Abstract
Background
When a bloodstream infection is suspected, the preliminary and definitive results of culture-based microbiological testing arrive too late to have any influence on the initial choice of empirical antibiotic treatment.
Methods
This review is based on pertinent publications retrieved by a selective search of the literature and on the authors’ clinical and scientific experience.
Results
A number of technical advances now enable more rapid microbiological diagnosis of bloodstream infections. DNA-based techniques for the direct detection of pathogenic organisms in whole blood have not yet become established in routine use because of various limitations. On the other hand, matrix-assisted laser desorption/ionization—time of flight (MALDI-TOF) mass spectrometry (MS) has become available for routine use in clinical laboratories and has markedly shortened the time to diagnosis after blood samples that have been cultured in automated blood-culture systems turn positive. Further developments of this technique now enable it to be used directly for blood cultures that have been flagged positive, as well as for subcultures that have been incubated for only a short time on a solid nutrient medium. The microbial biomass of the subculture can also be used in parallel for more rapid susceptibility testing with conventional methods, or, in future, with MALDI-TOF MS.
Conclusion
The potential of all of these new techniques will only be realizable in practice if they are optimally embedded in the diagnostic process and if sufficient attention is paid to pre-analytical issues, particularly storage and transport times.
From the very beginning, the task of diagnostic microbiology was rapid, confident identification and characterization of pathogens. This is particularly true for bloodstream infections, which—especially in the case of sepsis—acutely endanger the patient (e1).
Rapid detection is especially important in light of the virulence of many pathogens and the existence of resistent phenotypes. Moreover, we are confronted by a population increasingly vulnerable to infection, with ever more elderly, multimorbid, and immunocompromised individuals together with growing numbers of patients with implants and other foreign bodies (e2).
Pathogen diagnosis.
Rapid diagnosis is important due to pathogen virulence and the existence of resistant pathogen phenotypes, and also because the population structure is increasingly vulnerable to infections, with ever more elderly, multimorbid, and immunocompromised patients and/or those with implants and other foreign bodies.
Polymerase chain reaction (PCR).
With the development of the PCR technique, pathogen detection was accelerated and higher sensitivity and specificity were achieved for pathogens that could be cultured only with difficulty or not at all.
The rapidity of pathogen detection in culture is limited by the rate of division of the microorganisms concerned. Traditionally, there were “overnight” cultures for each culture-based step of diagnosis (primary culture, isolation of mixed cultures, biochemical identification, and susceptibility testing), often resulting in diagnosis times of 2 to 3 days or longer. The advent of nucleic acid amplification tests (NAT) in the form of the polymerase chain reaction (PCR) therefore amounted to a revolution: pathogen detection was accelerated, and higher sensitivity and specificity were achieved for the detection of fastidious or non-cultivable pathogens (1, e3, e4). More recently, mass spectrometry (MS) techniques have again revolutionized pathogen identification and in many institutions already form part of the routine diagnostic work-up. Further far-reaching changes can be expected from whole genome/microbiome sequencing and from automation of culture-based procedures.
Learning goals
After reading this article, the reader should be:
Familiar with the essential aspects of recent and coming developments in pathogen detection
Able to assess both the advantages and the disadvantages of these procedures
Acquainted with the use of recently developed techniques as they apply to the diagnosis of bloodstream infections
Methods
This review article is based on a selective literature survey and on our own diagnostic and research experience in the use of NAT and matrix-assisted laser desorption/ionization—time of flight (MALDI-TOF) MS for microbiological diagnosis of bloodstream infections.
Principles and overview
PCR and culture techniques.
Depending on the indication, PCR and culture can contribute synergistically to valid identification of the pathogen, determination of its susceptibility, and definition of further characteristics.
Nowadays PCR, other NAT, and sequencing techniques belong to the standard repertoire of microbiological diagnosis. These procedures and developments thereof are widely used for direct detection of pathogens or for molecular identification of cultured microorganisms, for detection of resistance genes or virulence factors, and for genotyping (e5– e8). On the other hand, it is being increasingly appreciated that culture procedures are in themselves highly sensitive (one microbial cell capable of multiplying in culture suffices), and only a culture isolate permits determination of the phenotype, which is more than the sum of all resistance-coding genes or mutations. Innovations in culture-based diagnosis also serve to detect pathogens with greater speed, sensitivity, and specificity.
Molecular genetic tests and phenotypic characterization based on the cultured isolate both have method-inherent limitations, but both also possess particular diagnostic advantages. Depending on the indication, these two diagnostic strategies can contribute synergistically to valid pathogen identification and determination of susceptibility, as well as closer characterization of the pathogen.
With regard to its influence on the quality and speed of pathogen identification, the introduction of MS techniques to the routine microbiological diagnostic work-up is comparable with the “PCR revolution”. In only a short time, MALDI-TOF MS systems have almost completely replaced conventional biochemical identification of pathogens in many laboratories and are well on the way to being employed for antibiotic susceptibility testing (e9– e12).
Pathogen detection by means of nucleic acid diagnostic tests
MALDI-TOF mass spectrometry systems.
In only a short time, MALDI-TOF MS systems have almost completely replaced conventional biochemical identification of pathogens in many laboratories and are well on the way to being employed for antibiotic susceptibility testing.
After the introduction of PCR and other NAT (figure 2) in 1985, their lack of dependence on culture and their high sensitivity and specificity swiftly led to acceptance of nucleic acid-based diagnostic techniques (e3). Typically, these procedures achieve analytic sensitivity of 1 to 100 genome equivalents of bacterial or fungal DNA. Specimens that are complex or exhibit inhomogeneous distribution of pathogen (e.g., tissue, blood, and stool) may lead to drastic reductions in sensitivity and specificity. The specificity of NAT depends heavily on the choice of target structures, the composition of primers and probes, and the PCR/microarray reaction conditions. The obvious advantages of NAT often lead to thoughtless and insufficiently validated employment of these techniques (e13). Among other errors, the influence of pre-analytic factors, the importance of optimal nucleic acid extraction, and the risk of contamination are frequently underestimated (e14– e16). To avoid similar “traps” when using recently developed innovative techniques, it is essential to consider the findings in conjunction with those of other methods and view them critically (box 1).
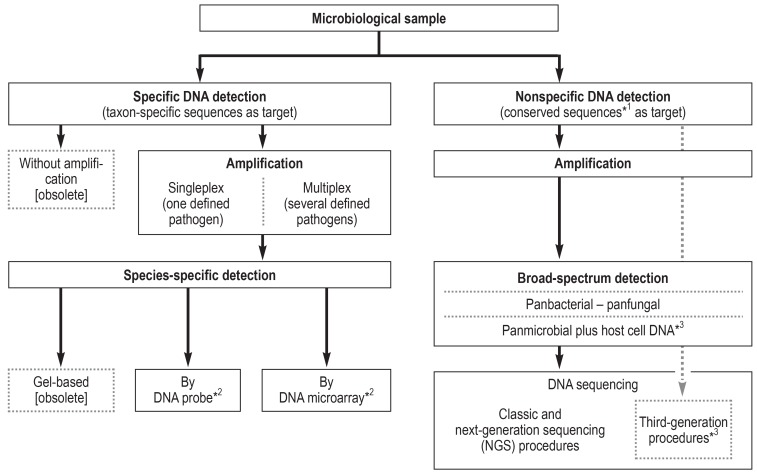
Figure 2.
Specific and nonspecific pathogen detection by means of DNA-based methods
*1 Framing taxon-specific sequence segments; *2 or by alternative DNA-based methods; *3 currently in development
BOX 1. Questions on assessment of NAT findings.
Can a positive NAT finding in itself explain the etiology of a suspected disease?
Does a positive NAT result really reflect the pathogen of the suspected infectious disease, or may it represent contamination of the specimen, e.g., by microorganisms that colonize the skin?
May the NAT findings contradict the results of culture-based or other investigations?
Can inappropriate pre-analytic handling (sampling and transport) be a cause of false-positive or false-negative results?
Is detection of resistance genes by NAT of patient samples associated with the suspected infectious pathogen, or do the genes come from other microorganisms (e.g., detection of the mecA gene from coagulase-negative staphylococci also present in the sample)?
Specificity of nucleic acid amplification testing (NAT).
The specificity of NAT depends heavily on the choice of target structures, the composition of primers and probes, and the PCR/microarray reaction conditions.
If a given patient’s infection cannot be attributed to the presence of defined pathogens by means of conventional diagnostic tests and species-specific NAT, broad-range nucleic acid-based methods can be used (Figure 2, eSupplement 1) (2, 3, e16– e19).
Whole-genome sequencing (WGS) and whole-metagenome sequencing (WMS) procedures and so-called next-generation sequencing (NGS) techniques are close to being ready for routine application (eSupplement 1) (4– 6, e20– e22).
In view of the differences among various NAT systems with regard to detection range, potential limitations of applicability for different specimens, and quality criteria (sensitivity, specificity, predictive value), close cooperation is advisable between the clinicians and the microbiology lab in order to select the techniques best suited to meet the clinical requirements. This is particularly true when NAT procedures are used in addition to conventional diagnostic techniques.
Evaluation of results.
To avoid the danger of misinterpretation when using recently developed innovative techniques, it is essential to consider the findings in conjunction with those of other methods and view them critically.
MALDI-TOF MS
The use of MS for pathogen identification was first proposed in the 1970s, but not until later did the German biophysicists Franz Hillenkamp and Michael Karas develop a technique, MALDI-TOF MS, suitable for routine application (for explanation of the principles of this method, please see eSupplement 1) (e23, e24). MALDI-TOF MS breaks down microbial, mainly ribosomal, proteins, using the data thus acquired to identify bacterial and fungal microorganisms. The main advantages of the technique are its extreme rapidity (a matter of minutes) and the cost efficiency of individual analyses with high specificity. Although MALDI-TOF MS is a phenotypic procedure, its high specificity, almost equal to that of NAT, is due to the high proportion of ribosomal proteins in the spectrum generated, practically reflecting the ribosomal nucleic acid sequences in entirety.
Owing to improvements in analytical methodology, analysis software, and the convenience of the devices used, MALDI-TOF MS has gained swift acceptance in the field of clinical microbiological diagnosis, to the point where it is now the standard for the identification of bacteria and yeasts and has led to considerable changes in diagnostic routine (7, e25, e26). The shortening of pathogen identification times achieved by MALDI-TOF MS may, for example, lead to improvement in clinical outcomes (e.g., survival rates) (8– 10).
Premises of microbiological diagnosis of bloodstream infections
Bloodstream infections, including sepsis and particularly septic shock, represent one of the most difficult challenges to diagnosis and treatment. The advances made in the past 25 years have, however, transformed our understanding of the nature of sepsis and led to altered definitions (most recently the Third International Consensus Definitions for Sepsis and Septic Shock—Sepsis-3) (e1, e27). Microbiological methods for diagnosis are still not sensitive enough to confidently detect the pathogen in all cases of bloodstream infection, and in particular they remain too slow to be of use in deciding on the initial empirical antibiotic treatment. They are indispensable, however, for decisions on specific antibiotic treatment after pathogen identification and susceptibility testing, as well as indirectly in compiling a database for use in future empirical treatment. A modern microbiological diagnostic work-up must be judged by how far it increases the sensitivity for detection of bloodstream infections and by how close it can come to the so-called “golden hour” of initial antimicrobial therapy.
MALDI-TOF MS.
The main advantages of MALDI-TOF MS are its extreme rapidity (a matter of minutes) and the cost efficiency of individual analyses with high specificity.
Sepsis-related mortality can be greatly reduced by early initiation of adequate antimicrobial treatment. The first hour after onset of the symptoms is crucial for the prognosis (11). Although there are no randomized clinical trials with significant conclusions regarding mortality and length of hospital stay, more rapid microbiological diagnosis could be of considerable benefit for the treatment of bloodstream infections (12, e28– e30). Early studies have confirmed a significant influence of rapid pathogen identification and susceptibility testing on mortality, length of stay in critical care, and costs (e30).
