Abstract
ZMIZ1 is a coactivator of several transcription factors, including p53, the androgen receptor, and NOTCH1. Here, we report 19 subjects with intellectual disability and developmental delay carrying variants in ZMIZ1. The associated features include growth failure, feeding difficulties, microcephaly, facial dysmorphism, and various other congenital malformations. Of these 19, 14 unrelated subjects carried de novo heterozygous single-nucleotide variants (SNVs) or single-base insertions/deletions, 3 siblings harbored a heterozygous single-base insertion, and 2 subjects had a balanced translocation disrupting ZMIZ1 or involving a regulatory region of ZMIZ1. In total, we identified 13 point mutations that affect key protein regions, including a SUMO acceptor site, a central disordered alanine-rich motif, a proline-rich domain, and a transactivation domain. All identified variants were absent from all available exome and genome databases. In vitro, ZMIZ1 showed impaired coactivation of the androgen receptor. In vivo, overexpression of ZMIZ1 mutant alleles in developing mouse brains using in utero electroporation resulted in abnormal pyramidal neuron morphology, polarization, and positioning, underscoring the importance of ZMIZ1 in neural development and supporting mutations in ZMIZ1 as the cause of a rare neurodevelopmental syndrome.
Keywords: ZMIZ1, intellectual disability, neurodevelopmental disorder, neuronal positioning, transcriptional coactivation
Main Text
Intellectual disability (ID) defines a diverse group of neurodevelopmental disorders that have a global prevalence of approximately 1%.1 ID is highly heterogeneous and can be categorized into syndromic and nonsyndromic forms. More than 1,000 underlying genetic causes of ID have been identified to date.2, 3, 4 Some of the genes previously associated with ID encode transcriptional regulators and are structured within a group of conditions called “transcriptomopathies.”5, 6, 7 Subjects with these syndromes may share a number of clinical features in addition to ID and developmental delay (DD), including growth restriction and facial dysmorphisms.
ZMIZ1 (also known as ZIMP10 or RAI17 [MIM: 607159]) is a transcriptional coregulator of the Protein Inhibitor of Activated STAT (PIAS)-like family. PIAS-like proteins do not bind DNA directly but regulate other DNA-binding transcription factors, generally through sumoylation.8 In addition, ZMIZ1 enhances the transcriptional activity of the tumor suppressor protein p539 and the androgen receptor (AR).10 ZMIZ1 is also involved in the transcriptional activation of a subset of NOTCH1 target genes, including MYC (MIM: 190080). Through its direct interaction with NOTCH1, ZMIZ1 plays a role in T lymphocyte development.11, 12 As NOTCH1 is directly involved in the regulation of cell fate during both neuronal and glial cell developments,13, 14, 15 a role for ZMIZ1 in neural development is equally plausible. Like other PIAS, ZMIZ1 is predicted to function as an E3 SUMO ligase. Sharma et al.10 indeed demonstrated that ZMIZ1 co-localizes with AR and SUMO-1 in the nucleus and forms a protein complex at replication foci. They also showed that ZMIZ1 enhances sumoylation of AR and that the increase of AR activity by ZMIZ1 is dependent on the sumoylation of the receptor using AR mutated at sumoylation sites.10 In 2015, Cordova-Fletes et al. reported a girl with ID and neuropsychiatric symptoms with a de novo balanced translocation, t(10;19)(q22.3;q13.33), that resulted in gene fusion between ZMIZ1 (chr10) and PRR12 (chr19 [MIM: 616633]), thereby disrupting the zinc-finger motif of ZMIZ1 (however, this study was by nature unable to attribute the observed phenotype to either ZMIZ1 or PRR12).16 More recently, Liu et al. showed that an enhancer of ZMIZ1 harbors recurrent single-nucleotide variations (SNVs) in autism spectrum disorder.17 Finally, animal models also suggest a role for ZMIZ1 in embryonic development, as mice homozygous for a null mutation in ZMIZ1 display embryonic lethality during organogenesis, with yolk sac vascular remodeling failure and abnormal embryonic vascular development.18
