Abstract
In Escherichia coli, after DNA damage, the SOS response increases the transcription (and protein levels) of approximately 50 genes. As DNA repair ensues, the level of transcription returns to homeostatic levels. ClpXP and other proteases return the high levels of several SOS proteins to homeostasis. When all SOS genes are constitutively expressed and many SOS proteins are stabilized by the removal of ClpXP, microscopic analysis shows that cells filament, produce mini-cells and have branching protrusions along their length. The only SOS gene required (of 19 tested) for the cell length phenotype is recN. RecN is a member of the Structural Maintenance of Chromosome (SMC) class of proteins. It can hold pieces of DNA together and is important for double-strand break repair (DSBR). RecN is degraded by ClpXP. Overexpression of recN+ in the absence of ClpXP or recN4174 (A552S, A553V), a mutant not recognized by ClpXP, produce filamentous cells with nucleoid partitioning defects. It is hypothesized that when produced at high levels during the SOS response, RecN interferes with nucleoid partitioning and Z-Ring function by holding together sections of the nucleoid, or sister nucleoids, providing another way to inhibit cell division.
Keywords: SOS response, homologous recombination, Proteases, DNA repair, DNA replication
Graphical Abstract
INTRODUCTION
Maintenance of genomic integrity is essential for continuation of a species. DNA is routinely subject to damage by many mechanisms that originate both internally and externally to the cell (Lindahl, 1993; Lindahl and Wood, 1999; Cox et al., 2000; Zeman and Cimprich, 2014). Organisms have multiple DNA repair systems (i.e., recombinational, nucleotide excision, base excision, non-homologous end-joining, translesion DNA synthesis). Some are constitutively expressed and some respond to DNA damage by specifically inducing the transcription of DNA repair genes (Erill et al., 2007; Ciccia and Elledge, 2010). The archetypal example of an inducible DNA repair system is the SOS response of Escherichia coli (reviewed in (Little and Mount, 1982; Walker, 1984; Bridges, 2005; Janion, 2008)). This system was first hypothesized by Witkins (Witkin, 1967) and then expanded upon by Radman (Radman, 1975). In subsequent years, it has been shown that RecA is the sensor of the SOS response monitoring the level of ssDNA in the cell. When the amount of ssDNA rises due to DNA damage, RecA binding to ssDNA and catalyzes the auto-proteolysis of the transcriptional repressor of the SOS response, LexA. This in turn increases the level of expression of at least 50 genes (Courcelle et al., 2001). These genes produce proteins that repair, recombine, mutagenize DNA and inhibit cell division. As DNA repair ensues, the level of ssDNA decreases and the concentration of LexA increases, returning the transcription of the SOS genes to homeostatic levels. Additionally, it has been shown that ClpXP, Lon and HslUV proteases help return the level of several SOS proteins to pre-DNA damage, homeostatic levels (Mizusawa and Gottesman, 1983; Frank et al., 1996; Khattar, 1997; W F Wu et al., 1999; Seong et al., 2000; Neher et al., 2003; Neher et al., 2006; Nagashima et al., 2006; Pruteanu and Baker, 2009a; Pruteanu and Baker, 2009b). One study revealed that many SOS proteins were preferentially found in a ClpXP “trap” after DNA damage (Neher et al., 2006). The ClpXP protease complex is composed of two stacked rings of seven ClpP subunits each with a hexamer of ClpX subunits at each end. ClpX is a chaperone/unfoldase that recognizes specific peptide sequence, unfolds the polypeptide, translocates the unfolded chain into the ClpP chamber (reviewed in (Sauer and Baker, 2011)). ClpXP can associate with other proteins. These include SspB, an adaptor protein that aids in the identification and processing of certain proteins to be degraded by ClpXP (Levchenko et al., 2000; Dougan et al., 2003; Chowdhury et al., 2010) and ClpA, an alternate chaperone/unfoldase that can also associate with ClpP to form the ClpAP protease (Katayama et al., 1988).
The placement of the cell division septum is determined by several factors (reviewed in (Buddelmeijer and Beckwith, 2002; Harry et al., 2006; Vicente et al., 2006; Barák and Wilkinson, 2007; Rowlett and Margolin, 2015; Blaauwen et al., 2017)). Initiating the process is the placement of the Z-ring. The Z-ring is spatially oriented by two systems: the Min System and Nucleoid Occlusion (reviewed in (Lutkenhaus, 2007; Ling Juan Wu and Errington, 2011; Di Ventura et al., 2013)). The Min system, comprised of the MinC, MinD, and MinE proteins, collectively prevent assembly at the poles of the cells through an oscillating mechanism (Raskin and de Boer, 1999; Hu and Lutkenhaus, 1999). Deletions of components of the Min system result in a characteristic phenotype in which dividing cells place their septum incorrectly, resulting in long cells and mini-cells (extremely small cells with no DNA) (Adler et al., 1967). Nucleoid Occlusion is a second mechanism that helps to correctly place the Z-ring between partitioned nucleoids. This mechanism is facilitated by slmA, a gene found in a synthetic lethal screen with minCDE mutants. SlmA binds to sites in the oriC half of the chromosome such that it interferes with Z-ring formation in these regions thereby facilitating Z-ring formation in between chromosomes and preventing chromosome guillotining (Bernhardt and de Boer, 2005; Cho and Bernhardt, 2013). It has been found that ClpXP can help to further regulate the assembly of the Z-Ring by degrading FtsZ (Weart et al., 2005; Camberg et al., 2009; Camberg et al., 2011; Camberg et al., 2014; Viola et al., 2017). Additional work has shown that ClpX, acting as a chaperone, can remodel the cytoskeleton by inhibiting polymerization of FtsZ (Sugimoto et al., 2010). ClpXP also degrades ZapC (Buczek et al., 2016) and ZipA is known to protect FtsZ from ClpXP proteolysis (Pazos et al., 2013). Lastly, MinD is also found in the ClpXP trap after DNA damage (Neher et al., 2006).
In this study, we have tried to maximize the level of as many SOS proteins as possible by simultaneously inactivating the LexA repressor and the ClpXP protease. We show that the lexA51 ΔclpP double mutant shows a unique mixture of cell division and cell shape phenotypes. The cell division phenotype resembles the classic “mini-cell” phenotype with three cells types: normal cells, long cells and mini-cells. The cell shape phenotype shows that some of the long cells have branching protrusions or “bumps” along their length. Here, we show that the RecN protein is required for the novel cell division phenotype in lexA51 ΔclpP mutants and that overproduction of RecN from the chromosome (recNop) in a ΔclpP mutant or a mutant of RecN that is likely resistant to ClpXP is sufficient to cause the long cell phenotype and a defect in chromosome partitioning. The mini-cell/bumps phenotype is more complicated, correlating with the absence of ClpXP and or LexA, although the absence of either alone is insufficient to cause these phenotypes. Lastly we show that the C-terminus of RecN, known to be critical for recognition by ClpXP, is also critical for normal RecN function in DNA repair as well as for the long cell/Par− phenotype. We propose that overproduction of RecN, a structural maintenance of chromosomes (SMC) protein (Graumann and Knust, 2009; Pellegrino et al., 2012; Upton and Sherratt, 2013), unchecked by the ClpXP protease in a lexA51 strain, forms structures with the nucleoids that inhibit the correct assembly, placement or function of the Z-ring.
EXPERIMENTAL PROCEDURES
Bacterial strains and media:
All bacterial strains used in this work are derivatives of Escherichia coli K-12 and are described in Table S1. The strains in the background JC13509 have the mutation sulB103, an allele of ftsZ that does not respond to the cell division inhibition caused by SulA during SOS (Bi and Lutkenhaus, 1990). Therefore, any cellular filamentation is due to the mutations in the strain and not due to induction of SOS. Strains constructed in the BW25113 background have a deletion of sulA. Some strains also carry a translational fusion of hupA to the fluorescent protein mCherry to visualize nucleoids directly without fixing or staining the cells. This mutant has been described elsewhere and used in different situations (Marceau et al., 2011; Pelletier et al., 2012). To the best of our knowledge, no phenotype has yet been found for this fusion gene. Most strains were generated using P1 transduction. The protocol for P1 transduction has been described previously (Willetts et al., 1969). All transductants were selected on 2% agar plates containing either Luria broth (LB) or 56/2 minimal medium containing 0.2% glucose, 0.001% thiamine, and specified amino acids (Willetts et al., 1969). Antibiotics used for selection included kanamycin (50 μg/mL), chloramphenicol (25 μg/mL), or tetracycline (10 μg/mL). Transductants were purified on the same type of media on which they were selected.
Creating mutations in recN:
To overproduce RecN, a strong promoter and optimized ribosome binding site was inserted directly in front of the recN start codon. This promoter and RBS site has been described previously (Massoni et al., 2012). Oligonucleotide primers (Table S2) were used to amplify the “overexpression” promoter from the radAop construct (Massoni et al., 2012) with ends homologous to sequences upstream (prSJS1400) and downstream (prSJS1401) of the recN ATG start codon. This fragment (also encoding chloramphenicol resistance between two flp sites) was then recombined onto the chromosome before the recN ATG by linear transformation using standard recombineering methods (Datsenko and Wanner, 2000). Chloramphenicol resistance was selected. The construct was verified by DNA sequencing. This construct allows for SOS-independent, constitutive RecN production. RecNop is also called recA4171.
Mutations (recN4172 (A552D, A553D), del(recN)4173, recN4174 (A552S, A553V), in the terminal 3’ end of the gene were introduced using a similar method to the above. Oligonucleotide primers (Table S2) were designed with homology to the end of the cat cassette (just after the flp site) used above, followed by the appropriate sequence to introduce the mutations in the recN gene, followed by sequences just upstream of these changes (prSJS1599, prSJS1607 and prSJS1608 for mutations recN4172, del(recN)4173 and recN4174 respectively). These were then combined with an oligonucleotide primer (prSJS1601) with homology to the other end of the cat cassette (such that it bound just upstream of the recN promoter sequences) and to sequences just after the recN stop codon. Using these primer pairs, a fragment of DNA containing homology to the 3’ end of the recN with one of the three mutations, the cat gene and the region just after the recN stop codon was amplified. The regions of homology were then extended in a second PCR reaction using prSJS1600 and prSJS1594. The fragments of DNA were then used to transform cells with either the normal recN promoter region or the recNop mutant (described above), using the recombineering method (described above) to generate CatR strains with the appropriate recN mutations on the chromosome. Note that these recN mutants have a non-flippable cat gene just after their stop codons.