It must be emphasized that both classic pathogen culture and molecular procedures, in themselves, do no more than demonstrate the presence of a microorganism (bacteremia or fungemia) or its DNA in a sample. The confident classification of a microbiologically detected organism as clinical pathogen of a bloodstream infection requires knowledge of the clinical context and acquaintance with all infection-relevant parameters.
Sepsis-related mortality.
Sepsis-related mortality can be greatly reduced by early initiation of adequate antimicrobial treatment. The first hour after onset of the symptoms is crucial for the prognosis.
Despite partial successes in reducing methicillin-resistant Staphylococcus aureus (MRSA) in particular countries or regions, the rates of multidrug-resistant pathogens are rising worldwide (e31– e35). For this reason, and in light of new commercially available antibiotics, (e36, e37), proper selection of antimicrobial treatment is decisive not only for the individual patient, but also for lowering selective pressure for the development of resistance.
The goal must be a swift transition to targeted treatment, based on the results of pathogen identification and susceptibility testing, in the interests of diagnostic and antibiotic stewardship.
Advances in the diagnosis of bloodstream infections
The conventional diagnosis of bloodstream infections consists of three steps:
Inoculation of the patient’s blood into a liquid medium (blood culture bottles) for primary pathogen culture
Subculture on solid growth media
Identification and antimicrobial susceptibility testing (figure 3)
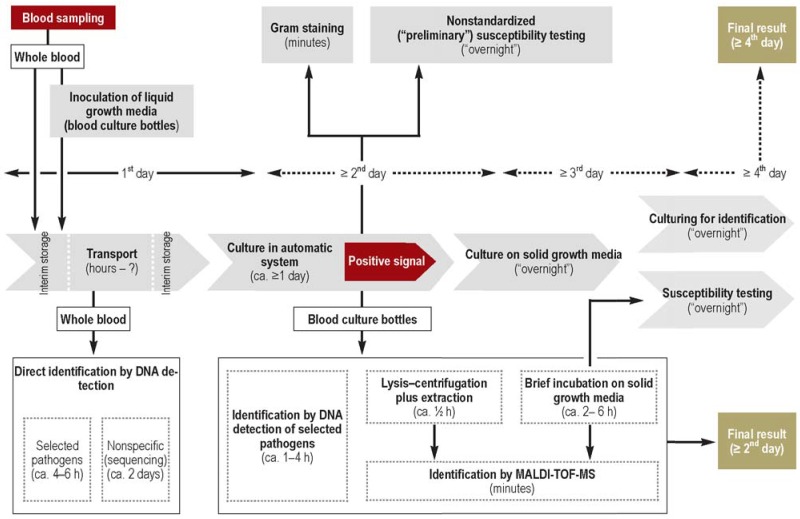
Figure 3.
Processes and times for classic pathogen diagnosis of bloodstream infections (gray) and modern means of accelerating diagnosis (white)
This procedure generally takes 3 to 4 days, perhaps longer in the case of compromised (previous antibiotic therapy) or slowly growing pathogens, and is therefore too slow to serve as a basis for early decisions on treatment (11).
For some decades blood culture bottles have been incubated in devices that automatically monitor microbial growth and emit a signal when a threshold value is attained (e38). For species determination and susceptibility testing, the sample is usually transferred from liquid growth medium to solid growth media for overnight incubation to produce individual colonies. In parallel to culture on solid growth media, a Gram stain is prepared and examined under the microscope (figure 3). The microscopy findings will ideally be delivered to the clinician 20 to 30 min after positive flagging of the blood culture and used to adjust the antibiotic regimen.
Up to a short time ago, the subsequent identification of individual colonies of microorganisms was based almost exclusively on biochemical methods, which required renewed microbial growth over a period of several hours and often could not be evaluated until after a further overnight incubation. Potential ways of accelerating the diagnosis of pathogens responsible for bloodstream infections rest either on more rapid pathogen identification from blood culture bottles flagged as positive or on NAT-based direct detection of the pathogen from blood samples as a complement to culture (figure 3).
Classification of findings.
The confident classification of a microbiologically detected organism as clinical pathogen of a bloodstream infection requires knowledge of the clinical context and acquaintance with all infection-relevant parameters.
Faster pathogen identification from positive blood culture bottles
DNA-based procedures
Recent years have seen the development of commercial multiplex PCR- or microarray-based procedures that enable detection and identification of pathogens from the liquid growth medium of blood cultures flagged as positive within 1 to 4 h (e39– e44).
A randomized controlled study showed that this method can help to optimize the antimicrobial treatment of bloodstream infections; no advantage could be demonstrated, however, with regard to mortality or duration of hospital stay (13). The counterarguments are the high costs, the additional workload, the lack of clarity concerning selection criteria for the samples to be investigated, the limited number of pathogens that can be detected, and, above all, the inauguration of MALDI-TOF MS as a procedure suitable for routine application (14).
MS procedures
Direct pathogen identification by means of MALDI-TOF MS from positive blood culture bottles has to be preceded by sample preparation using procedures such as lysis–centrifugation followed by ethanol/formic acid extraction. This quick technique—available either as an in-house adaptation or a commercial method—has shown identification rates of up to 80% and more within circa 20 to 40 min for detection of bacterial pathogens (7, 15– 17). In the case of yeasts, direct identification was possible in 62.5% of the samples (18).
Conventional diagnosis of bloodstream infections.
Inoculation of the patient’s blood into a liquid medium (blood culture bottles) for primary pathogen culture
Subculture on solid growth media
Identification and antimicrobial susceptibility testing
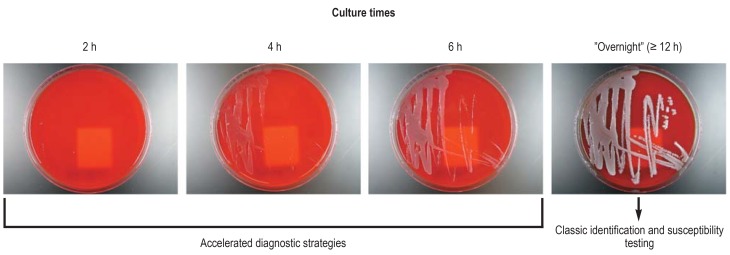
However, this technique has not achieved universal acceptance. The reasons for this lie in the relatively large increase in complexity, the increased reagent costs, the moderate identification rates, and its categorization as an add-on diagnostic test, because it does not replace subculture on solid growth media. This led to samples being processed in batches (e.g., twice daily or less), for organizational reasons, so that the anticipated benefit of rapid results largely failed to materialize (10, 16, 19). A more economical procedure better suited to integration into daily routine thus had to be found (14, 20). Instead of the usual “mature” colonies after overnight incubation, subcultures from positive blood cultures were incubated very briefly on solid growth media and then used (7). The growth on freshly inoculated solid growth media was monitored closely. On the first visual determination of microbial biomass (a fine “haze”), MALDI-TOF MS analysis was carried out (figure 1). It emerged that microbial colony material can be used for MALDI-TOF MS as soon as the first faint growth becomes visible on solid media. The mean incubation time to successful species identification for Gram-negative rods was only 2.0 h; for Gram-positive cocci it was 5.9 h (3.1 h with preceding protein extraction). The identification results were always the same as for cultures incubated for 24 h (7). The procedure can readily be integrated into the routine of a microbiology lab and requires neither additional materials nor more time to perform. Subsequent studies confirmed the benefits (21– 24). Furthermore, this identification technique can be advantageously combined with rapid susceptibility testing (25).
Figure 1.
Enterobacter cloacae cultures incubated for various lengths of time on a solid growth medium (blood agar plates) and their use in diagnosis
Direct DNA-based pathogen identification from whole blood
Direct pathogen identification.
Direct pathogen identification by means of MALDI-TOF MS from positive blood culture bottles has to be preceded by sample preparation using procedures such as lysis–centrifugation followed by ethanol/formic acid extraction.
Various experimental—later also commercial—species-specific and broad-spectrum NAT procedures were introduced to greatly shorten the time required for microbiological diagnosis of bloodstream infections directly from whole blood samples (26, e45– e47). Depending on the test system used, multiplex PCR can produce a result after circa 3.5 to 8 h. Broad-spectrum NAT takes much longer (roughly an extra working day) owing to the necessary sequencing of the PCR amplicon. In contrast to culture-based procedures, NAT can be positive as soon as microbial DNA is circulating in the bloodstream (“DNAemia”) and also in the case of microorganisms that are non-viable or whose status is viable but nonculturable (VBNC). While this may be disadvantageous in the event of contamination, it can lead to improved sensitivity in the presence of pathogens previously treated with antibiotics.
Haze-like growth.
Microbial biomass can be used for MALDI-TOF MS as soon as the first faint (haze-like) growth becomes visible on solid media.
Depending on their spectrum, commercially available tests for direct detection of pathogens in whole blood are variably effective for detection of bacteria and yeasts responsible for bloodstream infections as well as, in some cases, marker genes for some multidrug-resistant pathogens (26, 27, e48). These multiplex PCR systems for detection of specific pathogens are limited to the microorganisms that most frequently cause bloodstream infections (circa 25 to 90 pathogens, depending on the test system used) and yield false-negative results for rarer pathogens. Lowered analytical cutoff values to artificially decrease sensitivity have been introduced for some coagulase-negative staphylococci and nonhemolytic streptococci, which may be present as contaminants on the skin. These lowered amplification cycle number cutoffs may lead to under-reporting of infection-related bacteremia involving these pathogens in vulnerable groups (e.g., neutropenic patients and premature neonates) (28).
The broad-spectrum PCR procedures require additional time for analysis of the amplicon. They are not restricted to individual pathogens and also enable identification of fastidious or non-cultivable pathogens, such as Tropheryma whipplei, bartonellae, mycobacteria, or molds. They are also indicated for use in situations where their swiftness in yielding a result, compared with culture-based techniques, is beneficial for treatment or prevention of infections.
Direct DNA-based pathogen identification from whole blood.
Depending on the test system used, multiplex PCR can produce a result after circa 3.5 to 8 h. Broad-spectrum NAT takes much longer owing to the necessary sequencing of the PCR amplicon.
Recent years have seen the publication of a large number of studies comparing the identification results of PCR and culture techniques (27, 29– 31, e49– e52). Early investigations into clinical benefit were limited to data from retrospective evaluations (32– 34, e50, e53) or from studies that were prospective but not randomized (35– 37). The first two randomized controlled trials examined the clinical benefit of direct multiplex PCR from whole blood (38, 39). In hematology patients with febrile neutropenia, the median time before the switch to targeted antimicrobial treatment was 21.4 h in the study group (multiplex PCR in addition to blood cultures) and 42.5 h in the control group (blood cultures only); p = 0.018 (38). A positive PCR result led to a change in treatment in 33% of cases. A study in critical care patients with sepsis showed that the additional use of multiplex PCR from whole blood achieved a significant reduction in the time from sampling to communication of the result compared with blood culture alone (15.9 h versus 38.1 h) (39). The mean time that elapsed between blood sampling and the switch from empirical to specific antimicrobial treatment was 18.8 h in the study group and 38.3 h in the control group.
Broad-spectrum PCR techniques.
These are not restricted to individual pathogens and also enable identification of fastidious or non-cultivable pathogens, such as Tropheryma whipplei, bartonellae, mycobacteria, or molds.
Modern NAT procedures are possessed of high diagnostic accuracy and can lead to faster detection of the pathogen and thus to earlier administration of specific treatment. If difficult or nonculturable pathogens are suspected, broad-spectrum NAT—as well as antibody-detecting techniques, not discussed here—can contribute to their identification. However, various limitations have prevented wide application of the NAT techniques beyond the study setting (box 2). The principal disadvantages are the absence of information on antibiotic resistance phenotype and the lacking potential for close characterization of isolates. Efforts are being made to counter the sensitivity problem of the NAT procedures, e.g., by selective concentration of microbial DNA (e54).