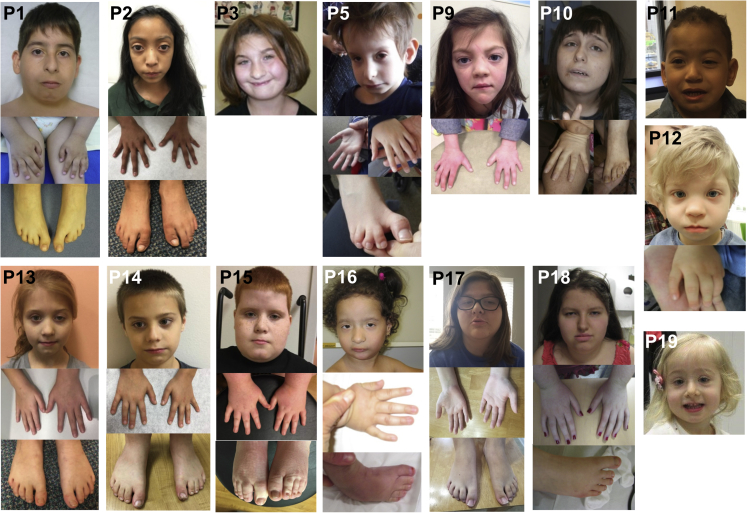
Here, we report 19 subjects (16 unrelated) with a syndromic form of ID/DD due to variants in ZMIZ1, which were identified by whole-exome or genome sequencing. The compilation of this series of subjects resulted from a transatlantic collaborative effort between University of Strasbourg (Strasbourg, France), Nantes University Hospital (Nantes, France), Dijon University Hospital (Dijon, France), Rouen University Hospital (Rouen, France), Necker Hospital/Imagine Institute (Paris, France), Lyon University Hospitals (Lyon, France), Baylor Genetics Laboratories (Houston, TX, USA), ARUP Laboratories (Salt Lake City, UT, USA), HudsonAlpha Institute for Biotechnology (Huntsville, AL, USA), Princess Anne Hospital (Southampton, UK), Medical University of Warsaw (Warsaw, Poland), Boston Children’s Hospital (Boston, MA, USA), University of Washington (Seattle, WA, USA), and GeneDx (Gaithersburg, MD, USA). The study was also partly facilitated by the web-based tools GeneMatcher,19 MyGene2 (see Web Resources), DECIPHER,20 and Matchmaker Exchange.21 All subjects were clinically assessed by at least one experienced clinical geneticist from the participating centers. All sequencing (whole-exome, genome, and Sanger) was performed after written informed consent for either clinical sequencing and/or center-specific institutional review board-approved research sequencing. Consent for published images (Figure 1) were obtained from all parents or legal guardians. All procedures were performed in accordance with the Helsinki Declaration. The main clinical features of the cohort are summarized in Table 1. More detailed clinical information for all subjects is provided in Table S1 and Supplemental Note.
Figure 1.
Topographical Images of Subjects with ZMIZ1 Variants
Table 1.
Summary of the Major Phenotypic Features of Subjects with ZMIZ1 Variants
| Subjects | #1 | #2 | #3 | #4 | #5 | #6 | #7 | #8 | #9 | #10 | #11 | #12 | #13 | #14a | #15a | #16 | #17 | #18 | #19 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Enrollment center | Nantes, France | Houston, USA | Houston, USA | Houston, USA | Dijon, France | Huntsville, USA | Huntsville, USA | Salt Lake City, USA | Salt Lake City, USA | Southampton, UK | Boston, USA | Warsaw, Poland | Houston, USA | Houston, USA | Houston, USA | Paris, France | Seattle, USA | Rouen, France | Paris, France |
| ZMIZ1 variantb | c.899C>T (p.Thr300Met) | c.859G>A (p.Ala287Thr) | c.3112dupA (p.Thr1038Asnfs∗4) | c.899C>T (p.Thr300Met) | c.893C>T (p.Thr298Ile) | c.2752dupC (p.Gln920Profs ∗34) | c.272A>G (p.Lys91Arg) | c.2610C>T (p.Ser870Ser) | c.887C>A (p.Thr296Lys) | c.1386dupC (p.Thr463Hisfs ∗14) | c.3097−2A>G (p.?) | c.3021delC (p.Phe1008Leufs∗7) | c.1386dupC (p.Thr463Hisfs ∗14) | c.1386dupC (p.Thr463Hisfs ∗14) | c.1386dupC (p.Thr463Hisfs ∗14) | c.887C>T (p.Thr296Ile) | c.2835delT (p.Met946Cysfs∗61) | t(X;10) (q27.3; q22.3) | t(10,12)(q22.2;q24.3) |
| Inheritance | de novo | de novo | de novo | de novo | de novo | de novo | de novo | de novo | de novo | de novo | de novo | de novo | unknown | unknown | unknown | de novo | de novo | de novo | de novo |
| Gender | male | female | female | female | male | female | female | male | female | female | male | male | female | male | male | female | female | female | female |
| Age at last visit | 11 y, 8 m | 12 y, 3 m | 12 y, 3 m | 1 y, 6 m | 5 y, 9 m | 20 y | 3 y | 6 y, 7 m | 8 y, 6 m | 19 y | 1 y, 4 m | 3 y, 6 m | 7 y | 10 y | 11 y | 7 y | 12 y, 17 m | 17 y | 2 y, 9 m |
