Abstract
Background
Previous studies have shown that foamy viral (FV) vectors are a promising alternative to gammaretroviral and lentiviral vectors and insulators can improve FV vector safety. However, in a previous analysis of insulator effects on FV vector safety, strong viral promoters were used to elicit genotoxic events. Here we developed and analyzed the efficacy and safety of a high-titer, clinically relevant FV vector driven by the housekeeping promoter elongation factor-1α and insulated with an enhancer blocking A1 insulator (FV-EGW-A1).
Methods
Human CD34+ cord blood cells were exposed to an enhanced green fluorescent protein expressing vector, FV-EGW-A1, at a multiplicity of infection of 10 and then maintained in vitro or transplanted into immunodeficient mice. Flow cytometry was used to measure engraftment and marking in vivo. FV vector integration sites were analyzed to assess safety.
Results
FV-EGW-A1 resulted in high-marking, multi-lineage engraftment of human repopulating cells with no evidence of silencing. Engraftment was highly polyclonal with no clonal dominance and a promising safety profile based on integration site analysis.
Conclusions
An FV vector with an elongation factor-1α promoter and an A1 insulator is a promising vector design for use in the clinic.
Keywords: gene therapy, viral vector, hematopoietic stem cell
Introduction
Hematopoietic stem cell (HSC) gene therapy has shown great promise in several clinical trials. Gammaretroviral and lentiviral (LV) vectors have provided sustained therapeutic benefit to patients with severe combined immunodeficiency (SCID)1,2, Wiskott-Aldrich Syndrome (WAS)3 and chronic granulomatous disease (CGD).4 However, in early clinical trials using gammaretroviral vectors some patients experienced severe adverse events such as vector mediated leukemia post-treatment1,5,6. Leukemia was caused by gammaretroviral vector proviruses inserting near proto-oncogenes resulting in clonal expansion.7 Following these adverse events the gene therapy field has moved to using LV vectors8, which have an improved safety profile relative to gammaretroviral vectors. However, clonal expansion has been observed in patients treated with LV vectors.9 Vector modifications such as the incorporation of housekeeping promoters and insulator elements10,11 have the potential to further improve the safety of HSC gene therapy.
Foamy virus (FV) vectors are a promising alternative to gammaretroviral and LV vectors. FV vectors have a broad tissue tropism eliminating the need for pseudotyping.12 They also have a large transgene capacity13 making them useful for diseases that require either large or multiple transgenes. Also, FV vectors can be produced at high titers14,15 making them clinically relevant for a wide range of diseases. In terms of safety, FV vectors have a favorable integration profile16 and have been shown to have less read-through transcription than gammaretroviral vectors or LV vectors.17 FV vectors have been used to successfully treat canine leukocyte adhesion deficiency in dogs maintaining a polyclonal distribution of gene-marked cells.18,19 When FV and LV vectors were compared in human repopulating cells in an immunodeficient mouse model, FV vectors were shown to have a more polyclonal distribution with fewer total integrations near or within proto-oncogenes.20 In this study LV vectors had 5 dominant clones with integrations in or near proto-oncogenes whereas FV vectors had zero dominant clones near or in proto-oncogenes, suggesting that FV vectors may be a safer alternative to LV vectors.20
Strong viral promoters, such as the spleen focus forming virus (SFFV) promoter, have the potential to activate nearby genes leading to clonal expansion and ultimately malignancy.21–23 Cellular promoters derived from human housekeeping genes, such as elongation factor-1α (EF1α) or phosphosglycerate kinase (PGK), can reduce the genotoxic risk of retroviral vectors. Vectors containing PGK had reduced tumor acceleration in comparison to vectors containing the SFFV promoter.23 The EF1α promoter drives persistent, therapeutic levels of transgene expression in several hematopoietic lineages.24,25 Salmon et al. showed that the EF1α promoter maintained higher expression than PGK in differentiated hematopoietic cells and showed no evidence of silencing.24 Together these studies highlight that EF1α is an efficient promoter in hematopoietic repopulating cells with the potential for use in the clinic.
