Abstract
Hsp70 chaperones are central hubs of the protein quality control network and collaborate with co-chaperones having a J-domain (an ∼70-residue–long helical hairpin with a flexible loop and a conserved His–Pro–Asp motif required for ATP hydrolysis by Hsp70s) and also with nucleotide exchange factors to facilitate many protein-folding processes that (re)establish protein homeostasis. The Hsp70s are highly dynamic nanomachines that modulate the conformation of their substrate polypeptides by transiently binding to short, mostly hydrophobic stretches. This interaction is regulated by an intricate allosteric mechanism. The J-domain co-chaperones target Hsp70 to their polypeptide substrates, and the nucleotide exchange factors regulate the lifetime of the Hsp70–substrate complexes. Significant advances in recent years are beginning to unravel the molecular mechanism of this chaperone machine and how they treat their substrate proteins.
Keywords: molecular chaperone, 70-kilodalton heat shock protein (Hsp70), protein folding, allosteric regulation, chaperone DnaK (DnaK), chaperone DnaJ (DnaJ), nanomachine, J-domain, protein homeostasis
Hsp70 function and allosteric cycle
The 70-kDa heat shock proteins (Hsp70) are universal tools of cellular protein folding. They are able to interact with many types of protein conformers as follows: with extended polypeptide segments during de novo folding of nascent chains at the ribosome or at the translocation pores (1, 2); with aggregation-prone folding intermediates and stress-denatured protein conformers, during reactivation of misfolded proteins (3); with aggregated and even amyloidic protein states during solubilization of protein aggregates and breaking of amyloid fibrils (4); and with natively folded proteins, during control of stability and activity of regulatory proteins like transcription factors (5) and assembly and disassembly of protein complexes like clathrin cages (6) or DNA replication initiation complexes (Fig. 1a) (7).
Figure 1.
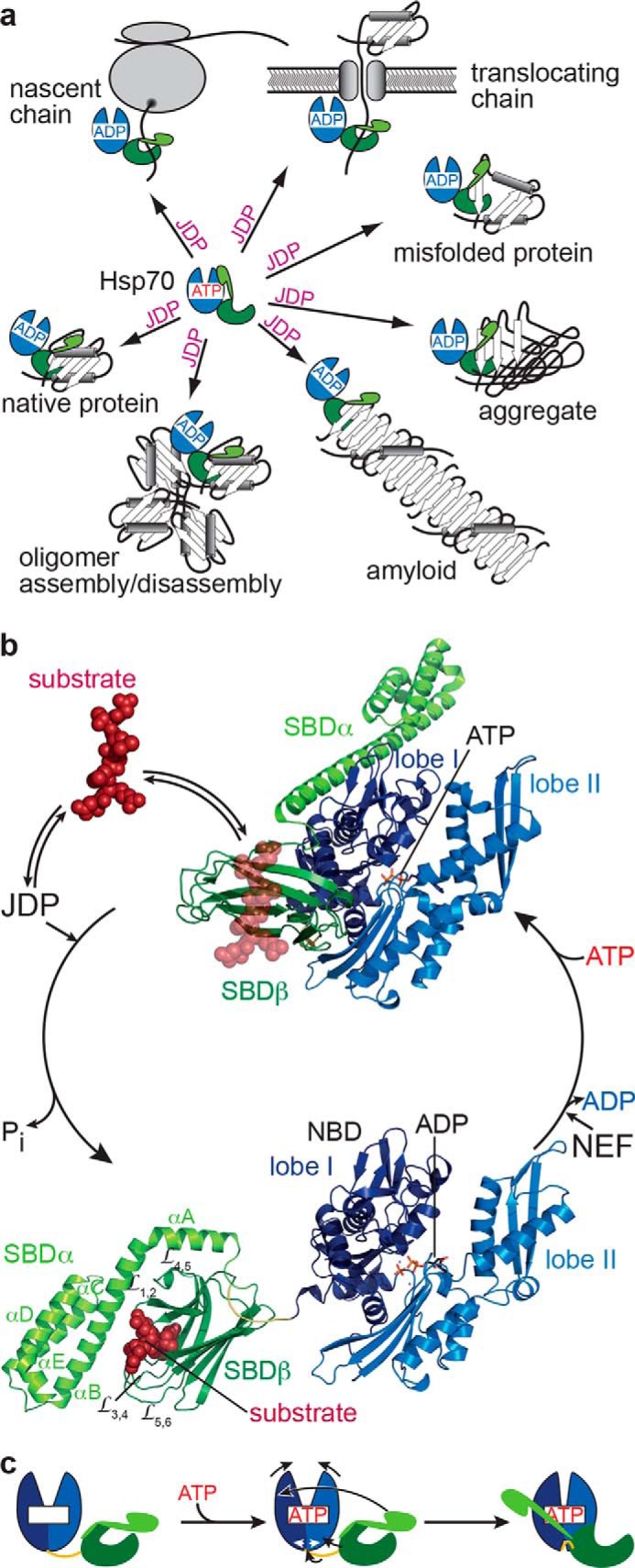
Hsp70 functions and ATPase cycle. a, JDPs target Hsp70s to a diverse set of substrates, including nascent polypeptide chains at the ribosome or translocation pore, to misfolded, aggregated, or amyloidic proteins, to oligomeric protein complexes, and to certain native proteins. b, structures of E. coli DnaK in the ATP-bound open conformation (PDB code 4B9Q, (16)) and the ADP-bound closed conformation (PDB code 2KHO (13)) with NBD lobe I in dark blue, lobe II in light blue, the conserved interdomain linker in yellow, SBDβ in dark green, and SBDα in light green. Substrates (dark red) targeted by JDPs bind with high association rates to the open conformation of Hsp70·ATP (indicated as transparent peptide from the aligned 1DKX structure (10)) and in synergism with the JDP trigger ATP hydrolysis and transition to the ADP-bound closed high-affinity state. NEFs accelerate ADP release, and rebinding of ATP converts Hsp70 back to the low-affinity ATP-bound state with subsequent substrate release. c, schematic illustrating ATP-induced conformational changes in Hsp70s; colors as in b.
Three key features of Hsp70s contribute to their versatility. 1) With their tweezers-like substrate-binding domain (SBD)2 Hsp70s interact with short degenerative sequence motifs, frequently found in practically all polypeptides, apart from intrinsically disordered proteins. This mode of action allows interaction with polypeptides and protein complexes independent of their size. In addition, the SBD also can interact with folding intermediates and folded parts of proteins and stabilize these structural entities (8). 2) The interaction of Hsp70s with folding proteins is transient and regulated by a sophisticated allosteric mechanism that couples ATP hydrolysis and ATP rebinding in their nucleotide-binding domain (NBD) with binding and release of substrate polypeptides. 3) Hsp70s do not act alone, but J-domain proteins (JDPs) and nucleotide-exchange factors (NEFs) tweak the allosteric mechanism of Hsp70s and thus control substrate binding and release (Fig. 1b).
Much has been learned about the molecular mechanism of allostery of Hsp70s through X-ray crystallography, NMR, and structure-guided mutagenesis. Although genetics and the analysis of the structures of individual domains already revealed a number of residues involved in the allosteric mechanism, only the recent elucidation of an ATP-bound Hsp70 in the open conformation allowed us to integrate these and additional residues into a functional network.
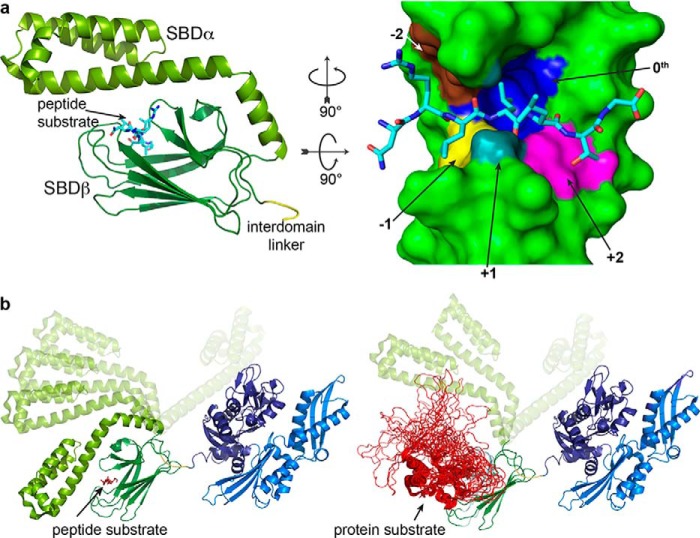
The first structural study on Hsp70s revealed their NBD as a structural relative to actin, consisting of two lobes with a deep cleft between them and each lobe subdivided into two subdomains with each of the four subdomains contributing to ATP binding (9). Later, the SBD was shown to have a unique fold, consisting of a twisted β-sandwich domain (SBDβ), an α-helical lid domain (SBDα), and a C-terminal unstructured region of largely unknown function (Figs. 1b and 2a) (10, 11). The SBDβ harbors the polypeptide-binding cleft with a deep pocket tailored for a single hydrophobic amino acid side chain. The cleft is built up by two pairs of β-strands that form a scissors-like opening and two concentric sets of upward protruding loops that enclose the peptide backbone. The SBDα restricts access to this cleft by docking onto two of the loops and forming a latch with the other two. The structure of the isolated SBD was consistent with a high affinity for peptide substrates and low substrate association and dissociation rates, a property that this domain has in the nucleotide-free and ADP-bound state. NMR studies suggested that in the ADP-bound state, NBD and SBD tumble largely independent of each other, only connected through the highly conserved and flexible linker (12, 13). In contrast, ATP binding to the NBD induces a dramatic rearrangement of the entire structure, leading to an increase of substrate association and dissociation rates by 100- and 1000-fold, respectively, and a decrease in affinity for peptides by 10–50-fold (14, 15). The structure of the ATP-bound open conformation of the Escherichia coli Hsp70 DnaK revealed these dramatic structural rearrangements (Fig. 1, b and c). The lobes of the NBD rotate relative to each other, leading to the opening of a lower crevice into which the linker binds, and SBDα and SBDβ dissociate from each other and dock onto two faces of the NBD (16, 17). The docking of the SBDβ to the NBD has two consequences: first, the substrate-binding cleft opens, consistent with high-substrate dissociation rates; and second, the NBD is locked in a lobe-rotated state, incapable of hydrolyzing ATP, explaining the very low intrinsic ATP hydrolysis rate of Hsp70s, a state that was described as a tug of war (18). Not surprisingly, such a mechanism requires a large number of residues to interact with each other and to coevolve as was revealed by evolutionary coupling analysis (19, 20). This transition between ADP- and ATP-bound states constitutes one key feature of Hsp70s: the association to substrate polypeptides occurs at the high rates of the ATP state, and concomitant ATP hydrolysis leads to trapping and tight binding in the ADP state. The coupling of polypeptide association and ATP hydrolysis is guaranteed by the fact that substrate polypeptides are able to stimulate the intrinsic ATPase rate through a series of residues that sense insertion of an amino acid side chain into the hydrophobic pocket in the SBDβ and transmits the conformational changes to the NBD (Fig. 3) (21). The efficient trapping of substrates requires the presence of a JDP that couples polypeptide binding to Hsp70s with ATP hydrolysis (22, 23). This nonequilibrium mode of substrate binding has as a consequence an extremely high affinity for substrates, which was coined ultra-affinity (24).
