Abstract
Polyphosphate (polyP) consists of a linear arrangement of inorganic phosphates and defies its structural simplicity with an astounding number of different activities in the cell. Already well known for its ability to partake in phosphate, calcium, and energy metabolism, polyP recently gained a new functional dimension with the discovery that it serves as a stabilizing scaffold for protein-folding intermediates. In this review, we summarize and discuss the recent studies that have identified polyP not only as a potent protein-like chaperone that protects cells against stress-induced protein aggregation, but also as a robust modifier of amyloidogenic processes that shields neuronal cells from amyloid toxicity. We consider some of the most pressing questions in the field, the obstacles faced, and the potential avenues to take to provide a complete picture about the working mechanism and physiological relevance of this intriguing biopolymer.
Keywords: chaperone, protein aggregation, amyloid, protein folding, stress, neurodegeneration, polyphosphate, proteostasis
Introduction
Polyphosphate (polyP)2 consists of a minimum of three up to 1000 inorganic phosphates, connected via high-energy phosphoanhydride bonds (Fig. 1). PolyP originally emerged during volcanic eruptions and is thought to have served as one of the first energy-rich molecules on earth (1). Early work revealed that some microorganisms accumulate polyP in dense, so-called metachromatic granules, which stain red with toluidine blue and are easily visible with a light microscope (2). However, few studies reported on the potential physiological role(s) of polyP in bacteria or any other organism, causing the polyP field to lay dormant for much of the 20th century. This situation changed, however, once polyP caught the attention of Sylvy Kornberg and later that of her husband Arthur Kornberg, who spent the last 15 years of his career investigating the “forgotten polymer” (3). Kornberg and his team developed many of the currently available polyP detection and quantification methods (4, 5) and, most importantly, identified the enzyme responsible for polyP synthesis in bacteria (6, 7). This breakthrough discovery enabled them, for the first time, to genetically manipulate polyP synthesis in a living organism and hence to systematically evaluate the physiological consequences of polyP depletion. It turned out that polyP fulfills a number of different functions in bacteria: polyP production was found to increase stress resistance, motility, and biofilm formation (5, 8–12) as well as to contribute to sporulation, quorum-sensing, and virulence (11–13). In eukaryotes, polyP appears to play an equally large number of diverse roles, ranging from stimulating blood clotting through the activation of factor XII to chelating calcium for bone mineralization, activating mTOR, and triggering apoptosis (14–17). This amalgam of seemingly unrelated functions (Fig. 1) poses new and even more intriguing questions as to how such a simple polyanion like polyP can fulfill all these different roles (Table 1). We will review the many known facets of polyP activity in pro- and eukaryotic organisms and explore whether polyP's ability to function as a protein-stabilizing scaffold might serve as one of the unifying principles of polyP action.
Figure 1.
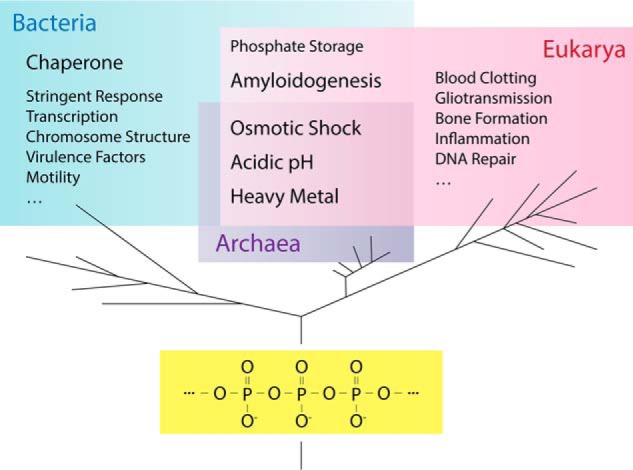
PolyP, a simple, universal, and highly multifunctional polyanion. The linear polyP chain consists of minimally three and maximally up to about 1000 orthophosphates linked by high-energy phosphoanhydride bonds. PolyP has been identified in all organisms tested so far, where it fulfills an astonishing array of different functions.
Table 1.
The physiological functions of polyP
| Species | Function | Subcellular localization (if described) | Refs. |
|---|---|---|---|
| E. coli | Stringent response | 4, 5 | |
| Stationary phase survival | 8, 26 | ||
| Stress resistance (heat, oxidants, osmotic shock, acidic pH, and heavy metal) | 8, 24–26, 28, 79 | ||
| Biofilm formation | 28, 54 | ||
| Persistence | 28 | ||
| Transcription | 22, 28, 31 | ||
| Translation | Ribosome | 77 | |
| DNA entry | Membrane | 118 | |
| Pseudomonas aeruginosa | Cellular structures | Nucleoid | 29 |
| Motility | 9, 10, 29 | ||
| Stress resistance (oxidants, desiccation) | 27, 29 | ||
| Biofilm formation | 12, 29 | ||
| Virulence | 12 | ||
| Quorum sensing | 12 | ||
| Colonization | 28 | ||
| S. cerevisiae | DNA synthesis and repair | Nucleus | 40, 119 |
| Post-translational modification (polyphosphorylation) | Nucleus/nucleolus | 58, 120 | |
| Phosphate homeostasis | Vacuole | 38 | |
| Arginine sequestration | Vacuole | 43 | |
| Stress resistance (heavy metal) | Cytoplasm | 41, 42 | |
| D. discoideum | Development and germination | 121, 122 | |
| Predation | 121 | ||
| Cytokinesis | 123 | ||
| Trypanosoma brucei Trypanosoma cruzi | Stress resistance (osmotic shock) | 124–126 | |
| Cytokinesis | 124 | ||
| Infection | 125, 127 | ||
| Homo sapiens | Ion channel activation | Plasma membrane | 59, 128 |
| Signaling (mTOR, FGF2, integrin β1, P2Y1) | Plasma membrane | 16, 50, 60, 61 | |
| Mitochondrial activity (mPTP opening, Ca2+ homeostasis, energy metabolism) | Mitochondria | 17, 57 | |
| Nucleolar function (rRNA transcription) | Nucleolus | 47 | |
| DNA repair | Nucleus | 120 | |
| Blood clotting | Secreted from polyP granules | 14 | |
| Inflammation | Secreted from polyP granules | 14, 48, 129–132 | |
| Bone mineralization | Secreted from polyP granules | 15 | |
PolyP biosynthesis and regulation in bacteria
Many bacteria, including numerous pathogens, encode the nonessential enzyme polyphosphate kinase (PPK1), which catalyzes the reversible transfer of the terminal γ-phosphate of ATP to polyP (the in vivo starting molecule is not yet known) (6). In addition to PPK1, some microorganisms encode a second, structurally unrelated kinase, PPK2, which catalyzes the transfer of terminal Pi from polyP to GDP to form GTP (7, 18, 19). Most, if not all, of these bacteria also encode the exopolyphosphatase PPX, which hydrolyzes polyP into individual phosphates, thereby indirectly utilizing the cellular ATP pool to maintain phosphate homeostasis (20).
Steady-state concentrations of polyP in the bacterial cytosol are typically in the low micromolar range, even in mutant strains that lack the polyphosphatase PPX (21). These results suggest that the levels and/or activity of PPK must be tightly regulated, a conclusion that makes energetic sense given that polyP synthesis draws from the cellular ATP pool. Yet, upon nutrient shift (4, 22) or exposure to osmotic changes (4), acidic pH (23), oxidants such as hypochlorous acid (24), or very high temperatures (25), bacteria rapidly accumulate high levels of polyP. In fact, under some of these stress conditions, bacteria have been shown to convert millimolar amounts of ATP into extremely (>1000 Pi) long chains of polyP, decreasing their cellular ATP pool by up to 30–50% in the process (24). Not surprisingly, preventing bacteria from synthesizing polyP (i.e. by deleting ppk) makes them exquisitely sensitive to nutrient deprivation, oxidants, or high temperature (24–28). In addition, deletion of the ppk gene has been shown to decrease bacterial pathogenicity in a comprehensive manner, i.e. by reducing motility, colonization, virulence factor production, and biofilm formation (Table 1) (28, 29).
Much of Kornberg's research focused on how bacterial polyP synthesis is regulated and which potential transcriptional regulators might control ppk gene expression (4, 5, 22). These studies revealed that the ppk gene is a direct target of σ38, the master transcriptional regulator for late stationary phase genes (30). PolyP, in turn, induces transcription of rpoS (the gene encoding σ38) and further amplifies its own synthesis (22, 28, 31). Although these results nicely explained the increased PPK and polyP levels observed in stationary phase bacteria, they failed to reveal how polyP synthesis is regulated during rpoS-independent stress conditions, such as oxidative or heat stress. As it turns out, in contrast to many stress-induced transcriptional responses, polyP synthesis appears to be primarily regulated on a post-transcriptional and/or post-translational level. In Pseudomonas fluorescens, for instance, mRNA transcripts of the ppk gene have been shown to be targeted by antisense RNA, which fine-tunes PPK synthesis and hence regulates polyP abundance (32). In Escherichia coli and other tested Gram-negative bacteria, nutrient shift–induced up-regulation of polyP has been proposed to be directly mediated by ppGpp, a secondary metabolite that quickly accumulates upon nutrient deprivation (4, 5, 33). This metabolite is thought to directly inhibit PPX, thereby preventing polyP hydrolysis (34). Similarly, our studies in HOCl-treated E. coli cells revealed that polyP accumulation is, at least in part, mediated by the reversible inactivation of PPX. In this case, reversible oxidation of a critical cysteine, located in the polyP-binding site, directly inactivates PPX until reducing conditions are restored (24). Despite these insights, it is clear that inactivation of PPX is only part of the polyP accumulation story. Because ppx deletion strains do not accumulate polyP in the absence of stress and PPK levels do not seem to significantly increase upon nutrient shift or other stress conditions in E. coli (21, 35, 36), it is highly likely for PPK to be post-translationally regulated, either directly or through stress-sensitive regulators. Once PPK is activated and polyP is synthesized, however, transient inactivation of PPX will guarantee that polyP levels remain high until nonstress conditions are restored.
