Abstract
The endoplasmic reticulum (ER) represents the entry point into the secretory pathway where nascent proteins encounter a specialized environment for their folding and maturation. Inherent to these processes is a dedicated quality-control system that detects proteins that fail to mature properly and targets them for cytosolic degradation. An imbalance in protein folding and degradation can result in the accumulation of unfolded proteins in the ER, resulting in the activation of a signaling cascade that restores proper homeostasis in this organelle. The ER heat shock protein 70 (Hsp70) family member BiP is an ATP-dependent chaperone that plays a critical role in these processes. BiP interacts with specific ER-localized DnaJ family members (ERdjs), which stimulate BiP's ATP-dependent substrate interactions, with several ERdjs also binding directly to unfolded protein clients. Recent structural and biochemical studies have provided detailed insights into the allosteric regulation of client binding by BiP and have enhanced our understanding of how specific ERdjs enable BiP to perform its many functions in the ER. In this review, we discuss how BiP's functional cycle and interactions with ERdjs enable it to regulate protein homeostasis in the ER and ensure protein quality control.
Keywords: heat shock protein (HSP), endoplasmic reticulum (ER), endoplasmic reticulum–associated protein degradation (ERAD), GRP78, ATPase, molecular chaperone, protein misfolding, protein folding, stress response, unfolded protein response (UPR), BiP, ER-localized DnaJ proteins
Overview
One-third of the mammalian genome encodes proteins that will be synthesized by membrane-bound ribosomes and translocated into the endoplasmic reticulum (ER)3 where they will undergo post-translational modifications, oxidative folding, and maturation to their functional tertiary or quaternary state. These error-susceptible steps in protein biosynthesis are both aided and monitored by resident ER chaperones and their cofactors in a process termed ER quality control (1, 2). Two major chaperone families exist in the ER that interact with a wide variety of clients: the lectin chaperones, which generally recognize incompletely folded glycosylated proteins, and the Hsp70 family member, BiP, which can interact with both nonglycosylated and glycosylated proteins and is the focus of this review. In addition to playing a major role in chaperoning newly synthesized proteins, BiP is also responsible for maintaining the permeability barrier of the ER during protein translocation, targeting misfolded proteins for retrograde translocation so they can be degraded by the proteasome, contributing to ER calcium stores, and sensing conditions of stress in this organelle to activate the mammalian unfolded protein response (UPR). The binding of adenosine nucleotides by BiP plays a vital role in all of these activities except calcium binding, and its interaction with specific ER-localized DnaJ (ERdj) family members allows it to contribute to diverse cellular functions. Recent advances have been made in understanding how nucleotide-induced structural changes in BiP are transduced through the molecule to regulate client binding. Similarly, studies on a number of the ERdjs provide new insights into how BiP functions in the cell. In this review, we focus on how these new data shape our thinking of BiP's functional cycle allowing it to regulate ER homeostasis and protein quality control.
Nucleotide-bound state of BiP regulates its interaction with clients
Like all Hsp70 proteins (3, 4), BiP's interaction with clients is regulated by its nucleotide-bound state. It possesses a highly conserved N-terminal nucleotide-binding domain (NBD), a substrate-binding domain (SBD) composed of eight β-strands with a helical lid, and a linker between them that controls the allosteric interaction between these two domains. Experiments conducted over several decades by multiple groups have provided an understanding of key aspects of the Hsp70 ATPase cycle that are shared by family members present in organisms ranging from bacteria to humans (Fig. 1A, steps 1–3). In the ATP-bound form, the SBD, with its lid open, is docked on the NBD resulting in a high on/off rate for its interaction with clients. ATP hydrolysis results in a conformational change between the two domains that results in their undocking and the closing of the lid over the SBD providing a high-affinity state for the bound client with both slow binding and release rates. This cycle is further regulated by DnaJ proteins, which in some cases interact directly with unfolded proteins and transfer them to the ATP-bound form of an Hsp70, while simultaneously triggering ATP hydrolysis and closing the lid on the client (5, 6). Nucleotide-exchange factors release ADP allowing ATP to rebind, thus allowing the client to be released and folded or targeted for degradation (7).
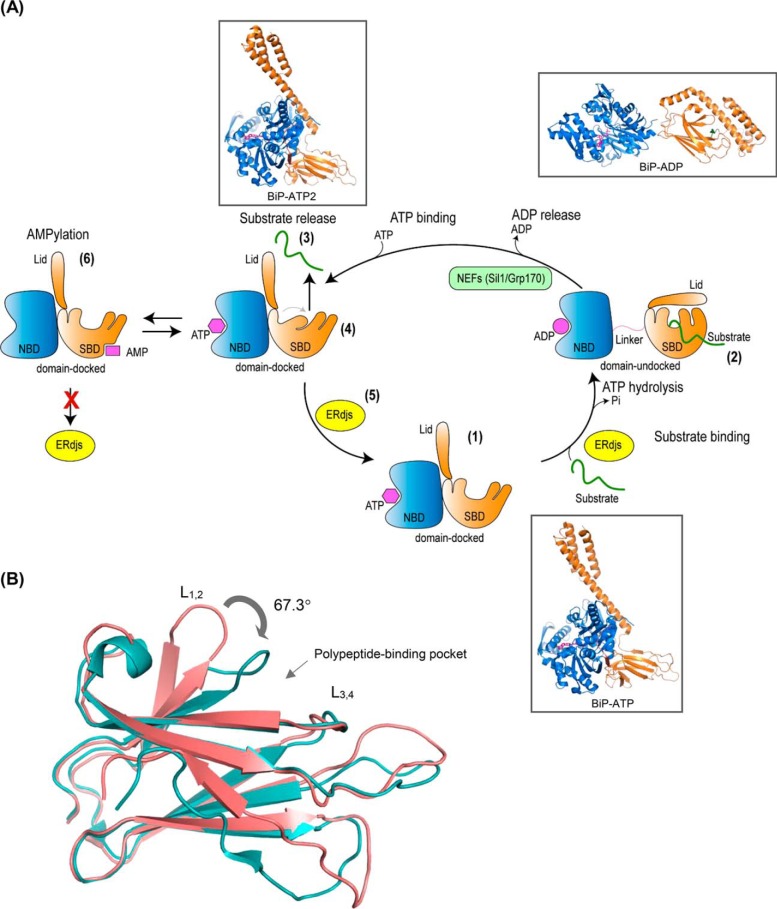
Figure 1.
A, BiP ATPase cycle. Step 1, in the ATP-bound form, the nucleotide-binding domain (NBD) (blue) and the substrate-binding domain (SBD) (orange), with its lid open, are docked to each other resulting to a form with high-substrate binding and release kinetics and low-substrate affinity. Step 2, upon ATP hydrolysis, the NBD and SBD become undocked, and the lid of the SBD closes providing a form that has high-substrate affinity but slow binding and release rates. This cycle is regulated by ER-localized DnaJ cofactors (ERdjs) that interact with unfolded proteins and transfer them to the ATP-bound form of BiP, while simultaneously triggering ATP hydrolysis. Step 3, substrate is released with the help of nucleotide-exchange factors (NEFs) that stimulate the release of ADP. Step 4, binding of ATP causes a conformational change in the SBD resulting in a more tightly compacted conformation that is thought to “squeeze” the substrate out. Step 5, interaction with ERdjs reorders the polypeptide-binding pocket of the BiP–ATP2 SBD, readying it to interact with another substrate. Step 6, BiP is post-translationally modified through AMPylation, and this causes the protein to be inactive. AMPylated BiP adopts a “domain-docked” structure similar to that of the ATP-bound state even in the apo- or ADP-bound state and is unable to interact productively with ERdjs. Ribbon representations of the structures (insets): ATP-bound BiP with the polypeptide-binding pocket open (BiP-ATP) (PDB 5e84 (8)); ADP-bound BiP from the structures of the isolated NBD (PDB 5evz (102)); SBD (PDB 5e85 (8)); and ATP-bound BiP with the polypeptide-binding pocket fully closed (BiP-ATP2) (PDB 6asy (8)). B, overlay of the SBD of BiP–ATP and BiP–ATP2. Comparison of SBDβ of BiP–ATP (deep salmon, PDB 5e84) and BiP–ATP2 (teal, PDB 6asy) with their superposition based on Cαs of β3–β7. The peptide-binding loops L1, 2 and L3, 4 are shown. The main difference of the two conformations is that the L1, 2 in BiP–ATP2 is rotated more than 60° compared with that of BiP–ATP resulting in a fully closed polypeptide-binding pocket.
