Abstract
High-throughput sequencing can be used to measure changes in tumor composition across space and time. Specifically, comparisons of pre- and post-treatment samples can reveal the underlying clonal dynamics and resistance mechanisms. Here, we discuss evidence for distinct modes of tumor evolution and their implications for therapeutic strategies. Additionally, we consider the utility of spatial tissue sampling schemes, single cell analysis, and circulating tumor DNA to track tumor evolution and the emergence of resistance, as well as approaches that seek to forestall resistance by targeting tumor evolution. Ultimately, characterization of the (epi)genomic, transcriptomic, and phenotypic changes that occur during tumor progression, coupled with computational and mathematical modeling of tumor evolutionary dynamics may inform personalized treatment strategies.
Keywords: clonal dynamics, computational modeling, therapeutic resistance, tissue correlative studies, tumor evolution
Missing the target by focusing on targeted approaches alone
The past two decades have brought about a bevy of targeted cancer therapies, including monoclonal antibodies and small molecules, that block immune checkpoints, interfere with cancer signaling pathways [1], or affect specific genetic vulnerabilities in tumors (e.g. PARP inhibitors [2]). The decision to treat with targeted therapy is typically based on the presence of a biomarker (such as a gene mutation or amplification) in a single tumor specimen. For certain cancers, targeted therapies have revolutionized patient care. Imatinib yields five-year survival rates of 95% for patients with chronic myeloid leukemia (CML) [3]. Unfortunately, for many solid tumors, response to targeted therapies is often 50% or lower [4]. Resistance to treatment can be classified as primary, describing patients who exhibit no response to treatment at all, or secondary, describing patients who initially respond to treatment, but later develop resistance, as the cells that are sensitive to treatment die and the resistant cell population continues to grow. As an example of secondary resistance, in BRAF mutant (V600E) melanoma, treatment with the tyrosine kinase inhibitor vemurafenib yields dramatic initial responses, but most patients eventually relapse with drug-resistant, deadly disease [5]. Similarly, HER2 amplified and/or over-expressing breast cancer tumors treated with the HER2-targeted monoclonal antibody trastuzumab also commonly exhibit resistance [6] and many patients who initially respond subsequently exhibit disease progression [7]. In colorectal cancer patients with EGFR-mutant, wild type-KRAS tumors, treatment with cetuximab yields a dismal objective tumor response rate of less than 15% [8], indicating that many tumors do not respond to treatment and hence, had primary resistance. Efforts to understand the complex mechanisms of resistance in pre-clinical models [9] and via tissue correlative studies highlight the many paths to resistance [10–13], which may not be fully elucidated by traditional, single-sample diagnostic tissue analysis.
The role of intratumor heterogeneity and non-genetic factors in resistance to targeted therapy
There is growing evidence that (epi)genetic and phenotypic heterogeneity within a tumor (intratumor heterogeneity, ITH, see glossary) contributes to resistance [14]. High ITH is associated with poor prognosis in head and neck cancer [15], lung cancer [16], ovarian cancer [17], and in pan-cancer analyses [18]. High ITH implies that the tumor is more likely to harbor a rare pre-existing resistant subclone and increases the likelihood that only a subset of tumor cells has the specific molecular aberration targeted by the therapy. Treatment targeting subclonal driver mutations has been associated with resistance and recurrence in CML [19] and multiple myeloma [20]. Similarly, treatments targeting subclonal copy number gains (versus clonal high-level gains) yielded suboptimal clinical benefit in gastric cancer patients treated with an FGFR inhibitor [21]. A better understanding of genetic ITH may explain why some patients who express specific molecular markers (at least in a single diagnostic sample) exhibit poor responses. There is a critical need to understand resistance as it arises in heterogeneous tumors and develop strategies to circumvent it.
Genetic heterogeneity provides a rich substrate for the emergence of resistance under treatment selective pressures, but it is not the only factor. While most targeted therapies are directed towards genetic alterations, epigenetic and microenvironmental causes of resistance are gaining greater recognition. Such epigenetic alterations can arise on relatively fast time scales due to varied microenvironmental influences resulting in heterogeneous gene expression patterns. This rapid change in gene expression can cause secondary resistance and will confound treatment decisions that are based solely on genotype [22]. For example, in melanoma cells, transcription-level variability, in the absence of corresponding genetic alterations, has been shown to cause resistance. Rare, synchronous, high-level transcription of multiple resistance markers combined with drug-induced epigenetic reprogramming led to preservation of the resistance-inducing transcriptional state [23]. Similarly, in NSCLC treated with anti-PD-1 checkpoint inhibitors, epigenetic upregulation of alternative immune checkpoints led to resistance [24], providing further evidence for non-genetic mechanisms of resistance. The surrounding tumor microenvironment is also known to play a role in cancer resistance, by protecting cancer cells from the full effects of infiltrating drugs. This limits cancer cell death and can lead to secondary resistance as the longer-surviving cancer cells continue to evolve under altered selective pressures [4]. In addition to providing a physical barrier, the surrounding tumor microenvironment may also release paracrine signaling factors that can alter tumor cell survival. In a murine model of Burkitt’s lymphoma, interleukin-6 and metalloproteinase 1 from the surrounding microenvironment affected cell survival following chemotherapy treatment [25]. While their presence and role are still somewhat debated, cancer stem cells, which are innately resistant to many treatments, are another potential cause of poor treatment response [4].
Towards a broader target
Comparing a tumor to a forest, current treatment approaches focus on individual trees (targetable genetic alterations present in a single sample). To combat resistance, an aerial view of the entire forest (tumor cells and their ecosystem/microenvironment) is beneficial — ideally with repeated longitudinal measurements to capture adaptive evolution. A spatially- and temporally-resolved characterization of tumor evolution (Figure 1) would include measurements of somatic alterations, as well as transcriptional and proteomic changes, using techniques that preserve tissue architecture. Changes in the tumor epigenetic landscape (DNA methylation and chromatin accessibility [26]) are also useful to trace. Although spatial profiling techniques are in their relative infancy, methods for in situ gene expression profiling such as MERFISH [27] and FISSEQ [28] have been used in developmental biology studies with extension to cancer. Additionally, a MasSpecPen has recently been developed and allows for nondestructive mass spectrometry analysis of ex vivo and in vivo cancer tissue [29]. Techniques such as MIBI [30] enable multiplexed proteomic analysis of formalin-fixed paraffin embedded archival tumor tissues. A multiscale, spatially and longitudinally resolved view of tumor progression will allow for a more complete understanding of the molecular and evolutionary determinants of resistance. Moreover, analysis of the resultant measurements within an ecological and evolutionary grounded framework may inform principled therapeutic strategies [31]. Previously, spatial statistics developed by ecologists have been used for the analysis of spatially resolved pathologic samples with phenotypic (cell type) information. The ecological statistics can be used to identify patterns of interactions that occur among cancer cells and their microenvironment (at the resolution of cell type) [32]. Such statistics can be similarly applied to genomic and proteomic data to delineate heterogeneity and interactions of resistant cell populations as they develop over time.