Chromatin insulators can be incorporated into retroviral vectors as an additional safety feature.26 The insulator is typically inserted into the U3 region of the 3’ long terminal repeat (LTR) and during reverse transcription the insulator is copied to the 5’ LTR. This results in two insulators that flank the transgene. Insulators can block nearby gene transactivation by enhancer elements within the vector provirus. They can also potentially reduce silencing of the transgene due to encroaching methylation.27 Our lab recently compared uninsulated FV vectors with FV vectors insulated with either the cHS4 insulator or the A1 insulator in SCID repopulating cells.28 In this previous study vectors contained a strong SFFV promoter in order to elicit genotoxicity, thereby improving the ability to detect the effects of insulators. In human repopulating cells FV vectors with A1 insulators had a more polyclonal distribution and were observed less frequently within 50 kb of proto-oncogenes than uninsulated FV vectors. A1 insulated vectors also had significantly less proviral integrations near the transcription start sites (TSS) of genes.28 FV vectors have a strong CTCF insulator in the LTR29, but the A1 insulator improved safety when stressed with a genotoxic SFFV promoter.28
The SFFV promoter is a useful tool to enhance genotoxicity for evaluating alternate vector designs including insulated vectors. However, SFFV would not be used in a clinical setting. Thus, in the present study, our goal was to perform a pre-clinical evaluation of a FV vector with an internal EF1α internal promotor and an A1 chromatin insulator. We evaluated an EF1α-driven, A1 insulated vector using cord blood CD34+ human repopulating cells in the NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mouse model. FV-EGW-A1 resulted in efficient multi-lineage transgene expression and the integration profile compared favorably to vectors with a strong SFFV promoter. This novel pre-clinical data supports the evaluation of EF1α-driven A1 insulated FV vectors in a clinical setting.
Materials and methods
Construction of FV-EGW-A1 vector
The EF1α promoter sequence used corresponds to the human genome draft genome hg38 on chromosome 6 from base pair 73,520,999 to 73,521,240 using the University of California-Santa Cruz BLAT tool.30 The promoter was synthesized and cloned into a FV backbone using standard molecular biology techniques. FV vector FV-EGW-A1 was constructed by cloning the A1 insulator fragment11,31 into the 3’ LTR of FV-EGW using the cut sites SalI and DrdI.
Vector preparation
FV vectors were produced by transient transfection of vector and helper plasmids into human embryonic kidney 293T cells using polyethylenimine (Polysciences, Warrinton, PA) and titered on HT1080 cells as previously described.32
Transduction of CD34+ human cord blood cells and progenitor assay
CD34+ human cord blood cells (StemCell Technologies, Rosalind, CA) were thawed, pooled and prestimulated as previously described.33 Prestimulation media consisted of Iscove’s Modified Dulbecco Medium with 10% heat inactivated fetal bovine serum, 5,000 U penicillin/streptomycin, and 100 ng/mL of the recombinant human cytokines interleukin 3, interleukin 6, stem cell factor, thrombopoietin, Fms-related tyrosine kinase 3 ligand, and granulocyte colony stimulating factor (ProSpec, Brunswick, NJ). Cells were transduced with FV vectors at a multiplicity of infection (MOI) of 10, or mock transduced with prestimulation media. Cells were incubated for 19 hours at 37°C. Cells were then harvested and used for either transplant, liquid culture or the colony forming unit (CFU) assay, as previously described.34
Xenotransplants of human cord blood CD34+ cells
All animal protocols were approved by the Institutional Animal Care and Use Committee of Washington State University.
4-week-old NSG mice (The Jackson Laboratory, Bar Harbor, ME) were housed in sterile microisolater cages and given water supplemented with tetracycline for 4 weeks. Mice received 25 mg/kg of busulfan (Sigma-Aldrich, St. Louis, MO) in a split dose 48 and 24 hours before transplantation. Busulfan doses were prepared in a 1:3 ratio of dimethyl sulfoxide and Roswell Park Memorial Institute Medium and delivered via intraperitoneal injections. 24 hours after conditioning, 1 × 105 FV vector exposed cells were transplanted into the mice via tail vein injections. At 3 week intervals, from 6 to 27 weeks post-transplant, 100 μl of peripheral blood was collected from the saphenous vein of each mouse to evaluate engraftment and marking. Mice were euthanized at 27 weeks post-transplant and whole blood, whole femurs were collected. Blood was collected via cardiac puncture and bone marrow was harvested from both the right and left femurs.
