Abstract
Ribosomal (r)-proteins are generally viewed as ubiquitous, constitutive proteins that simply function to maintain ribosome integrity. However, findings in the past decade have led to the idea that r-proteins have evolved specialized functions beyond the ribosome. For example, the 40S ribosomal protein uS5 (RPS2) is known to form an extraribosomal complex with the protein arginine methyltransferase PRMT3 that is conserved from fission yeast to humans. However, the full scope of uS5's extraribosomal functions, including whether uS5 interacts with any other proteins, is not known. In this study, we identify the conserved zinc finger protein 277 (ZNF277) as a new uS5-associated protein by using quantitative proteomics approaches in human cells. As previously shown for PRMT3, we found that ZNF277 uses a C2H2-type zinc finger domain to recognize uS5. Analysis of protein–protein interactions in living cells indicated that the ZNF277–uS5 complex is found in the cytoplasm and the nucleolus. Furthermore, we show that ZNF277 and PRMT3 compete for uS5 binding, because overexpression of PRMT3 inhibited the formation of the ZNF277–uS5 complex, whereas depletion of cellular ZNF277 resulted in increased levels of uS5-PRMT3. Notably, our results reveal that ZNF277 recognizes nascent uS5 in the course of mRNA translation, suggesting cotranslational assembly of the ZNF277–uS5 complex. Our findings thus unveil an intricate network of evolutionarily conserved protein–protein interactions involving extraribosomal uS5, suggesting a key role for uS5 beyond the ribosome.
Keywords: zinc finger, ribosome, proteomics, protein–protein interaction, protein complex, PRMT3, ribosomal protein, RPS2, uS5, ZNF277
Introduction
The ribosome is a large and complex molecular machine responsible for coordinating protein synthesis. In eukaryotes, the mature 80S ribosome is composed of two independent subunits: the small (40S) and large (60S) subunits that individually consist of an elaborated assortment of rRNA and ribosomal (r)-proteins.2 Specifically, the 40S subunit contains 33 r-proteins assembled around the 18S rRNA, whereas the 60S subunit is constituted of three different rRNAs (28S, 5.8S, and 5S) along with 46 r-proteins (1). Thanks to recent advances in protein structure determination approaches, unprecedented insights into how individual r-proteins and rRNAs are arranged into this exceptionally complex RNA–protein structure have been disclosed, helping to further our understanding of mRNA translation (2).
Production of a ribonucleoprotein particle of such complexity is one of the most energetically demanding process in eukaryotic cells, involving the activity of all three RNA polymerases (RNAPI, II, and III), as well as over 300 different proteins that function in ribosome assembly (3, 4). Throughout ribosome production, genes encoding r-proteins and maturation factors are transcribed by RNAPII, and the resulting mRNAs are exported out of the nucleus for translation in the cytoplasm; however, most of the translated protein products need to be imported back into the nucleus for assembly into pre-40S and pre-60S subunits (1, 3, 4). In parallel, as part of ribosomal subunit assembly in the nucleolus, transcription of rDNA loci by RNAPI produces a primary rRNA transcript (47S) that is cotranscriptionally processed into mature 28S, 18S, and 5.8S rRNAs via various endonucleolytic and exonucleolytic RNase machineries alongside hundreds of maturation factors and r-proteins (5, 6). Ultimately, 40S and 60S precursor particles are exported out of the nucleus via independent export pathways for final maturation steps in the cytoplasm (1).
During ribosome assembly, millions of nascent r-proteins need to be imported into the nucleus for incorporation into preribosomal particles (7). Although importin β-like receptors have been shown to be involved in r-protein nuclear import (8), the mechanistic details underlying the recognition of the 79 r-proteins by a few nuclear import receptors has remained poorly understood. Recently, it has been demonstrated that r-proteins use dedicated chaperones that are recruited in the cytoplasm to coordinate their folding, nuclear import, and incorporation into ribosomal subunit precursors (9–12). In Saccharomyces cerevisiae, Syo1 has been shown to mediate the coimport of r-protein L5 (uL18) and r-protein L11 (uL5) via the import receptor Kap104 (10, 13). Similarly, the Ankyrin-repeat protein Yar1 is cotranslationally recruited to nascent r-protein S3 (uS3) and protects uS3 from aggregation as well as escorts uS3 into the nucleus (10, 14, 15). The necessity of dedicated chaperones to escort nascent r-proteins appears to be evolutionarily conserved, as demonstrated by binding of Bcp1 and its human homolog, BCCIPβ, to r-protein L23 (uL14) in S. cerevisiae and human cells, respectively (16, 17). As yet, however, only a few dedicated r-protein chaperones have been identified, mostly in the budding yeast S. cerevisiae (18).
In addition to their role in the ribosome, r-proteins have been reported to form complexes outside the ribosome (19, 20). To date, most of the documented extraribosomal functions are involved in either autoregulation of r-protein production or surveillance of ribosome synthesis (20). In mammalian cells, several r-proteins, including uL5, uL11, and uL14, have been shown to associate with the murine double minute 2 (MDM2) protein, which can bind and promote p53 ubiquitination, ensuring rapid p53 degradation. Accordingly, growth conditions that affect ribosome assembly cause the accumulation of free uL5, uL11, and uL14, which interfere with p53 degradation by sequestering MDM2, resulting in cell cycle arrest or apoptosis (21–25). In addition, there are clear indications that implicate r-proteins in extraribosomal complexes, but for which the function remains elusive. This is the case for uS5, which has been shown to form a complex with the protein arginine methyltransferase 3 (PRMT3) in the fission yeast Schizosaccharomyces pombe and mammalian cells (26, 27). The existence of a similar complex in flies is also supported by results from a high-throughput two-hybrid screen, showing interactions between Drosophila homologs of PRMT3 (Art3) and uS5 (Sop) (28). Recently, we have reported that the extraribosomal PRMT3–uS5 complex can also recruit the programmed cell death protein 2 (PDCD2) and PDCD2-like (PDCD2L) proteins (29). Specifically, biochemical analysis revealed that PDCD2 and PDCD2L form distinct complexes with uS5-PRMT3, because PDCD2L was not detected in a PDCD2 purification, and reciprocally, PDCD2 was not identified as a PDCD2L-associated protein (29). Interestingly, the uS5-PDCD2 interaction has been also observed in Drosophila (30).
With the goal of getting a comprehensive view of the human uS5 protein interaction network, we used quantitative proteomics to identify additional uS5-associated proteins. Notably, we identified a conserved zinc finger protein, ZNF277, as a novel uS5-associated protein that directly interacts with uS5. Bimolecular fluorescence complementation assays revealed that the uS5–ZNF277 complex is located in the cytoplasm and the nucleolus. We also found that the most C-terminal zinc finger domain of ZNF277 is critical for a stable interaction with uS5. Importantly, our data indicate that ZNF277 is recruited cotranslationally by nascent uS5 and that ZNF277 and PRMT3 compete for uS5 binding in human cells. Our findings uncover ZNF277 as a novel protein associated with extraribosomal uS5, pointing to an emerging role for uS5 beyond the 40S ribosomal subunit.
Results
ZNF277 is a new uS5-interacting protein
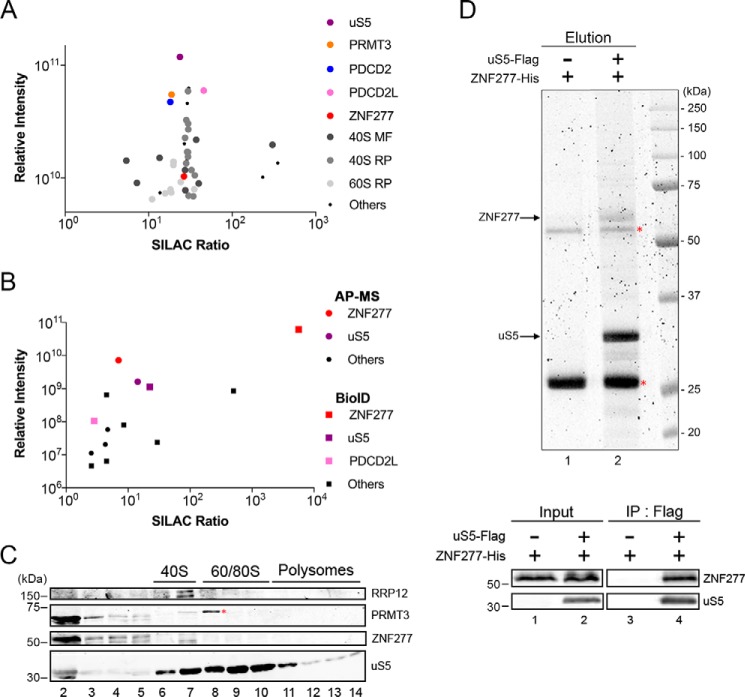
To get a comprehensive view of the protein interaction network of human uS5, we generated a HEK 293 cell line expressing a tetracycline-inducible GFP-tagged version of uS5 for affinity purification coupled to MS (AP–MS) analysis. Specific uS5-associated proteins were determined by a quantitative proteomics approach, stable isotope labeling by amino acids in cell culture (SILAC), that classifies interactions on the basis of specificity (ratio of peptide intensities between the GFP–uS5 pulldown and the control purification) and protein abundance, as estimated by the sum of peptide signal intensities of a given protein normalized to its molecular mass (31). In total, 282 proteins showed at least a 5-fold enrichment of the SILAC ratio in the GFP–uS5 purification relative to the control (Table S1). As expected for a component of the small ribosomal subunit, several r-proteins and 40S maturation factors were identified among the top 15% of uS5-associated proteins (Fig. 1A and Table S1). Proteins that were previously shown to form extraribosomal complexes with uS5, including PRMT3, PDCD2, and PDCD2L (29), were clearly enriched in the GFP–uS5 purification (Fig. 1A). Interestingly, a zinc finger protein, ZNF277, was also detected among the top 15% of uS5-copurifying proteins (Fig. 1A, red dot, and Table S1). ZNF277 is an evolutionarily conserved protein with clusters of C2H2-type zinc finger domains whose function remains unknown. Interestingly, microdeletions in ZNF277 were recently linked to an increased risk of language impairments (32). To validate the ZNF277–uS5 association and begin to explore the functional role of ZNF277 in human cells, we performed a reciprocal SILAC-based AP–MS analysis of ZNF277, as well as a complementary proximity-dependent biotinylation assay using BioID (33), also using SILAC-based quantitative proteomics. In total, we identified five and nine ZNF277-associated proteins using the AP–MS and BioID approaches, respectively (Tables S2 and S3). Importantly, as shown in Fig. 1B, uS5 was found to be the strongest ZNF277-interacting protein in both AP–MS and BioID analyses. PDCD2L was also identified in the BioID assay of ZNF277 but showed lower enrichment and abundance as compared with uS5 (Fig. 1B). As we have previously shown for the association between PRMT3 and PDCD2L (29), the copurification of PDCD2L with ZNF277 requires uS5 (Fig. S1), suggesting that uS5 bridges/stabilizes the association between ZNF277 and PDCD2L. Our results thus define ZNF277 as a new uS5-associated protein.
