Abstract
Intraductal papillary mucinous neoplasms (IPMNs) are precursors to pancreatic cancer; however, little is known about genetic heterogeneity in these lesions. The objective of this study was to characterize genetic heterogeneity in IPMNs at the single-cell level. We isolated single cells from fresh tissue from ten IPMNs, followed by whole genome amplification and targeted next generation sequencing of pancreatic driver genes. We then determined single-cell genotypes using a novel multi-sample mutation calling algorithm. Our analyses revealed that different mutations in the same driver gene frequently occur in the same IPMN. Two IPMNs had multiple mutations in the initiating driver gene KRAS that occurred in unique tumor clones, suggesting the possibility of polyclonal origin or an unidentified initiating event preceding this critical mutation. Multiple mutations in later-occurring driver genes were also common and were frequently localized to unique tumor clones, raising the possibility of convergent evolution of these genetic events in pancreatic tumorigenesis. Single-cell sequencing of IPMNs demonstrated genetic heterogeneity with respect to early and late occurring driver gene mutations, suggesting a more complex pattern of tumor evolution than previously appreciated in these lesions.
Keywords: single-cell sequencing, pancreatic cancer precursor lesion, intraductal papillary mucinous neoplasm, genetic heterogeneity, somatic mutation
INTRODUCTION
Pancreatic cancer is a deadly disease with a dismal prognosis, predicted to be the second leading cause of cancer death in the United States before 2030 [1]. However, pancreatic cancer arises through non-invasive precursor lesions that are curable if detected early enough, representing critical target for early detection approaches. Although the majority of pancreatic ductal adenocarcinomas (PDACs) are thought to arise from microscopic pancreatic intraepithelial neoplasia (PanIN), a significant minority arise through large cystic precursors, most commonly intraductal papillary mucinous neoplasm (IPMN) [2]. Because of their size and resultant earlier diagnosis, IPMNs represent an ideal system in which to study the biology and genetics of pancreatic cancer precursor lesions.
IPMNs are mucin-producing epithelial neoplasms in the pancreatic duct system and are distinguished from PanINs based on their size. IPMNs can be classified based on anatomic location and histologic subtype, but perhaps the most important classification is based on grade of dysplasia: IPMNs are classified into low-grade and high-grade dysplasia based on the degree of architectural and cytological atypia [2]. Multifocal IPMNs are common, and recent data show that the majority of these are independent neoplasms, highlighting the importance of multifocal neoplasia in the pancreas [3].
Decades of cancer research have demonstrated that cancer is a genetic disease caused by the accumulation of somatic mutations in oncogenes and tumor suppressor genes. Previous studies identified six crucial driver genes in pancreatic ductal neoplasia: KRAS, CDKN2A, TP53, and SMAD4, shared in PDACs and IPMNs, as well as GNAS and RNF43 in the IPMN pathway specifically [4]. In IPMNs, previous studies have demonstrated that KRAS and GNAS are likely to be the earliest genetic alterations, while mutations in the remaining genes occur at later stages of tumor progression [4]. Intratumor genetic heterogeneity of somatic mutations has been reported in a number of cancer types [5]. In contrast, recent studies have demonstrated remarkable homogeneity in driver gene alterations in advanced pancreatic cancers, suggesting that intratumor genetic heterogeneity (at least with respect to driver genes) is not a key factor in the later evolution of pancreatic cancer [6]. However, few studies to date have examined genetic heterogeneity in precursor lesions, and fewer still have focused on precursor lesions in the pancreas. Wu et al. assayed somatic mutations in fluid aspirated from IPMNs; they demonstrated that 13% of the IPMNs contained multiple KRAS mutations, and 4% had two different GNAS mutations [7]. This result raises the possibility of a polyclonal origin in a subset of IPMNs, in which an IPMN represents two independent clones that do not share a recent common progenitor. However, analysis of DNA shed into cyst fluid does not allow genotypes to be assigned to specific regions or cells in the IPMNs. Kanda et al. also found that a small percentage of PanINs also have multiple KRAS mutations [8], demonstrating that this genetic heterogeneity is a common feature of pancreatic precursor lesions. A deeper understanding of genetic heterogeneity in precursor lesions will provide insights into the earliest stages of neoplastic evolution in the pancreas, allowing perspectives not possible from the study of advanced cancers.
Computational modeling has previously been applied to infer the clonal composition of tumors, based on variant allele frequency and copy number from next generation sequencing of bulk tumor samples [9]. However, single-cell sequencing resolves individual cellular genotypes and definitively assigns mutations to each clone, providing a more complete understanding of intratumor genetic heterogeneity. Multiple recent studies have reported reliable single-cell sequencing of human cancer specimens [10–13]. However, to our knowledge, the genotypes of individual cells in pancreatic precursor lesions have yet to be examined. In this study, we assess genetic heterogeneity in IPMNs by identifying somatic mutations in single neoplastic cells from fresh IPMN tissue from surgical resections, providing new insights into the clonal structure of these pancreatic cancer precursor lesions.