Engraftment and marking analysis in the peripheral blood and bone marrow
50 μL of peripheral blood or ~5×105 bone marrow cells were suspended in flow buffer (phosphate-buffered saline with 10% heat inactivated fetal bovine serum) and were used for engraftment and marking analysis. To block non-specific staining, rat anti-mouse CD16/CD32 Fc block was added to each sample and incubated for 15 minutes on ice. Samples were then stained with fluorophore conjugated antibodies and incubated on ice for 30 minutes, as previously described.33 Red blood cells were lysed using 2 mL of 1X Red Blood Cell lysis buffer (Santa Cruz Biotechnology, Dallas, TX) per sample, mixed by inversion and incubated for 10 minutes at room temperature in the dark. Then 2 mL of flow buffer was added, cells were centrifuged for 5 minutes at 500 g and washed with 2 mL of flow buffer. Centrifugation was repeated and cells were resuspended in 200 μL of flow buffer. Samples were than analyzed using the Beckman-Coulter Gallios flow cytometer and analyzed using Kaluza software (Beckman-Coulter, Indianapolis, Indiana). Analysis for blood was based on 5,000 events from the live cell gate and analysis for bone marrow was based on 10,000 events from the live cell gate.
Analysis of integration profiles
Genomic DNA was extracted from bone marrow samples (Gentra puregene blood kit, cat # 158445, Qiagen). 3 μg of genomic DNA from each mouse was processed using modified genomic sequencing (MGS)-PCR as previously described35 and sequenced on an Illumina MiSeq (Genomic Sequencing and Analysis Facility, University of Texas-Austin, Austin, Texas). Sequences were then paired with Paired-End read merger software36 and processed with the Vector Integration Site Analysis (VISA) server as previously described.37 For cancer gene analysis, we used 1,571 cancer genes that had corresponding RefSeq mRNAs from the Network of Cancer Genes 5.0 as of January 4th, 2016.38 This list contains both proto-oncogenes and tumor suppressor genes. An integration site was considered to be within an oncogene when it was located within the oncogene transcript.
Results
Our goal was to evaluate the ability of a FV vector with a housekeeping EF1α promoter and an A1 insulator to provide multi-lineage gene marking in human CD34+ repopulating cells in the NSG model, and to evaluate safety by performing retroviral integration site (RIS) analysis. We first modified the previously described28 A1 insulated FV vector to contain the EF1α promoter, resulting in the vector, FV-EGW-A1 (Figure 1). FV-EGW-A1 could be produced at high titer, greater than 2×108 EGFP TU/mL (Figure S1).
Figure 1.
Vector design of FV-EGW-A1. EF1α, human elongation factor-1 alpha; EGFP, enhanced green fluorescent protein; W, woodchuck hepatitis virus post-transcriptional element; A1, A1 enhancer-blocking insulator element; pA, polyadenylation site (polyadenylation occurs from the vector LTR).
Efficient marking in human cord blood CD34+ progenitors
Human cord blood CD34+ cells were exposed to FV-EGW-A1 at an MOI of 10. Cells were then maintained in liquid culture, plated in methylcellulose for CFU analysis or transplanted into NSG mice. At 10 days post vector exposure, FV-EGW-A1 had a transduction frequency of 18% with a mean fluorescence intensity of 61.7 (Figure S1). The gene marking in progenitors was 30% as determined by enhanced green fluorescent protein (EGFP) expression in CFUs, with a plating efficiency of 42%.
Next we assessed 5,133 RIS retrieved from CD34+ cord blood cells that were maintained in liquid culture for ten days post vector exposure. The percentages of RIS integrating within genes and near TSS of genes was also similar to what we have observed for FV vectors in previous studies (Table 1).
Table 1.
