Abstract
The human 6-O-endosulfatases HSulf-1 and -2 catalyze the region-selective hydrolysis of the 6-O-sulfate group of the glucosamine residues within sulfated domains of heparan sulfate, thereby ensuring a unique and original post-biosynthetic modification of the cell surface proteoglycans. While numerous studies point out the role of HSulf-2 in crucial physiological processes as well as in pathological conditions particularly in cancer, its structural organization in two chains and its functional properties remain poorly understood. In this study, we report the first characterization by mass spectrometry (MS) of HSulf-2. An average molecular weight of 133,115 Da was determined for the whole enzyme by MALDI-TOF MS, i.e. higher than the naked amino acid backbone (98,170 Da), highlighting a significant contribution of post-translational modifications. The HSulf-2 protein sequence was determined by Nano-LC-MS/MS, leading to 63% coverage and indicating at least four N-glycosylation sites at Asn 108, 147, 174 and 217. These results provide a platform for further structural investigations of the HSulf enzymes, aiming at deciphering the role of each chain in the substrate binding and specificities and in the catalytic activities.
Keywords: HSulf-2, 6-O-Endosulfatase, Sulfatase, Heparan sulfate, Formylglycine, Mass spectrometry
Highlights
-
•
First characterization of HSulf-2 by mass spectrometry.
-
•
The Hsulf-2 protein sequence was covered.
-
•
A molecular weight of 133,115 Da was determined for the whole enzyme protein.
-
•
The sequence analysis located the catalytic residue formylglycine at cysteine 64.
-
•
Post-translational modifications notably contribute to HSulf-2 molecular weight.
1. Introduction
The human heparan sulfate 6-O-endosulfatases HSulf-1 and HSulf-2 (HSulfs, EC 3.1.6.14) catalyze the regioselective hydrolysis of the 6-O-sulfate groups within heparan sulfate (HS) chains present on cell surface proteoglycans and extracellular matrix [1,2]. HSulfs exhibit two unique features among human sulfatases, since to date they are the only known sulfatases to be secreted in the extracellular medium, and to be active at neutral pH and at the polymer level [3]. They belong to HS 6-O-endosulfatase family, which was firstly discovered in early 2000's in quail embryo [4,5], and then in chick [6], Xenopus laevis [7,8], sea urchin [9], zebrafish [10], Drosophila [11] and mammals [3,[12], [13], [14]]. Sulf enzymes were initially found during quail embryo development, highlighting their roles in modulating the binding of the morphogen Wnt to HS and thereby the Wnt signaling [8,15]. Since then, Sulfs have been shown to be involved in key developmental and tumoral processes through an unprecedented post-biosynthetic editing mechanism of the 6-O-sulfate pattern of HS [[16], [17], [18], [19], [20]]. Being confined to the highly sulfated domain (NS) of HS, HSulf-catalyzed 6-O-desulfation process leads to a modest decrease in the polysaccharide 6-O-sulfation content, and thus to a limited impact on its overall sulfation (4–5% sulfate loss) [21]. Remarkably, such moderate but subtle changes in HS sulfation pattern lead to massive alteration of its biological activities, affecting the polysaccharide modulatory properties towards a large number of growth factors, morphogens and chemokines [15,22,23].
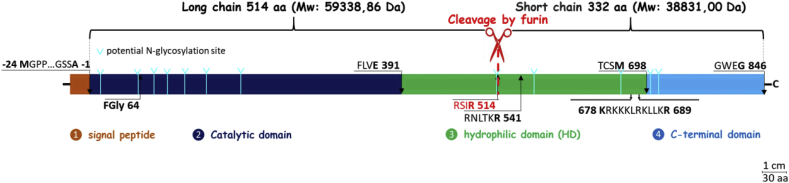
Despite the key role of HSulfs in the cellular glycomic machinery, these enzymes remain elusive protein objects as regard their structure and functions. In one hand, much effort has been devoted to the understanding of the enzyme reaction specificities, revealing a narrow specificity of HSulfs for highly sulfated disaccharide constituents within the NS domain of HS, and a processive oriented 6-O-desulfation starting from the non-reducing end [3,24]. In the other hand, much less is known about the structural organization at a molecular level, and no crystallographic data have been reported to date. Main structural insights arise from gene-derived sequence analysis and from mutation/deletion experiments [3,25,26]. HSulfs are biosynthesized as single polypeptide chain pro-enzymes (871 and 870 amino-acids for HSulf-1 (Q8IWU6) and HSulf-2 (Q8IWU5), respectively) comprising a signal peptide, followed by a catalytic domain featuring the formylglycine residue of prototype sulfatase active site, a hydrophilic domain (HD), and a C-terminal domain presenting a significant homology with that of the lysosomal glucosamine-6-sulfatase [3,4,16]. A maturation process including the removal of the signal peptide and the cleavage within the HD domain by a furin protease yields the mature enzyme as a two-chain protein likely joined by disulfide bonds [3,16,26]. As regard for HSulf-2, it results in a maturated active heterodimer, which consists in a longer chain comprising the catalytic domain (Phe 1 to Glu 391) and part of the HD domain cleaved at Arg 514, and a shorter chain composed of the remaining part of the HD domain and the C-terminal domain (Scheme 1).