MATERALS AND METHODS
This study was approved by the Institutional Review Board at The Johns Hopkins Hospital. We obtained fresh neoplastic tissue from ten surgically resected IPMNs. Fragments of cyst wall from one to four different regions of each IPMN were minced, digested, and filtered to make a single-cell suspension. Single cells were then isolated into individual wells of a 96-well plate by fluorescence activated cell sorting (FACS) on a MoFlo Legacy cell sorter (Beckman Coulter, Inc., Miami, FL, USA). We selected epithelial cells through labeling with an anti-EpCam antibody (Abcam, ab112068, 1.5μg/ml, Cambridge, UK), and we selected live cells through exclusion of cells labeling with propidium iodide (PI) (Sigma Aldrich, P4864, 1μg/ml, St. Louis, MO, USA). In addition, as first reported by Wang and colleagues [10], we specifically selected cells that had replicated their DNA to improve the efficiency of the subsequent whole genome amplification – this was accomplished with DyeCycle Violet (Life Technologies, V35003, 5μM, Carlsbad, CA, USA) staining and selection of cells in G2/M phase. The gating scheme for single cell sorting is shown in supplementary material, Figure S1. DNA from each single cell was then separately amplified by multiple displacement amplification (MDA) using isothermal random priming and extension with φ29 polymerase with the REPLI-g Single Cell kit (Qiagen, Hilden, North Rhine-Westphalia, Germany) – this WGA approach leads to higher coverage and lower error rate compared to other amplification approaches [14, 15]. Amplified DNA was quantified by qPCR, and amplification quality was estimated by multiplex PCR for ten separate genomic loci – only cells with successful amplification of 8/10 loci were used for next generation sequencing (supplementary material, Tables S1 and S2) [11, 16, 17]. DNA from each passing single cell was then analyzed by targeted next generation sequencing on the IonTorrent PGM (Life Technologies) following library preparation with a custom AmpliSeq panel incorporating the hotspot or coding regions of 11 key pancreatic driver genes (ARID1A, BRAF, CDKN2A, GNAS, KRAS, MAP2K4, PIK3CA, RNF43, SMAD4, TGFBR2, TP53), as well as the locations of nearby SNPs with high minor allele frequency. Technical controls were processed using the same cell sorting and amplification pipeline, followed by targeted next generation sequencing using the Ion AmpliSeq Cancer Hotspot Panel v2 (Pa01 cell line) (Life Technologies) or our Ion AmpliSeq custom panel of pancreatic drivers described above (normal cells, intraductal oncocytic papillary neoplasm).
We implemented a novel multi-sample somatic mutation calling pipeline that leveraged information from multiple single cells and bulk sequencing and reduced errors due to amplification bias, requiring the combination of several existing tools and development of a new clustering and imputation algorithm to refine results. This custom pipeline was used to call somatic mutations in control and IPMN single-cell and bulk samples and germline variants in normal bulk samples. Details are provided in the supplementary material, Supplementary materials and methods and in supplementary material, Figures S2 and S3. We first applied a standard variant calling method [18–20] to all samples from each case, followed by detailed inspection of the sequencing reads aligned to each site in each sample. Initial variant calls in all samples were made by GATK’s HaplotypeCaller (HC) (Broad Institute, Cambridge, MA, USA) with default parameters [19]. For each IPMN, alternate read count and VAF estimation at all sites where HC detected a variant in any sample was done with the mpileup tool independently for each single-cell sample (supplementary material, Table S3). In each single-cell sample, each variant site was classified as either harboring a somatic mutation (IPMN VAF≥5% and alternate read count ≥ 5, and normal bulk VAF<2% and alternate read count <4, provided that normal bulk coverage ≥100X), consistent with reference (coverage≥100 and VAF<0.2% and alternate read count <2), or indeterminate. We then applied an iterative single-cell genotype clustering algorithm using minimum distance linkage to impute the status of indeterminate mutations [21]. The in-house iterative imputation algorithm that was used to reclassify indeterminate sites leveraged information from multiple samples from the same IPMN (supplementary material, Figure S4). In addition to the coding regions of eleven pancreatic cancer driver genes, our targeted next generation sequencing panel included nearby SNPs with high minor allele frequency, thus likely to be heterozygous in the germline in a large proportion of patients. After determining the germline status of each SNP in the patient’s normal DNA, we removed single cells with fewer than 51% heterozygous SNPs correctly called as heterozygous (supplementary material, Tables S4 and S5). The 51% threshold was determined by standard box-plot techniques (supplementary material, Supplementary materials and methods, supplementary material, Figure S5) [22]. The average allelic dropout (ADO) rate across all single cells was 13% after outlier removal. This process excluded samples with incomplete or inefficient WGA. In addition, cells with no somatic mutations were also excluded, as they likely represented contaminating non-neoplastic cells (supplementary material, Table S1).
RESULTS
Clinicopathological data
Clinicopathological data of the ten cases are summarized in Table 1. All patients had a histologically confirmed diagnosis of IPMN. Of the ten IPMNs analyzed, seven cases were non-invasive (three with low-grade dysplasia and four with high-grade dysplasia), while three had adjacent invasive carcinomas (two invasive ductal adenocarcinomas and one invasive colloid carcinoma). The IPMNs were classified as gastric (7 cases), intestinal (2 cases), and oncocytic (1 case) histological subtypes.
Table 1.
