Abstract
Small cell lung cancer (SCLC) accounts for 15% of lung cancer and is almost always linked to inactivating RB1 and TP53 mutations. SCLC frequently responds, albeit briefly, to chemotherapy. The canonical function of the RB1 gene product, pRB, is to repress the E2F transcription factor family. pRB also plays both E2F-dependent and E2F-independent mitotic roles. We performed a synthetic lethal CRISPR/Cas9 screen in an RB1−/− SCLC cell line that conditionally expresses RB1 to identify dependencies that are caused by pRB loss and discovered that RB1−/− SCLC cell lines are hyperdependent on multiple proteins linked to chromosomal segregation, including Aurora B kinase. Moreover, we show that an Aurora B kinase inhibitor is efficacious in multiple preclinical SCLC models at concentrations that are well tolerated in mice. These results suggest that pRB loss is a predictive biomarker for sensitivity to Aurora B kinase inhibitors in SCLC and perhaps other RB1−/− cancers.
Keywords: pRB, RB1, retinoblastoma gene, retinoblastoma protein, Aurora B Kinase, AURKB, Synthetic Lethal, SCLC, small cell lung cancer
Introduction
Small cell lung cancer (SCLC) is a high-grade neuroendocrine cancer that accounts for 15% of lung cancer, with 33,000 new SCLC cases in the U.S. each year [1]. Although 60–70% of patients with extensive-stage (i.e. metastatic) SCLC initially respond to conventional chemotherapy (cisplatin and etoposide), these responses are typically short-lived, and median overall survival is only 9–11 months [1]. There are no approved targeted therapies for SCLC.
Identifying therapeutic targets in SCLC has been challenging, partly because driver mutations in SCLC are primarily loss of function (LOF), typically involving the tumor suppressor genes RB1 and TP53 [2–4]. The importance of these two tumor suppressors in SCLC is underscored by genetically engineered mouse studies, where inactivation of Rb1 and Trp53 in the lung causes SCLC [5, 6]. Although TP53 is highly mutated in many types of human cancer, RB1 is only frequently mutated in SCLC amongst adult cancers [2–4, 7].
The canonical function of pRB is to repress E2F-dependent transcription [8]. pRB operates in a pathway that includes its upstream regulators p16, Cyclin D1, and CDK4, which control pRB phosphorylation and consequently its ability to repress E2F [9]. Many types of cancer have pRB pathway mutations without a strong bias toward any individual pathway component. In contrast, almost all SCLCs harbor RB1 mutations, whereas CDKN2A (p16), CCND1 (Cyclin D1), and CDK4 mutations are conspicuously rare. This suggests a specific, perhaps E2F-independent, role for pRB loss in SCLC pathogenesis that is not shared by its upstream regulators or that loss of these upstream regulators is antithetical to SCLC pathogenesis. With regards to the former possibility, a number of E2F-independent functions have been ascribed to pRB, including a role in maintaining mitotic fidelity [10–13].
Synthetic lethality provides a paradigm for targeting cancers that have sustained loss of function mutations in tumor suppressor genes. In applying this paradigm one looks for specific vulnerabilities that are created upon loss of the gene of interest. The classic example of a successful synthetic lethal approach in cancer is the use of PARP inhibitors to target BRCA-deficient tumors [14]. BRCA-deficient tumor have defects in homologous recombination and become hyperdependent on PARP-dependent collateral DNA repair mechanisms [15]. PARP inhibitors are now approved for treatment of recurrent ovarian cancers and metastatic breast cancers harboring loss of function BRCA mutations [16–18]. It is unknown whether pRB loss confers dependencies (i.e. synthetic lethality) in SCLC. Herein, we used an isogenic cell system and Cas9/CRISPR to identify synthetic lethal targets that result from pRB loss in SCLC.
Results
pRB Loss is Synthetic Lethal with Multiple Genes that Regulate Chromosomal Segregation in SCLC
To identify synthetic lethal interactors with RB1 in SCLC, we first infected two RB1−/− SCLC cell lines (NCI-H82 and NCI-H69) with a lentivirus that expresses pRB in the presence of doxycycline (DOX-On pRB) or with the corresponding empty vector (DOX-On EV). In all our experiments virally infected cells were maintained as polyclonal pools under drug selection suitable for the corresponding virus.
As expected [19, 20], pRB, once reintroduced into NCI-H69 cells, was unphosphorylated and suppressed cell proliferation (Fig. 1A-C). In contrast, exogenous pRB was partially phosphorylated in NCI-H82 cells and did not grossly suppress cell proliferation (Fig. 1A,B and D). We then lentivirally infected the DOX-On pRB and DOX-On EV NCI-H82 cells to express Cas9 and confirmed their ability to edit a GFP reporter plasmid within 13 days of receiving an sgGFP (Supplementary Fig. S1A,B).
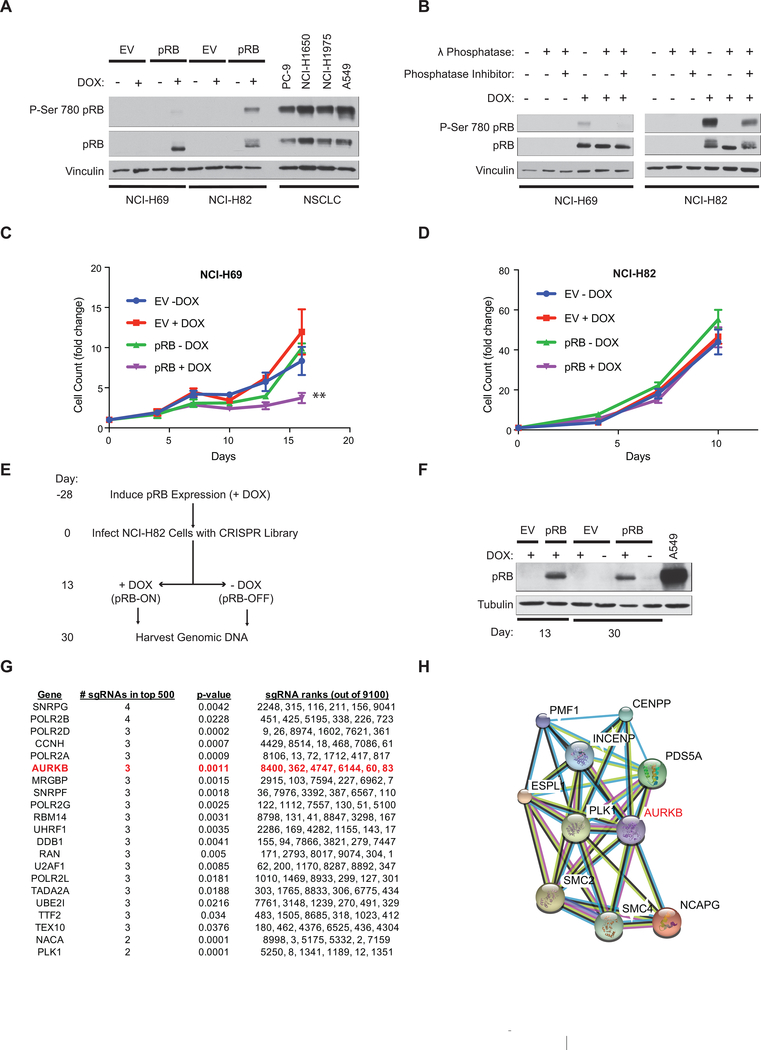
Figure 1: RB1 is synthetic lethal with multiple genes that regulate chromosomal segregation.
A, Immunoblot analysis of NCI-H69 and NCI-H82 RB1−/− small cell lung cancer lines (SCLC) that were infected with a DOX-On pRB or DOX-On EV and then grown in the presence or absence of DOX, as indicated, for 48 hours. Extracts of RB1+/+ NSCLC cell lines (PC-9, NCI-H1650, NCI-H1975, and A549) were included for comparison. B, Immunoblot assays of NCI-H69 (left) and NCI-H82 (right) cells grown as in A. Cell extracts were then treated with λ phosphatase in the presence of absence of a phosphatase inhibitor as indicated. C, D, Cell proliferation of NCI-H69 (C) and NCI-H82 (D) cells grown as in A. The values for each cell line were normalized to a day 0 value of 1. Where indicated DOX was added on day 0. **=p<0.01. E, Schema for the synthetic lethal CRISPR screen. An identical screen was performed with NCI-H82 cells infected with the DOX-On EV as a control. n=3 biological replicates. F, Immunoblot analysis of NCI-H82 cells subjected to the protocol depicted in E. Note pRB reexpression at day 13 before randomization to DOX or NO DOX and persistent pRB expression at day 30 for DOX-On pRB cells maintained in DOX. G, Top 21 synthetic lethal genes as determined by RIGER-E log fold change 2nd best analysis followed by sorting based on the number of sgRNAs that scored in the top 500 of all sgRNAs (9100) in the library. H, Protein-protein interaction network analysis (http://string-db.org/) of statistically significant synthetic lethal hits linked to chromosomal segregation.
Given that pRB reexpression had no gross effect on cell proliferation in NCI-H82 cells, we used the DOX-On pRB NCI-H82 cells to perform a pRB synthetic lethal screen using CRISPR/Cas9-based gene editing. Cas9-positive DOX-On pRB and DOX-On EV cells were grown in the presence of DOX for 28 days and then infected (“Day 0”) (MOI of 0.3) with a lentiviral sgRNA library targeting 1350 epigenetic, cell cycle, and cancer-relevant genes (6 sgRNAs per gene) (Fig. 1E, Supplementary Table S1). The library also contained 1000 non-targeting control sgRNAs. Thirteen days later the cells were split (50:50) into media that did or did not contain DOX, and passaged until day 30. Cell aliquots were removed for genomic DNA (gDNA) isolation at day 13 and at day 30. pRB expression was maintained in the cells continuously exposed to DOX (Fig. 1F).
We used deep sequencing of the gDNA samples and the RIGER-E Log Fold Change Second Best Scoring Algorithm to score the relative depletion or enrichment of the individual sgRNAs in the day 30 samples (plus and minus DOX) compared to the corresponding shared day 13 sample. Multiple sgRNAs targeting genes known to interact with RB1 [CDKN2A, E2F8, EID1, and RB1 itself] were enriched over time in the DOX-On pRB cells (+DOX), but not the DOX-On EV cells (+DOX) (Supplementary Fig. S1C, D and Supplementary Tables S2 & S3, see Methods) implying that the pRB induced by DOX was at least partially active and that our screening metholodogy was technically sound. The recovery of these well annotated pRB-interactors [21, 22] suggest that many of the other genes that scored in this assay are likewise required for SCLC suppression by pRB.
Using this strategy, we also identified 104 genes that were synthetic lethal with RB1, based on sgRNA depletion in the pRB-deficient cells (DOX-On pRB; No DOX) compared to the pRB-proficient cells (DOX-On pRB; +DOX), using a p value cut-off of p<0.05 (Fig. 1G and Supplementary Tables S2 & S4). Many of the top scoring genes (p<0.01) (hereafter called “hits”) had 3 or 4 of their 6 sgRNAs in the library score in the top 500 (~top 5%) of differentially depleted sgRNAs (Fig. 1G). These hits were unlikely to be ‘noise’ as very few non-targeting control sgRNAs were similarly depleted and there were very few hits in the analogous DOX-On EV cell screen (Supplementary Fig. S1E, F). We obtained a very similar list of synthetic lethal hits using STARS [23], which is an orthogonal screen analysis algorithm, suggesting that our hit list was robust and independent of the algorithm chosen for analysis (Supplementary Table S4). Interestingly, many of the hits encode regulators of chromosomal segregation that functionally interact, including components of condensin complexes [SMC2, NCAPG, SMC4] and their upstream regulators (AURKB, PLK1, INCENP) [24–28] (Fig. 1H).
