Abstract
The chromosomal passenger complex (CPC) is a master regulator of mitosis. CPC consists of inner centromere protein (INCENP), Survivin, Borealin, and the kinase Aurora B and plays key roles in regulating kinetochore–microtubule attachments and spindle assembly checkpoint signaling. However, the role of CPC in sister chromatid cohesion, mediated by the cohesin complex, remains incompletely understood. Here, we show that Aurora B kinase activity contributes to centromeric cohesion protection partly through promoting kinetochore localization of the kinase Bub1. Interestingly, disrupting the interaction of INCENP with heterochromatin protein 1 (HP1) in HeLa cells selectively weakens cohesion at mitotic centromeres without detectably reducing the kinase activity of Aurora B. Thus, through this INCENP–HP1 interaction, the CPC also protects centromeric cohesion independently of Aurora B kinase activity. Moreover, the requirement for the INCENP–HP1 interaction in centromeric cohesion protection can be bypassed by tethering HP1 to centromeres or by depleting the cohesin release factor Wapl. We provide further evidence suggesting that the INCENP–HP1 interaction protects centromeric cohesion by promoting the centromere localization of Haspin, a protein kinase that antagonizes Wapl activity at centromeres. Taken together, this study identifies Aurora B kinase activity–dependent and –independent roles for the CPC in regulating centromeric cohesion during mitosis in human cells.
Keywords: centromere, mitosis, kinetochore, cell cycle, histone modification, chromosomal passenger complex, cohesin, Haspin, heterochromatin protein 1, sister chromatid cohesion, Aurora B, INCENP
Introduction
Chromosomal instability resulting from chromosome missegregation during mitosis is a hallmark of cancer cells and contributes to tumorigenesis (1, 2). The fidelity of chromosome segregation is guaranteed by a sophisticated mechanism that ensures timely resolution of sister chromatid cohesion, correct attachment of kinetochores to microtubules, and proper response of spindle assembly checkpoint (SAC)3 monitoring the status of kinetochore–microtubule (KT–MT) attachments, which allows chromosome segregation only when all chromosomes have completed biorientation on the metaphase plate (3).
The chromosomal passenger complex (CPC) is a master regulator of mitosis (4). The CPC consists of the kinase subunit Aurora B, the scaffold subunit inner centromere protein (INCENP), and the two regulatory subunits Survivin and Borealin. These proteins play specific roles in controlling the localization and function of the CPC. During early mitosis, Survivin and Borealin associate with the N-terminal CEN domain of INCENP and mediate the centromeric localization of CPC by directly binding to phosphorylated histone tails (5–7). The C terminus of INCENP binds to Aurora B through an IN-box domain, leading to allosteric activation of Aurora B (8–11). Heterochromatin protein 1 (HP1), which binds to INCENP or Borealin and is distributed along with the CPC to various locations throughout mitosis, can be considered the fifth subunit of the CPC. However, the functional significance of HP1 in the CPC remains incompletely understood.
Aurora B kinase activity plays a key role in the correction of erroneous KT–MT attachments. Aurora B–mediated phosphorylation of multiple kinetochore substrates in the KMN network, which consists of the KNL1, Mis12, and Ndc80 complexes, has been implicated in releasing incorrect attachments (12–15). Aurora B kinase is also required for SAC activation. This is achieved both through releasing improperly attached microtubules to generate unattached kinetochore and through phosphorylating substrates in the kinetochore to directly or indirectly recruit spindle checkpoint proteins (16). Compared with these well-established functions, the exact role of the CPC in the regulation of sister chromatid cohesion is unclear.
Sister chromatid cohesion is mediated by a multiple-subunit cohesin complex composed of Smc1, Smc3, Scc1, SA2 (or SA1), and the accessory subunit Pds5B (or Pds5A) (17). Sororin and Wapl competitively interact with Pds5A/B to positively and negatively regulate the association of cohesin with chromatin, respectively (18–22). Through antagonizing Wapl activity in cohesin release, Sororin ensures DNA replication–dependent sister chromatid cohesion and its maintenance in the subsequent G2 phase (22–24). During early mitosis, although cohesin at centromeres is protected by a complex mechanism involving the Wapl antagonists Sgo1, Sororin, and Haspin until the proteolytic cleavage of Scc1 by the protease Separase at anaphase onset (25), most cohesin is removed by Wapl from chromosome arms in a cleavage-independent manner (18, 19, 26–28). This prophase pathway of cohesin removal requires phosphorylation of SA2 (29–31) and Sororin (22, 32). Phosphorylation of Sororin by Cyclin-dependent kinase 1 and Aurora B destabilizes its interaction with Pds5A/B and causes Wapl activation and cohesin release (33), enabling the resolution of chromosome arms in prophase and prometaphase. Therefore, Aurora B kinase activity negatively regulates sister chromatid cohesion along the chromosome arms in early mitosis.
In this study, we report that Aurora B kinase activity positively regulates sister chromatid cohesion at mitotic centromeres at least partly through promoting the kinetochore localization of Bub1 in human cells. Importantly, through the interaction of its INCENP subunit with HP1, the CPC also plays an Aurora B kinase activity–independent role in the protection of centromeric cohesion.
Results
Inhibiting Aurora B kinase activity causes centromeric cohesion defects, which can be rescued by centromere-tethered Bub1
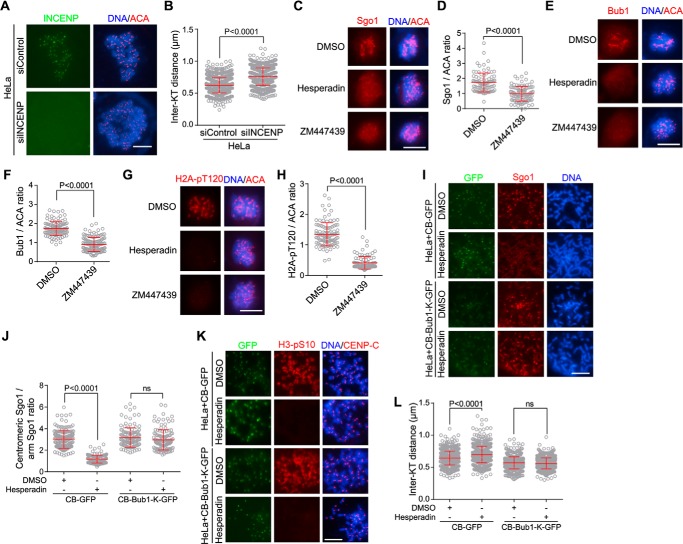
To dissect the role of CPC in the regulation of sister chromatid cohesion, we depleted its scaffold subunit INCENP by small interfering RNA (siRNA) and analyzed chromosomes prepared from HeLa cells that were arrested in mitosis with the microtubule destabilizer nocodazole. Immunofluorescence microscopy showed that the average interkinetochore (inter-KT) distance on chromosomes prepared from INCENP-depleted cells was 23% further apart than the control (Fig. 1, A and B), which is indicative of weakened centromeric cohesion. Similarly, inhibition of Aurora B kinase activity by the small-molecule inhibitor Hesperadin (34), as evidenced by loss of H3 serine 10 phosphorylation (H3-pS10) on chromatin (35), significantly increased the inter-KT distance (Fig. S1, A and B). As previously shown (28), we also noticed that Aurora B inhibition impaired the resolution of chromosome arms in nocodazole-arrested mitotic cells (Fig. S1A). These results indicate that, in contrast to its role in releasing cohesin from chromosome arms, Aurora B kinase activity is required to protect proper strength of sister chromatid cohesion at mitotic centromeres.
Figure 1.
Inhibiting Aurora B kinase activity causes centromeric cohesion defects, which can be rescued by centromere-tethered Bub1. A and B, HeLa cells transfected with control or INCENP siRNA were treated with nocodazole for 3 h. Mitotic cells collected by shake-off were spun on slides and stained with INCENP antibodies, ACA, and DAPI (A). The inter-KT distance was measured on over 886 chromosomes in 20 cells (B). C–H, HeLa cells were treated for 1 h with nocodazole and MG132 together with DMSO or the indicated Aurora B inhibitors. Cells were then stained with DAPI, ACA, and antibodies for Sgo1 (C), Bub1 (E), and H2A-pT120 (G). The immunofluorescence intensity ratios of centromeric Sgo1/ACA (D), Bub1/ACA (F), and H2A-pT120/ACA (H) were determined on 90–123 chromosomes in 20 cells. I–L, HeLa cells stably expressing CB-GFP or CB-Bub1-K-GFP were treated for 1 h with nocodazole and MG132 together with DMSO or Hesperadin. Mitotic chromosome spreads were stained with DAPI and antibodies for Sgo1, H3-pS10, and CENP-C. Example images are shown (I and K). The immunofluorescence intensity ratio of centromeric Sgo1/arm Sgo1 was determined on at least 124 chromosomes in 20 cells (J). The inter-KT distance was measured on over 759 chromosomes in 20 cells (L). Means and error bars representing S.D. are shown (unpaired t test). Scale bars, 10 μm. See also Fig. S1. ns, not significant.
We also found that inhibition of Aurora B kinase activity by Hesperadin and ZM447439 (Fig. S1C), another small-molecule inhibitor of Aurora B kinase (36), significantly reduced the centromeric localization of Sgo1 (Fig. 1, C and D). Consistently, Aurora B inhibition caused a strong reduction in the kinetochore localization of Bub1 kinase (Fig. 1, E and F) as well as Bub1-mediated phosphorylation of histone H2A at threonine 120 (H2A-pT120) (Fig. 1, G and H) (37), which is a direct binding site for Sgo1 at mitotic centromeres (38, 39). We therefore speculated that Aurora B kinase activity ensures full enrichment of Sgo1 at mitotic centromeres at least partly through promoting the kinetochore localization of Bub1 kinase. To test this hypothesis, we expressed a CENP-B (CB) fusion protein, CB-Bub1-K-GFP, in which Bub1 kinase domain is fused to the centromeric targeting domain of CENP-B (40, 41), which binds a 17-bp CENP-B–box motif within the α-satellite repeats of human centromeres (42–44). As expected, treatment of cells stably expressing CB-GFP with Hesperadin caused strong delocalization of Sgo1 from mitotic centromeres (Fig. 1, I and J). Interestingly, centromeric localization of Sgo1 was largely retained when cells stably expressing CB-Bub1-K-GFP were treated with Hesperadin, consistent with the role of Bub1-mediated H2A-pT120 in recruiting Sgo1 to centromeres. Moreover, the inter-KT distance in cells expressing CB-Bub1-K-GFP was not sensitive to Aurora B inhibition by Hesperadin (Fig. 1, K and L). Thus, the need for Aurora B kinase activity in maintaining proper inter-KT distance can be bypassed by tethering Bub1 to centromeres. These results are also consistent with the role of Sgo1 in protecting centromeric cohesion (30, 45–47). Previous studies reported that Aurora B kinase activity is required for efficient localization and activation of Mps1 at kinetochores (48–53) and that Mps1-mediated phosphorylation of KNL1 recruits Bub1 to kinetochores (54–57). In line with this, we found that treatment of cells with reversine and AZ3146, small-molecule inhibitors of Mps1 that strongly reduce the centromeric accumulation of Bub1 and Sgo1 (37, 51, 58), significantly increased the inter-KT distance (Fig. S1, D and E). Moreover, Mps1 inhibition significantly increased the inter-KT distance in cells stably expressing CB-GFP but not CB-Bub1-K-GFP, indicating that the requirement for Mps1 kinase activity in maintaining proper centromeric cohesion can be bypassed by centromere-tethered Bub1 (Fig. S1, F and G). Taken together, these results indicate that Aurora B kinase activity protects centromeric cohesion at least partly through promoting the localization of Bub1 kinase at kinetochores.
