Abstract
The variant histone H3.3 is incorporated into the genome in a transcription-dependent manner. This histone is thus thought to play a role in epigenetic regulation. However, our understanding of how H3.3 controls gene expression and epigenome landscape has remained incomplete. This is partly because precise localization of H3.3 in the genome has been difficult to decipher particularly for cells in vivo. To circumvent this difficulty, we generated knockin mice, by homologous recombination, to replace both of the two H3.3 loci (H3f3a and H3f3b) with the hemagglutinin-tagged H3.3 cDNA cassette, which also contained a GFP gene. We show here that the hemagglutinin-tagged H3.3 and GFP are expressed in the majority of cells in all adult tissues tested. ChIP-seq data, combined with RNA-seq, revealed a striking correlation between the level of transcripts and that of H3.3 accumulation in expressed genes. Finally, we demonstrate that H3.3 deposition is markedly enhanced upon stimulation by interferon on interferon-stimulated genes, highlighting transcription-coupled H3.3 dynamics. Together, these H3.3 knockin mice serve as a useful experimental model to study epigenome regulation in development and in various adult cells in vivo.
Keywords: interferon, transcription, epigenetics, chromatin, ChIP-sequencing (ChIP-seq), H3.3, Histone variant
Introduction
H3.3 is a conserved variant histone distinct from the canonical histone H3 (H3.1 and H3.2). Although it differs only in four or five amino acids from the canonical H3, H3.3 has an activity and mode of action different from H3.1 and H3.2 (1–3). Unlike the canonical histone H3, which is synthesized and deposited into nucleosomes during DNA replication, H3.3 is expressed throughout cell cycle and is incorporated into chromatin along with transcription. Its deposition is in some cases induced by transcriptional activation triggered by external signals, such as interferons, and linked to transcription elongation (4–7). Once deposited, H3.3 remains for a long time, although with varying turnover kinetics (8). For these reasons, H3.3 is implicated to play a role in epigenetic regulation (9). Although its deposition has been generally associated with expressed genes, H3.3 is deposited in bivalent genes in ES cells, as well as heterochromatin and telomeres (10–12). The H3.3 specific histone chaperon, HIRA mediates H3.3 deposition in expressed genes, bivalent genes, and damaged DNA sites (10, 11, 13, 14). ATRX and DAXX, on the other hand, direct H3.3 incorporation into heterochromatin and telomeres (11, 15–18).
H3.3 is encoded by two independent loci in both humans and mice (H3f3a and H3f3b), which produce a protein with the identical amino acid sequences. Targeted knockout of H3f3b shows the requirement of this gene in early embryogenesis and sperm development (19, 20). Conditional knockout of both H3f3a and Hf3b also results in defective embryonic development and abnormal cell growth (21). Furthermore, mutations in the H3F3A gene and genes in the ATRX pathway have been linked to pediatric brain cancer (22–25). Recently a chromatin reader, ZMYND11, that recognizes H3.3 methylated at lysine 36 has been identified (26, 27). These lines of studies demonstrate the fundamental role of H3.3 in the life of mammals.
Despite the well-acknowledged importance, our understanding of H3.3 has been hampered considerably, because of the difficulties in distinguishing H3.3 from the canonical histone H3. Thus, regulated expression of endogenous H3.3 proteins has remained elusive, including regulated distribution of H3.3 in the genome. Further, it is largely unknown how H3.3 responds to physiological changes in the cells. Exogenous introduction of a tagged H3.3, although useful in some cases, does not substitute for the endogenous H3.3 genes. It is of note that Wen et al. (30) reported construction of H3f3b-HA4 mice using a zinc finger genome-editing method. However, there is no mouse model available for the H3.3f3a gene. Because of the lack of a suitable in vivo models, our knowledge on the status and the role of H3.3 in terminally differentiated cells and small populations of specialized cells is very scarce in mammals.
To study the activity of endogenous H3.3 in vivo, we generated, by homologous recombination, knockin mice that express HA-tagged H3.3 from the H3f3a and H3f3b loci. We report here that H3f3a-HA and H3f3b-HA proteins are expressed broadly in all adult tissues examined and mostly at similar levels, including various cell types in the immune system. ChIP-seq analysis revealed that H3.3 is enriched in expressed genes in a manner closely recapitulating levels of mRNA expression, its localization found from the promoter, the transcription start site (TSS) to the gene body, and the transcription end site (TES). Finally, we demonstrate that interferon stimulation causes rapid H3.3 incorporation within interferon-stimulated genes, highlighting a dynamic nature of H3.3 deposition. Together, these knockin mice would serve as a valuable model for studying the mode and mechanisms of H3.3 activity in cells in vivo, in heath and disease.
Results
Construction of knockin mice expressing HA-tagged histone H3.3
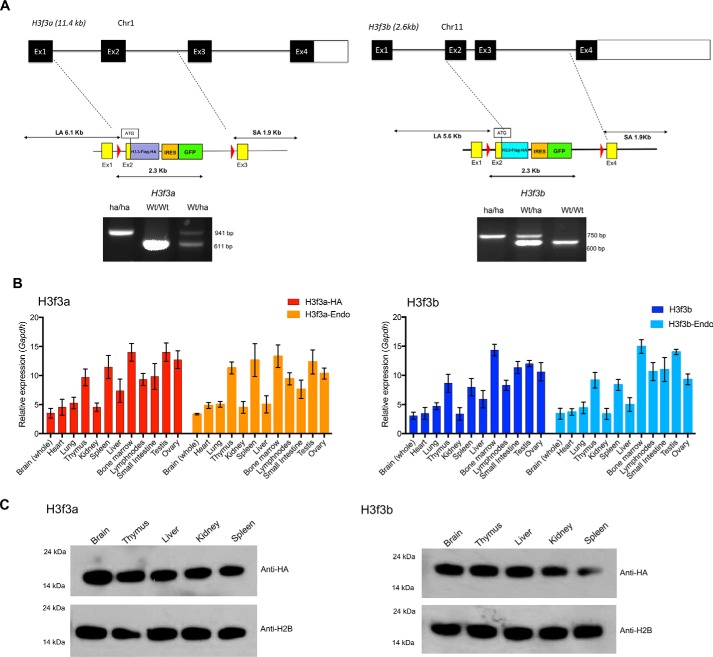
With the aim of developing new mouse strains expressing tagged H3.3 from the endogenous loci, we replaced part of the H3f3a and H3f3b genes with an HA-tagged H3.3 cDNA cassette by homologous recombination. Fig. 1A and Fig. S1A schematically depict the strategy used. The exon 2 containing the ATG through exon 3 of the H3f3a and H3f3b loci were replaced by the cassette containing H3.3 cDNA fused to the HA and FLAG tag (hereafter H3f3A-HA and H3f3b-HA). An IRES-EGFP was inserted at the 3′ end, enabling independent monitoring of free EGFP expression from each locus. In addition, loxP sites were placed flanking the cassettes for potential conditional deletion. At the 3′ end of EGFP cDNA, drug selection marker, neomycin- or hygromycin-resistant gene flanked by FRT was placed in the H3f3a and H3fb targeting vector, respectively. The drug makers were later removed by crossing with FLP mice. The inserted HA-FLAG tag sequence is shown in Fig. S1B. Mice with germ line transmission were genotyped by PCR (Fig. 1A, lower panel, and Fig. S1C). Thus, the recombination at either locus did not alter the regulatory regions, promoters, 5′- and 3′-UTRs and introns (except a sequence between exon 2 and exon 3). Heterozygous mice were born as expected and grew with a comparable weight increase with WT mice. In addition, both males and females were fertile. Because the replacement of a single allele was sufficient to detect HA-tagged H3.3 and GFP expression, all experiments in this work were carried out with heterozygous mice. The number of homozygous H3f3a-HA and H3f3b-HA pups appeared somewhat lower than expected. In addition, the number of double knockin mice carrying both H3f3a-HA and H3f3b-HA pups appeared lower. The precise ratios of homozygous mice and double knockin mice await results of additional, mating experiments.