Figure 2.
Hsp70–substrate interaction and helical lid dynamics. a, on the left is shown the substrate-binding domain of the E. coli Hsp70, DnaK, with the model peptide NRLLLTG (in cyan) bound to the binding cleft (10) (PDB code 1DKZ). This side view of the crystal structure shows how the β-subdomain cradles the model substrate in an extended conformation, with the helical lid serving as a cover over the peptide substrate. On the right is shown a top view of the SBD–NRLLLTG complex (with the helical lid removed) to illustrate the five pockets that comprise the canonical binding site (numbered). The structure shows the extended backbone of the peptide substrate model and the fit of side chains into the pockets on the chaperone. b, on the left is shown the ADP- and substrate-bound conformation of DnaK (PDB code 2KHO (13)), including a bound model peptide substrate (NRLLLTG) (in red), with a schematic representation of the potential movements of the helical lid that have been observed to occur by a variety of methods (34–36). On the right is shown how the lifting of the helical lid away from the β-subdomain enables the binding of a partially folded client protein to the canonical binding groove in the SBD. The protein depicted here, a mutant form of apoflavodoxin, illustrates a representative ensemble of unfolded states as determined by NMR (PDB code 2KQU) (121).
Figure 3.
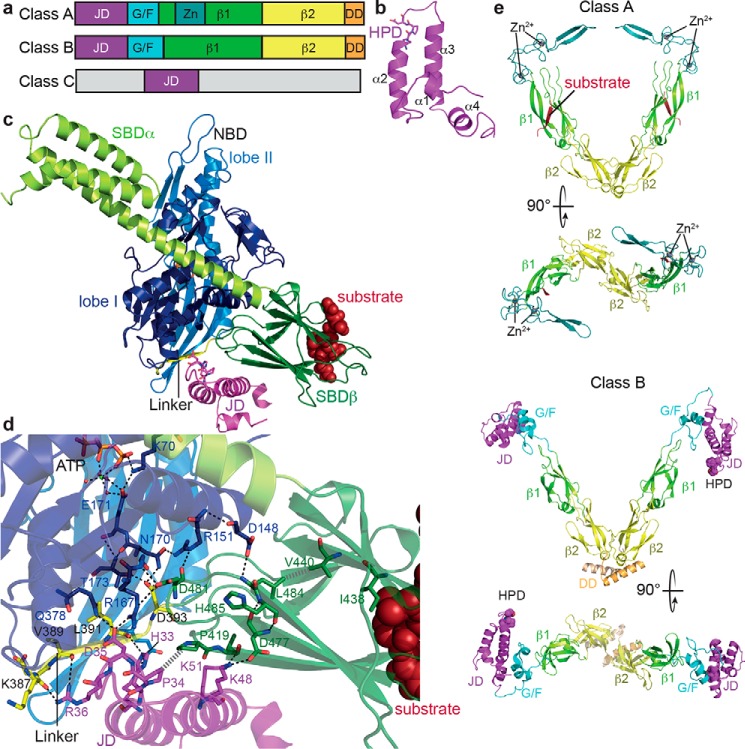
Structure and function of JDPs. a, domain organization of JDPs; JD, J-domain; G/F, glycine-phenylalanine–rich region; Zn, zinc finger; β1, first β-sandwich domain; β2, second β-sandwich domain; DD, dimerization domain. b, NMR structure of the J-domain of E. coli DnaJ (PDB code 1XBL (122)) indicating the four helices of the helical hairpin and the HPD motif. c, structure of the ATP-bound open conformation of E. coli DnaK in complex with the J-domain of E. coli DnaJ (PDB code 5NRO (82)); NBD lobe I, dark blue; NBD lobe II, light blue; linker, yellow; SBDβ, dark green; SBDα, light green; J-domain, purple. The SBDβ was aligned with the structure of the SBD in complex with a substrate peptide (PDB code 1DKX (10)) to show how peptides (dark red) bind to the ATP-bound open conformation. d, zoom into the structure presented in c showing residues involved in J-domain–DnaK interactions and in substrate and DnaJ-mediated stimulation of the ATPase activity as sticks, polar contacts as black dashed lines, and hydrophobic interactions as gray hatched lines. e, cartoon representation of the β-sandwich domains and the zinc finger of the class A JDP Saccharomyces cerevisiae Ydj1 (PDB code 1NLT (86); dimerization domain is missing in the structure) and the full structure of the class B JDP T. thermophilus DnaJ (PDB code 4J80 (123); J-domain, purple; G/F region, cyan; zinc, dark teal; β1, light green; β2, yellow; dimerization domain, orange.
Interaction of Hsp70s and their substrates
The functions of Hsp70s, although impressively diverse, have at their center a common mechanism that is based on the binding and subsequent ATP-mediated release of substrates. Generally, Hsp70 substrates are partially folded and thus expose sequences that would normally be intimately packed into the natively folded client and inaccessible. In some cases, clients display sequences that are accessible expressly for the purpose of chaperone recognition for specific functions (25). As discussed in more detail below, JDPs mediate the productive delivery of substrates to Hsp70s and may play critical roles in how substrates first interact with an Hsp70. Moreover, a key aspect of the action of Hsp70s on their substrates is the timing of the cycle of binding and release, which the JDP strongly influences by stimulation of the Hsp70 ATPase activity, as does the NEF (see below for deeper discussions of the co-chaperone functions). Although the chaperone system functions with these co-chaperones, the Hsp70 itself has a capacity to bind a wide array of clients with nucleotide-modulated affinity.
Hsp70s are selectively promiscuous in their binding to substrates
Peptide array data have shown that the E. coli Hsp70, DnaK, which has served as a paradigm for the Hsp70 family, has a strong preference to bind hydrophobic sequences (consistent with the preference for partially unfolded substrates) with some positive residues flanking the bound region (26, 27). Two algorithms that predict possible substrate-binding sites of DnaK have been developed, largely based on the peptide array data: one is from Bukau and co-workers (26), and the other, called Limbo, is from Rousseau and Schymkowitz and co-workers (27). Application of these prediction algorithms reveals that there are potential DnaK-binding sites with high frequency (one every 40 residues) in the E. coli proteome. Analogous predictive algorithms have been developed for the eukaryotic Hsp70, BiP, using a combination of phage display and peptide array data (28, 29). The results show that there may be some chaperone-to-chaperone variation in substrate preferences, but the ability to bind many sequences is a common Hsp70 property. The binding function of Hsp70s thus displays promiscuity in that many clients can bind, and selectivity, in that Hsp70s do not bind all proteins or all sequences in the proteome. It is fascinating to explore the origins of this selective promiscuity.
Client binding to Hsp70s is mediated primarily by a channel in the SBD. Most of our understanding of the structural details of Hsp70–substrate binding has been derived from studies of peptide models (25). A number of structures from crystallography (10, 30, 31) and NMR (32, 33) show that these peptide substrate models bind in an extended conformation to a site in the SBDβ, with the helical subdomain as a lid (Fig. 2a). The helical lid must dissociate before the substrate can bind and be released, which explains why binding and release from ADP-bound Hsp70s is very slow. Nonetheless, the helical lid has dynamic character even in ADP-bound Hsp70s (Fig. 2b) (34–36).
Inspection of available crystal structures of the SBD bound to peptide models provides a detailed picture of the binding interaction. The binding cleft is made up of five pockets formed by the β-sandwich subdomain, suggesting that the core binding site for an Hsp70 substrate is five residues long (Fig. 2a, right panel). The residue occupying the central pocket of the binding site, called the “0th position,” confers the highest sequence specificity in Hsp70–substrate binding (37). Leu fits optimally in this pocket, and Ile, Val, and Phe fit less well. The ability of the other pockets to accommodate a range of amino acids underlies the promiscuity, whereas the stringent fit to the central pocket and the preference for positively charged sequences account for selectivity.
Their selective promiscuity suggests that Hsp70s could bind most proteins in the proteome, if binding sites were accessible. This raises the intriguing question of how many clients interact with Hsp70s in vivo? Depletion of DnaK in E. coli caused widespread aggregation of cytosolic proteins associated with a wide array of functions, particularly when combined with the deletion of a trigger factor (38, 39). A complementary study identified in vivo substrates of DnaK by treating cells with apyrase to lock substrates into complex with the chaperone high-affinity state, followed by immunoprecipitation and identification of the bound substrates by MS (40). Of the ∼700 DnaK substrates identified, most were cytosolic. There were some properties in common with these substrates: increased aggregation propensity (as demonstrated in an in vitro expression system by Niwa et al. (41)), relatively low abundance, complex topologies, and the presence of predicted DnaK-binding sites. Both newly synthesized and pre-existing proteins were among the DnaK substrates, as expected given the roles of DnaK both early in biosynthesis and later in maintenance of the health of the proteome. The authors concluded that DnaK is a key hub in protein homeostasis and that a large fraction of expressed proteins may flux through DnaK.
Varied nature of Hsp70 complexes with protein substrates
Proteins could be envisioned to bind to Hsp70s in a variety of ways. Accessible, unstructured regions of highly unfolded substrates may bind very much like peptides. Alternatively, partially folded proteins could bind in a compact, molten globule-like state. Finally, a structured protein substrate may display an unfolded accessible sequence, for example at one of its termini. Although we lack a detailed structural picture of a protein bound to an Hsp70, there are studies supporting each of these paradigms (8, 42, 43). A recent in-depth nuclear magnetic resonance (NMR) study (44) examined how DnaK bound two model substrate proteins, one with a helical native structure and one with a largely β-sheet native fold. Both of these substrates were presented to the chaperone as ensembles containing some partially folded species and some unfolded species. Chaperone binding occurred via conformational selection of unfolded states rather than by “unfolding” of partially folded species, and the mode of binding was like that of peptide models. In other cases, the Hsp70-bound substrate has been found to exist in a compact non-native or near-native state. For example, a recent single molecule optical tweezer study of the impact of the DnaK chaperone system on the folding and unfolding of maltose-binding protein (8) showed that the helical lid and the binding cleft were implicated in binding. The helical lid lifted off the β-subdomain when the chaperone bound to a partially folded state of the client (depicted in Fig. 2b, right panel). The third binding mode has been described when the constitutively expressed eukaryotic Hsp70, Hsc70, recognizes clathrin via a short terminal sequence that is unfolded and accessible while the rest of the protein is folded (see more description of this system below) (45, 46). These studies point out how the different substrate-binding modes of Hsp70s may underlie the functional diversity of this family of chaperones.
How does Hsp70 binding affect its client protein and how does this relate to Hsp70 functions?