Scheme 1.
Schematic view of the domain organization of HSulf-2.
Domains from N— to C— terminus are displayed with their four ending residues: signal peptide (orange, 24 aa); catalytic domain (dark blue, 391 aa); hydrophilic domain (green, 307 aa); C-terminus (blue, 148 aa). The following sequence key points are indicated: Fgly 64, active site formylglycine at Cys64; RSIR 514, main furin cleavage site in HD; RNLTKR 541, secondary furin cleavage site in HD; 678 KRKKKLRKLLKR 689, HD C-terminus basic residues cluster.
It is worth noting that HSulfs were most often indirectly detected in previous studies, generally by western-blotting and/or through the monitoring of its catalytic activity. We report here the first mass spectrometry characterization of HSulf-2, allowing its direct detection at the protein level and the coverage of the full protein sequence by a bottom-up proteomics approach.
2. Materials and methods
2.1. Materials
All reagents were of analytical grade. DL-dithiothreitol (DTT), iodoacetamide (IAA), ammonium bicarbonate (NH4HCO3), and urea were all purchased from Sigma-Aldrich (Saint-Quentin Fallavier, France). Acetonitrile and formic acid were obtained from Fluka (France); sequencing-grade trypsin, Arg-C, Chymotrypsin, Asp-N, Trypsin/Lys-C Mix and PNGase F were from Promega (France). Purified recombinant HSulf-2 was prepared according to a procedure described in a forthcoming report. Briefly, the enzyme was purified from the conditioned medium of HSulf-2 transfected HEK293F cells, using cation exchange and size exclusion chromatography, successively (A. Seffouh et al., Cell Mol. Life Sci. in press). The purified active enzyme was stored at −80 °C in 50 mM Tris buffer, 300 mM NaCl, 5 mM MgCl2, 5 mM CaCl2 pH 7.5. All buffers and solutions were prepared using ultra-pure water from a MilliQ apparatus (Millipore, Merck, France).
2.2. N-deglycosylation of HSulf-2
N-deglycosylation of HSulf-2 (3 μg) by PNGase F was carried out under denaturing conditions in a 20 μL final volume according to the provider instructions (Promega). The deglycosylation of HSulf-2 was carried out by the addition of 2 μL of recombinant PNGase F and incubation at 37 °C for 1–3 h.
2.3. Desalting of HSulf-2
Desalting of HSulf-2 was performed by diafiltration using Microcon® DNA Fast Flow 100 kDa (Millipore) previously washed with 300 μL water. HSulf-2 sample (30 μg, 40 μL) supplemented to 200 μL with water was centrifuged in Microcon at 500 g for 4 min at 4 °C. Then, 200 μL of water were added and centrifugation was repeated for few min to get about 30 μL final volume. Finally, the Microcon was returned to recover the HSulf-2 sample in 30 μL water retentate.
2.4. MALDI-TOF mass spectrometry
MS experiments were carried out on a MALDI Autoflex speed TOF/TOF MS instrument (Bruker Daltonics, Germany), equipped with a SmartBeam II™ laser pulsed at 1 kHz. The spectra were recorded in the positive linear mode (delay: 600 ns; ion source 1 (IS1) voltage: 19.0 kV; ion source 2 (IS2) voltage: 16.6 kV; lens voltage: 9.5 kV). MALDI data acquisition was carried out in the mass range 5000–150000 Da, and 10000 shots were summed for each spectrum. Mass spectra were processed using FlexAnalysis software (version 3.3.80.0, Bruker Daltonics). The instrument was calibrated using mono- and multi-charged ions of BSA (BSA Calibration Standard Kit, AB SCIEX, France). MALDI-TOF MS analysis was achieved by mixing 1.5 μL of sinapinic acid matrix at 20 mg/mL in acetonitrile/water (50/50; v/v), 0.1% TFA, with 1.5 μL of the desalted protein solution (0.71 mg/mL).