Figure 1.
Identification of a human uS5–ZNF277 complex. A, scatterplot showing results from GFP–uS5 AP–MS plotted by SILAC ratio on the x axis (peptide intensity originating from the GFP–uS5 purification versus the GFP control), which reflects specificity; and relative peptide intensity up the y axis (total peptide intensity for each protein), reflecting the relative abundance of each protein in the purification. Note that the y axis starts with intensity levels above 5 × 109 to emphasize the top 15% most abundant proteins. B, scatterplot showing results from GFP–ZNF277 AP–MS (circles) and BirA–ZNF277 BioID (square) assays coupled to SILAC-based quantifications, as described in Fig. 1A. C, Western blotting analysis of the indicated proteins using fractions of centrifuged sucrose gradients that were prepared using extracts of HEK 293 cells. The positions of the 40S, 60S/80S, and polysomes sedimentation are indicated on top. The red asterisk indicates the presence of a nonspecific protein detected using the PRMT3 antibody. D, top panel, SYPRO Ruby-stained SDS-PAGE of anti-FLAG precipitates prepared using extracts of E. coli that coexpressed ZNF277-His and uS5-FLAG (lane 2) or that expressed ZNF277-His alone (lane 1). The red asterisks indicate the position of the IgG heavy and light chains. Bottom panel, Western blotting analysis of total extracts (input, lanes 1 and 2) and anti-FLAG precipitates (IP, lanes 3–4) prepared from E. coli that coexpressed ZNF277 and uS5 (lanes 2 and 4) or that expressed ZNF277 alone (lanes 1 and 3).
We next addressed whether ZNF277 associated with free uS5 or 40S-incorporated uS5 by examining the distribution of ZNF277 after velocity sedimentation on sucrose gradients. As shown in Fig. 1C, the majority of ZNF277 was distributed in the low-density fractions (fractions 2–4), showing a similar distribution as PRMT3, which forms an extraribosomal complex with free uS5 (29). In contrast, 40S ribosomal subunits, 80S monosomes, and polysomes were detected in fractions 6–14, where ZNF277 was not detected (Fig. 1C). Importantly, detection of the 40S maturation factor RRP12 in fractions 6 and 7 (Fig. 1C) confirmed that our procedure could detect transient associations between a maturation factor and ribosomal subunit precursors. Together with the lack of additional r-proteins in affinity purification assays of ZNF277 (Fig. 1B), the sucrose gradient sedimentation analysis supports a model in which ZNF277 primarily associates with free uS5.
To examine whether uS5 interacts directly with ZNF277, we tested whether a uS5–ZNF277 complex could be reconstituted in vitro using recombinant versions of both proteins. Notably, coexpression of uS5 and ZNF277 in Escherichia coli was able to reconstitute a stable complex as demonstrated by the detection of uS5 and ZNF277 as the two main proteins recovered after affinity purification of uS5-FLAG under stringent conditions (Fig. 1D, lane 2). As a control, anti-FLAG affinity purification using extracts of E. coli that did not express uS5-FLAG did not recover ZNF277 (Fig. 1D, lane 1), despite the presence of ZNF277 in the extract (Fig. 1D, bottom panel, lanes 1 and 2). From these data, we conclude that ZNF277 directly interacts with uS5.
C2H2 zinc finger domains of ZNF277 are important for the association with uS5
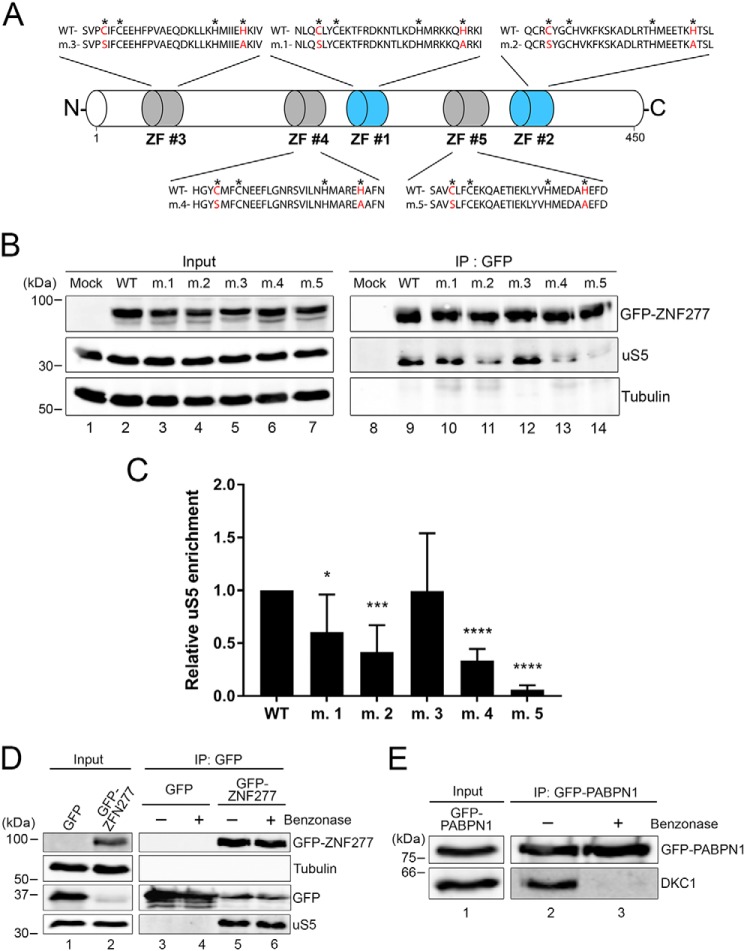
The single C2H2 zinc finger domain of PRMT3 is critical for binding to uS5 in both yeast and human cells (27, 34). Inspection of ZNF277 annotation using UniProt (35) indicated the presence of two typical C2H2 zinc finger domains with the consensus amino acid sequence pattern: C-X(2,4)-C-X(3)-[LIVMFYWC]-X(8)-H-X(3,5)-H (Fig. 2A, ZF#1 and ZF#2). However, further inspection of ZNF277 amino acid sequence across multiple eukaryotic species revealed the presence of three additional atypical C2H2 zinc finger domains that slightly deviate from the consensus pattern (Fig. 2A, ZF#3–ZF#5, and Fig. S2). To determine the functional relevance of the different ZNF277 zinc finger domains for uS5 association, we substituted the first cysteine for a serine and the last histidine for an alanine in each zinc finger domain, generating zinc finger mutants 1–5 (m.1–m.5; Fig. 2A). We next assessed the ability of WT and mutant versions of GFP–ZNF277 to associate with endogenous uS5 by coimmunoprecipitation assays. As shown in Fig. 2B, uS5 was efficiently recovered in anti-GFP precipitates prepared from extracts of cells that expressed the WT version of ZNF277 (see lane 9). In contrast, a control purification prepared from extracts of cells that expressed GFP alone did not copurify uS5 (Fig. 2B, lane 8). Analysis of uS5 recovery using ZNF277 zinc finger mutants indicated that the integrity of ZF#2, ZF#4, and ZF#5 was most important for association with uS5 (Fig. 2B, lanes 9–14). Notably, ZF#5 was found to be absolutely required for the stable association between uS5 and ZNF277 (see lane 14). Quantification of multiple coimmunoprecipitation experiments confirmed that the integrity of ZF#5 in ZNF277 was critical for uS5 binding (Fig. 2C). We conclude that, as for PRMT3, the integrity of a C2H2 zinc finger domain in ZNF277 is essential for stable association with uS5, suggesting a similar mode of uS5 recognition between PRMT3 and ZNF277.
Figure 2.
The integrity of zinc finger domains in ZNF277 is important for uS5 binding. A, schematic of ZNF277 primary structure showing its five zinc finger (ZF) domains. The typical C2H2 zinc fingers ZF#1 and ZF#2 are shown in blue, whereas atypical C2H2 zinc fingers ZF#3-ZF#5 are in gray (see also Fig. S2). Asterisks are present above the canonical cysteine and histidine residues of the zinc finger motif. Cysteine and histidine residues shown in red indicate the various substitutions introduced to generate ZNF277 variants m.1–m.5. B, Western blotting analysis of total extracts (lanes 1–7) and anti-GFP precipitates (lanes 8–14) prepared from HEK 293 cells that were transiently transfected for 48 h with the indicated versions of ZNF277. Mock refers to HEK 293 cells transfected with a GFP control plasmid. C, quantification of uS5 levels recovered in GFP immunoprecipitates normalized to the levels of GFP–ZNF277. The values were expressed relative to WT ZNF277, which were set to 1.0. The data and error bars represent the averages and standard deviation from six independent experiments. *, p ≤ 0.05; ***, p ≤ 0.001; ****, p ≤ 0.0001; Student's t test. D, Western blotting analysis of total extracts (lanes 1 and 2) and anti-GFP purifications (lanes 3–6) prepared from cells that stably expressed GFP (lanes 1, 3, and 4) and GFP–ZNF277 (lanes 2, 5, and 6). Extracts were treated (+, lanes 4 and 6) or not treated (−, lanes 3 and 5) with Benzonase before GFP immunoprecipitation. E, the RNA-dependent interaction between PABPN1 and Dyskerin (DKC1) was lost after the Benzonase treatment (compare lanes 2 and 3). IP, immunoprecipitation.