Figure 1.
Knockin mice expressing HA-tagged H3.3. A, top panel, targeting strategy for replacing the endogenous H3f3a and H3f3b loci with the H3.3-HA cDNA cassettes. The exon–intron organization of the endogenous H3f3a and H3f3 loci. Below are the schematic structure of targeting vector and the 5′ and 3′ site arms. The targeting vectors and H3.3 cDNA fused to FLAG-HA, free EGFP, and drug selection markers. Bottom panels, identification homozygous knockin (ha/ha), heterozygous (ha/Wt), and WT (Wt/Wt) pups. Quantitative PCR–based genotyping was performed with tail DNA using primers in Fig. S1C. B, expression of endogenous H3.3 and knockin H3.3-HA mRNA in heterozygous knockin mouse. qRT-PCR detection of mRNA from the H3f3a(ha/wt) (left panel) and H3f3b(ha/wt) loci (right panel) in indicated adult tissues. H3f3a-HA and H3f3b-HA are transcripts from knockin loci, and H3f3a-Endo and H3f3b-Endo are transcripts from endogenous loci. Expression levels were normalized to Gapdh. The values represent the averages of three independent analysis using tissues from 8-week-old mice. C, immunoblot detection of H3.3-HA proteins, from knockin mice. 500 ng of histones prepared from 8-week-old heterozygous mice (H3f3a-HA on the left and H3f3b-HA on the right) were tested by immunoblot using anti-HA or anti-H2B antibody.
In Fig. 1B, expression of WT or HA-tagged H3.3 mRNA was compared by qRT-PCR. H3f3a-HA and H3f3b-HA mRNA were expressed at varying levels in all adult tissues examined, closely paralleling that of WT mRNA for both loci. The expression was generally higher in the reproductive organs and tissues of the immune system but lower in liver, heart, and kidney. Expression of HA-tagged H3.3 protein was detected in various adult tissues by immunoblot analysis using anti-HA antibody (Fig. 1C). H3.3A-HA and H3.3B-HA both migrated between 14- and 24-kDa molecular masses as expected and found in all tissues at similar levels.
H3.3A-HA and H3.3B-HA are expressed in adult tissues: immunohistochemical analysis
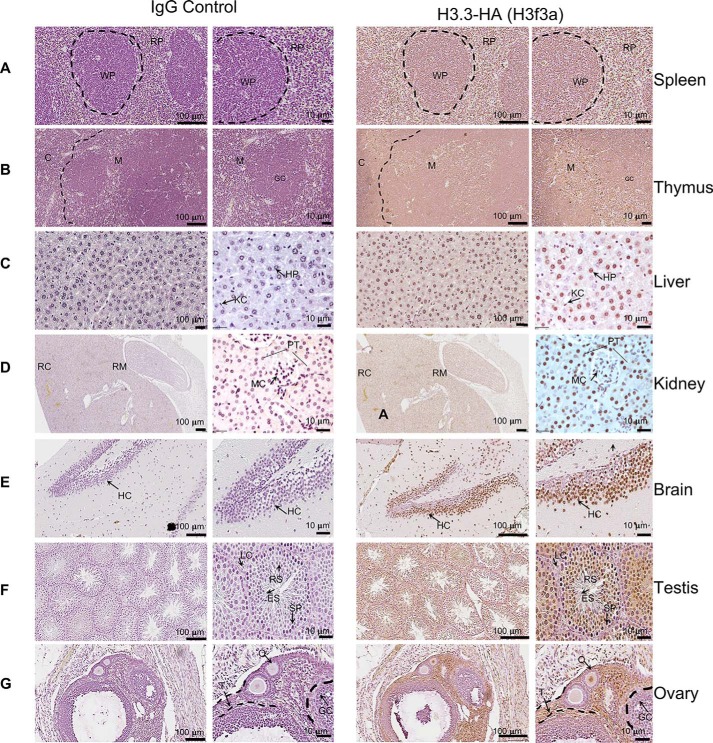
To examine detailed expression of HA-tagged H3.3, we performed immunohistochemistry (IHC) for adult tissues. spleen, thymus, liver, kidney, brain, testis, and ovary. Our purpose was to assess whether H3.3 expression varies among tissues and among cells within a given tissue. In Fig. 2 (A–D), 5-μm sections from indicated organs from H3f3a-HA mice were stained with anti-HA antibody (right panel). Matching sections were stained with control IgG, counterstained with hematoxylin to localize the nuclei (left panel). Images acquired at low and high magnifications are shown. In spleen, most lymphocytes, residing in the white pulp and red pulp displayed robust HA stain. Similarly, most cells in thymus, both in medulla and cortex were positive for HA signals. In liver, HA stain was found widely in hepatocytes and Kupffer cells. Likewise, in kidney most cells, including those in the glomerular tubules were stained with anti-HA antibody. In the brain, again, neurons clustered in the hippocampus area exhibited strong HA signals, showing exclusive localization in the nuclei. In testis, spermatocytes and round spermatids were stained along with non–germ line Leydig cells and granulosa. In ovary, HA stain was found mostly in the nuclei of theca cells and oocytes but more sparsely in granulosa cells. Essentially, the same results were obtained from IHC analyses from the H3f3b-HA mice (Fig. S2). Thus, H3.3-HA is expressed broadly in most, if not all types of cells, in all adult tissues tested.
Figure 2.
