Abstract
Background & Aims
A generalized human pacemaking syndrome, chronic atrial and intestinal dysrhythmia (CAID) (OMIM 616201), is caused by a homozygous SGO1 mutation (K23E), leading to chronic intestinal pseudo-obstruction and arrhythmias. Because CAID patients do not show phenotypes consistent with perturbation of known roles of SGO1, we hypothesized that noncanonical roles of SGO1 drive the clinical manifestations observed.
Methods
To identify a molecular signature for CAID syndrome, we achieved unbiased screens in cell lines and gut tissues from CAID patients vs wild-type controls. We performed RNA sequencing along with stable isotope labeling with amino acids in cell culture. In addition, we determined the genome-wide DNA methylation and chromatin accessibility signatures using reduced representative bisulfite sequencing and assay for transposase-accessible chromatin with high-throughput sequencing. Functional studies included patch-clamp, quantitation of transforming growth factor-β (TGF-β) signaling, and immunohistochemistry in CAID patient gut biopsy specimens.
Results
Proteome and transcriptome studies converge on cell-cycle regulation, cardiac conduction, and smooth muscle regulation as drivers of CAID syndrome. Specifically, the inward rectifier current, an important regulator of cellular function, was disrupted. Immunohistochemistry confirmed overexpression of Budding Uninhibited By Benzimidazoles 1 (BUB1) in patients, implicating the TGF-β pathway in CAID pathogenesis. Canonical TGF-β signaling was up-regulated and uncoupled from noncanonical signaling in CAID patients. Reduced representative bisulfite sequencing and assay for transposase-accessible chromatin with high-throughput sequencing experiments showed significant changes of chromatin states in CAID, pointing to epigenetic regulation as a possible pathologic mechanism.
Conclusions
Our findings point to impaired inward rectifier potassium current, dysregulation of canonical TGF-β signaling, and epigenetic regulation as potential drivers of intestinal and cardiac manifestations of CAID syndrome. Transcript profiling and genomics data are as follows: repository URL: https://www.ncbi.nlm.nih.gov/geo; SuperSeries GSE110612 was composed of the following subseries: GSE110309, GSE110576, and GSE110601.
Keywords: CAID Syndrome (Chronic Atrial and Intestinal Dysrhythmia), Chronic Intestinal Pseudo-obstruction, TGF-β Signaling, Epigenetics
Abbreviations used in this paper: ATAC, assay for transposase-accessible chromatin with high throughput; CAID, chronic atrial and intestinal dysrhythmia; CIPO, chronic intestinal pseudo-obstruction; DAB, 3,3′-diaminobenzidine tetra hydrochloride; ERK, extracellular signal–regulated kinase; FDR, false discovery rate; GO, Gene Ontology; IK1, inward rectifying potassium current; JNK, c-Jun-N-terminal kinase; MAPK, mitogen-activated protein kinase; mRNA, messenger RNA; p, passage number; PCR, polymerase chain reaction; RMP, resting membrane potential; RRBS, reduced representative bisulfite sequencing; SILAC, stable isotope labeling by amino acids in cell culture; TAGLN, Transgelin; TGF-β, transforming growth factor-β; TPM, tropomyosin
Graphical abstract
Summary.
CAID syndrome is a genetic disease in which failure of pacemakers in the entire human body leads to loss of all bowel movements. We here identify unknown mechanisms by which CAID syndrome causes this intestinal disease.
Lifelong rhythmic contractions are the hallmark of the human heart and gut. Both organs contain pacemaker cells that spontaneously can generate an action potential through a shared polarization mechanism involving ion channels. In the heart, the sinoatrial node is responsible for this mechanism, whereas in the gut, the network of interstitial cells of Cajal and the autonomous enteric nervous system depolarize the myocytes and allow the induction of electric signals.
Disorders of pacemaking are a major cause of gastrointestinal and cardiac diseases. Chronic intestinal pseudo-obstruction (CIPO) is a rare and severe disorder of gastrointestinal motility in which intestinal obstruction occurs in the absence of a mechanical obstacle. CIPO is one of the most important causes of intestinal insufficiency in pediatric (15%) and adult cases (20%).1, 2, 3 CIPO can result from the insufficiency of pacemaker cells and the enteric nervous system to generate or propagate electric signals and stimulate subsequent contraction of the smooth muscles, leading to contraction defects (neurogenic CIPO). It also can result from primary smooth muscle contraction defects (myogenic CIPO). CIPO is a devastating and life-long disease that often requires surgical intestinal decompression as well as total parenteral nutrition. Even though X-linked, autosomal-dominant, and recessive mutations have been identified in FLNA, ACTG2, TYMP, POLG1, and RAD21, most cases are sporadic and leave the causes and pathomechanisms of CIPO poorly defined.4, 5, 6, 7, 8
We recently identified a generalized human pacemaking syndrome, encompassing both gastrointestinal and cardiac dysrhythmias. We have termed this condition chronic atrial and intestinal dysrhythmia (CAID) syndrome (OMIM 616201). We identified a recessive point mutation in the cohesion regulator SGO1, namely K23E, as the cause of CAID syndrome.3 CAID patients appeared normal at birth, and no patient had any other congenital anomalies. Gastrointestinal symptoms usually preceded cardiac symptoms. In different individuals with CAID, CIPO was found to be of neurogenic, myogenic, or mixed origin.3 Representative manometries of CAID patients showed visceral neuropathy with abnormal response to intravenous erythromycin and colonic manometries showed visceral myopathy with low-amplitude propagated contractions after stimulation.3 Gastrointestinal symptoms were also the main determinant of morbidity and mortality.
Karyotypes in affected individuals showed the typical railroad appearance of a centromeric cohesion defect, identifying CAID as a novel cohesinopathy. Although some clinical manifestations overlap between CAID and other cohesinopathies, CAID syndrome is set apart in that it does not cause intellectual and growth retardation. In addition, there are no clinical phenotypes directly associated with the canonical role of SGO1 in centromeric cohesion and sister chromatid segregation, such as premature aging or cancer. Interestingly, fibroblasts from CAID patients showed accelerated cell-cycle progression, a higher rate of senescence, and enhanced activation of transforming growth factor-β (TGF-β) signaling at late passage,3 identifying clinical cross-talk between the TGF-β and cohesin cascades. Unfortunately, homozygous Sgo1 knockout mice and homozygous K23E knock-in mice are embryonic lethal (data not shown), leaving only cell-based models for the study of this condition.9
The clinical observations raise essential pathophysiological questions about the role of SGO1 and have prompted us to perform unbiased screens to identify the molecular signature of CAID syndrome in vivo along all steps of the central dogma. Here, we investigated how the SGO1 mutation affects regulation on epigenetic RNA expression and proteomic levels using reduced representative bisulfite sequencing (RRBS), assay for transposase-accessible chromatin with high throughput (ATAC) sequencing, RNA sequencing, and stable isotope labeling with amino acids in cell culture (SILAC) on CAID patient fibroblasts. Because the first symptoms of CAID pathology are not present at birth and appear from 5 years of age onward, the phenotypic manifestations seem to be associated with aging. Thus, the screens were performed on early (passage [p]8) and late (p14) passages. We report differential expression of several genes involved in the cohesin complex and cell-cycle regulation, cardiac conduction, and smooth muscle identity and contraction, as well as perturbation of potassium currents in CAID syndrome, canonical TGF-β signaling, DNA methylation, and chromatin compaction. These results expose unanticipated noncanonical roles for SGO1 in CAID pathogenesis and raise important questions for other disorders of gastrointestinal motility.
Results
Transcriptome Profiling of CAID Syndrome
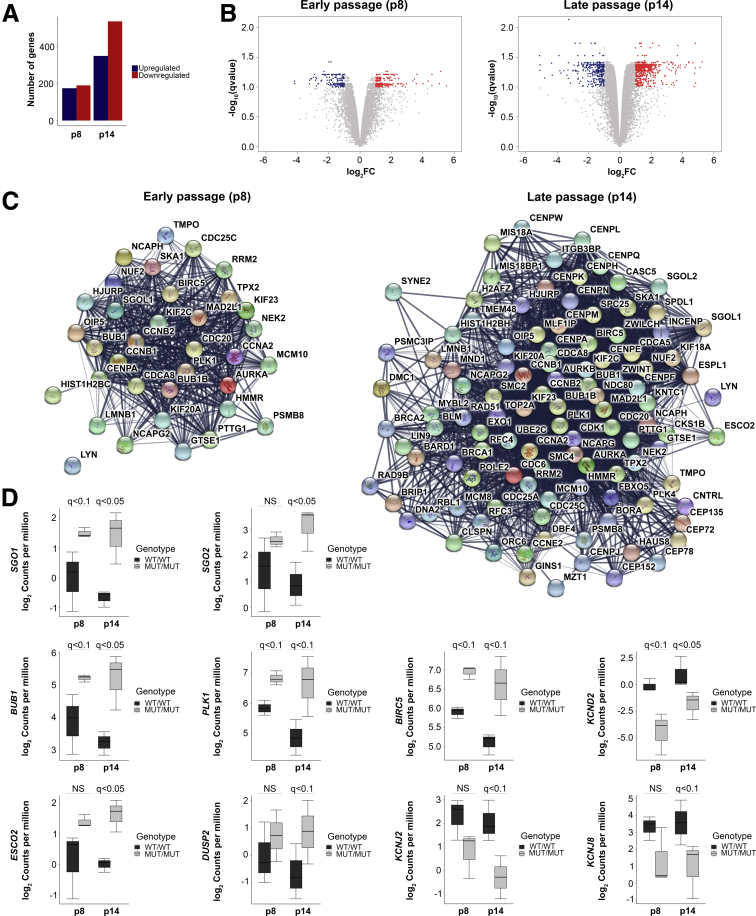
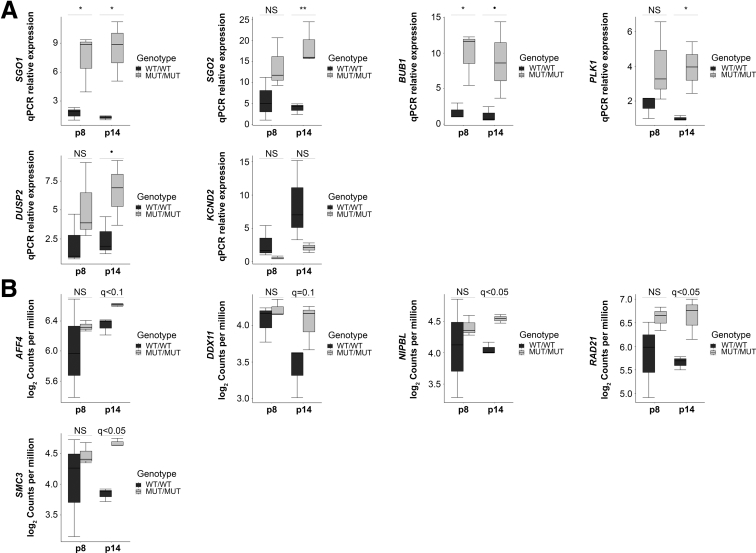
To characterize the transcriptomic profile of CAID patients homozygous for the SGO1 K23E mutation (MUT/MUT) vs wild-type SGO1 controls (WT/WT), we performed paired-end RNA sequencing on human dermal fibroblasts from 3 cases and 3 controls at early (p8) and late passages (p14). Consistent with the more pronounced phenotype with aging, we identified 173 down-regulated and 188 up-regulated genes at p8, whereas at p14, we identified 346 down-regulated and 531 up-regulated genes (Figure 1A, Supplementary Table 1). Volcano plots of differentially expressed genes at p8 and p14 showed that the vast majority of messenger RNA (mRNA) expression changes observed between CAID patients and controls are less than 4-fold (Figure 1B). Biological functions associated with differentially expressed genes at p8 also were associated with gene expression differences at p14. However, additional biological functions including heart development and contraction, muscle contraction, chromatin compaction, behavior, and potassium channels also were identified at p14 (Tables 1 and 2, Supplementary Table 2). At p8, we identified a specific cluster of up-regulated genes (35 genes) associated with the cell cycle, and this cluster was considerably larger at p14 (109 genes). Of note, we detected up-regulation of the following genes at different levels of significance: SGO1 itself; SGO2, the second member of the shugoshin family involved mainly in meiosis; and Budding Uninhibited By Benzimidazoles 1 (BUB1) and PLK1, required for SGO1 localization at the centromere (Figures 1D and 2A). At early passage (p8), SGO1, SGO2, BUB1, and PLK1 mRNA levels were higher in CAID patients compared with controls, and at late passage (p14) their levels were maintained or up-regulated in CAID patients and decreased in controls. This correlates with overexpression of BUB1 in CAID patients at the protein level (Figure 3C and D). Thus, the SGO1 K23E mutation does not abolish normal SGO1 function in mitosis,3 but may be associated with causal or compensatory overexpression of SGO1, SGO2, BUB1, and PLK1. Interestingly, we also a noticed a significant up-regulation of ESCO2, a gene involved in Roberts syndrome, another cohesinopathy (Figure 1D). This led us to investigate whether other cohesin components are dysregulated as well, and we found a trend toward up-regulation for AFF4, DDX11, NIPBL, RAD21, and SMC3 in CAID patients, although the log2 fold changes and/or q values of the mRNA expression levels were below the threshold (Figure 2B). Interestingly, we also found a significant up-regulation of BIRC5, an anti-apoptotic gene that binds an amino-terminal motif of SGO1 (Figure 1D).10 BIRC5 contributes to the control of the total cardiomyocyte number and its cardiac-specific knock-out leads to conduction defects and sino-atrial node dysfunction.11, 12 We also identified down-regulation of potassium voltage-gated channels KCNJ2 (Kir2.1), KCNJ8 (Kir6.1), and KCND2 (Kv.4.2), suggesting altered potassium channel currents in CAID syndrome (Figure 1D). Taken together, these data suggest that cell-cycle regulation, cardiac conduction, chromatin compaction, and potassium channels are key pathways involved in CAID syndrome.