Understanding how the interaction of Hsp70s affects the conformational ensemble of their clients will be needed to understand the physiological roles of Hsp70s, but the evidence accumulated so far indicates that the action of Hsp70s may differ from substrate to substrate. NMR experiments showed that DnaK immobilizes its substrates in the region of the binding sites but causes only a modest effect on the conformations sampled by the sequences surrounding them (47, 48). This constitutes a holding model and ascribes a fairly passive role to the Hsp70 chaperone with the binding and hydrolysis of ATP acting predominantly as a timing mechanism.
An alternative result was reported using single molecule fluorescence (49). The authors found that the denatured rhodanese was markedly expanded upon interaction with DnaK, and a model was proposed where multiple DnaKs bind simultaneously to the client. This single molecule fluorescence result is consistent with an “unfoldase” role of the chaperone, described as “entropic pulling,” in which the binding of chaperones is postulated to exert a force on the client in such a way that regions apart from the binding site are unfolded (24, 50, 51). In this model, the energy from ATP is used to re-route an Hsp70 substrate from a misfolding pathway to a productive pathway to native protein by stabilizing a nonequilibrium state of the protein. Quantitative analysis based on this model can account for the energy demand of Hsp70s.
Although the holding/unfolding models and exact description of how or whether Hsp70s alter the folding of their substrates remain a subject of debate and will require more experimentation, chaperone action is likely to be substrate-dependent, and there is widespread agreement that regardless of the detailed mechanism, the action of Hsp70s reduces the likelihood of aggregation of its substrates. Many examples showing inhibition of aggregation by Hsp70s have been reported (52–54).
Examples of the interaction of Hsp70s with physiological clients
One of the first physiological clients of Hsp70s to be examined was σ32 (55). This transcription factor is a master regulator of the heat shock response in E. coli, binding to the RNA polymerase core enzyme in place of the σ70 subunit and targeting the polymerase to the heat-shock promoters (56). In the absence of cellular stress, the DnaK system (with its co-chaperones DnaJ and GrpE) inactivates σ32 by increasing its degradation by FtsH. Thus, expression of genes under control of heat-shock promoters is high when the concentration of the DnaK system components is lowered by their action on unfolded proteins and low when the DnaK system is free to bind σ32. Detailed analysis of the binding has revealed that there is one DnaK-binding site in native σ32 in an unstructured region within the 284-amino acid-long σ32 molecule. Binding of DnaK was shown by hydrogen exchange to cause a conformational change in the N-terminal region of σ32, which presumably renders the transcription factor amenable to proteolytic degradation. Mutants of σ32 that display longer half-lives and thus lose regulation of the heat-shock response by chaperone levels are often poor DnaK binders. Altogether, this system is emblematic of the physiological exploitation of the functions of the Hsp70 chaperone.
Ensuring that the synthesis and folding of the light and heavy chains of Igs are coordinated properly was recognized early on to be a challenge in cells. This led to the discovery of the ER resident Hsp70, BiP, the heavy-chain binding protein, the role of which was proposed to be binding to the newly synthesized heavy chains so that they are protected until they form a complex with their respective light chain (LC) (57). In-depth in vitro and in vivo studies from the Hendershot and Buchner labs have led to a model in which the function of BiP is to recognize incompletely folded states of the heavy chain CH1 domain which remain reduced until they coordinately fold, oxidize, and assemble with the corresponding LC (58). Thus, BiP binding occurs to an unfolded CH1 domain, and the recognition is mediated by linear sequences, well mimicked by peptides but not entirely as the lid does not close over the peptide-binding pocket (43). The timing and coordination of the assembly of the full IgG is complex and influenced by co-chaperones as well as other factors (59). Again, this system exemplifies the exploitation of Hsp70's chaperone functions for a physiological need.
The earliest descriptions of the eukaryotic Hsp70, Hsc70, was as the clathrin-uncoating ATPase (60), and although this abundant cytoplasmic chaperone performs many functions, its role as a specialized disassembler of clathrin coats is a fascinating example of physiological reliance on its capabilities. A number of studies have provided a consistent picture of the way Hsc70 recognizes and facilitates disassembly of clathrin triskelions from coated vesicles (6, 46, 61). The Hsc70-binding sites on clathrin (QLMLT sequences) are at the extreme flexible C-terminal tails of each clathrin molecule. Following the canonical mechanism of Hsp70s, the combination of substrate and JDP activates the ATPase of Hsc70, shifting it to the high-affinity ADP–bound state and leading to a stable complex of clathrin triskelion with Hsc70s. The disassembly mechanism may rely on steric interference of the chaperone-bound triskelions with one another in the coat architecture, thus favoring their free, disassembled state (which in turn remain bound to the Hsc70–ADP chaperone). An alternative model invokes a more active entropic pulling mechanism for Hsc70-mediated coat disassembly (62).
Hsp70 partners with other chaperones
Many functions of Hsp70s rely on their partnerships with other classes of chaperones. The hand-off of substrates from the bacterial Hsp70, DnaK, to the chaperonin GroEL is a classic demonstration of a chaperone team (63). In the case of nascent chains, DnaK may interact while a chain is translating, usually after the ribosome-associated chaperone, trigger factor (64). Alternatively, DnaK may bind to fully synthesized stress-unfolded substrates in the cytosol (65). In either case, upon chain release from DnaK triggered by ATP binding, the partially folded substrate may then be bound by GroEL, where its folding to the native state is facilitated by cycles of action of GroEL, GroES, and ATP. There are similar hand-offs and roles of Hsp70s in eukaryotes (66), including eukaryotic substrates such as the von Hippel-Lindau tumor suppressor complex (67), which follow a pathway from Hsp70 to the chaperonin TRiC to achieve their complex folds. The Hsp70 role in these cascades of chaperone is to prevent aggregation of early folding states.
Early biochemical studies showed the cooperative action of the Hsp70 and Hsp90 chaperones in folding and assembly in eukaryotes, along with a dedicated co-chaperone Hop, which facilitates substrate transfer from the Hsp70 to the Hsp90 (69, 70). The hormone receptor maturation pathway is a seminal physiological example of how Hsp70s function as intermediaries in the multichaperone, Hsp70/Hsp90 system (71). Structural data suggest that this handoff involves a multicomponent complex (5). Analogous to its role with the chaperonins, the Hsp70 appears to hold the client in a significantly and incompletely folded state, prevent its aggregation, and then through the co-chaperones hand it off for maturation on the Hsp90. Exciting new biochemical and structural insights about the interaction of the Hsp70/Hsp90 chaperone team with clients is emerging recently (71, 72). An interesting twist on the various proposals for the impact of Hsp70 binding on its clients has recently been articulated by Rüdiger and co-workers (73), who tested the partnership between Hsp70 and Hsp90 in the refolding of luciferase. They demonstrate in a reconstituted system that the Hsp70 interaction leads to a dead-end trapping of the substrate under physiological conditions and that rescue requires intervention by an Hsp90 partner (73).
Another important function of Hsp70 is as part of a disaggregation machinery in cells. This topic has been reviewed quite recently (74), and so we only briefly describe it here. Protein aggregates are hallmarks of misfolding diseases, although the etiology of their role in pathology remains unclear. Nonetheless, all organisms have developed machinery that has the capacity to disassemble protein aggregates. In bacteria, this would be DnaK working with its J-domain partner DnaJ and its NEF GrpE, and the AAA+ ATPase, ClpB. In yeast, the cytoplasmic disaggregation machinery comprises an Hsp70 (Ssa1–4), a JDP (either Ydj1 or Sis1), an intriguing class of NEF, Hsp110 (here called Sse1/2), and the specialized disaggregation chaperone Hsp104. In the metazoan cytoplasm, an Hsp70 (either HspA1/Hsp70 or HspA8/Hsc70) partners with the JDPs DnaJA2 and DnaJB1 and a member of the Hsp110 family, HspH1–3, to make up the disaggregation machinery. In each of these systems, small heat shock proteins join the party, helping disaggregation by stabilizing smaller aggregates during aggregate formation and sequestering denaturing/misfolding proteins in a near-native state (75). In the presence of ClpB/Hsp104, the role of the Hsp70s in disaggregation is to recognize stretches of accessible polypeptide early on and to activate the downstream disaggregation machines by handing off the substrates. In metazoans where the AAA+ disaggregases are absent, the role of Hsp70 comprises both recognition of accessible polypeptide stretches and disentangling of the aggregates.
Hsp70s work with a team of quality control chaperone and protease partners to play a key role when the cellular decision is made that a substrate must be degraded, either because it has no hope of folding on the time scale needed by the cell or because its lifetime is regulated as part of its cellular function. The Hsp70s' action on substrates in degradation decisions is dual: recognition of substrates fated for degradation and preparation of these substrates to interact with the downstream partners in degradation. Both prokaryotic and eukaryotic degradation systems have been the focus of considerable research, and a recent review summarizes how Hsp70s participate in these systems (76). Therefore, here we will describe just one example to illustrate the general role of the Hsp70 in these systems. In eukaryotes, the chaperone-assisted ubiquitin–proteasome pathway exploits the canonical substrate-binding role of the Hsp70 SBD, coupled with interaction of the Hsp70 NBD with a specialized NEF, Bag1, that also interacts with the proteasome, and the co-chaperone CHIP (C terminus of Hsp70-interacting protein) that acts as an E3 ubiquitin ligase and also binds the C-terminal EEVD of the Hsp70 and its partner Hsp90. The whole complex is a nanomachine capable of recognizing substrates to be degraded, facilitating their tagging with ubiquitin, and handing them off to the proteasome.
Additional roles of Hsp70s are being described in a growing number of cellular systems (77–80), and we anticipate that this trend will continue. These chaperones are central machines of Nature that have been harnessed throughout evolution in cases where relatively generic recognition motifs signal the exposure of sites that ready a client for subsequent steps, and where the subsequent steps need help: protection from competing processes like aggregation, productive hand-off to downstream interaction partners (Hsp70 to Hsp90), or timing of release to tune a cellular process (triage decisions for degradation or not) (81).