Immunohistochemical detection of H3. 3-HA in adult tissues. Indicated tissue sections from y8-week-old H3f3a-HA mice were stained with control IgG counter stained with hematoxylin to identify nuclei (left panels) or stained with anti-HA antibody and counterstained with diamino benzidine (right panels). The images are viewed at two different magnifications (indicted in each panel). Representative structures and cell types are marked by capital letters. WP, white pulp; RP, red pulp in spleen; C, cortical zone; M, medulla in thymus; HP, hepatocytes; KC, Kupffer cells in liver; RM, renal medulla; RC, renal cortex; RT, renal tubules in kidney; HC, hippocampus in brain; LC, Leydig cells; ES, elongating spermatid; RS, round spermatid; SP, spermatid in testis; T, theca; GC, germinal center; O, developing oocyte; PT, proximal tubules; MC, mesangial cells. See similar staining patterns in the H3f3b-HA mouse tissues in Fig. 2.
H3.3-driven GFP is expressed in most of immune cell types: flow cytometry analysis
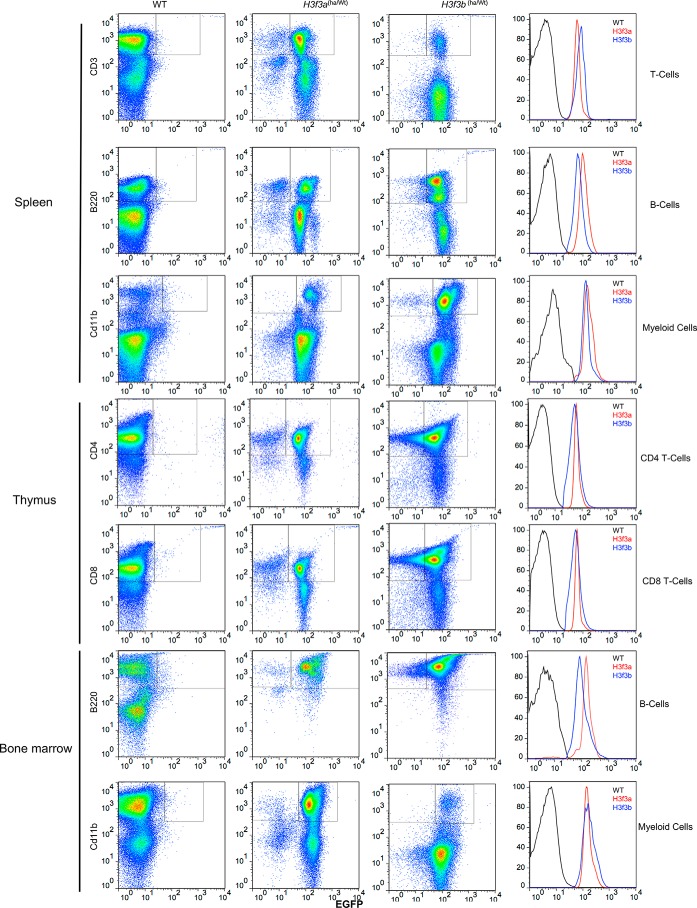
To assess H3.3 expression in different cell types in further detail, we opted for monitoring GFP signals by flow cytometry. H3.3 expression was tested for T lymphocytes (CD3+), B lymphocytes, and myeloid cells from thymus, spleen, and bone marrow. Fig. 3 shows flow cytometry profiles of GFP-positive cells with CD3, B220, and CD11b markers. The cells from both H3f3a-HA and H3f3b-HA mice were tested, with cells from WT mice serving as a negative control for GFP. A large majority of cells in spleen (T cells, B lymphocytes, and myeloid cells) were GFP-positive in both H3f3a-HA and H3f3b-HA mice. Similarly, most of the thymic T cells (CD4+ and CD8+) expressed GFP. Likewise, most of B cells and myeloid cells residing in bone marrow were positive for GFP. The histograms in Fig. 3 (right panels) showed that GFP from H3f3a-HA and H3f3b-HA mice are expressed at comparable levels in each cell type in these tissues. Furthermore, in all cases GFP signals exhibited a narrow monophasic peak, indicating that H3.3 expression levels are largely uniform within each cell type. These results were reproducibly observed with samples from additional four mice aged from 6 to 10 weeks. We also found that the majority of macrophages, dendritic cells, and NK cells in spleen expressed GFP signals (Fig. S3).
Figure 3.
Flow cytometry analysis of immune cells expressing EGFP from H3f3a-HA or H3f3b-HA mice. Cells from spleen, thymus, or bone marrow from 9-week-old H3f3a-HA or H3f3b-HA mice were stained with antibodies for B cells (B220), T cells (CD3 and CD4), or myeloid cells (Cd11b). Cells with these markers (y axis) were tested for EGFP signals (x axis). Histograms in the right panels depict EGFP mean fluorescent intensity.
Genic deposition of H3.3 recapitulates the levels of transcript expression: ChIP-seq and RNA-seq analyses
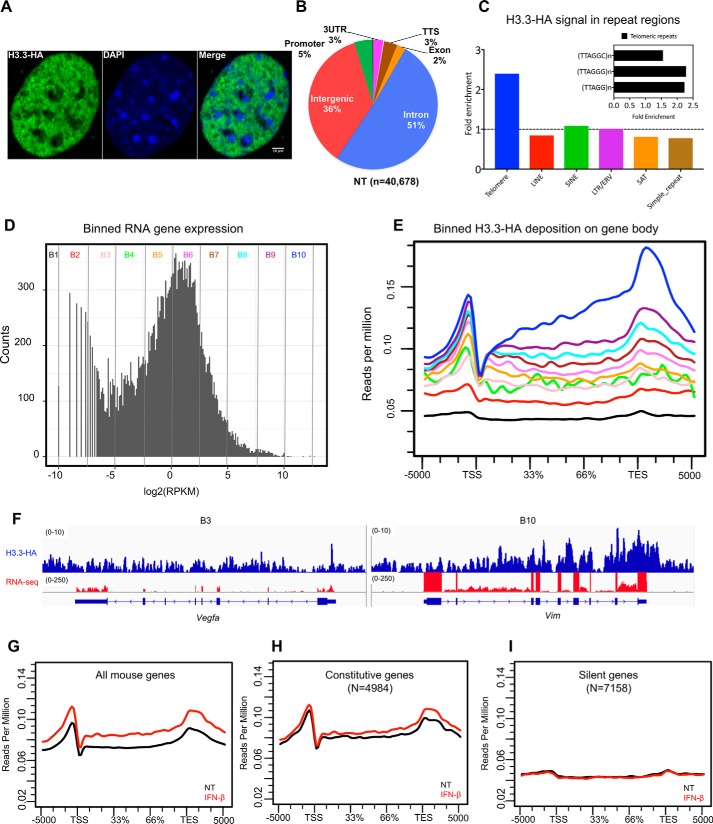
H3.3 deposition has traditionally been investigated by using exogenously introduced epitope-tagged H3.3 as a surrogate vector. The use of exogenous H3.3 vector has been accredited, because exogenous H3.3 was shown to be incorporated into nucleosomes and deposited in chromatin. Nevertheless, the use of exogenous vector has a potential caveat, in that regulated expression of endogenous H3.3 is not easily reproduced by an exogenous H3.3. The use of the knockin mice would obviate this caveat, presumably allowing more accurate assessment of H3.3 deposition. We thus examined genome-wide distribution of H3.3 in mouse embryonic fibroblasts (MEFs) established from H3f3b-HA embryos. Confocal image of H3f3b-HA MEFs showed that H3.3-HA immunoreactivity was found predominantly in the “euchromatic” regions of the nucleus (Fig. 4A). On the other hand HA signals were generally low in 4′,6′-diamino-2-phenylindole–dense “heterochromatic” regions, consistent with the results with exogenous H3.3 (5, 6). No HA stain was found in WT MEFs. ChIP-seq analysis was performed for growing MEFs using anti-HA antibody. We found a total of ∼40,700 H3.3-HA peaks, which were localized to three genomic regions, genic (blue), intergenic (red), and regulatory/promoter (green and other colors) (Fig. 4B). Nearly 60% of H3.3 signals were found in the genic regions (TSS, exons, and introns). Goldberg et al. (11) have shown that H3.3 is enriched in telomere repeats in mouse embryonic stem cells. We thus tested whether H3.3 is enriched in telomeres in MEFs as well. We found that H3.3 signals were enriched in the consensus telomere repeats TTAGGGn and closely related telomeric repeats TTAGGn and TTAGGCn. As shown in Fig. 4C, H3.3 signals were significantly enriched in telomere repeats. On the other hand, H3.3 signals on other repeat sequences, LINE, SINE, LTR, SAT, or simple repeats were near or below the background. These results closely resemble those of Goldberg et al.