Because C2H2-type zinc fingers are capable of making interactions with DNA and RNA, we addressed whether the interaction between ZNF277 and uS5 depended on nucleic acids. We therefore pretreated total extracts with Benzonase, a robust nuclease that degrades all form of RNA and DNA, before affinity purification of ZNF277. Notably, pretreatment with Benzonase did not affect the interaction between ZNF277 and uS5 (Fig. 2D, compare lanes 5 and 6). As a control, the RNA-dependent interaction between PABPN1 and Dyskerin/DKC1 (36) was lost after the Benzonase treatment (Fig. 2E, compare lanes 2 and 3), confirming that the nuclease treatment effectively degraded nucleic acids present in the extract. These results thus support a direct protein–protein interaction between ZNF277 and uS5.
The ZNF277–uS5 complex is found in the cytoplasm and nucleolus
A proteome-wide analysis of subcellular localization in different human cell types indicates that ZNF277 is mainly localized to the nucleus with some cytoplasmic staining (37). Consistent with these results, a U-2 OS cell line stably expressing a GFP-tagged version of ZNF277 showed fluorescence signal in both the nucleus and the cytoplasm (Fig. 3A, panels c and d). Interestingly, the nuclear distribution of ZNF277 in U-2 OS cells showed concentrated signal reminiscent of nucleolar staining (Fig. 3A, panel d), which are sites of ribosome biogenesis. To test this possibility and further characterize the subcellular localization of ZNF277, the fluorescence analysis was combined with an immunostaining procedure for endogenous fibrillarin, which is a nucleolar marker protein. Comparison of the different fluorescence signal showed that a fraction of GFP–ZNF277 was concentrated in nuclear regions that colocalized with anti-fibrillarin staining (Fig. 3B, panels a–d). In contrast, U-2 OS cells that stably expressed GFP did not show nucleolar staining (Fig. 3A, panels a and b).
Figure 3.
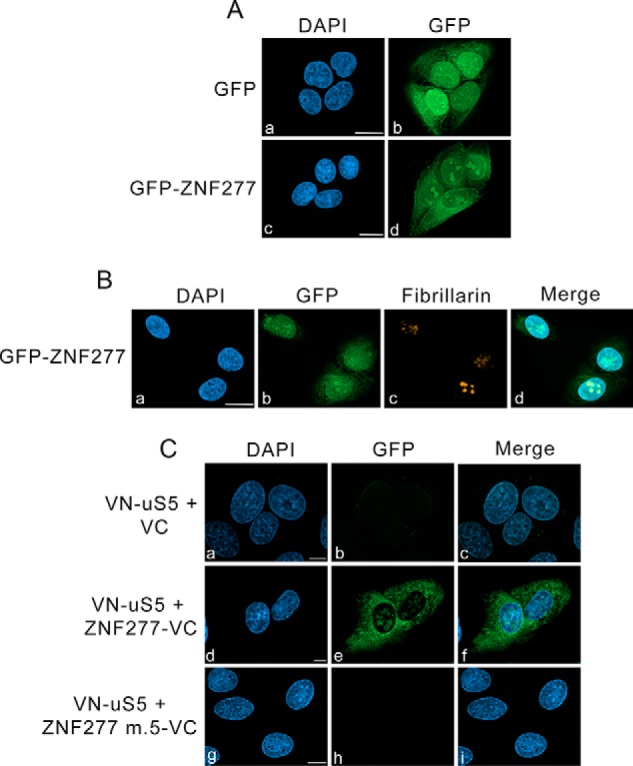
ZNF277–uS5 complexes localize to the cytoplasm and nucleolus. A, U-2 OS cells induced to expressed GFP (panels a and b) and GFP–ZNF277 (panels c and d) were fixed and analyzed by direct fluorescence. DNA staining with 4′,6-diamidino-2-phenylindole (DAPI) shows the nucleus of each cell (panels a and c). Bar, 10 μm. B, U-2 OS cells induced to express GFP–ZNF277 were simultaneously analyzed by direct GFP fluorescence (panel b) and immunostaining for the nucleolar marker fibrillarin (panel c). DNA staining with DAPI shows the nucleus of each cell (panel a). Bar, 10 μm. C, representative BiFC images showing interaction between VN-uS5 and ZNF277-VC in living human cells. U-2 OS cells that coexpressed VN-uS5 with either the VC control (panels a–c), WT ZNF277-VC (panels d–f), and ZF#5 mutant ZNF277-VC (panels g–i) were fixed and analyzed by direct fluorescence. DNA staining with DAPI shows the nucleus of each cell (panels a, d, and g). Bar, 10 μm.
We next used the bimolecular fluorescence complementation (BiFC) assay to address where in the cell the uS5 and ZNF277 physical interaction existed. BiFC consists of fusing two nonfluorescent fragments of the yellow fluorescent protein (YFP) to two proteins from a stable complex leading to restoration of fluorescence within a cell by reconstituting the split YFP, thereby providing indication about the cellular localization of a complex (38). We used an improved version of YFP named Venus (39) to create fusions with uS5 and ZNF277: the N-terminal fragment of Venus (VN) was fused to the N terminus of uS5, whereas the C-terminal fragment of Venus (VC) was fused to the C terminus of both WT and ZF#5 mutant versions of ZNF277. Next, the VN-uS5 and ZNF277-VC constructs were cotransfected in U-2 OS cells, and 24 h post-transfection, the cells were fixed and visualized by fluorescence microscopy. Consistent with our biochemical studies, uS5 and ZNF277 interacted with each other, resulting in a reconstituted Venus signal that showed nucleolar localization as well as a diffuse cytoplasmic distribution (Fig. 3C, panels d–f). In contrast, despite confirmation that each fusion protein was expressed (Fig. S3), combining fusions of uS5 and ZNF277 mutant ZF#5 produced background fluorescence (Fig. 3C, panels g–i), consistent with biochemical results indicating that a functional ZF#5 is required for ZNF277 to interact with uS5 (Fig. 2). Together, these results confirm a direct interaction between uS5 and ZNF277 that occurs in the cytoplasm and the nucleolus.
ZNF277 and PRMT3 compete for uS5 binding
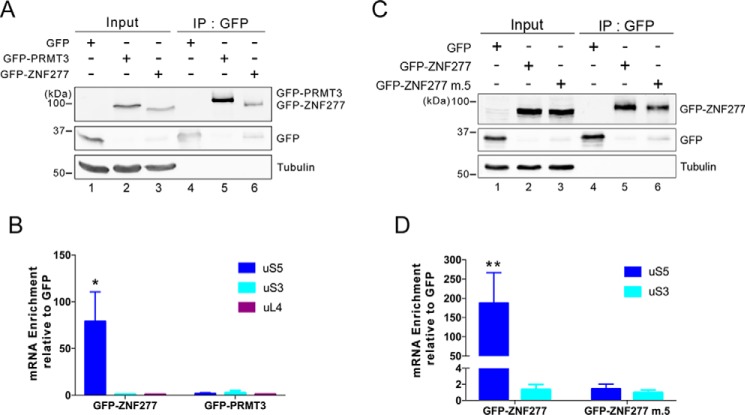
The observation that ZNF277 and PRMT3 both require a functional C2H2-type zinc finger domain to form a stable complex with extraribosomal uS5 suggested a similar mode of uS5 recognition by PRMT3 and ZNF277. Furthermore, it is noteworthy that PRMT3 was not detected in a ZNF277 purification (Fig. 1B), and reciprocally, ZNF277 was not identified as a PRMT3-associated protein (29). Collectively, these observations suggested a model in which PRMT3 and ZNF277 are mutually exclusive partners of uS5, using their C2H2 zinc finger domains to recognize a similar binding site on uS5. To test this possibility, we examined whether increased levels of PRMT3 impaired the formation of the uS5–ZNF277 complex in human cells. Notably, increased dosage of PRMT3 resulted in a marked decrease in the level of uS5 that was copurified with a GFP-tagged version of ZNF277 as compared with cells that overexpressed a control protein (Fig. 4A, compare lanes 4 and 5). To confirm that the impairment in ZNF277–uS5 complex formation was due to increased levels of PRMT3–uS5 complex, we used a version of PRMT3 that no longer binds to uS5 as a result of a substitution (cysteine 50 to serine) that disrupts the C2H2 zinc finger domain of PRMT3 (27, 34). Consistent with the idea that PRMT3 and ZNF277 compete for uS5 binding, overexpression of the PRMT3 C50S zinc finger mutant did not outcompete the formation of the ZNF277–uS5 complex (Fig. 4A, lane 6).
Figure 4.