BOX 2. Limitations of direct detection of pathogens from whole blood.
-
False-positive results owing to:
Detection of DNA from non-viable pathogens or from microorganisms that have already been adequately treated
Detection of DNA from contaminants (e.g., from transient bacteremia/fungemia or from a skin cylinder punched out in the course of sampling)
-
False-negative results owing to:
Low sensitivity compared with culture (e.g., due to artificial adjustment of detection thresholds)
Interference with primers and probes by high amounts of nonmicrobial host DNA
Presence of inhibitors (e.g., heparin, iron, and immunoglobulins)
Discrepancies with the result from conventional blood culture (no pathogen detected, different pathogen detected, simultaneous detection of two or more pathogens)
Comparatively high costs (apparatus, kits, staff time)
No or only limited (restricted to a few [known] resistance genes) information about antibiotic susceptibility
No possibility of typing
Currently limited research data available on most of the test systems
Antibiotic susceptibility testing and resistance gene detection
Although species identification in itself can yield information helpful for treatment decisions, it is the results of antimicrobial susceptibility that are crucial in guiding the final choice of targeted therapy. For this reason it is important to produce the susceptibility results as soon as possible in the presence of critical infections.
Conventionally, individual colonies from solid growth media are employed for susceptibility testing. In the diagnosis of bloodstream infections, this entails considerable delay owing to the incubation time needed after subcultivation from positive blood culture bottles.
Acceleration of susceptibility testing by changing the method of culturing
With the direct inoculation of test systems from positive blood culture bottles, one does not wait for the growth of subcultured colonies on solid growth media. Cell pellets produced by centrifugation of positive blood culture bottles are used for direct inoculation into susceptibility testing systems. For yeasts, the time to availability of results was shortened by 15.1 h (18). While the results for Gram-negative rods showed good agreement with conventional procedures, no satisfactory correspondence was achieved for Gram-positive cocci and yeasts (18, 40) (e55).
Modern NAT techniques.
Modern NAT procedures possess high diagnostic accuracy and can lead to faster detection of the pathogen and thus to earlier administration of specific treatment.
One elegant and cost-efficient strategy is based on using the biomass of briefly incubated subcultures. The subcultures used for susceptibility testing are incubated on solid growth media for only a very short time. The agreement with standard testing was found to be over 99 % (25). The mean incubation period for the subcultures was only 3.8 h for Gram-positive cocci and 2.4 h for Gram-negative rods. The time saved was thus approximately one day, without any added costs or complexity. This method can readily be combined with rapid identification by means of MALDI-TOF MS from subcultures incubated briefly on solid growth media (14).
Use of MALDI-TOF MS for accelerated detection of resistance and for susceptibility testing
The time savings and lower costs associated with MALDI-TOF MS have prompted various efforts to adapt this technique for the detection of individual resistance mechanisms and also, recently, as an alternative for the universal susceptibility testing. For instance, antibiotic-related alterations of the pathogen’s proteome may result in changes in mass that can be demonstrated by MALDI-TOF MS (e56). Another approach rests on changes in mass of the antibiotics used that can be caused by microbial resistance mechanisms. An example is hydrolysis of betalactam antibiotics by bacterial betalactamases (e57). These methods take around 0.5 to 4 h but are restricted to just a few resistance mechanisms and involve greater test complexity.
A new approach to the universal application of MALDI-TOF MS for rapid susceptibility rests on growth-based differentiation into “susceptible” and “resistant”—similar to conventional susceptibility testing. Antibiotics are added in breakpoint concentrations, and MALDI-TOF MS is used to find out whether microbial growth of the isolates being tested has taken place (=resistant) or not (=susceptible) (e58). This procedure was recently shortened and simplified still further by the design of a method involving incubation of microdroplets of the isolates directly on the MALDI-TOF MS targets (direct-on-target microdroplet growth assay, DOT-MGA) (e59). In a pilot study, this technique—still in development but potentially suitable for routine application—took 4 to 5 h to complete susceptibility testing for several antibiotics with sensivity and specificity similar to those of the current standard procedure (e59). Moreover, DOT-MGA offers the advantage of parallel identification and susceptibility testing in one assay and can also be employed for susceptibility testing directly from blood cultures (e59, e60).
Direct DNA-based detection of resistance genes in whole blood samples
Efficiency of direct susceptibility testing.
While the results for Gram-negative rods have shown good agreement with conventional procedures, no sufficiently good correspondence has been achieved for Gram-positive cocci and yeasts.
Ideally, in monoinfections, the microbial DNA isolated directly from samples of whole blood could be used not only for pathogen identification, but also to detect the genes or, if mutation related, the DNA segments determining the antibiotic resistance. However, the PCR multiplex systems in current use are stretched to their technical limits by the detection of the pathogens most frequently causing bloodstream infections, so that one either dispenses with molecular detection of resistance or includes only individual resistance genotypes. In the latter case, detection ensues either in parallel, in one assay, or, if the primary PCR points to a corresponding pathogen, in a second PCR. The commercial test systems available so far extend to the detection of the mecA/mecC genes as a sign of MRSA isolates or demonstration of the vanA-/vanB genes as a sign of vancomycin-resistant enterococci (VRE).
Pre-analytics, standardization and quality assurance
Pre-analytic factors (eSupplement 2) (12, 38, e61, e62) and aspects of standardization and quality assurance (eSupplement 2) exert considerable influence on the quality of the results of microbiological diagnosis of bloodstream infections (e63).
Summary
The modern microbiological diagnosis of bloodstream infections is moving close to the time of clinical suspicion of the presence of infection, but is still far from having any direct influence on the decision regarding initial antibiotic treatment. MALDI-TOF MS, combined with the use of briefly incubated solid growth media, has contributed to a massive shortening of the time needed for pathogen identification and susceptibility testing. PCR-based rapid tests are helpful in the event of unsuccessful culture of bloodstream infection pathogens, but cannot replace culture. The benefits of the new techniques can take effect only if pre-analytics, the workflow, the times required for each step, and quality management are all optimally adjusted and if the principal indications and limitations of the individual diagnostic procedures are known to the requesting physician.
Supplementary Material
eSUPPLEMENT 1
Basic principles of DNA-based and mass spectrometry methods
Broad-spectrum nucleic acid diagnostics
If a patient’s medical history and clinical presentation suggest no clear initial diagnosis and conventional diagnostic procedures (culture, serology) do not attribute the infection event to the presence of one or more pathogenic microorganisms, the treating physician can request so-called “broad-spectrum” nucleic acid-based diagnostics, consisting of DNA amplification followed by sequencing or probe identification of the amplicons (figure 2). These NAT detection methods, also referred to as “group-specific,” “broad-range,” “eu-/panbacterial,” or “panfungal,” rest on the presence of certain genes or gene segments which, in at least parts of their nucleic acid sequence, have been conserved by “evolutionary pressure” and therefore display a high degree of homology among various species.
These gene sequences represent a certain gold standard in the definition of bacterial, fungal, and parasite species and their phylogenetic classification, and their high diagnostic potential can also be utilized for broad-spectrum pathogen detection. Some examples of systematically investigated “marker genes” are hsp65 (65-kDa heat-shock protein), rpoB (B subunit of RNA polymerase), and the genes for the ribosomal subunits (bacteria: 16S rRNA, 23S rRNA genes; fungi: 18S rRNA, 28S rRNA genes) or interposed sequence regions known as internal transcribed spacer (ITS) regions.
The ribosomal genes and ITS regions represent ideal target sequences for molecular biological species identification of cultured isolates and also—under certain experimental conditions—for direct detection from the clinical sample (2). The amplicons are subjected to automated DNA sequencing, and the nucleic acid sequences determined are compared with databases, whereby the quality of the database contents is decisive for the interpretation of the sequencing findings (e17, 64). The results are communicated either as clear concordance with reference sequences from a given species or, as observed more frequently in practice for the investigation of clinical specimens, a list of several phylogenetically related species in ascending order of sequence homology. This enables clear species attributions in many cases, sometimes with the aid of existing culture results or molecular genetic analysis of further cross-species target sequences.
Many in-house techniques have been developed for broad-spectrum nucleic acid diagnostics; to date, commercial procedures are available for detection of pathogens responsible for sepsis.
If the sample for investigation consists of normally sterile material (e.g., blood, but also cerebrospinal fluid, joint fluid, or tissue), amplicon sequence analysis usually yields clear species identification even in the case of fastidious or non-cultivable pathogens and previously unknown pathogens. This type of analysis fails, however, in the presence of (the DNA of) various microorganisms in the same sample, in which case overlapping of the different species-specific sequence signals hampers interpretation. The indication for these specialized investigations therefore depends on certain preconditions and constellations (box 2) (3).
The greatest problem in the technical implementation of broad-spectrum NAT techniques in the microbiology lab is the potential for contamination from the ubiquitous ambient bacterial and fungal DNA (in the air and on surfaces) and the physiological microbiotic flora of patients and staff (e16). Even reaction vessels and reagents employed for DNA isolation and amplification can become contaminated to a varying degree with bacterial or fungal DNA during production or use (e18). With sensitive broad-spectrum test systems, for example, DNA from Pseudomonas spp. can be demonstrated in many batches of aqueous reagents approved for PCR (e19).
Until a complete palette of “DNA-free” sample vessels and other reaction components are commercially available, together with closed systems for contamination-free processing and analysis, accreditable and thus reliable performance of broad-spectrum NAT investigations will necessarily be restricted to a small number of ready-made test kits from certain manufacturers (e.g., those producing sepsis PCR kits) and to specialized laboratories that conduct elaborate batch testing for individual reaction components and possess the necessary lab space and equipment.
Therefore, before ordering broad-spectrum NAT investigations the attending physician must always weigh the relatively high demands on technology, quality management, special reagents, and working time (and thus costs) against the anticipated benefit for the individual patient.
Whole-genome sequencing and whole-metagenome sequencing
The technical accomplishment of whole-genome sequencing (WGS) and whole-metagenome sequencing (WMS) has been made possible by so-called next-generation sequencing (NGS), so that these methods are well on the way to becoming routine (4, 5, e20). Their advantage lies in the potential for rapid (hours to a few days) simultaneous identification, genotyping, and determination of genetically coded traits of virulence and resistance. Even parallel analysis of the host genome is possible if the sample contains human DNA.
These procedures, starting with the pyrosequencing technology in 2005, permit complete presentation of the genetic information for individual pathogens (by WGS) or complete microbiota (by WMS) in a relatively short time (e65). Current NGS platforms yield a throughput of 0.1 to 6000 Gb, depending on the device used (for comparison: 0.0003 Gb for the conventional, so-called Sanger sequencing [e66]). However, the current reading lengths of around 50 to 400 bp for “short-read” techniques are still much lower than for the first-generation sequencing methods (Sanger), which achieved reading lengths of 0.5 to 1 kb. Massive parallel sequencing following amplification results in a large number of small portions which, after assembly, provide multiple coverage or reading depth of the genome. New “third-generation” sequencing methods (single-molecule real-time sequencing techniques, biological nanopore-based techniques), which have single DNA molecules as target and thus require no amplification, are capable of achieving DNA fragments of = 20 kb (e67– e69). Depending on procedure, particularly for the long-read techniques, the error rates may be relatively high (10 bis >30%), especially in complex areas of the genome with many repeats or phage insertions. For this reason, sufficiently high coverage and efficient algorithms for assembly and error correction are necessary to pinpoint errors and close gaps (e70).