(11) and confirm H3.3 localization on telomeres. To correlate H3.3 deposition with transcription, we performed RNA-seq for the MEFs. In Fig. 4D, RNA-seq peak signals found on all annotated mouse genes were fractioned into 10 bins according to increasing transcripts levels (x axis), where the number of genes in each bin is shown on the y axis. Bin 1 represented 7,158 silent genes, and bin 10 denoted most highly expressed genes. Majority of expressed genes were in bin 5 to bin 7, peaking at bin 6. We next plotted average H3.3 deposition for genes in each bin (Fig. 4E). Strikingly, the amount of H3.3 deposition closely paralleled that of mRNA levels, i.e. the higher the RNA expression, the higher the H3.3 deposition and vice versa. It is interesting to note that for genes expressed at moderate levels (bins 3–6), H3.3 deposition peaked at/near the TSS, with fewer H3.3 signals over the gene body. This pattern of H3.3 accumulation has been reported as a general feature of H3.3 accumulation in many studies (11, 13, 28, 29). However, for genes expressed at higher levels (bins 7–10), H3.3 signals increased more significantly over the gene body, with the greatest rise near or at the TES and beyond. H3.3 signals were virtually absent in silent genes (bin 1). Fig. 4F provides IGV screenshots of H3.3 deposition and RNA peaks for a low (bin 3: Vegfa) and high expressed genes (bin 10: Vim). Heat maps in Fig. S4A show H3.3 distribution on individual genes in each bin. Fig. 4 (F–I) shows average H3.3 deposition patterns over all genes, constitutively expressed genes and silent genes, respectively. Detailed heat maps for constitutive and silent genes Fig. 4 (H–I) are shown in Fig. S4 (B–D).
Figure 4.
Genome-wide H3.3 distribution: ChIP-seq analysis with anti-HA antibody. A, confocal image of H3.3 distribution in the MEF nucleus. MEFs from H3f3b-HA mice were stained with HA antibody (green) and counterstained with 4′,6′-diamino-2-phenylindole (blue). The white vertical bar indicates 10 μm. B, pie chart depicting genomic distribution of H3.3 peaks (%). TTS, transcription termination site; NT, no treatment. C, H3.3-HA enrichment at various repetitive elements. The levels of H3.3 at the telomeric regions (TTAGGG)n, long interspersed elements (LINE), short interspersed elements (SINE), long terminal repeats (LTR), satellite regions (SAT), and simple repeats were calculated by dividing the reads of HA immunoprecipitated sample and the corresponding input sample. The enrichments were calculated using the RepEnrich2 software (36) (https://github.com/nerettilab/RepEnrich2). (Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.) The fold enrichment on the y axis represents the number of reads in the HA sample over the input ChIP. The dotted line shows H3.3 signals that are more than 1-fold and above. The inset shows three different subclasses of telomeric repeat sequences and their fold enrichment over input samples. ERV, endogenous retrovirus. D, RNA-seq analysis. All annotated mouse genes were divided into 10 bins according to the transcript levels (log2, RPKM), and each bin contained equal number of genes (∼3000 genes) with the exception of B1 and B2, which had ∼7,000 genes and plotted as histogram. The y axis (counts) represents how many times mRNA expression occurred within the specified range of RPKM in each bin (Table S1 (Binned genes)). E, ChIP-seq analysis. Average H3.3 accumulation was plotted for genes in 10 bins for the genic regions (TSS, gene body, TES ± 5 kb). Numbers on the y axis represent reads per million. F, IGV screenshots for ChIP-seq peaks (blue) and RNA peaks (red). G–I, average H3.3 distribution patterns for all genes (G), constitutively expressed genes (H), and silent genes (I) are shown.