| Weight (g) birth (SD) | 3,520 (+0.4) | ND | ND | ND | 2,185 (−2.8) | 3,090 (−0.2) | ND | ND | 2,135 (−2.7) | 2,380 (−2) | 2,780 (−1.4) | 3,300 (0) | ND | ND | 3,165 (−0.4) | 2,200 (−2.5) | 2,899 (−0.4) | 2,510 (−1.8) | 2,560 (−1.8) |
| Length (cm) birth (SD) | 51 (+0.5) | ND | ND | ND | 44 (−3) | ND | ND | ND | 47 (−1.1) | ND | 47 (−1.5) | 55 (+2.6) | ND | ND | ND | 45 (−2.2) | 45 (−2.2) | 47 (−1.1) | 47.5 (−1.1) |
| OFC (cm) birth (SD) | 35 (+0.4) | ND | ND | ND | 30 (−3.5) | ND | ND | ND | 30.5 (−2.9) | ND | 33.5 (−1.3) | 34 (−0.4) | ND | ND | ND | 32.5 (−1.2) | 33 (−1.2) | 32.5 (−1.2) | 32 (−1.2) |
| Growth | |||||||||||||||||||
| Growth failure yes/no | no | yes (weight) | no | ND | yes | yes (weight) | NA | no | yes | yes | no | no | yes | yes | yes | yes | yes | no | no |
| Weight (kg) last visit (SD) | 48.7 (+2.0) | 32.7 (−1.5) | 56.7 (+1.2) | ND | 15.3 (−2.4) | 56.7 (−0.2) | 12.25 (−1.0) | 25.9 (+1.0) | 20.0 (−2.1) | 41.45 (−2.6) | 11 (−0.9) | 17 (+0.9) | 22.5 (−0.1) | 27.4 (−0.9) | 45.9 (+1.1) | 20 (−0.5) | 63.1 (+1.4) | 83.3 (>+4) | 11.5 (−1) |
| Height (cm) last visit (SD) | 148.5 (−1.0) | 150.7 (−0.3) | 152.25 (−0.1) | ND | 99 (−3.0) | 139.7 (−3.6) | 86.37 (−2.1) | 116.7 (−0.5) | 121.5 (−1.6) | 137.6 (−4.3) | 81 (−1.0) | 106 (+1.7) | 115 (−1.3) | 126.8 (−1.8) | 121.6 (−3.4) | 114 (−1.0) | 158.1 (+0.3) | 157 (−0.5) | 89 (−0.6) |
| OFC (cm) last visit (SD) | 53 (−1.0) | 55.7 (+1.4) | 50.25 (−2.2) | ND | 47.7 (−3.0) | 54 (−0.3) | NA | 52 (0.0) | 49.0 (−2.2) | 53 (−1.8) | 46 (−1.6) | 51 (+0.1) | 49.4 (−1.5) | 50.3 (−1.8) | 52.1 (−0.9) | 48.5 (−2.0) | 53.7 (0.0) | 56 (+1) | 46 (−2) |
| Feeding difficulties | − | − | ND | + | + | + | + | ND | + | + | + | + | − | − | − | + | − | − | − |
| Neurological Abnormalities | |||||||||||||||||||
| Intellectual disability | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Motor delay | + | − | − | + | + | − | + | + | + | + | + | + | ND | ND | ND | + | − | + | + |
| Speech delay | + | − | − | ND | + | + | + | + | + | + | + | + | + | + | ND | + | + | + | + |
| Abnormal behavior | − | + | + | ND | + | + | ND | + | + | + | + | + | − | + | ND | + | + | + | − |
| Seizures | − | − | ND | + | − | − | − | + | − | − | − | − | − | − | − | − | + | − | − |
| Hypotonia | + | − | − | ND | + | − | ND | ND | − | − | + | + | + | − | + | + | + | + | + |
| Hearing loss | − | − | + | − | + | − | − | − | − | − | − | − | + | − | − | + | − | − | − |
| Brain abnormalities MRI | + | − | − | − | ND | + | − | − | ND | + | ND | ND | + | ND | + | + | + | − | − |
| Congenital Malformations | |||||||||||||||||||
| Cardiac | − | + | − | ND | + | − | ND | + | − | − | − | − | − | − | − | − | + | − | − |
| Urogenital/kidney | − | + | − | + | ND | + | ND | ND | + | − | + | + | − | − | + | − | − | + | − |
| Eye | − | + | + | + | + | + | ND | ND | + | + | + | − | + | + | − | − | + | − | − |
| Craniofacial dysmorphism | + | + | + | + | + | − | ND | + | + | + | + | + | + | + | + | + | + | + | − |
| Others | − | − | − | − | − | − | − | − | − | + | + | + | − | − | − | − | + | + | − |
| Skeletal Abnormalities | |||||||||||||||||||
| Joint hypermobility | − | + | − | + | − | − | ND | + | − | + | − | − | + | − | + | + | − | + | − |
| Pectus deformity | − | + | − | ND | − | − | ND | − | − | − | − | − | − | − | − | − | − | − | − |
| Thorax | + | + | − | ND | − | − | ND | − | + | − | − | − | − | − | − | − | + | − | − |
| Hands | + | + | − | ND | − | − | ND | + | + | + | − | + | − | − | + | + | − | + | − |
| Feet | + | + | + | + | + | − | ND | + | + | + | − | − | − | − | − | + | + | + | − |
| Palate | − | − | ND | + | ND | − | ND | ND | + | − | + | − | − | − | − | + | − | − | − |
Abbreviations are as follows: y, years; m, months; ND, not determined; SD, standard deviation; OFC, occipitofrontal circumference; and MRI, magnetic resonance imaging. More comprehensive information regarding clinical features can be found in Table S1 and Supplemental Note.