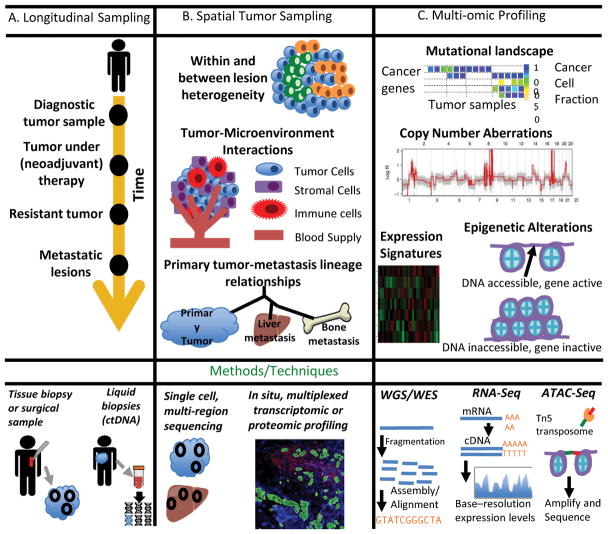
Figure 1 (Key Figure). Characterizing the spatio-temporal dynamics and mechanisms of resistance.
A. Longitudinal sampling can be used to track molecular changes during disease progression. In addition to solid tumor samples, liquid biopsies are particularly useful for studying temporal evolution. B. Spatial sampling can be used to characterize within and between lesion heterogeneity and to define tumor-immune cell interactions, all of which play a role in resistance. Single-cell sequencing methods examine heterogeneity at an extremely local level. In situ, multiplexed proteomic techniques, such as Nanostring digital spatial profiling, allow for the study of the co-evolution of the tumor and immune microenvironment during treatment. C. Multi-scale profiling at the genomic, transcriptomic, proteomic, and epigenomic levels can provide a complete picture of functional and non-functional heterogeneity. At the genomic level, tumor tissue can be characterized via the mutational landscape and copy number aberrations. ATAC-seq can be used to study epigenetic alterations.
Unwanted tumor evolution breeds resistance
The theory of clonal evolution described by Peter Nowell over 40 years ago by has been foundational for understanding tumor initiation and progression [33]. The clonal evolution model posits that after transformation of a single founding neoplastic cell, tumors evolve through an iterative and dynamic process as they continuously accumulate somatic mutations, some of which confer selective growth advantages. In the classic linear progression model, stringent positive selection for phenotypic traits results in selective sweeps throughout the course of tumor progression (see [34, 35] for recent reviews). An implication of ongoing selection is that multiple subclonal drivers may need to be targeted in addition to clonal alterations in order to achieve effective disease control.
In recent years, other tumor progression models have been proposed, including effectively neutral evolution and punctuated evolution. Under the Big Bang model of colorectal tumor growth, after malignant transformation, the tumor grows as a single (terminal) expansion composed of effectively equally fit clones [36], compatible with effectively neutral evolution. In this model, the timing of a mutation is the fundamental determinant of its frequency in the final tumor, rather than stringent selection for additional driver mutations. Subsequent work has corroborated neutral evolution in other solid tumors [37–39], suggesting that it may be relatively common. Importantly, under neutral evolution early clonal alterations (present in all tumor cells) correspond to the key drivers of progression and may therefore represent ideal therapeutic targets. However, the many subclones that subsequently diversify provide a potentially rich substrate for the emergence of resistance in the context of treatment selective pressure. Punctuated evolution due to mutational bursts or cataclysmic genomic rearrangements has also been described in multiple tumor types, including breast, prostate and pancreatic cancers [40–42]. The sudden accrual of a multitude of genomic changes represents another source of alterations that may contribute to resistance.
Irrespective of the mode of evolution in treatment-naive tumors, the application of therapy imposes selective pressure, resulting in the expansion of a pre-existing (potentially undetectable) resistant subclone. Additionally, when cancer recurs due to treatment failure or resistance, it often presents at organ sites beyond the primary tumor [43], suggesting the importance of delineating the evolution of metastases in addition to primary tumors. As progression of disease and treatment provide an expanding substrate for the selection of resistance, early detection of cancer is likely to provide a much more tractable target. Indeed, at early time points, surgery and/or radiotherapy without systemic therapy may be effective.
Mathematical and computational modeling of tumor progression and resistance
Mathematical modeling of tumor evolutionary processes, including progression, metastasis, and treatment, has become increasingly prevalent and provides a powerful tool to elucidate complex biological processes when used in tandem with ground-truth experimental or clinical data [44]. Several mathematical models describing tumor evolutionary dynamics have provided important insights into resistance (see [45] for a comprehensive review). These models generally make the simplifying assumption that tumors are composed of well-mixed cell populations [46]. However, spatial structure is a defining feature of solid tumors and can influence tumor dynamics [47] and computational models that account for this may be particularly informative and reveal distinct insights. Additionally, local mutational geography and ITH can impact clonal dynamics and should be incorporated when simulating tumor evolution, although this introduces additional complexity. Several spatial computational models of primary tumor growth have been described. While these models focus on important aspects of tumor progression such as angiogenesis (including delivery of chemotherapeutic agents to a tumor) [48] and tumor invasion in context of the surrounding microenvironment [49], none explicitly model treatment.