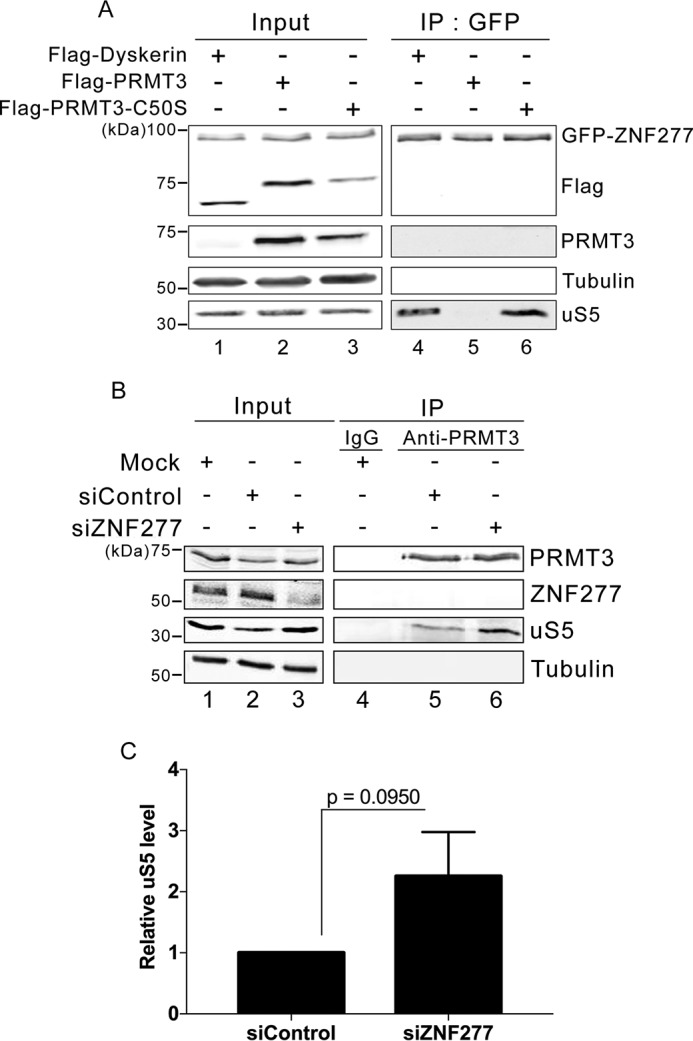
ZNF277 and PRMT3 compete for uS5 binding. A, Western blotting analysis of total extracts (lanes 1–3) and anti-GFP precipitates (lanes 4–6) prepared from a HEK 293 cell line that stably expresses GFP–ZNF277 and that were previously transfected with DNA constructs expressing FLAG–Dyskerin (lanes 1 and 4), FLAG–PRMT3 (lanes 2 and 5), and a zinc finger mutant (C50S) of FLAG–PRMT3 (lanes 3 and 6). B, Western blotting analysis of total extracts (lanes 1–3) and immunoprecipitates (lanes 4–6) prepared from HEK 293 cells that were previously treated with ZNF277-specific (lanes 3 and 6) and control nontarget (lanes 2 and 5) siRNAs. Control IgG (lane 4) and PRMT3-specific (lanes 5 and 6) antibodies were used for immunoprecipitations. C, quantification of uS5 levels recovered in anti-PRMT3 immunoprecipitates normalized to the levels of endogenous PRMT3. The values are expressed relative to cells treated with control siRNAs, which were set to 1.0. The data and error bars represent the averages and standard deviation from three independent experiments. The p value was calculated using a Student's t test.
Next, we assessed whether we could reciprocally affect the levels of endogenous PRMT3–uS5 complex by reducing the cellular abundance of ZNF277. For this, we depleted ZNF277 from human HEK 293 cells using siRNAs (Fig. 2B, compare lane 3 with lanes 1 and 2) and analyzed the level of uS5 that was copurified with endogenous PRMT3 after immunoprecipitation using a PRMT3-specific antibody. As shown in Fig. 4B, a deficiency in ZNF277 resulted in increased levels of uS5 in PRMT3 immunoprecipitates as compared with cells treated with a control siRNA (compare lane 6 to lane 5; see quantification in Fig. 4C). We conclude that PRMT3 and ZNF277 can compete for association with uS5 by forming mutually exclusive ZNF277–uS5 and PRMT3–uS5 complexes.
Normal uS5 levels are required for the cellular accumulation of ZNF277 and PRMT3
We noted that zinc finger mutants of ZNF277 that fail to associate with uS5 were expressed at lower levels than the WT protein (Fig. 2), as previously observed for PRMT3 (34). We therefore addressed whether free uS5 is needed for the proper accumulation of PRMT3 and ZNF277 in human cells. Using stable cell lines engineered to induce uS5-specific shRNAs after addition of doxycycline to the culture medium, we depleted roughly 50% of the total level of uS5 in a doxycycline-dependent manner (Fig. 5A, compare lanes 5 and 6 with lanes 2 and 3). Notably, depletion of uS5 using two independent clones that expressed different uS5-specific shRNAs resulted in a significant reduction of ZNF277 and PRMT3 protein accumulation (Fig. 5, A and B) but had no effect on the levels of PRMT3 and ZNF277 mRNAs (Fig. 5C). Interestingly, the effect of uS5 deficiency on PRMT3 and ZNF277 accumulation was not generalized to all uS5-associated proteins, because levels of PDCD2 remained unchanged after uS5 depletion (Fig. 5, A and B). Thus, loss of free uS5 caused the concomitant loss of PRMT3 and ZNF277, indicating that they need to be associated with uS5 for cellular accumulation.
Figure 5.
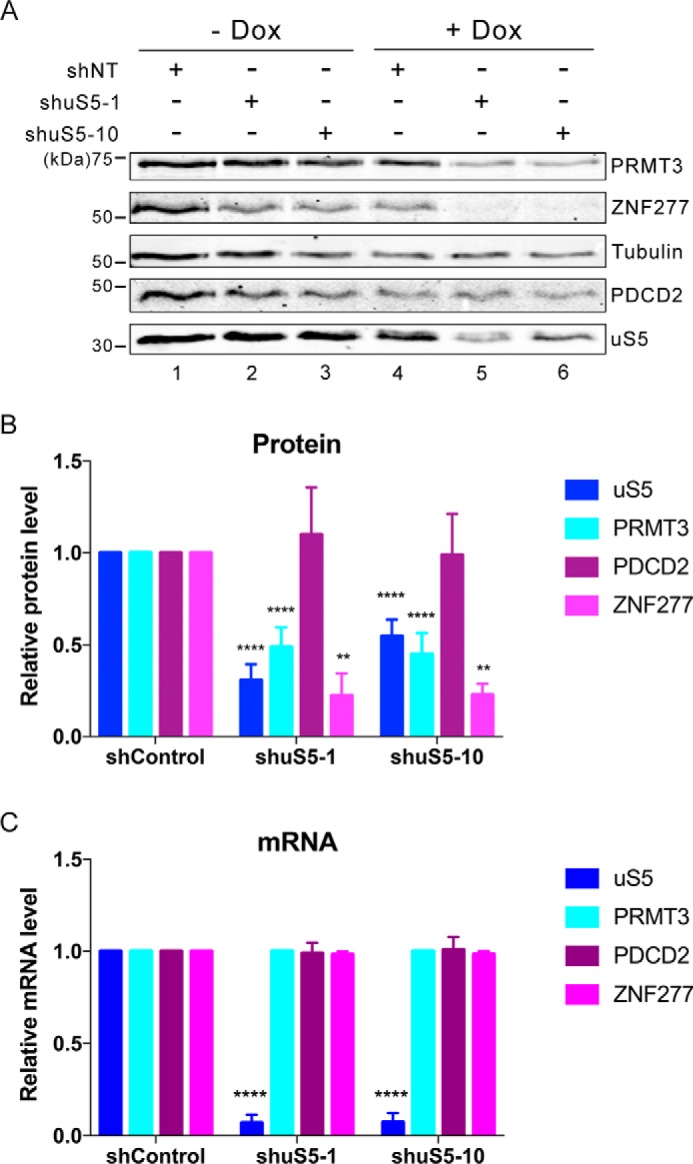
uS5 is required for the cellular accumulation of ZNF277 and PRMT3. A, Western blotting analysis of the indicated proteins using extracts prepared from HEK 293 cells that stably express inducible version of a control shRNA (lanes 1 and 4) and uS5-specific shRNAs (lanes 2, 3, 5, and 6). The cells were grown in the absence (lanes 1–3) or presence (lanes 4–6) of doxycycline (Dox) to induce the expression of the different shRNAs. B and C, quantification of protein (B) and mRNA (C) levels for the indicated genes as determined by immunoblotting (B) and RT-qPCR (C), respectively, using extracts of control (shControl) and uS5-deficient (shuS5-1 and shuS5-10) cells cultured in the presence of doxycycline. Protein levels (B) were normalized to tubulin and expressed relative to shControl cells, whereas mRNA levels (C) were normalized to the GAPDH mRNA and expressed relative shControl cells. The data and error bars represent the averages and standard deviation from at least three independent experiments. **, p ≤ 0.01; ****, p ≤ 0.0001; Student's t test.
Cotranslational recruitment of ZNF277 to nascent uS5
Given that uS5 was identified as the main ZNF277-associated protein as determined by both AP–MS and BioID approaches (Fig. 1), we explored the possibility that ZNF277 could play a chaperoning function toward uS5. Accordingly, an increasing amount of r-proteins have been shown to require dedicated chaperones for soluble expression and delivery to assembly sites (18). One frequent observation of dedicated r-protein chaperone is the cotranslational capture of the nascent r-protein (10). To test whether ZNF277 is recruited to uS5 in a cotranslational manner, we affinity-purified GFP, GFP–PRMT3, and GFP–ZNF277 from extracts of HEK 293 cells (Fig. 6A) that were previously treated with cycloheximide to block translation elongation and thereby maintain ribosome–mRNA associations. RNA was subsequently isolated from the GFP-Trap beads and analyzed by RT-qPCR for the specific enrichment of the uS5 mRNA. The data were normalized to the GAPDH mRNA to control for experimental variation, and the values were set to 1.0 for the control GFP purification. As shown in Fig. 6B, we observed a significant enrichment of the uS5 mRNA in GFP–ZNF277 precipitates, whereas the uS5 mRNA did not selectively copurify with GFP–PRMT3. Importantly, the enrichment of the uS5 mRNA in GFP–ZNF277 pulldowns was specific, because mRNAs encoding uS3 (RPS3) and uL4 (RPL4) were not found to be selectively enriched with GFP–ZNF277 (Fig. 6B). Next, we examined whether the ZF#5 mutant of ZNF277, which does not associate with uS5 (Fig. 2), can copurify with the uS5 mRNA. Notably, affinity purification of the ZNF277 mutant containing substitutions in ZF#5 failed to show any enrichment of the uS5 mRNA (Fig. 6, C and D), suggesting that a ZNF277–uS5 protein–protein interaction is required for the copurification of the uS5 mRNA with ZNF277. We conclude that ZNF277 has the capacity to recognize uS5 in a cotranslational manner.