Recent structural data and single molecule studies have provided new insights into the nucleotide-dependent, allosteric regulation of client binding and release by BiP; some that may be transferable to other Hsp70 (Fig. 1A, steps 4–5), as well as some that are clearly unique to the ER family member (Fig. 1A, step 6). Full-length structures of BiP with ATP bound were solved by crystallography (8) and NMR (9) utilizing T229A or T229G mutants, respectively, that were previously shown to have significantly reduced rates of ATP hydrolysis (10). Clear similarities were found with structures of Escherichia coli Hsp70, DnaK-ATP using a corresponding T199A mutant that was defective in ATP hydrolysis (11, 12), despite mammalian BiP having <50% homology to DnaK and possessing several different biochemical properties (13, 14). Because the ATP-bound forms of Hsp70 proteins are characterized by a “docked” SBD, these new structures also revealed a distinct NBD–SBD interface compared with DnaK. The interface in crystallized BiP was characterized by more hydrophobic interactions involving residues that are highly conserved in other eukaryotic Hsp70s irrespective of their resident organelle, but are missing in DnaK and mitochondrial Hsp70s (8). The “tighter docking” of the SBD to the NBD in this structure is consistent with a substantially slower interconversion between “docked” and “undocked” conformers of BiP in the NMR studies (9) compared with DnaK (15). The functional significance of this “tighter docking” of the SBD to NBD in eukaryotic Hsp70s will require more studies.
One additional BiP–ATP structure was obtained, this time with a WT NBD and only a small deletion in the loop between β-strands 3 and 4 (L3, 4) of the SBD, using a crystallization buffer containing high concentrations of phosphate to inhibit ATP hydrolysis (16). Importantly, this represents the first ATP–Hsp70 structure with a WT NBD. This BiP–ATP structure (BiP–ATP2) also has a docked SBD with an open lid, but in this case the L1, 2 loop has shifted 67.3° toward the β4 strand, occupying the peptide-binding pocket and making it more tightly compacted than either the previously described ATP- or ADP-bound forms (Fig. 1B). The authors suggest that in addition to opening the lid on the SBD, ATP binding can then serve to “squeeze” the bound client from the pocket of the SBD. These structures now suggest a much more active mechanism for client release than the two-state model that has long served as a paradigm for all Hsp70s. The identification of a second BiP–ATP structure is supported by the NMR data on the BiP hydrolysis mutant, which revealed two conformations (9), arguing for more flexibility in the ATP-bound state of BiP than observed for DnaK (9). Based on the high degree of homology and evidence for functional conservation in client binding and release, it is reasonable to posit that this three-state model could extend to some of the other eukaryotic Hsps.
A critical question in this scenario is whether or not this novel BiP–ATP2 form represents a functional intermediate in the cycle. To this end, Liu and co-workers (16) demonstrated that the binding of the J domain–containing protein ERdj3 was able to reopen the tightly compacted SBD of the BiP–ATP2 form for interaction with another client. The ability of ERdj3 to modulate the conformation of the SBD was also observed with a single-molecule study that examined the interaction of BiP with a protein client instead of a peptide. They found that binding of ERdj3 triggered a more expanded configuration of lid over the SBD, allowing it to accommodate the CH1 domain of an antibody heavy chain (14). ERdj3 remained stably associated with the ADP–BiP/client complex and prevented nucleotide-exchange factors from releasing BiP from the CH1 domain. This is reminiscent of the inability to detect BiP cycling from Ig heavy chains in vivo when ERdj3 is also bound to the heavy chain (17). Together, these studies revealed much more conformational variability in the SBD than previously detected.
Unlike other Hsp70 proteins, BiP is subject to a reversible, post-translational modification, which is rapidly removed when unfolded proteins accumulate and the demand for BiP increases (18–20). Through a combination of in vitro, cell culture-based, in vivo, and genetic studies conducted over the past decade, significant progress has been made in understanding this modification and the enzyme responsible. Whereas BiP was originally thought to be ADP-ribosylated under conditions of low demand, more recent studies in mammals (21–23) and flies (24) have demonstrated the modification to be AMPylation and the responsible enzyme to be FICD (filamentation induced by cAMP domain), which is also known as HYPE (Huntington yeast-interacting protein E). FICD attaches a single molecule of AMP to threonine 518 in loop L7, 8 in the SBD of BiP (22). This modification enhances peptide release from recombinant BiP and also prevents the stimulation of ATP hydrolysis by a J domain–containing protein. Structural data revealed that even when in the apo-form, which normally assumes an ADP-bound conformation, AMPylated BiP adopts a “domain-docked” structure that is more characteristic of the ATP-bound state and inhibits J domain–assisted transfer of a peptide client to BiP (9, 25). Together, these data provide insights into why modified BiP is not detected with clients (20) and further argue that modified BiP represents an inactive pool of BiP that can be readily reactivated by removing this modification when demand for BiP arises. A separate study by this group argues that FICD may also be capable of de-AMPylating BiP when unfolded proteins accumulate (23). However, the mechanism by which unfolded proteins convert FICD to a de-AMPylating enzyme remains unknown.
Diverse functions of BiP in the ER are assisted by DnaJ-like cofactors
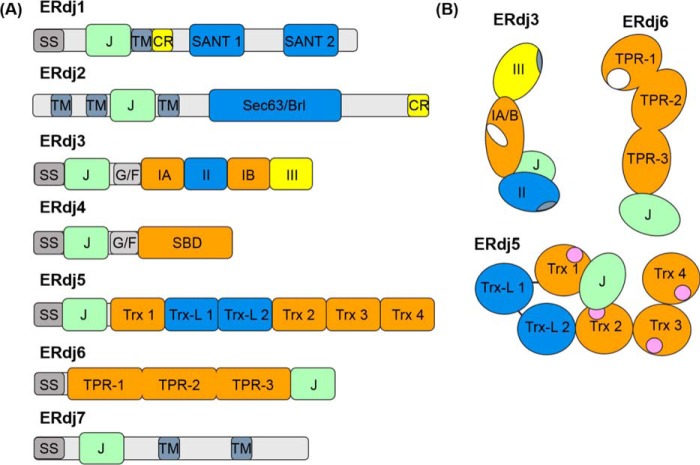
In the cell, BiP performs its functions with the assistance of ER-localized DnaJ cofactors (ERdjs). To date, seven DnaJ family members have been reported with the J domain oriented to the ER lumen where they can interact with BiP. Four of these (ERdj3–6) appear to interact directly with a variety of clients (Table 1 and Fig. 2). Our focus will be on these, but we will briefly discuss recent studies that shed light on the functions of the other three as well. DnaJ family members are divided into three subtypes based on their domain structure (26, 27). Type I (DNAJA) DnaJ proteins have all of the domains present in E. coli DnaJ, the founding member of this superfamily, including an N-terminal J domain, followed by a Gly/Phe-rich region that is thought to provide a flexible linker to the substrate-binding domain, which often includes a cysteine-rich zinc-binding domain, and which terminates with a dimerization domain. Type II (DNAJB) proteins possess all the same domains and in the same order as type I proteins, except that the cysteine-rich domain is missing. Both type I and II DnaJ family members can bind directly to clients. Type III (DNAJC) proteins form the largest group of DnaJs. This subtype only contains the J domain, which can occur anywhere in the protein. Some of the type III DnaJ proteins bind clients directly, whereas others do not appear to do so. The ER has DnaJ family members (ERdjs) representing all three subclasses, which were numbered according to their discovery. The past decade has provided a wealth of information on their structures, regulation, and client binding. Although ER functions are becoming available, we still lack clear insights into how the ERdjs compete for interaction with BiP to regulate its various functions.
Table 1.
Structural, functional, and substrate interaction properties of the ERdj family
The following abbreviations are used: M, membrane; L, lumen; NB, nonbinder of nascent secretory protein substrate; NA, not applicable.
| Protein size | DnaJ classification | ER location | Substrate-binding domain | In vivo sequence-binding profilea | Secretory protein processing role | |
|---|---|---|---|---|---|---|
| kDa | ||||||
| ERdj1 | 63.9 | Type III | M | NB | NA | Translation regulator |
| ERdj2 | 88.0 | Type III | M | NB | NA | Translocation regulator |
| ERdj3 | 40.5 | Type I | L | Domain I | Diverse sequences | Pro-folding or ERAD? |
| ERdj4 | 25.5 | Type II | L | Trx 1–4 | Aggregation-prone | ERAD, UPR regulator |
| ERdj5 | 91.0 | Type III | L | SBD | Aggregation-prone | Disulfide reductase, pro-folding, ERAD |
| ERdj6 | 57.6 | Type III | L | TPR-1 | Untested | Pro-folding? |
| ERdj7 | 42.4 | Type III | M | NB | NA | Unknown |
a In vivo binding profiles have been assessed with immunoglobulin substrates (62).
Figure 2.