Aurora B Kinase is Synthetic Lethal with pRB in NCI-H82 Cells
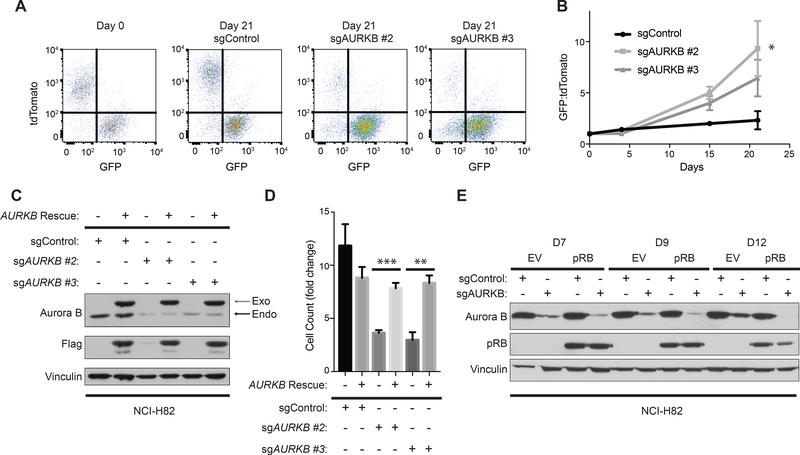
We focused on AURKB because it was the highest scoring “druggable” hit. To test whether AURKB was a true positive synthetic lethal hit, we first performed competition experiments with NCI-H82 cells that were infected to express both pRB and GFP or to express tdTomato alone and then mixed 1:1 (Supplementary Fig. S2A,B). Treating this mixture with a lentivirus expressing one of two effective AURKB sgRNAs caused a dramatic outgrowth of the GFP positive (pRB-proficient) cells compared to the irrelevant sgRNA controls (Fig. 2A,B, Supplementary Fig. S2C, D, E). These effects were on-target because growth inhibition of the parental NCI-H82 cells by the AURKB sgRNAs was completely reversed by a DOX-inducible sgRNA-resistant AURKB variant (Fig. 2C,D).
Figure 2: Genetic inactivation of AURKB is synthetic lethal with RB1 loss in NCI-H82 cells.
A, FACS analysis of NCI-H82 cells infected to produce both pRB and GFP (infected with pRB-IRES-GFP lentivirus) or to produce tdTomato (infected with EV-IRES-tdTomato lentivirus) after being mixed at a 1:1 ratio and infected with a lentivirus expressing Cas9 and the indicated sgRNA. B, Quantification of the GFP:tdTomato ratio at the indicated timepoints of cells treated as in A. n=5 biological replicates. *=p<0.05 of sgAURKB #2 at day 21 compared to sgControl. C, Immunoblot analysis, and D, cellular proliferation assays [based on cell count (fold change relative to day 0)] of NCI-H82 cells that were infected with a Dox-inducible sgRNA-resistant Aurora B kinase cDNA and superinfected with the indicated sgRNAs. The cells were selected in the presence of DOX (see Methods) to maintain expression of the sgRNA-resistant Aurora B Kinase and then grown in the presence or absence of DOX to either maintain or withdraw expression of exogenous Aurora B kinase for 5 days. n=4 biological replicates. **=p<0.01, ***=p<0.001. In C, gray arrow shows exogenous Aurora B kinase and black arrow shows endogenous Aurora B kinase. E, Immunoblot analysis of NCI-H82 cells expressing Cas9 that were superinfected with DOX-On pRB or DOX-On EV (see Supplementary Fig. S1A), grown in the presence of DOX, and then infected with a lentivirus expressing the indicated sgRNA. Cell extracts were harvested at the times indicated after introducing the sgRNAs.
In another variation of this experiment, the Cas9-positive DOX-On pRB and DOX-On EV NCI-H82 cells were grown in the presence of DOX and infected with a lentivirus expressing an AURKB sgRNA or control sgRNA. Notably, there was progressive enrichment of cells that had failed to edit AURKB amongst the EV (pRB-deficient) cells compared to the pRB-proficient cells (Fig. 2E).
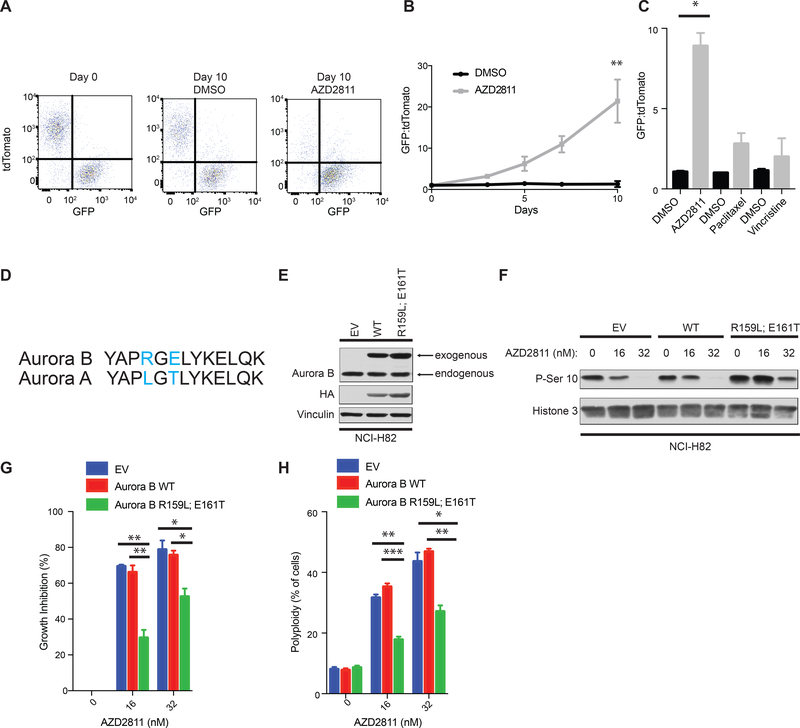
In a complementary set of experiments the GFP (pRB-proficient) and tdTomato (pRB-deficient) cells were again mixed at approximately 1:1 and then treated with the Aurora B kinase specific inhibitor AZD2811 (Supplementary Fig. S3A, B). Treatment with AZD2811 also caused a dramatic enrichment of GFP positive (pRB-proficient) cells compared to the cells treated with DMSO (Fig. 3A,B). Furthermore, AZD2811 did not cause progressive enrichment of GFP positive cells when neither the GFP nor tdTomato cells expressed pRB (Supplementary Fig. S3C) and other antimitotic drugs including paclitaxel, vincristine, the PLK1 inhibitor BI 6727, and the Mps1 inhibitor CFI-402257 did not phenocopy AZD2811 (Fig. 3C and Supplementary Fig. S3D,E, I-L). The targeted sgRNA library we used for our screen did not contain sgRNAs targeting the Aurora B kinase paralog Aurora A kinase. The Aurora A kinase specific inhibitor MK-5108 [29] (Supplementary Fig. S3B, M) selectively depleted RB1−/− cells in both short-term assays and long-term mixing assays (Supplementary Fig. S3N,O), although in competition experiments its effects were less pronounced than the Aurora B kinase inhibitor AZD2811 (compare Fig. 3B to Fig. S3N).
Figure 3: Pharmacological inhibition of Aurora B kinase is synthetic lethal with RB1 loss in NCI-H82 cells.
A, FACS analysis of NCI-H82 cells infected to produce both pRB and GFP or to produce tdTomato after being mixed at a 1:1 ratio and treated with 16 nM AZD2811or DMSO for the indicated number of days. B, Quantification of the GFP:tdTomato ratio at the indicated timepoints of cells treated as in A. n=5 biological replicates. **=p<0.01. C, Quantitation of the GFP:tdTomato ratio on day 8 of NCI-H82 cells as in A treated with either 16 nM AZD2811, 8 nM paclitaxel, 2 nM vincristine, or DMSO. n=3 biological replicates. *=p<0.05. D, Alignment of human Aurora B kinase residues 156–168 with the corresponding region of Aurora A kinase. E161 in Aurora B (blue) forms a hydrogen bound with AZD2811 and is not conserved in Aurora A [30]. A nearby residue R159 in Aurora B (blue) is also not conserved in Aurora A. Given the high selectivity of AZD2811 for Aurora B relative to Aurora A, we mutated both of these residues in Aurora B to the residues found in Aurora A and made an AZD2811 drug-resistant Aurora B kinase mutant (R159L; E161T). E, F, Immunoblot analysis of NCI-H82 infected with a lentivirus that constitutively expresses Aurora B kinase [wild-type (WT)], Aurora B kinase (R159L; E161T), or the empty vector. In F, the cells were treated with the indicated concentrations of AZD2811 for 24 hours. G, Growth inhibition (%) based on viable cell numbers relative to untreated (DMSO) controls and H, Quantification of % polyploidy as determined by FACS of propidium iodide stained cells as in E, that were treated with AZD2811 for 72 hours. n=3 biological replicates. *=p<0.05, **=p<0.01.
To ask if the effects of AZD2811 were on-target, we exploited the fact that the biochemical and cellular IC50s for AZD2811 against Aurora A are 1000-fold higher than the corresponding values for Aurora B [30]. We then changed two residues in Aurora B that differ with Aurora A in the region that contacts AZD2811 [30] (Fig. 3D). The resulting variant, Aurora B R159L; E161T, but not wild-type Aurora B, partially rescued growth inhibition and polyploidy induced by AZD2811 in NCI-H82 cells (Fig. 3E-H; see also Supplementary Fig. S7F-H). Similar results were seen in NCI-H82 cells that were engineered to lack endogenous Aurora B using CRISPR/Cas9 (Supplementary Fig. S3F-H), arguing that the Aurora B R159L; E161T variant was not simply acting as a decoy for endogenous Aurora B. Together, these data show that Aurora B kinase inhibition is synthetic lethal with pRB loss in NCI-H82 cells.
pRB is synthetic lethal with Aurora B Kinase in other SCLC, NSCLC, and breast cancer cell lines
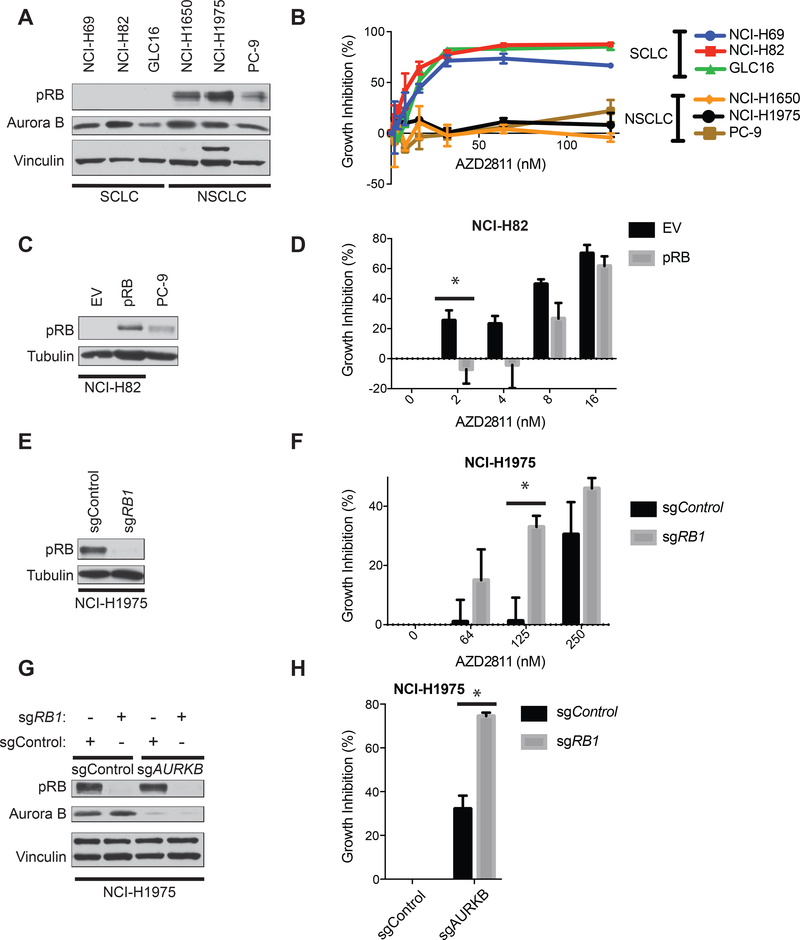
The ability of NCI-H82 cells to proliferate after restoration of pRB function raised the question of whether pRB and Aurora B kinase would be synthetic lethal in other celluar contexts. We therefore asked whether inhibition of AURKB is synthetic lethal with RB1 loss in other SCLC, NSCLC, and breast cancer cell lines. Consistent with our findings in NCI-H82 cells, 3 different RB1−/− SCLC lines (NCI-H69, NCI-H82, and GLC16 cells) were highly sensitive to AZD2811 (IC50 < 50 nM) compared to 3 different RB1+/+ NSCLC lines (NCI-H1650, NCI-1975, and PC-9 cells) (Fig. 4A,B). This differential sensitivity was not due to gross differences in the cell-cycle distribution amongst the 6 cell lines (Supplementary Fig. S4A,D). The pharmacodynamic IC50 values for AZD2811 were lower in RB1−/− SCLC cells than in RB1+/+ NSCLC cells, which likely contributes to the increased sensitivity of the former to AZD2811 in cellular fitness assays (Supplementary Fig. S4E, F). Nonetheless, 125 nM AZD2811 caused nearly complete inhibition of Aurora B kinase in NCI-H1975 and PC-9 cells without suppressing their proliferation (Figure 4B), suggesting that pharmacodynamic differences do not fully account for the striking differential sensitivity of these cell lines to AZD2811.
Figure 4: RB1 is synthetic lethal with AURKB in multiple SCLC and NSCLC cell lines.