Subjects #14 and #15 are siblings of subject #13.
Positions refer to GenBank: NM_020338.3. Nucleotide numbering uses +1 as the A of the ATG translation initiation codon in the reference sequence, with the initiation codon as codon 1.
All subjects from the series exhibited ID/DD (n = 19). Growth failure and feeding difficulties were each reported in 10 and 9 subjects, respectively. Five subjects had microcephaly (subjects #3, #5, #9, #16, and #19 with SD −2.2, −3.0, −2.2, −2.0, and −2.0, respectively). All subjects had additional neurological features, including motor delay (n = 12), speech delay (n = 15), abnormal behavior (n = 13), seizures (n = 3), hypotonia (n = 10), and hearing loss (n = 4). Thirteen subjects had other congenital malformations, including ventricular septal defects (n = 2), abnormal mitral valve (n = 1), patent ductus arteriosus (n = 1), renal pelviectasis (n = 1), cryptorchidism (n = 2), vesicoureteral reflux (n = 3), and atrophic kidney (n = 1). Eleven subjects had additional ophthalmologic anomalies, i.e., decreased vision (n = 1), ptosis (n = 3), glaucoma (n = 1), hypermetropia (n = 2), astigmatism (n = 1), myopia (n = 1), Duane syndrome (n = 1), nasolacrimal duct stenosis (n = 1), coloboma of the retina (n = 1), and amblyopia (n = 1). Nine subjects had skeletal defects including joint hypermobility (n = 8), pectus excavatum (n = 1), scoliosis (n = 3), spondylolisthesis (n = 1), and finally, abnormalities of the feet (bilateral 2/3 toe syndactyly n = 6; cone-shaped epiphyses n = 2; bilateral clubfeet n = 2; short toes n = 1), hands (short fingers n = 6; long fingers n = 1, tapered fingers n = 1; brachydactyly n = 1), and palate (n = 3). Craniofacial dysmorphisms were observed in 16 subjects (see Figure 1 as well as Table S1 and Supplemental Note for details).
All ZMIZ1 variants and genomic alterations reported in this work are detailed in Table 2. Protocols for single- or trio-based whole-exome/genome sequencing and Sanger sequencing were previously described4, 26, 27, 28, 29, 30, 31 (see also Supplemental Material and Methods). None of the variants identified here were present in gnomAD,32 the Exome Variant Server (see Web Resources), or an internal database of 550 exomes. Aside from one family (subjects #13, #14, and #15) for whom parental samples were unavailable, all variants were shown to be de novo (Figure S1). The variants were all predicted to be disease causing by various bioinformatics tools and to affect specific functional domains of the protein (Table 2; Figure 2A). Two additional subjects (#18 and #19) with balanced de novo translocations between chromosomes X and 10 (46,XX,t(X;10)(q27;q23)) and between chromosomes 10 and 12 (46,XX,t(10;12)(q22.2;q24.3)), respectively, were identified by blood karyotyping. The breakpoints on the X chromosome (Xq27.3, chrX:143274164_143274161, GRCh37) and on chromosome 10 (10q22.3, chr10:80552343_80552344, GRCh37) for the t(X;10)(q27;q23) translocation, and on chromosome 12 (12q24.32, chr12:128533508_128533517, GRCh37) and chromosome 10 (10q22.3, chr10:81010865_81010871, GRCh37) for t(10;12)(q22.2;q24.3) translocation were defined by whole-genome sequencing (Table 2). While the t(10;12)(q22.2;q24.3) translocation of subject #19 disrupts ZMIZ1, no gene is disrupted by t(X;10)(q27;q23) translocation in subject #18. However, the breakpoint on chromosome 10 in this subject is located 276 kb upstream of ZMIZ1, where it disrupts a distal enhancer (position chr10:80559750–80568675) known to harbor recurrent SNVs in autism spectrum disorder subjects and predicted to interact with the ZMIZ1 promoter.17 Moreover, as detailed below, the deleterious effect of the translocations on ZMIZ1 expression was further confirmed by quantitative RT-PCR analysis of ZMIZ1 transcripts.
Table 2.
