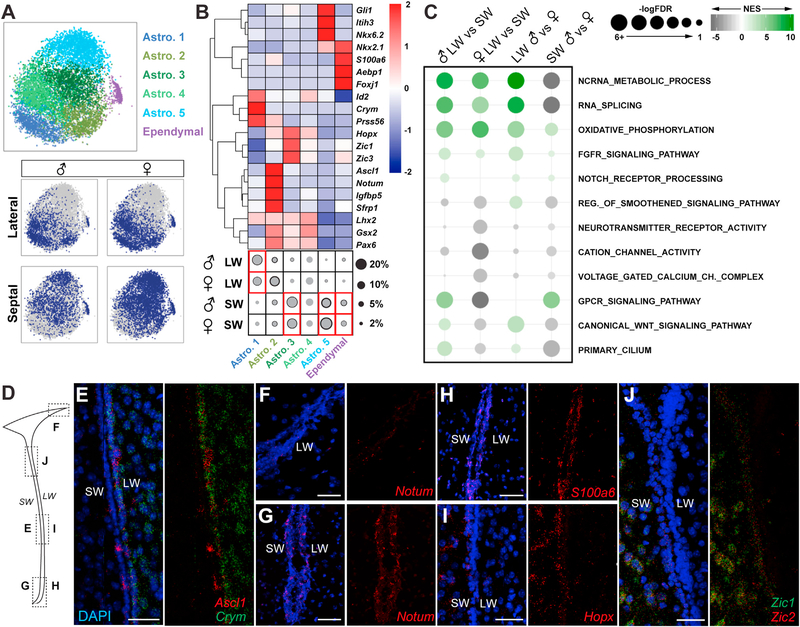
Figure 2. Molecular Characterization of Astrocyte Clusters.
(A) t-SNE projections of five major astrocyte subclusters and ependymal cells and their distribution in different samples.
(B) Heatmap (scaled by row) showing average expression of markers, including TFs and subtype identifiers (FDR < 0.05). The fraction of each subtype is represented with circles (gray, mean; black, mean + SD; two biological replicates). Red box indicates regional enrichment. Astro.4 was not detected in the second replicate due to the lack of subcluster-specific markers, and therefore, black circle was not drawn. LW, lateral wall; SW, septal wall.
(C) GSEA showing significantly regulated molecular processes relevant to V-SVZ NSC biology across four comparisons. NES, normalized enrichment score.
(D) Schema showing coronal section of lateral ventricle and location of RNAscope images in (E)–(J).
(E–J) RNAscope images, with and without DAPI, of (E) Ascl1 (red) and Crym (green), Notum (red) in the (F) dorso-lateral V-SVZ and (G) ventral V-SVZ, (H) S100a6 (red), (I) Hopx (red), and (J) Zic1 (green) and Zic2 (red). DAPI is in blue.
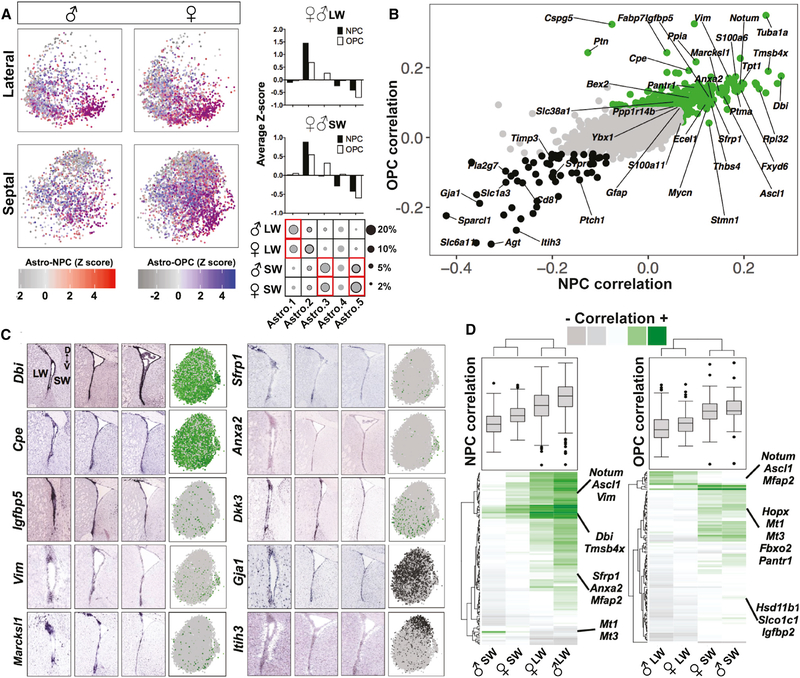
Interestingly, V-SVZ astrocyte subclusters were differentially distributed in the lateral and septal preparations (Figures 2A and 2B). Astro.1 was more abundant in samples of lateral wall, whereas Astro.3 and Astro.5 were more abundant in those from the septal wall. In contrast, Astro.2 and Astro.4 were more evenly distributed in both lateral and septal walls. Binomial specificity analysis revealed molecular markers enriched in astrocyte subclusters (Figures 2B and S3A; Table S2), including transcription factors, which demarcate different spatial domains in the V-SVZ, or involved in activation of quiescent NSCs (Kohwi et al., 2005; López-Juárez et al., 2013; Merkle et al., 2007, 2014; Samanta et al., 2015; Urbán et al., 2016). Astro.1 was enriched in Crym, Prss56, and Id2; Astro.2 Notum, Sfrp1, Igfbp5, S100a6, Thbs4, and Ascl1; Astro.3 Hopx and Zic family genes; and Astro.5 Agt, Gli1, Itih3, and Nkx6–2 (Figures 2B and S3A; Table S2). Although Gsx2, Pax6, and Lhx2 were more uniformly expressed across multiple astrocyte subclusters, they were depleted in Astro.5 (Figures 2B and S3A; Table S2).
We performed RNAscope to validate the expression patterns of selected genes in astrocyte subclusters. Crym (Astro.1) was highly enriched in the lateral V-SVZ and striatum (Figures 2D, 2E, S3B, and S3D) in contrast to Hopx, Zic1, and Zic2 (Astro.3), which were predominantly localized in the V-SVZ septal wall and septum (Figures 2D, 2I, 2J, S3B, S3E, and S3F), confirming that Astro.1 and Astro.3 are enriched in the lateral and septal wall, respectively. Interestingly, Astro.2 was enriched in genes predominantly localized to the V-SVZ, such as Notum, S100a6, and Ascl1 (Figures 2E–2H, S3C, S3D, and S3G). Importantly, RNAscope analysis also revealed that, within the V-SVZ itself, some genes exhibited regional differences along the dorsoventral axis. Notum (Astro.2) was enriched in the ventral as compared to dorsal V-SVZ (Figures 2F, 2G, and S3C), and Zic1 and Zic2 (Astro.3) were enriched in the dorsal septal wall and septum at rostral levels (Figures 2J and S3E), highlighting smaller domains of V-SVZ astrocytes with different molecular identities within larger regions of the V-SVZ.
The distribution of astrocyte subclusters showed clear regional patterns, but not major compositional sex differences. However, comparison of male and female lateral and septal wall astrocytes at the gene expression level highlighted many differentially expressed genes between walls and sexes, including transcription factors and receptors (Table S3). Using GSEA, we identified Gene Ontology gene sets preferentially associated with lateral or septal V-SVZ and sex (Figure 2C; Table S3). Several molecular and metabolic processes important for stem cell self-renewal, maintenance, and proliferation, such as Fgfr and Notch signaling and oxidative phosphorylation, showed lateral wall and male enrichment (Engler et al., 2018; Kawai et al., 2017; Llorens-Bobadilla et al., 2015; Zheng et al., 2004). Wnt and Smoothened signaling, as well as primary cilium pathways (Ahn and Joyner, 2005; Azim et al., 2014; Ihrie et al., 2011), were up-regulated in male lateral and female septal samples. In contrast, G-protein coupled receptor (GPCR) signaling associated with NSC quiescence (Codega et al., 2014) was enriched in the male septal wall (Figure 2C; Table S3). Interestingly, female astrocytes were enriched in neurotransmitter receptor activity and ion channel pathways. Lastly, non-coding RNA processing and RNA splicing showed lateral wall enrichment, consistent with the importance of non-coding RNAs in V-SVZ neurogenesis (Ramos et al., 2015; Figure 2C; Table S3). In sum, the lateral and septal walls have different V-SVZ astrocyte compositions, which exhibit significant regional and sex differences at the gene expression level.