Distribution of integration sites in transduced human cord blood 10 days post vector exposure
| Vector | Unique RIS | Within genes | Within 50 kbp of TSS | In proto-oncogenes | Within 50 kbp of proto-oncogene TSS |
|---|---|---|---|---|---|
| FV-EGW-A1 | 5,133 | 44.6% | 51.2% | 8.3% | 7.9% |
| FV-SGW-A11 | 6,489 | 41.8% | 49.0% | 8.2% | 7.6% |
| FV-SGW1 | 1,594 | 43.0% | 53.5% | 8.8% | 7.9% |
data from Browning et al. 2017
RIS is retroviral integration sites, TSS is transcription start sites
Efficient engraftment and stable, multi-lineage transgene expression of SCID repopulating cells
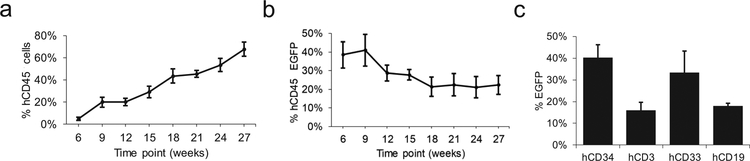
Human cord blood CD34+ cells were exposed to FV-EGW-A1 at an MOI of 10 and then transplanted into myeloablated, 4-week-old NSG mice. Engraftment of human CD45+ (hCD45) cells steadily increased from an average of 5% to an average of 72% at 27 weeks post-transplant (Figure 2a). Marking in the peripheral blood stabilized at 18 weeks post-transplant (Figure 2b), and was similar to marking in the bone marrow at 27 weeks (22% EGFP hCD45+ cells). Multi-lineage marking was observed in all of the transplanted mice with no lineage-specific effects (Figure 2c). Thus, FV vectors with a clinically relevant design using the EF1α promoter and A1 insulator provided sustained expression in a high percentage of repopulating cells with multi-lineage engraftment and with no evidence of silencing.
Figure 2.
Engraftment of human hematopoietic repopulating cells and vector marking in peripheral blood in vivo. (a) Engraftment of human hematopoietic repopulating cells in peripheral blood collected 6–27 weeks post-transplant. (b) Proportion of human hematopoietic repopulating cells expressing EGFP. (c) Multi-lineage marking in peripheral blood. Marking was quantified by expression of enhanced green fluorescent protein (EGFP).
Polyclonal distribution of SCID repopulating cells with no dominant clones in vivo
The development of vector-mediated malignancies can take years to develop in patients. The vector-mediated malignancies observed in HSC gene therapy patients have been the result of vectors integrating near proto-oncogenes. These integrations cause the proto-oncogenes to be dysregulated and give the clones a competitive advantage, resulting in clonal dominance. Thus, we carefully evaluated the contribution and distribution of RIS in the bone marrow of the transplanted mice to screen for potential indicators of genotoxicity.
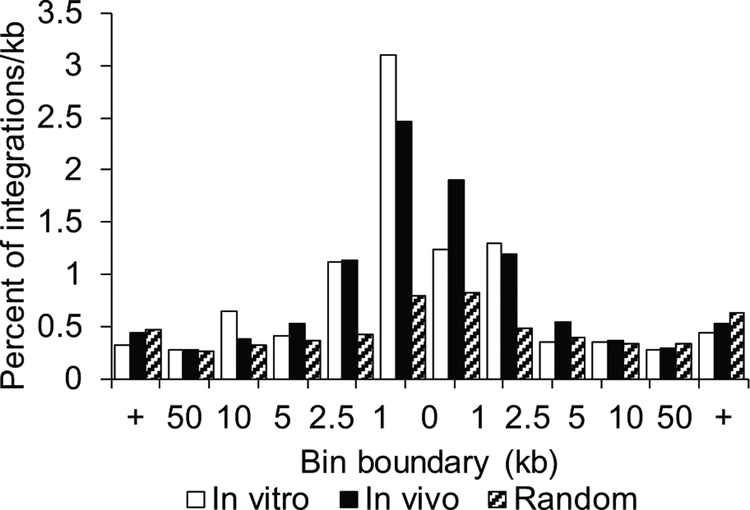
A total of 809 unique RIS were captured from the bone marrow of the 6 mice at 27 weeks post transplantation. The A1 insulated FV vector driven by EF1α integrated in a nearly random distribution with a slight increase of RIS 1 kb upstream from TSS. We compared this to the 5,133 RIS retrieved from the CD34+ cord blood cells that were maintained in liquid culture for ten days post vector exposure and saw no large differences from the integration profile (Figure 3).
Figure 3.
Retroviral integration sites of foamy viral vector FV-EGW-A1 in vitro and in vivo. Percentage of integrations per kb in relation to transcription start sites of RefSeq genes.
For FV-EGW-A1, there were a similar number of RIS within genes and within 50 kb of TSS of genes to FV-SGW-A1 (Table 2). As expected, the number of RIS within 50 kb of TSS was significantly lower than previously seen in the uninsulated FV and LV vectors with the SFFV promoter, FV-SGW and LV-SGW. While the frequency of integrations within proto-oncogenes was lower for FV-EGW-A1 the number of integrations within 50 kb of TSS of proto-oncogenes was the same (Table 2).
Table 2.