INCENP interacts with the chromo shadow domain (CSD) of HP1α and HP1γ
Previous studies showed that INCENP interacts with HP1 (59–62) and that loss of HP1α and/or HP1γ weakens centromeric cohesion (63–65). We were thus prompted to examine whether the INCENP–HP1 interaction might contribute to the role of CPC in regulating sister chromatid cohesion.
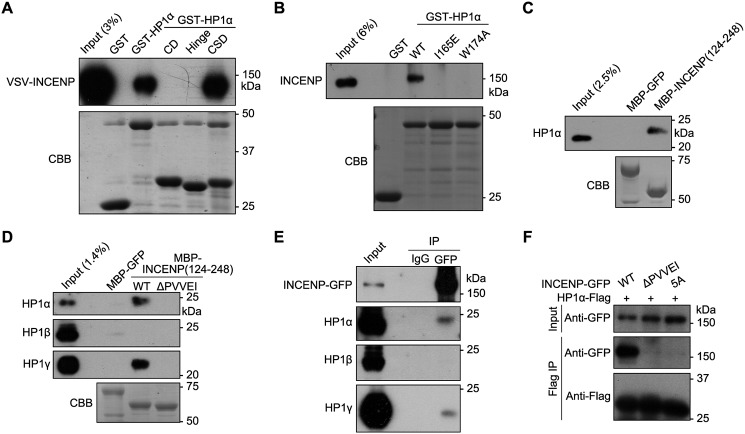
We found that GST-fused full-length HP1α or the CSD, but not CD and Hinge, selectively pulled down VSV-tagged INCENP (VSV-INCENP) transiently expressed in HEK-293T cells (Fig. 2A), indicating that CSD is sufficient to bind INCENP. Moreover, endogenous INCENP in mitotic HeLa cell lysates was efficiently pulled down by GST-HP1α (Fig. 2B) but not by GST-HP1α containing the mutations I165E and W174A that disrupt the formation of CSD dimer and hydrophobic pocket, respectively (66–68). This implies that INCENP utilizes a consensus (P/L)XVX(M/L/V) motif (PXVXL in short; X represents any amino acid) to interact with the hydrophobic pocket of CSD dimer (68, 69). We found that endogenous HP1α and HP1γ, but not HP1β, in mitotic HeLa cell lysates was pulled down by MBP-fused INCENP fragment encompassing residues 124–248 (MBP-INCENP(124–248)) but not by the MBP-INCENP-ΔPVVEI mutant lacking the highly conserved PVVEI motif (Fig. 2, C and D). We then carried out coimmunoprecipitation experiments and found that INCENP-GFP associated with HP1α and HP1γ, but not HP1β, in mitotic HeLa cell lysates (Fig. 2E). Moreover, FLAG-tagged HP1α (HP1α-FLAG) associated with INCENP-GFP but not with INCENP-ΔPVVEI-GFP, which lacks the PVVEI motif, or with the INCENP mutant in which the PVVEI motif was mutated to five alanines (INCENP-5A-GFP) (Fig. 2F). We thus conclude that INCENP interacts with the CSD of HP1α through a PVVEI motif.
Figure 2.
INCENP interacts with the CSD of HP1α and HP1γ. A, lysates of nocodazole-arrested mitotic HeLa cells transiently expressing VSV-INCENP were subjected to pulldown by GST or the indicated GST fusion proteins followed by anti-VSV immunoblotting and Coomassie Brilliant Blue (CBB) staining. B, lysates of nocodazole-arrested mitotic HeLa cells were subjected to pulldown by GST or the indicated GST fusion proteins followed by anti-INCENP immunoblotting and Coomassie Brilliant Blue staining. C and D, lysates of nocodazole-arrested mitotic HeLa cells were subjected to pulldown by MBP or the indicated MBP fusion proteins followed by anti-HP1α (C and D), -HP1β, or -HP1γ (D) immunoblotting and Coomassie Brilliant Blue staining. E, lysates of nocodazole-arrested mitotic HeLa cells stably expressing INCENP-GFP were subjected to anti-GFP immunoprecipitation (IP) followed by immunoblotting with the indicated antibodies. F, lysates of nocodazole-arrested mitotic HeLa cells transiently expressing HP1α-FLAG and INCENP-GFP (WT or the indicated mutants) were subjected to anti-FLAG immunoprecipitation followed by anti-FLAG or anti-GFP immunoblotting.
Disrupting the INCENP–HP1 interaction delocalizes HP1 from mitotic centromeres and increases chromosome missegregation
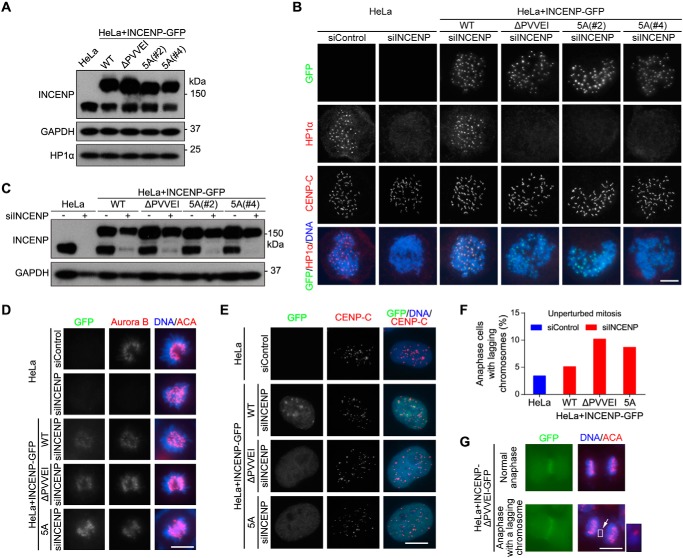
We stably expressed WT INCENP-GFP and the HP1 binding–deficient mutants INCENP-ΔPVVEI-GFP and INCENP-5A-GFP in HeLa cells (Fig. 3A). Immunofluorescence microscopy showed concentrated localization of HP1α and HP1γ at inner centromeres of mitotic chromosomes, which was abolished by transfection with siRNA targeting the 3′-UTR of endogenous INCENP (Figs. 3, B and C, and S2A). Interestingly, exogenous expression of INCENP-GFP, but not INCENP-ΔPVVEI-GFP and INCENP-5A-GFP, supported the centromeric localization of HP1α and HP1γ in the absence of endogenous INCENP. Moreover, INCENP-GFP (WT or the mutants) and endogenous Aurora B localized normally to centromeres in prometaphase and metaphase and subsequently to the anaphase spindle midzone (Figs. 3, B and D, and S2, B and C).
Figure 3.
Disrupting the INCENP–HP1 interaction delocalizes HP1 from mitotic centromeres and increases chromosome missegregation. A, lysates of nocodazole-arrested mitotic HeLa cells with or without stable expression of the indicated exogenous INCENP-GFP proteins were immunoblotted. B and C, HeLa cells and the indicated stable cell lines transfected with control or INCENP siRNA were treated with nocodazole for 3 h. Mitotic cells collected by shake-off were spun on slides and stained with DAPI and antibodies for GFP, HP1α, and CENP-C (B). The mitotic cell lysates were immunoblotted (C). D and E, asynchronous HeLa cells and the indicated stable cell lines transfected with control or INCENP siRNA were stained with DAPI and antibodies for Aurora B, CENP-C, or ACA. Example cells in prometaphase (D) and interphase (E) are shown. F and G, asynchronous HeLa cells and the indicated stable cell lines were fixed and stained with ACA and DAPI. The percentage of cells with lagging chromosomes was determined in 100 anaphase cells (F). Example images are shown (G). Scale bars, 10 μm. See also Fig. S2.
In addition, we further noticed that the mutant forms of INCENP-GFP were defective in localizing to the heterochromatic loci in interphase cells (Fig. 3E). Similar defects were observed when the INCENP mutant proteins were transiently expressed in mouse NIH 3T3 cells (Fig. S2D). These results indicate that although the INCENP–HP1 interaction is dispensable for the centromeric localization of CPC during mitosis it is required for CPC localization to heterochromatin in interphase.
Moreover, we found that the frequency of anaphases with lagging chromosomes was obviously higher in cells expressing the mutant forms of INCENP-GFP than in control cells (Fig. 3, F and G). An increased rate of chromosome missegregation was also observed when cells were released from transient mitotic arrest with S-trityl-l-cysteine (STLC) (Fig. S2E), an Eg5 inhibitor that reversibly blocks cells in prometaphase with monopolar spindles.
Taken together, these results indicate that INCENP targets HP1 to mitotic centromeres, but not vice versa, and that the INCENP–HP1 interaction is required to prevent chromosome missegregation during mitosis. In line with this, HP1β, which is defective in binding INCENP (Fig. 2, D and E), is not enriched at centromeres in mitosis (63).
Disrupting the INCENP–HP1 interaction does not compromise Aurora B kinase activity
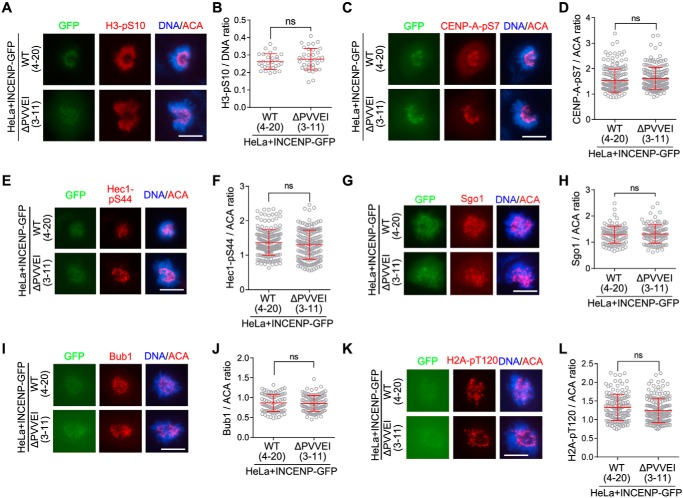
We next examined whether Aurora B kinase activity is compromised in cells expressing the HP1 binding–deficient mutants of INCENP-GFP. Aurora B phosphorylates multiple substrates, including histone H3 serine 10 on chromatin (35) and CENP-A at serine 7 at centromeres (70) (Fig. S1C) and KMN network proteins at kinetochores (12–15). Immunofluorescence staining showed that the levels of H3-pS10 were comparable in cells expressing the WT or mutant forms of INCENP-GFP (Figs. 4, A and B, and S3, A and B). This indicates that global Aurora B kinase activity is not compromised in the absence of INCENP–HP1 interaction. We further found that phosphorylation of CENP-A at serine 7 (CENP-A-pS7) and phosphorylation of Hec1 at serine 44 (Hec1-pS44), the readout of Aurora B kinase activity at centromeres and outer kinetochores, respectively, were insensitive to the loss of INCENP–HP1 interaction (Figs. 4, C–F, and S3, C and D).