Adult V-SVZ Lateral Wall Is More Proliferative and Neurogenic Than the Septal Wall
Our large-scale single-cell profiling of the adult V-SVZ from regionally dissected walls allowed us to compare neurogenesis and gliogenesis in the septal and lateral walls. We first focused on the neuronal lineage. Lineage trajectories generated using all astrocyte and neuronal progenitor clusters revealed that non-dividing astrocytes were connected to aNSCs via a narrow path of activation representing transitional cells (Figure 3A). These transitional cells clustered with Astro.2 and were in a continuous trajectory that progressed via two sequential stages of aNSCs (aNSC1 and aNSC2/TAC) to neuroblasts, suggesting a key role for Astro.2 in NSC activation (Figure 3A). GSEA revealed a clear correspondence between our neuronal lineage trajectory and previously published studies with lower throughput (Figures S2B and S2C; Dulken et al., 2017; Llorens-Bobadilla et al., 2015). Interestingly, expression of proliferation markers (e.g., mKi67, Mcm2, and Top2a) in the neuronal lineage peaked in aNSC1s, aNSC2s, and TACs and decreased in Dcx+ NBs (Figure 3C; Table S2). Moreover, transition cells upregulated Ascl1 and Egfr prior to canonical cell cycle genes (e.g., mKi67, Mcm2, and Top2a), revealing that stem cell activation and cell proliferation can be uncoupled (Figure 3C). Importantly, activated neuronal progenitors and NBs were more enriched in the lateral wall than the septal wall (Figures 3A and 3B). We confirmed in vivo that the septal wall has fewer dividing cells and NBs than the lateral wall, except at rostral levels of the V-SVZ, by immunostaining and quantification for Ki67 and doublecortin (DCX) (Figures 3D, 3E, and S4A–S4C).
To investigate the transcriptional regulatory steps defining neuronal lineage differentiation, we calculated the co-expression probability ratios for each transcription factor (TF) expressed in the neuronal lineage in individual cells and isolated TFs forming significant clusters (Figure S5A; Table S4) to identify TF co-expression modules and TFs with mutually exclusive expression profiles. Plotting the average expression of the modules across each cell and its nearest neighbors on force-directed graphs revealed TF modules with distinct distributions along the neuronal lineage trajectory (Figures 3F and S5A; Table S4). In addition to the classical markers of neuronal progenitor (NPC) regulation (e.g., Ascl1, Dlx2, and Sp8), the neuronal lineage modules included TFs implicated in cell proliferation and differentiation of stem cells in various tissues (e.g., Phf5a and Bcl11a) and of cancer stem cells (e.g., Hdgf and Ybx1; Fotovati et al., 2011; Luc et al., 2016; Strikoudis et al., 2016; Zhao et al., 2013) as well as TF co-expression modules in astrocyte subtypes (e.g., Hopx-Etv4, Gli1-Gli3, and Nr1d1-Id3). This multi-step progression reveals an unappreciated intricacy in neuronal lineage differentiation downstream of qNSC to aNSC transition and TF co-expression profiles that target transcriptionally discrete stages of NSC differentiation.
Oligodendrocyte Progenitors Are Enriched in the Male Septal Wall
Little is known about oligodendrocyte lineage progression in the adult V-SVZ and whether there are regional differences between lateral and septal walls. To address this, we first performed clustering analyses on oligodendrocyte lineage cells, revealing cell types at different stages of oligodendrocyte differentiation corresponding to OPCs, COPs, newly formed and myelin-forming oligodendrocytes (NFOL/MFOLs), and mature oligodendrocytes (MOLs). We next generated a force-directed visualization of the oligodendrocyte lineage k-nearest neighbors graph to define their lineage order (Figure 4A), which recapitulated the predicted lineage order previously reported from single cell RNA (scRNA)-seq profiles of oligodendrocyte differentiation (Marques et al., 2016; Figure S2E).
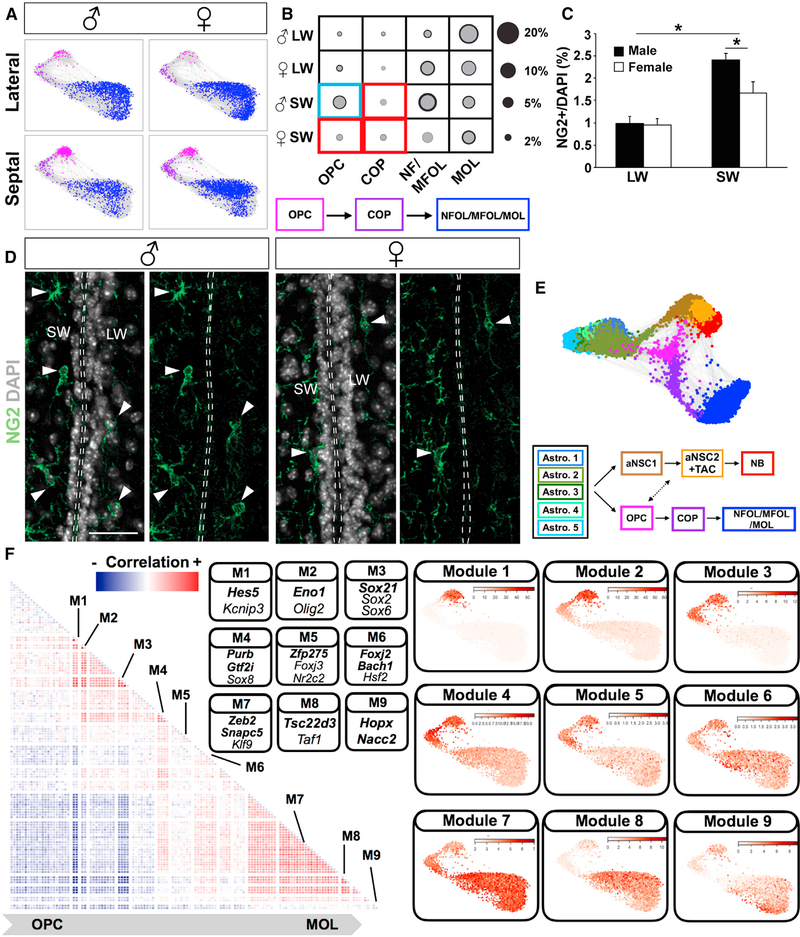
Figure 4. Regional and Sex Differences in Oligodendrocyte Lineage.
(A) Force-directed graphs showing oligodendrocyte lineage progression from OPCs to MOLs (MOL, mature oligodendrocytes; NFOL/MFOL, newly formed and myelin-forming oligodendrocytes) in four samples and a schematic summary of the lineage differentiation order.
(B) The fraction of each cell type in different samples is represented with circles (gray, mean; black, mean + SD; two biological replicates). Red box indicates regional enrichment and blue box indicates both regional and sex-specific enrichment.
(C) Quantification of NG2+ soma demonstrating sex- and region-specific differences in OPC density (%NG2/DAPI; p < 0.05; n = 3; mean + SEM). Note fibers were not quantified.
(D) Representative images of NG2 immunostainings in the V-SVZ from male and female mice. Scale bar: 50 μm.
(E) Force-directed graphs with both neuronal and oligodendrocyte lineages and a schematic summary of the differentiation order and branching point of these lineages. See also Figure S4D and Video S1.
(F) (Left) Heatmap showing correlation matrix for significant TF modules. Both axes show TFs with the same dimensionality. The correlation between the same TF was set to 0 for visualization purposes. (Right) Significant TF clusters (Figure S5B; Table S4) were ordered based on their dispersion on force-directed graphs.
The average expressions of TF modules in each cell were plotted on force-directed graphs. Modules with distinct expression patterns were separated with white lines in the heatmap. Representative TFs from different modules are shown in the panel, and TFs associated with previously undescribed co-expression patterns in each module are highlighted in bold.
To investigate whether neuronal and oligodendrocyte line-ages are related in the adult V-SVZ, we combined neuronal and oligodendrocyte lineage trajectories with V-SVZ astrocytes (Video S1; Figures 4E and S4D). Importantly, astrocyte clusters exhibit different connectivity within the trajectory. For example, Astro.2 is connected to both OPCs and neuronal progenitors, whereas Astro.3, which is enriched in the septal wall, is connected only with OPCs (Video S1). Interestingly, oligodendrocyte progenitors were also connected to aNSCs in the neuronal trajectory, supporting a model for two lineages of oligodendrocytes.
Unexpectedly, the most prominent sex difference we detected in the V-SVZ was among oligodendrocyte progenitors. OPCs and COPs were significantly enriched in the septal wall over the lateral wall in both sexes, with OPCs more enriched in males as compared to females (Figures 4A and 4B). To independently validate this finding in vivo, we performed immunostaining for NG2, an OPC marker, and quantified OPCs in the septal and lateral V-SVZ in both sexes. This confirmed that OPCs were enriched in the septal V-SVZ and, furthermore, that they were more abundant in the septal V-SVZ in males than in females (Figures 4C and 4D). In sum, the higher number of neuronal lineage progenitors in the lateral wall, in contrast to the enrichment of oligodendrocyte progenitors in the septal wall, point to important regional differences in the V-SVZ regarding neurogenesis and gliogenesis (Figure S4E).
