Abstract
Inheritance of the single mitochondrial nucleoid (kinetoplast) in the trypanosome requires numerous proteins many of whose precise roles are unclear. By considering kinetoplast DNA (kDNA) as a template for cleavage into two equal-size networks, we predicted sets of mutant kinetoplasts associated with defects in each of five steps in the kinetoplast cycle. Comparison of these kinetoplasts with those obtained after gene knockdowns enabled assignment of proteins to five classes - kDNA synthesis, site of scission selection, scission, separation, and partitioning. These studies highlight how analysis of mutant kinetoplast phenotypes may be used to predict functional categories of proteins involved in biogenesis of kinetoplasts.
Keywords: Trypanosome, Trypanosoma brucei, kinetoplast, mitochondrial DNA, segregation, kDNA
Kinetoplast biogenesis and inheritance cycle
The single-cell eukaryote Trypanosoma brucei causes human African trypanosomiasis (HAT) and the cattle disease nagana in regions of sub-Saharan Africa. T. brucei is spread to vertebrates through the bite of an infected tsetse fly which harbors insect stage (procyclic) trypanosomes. The mitochondrial genome of T. brucei is comprised of circular double-stranded DNAs (minicircles and maxicircles) catenated into a disk-like network in a single “kinetoplast” (see Glossary). Loss of kinetoplast DNA (kDNA) disrupts mitochondrial functions in bloodstream T. brucei and interferes with development in the tsetse fly, breaking vector-to-mammal transmission that is needed to spread disease [1, 2]. Bloodstream trypanosomes may lose kDNA, becoming dyskinetoplastic, in which case their long-term survival is only possible if they acquire a mutation in the γ-subunit of ATP synthase [3]. Naturally-occurring dyskinetoplastic Trypanosoma equiperdum and T. evansi strains are known (reviewed in [1]). Trypanosome basal bodies are “modified” centrioles found near kinetoplasts. The region between basal bodies and the kinetoplast constitutes a tripartite attachment complex (TAC) [4]. We term proteins that associate with TAC “TAC-associated proteins (TACAPs)” (reviewed in [5]).
“Segregation” is widely-used in the field to describe aspects of the kinetoplast cycle. Given our current state of knowledge about proteins involved in biogenesis and inheritance of kinetoplasts, the meaning of the word “segregation” is ambiguous, at best. Originally used to describe scission of kDNA networks, inferred from examination of electron micrographs [6], “segregation” has also been used to describe movement and/or duplication of kinetoplasts [4, 7].
Biogenesis of kinetoplasts is coordinated with the cell cycle. Trypanosomes in G1 have a single kinetoplast (K) and one nucleus (N) (1K1N, Figure 1). kDNA is synthesized in S-phase [8, 9]. Two kinetoplasts per cell are observed in G2 in 2K1N trypanosomes [10]. Kinetic analysis using pre-S phase enriched trypanosomes indicates that duplication of kinetoplasts peaks 1 h after termination of kDNA synthesis [11]. The period between termination of kDNA synthesis and division of kinetoplasts may be termed kinetoplast G2 (G2K). Nuclei of 2K1N trypanosomes divide to generate 2K2N cells that produce two 1K1N cells after cytokinesis [12]. Restriction of kinetoplast duplication to G2 indicates that a kinetoplast is “licensed” for scission or separation at this stage of the cell cycle.
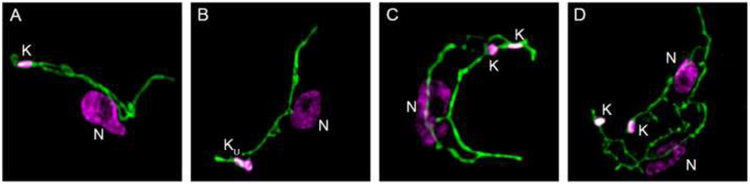
Figure 1. Kinetoplasts inside mitochondria.
Mitochondria of bloodstream trypanosomes were labeled with mitotracker (green) [46], and DNA in the nucleus (N) or kinetoplast (K) labeled with DAPI (4,6-diamino-2-phenylindole). Images were captured with a fluorescence microscope. Panel A, 1K1N; B, 1KU1N trypanosome; C, 2K1N trypanosome; D, 2K2N trypanosome.
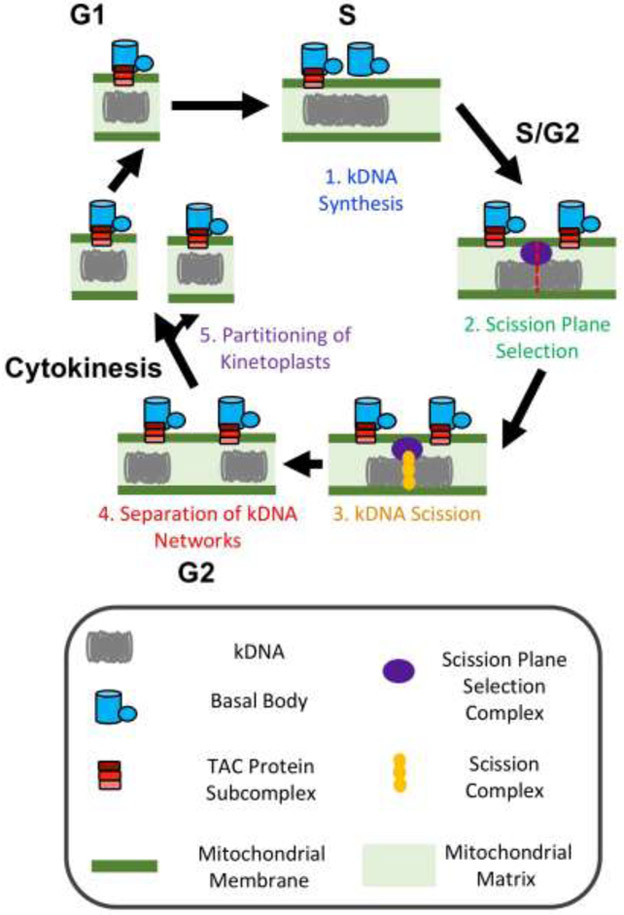
A long-term goal of the field is to provide a molecular description of the kinetoplast cycle (Figure 2) [11, 13-18]. That objective calls for assignment of roles to proteins at specific steps of the kinetoplast cycle. In this context, we suggest that investigators try to avoid the term “segregation” because of the ambiguity in its meaning. We offer terminology that the field may consider using to describe the major steps in the duplication cycle of kinetoplasts. Further, we provide guidelines for analysis of mutant kinetoplasts from gene knockdowns that allows placement of proteins into one of five steps in the kinetoplast cycle (Figure 2).