Preparation of cells for microscopy
Cells were grown first as overnights in Luria broth. Then they were diluted 1:50 into minimal media and grown for approximately 2.5 hours into log phase. Then 3 μl of the minimal media grown culture was loaded onto a fresh 2% agarose pad made from minimal medium with low-melting agarose. A coverslip was added on top of the cells. Protocol for preparing agarose slides has been described elsewhere (Levin 2002). Cells on slides were incubated for 2–3 hours at 37°C. Images (phase contrast and fluorescent) were taken for at least 9 different fields of view (3 fields on 3 different days) and analyzed. A Nikon E600 microscope equipped with automated filter wheels, shutters, CoolLED light source and an ORCA-ER camera were used for image acquisition.
Analysis of microscopic images
Images were analyzed with the following software: I-Vision (BioVision Technologies, Inc.), OpenLabs 5.5.1 (Improvision, Inc.), SuperSegger (Stylianidou et al., 2016) and MatLab R2016a (Mathworks, Inc.). Individual cells were defined using SuperSegger and strains were quantified for number of cells, cell area, number of mini cells, bumps, etc, using specially written Matlab programs. Statistical analysis was completed on the cell area with the Student’s t-test. The cutoff p < 0.001 was used to determine significance. p-values are reported in footnotes for each Table. We report the total number of pixels in a cell for the cell area and use this as a proxy for cell length. The average cell width for 80–100 cells, in number of pixels, for the two extreme types of cells studied: lexA+ clpP+ cells and lexA51 ΔclpP, are 11.3 ± 1.9 and 11.4 ± 1.6 respectively, and thus are very similar. Given our microscopic parameters, one pixel is equal to 0.0625 microns. This validates our assumption that cell area in pixels is a good proxy for cell length.
Western Blots
Cells were grown in LB medium overnight. The absorbance at 600 nm of the cultures diluted four-fold were determined by spectrophotometry. 250 μl of the cultures were harvested by centrifugation at 13,000 rpm for 5 min at room temperature. Cell pellets were resuspended in 75 ul of 5x sample buffer (87.5 mM Tris-HCL ph 6.8, 25% glycerol, 0.5% bromphenol blue, 10% SDS, 5% β-mercaptoethanol) and boiled at 95°C for 5 min, then incubated for 10 min on ice. Crude cell lysates were further diluted five-fold with water and loaded on NuPAGE Novex 4–12% Bis-Tris Protein Gel (1.0 mm, 15-well, Thermo Fisher Scientific) in the amount normalized to the value of optical density of the sample with the lowest OD600, so that equal amount of lysed cells were loaded per well. Prestained protein standard was loaded in one of the wells (NEB, P7712S, 3 μl). SDS-PAGE was performed in MOPS running buffer at 100 V for 2 hours, following by transfer to PVDF membrane at 14 V overnight at 4°C. After blocking in 5% nonfat milk for 3 hours, the membrane was incubated with primary antibody for 1 hour, washed and incubated with horseradish peroxidase-conjugated secondary antibody for 1 hour. Immunodetection was performed with Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific). Images were acquired using ChemiDoc Touch Imaging System (Bio-Rad) and analyzed using ImageJ software. Blots were stained either with anti-RecN rabbit antisera (a kind gift from Ken Marians) diluted 1:10,000 and HRP-conjugated donkey anti-rabbit IgG (1:1,000, Amersham, NA934). Relative density values obtained from ImageJ for each sample were subjected to subtraction of the value corresponding to the non-specific band in SS5200 strain. These amounts were normalized to the OD600 of each cell culture to equalize the amount of cell lysate loaded per well. Data were presented as a mean adjusted relative density ± standard deviation.
RESULTS
To maintain viability when expressing the SOS response constitutively, it is necessary to have either a sulA or sulB mutation to prevent lethality due to cellular filamentation (Huisman et al., 1980; Lutkenhaus, 1983). Therefore, many strains used in this work have sulB103, an allele of ftsZ that does not respond to the SOS cell division inhibition mediated by SulA (Lutkenhaus, 1983; Bi and Lutkenhaus, 1990). Hence, any changes in cell size are likely due to the added mutations in the strains and not due to SOS filamentation. Furthermore, some strains have hupA::mcherry fusion gene in place of the wild type hupA gene to visualize nucleoids directly with fluorescence microscopy. This fusion gene has been described and characterized elsewhere (Marceau et al., 2011; Pelletier et al., 2012). The data collected in this work is in the form of phase contrast and fluorescent images of cells grown 2–3 hours on minimal media agarose pads at 37°C. These images were then quantitated using SuperSegger (Stylianidou et al., 2016) to identify the cells and Matlab programs to analyze the cell area and shape (see Materials and Methods). Statistical analyses of average cell area were done with the Student’s t-test. p values less than 0.001 are considered significant.
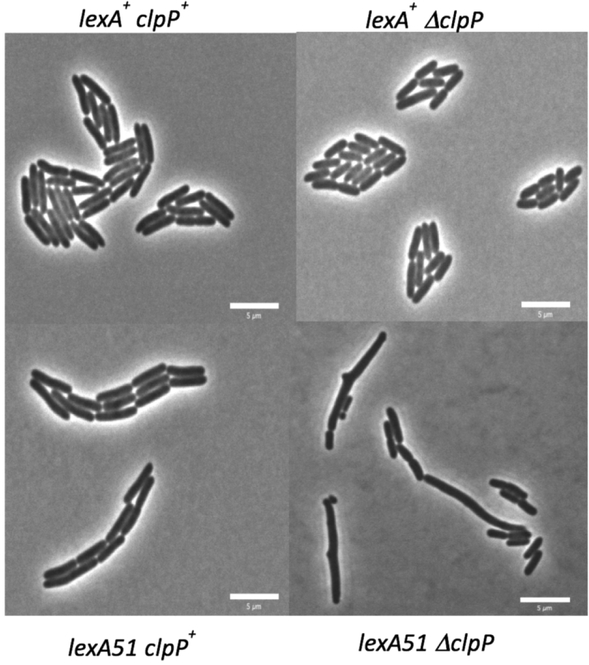
ΔclpP lexA51 and ΔclpX lexA51 mutants show a novel cell division phenotype
ClpXP protease specifically targets several SOS proteins after induction of the SOS response, implying an active role of proteolysis in returning the cell to a normal state following DNA damage repair (Neher et al., 2006). E. coli encodes other proteases, chaperones and adaptor proteins to help it maintain protein homeostasis, including ClpAP, Lon, DegP, HslUV, ClpB and SspB (Strauch et al., 1989; Sauer and Baker, 2011). ClpXP, Lon and HslUV have been shown to also degrade SOS proteins (see above). It was hypothesized that removing any of these proteolytic machines, chaperones or adaptors in an SOS constitutive strain might yield even higher levels of SOS proteins that in turn might impede cell function and create novel phenotypes. To explore this possibility, lon, hslU, degP, clpA, clpB, clpX, clpP, and sspB genes were deleted in a lexA51 strain and then these double mutant strains were visualized microscopically for phenotypes.
When initially characterizing these mutants, it was seen that some cells were longer than normal, some mini-cells were produced and some branching structures were seen. As mentioned above, mini-cells are very small cells without nucleoids that have arisen from septum formation at the poles. Branching phenotypes have been observed previously (Denome et al., 1999; Gullbrand et al., 1999; Nelson and Young, 2000; Nelson and Young, 2001; de Pedro et al., 2003; Potluri et al., 2012) and have been suggested to arise via the action of penicillin-binding proteins and are independent of cell division. Note that cell area is used as a proxy for cell length and the justification for this assumption is given in the Experimental Procedures section.
We determined average cell length and the frequency of mini-cells produced and branching structures per cell. As a different way to view cell area across the population from just the average cell area, we also binned the cell areas and reported the percentage of cells greater than 300 pixels (approximately the area of two cells). Table 1 summarizes this information for wild type, the lexA51 single mutant, the eight single mutants and the eight double mutants. It is shown that all protease, chaperone and adaptor single mutants have a minimal phenotype showing cell areas (even with binning) that are almost equal to wild type and very low frequencies of mini-cells and branching. The lexA51 single mutant shows cell area is greater on average and about 9% of the cells are greater than 300 pixels in area.
Table 1.
The effects of mutations in genes affecting protease, chaperone and adaptor functions on cellular size and shape1
| Strain | lexA | Other | Average Cell Area | %Cells (Area>300) | Bumps | Mini Cells |
|---|---|---|---|---|---|---|
| SS7117 | + | + | 160 | 1 | 0 | 0 |
| SS9921 | 51 | + | 220 | 9 | 0 | 0.01 |
| SS10329 | + | ΔclpA | 170 | 1 | 0 | 0 |
| SS10342 | 51 | ΔclpA | 230 | 15 | 0 | 0.02 |
| SS10325 | + | ΔclpB | 170 | 1 | 0 | 0 |
| SS10339 | 51 | ΔclpB | 240 | 16 | 0 | 0 |
| SS9189 | + | ΔclpP | 160 | 1 | 0.01 | 0.01 |
| SS9190 | 51 | ΔclpP | 330 | 28 | 0.04 | 0.19 |
| SS10328 | + | ΔclpX | 130 | 0 | 0.01 | 0 |
| SS10334 | 51 | ΔclpX | 450 | 38 | 0.08 | 0.05 |
| SS12407 | + | ΔdegP | 180 | 2 | 0.01 | 0 |
| SS9953 | 51 | ΔdegP | 260 | 25 | 0.01 | 0.01 |
| SS12408 | + | ΔhslU | 180 | 1 | 0 | 0 |
| SS9955 | 51 | ΔhslU | 250 | 19 | 0 | 0.01 |
| SS10987 | + | Δlon | 160 | 1 | 0 | 0 |
| SS9954 | 51 | Δlon | 530 | 35 | 0.04 | 0.02 |
| SS10326 | + | ΔsspB | 160 | 0 | 0 | 0 |
| SS10338 | 51 | ΔsspB | 340 | 27 | 0.01 | 0.03 |
1400–3200 cells were counted for each strain. Average cell area is given in number of pixels. Bumps and mini cells are given as a frequency per total number of cells. Cells were grown in as mentioned the Experimental Procedures. The average cell area of ΔclpP lexA51 (SS9190) is significantly different from the ΔclpP single mutant (SS9189) and the lexA51 single mutant (SS9921) with p < 0.001 by the Student’s t-test.