A, Immunoblot analysis of the indicated 3 RB1−/− SCLC cell lines and 3 EGFR-mutant RB1+/+ NSCLC cell lines. B, Growth inhibition (%), based on viable cell numbers relative to untreated controls, of the indicated cell lines treated with the AZD2811 for 72 hours. n=3 biological replicates. C, Immunoblot analysis of NCI-H82 cells infected with a lentivirus that constitutively expresses pRB or EV. D, Growth inhibition (%), based on viable cell numbers relative to untreated (DMSO) controls, of the cells in C, treated with the AZD2811 for 48 hours. n=3 biological replicates. *=p<0.05. E, Immunoblot analysis of NCI-H1975 RB1+/+ cells infected with a lentivirus that expresses Cas9 and an sgRNA targeting RB1 [or a non-targeting sgRNA (sgControl)]. F, Growth inhibition (%), based on viable cell numbers relative to untreated (DMSO) controls, of the cells in E, treated with the AZD2811 for 48 hours. n=4 biological replicates. *=p<0.05. G, Immunoblot analysis NCI-H1975 RB1+/+ cells that were first infected with a lentivirus that expresses Cas9 and an sgRNA targeting RB1 or a non-targeting sgRNA (sgControl) and then superinfected with an sgRNA targeting AURKB (sgAURKB) or a non-targeting sgRNA (sgControl) as indicated. H, Growth inhibition (%), based on viable cell numbers relative to sgControl cells in G, after growth in culture for 48 hours. n=2 biological replicates. *=p<0.05.
c-MYC amplification has been correlated with sensitivity of SCLC cell lines to Aurora B kinase inhibitors and the NCI-H82 cells we used in our screen are c-MYC amplified [31–34]. To ask whether c-MYC status was a confounder in our studies we identified multiple RB1−/− cancer cells lines, including NCI-H1876 SCLC cells, and MDA-MB-436 and MDA-MB-468 breast cancer cells, that have low c-MYC expression (Supplementary Fig. S5A) and, where studied, unamplified c-MYC. All of these cell lines were, like NCI-H82 cells, highly sensitive to AZD2811 (Supplementary Fig. S5B) demonstrating that RB1 loss is a marker for increased dependence on AURKB irrespective of c-MYC expression.
We then asked whether pRB status caused the differences in sensitivity to AZD2811 between RB1−/− SCLC lines and RB1+/+ NSCLC lines. Stable reintroduction of pRB into NCI-H82 and GLC16 RB1−/− SCLC cells conferred partial resistance to AZD2811 (Fig. 4C,D and Supplementary Fig. S4G,H), while inactivating RB1 using CRISPR/Cas9 in NCI-H1975 and PC-9 NSCLC cells had the opposite effect (Fig. 4E,F and Supplementary Fig. S4I,J), again without grossly altering cell-cycle distribution (Supplementary Fig. S4B,C). Furthermore, these differences were not caused by increases in MYC expression as a result of pRB loss (Supplementary Fig. S5C, D). As an orthogonal approach, we used CRISPR/Cas9 to generate NCI-H1975 cells deleted for RB1 alone, AURKB alone, or both (Fig. 4G). In keeping with our pharmacological results, RB1 inactivation enhanced the antiproliferative effects of deleting AURKB (Fig. 4H).
RB1-deficient breast cancer cell lines were likewise more sensitive than RB1+/+ breast cancer lines to genetic or pharmacological inhibition of Aurora B kinase (Supplementary Fig. S6A-F). Furthermore, inactivating RB1 in RB1+/+ breast cancer cell lines with an sgRNA or an shRNA increased their sensitivity to various Aurora kinase inhibitors (Supplementary Fig. S6G-J). Therefore RB1 and AURKB display a synthetic lethal relationship in multiple cancer cell lines of different lineages.
pRB Loss Exacerbates Mitotic Abnormalities Caused by Aurora B Kinase Inhibition
Aurora B kinase inhibition causes polyploidy and apoptosis in many cancer cell lines [35]. Two of the three RB1−/− lines (NCI-H82 and GLC16) became polyploid in the presence of low concentrations of AZD2811, while the third SCLC cell line (NCI-H69) arrested with 4N DNA content (Supplementary Fig. S7A), and all underwent time-dependent apoptosis at low concentrations of AZD2811 (Supplementary Fig. S7C). In contrast, these concentrations of AZD2811 did not affect the cell-cycle distribution of the RB1+/+ NSCLC lines and a 4-fold higher concentration of AZD2811 caused only modest effects (Supplementary Fig. S7B).
Strikingly, pRB reexpression in the DOX-On pRB NCI-H82 cells suppressed the polyploidy caused by AZD2811 (Fig. 5A,B & Supplementary Fig. S7G) and decreased AZD2811-induced apoptosis (Fig. 5C,D). This was not because pRB restored Aurora B kinase activity in the face of AZD2811 (Supplementary Fig. S7D,G), prevented cells from entering mitosis (Supplementary Fig. S7E, G), or increased expression of Aurora B or its paralog Aurora A (Supplementary Fig. S7F). Furthermore, the AZD2811-resistant Aurora B kinase (R159L; E161T) suppressed polyploidy to a comparable degree in both pRB-deficient (-DOX) and pRB-proficient (+DOX) cells (Supplementary Fig. S7H). Collectively these results argue that the ability of pRB to suppress the polyploidy caused by AZD2811 is not because pRB alters the ability of AZD2811 to inhibit Aurora B kinase or due to gross alterations in the fraction of cells in M-phase.
Figure 5: pRB loss exacerbates mitotic abnormalities caused by Aurora B kinase inhibition.
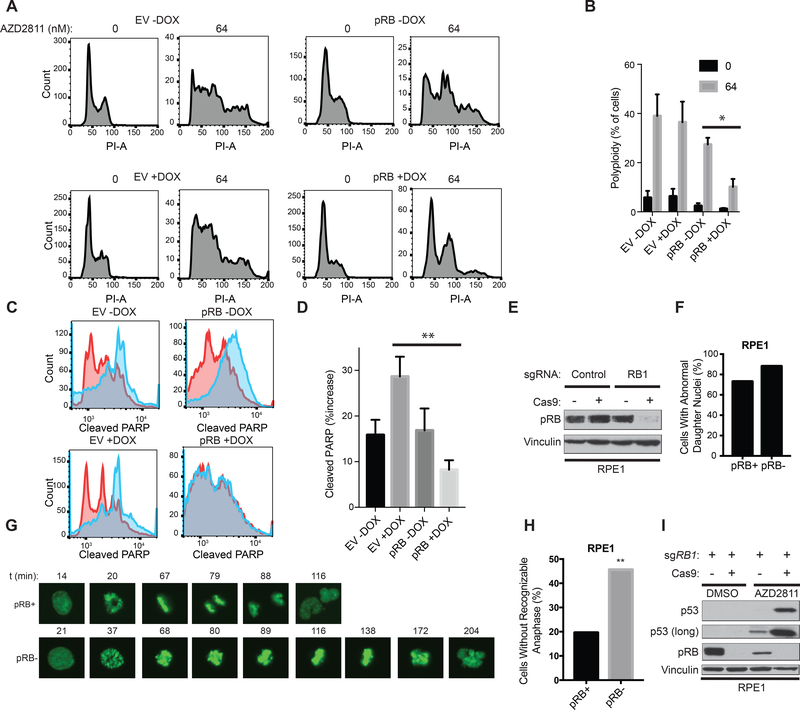
A-D, FACS analysis after propidium iodide (PI) staining (A) or staining with antibody against Cleaved PARP (C) of NCI-H82 RB1−/− SCLC cells that were infected with a DOX-On pRB or DOX-On EV, pretreated in the presence or absence of DOX, as indicted, for 48 hours, and then treated with AZD2811 [blue in (C)] or DMSO [red in (C)] for an additional 48 hours. B, Quantification of % polyploidy as determined by FACS of PI stained cells as in A. n=3 biological replicates. *=p<0.05 comparing pRB -DOX vs. pRB +DOX AZD2811 treated cells. D, Quantification of % increase in cleaved PARP as in C in AZD2811 treated cells relative to the DMSO controls. n=5 biological replicates. **=p<0.01 comparing EV +DOX vs. pRB +DOX AZD2811 treated cells. E, Immunoblot analysis of RPE1 cells expressing GFP-H2B and DOX-On Cas9 that were then infected with a lentivirus expressing an sgRNA targeting RB1 before being grown in the presence (pRB-) or absence (pRB+) of DOX as indicated for 24 hours. F, Quantitation of the % of cells from G that completed mitosis with an abnormally shaped daughter nuclei. G, Representative images from time-lapse fluorescence imaging of pRB+ and pRB- cells in E, that were synchronized in G2 by incubation with R0–3306 (9 μM) for 18 hours and then released into media containing AZD2811 (125 nM) for the times indicated. H, Quantification of % of cells that initiated mitosis, but failed to progress to anaphase, in cells treated as in G. n=51 mitotic cells (pRB+), n=58 mitotic cells (pRB-) from 2 biological replicates. **=p<0.01 using a two-sided chi-square test. I, Immunoblot analysis of RPE1 cells as in E, that were grown in AZD2811 (125 nM) or DMSO for 72 hours.
pRB loss in non-transformed retinal pigment epithelial (RPE1) cells causes chromosomal missegregation and aneuploidy [12, 13]. We next infected RPE1 cells that 1) stably express GFP-H2B to facilitate imaging of chromosomes and 2) express Cas9 in the presence of DOX (see Methods) with a lentivirus expressing an RB1 sgRNA or a control sgRNA (Fig. 5E). We then performed live cell imaging of RB1 sgRNA-infected cells that were grown in the presence (pRB-deficient) or absence (pRB-proficient) of DOX, synchronized in late G2/M using the CDK1 inhibitor R0–3306 [36], and then released into AZD2811 or DMSO. AZD2811 caused mitotic abnormalities in both pRB-proficient and pRB-deficient cells, as most daughter cell nuclei in both conditions were abnormally shaped (Fig. 5F). However, significantly more pRB-deficient cells treated with AZD2811 failed to enter an identifiable anaphase and instead underwent mitotic slippage and induced p53 (Fig. 5G-I).
pRB Status Affects Multiple Genes Involved in the Mitotic Spindle and Chromosomal Segregation in SCLC
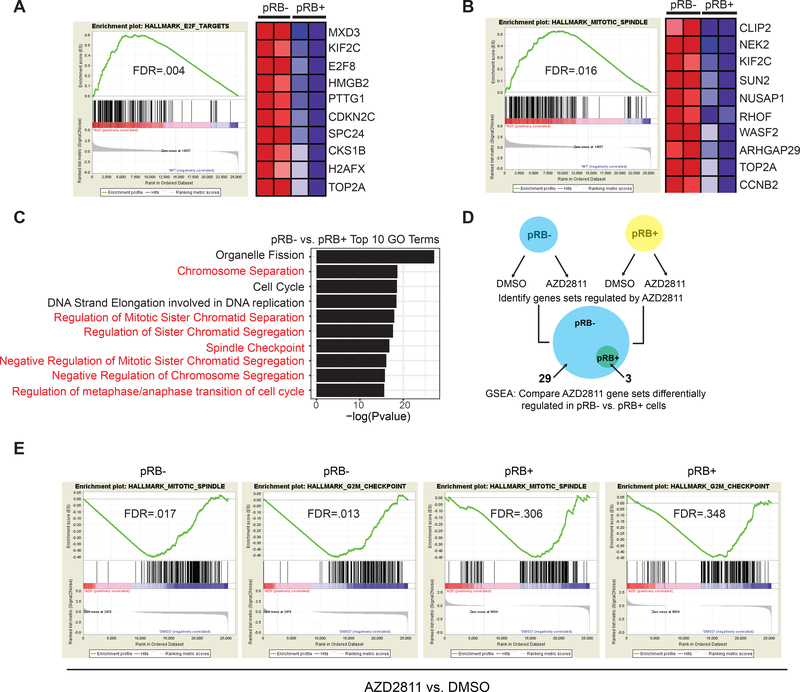
Given that pRB loss exacerbates the mitotic abnormalities caused by Aurora B kinase inhibition (Fig. 5), we next asked whether pRB status affects, directly or indirectly, genes linked to mitosis by performing RNA-Seq with DOX-On pRB NCI-H82 cells that were grown in the presence or absence of DOX for 96 hours (Supplementary Table S5). Despite the partial phosphorylation of exogenous pRB (and absence of an overt cell-cycle block) after restoring pRB expression in these cells, Gene Set Enrichment Analysis (GSEA) identified multiple pRB-regulated gene sets in the DOX-treated cells, including signature E2F target genes (Fig. 6A), consistent with our sgRNA enrichment data described above (Supplementary Fig. S1C). Interestingly, 7 of the 10 most upregulated gene sets in the pRB-deficient cells compared to pRB-proficient cells are linked to mitotic fidelity and chromosome segregation (Fig. 6B,C & Supplementary Table S5).