WGS techniques therefore represent the ultimate tool for genotyping of microbial isolates, because they are capable of detecting individual nucleotide exchanges between isolates. Using these techniques, the transmission pathways of pathogens can be elucidated in detail and their dissemination in a health facility, region, or beyond can practically be traced. In the area of hospital hygiene, prospective WGS can document infection pathways continuously and in real time, and achieves this at a lower cost than measures to prevent transmission (6, e21, e22).
The routine use of these techniques is currently limited particularly by the costs, the quality of databases, insufficient standardization, lack of IT capacity, and problems of “big-data” management.
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry
Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (MS) enables marker-free analysis of large biomolecules by cocrystallization of matrix material and analyte (=sample) (e24). For diagnostic characterization, material from microbial pure cultures is applied to a plate together with the matrix and the resulting cocrystals are irradiated by a pulsed laser in full vacuum. The absorbed laser energy leads to ionization and release of ions that are then accelerated by an electrode in the electrical field generated, detected by an ion detector, and transformed into electrical signals. The time of flight of the pathogen-specific composite ions depends on their mass and charge. The measured values are compared with reference spectra, so the results are dependent of the size and quality of the databases used.
The culture-based areas of application for routine MALDI-TOF MS in diagnostic microbiology include especially identification of prokaryotic (bacteria, archaea) and eukaryotic pathogens (fungi, single-and multi-celled parasites) at species level, and sometimes at subspecies or even strain level (8, e25, e26, e71). Detection of specific viral proteins (e.g., norovirus capsid protein) by this means has also been reported (e72). Moreover, applications for detection of bacterial toxins, e.g., for Staphylococcus enterotoxins and Panton–Valentine leukocidin (PVL) have been described (e73, e74). Further reported uses are direct detection of microorganisms from positive blood cultures and from samples of urine and foodstuffs (18, e75– e78). An application of MALDI-TOF MS that is well on the way to routine use is identification of resistance in bacteria and fungi, or antibiotic susceptibility testing of clinical isolates (e56– e59, e79– e84).
eSUPPLEMENT 2
Pre-analytics, standardization, and quality assurance
Pre-analytic processes and factors
Particularly if add-on tests are used, costs can increase sharply, due to the test itself and/or increased personnel requirements. Not least for this reason, close attention should be paid to the necessary practical and administrative/organizational processes in the course of pre-analytics (e85).
These include determination of the indications, patient preparation, selection of the nature and number of samples for the add-on test systems, any diagnostic algorithms, transport and storage times (figure 2), and the organization of working times and diagnostic procedures in the laboratory. For example, excessively long transport and interim storage times or too small inoculates may lead to false-negative results in automated blood culture diagnostics (38, e61, e62).
Standardization and quality assurance
The quality requirements formulated at the beginning of the PCR era regarding standardization, reproducibility, and validity still hold true, not only for DNA-based methods: “Uncritical use of PCR procedures on unproven specimen types or the application of these methods under inappropriate conditions amplifies the potential for error and leads to diagnostic misinterpretations” (translated from the original German in [1]). Naturally the same requirements apply to MALDI-TOF MS-based techniques.
With the growing acceptance, widening use, and especially the ever-increasing variety of closed analysis systems, ready-prepared commercial test kits, and in-house protocols for nucleic acid detection of relevant pathogens, there is an increasing need for suitable external quality assurance measures. These should preferably take the shape of centrally organized inter-laboratory ring trials, but there could also be bilateral or multilateral exchange of well-characterized sample materials between laboratories.
While after successful accreditation and/or certification of a diagnostic facility only a well-organized workflow and reproducible, standardized implementation of precisely predefined protocols can be assured (lab-internal quality management), external quality control measures are aimed mainly at testing the sensitivity and specificity of NAT protocols and the assurance of precise and above all comparable results in diagnostic routine. Thus, internal quality management and external quality control are complementary.
Diagnostic institutions profit in more than one way from inter-laboratory ring trials or applying comparable quality assurance measures, as in this way they formally comply with the demands of certification and accreditation guidelines and also fulfill the requirements of the Richtlinie der Bundesärztekammer zur Qualitätssicherung laboratoriumsmedizinischer Untersuchungen (German Medical Association Guideline on Quality Assurance in Medical Laboratory Examinations; RiLiBÄK, part 3) (e63). Moreover, professional societies are also striving to achieve quality assurance and standardization of examination techniques, as exemplified by the microbiological/infectiological quality standards (MIQ) of the Deutsche Gesellschaft für Hygiene und Mikrobiologie (German Society for Hygiene and Microbiology; DGHM) (e86), e.g., MIQ 30, Qualitätsmanagement im medizinisch-mikrobiologischen Laboratorium (Quality Management in the Medical–Microbiological Laboratory); the norms of DIN series 58959, Qualitätsmanagement in der medizinischen Mikrobiologie (Quality Management in Medical Microbiology) (e87); and the Checkliste Mikrobiologie und Hygiene – Molekularbiologie in der Infektionsdiagnostik (Checklist Microbiology and Hygiene—Molecular Biology in the Diagnosis of Infections) in the Handbuch für die Akkreditierung Medizinischer Laboratorien (Handbook for the Accreditation of Medical Laboratories) (e88). All of these documents are published in German.
The Society for Promoting Quality Assurance in Medical Laboratories (Gesellschaft zur Förderung der Qualitätssicherung in medizinischen Laboratorien e.V.; INSTAND) has set up a German-language journal entitled GMS Zeitschrift zur Förderung der Qualitätssicherung in medizinischen Laboratorien (GMS Journal for Promoting Quality Assurance in Medical Laboratories) (www.egms.de/static/de /journals/index.htm) (e89). Here the participants in INSTAND ring trials receive reports of potential sources of error and further information on current issues in quality assurance.
Further information on CME.
-
Participation in the CME certification program is possible only over the Internet: cme.aerzteblatt.de. This unit can be accessed until 3 March 2019. Submissions by letter, e-mail or fax cannot be considered.
The following CME units can still be accessed for credit:
„The Nomenclature, Definition and Distinction of Types of Shock“ (issue 45/2018) until 3 February 2019
”The neurophysiology and treatment of motion sickness” (issue 41/2018) until 6 January 2019
”The diagnosis and treatment of anxiety disorders” (issue 37/2018) until 9 December 2018
This article has been certified by the North Rhine Academy for Continuing Medical Education. Participants in the CME program can manage their CME points with their “uniform CME number” (einheitliche Fortbildungsnummer, EFN).
The EFN must be stated during registration on
www.aerzteblatt.de (“Mein DÄ”) or else entered in “Meine Daten,” and the participant must agree to communication of the results. The 15-digit EFN is found on the CME card (8027XXXXXXXXXXX).
CME credit for this unit can be obtained via cme.aerzteblatt.de until 3 March 2019.
Only one answer is possible per question. Please select the answer that is most appropriate.
Question 1
What advantage does the additional microbiological diagnosis of bloodstream infections by means of nucleic acid amplification testing (NAT) have over culture-based procedures?
Consistently higher sensitivity
No need for preparation of the sample
Can be carried out by untrained staff
No quality assurance measures needed
Faster test result
Question 2
When is the use of broad-spectrum NAT indicated?
For detection of fastidious or non-cultivable pathogens and/or when its speed offers benefits for treatment or prophylaxis of infections
Only for experimental purposes in clinical trials
Exclusively for the detection of yeasts and molds
Exclusively for reasons of prevention (e.g., MRSA screening)
When mixed bacterial and viral infection is suspected
Question 3
What modification of the culture technique can accelerate susceptibility testing?
More glucose is added to the culture medium to improve the conditions for growth.
Positive blood culture bottles are centrifuged and the pellet is used directly for susceptibility testing.
The incubation temperature is raised to 39° so that the blood cultures reach the logarithmic growth phase sooner.
Sample material is first inoculated onto a solid growth medium, then individual colonies of bacteria are subjected to susceptibility testing.
Ampicillin is added to the blood cultures to select for resistant pathogens from the outset.
Question 4
What method is used for the microbiological diagnosis of bloodstream infections?
Exclusively NAT techniques are used.
It is primarily culture-based; the cultured pathogens are identified by various means and tested for antibiotic susceptibility.
Primarily serological detection methods are applied.
Cell culture techniques based on human host cells are employed.
Exclusively Gram-stains are used.
Question 5
What limits the speed of culture-based procedures in microbiological diagnosis?
The technical parameters of the incubators required
Suboptimal growth conditions due to standardized culture conditions with negative effects on the division rate
The generation time of the microorganism
The high amount of work involved
Exposure of the cultures to UV light and daylight spectra (including wavelengths of artificial light sources)
Question 6
What is the functional principle of diagnostic MALDI-TOF mass spectrometry (MS)?
Analysis of microbial nucleic acids (DNA) by MS
Analysis of microbial RNA by MS
Analysis of microbial, primarily ribosomal proteins by MS
Analysis of microbial islands of pathogenicity or other virulence-related elements by MS
Analysis of large biomolecules used for the typing of bacteria and fungi by MS
Question 7
What organisms are routinely diagnosed with MALDI-TOF MS?
Prion proteins
Bacteria and yeasts
Retroviruses
Double-stranded DNA viruses
Transfer RNA
Question 8
What is the conventional procedure for the diagnosis of bloodstream infections following blood sampling?
Inoculation into serum tubes, subculture on solid growth medium, antibiotic susceptibility testing on agar plates
Inoculation into blood culture bottles, subculture on solid growth medium, Gram staining for antibiotic susceptibility testing
Inoculation onto solid growth medium, Gram staining for pathogen identification, blood culture bottles for antibiotic susceptibility testing
Inoculation into the liquid medium of blood culture bottles, subculture on solid growth medium, identification and antibiotic susceptibility testing
Inoculation into blood culture bottles with various antibiotics, subculture and final antibiotic testing on solid growth medium
Question 9
From when can microbial colony material be used for MALDI-TOF-MS analysis?
After brief (30 min) incubation on solid growth medium
After full development of typical colony morphology
After appearance of at least three separate colonies at least 3 mm in diameter
After incubation for 1 h in liquid growth medium and brief (1 h) incubation on solid growth medium
After first visual detection of (haze-like) growth on solid growth medium
Question 10
What can lead to false-positive results when using methods for direct pathogen detection from whole blood?
Absence of means of typing
Presence of inhibitors such as heparin, iron, or immunoglobulins
Interference with primers and probes by high proportions of nonmicrobial host DNA
Low sensitivity compared with culture
Detection of DNA from non-viable pathogens or from microorganisms that have already been treated adequately
►Participation is possible only via the Internet: cme.aerzteblatt.de
Acknowledgments
Translated from the original German by David Roseveare
Footnotes
Conflict of interest statement Prof. Becker is co-inventor of the patent application “Preparation of living microbial samples and microorganisms for subsequent measurement and analysis by mass spectrometry” (“Aufbereitung lebendiger, mikrobieller Proben und Mikroorganismen für anschließende massenspektrometrische Messung und Auswertung”) and the patent application “Device and method for treating fluids, particularly body fluids” (“Vorrichtung und Verfahren zum Aufbereiten von Flüssigkeiten, insbesondere Körperflüssigkeiten”). The patent applications were licensed by University Hospital Münster and the inventor’s due portion was transferred to Prof. Becker in accordance with the law. He has received payments for lectures, including reimbursement of travel costs, from Becton Dickinson, bioMérieux, Bruker, Daltonik, Hain, Lifescience, Roche Molecular Systems, and ThermoFisher.
PD Dr. Idelevich is co-inventor of the patent application “Preparation of living microbial samples and microorganisms for subsequent measurement and analysis by mass spectrometry” (Aufbereitung lebendiger, mikrobieller Proben und Mikroorganismen für anschließende massenspektrometrische Messung und Auswertung) and the patent application “Device and method for treating fluids, particularly body fluids” (Vorrichtung und Verfahren zum Aufbereiten von Flüssigkeiten, insbesondere Körperflüssigkeiten)“. The patent applications were licensed by University Hospital Münster and the inventor’s due portion was transferred to PD Dr. Idelevich in accordance with the law. He has received reimbursement of congress attendance charges and associated travel and accommodation costs from Astellas. He has been paid for giving lectures by Novartis, Pfizer, and Bruker.