Figure 1.
Transcriptomic profiling of CAID patient dermal fibroblasts. (A) Volcano plot of the differentially expressed genes at early (p8) and late stage (p14). Log2 fold change: blue, <-1; red, >1. (B) Number of up-regulated and down-regulated genes identified at early (p8) and late passage (p14). (C) Cluster of genes associated with cell cycle identified at p8 that became larger at p14. (D) mRNA expression of candidate genes at early (p8) and late passage (p14) showing up-regulation of SGO1, SGO2, BUB1, PLK1, ESCO2, DUSP2, and BIRC5 and down-regulation of KCND2, KCNJ2, and KCNJ8. Error bars signify SD. For each group, N = 3 independent biological replicates. P values were corrected for multiple testing using the Benjamini–Hochberg method (q value). FC, fold change; MUT, K23E; WT, wild-type.
Table 1.
Biological Functions (GO Analysis) Associated With Differentially Expressed Genes in RNA Sequencing at Early Passage (p8)
| Biological functions | P value | Genes, n |
|---|---|---|
| Up-regulated | ||
| Cell cycle | 2.46E-19 | 35 |
| Sister chromatid segregation | 2.47E-17 | 22 |
| Sister chromatid cohesion | 9.62E-10 | 12 |
| Extracellular matrix organization | 4.45E-04 | 10 |
| Actomyosin structure organization | 6.47E-04 | 7 |
| Chromatin remodeling at the centromere | 5.39E-03 | 3 |
| Down-regulated | ||
| ABC transporters | 8.15E-07 | 6 |
| Chemical synaptic transmission | 1.42E-04 | 14 |
| Behavior | 2.54E-04 | 13 |
| Actin-mediated cell contraction | 8.25E-03 | 4 |
Table 2.
Biological Functions (GO Analysis) Associated With Differentially Expressed Genes in RNA Sequencing at Late Passage (p14)
| Biological functions | P value | Genes, n |
|---|---|---|
| Up-regulated | ||
| Cell cycle | 8.46E-64 | 109 |
| Sister chromatid segregation | 1.60E-49 | 63 |
| Sister chromatid cohesion | 7.92E-30 | 37 |
| Chromatin remodeling | 1.40E-06 | 16 |
| Histones phosphorylation | 1.69E-05 | 7 |
| Cardiac muscle fiber development | 8.80E-04 | 3 |
| Actin filament-based movement | 3.21E-03 | 9 |
| Cardiac muscle hypertrophy in response to stress | 6.36E-03 | 4 |
| Developmental growth | 9.47E-03 | 23 |
| Down-regulated | ||
| Cardiac conduction | 1.87E-04 | 9 |
| Circulatory system process | 5.19E-04 | 18 |
| Behavior | 5.39E-04 | 19 |
| Muscle contraction | 7.45E-04 | 10 |
| Potassium ion transport | 2.20E-03 | 10 |
| ABC transporters | 3.72E-03 | 4 |
| Heart contraction | 5.63E-03 | 10 |
Figure 2.
Candidates validation and expression profile of cohesinopathies genes. (A) Reverse transcription quantitative PCR (qPCR) validations of SGO1, SGO2, BUB1, PLK1, DUSP2, and KCND2 expression at early (p8) and late (p14) passage. Error bars signify SD. For each condition, the experiment was performed on N = 3 independent biological replicates in technical replicates. Significance was calculated using 1-way analysis of variance with the Bonferroni post-test (•P < .1, *P < .05, and **P < .01). (B) mRNA expression levels of genes involved in cohesinopathies at early (p8) and late (p14) passage. AFF4, CHOPS syndrome; DDX11, Warsaw breakage syndrome; NIPBL, RAD21, and SMC3, Cornelia de Lange syndrome. Error bars signify SD. P values were corrected for multiple testing using the Benjamini–Hochberg method (q value). For each group, N = 3 independent biological replicates. MUT, K23E; WT, wild-type.
Figure 3.
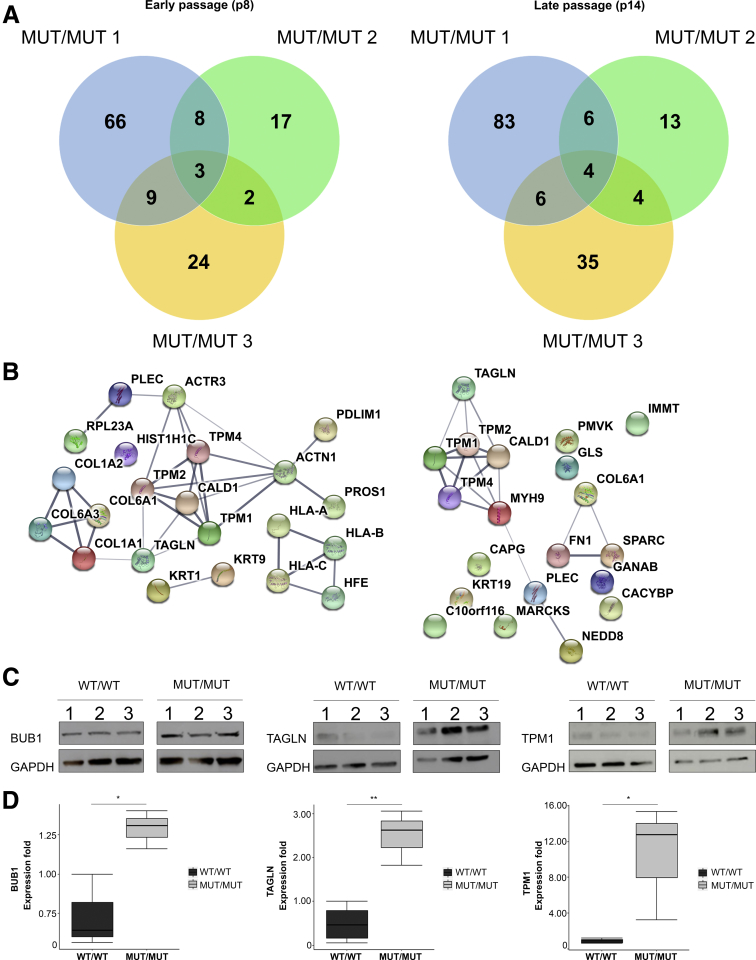
Proteomic profiling of CAID patient dermal fibroblasts. (A) Venn diagram of proteins differentially expressed in CAID patient/control duos at early (p10) and late passage (p14). (B) Protein network of differentially expressed proteins in CAID patients at early (p10) and late (p14) passage. N = 3 independent biological replicates were analyzed for each condition. P values were corrected for multiple testing using the Benjamini–Hochberg method (q value). (C) Western blot validation of TPM1, TAGLN, and BUB1 overexpression. (D) Densitometric quantification of BUB1, TAGLN, and TPM1 overexpression by Western blot. Error bars signify SD. For each group, N = 3 independent biological replicates (2 independent experiments). Significance was calculated by 1-way analysis of variance with the Bonferroni post-test (*P < .05, **P < .01). GAPDH, glyceraldehyde-3-phosphate dehydrogenase; MUT, K23E; WT, wild-type.
Proteome Profiling of CAID Syndrome
To investigate the effect of the SGO1 K23E mutation on protein expression, we performed SILAC on human dermal fibroblasts from 3 CAID patients and 3 controls. Cells were mixed together in a 1:1 ratio to form 3 duos composed of a wild-type control and a CAID patient. Because the global protein expression between duos was highly variable, we decided to look specifically at proteins expressed differentially in more than 1 duo (Figure 3A, Supplementary Table 3). At the earliest passage accessible by SILAC (p10), we detected 458 proteins, whereas at late passage (p14), 551 proteins were detected. At early passage (p10), 22 proteins were expressed differentially (q value ≤ 0.01) between duos, whereas at late passage (p14), we identified 20 proteins (Figure 3B). Gene ontology (GO) analysis showed that most of them are involved in extracellular matrix organization, regulation of actin cytoskeleton, and cardiac and smooth muscle contraction (Tables 3 and 4, Supplementary Table 4). We identified tropomyosin (TPM)1, TPM2, and TPM4, and Transgelin (TAGLN) as interesting biological candidates. TPMs code for tropomyosin family members, which play a major role in cardiac and smooth muscle contraction through CALD1 binding and subsequent actin cytoskeleton stabilization.13 TAGLN, also known as SM22α, is an actin-binding protein from the calponin family that has been suggested to regulate smooth muscle contractility through regulation of Ca2+-independent contractions.14 Overexpression of TPM1 and TAGLN in CAID patients also has been confirmed at the protein level using Western blot (Figure 3C and D). These results designate smooth muscle contraction through TPMs and TAGLN as a plausible pathway involved in the CAID gastrointestinal phenotype.
Table 3.
Biological Functions (GO Analysis) Associated With Differentially Expressed Proteins in SILAC at Early Passage (p8)
| Biological functions | P value | Genes, n |
|---|---|---|
| Assembly of collagen fibrils and other multimeric structures | 1.85E-09 | 5 |
| Smooth muscle contraction | 2.93E-08 | 4 |
| Supramolecular fiber organization | 1.94E-08 | 8 |
| Cardiac muscle contraction | 4.36E-05 | 3 |
| Hypertrophic cardiomyopathy | 5.26E-05 | 3 |
| Dilated cardiomyopathy | 6.69E-05 | 3 |
| Actin-mediated cell contraction | 1.22E-04 | 3 |
| Extracellular matrix organization | 2.00E-04 | 4 |
| Response to oxidative stress | 5.11E-04 | 4 |
| Muscle organ development | 4.85E-03 | 3 |
Table 4.
Biological Functions (GO Analysis) Associated With Differentially Expressed Proteins in SILAC at Late Passage (p14)
| Biological functions | P value | Genes, n |
|---|---|---|
| Smooth muscle contraction | 2.93E-08 | 4 |
| Muscle contraction | 2.93E-05 | 4 |
| ECM proteoglycans | 3.88E-05 | 3 |
| Cardiac muscle contraction | 4.36E-05 | 3 |
| Hypertrophic cardiomyopathy | 5.26E-05 | 3 |
| Dilated cardiomyopathy | 6.69E-05 | 3 |
| Striated muscle cell differentiation | 9.15E-05 | 4 |
| Actin-mediated cell contraction | 1.22E-04 | 3 |
| Extracellular matrix organization | 1.26E-04 | 4 |
| Adrenergic signaling in cardiomyocytes | 2.69E-04 | 3 |
ECM, extracellular matrix.