Domain structure of ERdj family members. A, domain arrangements of primary sequences of each of the known ERdj proteins. Abbreviations used are as follows: SS, signal sequence; J, J domain; TM, transmembrane region; CR, charged amino acid region; SANT, SANT domain; Sec63/Brl, Sec63/Brl domain; G/F, glycine/phenylalanine-rich flexible linker region; IA and IB, bifurcated substrate-binding domain; II, cysteine-rich domain; III, dimerization domain; SBD, substrate-binding domain; Trx, thioredoxin domains; Trx-L, thioredoxin-like, enzymatically inactive domain; TPR, tetratricopeptide repeat domain (each containing three subdomains). B, cartoons representing orientation of the various domains of ERdj5 (72, 73) and ERdj6 (93, 94) based upon known structures. ERdj3 layout was derived from a combination of the solved structure of domain I–III of the yeast homologue Ydj1 (38, 39) and negative strain EM of ERdj3 showing the relative orientation of the J domain (43). The structures of ERdj1, -2, -4, and -7 are unsolved. The following functional regions are also indicated: ERdj3 and ERdj6 substrate-binding sites (white), ERdj3 dimerization/tetramerization regions within domains III and II respectively (blue-gray), and ERdj5's redox-active Trx motifs (pink).
ERdj1/DNAJC1 and ERdj2/DNAJC23
These two integral membrane DnaJ family members link translating polysomes to the Sec61 translocon and to the lumenal Hsp70 machinery via their J domains. ERdj1, a type III family member (Fig. 2A), was shown to bind to the tunnel exit site on the 80S ribosome through a positively charged cytosolic domain and serves to regulate translation in a BiP-dependent manner to ensure BiP is available to engage nascent polypeptide chains as they enter the ER (28, 29). Recent data demonstrate that the interaction of BiP with the J domain of ERdj1 in turn serves to increase the affinity of the cytosolic portion of ERdj1 for ribosomes (30). A cryo-EM structure of ribosomes bound to the ribosome-associated chaperone complex (RAC) has been solved (31), which reveals that RAC binds to the ribosome exit tunnel very close to where ERdj1 has been found to bind (29). This may indicate a competitive mechanism for the binding of cytosolic versus ER chaperones that could aid in discriminatory targeting the nascent polypeptide chain. In addition, a portion of the cytosolic region of ERdj1 contains two SANT–homology regions (28), although their functional significance has yet to be determined.
ERdj2 is also a subtype III protein that was originally identified in yeast as Sec63, a multipass transmembrane protein that is a component of the Sec61 translocon (32). A charged region at its C terminus allows it to interact with Sec62 (33), another translocon protein. Recent data argue that ERdj2, together with BiP and Sec62, serves to facilitate Sec61 channel opening, allowing precursors with weak signal sequences to be translocated (34) and to assist in the initial stages of cotranslational import of proteins into to the ER lumen (35).
ERdj3/DNAJB11
ERdj3 is a subtype I protein and the sole ER homologue of E. coli DnaJ. It was originally detected through its interaction with Shiga toxin, which enters the cell by moving backward through the secretory pathway (36), and with unassembled immunoglobulin heavy chains (37). It is widely expressed with the greatest levels in tissues with a high-secretory load, a pattern that mirrors that of BiP. The conservation with other type I family members like Saccharomyces cerevisiae Ydj1 (38, 39) is so strong that ERdj3 can readily be modeled (40), with the exception of domain II, which is smaller and stabilized by disulfide bonds instead of zinc ions (41). Although the type I DnaJ proteins that have been studied are thought to act as dimers, both endogenous and transfected ERdj3s (40.5 kDa) migrate as much larger species on nondenaturing gels and have particularly slow mobility in living cells (42). This is consistent with a recent negative stain EM analysis performed on purified recombinant ERdj3 that revealed it forms tetramers (43). In addition to the previously identified dimerization domain in domain III, a second intermolecular interaction site was observed in domain II (Fig. 2B). Mutational disruption of the second interaction site in domain II yielded dimers that were less able to bind clients in vivo. Because other type I DnaJ proteins are characterized by the presence of a cysteine-rich domain II, this begs the question of whether tetramer formation is unique to ERdj3 or whether this is a characteristic of other type I proteins.
There has been progress on understanding the mechanism of ERdj3 binding and release from clients. ERdj3 directly interacts with the substrate via highly conserved hydrophobic residues in the substrate-binding domain I in a process that is greatly enhanced by domain III dimerization (40). The binding to clients that fold rapidly in cells appears to be transient and terminated by transfer of the substrate to BiP, which depends on the interaction of the conserved QPD motif located in its J domain with BiP (44, 45). However, as noted under “Nucleotide-bound state of BiP regulates its interaction with clients,” this might be context/client-specific, as both in vitro assays (14) and studies in cultured cell assays (46) reveal that BiP and ERdj3 bind simultaneously and stably to the CH1 domain of the Ig heavy chain, which remains unstructured prior to assembly with Ig light chains (47). The release of a single arm of the dimer is sufficient to free ERdj3 from a client in cells (46). It is not currently clear how tetrameric forms will fit into this cycle.
Currently, there are conflicting reports as to whether ERdj3 serves a pro-folding or pro-degradation role in the ER. ERdj3's interaction with the Sec61 translocon (42, 48), through which nascent proteins enter the ER, could be compatible with a role in either stabilizing nascent proteins in a folding-competent conformation or recognizing proteins entering the ER that fail at the early stages in folding and must be degraded. Similarly, the relatively modest up-regulation of ERdj3 in response to ER stress is compatible with both functions (44). A few studies have been conducted on ERdj3's interaction with individual clients that provide some additional insights, but are also conflicting. For instance, ERdj3 binds stably to unassembled IgG γ1 heavy chains that have a half-life of over 12 h in plasmacytomas (37), which does not support a role in targeting this client for degradation. ERdj3-depleted cells have been used to determine the effects on disease-associated variants of α1-antitrypsin (α1-AT) (49) and glucocerebrosidase (50). The Z variant of α1-AT turned over faster, while the half-life of glucocerebrosidase was extended. Both mutant variants are glycoproteins and were redirected to lectin chaperones in ERdj3-depleted cells: calreticulin in the case of α1-AT and calnexin for glucocerebrosidase. Thus, neither study directly defines ERdj3's role but instead indicates that utilizing a lectin chaperone instead of the ER Hsp70 system can have diverse client-specific effects.
Intriguingly, unlike most soluble ER resident proteins, ERdj3 lacks a canonical ER retention motif, and a portion of it can be secreted when overexpressed or in association with unfolded proteins during ER stress (51). This secreted ERdj3 appears to maintain chaperone activity, as cell supernatants from overexpressing cells can inhibit the formation of extracellular aggregates and attenuate vacuole formation in cells expressing toxic prion proteins. Thus, it is conceivable that ERdj3's function both inside and outside the cell is to bind unfolded proteins and maintain them in a soluble state. Such a function is also consistent with a role in both folding nascent proteins and retrotranslocation of misfolded clients for degradation (52).
ERdj4/DNAJB9
ERdj4 is only subtype II DnaJ protein to be identified thus far in the ER of mammalian cells (53). It contains an N-terminal J domain that binds to BiP, followed by a glycine/phenylalanine-rich region, and a poorly defined client binding domain that has no structural homology to any other protein in databases. Despite an enigmatic structure, there has been progress in understanding ERdj4's function at the molecular, cellular, and organismal levels. Unlike ERdj3, its expression is particularly low in normal tissues (54) and cultured cells in the absence of ER stress (55). However, it is significantly induced at the transcriptional level by a variety of developmental schema, including early hematopoietic stem cell differentiation (56), angiogenesis (57), macrophage activation, TNFα-induced proinflammatory responses (58), and plasma cell differentiation (59). Many of these physiological processes are known to activate at least some elements of the ER stress response, which is consistent with the observation that ERdj4 is potently up-regulated by agents that disrupt ER homeostasis (53). In keeping with a role for ERdj4 in normal physiological processes, disruption of the ERdj4 locus by gene trap mutagenesis resulted in hypomorphic mice with poor perinatal viability (60). Surviving mice showed multiple metabolic defects and evidence of an elevated stress response in many tissues. Further studies revealed abnormal hematopoietic development in these mice (61), and enforced overexpression of ERdj4 in WT murine cord blood cells led to increased populations of functional hematopoietic stem cells (56). Analysis of the fitness of the surviving B cell precursors in the ERdj4 gene-trap mice revealed an inability of these mice to mount a T-dependent antibody response (61). However, it is noteworthy that T-independent responses were normal, arguing that ERdj4 does not play a critical role in antibody assembly or secretion, although both Ig heavy and light chains are natural clients (62).