J-domain proteins: Hsp70 targeting factors
JDPs are a family of multidomain proteins characterized by the presence of the so-called J-domain: ∼70-residue helical hairpin with a flexible loop of variable length and the universally conserved His–Pro–Asp (HPD) motif (Fig. 3, a and b). The J-domain is necessary to trigger ATP hydrolysis in Hsp70s by binding with its HPD motif to the docked linker and interacting with NBD and with SBDβ (82). The histidine and the aspartate of the HPD motif access two networks of hydrogen bonds and hydrophobic interactions that converge onto the catalytic residues for ATP hydrolysis (Fig. 3, c and d). In addition, the J-domain is connected to the pathway that leads the substrate-binding signal from the hydrophobic pocket in the SBDβ to the interface between SBDβ and NBD. The current model for the substrate and J-domain–mediated synergistic stimulation of ATP hydrolysis proposes that substrate binding to the SBDβ triggers release of the SBDβ from the NBD to allow back-rotation of the two NBD lobes into a position optimal for ATP hydrolysis (18, 21). The J-domain supports this signal by increasing the coupling within the pathway. In addition, the J-domain prevents the linker from slipping out of the lower crevice to arrest the back rotating NBD lobes at the correct position. Furthermore, the J-domain tweaks the hydrogen bond network in the NBD to optimally convert the receiving signals into ATP hydrolysis. This action of the J-domain is the basis for its function in targeting Hsp70s to their substrates. The localization of a J-domain may allow Hsp70s to select one specific binding site out of several alternative sites or even to bind to a suboptimal site, for example during translocation of a polypeptide into mitochondria, where Hsp70 should bind every 20–30 residues to drive efficient translocation, but a high-affinity Hsp70–binding site may not be present in such regular distances (83). Thus, the presence of a J-domain in any protein is considered as certain indication for the functional involvement of an Hsp70, and in the course of evolution the addition of a J-domain to a protein allowed for the recruitment of the Hsp70 chaperone power. In the absence of an Hsp70 substrate, JDPs often trigger Hsp70s to bind to themselves to form Hsp70 oligomers or to bind instead to the JDP itself. Therefore, interaction of JDPs with Hsp70s measured for example by surface plasmon resonance spectroscopy in the presence of ATP reflects the J-domain targeted binding of Hsp70 to the JDP as a substrate and is absent in Hsp70 mutants that are defective in ATP hydrolysis (e.g. DnaK-T199A) or in allostery (e.g. DnaK-V389A,L390A,L391A,L392A) or in substrate binding (e.g. DnaK-V436F) (84).
JDPs are traditionally divided into three classes according to the number of additional domains they share with the name-giving prototype JDP, E. coli DnaJ (Fig. 3a). Class A shares all domains with DnaJ, the N-terminal J-domain followed by the glycine-phenylalanine (G/F)-rich region, two topologically similar twisted β-sandwich domains with a Zn2+ finger inserted into the first of the two domains and a C-terminal dimerization domain (Fig. 3e). Class B JDPs contain a J-domain, a G/F-rich region, and a C-terminal domain (Fig. 3e). The C-terminal domain of many class B JDPs shares similarity with the C-terminal domain of DnaJ and also ends in a dimerization domain but always miss the Zn2+ finger. Class C JDPs only share the J-domain with DnaJ and contain any number of protein interaction or protein localization motifs (85). Among the class A and class B JDPs are many members that are considered to be general JDPs able to interact promiscuously with virtually all nascent polypeptides or misfolded or aggregated proteins (Fig. 3c). Class C JDPs are believed to be specialists able to interact with one or a small subset of Hsp70 substrates or to be localized where Hsp70 substrates emerge, such as the ribosomal exit tunnel (e.g. zuotin) or translocation pores (e.g. Sec63 or Pam18).
How class A and class B JDPs bind substrates is still debated. The zinc finger and β-sandwich domains of yeast class A JDP Ydj1 were crystallized with a peptide that augmented one of the twisted β-sheets of the first β-sandwich domain (Fig. 3c) (86). However, the structure did not explain the known binding preference of class A JDPs and were not consistent with biophysical data (55, 87). A similar construct of the human class B JDP Hdj1/DNAJB1 was crystallized with four peptides comprising the C terminus of human Hsc70 (GPTIEEVD) augmenting both β-sheets of the first β-sandwich domain in a very similar way (88). The peptides bound in this structure also engage in hydrophobic interactions more consistent with the binding preference of class A and class B JDPs. These results suggest that there might be several low-affinity binding sites for simultaneous interaction with a polypeptide chain, explaining why many class A and class B JDPs bind proteins with much higher affinity than peptides.
Recently, it was shown that some class A and class B JDPs cooperate in protein disaggregation. This cooperation involves not only a division of labor with DNAJA2 preferentially targeting small aggregates and DNAJB1 large aggregates but even more a direct interaction between the class A and the class B JDP, suggesting the formation of a high order complex with Hsp70s (89, 90).
Nucleotide exchange factors: regulators of Hsp70–substrate complex lifetime
Basal nucleotide dissociation rates of Hsp70s are generally very low (0.02–0.2 s−1) and become rate-limiting for the ATPase cycle in the presence of substrates and JDPs. Biochemical, crystallographic, and NMR data suggest that the NBD of Hsp70 toggles between at least two conformations, one with the nucleotide-binding cleft closed and one with the nucleotide-binding cleft open, allowing nucleotide binding and release. The transition rate between the closed and open conformation, which dictates the nucleotide binding and release rates, depends on the occupancy of the nucleotide-binding pocket, with the rate being highest in the nucleotide-free state and decreasing more and more when ADP, ADP plus phosphate, and ATP are coordinated by all four NBD subdomains in the nucleotide-binding pocket (16, 91–93).
Currently, four different families of NEFs are known to accelerate nucleotide release in Hsp70 proteins: the GrpE-type NEFs in bacteria, mitochondria, and plastides; the Bag-domain family of proteins in the nuclear/cytoplasmic compartment of eukaryotic cells; the HspBP1-type proteins in the nucleus, cytosol, and ER; and the Hsp110/Hsp170 family of proteins in the cytosol and ER (94). Although the different NEFs are structurally unrelated and must have evolved independently, they all act on subdomain IIB of the NBD to open the nucleotide-binding cleft by tilting it outward by 13–27° (GrpE, Bag, and Hsp110) or by rotating it around its longitudinal axis (HspBP1-type) (Fig. 4).
Figure 4.
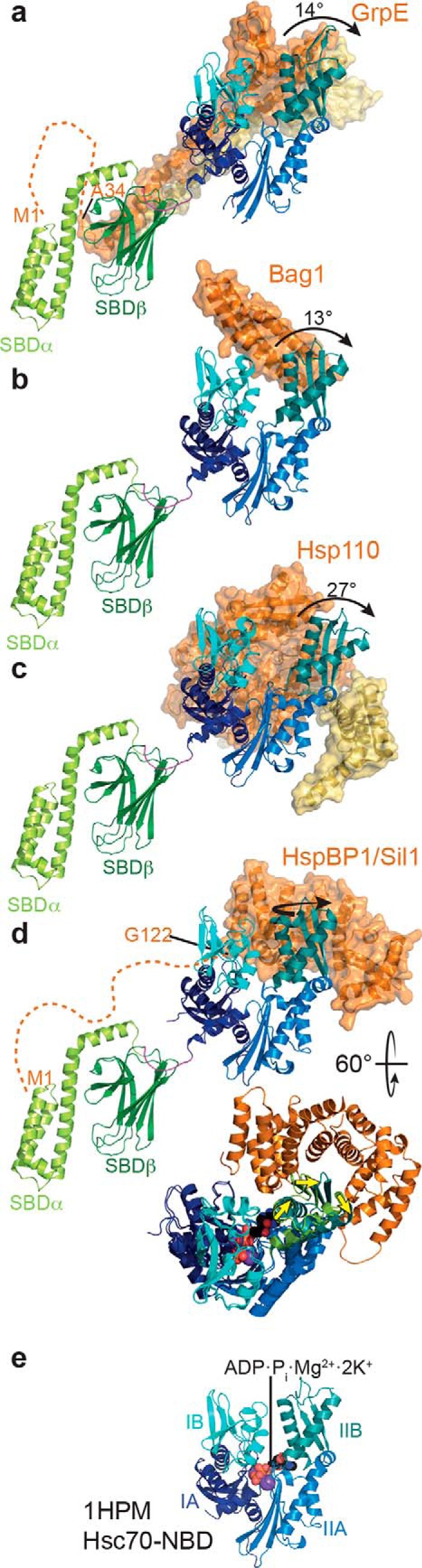
Mode of action of NEFs. Cartoon and surface representations of crystal structures of different NEFs (orange and yellow) in complex with an Hsp70 NBD (IA, dark blue; IB, cyan; IIA, light blue; IIB, dark teal), overlaid onto the full-length solution structure of DnaK (PDB code 2KHO (13), only SBD shown) to illustrate the position of the SBD. From top to bottom: E. coli GrpE in complex with E. coli DnaK–NBD (a, PDB code 1DKG (91)); Bag-domain of human Bag1 in complex with the NBD of bovine Hsc70 (b, PDB code 1HX1 (124)); yeast Sse1 in complex with the NBD of bovine Hsc70 (c, PDB code 3D2E (125)); yeast Sil1 in complex with the NBD of yeast Kar2 (d, PDB code 3QML (126)); yeast Sil1 in complex with the NBD of yeast Kar2 overlaid to bovine Hsc70 and rotated as indicated (yellow arrow indicates the displacement of subdomain IIB); e, NBD of bovine Hsc70 in complex with ADP, phosphate, Mg2+, and two K+ ions (PDB code 1HPM (68)). The NBDs of all structures are aligned to subdomains IA, IB, and IIA of the NBD of bovine Hsc70. Dashed lines at the N termini of GrpE and Sil1 indicate the unstructured regions not present in the crystal structure and proposed to bind into the substrate-binding pocket of Hsp70 (115, 116). The N-terminal methionine (M1) and the first residue in the structure are indicated.
Although E. coli GrpE is a dimer, it seems to interact with a single DnaK molecule in an asymmetric conformation through only one of its subunits, contacting subdomains IA, IB, and IIB of the DnaK NBD (Fig. 4a) (91). Therefore, its small β-sheet domain interacts with residues in the nucleotide-binding cleft, breaking a salt bridge (Arg-56–Glu-264) that crosses the nucleotide-binding cleft in the ADP-bound state and limits nucleotide dissociation (95). The GrpE dimer of Geobacillus kaustophilus was crystallized in an assembly with two DnaK molecules, seemingly functionally engaged with both as judged by the open nucleotide-binding cleft and interactions with subdomains IB and IIB of both NBDs, despite also forming an asymmetric dimer with only one protomer interacting in addition with subdomain IA of one of the DnaK molecules (96). Thus, it remains an open question whether all GrpE dimers could interact with two DnaKs that are in close proximity to accelerate nucleotide exchange simultaneously or in a rapid sequential order. Such a mechanism would explain why GrpE is a dimer, whereas the other NEFs, except for Bag2 (97), are monomeric. GrpE stimulates dissociation of ADP, ADP in the presence of 10 mm phosphate, and ATP with similar efficacy, suggesting that GrpE can interact with different conformations of the NBD and actively opens the nucleotide-binding cleft (93, 98).
The Bag-domain proteins are a family of modular multidomain proteins that share the ∼80-residue three-helix bundle Bag-domain that is necessary and sufficient for stimulating nucleotide release in eukaryotic cytosolic Hsp70 proteins (Fig. 4b). The Bag-domain interacts with NBD subdomains IB and IIB splaying open the nucleotide-binding cleft. Based on the significant difference in Bag1-stimulated nucleotide exchange for ADP, ADP plus phosphate, and ATP, it was proposed that Bag proteins act more passively and effect nucleotide exchange only by binding to and stabilizing the open conformation of Hsp70s' NBD (93).