These modeling frameworks can be further strengthened by using parameters derived from patient genomic data. For example, a spatial agent-based model was developed to simulate the growth of glandular epithelial (e.g. colon) tumors. This model employed statistical inference techniques to deduce patient-specific evolutionary parameters using genomic data [36]. Additionally, the spatial growth of primary tumors under different evolutionary “modes” (ranging from effectively neutral growth to strong selection) was simulated using genomic data in conjunction with the simulations. This method was able to classify the “modes” of evolution for individual patients by examining patterns of between-region genetic divergence [37]. Another spatial tumor model was developed that combines genetic evolution with spatial growth and migration to show that resistant subclones are almost always present in clinically detectable lesions [50]. Given the stochastic nature of resistance and the multiple evolutionary trajectories that can occur during tumor growth, spatial computational models informed by patient-derived tumor data can improve understanding of the dynamics and mechanisms of resistance. Comparing “virtual” tumors simulated under varied conditions can be used to evaluate the evolutionary trajectories and resultant genomic patterns for a given patient. Such models may also be used to assess therapeutic strategies and to inform study designs prior to in vitro and in vivo pre-clinical testing. In the future, a more comprehensive spatial modeling approach that incorporate ecosystem variables such as hypoxia, immune infiltration, and stromal cell activation with genomic variables could further improve predictions regarding the evolutionary trajectory of the cancer clones.
Tumor sampling strategies
Previous studies have shown that a single tumor sample is unlikely to capture the ITH present in the entire tumor cell population [51–53] (Figure 2A). In a study of 100 NSCLC cases, over 75% of the tumors had subclonal driver alterations, most of which would have appeared clonal in a single biopsy [51]. In a study of glioblastoma, most patients had multiple molecular subtypes of disease found in different regions within a single tumor [52]. In renal cancer, expression-based profiling of samples from different regions can yield drastically different prognoses [53]. These examples motivate approaches that account for spatial ITH when characterizing cancer genomic landscapes. When studying the impact of therapy using paired pre- and post-treatment samples, regional ITH can be particularly confounding as it is difficult to delineate the appearance of a novel treatment-induced clone from a clone that was present initially but missed due to inadequate sampling.
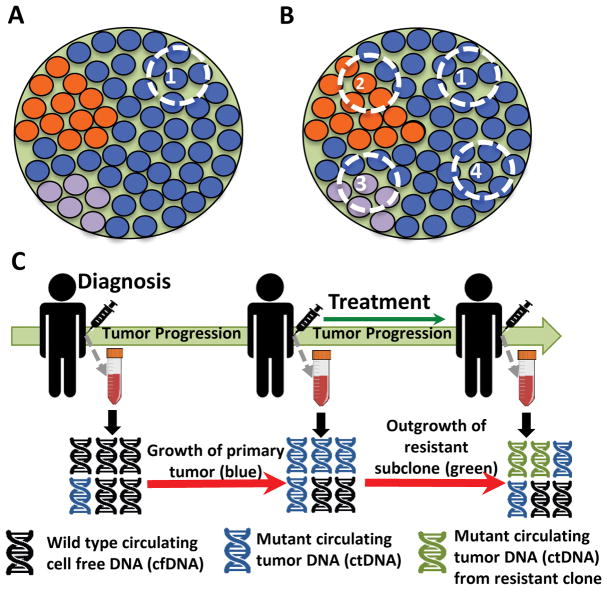
Figure 2. Sampling strategies to infer evolutionary dynamics.
For the tumor schematics in A and B, the green background indicates clonal alterations present in all tumor cells whereas other colors correspond to subclonal alterations that may be undetected due to spatial heterogeneity A. Sequencing of single tumor sample (e.g. diagnostic biopsy) illustrates that subclonal alterations and intratumor heterogeneity (ITH) may be overlooked. B. Multi-region sequencing (MRS) can better capture ITH and enables discovery of clonal versus subclonal alterations, which can, in turn, reveal the underlying evolutionary dynamics of the tumor. C. Liquid biopsies allow for ctDNA profiling, which provide a tractable method for the longitudinal characterization of clonal dynamics in the context of therapy and can be used to monitor the emergence of resistance prior to clinical manifestation on imaging.
Multi-region sampling
Multi-region sequencing (MRS) accounts for tumor spatial heterogeneity by sampling and profiling multiple regions from a single tumor specimen (Figure 2B). Compared to a single sample, MRS better captures ITH and enables more robust discrimination of clonal versus subclonal alterations, which in turn reflect the underlying evolutionary dynamics of the tumor. The computational framework developed by Sun et al. exploits MRS data to compute genetic divergence between samples in order to distinguish effectively neutral evolution from strong selection. Through simulation studies, they show that sampling additional tumor regions affords greater power for distinguishing between different modes of evolution as compared to deeper sequencing of fewer regions, with attendant implications for study design [37].
MRS can also illuminate functional ITH and potential convergent evolution, as originally demonstrated in renal cell carcinoma [53,54]. In a study of multiple synchronous lung cancers, MRS showed a high level of genomic heterogeneity (multiple distinct oncogenic alterations) between lesions from the same patient. However, these heterogeneous aberrations often mapped to a few key signaling pathways [55], suggesting convergent evolution and highlighting the utility of MRS to delineate functional ITH.
Single-cell sequencing
Single-cell profiling can characterize tumor heterogeneity at unprecedented resolution. Single-cell RNA-seq is becoming mainstream, but requires viable cells, making its application to primary human tumor samples relatively limited [56,57]. Throughput and cost are other key considerations for broader implementation of single-cell technologies. An additional challenge surrounding DNA-based single-cell assays is technical noise, which hinders accurate genotyping [58]. Current efforts have largely focused on single-cell copy number analyses to characterize clonal evolution [59,40], with notable exceptions that have performed clonal genotype inference [60]. Stochastic profiling, a related technique, provides an another approach for uncovering single cell molecular programs by identifying co-regulated, heterogeneously expressed genes within small cellular populations [61]. As technological and bioinformatic methods improve, single-cell approaches will increasingly be used to characterize tumor evolution, likely in conjunction with bulk or multi-region sequencing [62,63].
Liquid biopsies
Since it is often impractical to obtain repeat biopsies to monitor solid tumor progression, circulating tumor DNA (ctDNA) is useful for following longitudinal tumor progression. Non-invasive liquid biopsies (such as blood or urine) yield ctDNA or circulating tumor cells that can be used to characterize ITH and tumor growth dynamics [64] (Figure 2C). Studies of diverse tumor types, including breast cancer [64], lung cancer [65,66], and lymphoma [67] indicate that ctDNA can capture clonal evolution, although analyses are often restricted to small numbers of mutations. Liquid biopsies also provide a potential approach for earlier cancer detection [68]. This is particularly appealing since detection at a stage when surgical resection is still feasible and treatments are more effective would significantly reduce cancer related mortality.