Finally, to investigate the transcriptional regulatory steps defining oligodendrocyte lineage progression, we analyzed TF co-expression modules in each cell in the oligodendrocyte line-age trajectory. We identified TF regulatory modules that exhibit sequential progression along the oligodendrocyte lineage (Figures 4F and S5B; Table S4). These previously uncharacterized temporal modules comprised oligodendrocyte lineage progression TFs (e.g., Kcnip3, Olig2, Sox2, and Klf9; Maki et al., 2013; Marques et al., 2016; Tokumoto et al., 2017; Wu et al., 2012) and co-expression partners (e.g., Hes5, Eno1, Sox21, Purb, Zfp275, Foxj2, Zeb2, Tsc22d3, and Hopx; Figure 4F). Among these, Sox2 and Sox21 interaction are regulators of pluripotency and human glioma progression (Caglayan et al., 2013; Kuzmichev et al., 2012).
Functional Classification of Non-dividing V-SVZ Stem Cells
An important open question is whether individual cells are multi-potent or whether they exhibit commitment to neuronal or glial fates. We therefore examined whether V-SVZ astrocytes have neuronal or oligodendrocyte lineage potential at the single-cell level and whether this is enriched in the lateral or septal wall. To functionally classify the large astrocyte cluster in an unbiased manner, we calculated the correlation between scRNA-seq profiles of individual non-dividing astrocytes with aNSC1s (NPC Corr.) and OPCs (OPC Corr.) (STAR Methods). Only a modest number of cells in the astrocyte cluster were highly correlated with NPCs and/or OPCs but were present in each astrocyte sub-cluster (Figure 5A). Consistent with the putative lineage order in the force-directed graphs (Figures 3A and 4E), the Astro.2 population showed highest correlation with both NPCs and OPCs, although the septal V-SVZ-enriched Astro.3 was correlated with OPCs only (Figure 5A), in line with the higher proportion of OPCs in the septal wall (Figures 4B and 4C).
Figure 5. Functional Classification of Non-dividing V-SVZ Stem Cells and Their Spatial Organization.
(A) (Left) Individual cells were colored based on their Z-scored correlation values (red, NPC-correlated; blue, OPC-correlated; purple, correlated with both NPCs and OPCs) and plotted on t-SNE projections in each sample. (Right) The average Z scores of cells per cluster in each region. The circle plots are the same as those shown in Figure 2B.
(B) Scatterplot showing the correlation between each gene and NPCs or OPCs. Highly correlated genes are labeled in green and anti-correlated genes in black.
(C) In situ hybridization (ISH) data from Allen Mouse Brain Atlas (coronal sections) showing spatial distribution of selected NPC and/or OPC-correlated astrocyte markers at three different rostro-caudal levels and the scaled expression of each gene on t-SNEs (green, correlated genes; black, anti-correlated genes).
(D) Heatmap of gene correlation values for the top hundred positively correlated genes with highest variability across four samples and the boxplots showing the data distribution. Genes with high regional and sex differences are highlighted.
Next, we performed a gene level analysis to rank astrocyte genes based on their lineage potency (see STAR Methods), which revealed different correlation patterns with NPC and/or OPC potency (Table S5). Many NPC-correlated genes (Table S5) also had high correlation with OPCs (Table S5), indicating shared molecular mechanisms of potency. Top markers that correlated with both NPCs and OPCs included S100a6, Notum, Dbi, and Sfrp1, as well as known V-SVZ qNSC activation and injury-response markers, such as Ascl1, Vim, Thbs4, and Rpl32 (Benner et al., 2013; Dulken et al., 2017; Llorens-Bobadilla et al., 2015; Figure 5B). Some markers were correlated only with NPCs (e.g., Stmn1) or OPCs (e.g., Cspg5 and Dkk3; Table S5); moreover, our analysis also revealed anti-correlated markers, including classical astrocyte markers, such as Gja1, Slc1a3, and Sparcl1. As almost all V-SVZ astrocytes are Slc1a3+, the level of expression of these pan-astrocyte genes (e.g., Gja1 and Slc1a3) at the single-cell level may be important to assess the lineage potency of an individual astrocyte (Figure 5B). Finally, among known markers expressed by adult V-SVZ NSCs, aNSC markers were correlated with NPCs and OPCs (Ascl1, Nes, and Gsx2), whereas qNSC-enriched genes (Vcam1, Cdh2, Nr2e1, S1pr1, and Hes5) were not (Figure S5C). Together, our analysis identifies markers correlated with NPC and/or OPC potency and allows the ranking of previously proposed markers in terms of their lineage potency.
To gain insight into the spatial distribution of cells expressing NPC and/or OPC-correlated and anti-correlated markers, we used Allen Mouse Brain Atlas ISH data (Lein et al., 2007). Consistent with their high ranking in the lineage potency analysis, Dbi, Cpe, Igfbp5, Vim, Marcksl1, Sfrp1, Anxa2, and Dkk3 transcripts showed high enrichment in V-SVZ, including various regional distributions along the dorsal-ventral and rostro-caudal axis (Figure 5C). In contrast, highly NPC and OPC anti-correlated astrocyte markers, such as Gja1, showed high expression in both the V-SVZ and adjacent parenchymal cells and Itih3 was more enriched in non-SVZ cells (Figure 5C). These ISH patterns indicate the significance of V-SVZ enrichment in selecting salient markers for lineage potency.
Finally, when lineage potencies of non-dividing astrocytes were compared separately by wall and by sex, NPC- associated genes were enriched in lateral wall astrocytes and OPC-associated genes in septal wall astrocytes (Figure 5D; Table S5), consistent with the quantitative differences in NPCs or OPCs in these regions. Interestingly, several NPC and/or OPC-correlated genes also showed clear regional bias, such as Mt1 or Mt3, which were strong markers of lineage potency in septal astrocytes but did not correlate with either lineage in lateral astrocytes (Figure 5D). Similarly, we detected several genes with high correlation variability that depend on sex in male versus female comparisons, such as Igfbp2, Slco1c1, and Mfap2, further supporting male and female V-SVZ astrocyte functional gene expression level differences (Figure 5D; Table S5).
DISCUSSION
We performed large-scale, single-cell profiling to characterize the complexity of the adult V-SVZ NSC stem cell niche as well as regional differences between the lateral and septal walls. We identified the major V-SVZ cell types, including molecularly distinct subtypes. Our analysis revealed that lateral and septal walls have different lineage biases toward neuronal or glial fate at the single-cell level and exhibit some sex differences. Furthermore, we identified single-cell-level transcription factor co-expression modules in neurogenesis and oligodendrogenesis that track the transcriptional progression of these lineages with high resolution.
The V-SVZ exhibits regional and cell type heterogeneity at multiple scales: anatomical (different walls of the ventricle), large spatial domains within each wall and restricted microdomains that generate specific subtypes of neurons or glia (Chaker et al., 2016). Although transcriptome analysis in the postnatal V-SVZ has uncovered molecular differences between the dorsal and the lateral walls (Azim et al., 2015), all profiling studies in the adult V-SVZ, including recent low-throughput, single-cell studies, have focused only on the lateral wall and the neuronal lineage (Basak et al., 2018; Codega et al., 2014; Dulken et al., 2017; Llorens-Bobadilla et al., 2015). These FACS-based studies have provided important initial insight into stem cell quiescence and have highlighted sequential transitions that NSCs undergo during neuronal lineage progression (Basak et al., 2018; Codega et al., 2014; Dulken et al., 2017; Llorens-Bobadilla et al., 2015).
Here, we separately profiled cells from the entire lateral and septal wall of male and female adult mice without FACS purification, thereby capturing a broader repertoire of V-SVZ cell types, including cells at multiple stages of differentiation along both the neuronal and oligodendrocyte lineages as well as niche cells. Importantly, this provides unique insight into regional differences in the lateral and septal walls, including the less-explored oligodendrocyte lineage in the adult V-SVZ, and has allowed us to identify transcription factor co-expression modules at the single-cell level, which likely delineate transcriptional regulatory changes occurring during neuronal and oligodendrocyte differentiation.
We detected regional differences in the composition of neuronal and oligodendrocyte progenitors, suggesting that, although both walls can give rise to neurons and glia, the lateral wall is more enriched in neurogenesis and the septal wall in gliogenesis. The most striking sex difference was in OPCs, which were enriched in the septal wall and especially in males. This sex difference may be due to differences in the number and types of stem cells and progenitors in males and females as well as to differences in niche signals. Female and male oligodendrocyte levels and myelination are differentially sensitive to steroids, such as progesterone and dihydrotestosterone (Swamydas et al., 2009). Although the functional consequences of different OPC numbers are unclear, we speculate that adaptive myelination may exhibit sex differences.