Sequence Changes Identified
| Subject No. | Nucleotide Alterationa | Coding Sequence Alterationb | Amino Acid Alteration | SIFTc | LRTd | GERP++e | PhyloPf | PolyPheng | CADDh | MutationTaster |
|---|---|---|---|---|---|---|---|---|---|---|
| 1,4 | chr10:81052055C>T | c.899C>T | p.Thr300Met | 0 | 0.001 | 5.16 | 5.494 | 1 | 26.8 | disease-causing |
| 2 | chr10:81052015G>A | c.859G>A | p.Ala287Thr | 0 | 0.001 | 5.16 | 5.406 | 0.998 | 27.3 | disease-causing |
| 3 | chr10:81072414_81072415insA | c.3112dupA | p.Thr1038Asnfs∗4 | NA | NA | 5.16 | 5.572 | NA | 35 | disease-causing |
| 5 | chr10:81052049C>T | c.893C>T | p.Thr298Ile | 0.001 | 0.001 | 5.16 | 5.494 | 1 | 25.8 | disease-causing |
| 6 | chr10:81067244_81067245insC | c.2752dupC | p.Gln920Profs∗34 | NA | NA | 4.57 | 5.306 | NA | 35 | disease-causing |
| 7 | chr10:80976023A>G | c.272A>G | p.Lys91Arg | 0.032 | 0.004 | 5.3 | 4.442 | 0.956 | 27.1 | disease-causing |
| 8 | chr10:81066043C>T | c.2610C>Ti | p.Ser870Ser | NA | NA | 0.619 | −0.048 | NA | 6.092 | disease-causing |
| 9 | chr10:81052043C>A | c.887C>A | p.Thr296Lys | 0.001 | 0.001 | 5.16 | 5.494 | 1 | 25.6 | disease-causing |
| 10,13,14,15 | chr10:81056383_81056384insC | c.1386dupC | p.Thr463Hisfs∗14 | NA | NA | 4.96 | 2.061 | NA | 33 | disease-causing |
| 11 | chr10:81072397A>G | c.3097−2A>G j | ? | NA | NA | 5.16 | 4.614 | NA | 34 | disease-causing |
| 12 | chr10:81070866_81070866delC | c.3021delC | p.Phe1008Leufs∗7 | NA | NA | 4.72 | 4.438 | NA | 35 | disease-causing |
| 16 | chr10:81052043C>T | c.887C>T | p.Thr296Ile | 0.11 | 0.001 | 5.16 | 5.497 | 1 | 25.4 | disease-causing |
| 17 | chr10:81067328_81067329delTG | c.2835delT | p.Met946Cysfs∗61 | NA | NA | NA | NA | NA | 35 | disease-causing |
| 18 | t(X;10)(q27.3;q22.3) | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| 19 | t(10,12)(q22.2;q24.3) | NA | NA | NA | NA | NA | NA | NA | NA | NA |
Predictive analyses were performed with MutationTaster,22 PolyPhen-2 v2.2.2r398,23 GATK analysis toolkit,24 and ESEfinder.25 All variants were absent from the Exome Variant Server and the gnomAD database. NA, not applicable.
Positions refer to Hg19 (GRCh37).
Positions refer to GenBank: NM_020338.
Function prediction tool based on protein sequence conservation among homologs. Variants with scores between 0 and 0.05 are considered deleterious.
DNA sequence evolutionary model expressed as a p value.
GERP++NR score: DNA conservation score.
Vertebrate PhyloP scores. Values vary between −20 and +9.873. Sites predicted to be conserved are assigned positive scores, while sites predicted to be fast-evolving are assigned negative scores.
PolyPhen2_HDIV_score: variants with scores between 0.85 and 1.0 are predicted to be damaging with high confidence.
CADDv1.4 scores range from 1 to 99, with a higher score indicating greater deleteriousness.
Probable impact on splicing: part of CTCCCTA (ESE SF2/ASF) and CGTCTCC (ESE SRp40).
Acceptor splice site lost.
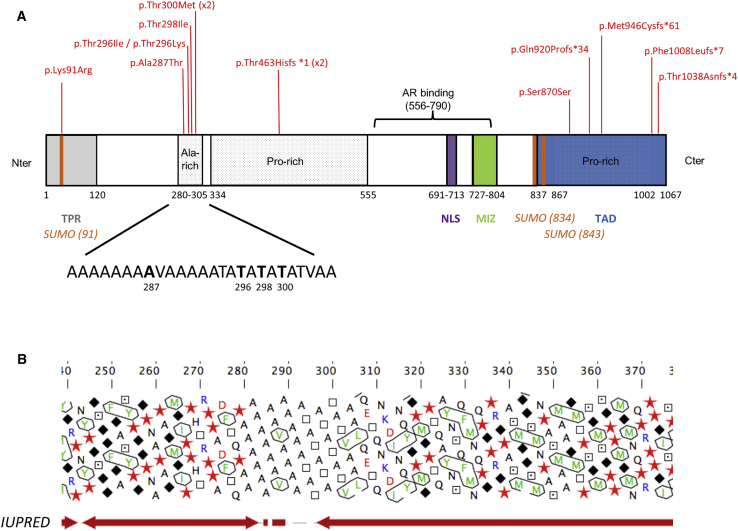
Figure 2.