Typical of type II DnaJ family members, it was anticipated that ERdj4 would bind directly to unfolded clients. The first evidence of this activity came through analysis of two disease-associated mutants of surfactant protein C (SP-C), which fail to fold in the ER and are retrotranslocated for rapid degradation by the proteasome (63). ERdj4 was shown to bind at significantly higher levels to the two mutant proteins than to WT SP-C, and siRNA-mediated decrease in ERdj4 expression led to a prolonged half-life for the mutant proteins, providing the first indication that ERdj4 might serve to support BiP's role in ER-associated degradation (ERAD). The effects of reduced or overexpressed ERdj4 on cellular pro-insulin (60) and epithelial sodium channel levels (64) are also consistent with it functioning in ERAD, as is its association with retrotranslocon components (55). Whether it serves as a functional part of the retrotranslocon or is actively being extracted with clients is not clear at this point. It is intriguing to speculate that the developmental defects observed upon ERdj4 loss are revealing the importance of a well-functioning ERAD system to these processes. Similar to ERdj3, ERdj4 also lacks an ER retention signal; however, these are no reports of ERdj4 being secreted or possessing extracellular activity.
A recent study indicates that UPR-transducer Ire1 may be an unexpected client of ERdj4 (65). BiP is known to bind Ire1 and to suppress its dimerization and activation during normal ER homeostasis (66), which begged the question of whether an ER-localized DnaJ protein recruited BiP to Ire1. Genetic disruption of both ERdj2 and ERdj4 in CHO cells led to constitutive Ire1 activation, although ERdj4 was more selective in its activation of Ire1 but not PERK (65). Conversely, an independent study found targeted disruption of ERdj2 in kidney tissue led to activation of Ire1 but not PERK (67). This raises the possibility of tissue-specific distinctions in the role of ERdj2 in regulating UPR transducers, but it is important to note that neither study directly examined ERdj2 binding to Ire1, which is complicated by the fact that unlike ERdj4, ERdj2 is an integral membrane protein. Exogenous expression of ERdj4 in the ΔERdj4 CHO cell line promoted BiP binding to an Ire1 construct, which depended on ERdj4 possessing a J domain capable of interacting with BiP (65). ERdj6 overexpression was unable to restore BiP binding to the Ire1 construct, demonstrating some specificity to ERdj requirements in regulating Ire1.
ERdj5/DNAJC10
ERdj5 is a subtype III DnaJ family member (68, 69) and has the unique property of also belonging to the PDI family of enzymes that serve to form, isomerize, or reduce disulfide bonds (70, 71). ERdj5 is ubiquitously expressed under normal physiological conditions and is only modestly induced by ER stress. In addition to possessing an N-terminal J domain, ERdj5 has six thioredoxin (Trx) domains, two of which are enzymatically inactive (Trx-like), and an ER retention sequence. Structural data show that the Trx domains are arranged in two clusters (72, 73). Each cluster possesses two Trx domains, but the N-terminal cluster also has the two catalytically inactive domains. Importantly, mutational inactivation of Trx domains in PDI, the founding member of this family, does not disrupt client binding or prevent their aggregation (74). Consistent with this property, ERdj5 can also bind to peptides lacking cysteines in vivo (62). The active sites of the four Trx domains were oriented in the same plane as the J domain in the first structure (72), likely situating them for interaction with clients bound to BiP. This structure also revealed a binding site for EDEM, a component of the ERAD machinery for glycosylated proteins, allowing ERdj5 to provide a physical lumenal link between ER quality control of BiP clients that are often unglycosylated and lectin clients. A second ERdj5 crystallization attempt produced a different three-dimensional arrangement of the N- and C-terminal clusters (73). This suggested more flexibility in the structure than had been anticipated, and single molecule studies revealed rapid movement between the two clusters. Mutants in which this flexibility was disrupted were less able to reduce disulfides in ERAD clients in vivo and aid in their degradation, arguing that the highly dynamic nature of ERdj5 is critical to its function and supports the involvement of multiple Trx domains in this activity.
Although all PDI family members can participate in disulfide relays, their redox equilibrium in the ER determines whether they are more likely to contribute to disulfide formation or reduction. The apparent equilibrium constant of recombinant ERdj5 was ∼100 times that of the ER redox state, making it the most reduced of the PDI family members and arguing that it acts as a reductase (75). In keeping with this, overexpression of ERdj5 in cells decreased the formation of mutant NHK α1-antitrypsin disulfide-linked dimers and inhibited the formation of mixed disulfide-bonded aggregates of the Ig J chain, confirming reductase activity in vivo. Subsequently, ERdj5 has been shown to enhance the degradation of SP-C (63) and rhodopsin mutants that are characterized by incorrect disulfide bond formation (76), substantiating a role for ERdj5 in reducing disulfide bonds in ERAD substrates that otherwise would hinder their passage through the retrotranslocon. In addition to participating in ERAD, ERdj5 is critical for the maturation of the low-density lipoprotein receptor, which requires the reduction of non-native disulfide bonds that are part of its normal folding trajectory (77).
There is mounting evidence that pathogens and toxins can hijack the ER quality control machinery to gain entrance to the cytosol where they exert their destructive capabilities (78). For instance, SV40 and BK PyV use the retrograde trafficking pathway to move from the cell surface to the ER and are then retrotranslocated to the cytosol using ERAD machinery where they establish an infection (79). ERdj5 was shown to cooperate with PDI to reduce pentameric VP1 into monomers that can be more readily delivered to the cytosol for the production of infectious particles (80). Similarly, ERdj5 assists in the retrotranslocation of the catalytic A chain of the cholera toxin to cytosol (81). The A chain is assembled from two subunits that are covalently linked via a disulfide bond that must be broken in the ER prior to retrotranslocation (82).
A critical element of ER function is the maintenance of calcium stores, which are essential to signaling cascades throughout the cell, as well as for the successful folding of proteins in this organelle. ER calcium levels are regulated by channels like the 1,4,5-trisphosphate and ryanodine receptors, which release calcium to the cytosol, and the sarco/endoplasmic reticulum Ca2+-ATPases (SERCA) that pump calcium back into the ER (83). The activity of SERCA2b, a ubiquitously expressed isoform, is negatively regulated by disulfide bond formation. At low calcium levels, ERdj5 reduces the regulatory bond thus activating SERCA2b. Conversely, high ER calcium levels induced ERdj5 oligomerization, which inhibited its reductase activity and thus its ability to regulate SERCA2b (84). These data provide an interesting link between ER redox, calcium levels, and protein homeostasis in this organelle. Given the importance of ER homeostasis to cellular and organismal functions, it is surprising that an ERdj5 knockout mouse was viable and showed no obvious defects other than a modest stress response in salivary glands (85). It is conceivable that either ERdj5 does not regulate other SERCA isoforms or that another PDI family member, like ERp57, which has also been shown to interact with SERCA2b (86), compensates.
ERdj6/DNAJC3
Like ERdj5, ERdj6 is a type III DnaJ protein that possesses additional functional domains not usually found in DnaJ family members. In this case, nine tandemly arranged tetratricopeptide repeat (TPR) motifs make up a significant portion of this protein (87). ERdj6 was originally identified as an inhibitor of the cytosolically localized, dsRNA-activated kinase (PKR) and was named p58IPK (88). Consequently, it was reported to also inhibit ER-localized PERK (89) and the cytosolic amino acid-regulated GCN2 (90) kinases, all of which target eIF2α to negatively regulate translation. As such, p58IPK was thought to be a cytosolically disposed, peripheral ER protein. The identification of an N-terminal ER-targeting signal sequence and protein orientation studies revealed that under normal physiological conditions p58IPK/ERdj6 is a lumenal ER resident protein that binds to BiP (91) and interacts with secretory pathway clients (92). However, ER stress reduces the efficiency of ERdj6 translocation into the ER leading to a portion of the protein accumulating in the cytosol (91). The cytosolic pool functions to modulate translation in response to cellular stresses, whereas in the ER, ERdj6 functions as a ubiquitously expressed co-chaperone of BiP.
ERdj6 is a monomer with a very elongated shape that positions the HPD motif of the J domain at the C terminus of the structure (93, 94). The TPR domain consists of three subdomains organized head-to-tail, and each subdomain contains three 34-amino acid TPR motifs. The sequence identity between the different TPR motifs is low despite a similar structural fold. TPR subdomain I has a binding site for unfolded recombinant rhodanese and luciferase, which is thought to localize to a conserved hydrophobic patch in this subdomain (93). Subdomains II and III are more hydrophilic, and the TPR6 motif in subdomain II is required for the inhibition of PKR suggesting this subdomain contains the binding site for PKR (95). Within the ER, ERdj6 can selectively bind misfolded, but not folded, vesicular stomatitis virus G protein and is released as the client folds, which depends on a functional interaction between ERdj6 and BiP (92). Only a few studies have been conducted to directly determine whether lumenal ERdj6 functions as a pro-folding or pro-degradation co-chaperone for BiP. For instance, the turnover of ectopically expressed TCRα chains in ERdj6 null cells was not affected, but a higher expression of this ERAD client during the pulse period led these investigators to suggest this reflected a role in co-translocational degradation due to the cytosolic form of ERdj6 recruiting Hsc70 (96). To address this possibility, another group engineered an ER-specific form of ERdj6 using the signal sequence of preprolactin, which is well-targeted even under stress conditions and a cytosolically expressed form that lacked the signal sequence (91). These investigators found that ectopic expression of the ER form, but not the cytosolic form, enhanced the levels of a secretory pathway client by assisting in translocation. Their studies further indicated that overexpression of the ER form of ERdj6 enhanced the proper maturation of one client, arguing for a possible role as a pro-folding co-chaperone. Clearly more isoform-specific analyses directed at understanding ERdj6's function in the ER as well as the cytosol are needed.