2.5. In-solution proteolytic digestions of HSulf-2
For trypsin digestion, 2 μL of HSulf-2 sample (3 μg) were diluted in 5.5 μL of 50 mM NH4HCO3 pH 8.0 to which 2.5 μL of 8 M urea were added (2 M final), and then incubated for 1 h at room temperature under moderate stirring. Samples were then reduced by addition of 1.1 μL DTT (5 mM final) for 1 h at 37 °C under moderate stirring. Samples were then alkylated by addition of 1 μL IAA (20 mM final) and left for 45 min at room temperature in the dark. Before performing trypsin digestion, 6.9 μL 50 mM NH4HCO3, pH 8.0, was added to the samples to reach a final concentration of 1 mM urea. Trypsin (1 μL of 0.4 μg.μL−1) was then added before overnight incubation at 37 °C. Conditions for digestion by Trypsin/Lys-C Mix, Asp-N, Chymotrypsin, and Arg-C are reported in Supplementary Materials.
2.6. In-gel proteolytic digestions of HSulf-2
HSulf-2 was analyzed in reducing conditions in 12% polyacrylamide gel. HSulf-2 treated/untreated with PNGase F (25/10 μL) was mixed with an equal volume of Laemmli SDS-sample buffer (60 mM Tris-Cl pH 6.8, 2% SDS, 10% glycerol, 5% β-mercaptoethanol, 0.01% bromophenol blue), heated for 5 min at 95 °C and then centrifuged and cooled at ambient temperature before to be loaded into wells.
The protein bands were excised, cut into small cubes, destained and washed with 50 μL of 0.1 M NH4HCO3, and then shrunk by dehydration in acetonitrile for 5 min (repeated 3 times). The gel pieces were next incubated in 50 μL 10 mM DTT for 35 min at 56 °C. After cooling to room temperature, the DTT solution was replaced by 50 μL acetonitrile for 5 min, then by 50 μL 0.1 M NH4HCO3 for 5 min followed by dehydration in acetonitrile for 5 min. Alkylation was carried out by addition of 50 μL of 50 mM IAA and incubation for 30 min at room temperature in the dark. The gel pieces were rinsed twice alternatively with 50 μL of acetonitrile, and with 50 μL of 0.1 M NH4HCO3 (5 min each), followed by a final dehydration step with acetonitrile. The gel pieces were then rehydrated in 40 μL of 0.1 M NH4HCO3, pH 8.0, containing 0.4 μg of sequencing grade trypsin. After overnight digestion at 37 °C, the supernatant was collected, the gel pieces were re-suspended in 50 μL of acetonitrile/water/formic acid (60:40:0.1, v/v/v) and sonicated for 10 min at 37 °C. Finally, the two resulting supernatants were gathered, vacuum-dried and stored at −80 °C until analysis.
2.7. Peptide preparation for LC-MS/MS analysis
Prior to NanoLC-MS/MS analysis, HSulf-2 peptides from in-gel digestion were re-suspended in 20 μL of solvent A (acetonitrile/water/formic acid 2:98:0.1, v/v/v). HSulf-2 peptides from in-solution digestion were desalted by two filtration cycles on ZipTip C18 (Millipore) according to the manufacturer procedure to remove urea. Eluted peptides (40 μL) were vacuum-dried and stored at −80 °C until analysis. HSulf-2 peptide digests were re-suspended in 30 μL of solvent A.
2.8. Reverse phase NanoLC-MS/MS analysis
Peptide mixtures were analyzed on a Dual Gradient Ultimate 3000 chromatographic system (Dionex) coupled to a LTQ-Orbitrap™ XL mass spectrometer (Thermo-Fisher Scientific).
5 μL of peptide solution in solvent A were injected at 20 μL/min flow rate onto a C18 pre-column (Acclaim PepMap C18, Dionex) to concentrate and desalt for 5 min in solvent A (water/acetonitrile/formic acid, 98:2:0.1, v/v/v). Then, peptide separation was carried out on a C18 capillary column (Acclaim PepMap C18, Dionex) at 300 μL/min flow rate according to the gradient: 0% solvent B (acetonitrile/water/formic acid 80/20/0,1; v/v/v) during 6 min, then 0–70% B over 49 min, then 70%–100% B over 2 min, 100% B during 10 min and finally decreasing to 0% B in 3 min. The column was finally re-equilibrated with 100% solvent A for 15 min. The LC eluent was sprayed into the MS instrument with a glass emitter tip (Pico-tip, New Objective, USA).