Figure 6: pRB status affects multiple genes involved in the mitotic spindle and chromosomal segregation in SCLC.
A, B, Gene Set Enrichment Analysis (GSEA) using the hallmarks gene sets of A, E2F Targets and B, Mitotic Spindle genes from an RNA-Seq experiment performed in DOX-On pRB NCI-H82 RB1−/− SCLC cells that were grown in the presence (pRB+) or absence (pRB-) of DOX for 96 hours. The accompanying heatmaps shows the top 10 enriched mRNAs (red) in the respective genes sets in pRB-deficient cells compared to the pRB-proficient cells. C, Top biological processes from Gene Ontology (GO) GSEA analysis enriched in pRB-deficient cells (pRB-) compared to pRB-proficient cells (pRB+) from the RNA-Seq experiment described above. The top 10 statistically significant GO gene sets are shown. Gene sets linked to chromosomal segregation are highlighted in red. For A-C, n=2 biological replicates. D, Schema for the RNA sequencing/GSEA experiment to identify gene sets regulated by AZD2811 in DOX-On NCI-H82 RB1−/− SCLC grown in the presence (pRB+, yellow) or absence (pRB-, blue) of DOX for 48 hours, and then treated with AZD2811 (32 nM) for an additional 48 hours. 29 hallmark gene sets were altered by AZD2811 in pRB- cells while only 3 hallmark gene sets were altered in pRB+ cells. E, GSEA showing behavior of two gene sets linked to mitosis (Mitotic Spindle and G2/M Checkpoint) after treatment with AZD2811 in pRB- compared to pRB+ cells are shown. n=2 biological replicates.
To explore the convergent effects of pRB and Aurora B kinase on mitosis further, we again used RNA-Seq followed by GSEA to assess transcriptional changes, whether direct or indirect, in DOX-On pRB NCI-H82 cells that were grown in the presence or absence of DOX and then treated with AZD2811 or DMSO (Fig. 6D and Supplementary Table S5). AZD2811 induced statistically significant changes in 29 gene sets in pRB-deficient cells. Remarkably, only 3 of these gene sets remained statistically significant in pRB-proficient cells and AZD2811 did not statistically alter any gene sets in pRB-proficient cells that it did not also affect in the pRB-deficient cells (Fig. 6D and Supplementary Table S5). The 26 gene sets that were selectively altered by AZD2811 in pRB-deficient cells included gene sets linked to the G2/M checkpoint and the mitotic spindle (Fig. 6E and Supplementary Table S5). These data show that pRB and Aurora B kinase have partially redundant roles with respect to the control of mitosis in SCLC, possibly explaining their synthetic lethal relationship.
Aurora B Kinase Inhibition has Efficacy in Mouse Models of pRB-Deficient Neuroendocrine Cancers in vivo
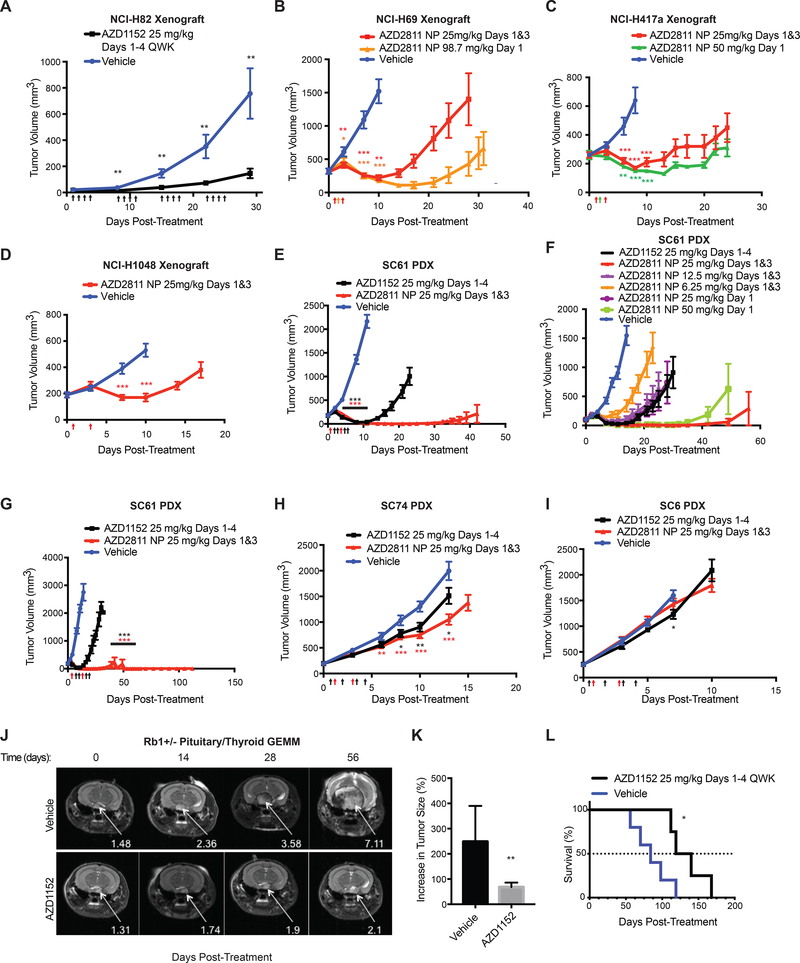
Next we treated several RB1−/− SCLC cell line xenograft models with AZD1152[37] or with AZD2811 encapsulated in a nanoparticle (AZD2811 NP) [38]. AZD2811 NP displays improved pharmacokinetics and efficacy in mouse models compared to AZD1152 [38]. Both AZD1152 and AZD2811 NP were highly active against NCI-H82, NCI-H69, NCI-H417a, and NCI-1048 xenografts (Fig. 7A-D, Supplementary Fig. S8A,B) without causing overt toxicity (Supplementary Fig. S8C-H). Treatment of NCI-H417a xenografts with AZD2811 NP decreased phospho-histone H3 levels, consistent with inhibition of Aurora B kinase activity in vivo, and increased polyploidy and apoptosis (Supplementary Fig. S9A-D). Importantly, NCI-H82 xenografts expressing Aurora B R159L; E161T, but not wild-type Aurora B, were resistant to AZD2811 NP (Supplementary Fig. S9E-G) demonstrating that AZD2811 NP’s anti-tumor effects in vivo were due specifically to Aurora B kinase inhibition.
Figure 7: Aurora B Kinase inhibitors are efficacious in mouse models of RB1−/− neuroendocrine cancers in vivo.
A, Caliper-determined tumor volumes of RB1−/− NCI-H82 xenografts grown subcutaneously in NCr nude mice treated with AZD1152 (25 mg/kg/day given by intraperitoneal (IP) injection on days 1–4, 8–11, 15–18, 22–25) or vehicle (30 mM Tris pH 9). Treatment was initiated when tumors were visible by eye (~20 mm3) (day 1). n=20 tumors (AZD1152) and 20 tumors (vehicle). B-D, Caliper-determined tumor volumes of RB1−/− (B) NCI-H69, (C) NCI-H417a, (D) NCI-H1048 xenografts grown subcutaneously in athymic nude mice treated with AZD2811 NP administered by tail vein injection at the doses indicated or vehicle (0.9% saline for NCI-H69 & NCI-1048, unloaded NP for NCI-H8417a). Treatment was initiated when tumors were ~200 mm3. For B, n=10 tumors per arm. For C, n=12 tumors (vehicle), 6 tumors (AZD2811 NP 25 mg/kg), 6 tumors (50 mg/kg). For D, n=10 tumors per arm. E-I, Caliper-determined tumor volumes of RB1−/− (E, F, G) SC61, (H) SC74, (I) SC6 patient-derived xenografts (PDXs) grown subcutaneously in athymic nude mice treated with AZD2811 NP or AZD1152 administered by tail vein injection at the doses indicated or vehicle (unloaded NP). Treatment was initiated when tumors were ~200 mm3. In G, treatment was a single cycle (2 doses for AZD2811 NP or 4 day osmotic mini-pump for AZD1152) and mice were monitored for 112 days. 8 of 10 mice treated with AZD2811 NP showed no regrowth of tumors during this time. For E,H,I, n=20 tumors per arm. For F, n=at least 12 tumors per arm. For G, n=10 tumors per arm. For A-I, upward arrows on the x-axis denote treatment days. For A-I, *=p<0.05, **=p<0.01, ,***=p<0.001 where indicated comparing AZD1152 or AZD2811 NP to vehicle. For E, p<0.001 for both AZD1152 and AZD2811 NP for days 4, 8, 11. J, Representative MRIs of pituitary tumors arising in Rb1+/− mice that were treated with AZD1152 (25 mg/kg/day given by IP injection given 4 days a week for 8 weeks) or vehicle (30 mM Tris pH 9) at the indicated number of days after initial treatment. Arrows show tumors. K, % increase in size of tumors (thyroid and pituitary) arising in Rb1+/− mice after treatment for 8 weeks with AZD1152 or vehicle. n= 8 tumors (AZD1152) or 6 tumors (vehicle). **p<0.01. L, Kaplan-Meier survival analysis of Rb1+/− mice bearing pituitary or thyroid tumors treated with AZD1152 or vehicle for 8 weeks. n= 4 mice (AZD1152) and 5 mice (vehicle). *=p<0.05.
AZD2811 NP, using a variety of doses and treatment schedules, was also highly active in the RB1−/− SC61 SCLC PDX model (Fig. 7E-G, Supplementary Fig. S8I, S10A-H, and Supplementary Table S6) and, in 8/10 cases, caused sustained complete remissions after only two doses of AZD2811 NP (25 mg/kg on days 1 and 3) (Fig. 7G). AZD2811 NP and AZD1152 also slowed the growth of RB1−/− SCLC SC74 PDX tumors, but were ineffective in the RB1−/− SCLC SC6 SCLC PDX model (Fig. 7H,I, Supplementary Fig. S8J, S10A-H, and Supplementary Table S6). Finally, AZD1152 was active against autochthonous pituitary and thyroid neuroendocrine tumors arising in Rb1+/− mice (Fig. 7J-L and Supplementary Fig. S8K,L,M) [39]. Together, these data show that Aurora B kinase inhibition has efficacy against RB1−/− SCLC cell line xenografts, RB1−/− SCLC PDXs, and autochthonous Rb1−/− neuroendocrine tumors.
Discussion
Our screen unexpectedly revealed differential dependence of pRB-deficient and pRB-proficient cells on genes, such as AURKB and PLK1, that are essential during embryogenesis and usually considered essential in somatic cells [40, 41]. This might have been due to CRISPR/Cas9’s ability to generate hypomorphic alleles, although the AURKB sgRNAs that scored in our primary screen targeted functionally important Aurora B subdomains and also profoundly lowered Aurora B kinase protein levels (Supplementary Fig. S1G). Another factor is that our screen measured relative dependence rather than absolute dependence. The AURKB sgRNAs were lost over time in the pRB-proficient NCI-H82 cells, but were lost more rapidly in the pRB-deficient cells. Thus, while pRB-proficient cells sustain a loss of fitness upon loss of Aurora B, the effect is far more pronounced in pRB-deficient cells. This translates into a quantitative difference in sensitivity to pharmacological inhibition of Aurora B.
Our screen was conducted with a focused sgRNA library and can now be expanded to other libraries. In this regard, our library did not target TSC2 and SKP2, which have been reported to be synthetic lethal with RB1 [42, 43], and did not target the AURKB paralog AURKA, which is also synthetic lethal with RB1 (companion paper by Plowman and coworkers and Supplementary Fig. S3M-O). Another limitation of our study is that the partial phosphorylation of the exogenous pRB in NCI-H82 cells might have masked certain differential dependencies.
pRB represses the transcription of E2F-responsive mitotic genes such as MAD2, CENPE and HEC1 [44]. pRB also directly promotes chromosomal condensation and cohesion and thereby affects chromosomal segregation [10–13, 45, 46]. For example, pRB regulates condensin II localization to chromosomes by binding to CAP-D3 [11], a regulatory subunit of the condensin II complex. pRB also binds to and promotes the activity of the H4K20 methytransferases Suv4–20h1 and Suv4–20h2 [10], which are necessary for binding of cohesin to chromosomes [13]. We documented transcriptional deregulation of multiple mitotic genes in pRB-deficient cells, which was exacerbated further by loss of Aurora B kinase activity. Whether these transcriptional changes are driven by E2F or are an indirect consequence of pRB’s biochemical functions during mitosis described above requires further study. Nonetheless, the fact that multiple mitotic genes scored as synthetic lethal with RB1 underscores the physiological relevance of pRB’s control of mitotic fidelity.