PolyP in eukaryotic organisms
Intriguingly, the polyP-synthesizing machinery is not universally conserved, and for most eukaryotic organisms, the mechanism by which polyP is synthesized remains unknown (37). One exception is the baker's yeast Saccharomyces cerevisiae, where the vacuolar transporter chaperone 4 (VTC4) was found to use ATP to generate polyP and concomitantly transport the polyanion into the vacuole (38, 39). Vacuolar polyP serves a central role in maintaining phosphate homeostasis, sequestering phosphate during growth in phosphate-rich environments (38), and releasing phosphate during the cell cycle to provide the building blocks for DNA replication (40). Moreover, polyP chelates metals, such as manganese and cadmium, thereby preventing metal-induced cellular damage (41, 42), and sequesters arginine to increase nitrogen storage without affecting the osmotic pressure (43). One other eukaryotic polyP-generating system has been proposed to exist in Dictyostelium discoideum, which, in addition to a bacterial PPK homologue, contains a tripartite complex of actin-related proteins that was found to synthesize polyP ex vivo (44). Unfortunately, however, the protein(s) responsible for the polyP-synthesizing activity have not yet been identified. No sequence or structural homology exists between the known polyP-synthesizing enzymes PPK1, PPK2, or Vtc4, and respective homologues have not been found in higher eukaryotic species, leaving the field for new discoveries wide open.
The earliest studies about the distribution of polyP in mammalian cells used subcellular fractionation and biochemical quantification assays. Kumble and Kornberg (45) reported that polyP is present in the nucleus, plasma membrane, cytoplasm, and intracellular organelles. Staining of cells and tissues for polyP using 4′,6-diamidino-2-phenylindole (DAPI), whose excitation and emission spectra shift significantly when bound to polyP instead of DNA (46) as well as recently developed polyP-specific probes, including fusion proteins with the polyP-binding domain of PPX (47, 48) or JC-D7/8 (49), supported these results. Moreover, they demonstrated that polyP is particularly enriched in the nucleolus, acidocalcisomes (organelles rich in protons, calcium, and phosphorus), and mitochondria. Moreover, polyP was found to be secreted from astrocytes and subsequently taken up by neurons, suggesting that it is present both inside and outside of the cell (50, 51). Apart from certain cell types such as thrombocytes and mast cells, which store up to 130 mm Pi in the form of medium-sized polyP chains in dense granules (14, 48, 52), eukaryotic polyP levels are generally in the micromolar range and the chain lengths range between 50 and 800 Pi (45). Brain tissue appears to contain some of the highest concentrations of polyP (∼100 μm) based on tissue-wide studies in rats and mice (45, 53). So far, no trigger for increased polyP synthesis has been reported in mammalian cells. However, brain polyP levels do seem to decline with age and disease state (i.e. Alzheimer's disease), suggesting that physiological events might affect polyP synthesis in mammalian species (54, 55).
Despite the challenges in elucidating the function of a molecule, whose abundance and/or activity cannot be directly manipulated via genetic approaches, we know a surprising amount about the processes that eukaryotic polyP appears to be involved in (Table 1). Much of this knowledge comes again from the Kornberg group, who generated the first mammalian cell line that stably expressed yeast PPX to degrade endogenous polyP (16). Pavlov and co-workers (17) applied a similar strategy to test the role of polyP in mitochondria, and Morrissey and co-workers (56) developed end-labeled polyP to monitor its dynamics and interactions. These studies produced ample evidence that polyP is either directly or indirectly involved in a myriad of different processes, including opening of the mitochondrial permeability transition pore (mPTP) and Ca2+ signaling (17, 57), activating plasma protease factor XII and forming fibrin fibers at pro-coagulant sites upon release from thrombocytes (14), and exerting pro-inflammatory responses upon release from mast cells (48). PolyP has also been proposed to function as a gliotransmitter in the autonomic nervous system (50) and to be involved in maintaining mitochondrial membrane potential (17). Finally, a variety of other proteins have been identified to stably or transiently interact with polyP, including mTOR, Nsr1, TRPM8, FGF-2, integrin β1, as well as several ribosomal proteins and glycosomal enzymes, resulting in altered activities and profound influences on cell growth and survival (16, 58–63). The underlying mechanism by which polyP interacts with these structurally unrelated proteins remains unknown.
PolyP—an inorganic polyanion with protein-like chaperone qualities
Molecular chaperones serve as the guardians of the proteome (64). Their primary functions are to support the de novo folding of newly translated polypeptides and prevent irreparable stress-induced misfolding of native proteins within the crowded and metastable cellular environment (65). Most chaperones are proteins, grouped into individual families that differ in structure, complexity, and regulation (66). The chaperone families range from energy-independent small proteins that function as monomeric (e.g. HdeA), dimeric (e.g. Hsp33), or high-molecular-weight oligomers (e.g. sHsps) to complex multichaperone machineries (e.g. TRIC, Hsp70, Hsp90) that use ATP binding and hydrolysis to regulate co-chaperone and substrate protein binding and release (67, 68). The structural and functional diversification among the individual chaperone families appears to contribute to their ability to cater to a wide variety of different client proteins that are present in vastly diverse folding environments (69). Given the highly sophisticated mechanisms that many of these chaperones employ, it came as a significant surprise when our lab discovered that the simple polyanion polyP exerts protein-like chaperone qualities (24). Like classical protein chaperones, we found that micromolar concentrations of polyP effectively prevented protein aggregation during a variety of different proteotoxic stress conditions, including oxidative stress (24) and high temperature (Fig. 2) (25). Deletion of the PPK gene led to the accumulation of aggregated proteins in vivo despite the bacteria's attempts to compensate for the lack of polyP with increased heat shock gene expression (24, 25). These results helped to explain previous reports about the exquisite sensitivity of ppk deletion strains toward proteotoxic stress conditions and suggested that the chaperone function of polyP plays a physiologically relevant role in stress protection (24, 27, 28). In addition, some of the stress-protective effects of polyP in vivo might also be due to polyP's ability to serve as a reservoir for high-energy phosphoanhydride bonds or its metal-chelating activity (24, 70). However, dissecting which of these functions might play the predominant cytoprotective role will be extremely challenging.
Figure 2.
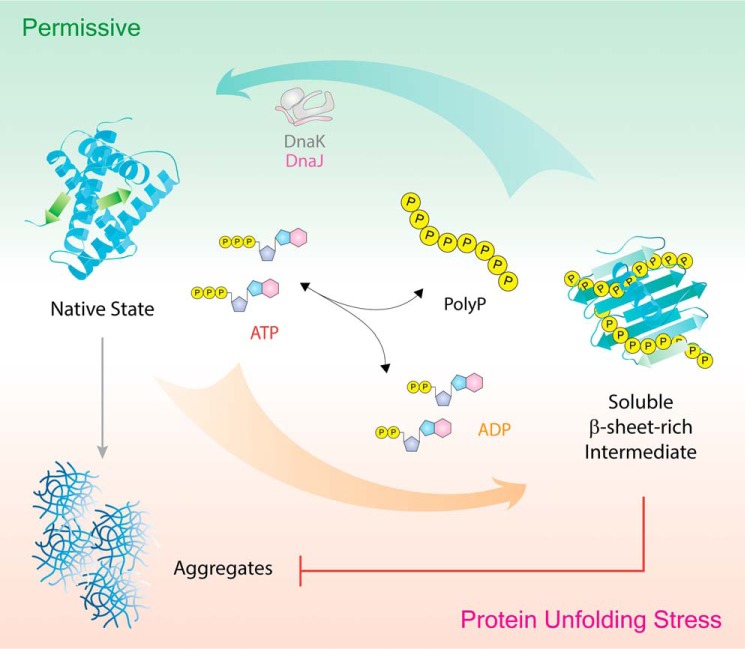
PolyP, an inorganic molecule with protein-like chaperone qualities. Under proteotoxic stress conditions, such as oxidative stress or heat shock, bacteria convert significant quantities of cellular ATP into long chains of polyP, which functions as a protein-stabilizing scaffold that binds protein unfolding intermediates and stabilizes them in a soluble β-sheet–rich conformation. This stable interaction effectively prevents protein aggregation both in vitro and in vivo. Once stress conditions are alleviated, chaperone foldases such as the DnaK system are able to refold polyP-bound client proteins and restore their native structure and function.