Distribution of integration sites in human repopulating cells
| Vector Type | Vector | Unique RIS | In Genes | Within 50 kb of TSS | In Proto-oncogenes | Within 50 kb of proto-oncogene TSS |
|---|---|---|---|---|---|---|
| Foamy | FV-EGW-A1 | 809 | 40.0% | 54.3% | 5.3% | 8.0% |
| FV-SGW-A11 | 913 | 38.6% | 54.4% | 8.1%a | 8.0% | |
| FV-SGW2 | 826 | 40.0% | 59.8%a | 9.5%a | 9.6% | |
| Lenti | LV-SGW2 | 461 | 74.0%a | 73.0%a | 12.3%a | 12.6%a |
| N/A | Random | 1000 | 43.8%b | 44.8%a | 6.7% | 6.8% |
data from Browning et al. 2017
data from Everson et al. 2016
significantly different than FV-EGW-A1, p<0.001
significantly different than FV-EGW-A1, p<0.05
RIS is retroviral integration sites, TSS is transcription start sites
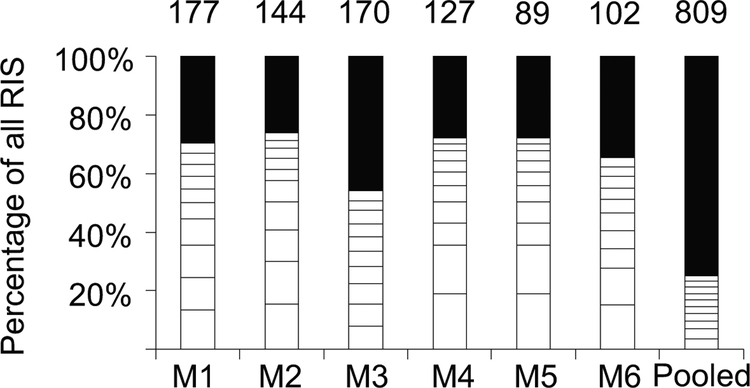
RIS in each mouse were analyzed using span count28 to detect clonal contribution and dominance. Clonal dominance is defined as a single integration accounting for over 20% of all RIS spans. SCID-repopulating cells were highly polyclonal and no clonal dominance was observed in any of the mice (Figure 4). Next, we analyzed the top ten clones from each mouse for proximity to proto-oncogenes. A top ten clone in one mouse (M6) had a clone accounting for 6.2% of the total spans with an integration 30 kb downstream from the proto-oncogene CYP2A6.
Figure 4.
Clonality analysis of foamy viral vector FV-EGW-A1 proviral integrations in vivo. Each bar represents the clonal composition in the bone marrow of individual mice. The last bar represents the combined analysis for the pooled integrations from all of the mice. The white boxes depict the top ten clones with the highest span counts for each mouse. The number on top of each bar represents the total number of unique integrations. RIS is retroviral integration site, M is mouse.
We also compared the RIS to a list of 38 lymphoid-specific proto-oncogenes.39 Two integrations were within 50 kb of proto-oncogenes on this list. One RIS (M1) was identified 2 kb upstream from the TSS of MLLT10 but only accounted for less than 1% of the total spans in this mouse. Another RIS (M3) integrated 11 kb upstream from the TSS of STIL but only accounted for less than 1% of the total spans in this mouse.
Discussion
HSC gene therapy has enormous potential for hematopoietic disorders and genetic diseases. Therefore it is important to develop vectors that reduce vector mediated adverse side effects. Here we provide novel pre-clinical data that supports the use of a FV vector with an EF1α promoter and an A1 insulator in patients.
Previously it was shown that the A1 insulated FV vector fulfilled the criteria for a promising insulated vector by providing significant insulating activity while maintaining high vector titer.28 However, the previous study utilized a strong SFFV promoter to evaluate the potential effects of the A1 insulated FV vector on the host genome. While SFFV is an effective tool for determining the genotoxic potential of new vectors, here we evaluated the safety of a clinically relevant A1 insulated FV vector. The A1 insulated FV vector driven by the housekeeping promoter EF1α was high titer and resulted in a highly polyclonal distribution of RIS, a reduced number of integrations near proto-oncogenes, and no clonal dominance in human cord blood CD34+ SCID repopulating cells.