Hsp110/Hsp170s are homologs of Hsp70s deviating from Hsp70s only by the linker between NBD and SBD, which is not conserved, and insertions of highly charged sequences of different lengths in the SBDβ between β-strands 7 and 8 and at the end of the SBDα. They bind with their NBD face-to-face to the NBD of Hsp70, interacting with subdomains IA, IB, and IIB and with their SBDα to the side of the NBD, interacting with subdomain IIA (Fig. 4c). Although Hsp110s have an ATPase activity (99), this ability is neither important for nucleotide exchange function nor for complementation of the deletion of the Hsp110 encoding genes sse1 and sse2 in yeast (100, 101). However, ATP binding to Hsp110 seems to be important for their nucleotide exchange function as ATP stabilizes their NBD (101, 102).
The members of the armadillo-repeat HspBP1 family of NEFs interact mainly with subdomain IIB of the Hsp70 NBD. These NEFs achieve the opening of the nucleotide-binding cleft by rotating subdomain IIB around its length axis rather than tilting it away from subdomain IB (Fig. 4d).
Why do Hsp70s need NEFs? As mentioned above, after JDP-mediated trapping of substrates nucleotide exchange becomes rate-limiting for substrate release, and NEFs regulate the lifetime of the Hsp70 substrate complex. However, Hsp70 could have evolved to have a higher nucleotide exchange rate. In general, eukaryotic Hsp70s already have a 10-fold higher ADP dissociation rate than the prokaryotic DnaK-type Hsp70s, because in DnaK two salt bridges across the nucleotide-binding cleft (Lys-55–Glu-267 and Arg-56–Glu-264) slow down its opening and thus ADP dissociation, whereas in eukaryotic Hsp70s only one salt bridge is retained (Lys-55–Glu-267) (95). The prokaryotic HscA has none of the two salt bridges and has a 700-fold higher nucleotide release rate than DnaK (95). Therefore, it seems that evolution could have tuned the Hsp70 chaperones optimally for the ATPase cycle. However, HscA seems to be a very specialized Hsp70 only interacting with one or a small set of substrates, whereas many Hsp70s like DnaK or the eukaryotic constitutive Hsc70 are highly promiscuous, interacting with a large number of proteins in the nascent or misfolded state. If different substrates would need to be in complex with Hsp70 for different lifetimes, then no optimal intrinsic ADP dissociation rate could have evolved, and NEFs become advantageous.
In addition, the optimal lifetime of the Hsp70–substrate complex might be different under different conditions like heat shock, and regulating the NEF activity might be easier than adapting the Hsp70 intrinsic ADP dissociation rate directly. E. coli and Thermus thermophilus GrpE were shown to unfold reversibly and lose NEF activity at heat shock temperatures, prolonging the lifetime of the Hsp70–substrate complex under conditions when refolding seems impossible (103–105). Also, regulating the concentration of NEFs could have a profound influence on Hsp70 chaperone activity as refolding of denatured luciferase and solubilization of amorphous protein aggregates and amyloid fibrils are accelerated by NEFs in vitro only in a very narrow concentration range (93, 98, 101, 106, 107). Moreover, not all NEFs work equally well in solubilization of protein aggregates, suggesting functions beyond nucleotide exchange (106, 107). When the prokaryotic Hsp70 DnaK cooperates with the Hsp100 AAA+ chaperone ClpB to solubilize protein aggregates, GrpE is not necessary for substrate transfer from DnaK to ClpB. In fact, ClpB and GrpE compete for the same interaction site on DnaK (108).
In eukaryotic cells, compartmentalization of NEFs could be another regulatory mechanism for Hsp70 activity. For example, in yeast the HspBP1-type NEF Fes1 exists in two splice variants: the major one is cytosolic and the minor one is nuclear (109). Under heat shock, the cytosolic one is even increased much more than the nuclear one. Similarly, human Hsp105 is mostly cytosolic under nonstress conditions and partially translocates to the nucleus after heat shock (110, 111). Overexpression of Hsp110 in the nucleus in certain cancer cells is linked to resistance to anti-cancer drugs and poor survival prognosis for the patient (112). Another aspect of subcellular localization is found in the Bag family of proteins. There are five different members of Bag proteins in human cells. These proteins mainly differ in their N-terminal sequences with a number of different protein–protein interaction domains that target the Bag proteins to different sites, for example the proteasome (Bag1) (113) or the cytosolic domain of the TNFα receptor (Bag4/SODD) (114). Such localization could lead to targeted nucleotide exchange of Hsp70 and, consequently, upon rebinding of ATP, to release of Hsp70 substrates.
NEFs can also have a function beyond accelerating nucleotide exchange. Yeast Fes1 and human HspBP1 have a long unstructured N-terminal sequence that was shown to bind into the substrate-binding pocket and to prevent rebinding of substrates after nucleotide exchange and ATP-binding–mediated release (115). This function is essential for Fes1 activity in yeast. A similar function seems also to be operational in GrpE, which has an unstructured N-terminal extension that was shown to compete with substrates for binding to the substrate-binding cleft, but does not actively accelerate substrate release in the absence of ATP (116).
Finally, Hsp110 NEFs are able to bind misfolded proteins (117). How this function contributes to the working of the Hsp70 chaperone machinery is currently unclear (118).
Current questions
Hsp70s have been known to form oligomeric states for a long time. More recently, different oligomeric structures in the ADP- and ATP-bound states were proposed (72, 119). How these states contribute to Hsp70 function is currently not clear. Also, as mentioned above class A and class B JDPs form complexes in vitro and in vivo boosting Hsp70 disaggregation activity (89, 90, 120). Because both class A and class B JDPs are dimers, potentially four J-domains are available for interacting with Hsp70s and could target several Hsp70s simultaneously to bind to aggregates in close proximity to each other. Whether higher order complexes of Hsp70s are formed here is not clear. Interestingly, class A and class B complexes do not increase the efficacy of Hsp70s in breaking and disassembling amyloid fibrils (107). The number of JDPs has increased significantly during evolution to multicellular complex organisms, and JDPs within the same cellular compartment deviate in their Hsp70 interaction surface despite seemingly interacting with the same Hsp70s. How such deviations in the interaction surface affect the Hsp70 chaperone activity will be interesting to explore.
This is the second article in the JBC Reviews series “Molecular chaperones and protein quality control.” The authors declare that they have no conflicts of interest with the contents of this article.
- SBD
- substrate-binding domain
- ER
- endoplasmic reticulum
- PDB
- Protein Data Bank
- NBD
- nucleotide-binding domain
- LC
- light chain
- JDP
- J-domain protein
- NEF
- nucleotide-exchange factor
- JDP
- J-domain protein.
References
- 1. Preissler S., and Deuerling E. (2012) Ribosome-associated chaperones as key players in proteostasis. Trends Biochem. Sci. 37, 274–283 10.1016/j.tibs.2012.03.002 [DOI] [PubMed] [Google Scholar]
- 2. Neupert W., and Brunner M. (2002) The protein import motor of mitochondria. Nat. Rev. Mol. Cell Biol. 3, 555–565 10.1038/nrm878 [DOI] [PubMed] [Google Scholar]
- 3. Mayer M. P., and Bukau B. (2005) Hsp70 chaperones: cellular functions and molecular mechanism. Cell. Mol. Life Sci. 62, 670–684 10.1007/s00018-004-4464-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nillegoda N. B., and Bukau B. (2015) Metazoan Hsp70-based protein disaggregases: emergence and mechanisms. Front. Mol. Biosci. 2, 57 10.3389/fmolb.2015.00057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wegele H., Müller L., and Buchner J. (2004) Hsp70 and Hsp90–a relay team for protein folding. Rev. Physiol. Biochem. Pharmacol. 151, 1–44 10.1007/s10254-003-0021-1 [DOI] [PubMed] [Google Scholar]
- 6. Sousa R., and Lafer E. M. (2015) The role of molecular chaperones in clathrin mediated vesicular trafficking. Front. Mol. Biosci. 2, 26 10.3389/fmolb.2015.00026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mayer M. P. (2005) Recruitment of Hsp70 chaperones: a crucial part of viral survival strategies. Rev. Physiol. Biochem. Pharmacol. 153, 1–46 10.1007/s10254-004-0025-5 [DOI] [PubMed] [Google Scholar]
- 8. Mashaghi A., Bezrukavnikov S., Minde D. P., Wentink A. S., Kityk R., Zachmann-Brand B., Mayer M. P., Kramer G., Bukau B., and Tans S. J. (2016) Alternative modes of client binding enable functional plasticity of Hsp70. Nature 539, 448–451 10.1038/nature20137 [DOI] [PubMed] [Google Scholar]
- 9. Flaherty K. M., DeLuca-Flaherty C., and McKay D. B. (1990) Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature 346, 623–628 10.1038/346623a0 [DOI] [PubMed] [Google Scholar]
- 10. Zhu X., Zhao X., Burkholder W. F., Gragerov A., Ogata C. M., Gottesman M. E., and Hendrickson W. A. (1996) Structural analysis of substrate binding by the molecular chaperone DnaK. Science 272, 1606–1614 10.1126/science.272.5268.1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bertelsen E. B., Zhou H., Lowry D. F., Flynn G. C., and Dahlquist F. W. (1999) Topology and dynamics of the 10 kDa C-terminal domain of DnaK in solution. Protein Sci. 8, 343–354 10.1110/ps.8.2.343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Swain J. F., Dinler G., Sivendran R., Montgomery D. L., Stotz M., and Gierasch L. M. (2007) Hsp70 chaperone ligands control domain association via an allosteric mechanism mediated by the interdomain linker. Mol. Cell 26, 27–39 10.1016/j.molcel.2007.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bertelsen E. B., Chang L., Gestwicki J. E., and Zuiderweg E. R. (2009) Solution conformation of wild-type E. coli Hsp70 (DnaK) chaperone complexed with ADP and substrate. Proc. Natl. Acad. Sci. U.S.A. 106, 8471–8476 10.1073/pnas.0903503106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schmid D., Baici A., Gehring H., and Christen P. (1994) Kinetics of molecular chaperone action. Science 263, 971–973 10.1126/science.8310296 [DOI] [PubMed] [Google Scholar]