Figure 2. Five steps in kinetoplast biogenesis and inheritance.
Major steps in the kinetoplast cycle are depicted. Shown are; (1) kDNA synthesis, (2) scission site selection, (3) scission (cleavage), (4) separation of cleaved kDNAs, and (5) partitioning (at cytokinesis).
Synthesis of kDNA refers to incorporation of nucleotides into the mitochondrial DNA network (Figure 1). “Scission” involves cleavage of kDNA into two networks. “Separation” denotes movement of cleaved kinetoplasts away from each other inside a single mitochondrial (Figure 1). “Partitioning” is the sorting of kinetoplasts into daughter trypanosomes (Figure 2). Minimally, the kinetoplast cycle has five steps (Figure 2): (1) Replication of kDNA; (2), selection of a site for scission of kDNA; (3) scission of kDNA; (4) separation of cleaved kinetoplasts; and (5) partitioning of kinetoplasts into two trypanosomes.
Five classes of mutants for kinetoplast biogenesis and inheritance
On the basis of the steps involved in the kinetoplast cycle (Figure 2), protein roles may be categorized as follow: First, identify and quantitate all trypanosome cell types produced within three cell division cycles after knockdown of a gene. Second, compare unusual cell types to those predicted after loss of genes at different steps of the kinetoplast cycle (Figure 2). Third, assign a gene to the class with the closest set of predicted abnormal trypanosomes (Table 1).
Table 1.
Sets of unusual kinetoplast combinations predicted for classes of mutants
| Category | Stage in kinetoplast cycle | Expected set of abnormal trypanosomes |
|---|---|---|
| Class I | kDNA synthesis | 1KS1N, KS/KS1N, KS/KS2N |
| Class II | Selection of Scission Site | KL/KS1N, KL/KS2N, 1KL1N, 1KS1N |
| Class III | Scission of kDNA network | 1KU1N, 1KU2N, 0K1N. Electron microscopy shows one uncleaved kDNA per kinetoplast |
| Class IV | Separation of Cleaved kDNA networks | 1KU1N, 1KU2N, 0K1N. Two cleaved kDNAs per kinetoplast, detected in electron microscopy analysis |
| Class V | Partitioning of kinetoplasts into new cells | 2K1N, 0K1N |
Different types of kinetoplasts and the expected sets appearing together are presented for mutants belonging to classes I through V. Kinetoplast designations are as follows; K, kDNA containing one genomes’ worth of minicircles and maxicircles; KS, kinetoplast contains less < 0.5 of one genome’s equivalent of kDNA; KL, kinetoplast contains > 0.5 < 1 of one genome’s equivalent of kDNA; KU, kDNA has two genome’s amount of minicircles and maxicircles.
Class I - kDNA synthesis mutants
Due to failure to synthesize a full component of mitochondrial DNA, kDNA content of a mutants is less than normal at the end of kinetoplast S-phase. This smaller kDNA intermediate (KL*) is cleaved into two, and the immediate product (1KS1N) has less kDNA than normal kinetoplasts. In contrast nuclei in the cells have normal content (N) compared to control cells (Figure 3A).
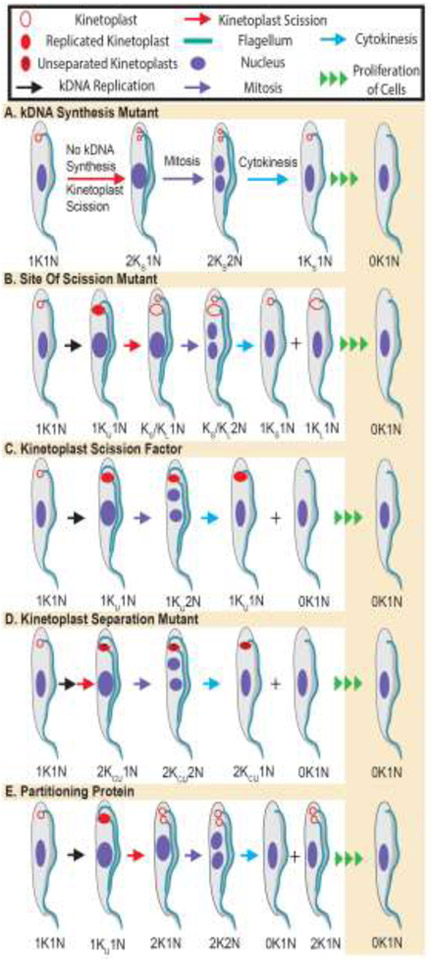
Figure 3. Predicted sets of kinetoplasts in mutant trypanosomes follwing failure of specified steps of the kinetoplast cycle.