The real potential of polyP as a protein-stabilizing scaffold became evident when we heated known thermolabile proteins, such as luciferase or lactate dehydrogenase in the presence of polyP, and found that the proteins remained fully soluble even upon heating to near-boiling temperatures (i.e. 85 °C) (24, 25). The effective polyP concentration was in the low millimolar range (per Pi) and hence at least 3 orders of magnitude below the working concentration of chemical chaperones (e.g. glycerol or trehalose), which are thought to exert their protein-stabilizing effects through nonspecific solvation effects (71, 72). Moreover, we found that the chaperone function of polyP is chain length-dependent, with longer polyP chains being disproportionally more effective in preventing protein aggregation in vitro than short-chain polyPs (<16-mer), excluding a “simple” charge effect (24, 25). In summary, these results suggested that polyP, which itself is redox-inert and thermostable, might have served as the primordial chaperone and now functions in the first line of defense under extreme stress conditions that might inactivate protein-based chaperones (Fig. 2) (24, 25).
Analysis of the fate of the bound client proteins revealed that polyP not only preserves their solubility but also maintains the client proteins in a folding-competent conformation, even upon incubation at near-boiling temperatures (25). Subsequent incubation of polyP–client complexes under nonstress conditions restored the enzymatic activity of the clients provided that the DnaK-refolding system was present (Fig. 2) (24, 25). In contrast, hydrolysis of polyP in the absence of other chaperone systems caused the rapid aggregation of the client proteins (24, 25). At this point, we do not know whether substrate release is simply triggered by the presence of chaperone foldases or whether polyP directly affects and regulates the activity of other chaperones as well. More research is clearly needed to gain detailed insights into the precise roles that polyP plays within the cellular proteostasis network.
For many years, it has been a well-accepted dogma that molecular chaperones bind client proteins through hydrophobic contact sites, thereby shielding aggregation-sensitive surfaces of clients against nonspecific interactions (73–75). This mechanism is clearly not compatible with polyP, which is one of the most highly negatively charged molecules known (54). Instead, it strongly suggests that the main driving force behind polyP–client interactions is of an electrostatic nature. Yet, how polyP is able to recognize and bind a wide range of different unfolding intermediates while discriminating against their native counterparts is not known. So far, no correlation has been observed between the preference of polyP and the isoelectric points (pI) of the potential clients, although a comprehensive analysis still needs to be performed. Moreover, polyP appears indiscriminative toward aggregation-sensitive endogenous E. coli proteins, which appear to be all similarly protected by polyP (24). Of course, it is unclear whether polyP interacts individually with all of the different proteins or broadens its effect by manipulating the activity of other chaperones that are present in these lysates. Finally, the chaperone activity of polyP appears to be chain length–dependent (24, 25), which suggests that certain structural elements in polyP rather than simply the presence of negative charges are responsible for the disproportionally higher activity of longer polyP chains. Our recent studies showed that polyP stabilizes a range of different thermally unfolded proteins in a β-sheet–rich, amyloid-like yet soluble conformation (24, 25). This is despite the fact that the proteins that we tested so far are predominantly α-helical in their native conformation. Because polyP does not affect the conformation of these proteins in their native state, we propose that polyP chains bind and stabilize β-sheet structures as they transiently populate during the unfolding process. More studies are clearly needed to investigate this idea (Fig. 2). It is of note that other polyanions, such as DNA and RNA, have now been shown to also moonlight as chaperones in vitro, with an efficiency that in some cases even exceeds that of polyP (76). We are now awaiting high-resolution structures, which will hopefully reveal how these polyanions interact with and stabilize proteins.
PolyP's other potential roles in cellular proteostasis
In addition to the chaperone function, polyP has several other features and functional activities that might contribute to maintaining proteostasis in the cell. For example, it has been proposed that polyP oversees the fidelity of protein translation by refining the magnesium-sensitive codon–anticodon pairing and/or by modifying the structure of the ribosome (77). By preventing the overproduction of error-prone polypeptides, polyP might thereby mitigate the burden on the quality control network. Moreover, it has been known for years that polyP is able to chelate heavy metals, such as cadmium, copper, and manganese, that are known to target protein folding (41, 78–82). Yet, another strategy to protect the proteome might utilize the osmotic properties of polyP to attain an optimal folding environment. For instance, the extremophile Methanosarcina mazei deliberately up-regulates polyP to maintain its normal osmolarity at a concentration of salt that would otherwise render the cytoplasm nearly desiccated (83). In summary, it appears that polyP, one of the simplest yet most versatile molecule known, is able to protect the proteome under vastly different and constantly changing conditions.
PolyP—a modifier of amyloidogenic processes in vitro
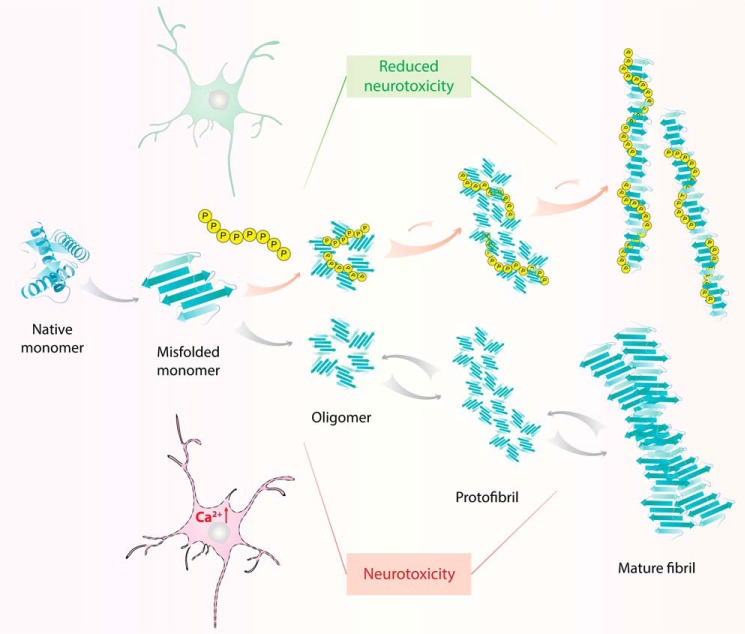
Inspired by the observation that protein-folding intermediates, once in complex with polyP, exert β-sheet–rich features that are reminiscent of soluble amyloid-like microaggregates (24, 25), we explored the role of polyP in amyloidogenic processes. All of the known amyloidogenic proteins start life as soluble proteins, typically with an intrinsically disordered or α-helix–rich structure (84). Using mechanisms and following in vivo kinetics that have yet to be defined for the majority of amyloids, the monomers eventually undergo conformational rearrangements into association-competent β-sheet structures, which form oligomers, proto-fibrils, and finally the mature fibrils (84, 85). Irrespective of the sequence and structure of the amyloidogenic monomers, the overall morphology and characteristics of the mature fibrils appear to be surprisingly similar: cross–β-sheet repeat structures, which are protease-resistant, SDS-insoluble, and thioflavin T-positive (86–88). It is important to note that the mature fibrils are not inert end products but are known to undergo shedding events that increase the number of oligomers and proto-fibrils, which will nucleate additional oligomerization processes (89–91).
Functional amyloid fibrils are found in bacteria where they constitute the structural framework of biofilms (92, 93), as well as in mammals, where they serve as a reservoir of pituitary hormones (94) or generate pigmentation in melanocytes (95). However, amyloidogenic proteins are even better known for their roles in protein-folding diseases (84, 85), particularly Aβ, tau, α-synuclein, and huntingtin, whose fibril depositions in the brain have been associated with Alzheimer's, Parkinson's, and Huntington's disease, respectively (96–98). Because age is the greatest risk factor for developing these neurodegenerative diseases (99), amyloid pathology has become exceedingly prevalent, and effective therapeutics are desperately needed to target these devastating diseases (100).
It is generally agreed upon that the rate-determining step in most amyloidogenic processes is the initial conformational change that converts soluble monomers into association-competent β-sheet–rich proteins (101). Hence, our finding that polyP stabilizes β-sheet–containing folding intermediates (24, 25) fueled the idea that polyP might increase the rate of fibril formation. At a minimum, this activity would explain the previous observation that polyP accelerates biofilm formation in bacteria (11, 12). Indeed, experiments with the bacterial amyloid CsgA revealed that polyP accelerates CsgA fibril (i.e. Curli) formation both in vitro and in vivo (54). Subsequent analysis with different disease-associated amyloids confirmed this activity and revealed, in some cases, an extraordinary acceleration of in vitro fibril formation through polyP (Fig. 3) (54). One extreme example is the human tau protein, whose lag phase is reduced from many months (102) to around 30 h simply by adding polyP at a 1:5 (polyP/tau) ratio (54). Another example is the tau fragment TauK19, which takes 24–48 h to form fibrils in the absence of polyP yet fibrillates within minutes in its presence (54). Depending on the amyloidogenic protein tested, polyP shortens the initial lag phase and/or expedites fibril elongation. Similar to the anti-aggregation function of polyP, the pro-aggregation function of polyP is also chain length–dependent, with longer chains being disproportionally more effective in nucleating fibril formation than shorter chains (54). This is a particularly intriguing aspect given that cells in the mammalian brain contain extremely long polyP chains (>800 Pi), and it makes one wonder whether or how fibril formation is avoided in cells that contain both amyloids and polyP. In addition to affecting nucleation and elongation, polyP binding also causes substantial structural changes in the mature fibrils (Fig. 3) (54). These structural rearrangements likely contribute to the fact that fibrils formed in the presence of polyP are less proteolytically stable, less able to shed, and less effective in seeding (54). Although the data appear convincing that polyP accelerates amyloid fibril formation, a better temporal resolution is needed to define when polyP binds and to determine which specific steps in the fibril formation are accelerated. Moreover, we need higher-resolution structural information to visualize how polyP interacts with fibrils. The most important aspect, however, is to understand the extent to which polyP contributes to in vivo fibril formation and how premature polyP-mediated fibril formation might be prevented in the human brain. With the advancement of super-resolution microscopy and in combination with novel labeling techniques, answers for these crucial questions will hopefully be found soon.