Gastrointestinal Histology of CAID Syndrome
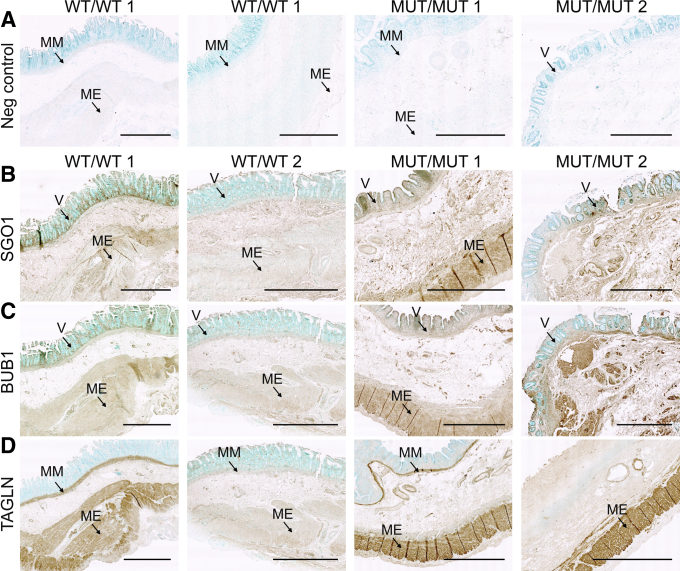
To confirm the results of unbiased screens of the CAID proteome and transcriptome, we performed immunohistochemistry by 3,3′-diaminobenzidine tetra hydrochloride (DAB) and fluorescence staining on gut biopsy specimens. We performed staining in the absence of primary antibody (negative control) to make sure that the signal was not caused by nonspecific tissue reactivity (Figures 4A and 5B). We further confirmed our results through an immunizing peptide blocking experiment to ensure the specificity of the antibodies used (Figure 5A). These experiments show BUB1 overexpression in the gut of CAID patients (Figure 4C), which is most striking in the muscularis externa and intestinal villi (Figure 5C). Similarly, SGO1 expression is higher in CAID patients compared with controls and this differential expression is enhanced in the muscularis externa and the villi (Figures 4B and 5C). Finally, we found TAGLN to be overexpressed specifically in the muscularis mucosa and muscularis externa of CAID patients (Figures 4D and 5D). We previously reported thinning of smooth muscle layers, disruption of fiber architecture of smooth muscles, and extensive fibrosis, a hallmark of chronic TGF-β activation, in CAID patient guts.3 Taking this into consideration, overexpression of TAGLN in muscular layers might be indicative of reparative changes. Such changes could indicate either a causal pathomechanism or compensatory processes in damaged tissues. In summary, these results suggest a possible role of SGO1 and BUB1 in CAID gastrointestinal phenotype.
Figure 4.
Gastrointestinal histology of CAID patients. Biopsy specimens (5 μm) from controls and individuals with CAID. All biopsy specimens are oriented to show the villi on top and the muscularis externa at the bottom. (A) Negative controls show that no signal was obtained in the absence of the primary antibody. (B–D) DAB staining was performed for SGO1, BUB1, and TAGLN. (B) SGO1 and (C) BUB1 show higher expression in the villi and muscularis externa of CAID patient intestines vs controls. (D) TAGLN shows higher expression in muscularis mucosa and muscularis externa in CAID patient intestines vs controls (N = 3, 2 shown). Experiment was performed 3 times independently on 3 biological replicates for each condition. The Scale bar is 1000 μm except MUT/MUT1 SGO1, BUB1, TAGLN, and MUT/MUT 2 TAGLN where Scale bar: 2000 μm. Images were acquired using a Zeiss AxioScan Z1 slide scanner and processed with Zen software. MUT, K23E; WT, wild-type.
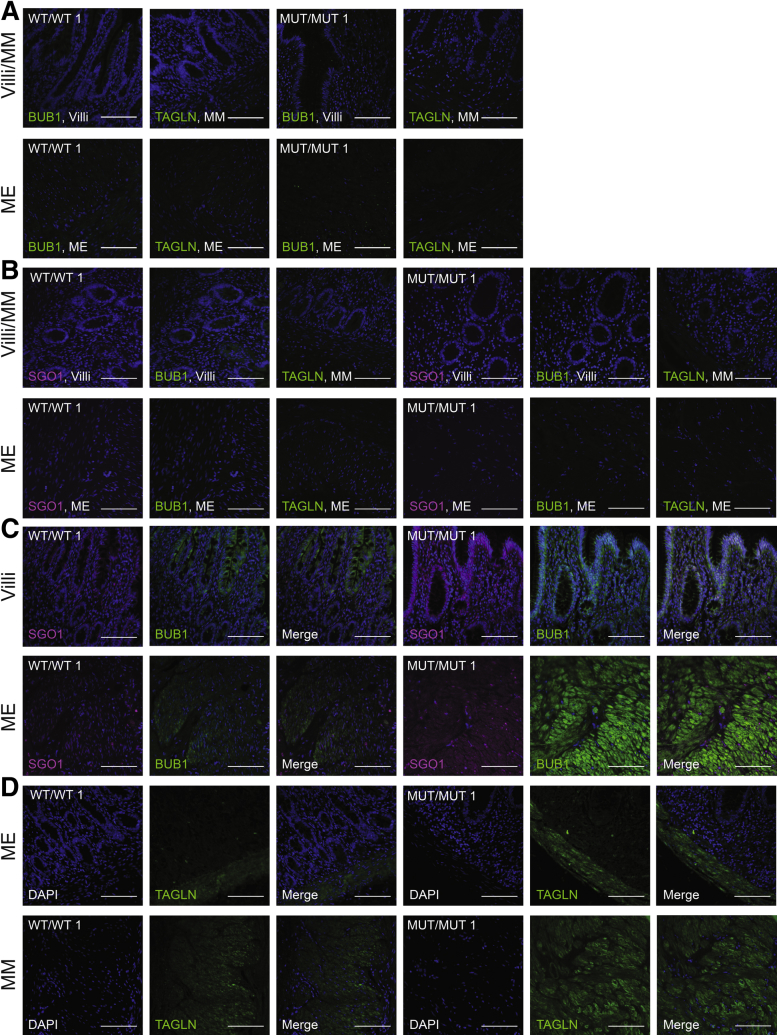
Figure 5.
Gastrointestinal fluorescent histology of CAID patients. Biopsy specimens (5 μm) from controls and individuals with CAID. All biopsy specimens are oriented to show the villi on top and the muscularis externa at the bottom. (A) Immunizing peptide blocking experiment showing the specificity of BUB1 and TAGLN antibodies. (B) Negative controls showing that no signal was obtained in the villi (SGO1 and BUB1), the muscularis mucosa (TAGLN), and the muscularis externa (SGO1, BUB1, and TAGLN) in the absence of the primary antibody. (C and D) Fluorescence staining was performed for SGO1, BUB1, and TAGLN. (C) SGO1 and BUB1 expression were higher in the villi and muscularis externa of CAID patient intestines vs controls. In controls, SGO1 expression was mainly nuclear whereas in CAID patients, SGO1 was delocalized to the cytoplasm. (D) TAGLN expression was higher in the muscularis mucosa and the muscularis externa in CAID patient intestines vs controls. V, villi; ME, muscularis externa; MM, muscularis mucosa (N = 3, 2 shown). Experiment was performed 3 times independently on 3 biological replicates for each condition. Scale bar: 100 μm. Images were acquired at 40× using a Leica DMi8 inverted fluorescence microscope and processed with Leica LAS X software. DAPI, 4′,6-diamidino-2-phenylindole. MUT, K23E; WT, wild-type.
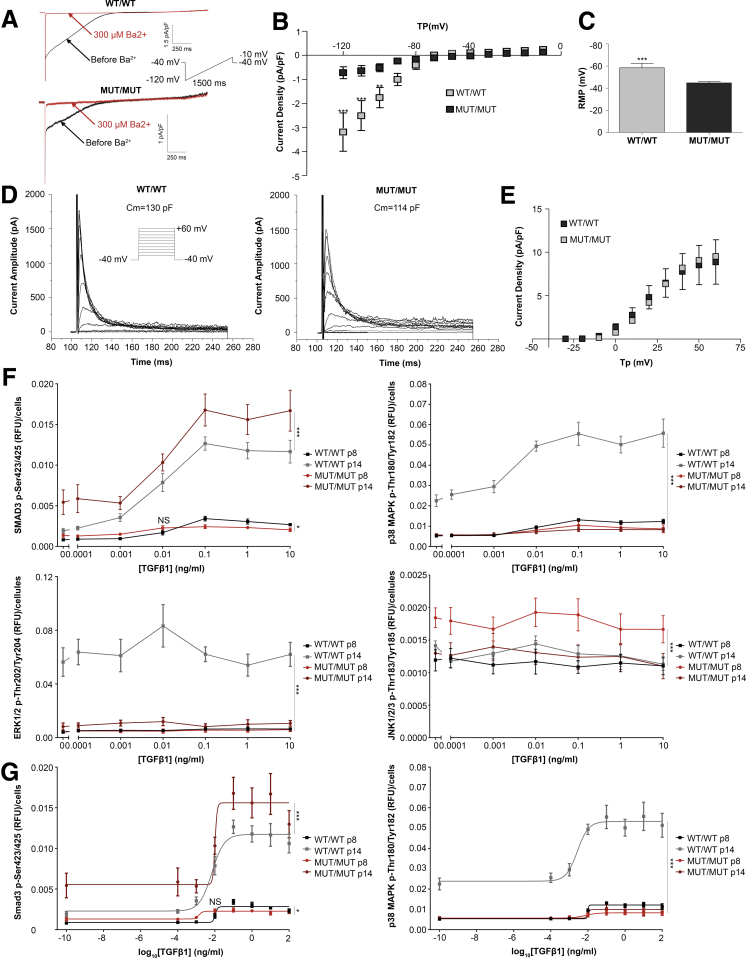
Electrophysiology of Potassium Channels in CAID Fibroblasts
Because we noticed a decrease of KCNJ2, KCNJ8, and KCND2 mRNA levels, we explored potential contributions of different potassium currents to the pathogenesis of CAID syndrome by performing electrophysiology on CAID patient dermal fibroblast cell lines. We identified the inward rectifying potassium current (IK1) to be reduced significantly in CAID patient cell lines vs controls (Figure 6A and B). The resting membrane potential (RMP), controlled by IK1, was decreased accordingly (Figure 6C). However, no difference was detected in the outward potassium current (including the Transient outward potassium current [Ito]) (Figure 6D and E). These findings suggest that IK1 is impaired in CAID syndrome and could lead to an imbalance in cell excitability.
Figure 6.
Electrophysiology of CAID patient dermal fibroblasts and analysis response of the canonical and noncanonical TGF-β pathways. (A) IK1 recordings (at 0.1 Hz) from controls and CAID patient human dermal fibroblasts. Typical response to Ba2+. (B) Means ± SEM Ba2+-sensitive current density vs voltage relationships in controls (n = 8 cells) vs CAID patients (n = 8 cells). Significance was calculated by 2-way analysis of variance with the Bonferroni post-test (**P < .01 and ***P < .001). (C) Means ± SEM RMP in controls (n = 8 cells) vs CAID patients (n = 13 cells). Significance was calculated using the Student t test with Bonferroni correction. (D) Time- and voltage-dependent outward currents in human fibroblasts. Outward currents were elicited by 200-ms voltage steps from a holding potential of −40 mV to membrane potentials between −30 to +60 mV. (E) Means ± SEM of current density vs voltage relationships for time-dependent outward currents in human fibroblasts. There were no significant differences between controls (n = 11 cells) and CAID patients (n = 18 cells). Significance was calculated by 2-way analysis of variance with the Bonferroni post-test (**P < .01 and ***P < .001). All experiments were replicated 10 times independently with a minimum of 8 cells each time, on N = 1 biological replicate for control, and N = 2 biological replicates for CAID patients. (F) Phosphorylation of SMAD3 was increased significantly at baseline and after stimulation with TGF-β1 in CAID patients (N = 3, n = 6) vs controls (N = 3, n = 6) at early (p8) (0–0.1 ng/mL, NS; 0.01–10 ng/mL, P < .001) and late passage (p14) (P < .001). Phosphorylation of p38 and ERK1/2 was increased significantly in controls (N = 3, n = 6) at p14, but not in CAID patients (N = 3, n = 6). At early passage (p8) controls and CAID patients were not sensitive to ligand stimulation. Phosphorylation analysis of JNK1/2/3 showed that controls (N = 3, n = 6) and CAID patients (N = 3, n = 6) were not sensitive to ligand stimulation for both passages studied. Significance was calculated by 2-way analysis of variance with the Bonferroni post-test (*P < .01, and ***P < .001). (G) Dose-response analysis of the canonical (SMAD3) and non-canonical (p38) TGF-β pathway. (N = 3, n = 6). Significance was calculated by 2-way analysis of variance with the Bonferroni post-test (∗P < .01, and ∗∗∗P < .001). All experiments were performed 4 times independently. MUT, K23E; WT, wild-type.