Clinicopathological and molecular data from analyzed IPMNs
| Case | Sex | Age | Location | Size (cm) IPMN(s); carcinoma | Predominant Histological Subtype | Final Diagnosis | Tissue Analyzed | Sections | Mutations identified |
|---|---|---|---|---|---|---|---|---|---|
| IP04 | M | 76 | Head | 4 | Intestinal | IPMN with HGD | IPMN | A: IPMN F: IPMN G: IPMN |
GNAS(p.R201C) |
| IP08 | F | 64 | Tail | 2; 0.8 | Gastric | IPMN with carcinoma (ductal) | IPMN, carcinoma | A: carcinoma B: IPMN C: IPMN |
KRAS(p.G12V), CDKN2A(p.G23Vfs), ARID1A(p.V1825L) |
| IP10 | F | 65 | Body | 3.7 | Intestinal | IPMN with carcinoma (colloid)* | IPMN | B: IPMN C: IPMN |
GNAS(p.R201C), RNF43(p.L12Dfs), CDKN2A(p.G55Afs) |
| IP11 | M | 78 | Head | 1.5 | Gastric | IPMN with LGD+ | IPMN | A: IPMN | KRAS(p.G12D), GNAS(p.R201C) |
| IP12 | M | 59 | Tail | 3.7 | Gastric | IPMN with LGD | IPMN | B: IPMN C: IPMN |
KRAS(p.G12D), ARID1A(p.P70Lfs, p.A826Efs, p.E1786Gfs) |
| IP16 | F | 82 | Head | 1.1, 1.8; 0.2 | Gastric | IPMN with carcinoma (ductal) | IPMN, carcinoma | A: IPMN B: IPMN C: carcinoma |
KRAS(p.G12D, p.G12V, p.G12R), TP53(p.175H, p.C229*), ARID1A(p.T1031I), TGFBR2(p.R232Q), |
| IP20 | F | 85 | Tail | 5.5 | Oncocytic | IPMN with HGD | IPMN | A: IPMN B: IPMN C: IPMN D: IPMN |
None |
| IP22 | M | 65 | Tail | 3.2 | Gastric | IPMN with LGD | IPMN | A: IPMN B: IPMN |
KRAS(p.G12V), GNAS(p.R201C), RNF43(p.G3Sfs, p.V31Dfs, p.C91Y, p.G166V, p.VD299VY, p.H420Pfs), SMAD4 (p.E205K), CDKN2A(p.H83D) |
| IP24 | F | 63 | Head | >1.4# | Gastric | IPMN with HGD+ | IPMN | A: IPMN B: IPMN |
KRAS(p.G12V), GNAS(p.R201C), RNF43(p.A11Lfs, p.T28I, p.G207D) |
| IP27 | M | 67 | Head | 3 | Gastric | IPMN with HGD | IPMN | A: IPMN B: IPMN |
KRAS(p.G12D, p.G12R), GNAS(p.R201H), ARID1A(p.M841Ifs) |
The carcinoma was not identified at the time of specimen processing and was not sampled for this study.
These IPMNs co-occurred with grossly distinct biliary carcinomas that were not sampled for this study.
The IPMN diffusely involved the entire pancreas microscopically – the size in the table reflects largest gross measurement.
Technical validation of single cell sequencing pipeline
In order to validate our single cell pipeline, we used three technical controls to calculate the false positive rate (FPR) (supplementary material, Supplementary materials and methods). In brief, we first performed single cell sorting followed by whole genome amplification and targeted next generation sequencing of the pancreatic cancer cell line Pa01 [23]. Because a normal tissue control was not available for this cell line, we designated all mutations called in the bulk samples as true positives, while any mutation present in the single cells but not the bulk was a false positive – this resulted in a FPR of 2.5 × 10−5 false positives per base pair sequenced. The use of a cell line for technical control has the advantage of genetic homogeneity; however, it does not account for potential errors induced by tissue processing steps. As such, we included two additional technical controls derived from resected human pancreatic samples. First, we isolated single cells from tissue samples from pancreata with pancreatic ductal adenocarcinoma. Due to the low neoplastic cellularity of these tumors, a significant proportion of the isolated cells were derived from normal epithelium. As such, after whole genome amplification and single cell sequencing, we identified single cells that lacked all somatic mutations present in the bulk sample – we deduced that such cells are contaminating normal epithelial cells. We then used the mutations called in these cells to calculate an independent FPR of 3.0 × 10−5 false positives per base pair sequenced. Finally, we used the data from the single cells from an intraductal oncocytic papillary neoplasm (IOPN) to calculate another independent FPR of 3.1 × 10−5 false positives per base pair sequenced. Because IOPNs lack mutations in previously characterized pancreatic driver genes, all mutations called in this lesion in our targeted panel were false positives. Importantly, the FPR is consistent across these three independent technical controls, suggesting that it is protocol-specific and does not vary substantially between different experimental samples. Moreover, the same false positive mutation was not called in multiple single cells in any of the technical controls, highlighting that the mutations identified in our experimental samples are unlikely to be technical artifacts (supplementary material, Table S6).
Somatic mutation calling
The number of somatic mutations called in the ten analyzed IPMNs ranged from 0 to 10 (Table 1, Figures 1–3, supplementary material, Table S7, Figures S4 and S6). Five IPMNs had different mutations in the same driver gene called in different single cells.