Functionally distinct quiescent stem cell pools located in either wall of the V-SVZ may underlie regional compositional differences in progenitor populations, as our cell- and gene-level analyses suggest that lateral and septal wall astrocytes exhibit higher NPC or OPC correlation, respectively. Indeed, spatially distinct pools of NSCs can sense and respond to different local and long-range physiological signals to generate different types of OB interneurons (Paul et al., 2017), and V-SVZ NSCs are promptly recruited upon injury or under pathological conditions (Chaker et al., 2016). In light of our findings, it will be interesting to carefully dissect regional V-SVZ responses to demyelinating insults and other injury paradigms in female and male mice separately in the future and isolate regional dormant cell subtypes that are lineally related to different injury response cells. Indeed, additional layers of heterogeneity among both stem cells and niche cells remain to be resolved. This may require more precise isolation of regional subtypes and optimized protocols to enrich for different cell types. Importantly, our study provides a platform to identify molecularly distinct pools of stem cells under homeostasis, in response to different physiological states, and that are recruited by injury in each sex.
Significant effort has been made to classify V-SVZ NSCs, yielding a handful of markers that also label non-SVZ astrocytes or show high enrichment in other cell types. We identify several lineage potency markers, including signaling molecules with high V-SVZ astrocyte enrichment. These markers exhibited different regional expression patterns within the V-SVZ walls, highlighting additional V-SVZ astrocyte heterogeneity still to be resolved. In addition to significant regional differences, we also found that several key molecular pathways crucial for NSC maintenance, self-renewal, and proliferation were differentially expressed in male versus female astrocytes, indicating differential effects of these cues and signaling pathways on male and female NSCs. In the future, it will be important to characterize regional cellular heterogeneity and sex differences in the adult human V-SVZ, which contains largely quiescent stem cells (Sanai et al., 2004).
Finally, our large-scale census of regionally dissected adult V-SVZ cells will provide a valuable resource for future functional studies, including identification and regulation of regionally distinct adult V-SVZ neural stem cells, their lineages, and interactions with different niche components. It will be useful for comparative analysis of NSCs in the developing brain and adult subgranular zone neural stem cell niche as well as for comparison with astrocytes, microglia, and vascular cells in different brain regions to probe regional heterogeneity and sex differences
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Fiona Doetsch (fiona.doetsch@unibas.ch).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
All experiments were performed in accordance with institutional and national guidelines for animal use and approved by IACUC at Columbia University and the cantonal veterinary office of Basel-Stadt. All mice (males and females) were group-housed conventionally with ad libitum food and water in 12-h light/12-h dark cycles, and sacrificed at the same time of day. B6.Cg-Tg(GFAP-cre/ERT2)505Fmv/J mice (hGFAP::CreERT2) were bred with B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J mice (R26RCAG-tdTomato) to generate hGFAP::CreERT2; R26RCAG-tdTomato mice. For single cell analysis, the tissue samples were microdissected from eight to ten week old hGFAP::CreERT2; R26RCAG-tdTomato and R26RCAG-tdTomato female and male mice. Females were in the estrus phase of the estrus cycle. Two biological replicates each were performed for male lateral wall, male septal wall, female lateral wall and female septal wall (Male Lateral Replicate 1: hGFAP::CreERT2; R26RCAG-tdTomato (2 mice); Male Septal Replicate 1: hGFAP:: CreERT2; R26RCAG-tdTomato (2 mice); Female Lateral Replicate 1: hGFAP::CreERT2; R26RCAG-tdTomato (3 mice); Female Septal Replicate 1: hGFAP::CreERT2; R26RCAG-tdTomato (3 mice); Male Lateral Replicate 2: hGFAP::CreERT2; R26RCAG-tdTomato (1 mouse); Male Septal Replicate 2: R26RCAG-tdTomato (3 mice); Female Lateral Replicate 2: hGFAP::CreERT2; R26RCAG-tdTomato (2 mice); Female Septal Replicate 2: hGFAP::CreERT2; R26RCAG-tdTomato (2 mice)).
METHOD DETAILS
V-SVZ dissection and dissociation
Whole-mounts of the lateral and septal walls of the V-SVZ were dissected separately as described (Mirzadeh et al., 2010). The whole-mount dissections included the entire length of the lateral and septal walls of the V-SVZ, as well as inevitably some adjacent striatal and septal tissue. Dissected whole mounts were minced into small pieces, digested with papain (Worthington, 6 mg per sample, 10 min at 37°C) in PIPES solution [120 mM NaCl, 5 mM KCl, 20 mM PIPES (Sigma), 0.45% glucose, 1x Antibiotic/Antimycotic (GIBCO), and phenol red (Sigma) in water; pH adjusted to 7.6] (Codega et al., 2014). Digested tissue was triturated to single cells in the presence of ovomucoid (Worthington, 0.7 mg/ml) and DNase (Worthington, 0.5 mg/ml) in DMEM/F12. The single cell suspension was layered on top of 22% Percoll (Sigma) and centrifuged for 10 mins at 4°C without brakes to remove debris and myelin. Cells were washed with 1% BSA/0.1% Glucose/Hank’s Balanced Salt Solution by centrifugation at 4°C. The single cell suspension was treated with red blood cell lysis buffer (Sigma) (1:1 dilution) for 1 minute, washed by two rounds of centrifugation (1300 rpm at 4°C) and resuspended in 1X Tris Buffered Saline. Cells were passed through a 40 μm cell strainer (Fisher) to remove any cell clumps before loading, and were stained with Calcein AM live stain dye (Fisher, 1:500) on ice for 15–30 minutes.
Single cell library preparation and sequencing
Single cell capture and reverse transcription (RT) were performed as previously described (Yuan and Sims, 2016). Cells were loaded in the microwell devices at a concentration of 600–1000 cells per μl. Cell capture, lysis (2-Mercaptoethanol: Fisher Scientific, Buffer TCL: QIAGEN), and reverse transcription (Maxima H Minus Reverse Transcriptase: ThermoFisher, SUPERaseIN: Thermofisher, Template switch oligo: IDT) were all performed on 150K microwell devices (microwell size: 58 μm diameter, 75 μm height) connected to an automated microfluidics system. The devices were scanned on a fluorescence microscope (Eclipse Ti-U, Nikon) during the RNA capture step, and checked for lysis efficiency. RT reactions were performed on Drop-seq beads (MACOSKO-2011–10, ChemGenes) also captured in the microwells. Beads were collected from the devices and pooled for cDNA amplification step. Prior to PCR, the beads were treated with Exo-I (New England Biolabs). Following PCR (KAPA HotStart ReadyMix, Kapabiosystems), purified cDNA was used as input for Nextera tagmentation reactions. cDNA and library quality was assessed using Qubit and Bioanalyzer (Agilent Technologies). All DNA purification steps were carried out using Ampure XP beads (Beckman). High quality samples were sequenced on a NextSeq 500 sequencer using a 75 cycle High Output Kit (Illumina, 1×75 bp).
RNA-scope
Brains were quick frozen, and stored at 80C. 18 mm coronal sections were cut using the cryostat. Sections were fixed in pre-chilled 4% PFA for 30 min at 4°C, washed rapidly 5 times in PBS, followed by 5 more washes in fresh PBS and dehydrated sequentially in 50% Ethanol, 70% Ethanol, and 100% Ethanol (two times) for 5 minutes each. Sections were processed for FISH using the RNAscope Multiplex Fluorescent Assay Kit (Advanced Cell Diagnostics). Sections were dried for 5 min at room temperature (RT), permeabilized for 20 min at RT using Protease IV solution (Cat No. 322336), washed and maintained in PBS until probe addition. Probes were pre-heated to 40°C, and added to sections in probe solutions for 2hrs at 40°C. Probes targeting Ascl1 (Cat No. 313291, NM_008553.4), Crym (Cat No. 466131-C3, NM_016669.1), Hopx (Cat No. 405161, NM_001159900.1), Zic1 (Cat No. 493121-C3, NM_009573.3), Zic2 (Cat No. 428211, NM_009574.3), Notum (Cat No. 428981, NM_175263.4) and S100a6 (Cat No. #412981,NM_011313.2) were used. Sections were washed 2 times 2 min in RNAscope Wash Buffer, incubated in RNAscope AMP-1 solution for 30 min at 40°C, washed 2 times 2 min in Wash Buffer, incubated in RNAscope AMP-2 solution for 15 min at 40°C, washed 2 times 2 min in Wash Buffer, incubated in RNAscope AMP-3 solution for 30min, washed 2 times 2min in Wash Buffer and incubated in AMP-4-FL solution for 15min, washed 2 times 2 min in Wash Buffer, incubated in DAPI solution for 30 s and mounted with AquaPolymount. Tile scans of the V-SVZ were acquired using an inverted Zeiss Confocal LSM700 and Zen Blue software. For RNA-scope signal intensity quantification, three sections per brain from three male and three female mice were analyzed for each gene using Fiji (Schindelin et al., 2012). RNA-scope signal intensities were normalized to DAPI intensity in the same region of interest to measure the relative abundance of a given gene.