The double mutants all show some increase relative to either of the single mutants. For clpA, clpB, degP and hslU, the double mutants (with lexA51), the increase is modest for cell area (changes from 220 to 230–260) and binning (changes from 9% to 15–25%) as compared to the lexA51 single mutant. Some mini-cells and branching structures are seen but these are either equal to the lexA51 cells or just above background (0 compared to 0.02). The other mutants ΔclpX lexA51, ΔclpP lexA51, ΔsspB lexA51, Δlon lexA51 all showed significant increases in cell area (changes from 220 to 330–530) and increases in the percent of cells above 300 pixels (changes from 9% to 27–38%) with significant mini-cell formation (changes from 0 to 0.02 – 0.19) and in most cases, significant branching (changes from 0 to 0.01 to 0.08).
It is concluded that while all the genes tested show some increase in phenotype, the largest synthetic increases are observed when lexA51 is combined with a deletion of clpX, clpP sspB and lon. Therefore, in log phase cells constitutively expressing the SOS response, and missing the ClpXP protease, the SspB adaptor protein and the Lon protease degrade (or help degrade) some protein(s), most likely an SOS protein, that when transcribed at SOS levels inhibits proper assembly, localization and or function of the Z-ring (we will collectively refer to this phenotype as a Z-ring problem since we will not report any experiments that distinguish between the three possibilities) for cell division, leading to the long/mini-cell phenotype. Since ClpX and ClpP form a complex that is often activated by having SspB deliver a protein to be degraded to ClpXP, we will pursue an explanation for the phenotypes associated with the ΔclpP lexA51 double mutant below (Figure 1). The lexA51 lon phenotype will be reported elsewhere.
Figure 1.
ΔclpP lexA51 produces a novel cell division phenotype that includes long cells, mini-cells and bumps. As explained in the Experimental Procedures section, the cells were grown on minimal media slabs of 2% low melting agarose at 37°C for 3–4 hours before imaging the cells with standard phase contrast microscopy. The pictures shown are examples of the types of cells seen with the following genotypes: A) SS7117 (clpP+ lexA+), B) SS9189 (ΔclpP lexA+), C) SS9921 (clpP+ lexA51) and D) SS9190 (ΔclpP lexA51).
The long/mini-cell phenotypes are additive between the lexA51 ΔclpP double mutant and the ΔminCDE single mutant
With the exception of the bumps, the phenotype of the ΔclpP lexA51 mutant was similar to that observed with the ΔminCDE mutant in that it shows both long cells and mini-cells (Alder et al. 1967). Table 2 shows that the average cell areas and the percentage of cells with area larger than 300 pixels is greater in the ΔminCDE mutants than in the ΔclpP lexA51 mutant and the frequency of mini-cell production was also 10-fold greater in the ΔminCDE mutant. Nonetheless, there is some similarity in the phenotype. Therefore, it was of interest to test if the long/mini-cell phenotypes observed in the ΔclpP lexA51 were occurring by the same or different pathway as that of the ΔminCDE. This was genetically tested by constructing a ΔclpP lexA51 ΔminCDE triple mutant and comparing it to the ΔminCDE single mutant and the ΔclpP lexA51 double mutant. Table 2 shows that the ΔclpP lexA51 ΔminCDE triple mutant revealed a highly significant, additive increase in average cell area compared to either of the starting mutants and an increase in the frequency of branching structures. There was no difference in mini-cell production. This suggests that additive increase in cell area and branching are likely occurring by different pathways in the two mutant strains. However, mini-cell production is occurring by the same pathway and this is dominated by the absence of MinCDE.
Table 2.
The effects of mutations in minCDE, clpP and lexA on cellular size and shape1
| Strain | lexA | clpP | minCDE | Average Cell Area | %Cells (Area>300) | Bumps | Mini cells |
|---|---|---|---|---|---|---|---|
| SS7117 | + | + | + | 160 | 1 | 0 | 0 |
| SS9190 | 51 | Δ | + | 330 | 28 | 0.04 | 0.19 |
| SS12699 | + | + | Δ | 570 | 75 | 0.02 | 2.06 |
| SS9194 | 51 | Δ | Δ | 840 | 70 | 0.14 | 2.06 |
850–3200 cells were counted for each strain. Average cell area is given in number of pixels. Bumps and mini-cells are given as a frequency per total number of cells. Cells were grown as mentioned the Experimental Procedures. The average cell area of ΔclpP lexA51 (SS9190) is significantly different (p < 0.001) from the ΔminCDE single mutant (SS12699) by the Student’s t-test. The differences in average cell area between both the ΔclpP lexA51(SS9190) double mutant and the ΔminCDE single mutant (SS12699) and the ΔclpP lexA51 ΔminCDE (SS9194) triple mutant are significant (p < 0.001).
recN is required for the long/mini-cell/branching phenotype in a ΔclpP lexA51 mutant
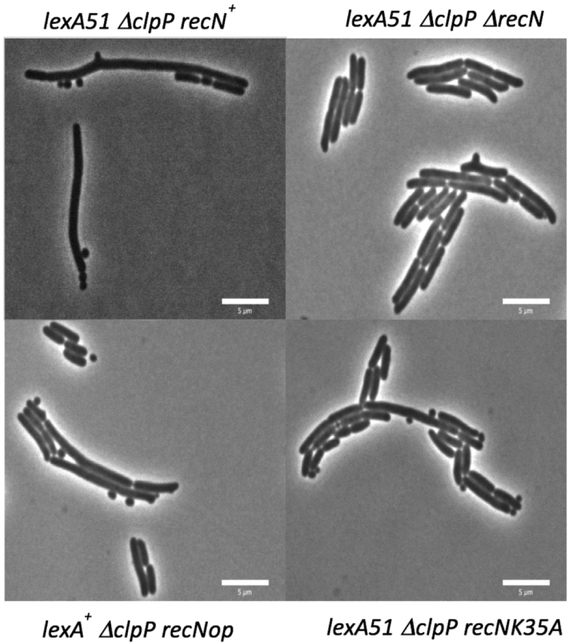
From the above experiments, it was hypothesized that the long/mini-cell/branching phenotype produced in the ΔclpP lexA51 strain was due to the accumulation of some SOS protein(s) that somehow cause a Z-ring problem in log phase cells. While many proteins are produced at high levels during the SOS response and it is possible that more than one protein may be responsible for this phenotype, we assumed that this long/mini-cell/branching phenotype was produced by a single protein. Therefore, to determine the protein required for this phenotype, we introduced deletion mutations into several SOS genes in the ΔclpP lexA51 strain and screened these triple mutants for suppression of the phenotype. The SOS genes tested were dinB, dinD, dinI, polB, recA, recF, recN, recQ, recX, ruvA, ruvB, sulA, umuC, umuD, uvrA, uvrB, uvrC, uvrD and yebG. Qualitative screening of the triple mutants microscopically revealed that only introduction of the ΔrecN mutation into the ΔclpP lexA51 mutant reverted the phenotype back to cells resembling the lexA51 single mutant in terms of average cell area, frequency of bumps and mini-cells (Figure 2). Quantitatively, Table 3 shows that ΔrecN suppresses the long cell phenotype (average cell area) of ΔclpP lexA51 from 330 pixels to 200 pixels (p <0.001) with an accompanying decrease in the binning (change from 28% to 5%), a large decrease in the frequency of mini-cell production (changes from 0.19 to 0.01) and a small, 2-fold, decrease in branching (change from 0.04 to 0.02). This implies that recN is required for a large portion of all three phenotypes.
Figure 2.
recN-dependence of the long cell/mini-cell and bumps phenotypes seen in the ΔclpP lexA51 double mutant. As explained in the Experimental Procedures section, the cells were grown on minimal media slabs of 2% low melting agarose at 37°C for 3–4 hours before imaging the cells with standard phase contrast microscopy. The pictures shown are examples of the types of cells seen with the following genotypes: A) SS9190 (ΔclpP lexA51 recN+), B) SS9198(ΔclpP lexA51 ΔrecN), C) SS10337 (ΔclpP lexA+ recNop) and D) SS10357 (ΔclpP lexA51 recNK35A).
Table 3.
The effects of mutations in lexA, clpP, the recN promoter and recN on cellular size and shape1
| Strain | lexA | clpP | recNp | recN | Average Cell Area | %Cells (Area>300) | Bumps | Mini cells |
|---|---|---|---|---|---|---|---|---|
| SS7117 | + | + | + | + | 160 | 1 | 0 | 0 |
| SS9921 | 51 | + | + | + | 220 | 9 | 0 | 0.01 |
| SS9189 | + | Δ | + | + | 160 | 1 | 0.01 | 0.01 |
| SS12334 | + | + | + | Δ | 180 | 3 | 0 | 0 |
| SS10336 | + | + | op | + | 150 | 1 | 0 | 0 |
| SS10337 | + | Δ | op | + | 280 | 20 | 0.03 | 0.04 |
| SS9190 | 51 | Δ | + | + | 330 | 28 | 0.04 | 0.19 |
| SS9198 | 51 | Δ | + | Δ | 200 | 5 | 0.02 | 0.01 |
| SS10357 | 51 | Δ | + | K35A | 210 | 7 | 0.02 | 0.04 |
1400–3200 cells were counted for each strain. Average cell area is given in number of pixels. Bumps and mini-cells are given as a frequency per total number of cells. Cells were grown as mentioned the Experimental Procedures. All statistical tests were done using the Student’s t-test. The difference in average cell area of ΔrecN ΔclpP lexA51 (SS9198) and the ΔclpP lexA51 mutant (SS9190) is significant (p < 0.001). The difference in cell area of recNK35A ΔclpP lexA51 (SS10357) is significant from the ΔclpP lexA51 mutant (SS9190) (p < 0.001). The difference in average cell area of recNK35A ΔclpP lexA51 (SS10357) was not found to be statistically different than the average cell area of ΔrecN ΔclpP lexA51 (p = 0.025). The difference in average cell area between recNop ΔclpP (SS10337) and the ΔclpP mutant (SS9190) or the recNop mutant (SS10336) are significant (p < 0.001).