ERdj7/DNAJC25
ERdj7 was discovered in a proteomic study conducted on canine pancreatic rough microsomes where it was demonstrated to be an integral membrane protein with a lumenally-oriented J domain (97). Mass spectrometric analyses indicated that ERdj7 is the most abundant ERdj family member in these rough microsomes, although it does not appear to be induced by ER stress. Although rice and Arabidopsis only have homologues of four of the mammalian ERdjs, both organisms express ERdj7 (98). Despite its abundance and conservation across kingdoms, the role of ERdj7 in ER functions remains a mystery.
Toward understanding the BiP/ERdj network
The ERdjs have largely been studied in isolation, and it has not been determined whether the various ERdjs usually act temporally or how they might compete for clients or for interaction with BiP. ERdj1 and ERdj2 are components of the translocon, and current data support a role for these proteins in nascent protein entry into the ER. As such, they should represent the most upstream cofactors of BiP, although there is no evidence that either of these ERdjs bind directly to clients. Conversely, ERdj3–6 bind to clients once they are in the ER. Although studies designed to understand the functions of these ERdjs in folding versus ERAD are limited, overall, they seem to implicate ERdj4 and ERdj5 in recognition or targeting of clients for degradation. Based on ERdj5 having reductase activity and ERdj4 binding to retrotranslocon components, it is appealing to hypothesize that ERdj5 acts upstream of ERdj4 to reduce clients, so they can move more readily into the channel, and then transfer them to ERdj4. However, this type of temporal experiment is currently lacking. It is noteworthy that SP-C mutants bind to both of these co-chaperones, and their overexpression in combination accelerates degradation of the mutants (63).
An in vivo peptide expression study was undertaken to identify binding preferences for ERdj3, ERdj4, and ERdj5 on a truncated Ig γ heavy chain and a nonsecreted light chain that are natural clients of all three ERdjs (62). ERdj3 binds to frequent sites spread throughout both clients that overlapped with those that associated with BiP. This is reminiscent of in vitro peptide-binding assays conducted on DnaK, the E. coli Hsp70, and DnaJ, which reveal a similar binding preference for these two proteins (99). Based on much earlier studies to identify BiP sequence binding preferences (100), ERdj3 likely interacts with short sequences that are more hydrophobic in nature. Conversely, ERdj4 and ERdj5 recognized the same sites on each client, and these sites were longer than those BiP bound, occurred less frequently, and were even absent in the domain of quality control focus for the heavy chain. Unlike the BiP-binding sites, they were readily disrupted or introduced by mutation and could be identified with the TANGO algorithm, which predicts regions with a high potential for β aggregate formation (101). Although these sites represent a distinct sequence preference type for DnaJ co-chaperones, this study did not provide insights into how these ERdj proteins compete for interaction with clients or how these ERdjs specifically recognize aggregation-prone regions, as their client-binding sites have not been determined, and they have no sequence or structural homology to each other.
Summary
Recent progress has provided a new paradigm for the allosteric regulation of BiP's ATPase cycle: some aspects that are unique to BiP and some that are likely to be applicable to other Hsp70 proteins. We have also witnessed the identification of multiple new ER-localized DnaJ family members and a large number of studies that provide insights into the functions and structures of some of these proteins. It is clear that more studies are needed to understand whether the ERdjs that bind clients play dedicated roles in folding versus ERAD or whether this outcome is client-specific. It is also unclear whether the client-binding ERdjs have distinct client specificities or if they are largely indiscriminate in their client preferences. The finding that some clients can associate with multiple ERdjs begs the questions of how common this is, whether there is an order to these interactions, and how transfer from one type of ERdj protein to another is achieved. This is all the more puzzling, because the basal levels of the different ERdj proteins vary widely during normal ER homeostasis and after UPR activation. The combined use of structural, single molecule, cell biology, and model system methodologies is likely to provide answers to many of these questions in the coming decade.
This work was supported by the American Lebanese Syrian Associated Charities of St. Jude Children's Research Hospital and National Institutes of Health NIGMS Grant 5R01GM054068-20 (to L. M. H.). This is the third article in the JBC Reviews series “Molecular chaperones and protein quality control.” The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- ER
- endoplasmic reticulum
- BiP
- immunoglobulin-binding protein
- ERdj
- ER-localized DnaJ protein
- UPR
- unfolded protein response
- NBD
- nucleotide-binding domain
- SBD
- substrate-binding domain
- RAC
- ribosome-associated complex
- Ig
- immunoglobulin
- SP-C
- surfactant protein C
- ERAD
- ER-associated degradation
- PKR
- RNA-dependent protein kinase
- PERK
- PRK-like ER kinase
- PDI
- protein-disulfide isomerase
- Trx
- thioredoxin
- SERCA
- sarcoplasmic reticulum calcium ATPase
- PDB
- Protein Data Bank
- FICD
- filamentation induced by cAMP domain
- α1-AT
- α1-antitrypsin
- TPR
- tetratricopeptide repeat.
References
- 1. Ellgaard L., and Helenius A. (2003) Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 4, 181–191 10.1038/nrm1052 [DOI] [PubMed] [Google Scholar]
- 2. Braakman I., and Bulleid N. J. (2011) Protein folding and modification in the mammalian endoplasmic reticulum. Annu. Rev. Biochem. 80, 71–99 10.1146/annurev-biochem-062209-093836 [DOI] [PubMed] [Google Scholar]
- 3. Mayer M. P., and Kityk R. (2015) Insights into the molecular mechanism of allostery in Hsp70s. Front. Mol. Biosci. 2, 58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mayer M. P. (2018) Intra-molecular pathways of allosteric control in Hsp70s. Philos. Trans. R. Soc. Lond. B Biol. Sci. 373, 20170183 10.1098/rstb.2017.0183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Misselwitz B., Staeck O., and Rapoport T. A. (1998) J proteins catalytically activate Hsp70 molecules to trap a wide range of peptide sequences. Mol. Cell 2, 593–603 10.1016/S1097-2765(00)80158-6 [DOI] [PubMed] [Google Scholar]
- 6. Kityk R., Kopp J., and Mayer M. P. (2018) Molecular mechanism of J-domain-triggered ATP hydrolysis by Hsp70 chaperones. Mol. Cell 69, 227–237.e4 10.1016/j.molcel.2017.12.003 [DOI] [PubMed] [Google Scholar]
- 7. Behnke J., Feige M. J., and Hendershot L. M. (2015) Mechanisms of action and biological functions of BiP's nucleotide-exchange factors in the endoplasmic reticulum. J. Mol. Biol. 427, 1589–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yang J., Nune M., Zong Y., Zhou L., and Liu Q. (2015) Close and allosteric opening of the polypeptide-binding site in a human Hsp70 chaperone BiP. Structure 23, 2191–2203 10.1016/j.str.2015.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wieteska L., Shahidi S., and Zhuravleva A. (2017) Allosteric fine-tuning of the conformational equilibrium poises the chaperone BiP for post-translational regulation. Elife 6, e29430 10.7554/eLife.29430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wei J., Gaut J. R., and Hendershot L. M. (1995) In vitro dissociation of BiP:peptide complexes requires a conformational change in BiP after ATP binding but does not require ATP hydrolysis. J. Biol. Chem. 270, 26677–26682 10.1074/jbc.270.44.26677 [DOI] [PubMed] [Google Scholar]
- 11. Kityk R., Kopp J., Sinning I., and Mayer M. P. (2012) Structure and dynamics of the ATP-bound open conformation of Hsp70 chaperones. Mol. Cell 48, 863–874 10.1016/j.molcel.2012.09.023 [DOI] [PubMed] [Google Scholar]