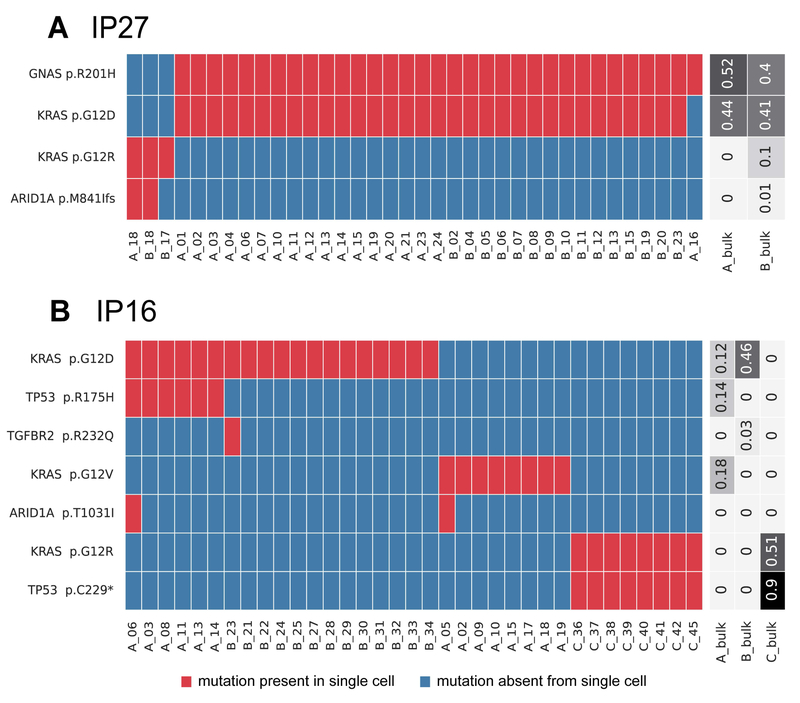
Figure 1. Somatic mutations identified in single cells from IPMNs with multiple KRAS mutations.
Somatic mutations are presented in heatmaps with each row representing a mutation and each column representing a single cell. Single cells are designated by their tissue section (A-C) and cell number. Cells and mutations were clustered with Euclidean distance bi-clustering. The colors indicate the mutation calls after imputation, with red indicating mutant and blue indicating wild-type. Variant allele frequencies of the identified mutations in bulk samples from each section are indicated on the right. Both depicted IPMNs have multiple unique KRAS mutations. The majority of cells in IP27 (A), a gastric-type IPMN with high-grade dysplasia, have p.G12D in KRAS (as well as p.R201H in GNAS), while a small subclone lacks these mutations and instead has p.G12R in KRAS. IP16 (B) represents a gastric-type IPMN (sections A and B) with an adjacent ductal adenocarcinoma (section C). In this case, the IPMN contained two unique and mutually exclusive KRAS mutations, while the cancer had a third KRAS mutation as well as a unique mutation in TP53.
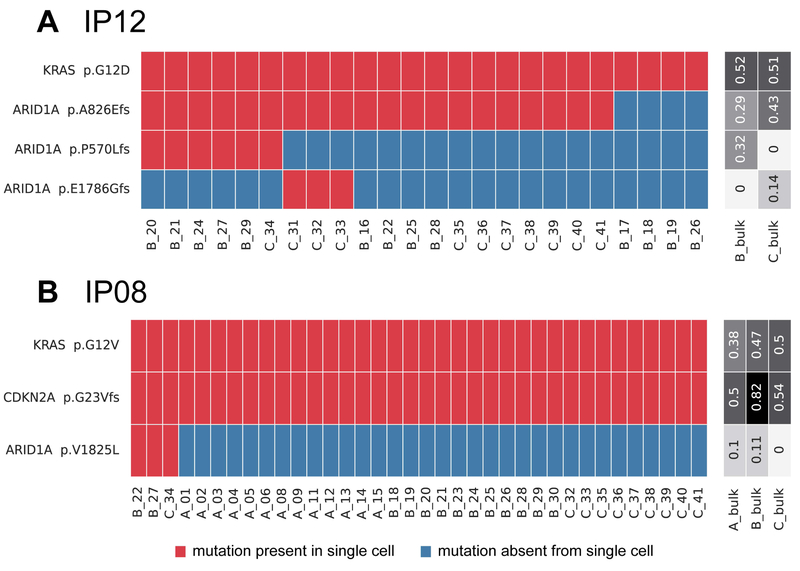
Figure 3. Somatic mutations identified in single cells from IPMNs with subclonal ARID1A mutations.
Somatic mutations are presented in heatmaps with each row representing a mutation and each column representing a single cell. Single cells are designated by their tissue section (A–C) and cell number. Cells and mutations were clustered with Euclidean distance bi-clustering. The colors indicate the mutation calls after imputation, with red indicating mutant and blue indicating wild-type. Variant allele frequencies of the identified mutations in bulk samples from each section are indicated on the right. Both depicted IPMNs have subclonal ARID1A mutations. In IP12 (A), a gastric IPMN with low-grade dysplasia, there was a subclonal inactivating mutation representing a first hit in this tumor suppressor gene as well as two mutually exclusive second hits, each present in non-overlapping subclones. In IP08 (B), an IPMN with high-grade dysplasia, there is a single subclonal ARID1A mutation present in only three single cells.