Prof. Reischl declares that no conflict of interest exists.
References
- 1.Becker K, Peters G. Moderne diagnostische Verfahren in der medizinischen Mikrobiologie. Internist (Berl) 1995;36:95–101. [PubMed] [Google Scholar]
- 2.Clarridge JE 3rd. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin Microbiol Rev. 2004;17:840–862. doi: 10.1128/CMR.17.4.840-862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reischl U. Thiemann F, Cullen PM, Klein HG, editors. Indikationen für die molekulare Diagnostik - Bakterien, Pilze, Eukaryonten Leitfaden Molekulare Diagnostik: Grundlagen, Gesetze, Tipps und Tricks. Weinheim: Wiley-VCH. 2006:175–183. [Google Scholar]
- 4.Quainoo S, Coolen JPM, van Hijum S, et al. Whole-genome sequencing of bacterial pathogens: the future of nosocomial outbreak analysis. Clin Microbiol Rev. 2017;30:1015–1063. doi: 10.1128/CMR.00016-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tagini F, Greub G. Bacterial genome sequencing in clinical microbiology: a pathogen-oriented review. Eur J Clin Microbiol Infect Dis. 2017;36:2007–2020. doi: 10.1007/s10096-017-3024-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mellmann A, Bletz S, Böking T, et al. Real-time genome sequencing of resistant bacteria provides precision infection control in an institutional setting. J Clin Microbiol. 2016;54:2874–2881. doi: 10.1128/JCM.00790-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Idelevich EA, Schüle I, Grünastel B, Wüllenweber J, Peters G, Becker K. Rapid identification of microorganisms from positive blood cultures by MALDI-TOF mass spectrometry subsequent to very short-term incubation on solid medium. Clin Microbiol Infect. 2014;20:1001–1006. doi: 10.1111/1469-0691.12640. [DOI] [PubMed] [Google Scholar]
- 8.Bizzini A, Greub G. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry, a revolution in clinical microbial identification. Clin Microbiol Infect. 2010;16:1614–1619. doi: 10.1111/j.1469-0691.2010.03311.x. [DOI] [PubMed] [Google Scholar]
- 9.Perez KK, Olsen RJ, Musick WL, et al. Integrating rapid diagnostics and antimicrobial stewardship improves outcomes in patients with antibiotic-resistant Gram-negative bacteremia. J Infect. 2014;69:216–225. doi: 10.1016/j.jinf.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 10.Vlek AL, Bonten MJ, Boel CH. Direct matrix-assisted laser desorption ionization/time-of-flight mass spectrometry improves appropriateness of antibiotic treatment of bacteremia. PLoS One. 2012;7 doi: 10.1371/journal.pone.0032589. e32589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar A, Roberts D, Wood KE, et al. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med. 2006;34:1589–1596. doi: 10.1097/01.CCM.0000217961.75225.E9. [DOI] [PubMed] [Google Scholar]
- 12.Idelevich EA, Grünastel B, Peters G, Becker K. Direct blood culturing on solid medium outperforms an automated continuously monitored broth-based blood culture system in terms of time to identification and susceptibility testing. New Microbes New Infect. 2016;10:19–24. doi: 10.1016/j.nmni.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banerjee R, Teng CB, Cunningham SA, et al. Randomized trial of rapid multiplex polymerase chain reaction-based blood culture identification and susceptibility testing. Clin Infect Dis. 2015;61:1071–1080. doi: 10.1093/cid/civ447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Idelevich EA, Becker K. Identification and susceptibility testing from shortly incubated cultures accelerate blood culture diagnostics at no cost. Clin Infect Dis. 2016;62:268–269. doi: 10.1093/cid/civ824. [DOI] [PubMed] [Google Scholar]
- 15.Schubert S, Weinert K, Wagner C, et al. Novel, improved sample preparation for rapid, direct identification from positive blood cultures using matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry. J Mol Diagn. 2011;13:701–706. doi: 10.1016/j.jmoldx.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martiny D, Dediste A, Vandenberg O. Comparison of an in-house method and the commercial Sepsityper™ kit for bacterial identification directly from positive blood culture broths by matrix-assisted laser desorption-ionisation time-of-flight mass spectrometry. Eur J Clin Microbiol Infect Dis. 2012;31:2269–2281. doi: 10.1007/s10096-012-1566-1. [DOI] [PubMed] [Google Scholar]
- 17.Kok J, Thomas LC, Olma T, Chen SC, Iredell JR. Identification of bacteria in blood culture broths using matrix-assisted laser desorption-ionization Sepsityper and time of flight mass spectrometry. PLoS One. 2011;6 doi: 10.1371/journal.pone.0023285. e23285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Idelevich EA, Grunewald CM, Wüllenweber J, Becker K. Rapid identification and susceptibility testing of Candida spp from positive blood cultures by combination of direct MALDI-TOF mass spectrometry and direct inoculation of Vitek 2. PLoS One. 2014;9 doi: 10.1371/journal.pone.0114834. e114834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lagacé-Wiens PR, Adam HJ, Karlowsky JA, et al. Identification of blood culture isolates directly from positive blood cultures by use of matrix-assisted laser desorption ionization-time of flight mass spectrometry and a commercial extraction system: analysis of performance, cost, and turnaround time. J Clin Microbiol. 2012;50:3324–3328. doi: 10.1128/JCM.01479-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Culbreath K, Petti CA. Balancing enthusiasm for innovative technologies with optimizing value: an approach to adopt new laboratory tests for infectious diseases using bloodstream infections as exemplar. Open Forum Infect Dis. 2015;2 doi: 10.1093/ofid/ofv075. ofv075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kohlmann R, Hoffmann A, Geis G, Gatermann S. MALDI-TOF mass spectrometry following short incubation on a solid medium is a valuable tool for rapid pathogen identification from positive blood cultures. Int J Med Microbiol. 2015;305:469–479. doi: 10.1016/j.ijmm.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 22.Verroken A, Defourny L, Lechgar L, Magnette A, Delmee M, Glupczynski Y. Reducing time to identification of positive blood cultures with MALDI-TOF MS analysis after a 5-h subculture. Eur J Clin Microbiol Infect Dis. 2015;34:405–413. doi: 10.1007/s10096-014-2242-4. [DOI] [PubMed] [Google Scholar]
- 23.Zabbe JB, Zanardo L, Megraud F, Bessede E. MALDI-TOF mass spectrometry for early identification of bacteria grown in blood culture bottles. J Microbiol Methods. 2015;115:45–46. doi: 10.1016/j.mimet.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 24.Köck R, Wüllenweber J, Horn D, Lanckohr C, Becker K, Idelevich EA. Implementation of short incubation MALDI-TOF MS identification from positive blood cultures in routine diagnostics and effects on empiric antimicrobial therapy. Antimicrob Resist Infect Control. 2017;6 doi: 10.1186/s13756-017-0173-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Idelevich EA, Schüle I, Grünastel B, Wüllenweber J, Peters G, Becker K. Acceleration of antimicrobial susceptibility testing of positive blood cultures by inoculation of Vitek 2 cards with briefly incubated solid medium cultures. J Clin Microbiol. 2014;52:4058–4062. doi: 10.1128/JCM.02400-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liesenfeld O, Lehman L, Hunfeld KP, Kost G. Molecular diagnosis of sepsis: New aspects and recent developments. Eur J Microbiol Immunol (Bp) 2014;4:1–25. doi: 10.1556/EuJMI.4.2014.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lehmann LE, Hunfeld KP, Emrich T, et al. A multiplex real-time PCR assay for rapid detection and differentiation of 25 bacterial and fungal pathogens from whole blood samples. Med Microbiol Immunol. 2008;197:313–324. doi: 10.1007/s00430-007-0063-0. [DOI] [PubMed] [Google Scholar]
- 28.Reers Y, Idelevich EA, Pätkau H, et al. Multiplex PCR assay underreports true bloodstream infections with coagulase-negative staphylococci in hematological patients with febrile neutropenia. Diagn Microbiol Infect Dis. 2016;85:413–415. doi: 10.1016/j.diagmicrobio.2016.04.024. [DOI] [PubMed] [Google Scholar]
- 29.Bloos F, Hinder F, Becker K, et al. A multicenter trial to compare blood culture with polymerase chain reaction in severe human sepsis. Intensive Care Med. 2010;36:241–247. doi: 10.1007/s00134-009-1705-z. [DOI] [PubMed] [Google Scholar]
- 30.von Lilienfeld-Toal M, Lehmann LE, Raadts AD, et al. Utility of a commercially available multiplex real-time PCR assay to detect bacterial and fungal pathogens in febrile neutropenia. J Clin Microbiol. 2009;47:2405–2410. doi: 10.1128/JCM.00491-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Westh H, Lisby G, Breysse F, et al. Multiplex real-time PCR and blood culture for identification of bloodstream pathogens in patients with suspected sepsis. Clin Microbiol Infect. 2009;15:544–551. doi: 10.1111/j.1469-0691.2009.02736.x. [DOI] [PubMed] [Google Scholar]
- 32.Bravo D, Blanquer J, Tormo M, et al. Diagnostic accuracy and potential clinical value of the LightCycler SeptiFast assay in the management of bloodstream infections occurring in neutropenic and critically ill patients. Int J Infect Dis. 2011;15:e326–e331. doi: 10.1016/j.ijid.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 33.Lehmann LE, Alvarez J, Hunfeld KP, et al. Potential clinical utility of polymerase chain reaction in microbiological testing for sepsis. Crit Care Med. 2009;37:3085–3090. doi: 10.1097/CCM.0b013e3181b033d7. [DOI] [PubMed] [Google Scholar]
- 34.Maubon D, Hamidfar-Roy R, Courby S, et al. Therapeutic impact and diagnostic performance of multiplex PCR in patients with malignancies and suspected sepsis. J Infect. 2010;61:335–342. doi: 10.1016/j.jinf.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 35.Grif K, Fille M, Würzner R, et al. Rapid detection of bloodstream pathogens by real-time PCR in patients with sepsis. Wien Klin Wochenschr. 2012;124:266–270. doi: 10.1007/s00508-012-0159-4. [DOI] [PubMed] [Google Scholar]
- 36.Lodes U, Bohmeier B, Lippert H, König B, Meyer F. PCR-based rapid sepsis diagnosis effectively guides clinical treatment in patients with new onset of SIRS. Langenbecks Arch Surg. 2012;397:447–455. doi: 10.1007/s00423-011-0870-z. [DOI] [PubMed] [Google Scholar]
- 37.Wallet F, Nseir S, Baumann L, et al. Preliminary clinical study using a multiplex real-time PCR test for the detection of bacterial and fungal DNA directly in blood. Clin Microbiol Infect. 2010;16:774–779. doi: 10.1111/j.1469-0691.2009.02940.x. [DOI] [PubMed] [Google Scholar]
- 38.Idelevich EA, Silling G, Niederbracht Y, et al. Impact of multiplex PCR on antimicrobial treatment in febrile neutropenia: a randomized controlled study. Med Microbiol Immunol. 2015;204:585–592. doi: 10.1007/s00430-014-0385-7. [DOI] [PubMed] [Google Scholar]
- 39.Tafelski S, Nachtigall I, Adam T, et al. Randomized controlled clinical trial evaluating multiplex polymerase chain reaction for pathogen identification and therapy adaptation in critical care patients with pulmonary or abdominal sepsis. J Int Med Res. 2015;43:364–377. doi: 10.1177/0300060514561135. [DOI] [PubMed] [Google Scholar]
- 40.Ling TK, Liu ZK, Cheng AF. Evaluation of the VITEK 2 system for rapid direct identification and susceptibility testing of gram-negative bacilli from positive blood cultures. J Clin Microbiol. 2003;41:4705–4707. doi: 10.1128/JCM.41.10.4705-4707.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E1.Singer M, Deutschman CS, Seymour CW, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3) JAMA. 2016;315:801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E2.O‘Neill J. Review on Antimicrobial Resistance. London: 2014. Review on Antimicrobial Resistance Antimicrobial Resistance: Tackling a crisis for the health and wealth of nations. [Google Scholar]