recN expression correlates with the phenotypes seen in the ΔclpP and lexA51 strains
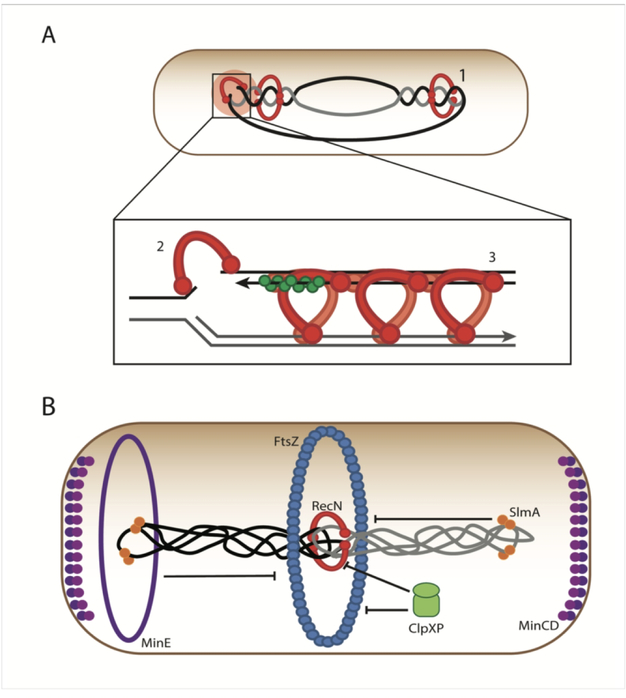
The RecN protein is a member of the Structural Maintenance of Chromosomes (SMC) family of proteins (Graumann and Knust, 2009; Pellegrino et al., 2012; Hirano, 2016) and has been shown to have roles in DNA repair, particularly in repair of double-strand breaks in E. coli and other bacteria (Meddows et al., 2005; Grove et al., 2009) and in Sister Chromosome Cohesion after DNA replication (Vickridge et al., 2017) (Figure 3A). More recently, RecN has been shown to stimulate RecA’s ability to catalyze D-loop formation in vitro (Uranga et al., 2017) (Figure 3A). Structurally, SMC complexes are dimers forming a “V” structure. The N- and C- terminal domains are separated by a large coiled-coil domain and a central hinge. The two terminal domains come together to form a larger globular domain that binds and hydrolyzes ATP. A general mechanism for the function of SMCs hypothesizes that the SMCs first bind to duplex DNA (loops or two duplexes either end to end or side by side), then the N- and C-terminal domains come together with DNA “caught” in the protein complex. This DNA/globular protein complex is then stabilized by binding ATP. Finally, ATP hydrolysis “releases” the structures. Many SMCs interact with other proteins to stabilize the DNA-globular domain structures (e.g., MukEF with MukB (Badrinarayanan et al., 2012; Bahng et al., 2016)). RecN seems to differ from other SMCs in that no accessory proteins have yet been suggested for RecN. Additionally, sequence comparisons and structural studies reveal that RecN coiled-coil region is shorter and stiffer than most other SMCs (Grove et al., 2009; Pellegrino et al., 2012). For these reasons RecN may use a similar, but not necessarily identical mechanism, for stabilizing DNA structures. Pellegrino and colleagues have suggested a spiral structure for RecN where two duplexes of DNA are stabilized Protein/DNA complex in a side by side arrangement (Pellegrino et al., 2012).
Figure 3.
Part A. RecN has been implicated in several roles in coordinating repair to DNA damage. (1) Normally, bidirectional replication leads to the formation of pre-catenanes, or inter-wound daughter strands. In the absence of DNA damage, the daughter strands are separated by Topoisomerase IV, allowing sister chromatid segregation. When DNA damage triggers the SOS Response, RecN may act to hold these precatenanes together, preventing segregation. This acts to keep the sister chromatids close, facilitating template-guided DNA damage repair (Vickridge et al., 2017). (2) RecN is also thought to perform an end-joining function during Double-Strand Break Repair (Meddows et al., 2005; Grove et al., 2009). (3) RecN may interact with RecA to facilitate homology search and strand invasion during homologous recombination of double-strand breaks (Uranga et al., 2017). Note that in all sections, whether RecN is diagrammed as part of a dimer or polymer, it is pictured as either an open circle or a helical structure as suggested by Pellegrino et al. (Pellegrino, Radzimanowski, de Sanctis, Boeri Erba, McSweeney, and Timmins, 2012b). Part B. Prior to cell division, the Z-ring forms via polymerization of FtsZ protein. This polymerization is dynamic and modulated by several proteins. It is known that the oscillating Min system prevents assembly of the Z-Ring at the poles via the action of MinC. The Nucleoid Occlusion system prevents the Z-Ring from forming prematurely over the nucleoid prior to complete of DNA replication via the protein SlmA. ClpXP has been shown to directly modulate the assembly of FtsZ polymers. During times of cell stress, the SOS response may be induced, triggering the expression of the protein RecN. RecN is known to coordinate double strand break repair. We postulate that early in an SOS Response, it acts as a cohesion-like protein to hold together the sister nucleoids. This provides an opportunity for the cell to repair DNA via homologous recombination using the newly replicated sister chromatid as a template. To ensure cell division does not progress before damage is repaired, high levels of RecN inhibit division. This provides a second way (in addition to SulA-dependent inhibition of FtsZ polymerization) for the cell to inhibit division while allocating resources to fix the damage. It is possible that RecN works with other SOS proteins in this function. This function is moderated by degradation by ClpXP. In the study above where RecN is overproduced in the absence of ClpXP, these extraordinarily high levels of RecN cause cell division defects.
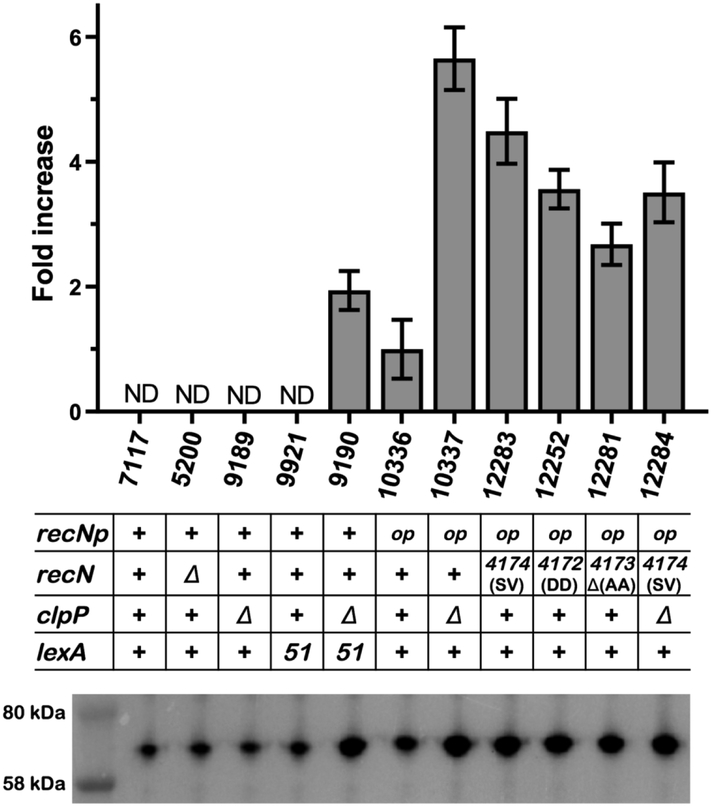
The above experiments suggest that RecN is required for the phenotypes. It is transcribed at high levels in a lexA defective strain and stabilized from protease digestion by removal of ClpP. Thus, the levels of RecN would increase in the ΔclpP lexA51 strain relative to either single mutant (they would be lower in either of the single mutants since they would be either transcribed at high levels and degraded (lexA51 clpP+) or transcribed at repressed levels and not degraded (lexA+ ΔclpP)). This was tested by performing Western Blots with those strains with antibodies raised against the RecN protein (provided by Dr. K. J. Marians, Sloan Kettering Institute). Figure 4 shows that the RecN antibodies detect a non-specific band of a protein in a recN deleted strain that migrates at the same position in the gel relative to RecN. This is seen in lane 3 of Figure 4 where the profile of a del(recN) strain is shown. To correct for this non-specific band, the total amount of intensity of the non-specific band is subtracted from the bands in the other strains. When this is done, the levels of RecN in wild type, lexA51 and ΔclpP strains are below the level of detection. However the amount of RecN in the lexA51 ΔclpP strain is detectable. This shows that increased levels of RecN protein correlates with the strains exhibiting the phenotypes.
Figure 4.
Western blot analysis of RecN expression. A typical Western blot is shown in the lower part of the figure. Quantitation of the Western blot is shown in the bar graph at the top of the Figure. The columns in the bar graph and the lanes in the westerns line up with one another and the strain number and genotype are listed in between. As explained in the Experimental Procedures section, whole cell lysates from overnight cultures were loaded on a precast polyacrylamide gel in the amount normalized to the values of OD600, proteins were separated by SDS-PAGE and detected using anti-RecN antibodies. Data are shown as a mean adjusted relative density of the bands ± standard deviation calculated from four independent experiments, where only three out of four values were taken into consideration. Strain numbers are indicated on the X axis. One of the analyzed blots is shown on the bottom. recN promoter is labeled as recNp. The amount of RecN in SS10336, after subtraction of the non-specific band in SS5200, is set to 1 and amounts higher are given by the fold increase given on the Y-axis. The RecN levels for strains SS7117, SS9189 and SS9921 were non-detectable (ND) above the non-specific band in SS5200.
While the above experiments show both the presence and amount of RecN correlate with the long cell phenotype in the ΔclpP lexA51 strain, it is possible that other proteins may also be required. To test if overproduction of RecN alone is sufficient for the phenotype, we constructed a strain that produced RecN at high levels constitutively and independently of SOS regulation. This was done by inserting a strong promoter and ribosome binding site (designated recNop for recN “overproducer”) in between the LexA-regulated recN promoter (recNp) and the start codon of the recN gene using a promoter sequence identical to the one used to overproduce the radA protein (Massoni et al., 2012). Figure 4 shows that in the presence of ClpP, the amount of RecN protein detectable is about 50% of the levels seen in a ΔclpP lexA51 strain. Figure 2 shows that the recNop clpP+ strain does not show the phenotype. It is possible, however, that the level of transcription attained with recNop is sufficient for the long/mini-cell phenotype but that the amount of RecN is not sufficient because the RecN protein is still degraded at a high enough rate by ClpXP such that the protein does not accumulate to produce the phenotype. If so, then the long/mini-cell phenotype should be visible in a lexA+ strain with recNop and ΔclpP. Figure 2 shows that the recNop ΔclpP strain produces a similar phenotype as the ΔclpP lexA51 strain. Table 3 shows that the average cell area of the recNop ΔclpP double mutant (280 pixels) is statistically higher than the average cell area of the recNop (150 pixels) or the ΔclpP (160 pixels) single mutants (for both comparisons, p < 0.001). It is not as great, however, as the ΔclpP lexA51 strain (330 pixels). Table 3 shows that the frequency of bump formation is also similar between the recNop ΔclpP and ΔclpP lexA51 strains, however the frequency of mini-cells is much higher in the ΔclpP lexA51 strain than the recNop ΔclpP. Figure 4 shows that the recNop ΔclpP strain has greatly increased levels of RecN (about 3-fold over a ΔclpP lexA51 strain). From this we conclude that high levels of RecN (overproduction of RecN constitutively in the absence of degradation by ClpXP protease) is sufficient to cause a Z-ring problem albeit not to the same level as seen in the ΔclpP lexA51 strain. We also see that there is not a strict correlation between the amount of RecN protein and the severity of the phenotype: the recNop ΔclpP strain has higher levels of RecN than the ΔclpP lexA51 strain, but the ΔclpP lexA51 strain has slightly stronger phenotype than the recNop ΔclpP strain. Therefore, it is possible that other factors contribute to the long cell/mini-cell phenotype. This will be discussed further below.