Early driver gene mutations
Of the ten IPMNs analyzed, nine harbored KRAS and/or GNAS mutations (Table 1). We identified two IPMNs with genetic heterogeneity with respect to these early driver gene mutations. One case (IP27) was diagnosed as an IPMN with high-grade dysplasia - two pieces of the cyst lining were harvested from the same grossly-evident cyst. Two different distinct KRAS mutations were identified in this neoplasm (Figure 1A). The majority of cells that harbored a GNAS p.R201H mutation also had a KRAS p.G12D mutation. Three cells lacked both the GNAS p.R201H and KRAS p.G12D mutations but had a different mutation in KRAS (p.G12R). These data suggest this neoplasm could have originated from two independent clones, each with unique mutations in early driver genes and without any shared genetic alterations. In the second IPMN (IP16), there were two adjacent grossly-evident cysts (samples A and B) as well as a distinct firm area (sample C). Histologic examination revealed IPMN with high-grade dysplasia that was microscopically contiguous between the two cysts (suggesting that they likely represented the same IPMN) and infiltrating ductal adenocarcinoma in the firm area. Two different clones with unique KRAS mutations were identified in the cysts – a clone with KRAS p.G12D was identified in both IPMN samples, while a clone with KRAS p.G12V was limited to a single IPMN sample. A mutation in TP53 (p.R175H) occurred in a subset of cells with KRAS p.G12D. In contrast, the invasive cancer harbored a different KRAS mutation (p.G12R) as well as a unique TP53 mutation (p.C229*) (Figure 1B). These results suggest two independent clones within the IPMNs, as well as a genetically distinct invasive adenocarcinoma, all in a relatively small area of this patient’s pancreas.
In the other seven cases, shared KRAS and/or GNAS mutations were present in the vast majority of neoplastic cells analyzed, suggesting that these were clonal mutations.
Mutations in other driver genes
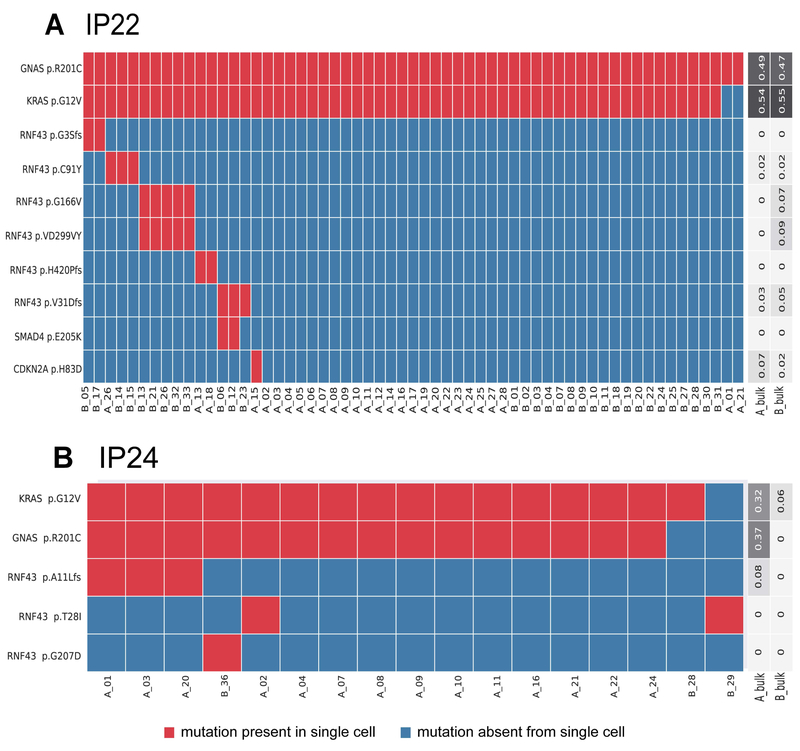
We identified mutations in RNF43 in three IPMNs (Table 1). The first RNF43-mutant case (IP22) was diagnosed as IPMN with low-grade dysplasia and harbored six different RNF43 mutations, including three frameshift insertion/deletion mutations, two missense mutations, and one binucleotide substitution. The binucleotide substitution (p.VD299VY) occurred in the same clone as one missense mutation (p.G166V), suggesting biallelic RNF43 alteration in this clone. The remaining RNF43 mutations were mutually exclusive, suggesting the presence of five separate clones with unique alterations in RNF43 (Figure 2A). In another IPMN with high-grade dysplasia (IP24), we identified three RNF43 mutations each present in different single cell populations: p.A11Lfs occurred in three single cells from section A, while p.G207D occurred in a single cell from section B and p.T28I occurred in one single cell from each tissue section (Figure 2B). In the third RNF43-mutant neoplasm (IP10), which was diagnosed as colloid carcinoma, the RNF43 mutation (p.L12Dfs) occurred with mutations in GNAS (p.R201C) and CDKN2A (p.G55Afs) in all cells analyzed from this IPMN (Supplementary Figure 6). However, we analyzed very few single cells in this case and thus cannot confidently determine the clonality of these mutations.
Figure 2. Somatic mutations identified in single cells from IPMNs with multiple RNF43 mutations.
Somatic mutations are presented in heatmaps with each row representing a mutation and each column representing a single cell. Single cells are designated by their tissue section (A-C) and cell number. Cells and mutations were clustered with Euclidean distance bi-clustering. The colors indicate the mutation calls after imputation, with red indicating mutant and blue indicating wild-type. Variant allele frequencies of the identified mutations in bulk samples from each section are indicated on the right. Both depicted IPMNs have multiple unique RNF43 mutations, with five mutually exclusive subclones with different RNF43 mutations in IP22 (A) and three mutually exclusive RNF43 mutations in IP24 (B).
We also identified an IPMN with low-grade dysplasia (IP12) that harbored three different inactivating ARID1A mutations in addition to a clonal KRAS mutation (Figure 3A). One mutation (p.A826Efs) occurred in 83% of single cells. The other two ARID1A mutations (p.P570Lfs and p.E1786Gfs) were mutually exclusive, occurring in in 25% and 13% of single cells, respectively, and were only present in cells with the first ARID1A alteration. Three other IPMNs, IP16, IP27 (Figure 1), and IP08 (Figure 3B), also had subclonal ARID1A mutations, each present in only two to three single cells.