Figure 4.
Disrupting the INCENP–HP1 interaction does not compromise Aurora B kinase activity. A and B, HeLa cells in which endogenous INCENP was stably replaced by exogenous INCENP-GFP (WT or the indicated mutant) were stained with DAPI, ACA, and the anti-H3-pS10 antibody (A). The immunofluorescence intensity ratio of H3-pS10/DNA was determined in around 30 cells (B). C–L, cells described in A were stained with DAPI, ACA, and antibodies for CENP-A-pS7 (C), Hec1-pS44 (E), Sgo1 (G), Bub1 (I), and H2A-pT120 (K). The immunofluorescence intensity ratios of centromeric CENP-A-pS7 (D), Hec1-pS44/ACA (F), Sgo1/ACA (H), Bub1/ACA (J), and H2A-pT120/ACA (L) were determined on 100–201 chromosomes in 20 cells. Means and error bars representing S.D. are shown (unpaired t test). Scale bars, 10 μm. See also Fig. S3. ns, not significant.
To test whether loss of INCENP–HP1 interaction influences Aurora B activation, we used conditions in which Aurora B is initially inhibited in mitotic cells, but reactivation is then allowed upon removal of Aurora B inhibitor (Fig. S3E). After treatment with Hesperadin, Hec1-pS44 signal at kinetochores was abolished as expected. After removal of Hesperadin, Hec1-pS44 at kinetochores was largely recovered within 60–90 min in cells expressing WT INCENP-GFP. When we repeated these experiments in cells expressing the mutant forms of INCENP-GFP, the kinetics of Hec1-pS44 recovery was not delayed (Fig. S3, F and G). These findings suggest that, in these experimental circumstances, the INCENP–HP1 interaction does not contribute to activation of Aurora B and phosphorylation of substrates at kinetochores. Moreover, in cells expressing either the wildtype or mutant forms of INCENP-GFP, we did not find obvious differences in centromeric enrichment of Sgo1 and H2A-pT120 as well as the kinetochore localization of Bub1 (Figs. 4, G–L, and S3, H–K). Taken together, these results indicate that loss of the INCENP–HP1 interaction does not detectably compromise Aurora B kinase activity in mitotic cells.
Disrupting the INCENP–HP1 interaction weakens centromeric cohesion
Inspection of chromosome spreads prepared from nocodazole-arrested mitotic cells showed that, upon depletion of endogenous INCENP by siRNA, cells expressing the mutant forms of INCENP-GFP did not have obvious loss of sister chromatid cohesion (Fig. S4, A and B). Similar results were obtained when cells were arrested in mitosis with STLC (Fig. S4C).
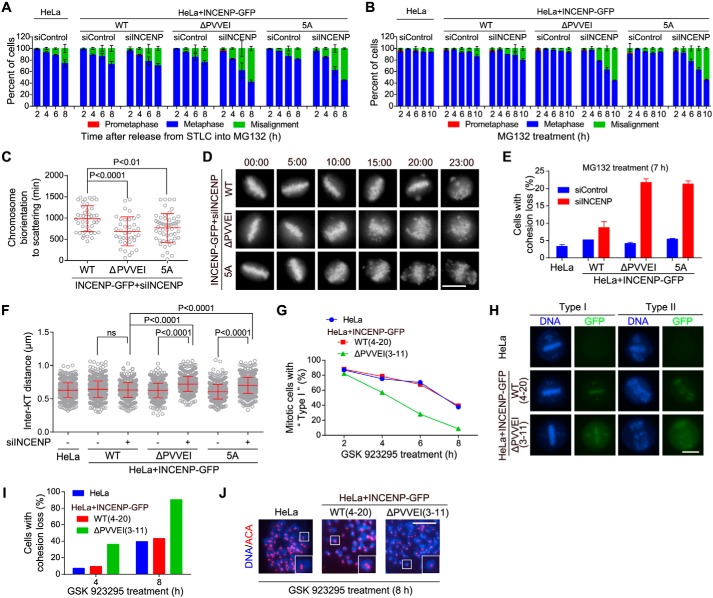
Next, we treated cells with STLC to accumulate monopolar mitoses and then released them into MG132, a proteasome inhibitor that arrests cells in metaphase, to allow bipolar spindle formation and chromosome alignment (71). Examination of fixed cells showed that cells expressing the mutant forms of INCENP-GFP were largely impaired in maintaining the alignment of chromosomes on the metaphase plate (Fig. 5A). Similar results were observed when these cells were directly subjected to MG132-induced metaphase arrest (Fig. 5B). Time-lapse live-cell imaging further showed that, upon MG132-induced metaphase arrest, cells expressing the mutant forms of INCENP-GFP began to exhibit irreversible chromosome scattering from the metaphase plate much earlier than control cells (Fig. 5, C and D).
Figure 5.
Disrupting the INCENP–HP1 interaction weakens centromeric cohesion. A, HeLa cells and the indicated stable cell lines transfected with control or INCENP siRNA were released from 5-h treatment with STLC into MG132-containing medium and then fixed at the indicated time points for DNA staining. The percentage of mitotic cells in prometaphase, metaphase, and metaphase with some misaligned chromosomes was determined in around 200 cells (n = 2). B, cells transfected as in A were exposed to MG132, then fixed at the indicated time points for DNA staining, and quantified in around 200 cells (n = 2). C and D, the indicated INCENP-GFP cell lines stably expressing H2B-GFP were exposed to MG132 and subjected to live imaging. The time from the achievement of metaphase chromosome alignment to chromosome scattering was determined in at least 37 cells (C). Selected frames of the movies are shown (D). Time stated in hours:minutes. E, cells transfected as in A were exposed to MG132 for 7 h. Using mitotic chromosome spreads, the percentage of cells with cohesion loss was determined in 100 cells (n = 2). Example images are shown in Fig. S4D. F, cells transfected as in A were treated with nocodazole for 3 h. Mitotic chromosome spreads were stained with ACA and DAPI. The inter-KT distance was measured on over 802 chromosomes in 20 cells. G and H, HeLa cells in which endogenous INCENP was stably replaced by exogenous INCENP-GFP (WT and the indicated mutant) were treated with GSK 923295 and then fixed at the indicated time points for DNA staining. The percentage of mitotic cells with mild (Type I) or severe (Type II) chromosome misalignment was determined in around 150 cells. I and J, mitotic cells treated as in G were collected to prepare chromosome spreads. The percentage of cells with cohesion loss was determined in around 100 cells (I). Example images are shown (J). Means and error bars representing S.D. are shown (unpaired t test). Scale bars, 10 μm. See also Fig. S4. ns, not significant.
These results prompted us to investigate whether sister chromatid cohesion was compromised in cells expressing the mutant forms of INCENP-GFP. Inspection of chromosome spreads prepared from cells arrested in metaphase with MG132 revealed a strong increase in premature chromatid separation (Figs. 5E and S4D). We also found that the inter-KT distance of mitotic chromosomes was around 20% further apart in mutant INCENP-GFP–expressing cells than that in the control cells (Fig. 5F), which is indicative of compromised cohesion at mitotic centromeres.
To further corroborate the contribution of INCENP–HP1 interaction to centromeric cohesion protection, we isolated two clones, 4-20 and 3-11, in which endogenous INCENP was depleted by CRISPR/Cas9-mediated genome editing (72) in cells stably expressing INCENP-GFP and INCENP-ΔPVVEI-GFP, respectively (Fig. S4E). Indeed, we found similar defects in maintaining chromosome alignment and sister chromatid cohesion during MG132-mediated metaphase arrest in INCENP-ΔPVVEI-GFP–expressing stable cell line (clone 3-11) (Fig. S4, F–H).
To rule out the possibility that these cohesion defects were only observed upon treatment with MG132, we treated cells with GSK 923295, a small-molecule inhibitor of the mitotic kinesin CENP-E (73). As expected, upon treatment with GSK 923295 for 2 h, control HeLa cells were arrested in a metaphase-like state (Type I) in which most chromosomes were aligned on the metaphase plate with only a few chromosomes scattered to the spindle pole–proximal area (Fig. 5, G and H). Over time, more chromosomes that had completed the alignment started to scatter from the metaphase plate (Type II). Inspection of chromosome spreads showed that these “Type II” cells were largely impaired in maintaining sister chromatid cohesion (Fig. 5, I and J). This indicates that cohesion loss largely accounts for the increased misalignment of chromosomes during the sustained inhibition of CENP-E. Importantly, we found that the defects in maintaining metaphase chromosome alignment (Fig. 5, G and H) and sister chromatid cohesion were much stronger in cells expressing the INCENP-ΔPVVEI-GFP mutant than in the control (Fig. 5, I and J).
We recently reported that double knockout of HP1α and HP1γ weakens sister chromatid cohesion at mitotic centromeres (63). In contrast, when HP1α and HP1β or HP1β and HP1γ were knocked out in combination (Fig. S4, I–K), cells were capable of maintaining metaphase chromosome alignment and sister chromatid cohesion (Fig. S4, L–N). Thus, HP1β, which is defective in binding INCENP and localizing at mitotic centromeres, is dispensable for centromeric cohesion protection. In sum, these results indicate that the INCENP–HP1 interaction is required to sustain proper strength of centromeric cohesion, which is particularly important for the maintenance of cohesion between sister chromatids on the metaphase plate.