- 12. Qi R., Sarbeng E. B., Liu Q., Le K. Q., Xu X., Xu H., Yang J., Wong J. L., Vorvis C., Hendrickson W. A., Zhou L., and Liu Q. (2013) Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP. Nat. Struct. Mol. Biol. 20, 900–907 10.1038/nsmb.2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bonomo J., Welsh J. P., Manthiram K., and Swartz J. R. (2010) Comparing the functional properties of the Hsp70 chaperones, DnaK and BiP. Biophys. Chem. 149, 58–66 10.1016/j.bpc.2010.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marcinowski M., Höller M., Feige M. J., Baerend D., Lamb D. C., and Buchner J. (2011) Substrate discrimination of the chaperone BiP by autonomous and cochaperone-regulated conformational transitions. Nat. Struct. Mol. Biol. 18, 150–158 10.1038/nsmb.1970 [DOI] [PubMed] [Google Scholar]
- 15. Zhuravleva A., Clerico E. M., and Gierasch L. M. (2012) An interdomain energetic tug-of-war creates the allosterically active state in Hsp70 molecular chaperones. Cell 151, 1296–1307 10.1016/j.cell.2012.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang J., Zong Y., Su J., Li H., Zhu H., Columbus L., Zhou L., and Liu Q. (2017) Conformation transitions of the polypeptide-binding pocket support an active substrate release from Hsp70s. Nat. Commun. 8, 1201 10.1038/s41467-017-01310-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vanhove M., Usherwood Y.-K., and Hendershot L. M. (2001) Unassembled Ig heavy chains do not cycle from BiP in vivo, but require light chains to trigger their release. Immunity 15, 105–114 10.1016/S1074-7613(01)00163-7 [DOI] [PubMed] [Google Scholar]
- 18. Carlsson L., and Lazarides E. (1983) ADP-ribosylation of the Mr 83,000 stress-inducible and glucose-regulated protein in avian and mammalian cells: modulation by heat shock and glucose starvation. Proc. Natl. Acad. Sci. U.S.A. 80, 4664–4668 10.1073/pnas.80.15.4664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hendershot L. M., Ting J., and Lee A. S. (1988) Identity of the immunoglobulin heavy-chain–binding protein with the 78,000-dalton glucose-regulated protein and the role of posttranslational modifications in its binding function. Mol. Cell. Biol. 8, 4250–4256 10.1128/MCB.8.10.4250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Freiden P. J., Gaut J. R., and Hendershot L. M. (1992) Interconversion of three differentially modified and assembled forms of BiP. EMBO J. 11, 63–70 10.1002/j.1460-2075.1992.tb05028.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sanyal A., Chen A. J., Nakayasu E. S., Lazar C. S., Zbornik E. A., Worby C. A., Koller A., and Mattoo S. (2015) A novel link between Fic (filamentation induced by cAMP)-mediated adenylylation/AMPylation and the unfolded protein response. J. Biol. Chem. 290, 8482–8499 10.1074/jbc.M114.618348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Preissler S., Rato C., Chen R., Antrobus R., Ding S., Fearnley I. M., and Ron D. (2015) AMPylation matches BiP activity to client protein load in the endoplasmic reticulum. Elife 4, e12621 10.7554/eLife.12621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Preissler S., Rato C., Perera L., Saudek V., and Ron D. (2017) FICD acts bifunctionally to AMPylate and de-AMPylate the endoplasmic reticulum chaperone BiP. Nat. Struct. Mol. Biol. 24, 23–29 10.1038/nsmb.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ham H., Woolery A. R., Tracy C., Stenesen D., Krämer H., and Orth K. (2014) Unfolded protein response-regulated Drosophila Fic (dFic) protein reversibly AMPylates BiP chaperone during endoplasmic reticulum homeostasis. J. Biol. Chem. 289, 36059–36069 10.1074/jbc.M114.612515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Preissler S., Rohland L., Yan Y., Chen R., Read R. J., and Ron D. (2017) AMPylation targets the rate-limiting step of BiP's ATPase cycle for its functional inactivation. Elife 6, e29428 10.7554/eLife.29428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cheetham M. E., and Caplan A. J. (1998) Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones 3, 28–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kampinga H. H., and Craig E. A. (2010) The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11, 579–592 10.1038/nrm2941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dudek J., Greiner M., Müller A., Hendershot L. M., Kopsch K., Nastainczyk W., and Zimmermann R. (2005) ERj1p has a basic role in protein biogenesis at the endoplasmic reticulum. Nat. Struct. Mol. Biol. 12, 1008–1014 10.1038/nsmb1007 [DOI] [PubMed] [Google Scholar]
- 29. Blau M., Mullapudi S., Becker T., Dudek J., Zimmermann R., Penczek P. A., and Beckmann R. (2005) ERj1p uses a universal ribosomal adaptor site to coordinate the 80S ribosome at the membrane. Nat. Struct. Mol. Biol. 12, 1015–1016 10.1038/nsmb998 [DOI] [PubMed] [Google Scholar]
- 30. Benedix J., Lajoie P., Jaiswal H., Burgard C., Greiner M., Zimmermann R., Rospert S., Snapp E. L., and Dudek J. (2010) BiP modulates the affinity of its co-chaperone ERj1 for ribosomes. J. Biol. Chem. 285, 36427–36433 10.1074/jbc.M110.143263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Leidig C., Bange G., Kopp J., Amlacher S., Aravind A., Wickles S., Witte G., Hurt E., Beckmann R., and Sinning I. (2013) Structural characterization of a eukaryotic chaperone–the ribosome-associated complex. Nat. Struct. Mol. Biol. 20, 23–28 10.1038/nsmb.2447 [DOI] [PubMed] [Google Scholar]
- 32. Brodsky J. L., and Schekman R. (1993) A Sec63p-BiP complex from yeast is required for protein translocation in a reconstituted proteoliposome. J. Cell Biol. 123, 1355–1363 10.1083/jcb.123.6.1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wittke S., Dünnwald M., and Johnsson N. (2000) Sec62p, a component of the endoplasmic reticulum protein translocation machinery, contains multiple binding sites for the Sec-complex. Mol. Biol. Cell 11, 3859–3871 10.1091/mbc.11.11.3859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hassdenteufel S., Johnson N., Paton A. W., Paton J. C., High S., and Zimmermann R. (2018) Chaperone-mediated Sec61 channel gating during ER import of small precursor proteins overcomes Sec61 inhibitor-reinforced energy barrier. Cell Rep. 23, 1373–1386 10.1016/j.celrep.2018.03.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lang S., Benedix J., Fedeles S. V., Schorr S., Schirra C., Schäuble N., Jalal C., Greiner M., Hassdenteufel S., Tatzelt J., Kreutzer B., Edelmann L., Krause E., Rettig J., Somlo S., et al. (2012) Different effects of Sec61α, Sec62, and Sec63 depletion on transport of polypeptides into the endoplasmic reticulum of mammalian cells. J. Cell Sci. 125, 1958–1969 10.1242/jcs.096727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yu M., Haslam R. H., and Haslam D. B. (2000) HEDJ, an Hsp40 co-chaperone localized to the endoplasmic reticulum of human cells. J. Biol. Chem. 275, 24984–24992 10.1074/jbc.M000739200 [DOI] [PubMed] [Google Scholar]
- 37. Meunier L., Usherwood Y. K., Chung K. T., and Hendershot L. M. (2002) A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol. Biol. Cell 13, 4456–4469 10.1091/mbc.e02-05-0311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li J., Qian X., and Sha B. (2003) The crystal structure of the yeast Hsp40 Ydj1 complexed with its peptide substrate. Structure 11, 1475–1483 10.1016/j.str.2003.10.012 [DOI] [PubMed] [Google Scholar]
- 39. Wu Y., Li J., Jin Z., Fu Z., and Sha B. (2005) The crystal structure of the C-terminal fragment of yeast Hsp40 Ydj1 reveals novel dimerization motif for Hsp40. J. Mol. Biol. 346, 1005–1011 10.1016/j.jmb.2004.12.040 [DOI] [PubMed] [Google Scholar]