Mutations in other frequently altered driver genes in pancreatic ductal neoplasia occurred uncommonly in our cohort. Two IPMNs (IP08 and IP10) had clonal mutations in CDKN2A, while IP22 had a subclonal mutation in this gene. Although our cohort included four cases of IPMN with high-grade dysplasia and three cases of IPMN with associated adenocarcinoma, we identified TP53 mutations in only one patient – unique TP53 mutations were identified in IPMN and adjacent carcinoma in IP16. In addition, we only identified single mutations in SMAD4 and in TGFBR2. Intriguingly, the SMAD4 mutation co-occurred with an RNF43 mutation in a subclone of IP22, which had only low-grade dysplasia, while the TGFBR2 mutation occurred in the component IP16 with high-grade dysplasia.
Of note, in the single case of intraductal oncocytic papillary neoplasm (IOPN) (IP20), we did not identify any mutations in the genes in our panel, consistent with recent reports that these neoplasms are genetically distinct from other subtypes of IPMN [24].
DISCUSSION
In this study, we provide the first single-cell genetic analysis of precursor lesions to invasive pancreatic cancer. In addition to demonstrating the feasibility of this type of analysis, these results provide insights into the genetic heterogeneity of early pancreatic tumorigenesis.
The majority of the IPMNs (seven of ten) had KRAS and/or GNAS mutations that were shared by the vast majority of analyzed cells. As the proportion of wild-type cells was similar to our ADO rate, the data suggest that these mutations are clonal in these IPMNs, that is, present in every neoplastic cell. In two of the ten IPMNs, we identified multiple clones with distinct KRAS mutations, suggesting the presence of independent neoplasms or neoplasms in which the shared genetic alteration does not occur in any of the known driver genes. In one IPMN (case IP16), the clone in the invasive carcinoma was genetically distinct from the two clones in the IPMN, consistent with an IPMN with concomitant rather than associated invasive adenocarcinoma [25]. The IPMNs in both IP16 and IP27 had multiple KRAS mutations within the same grossly defined IPMN. In these cases, the two clones were not spatially separated but instead were identified in the same small tissue fragment harvested from the wall of the IPMN. These data could suggest that these IPMNs were polyclonal, made up of multiple clones without a shared truncal genetic alteration. Although multiple KRAS mutations have been described in cyst fluid from IPMNs, this is the first report of single cell genotypes demonstrating this possible polyclonality. An alternative interpretation of these data is that these IPMNs were monoclonal but initiated by an unidentified alteration prior to the development of KRAS mutations. The existence of such an earlier initiating alteration in pancreatic tumorigenesis has not yet been described.
The results of these studies also provide insight into the timing of driver gene mutations in pancreatic tumorigenesis. Like previous studies, our data suggest that mutations in KRAS and GNAS occur very early, as they are clonal in the majority of neoplasms in our study. In the IPMNs in which mutations in these genes were subclonal, there were no clonal mutations in other driver genes, suggesting that KRAS and GNAS are the earliest known driver genes in the pancreas. In contrast, mutations in RNF43 and ARID1A are clearly subclonal in a subset of IPMNs, indicating that these mutations occur after the clonal KRAS or GNAS mutations. The persistence of cells without mutations in these genes highlights the heterogeneous clonal composition of IPMNs, perhaps suggesting that these driver gene mutations only provide slight selective advantage to certain cells within the tumor microenvironment. Of note, rare single cells (such as C_10 in IP10 and B_29 in IP24) lack mutations in KRAS and GNAS but have mutations in other driver genes. We interpret the lack of KRAS/GNAS mutations as a false negative result due to allelic drop out, a known artifact in single-cell sequencing data due to limited starting material. However, we cannot exclude that the identified genotypes are accurate, raising the possibility of a different sequence of driver gene mutations in a subset of cases.
In addition to mixtures of wild-type and mutant cells, our data also demonstrate that a subset of IPMNs consist of mixtures of neoplastic cells with different mutations in the same driver gene. In addition to clonal KRAS and GNAS mutations, IP22 also has five different clones, each with unique mutations in RNF43. Similarly, IP24 has three unique mutually exclusive RNF43 mutations. In contrast, IP12 has three different ARID1A mutations, two of which occurred as mutually exclusive second hits in small subclones of cells with the first ARID1A mutation. These IPMNs provide snapshots of the acquisition of tumor suppressor gene mutations in precursor lesions, suggesting a more complicated process than the sequential acquisition of two “hits”. Moreover, they suggest the presence of convergent evolution in at least a subset of IPMNs, in which mutations in a specific driver are strongly selected at a certain time point in tumorigenesis, resulting in multiple clones independently acquiring unique mutations in the selected gene. The identification of somatic mutations in ARID1A in IPMNs in also novel – although somatic mutations in this well-characterized tumor suppressor gene have been previously reported in PDAC, this is the first report of frequent ARID1A alterations in IPMNs [26].