Centromere-tethered HP1 and Wapl depletion bypass the requirement for INCENP–HP1 interaction in centromeric cohesion protection
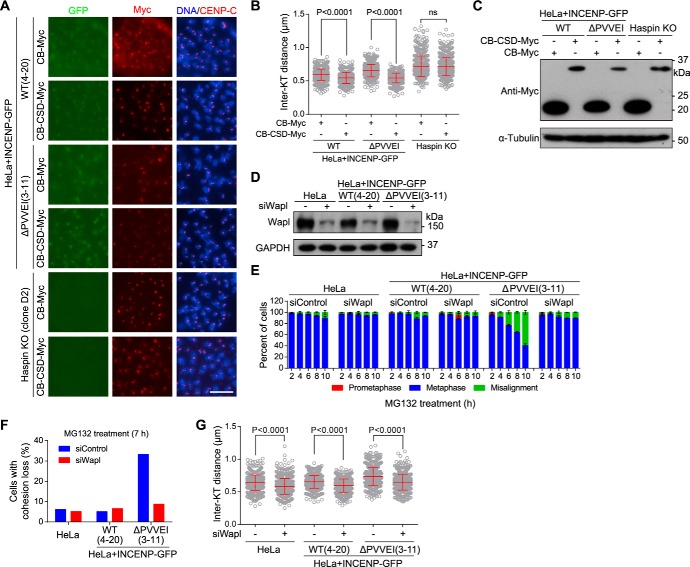
Given the requirement for INCENP in HP1 localization at mitotic centromeres, we next examined whether INCENP promotes centromeric cohesion at least partly through targeting HP1 to centromeres. We expressed the CSD of HP1α as a CENP-B fusion protein (CB-CSD-Myc). Immunofluorescence microscopy showed that expression of CB-CSD-Myc in Haspin-knockout (clone D2) cells was unable to shorten the inter-KT distance, which is in line with the requirement for Haspin in centromeric cohesion protection by HP1 (63). Importantly, expression of CB-CSD-Myc, but not CB-Myc, efficiently shortened the inter-KT distance in cells expressing either WT or the mutant form of INCENP-GFP (Fig. 6, A–C). Thus, targeting the CSD of HP1 to centromeres rescues sister chromatid cohesion defects in cells lacking the INCENP–HP1 interaction. These results are in line with our previous observation that stable expression of the CSD of HP1α as a CENP-B fusion protein (CB-CSD-GFP) is sufficient to support proper centromeric cohesion in the absence of endogenous HP1α and HP1γ (63). Moreover, we found that inhibition of Aurora B kinase activity by Hesperadin treatment was still capable of significantly increasing the inter-KT distance in HP1α and HP1γ double-knockout cells stably expressing CB-CSD-GFP (Fig. S5, A and B), suggesting that Aurora B kinase activity can protect centromeric cohesion in a way that is independent of the centromeric localization of HP1.
Figure 6.
Centromere-tethered HP1 and Wapl depletion bypass the requirement for INCENP–HP1 interaction in centromeric cohesion protection. A–C, the indicated stable cell lines were transfected with CB-Myc or CB-CSD-Myc and treated with nocodazole for 3 h. Mitotic chromosome spreads were stained with DAPI and antibodies for Myc and CENP-C. Example images are shown (A). The inter-KT distance was measured on over 616 chromosomes in 20 cells (B). The asynchronous cell lysates were immunoblotted (C). D–G, HeLa cells and the indicated stable cell lines transfected with control or Wapl siRNA were immunoblotted (D); exposed to MG132, fixed at the indicated time points for DNA staining, and quantified in around 200 cells (n = 2) (E); used to prepare chromosome spreads after 7-h treatment with MG132 followed by determining the percentage of cells with cohesion loss in around 60 cells (F); or treated with nocodazole for 3 h to make chromosome spreads and stained with ACA and DAPI followed by measuring the inter-KT distance on over 749 chromosomes in over 30 cells (G). Means and error bars representing S.D. are shown (unpaired t test). Scale bars, 10 μm. See also Fig. S5. ns, not significant.
Whereas the majority of cohesin is released from chromosome arms by Wapl in early mitosis to allow sister-chromatid resolution (18, 19), Wapl activity is normally inhibited at mitotic centromeres (25). We wondered whether aberrantly increased local Wapl activity might account for the centromeric cohesion defects in cells lacking the INCENP–HP1 interaction. We found that Wapl depletion by siRNA effectively prevented metaphase chromosome misalignment in INCENP-ΔPVVEI-GFP–expressing cells during MG132-induced metaphase arrest (Fig. 6, D and E). Consistently, these mutant cells were capable of maintaining sister chromatid cohesion in the absence of Wapl (Fig. 6F). Moreover, proper inter-KT distance was restored by Wapl depletion in cells expressing the INCENP-ΔPVVEI-GFP mutant (Fig. 6G). Thus, Wapl depletion bypasses the requirement for INCENP–HP1 interaction in centromeric cohesion protection.
The INCENP–HP1 interaction promotes centromeric localization of Haspin to antagonize Wapl
During mitosis, Haspin localizes predominantly to centromeres and phosphorylates histone H3 at threonine 3, which is particularly concentrated at centromeres (6, 7, 74). Given that INCENP targets HP1 to mitotic centromeres and that HP1 promotes the centromeric accumulation of Haspin (7, 63), we next examined whether Haspin localization at mitotic centromeres is compromised in cells lacking the INCENP–HP1 interaction.
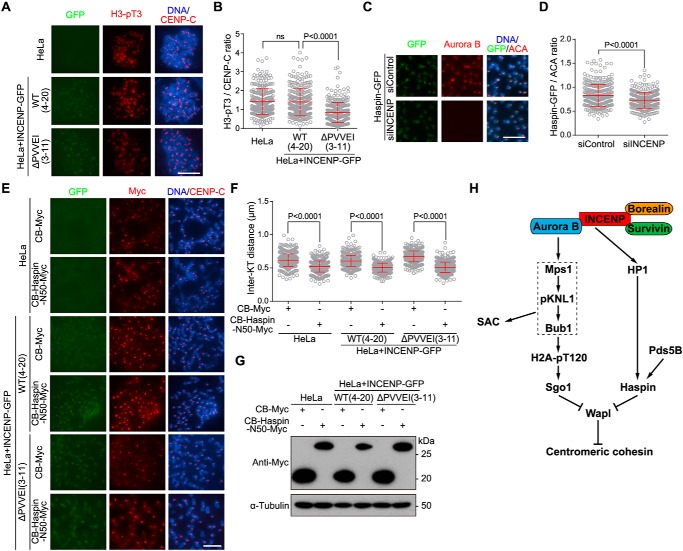
Immunofluorescence microscopy showed that the level of phosphorylation of histone H3 at threonine 3 (H3-pT3) at mitotic centromeres, the readout of Haspin activity, was significantly lower in INCENP-ΔPVVEI-GFP–expressing cells than in control cells (Fig. 7, A and B). Using HeLa cells stably expressing Haspin-GFP, we further found that depletion of INCENP by siRNA reduced the localization of Haspin-GFP at mitotic centromeres (Fig. 7, C and D). Thus, the INCENP–HP1 interaction is required for full enrichment of Haspin at centromeres in mitosis.
Figure 7.
The INCENP–HP1 interaction promotes centromeric localization of Haspin to antagonize Wapl. A and B, HeLa cells and the indicated stable cell lines were treated with nocodazole for 3 h. Mitotic cells were spun on slides and stained with DAPI and antibodies for H3-pT3 and CENP-C (A). The centromeric H3-pT3/CENP-C immunofluorescence intensity ratio was determined on 273–285 chromosomes in 20 cells (B). C and D, HeLa cells stably expressing Haspin-GFP were transfected with control or INCENP siRNA and treated with nocodazole for 3 h. Mitotic chromosome spreads were stained with DAPI and antibodies for GFP, Aurora B, and ACA (C). The centromeric GFP/CENP-C immunofluorescence intensity ratio was determined on around 320 chromosomes in 30 cells (D). E–G, HeLa cells and the indicated stable cell lines were transfected with CB-Myc or CB-Haspin-N50-Myc and treated with nocodazole for 3 h. Mitotic chromosome spreads were stained with DAPI and antibodies for Myc and CENP-C (E). The inter-KT distance was measured on 560–640 chromosomes in 25 cells (F). The asynchronous cell lysates were immunoblotted (G). H, model for the Aurora B kinase activity–dependent and –independent roles for the CPC in protecting sister chromatid cohesion at mitotic centromeres. Aurora B kinase activity–dependent localization of the Mps1–pKNL1–Bub1 signaling cascade to the kinetochore promotes centromeric cohesion through phosphorylating H2A and recruiting Sgo1 to centromeres. Aurora B kinase activity–independent INCENP–HP1 interaction protects centromeric cohesion through promoting the centromeric localization of Haspin. Both Sgo1 and Haspin antagonize Wapl activity in releasing cohesin from mitotic centromeres. Means and error bars representing S.D. are shown (unpaired t test). Scale bars, 10 μm. ns, not significant.
We previously showed that the Pds5-interacting motif (PIM) in the N terminus of Haspin competes with the PIM of Wapl for binding Pds5B and that centromeric targeting of the PIM-containing fragment of Haspin as a CENP-B fusion protein rescues the centromeric cohesion defect in cells lacking the Pds5B–Haspin interaction (75). We hypothesized that, if the INCENP–HP1 interaction protects centromeric cohesion through localizing Haspin, artificially targeting Haspin to centromeres might bypass the need for INCENP–HP1 interaction in cohesion protection. Indeed, expression of the N terminus (residues 1–50) of Haspin as a CENP-B fusion protein (CB-Haspin-N50-Myc), but not CB-Myc, strongly shortened the inter-KT distance in cells expressing the INCENP-ΔPVVEI-GFP mutant (Fig. 7, E–G), indicative of the restoration of proper centromeric cohesion. Taken together, these results indicate that INCENP targets HP1 to mitotic centromeres, which promotes the centromeric localization of Haspin to antagonize Wapl activity in cohesin release.
Discussion
Timely resolution of sister chromatid cohesion in a stepwise manner during mitosis is critical for precise chromosome segregation. Defects in this process lead to chromosomal instability in cancer cells (76). It has been known for a long time that Aurora B kinase activity promotes cohesin release from chromosome arms, thereby enabling the resolution of sister chromatids in prophase and prometaphase (27, 28). Previous studies also found that depletion of Survivin or Aurora B or inhibition of Aurora B kinase activity impaired centromeric cohesion during mitosis (77, 78). We confirmed these observations and further demonstrated that the requirement for Aurora B kinase activity in centromeric cohesion protection can be bypassed by centromere-tethered Bub1, whose kinase activity toward histone H2A is required to recruit Sgo1 to centromeres (38, 39, 41, 46, 79–81). Thus, Aurora B kinase activity–dependent localization of the Mps1–pKNL1–Bub1 (p represents phosphorylation) signaling cascade to kinetochores not only plays an important role in SAC signaling through recruiting spindle checkpoint proteins (82) but also is required for centromeric cohesion through phosphorylating H2A and recruiting Sgo1 to centromeres (see the working model in Fig. 7H).