The pRB tumor suppressor pathway includes p16, Cyclin D1, and CDK4, which control pRB phosphorylation and its ability to repress E2F [9]. Many types of cancer have pRB pathway mutations without a strong bias toward any individual pathway component. In stark contrast, SCLCs stereotypically mutate RB1 [2–4]. pRB’s mitotic functions are at least partially CDK-resistant and we found that inactivating RB1 in p16−/− NSCLC cells[47] (Supplementary Fig. S4K) made them hyperdependent on Aurora B. Perhaps pRB loss causes greater E2F derepression than upstream pRB pathway mutations and E2F activity must exceed a certain threshold for SCLC pathogenesis and Aurora B kinase hyperdependence. Alternatively, pRB’s CDK-resistant mitotic functions might suppress SCLC and dependence on Aurora B.
We demonstrated Aurora B hyperdependence in multiple pRB-deficient cell types. It remains possible, however, that the genetic interaction between RB1 and AURKB is influenced by other SCLC driver mutations. For example, c-MYC and Aurora B have a synthetic lethal relationship [34] and c-MYC amplification in SCLC correlates with increased Aurora B dependence [31–33]. Many SCLC tumors and cell lines, including NCI-H82 cells, are c-MYC amplified. However, we observed that RB1−/− cancer cells that did not overexpress c-MYC were still highly sensitive to Aurora B kinase inhibitors. Furthermore, AZD2811 NP was highly active in the SC61 SCLC PDX model, which is c-MYC unamplified, but had virtually no effect on the SC6 SCLC PDX model, which is c-MYC amplified (Supplementary Fig. S10A-H, Supplementary Table S6). Likewise, TP53 mutations, which are nearly universal in SCLC, increase Aurora kinase dependence in colorectal cancer cells [35]. Clearly additional studies will be needed to understand how genetic context influences the dependence of RB1−/− SCLC cells on Aurora B, in part to explain the heterogeneous responses of RB1−/− SCLC tumors we observed preclinically and that are likely to be encountered clinically.
Aurora B kinase inhibitors are bone marrow suppressive and display only modest activity in unselected cancer patients at their maximal tolerated doses. Nonetheless, a recent all-comers Phase 1 trial of the dual Aurora A/B kinase inhibitor Alisertib [35] demonstrated clinical activity in a subset of heavily pretreated SCLCs (20% response rate) and breast cancers[48]. Our findings suggest that the therapeutic index for Aurora B kinase inhibitors would be higher in patients with RB1−/− tumors. In addition to SCLC and pediatric retinoblastomas, RB1 mutations occur in a variety of cancers, including breast cancers, bladder cancers, prostate cancers, and sarcomas, and are also emerging a cause of acquired resistance to targeted agents such as AR antagonists, EGFR antagonists, and CDK4/6 inhibitors. Another way to improve their therapeutic index would be to optimize their biodistribution, such as is being tried with the AZD2811 NP[38].
Methods
Cell Lines and Cell Culture
WERI-Rb-1 (obtained in 6/2016), NCI-H1417 (6/2017), NCI-H1876 (11/2016), 293FT, and hTERT-RPE1 (1/2016) cells were originally obtained from American Type Culture Collection (ATCC). NCI-H69, NCI-H82, GLC16, NCI-H1650, and NCI-H1975 cells were a kind gift from Dr. Kwok-kin Wong’s laboratory (New York University) and were obtained in 8/2014. MDA-MB-436 and MDA-MB-468 cells were a kind gift from AVEO Oncology. PC-9 cells were a kind gift from Dr. Geoff Shapiro’s laboratory (Dana-Farber Cancer Institute) obtained in 11/2014. Cell line authentification was performed (prior to freezing initial early passage stocks) on NCI-H69, NCI-H82, NCI-H1650, and NCI-H1975 cells by Genetica DNA Laboratories in 9/2014 and were found to match the specifications listed in ATCC. NCI-H69, NCI-H82, GLC16, NCI-H1650, NCI-H1975, WERI-Rb-1, NCI-H1417, and PC-9 cells were maintained in RPMI-1640 media. 293FT, MDA-MB-436, and MDA-MB-468 cells were maintained in DMEM media. hTERT-RPE1 and NCI-H1876 cells were maintained in DMEM/F12 media. All media was supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 μg/mL streptomycin except NCI-H1876 cells where the media was supplemented with 5% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin, and HITES [10 nM hydrocortisone, Insulin-Transferrin-Selenium (Sigma), and 10 nM beta-estradiol]. Doubling times for cell lines in Fig. 4A are: NCI-H69 (68 hours), NCI-H82 (26 hours), GLC16 (35 hours), NCI-H1650 (35 hours), NCI-H1975 (41 hours), PC-9 (30 hours). MCF10A TP53–/– cells were purchased from Horizon Discovery and maintained in DMEM/F12 media supplemented with 5% horse serum, 20ng/mL EGF, 0.5mg/mL hydrocortisone, 100ng/mL cholera toxin and 10ug/mL insulin. All cell lines were maintained in 5% CO2 at 37˚C. All cell lines when initially obtained were tested for mycoplasma using the MycoAlert Mycoplasma Detection Kit (Lonzo #LT07–418) and were negative. Early passage cells of all parental cell lines listed above were frozen using Bambanker’s freezing media (Bulldog Bio). Cells were then maintained in culture for <4 months at which point a new early passage vials was thawed. Where indicated, the following chemicals (stored at −20˚C) were also added to the media as indicated in the text: doxycycline (stock 1 mg/mL in H20), AZD2811 (formely AZD1152-HQPA Selleck cat no. #S1147, stock 1 mM in DMSO), paclitaxel (Selleck #S1150, stock 10 mM in DMSO), vincristine (Selleck #S1241, stock 10 mM in DMSO), MK-5108 (Selleck #S2770, stock 10 mM in DMSO), BI 6727 (Selleck #S2235, stock 5 mM in DMSO) , CFI-402257 (MedChem Express #HY-101340, stock 5 mM in DMSO), aphidicolin (Sigma #A0781, stock 3 mM in DMSO), nocodazole (Sigma #M1404, stock 5mM in DMSO), and RO-3306 (Sigma #SML0569, stock 10 mM in DMSO).
Lentivirus and Retrovirus Production
Lentiviruses were made by Lipofectamine 2000-based cotransfection of 293FT cells with the respective lentiviral expression vectors and the packaging plasmids psPAX2 (Addgene #12260) and pMD2.G (Addgene #12259) in a ratio of 2:2:1. Virus-containing supernatant was collected at 48 and 72 hours after transfection, pooled together (15 mL total per 10 cm tissue culture dish), passed through a 0.45 μm filter, aliquoted, and frozen at −80 °C until use.
Retroviruses were made by Lipofectamine 2000 based cotransfection of 293FT with pBABE-H2BGFP plasmid (gift of Fred Dick, Addgene plasmid #26790) along with the pUMVC and pVSV-G packaging plasmids in a ratio of 4:3:1 according to manufacturer’s instructions.
Lentiviral and Retrovirus Infection
Suspension cells were counted using a Vi-Cell XR Cell Counter (Beckman Coulter) and resuspended in 1 mL lentivirus with 5 μg/mL polybrene at the following concentrations in individual wells of a 12 well plate: 1 X 106 cells/mL for NCI-H82 cells, or 2 X106 cells/mL for NCI-H69 and GLC16 cells. The plates were then centrifuged at 434 × g for 2 hours at 30° C. 12–16 hours later the virus was removed and cells were grown for 72 hours before being placed under drug selection.
100,000 adherent cells per well were plated in 6 well plates in 2 mL of media. The following day 500 μl of lentivirus was added together with 8 μg/mL polybrene and the plates were centrifuged as above. The lentivirus was removed the next day and the cells were grown for at least 24 hours before being placed under drug selection. Both suspension and adherent cells were selected by growth in puromycin (1 μg/mL), blasticidin (10 μg/mL), or G418 (800 μg/mL) and maintained in media containing puromycin (1 μg/mL) or blasticidin (10 μg/mL), or G418 (400 μg/mL), respectively.
To make the RPE-1 cell line expressing H2B-GFP, RPE-1 cells were infected with retroviruses encoding H2B-GFP for 24 hours in the presence of 10 μg/mL polybrene, washed, and allowed to recover for 24 h before selection by fluorescence cell sorting.
Cell Proliferation Assays
Cells were counted on day 0 using the Vi-Cell XR Cell Counter and plated in 12 well plates at 200,000 cells/mL in 1 mL of media per well for NCI-H69 cells or petri dishes at 32,500 cells/ml in 8 mLs of media for NCI-H82 cells. Cell counts were then determined using the Vi-Cell XR Cell Counter every 3 days and normalized to day 0.
Growth Inhibition Assays
Cells were plated in RPMI media supplemented with 10% FBS in 6 well plates containing 2 ml of media per well at the following cell densities: NCI-H69 (200,000/mL), NCI-H82 (100,000/mL), GLC16 (200,000/mL), NCI-H1650 (100,000/mL), NCI-H1975 (100,000/mL), PC-9 (100,000/mL), MDA-MB-436 (100,000/mL), MDA-MB-468 (100,000/mL), WERI-Rb-1 (200,000/mL), NCI-H1417 (200,000/mL), NCI-H1876 (500,000/mL). Viable cell counts at the indicated times were determined using a Vi-Cell XR Cell Counter. The percent growth inhibition was calculated as 100 × [1 – the ratio of the treatment sample value/control (DMSO or non-targeting sgRNA) sample value].
MCF10A cell viability assays
500 cells per well were seeded into 384-well plates. After 24 hours cells were exposed to small molecule inhibitors resuspended in DMSO using an Echo 550 liquid handler (Labcyte). Cells were incubated with the inhibitor for five days after which cell viability was estimated using Cell Titre-Glo (Promega). CellTitre-Glo luminescence data was normalized to the signal in DMSO exposed cells to calculate Surviving Fractions (SF). Graphpad Prism software was used to convert SF into four parameter logistic regression dose/response curves.
Lambda Phosphatase Assays
Cell extracts were prepared in EBC buffer without phosphatase inhibitors. 40 μg of protein lysate was treated with λ-phosphatase and/or a phosphatase inhibitor for 1 hour at 30°C using a Lambda Protein Phosphatase kit (NEB #P0753) according to the manufacturer’s instriuctions. Extracts were then boiled in sample buffer, resolved by SDS-PAGE, and analyzed by immunoblotting as described in Supplementary Methods.
GFP Reporter Assay for Cas9 Activity
NCI-H82 DOX-On pRB or EV cells previously infected Lenti-Cas9–2A-Blast were superinfected with a lentivirus (pXPR_011 (Addgene #59702)) expressing GFP that also encodes an sgRNA targeting GFP. Puromycin-resistant cells were analyzed by FACS for GFP. The GFP negative population in the cells transduced with Lenti-Cas9–2A-Blast reflects the % of cells that successfully CRISPR-edited GFP.
sgRNA Library Construction
Gene-targeting sgRNAs and appropriate controls were designed using the rule set described at the GPP portal (http://portals.broadinstitute.org/gpp/public/) [49]. Oligonucleotides were flanked by PCR primer sites and PCR was used to amplify DNA using NEBNext kits. The PCR products were purified using Qiagen PCR cleanup kits and cloned into pXPR_BRD003 using Golden Gate cloning reactions. Pooled libraries were amplified using electro-competent Stbl4 cells. Viruses were generated as outlined at the GPP portal. The sgRNA library targeted epigenetics regulators, cell-cycle genes, and cancer-relevant genes. It consisted of 9100 sgRNAs targeting 1350 genes (6 sgRNAs targeting each gene) and 1000 non-targeting sgRNAs.
Isogenic pRB Synthetic Lethal CRISPR Screen and Analysis
NCI-H82 cells that had been infected with pTripZ-RB1 (or with the corresponding empty vector) were maintained in G418 and then superinfected with Lenti-Cas9–2A-Blast and placed under Blasticidin selection. The successfully infected cells were grown in the presence of 0.5 μg/mL doxycycline (DOX), 10 μg/mL Blasticidin, and 400 μg/mL G418 for 30 days using tetracycline negative FBS (Gemini #100–800).
Prior to the initiation of the screen, a pilot experiment was performed to test the proliferation of DOX-On pRB cells under the conditions that would be used during the screen. Specifically, DOX-On pRB cells that were grown in the presence of DOX for 30 days were then split 50:50 and grown in the presence (pRB-ON) or absence (pRB-OFF) of DOX and cell counts were determined every 3 days over the course of 14 days using a Vi-Cell XR Cell Counter. Doubling times calculated over the 14 day proliferation assay were 40.27 hours for the pRB-ON cells and 40.63 hours for pRB-OFF cells. We concluded the proliferation rates of the 2 arms of the screen would be similar and that differences in proliferation were unlikely to confound our results.