Immunostaining
Adult male and female RjOrl:SWISS mice (2 months old) were transcardially perfused with 0.9% saline and 3.2% paraformaldehyde. Brains were fixed overnight and 20 μm coronal sections cut with a Leica VT1000S vibrating microtome. Brain sections were incubated in blocking solution (PBS with 2% bovine serum albumin (BSA) and 0.2% (Ki67 and DCX) or 0.05% (NG2) Triton X-100) for 60 minutes and then incubated in primary antibodies in blocking solution for 36 hours at 4°C. After washing, sections were revealed with Alexa Fluor-conjugated secondary antibodies (Life Technologies and Jackson ImmunoResearch Laboratories) for 1–2 hours at room temperature, counterstained with 4’,6-Diamidino-2-Phenylindole Dihydrochloride (DAPI), (1 μg/ml, 5 minutes, SIGMA D9542) and mounted with FluorSave™ (Millipore corporation). The following primary antibodies were used: anti-rabbit NG2 (Merck AB5320, 1:100), anti-rabbit Iba1 (Wako Cat. #019–19741, 1:100), anti-rabbit Ki67 (Abcam ab15580, 1:100), anti-goat doublecortin (Santa Cruz (C18): sc-8066, 1:150). Samples were imaged using an inverted LSM 700 confocal (Zeiss).
QUANTIFICATION AND STATISTICAL ANALYSIS
Definition of Significance
The term significance was used if the p value was smaller than 0.05. p values are reported in the figures, figure legends, and STAR Methods.
Single Cell Data Processing
As previously described, cell and molecular barcodes are contained in read 1 of our paired-end sequencing data, while all genomic information is contained in read 2 (Yuan and Sims, 2016). We trimmed read 2 to remove 3′ polyA tails (> 7 A’s), and discarded fragments with fewer than 24 remaining nucleotides. Trimmed reads were aligned to the Mus musculus genome and transcriptome annotation (GRCm38, Gencode annotation vM10) using STAR v.2.5.0 with parameters –sjdbOverhang 65–twopassMode Basic–outSAMtype BAM Unsorted (Dobin et al., 2013). Only reads with unique, strand-specific alignments to exons were kept for further analysis.
We extracted 12-nt cell barcodes (CBs) and 8-nt unique molecular identifies (UMIs) from read 1. Degenerate CBs containing either ‘N’s or more than four consecutive ‘G’s were discarded. Synthesis errors, which can result in truncated 11-nt CBs, were corrected similarly to a previously reported method (Shekhar et al., 2016). Briefly, we identified all CBs with at least twenty apparent molecules and for which greater than 90% of UMI-terminal nucleotides were ‘T’. These putative truncated CBs were corrected by removing their last nucleotide. This twelfth nucleotide became the new first nucleotide of corresponding UMIs, which were also trimmed of their last (‘T’) base. We note that one of second replicate experiments was conducted with commercial Drop-seq beads from Chemgenes with a modified structure. A ‘V’ base was added to the end of the UMI upstream of the poly(T) tail for quality-control purposes. As a result, we adjusted our analytical pipeline to recognize truncated CBs by the presence of excessive ‘T’ bases one nucleotide downstream of the position used in the previous pipeline.
All reads with the same CB, UMI, and gene mapping were collapsed to represent a single molecule. To correct for sequencing errors in UMI’s, we further collapsed UMI’s that were within Hamming distance 1 of another UMI with the same barcode and gene. To correct for sequencing errors in cell barcodes, we then collapsed CBs that were within Hamming distance one of another barcode, had at least 20 unique UMI-gene pairs a piece, and had at least 75% overlap of their UMI-gene pairs. Finally, we repeated UMI error correction and collapse using the error-corrected CBs. The remaining barcode-UMI-gene triplets were used to generate a digital gene expression matrix. The number of cells included in each sample was determined based on the inflection point in the cumulative histograms for molecules as previously described (Macosko et al., 2015; Yuan and Sims, 2016).
Clustering & Differential Expression Analyses
For unsupervised clustering, we first calculate k-nearest neighbors graphs from a cell by cell Spearman’s correlation matrix with k = 20. Spearman’s correlation was calculated from a subset of highly variable genes that were detected in fewer cells than expected given their apparent expression level (Satija et al., 2015). For the marker gene selection step only, molecular counts within each column of gene by cell expression matrices were normalized to sum to one. Genes were then ordered by their mean normalized value in the population and placed into bins of 50 genes. A gene’s detection frequency was calculated as the fraction of cells in which at least one molecule of a gene was detected, and its score was defined as the maximum detection frequency in its bin minus its detection frequency. Genes with scores greater than 0.15 were considered markers, and were used to compute Spearman’s correlation. Clustering was performed on the resulting k-nearest neighbors graph using Phenograph (Levine et al., 2015), a modularity-based clustering algorithm that produces robust clusterings on scRNA-seq data (Shekhar et al., 2016). Markers were re-selected for all subsequent sub-clusterings. Cluster specificity of each gene was measured using a binomial specificity test (Shekhar et al., 2016). The second replicates were clustered separately using the same settings as above to identify major cell types, their subclusters and cell compositions. All major cell clusters as well as subclusters were identified in the second replicates except Astro.4. The cell subclusters with the same identifying markers but that formed separate clusters were combined to maintain the compositional structure. SCDE analysis was used to identify differentially expressed genes across four astrocyte comparisons (Kharchenko et al., 2014).
Visualization of Single Cell Data
The similarity matrix described above was converted to a distance matrix, and used as input to tSNE for initial visualization (van der Maaten and Hinton, 2008). To visualize k-nearest neighbors graphs more directly and such that visualized distance was meaningful, we used force-directed graphs. Briefly, force-directed graphs model a k-nearest neighbors graph as a physical system with electrical and/or spring potentials between nodes, and visualize the system by minimizing its potential energy (Fruchterman and Reingold, 1991). Force-directed graph-based methods have previously been used to examine lineage relationships in single cell data (Weinreb et al., 2018). Layouts were generated from k-nearest neighbors graphs using the draw_networkx and spring_layout commands in the NetworkX v1.11 Python module with default parameters. All tSNE visualizations and force-directed graphs, differential expression analyses and gene expression plots from Figures 1, 2, 3, 4, and 5 were prepared using the larger first replicate dataset and the second replicate dataset was exclusively used for cell composition analysis.
Cell Filtering
Red blood cells (RBC) and doublets were removed after the initial clustering. Doublets were marked by significant expression of canonical markers for more than one cell type (e.g., Astrocyte-Oligodendrocytes, Astrocyte-Microglia) and formed separate clusters. To mitigate any effect these RBC and doublet populations might have had on the initial marker gene selection or clustering and to verify that no large clusters of doublets remained, data was re-clustered as described above without them. The second and/or third round of clustering revealed additional putative doublets expressing mature oligodendrocyte and aNSC-NB markers, and astrocyte and endothelial cell markers. As we were not able to validate such cells in the V-SVZ, they were also excluded from subsequent analysis (data not shown). In total, the putative doublets constituted cell barcodes ~7% of cell barcodes, in line with 4% doublets predicted by Poisson statistics (~70% bead loading efficiency). A small number of choroid plexus cells (4.5%) were detected in one of the four second replicate samples, and formed a separate cluster marked by high enrichment of Sostdc1, Kl, Wfdc2 and Ttr. This cluster was also excluded from downstream analyses and highly cross-talked marker Ttr in that sample was also removed from the expression matrix to improve clustering. The cell filtering resulted in the following cell numbers in each sample; Replicate 1: male lateral = 6649 cells, female lateral = 8676 cells, male septal = 4193 cells, female septal = 8889 cells; Replicate 2: male lateral = 1716 cells, female lateral = 3217 cells, male septal = 4897 cells, female septal = 3225 cells.
Gene Set Enrichment Analysis
To test for enrichment or depletion of previously reported gene sets, we first computed the average expression level of each gene in each cluster of cells identified using Phenograph (see above). Here, we defined the expression level as the number of UMI-corrected reads (molecules) for a specific gene divided by the total number of molecules detected in each cell. We then z-scored the log2-transformed the average expression level of each gene in each cluster, and used the z-scores to produce a ranked list for each cluster. GSEA was conducted for each gene set against each ranked list using the Java implementation (GSEA v2.2.2) available from http://software.broadinstitute.org/gsea/login.jsp and the following command: java -cp gsea2–2.2.2.jar -Xmx2512m xtools.gsea.GseaPreranked -gmx GMT_FILE_NAME -collapse false -mode Max_probe -norm meandiv -nperm 1001 -rnk RANK_FILE_NAME -scoring_scheme classic -rpt_label REPORT_FILE_NAME -include_only_symbols true -make_sets true -plot_top_x 20 -rnd_seed timestamp -set_max 6000 -set_min 15 -zip_report false -out . -gui false
C5 collection (Broad Institute) is utilized to identify GO biological processes, cellular components and molecular functions differentially regulated in four astrocyte comparisons (Figure 2C). SCDE output (Table S3) were pre-ranked based on effect size and inputted to Gsea with the following parameters: scoring_scheme: classic set_max: 500 set_min: 15 nperm: 1000
Transcription Factor Co-Expression Analysis
To assess co-expression and mutual exclusivity of TFs (GO Term: DNA binding transcription factor activity), we identified TFs with significant expression among cells (detected in > 80 cells) in the analyzed population. Homeo-domain containing, transcription cofactor Hopx was manually added to the original TF lists as it has highly regionalized expression. For each pair of TFs, we computed the log-transformed ratio of the joint probability of detecting the two TFs in the same cell to the product of their marginal detection probabilities:
Positive values of R indicate co-expression, and negative values indicate mutual exclusivity beyond what would be expected by chance. To display this analysis, we converted the matrix of R values to a Pearson correlation matrix and hierarchically clustered using the average linkage method. Significant clusters formed by these TFs were isolated using R (version 3) function pvclust(), which uses bootstrap resampling to compute p values (nboot = 2000). TFs forming significant clusters (alpha = 0.95) were defined as a TF module (Figure S5; Table S4).