Characterization of TGF-β Signaling in CAID Patient Fibroblasts
Because TGF-β signaling is clinically relevant in arrhythmias and plays a role in CAID syndrome,3, 15 we quantified the response to ligand stimulation of canonical and noncanonical TGF-β signaling pathways in CAID patients to further investigate the impact of the SGO1 K23E mutation. First, we confirmed the known activating effect of the SGO1 K23E mutation on the canonical pathway by measuring SMAD3 phosphorylation (Figure 6F). Independent of genotype, there was a significant increase of SMAD3 phosphorylation between early (p8) and late (p14) passage. At early passages (p8), there was no biologically meaningful difference in SMAD3 phosphorylation. At late passage (p14), SMAD3 phosphorylation was significantly higher in CAID patients, regardless of the TGF-β1 concentration used, even without stimulation. These results confirmed that SGO1 K23E increases Phospho-SMAD3 (pSMAD3) with and without stimulation and that this effect is enhanced with cell aging. At early passage (p8), the half-maximal effect (median effective concentration) is reached earlier in patients vs controls (0.0120 vs 0.0014 ng/mL), an effect that is lost at p14 (median effective concentration, 0.007 vs 0.010 ng/mL) (Figure 6G). This suggests that although pSMAD3 increases, the sensitivity to the TGF-β1 ligand is higher only at early passages. We analyzed the response of the noncanonical TGF-β pathway by means of Phosphop38 mitogen-activated protein kinase (MAPK), Phospho-extracellular signal–regulated kinase (ERK)1/2, and p-c-Jun-N-terminal kinase (JNK)1/2/3 measurements. This analysis showed that only wild-type fibroblasts, but not CAID cell lines, acquired a sensitivity to ligand stimulation at passage 14, but not at early passages for pp38 MAPK and pERK1/2 (Figure 6F and G). Interestingly, stimulation with TGF-β1 had no effect on the phosphorylation of JNK1/2/3 for both genotypes (Figure 6F). These results show a surprising uncoupling of the 2 pathways, potentially through a specific interaction with BUB1. In addition, at passage 14, we identified up-regulation of DUSP2, a dual-specificity phosphatase known to dephosphorylate ERK1/2, p38, and JNK (Figure 1D).16 Cumulatively, these analyses point to an up-regulation of canonical TGF-β signaling and uncoupling of canonical from noncanonical signaling in CAID pathology.
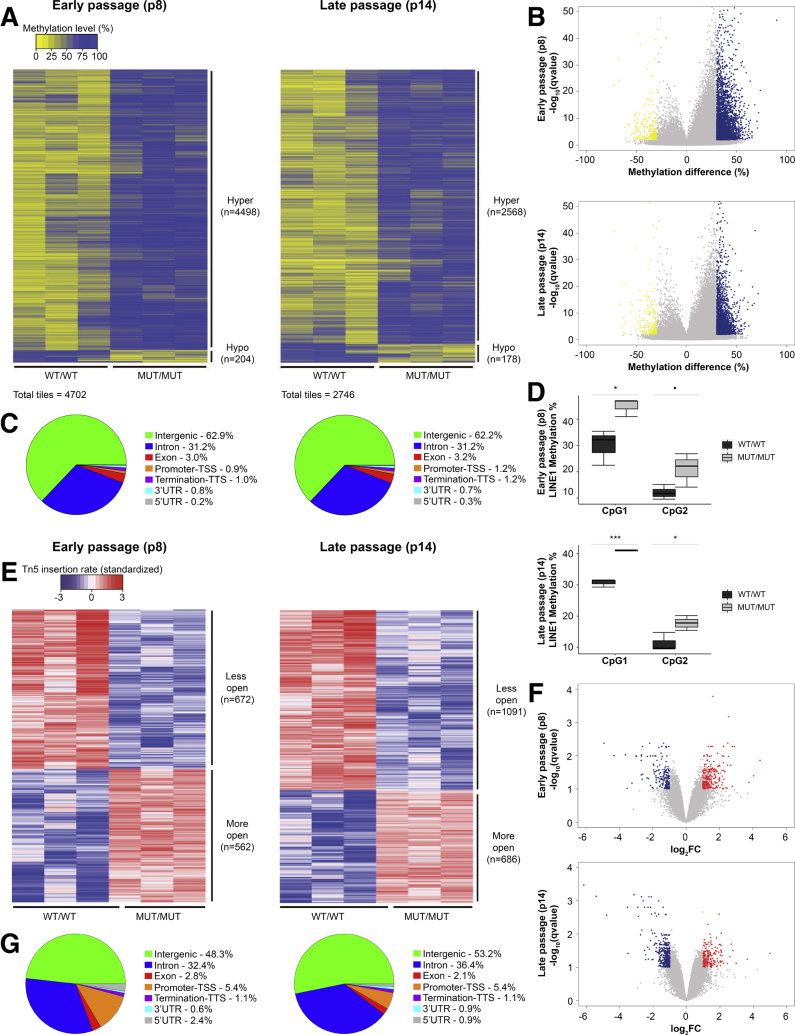
Epigenomics of CAID Syndrome
RNA sequencing analysis identified several differentially expressed genes as well as chromatin-related pathways in CAID patients, prompting us to study a potential role of SGO1 in epigenetic regulation through DNA methylation and chromatin compaction. To survey the methylation profile of CAID patients vs controls genome-wide at early (p8) and late (p14) passage, we used RRBS. A total of 375,002 tiles and 497,974 tiles, respectively, were interrogated at early (p8) and late passage (p14); tile refers to a 100-bp region containing a minimum of 2 CpG islands. A total of 4702 (4498 hypermethylated, 204 hypomethylated) and 2746 (2568 hypermethylated, 178 hypomethylated) differentially methylated tiles (≥30% methylation difference) were detected at early (p8) and late passage (p14), respectively (Figure 7A–C, Supplementary Table 5). Surprisingly, we found that CAID patients have significantly higher global methylation than controls for both passages. A total of 95.7% and 93.5% of the altered regions are hypermethylated in CAID patients at early (p8) and late passage (p14), respectively, pointing to global DNA hypermethylation as a possible pathomechanism in CAID syndrome. Differentially methylated tiles in genic regions (introns, exons, promoter–transcription starting site, termination -transcription termination site, 3’ untranslated region, and 5’ untranslated region) were associated with several biological pathways including heart and smooth muscle contraction, cellular potassium ion transport, as well as neuronal system process and behavior, features that are found commonly in other cohesinopathies (Tables 5 and 6, Supplementary Table 6). Of note, we identified regions associated with cellular potassium ion transport genes, supporting the hypothesis of a deregulation of potassium channels in CAID. However, no hypermethylation was observed in KCND2, KCNJ2, and KCNJ8 genic regions. The global hypermethylation pattern also was confirmed by pyrosequencing of 2 CpG sites in Long interspersed nuclear element (LINE-1) retrotransposons, which cover approximately 17% of the genome.17 We observed significantly higher LINE-1 methylation genome-wide in CAID patients for both passages studied (Figure 7D). At early passage (p8), we observed an increase in methylation percentage of approximately 15% and 5% for the CpG1 and the CpG2 sites, respectively. Similar results were obtained at late passage (p14), where an increase in methylation percentage of approximately 10% and 5% was observed for the CpG1 and the CpG2 sites, respectively.
Figure 7.
Epigenetic profile of CAID patients. (A) Heatmap representation of methylation levels in CAID patients vs controls among all tiles at early (p8) and late stage (p14) showed a global hypermethylation pattern. (B) Volcano plot of the methylation difference (%) at early (p8) and late stage (p14). Yellow ≥-30% and blue ≥30% methylation differences. (C) Tile proportions of differentially methylated regions at early (p8) and late stage (p14). For each group, N = 3 independent biological replicates. P values were corrected for multiple testing using the Benjamini–Hochberg method (q value). (D) Methylation percentage of LINE-1 CpG sites assessed by pyrosequencing. LINE-1 CpGs are significantly more methylated in CAID patients than in controls. Error bars signify SD. For each condition, the experiment was performed on N = 3 independent biological replicates in technical replicates. Significance was calculated by 1-way analysis of variance with the Bonferroni post-test (•P < .1, *P < .05, and ***P < .001). (E) Heatmap representation of chromatin compaction level in CAID patients vs controls among all tiles at early (p8) and late stage (p14). (F) Volcano plot of the log2 fold change of Transposase differential peaks at early (p8) and late stage (p14). Blue, log2 fold change <-1; red, log2 fold change >1. (G) Proportion of Transposase differential peaks at early (p8) and late stage (p14). For each group, N = 3 independent biological replicates. P values were corrected for multiple testing using the Benjamini–Hochberg method (q value). MUT, K23E; TSS, transcription starting site; TTS, transcription termination site; UTR, untranslated region; WT, wild-type.
Table 5.
Biological Functions (GO Analysis) Associated With Differentially Methylated Genes in RRBS at Early Passage (p8)
| Biological functions | P value | Genes, n |
|---|---|---|
| Hypermethylated | ||
| Neuronal system | 7.21E-14 | 73 |
| Regulation of membrane potential | 1.04E-06 | 60 |
| Behavior | 1.24E-06 | 77 |
| Regulation of heart contraction | 6.91E-05 | 37 |
| Muscle contraction | 9.72E-05 | 48 |
| Cellular potassium ion transport | 1.76E-04 | 31 |
| Midgut development | 8.59E-04 | 5 |
| Neuromuscular process | 2.58E-03 | 21 |
| Smooth muscle contraction | 3.42E-03 | 3 |
| Hypomethylated | ||
| Embryonic morphogenesis | 7.81E-06 | 15 |
| Heart morphogenesis | 3.24E-05 | 9 |
| Cardiovascular system development | 1.41E-04 | 15 |
| Heart looping | 6.29E-04 | 4 |
| Embryonic heart tube development | 1.63E-03 | 3 |
| Regulation of developmental growth | 4.80E-03 | 7 |
| Negative regulation of MAPK cascade | 5.78E-03 | 5 |
| Actin filament-based process | 6.78E-03 | 11 |
Table 6.
Biological Functions (GO Analysis) Associated With Differentially Methylated Genes in RRBS at Late Passage (p14)
| Biological functions | P value | Genes, n |
|---|---|---|
| Hypermethylated | ||
| Neuronal system | 4.82E-06 | 39 |
| Cardiac conduction | 2.49E-05 | 20 |
| Muscle contraction | 6.85E-05 | 24 |
| Behavior | 3.19E-04 | 47 |
| Regulation of membrane potential | 4.19E-04 | 36 |
| Cellular potassium ion transport | 8.25E-04 | 21 |
| Midgut development | 1.39E-03 | 4 |
| Actin filament-based process | 1.39E-03 | 50 |
| Hindgut development | 5.34E-03 | 3 |
| Hypomethylated | ||
| Embryonic morphogenesis | 4.52E-04 | 12 |
| Regulation of embryonic development | 1.33E-03 | 5 |
| Regulation of cellular response to stress | 1.38E-03 | 12 |
| Chromatin organization | 2.23E-03 | 7 |
| Negative regulation of response to wounding | 2.24E-03 | 4 |
| Heart morphogenesis | 4.98E-03 | 6 |
| Pathway-restricted SMAD phosphorylation | 2.47E-03 | 3 |
| Muscle structure development | 9.26E-03 | 10 |
SMAD, Mothers Against Decapentaplegic Homolog.
The striking global methylation signature of CAID prompted us to perform ATAC sequencing on 3 controls and 3 CAID patient cell lines at early (p8) and late passage (p14) to assess the chromatin accessibility profile (Transposase peaks). In parallel with these findings, we observed a mild chromatin closing signature at early (p8) passage, which was enhanced at late (p14) passage, suggesting that at late passage (p14) the effect of the mutation was stronger (Figure 7E–G, Supplementary Table 7). Transposase peaks near the gene transcription starting site were associated mainly within the same pathways identified by RNA sequencing and RRBS analysis (Tables 7 and 8, Supplementary Table 8). In aggregate, these data suggest that epigenetic modifications at methylation and chromatin configuration levels participate in CAID pathogenesis.
Table 7.
Biological Functions (GO Analysis) Associated With Transposase Peaks in ATAC Sequencing at Early Passage (p8)
| Biological functions | P value | Genes, n |
|---|---|---|
| Less open | ||
| Inwardly rectifying K+ channels | 5.80E-04 | 3 |
| Neuronal system | 3.49E-03 | 7 |
| Actin filament-based process | 3.88E-03 | 10 |
| Action potential | 6.38E-03 | 4 |
| Potassium ion transport | 8.71E-03 | 5 |
| More open | ||
| Regulation of nervous system development | 5.31E-05 | 13 |
| Apoptotic process involved in development | 5.30E-04 | 3 |
| Developmental growth | 1.85E-03 | 9 |
| Negative regulation of intracellular signal transduction | 3.20E-03 | 8 |
| Limb development | 8.81E-03 | 4 |
Table 8.