The LTQ-Orbitrap XL mass spectrometer (Thermo-Fisher Scientific) was operated in positive ionization mode. Singly charged species were excluded from fragmentation; dynamic exclusion of already fragmented precursor ions was applied for 300 s, with a repeat count of 1, a repeat duration of 30 s and an exclusion mass width of ± 1.5 m/z. The minimum MS signal for triggering MS/MS was set to 500.
One microscan was acquired for all scan modes. The Orbitrap cell recorded signals between 250 and 1600 m/z in profile mode with a resolution set to 60,000 in MS mode. The five most intense ions/scan were dissociated by CID (normalized collision energy 35%, precursor selection window 3 Da). During MS/MS scans, fragmentation and detection occurred in the linear ion trap analyser in centroid mode. The automatic gain control allowed accumulating up to 106 ions for FTMS scans and 104 ions for ITMSn scans. Maximum injection time was set to 500 ms for FTMS scans and 100 ms for ITMSn scans.
2.9. Database search
Raw data files were processed using Proteome Discover 1.4 (Thermo Fisher scientific) to obtain Mascot-compatible MGF files. Searches in the Swissprot Database were performed first in human taxonomy using the Mascot server (version 2.2.07, Matrix Science) with the following parameters: 1 missed cleavage, monoisotopic identification, tolerance on mass measurement 10 ppm for MS and 0.6 Da for MS2. To speed up the search, a homemade database was manually created containing the sequences of HSulf-2 chains. The MS/MS spectra were searched with semitryptic cleavage for trypsin, eight missed cleavage for chymotrypsin, and a maximum of two missed cleavages for the other proteases; no fixed modification was set; the following variable modifications were allowed: carbamidomethyl (C), carbamyl (K), carbamyl (N-term), deamidation (QN), formylGly (C), oxidation (M), and propionamide (C) without carbamyl (N-term, K) specifically for peptides resulting from in-gel digestion. Fragment types taken into account were those specified in the configuration “ESI-trap” for CID MS/MS. MS/MS spectra were all visually inspected to search for y/b discriminating ions and validate peptide sequences.
3. Results and discussion
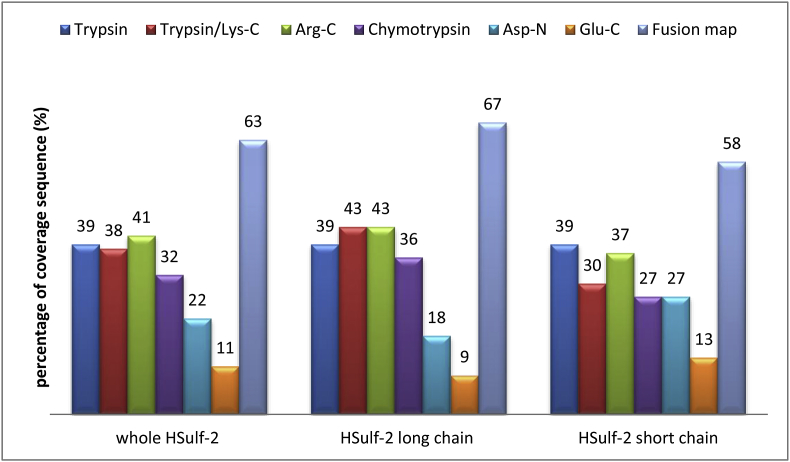
Recombinant HSulf-2 was overexpressed in the HEK293F cells, yielding an active enzyme on HS and heparin oligosaccharide substrate [24]. Trypsin digestion of HSulf-2 was carried out in solution in order to get complete sequence coverage of both short and long chains. However, this digestion performed in usual reducing/alkylating conditions led to poor sequence coverage (not shown), indicating a weak accessibility of the cleavage sites to trypsin. To increase proteolysis efficiency, proteolytic digestion was carried out in presence of urea. This protocol modification improved the digestion efficiency, since the sequence of both long and short chains of HSulf-2 was significantly covered for the first time (39% for each chain, Fig. 1). No significant coverage increase was observed following addition of various detergents (not shown), thus suggesting a particularly tight structural organization of HSulf-2.
Fig. 1.
Sequence coverage percentages obtained by in-solution digestion of HSulf-2 using the proteases trypsin, trypsin/Lys-C Mix, Arg-C, chymotrypsin, Asp-N and Glu-C.