Kinetoplast DNA (kDNA) (hollow red circle) was considered as a substrate for synthesis to produce a double-sized kinetoplast (KU). Cleavage of a kinetoplast precedes mitosis, and normally produces two equal-sized kDNAs and one nucleus (2K1N) in one trypanosome. Mitosis of 2K1N trypanosomes results in 2K2N cells that can go through cytokinesis and generate 1K1N cells. Early abnormal kinetoplasts predicted for each class of mutant in kinetoplast cycle is illustrated. In all cases sustained proliferation of early mutant trypanosomes leads to an increased fraction of dyskinetoplastic (0K1N) cells, but the path to this end result, as revealed by the panel of abnormal kinetoplasts that accompanies each mutation varies. A, Class I, kDNA Synthesis Mutants. After loss of a protein that is essential for synthesis of kDNA the total amount of DNA per kinetoplast decreases. Hence after scission of that kDNA the progeny has less than one kinetoplast’s equivalent (i.e., Ks) whereas the nucleus has a regular amount of DNA, making the cell a 2KS1N trypanosome. After mitosis, a 2KS2N trypanosome emerges which can go through cytokinesis. Following multiple rounds of division, the trypanosome gradually loses all of its kDNA and becomes dyskinetoplastic (i.e., 0K1N). B, Class II, Site of Scission Selection. After synthesis of kDNA in a normal cell a double-size uncleaved kinetoplast (KU) is the product in a 1KU1N cell: the kDNA is cleaved normally into two equal-sized progenies, before mitosis. Faulty choice of the site of scission resulting from knockdown of a factor that is import for identification of the scission site will cause cleavage of kDNA into two networks of unequal size (i.e., asymmetric scission of kinetoplast). The larger kinetoplast (KL) and the smaller kDNA (KS) can be found in premitotic (1KS/KL1N) cells or in post-mitotic 1KS/KL2N trypanosomes. Repeated asymmetric kinetoplast scission and proliferation of progeny eventually leads to a population of trypanosomes with a significant fraction of dyskinetoplastics (0K1N). C, Class III, Kinetoplast Scission Factors (KSFs). Cleavage of a double-size kinetoplast (1KU1N) into two networks is essential for inheritance of kinetoplasts. After knockdown of genes encoding KSFs, 1KU1N remains uncleaved in premitotic trypanosomes. However, failure of kDNA scission does not foil mitosis, so nuclear division results in 1KU2N trypanosomes that may accumulate in the population. Cytokinesis of 1KU2N cells produces 0K1N (dyskinetoplastic) and 1KU1N cells that after proliferation increase the fraction of dyskinetoplastic trypanosomes. D, Class IV, Separation of cleaved kinetoplasts. After scission of kinetoplasts the kDNA networks are detected by light microscopy as two entities when they move apart. Thus, in mutants where the separation of kinetoplasts does not take place, only one kDNA will be detectable although scission of kinetoplasts has occurred. Kinetoplasts in Class IV mutant 1K2N and 1K1N cells are only distinguishable from those of Class III (KSF) mutants in electron microscopy studies; they will reveal cleaved but adjacent kinetoplasts for separation factors, but show uncleaved kDNA in mutants of KSF’s. E, Class V, Partitioning of Kinetoplasts. During cytokinesis two kinetoplasts from mitotic 2K2N trypanosomes are sorted into two new progeny each of which has a 1K1N organelle content. If partitioning of the two kinetoplasts fails, the progeny trypanosomes are 2K1N and 0K1N. Thus, detection of 0K1N cells in absence of 1KU2N early after gene knockdown is diagnostic of partition mutants.
When 1KS1N nuclei divide, the products are 1KS2N. Thus, the early unusual products of this class of mutants are 2KS2N and 2KS1N. Should these cells proliferate, loss of kDNA continues until the nucleoid is no longer detectable and the trypanosomes are dyskinetoplastic (0K1N, Figure 3A) [19]. Class I mutants are exemplified by Pol1B [20], p38 [21], MIRF172 [22], TbKAP6 [23], mitochondrial Topo II [19], and Tb927.2.6100 [17].
Class II - site of scission selection mutants
During trypanosome proliferation, kinetoplasts are divided into two equal-size networks before partitioning into two cells (Figure 2). Thus, it stands to reason that trypanosomes have molecular machinery to recognize correct sites of scission on kinetoplasts. Erroneous selection of the site of scission would lead to cleavage of kDNA into two kinetoplasts of unequal size; one large (KL) and the other small (KS) in the same cell (KL/KS1N, i.e., asymmetric kinetoplast division [24], Figure 3B). After mitosis, KL/KS1N cells produce KL/KS2N trypanosomes. Thus, the abnormal early products of mutants in scission site selection are kinetoplasts that are either oversized (KL) or undersized (KS) (specifically, KL/KS2N, KL/KS1N, 1KLN and 1KS1N, Figure 3B). Sustained proliferation of these early cell types will produce dyskinetoplastic (0K1N) trypanosomes (Figure 3). Examples of Class II genes are TACAPs p166 [25], p197 (in procyclic T. brucei) [26], TAC65 [27], and TAC60 [28]. Acyl carrier protein (ACP) [29] is a Class II protein.