- E3.Saiki RK, Scharf S, Faloona F, et al. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985;230:1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- E4.Mullis KB, Faloona FA. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- E5.Krishna NK, Cunnion KM. Role of molecular diagnostics in the management of infectious disease emergencies. Med Clin North Am. 2012;96:1067–1078. doi: 10.1016/j.mcna.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E6.Sibley CD, Peirano G, Church DL. Molecular methods for pathogen and microbial community detection and characterization: current and potential application in diagnostic microbiology. Infect Genet Evol. 2012;12:505–521. doi: 10.1016/j.meegid.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E7.Jannes G, De Vos D. A review of current and future molecular diagnostic tests for use in the microbiology laboratory. Methods Mol Biol. 2006;345:1–21. doi: 10.1385/1-59745-143-6:1. [DOI] [PubMed] [Google Scholar]
- E8.Espy MJ, Uhl JR, Sloan LM, et al. Real-time PCR in clinical microbiology: applications for routine laboratory testing. Clin Microbiol Rev. 2006;19:165–256. doi: 10.1128/CMR.19.1.165-256.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E9.Charretier Y, Schrenzel J. Mass spectrometry methods for predicting antibiotic resistance. Proteomics Clin Appl. 2016;10:964–981. doi: 10.1002/prca.201600041. [DOI] [PubMed] [Google Scholar]
- E10.van Belkum A, Welker M, Erhard M, Chatellier S. Biomedical mass spectrometry in today‘s and tomorrow‘s clinical microbiology laboratories. J Clin Microbiol. 2012;50:1513–1517. doi: 10.1128/JCM.00420-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E11.Faron ML, Buchan BW, Ledeboer NA. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for use with positive blood cultures: methodology, performance, and optimization. J Clin Microbiol. 2017;55:3328–3338. doi: 10.1128/JCM.00868-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E12.Patel R. MALDI-TOF MS for the diagnosis of infectious diseases. Clin Chem. 2015;61:100–111. doi: 10.1373/clinchem.2014.221770. [DOI] [PubMed] [Google Scholar]
- E13.Bitter-Suermann D. Der Stellenwert der Polymerase-Kettenreaktion (PCR) für die klinische Diagnostik von Infektionskrankheiten. Dtsch Arztebl. 1993;90 A-3231. [Google Scholar]
- E14.Kwok S, Higuchi R. Avoiding false positives with PCR. Nature. 1989;339:237–238. doi: 10.1038/339237a0. [DOI] [PubMed] [Google Scholar]
- E15.Maurer JJ. Rapid detection and limitations of molecular techniques. Annu Rev Food Sci Technol. 2011;2:259–279. doi: 10.1146/annurev.food.080708.100730. [DOI] [PubMed] [Google Scholar]
- E16.Corless CE, Guiver M, Borrow R, Edwards-Jones V, Kaczmarski EB, Fox AJ. Contamination and sensitivity issues with a real-time universal 16S rRNA PCR. J Clin Microbiol. 2000;38:1747–1752. doi: 10.1128/jcm.38.5.1747-1752.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E17.Becker K, Harmsen D, Mellmann A, et al. Development and evaluation of a quality-controlled ribosomal sequence database for 16S ribosomal DNA-based identification of Staphylococcus species. J Clin Microbiol. 2004;42:4988–4995. doi: 10.1128/JCM.42.11.4988-4995.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E18.Klaschik S, Lehmann LE, Raadts A, Hoeft A, Stuber F. Comparison of different decontamination methods for reagents to detect low concentrations of bacterial 16S DNA by real-time-PCR. Mol Biotechnol. 2002;22:231–242. doi: 10.1385/MB:22:3:231. [DOI] [PubMed] [Google Scholar]
- E19.Mühl H, Kochem AJ, Disqué C, Sakka SG. Activity and DNA contamination of commercial polymerase chain reaction reagents for the universal 16S rDNA real-time polymerase chain reaction detection of bacterial pathogens in blood. Diagn Microbiol Infect Dis. 2010;66:41–49. doi: 10.1016/j.diagmicrobio.2008.07.011. [DOI] [PubMed] [Google Scholar]
- E20.Turaev D, Rattei T. High definition for systems biology of microbial communities: metagenomics gets genome-centric and strain-resolved. Curr Opin Biotechnol. 2016;39:174–181. doi: 10.1016/j.copbio.2016.04.011. [DOI] [PubMed] [Google Scholar]
- E21.Rossen JWA, Friedrich AW, Moran-Gilad J. ESCMID Study Group for Genomic and Molecular Diagnostics (ESGMD): Practical issues in implementing whole-genome-sequencing in routine diagnostic microbiology. Clin Microbiol Infect. 2018;24:355–360. doi: 10.1016/j.cmi.2017.11.001. [DOI] [PubMed] [Google Scholar]
- E22.Deurenberg RH, Bathoorn E, Chlebowicz MA, et al. Application of next generation sequencing in clinical microbiology and infection prevention. J Biotechnol. 2017;243:16–24. doi: 10.1016/j.jbiotec.2016.12.022. [DOI] [PubMed] [Google Scholar]
- E23.Anhalt JP, Fenselau C. Identification of bacteria using mass-spectrometry. Anal Chem. 1975;47:219–225. [Google Scholar]
- E24.Karas M, Bachmann D, Hillenkamp F. Influence of the wavelength in high-irradiance ultraviolet-laser desorption mass-spectrometry of organic molecules. Anal Chem. 1985;57:2935–2939. [Google Scholar]
- E25.Bizzini A, Greub G. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry, a revolution in clinical microbial identification. Clin Microbiol Infect. 2010;16:1614–1619. doi: 10.1111/j.1469-0691.2010.03311.x. [DOI] [PubMed] [Google Scholar]
- E26.Seng P, Drancourt M, Gouriet F, et al. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis. 2009;49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- E27.Weis S, Dickmann P, Pletz MW, Coldewey SM, Gerlach H, Bauer M. Sepsis 2017: Eine neue Definition führt zu neuen Konzepten. Dtsch Arztebl. 2017;114 A-1424. [Google Scholar]
- E28.Barenfanger J, Drake C, Kacich G. Clinical and financial benefits of rapid bacterial identification and antimicrobial susceptibility testing. J Clin Microbiol. 1999;37:1415–1418. doi: 10.1128/jcm.37.5.1415-1418.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E29.Trenholme GM, Kaplan RL, Karakusis PH, et al. Clinical impact of rapid identification and susceptibility testing of bacterial blood culture isolates. J Clin Microbiol. 1989;27:1342–1345. doi: 10.1128/jcm.27.6.1342-1345.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E30.Doern GV, Vautour R, Gaudet M, Levy B. Clinical impact of rapid in vitro susceptibility testing and bacterial identification. J Clin Microbiol. 1994;32:1757–1762. doi: 10.1128/jcm.32.7.1757-1762.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E31.Jurke A, Köck R, Becker K, et al. Reduction of the nosocomial meticillin-resistant Staphylococcus aureus incidence density by a region-wide search and follow-strategy in forty German hospitals of the EUREGIO, 2009 to 2011. Euro Surveill. 2013;18 doi: 10.2807/1560-7917.es2013.18.36.20579. pii=20579. [DOI] [PubMed] [Google Scholar]
- E32.Schaumburg F, Idelevich EA, Peters G, et al. Trends in antimicrobial non-susceptibility in methicillin-resistant Staphylococcus aureus from Germany (2004-2011) Clin Microbiol Infect. 2014;20:O554–O557. doi: 10.1111/1469-0691.12519. [DOI] [PubMed] [Google Scholar]
- E33.Meyer E, Schröder C, Gastmeier P, Geffers C. The reduction of nosocomial MRSA infection in Germany—an analysis of data from the Hospital Infection Surveillance System (KISS) between 2007 and 2012. Dtsch Arztebl Int. 2014;111:331–336. doi: 10.3238/arztebl.2014.0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E34.Tängdén T, Giske CG. Global dissemination of extensively drug-resistant carbapenemase-producing Enterobacteriaceae: clinical perspectives on detection, treatment and infection control. J Intern Med. 2015;277:501–512. doi: 10.1111/joim.12342. [DOI] [PubMed] [Google Scholar]
- E35.Walter J, Haller S, Blank HP, Eckmanns T, Abu Sin M, Hermes J. Incidence of invasive meticillin-resistant Staphylococcus aureus infections in Germany, 2010 to 2014. Euro Surveill. 2015;20(46) doi: 10.2807/1560-7917.ES.2015.20.46.30067. [DOI] [PubMed] [Google Scholar]
- E36.Roca I, Akova M, Baquero F, et al. The global threat of antimicrobial resistance: science for intervention. New Microbes New Infect. 2015;6:22–29. doi: 10.1016/j.nmni.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E37.Spellberg B, Bartlett J, Wunderink R, Gilbert DN. Novel approaches are needed to develop tomorrow‘s antibacterial therapies. Am J Respir Crit Care Med. 2015;191:135–140. doi: 10.1164/rccm.201410-1894OE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E38.Wilson ML, Weinstein MP, Reller LB. Automated blood culture systems. Clin Lab Med. 1994;14:149–169. [PubMed] [Google Scholar]
- E39.Altun O, Almuhayawi M, Ullberg M, Ozenci V. Clinical evaluation of the FilmArray blood culture identification panel in identification of bacteria and yeasts from positive blood culture bottles. J Clin Microbiol. 2013;51:4130–4136. doi: 10.1128/JCM.01835-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E40.Ledeboer NA, Lopansri BK, Dhiman N, et al. Identification of gram-negative bacteria and genetic resistance determinants from positive blood culture broths by use of the verigene gram-negative blood culture multiplex microarray-based molecular assay. J Clin Microbiol. 2015;53:2460–2472. doi: 10.1128/JCM.00581-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E41.Siu GK, Chen JH, Ng TK, et al. Performance evaluation of the verigene gram-positive and gram-negative blood culture test for direct identification of bacteria and their resistance determinants from positive blood cultures in Hong Kong. PLoS One. 2015;10 doi: 10.1371/journal.pone.0139728. e0139728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E42.Southern TR, VanSchooneveld TC, Bannister DL, et al. Implementation and performance of the BioFire FilmArray(R) Blood Culture Identification panel with antimicrobial treatment recommendations for bloodstream infections at a midwestern academic tertiary hospital. Diagn Microbiol Infect Dis. 2015;81:96–101. doi: 10.1016/j.diagmicrobio.2014.11.004. [DOI] [PubMed] [Google Scholar]
- E43.Bhatti MM, Boonlayangoor S, Beavis KG, Tesic V. Evaluation of FilmArray and Verigene systems for rapid identification of positive blood cultures. J Clin Microbiol. 2014;52:3433–3436. doi: 10.1128/JCM.01417-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E44.Ward C, Stocker K, Begum J, Wade P, Ebrahimsa U, Goldenberg SD. Performance evaluation of the Verigene(R) (Nanosphere) and FilmArray® (BioFire®) molecular assays for identification of causative organisms in bacterial bloodstream infections. Eur J Clin Microbiol Infect Dis. 2015;34:487–496. doi: 10.1007/s10096-014-2252-2. [DOI] [PubMed] [Google Scholar]
- E45.Davis TE, Fuller DD. Direct identification of bacterial isolates in blood cultures by using a DNA probe. J Clin Microbiol. 1991;29:2193–2196. doi: 10.1128/jcm.29.10.2193-2196.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E46.Peters RP, van Agtmael MA, Danner SA, Savelkoul PH, Vandenbroucke-Grauls CM. New developments in the diagnosis of bloodstream infections. Lancet Infect Dis. 2004;4:751–760. doi: 10.1016/S1473-3099(04)01205-8. [DOI] [PubMed] [Google Scholar]
- E47.Opota O, Croxatto A, Prod‘hom G, Greub G. Blood culture-based diagnosis of bacteraemia: state of the art. Clin Microbiol Infect. 2015;21:313–322. doi: 10.1016/j.cmi.2015.01.003. [DOI] [PubMed] [Google Scholar]
- E48.Opota O, Jaton K, Greub G. Microbial diagnosis of bloodstream infection: towards molecular diagnosis directly from blood. Clin Microbiol Infect. 2015;21:323–331. doi: 10.1016/j.cmi.2015.02.005. [DOI] [PubMed] [Google Scholar]