Figure 6.
ZNF277 binds to uS5 in a cotranslational manner. A, Western blotting analysis of total extracts (lanes 1–3) and anti-GFP precipitates (lanes 4–6) prepared from HEK 293 cells that stably express GFP (lanes 1 and 4), GFP–PRMT3 (lanes 2 and 5), and GFP–ZNF277 (lanes 3 and 6). B, analysis of mRNA enrichment (IP/input ratio) for the indicated genes in GFP, GFP–PRMT3, and GFP–ZNF277 immunoprecipitates was analyzed by RT-qPCR and normalized to a control housekeeping mRNA (GAPDH). The values were then set to 1.0 for the control GFP purification. *, p ≤ 0.05; Student's t test. C, Western blotting analysis of total extracts (lanes 1–3) and anti-GFP precipitates (lanes 4–6) prepared from HEK 293 cells that stably express GFP (lanes 1 and 4), GFP–ZNF277 WT (lanes 2 and 5), and GFP–ZNF277 with substitutions in ZF#5 (lanes 3 and 6). D, analysis of mRNA enrichment as described in B, but using GFP, GFP–ZNF277 WT, and GFP–ZNF277 m.5 immunoprecipitates. *, p ≤ 0.05; **, p ≤ 0.01; Student's t test. IP, immunoprecipitation.
A deficiency in ZNF277 does not affect ribosome biogenesis
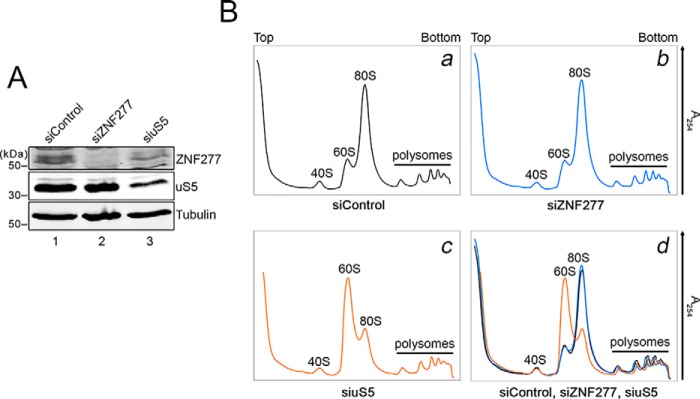
Cotranslational recognition of nascent uS5 by ZNF277 may support a chaperoning function to ZNF277. The idea of dedicated r-protein chaperones, specialized at accompanying and guarding highly basic and abundant r-proteins, is an emerging research area. Because mutants in r-protein chaperones usually result in ribosome biogenesis defects that resemble those observed upon depletion of their r-protein client, we compared ribosome profiles of cells deficient for ZNF277 and uS5. Depletion of over 90% of total ZNF277 (Fig. 7A, compare lanes 1 and 2) did not alter the levels of uS5 protein (Fig. 7A, compare lanes 1 and 2) and did not affect the ribosome profile detected from growing HeLa cells as compared with control cells (Fig. 7B, panels b and d). In contrast, a 35% depletion of total cellular uS5 (Fig. 7A, lane 3) caused a marked accumulation of free 60S subunit (Fig. 7B, panels c and d) as a result of a 40S subunit deficit, consistent with previous results obtained in fission yeast (40) and human cells (41). A deficiency in uS5 also caused a reduction in 80S monosomes (Fig. 7B, panels c and d). As shown in Fig. 5, the levels of ZNF277 were reduced in uS5-depleted cells (Fig. 7A, compare lanes 1 and 3). Together, these data argue that ZNF277 is unlikely to function as a dedicated chaperone involved in escorting uS5 from its synthesis in the cytoplasm to its assembly site in the preribosome.
Figure 7.
A deficiency in ZNF277 does not impair ribosome biogenesis. A, Western blotting analysis of total extracts prepared from HeLa cells that were transfected with siRNAs specific to ZNF277 (lane 2) and uS5 (lane 3) mRNAs as well as with nontarget control siRNAs (lane 1). Extracts were analyzed using antibodies specific for ZNF277, uS5, and tubulin. B, polysome profiles using extracts prepared from HeLa cells transfected with siRNAs specific to ZNF277 (panel b) and uS5 (panel c) mRNAs as well as nontarget control siRNAs (panel a) were separated using 5–50% sucrose gradients. Panel d shows an overlay of the ribosome profiles shown in panels a–c. The positions of free small (40S) and large (60S) ribosomal subunits, monosomes (80S), and polysomes are indicated.
Because our data indicate that ZNF277 and PRMT3 compete for uS5 association (Fig. 4), we tested the possibility that increased dosage of ZNF277, rather than a deficiency, could alter the ribosome profile. Transient expression of a GFP-tagged version of ZNF277 in HeLa cells resulted in a 10-fold increase relative to endogenous ZNF277 (Fig. S4A, lanes 3 and 4). Notably, increased levels of ZNF277 caused a small, but significant change in the ratio between small and large ribosomal subunits (Fig. S4, B and C): cells with increased levels of GFP–ZNF277 showed reduced levels of free 40S subunit concomitant with greater levels of free 60S subunit compared with cells that overexpressed GFP alone. Increased dosage of ZNF277 did not appear to affect monosome and polysome levels, however (Fig. S4B). These results suggest that excess ZNF277 in human cells can sequester uS5 and alter the balance between free ribosomal subunits.
Discussion
In this study, we report that human ZNF277 forms a stable extraribosomal complex with the 40S ribosomal protein uS5. Our results also distinguish two mutually exclusive extraribosomal complexes involving uS5: ZNF277–uS5 and PRMT3–uS5, both of which use a C2H2-type zinc finger domain to direct uS5 recognition. Interestingly, our data revealed that ZNF277 associates with nascent uS5 cotranslationally, whereas PRMT3 interacts with uS5 post-translationally. Collectively, our findings reveal an intricate network of evolutionarily conserved protein–protein interactions involving extraribosomal uS5, suggesting a key role for uS5 beyond the ribosome.
Previous work using fission yeast, Drosophila, and human cells (26–30) uncovered the existence of two independent protein complexes that involve an extraribosomal population of uS5 consisting of PRMT3–uS5–PDCD2L and PRMT3–uS5–PDCD2 (Fig. 8). The work described in the current study therefore adds a new layer to our understanding of extraribosomal uS5 by identifying the multizinc finger protein ZNF277 as a new uS5-associated protein. A complex between ZNF277 and uS5 (Fig. 1) is supported by independent studies that used high-throughput affinity purifications coupled to MS in human cells (42), as well as analysis of protein–protein interactions in Drosophila using high-throughput two-hybrid screening (28). Furthermore, genomic mediation analysis of trans-expression quantitative trait loci (trans-eQTL) in uS5 using multiomics data sets from the BXD mouse cohort identified Zfp277 (mouse homolog of human ZNF277 gene) as the potential regulator of uS5 (43). Together, all of these findings support the existence of evolutionarily conserved physical and functional connections between ZNF277 and uS5. What is the role of ZNF277 and the functional relevance of the ZNF277–uS5 interaction? Despite data consistent with recognition of nascent uS5 by ZNF277 in a cotranslational manner (Fig. 6), our results argue that ZNF277 is unlikely to function as a dedicated chaperone required to protect and escort free uS5 to 40S precursors. The line of evidence to support this conclusion is that a deficiency of ZNF277 affected neither uS5 protein levels nor ribosome biogenesis (Fig. 7), which contrasts with known dedicated r-protein chaperones (18). Independent of chaperoning functions, cotranslational formation of protein complexes has become an emerging concept of biology, which contributes to structural, spatial, and temporal aspects of complex assembly (44). We thus speculate that cotranslational recognition of uS5 by ZNF277 may provide a kinetic advantage to a potentially stronger interaction between uS5 and PRMT3.
Figure 8.
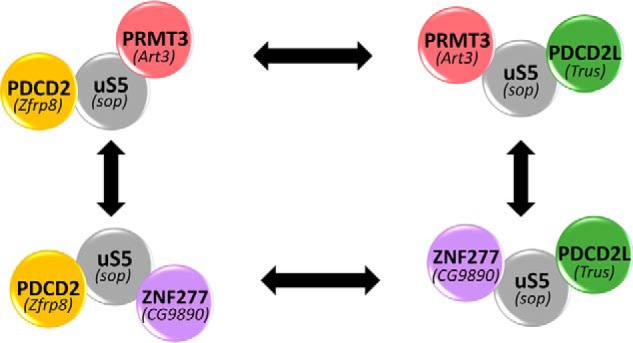
Protein complexes revolving around extraribosomal uS5. Shown is a schematic of independent PDCD2–uS5–PRMT3 (upper left) and PDCD2L–uS5-PRMT3 (upper right) complexes as we have described in humans (29). Mutually exclusive interactions of uS5 with ZNF277 and PRMT3 result in the formation of PDCD2–uS5–ZNF277 (lower left) and PDCD2L–uS5–ZNF277 (lower right) complexes. The existence of conserved complexes in Drosophila is supported by results from high-throughput two-hybrid screening and biochemical studies (28, 30), showing that homologs of PRMT3 (Art3), ZNF277 (CG9890), PDCD2 (Zfrp8), and PDCD2L (Trus) interact with uS5 (Sop).