Class III - kinetoplast scission mutants
Scission of a kinetoplast entails the conversion of one double-size kDNA into two networks of equivalent size. From kinetic analysis of chemically-synchronized trypanosomes, and the use of molecular markers [30], the earliest time when two kinetoplasts are detected in a population of trypanosomes is G2 [11]. Should scission of a kinetoplast fail, the immediate product is a 1KU1N trypanosome (Figure 3C) in which the kinetoplast contains two equivalents of kDNA. After mitosis, 1KU1N yields 1KU2N trypanosomes since failed kinetoplast division does not prevent mitosis (Figure 4). Documentation of this “failed kinetoplast division phenotype” is compelling when 1KU2N comprise 5% or more of the total trypanosome population. If 1KU2N trypanosomes divide, 1KL1N and 0K1N (dyskinetoplastic) cells are the earliest products (Figure 3C). Extended proliferation of these initial cells will produce more dyskinetoplastic trypanosomes (Figure 3C).
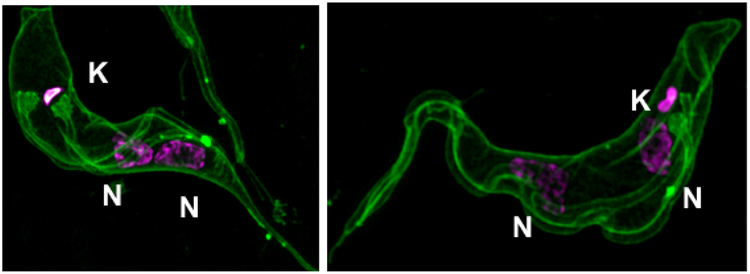
Figure 4. 1KU2N trypanosomes obtained after knockdown of a KSF.
Bloodstream T. brucei containing an RNAi construct were induced to knock down a KSF gene (TbCK1.2) [33]. After inducing knockdown of the gene with tetracycline, plasma membrane and flagella were stained with mCLING (green) [47], and kinetoplast (K) and nuclei (N) were detected with DAPI. Images were captured using a superresolution microscope as described [47].
Examples of gene knockdowns that led to accumulation of 1K2N are TbBB46, TbCEP57, KMP-11, TbUMSBP1/TbUMSBP2, TbCK1.2 and GCP3 (γ-tubulin) [31-35]. Electron microscopy of KU is predicted to show one undivided kDNA (larger than unit size) as observed after knockdown of UMSBP1/2 [32]. Genes that perturb flagellum biogenesis (e.g., produce detached or shortened flagella on 1K2N trypanosomes) are excluded from our list of potential Class III proteins, to limit digression into studies of proteins with multiple functions in T. brucei. We recognize that “asymmetric cytokinesis” of normal 2K2N cells can produce 1K2N and 1K0N (anucleate trypanosomes) [36, 37]. However, these kinetoplasts have normal amounts of kDNA; they are not 1KU that is easily identified by quantitation of kDNA content [18, 28, 38].
Class III genes encode kinetoplast scission factors (KSFs), a knockdown of which causes accumulation of 1KU2N trypanosomes. After division of 1KU2N cells, a new population of 1KU1N and dyskinetoplastic trypanosomes are produced. However, kDNA synthesis, basal body duplication, and flagellum biogenesis will be normal in cells lacking KSFs (Figure 4). Class III genes may encode polypeptides that (i) regulate trypanosome entry into G2 of the cell cycle, or (ii) cleave kDNA into two. Thus, for a better understanding of KSF function we need to learn whether 1KU2N trypanosomes obtained from knockdown of Class III genes are in G2 or M-phase of the cell cycle.
Finally, we considered a possibility that a protein could inhibit cleavage of kinetoplasts without accumulation of 1KU2N if trypanosomes proliferated efficiently. Such a scenario is only possible if loss of the protein has three concurrent effects; (i) blocks scission of a kinetoplast, (ii) accelerates nucleus mitosis, and (iii) promotes rapid cytokinesis, such that all traces of 1KU2N are lost from the population. There is no genetic support for this model.
Class IV - separation of kinetoplasts mutants
After cleavage of a double-size kinetoplast network into two, daughter kDNAs are presumed to be next to each other initially. Separated kinetoplasts are observed in G2 (Figure 1) [11, 30, 39], implying that daughter kDNAs move apart in G2 of the cell cycle. Based on these facts the initial products from mutation of a gene that is important for separation of kinetoplasts will include 1KU1N and 1KU2N that after dividing produce 1KU1N and 0K1N cells, similar to Class III genes. However, although kinetoplasts of 1KU2N in Class IV knockdowns appear to have one kDNA network in fluorescence microscopy, electron microscopy analysis of the nucleoids will reveal two kDNA disks juxtaposed to each other. Currently, we have not identified any gene for which convincing data is available for its classification as a Class IV mutant. However, we suspect that some genes producing 1K2N trypanosomes when knocked down might belong to this class.
Class V - partitioning mutants
Kinetoplasts in 2K2N trypanosomes are sorted into two 1K1N cells during cytokinesis. Mutations in genes encoding partitioning proteins or their regulatory factors will cause inappropriate sorting, producing 2K1N and 0K1N (dyskinetoplastic) trypanosomes as early abnormal trypanosomes (Figure 3D). After many rounds of proliferation, 0K1N trypanosomes become a major population in Class V mutants. Genes whose products qualify as partitioning factors include TACAPs p197 [18], TAC102 (in bloodstream T. brucei) [18], and TAC40 [40]. Protease PNT1 [14], and α-KDE2 [41] are also partitioning proteins.