Previously in a direct comparison of FV and LV vectors with identical transgene cassettes driven by the SFFV promoter, FV vectors had a significantly lower integration frequency within 50 kb of TSS of proto-oncogenes than LV vectors.20 However the SFFV promoter is not suitable for clinical applications. Here we showed a FV vector with a housekeeping promoter EF1α and an A1 insulator has a safe profile.
Clonal expansion is often a precursor to vector-mediated malignancies and occurs when a clone containing an RIS has a competitive advantage. Dominant clones can gain a growth or survival advantage from integrating near a proto-oncogene or proliferative gene. Although not all dominant clones will lead to clonal outgrowth and malignancy, the identification of dominant clones is informative as to the relative safety of retroviral vectors. LV and FV vectors are less prone to clonal dominance than gammaretroviral vectors16 and offer a promising alternative vector system for HSC gene therapy. A 2015 study by Negre et al. demonstrated that LV vectors corrected β-thalassemia in mice and reported that no evidence of clonal outgrowth was observed.40 Another group recently developed a self-inactivating LV vector carrying a codon-optimized human IL2RG cDNA driven by EF1α.41 This vector was shown to be effective at restoring T, B and NK cell counts in the bone marrow and peripheral blood of Il2rg−/−/Rag2−/− mice. They reported the mice to have polyclonal distributions with no signs of clonal dominance.41 In these two studies, the RIS from all of the mice were pooled showing a polyclonal/oligoclonal distribution.
One strength of our study is that we evaluated clonal dominance in individual transplanted mice. In studies where the RIS date is pooled from multiple mice, clonal dominance can be masked due to averaging the percentage of clonal abundance across multiple mice. With increases in the throughput of next generation sequencing and reductions in cost, we suggest that for future studies of RIS in SCID repopulating cells that data be reported per mouse to allow comparisons of vector genotoxicity between studies. Here we show that all of the FV-EGW-A1 mice individually maintain a polyclonal distribution of RIS with no evidence of clonal dominance. This approach is more sensitive for the identification of true dominant clones as evidenced by comparing the clonal contribution of individual mice to the pooled data (Figure 4).
The transactivation of proto-oncogenes remains a major concern for retroviral genotoxicity. Thus the enrichment of proviral integrations near the TSS of proto-oncogenes is of interest for therapeutic applications. FV proviruses are observed less frequently in hematopoietic repopulating cells near or within proto-oncogenes than commonly used vector system proviruses such as LV.16,20,42 Specifically, simple direct comparisons in both the dog model42 and human CD34+ cells in the NSG mouse model20 showed that FV proviruses integrate less frequently within 50 and 500 kb of proto-oncogene TSSs than LV proviruses. We previously showed that A1 insulated FV proviruses had reduced integrations near or within proto-oncogenes than uninsulated FV proviruses.28 In the current study we observed that the addition of the EF1α promoter to the A1 insulated FV vector design maintains a low number of integrations near and within proto-oncogenes and supports that EF1α does not negatively affect the integration distribution of FV vectors.
Two of the FV-EGW-A1 RIS integrated within 50 kb of proto-oncogenes associated with lymphocytic leukemias. RIS 21528954 integrated 2 kb upstream from MLLT10 and RIS 47325060 integrated 11 kb upstream from STIL. MLLT10 encodes a transcription factor and is involved in multiple chromosomal rearrangements that result in various leukemias. STIL encodes a cytoplasmic protein implicated in regulation of the mitotic spindle checkpoint. Chromosomal aberrations involving STIL may be a cause of some T-cell malignancies. Neither of these integrations showed signs of clonal outgrowth with both RIS accounting for less than 1% of the clonal contribution for each respective mouse. No FV vector integrations were observed near MDS1-EVI1 or LMO2.
In conclusion, FV-EGW-A1 allows for high marking, multi-lineage engraftment with no evidence of silencing. In a murine xenotransplantation model, FV-EGW-A1 resulted in a polyclonal population of SCID repopulating cells with no evidence of clonal dominance and a reduced number of proviral integrations within proto-oncogenes. Our study shows that the addition of the housekeeping promoter EF1α does not compromise the safety of an A1 insulated FV vector. Our data support the use of an EF1α-driven, A1 insulated FV vector in a clinical setting.
Supplementary Material
Acknowledgements
The authors would like to thank Joshua Wirtz for technical assistance. This work was supported by the National Institute of Health Grants AI097100 and AI102672 (G.D.T).
Footnotes
Conflict of Interest Statement
The authors declare no conflict of interest.