TF Module Visualization
To examine gradations of TF module expression in individual cells, we defined a cell’s neighborhood expression to be the sum of its expression with that of its k-nearest neighbors, and normalized each neighborhood expression profile to sum to the median number of molecules per neighborhood. A similar smoothing method was proposed in a recent study (Wagner et al., 2017). A TF module’s expression was then calculated as the mean expression of its genes in the neighborhood and visualized on force-directed graphs (see above). Normalizing each cell’s expression prior to calculating neighborhood expression or weighting the neighborhood sum so that the central cell made the greatest contribution yielded similar results.
Lineage Correlation Analysis
We use the Spearman’s correlation coefficients computed on a set of highly variable genes as described above to compare individual astrocytes to aNSC1 cells and OPCs (NPC and OPC, respectively). For each astrocyte, we computed the average over all pairwise correlations with aNSC1 cells and over all pairwise correlations with OPCs, yielding two correlation values for each cell. To identify markers of lineage potential, we computed the Spearman’s correlation coefficient between the normalized expression of each gene across astrocytes and the Spearman’s correlation coefficient with either NPCs or OPCs across astrocytes. These analyses were used to generate Figures 5A, 5B, and 5D.
Statistical Analysis of Immunostaining Data
Statistical details of experiments can be found in the figure legends. Data are presented as mean values ± standard error of the mean (SEM). n represents the number of evaluated animals. For quantification of immunostaining, four sections at different rostral/caudal levels from each animal were counted, from three males and three females. Cells throughout the entire dorsal-ventral extent of V-SVZ on both the lateral and septal walls were quantified using Fiji software (Schindelin et al., 2012). Data are presented as percentage of overall number of DAPI+ cells in each wall of the V-SVZ, or as cells/mm2. For Iba1 quantification, multipolar microglia were classified as “ramified” and round and unipolar microglia as “unramified.” Two-way statistical comparisons were conducted by two-tailed unpaired Student’s test.
DATA AND SOFTWARE AVAILABILITY
The accession number for the sequencing data reported in this paper is GEO: GSE109447.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-NG2 | Merck | Cat#5320; RRID:AB_11213678 |
| Rabbit polyclonal anti-Iba1 | Wako | Cat#019–19741; RRID:AB_839504 |
| Rabbit polyclonal anti-Ki67 | Abcam | Cat#ab15580; RRID:AB_443209 |
| Goat polyclonal anti-DCX | Santa Cruz Biotechnology | Cat#sc8066; RRID:AB_2088494 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Papain | Worthington | Cat#LS003119 |
| PIPES | Sigma | Cat#P1851 |
| Ovomucoid | Worthington | Cat#LS003087 |
| Percoll | Sigma | Cat#P1644 |
| Calcein AM | Invitrogen | Cat#C3100MP |
| 2-Mercaptoethanol | Fisher Scientific | Cat#BP176–100 |
| Critical Commercial Assays | ||
| Nextera XT DNA Library Preparation Kit | Illumina | Cat#FC-131–1024 |
| Qubit dsDNA HS Assay Kit | ThermoFisher | Cat#Q32854 |
| High Sensitivity DNA chips kit | Agilent Technologies | Cat#5067–4626 |
| NextSeq 500/550 High Output v2 kit (75 cycles) | Illumina | Cat#FC-404–2005 |
| Deposited Data | ||
| Raw and processed data | This manuscript | GEO: GSE109447 |
| Experimental Models: Organisms/Strains | ||
| Mouse: RjOrl:SWISS | JANVIER LABS | RjOrl:SWISS |
| Mouse: B6.Cg-Tg(GFAP-cre/ERT2)505Fmv/J | The Jackson Laboratory | JAX: 012849 |
| Mouse: B6;129S6-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze | The Jackson Laboratory | JAX: 007908 |
| Oligonucleotides | ||
| Template Switching Oligonucleotide AAGCAGTGGTATCA ACGCAGAGTGAAT rGrGrG |
Macosko et al., 2015 | IDT |
| Software and Algorithms | ||
| Phenograph | Levine et al., 2015 | https://github.com/jacoblevine/PhenoGraph |
| Force-directed graphs | Weinreb et al., 2018 | https://github.com/AllonKleinLab/SPRING/ |
| t- distributed stochastic neighbor embedding (t-SNE) | van der Maaten and Hinton, 2008 | https://lvdmaaten.github.io/tsne/ |
| Rstudio | https://www.rstudio.com | |
| Gene Set Enrichment Analysis | Subramanian et al., 2005 | http://software.broadinstitute.org/gsea/index.jsp |
| SCDE | Kharchenko et al., 2014 | http://hms-dbmi.github.io/scde/ |
| Other | ||
| DNase I | Worthington | Cat#LS002139 |
| Drop-seq beads | ChemGenes | Cat#MACOSKO-2011–10 |
| Maxima H Minus Reverse Transcriptase | ThermoFisher | Cat#EP0752 |
| Exo-I | New England Biolabs | Cat# M0293L |
| SUPERaseIN | ThermoFisher | Cat#AM2696 |
| KAPA HotStart ReadyMix | Kapabiosystems | Cat#KK2602 |
| Ampure XP beads | Beckman Coulter | Cat#A63881 |
| Buffer TCL | QIAGEN | Cat#1031576 |
| Red Blood Cell Lysing Buffer Hybri-Max | Sigma | Cat# R7757 |
| RNAscope Probe- Mm-Ascl1 | ACDBio | Cat No. 313291 |
| RNAscope Probe - Mm-Crym-C3 | ACDBio | Cat No. 466131-C3 |
| RNAscope Probe - Mm-Hopx | ACDBio | Cat No. 405161 |
| RNAscope Probe - Mm-Notum | ACDBio | Cat No. 428981 |
| RNAscope Probe - Mm-S100a6 | ACDBio | Cat No. 412981 |
| RNAscope Probe - Mm-Zic1-C3 | ACDBio | Cat No. 493121-C3 |
| RNAscope Probe - Mm-Zic2 | ACDBio | Cat No. 428211 |
| RNAscope Protease III & Protease IV Reagents | ACDBio | Cat No. 322340 |
| RNAscope Fluorescent Multiplex Reagent Kit | ACDBio | Cat No. 320850 |
| RNAscope Wash Buffer Reagents | ACDBio | Cat No. 310091 |
Highlights.
Single-cell transcriptomics of >41,000 cells from adult V-SVZ neural stem cell niche
Regional and sex differences in lateral and septal adult V-SVZ in males and females
Identify transcription factor co-expression modules and lineage potency markers
Regional biases for neuronal and oligodendrocyte lineages in the adult V-SVZ
ACKNOWLEDGMENTS
F.D. was supported by NIH NINDS R01 NS074039, United States; Swiss National Science Foundation 31003A_163088, Switzerland; and the University of Basel, Switzerland. P.A.S. was supported by NIH/NIBIB grant K01EB016071 and NIH/NCI grant R33CA202827, United States. D.M. was supported by an NRSA Ruth Kirschstein Postdoctoral Fellowship (NIH NINDS F32 NS090736), United States.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes five figures, one video, and five tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.12.044.
DECLARATION OF INTERESTS
Columbia University has filed a patent application on the microwell technology for single-cell RNA-seq. J.Y. and P.A.S. are listed as co-inventors on this patent application.