Although one could consider 2K0N and 0K2N (binucleate dyskinetoplastic) trypanosomes as products of severe defects in partitioning (Table 1), that decision is debatable. Knockdown of a protein involved in sorting of kinetoplasts should not affect mitosis. Therefore, 0K2N cells cannot be products of partitioning defects; they are products of asymmetric cytokinesis (aberrant cleavage furrow placement) of 2K2N cells [42], with 2K0N as the other abnormal product. One expects anucleate 2K0N cells to be short-lived and probably undetectable in an RNAi knockdown study, because it takes days for data to be collected. The considerations above illustrate the importance of examining a full panel of early abnormal trypanosomes from gene knockdowns before concluding that dyskinetoplastic cells originated from improper partitioning of kinetoplasts (Table 1).
Concluding Remarks
In the last four years the field has been seen excellent papers describing the involvement of proteins that localize between the kinetoplast and basal bodies (TACAPs) in inheritance of the nucleoid [14, 18, 22, 28, 40, 43]. It seems likely that TACAPs execute their functions by forming sub-complexes instead of acting together as one massive macromolecule complex comprised of all proteins that localize to TAC. Proteins (e.g., KSFs) that are not found on TAC can have profound effects on kinetoplast scission and/or separation. Several potential KSFs are encoded in the genome, from evaluation of published literature [31-35]. An effort to integrate KSFs into models of the machinery for mitochondrial genome scission and sorting will be well-received, since TACAPs are not essential for scission of kinetoplasts.
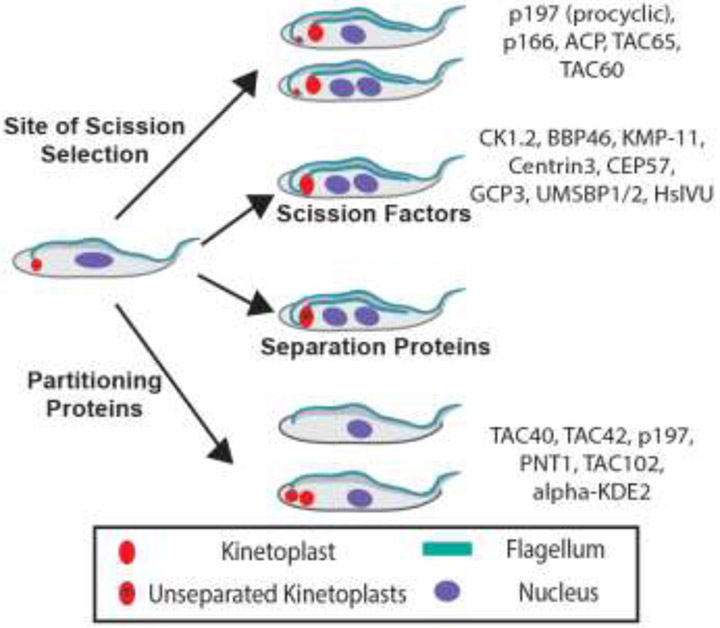
Our phenotypic analysis of early products from “loss of function” mutants allowed us to assign them to different steps in kinetoplast biogenesis and inheritance (Figure 5). This ordering of proteins enables investigators to develop new hypotheses on whether (or not) any subset of proteins function sequentially or interactively in vivo. Studies of possible interactions between TACAPs [18, 27, 28], as well as proteins that are not TAC-associated will be bolstered by functional categorization of genes. For example, do the postulated Partitioning Proteins TAC40, p197, PNT1, TAC102, and alpha-KDE2 interact physically? Similarly, do Site of Scission Selection Factors p197, p166, ACP, TAC65 and TAC60 form a sub-complex in trypanosomes (Figure 5)? Since techniques for testing protein interactions in vivo are available it should not be long before we obtain answers to some of these questions (see Outstanding Questions). We anticipate that the initial protein categorization summarized in Figure 5 will be expanded with new information provided by other investigators. It is possible that the initial assignment of proteins to different classes of mutants (Table 1) will be revised as more comprehensive data becomes available. These developments will help the field move closer to a more detailed molecular understanding of mitochondrial genome biogenesis and inheritance in the African trypanosome.
Figure 5. Distinction between functions of TAC-associated proteins and KSFs.
Early abnormal trypanosomes reported after knockdown of several proteins that affected kinetoplast cycle were analyzed as described (see Table 1 for summary). Genes were assigned to one of the five steps in kinetoplast biogenesis (see Figure 2), based on a comparison of published phenotypic data and the predicted of expected aberrant kinetoplasts (Table 1 and Figure 3).
Outstanding Questions.
What is the value of an initial assignment of genes to specific steps in the kinetoplast cycle through phenotypic analysis of mutant kinetoplasts?
How many kinetoplast scission factors are there, and what are their precise functions?
How are kinetoplasts attached to the machinery for scission or partition?
How are the functions of kinetoplast scission factors and TAC-associated proteins co-ordinated and/or integrated?
Concerning the systematic phenotypic analysis of gene knockdowns (Table 1, Figure 3), three issues are worth raising for discussion by the field. First, for a compelling quantitative description of the phenotypic observations, one should expect that a major diagnostic abnormal trypanosome cell type comprises at least 5 percent of the total population. Second, a Chi-square test of the statistical significance of the differences in distribution of cell types before and after gene knockdown returns a p-value < 5 × 10−3. The suggested higher threshold for the p-value is influenced by knowledge that most investigators use unsynchronized cells for these studies. Day-to-day variation of proportions for cell types in unsynchronized trypanosome populations can produce a distribution of cell types between the same control cells that gives p-values close to 5 × 10−2, a threshold that is frequently accepted for statistically significant differences. In addition, it will help if in work with kinetoplast biogenesis and inheritance, the trypanosomes are analyzed at fixed times after RNAi induction, so that changes in phenotypes and morphology can be documented as early as possible. Third, recognizing the power of microscopy images to sway readers to a particular point of view (e.g., proposed pathway of events), we kindly request, for the sake of reproducibility of observations between laboratories, that images of abnormal cells should only be presented in publications if those cell types constitute at least 5% of the total population of trypanosomes.