Interferon (IFN) stimulation triggers rapid H3.3 accumulation in IFN-stimulated genes (ISGs)
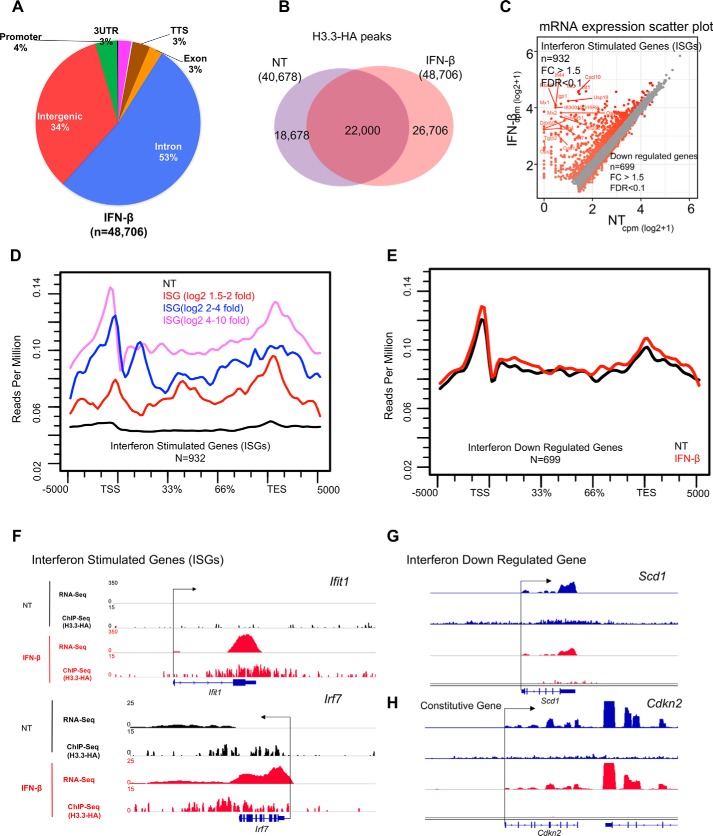
We previously showed that IFN stimulation causes marked accumulation of H3.3 in a number of genes transcriptionally activated by IFN. In these studies, we monitored the deposition of virally transduced H3.3-HA in fibroblasts (5, 6). To substantiate these observations for endogenous H3.3 and at a genome-wide level, ChIP-seq was performed for MEFs expressing H3f3b-HA stimulated by IFNβ for 6 h, in parallel with RNA-seq. We found ∼49,000 H3.3 peaks in IFN-treated cells, a significant increase over untreated cells, which had 40,700 peaks (Fig. 5A). This increase was presumably due to increased incorporation of the existing H3.3 into the chromatin rather than increased H3.3 expression (see H3f3a and H3f3b RNA peaks in Fig. S5A). Nevertheless, global pattern of H3.3 distribution in the genome was not substantially different between untreated and IFN-treated samples (pie chart in Fig. 5A). The Venn diagram in Fig. 5B shows that a large majority of H3.3 peaks were common in untreated and IFN-treated cells. RNA-seq found 932 genes up-regulated after IFN treatment (>1.5-fold change (FC) and FDR of 0.1), which were designated as ISGs (Fig. 5C and Table S1). Gene Ontology (GO) analysis showed enrichment in the type I IFN signaling, anti-viral, host defense, cytokine categories, as expected (Fig. S5B), whereas 699 genes were down-regulated by IFN treatment (Fig. 5C). Fig. 5D depicts H3.3 accumulation on ISGs, where H3.3 signals are plotted on three groups of ISGs, according to the degree of mRNA induction. The data showed that H3.3 deposition increased greatly in all ISGs. Interestingly, ISGs with the greatest induction (log2 FC > 4) showed the greatest increase in H3.3 deposition, those with intermediate induction (log2 FC 2–4) showed intermediate increase, and those with the least induction (log2 FC 1.5–2) showed the least increase in H3.3 accumulation. Conversely, H3.3 deposition was unchanged for genes down-regulated by IFN treatment both in amounts and patterns (Fig. 5E). The heat map in Fig. S6 depicts the rank order of H3.3-HA peak intensity on ISGs. Fig. 5 (E–H) provides snapshots of H3.3 deposition in typical ISGs, Ifit1 and Irf7; a down-regulated gene, Scd1; and a constitutively expressed Cdkn2 gene, supplemented with RNA peaks. Together, these data illustrate that H3.3 interacts with chromatin in a remarkably dynamic way and changes its localization in response to external signaling. Moreover, a direct correlation between the levels of ISG induction and that of H3.3 deposition is highly analogous with the correlation found for constitutively expressed genes in Fig. 4D, reinforcing the notion that H3.3 deposition is directly linked to transcriptional activity.
Figure 5.
IFN triggers H3. 3 accumulation on ISGs. A, pie chart depicting genomic distribution of H3.3-HA peaks in MEFs treated with IFN (6 h) shown as percentages (%). B, Venn diagram showing the overlap of peak coordinates of total H3.3 peaks in untreated (NT) and IFN-treated cells. C, scatter plot showing genes up-regulated (932 genes) or down-regulated (699 genes) after IFN treatment. Genes with >1.5-fold difference (FDR, 0.1) were identified as ISGs or genes down-regulated by IFN treatment. D, average H3.3 accumulation was plotted for ISGs according to activation levels (high, intermediate, or low). NT (black) represents average H3.3 accumulation in untreated for all ISGs or down-regulated IFN. E, average H3.3 accumulation was plotted for IFN down-regulated genes both during NT- (black) and IFN- (red) treated conditions. F–H, IGV screenshots of RNA expression (RPKM) and H3.3-HA sequence depth normalized tag density on ISGs (Ifit1, Irf7), down-regulated gene (Scd1), and constitutively expressed gene (Cdkn2) in untreated (black) and IFN-treated cells.
Discussion
We generated knockin mice in which the two H3.3 loci, H3f3a and H3f3b, were replaced with HA-tagged H3.3 by homologous recombination. Recently, Wen et al. (30) reported construction of H3f3b-HA mice using a zinc finger genome-editing method. We show here that our knockin mice allow sensitive detection of H3.3-HA protein expression in a variety of cells in vivo. These mice also proved useful for studying detailed, genome-wide distribution of H3.3. For these experiments, samples from heterozygous mice provided sufficient signals. Our knockin model is devoid of certain caveats inherent to experimental models that utilize exogenous H3.3, because introduced H3.3 does not readily recapitulate regulated expression of endogenous H3.3, and some cells are resistant to exogenous vector expression. IHC and flow cytometry analyses demonstrated that H3.f3a-HA and H3f3b-HA were expressed at comparable levels in most, if not all cells in various adult tissues, including brain and immune tissues. These tissues are rich in terminally differentiated, postmitotic cells, where our knowledge on the histone H3.3 is still scarce.
The use of anti-HA antibody to detect H3.3 proved particularly valuable for ChIP-seq experiments, because high-intensity H.3.3 peaks were observed with excellent resolution. This is noteworthy, given that commercially available anti-H3.3 antibodies are sometimes variable in quality. ChIP-seq analyses, combined with RNA-seq revealed a striking correlation between the amounts of H3.3 deposition and those of transcripts. Although the TSS-dominant H3.3 accumulation was found for the genes expressed moderately, genes expressed at higher levels showed greater H3.3 deposition over the gene body with even higher deposition toward the TES. This TES-biased pattern of deposition was also found in the majority of ISGs, where the levels of H3.3 deposition again correlated with the levels of transcriptional activation. These observations point to a complex mechanism of H3.3 accumulation that is presumably linked to transcription elongation and rate of RNA polymerase II passage through the gene body. In conclusion, these H3.3 knockin mice would serve as a useful model to study various aspects of H3.3 activity.