- 40. Jin Y., Zhuang M., and Hendershot L. M. (2009) ERdj3, a luminal ER DnaJ homologue, binds directly to unfolded proteins in the mammalian ER: identification of critical residues. Biochemistry 48, 41–49 10.1021/bi8015923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Marcus N. Y., Marcus R. A., Schmidt B. Z., and Haslam D. B. (2007) Contribution of the HEDJ/ERdj3 cysteine-rich domain to substrate interactions. Arch. Biochem. Biophys. 468, 147–158 10.1016/j.abb.2007.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Guo F., and Snapp E. L. (2013) ERdj3 regulates BiP occupancy in living cells. J. Cell Sci. 126, 1429–1439 10.1242/jcs.118182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen K. C., Qu S., Chowdhury S., Noxon I. C., Schonhoft J. D., Plate L., Powers E. T., Kelly J. W., Lander G. C., and Wiseman R. L. (2017) The endoplasmic reticulum HSP40 co-chaperone ERdj3/DNAJB11 assembles and functions as a tetramer. EMBO J. 36, 2296–2309 10.15252/embj.201695616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shen Y., and Hendershot L. M. (2005) ERdj3, a stress-inducible endoplasmic reticulum DnaJ homologue, serves as a cofactor for BiP's interactions with unfolded substrates. Mol. Biol. Cell 16, 40–50 10.1091/mbc.e04-05-0434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jin Y., Awad W., Petrova K., and Hendershot L. M. (2008) Regulated release of ERdj3 from unfolded proteins by BiP. EMBO J. 27, 2873–2882 10.1038/emboj.2008.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Otero J. H., Lizák B., Feige M. J., and Hendershot L. M. (2014) Dissection of structural and functional requirements that underlie the interaction of ERdj3 protein with substrates in the endoplasmic reticulum. J. Biol. Chem. 289, 27504–27512 10.1074/jbc.M114.587147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Feige M. J., Groscurth S., Marcinowski M., Shimizu Y., Kessler H., Hendershot L. M., and Buchner J. (2009) An unfolded CH1 domain controls the assembly and secretion of IgG antibodies. Mol. Cell 34, 569–579 10.1016/j.molcel.2009.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dejgaard K., Theberge J. F., Heath-Engel H., Chevet E., Tremblay M. L., and Thomas D. Y. (2010) Organization of the Sec61 translocon, studied by high resolution native electrophoresis. J. Proteome Res. 9, 1763–1771 10.1021/pr900900x [DOI] [PubMed] [Google Scholar]
- 49. Khodayari N., Marek G., Lu Y., Krotova K., Wang R. L., and Brantly M. (2017) Erdj3 has an essential role for Z variant α1-antitrypsin degradation. J. Cell. Biochem. 118, 3090–3101 10.1002/jcb.26069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tan Y. L., Genereux J. C., Pankow S., Aerts J. M., Yates J. R. 3rd., and Kelly J. W. (2014) ERdj3 is an endoplasmic reticulum degradation factor for mutant glucocerebrosidase variants linked to Gaucher's disease. Chem. Biol. 21, 967–976 10.1016/j.chembiol.2014.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Genereux J. C., Qu S., Zhou M., Ryno L. M., Wang S., Shoulders M. D., Kaufman R. J., Lasmézas C. I., Kelly J. W., and Wiseman R. L. (2015) Unfolded protein response-induced ERdj3 secretion links ER stress to extracellular proteostasis. EMBO J. 34, 4–19 10.15252/embj.201488896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nishikawa S. I., Fewell S. W., Kato Y., Brodsky J. L., and Endo T. (2001) Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J. Cell Biol. 153, 1061–1070 10.1083/jcb.153.5.1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shen Y., Meunier L., and Hendershot L. M. (2002) Identification and characterization of a novel endoplasmic reticulum (ER) DnaJ homologue, which stimulates ATPase activity of BiP in vitro and is induced by ER stress. J. Biol. Chem. 277, 15947–15956 10.1074/jbc.M112214200 [DOI] [PubMed] [Google Scholar]
- 54. Weitzmann A., Baldes C., Dudek J., and Zimmermann R. (2007) The heat shock protein 70 molecular chaperone network in the pancreatic endoplasmic reticulum–a quantitative approach. FEBS J. 274, 5175–5187 10.1111/j.1742-4658.2007.06039.x [DOI] [PubMed] [Google Scholar]
- 55. Lai C. W., Otero J. H., Hendershot L. M., and Snapp E. (2012) ERdj4 protein is a soluble endoplasmic reticulum (ER) DnaJ family protein that interacts with ER-associated degradation machinery. J. Biol. Chem. 287, 7969–7978 10.1074/jbc.M111.311290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. van Galen P., Kreso A., Mbong N., Kent D. G., Fitzmaurice T., Chambers J. E., Xie S., Laurenti E., Hermans K., Eppert K., Marciniak S. J., Goodall J. C., Green A. R., Wouters B. G., Wienholds E., and Dick J. E. (2014) The unfolded protein response governs integrity of the haematopoietic stem-cell pool during stress. Nature 510, 268–272 10.1038/nature13228 [DOI] [PubMed] [Google Scholar]
- 57. Pröls F., Mayer M. P., Renner O., Czarnecki P. G., Ast M., Gässler C., Wilting J., Kurz H., and Christ B. (2001) Upregulation of the cochaperone Mdg1 in endothelial cells is induced by stress and during in vitro angiogenesis. Exp. Cell Res. 269, 42–53 10.1006/excr.2001.5294 [DOI] [PubMed] [Google Scholar]
- 58. Berger B. J., Müller T. S., Buschmann I. R., Peters K., Kirsch M., Christ B., and Pröls F. (2003) High levels of the molecular chaperone Mdg1/ERdj4 reflect the activation state of endothelial cells. Exp. Cell Res. 290, 82–92 10.1016/S0014-4827(03)00316-1 [DOI] [PubMed] [Google Scholar]
- 59. Lee A. H., Iwakoshi N. N., and Glimcher L. H. (2003) XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell. Biol. 23, 7448–7459 10.1128/MCB.23.21.7448-7459.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fritz J. M., Dong M., Apsley K. S., Martin E. P., Na C. L., Sitaraman S., and Weaver T. E. (2014) Deficiency of the BiP cochaperone ERdj4 causes constitutive endoplasmic reticulum stress and metabolic defects. Mol. Biol. Cell 25, 431–440 10.1091/mbc.e13-06-0319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Fritz J. M., and Weaver T. E. (2014) The BiP cochaperone ERdj4 is required for B cell development and function. PLoS ONE 9, e107473 10.1371/journal.pone.0107473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Behnke J., Mann M. J., Scruggs F. L., Feige M. J., and Hendershot L. M. (2016) Members of the Hsp70 family recognize distinct types of sequences to execute ER quality control. Mol. Cell 63, 739–752 10.1016/j.molcel.2016.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Dong M., Bridges J. P., Apsley K., Xu Y., and Weaver T. E. (2008) ERdj4 and ERdj5 are required for endoplasmic reticulum-associated protein degradation of misfolded surfactant protein C. Mol. Biol. Cell 19, 2620–2630 10.1091/mbc.e07-07-0674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Buck T. M., Kolb A. R., Boyd C. R., Kleyman T. R., and Brodsky J. L. (2010) The endoplasmic reticulum-associated degradation of the epithelial sodium channel requires a unique complement of molecular chaperones. Mol. Biol. Cell 21, 1047–1058 10.1091/mbc.e09-11-0944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Amin-Wetzel N., Saunders R. A., Kamphuis M. J., Rato C., Preissler S., Harding H. P., and Ron D. (2017) A J-Protein co-chaperone recruits BiP to monomerize IRE1 and repress the unfolded protein response. Cell 171, 1625–1637.e13 10.1016/j.cell.2017.10.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bertolotti A., Zhang Y., Hendershot L. M., Harding H. P., and Ron D. (2000) Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2, 326–332 10.1038/35014014 [DOI] [PubMed] [Google Scholar]
- 67. Fedeles S. V., So J. S., Shrikhande A., Lee S. H., Gallagher A. R., Barkauskas C. E., Somlo S., and Lee A. H. (2015) Sec63 and Xbp1 regulate IRE1α activity and polycystic disease severity. J. Clin. Invest. 125, 1955–1967 10.1172/JCI78863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Cunnea P. M., Miranda-Vizuete A., Bertoli G., Simmen T., Damdimopoulos A. E., Hermann S., Leinonen S., Huikko M. P., Gustafsson J. A., Sitia R., and Spyrou G. (2003) ERdj5, an endoplasmic reticulum (ER)-resident protein containing DnaJ and thioredoxin domains, is expressed in secretory cells or following ER stress. J. Biol. Chem. 278, 1059–1066 10.1074/jbc.M206995200 [DOI] [PubMed] [Google Scholar]
- 69. Hosoda A., Kimata Y., Tsuru A., and Kohno K. (2003) JPDI, a novel endoplasmic reticulum-resident protein containing both a BiP-interacting J-domain and thioredoxin-like motifs. J. Biol. Chem. 278, 2669–2676 10.1074/jbc.M208346200 [DOI] [PubMed] [Google Scholar]
- 70. Ellgaard L., and Ruddock L. W. (2005) The human protein disulphide isomerase family: substrate interactions and functional properties. EMBO Rep. 6, 28–32 10.1038/sj.embor.7400311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ellgaard L., Sevier C. S., and Bulleid N. J. (2018) How are proteins reduced in the endoplasmic reticulum? Trends Biochem. Sci. 43, 32–43 10.1016/j.tibs.2017.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hagiwara M., Maegawa K., Suzuki M., Ushioda R., Araki K., Matsumoto Y., Hoseki J., Nagata K., and Inaba K. (2011) Structural basis of an ERAD pathway mediated by the ER-resident protein disulfide reductase ERdj5. Mol. Cell 41, 432–444 10.1016/j.molcel.2011.01.021 [DOI] [PubMed] [Google Scholar]