To further increase the sequence coverage of HSulf-2 in solution, various other proteases or protease mixture (trypsin/Lys-C, Arg-C, chymotrypsin, Asp-N, Glu-C) with different specificities were used. Slightly higher coverage percentages of the long chain sequence were obtained with Arg-C and the mix trypsin/Lys-C (43%), while trypsin remained the most efficient protease for the short chain (39%) (Fig. 1, Fig. S1). Overall, by merging the different peptides produced by the various proteases in a single sequence map, 63% of the whole HSulf-2 sequence was covered, the sequence coverage being slightly higher for the long chain (67%) than for the short one (58%) (Fig. 2). Arg-C was the only protease to produce a peptide from the long chain containing the residue Cys 64 detected mainly as a formylglycine residue (Fig. S2, Table S2). Formylglycine is a key feature of sulfatase active site, which is essential for the catalytic activity [27,28]. It worth noting that previous reported point mutation experiments abolishing the enzyme activity, targeted simultaneously Cys 64 and the adjacent residue Cys 65 [3,25]. For the first time, the catalytic residue formylglycine is thus precisely located at Cys 64 and directly identified at the protein level in HSulf-2, appearing as a major PTM occurring in HSulf-2.
Fig. 2.
Sequence coverage of the whole HSulf-2 enzyme. Merged sequence map of HSulf-2 determined by combining the covered sequences obtained by the proteases trypsin, trypsin/lys-C Mix, Arg-C, chymotrypsin, Asp-N and Glu-C. C: formylglycine in the catalytic domain; N: potential N-glycosylation site; RS: furin cleavage site.
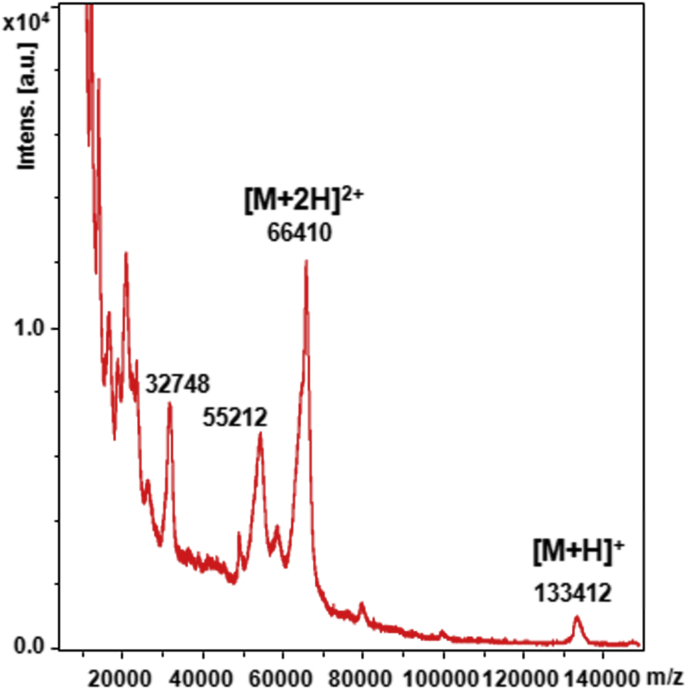
Until now, experimental molecular weight of HSulf-2 estimated to around 125 kDa relied on previously reported SDS-PAGE analysis, which showed the long chain at 75 kDa and the short chain hardly visible as a faint band at 50 kDa [26]. We confirmed this electrophoretic pattern (not shown), however, to provide an accurate mass value, HSulf-2 was submitted to MALDI-TOF MS analysis. Interestingly, the MALDI spectrum of the HSulf-2 showed in high m/z range a single low intensity peak at m/z 133,412, that we assigned to the mono-charged ion [M+H]+ of the whole HSulf-2 molecule (Fig. 3). A major peak at m/z 66,410 dominated the spectrum, which could be attributed to the doubly charged ion [M+2H]2+ of the entire HSulf-2 species. Based on these mono- and doubly-charged ions, an average experimental mass value of 133,115 ± 297 g mol−1 was determined for the first time by MS for the whole HSulf-2 enzyme. This experimental mass value was higher that the molecular weight deduced from the amino acid composition (98,170 g mol−1) suggesting a significant contribution of PTMs. An additional ion at m/z 55,212 was also detected, which could arise from possible variations in these modifications. The peak at m/z 66,410 exhibited a shoulder (at. m/z 65,420, and probably its doubly charged ion at m/z 32,748, Fig. 3) that remains unidentified. We cannot rule out that the observed shoulder along the doubly charged ion of the entire HSulf-2 species could be an isolated chain, which remains linked to the enzyme.
Fig. 3.