Proteins with clusters of C2H2 zinc fingers represent the largest class of human transcription factors (45). Accordingly, the mouse homolog of human ZNF277, Zfp277, was shown to function in the transcriptional repression of the CDKN2A (Ink4a/Arf) locus in a manner dependent on the Polycomb group protein Bmi1 (46). Depletion of ZNF277 in human HeLa cells did not result in a significant increase in mRNA levels expressed from the CDKN2A gene, however (data not shown). It is therefore possible that the ZNF277–CDKN2A regulatory circuit is inactive in immortalized human cancer cells, because the transcriptional role of mouse Zfp277 was demonstrated in nonimmortalized embryonic fibroblasts (46). Recently, a novel proteomics approach to identify proteins that bind RNA sequences of interest in living cells identified human ZNF277 as a protein recognizing the Nanos response element of the p38–MAPK14 mRNA (47), thus raising the possibility that ZNF277 can also bind RNA. It will therefore be interesting to learn whether extraribosomal uS5 functions in the regulation of the DNA- and/or RNA-binding activities of ZNF277.
Our findings suggest an attractive mode of cross-regulation between the ZNF277–uS5 and PRMT3–uS5 complexes. This conclusion is supported by data showing that overexpression of PRMT3 limits uS5 association with ZNF277, whereas a deficiency in ZNF277 increases the proportion of uS5-associated PRMT3 (Fig. 4). Depletion of uS5 also caused the concomitant loss of PRMT3 and ZNF277 (Fig. 5), a behavior that is frequently observed for constitutively interacting proteins of a complex (48). Interestingly, ZNF277 overexpression is associated with improved prognosis of human cancers according to the Human Protein Atlas Project analysis of available human tumors in the repository (49). Conversely, PRMT3 overexpression, which decreases the amount of ZNF277–uS5 complex, is associated with poor prognosis of human cancers (49). The opposing effects of ZNF277 and PRMT3 expression on human cancer prognosis thus suggest that extraribosomal uS5 could be targeted to reduce the growth of human cancers.
Our BioID analysis of ZNF277 also identified PDCD2L (Figs. 1B and 8), a protein that directly interacts with uS5 (29). The copurification of PDCD2L with ZNF277 is consistent with previous results from proteomic analyses of PDCD2- and PDCD2L-associated proteins, which identified ZNF277 among the significant hits (29). Our data also revealed that copurification of ZNF277 and PDCD2L depends on uS5 (Fig. S1), as previously shown for the copurification of PRMT3 and PDCD2/PDCD2L (29), thus supporting a model in which uS5 bridges or stabilizes the association between ZNF277 and PDCD2L (Fig. 8). It remains unclear why PDCD2 was not detected in our MS-based analysis of ZNF277-associated proteins despite the fact that endogenous ZNF277 was a top hit in PDCD2 purifications (29). One possibility is that the use of ZNF277 fusion proteins repelled the association between PDCD2 and uS5 in the context of the ZNF277–uS5–PDCD2 complex. Collectively, our findings suggest the occurrence of four independent complexes revolving around extraribosomal uS5 (Fig. 8). Because uS5 is overexpressed in diverse cancers (50–53) and is reported as a potential therapeutic target in colorectal (51) and prostate (53) cancers, elucidating the biological role of extraribosomal uS5 and its evolutionarily conserved associated proteins should provide significant insights into the emerging concept of ribosomal protein specialization and could ultimately be exploited to design strategies aimed at targeting extraribosomal uS5 in tumor cells.
Experimental procedures
Cell culture
HEK 293-FT, U-2 OS, and HeLa cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% of tetracycline-free fetal bovine serum. Inducible expression of GFP, GFP–PRMT3, GFP–ZNF277, GFP–ZNF277 mutant #5, BirA, and BirA–ZNF277 was achieved by flippase-mediated recombination in HEK 293-FT and/or in U-2 OS-FT cells, as previously described (54). Inducible lentiviral shRNA-expressing cells were generated using the pTRIPZ lentiviral inducible vector (GE Healthcare). Induction of GFP-tagged proteins was achieved with 500 ng/ml of doxycycline for 48 h, whereas the induction shRNA cells lines was achieved with a 72–96-h incubation with doxycycline. Induction of BirA-tagged proteins was achieved with 50 μm biotin for 24h. siRNAs were transfected with Lipofectamine 2000 at a final concentration of 25 nm (siControl and siZNF277) or 32 nm (siuS5) for 72 h.
SILAC, BioID, and immunoprecipitation assays
For SILAC and BioID experiments, proteins were metabolically labeled with stable isotopes of arginine and lysine in cell culture, as previously described (29). Briefly, HEK 293-FT cells expressing GFP- or BirA-tagged versions of proteins were grown in media containing labeled amino acids. 24–48 h after induction with doxycycline (SILAC) or biotin (BioID) cells were collected in lysis buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.1% Triton X-100, 10% glycerol, 2 mm MgCl2, 1 mm DTT, and 1× Complete protease inhibitor mixture (Roche)) and incubated at 4 °C for 20 min. Lysates were centrifuged for 10 min at 13,000 rpm at 4 °C, and equal amount of proteins were incubated with GFP-trap agarose beads from ChromoTek (Martinsried, Germany) or Dynabeads M-270 streptavidin (Thermo Fisher) for 3 h at 4 °C. The beads were then washed five times with lysis buffer, and proteins were subjected to two rounds of elution by adding 100 μl of denaturing buffer (50 mm Tris-HCl, pH 6.8, 2% SDS, 0.1 mm DTT) for 10 min at 90 °C. SILAC eluates were vacuum-concentrated in a SpeedVac and resuspended in reducing buffer (125 mm Tris-HCl, pH 8.0, 1.25% SDS, 37.5% glycerol, 60 mm DTT, 0.025% bromphenol blue). Gel electrophoresis, in-gel digestions, LC–MS/MS, and analysis of SILAC ratios were performed as described previously (29). BioID used on-bead digestion with trypsin followed by LC–MS/MS. To assess the requirement of nucleic acids for protein–protein interactions, extracts of HEK293 extracts were treated with 167 units/ml of Benzonase (Sigma, E1014) for 20 min at 4 °C. Benzonase-treated extracts were then centrifuged to remove precipitates, and supernatants were used for affinity purifications, as described.
Sucrose gradient analysis
To analyze the sedimentation pattern of ZNF277, sucrose gradient centrifugation was performed as previously described (26, 29). Briefly, cycloheximide-treated cells were washed twice with PBS, and lysis buffer (10 mm Tris-HCl, pH 7.4, 100 mm KCl, 10 mm MgCl2, 1 mm DTT, 1% Triton X-100, Complete protease inhibitor mixture (Roche), 40 units/ml RNase OUT (Life Technologies), and 50 μg/ml cycloheximide (Sigma–Aldrich)) was added directly to a 15-cm dish. The cells were scraped, incubated 15 min at 4 °C, and centrifuged for 10 min at 4 °C. Of the supernatant, 5% was kept for Western blotting analysis of the input, and the remainder (5–10 mg of total protein) was loaded onto a 5–50% sucrose gradient and centrifuged for 3 h at 40,000 rpm in a SW41 rotor (Beckman Coulter). The gradient was then fractionated by upward displacement with 55% (w/v) sucrose using a gradient fractionator (Brandel Inc.) connected to a UA-6 UV monitor (Teledyne Isco) for continuous measurement of the absorbance at 254 nm. 0.6-ml fractions were collected, proteins were precipitated with TCA (15% final) and analyzed by Western blotting. 40S, 60S, and 80S subunits curves were reproduced using WinDaq system, and the area under the curve was calculated.
Protein analysis and antibodies
Proteins were separated by SDS-PAGE, transferred to nitrocellulose membranes, and analyzed by immunoblotting using the following primary antibodies: anti-tubulin and anti-FLAG (T5168, and F1804, respectively; Sigma–Aldrich); anti-PRMT3, anti-PDCD2L, anti-PDCD2 (A302-526A, A303-783A, and A303-599A), respectively; Bethyl); anti-PDCD2L (A303-783A; Bethyl); anti-hRRP12 (sc-139043; Santa Cruz Biotechnology); anti-RPL17 (GTX111934; GeneTex), and anti-uS5 (RPS2) (a generous gift from Dr. Mark Bedford). Membranes were then probed with either a donkey anti-rabbit antibody conjugated to IRDye 800CW (926-32213; LiCOR) or a goat anti-mouse antibody conjugated to Alexa Fluor 680 (A-21057; Life Technologies). Proteins detection was performed using an Odyssey IR imaging system (LiCOR).
Recombinant proteins
For the coexpression of uS5 and ZNF277 in E. coli, we used the pET-duet-1 vector (Novagen) in BL21 DE3 cells. Construct pFB1329 contained the uS5 cDNA with sequence for a C-terminal FLAG tag in the first multiple cloning site (MCS) and the ZNF277 cDNA with sequence for a C-terminal His tag in the second MCS. pFB1340 only contained the ZNF277-His cDNA in the second MCS. Precultures of BL21 DE3 cells containing either pFB1329 or pFB1340 were diluted in a 1:10 ratio into fresh medium supplemented with 10 μm ZnCl2 and incubated for 2–3 h at 37 °C until a A600 nm of 0.5. The cultures were then supplemented with 0.5 mm isopropyl β-d-thiogalactopyranoside and 2% EtOH and incubated with shaking at 18 °C overnight. After a 20-min centrifugation at 4000 rpm, the cells were resuspended in radioimmune precipitation assay lysis buffer (10 mm Tris-Cl, pH 8.0, 350 mm NaCl, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, Complete protease inhibitor mixture (Roche)) and were treated with lysozyme at a final concentration of 1 mg/ml. Total extracts and nonlysed cells were then sonicated at 4 °C by intervals of 5 s on, 5 s off, for a total of 20 times at an amplitude of 19%. After sonication, Triton X-100 was added to a final concentration of 1%, incubated 30 min at 4 °C, and centrifuged for 30 min at 7000 rpm. 10% of the lysate was kept as an input for protein analysis. The remainder of the lysate was incubated with FLAG M2 affinity gel (Sigma–Aldrich) for 3 h at 4 °C. The beads were then washed five times using radioimmune precipitation assay buffer. The beads were then subjected to either an elution using FLAG wash buffer (50 mm Tris, pH 8.0, 250 mm NaCl, 0.1% Triton X-100, 10 μm ZnCl2) with 200 μg/μl of FLAG peptide or treated with 2× SDS-PAGE loading dye and heated at 95 °C for 10 min.