Figure 3.
PolyP, a modifier of amyloidogenic processes. Amyloidogenic proteins undergo conformational changes from their native monomeric state into a misfolded, β-sheet–rich aggregation-prone state, which can form insoluble oligomers, protofibrils, and mature fibrils with characteristic cross–β-sheet conformation. Shedding of mature fibrils into protofibrils and/or oligomers promotes further fibril formation. The prevalent model of amyloid toxicity identifies the oligomers and protofibrils as the major cytotoxic species, which disrupts the integrity of cellular membranes and destroys calcium homeostasis. PolyP accelerates the fibrillation of both functional amyloids (e.g. bacterial CsgA) as well as disease-associated amyloids, including Aβ-peptides, α-synuclein, and tau, and triggers morphological changes in mature fibrils. Cytotoxicity assays using neuronal cell lines revealed that the fibrils, formed in the presence of polyP, are no longer cytotoxic, suggesting that polyP might be a physiologically relevant modifier in amyloidogenic processes.
PolyP—a potential novel player in neurodegenerative diseases
Studies with several different neuronal cell cultures, including differentiated human neuroblastoma cells and PC12 cells, revealed that the presence of polyP during the fibril formation abrogated amyloid cytotoxicity. A similar cytoprotective effect was elicited by simply adding polyP to cells shortly before adding pre-formed amyloid fibrils (54, 103). Because polyP is both secreted and taken up by neuronal cells (50, 104), these results suggest that the presence of polyP in mammalian brains might influence amyloid toxicity, propagation, and/or disease progression.
At this point, it is unclear how polyP protects cells against the toxic effects of amyloids. The current model of amyloid toxicity proposes that the process of amyloid formation but not the mature fibrils elicits toxicity (105, 106). Toxic oligomers and protofibrils are thought to cause loss of membrane integrity and disruption of calcium homeostasis that ultimately leads to aberrant Ca2+ signaling and eventually cell death (107, 108). Hence, it is possible that by simply speeding up fibril formation, suppressing shedding, and preventing seeding, polyP decreases the effective concentration of toxic oligomers and protofibrils. It is also possible that polyP-associated fibrils are no longer recognized by receptor proteins responsible for the uptake of amyloid fibrils (109) or that the association with polyP increases the turnover of fibrils or triggers sequestration of the fibrils into inert structures. Finally, it has been shown that depletion of mitochondrial polyP blocks mPTP opening and the subsequent initiation of apoptosis induced by β-amyloid peptide (17). Therefore, polyP might serve as an indirect regulator of amyloid toxicity. More research is clearly needed to investigate these effects and to test whether the age-related decline in brain polyP levels that has previously been observed (55) might correlate with or potentially even contribute to disease onset and progression.
Summary and outlook
The goal of this review is to invigorate interest into a molecule, whose simple structure clearly defies its stunning functional versatility (Table 1). In addition to its fundamental roles as a Pi donor and energy source, the ability of polyP to serve as a stabilizing scaffold for countless soluble and amyloidogenic proteins has added a new dimension to its activities in the cell (24, 54, 70). It is intriguing that polyP can serve both as an anti-aggregation molecule to keep proteins soluble during stress-induced unfolding, as well as a pro-aggregation molecule that accelerates amyloid fibril formation. Based on some recent studies, it appears that the fundamental mechanism for both activities is the same: polyP binds and stabilizes proteins in a β-sheet–rich, amyloid-like conformation; however, the functional consequences appear to be determined by the intrinsic properties of the clients (24, 54). For nonamyloidogenic proteins such as luciferase and citrate synthase, association with polyP means solubility and the ability to refold if conditions permit. For amyloidogenic proteins exemplified by CsgA, Aβ, tau, and α-synuclein, association with polyP stimulates fibril formation and alters fibril morphology. Our findings that polyP aids bacteria in forming protective biofilms and shields mammalian cells against amyloid toxicity support the idea that polyP's stimulatory effects on fibril formation is indeed a desired cellular event. However, it now remains to be determined whether and how polyP alters and affects the in vivo fate of disease-associated amyloid fibrils, and whether age-mediated losses in cellular polyP levels contribute to the observation that age is one of the largest risk factors of amyloid-related neurodegenerative diseases.
We also want to point the readers to one of the biggest remaining puzzles in the field: the question of how polyP is synthesized in mammals. Unlike many other conserved molecules that are known in biology, polyP is not synthesized via the same conserved pathway but is derived from seemingly unrelated ATP-fueled engines that show no sequence or structural homology and have no homologues in higher eukaryotes! Even if one focuses only on the prokaryotic branch of the phylogenetic tree, clear homologues of E. coli PPK are absent in a large number of polyP-synthesizing species (110). During the search for alternative polyP-synthesizing machineries, a few enzymes have been recognized for their ability to use polyP as a phosphate donor (110, 111). Presumably, these reactions can be reversed by the presence of excess substrate; however, it is unknown whether such conditions can ever be achieved in vivo. Another hypothesis is that polyP may be generated by the proton-motive force without a designated enzyme (37). In mitochondria, complete depolarization of the membrane results in decreased polyP production, which suggests that a robust proton gradient is involved (112). This result also explains reports from several labs that found that cell lysis robustly kills polyP-synthesizing activity (45, 113). Yet, there is no direct evidence that polyP synthesis is indeed a spontaneous process that does not need catalysis. Finally, another group of phosphate species, inositol phosphates, has been implicated in the metabolism of polyP. Yeast, trypanosoma, and mouse strains that lack the enzyme to synthesize highly phosphorylated inositols show significantly reduced levels of polyP (114–117). Because the inositol phosphate pathway is well characterized, it is compelling to resolve the connection between inositol phosphates and polyP. This might serve as the entry point to gain a more detailed picture about polyP regulation. Finding the enzyme(s) that synthesize and regulate polyP synthesis in mammals is key; once this is achieved, many of the remaining doors will open and hopefully reveal how this ancient biomolecule works today.
Acknowledgments
We thank Justine Lempart for critically reading this manuscript and members of the Jakob lab for many helpful discussions.
This work was supported by National Institutes of Health Grants R35GM122506 and R21AG055090. This is the tenth article in the JBC Reviews series “Molecular chaperones and protein quality control.” The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- polyP
- polyphosphate
- mTOR
- mechanistic target of rapamycin
- PPX
- polyphosphatase
- PPK
- polyphosphate kinase
- mPTP
- mitochondrial permeability transition pore.