Previous studies have indicated that HP1 is a CPC-binding protein (59–61, 83) and that the INCENP–HP1 interaction assists Aurora B kinase activity to prevent chromosome segregation errors, particularly in nontransformed cells (62). Our finding that the INCENP–HP1 interaction is required for CPC localization to heterochromatic loci in G2 phase cells, together with a recent study (84), confirms the observation in a previous study (61). Interestingly, we found that loss of INCENP–HP1 interaction in HeLa cells does not seem to compromise Aurora B kinase activity at mitotic centromeres, likely because the amount of HP1 associated with CPC in cancer cells is below the threshold that is sufficient to cause an allosteric activation of Aurora B (62).
Importantly, we found that the INCENP–HP1 interaction is required to maintain sister chromatid cohesion at mitotic centromeres. This is in line with a recent study that infers that the CPC is localized at the inner centromere to sustain centromere cohesion on bioriented chromosomes with an unknown mechanism (85). At first glance, our results appear inconsistent with the previous observation that mitotic centromere targeting of HP1 is dispensable for sister chromatid cohesion in a HeLa-derived Tet-On cell line (61). However, it should be noted that the status of sister chromatid cohesion in the absence of INCENP–HP1 interaction was measured by Kang et al. (61) using chromosome spreads prepared from nocodazole-arrested mitotic cells. Consistently, we did not observe obvious cohesion defects in cells arrested in mitosis with either nocodazole or STLC. Intriguingly, we found that the INCENP–HP1 interaction is particularly important to maintain cohesion between sister chromatids on the metaphase plate, a situation where the kinetochore is under the sustained spindle pulling forces. The cohesion defects observed in cells lacking the INCENP–HP1 interaction are reminiscent of the “cohesion fatigue” phenotype (86–88). We demonstrated that Wapl depletion restores proper strength of centromeric cohesion in the absence of INCENP–HP1 interaction. In contrast, a recent study showed that Wapl-mediated opening of cohesin rings is not required after metaphase arrest to separate sister chromatid in cohesion fatigue (89). Thus, it seems that the sister chromatid cohesion defects in cells lacking INCENP–HP1 interaction are not simply an accelerated cohesion fatigue defect.
Although we cannot fully rule out the possibility that the INCENP–HP1 interaction protects centromeric cohesion through an additional unknown mechanism, we favor the idea that this interaction promotes the centromeric localization of Haspin, thereby antagonizing Wapl activity in cohesin release from mitotic centromeres (Fig. 7H) (63, 75, 90–92). Because both INCENP and Haspin use the PXVXL motif to bind HP1 (61, 63), it remains to be determined in the future how HP1 brought to centromeres by INCENP can also bind and recruit Haspin. This could be due to the dynamic nature of the interaction between the CSD dimer and PXVXL motif as well as the fact that INCENP is much more abundant than Haspin in somatic cells.
Moreover, we found that loss of the INCENP–HP1 interaction causes an elevated rate of chromosome missegregation. Then how might chromosome segregation errors occur in these cells with normal Aurora B kinase activity? It is possible that weakened cohesion at centromeres of metaphase chromosomes alters the kinetochore geometry and/or chromosome oscillation, thereby increasing errors in the KT–MT attachments.
Taken together, our results indicate that the CPC regulates sister chromatid cohesion at mitotic centromeres in ways that are both Aurora B kinase activity–dependent and –independent, thereby ensuring the maintenance of centromeric cohesion to the full extent in mitosis. This study provides important insight into the complex mechanism that ensures the fidelity of chromosome segregation during mitosis in human cells.
Experimental procedures
Cell culture, plasmids, siRNAs, transfection, and drug treatments
All cells were cultured in Dulbecco's modified Eagle's medium supplemented with 1% penicillin/streptomycin and 10% fetal bovine serum (Gibco) and maintained at 37 °C with 5% CO2. Cells stably expressing CB-GFP, H2B-GFP, INCENP-GFP, CB-Bub1-K-GFP, or Haspin-GFP were isolated and maintained in 3.0 and 2.0 μg/ml blasticidin (Sigma), respectively. To make pBos-CB-GFP, the H2B fragment in pBos-H2B-GFP (Clontech) was replaced with the KpnI/BamHI-digested PCR fragments encoding the centromere-targeting domain (residues 1–163) of CENP-B. The plasmid pBos-INCENP-GFP was constructed similarly. To make pBos-CB-Bub1-K-GFP, the PCR fragments encoding Bub1 kinase domain (residues 630–1085) were subcloned into the BamHI site of pBos-CB-GFP. To make pBos-CB-Myc, GFP sequence in pBos-CB-GFP was digested with BamHI and NotI and replaced with the Myc tag. Then the PCR fragments encoding HP1 CSD (residues 121–179) and Haspin N terminus (residues 1–50) were subcloned into the BamHI site of pBos-CB-Myc, resulting in pBos-CB-CSD-Myc and pBos-CB-Haspin-N50-Myc, respectively. All point mutations were introduced with the QuikChange II XL site-directed mutagenesis kit (Agilent Technologies). All plasmids were sequenced to verify desired mutations and absence of unintended mutations.
The siRNA targeting the 3′-UTR of INCENP (5′-GGCUUGGCCAGGUGUAUAUdTdT-3′), Wapl siRNA (5′-CGGACUACCCUUAGCACAAdTdT-3′), and control siRNA were ordered from RiboBio. Plasmid and siRNA transfections were done with FuGENE 6 (Promega) and Oligofectamine or Lipofectamine RNAiMAX (Invitrogen), respectively. Cells were arrested in mitosis with 0.33 μm nocodazole (Selleckchem). Other drugs used in this study were STLC (5 μm; Tocris Bioscience), MG132 (5–20 μm; MCE), GSK 923295 (1 μm; Selleckchem), Hesperadin (20–50 nm; Selleckchem), ZM447439 (2–3 μm; Selleckchem), Reversine (0.5 μm; MedChem Express), and AZ3146 (3 μm; MedChem Express). Mitotic cells were collected by selective detachment with “shake-off.”
CRISPR/Cas9 gene editing
Single-guide RNA (sgRNA) for human INCENP gene was ordered as oligonucleotides from Integrated DNA Technology, annealed, and cloned into the BbsI site of dual Cas9 and sgRNA expression vector pX330 (Addgene). The plasmids were transfected into HeLa cells stably expressing INCENP-GFP or INCENP-ΔPVVEI-GFP. After 48-h incubation, cells were split individually to make a clonal cell line with selection using 1 μg/ml puromycin for 2–3 days. The sgRNA targeting sequence (5′-TGCCTTCACCAGACAGAGCC-3′) is in an intron 3 bases upstream of the start codon of INCENP gene. A total of 96 clones (48 clones for each) were isolated and analyzed by immunoblotting to confirm the loss of endogenous INCENP.
The HeLa cell–derived HP1β-KO clone was described previously (63). HP1α was knocked out in HP1β-KO cells by means of double nicking with the pX462 (Addgene)-expressed Cas9-D10A mutant and a pair of sgRNAs targeting two sequences (5′-AGAACTGTCAGCTGTCCGCTtgg-3′, 17 bases downstream of the start codon, and 5′-GGATGAGGAGGAGTATGTTGtgg-3′; the PAM sequence is lowercase). Similarly, HP1γ was knocked out in HP1β-KO cells with the Cas9-D10A mutant and a pair of sgRNAs targeting two sequences (5′-TTTTCCACGACAAATTCTTCagg-3′, 78 bases downstream of the start codon, and 5′-CTAGATCGACGTGTAGTGAAtgg-3′). Clones with loss of HP1 proteins were isolated by immunostaining and confirmed by immunoblotting.
Antibodies
Rabbit polyclonal antibodies used were to H3-pT3 (B8633; a gift from Dr. Jonathan Higgins), GFP (A11122, Invitrogen), GAPDH (14C10, Cell Signaling Technology), H2A-pT120 (Active Motif), INCENP (2807, Cell Signaling Technology), CENP-A-pS7 (04-792, Millipore), VSV (V-4888, Sigma), and Wapl (A300-268A, Bethyl Laboratories). Rabbit anti-Bub1 and anti-Hec1-pS44 polyclonal antibodies were produced by immunization with the synthetic peptides NYGLPQPKNKPTGAR and PTFGKLpSINKPTSE (where pS is phosphoserine), respectively. Mouse monoclonal antibodies used were to HP1α (MAB3446 for immunoblotting; MAB3584 for immunostaining), HP1β (MAB3448), and HP1γ (MAB3450) (all from Millipore); FLAG tag (M2, Sigma); Myc tag (4A6, Millipore); H3-pS10 (6G3, Cell Signaling Technology); Aurora B (AIM-1, BD Biosciences); α-Tubulin (T-6047, Sigma); and Sgo1 (3C11, Abnova). Guinea pig polyclonal antibodies against CENP-C were from MBL (PD030). Anti-human centromere autoantibody (ACA) was from Immunovision. Secondary antibodies for immunoblotting were goat anti-rabbit or horse anti-mouse IgG–horseradish peroxidase (Cell Signaling Technology). Secondary antibodies for immunostaining were donkey anti-rabbit IgG–Alexa Fluor 488 or Cy3 (Jackson ImmunoResearch Laboratories), anti-mouse IgG–Alexa Fluor 488 or 546 (Invitrogen), anti-human IgG–Alexa Fluor 647 (Jackson ImmunoResearch Laboratories), and goat anti-guinea pig IgG–Alexa Fluor 647 (Invitrogen).
Fluorescence microscopy, time-lapse live-cell imaging, and statistical analysis
Cells cultured on coverslips were fixed with 2% paraformaldehyde in PBS for 10 min followed by extraction with 0.5% Triton X-100 in PBS for 5 min. To stain Hec1-pS44, cells were extracted for 5 min in PHEM (60 mm Pipes, 25 mm Hepes, 10 mm EGTA, and 2 mm MgCl2, pH 6.9) plus 1% Triton X-100 and fixed at room temperature for 20 min in PHEM plus 4% formaldehyde. Mitotic cells collected by shake-off were spun on slides by Cytospin (Cytospin 4, Thermo Scientific) at 1500 rpm for 5 min. To produce chromosome spreads, mitotic cells obtained by selective detachment were incubated in 75 mm KCl for 10 min. After attachment to glass coverslips by Cytospin, chromosome spreads were fixed with 2% paraformaldehyde in PBS for 10 min followed by extraction with 0.5% Triton X-100 in PBS for 5 min. Fixed cells and chromosome spreads were stained with primary antibodies for 1–2 h and secondary antibodies for 1 h, all with 3% BSA in PBS with 0.5% Triton X-100 and at room temperature. DNA was stained for 10 min with DAPI. Fluorescence microscopy was carried out at room temperature using a Nikon ECLIPSE Ni microscope with a Plan Apo Fluor 60× oil (numerical aperture, 1.4) objective lens and a Clara charge-coupled device (Andor Technology). The inter-KT distance was measured using the inner kinetochore marker CENP-C or centromere marker ACA on over 15 kinetochores per cell in at least 10 cells. Distance was determined by drawing a line from the outer kinetochore extending to the outer edge of its sister kinetochore. The length of the line was calculated using the imaging software of NIS-Elements BR (Nikon). Quantification of fluorescence intensity was carried out with NIH ImageJ using images obtained at identical illumination settings. Briefly, on chromosome spreads, the average pixel intensity of Sgo1, Bub1, H2A-pT120, CENP-A-pS7, Hec1-pS44, H3-pT3, Haspin-GFP, CENP-C, or ACA staining at centromeres, defined as circular regions including paired centromeres, or on chromosome arms, was determined using ImageJ. After background correction, the ratio of kinetochore or centromeric staining intensity versus CENP-C or ACA or centromeric Sgo1/arm Sgo1 was calculated for each centromere.