On day 0 (day of infection), ~3 X 107 cells (~3000 cells/sgRNA) were resuspended in complete media with 10% FBS, 5 μg/mL polybrene at concentration of 1 × 106 cells/mL in 50 mL conical tubes and the lentiviral sgRNA expression library described above was added at an multiplicity of infection (MOI) of 0.3. The cells were then distributed onto low adherence 6 well plates at a density of 2 X 106 cells per well and the plates were centrifuged at 434 × g for 2 hours. The following morning (day 1), the virus was removed and the cells were transferred to low adherence 10 cm plates at a concentration of 0.4 X 106 cells/mL and maintained in DOX. On day 3, the cells were transferred to t175 cm non-tissue culture treated flasks at a concentration of 0.2 X 106 cells/mL and maintained in DOX. On day 5, the cells were plated in fresh media in the presence of DOX and selected with 1 μg/mL puromycin for 72 hours.
A parallel experiment was performed on day 5 to determine the MOI. To do this, the cells infected with the sgRNA library and mock-infected cells were plated at 0.2 X 106 cells/mL in low adherence 6-well plates in the presence or absence of puromycin. After 72 hours, cells were counted using the Vi-Cell XR Cell Counter and the MOI was calculated using the following equation: (# of puromycin-resistant cells infected with the sgRNA library/# total cells infected with the sgRNA infected cells) – (# of puromycin-resistant mock-infected cells/ # total mock-infected cells).
After puromycin-selection (day 8), puromycin-resistant cells were then replated with fresh media and grown in the presence of DOX until day 13 after infection. On day 13, 2 X 107 cells were collected, washed in PBS, and frozen for genomic DNA isolation for the initial timepoint prior DOX withdrawal. Cells were then split 1:1 to be grown in the presence (pRB-proficient) or absence (pRB-deficient) of DOX using RPMI supplemented with tetracycline negative FBS and maintaining 3 X 107 cells per condition. Going forward, cells were passaged every 48 hours in the presence or absence of DOX and a minimum of 3 X 107 cells per arm were maintained. At 30 days after infection, 2 X 107 cells were collected, washed in PBS, and frozen for genomic DNA isolation for the final timepoint. An identical screen was performed with NCI-H82 cells expressing Cas9 and an insertless version of the DOX-On lentivirus (pTripZ EV) to control for non-specific effects of our screening strategy. The screen was performed in 3 biological replicates.
Following completion of the screen, genomic DNA (gDNA) was isolated using a Qiagen Genomic DNA midi prep kit (Cat No. #51185) according to the manufacturer’s protocol. PCR of gDNA and pDNA (sgRNA plasmid pool used to generate virus) was performed as previously described [23]. Raw Illumina reads were normalized between samples using: Log2[(sgRNA reads/total reads for sample) ×1e6)+1]. Log fold change (LFC) calculations between isogenic pairs were generated for subsequent RIGER-E analysis. The initial timepoint (day 13) was then subtracted from the end timepoint to determine the relative fold depletion or enrichment of each individual sgRNA after DOX withdrawal. RIGER-E Log Fold Change 2nd Best Algorithm was then used to compare the relative fold-depletion or fold-enrichment for each 2 class comparison.
For the synthetic lethal comparison in Figure 1G and Supplementary Table S4, the log fold change values for pRB-deficient cells (-DOX) at day 30 were compared to the log fold change values for pRB-proficient cells (+DOX) at day 30, and the depletion of sgRNAs was analyzed using the RIGER-E Log Fold Change 2nd Best Algorithm, which ranks the list in order of p-value. For the analysis for enrichment of sgRNAs in pRB-proficient cells in Supplementary Fig. S1C and Supplementary Table S3, the log fold change values for pRB-proficient cells (+DOX) at day 30 were compared to day 13, and the enrichment of sgRNAs was analyzed using the RIGER-E Log Fold Change 2nd Best Algorithm. Although sgRNAs targeting genes that functionally interact with RB1 (Supplementary Fig. S1C) were enriched over time, the magnitude of the log2 fold enrichment for these sgRNAs was low [59 out of 60 sgRNAs of the 10 genes had a log2 fold enrichment <1 over 17 days and only 1 sgRNA out of 60 sgRNAs fell 3 standard deviations above the mean (which is 1.14) of the 1000 non-targeting sgRNAs in the screen]. This result speaks more to the sensitivity of the CRISPR screening technology and demonstrates that our screening technology was able to detect subtle differences in enrichment over time. In particular, our methodology could detect enrichment of sgRNAs that negated pRB’s antiproliferative effects even in a cellular system where pRB did not cause gross antiproliferative effects (see Fig. 1D, 2B, 3B).
For both analyses, the list of hits were then further sorted for the number of sgRNAs (out of 6 sgRNAs in total for each gene) that scored in the top 5%. A p-value cutoff of <0.05 was used to call hits (please see Statistical Analysis section in Methods for how p-values are calculated in RIGER-E). To determine whether the synthetic lethal hits we identified were independent of the algorithm used for screen analysis, we also used the STARS algorithm [23] to reanalyze the data (Supplementary Table S4). The STARS algorithm generated a hit list that was very similar to the hit list we obtained using the RIGER-E Log Fold Change 2nd Best Algorithm. In fact, AURKB scored as the #1 hit in the screen when using the STARS algorithm to analyze median values. The data from all 3 biological replicates was used for all analyses.
The sgRNA library contained 6 sgRNAs targeting AURKB. There were 3 sgRNAs that scored in the top 500 (~top 5%) in the synthetic lethal analysis, which were then labeled as sg1, sg2, sg3. sg2 and sg3 were used for all validation experiments and an Aurora B Kinase sgRNA-resistant cDNA was able to completely rescue the proliferative defects caused by sg2 and sg3, indicating that their anti-proliferative effects were on-target. The other 3 sgRNAs (sg4, sg5, sg6) dropped out in both the presence or absence of exogenous pRB and therefore did not score as synthetic lethal. sg1 (CCAAACTTGCCTTTGCCCAG) targets AURKB nucleotides 247–266 in the catalytic domain and scored at 362 of 9100 sgRNAs, sg2 (TCTAGAGTATGCCCCCCGCG) targets nucleotides 459–478 in the catalytic domain and scored at 60 of 9100 sgRNAs, and sg3 (TTGCCCCTCCCAGATCATGG) targets nucleotides at the intron/exon junction at exon 7 around nucleotide 538 in the catalytic domain and scored at 83 of 9100 sgRNAs. sg4 (ATTCTAGAGTATGCCCCCCG) targets AURKB nucleotides 457–476 in the catalytic domain and scored at 8400 of 9100 sgRNAs, sg5 (CCCTTGCGCCAGTTACCTGT) targets the end of exon 3 in the regulatory domain and scored at 4747 of 9100 sgRNAs, and sg6 (GCTCCTTGTAGAGCTCCCCG) targets nucleotides 477–496 in catalytic domain and scored at 6144 of 9100 sgRNAs.
Pharmacodynamic Studies of AZD2811, MK-5108, BI 6727, and CFI-402257 in vitro
NCI-H82 cells were plated at 100,000 cells/mL on a petri dish and treated with AZD2811, MK-5108, BI 6727, or CFI-402257 at the indicated concentrations for 24 hours. Histones or soluble extracts were prepared and immunoblot analysis was performed as Supplementary Methods.
FACS-based Direct Competition Assay
NCI-H82 cells were infected with pLX304-RB1-IRES-GFP or pLX304-EV-GFP or pLX304-EV-tdTomato lentiviruses, selected with 10 μg/mL of Blasticidin, and then FACS sorted for GFP or tdTomato positive cells. For the competition assay performed with pharmacological inhibitors, cells were mixed at 1:1 ratios (pLX304-EV-tdTomato:pLX304-RB1-GFP ratio) on day 0 (with exact proportions of cells after mixing determined by repeat FACS analysis), and plated on petri dishes at 100,000 cells/mL in 8 mLs of media. Drugs were added to the cells at the concentrations indicated: AZD2811 (16 nM), paclitaxel (8 nM), vincristine (2 nM), or DMSO. The AZD2811, paclitaxel, and vincristine drug concentrations were chosen based on the pharmacodynamic and EC50 assays in Supplementary Fig. S3A, B, D, E. MK-5108, BI 6727, or CFI-402257 were used at the concentrations indicated in Supplementary Fig. S3. 200,000 cells were then harvested every 2–3 days for FACS analysis and the cells were replated in fresh drug in petri dishes again at 100,000 cells/mL.
For the competition assay using CRISPR genetic inhibition, pLX304-RB1-IRES-GFP and pLX304-EV-tdTomato were mixed at 1:1 ratios on day 0 (with exact proportions of cells after mixing determined by repeat FACS analysis), infected with pLentiCRISPR lentiviruses expressing the indicated sgRNAs (sgAURKB #2, sgAURKB #3, or a non-targeting sgRNA), selected with puromycin at 1 μg/mL for 72 hours, and FACS analysis was performed and cells were replated as above.
All samples were analyzed by FACS using a BD-Fortessa. For analysis, at least 10,000 cells were analyzed per sample. Live cells were first gated, doublets were then excluded, and the % of GFP-positive and tdTomato-positive cells were analyzed. The GFP:tdTomato ratio was calculated as a measure of pRB-proficient:pRB-deficient cells in the population. The GFP:tdTomato ratio for each timepoint was normalized to the GFP:tdTomato ratio at day 0 for each experiment. FlowJo was used for analysis.
FACS-based Propidium Iodide (PI) Cell Cycle Analysis
Cells were plated in RPMI media supplemented with 10% FBS at the following cell densities: NCI-H69 (200,000/ml), NCI-H82 (100,000/mL), GLC16 (200,000/mL), NCI-H1650 (50,000/mL), NCI-H1975 (50,000/mL), PC-9 (50,000/mL). 48 hours later, cells were washed once in ice-cold PBS and then fixed in ice-cold 80% ethanol (added dropwise) for at least 2 hours at −20°C. The cells were then centrifuged at 400 × g, washed once in PBS, centrifuged again at 400 × g, and then washed again in PBS containing 2% FBS. Finally, cells were centrifuged at 400 × g and stained with Propodium Iodide (PI) (BD # 550825) for 15 minutes at room temperature. FACS analysis for PI was then performed. For cell cycle experiments in Supplementary Fig. S4A-D, ModFIT was used for quantitative cell-cycle analysis. For cell-cycle experiments following treatment with AZD2811 (Fig. 5 and Supplementary Fig. S7), analysis was performed using FlowJo. % polyploidy was determined by gating on cells with >4N content.
FACS-based Cleaved PARP Apoptosis Analysis
NCI-H82 pTripZ-RB1 or pTripZ-EV cells were plated at 100,000 cells/mL and grown in the presence or absence of DOX for 48 hours. Cells were then treated with 64 nM AZD2811 while being maintained in the presence or absence of DOX for another 48 hours. In Supplementary Fig. S7C, NCI-H69, NCI-H82, GLC16 were plated at 100,000 cells/mL and grown in the presence of 64 nM AZD2811 for the times indicated. Cells were then washed once in ice-cold PBS and then fixed in ice-cold 80% ethanol (added dropwise) for at least 2 hours at −20°C. The cells were then centrifuged at 400 × g, washed once in PBS, centrifuged again at 400 × g, and then washed again in PBS containing 2% FBS. Cells were then incubated with Alexa-647 conjugated cleaved PARP antibody (Cell Signaling, Asp214, D64E10, #6987) per the manufacturer’s instructions at a dilution of 1:50 for 1 hour at room temperature, and then washed twice in PBS containing 2% FBS. FACS for Alexa-647 was then performed and analysis was performed using FlowJo. % increase in cleaved PARP was calculated by subtracting cleaved PARP positive cells in the DMSO treated samples from the AZD2811 treated samples.
FACS-based phospho-histone H3 (Ser10) and Polyploidy Analysis
NCI-H82 pTripZ-RB1 (Dox-On) cells were superinfected with pLX304-CMV-EV, pLX304-CMV-AURKB WT, or pLX304-CMV-AURKB R159L; E161T and selected with blasticidin. Cells were plated at 200,000 cells/mL and grown in the presence or absence of DOX for 48 hours. The cells were then treated with the indicated concentrations of AZD2811 while being maintained in the presence or absence of DOX for another 24 hours. The cells were then washed once in room temperature PBS and then fixed in 4% paraformaldehyde for 15 minutes at room temperature. The cells were then centrifuged at 400 × g at room temperature, washed once in PBS, centrifuged again at 400 × g, and then permeabilized with ice cold methanol at 4° C for 30 minutes. The cells were then washed again in PBS and then incubated with Alexa-647 conjugated phospho-histone H3 (Ser10) antibody (Cell Signaling, #3458) at a dilution of 1:100 for 1 hour at room temperature, then washed once in PBS containing 0.5% BSA, centrifuged at 400 × g, and then stained with Propodium Iodide (PI) (BD # 550825) for 15 minutes at room temperature. FACS analysis was then performed to determine the % positive phospho-histone H3 (Ser 10) cells and cell-cycle distribution. % polyploidy was determined by gating of cells with >4N content and % increase in polyploidy was calculated by subtracting % polyploidy in the DMSO treated samples from % polyploidy in the AZD2811 treated samples.