MALDI-TOF mass spectrometry analysis of HSulf-2. Mass spectrum of HSulf-2 in positive ionization mode (100 kDa-filtrated Hsulf-2, mixed with sinapinic acid matrix, linear mode).
A shift of the HSulf-2 bands on SDS-PAGE was previously reported after treatment by the N-glycosidase PNGase F [3,25], indicating the presence of N-glycans in agreement with the potential N-glycosylation sites along the HSulf-2 sequence (Scheme 1). We performed the trypsin in-gel digestion and nanoLC-ESI-MS/MS analysis of the resulting peptides on the most visible band attributed to the long chain, before and after treatment of HSulf-2 by PNGase F. The detected peptides belonged to the long chain (residues Phe 1 to Arg 514), ascertaining thus the identity of the main band on SDS-PAGE (Fig. S3). 46% of the long chain sequence was covered (i.e. 71% of the theoretical coverage expected by using trypsin), while the sequence coverage reached 66% (i.e. 100% of the theoretical coverage expected by using trypsin) after N-deglycosylation by PNGase F. The increased sequence coverage obtained after PNGase F treatment indicates that some cleavage sites in the long chain are protected from trypsin by N-glycan chains. Among the seven potential N-glycosylation sites found within the long chain (Asn 41, 88, 108, 125, 147, 174, 217), five on Asn 88, 108, 147, 174 and 217 were detected only in peptides obtained after N-deglycosylation. Deamidation introduced by PNGase F at Asn glycosylation site was observed on MS/MS spectra of de-glycosylated peptides (Fig. S4, Table S4). Conversely, Asn 125 is probably not a N-glycosylation site as it was detected in both glycosylated and N-deglycosylated forms of the long chain. The N-terminal first forty amino acids containing a potential N-glycosylation site were not covered. We assume that the various trypsin cleavage sites present within this N-terminal region may yield either too small peptides (≤6 residues, m/z above the chosen MS cutoff at 250) or the too acidic long peptide (PNIILVLTDDQDVELGSMQVMNK, with the potential site of glycosylation at Asn41) to be detected.
This study provides the first detailed structural data at the molecular level of the human endosulfatase HSulf-2 by mass spectrometry. The entire enzyme molecule was detected by MALDI-TOF allowing the determination of its average experimental mass at 133,115 Da, which indicated a significant contribution of post-translational modifications to the whole molecular mass. The protein sequence was covered by bottom-up ESI-MS/MS, evidencing the presence of the sulfatase specific catalytic residue formylglycine at Cys 64. The five conserved signatures of formylglycine-dependent sulfatase [29] are well identified in HSulf-2, among them the two catalytic signatures C88 to G98 and G135 to K143, and the two signatures comprising the sequences D52 to D53 and D317 to H318 involved in the coordination of a calcium ion. The increased protein sequence coverage upon N-deglycosylation by PNGase F support the presence of numerous N-glycans attached to HSulf-2 in agreement with previously reported apparent molecular mass shift on SDS-PAGE, particularly at Asn 108, 147, 174 and 217 within the long chain. While the in-gel digestion experiments of the long chain suggested Asn 88 as potentially N-glycosylated, this residue was actually identified in several peptide sequences formed under in-solution digestion conditions, indicating that Asn 88 was either not N-glycosylated like Asn 125 or could be a heterogeneous glycosylation site. Further characterization of Hsulf-2 N-glycans and whole protein top-down analysis are currently under way. The weak MS ionization efficiency of the protein and the need for multiple proteases to achieve the full sequence coverage in solution make HSulf-2 a tough protein to analyze and suggest tights folding and/or additional unusual modification of the protein backbone. This study provides the basis for further mass spectrometry investigations of the human endosulfatases.
Conflicts of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgements
We would like to thank V. Legros, D. Lebeau and W. Buchmann for help with running the Orbitrap and MALDI analysis. This work was supported in part by the CNRS and the GDR GAG (GDR 3739), the French Infrastructure for Integrated Structural Biology (FRISBI) ANR-10-INSB-05-01, the “Investissements d’avenir” program Glyco@Alps (ANR-15-IDEX-02), and by grants from the Agence Nationale de la Recherche (ANR-12-BSV8-0023 and ANR-17-CE11-0040) and Université Grenoble Alpes (UGA AGIR program).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2019.01.010.