Finally, how could TACAPs regulate scission site selection or partitioning of kinetoplasts when they lack DNA-binding domains (Figure 5)? The answer to the question may lie in three kinetoplast-binding proteins highlighted in our analysis, namely protease PNT1 [14] and universal minicircle-binding proteins UMSBP1 and UMSBP2 [32]. PNT1 is a partitioning protein (Figure 5), whereas UMSBP1 and UMSBP2, together, are KSFs (Figure 5) [32]. We hypothesize that PNT1 and UMSBP1/UMSBP2 have “moonlighting” functions [44, 45] as scaffolding proteins that anchor kinetoplasts to protein complexes needed for scission and partitioning of kDNA. Hence, two experimentally testable hypotheses from our phenotypic analyses of kinetoplasts and placement of genes in the kinetoplast cycle are (i) PNT1 is a scaffold for a partitioning complex on kinetoplasts (Figure 5), and (ii) UMSBP1/UMSBP2 anchors kinetoplast scission complexes to kDNA (Figure 5). We look forward to reading about answers to some of these questions in the near future.
Highlights.
Kinetoplasts (mitochondrial genome nucleoids) are important in bloodstream trypanosomes for establishment of mitochondrial membrane potential.
- Many proteins involved in segregation of kinetoplasts have been identified.
- A region between a kinetoplast and basal bodies is described as a tripartite attachment complex (TAC)
- A set of TAC-associated proteins (TACAPs) has been proposed as the machinery for kinetoplast segregation
- Sub-complexes of TACAPs that form in vivo have been described
Several proteins that do not associate with TAC are involved in maintenance of the kinetoplast. New kinetoplast-associated proteins have been identified
We are approaching an exciting period in the field when a molecular understanding of how all aspects of kinetoplast biogenesis is executed seems achievable.
Glossary
- Basal body
Microtubule organizing center for flagellum axoneme. Movements of basal bodies are associated with division of kinetoplasts. Some proteins that are found between basal bodies and kinetoplasts (i.e., TAC-associated proteins) are important in aspects of the kinetoplast biogenesis cycle.
- Kinetoplast
Mitochondrial nucleoid consisting of kDNA and associated proteins. During trypanosome proliferation, a second copy of the kinetoplast is detected by light microscopy in the G2 stage of the cell cycle. At cytokinesis two kinetoplasts are sorted, one each, into two daughter trypanosomes.
- Kinetoplast DNA (kDNA)
Mitochondrial DNA network in trypanosomes. Composed of interlocked (i.e., catenated) circular minicircles and maxicircles. Maxicircles encode the mitochondrial genome. Many genes encoded by maxicircles are incomplete until edited at the RNA level to incorporate (or remove) nucleotides. Information (guide RNAs) for editing maxicircle transcripts are found predominantly in minicircles. Edited pre-mRNAs can be translated to proteins by ribosomes.
- Kinetoplast scission factor (KSF)
Protein required for cleavage of kinetoplast DNA networks, generating two daughter kDNA networks each of which retains catenated minicircles and maxicircles.
- Partitioning
Distribution of cleaved kinetoplasts into two daughter trypanosomes during cytokinesis. Scission of a double-size kinetoplast with replicated kDNA is required ahead of partitioning the nucleoid.
- Scission (cleavage) of kinetoplast
Endonuclease cleavage of a double-size (i.e., replicated) kDNA network into two networks of approximately equal size. Daughter kDNAs retain catenated minicircles and maxicircles but they are not substrates for further scission reactions.
- Separation
Movement of cleaved kinetoplasts away from each other inside a single mitochondrion.