Liquid biopsies can be used to monitor treatment response and the clonal dynamics that lead to resistance. In NSCLC, longitudinal ctDNA from urine specimens was used to detect changes in the frequency of EGFR activating and resistance mutations [69]. In chronic lymphocytic leukemia patients who ultimately progressed following ibrutinib treatment, liquid biopsies were used to detect the treatment-associated patterns of clonal evolution. The authors showed that the kinetics of resistance depended on the pre-treatment size and relative fitness of the resistant subclone [13]. In AML, ultra-deep amplicon resequencing of serial blood samples similarly highlighted the impact of treatment on clonal heterogeneity [70]. Thus, liquid biopsies can capture the selective pressures imposed by treatment, track the outgrowth of resistant subclones, and may also inform adaptive therapeutic strategies that target the tumor’s changing composition.
While ctDNA allows for non-invasive longitudinal sampling, there are limitations to its use in monitoring the emergence of resistance. In particular, liquid biopsies do not provide spatial information (i.e. organ location, primary tumor versus metastasis) about the variants that may cause resistance. It is also unknown whether all tumor types shed ctDNA at a similar rate [71]. Thus, while ctDNA represents an important tool to survey the kinetics of resistance and minimal residual disease, at present it is most informative when combined with tissue profiling.
Exploiting tumor evolution to forestall resistance
Suitable sampling strategies combined with computational models to simulate growth and clonal dynamics under therapy will allow for treatment advances and may inform the development of therapies that prevent the outgrowth of resistant subclones. Below we discuss current treatment strategies that acknowledge the role of tumor evolution in both the development and prevention of resistance.
Combination Therapy
Combination therapy, the use of multiple drugs simultaneously, has been used to successfully combat resistance in the context of antimicrobials [72] and HIV therapies [73]. Since heterogeneous tumors have multiple subclones that may be targeted with distinct therapies, combination therapy has been considered in this context in order to prevent the outgrowth of resistant subclones (Figure 3A). A mathematical model was used to compare combination (simultaneous) versus sequential therapy in solid tumors and it was found that combination therapy results in longer-term disease control. It was further noted that advanced cancers with greater disease burden often require simultaneous treatment with three agents. Even with three drugs, recurrence is inevitable in large tumors if there is the possibility of a mutation conferring cross-resistance [46]. By constraining a tumor’s evolutionary trajectory and preventing the development of more aggressive, resistant subclones, combination therapy can in principle impede the evolution of tumor cells that would be resistant to future therapies. While these results are encouraging, they depend on the number of potential resistance mutations that exist in the cancer genome, which may vary considerably by therapy, tumor type, and across patients. Additionally, mechanisms of resistance can extend beyond point mutations to copy number aberrations, epigenetic, transcriptional, and proteomic alterations, implying manifold paths to resistance, that are yet to be fully recognized.
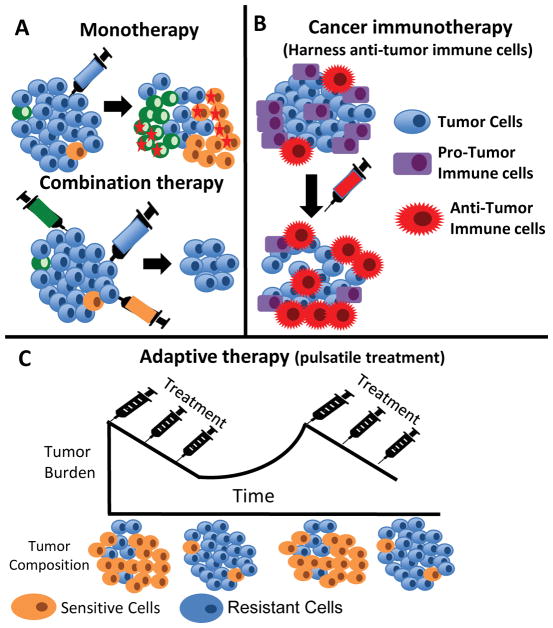
Figure 3. Approaches to forestall resistance.
A. Tumor schematic comparing the effects of monotherapy and combination therapy. Combination therapy treats a tumor with multiple drugs (different colored syringes) at the same time. This therapeutic strategy constrains the evolutionary trajectory of the tumor and prevents the development of more aggressive clones (orange and green cells marked with red stars) that would already be resistant to the second or third-line agents. B. Tumor schematic describing treatments targeting tumor-immune cell interactions. Cancer immunotherapy leverages the relationship between the tumor and its surrounding microenvironment to activate anti-tumor immune cells (eg CD4+ cells, NK cells, and B cells) and downregulate pro-tumor immune cells such as Tregs. C. Tumor schematic describing adaptive therapy. Adaptive therapy constrains the evolution of tumors with multiple competing subclones, some of which are resistant to therapy. In this schema, the blue cells are resistant to the treatment while the orange cells are sensitive. Treatment is given in a pulsatile manner such that the sensitive and resistant cell populations grow in the “off” and “on” phases of treatment, respectively. Ideally, both populations are maintained, but growth of the resistant subclone(s) is competitively constrained by neighboring sensitive cells.
In complementary work, combination therapy strategies were explored to treat heterogeneous tumors by using a computational optimization approach based on random sampling of RNAi based perturbations. It was found that optimal drug combinations depend on the level of genomic ITH and emphasize the inclusion of drugs with robust effects on all subpopulations rather than superior efficacy in targeting a single clone [74]. For a thorough review on considerations for modeling tumor dynamics towards the development of rational therapeutic strategies see ref [75]. In practice, combination targeted therapy has shown modest success in a clinical trial studying patients with BRAF V600E mutant colorectal cancer [76]. Additionally, combination therapy may be limited by the increased toxicity that accompanies simultaneous treatments. An additional concern is that without enumerating subclones a priori and targeting them, combination therapy may still leave patients vulnerable to resistance. Nonetheless, the strategies outlined above provide guiding principles for evaluating their efficacy and generalizability in pre-clinical models.