recNop ΔclpP strains are defective in their ability to partition their nucleoids
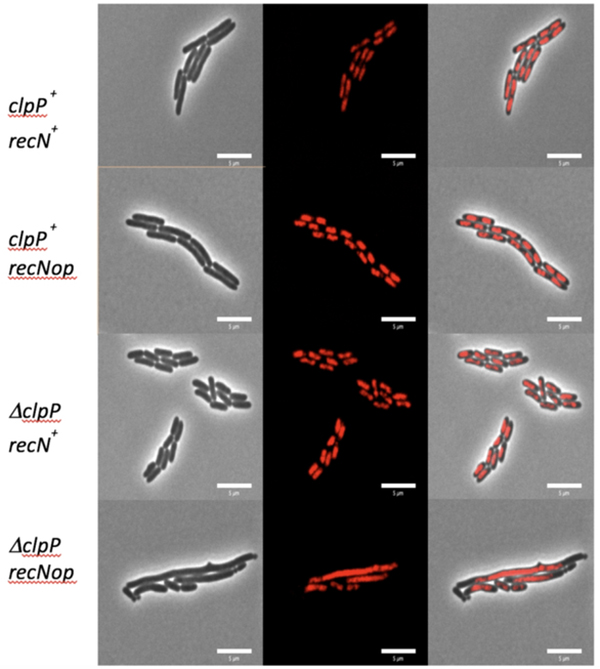
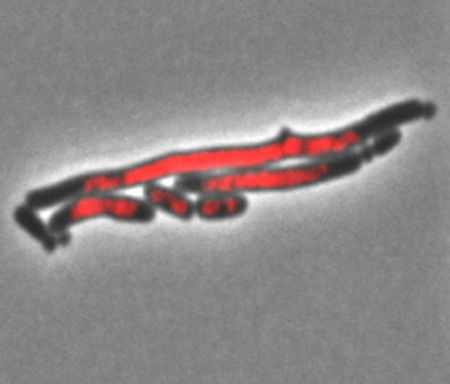
SMC proteins provide a method to stabilize higher order DNA structures in a non-covalent, possibly temporary way. These DNA structures could be ends of broken DNA, loops of DNA in same nucleoids or DNA between nucleoids. Strains deleted for RecN with I-Sce I generated DSBs (Meddows et al., 2005), treatments with mitomycin C (Keyamura et al., 2013; Vickridge et al., 2017) or UV (Odsbu and Skarstad, 2014) show nucleoids with morphological differences from wild type. We hypothesized that RecN overproduction caused the long/mini-cell phenotype by holding together different nucleoids, possibly recently replicated sister chromosomes, through loops of DNA on each nucleoid such that a Z-ring would not form between the nucleoids, leading to longer cells. If so, strains overproducing RecN should also have a defect in the cell’s ability to segregate or partition two sister chromosomes after DNA replication before cell division. To test this, hupA::mCherry, a fusion protein that allows for direct visualization of the nucleoids without fixing or staining the cells, was combined with the wild-type, ΔclpP, recNop and recNop ΔclpP mutant strains. The wild-type, ΔclpP and recNop single mutants showed one (or two) distinct, condensed nucleoid per cell (Figure 5). The recNop ΔclpP double mutants on the other hand had a partitioning defective phenotype where many nucleoids are diffuse and spread across the entirety of long cells (Figure 5). Table 4a shows that the presence of the hupA-mcherry has no significant effect on the any of the phenotypes measured in wild type, recNop, ΔclpP or recNop ΔclpP strains. This supports the idea that overproduction of RecN in the absence of ClpP causes more cohesion between parts of sister nucleoids, leading to poor segregation, and this in turn inhibits proper Z-ring assembly, placement and or function between the nucleoids and ultimately problems in cell division.
Figure 5.
Visualization of the partition defective phenotype seen in the recNop ΔclpP double mutants. Each strain contains a hupA-mCherry fusion protein that allows for direct visualization of the nucleoids. The three columns down show the phase contrast image (left column), red nucleoid image (middle column) and the merged image (right column). Down the figure, the rows are different strains and genotypes: A) SS6321 (clpP+ recN+), B) SS10346 (clpP+ recNop), C) SS10350 (ΔclpP recN+) and D) SS10353 (ΔclpP recNop). Only the double mutant (ΔclpP recNop) shows long cells, mini cells, and bumps with diffuse nucleoid in the long cells.
Table 4a.
The effects of amino acids changes in the last two codons of RecN on log/mini-cell phenotype and the ability to partition the nucleoids when overproducing the RecN protein1
| Strain | recNp | recN | clpP | hupA | Average Cell Area | %Cells (Area>300) |
Bumps | Mini cells | Partitioning Ability2 |
|---|---|---|---|---|---|---|---|---|---|
| SS7117 | + | + | + | + | 160 | 1 | 0 | 0 | ND |
| SS9189 | + | + | Δ | + | 160 | 1 | 0.01 | 0.01 | ND |
| SS10336 | op | + | + | + | 160 | 1 | 0 | 0 | ND |
| SS10337 | op | + | Δ | + | 280 | 20 | 0.03 | 0.04 | ND |
| SS12691 | op | 4172 (DD) | + | + | 180 | 3 | 0 | 0 | ND |
| SS12692 | op | Δ4173 (AA) | + | + | 180 | 3 | 0 | 0 | ND |
| SS12693 | op | 4174 (SV) | + | + | 320 | 20 | 0.01 | 0.01 | ND |
| SS6321 | + | + | + | mCherry | 170 | 1 | 0 | 0 | ++ |
| SS10350 | + | + | Δ | mCherry | 160 | 1 | 0.01 | 0 | ++ |
| SS10346 | op | + | + | mCherry | 180 | 2 | 0 | 0 | ++ |
| SS10353 | op | + | Δ | mCherry | 280 | 18 | 0.04 | 0.05 | − |
| SS12252 | op | 4172 (DD) | + | mCherry | 190 | 4 | 0 | 0 | ++ |
| SS12281 | op | Δ4173 (AA) | + | mCherry | 210 | 5 | 0 | 0 | ++ |
| SS12283 | op | 4174 (SV) | + | mCherry | 310 | 21 | 0.01 | 0.01 | − |
The letters in parentheses after the allele number represent the amino acids changed or deleted. 1900–4000 cells were counted for each strain. Average cell area is given in number of pixels. Cells were grown as mentioned the Experimental Procedures. All statistical tests were done using the Student’s t-test. The p value for the significance in the difference in average cell area between recNop recN4172 (A552D, A553D), recNop del(recN)4173, and recNop recN4174 (A552S, A553V), (SS12691, SS12692, and SS12693, respectively, or SS12252, SS12281, and SS12283, respectively) and recNop mutant (SS10336 or SS10346) is highly significant (p<0.001).
Example of cells that are properly partitioning their chromosome and ones that are not are shown in Figure 5.
The C-terminal alanines are critical for both RecN’s normal role in DNA repair and its role in the production of the long/mini-cell overproduction phenotype
Inactivation of ClpXP is required for recNop to cause the overproduction phenotypes. It is possible that the requirement for the absence of ClpXP is necessary either for RecN to accumulate to high levels or that some other protein(s) that is normally degraded by ClpXP is also required for the long/mini-cell phenotype or both. To test between these alternatives, we mutated the end of recN such that RecN is no longer recognized by ClpXP. Others has shown that the C-terminal amino acid sequence of RecN (LAA) is critical for ClpXP to recognize the RecN protein and degrade it (Nagashima et al., 2006). These authors showed that mutating the terminal two alanine residues (AA) to aspartic acid residues (DD) inhibits the ability of ClpXP to recognize RecN and degrade it; and that this protein, expressed from a plasmid, is sufficient to complement normal RecN function for survival after exposure to Mitomycin C. We hypothesized that changing the last two amino acids residues from alanine to aspartic acid in the context of the constitutive promoter (hereafter called recNop recN4172 (A552D, A553D)) would stabilize RecN to the ClpXP protease and produce the long/mini-cell phenotype in a clpP+ strain. Figure 4 shows that the recNop recN4172 (A552D, A553D) clpP+ strain produces slightly more RecN (almost 2-fold) than the lexA51 ΔclpP strain, but not quite as much as the recNop ΔclpP strain. Interestingly however, Table 4a shows that recNop recN4172 (A552D, A553D) clpP+ strain increased the average cell size minimally (from 160 pixels in SS10336 to 180 pixels in SS12691). This did not correspond to the increase in average cell size seen for recNop ΔclpP (280 pixels in SS10337). It also did not produce the partitioning defective phenotype (Table 4a -- SS12252). We conclude that placement of the two aspartic acid residues at the last two codons of recN did stabilize the protein from the effects of ClpXP, they were not optimal, however, to produce the long/mini-cell/Par− phenotype.
Given the above hypothesis and result, it would seem that some other protein normally degraded by ClpXP is also required required for the long cell phenotype. However, there is another possibility: the last two amino acids, critical for recognition by ClpXP, are also critical for the long/mini-cell phenotype; recN4172 (A552D, A553D) introduced two negative charges in the C-terminus and changed the hydrophobic character of the C-terminal end from hydrophobic to polar. Either or both of these characteristics may be critical for the long/mini-cell phenotypes. To test this, we made use of previous studies of model proteins recognized by ClpXP and having the sequence LAA at its C-terminal end (Flynn et al., 2003; McGinness et al., 2006). We made two mutants that we hypothesized would eliminate ClpXP recognition and maintain the hydrophobic nature of the C-terminus as much as possible. One mutant deleted the final two alanines (called del(recN)4173) and the other mutant replaced the two alanines with serine and valine (called recN4174 (A552S, A553V)). Figure 4 shows that in both cases, these mutations stabilized the RecN protein relative to the effects of ClpXP about as well as the recN4172 (A552D, A553D) mutations. Table 4a showed that removal of the two alanines in the recNop del(recN)4173 mutant had a much smaller average cell area compared with the recNop ΔclpP (180 pixels in SS12692 vs. 280 pixels in SS10337). It also did not show appreciable levels of long/mini-cells, bumps or the Par− phenotype. However, changing the two amino acids from alanine to serine and valine as in the recNop recN4174 (A552S, A553V) mutant, produced cells that were on average slightly larger than the recNop ΔclpP (320 pixels in SS12693 vs. 280 pixels in SS10337) strain and had the characteristic Par− phenotype. While recNop recN4174 did show some mini-cells and some branched structures (Table 4a), the frequency of this was below that detected in the recNop ΔclpP strain. We tentatively conclude that the amino acid changes in recN4174 (A552S, A553V) did not allow the degradation by ClpXP sufficiently that it was able to produce the long cell/Par− phenotypes. This overexpression, however, of the mutant protein in a ClpXP+ strain was not sufficient to produce the mini/branched-cell phenotypes to the same levels as the recNop ΔclpP strain.