Experimental procedures
Construction of H3f3a and H3f3b knockin mice
All the animal studies were performed as per the approved protocols of NICHD Animal Care and Use Committee (Animal Study Proposal (ASP) 14-044). The cDNA cassette human H3.3 cDNA fused to HA and FLAG tag and IRES-driven EGFP cassette were cloned into the targeting vectors TAM1 and TAM2 for H3f3a and H3fb3, respectively, along with the neomycin resistance gene or hygromycin resistance gene (Fig. 1A). Mouse ES cells (129/Sv ESC) replacing H3f3a and H3fb loci with the targeting vector were generated by homologous recombination. The complete sequence information for TAM1 and TAM2 vectors is available in the Mouse Genome Database. Isolation of ES clones, injection into pseudo pregnant C57BL/6 mice, identification of chimeric mice, and construction of mice with germ line transmission, as well as removal of the drug resistance genes by FRT recombination were performed by GenOway. TAM1 and TAM2 heterozygous mice were propagated in this laboratory and genotyped by specific primers Fig. S1C. Heterozygous mice were bred with C57BL/6 mice for at least six generations. For genotyping, genomic DNA was extracted from adult mouse tails by using DirectPCR Lysis Reagent (Viagen) supplemented with 1 mg/ml of proteinase K (Ambion) by incubating at 56 °C overnight and heat inactivating the proteinase K at 85 °C for 45 min. The lysate was spun down, and 2 μl of supernatant was used for PCR genotyping. A homozygous H3f3a knockin mice carried a 941-bp amplicon, WT mice had a 611-bp amplicon, and heterozygous mice displayed both bands. A homozygous H3f3b knockin mice had a 750-bp amplicon, WT mice had a 600-bp piece, and heterozygous mice carried both bands (Fig. 1A). The H3f3a and H3f3b mouse are available under the following accession numbers from Mouse Genome Informatics: H3f3a<tm1.1Koza> (accession number 6195526) and H3f3b<tm1.1Koza> (accession number 6195527).
Mouse embryonic fibroblasts
Tissues from embryos of heterozygous H3f3a-HA and H3f3b-HA mice were collected after removal of heads, tails, limbs, and internal organs and minced, trypsinized for 20 min at 37 °C, and washed. Single cells were cultured in Dulbecco's modified Eagle's medium (Mediatech) supplemented with 10% fetal bovine serum (Atlanta Biologicals) and 100 units/ml penicillin/streptomycin and split at 1:2 ratios when freshly confluent. MEFs were passaged 15 consecutive times, passing a growth crisis, and attained morphologically homogeneous cells.
Immunohistochemistry
Tissues from indicated adult organs were fixed in 10% formaldehyde (pH 7.0) overnight and embedded in paraffin. Tissues were sectioned at a 5-μm thickness, deparaffinized on slides. The slides were permeabilized with 2% Triton X-100 for 30 min and incubated with anti-HA (Abcam, ab9010) antibody diluted in TBS buffer containing 1% BSA at room temperature for 120 min, followed by blocking with 10% normal goat serum. The slides were then washed with 0.025% Triton X-100 in TBS and incubated with horseradish peroxidase (HRP)–conjugated secondary antibodies for 30 min at room temperature. The slides were developed using diamino benzidine substrate followed by two counterstaining with hematoxylin. The slides were imaged on Zeiss Axioplan 2 microscope.
Flow cytometry analysis
Single-cell suspensions from spleens, thymus, and bone marrow from 7–8-week-old heterozygous H3f3a-ha or H3f3b-ha mice or WT mice in RPMI 1640. After removal of red blood cells by lysis buffer (BD Biosciences), cell viability was assessed by Aqua (Invitrogen, LIVE/DEADTM fixable aqua dead cell stain kit). Following treatment with 0.5 μg of Fc Block (BD Biosciences) for 10 min, the cells were incubated with the following antibodies, phycoerythrin (PE)-conjugated CD3 (antibody 100206), B220 (antibody 103258), F4/80 (antibody 123146), Cd11b (antibody 101208), Cd11c (antibody 117308), and NK1.1 (antibody 108708) (Biolegend) diluted in PBS with 5% FBS and 0.09% sodium azide in the dark for 30 min at 4 °C. Surface fluorescence signals were detected in BD FACSCalibur (BD Biosciences) equipped with 488- and 633-nm lasers, and data were acquired by BD Cell Quest Pro software (BD Biosciences) and analyzed by FlowJo software.
Histone preparation and immunoblot analysis
Tissues and organs were homogenized in Dounce homogenizer in homogenization buffer (50 mm Tris-HCl, pH 7.5, 25 mm KCl, 250 mm sucrose, 2 mm sodium butyrate, 1 mm sodium orthovanadate, 0.5 mm phenylmethylsulfonyl fluoride, 1× protease inhibitor mixture) and centrifuged. The pellets were incubated in Triton extraction buffer (PBS containing 0.5% Triton X-100 (v/v), 2 mm phenylmethylsulfonyl fluoride, 0.02% (w/v) NaN3), centrifuged, and resuspended in 0.2 n HCl at a density of 4 × 107 nuclei/ml, and extraction was continued overnight at 4 °C. Extracts were centrifuged at 6,500 × g for 10 min at 4 °C to remove debris and stored at −20 C. Protein contents in the supernatants were measured by the Bradford assay. 0.5 μg of extracted proteins was separated on NuPAGE gels (4–12%) (Invitrogen) and transferred to a polyvinylidene difluoride membrane Immobilon-P (Millipore). The membrane was incubated with antibodies for HA (Abcam ab9110) and histone H2B (Abcam ab1790) at 1:2000 dilution followed by HRP–conjugated donkey anti-rabbit IgG at 1:5000 dilutions (Amersham Biosciences). The signal was visualized by the SuperSignal West Pico kit (Pierce) according to the manufacturer's protocols.
qRT-PCR
Total RNA was extracted with TRIzol (Life Technologies), treated with DNase I (Promega), and reverse-transcribed with SuperScriptTM II reverse transcriptase (Invitrogen) and mRNA levels for indicated genes was detected as described using primers listed in Table S3 (6). mRNA levels were normalized by Gapdh. Quantitative differences in H3f3a and H3f3b in various tissues were calculated using the 2−ΔΔCT method relative to Gapdh (31).
RNA-seq
RNA-seq library was prepared with Illumina stranded mRNA-seq kit, following the manufacturer's instructions. Total RNA was extracted from MEFs using RNeasy columns (Qiagen). RNA samples (500 ng) were used for library construction. Pooled libraries were sequenced by Illumina HiSeq 2500 sequencer. The RNA-seq Fastq files were aligned to the UCSC mm9 genome using TopHat2 (32) and Bowtie2 (33). Read counts per gene were generated using HTSeq-count, and differential expression analysis was performed using edgeR (34). Normalized bigWig files for IGV genome browser tracks were generated using deepTools (35).