In addition to the simultaneous combination therapy strategies discussed above, treatment regimens that use multiple drugs sequentially have also been explored computationally and experimentally. Tumor clonal evolution occurs in stages and each stage may have unique therapeutic vulnerabilities to be exploited. Resistant cells that develop over the course of tumor evolution can be collaterally sensitive to other drugs. Computationally generated fitness landscapes can be used to predict secondary resistance to front-line therapy, which can then be addressed by administering a second drug sequentially [77]. Experimentally, sequential treatment strategies and the role of collateral sensitivity have been explored in lung cancer cell lines treated with ALK inhibitors. The authors found that the length of time between sequential treatments may impact the evolving patterns of collateral sensitivity and cross-resistance [78]. Sequential drug therapy has also been explored as a way to combat antibiotic resistance and such studies highlight issues relevant to cancer treatment. As with cancer therapy, the mechanisms of resistance to beta-lactam antibiotics are many; in one study, even with the sequential use of 2–4 beta-lactam antibiotics, E. coli resistance was observed in over 70% of cases [79]. For both simultaneous and sequential combination therapy, the number of potential resistance mechanism that need to be targeted is immense and makes this task challenging.
Treatments targeting tumor-immune cell interactions
Tumors exist as part of complex ecosystems that contain fibroblasts, endothelial cells, and immune cells, in addition to the malignant tumor cells. Tumor evolution and continued mutational processes lead to the development of tumor neoantigens (often functional, non-synonymous mutations) that are recognized by T-cells, activating an anti-tumor immune response [80]. Unfortunately, by the time tumors are detectable in the clinic, they have evolved mechanisms to evade the immune system, through the downregulation of T-cell responses. Cancer immunotherapy capitalizes on the interactions between the tumor and surrounding immune cells by targeting the tumor’s evasion mechanisms and reactivating the immune system to attack the tumor (Figure 3B). Many of the canonical immunotherapeutic agents, such as anti-CTLA-4 therapy and anti-PD-1 therapy, are antibodies that block immune checkpoint proteins on the cell surface. These checkpoint proteins are normally responsible for inhibiting the immune system response. Hence, antibody-based checkpoint blockade results in increased activation of T-cells and a stronger anti-tumor immune response [80]. Despite the promise of immunotherapy, durable responses are limited to a subset of patients [81,82], with changes in intratumor heterogeneity and evolution of the tumor and surrounding microenvironment during immunotherapy treatment [83] being potential reasons for treatment failure. Heterogeneity of neoantigens provides another explanation for the observed variability in response to checkpoint blockade immunotherapy [80]. A high burden of putative neoantigens (as is seen in melanoma and NSCLC [81, 82]) was associated with a stronger anti-tumor cytotoxic T cell response and increased survival in a pan-cancer analysis [84]. There is a clear need to understand which patients have immunologically “cold” tumors that will never respond or will develop resistance, versus patients with tumors can be made immunologically “hot”. Beyond the need for improved patient stratification, enthusiasm should be tempered by the potential for severe off-target immune toxicity (the generalized activation of the immune system can result in widespread autoimmunity [81]). An improved understanding of the co-evolution of the tumor and immune microenvironment before and during immunotherapy is needed (potentially using in situ spatial profiling techniques, such as MIBI [30] and Nanostring digital spatial profiling [85]), and may reveal mechanisms of response and resistance [83,84].
Adaptive Containment Strategies
Adaptive therapy seeks to constrain evolution in tumors with multiple competing subclones, some of which are resistant to therapy. A key tenant of adaptive therapy is that resistance to treatment comes at a fitness cost. For example, cells with membrane efflux pumps required to expel chemotherapeutic agents have an energetic cost to the resistant clone [86]. Under adaptive therapy, treatments are given in a pulsatile manner such that the sensitive and resistant cell populations grow in the “off” and “on” phases of treatment, respectively (Figure 2C). Ideally, both cell populations are maintained and growth is competitively constrained by one another, preventing expansion of the resistant subclone(s).
Preclinical studies have provided support for adaptive therapy. Adaptive therapy was shown to increase the time to progression 2- to 10-fold in in vitro breast cancer models [87] and was also demonstrated to control growth of malignant melanoma xenografts treated with pulsatile vemurafenib [5]. While adaptive therapy is yet not widespread in patient care, some early successes have been achieved. Four melanoma patients who had developed resistance to BRAF inhibitors showed significant clinical response to the same agents after a median treatment-free period (drug holiday) of 8 months, although these responses were not lasting [88]. In a recent clinical trial of 11 patients with metastatic castrate-resistant prostate cancer, adaptive therapy maintained stable oscillations of tumor burdens in 10 patients with reduced cumulative drug (less than 50% of standard Abiraterone dosing) [89]. The adaptive therapy treatment regimen used in this clinical trial was based on a mathematical model, which considered the eco-evolutionary interactions between subclones in the context of evolutionary game theory, again highlighting the utility of modeling to guide initial hypotheses for testing in vivo. These case studies highlight the importance of understanding the evolution of both resistant and sensitive subclones on and off treatment. Adaptive therapy requires continual monitoring of disease burden, which is traditionally performed via imaging, but could also be profiled using ctDNA to quantify tumor burden and track tumor subclones simultaneously.
Concluding Remarks and Future Perspectives
Tumor progression is inherently an ecological and evolutionary process, which provides a valuable and established theoretical basis upon which to study the dynamics of tumor growth and resistance. To date, most studies have characterized tumors at a single timepoint (often diagnosis or surgery), using bulk or dissociative methods. The resultant data, in turn, necessitate the bioinformatic deconvolution of cellular populations [90,91] and the inference of dynamics, rather than direct measurements.
Improved spatial and temporal monitoring of tumor evolution has the potential to yield far greater resolution on these processes, but has been challenging for practical and technological reasons (see Outstanding Questions). Newly developed techniques that measure in situ single-cell gene and protein expression at high-throughput will be necessary for such efforts. These methods preserve tissue architecture so that cells can be studied in context and should facilitate the identification of rare resistant cell populations in lesions that are otherwise challenging to characterize using dissociative methods requiring large amounts of input material. Moreover, such methods may delineate tumor microenvironmental differences between responders and non-responders. Multiplexed proteomic approaches, in particular, hold the tantalizing promise of revealing the canalization of resistance mechanisms at a signaling level (amidst the vast genomic heterogeneity) with accompanying strategic therapeutic implications.
In tandem, patient-derived organoid models [92] and xenografts [93] have been shown to preserve the molecular and morphological characteristics of primary patient samples while yielding a renewable source of high-quality material. As such, these represent powerful platforms for drug screening [93], studying treatment response [94, 95], and improving our understanding of the dynamics and mechanisms of resistance. Moreover, they are amenable to facile genetic manipulations, lineage tracing, and rapid functional and phenotypic readouts. These experimental systems complement tissue correlative studies in primary patient samples and when coupled with iterative computational modeling, will contribute to a systematic understanding of the evolutionary dynamics and mechanisms of tumor progression and therapeutic resistance. A long-range goal of such efforts will be to develop strategies to forestall resistance and to predict the future course of disease for individual patients.