Lastly, we tested whether the recN4174 mutations were compatible with normal RecN function by combining that allele into a recB21 recC22 sbcB15 sbcC201 strain where RecN has been shown previously to be required for full UV resistance (Lloyd et al., 1983). Table 4b shows that in the absence of RecN, the del(recN) strain suffered a 10-fold decrease in UV resistance. In the presence of a normal recN promoter, recN4172 (A552D, A553D) showed full ability to repair DNA after DNA damage as expected (see above). Full UV resistance was also seen recN4174 (A552S, A553V). Table 4b also showed that del(recN)4173 was 10-fold less UV resistance than wild type and behaved like a recN null mutant for this phenotype. We conclude that recN4174 is able to function normally in its ability to repair DNA and produce the long cell phenotype. Deletion, however, of the last two amino acids as in del(recN)4173 removes its ability to produce the overexpression phenotypes to a large degree and to function normally in DNA repair.
Table 4b.
The effects of amino acids changes in the last two codons of RecN on surviving UV irradiation1
| Strain | recNp | recN | Fraction surviving 40 J |
|---|---|---|---|
| JC7623 | + | + | 0.39 ± 0.17 |
| SS12613 | + | Δ100::kan | 0.039 ± 0.032 |
| SS12614 | + | 4172 (DD) | 0.36 ± 0.01 |
| SS12615 | + | Δ4173(AA) | 0.031 ± 0.01 |
| SS12616 | + | 4174 (SV) | 0.61 ± 0.06 |
The letters in parentheses after the allele number represent the amino acids changed or deleted. Cells were grown in minimal media and treated as previously described(Sandler and Clark, 1994). The results presented above are the averages (and Standard Deviations) of 2–3 independent experiments. These strains have the additional genotype of recB21 recC22 sbcB15 sbcC201 in the AB1157 background as given in Table S1.
The ATPase activity of RecN is required for the long cell phenotype but not the mini-cell/bump phenotype
The RecN from E. coli has been shown to be difficult to purify due to its poor solubility properties (Grove et al., 2009). Nonetheless, RecN from several other bacteria has been purified and studied. Haemophilus influenzae RecN (50% amino acid identity with E. coli RecN) is able to perform normally in DSBR when replacing the chromosomal copy of recN in E. coli (Grove et al., 2009). As with other SMC proteins, RecN from Haemophilus influenzae (Grove et al., 2009) and Deinococcus radiodurans (Reyes et al., 2010) have been shown to have a weak ATPase activity. The ATPase activity in RecN from D. radiodurans is dsDNA-dependent (Reyes et al., 2010). It has been shown that the recNK35A mutation, removing ATPase activity, also removes the ability of mutant recN protein to function in DSBR (Grove et al., 2009; Keyamura et al., 2013). At least two biochemical studies use DNA ligation as an assay to study RecN activity (Reyes et al., 2010; Pellegrino et al., 2012). The D. radiodurans RecN(K67A) (equivalent mutation in E. coli is RecN(K35A)) mutant no longer has ATPase activity, but is still able to bind DNA and ATP and is still able to function to a large degree in the ligation assay. GFP-RecN and GFP-RecN(K35A) fusion proteins expressed from a plasmid in a ΔrecN strain, after mitomycin C treatment, show accumulation of nucleoid associated foci in both strains. These foci, however, decrease in number over time in a GFP-RecN+ strain and not in GFP-RecN(K35A) (Keyamura et al., 2013). Taken together, it is clear that release of DNA and RecN from DNA/RecN complexes is dependent on RecN’s ATPase activity, however, the formation of these structures is not.
To test if the ATPase activity of RecN was required to produce the long cell/mini-cell and bumps phenotypes of the ΔclpP lexA51 strain, recNK35A was introduced into the strain. Figure 2 shows that to a large degree, the phenotype of recNK35A ΔclpP lexA51 strain is similar to that of ΔrecN ΔclpP lexA51. Table 3 shows that average cell area of the recNK35A ΔclpP lexA51 strain is statistically smaller than of the ΔclpP lexA51 strain and similar to that of ΔrecN ΔclpP lexA51. The recNK35A ΔclpP lexA51 triple mutant also has a number of bumps and mini-cells like the ΔrecN ΔclpP lexA51 strain. These observations suggest that recN’s ATPase activity is required to cause a Z-ring problem and that the cell division phenotype is likely not due to just RecN binding the DNA.
Long/mini-cell and bumps phenotypes are independent of sulB103 or strain background
The observations above suggest that the long cell, mini-cell and bumps phenotypes are due to a Z-ring problem in strains that have high levels of RecN due to the presence of ΔclpP, lexA51 or recNop alleles. Since the placement of the septum depends heavily on FtsZ and the strains used in this work contain a mutant allele of ftsZ (sulB103) to prevent SOS filamentation, it is possible that the presence of sulB103 somehow helps to cause the observed phenotype. Additionally, it is formally possible that the observed phenotypes are due to some other mutation specific to the JC13509 background and are not representative of a general phenomenon. To test these two possibilities simultaneously, some of the above mutations (ΔclpP, lexA51 and recNop) were reconstructed in the BW25113 background with a ΔsulA mutation. Table 5 shows that the combinations of ΔclpP and lexA51 on one hand, and ΔclpP and recNop on the other, both combine to produce the same types of phenotypes seen above in the JC13509 background with sulB103 although the quantitation shows that the size of the wild type cells is much smaller and the amount of the changes are different. From this we conclude that the trends seen in the JC13509 strains with changes in clpP, lexA and recN do not depend on sulB103 or the JC13509 background.
Table 5.
ΔclpP lexA51 recN-dependence of long/mini-cell and bumps phenotype in ΔsulA sulB+ (ftsZ+) strains with a BW25113 background1
| Strain | lexA | clpP | recNp | recN | Average Cell Area | %Cells (Area>200) | Bumps | Mini cells |
|---|---|---|---|---|---|---|---|---|
| SS10044 | + | + | + | + | 100 | 0 | 0 | 0 |
| SS10047 | 51 | + | + | + | 140 | 8 | 0 | 0.02 |
| SS10352 | + | Δ | + | + | 100 | 0 | 0 | 0 |
| SS12698 | + | + | + | Δ | 100 | 0 | 0 | 0 |
| SS10064 | 51 | Δ | + | + | 270 | 50 | 0.01 | 0.43 |
| SS10354 | + | + | op | + | 100 | 0 | 0 | 0 |
| SS10355 | + | Δ | op | + | 160 | 15 | 0.02 | 0.05 |
The strains used in this table are all derivatives of BW25113. They have a ΔsulA mutation instead of a sulB103 to prevent SOS filamentation. 2300–3700 cells were counted for each strain. Average Cell Area is given in number of pixels. Bumps and mini-cells are given as a frequency per total number of cells. Cells were grown in as mentioned the Experimental Procedures. All statistical tests were done using the Student’s t-test. The difference in average cell area of ΔclpP lexA51 (SS10064) and either of the single mutants (SS10047 and SS10352) is significant (p < 0.001). Likewise, the difference in average cell area of recNop ΔclpP (SS10355) and either of the single mutants (SS10354 and SS10352) is significant (p < 0.001).
DISCUSSION
The SOS response is a temporal response to damage in the cell’s DNA. Initially, SOS genes are repressed at the level of transcription by LexA. This repression can mean low levels of expression for some proteins (e.g., SulA – 300 per cell), mid-range levels for some (e.g., LexA -- 3,000 per cell) while others are produced at very high levels (e.g., RecA --10,000 copies per cell) (Li et al., 2014). Regardless of the initial levels, after detection of DNA damage, the rate of all SOS genes, by definition, are increased at the level of transcription (Courcelle et al., 2001). Thus, the level of many proteins increases dramatically and in unison. This is necessary to accomplish their tasks in the cell to repair the DNA (recA, ruvABC, uvrABC and recN), to inhibit cell division (sulA) and or increase the frequency of mutagenesis (umuCD and dinB). As many of these proteins may be detrimental to the cell under normal growth conditions, the levels of these proteins must be decreased to homeostatic levels after the damage has been repaired. E. coli has at least two mechanisms to reset the level of expression: a decrease in RecA-DNA filaments leads to increased concentrations of LexA, which begins repression again at the level of transcription, and the ClpXP, Lon and HslUV proteases degrade many SOS proteins.
We have studied the SOS response where all the SOS genes are transcribed at the highest level by removing the LexA repressor and have increased the stability of some SOS proteins by removing the ClpXP protease. The resulting cell has as its normal physiology a state that is typically transient: all of the SOS proteins expressed at the highest possible level. In order to construct this cell one must include either a sulA or sulB mutation to inhibits SOS induced inhibition of cell division. While this triple mutant (lexA51 ΔclpP sulB103) is viable, it is not the exact situation that would occur during the SOS response. There is no DNA damage with which the cell would have to contend and other proteins, not SOS proteins, but normally degraded by the ClpXP protease, would also increase in levels in the cell. These other proteins may contribute to the phenotypes seen.
We have found that the chronic situation in the lexA51 ΔclpP strain leads to problems in cell division that can be monitored by increased cell length, mini-cells and bumps. These phenotypes were “synthetic” requiring both high levels of transcription and the absence of the ClpXP protease. This point must be stressed, since in no case for any of the strains studied here, was any phenotype seen when only one condition was applied. Four phenotypes were measured: the average cell length, the frequency of mini-cells, branching and a partitioning defective phenotype. The Par- phenotype was found in the recNop ΔclpP and recNop recN4174 (A552S A553V) strains (Figure 5 and Table 4) and the lexA51 ΔclpP strain (Figure S1).