- Tripartite attachment complex (TAC)
A region of the trypanosome between the kinetoplast and basal body. Some proteins that associate with TAC affect kinetoplast inheritance. However, not all TAC-associated proteins are important for kinetoplast scission and/or partitioning.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Schnaufer A, Domingo GJ and Stuart K (2002) Natural and induced dyskinetoplastic trypanosomatids: how to live without mitochondrial DNA. International journal for parasitology. 32, 1071–1084 [DOI] [PubMed] [Google Scholar]
- 2.Dewar CE, MacGregor P, Cooper S, Gould MK, Matthews KR, Savill NJ and Schnaufer A (2018) Mitochondrial DNA is critical for longevity and metabolism of transmission stage Trypanosoma brucei. PLoS pathogens. 14, e1007195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dean S, Gould MK, Dewar CE and Schnaufer AC (2013) Single point mutations in ATP synthase compensate for mitochondrial genome loss in trypanosomes. Proceedings of the National Academy of Sciences of the United States of America. 110, 14741–14746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ogbadoyi EO, Robinson DR and Gull K (2003) A high-order trans-membrane structural linkage is responsible for mitochondrial genome positioning and segregation by flagellar basal bodies in trypanosomes. Molecular biology of the cell. 14, 1769–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schneider A and Ochsenreiter T (2018) Failure is not an option - mitochondrial genome segregation in trypanosomes. Journal of cell science. 131. [DOI] [PubMed] [Google Scholar]
- 6.Hoeijmakers JH and Weijers PJ (1980) The segregation of kinetoplast DNA networks in Trypanosoma brucei. Plasmid. 4, 97–116 [DOI] [PubMed] [Google Scholar]
- 7.Robinson DR and Gull K (1991) Basal body movements as a mechanism for mitochondrial genome segregation in the trypanosome cell cycle. Nature. 352, 731–733 [DOI] [PubMed] [Google Scholar]
- 8.Woodward R and Gull K (1990) Timing of nuclear and kinetoplast DNA replication and early morphological events in the cell cycle of Trypanosoma brucei. Journal of cell science. 95 ( Pt 1), 49–57 [DOI] [PubMed] [Google Scholar]
- 9.Gluenz E, Povelones ML, Englund PT and Gull K (2011) The kinetoplast duplication cycle in Trypanosoma brucei is orchestrated by cytoskeleton-mediated cell morphogenesis. Molecular and cellular biology. 31, 1012–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siegel TN, Hekstra DR and Cross GA (2008) Analysis of the Trypanosoma brucei cell cycle by quantitative DAPI imaging. Molecular and biochemical parasitology. 160, 171–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sullenberger C, Pique D, Ogata Y and Mensa-Wilmot K (2017) AEE788 Inhibits Basal Body Assembly and Blocks DNA Replication in the African Trypanosome. Molecular pharmacology. 91, 482–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sherwin T and Gull K (1989) The cell division cycle of Trypanosoma brucei brucei: timing of event markers and cytoskeletal modulations. Philosophical transactions of the Royal Society of London. Series B, Biological sciences. 323, 573–588 [DOI] [PubMed] [Google Scholar]
- 13.Pena-Diaz P, Vancova M, Resl C, Field MC and Lukes J (2017) A leucine aminopeptidase is involved in kinetoplast DNA segregation in Trypanosoma brucei. PLoS pathogens. 13, e1006310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grewal JS, McLuskey K, Das D, Myburgh E, Wilkes J, Brown E, Lemgruber L, Gould MK, Burchmore RJ, Coombs GH, Schnaufer A and Mottram JC (2016) PNT1 Is a C11 Cysteine Peptidase Essential for Replication of the Trypanosome Kinetoplast. The Journal of biological chemistry. 291, 9492–9500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, Pappas-Brown V, Englund PT and Jensen RE (2014) TbKAP6, a mitochondrial HMG box-containing protein in Trypanosoma brucei, is the first trypanosomatid kinetoplast-associated protein essential for kinetoplast DNA replication and maintenance. Eukaryotic cell. 13, 919–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Povelones ML (2014) Beyond replication: Division and segregation of mitochondrial DNA in kinetoplastids. Molecular and biochemical parasitology [DOI] [PubMed] [Google Scholar]
- 17.Beck K, Acestor N, Schulfer A, Anupama A, Carnes J, Panigrahi AK and Stuart K (2013) Trypanosoma brucei Tb927.2.6100 is an essential protein associated with kinetoplast DNA. Eukaryotic cell. 12, 970–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffmann A, Kaser S, Jakob M, Amodeo S, Peitsch C, Tyc J, Vaughan S, Zuber B, Schneider A and Ochsenreiter T (2018) Molecular model of the mitochondrial genome segregation machinery in Trypanosoma brucei. Proceedings of the National Academy of Sciences of the United States of America. 115, E1809–E1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z and Englund PT (2001) RNA interference of a trypanosome topoisomerase II causes progressive loss of mitochondrial DNA. The EMBO journal. 20, 4674–4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bruhn DF, Mozeleski B, Falkin L and Klingbeil MM (2010) Mitochondrial DNA polymerase POLIB is essential for minicircle DNA replication in African trypanosomes. Molecular microbiology. 75, 1414–1425 [DOI] [PubMed] [Google Scholar]
- 21.Liu B, Molina H, Kalume D, Pandey A, Griffith JD and Englund PT (2006) Role of p38 in replication of Trypanosoma brucei kinetoplast DNA. Molecular and cellular biology. 26, 5382–5393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amodeo S, Jakob M and Ochsenreiter T (2018) Characterization of the novel mitochondrial genome replication factor MiRF172 in Trypanosoma brucei. Journal of cell science. 131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J, Pappas-Brown V, Englund PT and Jensen RE (2014) TbKAP6, a Mitochondrial HMG box-Containing Protein in Trypanosoma brucei, Is the First Trypanosomatid KAP protein Essential for kDNA Replication and Maintenance. Eukaryotic cell [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z, Drew ME, Morris JC and Englund PT (2002) Asymmetrical division of the kinetoplast DNA network of the trypanosome. The EMBO journal. 21, 4998–5005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao Z, Lindsay ME, Roy Chowdhury A, Robinson DR and Englund PT (2008) p166, a link between the trypanosome mitochondrial DNA and flagellum, mediates genome segregation. The EMBO journal. 