A few technical considerations in our study are important to note. First, like Wang et al, we specifically isolated cells with replicated DNA (G2/M cells) in order to provide more template and thus improve the efficiency of our single-cell whole genome amplification [10]. Although this approach improves the technical success of our assay, selection of this subset does bias our analysis to proliferating cells. Even with this caveat, our data show that these cells represent a broad spectrum of clones with varying driver gene mutations. Comparison of mutation calls from the bulk and single-cell analyses also provides important insights into the utility of detailed single-cell analysis. Some of the subclonal mutations in RNF43 in IP22 and IP24 were absent from the bulk sequencing data for the respective tissue fragments, despite sequencing at an average coverage of almost 700X in bulk samples. Although our data suggest that single-cell sequencing has an increased sensitivity to detect rare clones, it is possible that ultra-deep sequencing (>1000X) of bulk tissue could achieve a similar sensitivity. Still, an ultra-deep bulk approach cannot assign rare mutations with similar frequency to specific clones, which is the true strength of single-cell analyses. There was also a mutation that was identified in a bulk sample but not in any single cells from that section (p.V31Dfs in RNF43 in section A of IP22). This mutation had very low variant allele frequency in the bulk sample, suggesting that the number of single cells analyzed was likely not adequate to identify the rare cells with this mutation; such sensitivity issues will decrease as technical improvements allow analysis of larger numbers of single cells per sample. Overall, these findings highlight the strength of paired single-cell and bulk analysis of tissue samples, as the approaches are complementary. Finally, our data highlight the importance of assays to evaluate the quality of single-cell DNA amplification. Through our two-step filtering procedure (multiplex PCR, analysis of heterozygous germline SNPs), we restricted our analyses to only the most robustly amplified cells. Although this filtering excluded a significant proportion of the initially sorted cells (57%), it provides confidence in the quality of the data that passed these rigorous filters. In particular, we report a low ADO of 13% in analyzed cells, likely due to our analysis of only the most robustly amplified cells. Moreover, our FPR is consistent with those reported in other single-cell studies, and none of our control samples had false positive mutations that occurred in more than one single cell. This strongly suggests that even the rare subclones identified in our samples are unlikely to be artifacts caused by the extensive amplification required for single-cell sequencing.
Single-cell mutation calling remains an active research area in computational genomics, and the pipeline developed for this data set is novel. We found that currently available protocols to call mutations were not sufficiently optimized to take advantage of our sequencing data, which included IPMN single-cell samples, IPMN bulk samples, and matched normal bulk samples for each case in our study. The analysis pipeline developed in this work combines a standard variant caller designed for high specificity in bulk tissues and enhancements to handle single-cell amplification bias and increase caller sensitivity through multi-sample information pooling. The final set of mutation calls were a product of multiple tools and empirically selected thresholds; slight variations of these thresholds yielded stable results. We also utilized imputation to infer genotypes of cells for which sequencing data was indeterminate. Of note, imputation was not used in our study to identify new mutations but only to resolve indeterminate calls of mutations that had already been convincingly identified in other cells. Thus, this algorithm could only change the proportion of cells within an IPMN with a particular mutation.
Overall, our data provide the first insights into genetic heterogeneity of pancreatic cancer precursors at the single-cell level. Because our study encompassed a limited sample size of ten IPMNs, it is not possible to definitively determine the prevalence of this heterogeneity in patients with IPMN. Analysis of more cells and more lesions will be required to systematically catalogue this genetic heterogeneity and to correlate it with clinical features, such as grade of dysplasia and risk of malignant progression. Moreover, most of the IPMNs analyzed in our study were gastric-type, so our study provides limited insights into heterogeneity in other IPMN subtypes – inclusive cohorts with broad representation of all histological subtypes will be critical in future studies to comprehensively describe the nature and extent of this single-cell genetic heterogeneity in IPMNs. Still, our studies suggest complex patterns of clonal evolution in preinvasive lesions. In addition, more extensive sequencing (such as whole exome sequencing) and identification of different types of alterations (such as copy number changes) will provide a more complete picture of clonal evolution in IPMNs, but our driver-focused approach provides key insights into heterogeneity of alterations that drive pancreatic tumorigenesis. Genetic heterogeneity with respect to these driver genes is most likely to have functional consequences in the heterogeneous clones and thus is likely to have biological importance, providing novel insights into pancreatic tumorigenesis.
Supplementary Material
Figure S1. Gating scheme used for single cell sorting by fluorescence activated cell sorting (FACS)
Figure S2. Computational workflow for mutation calling and error estimation
Figure S3. Thresholds applied for determining mutation status
Figure S4. Somatic mutation calls in single IPMN cells before and after imputation
Figure S5. Outlier analysis of single cell ADO data
Figure S6. Somatic mutations identified in single cells from IP04, IP10, IP11
Table S1. Number of single cells analyzed from each IPMN
Table S2. Primers for multiplex PCR assay to evaluate WGA efficiency
Table S3. Somatic mutations identified in single IPMN cells
Table S4. Germline variant calls utilized for calculation of ADO
Table S5. ADO in each single IPMN cell
Table S6. False-positive mutations identified in single cells from control samples
Table S7. Somatic mutations identified in each IPMN section
Acknowledgements
The authors acknowledge the following sources of funding:
NIH/NCI P50 CA62924; NIH/NIDDK K08 DK107781; Sol Goldman Pancreatic Cancer Research Center; Buffone Family Gastrointestinal Cancer Research Fund; Kaya Tuncer Career Development Award in Gastrointestinal Cancer Prevention; AGA-Bernard Lee Schwartz Foundation Research Scholar Award in Pancreatic Cancer; Sidney Kimmel Foundation for Cancer Research Kimmel Scholar Award; AACR-Incyte Corporation Career Development Award for Pancreatic Cancer Research; Rolfe Pancreatic Cancer Foundation; Joseph C Monastra Foundation; The Gerald O Mann Charitable Foundation (Harriet and Allan Wulfstat, Trustees); Sigma Beta Sorority
Footnotes
Conflict of Interest
LDW is a paid consultant for Personal Genome Diagnostics. EDT receives salary support from Bristol-Meyers Squibb. The other authors report no conflict of interest.