- E49.Lamoth F, Jaton K, Prod‘hom G, et al. Multiplex blood PCR in combination with blood cultures for improvement of microbiological documentation of infection in febrile neutropenia. J Clin Microbiol. 2010;48:3510–3516. doi: 10.1128/JCM.00147-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E50.Dierkes C, Ehrenstein B, Siebig S, Linde HJ, Reischl U, Salzberger B. Clinical impact of a commercially available multiplex PCR system for rapid detection of pathogens in patients with presumed sepsis. BMC Infect Dis. 2009;9 doi: 10.1186/1471-2334-9-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E51.Lucignano B, Ranno S, Liesenfeld O, et al. Multiplex PCR allows rapid and accurate diagnosis of bloodstream infections in newborns and children with suspected sepsis. J Clin Microbiol. 2011;49:2252–2258. doi: 10.1128/JCM.02460-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E52.Rath PM, Saner F, Paul A, et al. Multiplex PCR for rapid and improved diagnosis of bloodstream infections in liver transplant recipients. J Clin Microbiol. 2012;50:2069–2071. doi: 10.1128/JCM.00745-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E53.Avolio M, Diamante P, Zamparo S, et al. Molecular identification of bloodstream pathogens in patients presenting to the emergency department with suspected sepsis. Shock. 2010;34:27–30. doi: 10.1097/SHK.0b013e3181d49299. [DOI] [PubMed] [Google Scholar]
- E54.Sachse S, Straube E, Lehmann M, Bauer M, Russwurm S, Schmidt KH. Truncated human cytidylate-phosphate-deoxyguanylate-binding protein for improved nucleic acid amplification technique-based detection of bacterial species in human samples. J Clin Microbiol. 2009;47:1050–1057. doi: 10.1128/JCM.02242-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E55.Kerremans JJ, Goessens WH, Verbrugh HA, Vos MC. Accuracy of identification and susceptibility results by direct inoculation of Vitek 2 cards from positive BACTEC cultures. Eur J Clin Microbiol Infect Dis. 2004;23:892–898. doi: 10.1007/s10096-004-1247-9. [DOI] [PubMed] [Google Scholar]
- E56.Kostrzewa M, Sparbier K, Maier T, Schubert S. MALDI-TOF MS: an upcoming tool for rapid detection of antibiotic resistance in microorganisms. Proteomics Clin Appl. 2013;7:767–778. doi: 10.1002/prca.201300042. [DOI] [PubMed] [Google Scholar]
- E57.Sparbier K, Schubert S, Weller U, Boogen C, Kostrzewa M. Matrix-assisted laser desorption ionization-time of flight mass spectrometry-based functional assay for rapid detection of resistance against beta-lactam antibiotics. J Clin Microbiol. 2012;50:927–937. doi: 10.1128/JCM.05737-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E58.Sparbier K, Schubert S, Kostrzewa M. MBT-ASTRA: A suitable tool for fast antibiotic susceptibility testing? Methods. 2016;104:48–54. doi: 10.1016/j.ymeth.2016.01.008. [DOI] [PubMed] [Google Scholar]
- E59.Idelevich EA, Sparbier K, Kostrzewa M, Becker K. Rapid detection of antibiotic resistance by MALDI-TOF mass spectrometry using a novel direct-on-target microdroplet growth assay. Clin Microbiol Infect. 2018;24:738–743. doi: 10.1016/j.cmi.2017.10.016. [DOI] [PubMed] [Google Scholar]
- E60.Idelevich EA, Storck LM, Sparbier K, Drews O, Kostrzewa M, Becker K. Rapid direct susceptibility testing from positive blood cultures by the matrix-assisted laser desorption ionization-time of flight mass spectrometry-based direct-on-target microdroplet growth assay. J Clin Microbiol. 2018;56 doi: 10.1128/JCM.00913-18. pii: e00913-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E61.Rönnberg C, Mildh M, Ullberg M, Özenci V. Transport time for blood culture bottles: underlying factors and its consequences. Diagn Microbiol Infect Dis. 2013;76:286–290. doi: 10.1016/j.diagmicrobio.2013.03.031. [DOI] [PubMed] [Google Scholar]
- E62.Kerremans JJ, van der Bij AK, Goessens W, Verbrugh HA, Vos MC. Needle-to-incubator transport time: logistic factors influencing transport time for blood culture specimens. J Clin Microbiol. 2009;47:819–822. doi: 10.1128/JCM.01829-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E63.Bundesärztekammer. Neufassung der „Richtlinie der Bundesärztekammer zur Qualitätssicherung laboratoriumsmedizinischer Untersuchungen - Rili-BÄK“ - Richtlinie der Bundesärztekammer zur Qualitätssicherung laboratoriumsmedizinischer Untersuchungen. Dtsch Arztebl. 2014;111 A-1583. [Google Scholar]
- E64.Turenne CY, Tschetter L, Wolfe J, Kabani A. Necessity of quality-controlled 16S rRNA gene sequence databases: identifying nontuberculous Mycobacterium species. J Clin Microbiol. 2001;39:3637–3648. doi: 10.1128/JCM.39.10.3637-3648.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E65.Margulies M, Egholm M, Altman WE, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E66.Besser J, Carleton HA, Gerner-Smidt P, Lindsey RL, Trees E. Next-generation sequencing technologies and their application to the study and control of bacterial infections. Clin Microbiol Infect. 2018;24:335–341. doi: 10.1016/j.cmi.2017.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E67.Rhoads A, Au KF. PacBio Sequencing and its applications. Genomics Proteomics Bioinformatics. 2015;13:278–289. doi: 10.1016/j.gpb.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E68.Schadt EE, Turner S, Kasarskis A. A window into third-generation sequencing. Hum Mol Genet. 2010;19 doi: 10.1093/hmg/ddq416. R227-40. [DOI] [PubMed] [Google Scholar]
- E69.Stoddart D, Heron AJ, Mikhailova E, Maglia G, Bayley H. Single-nucleotide discrimination in immobilized DNA oligonucleotides with a biological nanopore. Proc Natl Acad Sci USA. 2009;106:7702–7707. doi: 10.1073/pnas.0901054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E70.Chin CS, Alexander DH, Marks P, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- E71.Wieser A, Schneider L, Jung J, Schubert S. MALDI-TOF MS in microbiological diagnostics-identification of microorganisms and beyond (mini review) Appl Microbiol Biotechnol. 2012;93:965–974. doi: 10.1007/s00253-011-3783-4. [DOI] [PubMed] [Google Scholar]
- E72.Colquhoun DR, Schwab KJ, Cole RN, Halden RU. Detection of norovirus capsid protein in authentic standards and in stool extracts by matrix-assisted laser desorption ionization and nanospray mass spectrometry. Appl Environ Microbiol. 2006;72:2749–2755. doi: 10.1128/AEM.72.4.2749-2755.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E73.Bernardo K, Fleer S, Pakulat N, Krut O, Hunger F, Krönke M. Identification of Staphylococcus aureus exotoxins by combined sodium dodecyl sulfate gel electrophoresis and matrix-assisted laser desorption/ ionization-time of flight mass spectrometry. Proteomics. 2002;2:740–746. doi: 10.1002/1615-9861(200206)2:6<740::AID-PROT740>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- E74.Bittar F, Ouchenane Z, Smati F, Raoult D, Rolain JM. MALDI-TOF-MS for rapid detection of staphylococcal Panton-Valentine leukocidin. Int J Antimicrob Agents. 2009;34:467–470. doi: 10.1016/j.ijantimicag.2009.03.017. [DOI] [PubMed] [Google Scholar]
- E75.Christner M, Rohde H, Wolters M, Sobottka I, Wegscheider K, Aepfelbacher M. Rapid identification of bacteria from positive blood culture bottles by use of matrix-assisted laser desorption-ionization time of flight mass spectrometry fingerprinting. J Clin Microbiol. 2010;48:1584–1591. doi: 10.1128/JCM.01831-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E76.Ochoa ML, Harrington PB. Immunomagnetic isolation of enterohemorrhagic Escherichia coli O157:H7 from ground beef and identification by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and database searches. Anal Chem. 2005;77:5258–5267. doi: 10.1021/ac0502596. [DOI] [PubMed] [Google Scholar]
- E77.La Scola B, Raoult D. Direct identification of bacteria in positive blood culture bottles by matrix-assisted laser desorption ionisation time-of-flight mass spectrometry. PLoS One. 2009;4 doi: 10.1371/journal.pone.0008041. e8041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E78.Ferreira L, Sanchez-Juanes F, González-Avila M, et al. Direct identification of urinary tract pathogens from urine samples by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2010;48:2110–2115. doi: 10.1128/JCM.02215-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E79.Burckhardt I, Zimmermann S. Using matrix-assisted laser desorption ionization-time of flight mass spectrometry to detect carbapenem resistance within 1 to 25 hours. J Clin Microbiol. 2011;49:3321–3324. doi: 10.1128/JCM.00287-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E80.Burckhardt I, Zimmermann S. Susceptibility testing of bacteria using MALDI-TOF mass spectrometry. Front Microbiol. 2018;9 doi: 10.3389/fmicb.2018.01744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E81.Sparbier K, Lange C, Jung J, Wieser A, Schubert S, Kostrzewa M. MALDI biotyper-based rapid resistance detection by stable-isotope labeling. J Clin Microbiol. 2013;51:3741–3748. doi: 10.1128/JCM.01536-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E82.Lange C, Schubert S, Jung J, Kostrzewa M, Sparbier K. Quantitative matrix-assisted laser desorption ionization-time of flight mass spectrometry for rapid resistance detection. J Clin Microbiol. 2014;52:4155–4162. doi: 10.1128/JCM.01872-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E83.Sanguinetti M, Posteraro B. Mass spectrometry applications in microbiology beyond microbe identification: progress and potential. Expert Rev Proteomics. 2016:;13:965–977. doi: 10.1080/14789450.2016.1231578. [DOI] [PubMed] [Google Scholar]
- E84.Vella A, De Carolis E, Mello E, et al. Potential use of MALDI-ToF mass spectrometry for rapid detection of antifungal resistance in the human pathogen Candida glabrata. Sci Rep. 2017;7 doi: 10.1038/s41598-017-09329-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E85.Miller JM, Binnicker MJ, Campbell S, et al. A Guide to Utilization of the microbiology laboratory for diagnosis of infectious diseases: 2018 update by the Infectious Diseases Society of America and the American Society for Microbiology. Clin Infect Dis. 2018;67:813–816. doi: 10.1093/cid/ciy584. [DOI] [PubMed] [Google Scholar]
- E86.Podbielski A, Abele-Horn M, Becker K, et al. Jena: Urban & Fischer/Elsevier; München: 2016. Mikrobiologisch-infektiologische Qualitätsstandards (MiQ) Qualitätsstandards in der mikrobiologischen-infektiologischen Diagnostik. Im Auftrag der Deutschen Gesellschaft für Hygiene und Mikrobiologie (DGHM) [Google Scholar]
- E87.Deutsches Institut für Normung/Normenausschuss Medizin. Beuth. Berlin: 1997. DIN 58959 „Qualitätsmanagement in der medizinischen Mikrobiologie“. [Google Scholar]
- E88.Arbeitsgemeinschaft Medizinische Laboratoriumsdiagnostik (AML), Zentralstelle der Länder für Gesundheitsschutz bei Medizinprodukten (ZLG) Paschke. Berlin: 1997. Handbuch für die Akkreditierung medizinischer Laboratorien. [Google Scholar]
- E89.Reinauer H. Editorial - GMS Zeitschrift zur Förderung der Qualitätssicherung in medizinischen Laboratorien. GMS Z Forder Qualitatssich Med Lab. 2009;1 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eSUPPLEMENT 1
Basic principles of DNA-based and mass spectrometry methods
Broad-spectrum nucleic acid diagnostics
If a patient’s medical history and clinical presentation suggest no clear initial diagnosis and conventional diagnostic procedures (culture, serology) do not attribute the infection event to the presence of one or more pathogenic microorganisms, the treating physician can request so-called “broad-spectrum” nucleic acid-based diagnostics, consisting of DNA amplification followed by sequencing or probe identification of the amplicons (figure 2). These NAT detection methods, also referred to as “group-specific,” “broad-range,” “eu-/panbacterial,” or “panfungal,” rest on the presence of certain genes or gene segments which, in at least parts of their nucleic acid sequence, have been conserved by “evolutionary pressure” and therefore display a high degree of homology among various species.