Microscopy
Visual analysis of GFP-tagged proteins in human cells was as previously described (55). U-2 OS cells were washed twice with PBS, fixed with 4% paraformaldehyde for 15 min at room temperature, and washed twice again with PBS. Fixed cells were permeabilized using a 0.5% Triton X-100/PBS solution for 10 min and washed three times with PBS. For Fig. 3B, cells were equilibrated for 20 min in PBS with 1% BSA followed by a 1-h incubation with primary antibody dilution in PBS with 1% BSA (anti-fibrillarin 1/50 (Santa Cruz)). The cells were washed three times with PBS with 1% BSA and incubated with a secondary antibody dilution (mouse Alexa Fluor 568 1/500 (Invitrogen), in PBS with 1% BSA) for 1 h. For the BiFC experiments shown in Fig. 3C, U-2 OS cells were cotransfected with Venus vector N (VN) or Venus vector C (VC) fusions and fixed 20–24 h post-transfection for visual analysis as described above. For all microscopy experiments, three final washes with either PBS alone or PBS with 1% BSA were performed before slides were mounted on a coverslip with SlowFade Gold antifade solution (Life Technologies). Images were captured by Zeiss Axio Observer microscope using a 63× oil objective.
RNA coimmunoprecipitation assay
15-cm dishes of HEK 293-FT conditionally expressing GFP, GFP–PDCD2, GFP–ZNF277, or GFP–ZNF277 mutant #5 were induced using doxycycline for 48 h. The cells were washed twice with PBS, and 5% of cells were kept for total RNA extraction (input fraction). The cells were then resuspended in lysis buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.1% Triton X-100, 10% glycerol, 2 mm MgCl2, 1 mm DTT, Complete protease inhibitor mixture (Roche), and 40 units/ml RNase OUT (Life Technologies)) and incubated 15 min at 4 °C. Lysate was centrifuged at 13,000 rpm for 10 min at 4 °C, and 5% of the lysate was kept for protein analysis. The remainder of the lysate was incubated with GFP-Trap beads for 3 h at 4 °C. The beads were washed five times with lysis buffer, and 10% of the beads were kept for protein analysis. RNA was extracted from the remainder of the beads using TRIzol reagent (Life Technologies) and analyzed by RT-qPCR using the following set of primers: uS5, 5′-TATGCCAGTGCAGAAGCAGACC-3′/5′-CCTCCTTGGAGCACTTAACAC-3′ or 5′-GATGCCCGTCACCAAGTT-3′/5′-CTGATTCCTTAATAGGCAGGG-3′; uS3, 5′-CAAGAAGAGGAAGTTTGTCGC-3′/5′-GAACATTCTGTGTTCTGGTGG-3′; uL4, 5′-GCAGGTCATCAGACTAGTGC-3′/5′-GGTTTTGGTTGGTGCAAAC-3′; 18S, 5′-AAACGGCTACCACATCCAAG-3′/5′-CCTCCAATGGATCCTCGTTA-3′; and GAPDH, 5′-GTCAGCCGCATCTTCTTTTG-3′/5′-GCGCCCAATACGACCAAATC-3′).
Author contributions
K. L. D., D. B., A.-M. L.-V., and F. B. conceptualization; K. L. D., D. B., A.-M. L.-V., and F. B. formal analysis; K. L. D., D. B., A.-M. L.-V., and F. B. investigation; K. L. D., D. B., A.-M. L.-V., and F. B. methodology; K. L. D. and F. B. writing-original draft; F. B. resources; F. B. supervision; F. B. funding acquisition; F. B. project administration.
Supplementary Material
Acknowledgments
We are grateful to members of the Bachand lab for critical reading of the manuscript.
This work was supported by Canadian Institutes of Health Research Grant MOP-273292 (to F. B.) and by a graduate scholarship from the Natural Sciences and Engineering Research Council of Canada (to A.-M. L.-V.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1–S3 and Figs. S1–S4.
- r-protein
- ribosomal protein
- ZNF277
- zinc finger protein 277
- PRMT3
- protein arginine methyltransferase 3
- uS5/RPS2
- 40S ribosomal protein S2
- ZF
- zinc finger
- SILAC
- stable isotope labeling by amino acids in cell culture
- PDCD2
- programmed cell death protein 2
- PDCD2L
- PDCD2-like protein
- AP
- affinity purification
- BiFC
- bimolecular fluorescence complementation
- YFP
- yellow fluorescent protein
- qPCR
- quantitative PCR
- MCS
- multiple cloning site.
References
- 1. Nerurkar P., Altvater M., Gerhardy S., Schütz S., Fischer U., Weirich C., and Panse V. G. (2015) Eukaryotic ribosome assembly and nuclear export. Int. Rev. Cell Mol. Biol. 319, 107–140 10.1016/bs.ircmb.2015.07.002 [DOI] [PubMed] [Google Scholar]
- 2. Frank J. (2017) The mechanism of translation. F1000Res 6, 198 10.12688/f1000research.9760.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Henras A. K., Plisson-Chastang C., O'Donohue M. F., Chakraborty A., and Gleizes P. E. (2015) An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip. Rev. RNA 6, 225–242 10.1002/wrna.1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Peña C., Hurt E., and Panse V. G. (2017) Eukaryotic ribosome assembly, transport and quality control. Nat. Struct. Mol. Biol. 24, 689–699 10.1038/nsmb.3454 [DOI] [PubMed] [Google Scholar]
- 5. Thomson E., Ferreira-Cerca S., and Hurt E. (2013) Eukaryotic ribosome biogenesis at a glance. J. Cell Sci. 126, 4815–4821 10.1242/jcs.111948 [DOI] [PubMed] [Google Scholar]
- 6. Woolford J. L. Jr., and Baserga S. J. (2013) Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 195, 643–681 10.1534/genetics.113.153197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Warner J. R., Vilardell J., and Sohn J. H. (2001) Economics of ribosome biosynthesis. Cold Spring Harb. Symp. Quant. Biol. 66, 567–574 10.1101/sqb.2001.66.567 [DOI] [PubMed] [Google Scholar]
- 8. Jäkel S., and Görlich D. (1998) Importin β, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 17, 4491–4502 10.1093/emboj/17.15.4491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mitterer V., Gantenbein N., Birner-Gruenberger R., Murat G., Bergler H., Kressler D., and Pertschy B. (2016) Nuclear import of dimerized ribosomal protein Rps3 in complex with its chaperone Yar1. Sci. Rep. 6, 36714 10.1038/srep36714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pausch P., Singh U., Ahmed Y. L., Pillet B., Murat G., Altegoer F., Stier G., Thoms M., Hurt E., Sinning I., Bange G., and Kressler D. (2015) Co-translational capturing of nascent ribosomal proteins by their dedicated chaperones. Nat. Commun. 6, 7494 10.1038/ncomms8494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pillet B., García-Gómez J. J., Pausch P., Falquet L., Bange G., de la Cruz J., and Kressler D. (2015) The dedicated chaperone Acl4 escorts ribosomal protein Rpl4 to its nuclear pre-60S assembly site. PLoS Genet. 11, e1005565 10.1371/journal.pgen.1005565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stelter P., Huber F. M., Kunze R., Flemming D., Hoelz A., and Hurt E. (2015) Coordinated ribosomal L4 protein assembly into the pre-ribosome is regulated by its eukaryote-specific extension. Mol. Cell 58, 854–862 10.1016/j.molcel.2015.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kressler D., Bange G., Ogawa Y., Stjepanovic G., Bradatsch B., Pratte D., Amlacher S., Strauss D., Yoneda Y., Katahira J., Sinning I., and Hurt E. (2012) Synchronizing nuclear import of ribosomal proteins with ribosome assembly. Science 338, 666–671 10.1126/science.1226960 [DOI] [PubMed] [Google Scholar]
- 14. Holzer S., Ban N., and Klinge S. (2013) Crystal structure of the yeast ribosomal protein rpS3 in complex with its chaperone Yar1. J. Mol. Biol. 425, 4154–4160 10.1016/j.jmb.2013.08.022 [DOI] [PubMed] [Google Scholar]
- 15. Koch B., Mitterer V., Niederhauser J., Stanborough T., Murat G., Rechberger G., Bergler H., Kressler D., and Pertschy B. (2012) Yar1 protects the ribosomal protein Rps3 from aggregation. J. Biol. Chem. 287, 21806–21815 10.1074/jbc.M112.365791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ting Y. H., Lu T. J., Johnson A. W., Shie J. T., Chen B. R., Kumar S. S., and Lo K. Y. (2017) Bcp1 is the nuclear chaperone of Rpl23 in Saccharomyces cerevisiae. J. Biol. Chem. 292, 585–596 10.1074/jbc.M116.747634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wyler E., Wandrey F., Badertscher L., Montellese C., Alper D., and Kutay U. (2014) The β-isoform of the BRCA2 and CDKN1A(p21)-interacting protein (BCCIP) stabilizes nuclear RPL23/uL14. FEBS Lett. 588, 3685–3691 10.1016/j.febslet.2014.08.013 [DOI] [PubMed] [Google Scholar]
- 18. Pillet B., Mitterer V., Kressler D., and Pertschy B. (2017) Hold on to your friends: dedicated chaperones of ribosomal proteins: dedicated chaperones mediate the safe transfer of ribosomal proteins to their site of pre-ribosome incorporation. Bioessays 39, 1–12 10.1002/bies.201670013 , , , , , , , , , , , [DOI] [PubMed] [Google Scholar]
- 19. Lindström M. S. (2009) Emerging functions of ribosomal proteins in gene-specific transcription and translation. Biochem. Biophys. Res. Commun. 379, 167–170 10.1016/j.bbrc.2008.12.083 [DOI] [PubMed] [Google Scholar]
- 20. Warner J. R., and McIntosh K. B. (2009) How common are extraribosomal functions of ribosomal proteins? Mol. Cell 34, 3–11 10.1016/j.molcel.2009.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dai M. S., and Lu H. (2004) Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J. Biol. Chem. 279, 44475–44482 10.1074/jbc.M403722200 [DOI] [PubMed] [Google Scholar]