Aurora B Kinase sgRNA Rescue Experiments
NCI-H82 cells were first infected with a DOX-On pTripZ lentiviral sgRNA-resistant Aurora B Kinase cDNA expression vector, selected with G418, and then superinfected with pLentiCRISPR V2 puromycin-based lentivirus encoding Cas9, a puromycin-resistance gene, and either 1 of 2 independent sgRNAs targeting AURKB (sgAURKB #2 or sgAURKB #3) or a non-targeting sgRNA (control sgRNA). The puromycin-resistant cells were selected with puromycin while being maintained in the presence of DOX at 1 μg/mL for 21 days. The cells were then grown in the presence or absence of DOX using RPMI with tetracycline negative FBS (Gemini #100–800). Immunoblot analysis and cell counts were performed 5 days after DOX withdrawal.
Aurora B Kinase AZD2811 Drug-Resistant Rescue Experiments
Given the high selectivity of AZD2811 for Aurora B relative to Aurora A, we mutated two residues in Aurora B that are near the AZD2811 binding pocket (R159 and E161) to the residues found in Aurora A and made an AZD2811 drug-resistant Aurora B kinase mutant (R159L; E161T). For the experiments performed in Figs. 3E-H, NCI-H82 cells were then first infected with pLenti-EF1α-AURKB R159L; E161T, pLenti-EF1α-AURKB WT or the empty pLenti-EF1α vector lentiviruses and selected with puromycin. These cells were then used for the in vivo xenograft experiments in Supplementary Fig. S9E-G. For the experiments performed in Supplementary Figs. S3F-H, NCI-H82 cells were then first infected with pLX304-CMV-AURKB WT or pLX304-CMV-AURKB R159L; E161T and selected with blasticidin. Blasticidin-resistant cells were then superinfected with pLentiCRISPR expressing the indicated sgRNAs (sgAURKB #3 targets an intron-exon junction in AURKB and therefore only targets endogenous and not exogenous AURKB) and selected with puromycin. Growth inhibition and cell cycle experiments were performed 72 hours after treatment with AZD2811. % polyploidy was determined by gating on cells with >4N content. Pharmacodynamic experiments were performed 24 hours after treatment with AZD2811.
Time-lapse Microscopy
hTERT-RPE-1 cells expressing Dox-inducible Cas9 (a kind gift of Dr. Iain Cheeseman [50]) were superinfected with a retrovirus encoding H2B-GFP and sorted for GFP positive cells by FACS. RPE-1 cells expressing Doxycycline-inducible Cas9 and H2B-GFP were then superinfected with pLentiguide-Blast (a kind gift from Dr. Samuel McBrayer) containing an sgRNA targeting RB1 or a non-targeting sgRNA. Blasticidin-resistant cells were treated with 1 μg/mL DOX for 24 hours and after an additional 72 hours, pRB knockdown was confirmed by immunoblot analysis. Cells were then plated at a density of 30,000 cells per well in a 12 well MatTek plate containing glass coverslips and allowed to adhere overnight. The cells were then incubated in RO3306 (9μM) for 18 hours to synchronize cells in late G2 phase of the cell cycle [36]. The cells were released from RO-3306 by washing cells 5 times in phenol red free DMEM/F12 media for 1 minute per wash and then placed in phenol red free DMEM/F12 media containing either AZD2811 (125 nM) or DMSO. Live cell imaging was performed on a Nikon TI-E inverted widefield microscope equipped with a perfect focus and enclosed within a temperature- and CO2-controlled environment that maintained an atmosphere of 37° C and 5% humidified CO2. GFP Fluorescence and differential interference contrast (DIC) images were captured using Zyla sCMOS camera every 3 minutes with a 20X/0.45 Plan Fluor objective for at least 3 hours or until most cells had completed mitosis. Two independent wells per condition were imaged for each experiment and two biological replicates were performed. Images were analyzed using H2B-GFP to visualize chromosomes for the timing of metaphase and anaphase onset, the presence of recognizable anaphase, and morphological appearance of daughter nuclei.
RNA Sequencing and Gene Set Enrichment Analysis (GSEA)
For the pRB reexpression RNA Sequencing experiment, NCI-H82 pTripZ RB1 cells were plated at 100,000 cells/mL in 8 mls of complete media in petri dishes and grown in the presence or absence of DOX (at 0.5 μg/mL) for 96 hours on petri dishes changing the media and DOX at 48 hours. For the AZD2811 experiment, NCI-H82 pTripz RB1 cells were plated at 100,000 cells/mL and grown in the presence or absence of DOX (at 0.5 μg/ml) for 48 hours and then treated with AZD2811 (32 nM) or DMSO for an additional 48 hours. After 96 hours, RNA was harvested using RNeasy mini kit (Qiagen #74106) and RNA sequencing was performed.
Libraries were prepared using Illumina TruSeq Stranded mRNA sample preparation kits from 500 ng of purified total RNA according to the manufacturer’s protocol. The finished dsDNA libraries were quantified by Qubit fluorometer, Agilent TapeStation 2200, and RT-qPCR using the Kapa Biosystems library quantification kit according to manufacturer’s protocols. Uniquely indexed libraries were pooled in equimolar ratios and sequenced on an Illumina NextSeq500 with single-end 75bp reads by the Dana-Farber Cancer Institute Molecular Biology Core Facilities.
Sequenced reads were aligned to the UCSC hg19 reference genome assembly and gene counts were quantified using STAR (v2.5.1b). Differential expression testing was performed by DESeq2 (v1.10.1) as part of the VIPER analysis pipeline (https://bitbucket.org/cfce/viper/). Normalized read counts (RPKM) were calculated using cufflinks (v2.2.1).
For Gene Set Enrichment Analysis (GSEA), software was downloaded from the Gene Set Enrichment Analysis website [http://www.broad.mit.edu/gsea/downloads.jsp]. GSEA was performed using the ‘Gene-Ontology’ or ‘Hallmark’ gene sets for identification of enriched/depleted signatures. Gene Sets with an FDR<0.25 and a nominal p-value of <0.05 were considered significant.
Generation of SCLC Patient-Derived Xenograft (PDX) Models and Treatment Studies
To generate the SC61 PDX model (derived from a previously untreated primary oat cell SCLC), SC74 PDX model (derived from a previously treated SCLC lymph node metastasis), and SC6 PDX model (derived from a previously untreated SCLC lymph node metastasis), tumors of the same passage were transplanted subcutaneously into 5–10 athymic nude donor mice [51]. When these tumors reached 1000 to 2000 mm3, donor tumors were aseptically excised and viable tumor cut into fragments measuring approximately 20 mm3 and transplanted subcutaneously into experimental athymic nude mice. Growing tumors were recruited to study and allocated to treatment groups [vehicle (placebo nanoparticles (NP))], AZD1152, and/or AZD2811 NP groups) when tumors were in the range 62.5–405 mm3 with 6–10 mice per group. The nanoparticles were diluted to required concentration in 0.9% physiological saline. AZD1152 was diluted to required concentration in 30 mM Tris buffer, pH9. All agents were administered by slow intravenous tail vein injection.
SCLC Cell Line Xenograft Studies
For NCI-H82 xenograft experiments, NCI-H82 parental cells were grown to 108 cells, washed 3 times in sterile PBS, and resuspended at 5 X 107 cells/mL in PBS. The mice were anesthesized with isoflurane and 5 X 106 cells were injected subcutaneously into bilateral flanks of 8 week old NCr nude female mice (Taconic #NCRNU). The mice were monitored daily and when flank tumors were visible by eye (approximately 2 weeks after injection), mice were randomized to treatment with either AZD1152 (Apex Bio #A3214) or vehicle (30 mM Tris pH 9). Intraperitoneal (IP) injections were performed with AZD1152 at 25 mg/kg or vehicle for 4 days a week for 4 weeks. Tumor diameters were measured once a week using calipers during the 4 weeks of treatment at the times indicated and tumor volume was calculated: tumor volume (mm3) = (width)2 × length/2.
For the Aurora B Kinase AZD2811 Drug-Resistant Rescue Experiments in Supplementary Fig. S9E-G, NCI-H82 cells that were infected with pLenti-EF1α-AURKB R159L; E161T or pLenti-EF1α-AURKB WT were injected into 8 week old NCr nude female mice as described above. Once mice developed visible flank tumors, they were randomized to receive either 25 mg/kg of AZD2811 NP or Placebo NP administered by tail vein injection on days 1, 8, 15. Tumors were measured once a week for the 3 weeks of treatment ending on day 22 and tumor volumes were calculated as described above. The tumor volume fold increase at the endpoint on day 22 relative to the day 1 measurement for that tumor was reported.
For the NCI-H417a, NCI-H69 and NCI-H1048 xenograft experiments, cells were cultured in RPMI 1640 (Invitrogen) supplemented with 10% fetal calf serum (Invitrogen) and 1% L-glutamine (Invitrogen). All cell lines were cultured at 37°C in a humidified incubator with 7.5% CO2 for NCI-H417a and NCI-H1048 or 5% CO2 for NCI-H69 cells. NCI-H69 and NCI-H417a are suspension cell lines and NCI-H1048 is an adherent cell line detached using trypsin. All cells were then washed once with PBS and re-constituted in serum-free and glutamine-free RPMI 1640. Cells are injected subcutaneously into the left flank of female athymic nude mice. Cell inoculum was 1 × 107 (NCI-H69), 5 × 106 (NCI-H1048), 1 × 106 (NCI-H417a) in 50% Matrigel. The mice were monitored daily and tumor diameter measured using calipers at the times indicated and tumor volume was calculated: tumor volume (mm3) = (width)2 × length/2. Mice with growing tumors (NCI-69: 195–606 mm3, NCI-H1048: 114–329 mm3, NCI-417a: 165–361 mm3) were randomized to vehicle (placebo NP), AZD1152, and/or AZD2811 NP groups and treatment was administered at the times indicated as described in the PDX method section.
All mouse experiments using NCI-H82 xenografts and Rb1+/− mice that formed pituitary and thyroid tumors complied with National Institutes of Health guidelines and were approved by Dana-Farber Cancer Institute Animal Care and Use Committee (DFCI, protocol 03–105). All mouse experiments using NCI-H417a, NCI-H69 and NCI-H1048 xenografts and SC6, SC61, and SC74 SCLC PDX models were conducted in accordance with U.K. Home Office legislation, the Animal Scientific Procedures Act 1986, as well as the AstraZeneca Global Bioethics policy.
Rb1+/− Genetically-Engineered Mouse Model Treatment Study
Rb1+/− mice (Jackson Laboratory stock number 002102) underwent monthly MRIs beginning at 9 months of age. Once tumors were detected, mice were randomized to treatment with AZD1152 or vehicle (30 mM Tris pH 9) dosed at 25 mg/kg 4 days a week for 8 weeks by IP injection. MRIs were performed every 2 weeks during the treatment course and every 4 weeks thereafter until the mice had significant weight loss (15% or more) or were moribund or distressed at which point they were euthanized. Tumors <2 mm at the start of treatment were included in the final analysis. Median overall survival was determined using the Kaplan-Meier estimate.
Statistical Analysis
For the pRB synthetic lethal CRISPR screen, RIGER-E second best log fold change algorithm was used to perform 2-class comparisons (e.g. pRB-ON vs. pRB-OFF) and determine a rank list of synthetic lethal genes ranked by p-value where statistical significance is p< 0.05. The p-value was assessed empirically against a null distribution created by scrambling the mapping of sgRNAs to genes. The actual score for each gene from the unscrambled real data was compared to the null score distribution; the fraction of scores from the scrambled null distribution that were as good as or better than the observed real score was used to determine the p-value. Please see https://github.com/broadinstitute/rigerj for additional details on RIGER.
For the time-lapse imaging experiments, statistical significance was calculated using a two-sided chi-squared test. For the RNA sequencing experiments, statistical significance with calculated using FDR corrected for multiple hypothesis testing where <0.25 is considered statistically significant.
For the in vivo efficacy experiments in Fig. 7B-I, tumor volume data was analyzed using the Astrazeneca regression tool. Tumor growth inhibition (TGI) from the start of treatment was assessed by comparison of the geometric mean change in tumor volume for the control and treated groups. Tumor regression was calculated as the percentage reduction in tumor volume from the pre-treatment value: % Regression = (1-RTV) × 100% where RTV is the geometric mean relative tumour volume. Statistical significance was evaluated using a one-tailed Student’s t test.