These gene sequences represent a certain gold standard in the definition of bacterial, fungal, and parasite species and their phylogenetic classification, and their high diagnostic potential can also be utilized for broad-spectrum pathogen detection. Some examples of systematically investigated “marker genes” are hsp65 (65-kDa heat-shock protein), rpoB (B subunit of RNA polymerase), and the genes for the ribosomal subunits (bacteria: 16S rRNA, 23S rRNA genes; fungi: 18S rRNA, 28S rRNA genes) or interposed sequence regions known as internal transcribed spacer (ITS) regions.
The ribosomal genes and ITS regions represent ideal target sequences for molecular biological species identification of cultured isolates and also—under certain experimental conditions—for direct detection from the clinical sample (2). The amplicons are subjected to automated DNA sequencing, and the nucleic acid sequences determined are compared with databases, whereby the quality of the database contents is decisive for the interpretation of the sequencing findings (e17, 64). The results are communicated either as clear concordance with reference sequences from a given species or, as observed more frequently in practice for the investigation of clinical specimens, a list of several phylogenetically related species in ascending order of sequence homology. This enables clear species attributions in many cases, sometimes with the aid of existing culture results or molecular genetic analysis of further cross-species target sequences.
Many in-house techniques have been developed for broad-spectrum nucleic acid diagnostics; to date, commercial procedures are available for detection of pathogens responsible for sepsis.
If the sample for investigation consists of normally sterile material (e.g., blood, but also cerebrospinal fluid, joint fluid, or tissue), amplicon sequence analysis usually yields clear species identification even in the case of fastidious or non-cultivable pathogens and previously unknown pathogens. This type of analysis fails, however, in the presence of (the DNA of) various microorganisms in the same sample, in which case overlapping of the different species-specific sequence signals hampers interpretation. The indication for these specialized investigations therefore depends on certain preconditions and constellations (box 2) (3).
The greatest problem in the technical implementation of broad-spectrum NAT techniques in the microbiology lab is the potential for contamination from the ubiquitous ambient bacterial and fungal DNA (in the air and on surfaces) and the physiological microbiotic flora of patients and staff (e16). Even reaction vessels and reagents employed for DNA isolation and amplification can become contaminated to a varying degree with bacterial or fungal DNA during production or use (e18). With sensitive broad-spectrum test systems, for example, DNA from Pseudomonas spp. can be demonstrated in many batches of aqueous reagents approved for PCR (e19).
Until a complete palette of “DNA-free” sample vessels and other reaction components are commercially available, together with closed systems for contamination-free processing and analysis, accreditable and thus reliable performance of broad-spectrum NAT investigations will necessarily be restricted to a small number of ready-made test kits from certain manufacturers (e.g., those producing sepsis PCR kits) and to specialized laboratories that conduct elaborate batch testing for individual reaction components and possess the necessary lab space and equipment.
Therefore, before ordering broad-spectrum NAT investigations the attending physician must always weigh the relatively high demands on technology, quality management, special reagents, and working time (and thus costs) against the anticipated benefit for the individual patient.
Whole-genome sequencing and whole-metagenome sequencing
The technical accomplishment of whole-genome sequencing (WGS) and whole-metagenome sequencing (WMS) has been made possible by so-called next-generation sequencing (NGS), so that these methods are well on the way to becoming routine (4, 5, e20). Their advantage lies in the potential for rapid (hours to a few days) simultaneous identification, genotyping, and determination of genetically coded traits of virulence and resistance. Even parallel analysis of the host genome is possible if the sample contains human DNA.
These procedures, starting with the pyrosequencing technology in 2005, permit complete presentation of the genetic information for individual pathogens (by WGS) or complete microbiota (by WMS) in a relatively short time (e65). Current NGS platforms yield a throughput of 0.1 to 6000 Gb, depending on the device used (for comparison: 0.0003 Gb for the conventional, so-called Sanger sequencing [e66]). However, the current reading lengths of around 50 to 400 bp for “short-read” techniques are still much lower than for the first-generation sequencing methods (Sanger), which achieved reading lengths of 0.5 to 1 kb. Massive parallel sequencing following amplification results in a large number of small portions which, after assembly, provide multiple coverage or reading depth of the genome. New “third-generation” sequencing methods (single-molecule real-time sequencing techniques, biological nanopore-based techniques), which have single DNA molecules as target and thus require no amplification, are capable of achieving DNA fragments of = 20 kb (e67– e69). Depending on procedure, particularly for the long-read techniques, the error rates may be relatively high (10 bis >30%), especially in complex areas of the genome with many repeats or phage insertions. For this reason, sufficiently high coverage and efficient algorithms for assembly and error correction are necessary to pinpoint errors and close gaps (e70).
WGS techniques therefore represent the ultimate tool for genotyping of microbial isolates, because they are capable of detecting individual nucleotide exchanges between isolates. Using these techniques, the transmission pathways of pathogens can be elucidated in detail and their dissemination in a health facility, region, or beyond can practically be traced. In the area of hospital hygiene, prospective WGS can document infection pathways continuously and in real time, and achieves this at a lower cost than measures to prevent transmission (6, e21, e22).
The routine use of these techniques is currently limited particularly by the costs, the quality of databases, insufficient standardization, lack of IT capacity, and problems of “big-data” management.
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry
Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (MS) enables marker-free analysis of large biomolecules by cocrystallization of matrix material and analyte (=sample) (e24). For diagnostic characterization, material from microbial pure cultures is applied to a plate together with the matrix and the resulting cocrystals are irradiated by a pulsed laser in full vacuum. The absorbed laser energy leads to ionization and release of ions that are then accelerated by an electrode in the electrical field generated, detected by an ion detector, and transformed into electrical signals. The time of flight of the pathogen-specific composite ions depends on their mass and charge. The measured values are compared with reference spectra, so the results are dependent of the size and quality of the databases used.
The culture-based areas of application for routine MALDI-TOF MS in diagnostic microbiology include especially identification of prokaryotic (bacteria, archaea) and eukaryotic pathogens (fungi, single-and multi-celled parasites) at species level, and sometimes at subspecies or even strain level (8, e25, e26, e71). Detection of specific viral proteins (e.g., norovirus capsid protein) by this means has also been reported (e72). Moreover, applications for detection of bacterial toxins, e.g., for Staphylococcus enterotoxins and Panton–Valentine leukocidin (PVL) have been described (e73, e74). Further reported uses are direct detection of microorganisms from positive blood cultures and from samples of urine and foodstuffs (18, e75– e78). An application of MALDI-TOF MS that is well on the way to routine use is identification of resistance in bacteria and fungi, or antibiotic susceptibility testing of clinical isolates (e56– e59, e79– e84).
eSUPPLEMENT 2
Pre-analytics, standardization, and quality assurance
Pre-analytic processes and factors
Particularly if add-on tests are used, costs can increase sharply, due to the test itself and/or increased personnel requirements. Not least for this reason, close attention should be paid to the necessary practical and administrative/organizational processes in the course of pre-analytics (e85).
These include determination of the indications, patient preparation, selection of the nature and number of samples for the add-on test systems, any diagnostic algorithms, transport and storage times (figure 2), and the organization of working times and diagnostic procedures in the laboratory. For example, excessively long transport and interim storage times or too small inoculates may lead to false-negative results in automated blood culture diagnostics (38, e61, e62).
Standardization and quality assurance
The quality requirements formulated at the beginning of the PCR era regarding standardization, reproducibility, and validity still hold true, not only for DNA-based methods: “Uncritical use of PCR procedures on unproven specimen types or the application of these methods under inappropriate conditions amplifies the potential for error and leads to diagnostic misinterpretations” (translated from the original German in [1]). Naturally the same requirements apply to MALDI-TOF MS-based techniques.
With the growing acceptance, widening use, and especially the ever-increasing variety of closed analysis systems, ready-prepared commercial test kits, and in-house protocols for nucleic acid detection of relevant pathogens, there is an increasing need for suitable external quality assurance measures. These should preferably take the shape of centrally organized inter-laboratory ring trials, but there could also be bilateral or multilateral exchange of well-characterized sample materials between laboratories.
While after successful accreditation and/or certification of a diagnostic facility only a well-organized workflow and reproducible, standardized implementation of precisely predefined protocols can be assured (lab-internal quality management), external quality control measures are aimed mainly at testing the sensitivity and specificity of NAT protocols and the assurance of precise and above all comparable results in diagnostic routine. Thus, internal quality management and external quality control are complementary.
Diagnostic institutions profit in more than one way from inter-laboratory ring trials or applying comparable quality assurance measures, as in this way they formally comply with the demands of certification and accreditation guidelines and also fulfill the requirements of the Richtlinie der Bundesärztekammer zur Qualitätssicherung laboratoriumsmedizinischer Untersuchungen (German Medical Association Guideline on Quality Assurance in Medical Laboratory Examinations; RiLiBÄK, part 3) (e63). Moreover, professional societies are also striving to achieve quality assurance and standardization of examination techniques, as exemplified by the microbiological/infectiological quality standards (MIQ) of the Deutsche Gesellschaft für Hygiene und Mikrobiologie (German Society for Hygiene and Microbiology; DGHM) (e86), e.g., MIQ 30, Qualitätsmanagement im medizinisch-mikrobiologischen Laboratorium (Quality Management in the Medical–Microbiological Laboratory); the norms of DIN series 58959, Qualitätsmanagement in der medizinischen Mikrobiologie (Quality Management in Medical Microbiology) (e87); and the Checkliste Mikrobiologie und Hygiene – Molekularbiologie in der Infektionsdiagnostik (Checklist Microbiology and Hygiene—Molecular Biology in the Diagnosis of Infections) in the Handbuch für die Akkreditierung Medizinischer Laboratorien (Handbook for the Accreditation of Medical Laboratories) (e88). All of these documents are published in German.
The Society for Promoting Quality Assurance in Medical Laboratories (Gesellschaft zur Förderung der Qualitätssicherung in medizinischen Laboratorien e.V.; INSTAND) has set up a German-language journal entitled GMS Zeitschrift zur Förderung der Qualitätssicherung in medizinischen Laboratorien (GMS Journal for Promoting Quality Assurance in Medical Laboratories) (www.egms.de/static/de /journals/index.htm) (e89). Here the participants in INSTAND ring trials receive reports of potential sources of error and further information on current issues in quality assurance.