Biological Functions (GO Analysis) Associated With Transposase Peaks in ATAC Sequencing at Late Passage (p14)
| Biological functions | P value | Genes, n |
|---|---|---|
| Less open | ||
| Protein kinase A signaling | 1.45E-04 | 4 |
| Actin filament-based process | 1.31E-03 | 14 |
| Heart development | 3.53E-03 | 11 |
| Cellular potassium ion transport | 5.36E-03 | 6 |
| Neuronal system | 9.25E-03 | 8 |
| More open | ||
| Vasculogenesis | 5.64E-06 | 6 |
| Cardiovascular system development | 9.39E-04 | 12 |
| Developmental growth | 8.39E-03 | 9 |
| Muscle tissue development | 8.55E-03 | 7 |
| Cardiac muscle tissue development | 9.31E-03 | 5 |
Discussion
CAID syndrome is part of the cohesinopathy family, alongside Cornelia de Lange syndrome (NIPBL, RAD21, SMC1A, SMC3, and HDAC8), Robert syndrome (ESCO2), Warsaw breakage syndrome (DDX11), and others that result from mutations in cohesin complex components. Although there is some phenotypic overlap between CAID and other cohesinopathies (valve defects and gastrointestinal defects), CAID patients do not show intellectual or growth retardation. In addition, the clinical manifestations observed in CAID syndrome are difficult to reconcile with perturbation of canonical roles of SGO1 in cell cycling, suggesting that novel noncanonical roles of SGO1 are involved in CAID pathology. It has been shown over recent years that components of the cohesin complex play important roles in long-range transcriptional regulation.18, 19 Indeed, RAD21, SMC1A, and other interaction partners of SGO1 are involved in transcription regulation.20, 21 Thus, a noncanonical role of SGO1, possibly through 11-Zinc Finger Protein (CTCF) loading, chromatin remodeling, and epigenetic marks, in maintaining transcriptional modules during cardiac and intestinal development therefore is likely.21, 22 The early embryonic lethality of most cohesin component knockouts makes it impossible to precisely replicate cohesinopathies in mice without use of more sophisticated models. Homozygous Sgo1 knockout mice and homozygous K23E knock-in mice are embryonic lethal, leaving dermal fibroblasts and intestinal biopsy specimens from CAID patients as the only proxy available for characterization and understanding of the mechanisms involved in CAID syndrome9 (and data not shown).
In this study, we therefore follow all steps of the transcriptional cascade from epigenetics to proteomics to define a molecular signature of CAID syndrome. Although we did not identify a single driver pathway in CAID pathogenesis, multiple cumulative hits within related cascades seem to converge on a transcriptional dysregulation influenced by epigenetic and TGF-β–related events. At the mRNA level, especially at late passage (p14), the most striking finding is up-regulation of SGO1 and SGO2, along with many other genes involved in cell cycling or the cohesin complex, including direct interactors such as BUB1, PLK1, and BIRC5. Given that CAID fibroblasts show centromeric cohesion defects with only mild acceleration of cell cycling and without any impact on fidelity of mitosis or integrity of the genome,3 the observed effects of the SGO1 K23E mutation may be mainly compensatory. In fact, it is conceivable that overexpression of SGO1, SGO2, BUB1, and PLK1 can attenuate the direct effects of SGO1 K23E. Future studies should be directed at dissecting whether this overexpression is compensatory or causal.
In addition, recent publications have described noncanonical postmitotic cohesin functions. The serine/threonine kinase BUB1, previously recognized to play an important role in establishing the mitotic spindle checkpoint, now has been found to be required for canonical and noncanonical TGF-β signaling through type Transforming Growth Factor Beta Receptor 1 (TGFBRI) recruitment.23 This raises the possibility that enhanced TGF-β signaling in CAID patient cell lines is the result of overexpression of BUB1. Furthermore, we show here that SGO1 K23E uncouples canonical and noncanonical TGF-β signaling: while canonical TGF-β signaling is enhanced in CAID patient fibroblasts at early passage and increases further with late passage, a concomitant increase of noncanonical signaling occurs only in wild-type fibroblasts, but not CAID cell lines. This surprising uncoupling of the 2 TGF-β signaling pathways suggests that the SGO1 K23E mutation favors the canonical pathway through an unknown mechanism, possibly through a specific interaction with BUB1. This hypothesis is strengthened by the overexpression and the similar localization pattern of SGO1 and BUB1 in the villi and the muscularis externa from CAID patient intestines. On the other hand, the up-regulation of DUSP2 correlates with the inhibition of the noncanonical ERK1/2 MAPK, p38, and JNK signaling in CAID. All these results point to canonical TGF-β signaling as a key cascade involved in the intestinal manifestations of CAID syndrome and is a clinical identification of cross-talk between the TGF-β and cohesin cascades.
Interestingly, we also identified a trend toward up-regulation for genes involved in other cohesinopathies, with the most drastic changes for ESCO2, RAD21, and SMC3. Autosomal-dominant mutations in RAD21 and SMC3 have been associated with Cornelia de Lange syndrome, whereas recessive mutation in RAD21 has been found in CIPO. An analysis of RAD21 mutations and depletion in zebrafish models showed impaired cellular DNA damage response, disrupted transcription and intestinal transit, and the development of enteric neurons, similar to patients with CIPO, suggesting that these manifestations result from a loss-of-function of RAD21, while in our case, we found an up-regulation of RAD21, potentially representing a causal or compensatory mechanism.8, 24 In most cases, SMC3 and ESCO2 mutations do not cause gastrointestinal defects.
Even with recent advances in the proteomic field, characterization of the CAID patient proteome was challenging. Because the dynamic range of SILAC for protein identification and quantification in human beings can vary between 1000 and 5000, the whole proteome is not covered.25 The workflow of SILAC performs a single comparison between a control and a CAID patient without a cross-link between the different samples, giving rise to a lot of variations between duos. Furthermore, SILAC cannot detect proteins expressed at low levels, as was the case of several candidate genes identified by RNA sequencing. However, investigation of the proteome in CAID syndrome showed valuable insight into CAID pathomechanisms. We identified TPM1 and TAGLN as important genes overexpressed in CAID syndrome. Because TPM1 and CALD1 regulate smooth muscle contractions through Ca2+-dependent actin stabilization, it is conceivable that overexpression of TPM1 perturbs smooth muscle contractions, either through contraction stalling or insufficient compensation (myogenic CIPO).13 TGF-β signaling positively regulates both TPM1 and TPM2, and increases smooth muscle contractile force and tissue remodeling.26 TAGLN is a canonical TGF-β–inducible gene acting as an actin–cross-linking/gelling protein of the calponin family, which can regulate actin contractile function through Ca2+-independent contraction.14 Studies on TAGLN-deficient mice showed that TAGLN knock-out significantly inhibited Ca2+ independent but not Ca2+ dependant vascular contractility.27 Thus, overexpression of TAGLN might be a causal or compensatory process, either to balance Ca2+-dependent and -independent smooth muscle contractions or to bypass the perturbation of contractions secondary to the disruption and fibrosis of smooth muscle observed in CAID patients.
We found that IK1 was reduced significantly in CAID patient cell lines vs controls. This observation directly correlated with the down-regulation of KCNJ2, the ion-channel subunit that carries IK1. The RMP was decreased accordingly. IK1 is critically important to establish the resting membrane potential and thereby control excitability of a wide range of cells.28 The large changes in IK1 that we observed in CAID patient cells were strong candidates to underlie abnormalities in fibroblast, cardiomyocyte, and colonic functions.29, 30, 31 Further work is needed to relate this ion-channel dysfunction to specific components of CAID pathophysiology.
The results of this study also support the idea that CAID syndrome is a transcriptional dysregulation disorder. In addition to the unique differentially expressed gene network identified by RNA sequencing, the global methylation pattern observed in patients as well as the chromatin compaction signature suggest that epigenetic regulation has pathogenetic significance in CAID syndrome. This assertion was strengthened further by the significantly higher methylation of LINE-1 retrotransposons in CAID patients. Indeed, pyrosequencing of LINE-1 retrotransposons, representing approximately 17% of the human genome, is a well-established technique to survey global DNA methylation.17 Although DNA methylation is commonly associated with gene silencing, such a correlation was not found in our study. This is in line with recent studies showing that methylation can maintain gene expression through promoter blocking, and that gene down-regulation results from DNA methylation-independent mechanisms.32, 33 As an example, increased DNA methylation in the gene body can result in increased gene expression.34, 35 Although RRBS provides a large (∼1.5 million CpG) but partial view of the 30 million CpGs present in the genome, only a portion of indicative regions regulating gene expression (ie, promoters and enhancers) were evaluated. Nevertheless, RNA sequencing, RRBS, and ATAC sequencing analysis converged on common pathways in CAID and underscored a noncanonical role of SGO1 in transcriptional regulation through chromatin remodeling and epigenetic marks.
In conclusion, intestinal manifestations of CAID syndrome most likely result from several changes with a cumulative effect, and not a single dysregulated gene. We point out the dysregulation of canonical TGF-β signaling, DNA hypermethylation, chromatin compaction, and a decrease of IK1 as a vicious cycle producing the striking intestinal manifestations of CAID syndrome. Dysregulation of druggable pathways such as the TGF-β cascade and methylation may provide targets toward the identification of medical therapies for CAID syndrome, but also for study in other gastrointestinal conditions such as CIPO and irritable bowel syndrome, which affect patients worldwide.
Materials and Methods
All authors had access to the study data and reviewed and approved the final manuscript.
Skin Biopsy Specimens and Cell Lines
Human tissue acquisition from CAID patients was approved by the ethical committees at all participating institutions. Primary human dermal fibroblasts were derived from skin biopsy specimens from 3 wild-type controls and 3 CAID patients. The study was conducted after patients provided informed consent. All assays were performed at early (p8–p10) and a late (p14) passage. Primary dermal fibroblast; normal, human, adult (ATCC PCS201012, lots 61447289 and 61683453) and BJ (ATCC CRL-2522) have been used as control cell lines for SILAC experiments. All cell lines routinely were tested for the absence of mycoplasma infection.
Transcriptomic Studies
RNA extraction, library preparation, and RNA sequencing
RNA extraction from human dermal fibroblasts (3 controls and 3 CAID patients) at p8 and p14 was performed using the miRNAeasy mini kit (217004; Qiagen, Hilden, Germany) on an automated QIAcube (Qiagen) according to the manufacturer’s instructions. The complementary DNA libraries were generated on 500 ng of RNA with RNA integrity number greater than 9 using the Illumina TruSeq Stranded Total RNA Sample preparation kit (20020596; Illumina, San Diego, CA). Libraries were sequenced over multiple flow cells on the paired-end, 100-bp Illumina Hiseq4000.
RNA sequencing data processing and analysis
Trimmomatic (v.0.32; Usadel lab, Jülich and Aachen, Germany) was used to remove Illumina adapters from the raw reads.36 Reads with a quality score less than 30 and smaller than 32 bp were eliminated. The remaining reads were mapped to the human genome 19 (hg19) reference using Bowtie2 (v.2.1.0; Johns Hopkins University, Baltimore, MD) and TopHat (v.2.1.0; Johns Hopkins University).37, 38 FeatureCounts (v.1.4.4 Walter and Eliza Hall Institute of Medical Research, Parkville, Australia) was used to count mapped reads.39 The weighted trimmed mean of M values algorithm in the edgeR package (Bioconductor; Roswell Park Comprehensive Cancer Center, Buffalo, NY) was used to normalize gene expression levels across samples.40 The data were log-transformed, and we applied voom function in the limma package (Bioconductor) to get precision-weights data.41 Limma also was used to perform a weighted fit on the weighted data obtained from voom using the lmFit function. To investigate genotype effects on gene expression in dermal fibroblasts at passages 8 and 14, we used a nested linear model with the following design: gene expression ∼ sex + biopsy_age + genotype:passage. Nominal P values provided by lmFit were corrected for multiple testing using the Benjamini–Hochberg method. Sex chromosomal or low-count genes with an average read count less than 10 across all samples were excluded, resulting in 13,920 genes in total. Genes with a log2 fold change greater than 1 and less than -1 were considered as up-regulated and down-regulated, respectively. False discovery rate (FDR)-adjusted P value (q value) less than .1 was considered significant. The complete list of differentially expressed genes can be found in Supplementary Table 1. GO analysis was performed using Metascape (http://metascape.org).42 The complete GO analysis results can be found in Supplementary Table 2.
Real-time polymerase chain reaction validations
RNA was reverse-transcribed into complementary DNA using SuperScript IV Reverse Transcriptase (18090050; Invitrogen, Carlsbad, CA). Real-time polymerase chain reaction (PCR) was performed with FastStart Universal SYBR Green Master (11340500; Roche, Bâle, Suisse) using a LightCycler 96 Real-Time PCR system (Roche). Transcripts were normalized to GAPDH (Table 9).
Table 9.
Primers List for Reverse-Transcription PCR Validations
| Gene ID | Forward 5' primer sequence | Reverse 3' primer sequence |
|---|---|---|
| BUB1 | TCAGCCTGGTTTTGAATTCCTC | TACCACAATGACCCAAGATTCA |
| DUSP2 | GTAGCGGAAAAGGCCCTCAA | GTCCCGATCTGTGCTCTGAG |
| KCND2 | CTGGGGTACACACTGAAGAGTT | ACTCAGCGAACAGATAGAACCA |
| PLK1 | CCAACCATTAACGAGCTGCTTA | ACCTGTCTCTCGAACCACTG |
| SGO1 | CTGGAGCTGTCATCACTATTGG | CCAAAGTGAAAGAAGCCCAAGA |
| SGO2 | GAGTGCCCAGTGATGGAAAC | AATGCCCTGTTGTTGTGCTTTA |
Proteomic Studies
Cell culture and SILAC
SILAC Dulbecco's modified Eagle medium without L-arginine and L-lysine (88364; Thermo Fisher Scientific, Waltham, MA) was supplemented with 13C6, 15N4 L-arginine (20104102; Silantes, München, Germany), 13C6, and 15N2 L-lysine (2111604102; Silantes), or unlabeled 12C614N4 L-arginine (A5006; Sigma Aldrich, St. Louis, MO) and 12C6L-lysine (L5501; Sigma Aldrich) to produce heavy and light SILAC media, respectively. L-proline (P5607; Sigma Aldrich) also was added to a final concentration of 100 μg/mL to prevent amino acid conversion. A total of 500,000 fibroblasts from 3 controls and 3 CAID patients were seeded and grown in heavy and light medium, respectively, for at least 7 doubling times to ensure complete amino acid incorporation.