Figure 4 shows the level of RecN expression in several strains. The sensitivity of this measurement, however, was low because the antibodies used also detected a non-specific band that migrated at the same level as RecN using SDS-PAGE. Hence, detection of low levels of RecN (below the level of the non-specific band) was problematic. Nonetheless, it is clear that for high levels of RecN, like those equal or above the level produced in the lexA51 ΔclpP strain, one sees the overexpression phenotype (but one does not see them for the recNop strain even though its level of RecN expression was high enough for detection). There are some qualifications to that statement. The first is that the RecN protein must be wild type (and also must be deleted for clpP) or have at its C-terminus only certain amino acids that allow for the overproduction phenotype. RecN4174 (A552S A553V) is allowed while RecN (A552D A553D) or del(recN)4173 (deletion of the last two amino acids) are not (Table 4a). The second is that the severity of phenotype, particularly the bumps and mini-cells, is greatest in the lexA51 ΔclpP strains. Since lexA51 and ΔclpP affect a large number of proteins including RecN and the double mutant is able to produce more severe phenotypes than the recN4174 mutant (at slightly lower RecN levels), it is possible that other SOS proteins and ClpXP sensitive proteins (or combinations thereof) contribute the severity of the phenotypes.
The long cell and Par- phenotypes could be largely recapitulated with overexpression of a mutant RecN protein (recNop recN4174). We see that the mini-cell and bumps phenotypes seem to correlate better with the absence of ClpXP (and not so much with the overexpression of recN), suggesting that this leads to the imbalance of other cell division proteins in the cell. Two possibilities for these proteins are MinD and FtsZ. It is known that their balance is critical for proper placement of the cell division septum (see Introduction), both are known to be degraded by ClpXP (Camberg et al., 2009) or found in a ClpXP trap (Flynn et al., 2003; Neher et al., 2006) and both are known to produce mini-cells and or filaments when overproduced at modest levels (Ward and Lutkenhaus, 1985; Raskin and de Boer, 1999). The observation that removal of RecN or just RecN’s ATPase activity decreased the long cell phenotype and had a much smaller effect on the mini-cell and branching phenotypes is consistent with the above suggestion.
Several aspects of the mechanism of the lexA51 ΔclpP strain’s mutant phenotypes were examined. First, the long/mini-cell phenotype was reminiscent of phenotypes that has been associated with the removal of the minCDE genes. These proteins inhibit inappropriate Z-ring formation at the poles of cells. We show above the lexA51 ΔclpP double mutation is additive with cell length and mini-cell producing phenotypes of ΔminCDE. This is consistent with the idea that part of the long/mini-cell phenotype was due to overproduction of RecN. Based on the above results, one would suspect that ΔslmA would be viable with the lexA51 ΔclpP double mutation because the strain is wild type for minCDE. However, contrary to this idea, we saw that this combination (lexA51 ΔclpP ΔslmA) was lethal (data not shown). If, however, one focused on SlmA’s role in inhibiting Z-ring formation on the top of nucleoids, then the Par- phenotype of this strain (lexA51 ΔclpP) may better explain the lexA51 ΔclpP ΔslmA synthetic lethality.
It was found, in agreement with previous studies, that amino acids at the C-terminus are critical for the recognition of RecN by ClpXP. For the first time, however, it was shown that the terminal pair of alanines are also critical for RecN’s role in DNA repair. They may be changed, but they may not be deleted. Further studies will be necessary to provide information of the specific role of the C-terminus in RecN’s function in DNA repair.
Lastly, we tested whether these phenotypes were dependent on a particular allele of ftsZ (sulB103) used routinely to prevent SOS filamentation or if these phenotypes were the result of an unknown background mutation by testing for these phenotypes in the MG1655 (BW25113) background with a deletion of sulA. In both sets of strains and mutations, similar phenotypes were seen, albeit at not exactly the same levels, arguing that these phenotypes represent a general phenomenon.
Our model to explain the above results stems from other’s observations that RecN is an SMC that is implicated in nucleoid condensation (Odsbu and Skarstad, 2014) and sister chromosome cohesion (Vickridge et al., 2017) after DNA damage (Figure 3B). Both of these postulate a function for RecN in keeping together duplexes of DNA. These roles occur early in the SOS response when levels are still relatively low (but increasing). The phenotype reported here may mimic conditions that occur late in the SOS response when RecN levels have reached their highest point. We postulate that RecN starts to loop together different nucleoids, preventing FtsZ from forming a Z-ring between these nucleoids. It is possible that this mechanism represents yet another way that the SOS response inhibits cell division before it finishes with its DNA repair. It should be stressed that if this mechanism is indeed part of the SOS response, it is a transient, late phase of the response.
In closing, it is interesting that to point out that strains that were evolved to be highly resistance to ionizing radiation were found to have defects in ClpXP protease (Harris et al., 2009). Such mutations would give higher levels of RecN during the SOS response and could be a contributing factor to the suite of mutations offering high levels of resistance to double strand breaks.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by GM098885 from the National Institutes of Health. We would like to thank Robert G. Lloyd for sending strains and Ken Marians for supplying the RecN antibody. We would like to thanks Jodie Camberg and Peter Chien for reading the manuscript before submission and offering insightful comments. We have no conflicts of interest to report.
REFERENCES
- Adler HI, Fisher WD, Cohen A, and Hardigree AA (1967) Miniature Escherichia coli cells deficient in DNA. Proc Natl Acad Sci USA 57: 321–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badrinarayanan A, Reyes-Lamothe R, Uphoff S, Leake MC, and Sherratt DJ (2012) In vivo architecture and action of bacterial structural maintenance of chromosome proteins. Science 338: 528–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahng S, Hayama R, and Marians KJ (2016) MukB-mediated Catenation of DNA Is ATP and MukEF Independent. J Biol Chem 291: 23999–24008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barák I, and Wilkinson AJ (2007) Division site recognition in Escherichia coli and Bacillus subtilis. FEMS Microbiol Rev 31: 311–326. [DOI] [PubMed] [Google Scholar]
- Bernhardt TG, and de Boer PAJ (2005) SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over Chromosomes in E. coli. Mol Cell 18: 555–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E, and Lutkenhaus J (1990) Analysis of ftsZ mutations that confer resistance to the cell division inhibitor SulA (SfiA). J Bacteriol 172: 5602–5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaauwen, den T, Hamoen LW, and Levin PA (2017) The divisome at 25: the road ahead. Curr Opin Microbiol 36: 85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges BA (2005) Error-prone DNA repair and translesion DNA synthesis. II: The inducible SOS hypothesis. DNA Repair (Amst) 4: 725– 6– 739. [DOI] [PubMed] [Google Scholar]
- Buczek MS, Cardenas Arevalo AL, and Janakiraman A (2016) ClpXP and ClpAP control the Escherichia coli division protein ZapC by proteolysis. Microbiology (Reading, Engl) 162: 909–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buddelmeijer N, and Beckwith J (2002) Assembly of cell division proteins at the E. coli cell center. Curr Opin Microbiol 5: 553–557. [DOI] [PubMed] [Google Scholar]
- Camberg JL, Hoskins JR, and Wickner S (2009) ClpXP protease degrades the cytoskeletal protein, FtsZ, and modulates FtsZ polymer dynamics. Proc Natl Acad Sci USA 106: 10614–10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camberg JL, Hoskins JR, and Wickner S (2011) The interplay of ClpXP with the cell division machinery in Escherichia coli. J Bacteriol 193: 1911–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camberg JL, Viola MG, Rea L, Hoskins JR, and Wickner S (2014) Location of dual sites in E. coli FtsZ important for degradation by ClpXP; one at the C-terminus and one in the disordered linker. PLoS ONE 9: e94964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, and Bernhardt TG (2013) Identification of the SlmA active site responsible for blocking bacterial cytokinetic ring assembly over the chromosome. PLoS Genet 9: e1003304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury T, Chien P, Ebrahim S, Sauer RT, and Baker TA (2010) Versatile modes of peptide recognition by the ClpX N domain mediate alternative adaptor-binding specificities in different bacterial species. Protein Sci 19: 242–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, and Elledge SJ (2010) The DNA damage response: making it safe to play with knives. Mol Cell 40: 179–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle J, Khodursky A, Peter B, Brown PO, and Hanawalt PC (2001) Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics 158: 41–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, and Marians KJ (2000) The importance of repairing stalled replication forks. Nature 404: 37–41. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, and Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97: 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pedro MA, Young KD, Höltje J-V, and Schwarz H (2003) Branching of Escherichia coli cells arises from multiple sites of inert peptidoglycan. J Bacteriol 185: 1147–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denome SA, Elf PK, Henderson TA, Nelson DE, and Young KD (1999) Escherichia coli mutants lacking all possible combinations of eight penicillin binding proteins: viability, characteristics, and implications for peptidoglycan synthesis. J Bacteriol 181: 3981–3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Ventura B, Knecht B, Andreas H, Godinez WJ, Fritsche M, Rohr K, et al. (2013) Chromosome segregation by the Escherichia coli Min system. Mol Syst Biol 9: 686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan DA, Weber-Ban E, and Bukau B (2003) Targeted delivery of an ssrA-tagged substrate by the adaptor protein SspB to its cognate AAA+ protein ClpX. Mol Cell 12: 373–380. [DOI] [PubMed] [Google Scholar]
- Erill I, Campoy S, and Barbé J (2007) Aeons of distress: an evolutionary perspective on the bacterial SOS response. FEMS Microbiol Rev 31: 637–656. [DOI] [PubMed] [Google Scholar]
- Flynn JM, Neher SB, Kim YI, Sauer RT, and Baker TA (2003) Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol Cell 11: 671–683. [DOI] [PubMed] [Google Scholar]
- Frank EG, Ennis DG, Gonzalez M, Levine AS, and Woodgate R (1996) Regulation of SOS mutagenesis by proteolysis. Proc Natl Acad Sci USA 93: 10291–10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann PL, and Knust T (2009) Dynamics of the bacterial SMC complex and SMC-like proteins involved in DNA repair. Chromosome Res 17: 265–275. [DOI] [PubMed] [Google Scholar]