Domain Organization of ZMIZ1 and Its Variants from Affected Subjects
(A) Schematic representation of ZMIZ1 and 10 variants identified in 12 families. See Table 2 for DNA sequence changes in the subjects. The splice variant c.3097−2A>G and the t(10;19) (q22.3;q13.33) translocation are not represented here. Abbreviations: TPR, tetratricopeptide repeat; SUMO, SUMO acceptor site; NLS, nuclear localization signal; MIZ, SP-RING/MIZ domain; TAD, transactivation domain.
(B) Hydrophobic cluster analysis (HCA) of the alanine-rich region with predictions of intrinsically disordered regions from IUPRED33 performed with the Medor Metaserver.34 Symbols are used to represent amino acids with peculiar structural properties (star for proline, black diamond for glycine, square for threonine, and dotted square for serine, which may be either exposed or buried). Phase transition or phase separation has been put forward as a general mechanism for the transient formation of cellular bodies that behaves as liquid droplet. The occurrence of stochastic liquid-liquid phase transitions within a cell nucleus provides an interesting framework to explain recent experimental observations on the transcriptional regulation in higher eukaryotes.35
The ZMIZ1 protein sequence contains both folded domains and low-complexity regions predicted to be intrinsically disordered (Figure 2A). The folded domains include a tetratricopeptide repeat at the N terminus extremity (1–120), which is involved in the interaction with NOTCH1, and an MIZ/SP-RING finger (727–804) acting as an adaptor for sumoylation.36 Disordered regions include two large proline-rich domains and an alanine-rich motif (280–305). Of note, most variants were found in these low-complexity regions, with the exception of p.Lys91Arg (c.272A>G), which colocalizes with a SUMO acceptor site (Figure 2A). ZMIZ1 is, indeed, known to enhance AR sumoylation by its E3 SUMO ligase activity, which is essential for its coregulatory function.10, 37 The alanine-rich domain 280–305 contains five variants (p.Ala287Thr [c.859G>A], p.Thr296Lys [c.887C>A], p.Thr296Ile [c.887C>T], p.Thr298Ile [c.893C>T], and p.Thr300Met [c.899C>T] present in six unrelated subjects). The sequence of this domain (AAAAAAAAVAAAAATATATATATVAA) is evolutionarily conserved and specific to the ZMIZ1 protein (Figure S2). The p.Thr296Lys, p.Thr296Ile, p.Thr298Ile, and p.Thr300Met variants suggest that a strict alternation of alanine-threonine repeats is required for the physiological function of ZMIZ1. This motif is flanked by regular motifs of glycine and proline residues, suggesting a possible role in phase transition (Figure 2B). Phase transitions due to sequences of low complexity are an emerging mechanism of transcriptional regulation, and disease-related variants in such regions have been shown to disrupt transcription.38, 39, 40, 41 Disruption of the alanine-threonine repeat by variants may therefore have a direct impact on the coregulation of transcription mediated by ZMIZ1. Another series of seven variants is predicted to shorten or remove the C-terminal transactivation domain of the protein: (1) the four frameshift variants p.Gln920Profs∗34 (c.2752dupC), p.Met946Cysfs∗61 (c.2835delT), p.Phe1008Leufs∗7 (c.3021delC), and p.Thr1038Asnfs∗4 (c.3112dupA) each lead to a premature stop codon, (2) p.Ser870Ser (c.2610C>T) is silent at the protein level but is part of an exonic splicing enhancer site for SF2/ASF (C[C/T]CCCTA) and SRp40 (CGTC[C/T]CC), (3) the c.3097−2A>G splice-site variant leads to the loss of an acceptor site (AG>GG) in the last intron of the gene, resulting in two alternative transcripts (Figure S3), and finally, (4) the p.Thr463Hisfs∗1 (c.1386dupC) variant affects the proline-rich domain 334–555 and is predicted to be translated into a protein that is shortened by more than half of its length (464 amino acids instead of 1067 amino acids). It is highly probable that these variants result in truncated proteins with an absent or shortened transactivation domain and, thus, reduced cotranscriptional activity.
We further assessed the expression of ZMIZ1 at the mRNA level in peripheral blood from five subjects with point mutations (#1, #5, #8, #9, and #11) in an EBV-transformed lymphoblastoid cell line of subjects #18 with the t(X;10)(q27;q23) translocation and in peripheral blood of subject #19 with the t(10;12)(q22.2;q24.3) translocation. The expression level of ZMIZ1 in subjects with point mutations was found to be similar to that in healthy controls (Figure S4A). This observation was true for subjects with missense variants (subject #1, c.899C>T; subject #5, c.893C>T; and subject #9, c.887C>A) as well as for those with suspected or confirmed splice-site variants (subject #8, c.2610C>T and subject #11, c.3097−2A>G). In subject #18, the mRNA level was decreased compared to that in control subjects, supporting the presence of a position effect (Figure S4B). At the protein level, transfection of HeLa cells with wild-type or mutant (p.Lys91Arg, p.Thr300Met, or p.Thr1038fs) ZMIZ1 plasmids indicated nuclear localization (Figure S5).