- 15. Mayer M. P., Schröder H., Rüdiger S., Paal K., Laufen T., and Bukau B. (2000) Multistep mechanism of substrate binding determines chaperone activity of Hsp70. Nat. Struct. Biol. 7, 586–593 10.1038/76819 [DOI] [PubMed] [Google Scholar]
- 16. Kityk R., Kopp J., Sinning I., and Mayer M. P. (2012) Structure and dynamics of the ATP-bound open conformation of Hsp70 chaperones. Mol. Cell 48, 863–874 10.1016/j.molcel.2012.09.023 [DOI] [PubMed] [Google Scholar]
- 17. Qi R., Sarbeng E. B., Liu Q., Le K. Q., Xu X., Xu H., Yang J., Wong J. L., Vorvis C., Hendrickson W. A., Zhou L., and Liu Q. (2013) Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP. Nat. Struct. Mol. Biol. 20, 900–907 10.1038/nsmb.2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhuravleva A., Clerico E. M., and Gierasch L. M. (2012) An interdomain energetic tug-of-war creates the allosterically active state in Hsp70 molecular chaperones. Cell 151, 1296–1307 10.1016/j.cell.2012.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Smock R. G., Rivoire O., Russ W. P., Swain J. F., Leibler S., Ranganathan R., and Gierasch L. M. (2010) An interdomain sector mediating allostery in Hsp70 molecular chaperones. Mol. Syst. Biol. 6, 414 10.1038/msb.2010.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Malinverni D., Marsili S., Barducci A., and De Los Rios P. (2015) Large-scale conformational transitions and dimerization are encoded in the amino-acid sequences of Hsp70 chaperones. PLoS Comput. Biol. 11, e1004262 10.1371/journal.pcbi.1004262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kityk R., Vogel M., Schlecht R., Bukau B., and Mayer M. P. (2015) Pathways of allosteric regulation in Hsp70 chaperones. Nat. Commun. 6, 8308 10.1038/ncomms9308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Karzai A. W., and McMacken R. (1996) A bipartite signaling mechanism involved in DnaJ-mediated activation of the Escherichia coli DnaK protein. J. Biol. Chem. 271, 11236–11246 10.1074/jbc.271.19.11236 [DOI] [PubMed] [Google Scholar]
- 23. Laufen T., Mayer M. P., Beisel C., Klostermeier D., Mogk A., Reinstein J., and Bukau B. (1999) Mechanism of regulation of hsp70 chaperones by DnaJ cochaperones. Proc. Natl. Acad. Sci. U.S.A. 96, 5452–5457 10.1073/pnas.96.10.5452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. De Los Rios P., and Barducci A. (2014) Hsp70 chaperones are non-equilibrium machines that achieve ultra-affinity by energy consumption. Elife 3, e02218 10.7554/eLife.02218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Clerico E. M., Tilitsky J. M., Meng W., and Gierasch L. M. (2015) How hsp70 molecular machines interact with their substrates to mediate diverse physiological functions. J. Mol. Biol. 427, 1575–1588 10.1016/j.jmb.2015.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rüdiger S., Germeroth L., Schneider-Mergener J., and Bukau B. (1997) Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 16, 1501–1507 10.1093/emboj/16.7.1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Van Durme J., Maurer-Stroh S., Gallardo R., Wilkinson H., Rousseau F., and Schymkowitz J. (2009) Accurate prediction of DnaK-peptide binding via homology modelling and experimental data. PLoS Comput. Biol. 5, e1000475 10.1371/journal.pcbi.1000475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Blond-Elguindi S., Cwirla S. E., Dower W. J., Lipshutz R. J., Sprang S. R., Sambrook J. F., and Gething M. J. (1993) Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell 75, 717–728 10.1016/0092-8674(93)90492-9 [DOI] [PubMed] [Google Scholar]
- 29. Schneider M., Rosam M., Glaser M., Patronov A., Shah H., Back K. C., Daake M. A., Buchner J., and Antes I. (2016) BiPPred: Combined sequence- and structure-based prediction of peptide binding to the Hsp70 chaperone BiP. Proteins 84, 1390–1407 10.1002/prot.25084 [DOI] [PubMed] [Google Scholar]
- 30. Zahn M., Berthold N., Kieslich B., Knappe D., Hoffmann R., and Sträter N. (2013) Structural studies on the forward and reverse binding modes of peptides to the chaperone DnaK. J. Mol. Biol. 425, 2463–2479 10.1016/j.jmb.2013.03.041 [DOI] [PubMed] [Google Scholar]
- 31. Zhang P., Leu J. I., Murphy M. E., George D. L., and Marmorstein R. (2014) Crystal structure of the stress-inducible human heat shock protein 70 substrate-binding domain in complex with peptide substrate. PLoS ONE. 9, e103518 10.1371/journal.pone.0103518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pellecchia M., Montgomery D. L., Stevens S. Y., Vander Kooi C. W., Feng H. P., Gierasch L. M., and Zuiderweg E. R. (2000) Structural insights into substrate binding by the molecular chaperone DnaK. Nat. Struct. Biol. 7, 298–303 10.1038/74062 [DOI] [PubMed] [Google Scholar]
- 33. Stevens S. Y., Cai S., Pellecchia M., and Zuiderweg E. R. (2003) The solution structure of the bacterial HSP70 chaperone protein domain DnaK(393–507) in complex with the peptide NRLLLTG. Protein Sci. 12, 2588–2596 10.1110/ps.03269103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mapa K., Sikor M., Kudryavtsev V., Waegemann K., Kalinin S., Seidel C. A., Neupert W., Lamb D. C., and Mokranjac D. (2010) The conformational dynamics of the mitochondrial Hsp70 chaperone. Mol. Cell 38, 89–100 10.1016/j.molcel.2010.03.010 [DOI] [PubMed] [Google Scholar]
- 35. Lai A. L., Clerico E. M., Blackburn M. E., Patel N. A., Robinson C. V., Borbat P. P., Freed J. H., and Gierasch L. M. (2017) Key features of an Hsp70 chaperone allosteric landscape revealed by ion-mobility native mass spectrometry and double electron-electron resonance. J. Biol. Chem. 292, 8773–8785 10.1074/jbc.M116.770404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Banerjee R., Jayaraj G. G., Peter J. J., Kumar V., and Mapa K. (2016) Monitoring conformational heterogeneity of the lid of DnaK substrate-binding domain during its chaperone cycle. FEBS J. 283, 2853–2868 10.1111/febs.13769 [DOI] [PubMed] [Google Scholar]
- 37. Marcinowski M., Rosam M., Seitz C., Elferich J., Behnke J., Bello C., Feige M. J., Becker C. F., Antes I., and Buchner J. (2013) Conformational selection in substrate recognition by Hsp70 chaperones. J. Mol. Biol. 425, 466–474 10.1016/j.jmb.2012.11.030 [DOI] [PubMed] [Google Scholar]
- 38. Deuerling E., Schulze-Specking A., Tomoyasu T., Mogk A., and Bukau B. (1999) Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature 400, 693–696 10.1038/23301 [DOI] [PubMed] [Google Scholar]
- 39. Deuerling E., Patzelt H., Vorderwülbecke S., Rauch T., Kramer G., Schaffitzel E., Mogk A., Schulze-Specking A., Langen H., and Bukau B. (2003) Trigger factor and DnaK possess overlapping substrate pools and binding specificities. Mol. Microbiol. 47, 1317–1328 10.1046/j.1365-2958.2003.03370.x [DOI] [PubMed] [Google Scholar]
- 40. Calloni G., Chen T., Schermann S. M., Chang H.-C., Genevaux P., Agostini F., Tartaglia G. G., Hayer-Hartl M., and Hartl F. U. (2012) DnaK functions as a central hub in the E. coli chaperone network. Cell Rep. 1, 251–264 10.1016/j.celrep.2011.12.007 [DOI] [PubMed] [Google Scholar]
- 41. Niwa T., Kanamori T., Ueda T., and Taguchi H. (2012) Global analysis of chaperone effects using a reconstituted cell-free translation system. Proc. Natl. Acad. Sci. U.S.A. 109, 8937–8942 10.1073/pnas.1201380109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schlecht R., Erbse A. H., Bukau B., and Mayer M. P. (2011) Mechanics of Hsp70 chaperones enables differential interaction with client proteins. Nat. Struct. Mol. Biol. 18, 345–351 10.1038/nsmb.2006 [DOI] [PubMed] [Google Scholar]
- 43. Marcinowski M., Höller M., Feige M. J., Baerend D., Lamb D. C., and Buchner J. (2011) Substrate discrimination of the chaperone BiP by autonomous and cochaperone-regulated conformational transitions. Nat. Struct. Mol. Biol. 18, 150–158 10.1038/nsmb.1970 [DOI] [PubMed] [Google Scholar]
- 44. Sekhar A., Velyvis A., Zoltsman G., Rosenzweig R., Bouvignies G., and Kay L. E. (2018) Conserved conformational selection mechanism of Hsp70 chaperone-substrate interactions. Elife 7, e32764 10.7554/eLife.32764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Böcking T., Aguet F., Harrison S. C., and Kirchhausen T. (2011) Single-molecule analysis of a molecular disassemblase reveals the mechanism of Hsc70-driven clathrin uncoating. Nat. Struct. Mol. Biol. 18, 295–301 10.1038/nsmb.1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xing Y., Böcking T., Wolf M., Grigorieff N., Kirchhausen T., and Harrison S. C. (2010) Structure of clathrin coat with bound Hsc70 and auxilin: mechanism of Hsc70-facilitated disassembly. EMBO J. 29, 655–665 10.1038/emboj.2009.383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lee J. H., Zhang D., Hughes C., Okuno Y., Sekhar A., and Cavagnero S. (2015) Heterogeneous binding of the SH3 client protein to the DnaK molecular chaperone. Proc. Natl. Acad. Sci. U.S.A. 112, E4206–E4215 10.1073/pnas.1505173112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sekhar A., Rosenzweig R., Bouvignies G., and Kay L. E. (2015) Mapping the conformation of a client protein through the Hsp70 functional cycle. Proc. Natl. Acad. Sci. U.S.A. 112, 10395–10400 10.1073/pnas.1508504112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kellner R., Hofmann H., Barducci A., Wunderlich B., Nettels D., and Schuler B. (2014) Single-molecule spectroscopy reveals chaperone-mediated expansion of substrate protein. Proc. Natl. Acad. Sci. U.S.A. 111, 13355–13360 10.1073/pnas.1407086111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Goloubinoff P., Sassi A. S., Fauvet B., Barducci A., and De Los Rios P. (2018) Chaperones convert the energy from ATP into the nonequilibrium stabilization of native proteins. Nat. Chem. Biol. 14, 388–395 10.1038/s41589-018-0013-8 [DOI] [PubMed] [Google Scholar]
- 51. Sharma S. K., De los Rios P., Christen P., Lustig A., and Goloubinoff P. (2010) The kinetic parameters and energy cost of the Hsp70 chaperone as a polypeptide unfoldase. Nat. Chem. Biol. 6, 914–920 10.1038/nchembio.455 [DOI] [PubMed] [Google Scholar]