Two-way ANOVA analysis was performed on dose-response curves in Supplementary Fig. S6 and Supplementary Fig. S9A-D.
For all other experiments, statistical significance was calculated using unpaired, two-tailed Students t-test. p-values were considered statistically significant if the p-value was <0.05. For all figures, * indicates p-value <0.05, ** indicates p-value <0.01, *** indicates p-value <0.001, and **** indicates p-value <0.0001. Error bars represent SEM unless otherwise indicated.
Data and Materials Availability
Data generated from the RB1 synthetic lethal CRISPR screen are provided in Supplementary Tables S1–4. Data generated from the RNA-Seq and GSEA analyses are provided in Supplementary Table S5. Mutational/copy number variation status of oncogenic drivers (RB1, TP53, and MYC) in SCLC cell lines and SCLC PDX models are provided in Supplementary Table S6 and Supplementary Fig. S10. All other data and materials can be requested from the corresponding author upon reasonable request.
Supplementary Material
Significance.
Small cell lung cancer is rarely associated with actionable protooncogene mutations. We did a CRISPR/Cas9-based screen that showed that RB1−/− SCLC are hyperdependent on AURKB, likely because both genes control mitotic fidelity, and confirmed that Aurora B kinase inhibitors are efficacious against RB1−/− SCLC tumors in mice at non-toxic doses.
Acknowledgements
W.G. Kaelin is supported by HHMI, the Breast Cancer Research Foundation, and an NCI/NIH R35 grant (no. R35CA210068). M.G. Oser is supported by an NCI/NIH K08 grant (no. K08CA222657) and the Lung Cancer Research Foundation. C.J. Lord is supported by Programme Funding from Breast Cancer Now and Cancer Research UK. C.J. Ryan is a Sir Henry Wellcome Fellow, funded by the Wellcome Trust. D. Pellman is an HHMI investigator and is supported by an NIH grant (no. CA213404–20). A. Spektor is supported by an NCI/NIH K08 grant (no. K08CA208008–01) and the Burroughs Wellcome Fund Career Award for Medical Scientists (CAMS). E. Buss was an HHMI Medical Research Fellow. Special thanks to Xentech for technical help with the mouse experiments, Vidyasagar Koduri, Wenhua Gao, and Gang Lu for generation of destination vectors used for recombination cloning, Zach Herbert and the Molecular Biology Core Facility at DFCI for RNA-Seq analysis, Neil Umbreit for thoughtful discussions, and members of the Kaelin laboratory for critical reading of the manuscript.
Footnotes
Conflict of Interest Statement: W.G. Kaelin is a Lilly, Inc. board director (who is developing Aurora Kinase inhibitors), founder at Tango Therapeutics and Cedilla Therapeutics, and scientific advisor at Nextech Invest. P. Taylor, E.J. Pease., K. Maratea, J. Travers, S. Ashton, and S.T. Barry are Astra-Zeneca employees, who is developing Aurora Kinase inhibitors. M.G. Oser and W.G. Kaelin have a sponsored research agreement with AstraZeneca for future studies inspired by the findings in this manuscript.
References
- 1.Kalemkerian GP, et al. , Small cell lung cancer. J Natl Compr Canc Netw, 2013. 11(1): p. 78–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.George J, et al. , Comprehensive genomic profiles of small cell lung cancer. Nature, 2015. 524(7563): p. 47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peifer M, et al. , Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet, 2012. 44(10): p. 1104–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rudin CM, et al. , Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet, 2012. 44(10): p. 1111–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meuwissen R, et al. , Induction of small cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model. Cancer Cell, 2003. 4(3): p. 181–9. [DOI] [PubMed] [Google Scholar]
- 6.Schaffer BE, et al. , Loss of p130 accelerates tumor development in a mouse model for human small-cell lung carcinoma. Cancer Res, 2010. 70(10): p. 3877–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao J, et al. , Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal, 2013. 6(269): p. pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dick FA and Rubin SM, Molecular mechanisms underlying RB protein function. Nat Rev Mol Cell Biol, 2013. 14(5): p. 297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dyson NJ, RB1: a prototype tumor suppressor and an enigma. Genes Dev, 2016. 30(13): p. 1492–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalo S, et al. , Role of the RB1 family in stabilizing histone methylation at constitutive heterochromatin. Nat Cell Biol, 2005. 7(4): p. 420–8. [DOI] [PubMed] [Google Scholar]
- 11.Longworth MS, et al. , RBF1 promotes chromatin condensation through a conserved interaction with the Condensin II protein dCAP-D3. Genes Dev, 2008. 22(8): p. 1011–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manning AL, Longworth MS, and Dyson NJ, Loss of pRB causes centromere dysfunction and chromosomal instability. Genes Dev, 2010. 24(13): p. 1364–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manning AL, et al. , Suppression of genome instability in pRB-deficient cells by enhancement of chromosome cohesion. Mol Cell, 2014. 53(6): p. 993–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farmer H, et al. , Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature, 2005. 434(7035): p. 917–21. [DOI] [PubMed] [Google Scholar]
- 15.Rouleau M, et al. , PARP inhibition: PARP1 and beyond. Nat Rev Cancer, 2010. 10(4): p. 293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Audeh MW, et al. , Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet, 2010. 376(9737): p. 245–51. [DOI] [PubMed] [Google Scholar]
- 17.Domchek SM, et al. , Efficacy and safety of olaparib monotherapy in germline BRCA1/2 mutation carriers with advanced ovarian cancer and three or more lines of prior therapy. Gynecol Oncol, 2016. 140(2): p. 199–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olaparib for Metastatic Breast Cancer in Patients with a Germline BRCA Mutation. N Engl J Med, 2017. 377(17): p. 1700. [DOI] [PubMed] [Google Scholar]
- 19.Kratzke RA, et al. , RB-mediated tumor suppression of a lung cancer cell line is abrogated by an extract enriched in extracellular matrix. Cell Growth Differ, 1993. 4(8): p. 629–35. [PubMed] [Google Scholar]
- 20.Ookawa K, et al. , Reconstitution of the RB gene suppresses the growth of small-cell lung carcinoma cells carrying multiple genetic alterations. Oncogene, 1993. 8(8): p. 2175–81. [PubMed] [Google Scholar]
- 21.Lai A, et al. , RBP1 recruits the mSIN3-histone deacetylase complex to the pocket of retinoblastoma tumor suppressor family proteins found in limited discrete regions of the nucleus at growth arrest. Mol Cell Biol, 2001. 21(8): p. 2918–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyake S, et al. , Cells degrade a novel inhibitor of differentiation with E1A-like properties upon exiting the cell cycle. Mol Cell Biol, 2000. 20(23): p. 8889–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doench JG, et al. , Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol, 2016. 34(2): p. 184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kagami Y, Ono M, and Yoshida K, Plk1 phosphorylation of CAP-H2 triggers chromosome condensation by condensin II at the early phase of mitosis. Sci Rep, 2017. 7(1): p. 5583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaitna S, et al. , Incenp and an aurora-like kinase form a complex essential for chromosome segregation and efficient completion of cytokinesis. Curr Biol, 2000. 10(19): p. 1172–81. [DOI] [PubMed] [Google Scholar]
- 26.Lipp JJ, et al. , Aurora B controls the association of condensin I but not condensin II with mitotic chromosomes. J Cell Sci, 2007. 120(Pt 7): p. 1245–55. [DOI] [PubMed] [Google Scholar]
- 27.Shao H, et al. , Spatiotemporal dynamics of Aurora B-PLK1-MCAK signaling axis orchestrates kinetochore bi-orientation and faithful chromosome segregation. Sci Rep, 2015. 5: p. 12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thadani R, Uhlmann F, and Heeger S, Condensin, chromatin crossbarring and chromosome condensation. Curr Biol, 2012. 22(23): p. R1012–21. [DOI] [PubMed] [Google Scholar]
- 29.Shimomura T, et al. , MK-5108, a highly selective Aurora-A kinase inhibitor, shows antitumor activity alone and in combination with docetaxel. Mol Cancer Ther, 2010. 9(1): p. 157–66. [DOI] [PubMed] [Google Scholar]
- 30.Sessa F and Villa F, Structure of Aurora B-INCENP in complex with barasertib reveals a potential transinhibitory mechanism. Acta Crystallogr F Struct Biol Commun, 2014. 70(Pt 3): p. 294–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Helfrich BA, et al. , Barasertib (AZD1152), a Small Molecule Aurora B Inhibitor, Inhibits the Growth of SCLC Cell Lines In Vitro and In Vivo. Mol Cancer Ther, 2016. 15(10): p. 2314–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mollaoglu G, et al. , MYC Drives Progression of Small Cell Lung Cancer to a Variant Neuroendocrine Subtype with Vulnerability to Aurora Kinase Inhibition. Cancer Cell, 2017. 31(2): p. 270–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sos ML, et al. , A framework for identification of actionable cancer genome dependencies in small cell lung cancer. Proc Natl Acad Sci U S A, 2012. 109(42): p. 17034–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang D, et al. , Therapeutic potential of a synthetic lethal interaction between the MYC proto-oncogene and inhibition of aurora-B kinase. Proc Natl Acad Sci U S A, 2010. 107(31): p. 13836–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marxer M, et al. , p53 deficiency enhances mitotic arrest and slippage induced by pharmacological inhibition of Aurora kinases. Oncogene, 2014. 33(27): p. 3550–60. [DOI] [PubMed] [Google Scholar]
- 36.Vassilev LT, et al. , Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc Natl Acad Sci U S A, 2006. 103(28): p. 10660–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilkinson RW, et al. , AZD1152, a selective inhibitor of Aurora B kinase, inhibits human tumor xenograft growth by inducing apoptosis. Clin Cancer Res, 2007. 13(12): p. 3682–8. [DOI] [PubMed] [Google Scholar]
- 38.Ashton S, et al. , Aurora kinase inhibitor nanoparticles target tumors with favorable therapeutic index in vivo. Sci Transl Med, 2016. 8(325): p. 325–17. [DOI] [PubMed] [Google Scholar]
- 39.Jacks T, et al. , Effects of an Rb mutation in the mouse. Nature, 1992. 359(6393): p. 295–300. [DOI] [PubMed] [Google Scholar]
- 40.Fernandez-Miranda G, et al. , Genetic disruption of aurora B uncovers an essential role for aurora C during early mammalian development. Development, 2011. 138(13): p. 2661–72. [DOI] [PubMed] [Google Scholar]
- 41.Lu LY, et al. , Polo-like kinase 1 is essential for early embryonic development and tumor suppression. Mol Cell Biol, 2008. 28(22): p. 6870–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li B, et al. , Specific killing of Rb mutant cancer cells by inactivating TSC2. Cancer Cell, 2010. 17(5): p. 469–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao H, et al. , Deletions of Retinoblastoma 1 (Rb1) and Its Repressing Target S Phase Kinase-associated protein 2 (Skp2) Are Synthetic Lethal in Mouse Embryogenesis. J Biol Chem, 2016. 291(19): p. 10201–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Manning AL and Dyson NJ, RB: mitotic implications of a tumour suppressor. Nat Rev Cancer, 2012. 12(3): p. 220–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coschi CH, et al. , Haploinsufficiency of an RB-E2F1-Condensin II complex leads to aberrant replication and aneuploidy. Cancer Discov, 2014. 4(7): p. 840–53. [DOI] [PubMed] [Google Scholar]
- 46.Coschi CH, et al. , Mitotic chromosome condensation mediated by the retinoblastoma protein is tumor-suppressive. Genes Dev, 2010. 24(13): p. 1351–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tam KW, et al. , CDKN2A/p16 inactivation mechanisms and their relationship to smoke exposure and molecular features in non-small-cell lung cancer. J Thorac Oncol, 2013. 8(11): p. 1378–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Melichar B, et al. , Safety and activity of alisertib, an investigational aurora kinase A inhibitor, in patients with breast cancer, small-cell lung cancer, non-small-cell lung cancer, head and neck squamous-cell carcinoma, and gastro-oesophageal adenocarcinoma: a five-arm phase 2 study. Lancet Oncol, 2015. 16(4): p. 395–405. [DOI] [PubMed] [Google Scholar]
- 49.Doench JG, et al. , Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol, 2014. 32(12): p. 1262–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McKinley KL and Cheeseman IM, Large-Scale Analysis of CRISPR/Cas9 Cell-Cycle Knockouts Reveals the Diversity of p53-Dependent Responses to Cell-Cycle Defects. Dev Cell, 2017. 40(4): p. 405–420 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rubio-Viqueira B, et al. , An in vivo platform for translational drug development in pancreatic cancer. Clin Cancer Res, 2006. 12(15): p. 4652–61. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.