Transparency document related to this article can be found online at https://doi.org/10.1016/j.bbrep.2019.01.010.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Transparency document
References
- 1.Uchimura K. The Sulfs: expression, purification, and substrate specificity. Methods Mol. Biol. 2015;1229:401–412. doi: 10.1007/978-1-4939-1714-3_31. [DOI] [PubMed] [Google Scholar]
- 2.Diez-Roux G., Ballabio A. Sulfatases and human disease. Annu. Rev. Genom. Hum. Genet. 2005;6:355–379. doi: 10.1146/annurev.genom.6.080604.162334. [DOI] [PubMed] [Google Scholar]
- 3.Morimoto-Tomita M., Uchimura K., Werb Z., Hemmerich S., Rosen S.D. Cloning and characterization of two extracellular heparin-degrading endosulfatases in mice and humans. J. Biol. Chem. 2002;277:49175–49185. doi: 10.1074/jbc.M205131200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dhoot G.K., Gustafsson M.K., Ai X., Sun W., Standiford D.M., Emerson C.P. Regulation of Wnt signaling and embryo patterning by an extracellular sulfatase. Science. 2001;293:1663–1666. doi: 10.1126/science.293.5535.1663. [DOI] [PubMed] [Google Scholar]
- 5.Ai X., Do A.-T., Kusche-Gullberg M., Lindahl U., Lu K., Emerson C.P. Substrate specificity and domain functions of extracellular heparan sulfate 6-O-endosulfatases, QSulf1 and QSulf2. J. Biol. Chem. 2006;281:4969–4976. doi: 10.1074/jbc.M511902200. [DOI] [PubMed] [Google Scholar]
- 6.Braquart-Varnier C., Danesin C., Clouscard-Martinato C., Agius E., Escalas N., Benazeraf B., Ai X., Emerson C., Cochard P., Soula C. A subtractive approach to characterize genes with regionalized expression in the gliogenic ventral neuroepithelium: identification of chick sulfatase 1 as a new oligodendrocyte lineage gene. Mol. Cell. Neurosci. 2004;25:612–628. doi: 10.1016/j.mcn.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 7.Winterbottom E.F., Pownall M.E. Complementary expression of HSPG 6-O-endosulfatases and 6-O-sulfotransferase in the hindbrain of Xenopus laevis. Gene Expr. Patterns. 2009;9:166–172. doi: 10.1016/j.gep.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 8.Freeman S.D., Moore W.M., Guiral E.C., Holme A.D., Turnbull J.E., Pownall M.E. Extracellular regulation of developmental cell signaling by XtSulf1. Dev. Biol. 2008;320:436–445. doi: 10.1016/j.ydbio.2008.05.554. [DOI] [PubMed] [Google Scholar]
- 9.Fujita K., Takechi E., Sakamoto N., Sumiyoshi N., Izumi S., Miyamoto T., Matsuura S., Tsurugaya T., Akasaka K., Yamamoto T. HpSulf, a heparan sulfate 6-O-endosulfatase, is involved in the regulation of VEGF signaling during sea urchin development. Mech. Dev. 2010;127:235–245. doi: 10.1016/j.mod.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 10.Gorsi B., Whelan S., Stringer S.E. Dynamic expression patterns of 6-O endosulfatases during zebrafish development suggest a subfunctionalisation event for sulf2. Dev. Dynam. 2010;239:3312–3323. doi: 10.1002/dvdy.22456. [DOI] [PubMed] [Google Scholar]
- 11.Wojcinski A., Nakato H., Soula C., Glise B. DSulfatase-1 fine-tunes Hedgehog patterning activity through a novel regulatory feedback loop. Dev. Biol. 2011;358:168–180. doi: 10.1016/j.ydbio.2011.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohto T., Uchida H., Yamazaki H., Keino-Masu K., Matsui A., Masu M. Identification of a novel nonlysosomal sulphatase expressed in the floor plate, choroid plexus and cartilage. Genes Cells. 2002;7 doi: 10.1046/j.1356-9597.2001.00502.x. 173–85. [DOI] [PubMed] [Google Scholar]
- 13.Nagamine S., Koike S., Keino-Masu K., Masu M. Expression of a heparan sulfate remodeling enzyme, heparan sulfate 6-O-endosulfatase sulfatase FP2, in the rat nervous system. Brain Res. Dev. Brain Res. 2005;159:135–143. doi: 10.1016/j.devbrainres.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 14.Nagamine S., Keino-Masu K., Shiomi K., Masu M. Proteolytic cleavage of the rat heparan sulfate 6-O-endosulfatase SulfFP2 by furin-type proprotein convertases. Biochem. Biophys. Res. Commun. 2010;391:107–112. doi: 10.1016/j.bbrc.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 15.Ai X., Do A.