SILAC cell lysis and digestion
The heavy and light labeled cells were mixed together in a 1:1 ratio to form 3 duos composed of a wild-type control and a CAID patient. The mixed cells were centrifuged at 14,000 rpm at 4°C for 15 minutes. The cell pellets were resuspended in cold lysis buffer (1% sodium deoxycholate [SDC] in 50 mmol/L NH4HCO3) and sonicated with the Fisher Scientific Sonic Dismembrator Model 100 to lyse the cells. The samples then were transferred immediately to an Eppendorf Thermomixer R (Eppendorf, Hambourg, Allemagne) and incubated at 99°C for 5 minutes with shaking at 1400 rpm. The samples then were centrifuged at 40,000g for 10 minutes at 4°C. The supernatant was harvested, dithiothreitol was added to obtain a (T9284, Sigma Aldrich) final concentration of 5 mmol/L, and the samples were shaken at 800 rpm for 30 minutes at 56°C. After being returned to room temperature, the iodoacetamide was added to a final concentration of 15 mmol/L and the samples were incubated for 30 minutes in the dark. Trypsin was finally added at a final concentration of 5–10 ng/μL with a protein:trypsin ratio of approximately 50:1 to 200:1, and the samples were incubated at 37°C overnight. The enzymatic digestion was stopped by adding 30% Trifluoroacetic acid (TFA) to obtain a final pH of approximately 2 and centrifuged at 13,000g for 5 minutes. Peptide digestion (supernatant) was transferred to a new tube and the pellet was washed with 0.1% TFA and then centrifuged again. The supernatant was added to the peptide digestion.43
SILAC desalting, concentration, and mass spectrometry data acquisition
Samples were loaded on a homemade C18 precolumn directly connected to the switching valve of a homemade reversed-phase column with a gradient from 10% to 60% acetonitrile (0.2% formic acid [FA]) and a 600 nL/min flow rate on a NanoLC-2D system (Eksigent, Dublin, CA) connected to an Q-Exactive Plus (Thermo Fisher Scientific). Each full mass spectrometry spectrum acquired with a 70,000 resolution was followed by 12 MS/MS spectra, in which the 12 most abundant multiply charged ions were selected for MS/MS sequencing.
SILAC data processing and analysis
Data processing was performed using Maxquant (v1.5.0.25; Max Planck Institue of Biochemistry, Planegg, Germany).44 Tolerances on precursors and fragments were 15 ppm and 0.01 daltons, respectively. Variable selected post-translational modifications were carbamidomethyl (C), oxidation (M), and deamidation (NQ). SILAC labels selected were R10 (13C6, 15N4) + K8 (13C6, 15N2) for the heavy label. Genes with a fold change greater than 1.5 and less than 0.66 were considered as overexpressed and underexpressed, respectively. FDR-adjusted P value (q value) ≤ .01 was considered significant. The complete list of differentially expressed proteins can be found in Supplementary Table 3. GO analysis was performed using Metascape (http://metascape.org).42 The complete GO analysis results can be found in Supplementary Table 4.
Western blot validations
Protein lysates were extracted from fibroblasts. A total of 25 μg of proteins were loaded on 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (4561034; Bio-Rad, Hercules, CA) and transferred to a nitrocellulose membrane. Membranes were probed against SGO1 (mouse monoclonal, ab58023; Abcam, Cambridge, UK), BUB1 (mouse monoclonal, sc-365685; Santa Cruz, Dallas, TX), SM22α (rabbit polyclonal, sc-50446; Santa Cruz), and TPM1 (rabbit polyclonal, sc-28543; Santa Cruz) primary antibodies diluted 1:200 followed by horseradish-peroxidase–conjugated secondary antibodies. The blots were developed using SuperSignal (34580; Thermo Scientific). Signal intensity was quantified using ImageJ software (v.1.48; National Institutes of Health, Bethesda, MD).45
Tissue Studies
Immunohistology on intestine
Paraffin-embedded sections (5-μm thick) were immunostained using the Ultravision LP Detection system horseradish-peroxidase polymer and DAB Plus Chromogen kit (TL-015-HD; Thermo Scientific) for DAB staining. The slides were rehydrated by successive baths (xylene, EtOH 100%, EtOH 90%, EtOH 70%, EtOH 50%, dH2O). Antigen retrieval was performed by heating at 68°C for 20 minutes in 1 mmol/L citrate (pH 6.0) and cooled down to room temperature. Slides then were incubated in 0.3% Triton (T9284, Sigma Aldrich) for 30 minutes and blocked in phosphate-buffered saline + 10% serum (goat serum for DAB staining and donkey serum for fluorescent staining). For immunostaining SGO1 (anti-mouse, ab58023; Abcam), BUB1 (mouse monoclonal, sc-365685; Santa Cruz, and rabbit polyclonal, Ab70372; Abcam) and TAGLN (rabbit polyclonal, sc-50446; Santa Cruz) were diluted 1:500 for DAB and 1:200 for fluorescence staining. The primary antibodies were targeted with IgG secondary antibody conjugated to horseradish peroxidase for DAB staining and donkey anti-mouse and anti-rabbit Alexa 555 and 647 for fluorescent staining (1:500 dilution; Molecular Probes/Invitrogen, Carlsbad, CA). DAB immunostainings were counterstained using methyl green and mounted with Permount medium (Thermo Fisher Scientific). Fluorescent immunostainings were mounted with 4′,6-diamidino-2-phenylindole incorporated anti-fade medium (P36962; Thermo Fisher Scientific). Immunohistochemistry control experiments were performed by excluding the primary antibody. Immunizing peptide blocking also was performed following the protocol described earlier. Five times excess of BUB1 (H00000699-Q01; Novus Biologicals, Littleton, CO), and TAGLN (NBP1-45267; Novus Biologicals) human recombinant peptides was added to the primary antibody by weight. Pixel intensity was corrected for noise by subtraction of the immunofluorescence background in negative controls. Imaging of DAB-stained tissues was performed using a Zeiss AxioScan Z1 slide scanner and analyzed with Zen software (Zeiss, Oberkochen, Germany). Fluorescent-stained tissues were imaged using a Leica DMi8 inverted fluorescence microscope (Leica, Wetzlar, Germany) with a 40× objective. Image processing was performed with Leica LAS X software.
Cell Culture Assays
Patch clamp: ionic current and RMP recording
All in vitro recordings were obtained at 37°C. The whole-cell perforated-patch technique was used to record RMP in current-clamp mode, and the tight-seal patch clamp was used to record IK1 in voltage clamp mode. Borosilicate glass electrodes filled with pipette solution were connected to a patch-clamp amplifier (Axopatch 200A; Axon Union City, California, USA). Electrodes had tip resistances of 6–8 MΩ. Nystatin-free intracellular solution was placed in the tip of the pipette by capillary action (≈30 s), and then pipettes were backfilled with nystatin-containing (600 μg/mL) pipette solution. IK1 was recorded as the 300-μmol/L Ba2+-sensitive current. Tyrode solution contained 136 mmol/L NaCl, 1.8 mmol/L CaCl2, 5.4 mmol/L KCl, 1 mmol/L MgCl2, 0.33 mmol/L NaH2PO4, 10 mmol/L dextrose, and 5 mmol/L HEPES, titrated to pH 7.3 with NaOH. The pipette solution for RMP, outward potassium current, and IK1 recording contained 0.1 mmol/L guanosine triphosphate, 110 mmol/L potassium aspartate, 20 mmol/L KCl, 1 mmol/L MgCl2, 5 mmol/L Magnesium ATP (mgATP), 10 mmol/L HEPES, 5 mmol/L sodium phosphocreatine, and 0.005 mmol/L EGTA (pH 7.4, KOH). A total of 10 mmol/L tetraethylammonium was used to block outward potassium current. Junction potentials between bath and pipette solutions averaged 10.5 mV and were corrected for RMP measurements only. Currents are expressed as densities (picoamperes per picofarad (pA/pF)) to control for differences in cell size/capacitance.
α-Screen TGF-β signaling assays
The phosphorylation state of SMAD3, p38, ERK1/2, and JNK1/2/3 was quantified using Amplified Luminescence Proximity Homogenous Assays (Perkin Elmer Waltham, Massachusetts, USA). A total of 12,000–15,000 cells/well were seeded in a 24-well plate. Forty-eight hours after plating, the cells were starved overnight in total absence of fetal bovine serum. They then were stimulated with different concentrations of TGF-β1 (0–100 ng/mL) for 1 hour, washed with phosphate-buffered saline, and lysed for 20 minutes at room temperature using 200 μL/well of AlphaScreen SureFire Ultra lysis buffer supplemented with protease inhibitors. The quantifications were performed in 384-well plates (Proxiplate; Perkin Elmer), according to protocol guidelines and measured with the EnVision 2104 plate reader (Perkin Elmer). For each condition, Six independent measurements were performed for each cell line and experiments were repeated four times.
Epigenetic Studies
Library preparation and RRBS
DNA extraction from human dermal fibroblasts (3 controls and 3 CAID patients) at p8 and p14 was performed using the QIAamp DNA Mini Kit (51304; Qiagen). A total of 500 ng of genomic DNA was digested with Msp1 overnight at 37°C in an Eppendorf Thermocycler. Without deactivating MspI and cleaning up the digestion reactions, DNA end repair and A-tailing was conducted by adding Klenow fragment (3'→5' exo-) (M0212M; New England Biolabs Ipswich, Massachusetts, USA) and dNTP mixture (10 mmol/L deoxyadenosine triphosphate (dATP), 1 mmol/L Deoxycytidine triphosphate (dCTP), and 1 mmol/L Deoxyguanosine triphosphate (dGTP)) directly into each sample in the strip tube. The reaction was incubated at 30°C for 20 minutes and then at 37°C for 20 minutes. Bead clean-up was conducted using AMPure XP beads (B37419AB; Beckman Coulter Brea, California, USA) to remove small fragments. Ligation of NEB methylated adaptors (E7535; New England Biolabs) to DNA samples was performed using the T4 DNA ligase enzyme (M0202S; New England Biolabs) at 20°C for 20 minutes. The USER enzyme (M5505S; New England Biolabs) was added, and the reaction was incubated at 37°C for 15 minutes. The reaction was cleaned up using AMPure XP beads (B37419AB; Beckman Coulter). Bisulfite conversion and clean up then was performed on purified DNA using the EpiTect Fast bisulfite conversion kit (59824; Qiagen). Libraries of the bisulfite-converted DNA were generated by PCR reaction using the PfuTurbo Cx Hotstart DNA Polymerase (600410; Agilent Santa Clara, California, USA), deoxynucleotide triphosphates (dNTPs) (10 mmol/L), NEB indexing primers and NEB universal primers (E7535L; New England Biolabs), coupled with SYBR Green (S7563; Thermo Fisher Scientific). The libraries were cleaned using AMPure XP beads (B37419AB; Beckman Coulter). Paired-end sequencing of the libraries was performed on a paired-end, 125 pb Illumina Hiseq 2500.
RRBS data processing and analysis
Bsmap (v.2.6 Dan L Duncan Cancer Center, Houston, Texas, USA) was used for initial data processing, methylation calls, and reads alignment.46 Analysis and statistics of differentially methylated regions was performed using MethylKit (v.0.5.3 Bioconductor), a software based on the Benjamini–Hochberg FDR procedure (P value threshold of q = 0.01).47 Specific parameters were chosen including 100-bp, step-wise tiling windows, 2 CpG minimum per tile, and a minimum 30× CpG coverage of each tile per sample. The methylation level of a 100-bp tile was the average of all single CpGs within the tile, and the methylation level reported for a sample was the average methylation level across replicates. To identify differentially methylated CpG tiles in CAID patients vs controls at each passage, we used the R package DSS (Bioconductor)48 with the following model: methylation ∼ sex + biopsy_age + genotype:passage. We defined differentially methylated tiles as those showing a significant difference of methylation at an FDR (q value) < 0.01, and an absolute mean methylation difference greater than 30% (375,003 and 497,975 tiles in p8 and p14, respectively). HOMER (v.3.51 Salk Institute for Biological Studies, La Jolla, California, USA) was used to annotate differentially methylated sequences.49 The complete list of differentially methylated regions can be found in Supplementary Table 5. GO analysis was performed using Metascape (http://metascape.org).42 The complete GO analysis results can be found in Supplementary Table 6.