ChIP and ChIP-seq
Quantitative ChIP was performed for immunoprecipitated chromatin from MEFs as described (6). ChIP-seq libraries were prepared using 10–50 ng of immunoprecipitated DNA using a NEBNext® UltraTM DNA library prep kit for Illumina® (Cat E7370) following a standard protocol, normalized by concentration and sequenced in Illumina Next-Seq 550 following standard protocol. The quality of the reads was assessed by FastQC (v.0.9.5). The ChIP-seq reads were mapped onto the Mus musculus (mm9) genome using bowtie/2–2.2.5 and Samtools/1.2. To directly compare different ChIP-seq data, files were binned into 10-bp bins and then converted to sequence depth normalized to bigWig files using deepTools (v2.5.0). The peak calling was performed using both SICER/v1.1 (window size, 200; genome size, 0.787; gap size, 600; FDR, <0.01) and HOMER peak calling programs (Table S2). We performed visual inspection of the data using a local installation of the IGV genome browser view (Broad Institute). The peak annotation was performed using both CEAS-cistrome package and Homer software tools. The average metagene profiles of constitutive, silent, and induced genes was plotted using ngsplot/2.6.1 tool. We obtained repetitive element annotation version mm9- Jul 2007 - RepeatMasker open-3.2.8 - Repeat Library 20090604 from the repeatmasker site (http://www.repeatmasker.org/species/mm.html) site.5 We used RepEnrich2 pipeline available at https://github.com/nerettilab/RepEnrich2; M (36) and obtained count data for various repetitive elements including telomeres.5 Input and H3.3-HA ChIP fastq files were aligned to the mm9 mouse annotation to obtain bam files using the default bowtie2 settings (bowtie2 -q -threads = $(( SLURM_CPUS_PER_TASK - 4)) -x genome -U ../Sample.fastq.gz samtools view -Sb - Sample.bam). RepEnrich2_subset.py (python ../RepEnrich2_subset.py Sample.bam 30 Sample-Name -pairedend FALSE) was run on the resulting bam files to output uniquely mapping and multimapping reads and finally RepEnrich2.py (python/RepEnrich2.py mm9_repeatmasker.txt Sample_Output_Folder-Input/Sample-Name setup_folder../Sample.gz Sample_unique.bam -cpus 16 -pairedend FALSE) was run to obtain count files for various classes of repeat regions. The fold enrichments and log2 cpm were calculated using exactTest available on edgeR Bioconductor package after normalizing for the number of uniquely mapped reads.
Author contributions
M. B. data curation; M. B. software; M. B., T. T., C. C., and A. N. formal analysis; M. B. validation; M. B. investigation; M. B. visualization; M. B., T. T., C. C., A. N., V. N., N. S., S. B. G., A. G., and R. K. methodology; M. B. and K. O. writing-original draft; M. B. project administration; M. B. writing-review and editing; T. T. and K. O. conceptualization; T. T., M. L. D., and K. O. supervision; C. C. and N. S. resources.
Supplementary Material
Acknowledgments
We thank L. Banaszynski and C. D. Allis for discussion. We thank David Landsman and Iris Zhu of NCBI for initial help and guidance for bioinformatics analysis, as well as members of the Division of Developmental Biology (DDB) and the Ozato lab for advice. We gratefully acknowledge the contribution of the late Matthew Smith to this paper. This work utilized the computational resources of the National Institutes of Health High Performing Computation (HPC) Biowulf cluster.
The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1–S3 and Figs. S1–S6.
The raw sequence reads from RNA-seq experiments have been submitted in the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) and are accessible through Gene Expression Omnibus series accession number GSE118926.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- HA
- hemagglutinin
- TSS
- transcription start site
- TES
- transcription end site
- qRT-PCR
- quantitative RT-PCR
- IHC
- immunohistochemistry
- MEF
- mouse embryonic fibroblast
- IFN
- interferon
- ISG
- IFN-stimulated gene
- FC
- fold change
- FDR
- false discovery rate
- EGFP
- enhanced GFP.
References
- 1. Talbert P. B., and Henikoff S. (2017) Histone variants on the move: substrates for chromatin dynamics. Nat. Rev. Mol. Cell Biol. 18, 115–126 10.1038/nrm.2016.148,10.1038/nrn.2016.167 [DOI] [PubMed] [Google Scholar]
- 2. Allis C. D., and Jenuwein T. (2016) The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 17, 487–500 10.1038/nrg.2016.59 [DOI] [PubMed] [Google Scholar]
- 3. Sitbon D., Podsypanina K., Yadav T., and Almouzni G. (2017) Shaping chromatin in the nucleus: the bricks and the architects. Cold Spring harbor Symp. Quant. Biol. 82, 1–14 10.1101/sqb.2017.82.033753 [DOI] [PubMed] [Google Scholar]
- 4. Schneiderman J. I., Orsi G. A., Hughes K. T., Loppin B., and Ahmad K. (2012) Nucleosome-depleted chromatin gaps recruit assembly factors for the H3.3 histone variant. Proc. Natl. Acad. Sci. U.S.A. 109, 19721–19726 10.1073/pnas.1206629109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tamura T., Smith M., Kanno T., Dasenbrock H., Nishiyama A., and Ozato K. (2009) Inducible deposition of the histone variant H3.3 in interferon-stimulated genes. J. Biol. Chem. 284, 12217–12225 10.1074/jbc.M805651200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sarai N., Nimura K., Tamura T., Kanno T., Patel M. C., Heightman T. D., Ura K., and Ozato K. (2013) WHSC1 links transcription elongation to HIRA-mediated histone H3.3 deposition. EMBO J. 32, 2392–2406 10.1038/emboj.2013.176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Maze I., Wenderski W., Noh K. M., Bagot R. C., Tzavaras N., Purushothaman I., Elsässer S. J., Guo Y., Ionete C., Hurd Y. L., Tamminga C. A., Halene T., Farrelly L., Soshnev A. A., Wen D., et al. (2015) Critical role of histone turnover in neuronal transcription and plasticity. Neuron 87, 77–94 10.1016/j.neuron.2015.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kraushaar D. C., Jin W., Maunakea A., Abraham B., Ha M., and Zhao K. (2013) Genome-wide incorporation dynamics reveal distinct categories of turnover for the histone variant H3.3. Genome Biol. 14, R121 10.1186/gb-2013-14-10-r121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ng R. K., and Gurdon J. B. (2008) Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat. Cell Biol. 10, 102–109 10.1038/ncb1674 [DOI] [PubMed] [Google Scholar]