Supplementary Material
Acknowledgments
This work was supported by awards from the NIH (R01CA182514), Susan G. Komen Foundation (IIR13260750), and the Breast Cancer Research Foundation (BCRF-16-032) to C.C.
Glossary
- Cancer Stem Cells
a small subset set of tumor cells (with stem-cell-like self-renewal properties) that have tumorigenic ability; see [4]; cancer stem cells are thought to be resistant to treatment due to their quiescent state (providing chemoresistance), and many express multidrug resistance transporters, anti-apoptotic proteins, pro-survival signaling molecules [4]
- Collateral Sensitivity
in adaptive evolution, a phenomenon where an increase in fitness in one environment (e.g. the resistant cells that survive during treatment) leads to fitness changes in other environments (e.g. the resistant cells have increased sensitivity to other drugs)
- Clone
a set of cells that descend from a common ancestor and thus share genetic features
- Driver mutation
a selectively advantageous mutation that confers a fitness (e.g. growth) benefit; in cancer, “driver” genes include gain-of-function oncogenes and loss-of-function tumor suppressors that ultimately contribute to hallmarks of cancers; while driver mutations can be context dependent, canonical drivers have been defined for different cancer types [96]; a variety of computational tools exist, but many driver genes are identified based on the over-representation of damaging mutations or copy number aberrations in the putative driver genes
- Drug-induced epigenetic reprogramming
drug treatment can cause specific changes in chromatin accessibility (via changes in processes such as DNA methylation or DNA histone modifications); these epigenetic changes can lead to altered gene expression that can change cell fate and/or characteristics
- FISSEQ
fluorescence in situ sequencing; a method for genome-wide RNA expression profiling of intact cells and tissues; in this method, spatial tissue architecture is preserved allowing for RNA localization studies
- Intratumor heterogeneity (ITH)
cellular, genotypic, or phenotypic variation amongst cells within a tumor; genetic variation is a necessary substrate for evolution and contributes to fitness differences amongst tumor cells
- MERFISH
multiplexed error-robust fluorescence in situ hybridization (FISH); an imaging technique that can measure thousands of RNA species in individual cells; a robust combinatorial encoding scheme is used for each RNA species of interest; each RNA species is identified by successive rounds of hybridization that allow for the detection and correction of readout errors
- Selection
natural selection operates on phenotypes, resulting in an increased fitness advantage relative to the resident population and subsequent outgrowth of clones harboring that trait; thwarting selection can impede evolution
- Selective sweep
the selective outgrowth (clonal expansion) of cells such that positively selected clones sweep to fixation, thereby replacing the resident population; such complete selective sweeps can only occur if there is sufficient time before the acquisition of the next driver mutation
- Subclone
a subpopulation of cells that share a common genetic feature that is not present in the rest of the population
References
- 1.Scott A, et al. Antibody therapy of cancer. Nat Rev Cancer. 2012;12:278–287. doi: 10.1038/nrc3236. [DOI] [PubMed] [Google Scholar]
- 2.Ledermann J, et al. Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: a preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol. 2014;15:852–861. doi: 10.1016/S1470-2045(14)70228-1. [DOI] [PubMed] [Google Scholar]
- 3.Druker B, et al. K-ras Mutations and Benefit from Cetuximab in Advanced Colorectal Cancer. N Engl J Med. 2006;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 4.Holohan C, et al. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer. 2013;13:714–726. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- 5.Das Thakur M, et al. Modelling vemurafenib resistance in melanoma reveals a strategy to forestall drug resistance. Nature. 2013;494:251–255. doi: 10.1038/nature11814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Viani G, et al. Adjuvant trastuzumab in the treatment of her-2-positive early breast cancer: a meta-analysis of published randomized trials. BMC Cancer. 2007;7:153. doi: 10.1186/1471-2407-7-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nahta R, et al. Mechanisms of disease: understanding resistance to HER2-targeted therapy in human breast cancer. Nat Clin Pract Oncol. 2006;3:269–280. doi: 10.1038/ncponc0509. [DOI] [PubMed] [Google Scholar]
- 8.Karapetis C, et al. Targeting EGFR in Colorectal Cancer. N Engl J Med. 2008;359:1834–1836. doi: 10.1056/NEJMe0806778. [DOI] [PubMed] [Google Scholar]
- 9.Corcoran R, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discovery. 2012;2:227–235. doi: 10.1158/2159-8290.CD-11-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giltnane J, et al. Genomic profiling of ER+ breast cancers after short-term estrogen suppression reveals alterations associated with endocrine resistance. Sci Transl Med. 2017;9:402. doi: 10.1126/scitranslmed.aai7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hugo W, et al. Non-genomic and Immune Evolution of Melanoma Acquiring MAPKi Resistance. Cell. 2015;162:1271–1285. doi: 10.1016/j.cell.2015.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thress K, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non- small cell lung cancer harboring EGFR T790M. Nat Med. 2015;21:560–562. doi: 10.1038/nm.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burger J, et al. Clonal evolution in patients with chronic lymphocytic leukaemia developing resistance to BTK inhibition. Nat Commun. 2016;7:11589. doi: 10.1038/ncomms11589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maruysk A, et al. Intra-tumour heterogeneity: a looking glass for cancer? Nat Rev Cancer. 2014;12:323–334. doi: 10.1038/nrc3261. [DOI] [PubMed] [Google Scholar]
- 15.Mroz E, et al. MATH, a novel measure of intra-tumor genetic heterogeneity, is high in poor-outcome classes of head neck squamous cell carcinoma. Oral Oncol. 2013;49:211–215. doi: 10.1016/j.oraloncology.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J, et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science. 2014;346:256–259. doi: 10.1126/science.1256930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwarz R, et al. Spatial and temporal heterogeneity in high-grade serous ovarian cancer: a phylogenetic analysis. PLoS Med. 2015;12:e1001789. doi: 10.1371/journal.pmed.1001789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andor N, et al. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat Med. 2016;22:105–113. doi: 10.1038/nm.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerlinger M, et al. How Darwinian models inform therapeutic failure initiated by clonal heterogeneity in cancer medicine. Br J Cancer. 2010;103:1139–1143. doi: 10.1038/sj.bjc.6605912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lohr J, et al. Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell. 2014;25:91–101. doi: 10.1016/j.ccr.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pearson A, et al. High-level clonal FGFR amplification and response to FGFR inhibition in a translational clinical trial. Cancer Discov. 2016;6:838–851. doi: 10.1158/2159-8290.CD-15-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown R, et al. Poised epigenetic states and acquired drug resistance in cancer. Nat Rev Cancer. 2014;14:747–753. doi: 10.1038/nrc3819. [DOI] [PubMed] [Google Scholar]