References
- 1. Brown M. R., and Kornberg A. (2004) Inorganic polyphosphate in the origin and survival of species. Proc. Natl. Acad. Sci. U.S.A. 101, 16085–16087 10.1073/pnas.0406909101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Widra A. (1959) Metachromatic granules of microorganisms. J. Bacteriol. 78, 664–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kornberg A. (1995) Inorganic polyphosphate: toward making a forgotten polymer unforgettable. J. Bacteriol. 177, 491–496 10.1128/jb.177.3.491-496.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ault-Riché D., Fraley C. D., Tzeng C. M., and Kornberg A. (1998) Novel assay reveals multiple pathways regulating stress-induced accumulations of inorganic polyphosphate in Escherichia coli. J. Bacteriol. 180, 1841–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rao N. N., Liu S., and Kornberg A. (1998) Inorganic polyphosphate in Escherichia coli: the phosphate regulon and the stringent response. J. Bacteriol. 180, 2186–2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ahn K., and Kornberg A. (1990) Polyphosphate kinase from Escherichia coli. Purification and demonstration of a phosphoenzyme intermediate. J. Biol. Chem. 265, 11734–11739 [PubMed] [Google Scholar]
- 7. Zhang H., Ishige K., and Kornberg A. (2002) A polyphosphate kinase (PPK2) widely conserved in bacteria. Proc. Natl. Acad. Sci. U.S.A. 99, 16678–16683 10.1073/pnas.262655199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rao N. N., and Kornberg A. (1996) Inorganic polyphosphate supports resistance and survival of stationary-phase Escherichia coli. J. Bacteriol. 178, 1394–1400 10.1128/jb.178.5.1394-1400.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rashid M. H., and Kornberg A. (2000) Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 97, 4885–4890 10.1073/pnas.060030097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rashid M. H., Rao N. N., and Kornberg A. (2000) Inorganic polyphosphate is required for motility of bacterial pathogens. J. Bacteriol. 182, 225–227 10.1128/JB.182.1.225-227.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shi X., Rao N. N., and Kornberg A. (2004) Inorganic polyphosphate in Bacillus cereus: motility, biofilm formation, and sporulation. Proc. Natl. Acad. Sci. U.S.A. 101, 17061–17065 10.1073/pnas.0407787101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rashid M. H., Rumbaugh K., Passador L., Davies D. G., Hamood A. N., Iglewski B. H., and Kornberg A. (2000) Polyphosphate kinase is essential for biofilm development, quorum sensing, and virulence of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 97, 9636–9641 10.1073/pnas.170283397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim K. S., Rao N. N., Fraley C. D., and Kornberg A. (2002) Inorganic polyphosphate is essential for long-term survival and virulence factors in Shigella and Salmonella spp. Proc. Natl. Acad. Sci. U.S.A. 99, 7675–7680 10.1073/pnas.112210499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Müller F., Mutch N. J., Schenk W. A., Smith S. A., Esterl L., Spronk H. M., Schmidbauer S., Gahl W. A., Morrissey J. H., and Renné T. (2009) Platelet polyphosphates are proinflammatory and procoagulant mediators in vivo. Cell 139, 1143–1156 10.1016/j.cell.2009.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Omelon S., Georgiou J., Henneman Z. J., Wise L. M., Sukhu B., Hunt T., Wynnyckyj C., Holmyard D., Bielecki R., and Grynpas M. D. (2009) Control of vertebrate skeletal mineralization by polyphosphates. PLoS ONE 4, e5634 10.1371/journal.pone.0005634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang L., Fraley C. D., Faridi J., Kornberg A., and Roth R. A. (2003) Inorganic polyphosphate stimulates mammalian TOR, a kinase involved in the proliferation of mammary cancer cells. Proc. Natl. Acad. Sci. U.S.A. 100, 11249–11254 10.1073/pnas.1534805100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Abramov A. Y., Fraley C., Diao C. T., Winkfein R., Colicos M. A., Duchen M. R., French R. J., and Pavlov E. (2007) Targeted polyphosphatase expression alters mitochondrial metabolism and inhibits calcium-dependent cell death. Proc. Natl. Acad. Sci. U.S.A. 104, 18091–18096 10.1073/pnas.0708959104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ishige K., Zhang H., and Kornberg A. (2002) Polyphosphate kinase (PPK2), a potent, polyphosphate-driven generator of GTP. Proc. Natl. Acad. Sci. U.S.A. 99, 16684–16688 10.1073/pnas.262655299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nocek B., Kochinyan S., Proudfoot M., Brown G., Evdokimova E., Osipiuk J., Edwards A. M., Savchenko A., Joachimiak A., and Yakunin A. F. (2008) Polyphosphate-dependent synthesis of ATP and ADP by the family-2 polyphosphate kinases in bacteria. Proc. Natl. Acad. Sci. U.S.A. 105, 17730–17735 10.1073/pnas.0807563105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Akiyama M., Crooke E., and Kornberg A. (1993) An exopolyphosphatase of Escherichia coli. The enzyme and its ppx gene in a polyphosphate operon. J. Biol. Chem. 268, 633–639 [PubMed] [Google Scholar]
- 21. Rudat A. K., Pokhrel A., Green T. J., and Gray M. J. (2018) Mutations in Escherichia coli polyphosphate kinase that lead to dramatically increased in vivo polyphosphate levels. J. Bacteriol. 200, e00697 10.1128/JB.00697-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shiba T., Tsutsumi K., Yano H., Ihara Y., Kameda A., Tanaka K., Takahashi H., Munekata M., Rao N. N., and Kornberg A. (1997) Inorganic polyphosphate and the induction of rpoS expression. Proc. Natl. Acad. Sci. U.S.A. 94, 11210–11215 10.1073/pnas.94.21.11210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mullan A., Quinn J. P., and McGrath J. W. (2002) Enhanced phosphate uptake and polyphosphate accumulation in Burkholderia cepacia grown under low pH conditions. Microb. Ecol. 44, 69–77 10.1007/s00248-002-3004-x [DOI] [PubMed] [Google Scholar]
- 24. Gray M. J., Wholey W. Y., Wagner N. O., Cremers C. M., Mueller-Schickert A., Hock N. T., Krieger A. G., Smith E. M., Bender R. A., Bardwell J. C., and Jakob U. (2014) Polyphosphate is a primordial chaperone. Mol. Cell 53, 689–699 10.1016/j.molcel.2014.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yoo N. G., Dogra S., Meinen B. A., Tse E., Haefliger J., Southworth D. R., Gray M. J., Dahl J. U., and Jakob U. (2018) Polyphosphate stabilizes protein unfolding intermediates as soluble amyloid-like oligomers. J. Mol. Biol. 430, 4195–4208 10.1016/j.jmb.2018.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Crooke E., Akiyama M., Rao N. N., and Kornberg A. (1994) Genetically altered levels of inorganic polyphosphate in Escherichia coli. J. Biol. Chem. 269, 6290–6295 [PubMed] [Google Scholar]
- 27. Groitl B., Dahl J. U., Schroeder J. W., and Jakob U. (2017) Pseudomonas aeruginosa defense systems against microbicidal oxidants. Mol. Microbiol. 106, 335–350 10.1111/mmi.13768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dahl J. U., Gray M. J., Bazopoulou D., Beaufay F., Lempart J., Koenigsknecht M. J., Wang Y., Baker J. R., Hasler W. L., Young V. B., Sun D., and Jakob U. (2017) The anti-inflammatory drug mesalamine targets bacterial polyphosphate accumulation. Nat. Microbiol. 2, 16267 10.1038/nmicrobiol.2016.267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fraley C. D., Rashid M. H., Lee S. S., Gottschalk R., Harrison J., Wood P. J., Brown M. R., and Kornberg A. (2007) A polyphosphate kinase 1 (ppk1) mutant of Pseudomonas aeruginosa exhibits multiple ultrastructural and functional defects. Proc. Natl. Acad. Sci. U.S.A. 104, 3526–3531 10.1073/pnas.0609733104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Maciag A., Peano C., Pietrelli A., Egli T., De Bellis G., and Landini P. (2011) In vitro transcription profiling of the σS subunit of bacterial RNA polymerase: re-definition of the σS regulon and identification of σS-specific promoter sequence elements. Nucleic Acids Res. 39, 5338–5355 10.1093/nar/gkr129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kusano S., and Ishihama A. (1997) Functional interaction of Escherichia coli RNA polymerase with inorganic polyphosphate. Genes Cells 2, 433–441 10.1046/j.1365-2443.1997.13203301320330.x [DOI] [PubMed] [Google Scholar]
- 32. Silby M. W., Nicoll J. S., and Levy S. B. (2012) Regulation of polyphosphate kinase production by antisense RNA in Pseudomonas fluorescens Pf0-1. Appl. Environ. Microbiol. 78, 4533–4537 10.1128/AEM.07836-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cashel M., and Gallant J. (1969) Two compounds implicated in the function of the RC gene of Escherichia coli. Nature 221, 838–841 10.1038/221838a0 [DOI] [PubMed] [Google Scholar]
- 34. Kuroda A., Murphy H., Cashel M., and Kornberg A. (1997) Guanosine tetra- and pentaphosphate promote accumulation of inorganic polyphosphate in Escherichia coli. J. Biol. Chem. 272, 21240–21243 10.1074/jbc.272.34.21240 [DOI] [PubMed] [Google Scholar]