REFERENCES
- Ahn S, and Joyner AL (2005). In vivo analysis of quiescent adult neural stem cells responding to Sonic hedgehog. Nature 437, 894–897. [DOI] [PubMed] [Google Scholar]
- Azim K, Fischer B, Hurtado-Chong A, Draganova K, Cantù C, Zemke M, Sommer L, Butt A, and Raineteau O (2014). Persistent Wnt/β-catenin signaling determines dorsalization of the postnatal subventricular zone and neural stem cell specification into oligodendrocytes and glutamatergic neurons. Stem Cells 32, 1301–1312. [DOI] [PubMed] [Google Scholar]
- Azim K, Hurtado-Chong A, Fischer B, Kumar N, Zweifel S, Taylor V, and Raineteau O (2015). Transcriptional hallmarks of heterogeneous neural stem cell niches of the subventricular zone. Stem Cells 33, 2232–2242. [DOI] [PubMed] [Google Scholar]
- Basak O, Krieger TG, Muraro MJ, Wiebrands K, Stange DE, Frias-Aldeguer J, Rivron NC, van de Wetering M, van Es JH, van Oudenaarden A, et al. (2018). Troy+ brain stem cells cycle through quiescence and regulate their number by sensing niche occupancy. Proc. Natl. Acad. Sci. USA 115, E610–E619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benner EJ, Luciano D, Jo R, Abdi K, Paez-Gonzalez P, Sheng H, Warner DS, Liu C, Eroglu C, and Kuo CT (2013). Protective astrogenesis from the SVZ niche after injury is controlled by Notch modulator Thbs4. Nature 497, 369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose S, Wan Z, Carr A, Rizvi AH, Vieira G, Pe’er D, and Sims PA (2015). Scalable microfluidics for single-cell RNA printing and sequencing. Genome Biol. 16, 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caglayan D, Lundin E, Kastemar M, Westermark B, and Ferletta M (2013). Sox21 inhibits glioma progression in vivo by forming complexes with Sox2 and stimulating aberrant differentiation. Int. J. Cancer 133, 1345–1356. [DOI] [PubMed] [Google Scholar]
- Campbell JN, Macosko EZ, Fenselau H, Pers TH, Lyubetskaya A, Tenen D, Goldman M, Verstegen AM, Resch JM, McCarroll SA, et al. (2017). A molecular census of arcuate hypothalamus and median eminence cell types. Nat. Neurosci 20, 484–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaker Z, Codega P, and Doetsch F (2016). A mosaic world: puzzles revealed by adult neural stem cell heterogeneity. Wiley Interdiscip. Rev. Dev. Biol 5, 640–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codega P, Silva-Vargas V, Paul A, Maldonado-Soto AR, Deleo AM, Pastrana E, and Doetsch F (2014). Prospective identification and purification of quiescent adult neural stem cells from their in vivo niche. Neuron 82, 545–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetsch F, Caillé I, Lim DA, García-Verdugo JM, and Alvarez-Buylla A (1999). Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell 97, 703–716. [DOI] [PubMed] [Google Scholar]
- Dulken BW, Leeman DS, Boutet SC, Hebestreit K, and Brunet A (2017). Single-cell transcriptomic analysis defines heterogeneity and transcriptional dynamics in the adult neural stem cell lineage. Cell Rep. 18, 777–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler A, Rolando C, Giachino C, Saotome I, Erni A, Brien C, Zhang R, Zimber-Strobl U, Radtke F, Artavanis-Tsakonas S, et al. (2018). Notch2 signaling maintains NSC quiescence in the murine ventricular-subventricular zone. Cell Rep. 22, 992–1002. [DOI] [PubMed] [Google Scholar]
- Fiorelli R, Azim K, Fischer B, and Raineteau O (2015). Adding a spatial dimension to postnatal ventricular-subventricular zone neurogenesis. Development 142, 2109–2120. [DOI] [PubMed] [Google Scholar]
- Fotovati A, Abu-Ali S, Wang PS, Deleyrolle LP, Lee C, Triscott J, Chen JY, Franciosi S, Nakamura Y, Sugita Y, et al. (2011). YB-1 bridges neural stem cells and brain tumor-initiating cells via its roles in differentiation and cell growth. Cancer Res. 71, 5569–5578. [DOI] [PubMed] [Google Scholar]
- Friedman BA, Srinivasan K, Ayalon G, Meilandt WJ, Lin H, Huntley MA, Cao Y, Lee SH, Haddick PCG, Ngu H, et al. (2018). Diverse brain myeloid expression profiles reveal distinct microglial activation states and aspects of Alzheimer’s disease not evident in mouse models. Cell Rep. 22, 832–847. [DOI] [PubMed] [Google Scholar]
- Fruchterman TMJ, and Reingold EM (1991). Graph drawing by force-directed placement. J. Softw 21, 1129–1164. [Google Scholar]
- Fuentealba LC, Rompani SB, Parraguez JI, Obernier K, Romero R, Cepko CL, and Alvarez-Buylla A (2015). Embryonic origin of postnatal neural stem cells. Cell 161, 1644–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutachi S, Miya H, Watanabe T, Kawai H, Yamasaki N, Harada Y, Imayoshi I, Nelson M, Nakayama KI, Hirabayashi Y, and Gotoh Y (2015). Slowly dividing neural progenitors are an embryonic origin of adult neural stem cells. Nat. Neurosci 18, 657–665. [DOI] [PubMed] [Google Scholar]
- Ihrie RA, Shah JK, Harwell CC, Levine JH, Guinto CD, Lezameta M, Kriegstein AR, and Alvarez-Buylla A (2011). Persistent sonic hedgehog signaling in adult brain determines neural stem cell positional identity. Neuron 71, 250–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Káradóttir RT, and Kuo CT (2018). Neuronal activity-dependent control of postnatal neurogenesis and gliogenesis. Annu. Rev. Neurosci 41, 139–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai H, Kawaguchi D, Kuebrich BD, Kitamoto T, Yamaguchi M, Gotoh Y, and Furutachi S (2017). Area-specific regulation of quiescent neural stem cells by Notch3 in the adult mouse subependymal zone. J. Neurosci 37, 11867–11880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettenmann H, Hanisch UK, Noda M, and Verkhratsky A (2011). Physiology of microglia. Physiol. Rev 91, 461–553. [DOI] [PubMed] [Google Scholar]
- Kharchenko PV, Silberstein L, and Scadden DT (2014). Bayesian approach to single-cell differential expression analysis. Nat. Methods 11, 740–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohwi M, Osumi N, Rubenstein JL, and Alvarez-Buylla A (2005). Pax6 is required for making specific subpopulations of granule and periglomerular neurons in the olfactory bulb. J. Neurosci 25, 6997–7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev AN, Kim SK, D’Alessio AC, Chenoweth JG, Wittko IM, Campanati L, and McKay RD (2012). Sox2 acts through Sox21 to regulate transcription in pluripotent and differentiated cells. Curr. Biol 22, 1705–1710. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. (2007). Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176. [DOI] [PubMed] [Google Scholar]
- Levine JH, Simonds EF, Bendall SC, Davis KL, Amir el-A.D., Tadmor MD, Litvin O, Fienberg HG, Jager A, Zunder ER, et al. (2015). Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell 162, 184–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorens-Bobadilla E, Zhao S, Baser A, Saiz-Castro G, Zwadlo K, and Martin-Villalba A (2015). Single-cell transcriptomics reveals a population of dormant neural stem cells that become activated upon brain injury. Cell Stem Cell 17, 329–340. [DOI] [PubMed] [Google Scholar]
- López-Juárez A, Howard J, Ullom K, Howard L, Grande A, Pardo A, Waclaw R, Sun YY, Yang D, Kuan CY, et al. (2013). Gsx2 controls region-specific activation of neural stem cells and injury-induced neurogenesis in the adult subventricular zone. Genes Dev. 27, 1272–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luc S, Huang J, McEldoon JL, Somuncular E, Li D, Rhodes C, Ma-moor S, Hou S, Xu J, and Orkin SH (2016). Bcl11a deficiency leads to hematopoietic stem cell defects with an aging-like phenotype. Cell Rep. 16, 3181–3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Coskun V, Liang A, Yu J, Cheng L, Ge W, Shi Z, Zhang K, Li C, Cui Y, et al. (2015). Single-cell transcriptome analyses reveal signals to activate dormant neural stem cells. Cell 161, 1175–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. (2015). Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki T, Liang AC, Miyamoto N, Lo EH, and Arai K (2013). Mechanisms of oligodendrocyte regeneration from ventricular-subventricular zone-derived progenitor cells in white matter diseases. Front. Cell. Neurosci 7, 275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques S, Zeisel A, Codeluppi S, van Bruggen D, Mendanha Falcão A, Xiao L, Li H, Häring M, Hochgerner H, Romanov RA, et al. (2016). Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science 352, 1326–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matcovitch-Natan O, Winter DR, Giladi A, Vargas Aguilar S, Spinrad A, Sarrazin S, Ben-Yehuda H, David E, Zelada González F, Perrin P, et al. (2016). Microglia development follows a stepwise program to regulate brain homeostasis. Science 353, aad8670. [DOI] [PubMed] [Google Scholar]