Time-lapse live-cell imaging was carried out with the GE DV Elite Applied Precision DeltaVision system (GE Healthcare) equipped with Olympus oil objectives of 40× (numerical aperture, 1.35) UApo/340, an API Custom Scientific complementary metal-oxide semiconductor camera, and Resolve3D softWoRx imaging software. Cells expressing H2B-GFP were plated in four-chamber glass-bottomed 35-mm dishes (Cellvis) coated with poly-d-lysine and filmed in a climate-controlled and humidified environment (37 °C and 5% CO2). Images were captured every 5 min. The acquired images were processed using Adobe Photoshop and Adobe Illustrator. Statistical analyses were performed with a two-tailed unpaired Student's t test in GraphPad Prism 6. A p value less than 0.05 was considered significant.
Immunoblotting, immunoprecipitation, protein purification, and GST/MBP pulldown
SDS-PAGE, immunoblotting, and immunoprecipitation were carried out using standard procedures. Cell lysates were prepared in standard SDS sample buffer for immunoblotting. For the immunoprecipitation, cells were lysed in “P150 buffer” containing 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.5% Nonidet P-40, 10% glycerol, protease inhibitor mixture (P8340, Sigma), 1 mm phenylmethylsulfonyl fluoride or phenylmethanesulfonyl fluoride, 0.1 μm okadaic acid (Calbiochem), 10 mm NaF, 20 mm β-glycerophosphate, and Benzonase (Merck). After removal of insoluble materials by high-speed centrifugation, lysates were precleared with GammaBind G-Sepharose (17-0885-02, GE Healthcare). Lysates were incubated with antibodies for 3 h at 4 °C before addition of GammaBind G-Sepharose for a further 1 h. Beads were washed several times with the lysis buffer, boiled in standard SDS sample buffer, and subjected to immunoblotting.
Plasmids expressing MBP- or GST-fused HP1 and INCENP proteins were constructed by subcloning PCR fragments into pGEX-MBP-10xHis or pGEX-4T1. The plasmids were transformed into BL21 competent cells (Stratagene). Cells were grown in LB broth under antibiotic selection at 37 °C until reaching an A600 of 0.6–0.7, and protein expression was induced with 0.4 mm isopropyl 1-thio-β-d-galactopyranoside at 16 °C for 16 h. Cells were lysed by sonication in buffer A (50 mm Tris-HCl, pH 8.0, and 300 mm NaCl; for MBP fusion proteins) or buffer B (20 mm Tris-HCl, pH 8.0, 100 mm NaCl, 1 mm EDTA, and 1% Triton X-100; for GST fusion proteins). The lysate was clarified by centrifugation and incubated with amylose resin (BioLab) or GSH-Sepharose 4B (GE Healthcare) in lysis buffer. The resins were washed with lysis buffer or eluted with 150 mm maltose or 150 mm GSH. For pulldown of proteins from lysates (GST pulldown as the example), HeLa cells were first lysed in P150 buffer except that NaCl was used at 500 mm, and then the lysates were diluted to achieve the final concentration of NaCl at 150 mm. The lysates were precleared with GSH-Sepharose 4B beads and then incubated with GST fusion proteins immobilized to GSH-Sepharose 4B beads for 2 h.
Author contributions
Q. Y. and Q. C. validation; Q. Y., Q. C., M. Z., and F. W. investigation; Q. Y., Q. C., H. Y., M. Z., C. L., X. X., X. P., and F. W. methodology; H. Y. and F. W. conceptualization; H. Y. and F. W. resources; H. Y. and F. W. writing-review and editing; F. W. data curation; F. W. formal analysis; F. W. supervision; F. W. funding acquisition; F. W. writing-original draft; F. W. project administration.
Supplementary Material
Acknowledgment
We thank members of our laboratory for discussions and the Life Sciences Institute core facility for technical assistance.
This work was supported by National Key Research and Development Program of China Grant 2017YFA0503600 (to F. W.); National Natural Science Foundation of China (NSFC) Grants 31571393, 31771499, 31561130155, 31322032, and 31371359 (to F. W.); Natural Science Foundation of Zhejiang Province Grants LZ19C070001 and LR13C070001 (to F. W.) and LY17C070003 (to H. Y.); Newton Advanced Fellowship NA140075; and Fundamental Research Funds for the Central Universities. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S5.
- SAC
- spindle assembly checkpoint
- CPC
- chromosomal passenger complex
- INCENP
- inner centromere protein
- CSD
- chromo shadow domain
- HP1
- heterochromatin protein 1
- KT–MT
- kinetochore–microtubule
- inter-KT
- interkinetochore
- CB
- CENP-B
- STLC
- S-trityl-l-cysteine
- KMN
- KNL1/Mis12 complex/Ndc80 complex
- H3-pS10
- H3 serine 10 phosphorylation
- H2A-pT120
- phosphorylation of histone H2A at threonine 120
- VSV
- vesicular stomatitis virus
- MBP
- maltose-binding protein
- CENP
- centromere protein
- CENP-A-pS7
- phosphorylation of CENP-A at serine 7
- Hec1-pS44
- phosphorylation of Hec1 at serine 44
- H3-pT3
- phosphorylation of histone H3 at threonine 3
- PIM
- Pds5-interacting motif
- sgRNA
- single-guide RNA
- KO
- knockout
- ACA
- anti-human centromere autoantibody
- DAPI
- 4′,6-diamidino-2-phenylindole.
References
- 1. Bakhoum S. F., Ngo B., Laughney A. M., Cavallo J. A., Murphy C. J., Ly P., Shah P., Sriram R. K., Watkins T. B. K., Taunk N. K., Duran M., Pauli C., Shaw C., Chadalavada K., Rajasekhar V. K., et al. (2018) Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 553, 467–472 10.1038/nature25432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bakhoum S. F., and Cantley L. C. (2018) The multifaceted role of chromosomal instability in cancer and its microenvironment. Cell 174, 1347–1360 10.1016/j.cell.2018.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Trivedi P., and Stukenberg P. T. (2016) A centromere-signaling network underlies the coordination among mitotic events. Trends Biochem. Sci. 41, 160–174 10.1016/j.tibs.2015.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Carmena M., Pinson X., Platani M., Salloum Z., Xu Z., Clark A., Macisaac F., Ogawa H., Eggert U., Glover D. M., Archambault V., and Earnshaw W. C. (2012) The chromosomal passenger complex activates Polo kinase at centromeres. PLoS Biol. 10, e1001250 10.1371/journal.pbio.1001250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kelly A. E., Ghenoiu C., Xue J. Z., Zierhut C., Kimura H., and Funabiki H. (2010) Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science 330, 235–239 10.1126/science.1189505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang F., Dai J., Daum J. R., Niedzialkowska E., Banerjee B., Stukenberg P. T., Gorbsky G. J., and Higgins J. M. (2010) Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science 330, 231–235 10.1126/science.1189435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yamagishi Y., Honda T., Tanno Y., and Watanabe Y. (2010) Two histone marks establish the inner centromere and chromosome bi-orientation. Science 330, 239–243 10.1126/science.1194498 [DOI] [PubMed] [Google Scholar]
- 8. Sessa F., Mapelli M., Ciferri C., Tarricone C., Areces L. B., Schneider T. R., Stukenberg P. T., and Musacchio A. (2005) Mechanism of Aurora B activation by INCENP and inhibition by hesperadin. Mol. Cell 18, 379–391 10.1016/j.molcel.2005.03.031 [DOI] [PubMed] [Google Scholar]
- 9. Bishop J. D., and Schumacher J. M. (2002) Phosphorylation of the carboxyl terminus of inner centromere protein (INCENP) by the Aurora B kinase stimulates Aurora B kinase activity. J. Biol. Chem. 277, 27577–27580 10.1074/jbc.C200307200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Honda R., Körner R., and Nigg E. A. (2003) Exploring the functional interactions between Aurora B, INCENP, and survivin in mitosis. Mol. Biol. Cell 14, 3325–3341 10.1091/mbc.e02-11-0769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Adams R. R., Wheatley S. P., Gouldsworthy A. M., Kandels-Lewis S. E., Carmena M., Smythe C., Gerloff D. L., and Earnshaw W. C. (2000) INCENP binds the Aurora-related kinase AIRK2 and is required to target it to chromosomes, the central spindle and cleavage furrow. Curr. Biol. 10, 1075–1078 10.1016/S0960-9822(00)00673-4 [DOI] [PubMed] [Google Scholar]
- 12. DeLuca J. G., Gall W. E., Ciferri C., Cimini D., Musacchio A., and Salmon E. D. (2006) Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell 127, 969–982 10.1016/j.cell.2006.09.047 [DOI] [PubMed] [Google Scholar]
- 13. Cheeseman I. M., Chappie J. S., Wilson-Kubalek E. M., and Desai A. (2006) The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 127, 983–997 10.1016/j.cell.2006.09.039 [DOI] [PubMed] [Google Scholar]
- 14. Welburn J. P., Vleugel M., Liu D., Yates J. R. 3rd, Lampson M. A., Fukagawa T., and Cheeseman I. M. (2010) Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol. Cell 38, 383–392 10.1016/j.molcel.2010.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. DeLuca K. F., Lens S. M., and DeLuca J. G. (2011) Temporal changes in Hec1 phosphorylation control kinetochore-microtubule attachment stability during mitosis. J. Cell Sci. 124, 622–634 10.1242/jcs.072629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. London N., and Biggins S. (2014) Signalling dynamics in the spindle checkpoint response. Nat. Rev. Mol. Cell Biol. 15, 736–747 10.1038/nrm3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nasmyth K., and Haering C. H. (2009) Cohesin: its roles and mechanisms. Annu. Rev. Genet. 43, 525–558 10.1146/annurev-genet-102108-134233 [DOI] [PubMed] [Google Scholar]
- 18. Gandhi R., Gillespie P. J., and Hirano T. (2006) Human Wapl is a cohesin-binding protein that promotes sister-chromatid resolution in mitotic prophase. Curr. Biol. 16, 2406–2417 10.1016/j.cub.2006.10.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kueng S., Hegemann B., Peters B. H., Lipp J. J., Schleiffer A., Mechtler K., and Peters J. M. (2006) Wapl controls the dynamic association of cohesin with chromatin. Cell 127, 955–967 10.1016/j.cell.2006.09.040 [DOI] [PubMed] [Google Scholar]