References
- 1.Hacein-Bey-Abina S, Garrigue A, Wang GP, et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest. 2008;118(9):3132–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiuti A, Cattaneo F, Galimberti S, et al. Gene therapy for immunodeficiency due to adenosine deaminase deficiency. N Engl J Med. 2009;360(5):447–458. [DOI] [PubMed] [Google Scholar]
- 3.Boztug K, Schmidt M, Schwarzer A, et al. Stem-cell gene therapy for the Wiskott-Aldrich syndrome. N Engl J Med. 2010;363(20):1918–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ott MG, Schmidt M, Schwarzwaelder K, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med. 2006;12(4):401–409. [DOI] [PubMed] [Google Scholar]
- 5.Braun CJ, Boztug K, Paruzynski A, et al. Gene Therapy for Wiskott-Aldrich Syndrome—Long-Term Efficacy and Genotoxicity. Sci Transl Med. 2014;6(27):13–7. [DOI] [PubMed] [Google Scholar]
- 6.Stein S, Ott MG, Schultze-Strasser S, et al. Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nature Medicine. 2010;16:198–204. [DOI] [PubMed] [Google Scholar]
- 7.Trobridge GD. Genotoxicity of retroviral hematopoietic stem cell gene therapy. Expert Opin Biol Ther. 2011;11(5):581–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Montini E, Cesana D, Schmidt M, et al. Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration. Nature Biotechnology. 2006;24(6):687–696. [DOI] [PubMed] [Google Scholar]
- 9.Cavazzana-Calvo M, Payen E, Negre O, et al. Transfusion independence and HMGA2 activation after gene therapy of human [bgr]-thalassaemia. Nature. 2010;467(7313):318–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Groth AC, Liu M, Wang H, Lovelett E, Emery DW. Identification and characterization of enhancer-blocking insulators to reduce retroviral vector genotoxicity. PLoS One. 2013;8(10):e76528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Browning DL, Collins CP, Hocum JD, Leap DJ, Rae DT, Trobridge GD. Insulated Foamy Viral Vectors. Human gene therapy. 2016;27(3):255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Russell DW, Miller AD. Foamy virus vectors. Journal of virology. 1996;70(1):217–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trobridge G, Josephson N, Vassilopoulos G, Mac J, Russell DW. Improved foamy virus vectors with minimal viral sequences. Mol Ther. 2002;6(3):321–328. [DOI] [PubMed] [Google Scholar]
- 14.Josephson NC, Russell DW. Production of foamy virus vector and transduction of hematopoietic cells. Cold Spring Harb Protoc. 2010;2010(9):pdb.prot5481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nasimuzzaman M, Kim Y-S, Wang Y-D, Persons DA. High-titer foamy virus vector transduction and integration sites of human CD34+ cell–derived SCID-repopulating cells. Molecular Therapy — Methods & Clinical Development. 2014;1:14020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trobridge GD, Miller DG, Jacobs MA, et al. Foamy virus vector integration sites in normal human cells. In: Proc Natl Acad Sci U S A. Vol 103.2006:1498–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hendrie PC, Huo Y, Stolitenko RB, Russell DW. A rapid and quantitative assay for measuring neighboring gene activation by vector proviruses. Mol Ther. 2008;16(3):534–540. [DOI] [PubMed] [Google Scholar]
- 18.Trobridge GD, Horn PA, Beard BC, Kiem HP. Large Animal Models for Foamy Virus Vector Gene Therapy. In: Viruses. Vol 4.2012:3572–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bauer TR, Tuschong LM, Calvo KR, et al. Long-Term Follow-up of Foamy Viral Vector-Mediated Gene Therapy for Canine Leukocyte Adhesion Deficiency. Molecular Therapy. 2013;21(5):964–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Everson EM, Olzsko ME, Leap DJ, Hocum JD, Trobridge GD. A comparison of foamy and lentiviral vector genotoxicity in SCID-repopulating cells shows foamy vectors are less prone to clonal dominance. Molecular Therapy — Methods & Clinical Development. 2016;3:16048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montini E, Cesana D, Schmidt M, et al. The genotoxic potential of retroviral vectors is strongly modulated by vector design and integration site selection in a mouse model of HSC gene therapy. The Journal of clinical investigation. 2009;119(4):964–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Modlich U, Navarro S, Zychlinski D, et al. Insertional transformation of hematopoietic cells by self-inactivating lentiviral and gammaretroviral vectors. Mol Ther. 2009;17(11):1919–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cesana D, Ranzani M, Volpin M, et al. Uncovering and dissecting the genotoxicity of self-inactivating lentiviral vectors in vivo. Mol Ther. 2014;22(4):774–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salmon P, Kindler V, Ducrey O, Chapuis B, Zubler RH, Trono D. High-level transgene expression in human hematopoietic progenitors and differentiated blood lineages after transduction with improved lentiviral vectors. Blood. 2000;96(10):3392–3398. [PubMed] [Google Scholar]