- 23.Shaffer S, et al. Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature. 2017;546:431–435. doi: 10.1038/nature22794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koyama S, et al. Adaptive resistance to therapeutic PD-1 blockade is associated with upregulation of alternative immune checkpoints. Nat Commun. 2016;7:10501. doi: 10.1038/ncomms10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilbert L, et al. DNA damage-mediated induction of a chemoresistant niche. Cell. 2010;143:355–366. doi: 10.1016/j.cell.2010.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buenrostro J, et al. ATAC-seq: A Method for Assaying Chromatin Accessibility Genome- Wide. Curr Protoc Mol Biol. 2015;109:21.29.1–21.29.9. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen KH, et al. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348:412. doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee JH, et al. Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nat Protoc. 2015;10:442–458. doi: 10.1038/nprot.2014.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J, et al. Nondestructive tissue analysis for ex vivo and in vivo cancer diagnosis using a handheld mass spectrometry system. Sci Trans Med. 2017;406:eaan3968. doi: 10.1126/scitranslmed.aan3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Angelo M, et al. Multiplexed ion beam imaging of human breast tumors. Nat Med. 2014;20:436–442. doi: 10.1038/nm.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maley C, et al. Classifying the evolutionary and ecological features of neoplasms. Nat Rev Cancer. 2017;17:605–619. doi: 10.1038/nrc.2017.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nawaz S, et al. Computational pathology: Exploring the spatial dimension of tumor ecology. Cancer Letters. 2016;380:296–303. doi: 10.1016/j.canlet.2015.11.018. [DOI] [PubMed] [Google Scholar]
- 33.Nowell P. The Clonal Evolution of Tumor Cell Populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 34.Greaves M, et al. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scott J, et al. Somatic clonal evolution: A selection-centric perspective. Biochim Biophys Acta. 2017;1867:139–150. doi: 10.1016/j.bbcan.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 36.Sottoriva A, et al. A Big Bang model of human colorectal tumor growth. Nature Genet. 2015;47:209–216. doi: 10.1038/ng.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun R, et al. Between-region genetic divergence reflects the mode and tempo of tumor evolution. Nature Genet. 2017;49:1015–1024. doi: 10.1038/ng.3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ling S, et al. Extremely high genetic diversity in a single tumor points to prevalence of non-Darwinian cell evolution. Proc Natl Acad Sci. 2015;112:E6496–6505. doi: 10.1073/pnas.1519556112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williams M, et al. Identification of neutral tumor evolution across cancer types. Nature Genet. 2016;48:238–244. doi: 10.1038/ng.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao R, et al. Punctuated copy number evolution and clonal stasis in triple-negative breast cancer. Nature Genet. 2016;48:1119–1130. doi: 10.1038/ng.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baca S, et al. Punctuated evolution of prostate cancer genomes. Cell. 2013;153:666–677. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Notta F, et al. A renewed model of pancreatic cancer evolution based on genomic rearrangement patterns. Nature. 2016;538:378–382. doi: 10.1038/nature19823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nguyen D, et al. Metastasis: from dissemination to organ-specific colonization. Nat Rev. 2009;9:274–284. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- 44.Altrock P, et al. The mathematics of cancer: integrating quantitative models. Nat Rev Cancer. 2015;15:730–745. doi: 10.1038/nrc4029. [DOI] [PubMed] [Google Scholar]
- 45.Bozic I, et al. Resisting Resistance. Ann Rev Cancer Biol. 2017;1:203–221. [Google Scholar]
- 46.Bozic I, et al. Evolutionary dynamics of cancer in response to targeted combination therapy. Elife. 2013;2:e00747. doi: 10.7554/eLife.00747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martens E, et al. Spatial structure increases the waiting time for cancer. New J Phys. 2011:13. doi: 10.1088/1367-2630/13/11/115014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McDougall S, et al. Mathematical modelling of dynamic adaptive tumour-induced angiogenesis: Clinical implications and therapeutic targeting strategies. Journal of Theoretical Biology. 2006;241:564–589. doi: 10.1016/j.jtbi.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 49.Anderson A, et al. Tumor Morphology and Phenotypic Evolution Driven by Selective Pressure from the Microenvironment. Cell. 2006;127:905–915. doi: 10.1016/j.cell.2006.09.042. [DOI] [PubMed] [Google Scholar]
- 50.Waclaw B, et al. A spatial model predicts that dispersal and cell turnover limit intratumour heterogeneity. Nature. 2015;525:261–264. doi: 10.1038/nature14971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jamal-Hanjani M, et al. Tracking the Evolution of Non Small-Cell Lung Cancer. N Engl J Med. 2017;376:2109–2121. doi: 10.1056/NEJMoa1616288. [DOI] [PubMed] [Google Scholar]
- 52.Sottoriva A, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci. 2013;110:4009–4014. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gerlinger M, et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nature Genet. 2014;46:225–233. doi: 10.1038/ng.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gerlinger M, et al. Intratumor Heterogeneity and Branched Evolution Revealed by Multiregion Sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ma P, et al. Simultaneous evolutionary expansion and constraint of genomic heterogeneity in multifocal lung cancer. Nat Commun. 2017;8:823. doi: 10.1038/s41467-017-00963-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Patel A, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Puram S, et al. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell. 2017;171:1611–1624. doi: 10.1016/j.cell.2017.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gawad C, et al. Single-cell genome sequencing: current state of the science. Nat Rev. 2016;17:175–188. doi: 10.1038/nrg.2015.16. [DOI] [PubMed] [Google Scholar]
- 59.Wang Y, et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature. 2014;512:155–160. doi: 10.1038/nature13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roth A, et al. Clonal genotype and population structure inference from single-cell tumor sequencing. Nat Methods. 2016;13:573–576. doi: 10.1038/nmeth.3867. [DOI] [PubMed] [Google Scholar]