- 20. Rankin S., Ayad N. G., and Kirschner M. W. (2005) Sororin, a substrate of the anaphase-promoting complex, is required for sister chromatid cohesion in vertebrates. Mol. Cell 18, 185–200 10.1016/j.molcel.2005.03.017 [DOI] [PubMed] [Google Scholar]
- 21. Schmitz J., Watrin E., Lénárt P., Mechtler K., and Peters J. M. (2007) Sororin is required for stable binding of cohesin to chromatin and for sister chromatid cohesion in interphase. Curr. Biol. 17, 630–636 10.1016/j.cub.2007.02.029 [DOI] [PubMed] [Google Scholar]
- 22. Nishiyama T., Ladurner R., Schmitz J., Kreidl E., Schleiffer A., Bhaskara V., Bando M., Shirahige K., Hyman A. A., Mechtler K., and Peters J. M. (2010) Sororin mediates sister chromatid cohesion by antagonizing Wapl. Cell 143, 737–749 10.1016/j.cell.2010.10.031 [DOI] [PubMed] [Google Scholar]
- 23. Lafont A. L., Song J., and Rankin S. (2010) Sororin cooperates with the acetyltransferase Eco2 to ensure DNA replication-dependent sister chromatid cohesion. Proc. Natl. Acad. Sci. U.S.A. 107, 20364–20369 10.1073/pnas.1011069107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ladurner R., Kreidl E., Ivanov M. P., Ekker H., Idarraga-Amado M. H., Busslinger G. A., Wutz G., Cisneros D. A., and Peters J. M. (2016) Sororin actively maintains sister chromatid cohesion. EMBO J. 35, 635–653 10.15252/embj.201592532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Morales C., and Losada A. (2018) Establishing and dissolving cohesion during the vertebrate cell cycle. Curr. Opin. Cell Biol. 52, 51–57 10.1016/j.ceb.2018.01.010 [DOI] [PubMed] [Google Scholar]
- 26. Waizenegger I. C., Hauf S., Meinke A., and Peters J. M. (2000) Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell 103, 399–410 10.1016/S0092-8674(00)00132-X [DOI] [PubMed] [Google Scholar]
- 27. Losada A., Hirano M., and Hirano T. (2002) Cohesin release is required for sister chromatid resolution, but not for condensin-mediated compaction, at the onset of mitosis. Genes Dev. 16, 3004–3016 10.1101/gad.249202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Giménez-Abián J. F., Sumara I., Hirota T., Hauf S., Gerlich D., de la Torre C., Ellenberg J., and Peters J. M. (2004) Regulation of sister chromatid cohesion between chromosome arms. Curr. Biol. 14, 1187–1193 10.1016/j.cub.2004.06.052 [DOI] [PubMed] [Google Scholar]
- 29. Hauf S., Roitinger E., Koch B., Dittrich C. M., Mechtler K., and Peters J. M. (2005) Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol. 3, e69 10.1371/journal.pbio.0030069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McGuinness B. E., Hirota T., Kudo N. R., Peters J. M., and Nasmyth K. (2005) Shugoshin prevents dissociation of cohesin from centromeres during mitosis in vertebrate cells. PLoS Biol. 3, e86 10.1371/journal.pbio.0030086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sumara I., Vorlaufer E., Stukenberg P. T., Kelm O., Redemann N., Nigg E. A., and Peters J. M. (2002) The dissociation of cohesin from chromosomes in prophase is regulated by Polo-like kinase. Mol. Cell 9, 515–525 10.1016/S1097-2765(02)00473-2 [DOI] [PubMed] [Google Scholar]
- 32. Dreier M. R., Bekier M. E. 2nd, and Taylor W. R. (2011) Regulation of sororin by Cdk1-mediated phosphorylation. J. Cell Sci. 124, 2976–2987 10.1242/jcs.085431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nishiyama T., Sykora M. M., Huis in 't Veld P. J., Mechtler K., and Peters J. M. (2013) Aurora B and Cdk1 mediate Wapl activation and release of acetylated cohesin from chromosomes by phosphorylating Sororin. Proc. Natl. Acad. Sci. U.S.A. 110, 13404–13409 10.1073/pnas.1305020110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hauf S., Cole R. W., LaTerra S., Zimmer C., Schnapp G., Walter R., Heckel A., van Meel J., Rieder C. L., and Peters J. M. (2003) The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 161, 281–294 10.1083/jcb.200208092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hsu J. Y., Sun Z. W., Li X., Reuben M., Tatchell K., Bishop D. K., Grushcow J. M., Brame C. J., Caldwell J. A., Hunt D. F., Lin R., Smith M. M., and Allis C. D. (2000) Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102, 279–291 10.1016/S0092-8674(00)00034-9 [DOI] [PubMed] [Google Scholar]
- 36. Ditchfield C., Johnson V. L., Tighe A., Ellston R., Haworth C., Johnson T., Mortlock A., Keen N., and Taylor S. S. (2003) Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 161, 267–280 10.1083/jcb.200208091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. van der Waal M. S., Saurin A. T., Vromans M. J., Vleugel M., Wurzenberger C., Gerlich D. W., Medema R. H., Kops G. J., and Lens S. M. (2012) Mps1 promotes rapid centromere accumulation of Aurora B. EMBO Rep. 13, 847–854 10.1038/embor.2012.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kawashima S. A., Yamagishi Y., Honda T., Ishiguro K., and Watanabe Y. (2010) Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science 327, 172–177 10.1126/science.1180189 [DOI] [PubMed] [Google Scholar]
- 39. Liu H., Qu Q., Warrington R., Rice A., Cheng N., and Yu H. (2015) Mitotic transcription installs Sgo1 at centromeres to coordinate chromosome segregation. Mol. Cell 59, 426–436 10.1016/j.molcel.2015.06.018 [DOI] [PubMed] [Google Scholar]
- 40. Meppelink A., Kabeche L., Vromans M. J., Compton D. A., and Lens S. M. (2015) Shugoshin-1 balances Aurora B kinase activity via PP2A to promote chromosome bi-orientation. Cell Rep. 11, 508–515 10.1016/j.celrep.2015.03.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ricke R. M., Jeganathan K. B., Malureanu L., Harrison A. M., and van Deursen J. M. (2012) Bub1 kinase activity drives error correction and mitotic checkpoint control but not tumor suppression. J. Cell Biol. 199, 931–949 10.1083/jcb.201205115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Earnshaw W. C., Sullivan K. F., Machlin P. S., Cooke C. A., Kaiser D. A., Pollard T. D., Rothfield N. F., and Cleveland D. W. (1987) Molecular cloning of cDNA for CENP-B, the major human centromere autoantigen. J. Cell Biol. 104, 817–829 10.1083/jcb.104.4.817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pluta A. F., Saitoh N., Goldberg I., and Earnshaw W. C. (1992) Identification of a subdomain of CENP-B that is necessary and sufficient for localization to the human centromere. J. Cell Biol. 116, 1081–1093 10.1083/jcb.116.5.1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Masumoto H., Masukata H., Muro Y., Nozaki N., and Okazaki T. (1989) A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J. Cell Biol. 109, 1963–1973 10.1083/jcb.109.5.1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Salic A., Waters J. C., and Mitchison T. J. (2004) Vertebrate shugoshin links sister centromere cohesion and kinetochore microtubule stability in mitosis. Cell 118, 567–578 10.1016/j.cell.2004.08.016 [DOI] [PubMed] [Google Scholar]
- 46. Tang Z., Sun Y., Harley S. E., Zou H., and Yu H. (2004) Human Bub1 protects centromeric sister-chromatid cohesion through Shugoshin during mitosis. Proc. Natl. Acad. Sci. U.S.A. 101, 18012–18017 10.1073/pnas.0408600102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kitajima T. S., Kawashima S. A., and Watanabe Y. (2004) The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature 427, 510–517 10.1038/nature02312 [DOI] [PubMed] [Google Scholar]
- 48. Zhu T., Dou Z., Qin B., Jin C., Wang X., Xu L., Wang Z., Zhu L., Liu F., Gao X., Ke Y., Wang Z., Aikhionbare F., Fu C., Ding X., et al. (2013) Phosphorylation of microtubule-binding protein Hec1 by mitotic kinase Aurora B specifies spindle checkpoint kinase Mps1 signaling at the kinetochore. J. Biol. Chem. 288, 36149–36159 10.1074/jbc.M113.507970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nijenhuis W., von Castelmur E., Littler D., De Marco V., Tromer E., Vleugel M., van Osch M. H., Snel B., Perrakis A., and Kops G. J. (2013) A TPR domain-containing N-terminal module of MPS1 is required for its kinetochore localization by Aurora B. J. Cell Biol. 201, 217–231 10.1083/jcb.201210033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Saurin A. T., van der Waal M. S., Medema R. H., Lens S. M., and Kops G. J. (2011) Aurora B potentiates Mps1 activation to ensure rapid checkpoint establishment at the onset of mitosis. Nat. Commun. 2, 316 10.1038/ncomms1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Santaguida S., Tighe A., D'Alise A. M., Taylor S. S., and Musacchio A. (2010) Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J. Cell Biol. 190, 73–87 10.1083/jcb.201001036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ji Z., Gao H., and Yu H. (2015) Kinetochore attachment sensed by competitive Mps1 and microtubule binding to Ndc80C. Science 348, 1260–1264 10.1126/science.aaa4029 [DOI] [PubMed] [Google Scholar]
- 53. Hiruma Y., Sacristan C., Pachis S. T., Adamopoulos A., Kuijt T., Ubbink M., von Castelmur E., Perrakis A., and Kops G. J. (2015) Competition between MPS1 and microtubules at kinetochores regulates spindle checkpoint signaling. Science 348, 1264–1267 10.1126/science.aaa4055 [DOI] [PubMed] [Google Scholar]
- 54. Yamagishi Y., Yang C. H., Tanno Y., and Watanabe Y. (2012) MPS1/Mph1 phosphorylates the kinetochore protein KNL1/Spc7 to recruit SAC components. Nat. Cell Biol. 14, 746–752 10.1038/ncb2515 [DOI] [PubMed] [Google Scholar]