-T., Lozynska O., Kusche-Gullberg M., Lindahl U., Emerson C.P. QSulf1 remodels the 6-O sulfation states of cell surface heparan sulfate proteoglycans to promote Wnt signaling. J. Cell Biol. 2003;162:341–351. doi: 10.1083/jcb.200212083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vivès R.R., Seffouh A., Lortat-Jacob H. Post-synthetic regulation of HS structure: the Yin and Yang of the sulfs in cancer. Front. Oncol. 2014;3:331. doi: 10.3389/fonc.2013.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosen S.D., Lemjabbar-Alaoui H. Sulf-2: an extracellular modulator of cell signaling and a cancer target candidate. Expert Opin. Ther. Targets. 2010;14:935–949. doi: 10.1517/14728222.2010.504718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kleinschmit A., Koyama T., Dejima K., Hayashi Y., Kamimura K., Nakato H. Drosophila heparan sulfate 6-O endosulfatase regulates Wingless morphogen gradient formation. Dev. Biol. 2010;345:204–214. doi: 10.1016/j.ydbio.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maltseva I., Chan M., Kalus I., Dierks T., Rosen S.D. The SULFs, extracellular sulfatases for heparan sulfate, promote the migration of corneal epithelial cells during wound repair. PLoS One. 2013;8 doi: 10.1371/journal.pone.0069642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y.-H., Beck C. Distinct patterns of endosulfatase gene expression during Xenopus laevis limb development and regeneration. Regen. 2015;2:19–25. doi: 10.1002/reg2.27. (Oxford, England) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.El Masri R., Seffouh A., Lortat-Jacob H., Vivès R.R. The “in and out” of glucosamine 6-O-sulfation: the 6th sense of heparan sulfate. Glycoconj. J. 2017;34:285–298. doi: 10.1007/s10719-016-9736-5. [DOI] [PubMed] [Google Scholar]
- 22.Uchimura K., Morimoto-Tomita M., Bistrup A., Li J., Lyon M., Gallagher J., Werb Z., Rosen S.D. HSulf-2, an extracellular endoglucosamine-6-sulfatase, selectively mobilizes heparin-bound growth factors and chemokines: effects on VEGF, FGF-1, and SDF-1. BMC Biochem. 2006;7:2. doi: 10.1186/1471-2091-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yue X., Lu J., Auduong L., Sides M.D., Lasky J.A. Overexpression of Sulf2 in idiopathic pulmonary fibrosis. Glycobiology. 2013;23:709–719. doi: 10.1093/glycob/cwt010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seffouh A., Milz F., Przybylski C., Laguri C., Oosterhof A., Bourcier S., Sadir R., Dutkowski E., Daniel R., van Kuppevelt T.H., Dierks T., Lortat-Jacob H., Vivès R.R. HSulf sulfatases catalyze processive and oriented 6-O-desulfation of heparan sulfate that differentially regulates fibroblast growth factor activity. FASEB J. 2013;27:2431–2439. doi: 10.1096/fj.12-226373. [DOI] [PubMed] [Google Scholar]
- 25.Frese M.-A., Milz F., Dick M., Lamanna W.C., Dierks T. Characterization of the human sulfatase Sulf1 and its high affinity heparin/heparan sulfate interaction domain. J. Biol. Chem. 2009;284:28033–28044. doi: 10.1074/jbc.M109.035808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang R., Rosen S.D. Functional consequences of the subdomain organization of the sulfs. J. Biol. Chem. 2009;284:21505–21514. doi: 10.1074/jbc.M109.028472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dierks T., Schmidt B., V Borissenko L., Peng J., Preusser A., Mariappan M., von Figura K. Multiple sulfatase deficiency is caused by mutations in the gene encoding the human C(alpha)-formylglycine generating enzyme. Cell. 2003;113:435–444. doi: 10.1016/s0092-8674(03)00347-7. [DOI] [PubMed] [Google Scholar]
- 28.Appel M.J., Bertozzi C.R. Formylglycine, a post-translationally generated residue with unique catalytic capabilities and biotechnology applications. ACS Chem. Biol. 2015;10:72–84. doi: 10.1021/cb500897w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barbeyron T., Brillet-Guéguen L., Carré W., Carrière C., Caron C., Czjzek M., Hoebeke M., Michel G. Matching the diversity of sulfated biomolecules: creation of a classification database for sulfatases reflecting their substrate specificity. PLoS One. 2016;11 doi: 10.1371/journal.pone.0164846. https://doi: 10.1371/journal.pone.0164846 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.