- 35. Gray M. J., Wholey W. Y., Parker B. W., Kim M., and Jakob U. (2013) NemR is a bleach-sensing transcription factor. J. Biol. Chem. 288, 13789–13798 10.1074/jbc.M113.454421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang S., Deng K., Zaremba S., Deng X., Lin C., Wang Q., Tortorello M. L., and Zhang W. (2009) Transcriptomic response of Escherichia coli O157:H7 to oxidative stress. Appl. Environ. Microbiol. 75, 6110–6123 10.1128/AEM.00914-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rao N. N., Gómez-García M. R., and Kornberg A. (2009) Inorganic polyphosphate: essential for growth and survival. Annu. Rev. Biochem. 78, 605–647 10.1146/annurev.biochem.77.083007.093039 [DOI] [PubMed] [Google Scholar]
- 38. Ogawa N., DeRisi J., and Brown P. O. (2000) New components of a system for phosphate accumulation and polyphosphate metabolism in Saccharomyces cerevisiae revealed by genomic expression analysis. Mol. Biol. Cell 11, 4309–4321 10.1091/mbc.11.12.4309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hothorn M., Neumann H., Lenherr E. D., Wehner M., Rybin V., Hassa P. O., Uttenweiler A., Reinhardt M., Schmidt A., Seiler J., Ladurner A. G., Herrmann C., Scheffzek K., and Mayer A. (2009) Catalytic core of a membrane-associated eukaryotic polyphosphate polymerase. Science 324, 513–516 10.1126/science.1168120 [DOI] [PubMed] [Google Scholar]
- 40. Bru S., Martínez-Laínez J. M., Hernández-Ortega S., Quandt E., Torres-Torronteras J., Martí R., Canadell D., Ariño J., Sharma S., Jiménez J., and Clotet J. (2016) Polyphosphate is involved in cell cycle progression and genomic stability in Saccharomyces cerevisiae. Mol. Microbiol. 101, 367–380 10.1111/mmi.13396 [DOI] [PubMed] [Google Scholar]
- 41. Trilisenko L., Kulakovskaya E., and Kulakovskaya T. (2017) The cadmium tolerance in Saccharomyces cerevisiae depends on inorganic polyphosphate. J. Basic Microbiol. 57, 982–986 10.1002/jobm.201700257 [DOI] [PubMed] [Google Scholar]
- 42. Andreeva N., Ryazanova L., Dmitriev V., Kulakovskaya T., and Kulaev I. (2013) Adaptation of Saccharomyces cerevisiae to toxic manganese concentration triggers changes in inorganic polyphosphates. FEMS Yeast Res. 13, 463–470 10.1111/1567-1364.12049 [DOI] [PubMed] [Google Scholar]
- 43. Durr M., Urech K., Boller T., Wiemken A., Schwencke J., and Nagy M. (1979) Sequestration of arginine by polyphosphate in vacuoles of yeast (Saccharomyces cerevisiae). Arch. Microbiol. 121, 169–175 10.1007/BF00689982 [DOI] [PubMed] [Google Scholar]
- 44. Gómez-García M. R., and Kornberg A. (2004) Formation of an actin-like filament concurrent with the enzymatic synthesis of inorganic polyphosphate. Proc. Natl. Acad. Sci. U.S.A. 101, 15876–15880 10.1073/pnas.0406923101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kumble K. D., and Kornberg A. (1995) Inorganic polyphosphate in mammalian cells and tissues. J. Biol. Chem. 270, 5818–5822 10.1074/jbc.270.11.5818 [DOI] [PubMed] [Google Scholar]
- 46. Aschar-Sobbi R., Abramov A. Y., Diao C., Kargacin M. E., Kargacin G. J., French R. J., and Pavlov E. (2008) High sensitivity, quantitative measurements of polyphosphate using a new DAPI-based approach. J. Fluoresc. 18, 859–866 10.1007/s10895-008-0315-4 [DOI] [PubMed] [Google Scholar]
- 47. Jimenez-Nuñez M. D., Moreno-Sanchez D., Hernandez-Ruiz L., Benítez-Rondán A., Ramos-Amaya A., Rodríguez-Bayona B., Medina F., Brieva J. A., and Ruiz F. A. (2012) Myeloma cells contain high levels of inorganic polyphosphate which is associated with nucleolar transcription. Haematologica 97, 1264–1271 10.3324/haematol.2011.051409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Moreno-Sanchez D., Hernandez-Ruiz L., Ruiz F. A., and Docampo R. (2012) Polyphosphate is a novel pro-inflammatory regulator of mast cells and is located in acidocalcisomes. J. Biol. Chem. 287, 28435–28444 10.1074/jbc.M112.385823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Angelova P. R., Agrawalla B. K., Elustondo P. A., Gordon J., Shiba T., Abramov A. Y., Chang Y. T., and Pavlov E. V. (2014) In situ investigation of mammalian inorganic polyphosphate localization using novel selective fluorescent probes JC-D7 and JC-D8. ACS Chem. Biol. 9, 2101–2110 10.1021/cb5000696 [DOI] [PubMed] [Google Scholar]
- 50. Holmström K. M., Marina N., Baev A. Y., Wood N. W., Gourine A. V., and Abramov A. Y. (2013) Signalling properties of inorganic polyphosphate in the mammalian brain. Nat. Commun. 4, 1362 10.1038/ncomms2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Angelova P. R., Iversen K. Z., Teschemacher A. G., Kasparov S., Gourine A. V., and Abramov A. Y. (2018) Signal transduction in astrocytes: localization and release of inorganic polyphosphate. Glia 66, 2126–2136 10.1002/glia.23466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ruiz F. A., Lea C. R., Oldfield E., and Docampo R. (2004) Human platelet dense granules contain polyphosphate and are similar to acidocalcisomes of bacteria and unicellular eukaryotes. J. Biol. Chem. 279, 44250–44257 10.1074/jbc.M406261200 [DOI] [PubMed] [Google Scholar]
- 53. Gabel N. W., and Thomas V. (1971) Evidence for the occurrence and distribution of inorganic polyphosphates in vertebrate tissues. J. Neurochem. 18, 1229–1242 10.1111/j.1471-4159.1971.tb00222.x [DOI] [PubMed] [Google Scholar]
- 54. Cremers C. M., Knoefler D., Gates S., Martin N., Dahl J. U., Lempart J., Xie L., Chapman M. R., Galvan V., Southworth D. R., and Jakob U. (2016) Polyphosphate: a conserved modifier of amyloidogenic processes. Mol. Cell 63, 768–780 10.1016/j.molcel.2016.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lorenz B., Münkner J., Oliveira M. P., Kuusksalu A., Leitão J. M., Müller W. E., and Schröder H. C. (1997) Changes in metabolism of inorganic polyphosphate in rat tissues and human cells during development and apoptosis. Biochim. Biophys. Acta 1335, 51–60 10.1016/S0304-4165(96)00121-3 [DOI] [PubMed] [Google Scholar]
- 56. Choi S. H., Collins J. N., Smith S. A., Davis-Harrison R. L., Rienstra C. M., and Morrissey J. H. (2010) Phosphoramidate end labeling of inorganic polyphosphates: facile manipulation of polyphosphate for investigating and modulating its biological activities. Biochemistry 49, 9935–9941 10.1021/bi1014437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Seidlmayer L. K., Gomez-Garcia M. R., Blatter L. A., Pavlov E., and Dedkova E. N. (2012) Inorganic polyphosphate is a potent activator of the mitochondrial permeability transition pore in cardiac myocytes. J. Gen. Physiol. 139, 321–331 10.1085/jgp.201210788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Azevedo C., Livermore T., and Saiardi A. (2015) Protein polyphosphorylation of lysine residues by inorganic polyphosphate. Mol. Cell 58, 71–82 10.1016/j.molcel.2015.02.010 [DOI] [PubMed] [Google Scholar]
- 59. Zakharian E., Thyagarajan B., French R. J., Pavlov E., and Rohacs T. (2009) Inorganic polyphosphate modulates TRPM8 channels. PLoS ONE 4, e5404 10.1371/journal.pone.0005404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Shiba T., Nishimura D., Kawazoe Y., Onodera Y., Tsutsumi K., Nakamura R., and Ohshiro M. (2003) Modulation of mitogenic activity of fibroblast growth factors by inorganic polyphosphate. J. Biol. Chem. 278, 26788–26792 10.1074/jbc.M303468200 [DOI] [PubMed] [Google Scholar]
- 61. Segawa S., Fujiya M., Konishi H., Ueno N., Kobayashi N., Shigyo T., and Kohgo Y. (2011) Probiotic-derived polyphosphate enhances the epithelial barrier function and maintains intestinal homeostasis through integrin-p38 MAPK pathway. PLoS ONE 6, e23278 10.1371/journal.pone.0023278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Negreiros R. S., Lander N., Huang G., Cordeiro C. D., Smith S. A., Morrissey J. H., and Docampo R. (2018) Inorganic polyphosphate interacts with nucleolar and glycosomal proteins in trypanosomatids. Mol. Microbiol. 110, 973–994 10.1111/mmi.14131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Azevedo C., Singh J., Steck N., Hofer A., Ruiz F. A., Singh T., Jessen H. J., and Saiardi A. (2018) Screening a protein array with synthetic biotinylated inorganic polyphosphate to define the human PolyP-ome. ACS Chem. Biol. 13, 1958–1963 10.1021/acschembio.8b00357 [DOI] [PubMed] [Google Scholar]
- 64. Hartl F. U., Bracher A., and Hayer-Hartl M. (2011) Molecular chaperones In protein folding and proteostasis. Nature 475, 324–332 10.1038/nature10317 [DOI] [PubMed] [Google Scholar]
- 65. Ellis R. J. (2007) In Molecular Aspects of the Stress Response: Chaperones, Membranes and Networks (Csermely P., and Vígh L., eds) Springer, New York, 1–13 [Google Scholar]
- 66. Saibil H. (2013) Chaperone machines for protein folding, unfolding and disaggregation. Nat. Rev. Mol. Cell Biol. 14, 630–642 10.1038/nrm3658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bardwell J. C., and Jakob U. (2012) Conditional disorder in chaperone action. Trends Biochem. Sci. 37, 517–525 10.1016/j.tibs.2012.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kim Y. E., Hipp M. S., Bracher A., Hayer-Hartl M., and Hartl F. U. (2013) Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 82, 323–355 10.1146/annurev-biochem-060208-092442 [DOI] [PubMed] [Google Scholar]