- Grove JI, Wood SR, Briggs GS, Oldham NJ, and Lloyd RG (2009) A soluble RecN homologue provides means for biochemical and genetic analysis of DNA double-strand break repair in Escherichia coli. DNA Repair (Amst) 8: 1434–1443. [DOI] [PubMed] [Google Scholar]
- Gullbrand B, Akerlund T, and Nordström K (1999) On the origin of branches in Escherichia coli. J Bacteriol 181: 6607–6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris DR, Pollock SV, Wood EA, Goiffon RJ, Klingele AJ, Cabot EL, et al. (2009) Directed evolution of ionizing radiation resistance in Escherichia coli. J Bacteriol 191: 5240–5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harry E, Monahan L, and Thompson L (2006) Bacterial cell division: the mechanism and its precison. Int Rev Cytol 253: 27–94. [DOI] [PubMed] [Google Scholar]
- Hirano T (2016) Condensin-Based Chromosome Organization from Bacteria to Vertebrates. Cell 164: 847–857. [DOI] [PubMed] [Google Scholar]
- Hu Z, and Lutkenhaus J (1999) Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol Microbiol 34: 82–90. [DOI] [PubMed] [Google Scholar]
- Huisman O, D’Ari R, and George J (1980) Further characterization of sfiA and sfiB mutations in Escherichia coli. J Bacteriol 144: 185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janion C (2008) Inducible SOS response system of DNA repair and mutagenesis in Escherichia coli. Int J Biol Sci 4: 338–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama Y, Gottesman S, Pumphrey J, Rudikoff S, Clark WP, and Maurizi MR (1988) The two-component, ATP-dependent Clp protease of Escherichia coli. Purification, cloning, and mutational analysis of the ATP-binding component. J Biol Chem 263: 15226–15236. [PubMed] [Google Scholar]
- Keyamura K, Sakaguchi C, Kubota Y, Niki H, and Hishida T (2013) RecA protein recruits structural maintenance of chromosomes (SMC)-like RecN protein to DNA double-strand breaks. J Biol Chem 288: 29229–29237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khattar MM (1997) Overexpression of the hslVU operon suppresses SOS-mediated inhibition of cell division in Escherichia coli. FEBS Lett 414: 402–404. [DOI] [PubMed] [Google Scholar]
- Levchenko I, Seidel M, Sauer RT, and Baker TA (2000) A specificity-enhancing factor for the ClpXP degradation machine. Science 289: 2354–2356. [DOI] [PubMed] [Google Scholar]
- Li G-W, Burkhardt D, Gross C, and Weissman JS (2014) Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157: 624–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T (1993) Instability and decay of the primary structure of DNA. Nature 362: 709–715. [DOI] [PubMed] [Google Scholar]
- Lindahl T, and Wood RD (1999) Quality control by DNA repair. Science 286: 1897–1905. [DOI] [PubMed] [Google Scholar]
- Little JW, and Mount DW (1982) The SOS regulatory system of Escherichia coli. Cell 29: 11–22. [DOI] [PubMed] [Google Scholar]
- Lloyd RG, Picksley SM, and Prescott C (1983) Inducible expression of a gene specific to the RecF pathway for recombination in Escherichia coli K12. Mol Gen Genet 190: 162–167. [DOI] [PubMed] [Google Scholar]
- Lutkenhaus J (2007) Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu Rev Biochem 76: 539–562. [DOI] [PubMed] [Google Scholar]
- Lutkenhaus JF (1983) Coupling of DNA replication and cell division: sulB is an allele of ftsZ. J Bacteriol 154: 1339–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marceau AH, Bahng S, Massoni SC, George NP, Sandler SJ, Marians KJ, and Keck JL (2011) Structure of the SSB-DNA polymerase III interface and its role in DNA replication. EMBO J 30: 4236–4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massoni SC, Leeson MC, Long JE, Gemme K, Mui A, and Sandler SJ (2012) Factors limiting SOS expression in log-phase cells of Escherichia coli. J Bacteriol 194: 5325–5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinness KE, Baker TA, and Sauer RT (2006) Engineering controllable protein degradation. Mol Cell 22: 701–707. [DOI] [PubMed] [Google Scholar]
- Meddows TR, Savory AP, Grove JI, Moore T, and Lloyd RG (2005) RecN protein and transcription factor DksA combine to promote faithful recombinational repair of DNA double-strand breaks. Mol Microbiol 57: 97–110. [DOI] [PubMed] [Google Scholar]
- Mizusawa S, and Gottesman S (1983) Protein degradation in Escherichia coli: the lon gene controls the stability of sulA protein. Proc Natl Acad Sci USA 80: 358–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima K, Kubota Y, Shibata T, Sakaguchi C, Shinagawa H, and Hishida T (2006) Degradation of Escherichia coli RecN aggregates by ClpXP protease and its implications for DNA damage tolerance. J Biol Chem 281: 30941–30946. [DOI] [PubMed] [Google Scholar]
- Neher SB, Sauer RT, and Baker TA (2003) Distinct peptide signals in the UmuD and UmuD” subunits of UmuD/D” mediate tethering and substrate processing by the ClpXP protease. Proc Natl Acad Sci USA 100: 13219–13224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neher SB, Villén J, Oakes EC, Bakalarski CE, Sauer RT, Gygi SP, and Baker TA (2006) Proteomic profiling of ClpXP substrates after DNA damage reveals extensive instability within SOS regulon. Mol Cell 22: 193–204. [DOI] [PubMed] [Google Scholar]
- Nelson DE, and Young KD (2000) Penicillin binding protein 5 affects cell diameter, contour, and morphology of Escherichia coli. J Bacteriol 182: 1714–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DE, and Young KD (2001) Contributions of PBP 5 and DD-carboxypeptidase penicillin binding proteins to maintenance of cell shape in Escherichia coli. J Bacteriol 183: 3055–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odsbu I, and Skarstad K (2014) DNA compaction in the early part of the SOS response is dependent on RecN and RecA. Microbiology (Reading, Engl). [DOI] [PubMed] [Google Scholar]
- Pazos M, Natale P, and Vicente M (2013) A specific role for the ZipA protein in cell division: stabilization of the FtsZ protein. J Biol Chem 288: 3219–3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrino S, Radzimanowski J, de Sanctis D, Boeri Erba E, McSweeney S, and Timmins J (2012) Structural and functional characterization of an SMC-like protein RecN: new insights into double-strand break repair. Structure 20: 2076–2089. [DOI] [PubMed] [Google Scholar]
- Pelletier J, Halvorsen K, Ha B-Y, Paparcone R, Sandler SJ, Woldringh CL, et al. (2012) Physical manipulation of the Escherichia coli chromosome reveals its soft nature. Proc Natl Acad Sci USA 109: E2649–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potluri L-P, de Pedro MA, and Young KD (2012) Escherichia coli low-molecular-weight penicillin-binding proteins help orient septal FtsZ, and their absence leads to asymmetric cell division and branching. Mol Microbiol 84: 203–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruteanu M, and Baker TA (2009a) Controlled degradation by ClpXP protease tunes the levels of the excision repair protein UvrA to the extent of DNA damage. Mol Microbiol 71: 912–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruteanu M, and Baker TA (2009b) Proteolysis in the SOS response and metal homeostasis in Escherichia coli. Res Microbiol 160: 677–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radman M (1975) SOS repair hypothesis: phenomenology of an inducible DNA repair which is accompanied by mutagenesis. Basic Life Sci 5A: 355–367. [DOI] [PubMed] [Google Scholar]
- Raskin DM, and de Boer PA (1999) Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc Natl Acad Sci USA 96: 4971–4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes ED, Patidar PL, Uranga LA, Bortoletto AS, and Lusetti SL (2010) RecN is a cohesin-like protein that stimulates intermolecular DNA interactions in vitro. J Biol Chem 285: 16521–16529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowlett VW, and Margolin W (2015) The bacterial divisome: ready for its close-up. Philos Trans R Soc Lond, B, Biol Sci 370: 20150028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler SJ, and Clark AJ (1994) RecOR suppression of recF mutant phenotypes in Escherichia coli K-12. J Bacteriol 176: 3661–3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer RT, and Baker TA (2011) AAA+ proteases: ATP-fueled machines of protein destruction. Annu Rev Biochem 80: 587–612. [DOI] [PubMed] [Google Scholar]
- Seong IS, Oh JY, Lee JW, Tanaka K, and Chung CH (2000) The HslU ATPase acts as a molecular chaperone in prevention of aggregation of SulA, an inhibitor of cell division in Escherichia coli. FEBS Lett 477: 224–229. [DOI] [PubMed] [Google Scholar]
- Strauch KL, Johnson K, and Beckwith J (1989) Characterization of degP, a gene required for proteolysis in the cell envelope and essential for growth of Escherichia coli at high temperature. J Bacteriol 171: 2689–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stylianidou S, Brennan C, Nissen SB, Kuwada NJ, and Wiggins PA (2016) SuperSegger: robust image segmentation, analysis and lineage tracking of bacterial cells. Mol Microbiol 102: 690–700. [DOI] [PubMed] [Google Scholar]
- Sugimoto S, Yamanaka K, Nishikori S, Miyagi A, Ando T, and Ogura T (2010) AAA+ chaperone ClpX regulates dynamics of prokaryotic cytoskeletal protein FtsZ. J Biol Chem 285: 6648–6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton AL, and Sherratt DJ (2013) Breaking symmetry in SMCs. Nat Struct Mol Biol 20: 246–249. [DOI] [PubMed] [Google Scholar]
- Uranga LA, Reyes ED, Patidar PL, Redman LN, and Lusetti SL (2017) The cohesin-like RecN protein stimulates RecA-mediated recombinational repair of DNA double-strand breaks. Nat Commun 8: 15282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente M, Rico AI, Martínez-Arteaga R, and Mingorance J (2006) Septum enlightenment: assembly of bacterial division proteins. J Bacteriol 188: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickridge E, Planchenault C, Cockram C, Junceda IG, and Espeli O (2017) Management of E. coli sister chromatid cohesion in response to genotoxic stress. Nat Commun 8: 14618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viola MG, LaBreck CJ, Conti J, and Camberg JL (2017) Proteolysis-Dependent Remodeling of the Tubulin Homolog FtsZ at the Division Septum in Escherichia coli. PLoS ONE 12: e0170505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker GC (1984) Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol Rev 48: 60–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JE, and Lutkenhaus J (1985) Overproduction of FtsZ induces minicell formation in E. coli. Cell 42: 941–949. [DOI] [PubMed] [Google Scholar]
- Weart RB, Nakano S, Lane BE, Zuber P, and Levin PA (2005) The ClpX chaperone modulates assembly of the tubulin-like protein FtsZ. Mol Microbiol 57: 238–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willetts NS, Clark AJ, and Low B (1969) Genetic location of certain mutations conferring recombination deficiency in Escherichia coli. J Bacteriol 97: 244–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkin EM (1967) The radiation sensitivity of Escherichia coli B: a hypothesis relating filament formation and prophage induction. Proc Natl Acad Sci USA 57: 1275–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LJ, and Errington J (2011) Nucleoid occlusion and bacterial cell division. Nat Rev Microbiol 10: 8–12. [DOI] [PubMed] [Google Scholar]
- Wu WF, Zhou Y, and Gottesman S (1999) Redundant in vivo proteolytic activities of Escherichia coli Lon and the ClpYQ (HslUV) protease. J Bacteriol 181: 3681–3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeman MK, and Cimprich KA (2014) Causes and consequences of replication stress. Nat Cell Biol 16: 2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.