- 10. Banaszynski L. A., Wen D., Dewell S., Whitcomb S. J., Lin M., Diaz N., Elsässer S. J., Chapgier A., Goldberg A. D., Canaani E., Rafii S., Zheng D., and Allis C. D. (2013) Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell 155, 107–120 10.1016/j.cell.2013.08.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Goldberg A. D., Banaszynski L. A., Noh K. M., Lewis P. W., Elsaesser S. J., Stadler S., Dewell S., Law M., Guo X., Li X., Wen D., Chapgier A., DeKelver R. C., Miller J. C., Lee Y. L., et al. (2010) Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140, 678–691 10.1016/j.cell.2010.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Elsaesser S. J., and Allis C. D. (2010) HIRA and Daxx constitute two independent histone H3.3-containing predeposition complexes. Cold Spring Harbor Symp. Quant. Biol. 75, 27–34 10.1101/sqb.2010.75.008 [DOI] [PubMed] [Google Scholar]
- 13. Pchelintsev N. A., McBryan T., Rai T. S., van Tuyn J., Ray-Gallet D., Almouzni G., and Adams P. D. (2013) Placing the HIRA histone chaperone complex in the chromatin landscape. Cell Rep. 3, 1012–1019 10.1016/j.celrep.2013.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Adam S., Polo S. E., and Almouzni G. (2013) Transcription recovery after DNA damage requires chromatin priming by the H3.3 histone chaperone HIRA. Cell 155, 94–106 10.1016/j.cell.2013.08.029 [DOI] [PubMed] [Google Scholar]
- 15. Ivanauskiene K., Delbarre E., McGhie J. D., Küntziger T., Wong L. H., and Collas P. (2014) The PML-associated protein DEK regulates the balance of H3.3 loading on chromatin and is important for telomere integrity. Genome Res. 24, 1584–1594 10.1101/gr.173831.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Orsi G. A., Algazeery A., Meyer R. E., Capri M., Sapey-Triomphe L. M., Horard B., Gruffat H., Couble P., Aït-Ahmed O., and Loppin B. (2013) Drosophila Yemanuclein and HIRA cooperate for de novo assembly of H3.3-containing nucleosomes in the male pronucleus. PLoS Genet. 9, e1003285 10.1371/journal.pgen.1003285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Delbarre E., Jacobsen B. M., Reiner A. H., Sørensen A. L., Küntziger T., and Collas P. (2010) Chromatin environment of histone variant H3.3 revealed by quantitative imaging and genome-scale chromatin and DNA immunoprecipitation. Mol. Biol. Cell 21, 1872–1884 10.1091/mbc.e09-09-0839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Voon H. P., Hughes J. R., Rode C., De La Rosa-Velázquez I. A., Jenuwein T., Feil R., Higgs D. R., and Gibbons R. J. (2015) ATRX plays a key role in maintaining silencing at interstitial heterochromatic loci and imprinted genes. Cell Rep. 11, 405–418 10.1016/j.celrep.2015.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yuen B. T., Bush K. M., Barrilleaux B. L., Cotterman R., and Knoepfler P. S. (2014) Histone H3.3 regulates dynamic chromatin states during spermatogenesis. Development 141, 3483–3494 10.1242/dev.106450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bush K. M., Yuen B. T., Barrilleaux B. L., Riggs J. W., O'Geen H., Cotterman R. F., and Knoepfler P. S. (2013) Endogenous mammalian histone H3.3 exhibits chromatin-related functions during development. Epigenetics Chromatin 6, 7 10.1186/1756-8935-6-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jang C. W., Shibata Y., Starmer J., Yee D., and Magnuson T. (2015) Histone H3.3 maintains genome integrity during mammalian development. Genes Dev. 29, 1377–1392 10.1101/gad.264150.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schwartzentruber J., Korshunov A., Liu X. Y., Jones D. T., Pfaff E., Jacob K., Sturm D., Fontebasso A. M., Quang D. A., Tönjes M., Hovestadt V., Albrecht S., Kool M., Nantel A., Konermann C., et al. (2012) Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482, 226–231 10.1038/nature10833 [DOI] [PubMed] [Google Scholar]
- 23. Wu G., Broniscer A., McEachron T. A., Lu C., Paugh B. S., Becksfort J., Qu C., Ding L., Huether R., Parker M., Zhang J., Gajjar A., Dyer M. A., Mullighan C. G., Gilbertson R. J., et al. (2012) Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat. Genet. 44, 251–253 10.1038/ng.1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Aihara K., Mukasa A., Gotoh K., Saito K., Nagae G., Tsuji S., Tatsuno K., Yamamoto S., Takayanagi S., Narita Y., Shibui S., Aburatani H., and Saito N. (2014) H3F3A K27M mutations in thalamic gliomas from young adult patients. Neuro-oncology 16, 140–146 10.1093/neuonc/not144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gielen G. H., Gessi M., Hammes J., Kramm C. M., Waha A., and Pietsch T. (2013) H3F3A K27M mutation in pediatric CNS tumors: a marker for diffuse high-grade astrocytomas. Am. J. Clin. Pathol. 139, 345–349 10.1309/AJCPABOHBC33FVMO [DOI] [PubMed] [Google Scholar]
- 26. Wen H., Li Y., Xi Y., Jiang S., Stratton S., Peng D., Tanaka K., Ren Y., Xia Z., Wu J., Li B., Barton M. C., Li W., Li H., and Shi X. (2014) ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression. Nature 508, 263–268 10.1038/nature13045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guo R., Zheng L., Park J. W., Lv R., Chen H., Jiao F., Xu W., Mu S., Wen H., Qiu J., Wang Z., Yang P., Wu F., Hui J., Fu X., et al. (2014) BS69/ZMYND11 reads and connects histone H3.3 lysine 36 trimethylation-decorated chromatin to regulated pre-mRNA processing. Mol. Cell 56, 298–310 10.1016/j.molcel.2014.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mito Y., Henikoff J. G., and Henikoff S. (2005) Genome-scale profiling of histone H3.3 replacement patterns. Nat. Genet. 37, 1090–1097 10.1038/ng1637 [DOI] [PubMed] [Google Scholar]
- 29. Ray-Gallet D., Woolfe A., Vassias I., Pellentz C., Lacoste N., Puri A., Schultz D. C., Pchelintsev N. A., Adams P. D., Jansen L. E., and Almouzni G. (2011) Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Mol. Cell 44, 928–941 10.1016/j.molcel.2011.12.006 [DOI] [PubMed] [Google Scholar]
- 30. Wen D., Noh K. M., Goldberg A. D., Allis C. D., Rosenwaks Z., Rafii S., and Banaszynski L. A. (2014) Genome editing a mouse locus encoding a variant histone, H3.3B, to report on its expression in live animals. Genesis 52, 959–966 10.1002/dvg.22827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Livak K. J., and Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔC(T) method. Methods 25, 402–408 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 32. Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., and Salzberg S. L. (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Langmead B., and Salzberg S. L. (2012) Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Robinson M. D., McCarthy D. J., and Smyth G. K. (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ramírez F., Ryan D. P., Grüning B., Bhardwaj V., Kilpert F., Richter A. S., Heyne S., Dündar F., and Manke T. (2016) deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Criscione S. W., Zhang Y., Thompson W., Sedivy J. M., and Neretti N. (2014) Transcriptional landscape of repetitive elements in normal and cancer human cells. BMC Genomics 15, 583 10.1186/1471-2164-15-583 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.