- 69. Koldewey P., Horowitz S., and Bardwell J. C. A. (2017) Chaperone-client interactions: non-specificity engenders multifunctionality. J. Biol. Chem. 292, 12010–12017 10.1074/jbc.R117.796862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gray M. J., and Jakob U. (2015) Oxidative stress protection by polyphosphate–new roles for an old player. Curr. Opin. Microbiol. 24, 1–6 10.1016/j.mib.2014.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Jain N. K., and Roy I. (2009) Effect of trehalose on protein structure. Protein Sci. 18, 24–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Vagenende V., Yap M. G., and Trout B. L. (2009) Mechanisms of protein stabilization and prevention of protein aggregation by glycerol. Biochemistry 48, 11084–11096 10.1021/bi900649t [DOI] [PubMed] [Google Scholar]
- 73. Clerico E. M., Tilitsky J. M., Meng W., and Gierasch L. M. (2015) How Hsp70 molecular machines interact with their substrates to mediate diverse physiological functions. J. Mol. Biol. 427, 1575–1588 10.1016/j.jmb.2015.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Li Y., Gao X., and Chen L. (2009) GroEL Recognizes an amphipathic helix and binds to the hydrophobic side. J. Biol. Chem. 284, 4324–4331 10.1074/jbc.M804818200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Saio T., Guan X., Rossi P., Economou A., and Kalodimos C. G. (2014) Structural basis for protein antiaggregation activity of the trigger factor chaperone. Science 344, 1250494 10.1126/science.1250494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Docter B. E., Horowitz S., Gray M. J., Jakob U., and Bardwell J. C. (2016) Do nucleic acids moonlight as molecular chaperones? Nucleic Acids Res. 44, 4835–4845 10.1093/nar/gkw291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. McInerney P., Mizutani T., and Shiba T. (2006) Inorganic polyphosphate interacts with ribosomes and promotes translation fidelity in vitro and in vivo. Mol. Microbiol. 60, 438–447 10.1111/j.1365-2958.2006.05103.x [DOI] [PubMed] [Google Scholar]
- 78. Andreeva N., Ryazanova L., Dmitriev V., Kulakovskaya T., and Kulaev I. (2014) Cytoplasmic inorganic polyphosphate participates in the heavy metal tolerance of Cryptococcus humicola. Folia Microbiol. (Praha) 59, 381–389 10.1007/s12223-014-0310-x [DOI] [PubMed] [Google Scholar]
- 79. Keasling J. D., and Hupf G. A. (1996) Genetic manipulation of polyphosphate metabolism affects cadmium tolerance in Escherichia coli. Appl. Environ. Microbiol. 62, 743–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Alvarez S., and Jerez C. A. (2004) Copper ions stimulate polyphosphate degradation and phosphate efflux in Acidithiobacillus ferrooxidans. Appl. Environ. Microbiol. 70, 5177–5182 10.1128/AEM.70.9.5177-5182.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Gomes F. M., Carvalho D. B., Peron A. C., Saito K., Miranda K., and Machado E. A. (2012) Inorganic polyphosphates are stored in spherites within the midgut of Anticarsia gemmatalis and play a role in copper detoxification. J. Insect Physiol. 58, 211–219 10.1016/j.jinsphys.2011.09.008 [DOI] [PubMed] [Google Scholar]
- 82. Tamás M. J., Sharma S. K., Ibstedt S., Jacobson T., and Christen P. (2014) Heavy metals and metalloids as a cause for protein misfolding and aggregation. Biomolecules 4, 252–267 10.3390/biom4010252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Pflüger K., Ehrenreich A., Salmon K., Gunsalus R. P., Deppenmeier U., Gottschalk G., and Müller V. (2007) Identification of genes involved in salt adaptation in the archaeon Methanosarcina mazei Go1 using genome-wide gene expression profiling. FEMS Microbiol. Lett. 277, 79–89 10.1111/j.1574-6968.2007.00941.x [DOI] [PubMed] [Google Scholar]
- 84. Chiti F., and Dobson C. M. (2017) Protein misfolding, amyloid formation, and human disease: a summary of progress over the last decade. Annu. Rev. Biochem. 86, 27–68 10.1146/annurev-biochem-061516-045115 [DOI] [PubMed] [Google Scholar]
- 85. Knowles T. P., Vendruscolo M., and Dobson C. M. (2014) The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 15, 384–396 10.1038/nrm3810 [DOI] [PubMed] [Google Scholar]
- 86. Nordstedt C., Näslund J., Tjernberg L. O., Karlström A. R., Thyberg J., and Terenius L. (1994) The Alzheimer α-β-peptide develops protease resistance in association with its polymerization into fibrils. J. Biol. Chem. 269, 30773–30776 [PubMed] [Google Scholar]
- 87. Selkoe D. J., Ihara Y., and Salazar F. J. (1982) Alzheimers disease–insolubility of partially purified paired helical filaments in sodium dodecyl-sulfate and urea. Science 215, 1243–1245 10.1126/science.6120571 [DOI] [PubMed] [Google Scholar]
- 88. Naiki H., Higuchi K., Hosokawa M., and Takeda T. (1989) Fluorometric-determination of amyloid fibrils in vitro using the fluorescent dye, thioflavine-T. Anal. Biochem. 177, 244–249 10.1016/0003-2697(89)90046-8 [DOI] [PubMed] [Google Scholar]
- 89. Martins I. C., Kuperstein I., Wilkinson H., Maes E., Vanbrabant M., Jonckheere W., Van Gelder P., Hartmann D., D'Hooge R., De Strooper B., Schymkowitz J., and Rousseau F. (2008) Lipids revert inert Aβ amyloid fibrils to neurotoxic protofibrils that affect learning in mice. EMBO J. 27, 224–233 10.1038/sj.emboj.7601953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Cremades N., Cohen S. I., Deas E., Abramov A. Y., Chen A. Y., Orte A., Sandal M., Clarke R. W., Dunne P., Aprile F. A., Bertoncini C. W., Wood N. W., Knowles T. P., Dobson C. M., and Klenerman D. (2012) Direct observation of the interconversion of normal and toxic forms of α-synuclein. Cell 149, 1048–1059 10.1016/j.cell.2012.03.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Tipping K. W., Karamanos T. K., Jakhria T., Iadanza M. G., Goodchild S. C., Tuma R., Ranson N. A., Hewitt E. W., and Radford S. E. (2015) pH-induced molecular shedding drives the formation of amyloid fibril-derived oligomers. Proc. Natl. Acad. Sci. U.S.A. 112, 5691–5696 10.1073/pnas.1423174112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Dueholm M. S., Petersen S. V., Sønderkaer M., Larsen P., Christiansen G., Hein K. L., Enghild J. J., Nielsen J. L., Nielsen K. L., Nielsen P. H., and Otzen D. E. (2010) Functional amyloid in Pseudomonas. Mol. Microbiol. 77, 1009–1020 [DOI] [PubMed] [Google Scholar]
- 93. Chapman M. R., Robinson L. S., Pinkner J. S., Roth R., Heuser J., Hammar M., Normark S., and Hultgren S. J. (2002) Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295, 851–855 10.1126/science.1067484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Maji S. K., Perrin M. H., Sawaya M. R., Jessberger S., Vadodaria K., Rissman R. A., Singru P. S., Nilsson K. P., Simon R., Schubert D., Eisenberg D., Rivier J., Sawchenko P., Vale W., and Riek R. (2009) Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science 325, 328–332 10.1126/science.1173155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Rochin L., Hurbain I., Serneels L., Fort C., Watt B., Leblanc P., Marks M. S., De Strooper B., Raposo G., and van Niel G. (2013) BACE2 processes PMEL to form the melanosome amyloid matrix in pigment cells. Proc. Natl. Acad. Sci. U.S.A. 110, 10658–10663 10.1073/pnas.1220748110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Glenner G. G., and Wong C. W. (1984) Alzheimers-disease–initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem. Biophys. Res. Commun. 120, 885–890 10.1016/S0006-291X(84)80190-4 [DOI] [PubMed] [Google Scholar]
- 97. Polymeropoulos M. H., Lavedan C., Leroy E., Ide S. E., Dehejia A., Dutra A., Pike B., Root H., Rubenstein J., Boyer R., Stenroos E. S., Chandrasekharappa S., Athanassiadou A., Papapetropoulos T., Johnson W. G., et al. (1997) Mutation in the α-synuclein gene identified in families with Parkinson's disease. Science 276, 2045–2047 10.1126/science.276.5321.2045 [DOI] [PubMed] [Google Scholar]
- 98. Gusella J. F., Wexler N. S., Conneally P. M., Naylor S. L., Anderson M. A., Tanzi R. E., Watkins P. C., Ottina K., Wallace M. R., and Sakaguchi A. Y. (1983) A polymorphic DNA marker genetically linked to Huntingtons-disease. Nature 306, 234–238 10.1038/306234a0 [DOI] [PubMed] [Google Scholar]
- 99. Alzheimer's Association (2016) 2016 Alzheimer's disease facts and figures. Alzheimers Dement. 12, 459–509 10.1016/j.jalz.2016.03.001 [DOI] [PubMed] [Google Scholar]
- 100. Eisele Y. S., Monteiro C., Fearns C., Encalada S. E., Wiseman R. L., Powers E. T., and Kelly J. W. (2015) Targeting protein aggregation for the treatment of degenerative diseases. Nat. Rev. Drug Discov. 14, 759–780 10.1038/nrd4593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Naiki H., and Gejyo F. (1999) Kinetic analysis of amyloid fibril formation. Methods Enzymol. 309, 305–318 10.1016/S0076-6879(99)09022-9 [DOI] [PubMed] [Google Scholar]
- 102. Goedert M., Jakes R., Spillantini M. G., Hasegawa M., Smith M. J., and Crowther R. A. (1996) Assembly of microtubule-associated protein tau into Alzheimer-like filaments induced by sulphated glycosaminoglycans. Nature 383, 550–553 10.1038/383550a0 [DOI] [PubMed] [Google Scholar]