27, 143–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gheiratmand L, Brasseur A, Zhou Q and He CY (2013) Biochemical Characterization of the Bi-lobe Reveals a Continuous Structural Network Linking the Bi-lobe to Other Single-copied Organelles in Trypanosoma brucei. The Journal of biological chemistry. 288, 3489–3499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaser S, Oeljeklaus S, Tyc J, Vaughan S, Warscheid B and Schneider A (2016) Outer membrane protein functions as integrator of protein import and DNA inheritance in mitochondria. Proceedings of the National Academy of Sciences of the United States of America. 113, E4467–4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaser S, Willemin M, Schnarwiler F, Schimanski B, Poveda-Huertes D, Oeljeklaus S, Haenni B, Zuber B, Warscheid B, Meisinger C and Schneider A (2017) Biogenesis of the mitochondrial DNA inheritance machinery in the mitochondrial outer membrane of Trypanosoma brucei. PLoS pathogens. 13, e1006808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clayton AM, Guler JL, Povelones ML, Gluenz E, Gull K, Smith TK, Jensen RE and Englund PT (2011) Depletion of mitochondrial acyl carrier protein in bloodstream-form Trypanosoma brucei causes a kinetoplast segregation defect. Eukaryotic cell. 10, 286–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benz C, Dondelinger F, McKean PG and Urbaniak MD (2017) Cell cycle synchronisation of Trypanosoma brucei by centrifugal counter-flow elutriation reveals the timing of nuclear and kinetoplast DNA replication. Scientific reports. 7, 17599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dang HQ, Zhou Q, Rowlett VW, Hu H, Lee KJ, Margolin W and Li Z (2017) Proximity Interactions among Basal Body Components in Trypanosoma brucei Identify Novel Regulators of Basal Body Biogenesis and Inheritance. MBio. 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Milman N, Motyka SA, Englund PT, Robinson D and Shlomai J (2007) Mitochondrial origin-binding protein UMSBP mediates DNA replication and segregation in trypanosomes. Proceedings of the National Academy of Sciences of the United States of America. 104, 19250–19255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones NG, Thomas EB, Brown E, Dickens NJ, Hammarton TC and Mottram JC (2014) Regulators of Trypanosoma brucei Cell Cycle Progression and Differentiation Identified Using a Kinome-Wide RNAi Screen. PLoS pathogens. 10, e1003886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou Q and Li Z (2015) gamma-Tubulin complex in Trypanosoma brucei: molecular composition, subunit interdependence and requirement for axonemal central pair protein assembly. Molecular microbiology. 98, 667–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Z and Wang CC (2008) KMP-11, a basal body and flagellar protein, is required for cell division in Trypanosoma brucei. Eukaryotic cell. 7, 1941–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu H, Zhou Q and Li Z (2015) SAS-4 Protein in Trypanosoma brucei Controls Life Cycle Transitions by Modulating the Length of the Flagellum Attachment Zone Filament. The Journal of biological chemistry. 290, 30453–30463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Signorell A, Gluenz E, Rettig J, Schneider A, Shaw MK, Gull K and Butikofer P (2009) Perturbation of phosphatidylethanolamine synthesis affects mitochondrial morphology and cell-cycle progression in procyclic-form Trypanosoma brucei. Molecular microbiology. 72, 1068–1079 [DOI] [PubMed] [Google Scholar]
- 38.Patel G, Karver CE, Behera R, Guyett PJ, Sullenberger C, Edwards P, Roncal NE, Mensa-Wilmot K and Pollastri MP (2013) Kinase scaffold repurposing for neglected disease drug discovery: discovery of an efficacious, lapatinib-derived lead compound for trypanosomiasis. Journal of medicinal chemistry. 56, 3820–3832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Janzen CJ, Hake SB, Lowell JE and Cross GA (2006) Selective di- or trimethylation of histone H3 lysine 76 by two DOT1 homologs is important for cell cycle regulation in Trypanosoma brucei. Molecular cell. 23, 497–507 [DOI] [PubMed] [Google Scholar]
- 40.Schnarwiler F, Niemann M, Doiron N, Harsman A, Kaser S, Mani J, Chanfon A, Dewar CE, Oeljeklaus S, Jackson CB, Pusnik M, Schmidt O, Meisinger C, Hiller S, Warscheid B, Schnaufer AC, Ochsenreiter T and Schneider A (2014) Trypanosomal TAC40 constitutes a novel subclass of mitochondrial beta-barrel proteins specialized in mitochondrial genome inheritance. Proceedings of the National Academy of Sciences of the United States of America. 111, 7624–7629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sykes SE and Hajduk SL (2013) Dual functions of alpha-ketoglutarate dehydrogenase E2 in the Krebs cycle and mitochondrial DNA inheritance in Trypanosoma brucei. Eukaryotic cell. 12, 78–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.May SF, Peacock L, Almeida Costa CI, Gibson WC, Tetley L, Robinson DR and Hammarton TC (2012) The Trypanosoma brucei AIR9-like protein is cytoskeleton-associated and is required for nucleus positioning and accurate cleavage furrow placement. Molecular microbiology. 84, 77–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trikin R, Doiron N, Hoffmann A, Haenni B, Jakob M, Schnaufer A, Schimanski B, Zuber B and Ochsenreiter T (2016) TAC102 Is a Novel Component of the Mitochondrial Genome Segregation Machinery in Trypanosomes. PLoS pathogens. 12, e1005586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ginger ML (2014) Protein moonlighting in parasitic protists. Biochemical Society transactions. 42, 1734–1739 [DOI] [PubMed] [Google Scholar]
- 45.Jeffery CJ (2018) Protein moonlighting: what is it, and why is it important? Philosophical transactions of the Royal Society of London. Series B, Biological sciences. 373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vassella E, Straesser K and Boshart M (1997) A mitochondrion-specific dye for multicolour fluorescent imaging of Trypanosoma brucei. Molecular and biochemical parasitology. 90, 381–385 [DOI] [PubMed] [Google Scholar]
- 47.Wiedeman J and Mensa-Wilmot K (2018) A fixable probe for visualizing flagella and plasma membranes of the African trypanosome. PloS one. 13, e0197541. [DOI] [PMC free article] [PubMed] [Google Scholar]