- 55. Shepperd L. A., Meadows J. C., Sochaj A. M., Lancaster T. C., Zou J., Buttrick G. J., Rappsilber J., Hardwick K. G., and Millar J. B. (2012) Phosphodependent recruitment of Bub1 and Bub3 to Spc7/KNL1 by Mph1 kinase maintains the spindle checkpoint. Curr. Biol. 22, 891–899 10.1016/j.cub.2012.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. London N., Ceto S., Ranish J. A., and Biggins S. (2012) Phosphoregulation of Spc105 by Mps1 and PP1 regulates Bub1 localization to kinetochores. Curr. Biol. 22, 900–906 10.1016/j.cub.2012.03.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Primorac I., Weir J. R., Chiroli E., Gross F., Hoffmann I., van Gerwen S., Ciliberto A., and Musacchio A. (2013) Bub3 reads phosphorylated MELT repeats to promote spindle assembly checkpoint signaling. eLife 2, e01030 10.7554/eLife.01030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hewitt L., Tighe A., Santaguida S., White A. M., Jones C. D., Musacchio A., Green S., and Taylor S. S. (2010) Sustained Mps1 activity is required in mitosis to recruit O-Mad2 to the Mad1-C-Mad2 core complex. J. Cell Biol. 190, 25–34 10.1083/jcb.201002133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ainsztein A. M., Kandels-Lewis S. E., Mackay A. M., and Earnshaw W. C. (1998) INCENP centromere and spindle targeting: identification of essential conserved motifs and involvement of heterochromatin protein HP1. J. Cell Biol. 143, 1763–1774 10.1083/jcb.143.7.1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nozawa R. S., Nagao K., Masuda H. T., Iwasaki O., Hirota T., Nozaki N., Kimura H., and Obuse C. (2010) Human POGZ modulates dissociation of HP1α from mitotic chromosome arms through Aurora B activation. Nat. Cell Biol. 12, 719–727 10.1038/ncb2075 [DOI] [PubMed] [Google Scholar]
- 61. Kang J., Chaudhary J., Dong H., Kim S., Brautigam C. A., and Yu H. (2011) Mitotic centromeric targeting of HP1 and its binding to Sgo1 are dispensable for sister-chromatid cohesion in human cells. Mol. Biol. Cell 22, 1181–1190 10.1091/mbc.e11-01-0009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Abe Y., Sako K., Takagaki K., Hirayama Y., Uchida K. S., Herman J. A., DeLuca J. G., and Hirota T. (2016) HP1-assisted Aurora B kinase activity prevents chromosome segregation errors. Dev. Cell 36, 487–497 10.1016/j.devcel.2016.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yi Q., Chen Q., Liang C., Yan H., Zhang Z., Xiang X., Zhang M., Qi F., Zhou L., and Wang F. (2018) HP1 links centromeric heterochromatin to centromere cohesion in mammals. EMBO Rep. 19, e45484 10.15252/embr.201745484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yamagishi Y., Sakuno T., Shimura M., and Watanabe Y. (2008) Heterochromatin links to centromeric protection by recruiting shugoshin. Nature 455, 251–255 10.1038/nature07217 [DOI] [PubMed] [Google Scholar]
- 65. Shimura M., Toyoda Y., Iijima K., Kinomoto M., Tokunaga K., Yoda K., Yanagida M., Sata T., and Ishizaka Y. (2011) Epigenetic displacement of HP1 from heterochromatin by HIV-1 Vpr causes premature sister chromatid separation. J. Cell Biol. 194, 721–735 10.1083/jcb.201010118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Brasher S. V., Smith B. O., Fogh R. H., Nietlispach D., Thiru A., Nielsen P. R., Broadhurst R. W., Ball L. J., Murzina N. V., and Laue E. D. (2000) The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J. 19, 1587–1597 10.1093/emboj/19.7.1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Maison C., and Almouzni G. (2004) HP1 and the dynamics of heterochromatin maintenance. Nat. Rev. Mol. Cell Biol. 5, 296–304 10.1038/nrm1355 [DOI] [PubMed] [Google Scholar]
- 68. Thiru A., Nietlispach D., Mott H. R., Okuwaki M., Lyon D., Nielsen P. R., Hirshberg M., Verreault A., Murzina N. V., and Laue E. D. (2004) Structural basis of HP1/PXVXL motif peptide interactions and HP1 localisation to heterochromatin. EMBO J. 23, 489–499 10.1038/sj.emboj.7600088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Smothers J. F., and Henikoff S. (2000) The HP1 chromo shadow domain binds a consensus peptide pentamer. Curr. Biol. 10, 27–30 10.1016/S0960-9822(99)00260-2 [DOI] [PubMed] [Google Scholar]
- 70. Zeitlin S. G., Shelby R. D., and Sullivan K. F. (2001) CENP-A is phosphorylated by Aurora B kinase and plays an unexpected role in completion of cytokinesis. J. Cell Biol. 155, 1147–1157 10.1083/jcb.200108125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lampson M. A., Renduchitala K., Khodjakov A., and Kapoor T. M. (2004) Correcting improper chromosome-spindle attachments during cell division. Nat. Cell Biol. 6, 232–237 10.1038/ncb1102 [DOI] [PubMed] [Google Scholar]
- 72. Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., and Zhang F. (2013) Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wood K. W., Lad L., Luo L., Qian X., Knight S. D., Nevins N., Brejc K., Sutton D., Gilmartin A. G., Chua P. R., Desai R., Schauer S. P., McNulty D. E., Annan R. S., Belmont L. D., et al. (2010) Antitumor activity of an allosteric inhibitor of centromere-associated protein-E. Proc. Natl. Acad. Sci. U.S.A. 107, 5839–5844 10.1073/pnas.0915068107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Dai J., Sultan S., Taylor S. S., and Higgins J. M. (2005) The kinase haspin is required for mitotic histone H3 Thr 3 phosphorylation and normal metaphase chromosome alignment. Genes Dev. 19, 472–488 10.1101/gad.1267105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhou L., Liang C., Chen Q., Zhang Z., Zhang B., Yan H., Qi F., Zhang M., Yi Q., Guan Y., Xiang X., Zhang X., Ye S., and Wang F. (2017) The N-terminal non-kinase-domain-mediated binding of Haspin to Pds5B protects centromeric cohesion in mitosis. Curr. Biol. 27, 992–1004 10.1016/j.cub.2017.02.019 [DOI] [PubMed] [Google Scholar]
- 76. Losada A. (2014) Cohesin in cancer: chromosome segregation and beyond. Nat. Rev. Cancer 14, 389–393 10.1038/nrc3743 [DOI] [PubMed] [Google Scholar]
- 77. Kawashima S. A., Tsukahara T., Langegger M., Hauf S., Kitajima T. S., and Watanabe Y. (2007) Shugoshin enables tension-generating attachment of kinetochores by loading Aurora to centromeres. Genes Dev. 21, 420–435 10.1101/gad.1497307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Tanno Y., Kitajima T. S., Honda T., Ando Y., Ishiguro K., and Watanabe Y. (2010) Phosphorylation of mammalian Sgo2 by Aurora B recruits PP2A and MCAK to centromeres. Genes Dev. 24, 2169–2179 10.1101/gad.1945310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Baron A. P., von Schubert C., Cubizolles F., Siemeister G., Hitchcock M., Mengel A., Schröder J., Fernández-Montalván A., von Nussbaum F., Mumberg D., and Nigg E. A. (2016) Probing the catalytic functions of Bub1 kinase using the small molecule inhibitors BAY-320 and BAY-524. eLife 5, e12187 10.7554/eLife.12187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Liu H., Jia L., and Yu H. (2013) Phospho-H2A and cohesin specify distinct tension-regulated Sgo1 pools at kinetochores and inner centromeres. Curr. Biol. 23, 1927–1933 10.1016/j.cub.2013.07.078 [DOI] [PubMed] [Google Scholar]
- 81. Kitajima T. S., Hauf S., Ohsugi M., Yamamoto T., and Watanabe Y. (2005) Human Bub1 defines the persistent cohesion site along the mitotic chromosome by affecting Shugoshin localization. Curr. Biol. 15, 353–359 10.1016/j.cub.2004.12.044 [DOI] [PubMed] [Google Scholar]
- 82. London N., and Biggins S. (2014) Mad1 kinetochore recruitment by Mps1-mediated phosphorylation of Bub1 signals the spindle checkpoint. Genes Dev. 28, 140–152 10.1101/gad.233700.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Liu X., Song Z., Huo Y., Zhang J., Zhu T., Wang J., Zhao X., Aikhionbare F., Zhang J., Duan H., Wu J., Dou Z., Shi Y., and Yao X. (2014) Chromatin protein HP1 interacts with the mitotic regulator borealin protein and specifies the centromere localization of the chromosomal passenger complex. J. Biol. Chem. 289, 20638–20649 10.1074/jbc.M114.572842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ruppert J. G., Samejima K., Platani M., Molina O., Kimura H., Jeyaprakash A. A., Ohta S., and Earnshaw W. C. (2018) HP1α targets the chromosomal passenger complex for activation at heterochromatin before mitotic entry. EMBO J. 37, e97677 10.15252/embj.201797677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hengeveld R. C. C., Vromans M. J. M., Vleugel M., Hadders M. A., and Lens S. M. A. (2017) Inner centromere localization of the CPC maintains centromere cohesion and allows mitotic checkpoint silencing. Nat. Commun. 8, 15542 10.1038/ncomms15542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Lara-Gonzalez P., and Taylor S. S. (2012) Cohesion fatigue explains why pharmacological inhibition of the APC/C induces a spindle checkpoint-dependent mitotic arrest. PLoS One 7, e49041 10.1371/journal.pone.0049041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Daum J. R., Potapova T. A., Sivakumar S., Daniel J. J., Flynn J. N., Rankin S., and Gorbsky G. J. (2011) Cohesion fatigue induces chromatid separation in cells delayed at metaphase. Curr. Biol. 21, 1018–1024 10.1016/j.cub.2011.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Stevens D., Gassmann R., Oegema K., and Desai A. (2011) Uncoordinated loss of chromatid cohesion is a common outcome of extended metaphase arrest. PLoS One 6, e22969 10.1371/journal.pone.0022969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Sapkota H., Wasiak E., Daum J. R., and Gorbsky G. J. (2018) Multiple determinants and consequences of cohesion fatigue in mammalian cells. Mol. Biol. Cell 29, 1811–1824 10.1091/mbc.E18-05-0315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Liang C., Chen Q., Yi Q., Zhang M., Yan H., Zhang B., Zhou L., Zhang Z., Qi F., Ye S., and Wang F. (2018) A kinase-dependent role for Haspin in antagonizing Wapl and protecting mitotic centromere cohesion. EMBO Rep. 19, 43–56 10.15252/embr.201744737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Goto Y., Yamagishi Y., Shintomi-Kawamura M., Abe M., Tanno Y., and Watanabe Y. (2017) Pds5 regulates sister-chromatid cohesion and chromosome bi-orientation through a conserved protein interaction module. Curr. Biol. 27, 1005–1012 10.1016/j.cub.2017.02.066 [DOI] [PubMed] [Google Scholar]
- 92. Dai J., Sullivan B. A., and Higgins J. M. (2006) Regulation of mitotic chromosome cohesion by Haspin and Aurora B. Dev. Cell 11, 741–750 10.1016/j.devcel.2006.09.018 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.