- Mathys H, Adaikkan C, Gao F, Young JZ, Manet E, Hemberg M, De Jager PL, Ransohoff RM, Regev A, and Tsai LH (2017). Temporal tracking of microglia activation in neurodegeneration at single-cell resolution. Cell Rep. 21, 366–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menn B, Garcia-Verdugo JM, Yaschine C, Gonzalez-Perez O, Rowitch D, and Alvarez-Buylla A (2006). Origin of oligodendrocytes in the subventricular zone of the adult brain. J. Neurosci 26, 7907–7918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle FT, Mirzadeh Z, and Alvarez-Buylla A (2007). Mosaic organization of neural stem cells in the adult brain. Science 317, 381–384. [DOI] [PubMed] [Google Scholar]
- Merkle FT, Fuentealba LC, Sanders TA, Magno L, Kessaris N, and Alvarez-Buylla A (2014). Adult neural stem cells in distinct microdomains generate previously unknown interneuron types. Nat. Neurosci 17, 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzadeh Z, Merkle FT, Soriano-Navarro M, Garcia-Verdugo JM, and Alvarez-Buylla A (2008). Neural stem cells confer unique pinwheel architecture to the ventricular surface in neurogenic regions of the adult brain. Cell Stem Cell 3, 265–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzadeh Z, Doetsch F, Sawamoto K, Wichterle H, and Alvarez-Buylla A (2010). The subventricular zone en-face: wholemount staining and ependymal flow. J. Vis. Exp., 1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nait-Oumesmar B, Decker L, Lachapelle F, Avellana-Adalid V, Bachelin C, and Baron-Van Evercooren A (1999). Progenitor cells of the adult mouse subventricular zone proliferate, migrate and differentiate into oligodendrocytes after demyelination. Eur. J. Neurosci 11, 4357–4366. [DOI] [PubMed] [Google Scholar]
- Nomaru H, Sakumi K, Katogi A, Ohnishi YN, Kajitani K, Tsuchimoto D, Nestler EJ, and Nakabeppu Y (2014). Fosb gene products contribute to excitotoxic microglial activation by regulating the expression of complement C5a receptors in microglia. Glia 62, 1284–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega F, Gascón S, Masserdotti G, Deshpande A, Simon C, Fischer J, Dimou L, Chichung Lie D, Schroeder T, and Berninger B (2013). Oligodendrogliogenic and neurogenic adult subependymal zone neural stem cells constitute distinct lineages and exhibit differential responsiveness to Wnt signalling. Nat. Cell Biol 15, 602–613. [DOI] [PubMed] [Google Scholar]
- Otsuki L, and Brand AH (2017). The vasculature as a neural stem cell niche. Neurobiol. Dis 107, 4–14. [DOI] [PubMed] [Google Scholar]
- Paez-Gonzalez P, Asrican B, Rodriguez E, and Kuo CT (2014). Identification of distinct ChAT+ neurons and activity-dependent control of postnatal SVZ neurogenesis. Nat. Neurosci 17, 934–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul A, Chaker Z, and Doetsch F (2017). Hypothalamic regulation of regionally distinct adult neural stem cells and neurogenesis. Science 356, 1383–1386. [DOI] [PubMed] [Google Scholar]
- Ramos AD, Andersen RE, Liu SJ, Nowakowski TJ, Hong SJ, Gertz C, Salinas RD, Zarabi H, Kriegstein AR, and Lim DA (2015). The long noncoding RNA Pnky regulates neuronal differentiation of embryonic and postnatal neural stem cells. Cell Stem Cell 16, 439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro Xavier AL, Kress BT, Goldman SA, Lacerda de Menezes JR, and Nedergaard M (2015). A distinct population of microglia supports adult neurogenesis in the subventricular zone. J. Neurosci 35, 11848–11861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta J, Grund EM, Silva HM, Lafaille JJ, Fishell G, and Salzer JL (2015). Inhibition of Gli1 mobilizes endogenous neural stem cells for remyelination. Nature 526, 448–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanai N, Tramontin AD, Quiñones-Hinojosa A, Barbaro NM, Gupta N, Kunwar S, Lawton MT, McDermott MW, Parsa AT, Manuel-García Verdugo J, et al. (2004). Unique astrocyte ribbon in adult human brain contains neural stem cells but lacks chain migration. Nature 427, 740–744. [DOI] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, and Regev A (2015). Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol 33, 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhar K, Lapan SW, Whitney IE, Tran NM, Macosko EZ, Kowalczyk M, Adiconis X, Levin JZ, Nemesh J, Goldman M, et al. (2016). Comprehensive classification of retinal bipolar neurons by single-cell transcriptomics. Cell 166, 1308–1323.e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Wang Y, Kokovay E, Lin G, Chuang SM, Goderie SK, Roysam B, and Temple S (2008). Adult SVZ stem cells lie in a vascular niche: a quantitative analysis of niche cell-cell interactions. Cell Stem Cell 3, 289–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Vargas V, Crouch EE, and Doetsch F (2013). Adult neural stem cells and their niche: a dynamic duo during homeostasis, regeneration, and aging. Curr. Opin. Neurobiol 23, 935–942. [DOI] [PubMed] [Google Scholar]
- Sohn J, Orosco L, Guo F, Chung SH, Bannerman P, Mills Ko E, Zarbalis K, Deng W, and Pleasure D (2015). The subventricular zone continues to generate corpus callosum and rostral migratory stream astroglia in normal adult mice. J. Neurosci 35, 3756–3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strikoudis A, Lazaris C, Trimarchi T, Galvao Neto AL, Yang Y, Ntziachristos P, Rothbart S, Buckley S, Dolgalev I, Stadtfeld M, et al. (2016). Regulation of transcriptional elongation in pluripotency and cell differentiation by the PHD-finger protein Phf5a. Nat. Cell Biol 18, 1127–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, and Mesirov JP (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swamydas M, Bessert D, and Skoff R (2009). Sexual dimorphism of oligodendrocytes is mediated by differential regulation of signaling pathways. J. Neurosci. Res 87, 3306–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazoie M, Van der Veken L, Silva-Vargas V, Louissaint M, Colonna L, Zaidi B, Garcia-Verdugo JM, and Doetsch F (2008). A specialized vascular niche for adult neural stem cells. Cell Stem Cell 3, 279–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokumoto Y, Tamaki S, Kabe Y, Takubo K, and Suematsu M (2017). Quiescence of adult oligodendrocyte precursor cells requires thyroid hormone and hypoxia to activate Runx1. Sci. Rep 7, 1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbán N, van den Berg DL, Forget A, Andersen J, Demmers JA, Hunt C, Ayrault O, and Guillemot F (2016). Return to quiescence of mouse neural stem cells by degradation of a proactivation protein. Science 353, 292–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maaten L, and Hinton G (2008). Visualizing data using t-SNE. J. Mach. Learn. Res 9, 2579–2605. [Google Scholar]
- Vanlandewijck M, He L, Mäe MA, Andrae J, Ando K, Del Gaudio F, Nahar K, Lebouvier T, Laviña B, Gouveia L, et al. (2018). A molecular atlas of cell types and zonation in the brain vasculature. Nature 554, 475–480. [DOI] [PubMed] [Google Scholar]
- Wagner F, Yan Y, and Yanai I (2017). K-nearest neighbor smoothing for high-throughput single-cell RNA-seq data. bioRxiv 10.1101/217737. [DOI]
- Weinreb C, Wolock S, and Klein AM (2018). SPRING: a kinetic interface for visualizing high dimensional single-cell expression data. Bioinformatics 34, 1246–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Hernandez M, Shen S, Sabo JK, Kelkar D, Wang J, O’Leary R, Phillips GR, Cate HS, and Casaccia P (2012). Differential modulation of the oligodendrocyte transcriptome by sonic hedgehog and bone morphogenetic protein 4 via opposing effects on histone acetylation. J. Neurosci 32, 6651–6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing YL, Röth PT, Stratton JA, Chuang BH, Danne J, Ellis SL, Ng SW, Kilpatrick TJ, and Merson TD (2014). Adult neural precursor cells from the subventricular zone contribute significantly to oligodendrocyte regeneration and remyelination. J. Neurosci 34, 14128–14146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SZ, Taylor MM, and Bordey A (2011). Neurotransmitters couple brain activity to subventricular zone neurogenesis. Eur. J. Neurosci 33, 1123–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, and Sims PA (2016). An automated microwell platform for large-scale single cell RNA-seq. Sci. Rep 6, 33883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamanian JL, Xu L, Foo LC, Nouri N, Zhou L, Giffard RG, and Barres BA (2012). Genomic analysis of reactive astrogliosis. J. Neurosci 32, 6391–6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Ma MZ, Ren H, Liu Z, Edelman MJ, Pan H, and Mao L (2013). Anti-HDGF targets cancer and cancer stromal stem cells resistant to chemo-therapy. Clin. Cancer Res 19, 3567–3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Nowakowski RS, and Vaccarino FM (2004). Fibroblast growth factor 2 is required for maintaining the neural stem cell pool in the mouse brain subventricular zone. Dev. Neurosci 26, 181–196. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the sequencing data reported in this paper is GEO: GSE109447.