- 61.Janes K, et al. Identifying single-cell molecular programs by stochastic profiling. Nat Methods. 2010;7:311–317. doi: 10.1038/nmeth.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leung ML, et al. Single-cell DNA sequencing reveals a late-dissemination model in metastatic colorectal cancer. Genome Res. 2017;27:1287–1299. doi: 10.1101/gr.209973.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Salehi S, et al. ddClone: joint statistical inference of clonal populations from single cell and bulk tumour sequencing data. Genome Biol. 2017;18:44. doi: 10.1186/s13059-017-1169-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Murtaza M, et al. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nat Commun. 2015;6:8760. doi: 10.1038/ncomms9760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Abbosh C, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature. 2017;545:446–451. doi: 10.1038/nature22364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chaudhuri A, et al. Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA profiling. Cancer Discov. 2017 doi: 10.1158/2159-8290.CD-17-0716. CD-17–0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Scherer F, et al. Distinct biological subtypes and patterns of genome evolution in lymphoma revealed by circulating tumor DNA. Sci Transl Med. 2016;364:155. doi: 10.1126/scitranslmed.aai8545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bettegowda C, et al. Detection of Circulating Tumor DNA in Early- and Late-Stage Human Malignancies. Sci Transl Med. 2014;6:224. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Husain H, et al. Monitoring Daily Dynamics of Early Tumor Response to Targeted Therapy by Detecting Circulating Tumor DNA in Urine. Clin Can Res. 2017;23:4716–23. doi: 10.1158/1078-0432.CCR-17-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ojamies P, et al. Monitoring therapy responses at the leukemic subclone level by ultra-deep amplicon resequencing in acute myeloid leukemia. Leukemia. 2017;31:1048–105. doi: 10.1038/leu.2016.286. [DOI] [PubMed] [Google Scholar]
- 71.Perakis S, et al. Emerging concepts in liquid biopsies. BMC Medicine. 2017;15:75. doi: 10.1186/s12916-017-0840-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Worthington R, et al. Combination approaches to combat multidrug-resistant bacteria. Trends in Biotech. 2013;31:177–184. doi: 10.1016/j.tibtech.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Arts E, et al. HIV-1 Antiretroviral Drug Therapy. Cold Spring Harb Perspect Med. 2:a007161. doi: 10.1101/cshperspect.a007161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhao B, et al. Intratumor heterogeneity alters most effective drugs in designed combinations. PNAS. 2014;111:10773–10778. doi: 10.1073/pnas.1323934111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhao B, et al. Modeling tumor clonal evolution for drug combinations design. Trends in cancer. 2016;2:144–158. doi: 10.1016/j.trecan.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Corcoran R, et al. Combined BRAF, EGFR, and MEK Inhibition in Patients With BRAF V600E-Mutant Colorectal Cancer. Cancer Discov. 2018;8:428–443. doi: 10.1158/2159-8290.CD-17-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhao B, et al. Exploiting Temporal Collateral Sensitivity in Tumor Clonal Evolution. Cell. 2016;165:234–246. doi: 10.1016/j.cell.2016.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dhawan A, et al. Collateral sensitivity networks reveal evolutionary instability and novel treatment strategies in ALK mutated non-small cell lung cancer. Sci Rep. 2017;7:1232. doi: 10.1038/s41598-017-00791-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nichol D, et al. Steering Evolution with Sequential Therapy to Prevent the Emergence of Bacterial Antibiotic Resistance. PLOS Computational Biology. 2015;11:e1004493. doi: 10.1371/journal.pcbi.1004493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schumacher T, et al. Neoantigens in cancer immunotherapy. Science. 2015;348:69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- 81.Robert C, et al. Pembrolizumab versus ipilimumab in advanced melanoma. N Engl J Med. 2015;372:2521–2532. doi: 10.1056/NEJMoa1503093. [DOI] [PubMed] [Google Scholar]
- 82.Reck M, et al. Pembrolizumab versus Chemotherapy for PD-L1–Positive Non–Small-Cell Lung Cancer. N Engl J Med. 2016;375:1823–1833. doi: 10.1056/NEJMoa1606774. [DOI] [PubMed] [Google Scholar]
- 83.Riaz N, et al. Tumor and Microenvironment Evolution during Immunotherapy with Nivolumab. Cell. 2017;171:934–949. doi: 10.1016/j.cell.2017.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brown S, et al. Neo-antigens predicted by tumor genome meta-analysis correlate with increased patient survival. Genome Res. 2014;24:743–750. doi: 10.1101/gr.165985.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Geiss G, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–25. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 86.Szakacs G, et al. Targeting the Achilles Heel of Multidrug-Resistant Cancer by Exploiting the Fitness Cost of Resistance. Chem Rev. 114:5753–5774. doi: 10.1021/cr4006236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Silva A, et al. Evolutionary Approaches to Prolong Progression-Free Survival in Breast Cancer. Cancer Res. 2012;72:6362–6370. doi: 10.1158/0008-5472.CAN-12-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rogiers A, et al. Dabrafenib plus trametinib rechallenge in four melanoma patients who previously progressed on this combination. Melanoma Res. 2017;27:164–167. doi: 10.1097/CMR.0000000000000320. [DOI] [PubMed] [Google Scholar]
- 89.Zhang J, et al. Integrating evolutionary dynamics into treatment of metastatic castrate-resistant prostate cancer. Nat Commun. 2017;8:1816. doi: 10.1038/s41467-017-01968-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Andor N, et al. EXPANDS: Expanding ploidy and allele frequency on nested subpopulations. Bioinformatics. 2014;30:50–60. doi: 10.1093/bioinformatics/btt622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Roth A, et al. PyClone: Statistical inference of clonal population structure in cancer. Nat Methods. 2014;11:396–398. doi: 10.1038/nmeth.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sachs N, et al. A Living Biobank of Breast Cancer Organoids Captures Disease Heterogeneity. Cell. 2017;172:373–386. doi: 10.1016/j.cell.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 93.Bruna A, et al. A Biobank of Breast Cancer Explants with Preserved Intra-tumor Heterogeneity to Screen Anticancer Compounds. Cell. 2017;167:260–274. doi: 10.1016/j.cell.2016.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vlachogiannis G, et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 2018;359:920–926. doi: 10.1126/science.aao2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jenkins R, et al. Ex Vivo Profiling of PD-1 Blockade Using Organotypic Tumor Spheroids. Cancer Discov. 2018;8:196–215. doi: 10.1158/2159-8290.CD-17-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bailey M, et al. Comprehensive Characterization of Cancer Driver Genes and Mutations. Cell. 2018;173:371–385. doi: 10.1016/j.cell.2018.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.