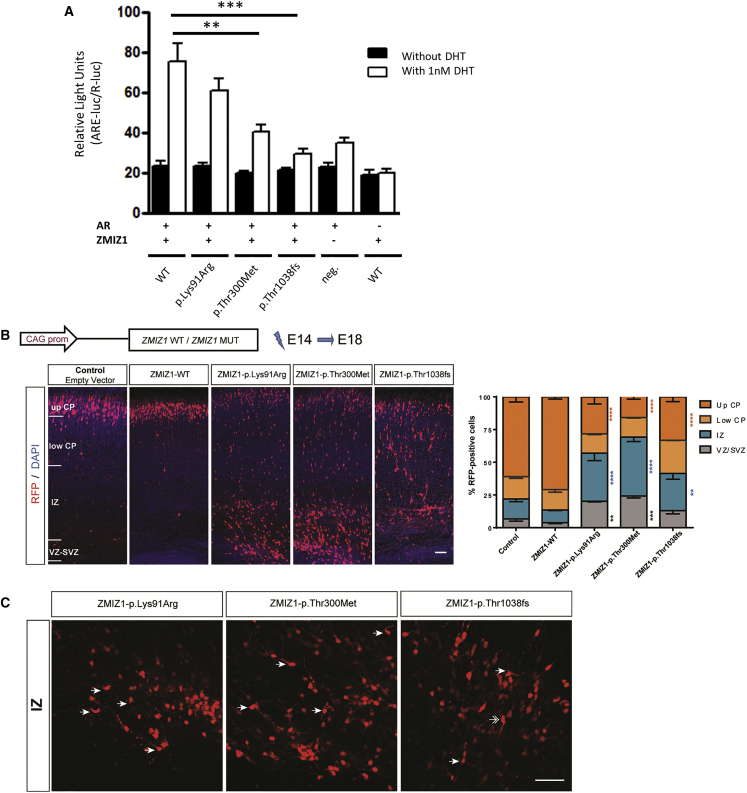
As ZMIZ1 is a well-known AR coactivator,10 we sought to analyze the effect of three representative ZMIZ1 mutations on this process. We cotransfected the HEK293T cell line with expression vectors containing the following cDNA constructs: ZMIZ1 (wild-type, c.272A>G, c.899C>T, or c.3112dupA), AR, renilla luciferase, and a consensus androgen response element (ARE)-driven luciferase reporter. Luciferase activity was determined with and without induction by the AR ligand dihydrotestosterone (DHT) (Figure 3A). Compared to wild-type ZMIZ1 protein, all three mutant proteins showed decreased induction of luciferase activity after the addition of DHT (although not statistically significant for the p.Lys91Arg mutant). These results indicate that ZMIZ1 variants impair the coactivation activity of the protein.
Figure 3.
In Vitro and In Vivo Effects of ZMIZ1 Variants
(A) Effect of ZMIZ1 variants on DHT-induced AR activity, as determined by the ARE-luciferase assay. HEK293T cells were cultivated with 1 nM DHT or without DHT (ethanol as vehicle) and transiently transfected with 30 ng of ZMIZ1 expression constructs in combination with 5 ng of pEGFP-ARwt (wild-type AR expression plasmid), 150 ng of pARE-luc (androgen response element (ARE)-driven luciferase reporter plasmid), and 20 ng of pRenilla-luc (renilla luciferase expression vector for normalization). The total amount of plasmids per well was kept constant by adding empty vectors as needed. Control transfections without the pEGFP-ARwt or the ZMIZ1 vectors were also performed. Transfection experiments were repeated three times in quadruplicate. Relative luciferase units (RLUs) are presented as the mean ± SEM. Two-tailed t test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(B) Effect of ZMIZ1 variants on neuronal migration and positioning. cDNA constructions were coelectroporated with an RFP-encoding reporter construction (dt-Tomato) in progenitor cells adjacent to the ventricle of E14.5 embryonic mouse cortices. Analyses were performed at E18.5. Images show coronal sections of E18.5 brains electroporated at E14.5 with either a control vector, WT ZMIZ1 or one of the three mutated forms of ZMIZ1. Sections were stained with 4′,6-diamidino-2-phenylindole (DAPI). Abbreviations: CP, cortical plate; IZ, intermediate zone; and VZ/SVZ, ventricular zone/subventricular zone. Scale bar, 50 μm. Histograms present the quantification of the percentage of fluorescent neurons in four different regions: upper and lower CP, IZ, and VZ/SVZ. Data are presented as the mean ± SEM, two-way ANOVA with Tukey’s multiple comparisons test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 compared to an empty vector control.
(C) Morphological features of cells accumulated in the IZ with the three ZMIZ1 variants. Arrow shows cells with long and abnormally oriented processes. Double arrow shows a bipolar cell with a leading process oriented toward the VZ. Scale bar, 50 μm.