- 73. Maegawa K. I., Watanabe S., Noi K., Okumura M., Amagai Y., Inoue M., Ushioda R., Nagata K., Ogura T., and Inaba K. (2017) The highly dynamic nature of ERdj5 is key to efficient elimination of aberrant protein oligomers through ER-associated degradation. Structure 25, 846–857.e4 10.1016/j.str.2017.04.001 [DOI] [PubMed] [Google Scholar]
- 74. Puig A., Lyles M. M., Noiva R., and Gilbert H. F. (1994) The role of the thiol/disulfide centers and peptide binding site in the chaperone and anti-chaperone activities of protein disulfide isomerase. J. Biol. Chem. 269, 19128–19135 [PubMed] [Google Scholar]
- 75. Ushioda R., Hoseki J., Araki K., Jansen G., Thomas D. Y., and Nagata K. (2008) ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science 321, 569–572 10.1126/science.1159293 [DOI] [PubMed] [Google Scholar]
- 76. Athanasiou D., Bevilacqua D., Aguila M., McCulley C., Kanuga N., Iwawaki T., Chapple J. P., and Cheetham M. E. (2014) The co-chaperone and reductase ERdj5 facilitates rod opsin biogenesis and quality control. Hum. Mol. Genet. 23, 6594–6606 10.1093/hmg/ddu385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Oka O. B., Pringle M. A., Schopp I. M., Braakman I., and Bulleid N. J. (2013) ERdj5 is the ER reductase that catalyzes the removal of non-native disulfides and correct folding of the LDL receptor. Mol. Cell 50, 793–804 10.1016/j.molcel.2013.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. He K., Ravindran M. S., and Tsai B. (2015) A bacterial toxin and a nonenveloped virus hijack ER-to-cytosol membrane translocation pathways to cause disease. Crit. Rev. Biochem. Mol. Biol. 50, 477–488 10.3109/10409238.2015.1085826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kartenbeck J., Stukenbrok H., and Helenius A. (1989) Endocytosis of simian virus 40 into the endoplasmic reticulum. J. Cell Biol. 109, 2721–2729 10.1083/jcb.109.6.2721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Inoue T., Dosey A., Herbstman J. F., Ravindran M. S., Skiniotis G., and Tsai B. (2015) ERdj5 reductase cooperates with protein disulfide isomerase to promote Simian virus 40 endoplasmic reticulum membrane translocation. J. Virol. 89, 8897–8908 10.1128/JVI.00941-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Williams J. M., Inoue T., Banks L., and Tsai B. (2013) The ERdj5-Sel1L complex facilitates cholera toxin retrotranslocation. Mol. Biol. Cell 24, 785–795 10.1091/mbc.e12-07-0522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Tsai B., Rodighiero C., Lencer W. I., and Rapoport T. A. (2001) Protein disulfide isomerase acts as a redox-dependent chaperone to unfold cholera toxin. Cell 104, 937–948 10.1016/S0092-8674(01)00289-6 [DOI] [PubMed] [Google Scholar]
- 83. Berridge M. J., Bootman M. D., and Roderick H. L. (2003) Calcium signalling: dynamics, homeostasis and remodelling. Nat. Rev. Mol. Cell Biol. 4, 517–529 10.1038/nrm1155 [DOI] [PubMed] [Google Scholar]
- 84. Ushioda R., Miyamoto A., Inoue M., Watanabe S., Okumura M., Maegawa K. I., Uegaki K., Fujii S., Fukuda Y., Umitsu M., Takagi J., Inaba K., Mikoshiba K., and Nagata K. (2016) Redox-assisted regulation of Ca2+ homeostasis in the endoplasmic reticulum by disulfide reductase ERdj5. Proc. Natl. Acad. Sci. U.S.A. 113, E6055–E6063 10.1073/pnas.1605818113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hosoda A., Tokuda M., Akai R., Kohno K., and Iwawaki T. (2009) Positive contribution of ERdj5/JPDI to endoplasmic reticulum protein quality control in the salivary gland. Biochem. J. 425, 117–125 [DOI] [PubMed] [Google Scholar]
- 86. Li Y., and Camacho P. (2004) Ca2+-dependent redox modulation of SERCA2b by ERp57. J. Cell Biol. 164, 35–46 10.1083/jcb.200307010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Lee T. G., Tang N., Thompson S., Miller J., and Katze M. G. (1994) The 58,000-dalton cellular inhibitor of the interferon-induced double-stranded RNA-activated protein kinase (PKR) is a member of the tetratricopeptide repeat family of proteins. Mol. Cell. Biol. 14, 2331–2342 10.1128/MCB.14.4.2331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Lee T. G., Tomita J., Hovanessian A. G., and Katze M. G. (1990) Purification and partial characterization of a cellular inhibitor of the interferon-induced protein kinase of Mr 68,000 from influenza virus-infected cells. Proc. Natl. Acad. Sci. U.S.A. 87, 6208–6212 10.1073/pnas.87.16.6208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Yan W., Frank C. L., Korth M. J., Sopher B. L., Novoa I., Ron D., and Katze M. G. (2002) Control of PERK eIF2α kinase activity by the endoplasmic reticulum stress-induced molecular chaperone P58IPK. Proc. Natl. Acad. Sci. U.S.A. 99, 15920–15925 10.1073/pnas.252341799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Roobol A., Roobol J., Bastide A., Knight J. R., Willis A. E., and Smales C. M. (2015) p58IPK is an inhibitor of the eIF2α kinase GCN2 and its localization and expression underpin protein synthesis and ER processing capacity. Biochem. J. 465, 213–225 10.1042/BJ20140852 [DOI] [PubMed] [Google Scholar]
- 91. Rutkowski D. T., Kang S. W., Goodman A. G., Garrison J. L., Taunton J., Katze M. G., Kaufman R. J., and Hegde R. S. (2007) The role of p58IPK in protecting the stressed endoplasmic reticulum. Mol. Biol. Cell 18, 3681–3691 10.1091/mbc.e07-03-0272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Petrova K., Oyadomari S., Hendershot L. M., and Ron D. (2008) Regulated association of misfolded endoplasmic reticulum lumenal proteins with P58/DNAJc3. EMBO J. 27, 2862–2872 10.1038/emboj.2008.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Tao J., Petrova K., Ron D., and Sha B. (2010) Crystal structure of P58(IPK) TPR fragment reveals the mechanism for its molecular chaperone activity in UPR. J. Mol. Biol. 397, 1307–1315 10.1016/j.jmb.2010.02.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Svärd M., Biterova E. I., Bourhis J. M., and Guy J. E. (2011) The crystal structure of the human co-chaperone P58(IPK). PLoS ONE 6, e22337 10.1371/journal.pone.0022337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Tang N. M., Ho C. Y., and Katze M. G. (1996) The 58-kDa cellular inhibitor of the double-stranded RNA-dependent protein kinase requires the tetratricopeptide repeat 6 and DnaJ motifs to stimulate protein synthesis in vivo. J. Biol. Chem. 271, 28660–28666 10.1074/jbc.271.45.28660 [DOI] [PubMed] [Google Scholar]
- 96. Oyadomari S., Yun C., Fisher E. A., Kreglinger N., Kreibich G., Oyadomari M., Harding H. P., Goodman A. G., Harant H., Garrison J. L., Taunton J., Katze M. G., and Ron D. (2006) Cotranslocational degradation protects the stressed endoplasmic reticulum from protein overload. Cell 126, 727–739 10.1016/j.cell.2006.06.051 [DOI] [PubMed] [Google Scholar]
- 97. Zahedi R. P., Völzing C., Schmitt A., Frien M., Jung M., Dudek J., Wortelkamp S., Sickmann A., and Zimmermann R. (2009) Analysis of the membrane proteome of canine pancreatic rough microsomes identifies a novel Hsp40, termed ERj7. Proteomics 9, 3463–3473 10.1002/pmic.200800722 [DOI] [PubMed] [Google Scholar]
- 98. Ohta M., and Takaiwa F. (2014) Emerging features of ER resident J-proteins in plants. Plant Signal. Behav. 9, e28194 10.4161/psb.28194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Rüdiger S., Schneider-Mergener J., and Bukau B. (2001) Its substrate specificity characterizes the DnaJ co-chaperone as a scanning factor for the DnaK chaperone. EMBO J. 20, 1042–1050 10.1093/emboj/20.5.1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Blond-Elguindi S., Cwirla S. E., Dower W. J., Lipshutz R. J., Sprang S. R., Sambrook J. F., and Gething M. J. (1993) Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell 75, 717–728 10.1016/0092-8674(93)90492-9 [DOI] [PubMed] [Google Scholar]
- 101. Fernandez-Escamilla A. M., Rousseau F., Schymkowitz J., and Serrano L. (2004) Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat. Biotechnol. 22, 1302–1306 10.1038/nbt1012 [DOI] [PubMed] [Google Scholar]
- 102. Hughes S. J., Antoshchenko T., Chen Y., Lu H., Pizarro J. C., and Park H. W. (2016) Probing the ATP Site of GRP78 with nucleotide triphosphate analogs. PLoS ONE 11, e0154862 10.1371/journal.pone.0154862 [DOI] [PMC free article] [PubMed] [Google Scholar]