- 52. Luk K. C., Mills I. P., Trojanowski J. Q., and Lee V. M. (2008) Interactions between Hsp70 and the hydrophobic core of α-synuclein inhibit fibril assembly. Biochemistry 47, 12614–12625 10.1021/bi801475r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Aprile F. A., Arosio P., Fusco G., Chen S. W., Kumita J. R., Dhulesia A., Tortora P., Knowles T. P., Vendruscolo M., Dobson C. M., and Cremades N. (2017) Inhibition of α-synuclein fibril elongation by Hsp70 is governed by a kinetic binding competition between α-synuclein species. Biochemistry 56, 1177–1180 10.1021/acs.biochem.6b01178 [DOI] [PubMed] [Google Scholar]
- 54. Kundel F., De S., Flagmeier P., Horrocks M. H., Kjaergaard M., Shammas S. L., Jackson S. E., Dobson C. M., and Klenerman D. (2018) Hsp70 inhibits the nucleation and elongation of tau and sequesters tau aggregates with high affinity. ACS Chem. Biol. 13, 636–646 10.1021/acschembio.7b01039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rodriguez F., Arsène-Ploetze F., Rist W., Rüdiger S., Schneider-Mergener J., Mayer M. P., and Bukau B. (2008) Molecular basis for regulation of the heat shock transcription factor σ32 by the DnaK and DnaJ chaperones. Mol. Cell 32, 347–358 10.1016/j.molcel.2008.09.016 [DOI] [PubMed] [Google Scholar]
- 56. Guisbert E., Herman C., Lu C. Z., and Gross C. A. (2004) A chaperone network controls the heat shock response in E. coli. Genes Dev. 18, 2812–2821 10.1101/gad.1219204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Munro S., and Pelham H. R. (1986) An Hsp70-like protein in the ER: identity with the 78 kd glucose-regulated protein and immunoglobulin heavy chain binding protein. Cell 46, 291–300 10.1016/0092-8674(86)90746-4 [DOI] [PubMed] [Google Scholar]
- 58. Feige M. J., Hendershot L. M., and Buchner J. (2010) How antibodies fold. Trends Biochem. Sci. 35, 189–198 10.1016/j.tibs.2009.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rosam M., Krader D., Nickels C., Hochmair J., Back K. C., Agam G., Barth A., Zeymer C., Hendrix J., Schneider M., Antes I., Reinstein J., Lamb D. C., and Buchner J. (2018) Bap (Sil1) regulates the molecular chaperone BiP by coupling release of nucleotide and substrate. Nat. Struct. Mol. Biol. 25, 90–100 10.1038/s41594-017-0012-6 [DOI] [PubMed] [Google Scholar]
- 60. Schlossman D. M., Schmid S. L., Braell W. A., and Rothman J. E. (1984) An enzyme that removes clathrin coats: purification of an uncoating ATPase. J. Cell Biol. 99, 723–733 10.1083/jcb.99.2.723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Böcking T., Aguet F., Rapoport I., Banzhaf M., Yu A., Zeeh J. C., and Kirchhausen T. (2014) Key interactions for clathrin coat stability. Structure 22, 819–829 10.1016/j.str.2014.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sousa R., Liao H.-S., Cuéllar J., Jin S., Valpuesta J. M., Jin A. J., and Lafer E. M. (2016) Clathrin-coat disassembly illuminates the mechanisms of Hsp70 force generation. Nat. Struct. Mol. Biol. 23, 821–829 10.1038/nsmb.3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Langer T., Lu C., Echols H., Flanagan J., Hayer M. K., and Hartl F. U. (1992) Successive action of DnaK, DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature 356, 683–689 10.1038/356683a0 [DOI] [PubMed] [Google Scholar]
- 64. Frydman J., Nimmesgern E., Ohtsuka K., and Hartl F. U. (1994) Folding of nascent polypeptide chains in a high molecular mass assembly with molecular chaperones. Nature 370, 111–117 10.1038/370111a0 [DOI] [PubMed] [Google Scholar]
- 65. Schröder H., Langer T., Hartl F. U., and Bukau B. (1993) DnaK, DnaJ and GrpE form a cellular chaperone machinery capable of repairing heat-induced protein damage. EMBO J. 12, 4137–4144 10.1002/j.1460-2075.1993.tb06097.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Albanèse V., Yam A. Y., Baughman J., Parnot C., and Frydman J. (2006) Systems analyses reveal two chaperone networks with distinct functions in eukaryotic cells. Cell 124, 75–88 10.1016/j.cell.2005.11.039 [DOI] [PubMed] [Google Scholar]
- 67. Melville M. W., McClellan A. J., Meyer A. S., Darveau A., and Frydman J. (2003) The Hsp70 and TRiC/CCT chaperone systems cooperate in vivo to assemble the von Hippel-Lindau tumor suppressor complex. Mol. Cell. Biol. 23, 3141–3151 10.1128/MCB.23.9.3141-3151.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wilbanks S. M., and McKay D. B. (1995) How potassium affects the activity of the molecular chaperone Hsc70. II. Potassium binds specifically in the ATPase active site. J. Biol. Chem. 270, 2251–2257 10.1074/jbc.270.5.2251 [DOI] [PubMed] [Google Scholar]
- 69. Schumacher R. J., Hansen W. J., Freeman B. C., Alnemri E., Litwack G., and Toft D. O. (1996) Cooperative action of Hsp70, Hsp90, and DnaJ proteins in protein renaturation. Biochemistry 35, 14889–14898 10.1021/bi961825h [DOI] [PubMed] [Google Scholar]
- 70. Smith D. F., and Toft D. O. (2008) Minireview: the intersection of steroid receptors with molecular chaperones: observations and questions. Mol. Endocrinol. 22, 2229–2240 10.1210/me.2008-0089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kirschke E., Goswami D., Southworth D., Griffin P. R., and Agard D. A. (2014) Glucocorticoid receptor function regulated by coordinated action of the hsp90 and hsp70 chaperone cycles. Cell 157, 1685–1697 10.1016/j.cell.2014.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Morgner N., Schmidt C., Beilsten-Edmands V., Ebong I.-O., Patel N. A., Clerico E. M., Kirschke E., Daturpalli S., Jackson S. E., Agard D., and Robinson C. V. (2015) Hsp70 forms antiparallel dimers stabilized by post-translational modifications to position clients for transfer to Hsp90. Cell Rep. 11, 759–769 10.1016/j.celrep.2015.03.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Morán Luengo T., Kityk R., Mayer M. P., and Rüdiger S. G. D. (2018) Hsp90 breaks the deadlock of the Hsp70 chaperone system. Mol. Cell 70, 545–552.e9 10.1016/j.molcel.2018.03.028 [DOI] [PubMed] [Google Scholar]
- 74. Mogk A., Bukau B., and Kampinga H. H. (2018) Cellular handling of protein aggregates by disaggregation machines. Mol. Cell 69, 214–226 10.1016/j.molcel.2018.01.004 [DOI] [PubMed] [Google Scholar]
- 75. Ungelenk S., Moayed F., Ho C.-T., Grousl T., Scharf A., Mashaghi A., Tans S., Mayer M. P., Mogk A., and Bukau B. (2016) Small heat shock proteins sequester misfolding proteins in near-native conformation for cellular protection and efficient refolding. Nat. Commun. 7, 13673 10.1038/ncomms13673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Fernández-Fernández M. R., Gragera M., Ochoa-Ibarrola L., Quintana-Gallardo L., and Valpuesta J. M. (2017) Hsp70-a master regulator in protein degradation. FEBS Lett. 591, 2648–2660 10.1002/1873-3468.12751 [DOI] [PubMed] [Google Scholar]
- 77. Jores T., Lawatscheck J., Beke V., Franz-Wachtel M., Yunoki K., Fitzgerald J. C., Macek B., Endo T., Kalbacher H., Buchner J., and Rapaport D. (2018) Cytosolic Hsp70 and Hsp40 chaperones enable the biogenesis of mitochondrial β-barrel proteins. J. Cell Biol. 217, 3091–3108 10.1083/jcb.201712029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Gorenberg E. L., and Chandra S. S. (2017) The role of co-chaperones in synaptic proteostasis and neurodegenerative disease. Front. Neurosci. 11, 248 10.3389/fnins.2017.00248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Tsuboyama K., Tadakuma H., and Tomari Y. (2018) Conformational activation of argonaute by distinct yet coordinated actions of the Hsp70 and Hsp90 chaperone systems. Mol. Cell 70, 722–729.e4 10.1016/j.molcel.2018.04.010 [DOI] [PubMed] [Google Scholar]
- 80. Cho H., and Shan S.-O. (2018) Substrate relay in an Hsp70-cochaperone cascade safeguards tail-anchored membrane protein targeting. EMBO J. 37, e99264 10.15252/embj.201899264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Stankiewicz M., Nikolay R., Rybin V., and Mayer M. P. (2010) CHIP participates in protein triage decisions by preferentially ubiquitinating Hsp70-bound substrates. FEBS J. 277, 3353–3367 10.1111/j.1742-4658.2010.07737.x [DOI] [PubMed] [Google Scholar]
- 82. Kityk R., Kopp J., and Mayer M. P. (2018) Molecular mechanism of J-domain–triggered ATP hydrolysis by Hsp70 chaperones. Mol. Cell 69, 227–237.e4 10.1016/j.molcel.2017.12.003 [DOI] [PubMed] [Google Scholar]
- 83. De Los Rios P., Ben-Zvi A., Slutsky O., Azem A., and Goloubinoff P. (2006) Hsp70 chaperones accelerate protein translocation and the unfolding of stable protein aggregates by entropic pulling. Proc. Natl. Acad. Sci. U.S.A. 103, 6166–6171 10.1073/pnas.0510496103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Mayer M. P., Laufen T., Paal K., McCarty J. S., and Bukau B. (1999) Investigation of the interaction between DnaK and DnaJ by surface plasmon resonance spectroscopy. J. Mol. Biol. 289, 1131–1144 10.1006/jmbi.1999.2844 [DOI] [PubMed] [Google Scholar]
- 85. Kampinga H. H., and Craig E. A. (2010) The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11, 579–592 10.1038/nrm2941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Li J., Qian X., and Sha B. (2003) The crystal structure of the yeast Hsp40 Ydj1 complexed with its peptide substrate. Structure 11, 1475–1483 10.1016/j.str.2003.10.012 [DOI] [PubMed] [Google Scholar]
- 87. Rüdiger S., Schneider-Mergener J., and Bukau B. (2001) Its substrate specificity characterizes the DnaJ co-chaperone as a scanning factor for the DnaK chaperone. EMBO J. 20, 1042–1050 10.1093/emboj/20.5.1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Suzuki H., Noguchi S., Arakawa H., Tokida T., Hashimoto M., and Satow Y. (2010) Peptide-binding sites as revealed by the crystal structures of the human Hsp40 Hdj1 C-terminal domain in complex with the octapeptide from human Hsp70. Biochemistry 49, 8577–8584 10.1021/bi100876n [DOI] [PubMed] [Google Scholar]