LINE-1 pyrosequencing
DNA extraction from human dermal fibroblasts (3 controls and 3 CAID patients) at p8 and p14 was performed using the QIAamp DNA Mini Kit (51304; Qiagen). Bisulfite conversion was performed as previously described in the RRBS methods section. Bisulfite converted DNA was amplified and enriched for LINE-1 conservative sequences using specific primers (LINE-1 forward and LINE-1 biotinylated reverse primer) designed with PyroMark Assay Design (Qiagen). The primers were designed to allow the coverage of 2 CpG sites. The methylation percentage of LINE-1 CpG sites was assessed using PyroMark Q24 (Qiagen) pyrosequencing technology (Table 10).
Table 10.
LINE-1 Pyrosequencing Primers
| Primer | Sequence |
|---|---|
| Forward 5’ | GGGTATAGGGAAGGGAATATTATATATTAG |
| Reverse 3’ | CCATCAACTCACCATCTACATT |
| Sequencing | AGGGAATATTATATATTAGGGT |
Library preparation and ATAC sequencing
ATAC sequencing libraries were generated from 100,000 human skin fibroblasts from 3 controls and 3 CAID patients. Cells were lysed with ice-cold hypotonic cell lysis buffer (0.1% wt/vol sodium citrate tribasic dihydrate and 0.1% vol/vol Triton) for 30 minutes on ice followed by a second 30-minute lysis step with ice-cold cell lysis buffer (10 mmol/L Tris-HCl, pH 7.4, 10 mmol/L NaCl, 3 mmol/L MgCl2, and 0.1% vol/vol Octylphenoxy poly(ethyleneoxy)ethanol, branched (IGEPAL) CA-630 NON999, Bioshop, Burlington, Ontario, Canada). The transposition reaction was performed in a 25-μL reaction volume using 5 μL tagmentation DNA enzyme in 2× Tagmentation DNA (TD) buffer (FC-121-1030; Illumina). EDTA (25 mmol/L) was added to the reaction mix before DNA recovery with MiniElute PCR Purification columns (28004; Qiagen). Paired-end sequencing of the libraries was performed on a paired-end, 125 pb Illumina Hiseq 2500.
ATAC sequencing data processing and analysis
ATAC sequencing reads were trimmed for adapter sequences and low-quality score bases (Phred score < 20) and were mapped to the human reference genome (GRCh37/hg19). Mapping was performed using Burrows-Wheeler Aligner (BWA-MEM) in (Source Forge, New York, USA) paired-end mode at default parameters.50 Only reads that were paired properly and had a unique alignment (mapping quality > 10) were retained. PCR duplicates were removed using Picard tools (http://broadinstitute.github.io/picard/). Peaks were first called on ATAC sequencing using the MACS2 (Liu lab, Cambridge, MA) software suite with the added parameters “-g hs -q 0.05 –keep-dup all –broad –nomodel –extsize 200 –nolambda.”51 For comparison between samples, all peaks from each sample were merged to provide 1 set of combined peaks. For quantification of the combined peaks, we used FeatureCount (v1.4.4) to count the number of overlapping reads for each sample.39 For all downstream analyses, we excluded sex chromosomal or low-count peaks with an average read count lower than 30 across all samples, resulting in 69,249 peaks in total. Chromatin accessibility levels across samples were normalized using the weighted trimmed mean of M values algorithm, implemented in edgeR R package.40 Afterward, we log-transformed the data and obtained precision-weights using the voom function in the limma package.41 Finally, we performed a weighted fit, using lmFit (also from limma) and the weights obtained from voom. To interrogate for genotype effects on chromatin accessibility in fibroblasts at passages 8 and 14, we used a nested linear model with the following design: chromatin accessibility ∼ sex + biopsy_age + genotype:passage. Nominal P values provided by lmFit were corrected for multiple testing using the Benjamini–Hochberg method. ATAC sequencing mapping statistics and the complete list of differentially methylated regions can be found in Supplementary Table 7. Peaks with a log2 fold change greater than 1 and less than -1 were considered as more open and less open, respectively. FDR-adjusted P value (q value) < .1 was considered significant. GO analysis was performed using Metascape (http://metascape.org).42 The complete GO analysis results can be found in Supplementary Table 8.
Statistical Analyses
P values from RNA sequencing, SILAC, RRBS, and ATAC sequencing experiments were corrected for multiple testing using the Benjamini–Hochberg method (q value). GO analysis P values were calculated based on accumulative hypergeometric distribution and corrected for multiple testing using the Benjamini–Hochberg procedure. Reverse-transcription quantitative PCR, Western blot, and LINE-1 retrotransposons methylation quantification significance was calculated using 1-way analysis of variance with the Bonferroni post-test. Multiple group statistical comparisons (electrophysiology and TGF-β signaling response) were obtained by 2-way analysis of variance with the Bonferroni post-test, and individual group mean differences (electrophysiology) were evaluated by the Student t test with Bonferroni correction.
Acknowledgments
The authors are profoundly thankful to all participating families and patients. The authors thank E. Bonneil and the Institut de recherche en immunologie et en cancérologie (IRIC) proteomic platform for expert assistance.
Members of the CoHeart Consortium
Gregor Andelfinger, Centre Hospitalier Universitaire Sainte Justine Research Center, Université de Montréal, Montréal, Québec, Canada.
Jeroen Bakkers, Hubrecht Institute-KNAW and University Medical Center Utrecht, Utrecht, Netherlands.
Bart Loeys, Cardiogenetics, Center for Medical Genetics, University of Antwerp/Antwerp University Hospital, Antwerp, Belgium.
Michel Pucéat, Université Aix-Marseille, InSERM UMR-1251, Marseille, France, and Laboratoire International Associé INSERM, Centre Hospitalier Universitaire Sainte Justine Research Center, Montréal, Québec, Canada.
Footnotes
Author contributions Jessica Piché and Gregor Andelfinger designed the study and wrote the paper; Gregor Andelfinger supervised the study and obtained funding; Philippe Chetaille obtained critical patient material; Jessica Piché, Natacha Gosset, Lisa-Marie Legault, Alain Pacis, Andrea Oneglia, Maxime Caron, Donghai Liu, Xioyan Qi, Mélanie Breton-Larrivée, and Séverine Leclerc provided technical support, performed experiments, and acquired or analyzed data; Serge McGraw, Luis Barreiro, and Stanley Nattel analyzed and interpreted data; and the CoHEART consortium provided critical revision.
Conflicts of interest The authors disclose no conflicts.
Funding Supported by doctoral awards from the Le Fonds de recherche du Québec – Santé (FRQS), the Foundation La Fondation du Grand défi Pierre Lavoie, and FESP (J.P.); by Chercheur-boursier Junior 1 from the FRQS (S.M.); by Chercheur-boursier Senior from the FRQS and the Banque Nationale Chaire de Recherche en Génétique Cardiovasculaire (G.A.); by an operating grant from the Canadian Institutes of Health Research 366129 (G.A. and S.N.), by an E-RARE CoHEART network grant (G.A.).
Contributor Information
Gregor Andelfinger, Email: gregor.andelfinger@recherche-ste-justine.qc.ca.
CoHEART Consortium:
Gregor Andelfinger, Jeroen Bakkers, Bart Loeys, and Michel Pucéat
Supplementary Material
References
- 1.Stanghellini V., Cogliandro R.F., de Giorgio R., Barbara G., Salvioli B., Corinaldesi R. Chronic intestinal pseudo-obstruction: manifestations, natural history and management. Neurogastroenterol Motil. 2007;19:440–452. doi: 10.1111/j.1365-2982.2007.00902.x. [DOI] [PubMed] [Google Scholar]
- 2.Stanghellini V., Cogliandro R.F., De Giorgio R., Barbara G., Morselli-Labate A.M., Cogliandro L., Corinaldesi R. Natural history of chronic idiopathic intestinal pseudo-obstruction in adults: a single center study. Clin Gastroenterol Hepatol. 2005;3:449–458. doi: 10.1016/s1542-3565(04)00675-5. [DOI] [PubMed] [Google Scholar]
- 3.Chetaille P., Preuss C., Burkhard S., Cote J.M., Houde C., Castilloux J., Piche J., Gosset N., Leclerc S., Wunnemann F., Thibeault M., Gagnon C., Galli A., Tuck E., Hickson G.R., El Amine N., Boufaied I., Lemyre E., de Santa Barbara P., Faure S., Jonzon A., Cameron M., Dietz H.C., Gallo-McFarlane E., Benson D.W., Moreau C., Labuda D., Zhan S.H., Shen Y., Jomphe M., Jones S.J., Bakkers J. Mutations in SGOL1 cause a novel cohesinopathy affecting heart and gut rhythm. Nat Genet. 2014;46:1245–1249. doi: 10.1038/ng.3113. [DOI] [PubMed] [Google Scholar]
- 4.Gargiulo A., Auricchio R., Barone M.V., Cotugno G., Reardon W., Milla P.J., Ballabio A., Ciccodicola A., Auricchio A. Filamin A is mutated in X-linked chronic idiopathic intestinal pseudo-obstruction with central nervous system involvement. Am J Hum Genet. 2007;80:751–758. doi: 10.1086/513321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lehtonen H.J., Sipponen T., Tojkander S., Karikoski R., Jarvinen H., Laing N.G., Lappalainen P., Aaltonen L.A., Tuupanen S. Segregation of a missense variant in enteric smooth muscle actin gamma-2 with autosomal dominant familial visceral myopathy. Gastroenterology. 2012;143:1482–14891.e3. doi: 10.1053/j.gastro.2012.08.045. [DOI] [PubMed] [Google Scholar]
- 6.Nishino I., Spinazzola A., Hirano M. Thymidine phosphorylase gene mutations in MNGIE, a human mitochondrial disorder. Science. 1999;283:689–692. doi: 10.1126/science.283.5402.689. [DOI] [PubMed] [Google Scholar]
- 7.Giordano C., Powell H., Leopizzi M., De Curtis M., Travaglini C., Sebastiani M., Gallo P., Taylor R.W., d'Amati G. Fatal congenital myopathy and gastrointestinal pseudo-obstruction due to POLG1 mutations. Neurology. 2009;72:1103–1105. doi: 10.1212/01.wnl.0000345002.47396.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonora E., Bianco F., Cordeddu L., Bamshad M., Francescatto L., Dowless D., Stanghellini V., Cogliandro R.F., Lindberg G., Mungan Z., Cefle K., Ozcelik T., Palanduz S., Ozturk S., Gedikbasi A., Gori A., Pippucci T., Graziano C., Volta U., Caio G., Barbara G., D'Amato M., Seri M., Katsanis N., Romeo G., De Giorgio R. Mutations in RAD21 disrupt regulation of APOB in patients with chronic intestinal pseudo-obstruction. Gastroenterology. 2015;148:771–782.e11. doi: 10.1053/j.gastro.2014.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamada H.Y., Yao Y., Wang X., Zhang Y., Huang Y., Dai W., Rao C.V. Haploinsufficiency of SGO1 results in deregulated centrosome dynamics, enhanced chromosomal instability and colon tumorigenesis. Cell Cycle. 2012;11:479–488. doi: 10.4161/cc.11.3.18994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeyaprakash A.A., Basquin C., Jayachandran U., Conti E. Structural basis for the recognition of phosphorylated histone h3 by the survivin subunit of the chromosomal passenger complex. Structure. 2011;19:1625–1634. doi: 10.1016/j.str.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 11.Levkau B., Schafers M., Wohlschlaeger J., von Wnuck Lipinski K., Keul P., Hermann S., Kawaguchi N., Kirchhof P., Fabritz L., Stypmann J., Stegger L., Flogel U., Schrader J., Fischer J.W., Hsieh P., Ou Y.L., Mehrhof F., Tiemann K., Ghanem A., Matus M., Neumann J., Heusch G., Schmid K.W., Conway E.M., Baba H.A. Survivin determines cardiac function by controlling total cardiomyocyte number. Circulation. 2008;117:1583–1593. doi: 10.1161/CIRCULATIONAHA.107.734160. [DOI] [PubMed] [Google Scholar]