- 25.Byun HM, Suh D, Jeong Y, et al. Plasmid vectors harboring cellular promoters can induce prolonged gene expression in hematopoietic and mesenchymal progenitor cells. Biochemical and biophysical research communications. 2005;332(2):518–523. [DOI] [PubMed] [Google Scholar]
- 26.Evans-Galea MV, Wielgosz MM, Hanawa H, Srivastava DK, Nienhuis AW. Suppression of Clonal Dominance in Cultured Human Lymphoid Cells by Addition of the cHS4 Insulator to a Lentiviral Vector. Mol Ther. 2007;15(4):801–809. [DOI] [PubMed] [Google Scholar]
- 27.Sun FL and Elgin SC. Putting Boundaries on Silence. Cell. 1999;99(5):459–62. [DOI] [PubMed] [Google Scholar]
- 28.Browning DL, Everson EM, Leap DJ, et al. Evidence for the in vivo safety of insulated foamy viral vectors. Gene Ther. 2017;24(3):187–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goodman MA, Arumugam P, Pillis DM, et al. Foamy Virus Vector Carries a Strong Insulator in Its Long Terminal Repeat Which Reduces Its Genotoxic Potential. J Virol. 2018;92(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12(4):656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu M, Maurano MT, Wang H, et al. Genomic discovery of potent chromatin insulators for human gene therapy. Nature biotechnology. 2015;33(2):198–203. [DOI] [PubMed] [Google Scholar]
- 32.Kiem HP, Wu RA, Sun G, von Laer D, Rossi JJ, Trobridge GD. Foamy combinatorial anti-HIV vectors with MGMTP140K potently inhibit HIV-1 and SHIV replication and mediate selection in vivo. Gene therapy. 2010;17(1):37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olszko ME, Adair JE, Linde I, et al. Foamy viral vector integration sites in SCID-repopulating cells after MGMTP140K-mediated in vivo selection. Gene therapy. 2015;22(7):591–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou S, Mody D, DeRavin SS, et al. A self-inactivating lentiviral vector for SCID-X1 gene therapy that does not activate LMO2 expression in human T cells. 2010;116(6):900–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rae DT, Collins CP, Hocum JD, Browning DL, Trobridge GD. Modified Genomic Sequencing PCR Using the MiSeq Platform to Identify Retroviral Integration Sites. Human gene therapy methods. 2015;26(6):221–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J, Kobert K, Flouri T, Stamatakis A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics (Oxford, England). 2014;30(5):614–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hocum JD, Battrell LR, Maynard R, et al. VISA--Vector Integration Site Analysis server: a web-based server to rapidly identify retroviral integration sites from next-generation sequencing. BMC bioinformatics. 2015;16:212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.An O, Dall’Olio GM, Mourikis TP, Ciccarelli FD. NCG 5.0: updates of a manually curated repository of cancer genes and associated properties from cancer mutational screenings. Nucleic Acids Res. 2016;44(D1):D992–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lab. B. Human Lymphoma Cancer Gene List. http://www.bushmanlab.org/links/genelists. Accessed April 4, 2018.
- 40.Negre O, Bartholomae C, Beuzard Y, et al. Preclinical evaluation of efficacy and safety of an improved lentiviral vector for the treatment of beta-thalassemia and sickle cell disease. Curr Gene Ther. 2015;15(1):64–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poletti V, Charrier S, Corre G, et al. Preclinical Development of a Lentiviral Vector for Gene Therapy of X-Linked Severe Combined Immunodeficiency. Molecular Therapy - Methods & Clinical Development. 2018;9(15):257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beard BC, Keyser KA, Trobridge GD, et al. Unique Integration Profiles in a Canine Model of Long-Term Repopulating Cells Transduced with Gammaretrovirus, Lentivirus, or Foamy Virus. Hum Gene Ther. 2007;18(5):423–34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.