- 89. Nillegoda N. B., Kirstein J., Szlachcic A., Berynskyy M., Stank A., Stengel F., Arnsburg K., Gao X., Scior A., Aebersold R., Guilbride D. L., Wade R. C., Morimoto R. I., Mayer M. P., and Bukau B. (2015) Crucial HSP70 co-chaperone complex unlocks metazoan protein disaggregation. Nature 524, 247–251 10.1038/nature14884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Nillegoda N. B., Stank A., Malinverni D., Alberts N., Szlachcic A., Barducci A., De Los Rios P., Wade R. C., and Bukau B. (2017) Evolution of an intricate J-protein network driving protein disaggregation in eukaryotes. Elife 6, e24560 10.7554/eLife.24560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Harrison C. J., Hayer-Hartl M., Di Liberto M., Hartl F., and Kuriyan J. (1997) Crystal structure of the nucleotide exchange factor GrpE bound to the ATPase domain of the molecular chaperone DnaK. Science 276, 431–435 10.1126/science.276.5311.431 [DOI] [PubMed] [Google Scholar]
- 92. Zhang Y., and Zuiderweg E. R. (2004) The 70-kDa heat shock protein chaperone nucleotide-binding domain in solution unveiled as a molecular machine that can reorient its functional subdomains. Proc. Natl. Acad. Sci. U.S.A. 101, 10272–10277 10.1073/pnas.0401313101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Gassler C. S., Wiederkehr T., Brehmer D., Bukau B., and Mayer M. P. (2001) Bag-1M accelerates nucleotide release for human Hsc70 and Hsp70 and can act concentration-dependent as positive and negative cofactor. J. Biol. Chem. 276, 32538–32544 10.1074/jbc.M105328200 [DOI] [PubMed] [Google Scholar]
- 94. Bracher A., and Verghese J. (2015) The nucleotide exchange factors of Hsp70 molecular chaperones. Front. Mol. Biosci. 2, 10 10.3389/fmolb.2015.00010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Brehmer D., Rüdiger S., Gässler C. S., Klostermeier D., Packschies L., Reinstein J., Mayer M. P., and Bukau B. (2001) Tuning of chaperone activity of Hsp70 proteins by modulation of nucleotide exchange. Nat. Struct. Biol. 8, 427–432 10.1038/87588 [DOI] [PubMed] [Google Scholar]
- 96. Wu C.-C., Naveen V., Chien C.-H., Chang Y.-W., and Hsiao C.-D. (2012) Crystal structure of DnaK complexed with nucleotide exchange factor GrpE in the DnaK chaperone system: insight into the intermolecular communication. J. Biol. Chem. 287, 21461–21470 10.1074/jbc.M112.344358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Xu Z., Page R. C., Gomes M. M., Kohli E., Nix J. C., Herr A. B., Patterson C., and Misra S. (2008) Structural basis of nucleotide exchange and client binding by the Hsp70 cochaperone Bag2. Nat. Struct. Mol. Biol. 15, 1309–1317 10.1038/nsmb.1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Packschies L., Theyssen H., Buchberger A., Bukau B., Goody R. S., and Reinstein J. (1997) GrpE accelerates nucleotide exchange of the molecular chaperone DnaK with an associative displacement mechanism. Biochemistry 36, 3417–3422 10.1021/bi962835l [DOI] [PubMed] [Google Scholar]
- 99. Raviol H., Bukau B., and Mayer M. P. (2006) Human and yeast Hsp110 chaperones exhibit functional differences. FEBS Lett. 580, 168–174 10.1016/j.febslet.2005.11.069 [DOI] [PubMed] [Google Scholar]
- 100. Raviol H., Sadlish H., Rodriguez F., Mayer M. P., and Bukau B. (2006) Chaperone network in the yeast cytosol: Hsp110 is revealed as an Hsp70 nucleotide exchange factor. EMBO J. 25, 2510–2518 10.1038/sj.emboj.7601139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Dragovic Z., Broadley S. A., Shomura Y., Bracher A., and Hartl F. U. (2006) Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 25, 2519–2528 10.1038/sj.emboj.7601138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Andréasson C., Fiaux J., Rampelt H., Mayer M. P., and Bukau B. (2008) Hsp110 is a nucleotide-activated exchange factor for Hsp70. J. Biol. Chem. 283, 8877–8884 10.1074/jbc.M710063200 [DOI] [PubMed] [Google Scholar]
- 103. Grimshaw J. P., Jelesarov I., Schönfeld H. J., and Christen P. (2001) Reversible thermal transition in GrpE, the nucleotide exchange factor of the DnaK heat-shock system. J. Biol. Chem. 276, 6098–6104 10.1074/jbc.M009290200 [DOI] [PubMed] [Google Scholar]
- 104. Groemping Y., and Reinstein J. (2001) Folding properties of the nucleotide exchange factor GrpE from Thermus thermophilus: GrpE is a thermosensor that mediates heat shock response. J. Mol. Biol. 314, 167–178 10.1006/jmbi.2001.5116 [DOI] [PubMed] [Google Scholar]
- 105. Grimshaw J. P., Jelesarov I., Siegenthaler R. K., and Christen P. (2003) Thermosensor action of GrpE. The DnaK chaperone system at heat shock temperatures. J. Biol. Chem. 278, 19048–19053 10.1074/jbc.M300924200 [DOI] [PubMed] [Google Scholar]
- 106. Rampelt H., Kirstein-Miles J., Nillegoda N. B., Chi K., Scholz S. R., Morimoto R. I., and Bukau B. (2012) Metazoan Hsp70 machines use Hsp110 to power protein disaggregation. EMBO J. 31, 4221–4235 10.1038/emboj.2012.264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Gao X., Carroni M., Nussbaum-Krammer C., Mogk A., Nillegoda N. B., Szlachcic A., Guilbride D. L., Saibil H. R., Mayer M. P., and Bukau B. (2015) Human Hsp70 disaggregase reverses Parkinson's-linked α-synuclein amyloid fibrils. Mol. Cell 59, 781–793 10.1016/j.molcel.2015.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Rosenzweig R., Moradi S., Zarrine-Afsar A., Glover J. R., and Kay L. E. (2013) Unraveling the mechanism of protein disaggregation through a ClpB-DnaK interaction. Science 339, 1080–1083 10.1126/science.1233066 [DOI] [PubMed] [Google Scholar]
- 109. Gowda N. K., Kaimal J. M., Masser A. E., Kang W., Friedländer M. R., and Andréasson C. (2016) Cytosolic splice isoform of Hsp70 nucleotide exchange factor Fes1 is required for the degradation of misfolded proteins in yeast. Mol. Biol. Cell 27, 1210–1219 10.1091/mbc.E15-10-0697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Ishihara K., Yasuda K., and Hatayama T. (1999) Molecular cloning, expression and localization of human 105 kDa heat shock protein, hsp105. Biochim. Biophys. Acta 1444, 138–142 10.1016/S0167-4781(98)00254-1 [DOI] [PubMed] [Google Scholar]
- 111. Gauley J., Young J. T., and Heikkila J. J. (2008) Intracellular localization of the heat shock protein, HSP110, in Xenopus laevis A6 kidney epithelial cells. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 151, 133–138 10.1016/j.cbpa.2008.06.010 [DOI] [PubMed] [Google Scholar]
- 112. Kimura A., Ogata K., Altan B., Yokobori T., Ide M., Mochiki E., Toyomasu Y., Kogure N., Yanoma T., Suzuki M., Bai T., Oyama T., and Kuwano H. (2016) Nuclear heat shock protein 110 expression is associated with poor prognosis and chemotherapy resistance in gastric cancer. Oncotarget 7, 18415–18423 10.18632/oncotarget.7821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Lüders J., Demand J., and Höhfeld J. (2000) The ubiquitin-related BAG-1 provides a link between the molecular chaperones Hsc70/Hsp70 and the proteasome. J. Biol. Chem. 275, 4613–4617 10.1074/jbc.275.7.4613 [DOI] [PubMed] [Google Scholar]
- 114. Jiang Y., Woronicz J. D., Liu W., and Goeddel D. V. (1999) Prevention of constitutive TNF receptor 1 signaling by silencer of death domains. Science 283, 543–546 10.1126/science.283.5401.543 [DOI] [PubMed] [Google Scholar]
- 115. Gowda N. K. C., Kaimal J. M., Kityk R., Daniel C., Liebau J., Öhman M., Mayer M. P., and Andréasson C. (2018) Nucleotide exchange factors Fes1 and HspBP1 mimic substrate to release misfolded proteins from Hsp70. Nat. Struct. Mol. Biol. 25, 83–89 10.1038/s41594-017-0008-2 [DOI] [PubMed] [Google Scholar]
- 116. Brehmer D., Gässler C., Rist W., Mayer M. P., and Bukau B. (2004) Influence of GrpE on DnaK–substrate interactions. J. Biol. Chem. 279, 27957–27964 10.1074/jbc.M403558200 [DOI] [PubMed] [Google Scholar]
- 117. Polier S., Hartl F. U., and Bracher A. (2010) Interaction of the Hsp110 molecular chaperones from S. cerevisiae with substrate protein. J. Mol. Biol. 401, 696–707 10.1016/j.jmb.2010.07.004 [DOI] [PubMed] [Google Scholar]
- 118. Garcia V. M., Nillegoda N. B., Bukau B., and Morano K. A. (2017) Substrate binding by the yeast Hsp110 nucleotide exchange factor and molecular chaperone, Sse1, is not obligate for its biological activities. Mol. Biol. Cell 28, 2066–2075 10.1091/mbc.e17-01-0070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Sarbeng E. B., Liu Q., Tian X., Yang J., Li H., Wong J. L., Zhou L., and Liu Q. (2015) A functional DnaK dimer is essential for the efficient interaction with heat shock protein 40 kDa (Hsp40). J. Biol. Chem. 290, 8849–8862 10.1074/jbc.M114.596288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Kirstein J., Arnsburg K., Scior A., Szlachcic A., Guilbride D. L., Morimoto R. I., Bukau B., and Nillegoda N. B. (2017) In vivo properties of the disaggregase function of J-proteins and Hsc70 in Caenorhabditis elegans stress and aging. Aging Cell 16, 1414–1424 10.1111/acel.12686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Ayuso-Tejedor S., Angarica V. E., Bueno M., Campos L. A., Abián O., Bernadó P., Sancho J., and Jiménez M. A. (2010) Design and structure of an equilibrium protein folding intermediate: a hint into dynamical regions of proteins. J. Mol. Biol. 400, 922–934 10.1016/j.jmb.2010.05.050 [DOI] [PubMed] [Google Scholar]
- 122. Pellecchia M., Szyperski T., Wall D., Georgopoulos C., and Wüthrich K. (1996) NMR structure of the J-domain and the Gly/Phe-rich region of the Escherichia coli DnaJ chaperone. J. Mol. Biol. 260, 236–250 10.1006/jmbi.1996.0395 [DOI] [PubMed] [Google Scholar]
- 123. Barends T. R., Brosi R. W., Steinmetz A., Scherer A., Hartmann E., Eschenbach J., Lorenz T., Seidel R., Shoeman R. L., Zimmermann S., Bittl R., Schlichting I., and Reinstein J. (2013) Combining crystallography and EPR: crystal and solution structures of the multidomain cochaperone DnaJ. Acta Crystallogr. D Biol. Crystallogr. 69, 1540–1552 10.1107/S0907444913010640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Sondermann H., Scheufler C., Schneider C., Hohfeld J., Hartl F. U., and Moarefi I. (2001) Structure of a Bag/Hsc70 complex: convergent functional evolution of Hsp70 nucleotide exchange factors. Science 291, 1553–1557 10.1126/science.1057268 [DOI] [PubMed] [Google Scholar]
- 125. Polier S., Dragovic Z., Hartl F. U., and Bracher A. (2008) Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding. Cell 133, 1068–1079 10.1016/j.cell.2008.05.022 [DOI] [PubMed] [Google Scholar]
- 126. Yan M., Li J., and Sha B. (2011) Structural analysis of the Sil–Bip complex reveals the mechanism for Sil1 to function as a nucleotide-exchange factor. Biochem. J. 438, 447–455 10.1042/BJ20110500 [DOI] [PubMed] [Google Scholar]