- 12.Schrickel J.W., Lickfett L., Lewalter T., Tiemann K., Nickenig G., Baba H., Heusch G., Schulz R., Levkau B. Cardiomyocyte-specific deletion of survivin causes global cardiac conduction defects. Basic Res Cardiol. 2012;107:299. doi: 10.1007/s00395-012-0299-8. [DOI] [PubMed] [Google Scholar]
- 13.Smith C.W., Pritchard K., Marston S.B. The mechanism of Ca2+ regulation of vascular smooth muscle thin filaments by caldesmon and calmodulin. J Biol Chem. 1987;262:116–122. [PubMed] [Google Scholar]
- 14.Elsafadi M., Manikandan M., Dawud R.A., Alajez N.M., Hamam R., Alfayez M., Kassem M., Aldahmash A., Mahmood A. Transgelin is a TGFbeta-inducible gene that regulates osteoblastic and adipogenic differentiation of human skeletal stem cells through actin cytoskeleton organization. Cell Death Dis. 2016;7:e2321. doi: 10.1038/cddis.2016.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gramley F., Lorenzen J., Koellensperger E., Kettering K., Weiss C., Munzel T. Atrial fibrosis and atrial fibrillation: the role of the TGF-beta1 signaling pathway. Int J Cardiol. 2010;143:405–413. doi: 10.1016/j.ijcard.2009.03.110. [DOI] [PubMed] [Google Scholar]
- 16.Wei W., Jiao Y., Postlethwaite A., Stuart J.M., Wang Y., Sun D., Gu W. Dual-specificity phosphatases 2: surprising positive effect at the molecular level and a potential biomarker of diseases. Genes Immun. 2013;14:1–6. doi: 10.1038/gene.2012.54. [DOI] [PubMed] [Google Scholar]
- 17.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Funke R., Gage D., Harris K., Heaford A., Howland J., Kann L., Lehoczky J., LeVine R., McEwan P., McKernan K., Meldrim J., Mesirov J.P., Miranda C., Morris W., Naylor J., Raymond C., Rosetti M., Santos R., Sheridan A., Sougnez C., Stange-Thomann Y., Stojanovic N., Subramanian A., Wyman D., Rogers J., Sulston J., Ainscough R., Beck S., Bentley D., Burton J., Clee C., Carter N., Coulson A., Deadman R., Deloukas P., Dunham A., Dunham I., Durbin R., French L., Grafham D., Gregory S., Hubbard T., Humphray S., Hunt A., Jones M., Lloyd C., McMurray A., Matthews L., Mercer S., Milne S., Mullikin J.C., Mungall A., Plumb R., Ross M., Shownkeen R., Sims S., Waterston R.H., Wilson R.K., Hillier L.W., McPherson J.D., Marra M.A., Mardis E.R., Fulton L.A., Chinwalla A.T., Pepin K.H., Gish W.R., Chissoe S.L., Wendl M.C., Delehaunty K.D., Miner T.L., Delehaunty A., Kramer J.B., Cook L.L., Fulton R.S., Johnson D.L., Minx P.J., Clifton S.W., Hawkins T., Branscomb E., Predki P., Richardson P., Wenning S., Slezak T., Doggett N., Cheng J.F., Olsen A., Lucas S., Elkin C., Uberbacher E., Frazier M., Gibbs R.A., Muzny D.M., Scherer S.E., Bouck J.B., Sodergren E.J., Worley K.C., Rives C.M., Gorrell J.H., Metzker M.L., Naylor S.L., Kucherlapati R.S., Nelson D.L., Weinstock G.M., Sakaki Y., Fujiyama A., Hattori M., Yada T., Toyoda A., Itoh T., Kawagoe C., Watanabe H., Totoki Y., Taylor T., Weissenbach J., Heilig R., Saurin W., Artiguenave F., Brottier P., Bruls T., Pelletier E., Robert C., Wincker P., Smith D.R., Doucette-Stamm L., Rubenfield M., Weinstock K., Lee H.M., Dubois J., Rosenthal A., Platzer M., Nyakatura G., Taudien S., Rump A., Yang H., Yu J., Wang J., Huang G., Gu J., Hood L., Rowen L., Madan A., Qin S., Davis R.W., Federspiel N.A., Abola A.P., Proctor M.J., Myers R.M., Schmutz J., Dickson M., Grimwood J., Cox D.R., Olson M.V., Kaul R., Raymond C., Shimizu N., Kawasaki K., Minoshima S., Evans G.A., Athanasiou M., Schultz R., Roe B.A., Chen F., Pan H., Ramser J., Lehrach H., Reinhardt R., McCombie W.R., de la Bastide M., Dedhia N., Blocker H., Hornischer K., Nordsiek G., Agarwala R., Aravind L., Bailey J.A., Bateman A., Batzoglou S., Birney E., Bork P., Brown D.G., Burge C.B., Cerutti L., Chen H.C., Church D., Clamp M., Copley R.R., Doerks T., Eddy S.R., Eichler E.E., Furey T.S., Galagan J., Gilbert J.G., Harmon C., Hayashizaki Y., Haussler D., Hermjakob H., Hokamp K., Jang W., Johnson L.S., Jones T.A., Kasif S., Kaspryzk A., Kennedy S., Kent W.J., Kitts P., Koonin E.V., Korf I., Kulp D., Lancet D., Lowe T.M., McLysaght A., Mikkelsen T., Moran J.V., Mulder N., Pollara V.J., Ponting C.P., Schuler G., Schultz J., Slater G., Smit A.F., Stupka E., Szustakowki J., Thierry-Mieg D., Thierry-Mieg J., Wagner L., Wallis J., Wheeler R., Williams A., Wolf Y.I., Wolfe K.H., Yang S.P., Yeh R.F., Collins F., Guyer M.S., Peterson J., Felsenfeld A., Wetterstrand K.A., Patrinos A., Morgan M.J., de Jong P., Catanese J.J., Osoegawa K., Shizuya H., Choi S., Chen Y.J., Szustakowki J. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 18.Deardorff M.A., Bando M., Nakato R., Watrin E., Itoh T., Minamino M., Saitoh K., Komata M., Katou Y., Clark D., Cole K.E., De Baere E., Decroos C., Di Donato N., Ernst S., Francey L.J., Gyftodimou Y., Hirashima K., Hullings M., Ishikawa Y., Jaulin C., Kaur M., Kiyono T., Lombardi P.M., Magnaghi-Jaulin L., Mortier G.R., Nozaki N., Petersen M.B., Seimiya H., Siu V.M., Suzuki Y., Takagaki K., Wilde J.J., Willems P.J., Prigent C., Gillessen-Kaesbach G., Christianson D.W., Kaiser F.J., Jackson L.G., Hirota T., Krantz I.D., Shirahige K. HDAC8 mutations in Cornelia de Lange syndrome affect the cohesin acetylation cycle. Nature. 2012;489:313–317. doi: 10.1038/nature11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidt D., Schwalie P.C., Ross-Innes C.S., Hurtado A., Brown G.D., Carroll J.S., Flicek P., Odom D.T. A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res. 2010;20:578–588. doi: 10.1101/gr.100479.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mannini L., Lamaze F., Cucco F., Amato C., Quarantotti V., Rizzo I.M., Krantz I.D., Bilodeau S., Musio A. Mutant cohesin affects RNA polymerase II regulation in Cornelia de Lange syndrome. Sci Rep. 2015;5:16803. doi: 10.1038/srep16803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wendt K.S., Yoshida K., Itoh T., Bando M., Koch B., Schirghuber E., Tsutsumi S., Nagae G., Ishihara K., Mishiro T., Yahata K., Imamoto F., Aburatani H., Nakao M., Imamoto N., Maeshima K., Shirahige K., Peters J.M. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 22.Rubio E.D., Reiss D.J., Welcsh P.L., Disteche C.M., Filippova G.N., Baliga N.S., Aebersold R., Ranish J.A., Krumm A. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci U S A. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nyati S., Schinske-Sebolt K., Pitchiaya S., Chekhovskiy K., Chator A., Chaudhry N., Dosch J., Van Dort M.E., Varambally S., Kumar-Sinha C., Nyati M.K., Ray D., Walter N.G., Yu H., Ross B.D., Rehemtulla A. The kinase activity of the Ser/Thr kinase BUB1 promotes TGF-beta signaling. Sci Signal. 2015;8:ra1. doi: 10.1126/scisignal.2005379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deardorff M.A., Wilde J.J., Albrecht M., Dickinson E., Tennstedt S., Braunholz D., Monnich M., Yan Y., Xu W., Gil-Rodriguez M.C., Clark D., Hakonarson H., Halbach S., Michelis L.D., Rampuria A., Rossier E., Spranger S., Van Maldergem L., Lynch S.A., Gillessen-Kaesbach G., Ludecke H.J., Ramsay R.G., McKay M.J., Krantz I.D., Xu H., Horsfield J.A., Kaiser F.J. RAD21 mutations cause a human cohesinopathy. Am J Hum Genet. 2012;90:1014–1027. doi: 10.1016/j.ajhg.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thakur S.S., Geiger T., Chatterjee B., Bandilla P., Frohlich F., Cox J., Mann M. Deep and highly sensitive proteome coverage by LC-MS/MS without prefractionation. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.003699. M110.003699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prunotto M., Bruschi M., Gunning P., Gabbiani G., Weibel F., Ghiggeri G.M., Petretto A., Scaloni A., Bonello T., Schevzov G., Alieva I., Bochaton-Piallat M.L., Candiano G., Dugina V., Chaponnier C. Stable incorporation of alpha-smooth muscle actin into stress fibers is dependent on specific tropomyosin isoforms. Cytoskeleton. 2015;72:257–267. doi: 10.1002/cm.21230. [DOI] [PubMed] [Google Scholar]
- 27.Je H.D., Sohn U.D. SM22alpha is required for agonist-induced regulation of contractility: evidence from SM22alpha knockout mice. Mol Cells. 2007;23:175–181. [PubMed] [Google Scholar]
- 28.Wang J., Huang Y., Ning Q. Review on regulation of inwardly rectifying potassium channels. Crit Rev Eukaryot Gene Express. 2011;21:303–311. doi: 10.1615/critreveukargeneexpr.v21.i4.10. [DOI] [PubMed] [Google Scholar]
- 29.Qi X.Y., Huang H., Ordog B., Luo X., Naud P., Sun Y., Wu C.T., Dawson K., Tadevosyan A., Chen Y., Harada M., Dobrev D., Nattel S. Fibroblast inward-rectifier potassium current upregulation in profibrillatory atrial remodeling. Circ Res. 2015;116:836–845. doi: 10.1161/CIRCRESAHA.116.305326. [DOI] [PubMed] [Google Scholar]
- 30.Barry D.M., Nerbonne J.M. Myocardial potassium channels: electrophysiological and molecular diversity. Ann Rev Physiol. 1996;58:363–394. doi: 10.1146/annurev.ph.58.030196.002051. [DOI] [PubMed] [Google Scholar]
- 31.Huang X., Lee S.H., Lu H., Sanders K.M., Koh S.D. Molecular and functional characterization of inwardly rectifying K(+) currents in murine proximal colon. J Physiol. 2018;596:379–391. doi: 10.1113/JP275234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clark S.J., Melki J. DNA methylation and gene silencing in cancer: which is the guilty party? Oncogene. 2002;21:5380–5387. doi: 10.1038/sj.onc.1205598. [DOI] [PubMed] [Google Scholar]
- 33.Jones P.A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 34.Hawkins R.D., Hon G.C., Lee L.K., Ngo Q., Lister R., Pelizzola M., Edsall L.E., Kuan S., Luu Y., Klugman S., Antosiewicz-Bourget J., Ye Z., Espinoza C., Agarwahl S., Shen L., Ruotti V., Wang W., Stewart R., Thomson J.A., Ecker J.R., Ren B. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang X., Han H., De Carvalho D.D., Lay F.D., Jones P.A., Liang G. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014;26:577–590. doi: 10.1016/j.ccr.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 40.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tripathi S., Pohl M.O., Zhou Y., Rodriguez-Frandsen A., Wang G., Stein D.A., Moulton H.M., DeJesus P., Che J., Mulder L.C., Yanguez E., Andenmatten D., Pache L., Manicassamy B., Albrecht R.A., Gonzalez M.G., Nguyen Q., Brass A., Elledge S., White M., Shapira S., Hacohen N., Karlas A., Meyer T.F., Shales M., Gatorano A., Johnson J.R., Jang G., Johnson T., Verschueren E., Sanders D., Krogan N., Shaw M., Konig R., Stertz S., Garcia-Sastre A., Chanda S.K. Meta- and orthogonal integration of influenza “OMICs” data defines a role for UBR4 in virus budding. Cell Host Microbe. 2015;18:723–735. doi: 10.1016/j.chom.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ong S.E., Mann M. A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC) Nat Protoc. 2006;1:2650–2660. doi: 10.1038/nprot.2006.427. [DOI] [PubMed] [Google Scholar]
- 44.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 45.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xi Y., Li W. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics. 2009;10:232. doi: 10.1186/1471-2105-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Akalin A., Kormaksson M., Li S., Garrett-Bakelman F.E., Figueroa M.E., Melnick A., Mason C.E. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012;13:R87. doi: 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feng H., Conneely K.N., Wu H. A Bayesian hierarchical model to detect differentially methylated loci from single nucleotide resolution sequencing data. Nucleic Acids Res. 2014;42:e69. doi: 10.1093/nar/gku154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.