SUPPLEMENTARY MATERIAL ONLINE
Supplementary materials and methods YES
Supplementary figure legends NO, because all legends are shown below their Suppl Figure in the single PDF.
References
- 1.Rahib L, Smith BD, Aizenberg R, et al. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res 2014; 74: 2913–2921. [DOI] [PubMed] [Google Scholar]
- 2.Basturk O, Hong SM, Wood LD, et al. A revised classification system and recommendations from the Baltimore consensus meeting for neoplastic precursor lesions in the pancreas. Am J Surg Pathol 2015; 39: 1730–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pea A, Yu J, Rezaee N, et al. Targeted DNA sequencing reveals patterns of local progression in the pancreatic remnant following resection of Intraductal Papillary Mucinous Neoplasm (IPMN) of the pancreas. Ann Surg 2017; 266: 133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Felsenstein M, Hruban RH, Wood LD. New developments in the molecular mechanisms of pancreatic tumorigenesis. Adv Anat Pathol 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGranahan N, Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell 2017; 168: 613–628. [DOI] [PubMed] [Google Scholar]
- 6.Makohon-Moore AP, Zhang M, Reiter JG, et al. Limited heterogeneity of known driver gene mutations among the metastases of individual patients with pancreatic cancer. Nat Genet 2017; 49: 358–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu J, Matthaei H, Maitra A, et al. Recurrent GNAS mutations define an unexpected pathway for pancreatic cyst development. Sci Transl Med 2011; 3: 92ra66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanda M, Matthaei H, Wu J, et al. Presence of somatic mutations in most early-stage pancreatic intraepithelial neoplasia. Gastroenterology 2012; 142: 730–733 e739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beerenwinkel N, Schwarz RF, Gerstung M, et al. Cancer evolution: mathematical models and computational inference. Syst Biol 2015; 64: e1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, Waters J, Leung ML, et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature 2014; 512: 155–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu X, Hou Y, Yin X, et al. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell 2012; 148: 886–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gawad C, Koh W, Quake SR. Dissecting the clonal origins of childhood acute lymphoblastic leukemia by single-cell genomics. Proc Natl Acad Sci U S A 2014; 111: 17947–17952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leung ML, Davis A, Gao R, et al. Single-cell DNA sequencing reveals a late-dissemination model in metastatic colorectal cancer. Genome Res 2017; 27: 1287–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Bourcy CF, De Vlaminck I, Kanbar JN, et al. A quantitative comparison of single-cell whole genome amplification methods. PLoS One 2014; 9: e105585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hou Y, Wu K, Shi X, et al. Comparison of variations detection between whole-genome amplification methods used in single-cell resequencing. Gigascience 2015; 4: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hou Y, Song L, Zhu P, et al. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell 2012; 148: 873–885. [DOI] [PubMed] [Google Scholar]
- 17.Rago C, Huso DL, Diehl F, et al. Serial assessment of human tumor burdens in mice by the analysis of circulating DNA. Cancer Res 2007; 67: 9364–9370. [DOI] [PubMed] [Google Scholar]
- 18.Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009; 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011; 43: 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997 2013.
- 21.Müllner D Modern hierarchical, agglomerative clustering algorithms. arXiv preprint arXiv:11092378 2011.
- 22.McGill R, Tukey JW, Larsen WA. Variations of Box Plots. The American Statistician 1978; 32: 12–16. [Google Scholar]
- 23.Jones S, Zhang X, Parsons DW, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 2008; 321: 1801–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Basturk O, Tan M, Bhanot U, et al. The oncocytic subtype is genetically distinct from other pancreatic intraductal papillary mucinous neoplasm subtypes. Mod Pathol 2016; 29: 1058–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamaguchi K, Kanemitsu S, Hatori T, et al. Pancreatic ductal adenocarcinoma derived from IPMN and pancreatic ductal adenocarcinoma concomitant with IPMN. Pancreas 2011; 40: 571–580. [DOI] [PubMed] [Google Scholar]
- 26.Sausen M, Phallen J, Adleff V, et al. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nat Commun 2015; 6: 7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Gating scheme used for single cell sorting by fluorescence activated cell sorting (FACS)
Figure S2. Computational workflow for mutation calling and error estimation
Figure S3. Thresholds applied for determining mutation status
Figure S4. Somatic mutation calls in single IPMN cells before and after imputation
Figure S5. Outlier analysis of single cell ADO data
Figure S6. Somatic mutations identified in single cells from IP04, IP10, IP11
Table S1. Number of single cells analyzed from each IPMN
Table S2. Primers for multiplex PCR assay to evaluate WGA efficiency
Table S3. Somatic mutations identified in single IPMN cells
Table S4. Germline variant calls utilized for calculation of ADO
Table S5. ADO in each single IPMN cell
Table S6. False-positive mutations identified in single cells from control samples
Table S7. Somatic mutations identified in each IPMN section