- 103. Muller W. E. G., Wang S., Ackermann M., Neufurth M., Steffen R., Mecja E., Munoz-Espi R., Feng Q., Schroder H. C., and Wang X. (2017) Rebalancing β-amyloid-induced decrease of ATP level by amorphous Nano/Micro polyphosphate: suppression of the neurotoxic effect of amyloid β-protein fragment 25–35. Int. J. Mol. Sci. 18, 2154–2171 10.3390/ijms18102154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Stotz S. C., Scott L. O., Drummond-Main C., Avchalumov Y., Girotto F., Davidsen J., Gómez-Garcia M. R., Rho J. M., Pavlov E. V., and Colicos M. A. (2014) Inorganic polyphosphate regulates neuronal excitability through modulation of voltage-gated channels. Mol. Brain 7, 42 10.1186/1756-6606-7-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Janson J., Ashley R. H., Harrison D., McIntyre S., and Butler P. C. (1999) The mechanism of islet amyloid polypeptide toxicity is membrane disruption by intermediate-sized toxic amyloid particles. Diabetes 48, 491–498 10.2337/diabetes.48.3.491 [DOI] [PubMed] [Google Scholar]
- 106. Roberts H. L., and Brown D. R. (2015) Seeking a mechanism for the toxicity of oligomeric α-synuclein. Biomolecules 5, 282–305 10.3390/biom5020282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Demuro A., Mina E., Kayed R., Milton S. C., Parker I., and Glabe C. G. (2005) Calcium dysregulation and membrane disruption as a ubiquitous neurotoxic mechanism of soluble amyloid oligomers. J. Biol. Chem. 280, 17294–17300 10.1074/jbc.M500997200 [DOI] [PubMed] [Google Scholar]
- 108. Verma M., Vats A., and Taneja V. (2015) Toxic species in amyloid disorders: oligomers or mature fibrils. Ann. Indian Acad. Neurol. 18, 138–145 10.4103/0972-2327.144284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Ihse E., Yamakado H., van Wijk X. M., Lawrence R., Esko J. D., and Masliah E. (2017) Cellular internalization of α-synuclein aggregates by cell surface heparan sulfate depends on aggregate conformation and cell type. Sci. Rep. 7, 9008 10.1038/s41598-017-08720-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Wang L., Yan J., Wise M. J., Liu Q., Asenso J., Huang Y., Dai S., Liu Z., Du Y., and Tang D. (2018) Distribution patterns of polyphosphate metabolism pathway and its relationships with bacterial durability and virulence. Front. Microbiol. 9, 782 10.3389/fmicb.2018.00782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Nahálka J., and Pätoprstý V. (2009) Enzymatic synthesis of sialylation substrates powered by a novel polyphosphate kinase (PPK3). Org. Biomol. Chem. 7, 1778–1780 10.1039/b822549b [DOI] [PubMed] [Google Scholar]
- 112. Pavlov E., Aschar-Sobbi R., Campanella M., Turner R. J., Gómez-Garcia M. R., and Abramov A. Y. (2010) Inorganic polyphosphate and energy metabolism in mammalian cells. J. Biol. Chem. 285, 9420–9428 10.1074/jbc.M109.013011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Kornberg A., Rao N. N., and Ault-Riché D. (1999) Inorganic polyphosphate: a molecule of many functions. Annu. Rev. Biochem. 68, 89–125 10.1146/annurev.biochem.68.1.89 [DOI] [PubMed] [Google Scholar]
- 114. Lonetti A., Szijgyarto Z., Bosch D., Loss O., Azevedo C., and Saiardi A. (2011) Identification of an evolutionarily conserved family of inorganic polyphosphate endopolyphosphatases. J. Biol. Chem. 286, 31966–31974 10.1074/jbc.M111.266320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Cordeiro C. D., Saiardi A., and Docampo R. (2017) The inositol pyrophosphate synthesis pathway in Trypanosoma brucei is linked to polyphosphate synthesis in acidocalcisomes. Mol. Microbiol. 106, 319–333 10.1111/mmi.13766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Ghosh S., Shukla D., Suman K., Lakshmi B. J., Manorama R., Kumar S., and Bhandari R. (2013) Inositol hexakisphosphate kinase 1 maintains hemostasis in mice by regulating platelet polyphosphate levels. Blood 122, 1478–1486 10.1182/blood-2013-01-481549 [DOI] [PubMed] [Google Scholar]
- 117. Hou Q., Liu F., Chakraborty A., Jia Y., Prasad A., Yu H., Zhao L., Ye K., Snyder S. H., Xu Y., and Luo H. R. (2018) Inhibition of IP6K1 suppresses neutrophil-mediated pulmonary damage in bacterial pneumonia. Sci. Transl. Med. 10, eaal4045 10.1126/scitranslmed.aal4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Pavlov E., Grimbly C., Diao C. T., and French R. J. (2005) A high-conductance mode of a poly-3-hydroxybutyrate/calcium/polyphosphate channel isolated from competent Escherichia coli cells. FEBS Lett. 579, 5187–5192 10.1016/j.febslet.2005.08.032 [DOI] [PubMed] [Google Scholar]
- 119. Bru S., Samper-Martin B., Quandt E., Hernandez-Ortega S., Martinez-Lainez J. M., Gari E., Rafel M., Torres-Torronteras J., Marti R., Ribeiro M. P. C., Jimenez J., and Clotet J. (2017) Polyphosphate is a key factor for cell survival after DNA damage in eukaryotic cells. DNA Repair (Amst) 57, 171–178 10.1016/j.dnarep.2017.08.001 [DOI] [PubMed] [Google Scholar]
- 120. Bentley-DeSousa A., Holinier C., Moteshareie H., Tseng Y. C., Kajjo S., Nwosu C., Amodeo G. F., Bondy-Chorney E., Sai Y., Rudner A., Golshani A., Davey N. E., and Downey M. (2018) A screen for candidate targets of lysine polyphosphorylation uncovers a conserved network implicated in ribosome biogenesis. Cell Rep. 22, 3427–3439 10.1016/j.celrep.2018.02.104 [DOI] [PubMed] [Google Scholar]
- 121. Zhang H., Gómez-Garcia M. R., Brown M. R., and Kornberg A. (2005) Inorganic polyphosphate in Dictyostelium discoideum: influence on development, sporulation, and predation. Proc. Natl. Acad. Sci. U.S.A. 102, 2731–2735 10.1073/pnas.0500023102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Livermore T. M., Chubb J. R., and Saiardi A. (2016) Developmental accumulation of inorganic polyphosphate affects germination and energetic metabolism in Dictyostelium discoideum. Proc. Natl. Acad. Sci. U.S.A. 113, 996–1001 10.1073/pnas.1519440113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Zhang H., Gómez-Garcia M. R., Shi X., Rao N. N., and Kornberg A. (2007) Polyphosphate kinase 1, a conserved bacterial enzyme, in a eukaryote, Dictyostelium discoideum, with a role in cytokinesis. Proc. Natl. Acad. Sci. U.S.A. 104, 16486–16491 10.1073/pnas.0706847104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Fang J., Rohloff P., Miranda K., and Docampo R. (2007) Ablation of a small transmembrane protein of Trypanosoma brucei (TbVTC1) involved in the synthesis of polyphosphate alters acidocalcisome biogenesis and function, and leads to a cytokinesis defect. Biochem. J. 407, 161–170 10.1042/BJ20070612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Galizzi M., Bustamante J. M., Fang J., Miranda K., Soares Medeiros L. C., Tarleton R. L., and Docampo R. (2013) Evidence for the role of vacuolar soluble pyrophosphatase and inorganic polyphosphate in Trypanosoma cruzi persistence. Mol. Microbiol. 90, 699–715 10.1111/mmi.12392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Ruiz F. A., Rodrigues C. O., and Docampo R. (2001) Rapid changes in polyphosphate content within acidocalcisomes in response to cell growth, differentiation, and environmental stress in Trypanosoma cruzi. J. Biol. Chem. 276, 26114–26121 10.1074/jbc.M102402200 [DOI] [PubMed] [Google Scholar]
- 127. Lander N., Ulrich P. N., and Docampo R. (2013) Trypanosoma brucei vacuolar transporter chaperone 4 (TbVtc4) is an acidocalcisome polyphosphate kinase required for in vivo infection. J. Biol. Chem. 288, 34205–34216 10.1074/jbc.M113.518993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Kim D., and Cavanaugh E. J. (2007) Requirement of a soluble intracellular factor for activation of transient receptor potential A1 by pungent chemicals: role of inorganic polyphosphates. J. Neurosci. 27, 6500–6509 10.1523/JNEUROSCI.0623-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Bae J. S., Lee W., and Rezaie A. R. (2012) Polyphosphate elicits pro-inflammatory responses that are counteracted by activated protein C in both cellular and animal models. J. Thromb. Haemost. 10, 1145–1151 10.1111/j.1538-7836.2012.04671.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Dinarvand P., Hassanian S. M., Qureshi S. H., Manithody C., Eissenberg J. C., Yang L., and Rezaie A. R. (2014) Polyphosphate amplifies proinflammatory responses of nuclear proteins through interaction with receptor for advanced glycation end products and P2Y1 purinergic receptor. Blood 123, 935–945 10.1182/blood-2013-09-529602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Hassanian S. M., Dinarvand P., Smith S. A., and Rezaie A. R. (2015) Inorganic polyphosphate elicits pro-inflammatory responses through activation of the mammalian target of rapamycin complexes 1 and 2 in vascular endothelial cells. J. Thromb. Haemost. 13, 860–871 10.1111/jth.12899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Smith S. A., Choi S. H., Davis-Harrison R., Huyck J., Boettcher J., Reinstra C. M., and Morrissey J. H. (2010) Polyphosphate exerts differential effects on blood clotting, depending on polymer size. Blood 116, 4353–4359 [DOI] [PMC free article] [PubMed] [Google Scholar]