- 22. Horn H. F., and Vousden K. H. (2008) Cooperation between the ribosomal proteins L5 and L11 in the p53 pathway. Oncogene 27, 5774–5784 10.1038/onc.2008.189 [DOI] [PubMed] [Google Scholar]
- 23. Jin A., Itahana K., O'Keefe K., and Zhang Y. (2004) Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol. Cell. Biol. 24, 7669–7680 10.1128/MCB.24.17.7669-7680.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lindström M. S., Jin A., Deisenroth C., White Wolf G., and Zhang Y. (2007) Cancer-associated mutations in the MDM2 zinc finger domain disrupt ribosomal protein interaction and attenuate MDM2-induced p53 degradation. Mol. Cell. Biol. 27, 1056–1068 10.1128/MCB.01307-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lohrum M. A., Ludwig R. L., Kubbutat M. H., Hanlon M., and Vousden K. H. (2003) Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell 3, 577–587 10.1016/S1535-6108(03)00134-X [DOI] [PubMed] [Google Scholar]
- 26. Bachand F., and Silver P. A. (2004) PRMT3 is a ribosomal protein methyltransferase that affects the cellular levels of ribosomal subunits. EMBO J. 23, 2641–2650 10.1038/sj.emboj.7600265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Swiercz R., Person M. D., and Bedford M. T. (2005) Ribosomal protein S2 is a substrate for mammalian PRMT3 (protein arginine methyltransferase 3). Biochem. J. 386, 85–91 10.1042/BJ20041466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Giot L., Bader J. S., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y. L., Ooi C. E., Godwin B., Vitols E., Vijayadamodar G., Pochart P., Machineni H., Welsh M., Kong Y., et al. (2003) A protein interaction map of Drosophila melanogaster. Science 302, 1727–1736 10.1126/science.1090289 [DOI] [PubMed] [Google Scholar]
- 29. Landry-Voyer A. M., Bilodeau S., Bergeron D., Dionne K. L., Port S. A., Rouleau C., Boisvert F. M., Kehlenbach R. H., and Bachand F. (2016) Human PDCD2L is an export substrate of CRM1 that associates with 40S ribosomal subunit precursors. Mol. Cell. Biol. 36, 3019–3032 10.1128/MCB.00303-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Minakhina S., Naryshkina T., Changela N., Tan W., and Steward R. (2016) Zfrp8/PDCD2 interacts with RpS2 connecting ribosome maturation and gene-specific translation. PLoS One 11, e0147631 10.1371/journal.pone.0147631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., and Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 10.1074/mcp.M200025-MCP200 [DOI] [PubMed] [Google Scholar]
- 32. Ceroni F., Simpson N. H., Francks C., Baird G., Conti-Ramsden G., Clark A., Bolton P. F., Hennessy E. R., Donnelly P., Bentley D. R., Martin H., IMGSAC, SLI Consortium, WGS500 Consortium, Parr J., et al. (2014) Homozygous microdeletion of exon 5 in ZNF277 in a girl with specific language impairment. Eur. J. Hum. Genet. 22, 1165–1171 10.1038/ejhg.2014.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Roux K. J., Kim D. I., Raida M., and Burke B. (2012) A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801–810 10.1083/jcb.201112098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Perreault A., Gascon S., D'Amours A., Aletta J. M., and Bachand F. (2009) A methyltransferase-independent function for Rmt3 in ribosomal subunit homeostasis. J. Biol. Chem. 284, 15026–15037 10.1074/jbc.M109.004812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. UniProt Consortium, T. (2018) UniProt: the universal protein knowledgebase. Nucleic Acids Res. 46, 2699 10.1093/nar/gky092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nguyen D., Grenier St-Sauveur V., Bergeron D., Dupuis-Sandoval F., Scott M. S., and Bachand F. (2015) A polyadenylation-dependent 3′ end maturation pathway is required for the synthesis of the human telomerase RNA. Cell Reports 13, 2244–2257 10.1016/j.celrep.2015.11.003 [DOI] [PubMed] [Google Scholar]
- 37. Thul P. J., Akesson L., Wiking M., Mahdessian D., Geladaki A., Ait Blal H., Alm T., Asplund A., Bjork L., Breckels L. M., Backstrom A., Danielsson F., Fagerberg L., Fall J., Gatto L., et al. (2017) A subcellular map of the human proteome. Science 356 [DOI] [PubMed] [Google Scholar]
- 38. Kerppola T. K. (2006) Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat. Protocols 1, 1278–1286 10.1038/nprot.2006.201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nagai T., Ibata K., Park E. S., Kubota M., Mikoshiba K., and Miyawaki A. (2002) A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20, 87–90 10.1038/nbt0102-87 [DOI] [PubMed] [Google Scholar]
- 40. Perreault A., Bellemer C., and Bachand F. (2008) Nuclear export competence of pre-40S subunits in fission yeast requires the ribosomal protein Rps2. Nucleic Acids Res. 36, 6132–6142 10.1093/nar/gkn625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. O'Donohue M. F., Choesmel V., Faubladier M., Fichant G., and Gleizes P. E. (2010) Functional dichotomy of ribosomal proteins during the synthesis of mammalian 40S ribosomal subunits. J. Cell Biol. 190, 853–866 10.1083/jcb.201005117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Huttlin E. L., Ting L., Bruckner R. J., Gebreab F., Gygi M. P., Szpyt J., Tam S., Zarraga G., Colby G., Baltier K., Dong R., Guarani V., Vaites L. P., Ordureau A., Rad R., et al. (2015) The BioPlex network: a systematic exploration of the human interactome. Cell 162, 425–440 10.1016/j.cell.2015.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li H., Wang X., Rukina D., Huang Q., Lin T., Sorrentino V., Zhang H., Bou Sleiman M., Arends D., McDaid A., Luan P., Ziari N., Velázquez-Villegas L. A., Gariani K., Kutalik Z., et al. (2018) An integrated systems genetics and omics toolkit to probe gene function. Cell Syst. 6, 90–102.e4 10.1016/j.cels.2017.10.016 [DOI] [PubMed] [Google Scholar]
- 44. Williams N. K., and Dichtl B. (2018) Co-translational control of protein complex formation: a fundamental pathway of cellular organization? Biochem. Soc. Trans. 46, 197–206 10.1042/BST20170451 [DOI] [PubMed] [Google Scholar]
- 45. Weirauch M. T., and Hughes T. R. (2011) A catalogue of eukaryotic transcription factor types, their evolutionary origin, and species distribution. Subcell. Biochem. 52, 25–73 10.1007/978-90-481-9069-0_3 [DOI] [PubMed] [Google Scholar]
- 46. Negishi M., Saraya A., Mochizuki S., Helin K., Koseki H., and Iwama A. (2010) A novel zinc finger protein Zfp277 mediates transcriptional repression of the Ink4a/arf locus through polycomb repressive complex 1. PLoS One 5, e12373 10.1371/journal.pone.0012373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ramanathan M., Majzoub K., Rao D. S., Neela P. H., Zarnegar B. J., Mondal S., Roth J. G., Gai H., Kovalski J. R., Siprashvili Z., Palmer T. D., Carette J. E., and Khavari P. A. (2018) RNA–protein interaction detection in living cells. Nat. Methods 15, 207–212 10.1038/nmeth.4601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wyant G. A., Abu-Remaileh M., Frenkel E. M., Laqtom N. N., Dharamdasani V., Lewis C. A., Chan S. H., Heinze I., Ori A., and Sabatini D. M. (2018) NUFIP1 is a ribosome receptor for starvation-induced ribophagy. Science 360, 751–758 10.1126/science.aar2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Uhlen M., Zhang C., Lee S., Sjostedt E., Fagerberg L., Bidkhori G., Benfeitas R., Arif M., Liu Z., Edfors F., Sanli K., von Feilitzen K., Oksvold P., Lundberg E., Hober S., et al. (2017) A pathology atlas of the human cancer transcriptome. Science 357 [DOI] [PubMed] [Google Scholar]
- 50. Cancer Genome Atlas Research Network (2008) Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068 10.1038/nature07385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Grade M., Hummon A. B., Camps J., Emons G., Spitzner M., Gaedcke J., Hoermann P., Ebner R., Becker H., Difilippantonio M. J., Ghadimi B. M., Beissbarth T., Caplen N. J., and Ried T. (2011) A genomic strategy for the functional validation of colorectal cancer genes identifies potential therapeutic targets. Int. J. Cancer 128, 1069–1079 10.1002/ijc.25453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liliensiek B., Rocha M., Umansky V., Benner A., Lin J., Ziegler R., Nawroth P. P., and Schirrmacher V. (1998) Identification of four genes in endothelial cells whose expression is affected by tumor cells and host immune status: a study in ex vivo-isolated endothelial cells. Blood 92, 3394–3404 [PubMed] [Google Scholar]
- 53. Wang M., Hu Y., and Stearns M. E. (2009) RPS2: a novel therapeutic target in prostate cancer. J. Exp. Clin, Cancer Res. 28, 6 10.1186/1756-9966-28-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bergeron D., Pal G., Beaulieu Y. B., Chabot B., and Bachand F. (2015) Regulated intron retention and nuclear pre-mRNA decay contribute to PABPN1 autoregulation. Mol. Cell. Biol. 35, 2503–2517 10.1128/MCB.00070-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mallet P. L., and Bachand F. (2013) A proline-tyrosine nuclear localization signal (PY-NLS) is required for the nuclear import of fission yeast PAB2, but not of human PABPN1. Traffic 14, 282–294 10.1111/tra.12036 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.