Next, we sought to investigate the effect of the three ZMIZ1 variants on neuronal positioning in vivo.42 We first validated that the wild-type and mutant proteins were stable (not degraded) when overexpressed in neuronal cells using the neuro2A cell line (Figure S6). We then used in utero electroporation to induce the overexpression of human ZMIZ1 variants under the control of a CAG promoter together with a pCAGGS-IRES-Tomato (RFP) reporter in progenitor cells in the ventricular zone. Mice cortices were electroporated at embryonic day 14 (E14.5), and the distribution of electroporated cells was analyzed at E18.5. The relative percentage of electroporated RFP-positive cells was calculated for four different regions of the cortical wall: the ventricular and subventricular zones (VZ/SVZ), the intermediate zone (IZ), and the upper and lower cortical plate (CP) (Figure 3B). In brain sections expressing the empty vector control, the majority of the electroporated neurons were positioned within the upper layers of the CP. Likewise, overexpression of human WT-ZMIZ1 showed a normal pattern of distribution, similar to that in the control. In contrast, in utero electroporation of ZMIZ1 pathogenic variants resulted in impaired neuronal positioning at this stage with an accumulation of electroporated cells in the VZ/SVZ and IZ and a corresponding depletion of these cells in the upper CP. The phenotype of two variants, p.Lys91Arg and p.Thr300Met, appeared more severe than that of p.Thr1038fs, with approximately 60% of the electroporated cells remaining in the IZ and VZ/SVZ. In the third variant p.Thr1038fs, approximately 40% of electroporated cells remained in these regions, and interestingly, 25% was found in the lower CP, suggesting possible differences in the severity of consequences from the variants. Rescue experiments consisting of co-electroporation of the WT ZMIZ1 plasmid along with different mutants showed a partial rescue of the WT phenotype only in the case of the p.Thr300Met variant (Figure S7). Moreover, a closer look at the arrested cells in the IZ revealed striking morphological features. The majority of cells in the IZ were round; however, the cells emitting processes appeared aberrantly long and abnormally oriented (Figure 3C). This phenotype is in contrast with the expected dynamics and morphological changes of migrating neurons. Indeed, in the IZ, migrating newborn neurons adopt a bipolar morphology and extend a leading process toward the CP.43 The presence of rounded cells and of cells with long, randomly oriented processes suggests the disruption of proper polarization. Abnormal morphology and polarization could underlie the inability of the cells to migrate properly. Together, these data suggest an important role for ZMIZ1 in early stages of the postmitotic positioning of pyramidal neurons in the developing cerebral cortex.
Taken together, our data indicate that ZMIZ1 plays an important role during embryonic development. High expression levels of ZMIZ1 in craniofacial tissue and brain18, 44 are consistent with the clinical phenotype of ID/DD and the craniofacial dysmorphism observed in 16 of the subjects included in this study. Moreover, in the brain, ZMIZ1 has been shown to interact with chromatin remodeling complex (nBAF) and activator protein 1 (AP-1), which regulate the activity of genes essential for normal synapse and dendrite development.45, 46 During mouse embryogenesis, the expression of ZMIZ1 was shown to be very dynamic, with a remarkable expression pattern in the limb buds; ZMIZ1 expression was restricted to the interdigital region at limb stage S8 (embryonic day 12),18 an observation that is in line with syndactyly, which was observed in 6 of our subjects (Figure 1, Tables 1 and S1 and Supplemental Note). Furthermore, we showed here that overexpression of ZMIZ1 mutants in mouse progenitor cells impaired neuronal migration. This result might be explained by an effect on the well-known partner of ZMIZ1, NOTCH1, which has been shown to regulate neurite outgrowth in neuroblastoma cells13 and to promote the acquisition of the first specialized cell type (i.e., radial glia) in the mouse forebrain during embryogenesis.14
In conclusion, we describe a neurodevelopmental disorder caused by de novo heterozygous point mutations or rearrangement of ZMIZ1. Further studies are needed to definitively conclude as to a mechanism of action of these variants.
Declaration of Interests
I.M.W., K.M., and R.E.S. are employees of GeneDx, Inc., a wholly owned subsidiary of OPKO Health, Inc. The other authors declare no competing interests.
Acknowledgments
We thank Drs. Olivier Pourquié and Clifford Tabin (Harvard Medical School) for discussions on limb development. We thank Dr. Kevin Bowling (HudsonAlpha Institute for Biotechnology) for computation of CADD scores. We are grateful to the families who participated in this study. This work was supported by grants from the Agence Nationale de la Recherche (ANR), the INSERM (UMR_S 1109) and the Institut Universitaire de France (IUF) to S.B.; the European regional development fund (European Union) INTERREG V program (project n°3.2 TRIDIAG) to R.C. and S.B.; MSDAvenir grant (Autogen project) to S.B.; the French Ministry of Health (DGOS) and the French National Agency for Research (ANR) (PRTS 2013) grants to C.S.-B.; the MSDAvenir (DevoDecode project) and ANR to J.A., C.T.G., and C.D.; and a grant from the National Human Genome Research Institute to G.M.C. (UM1HG007301). The DDD study presents independent research commissioned by the Health Innovation Challenge Fund (grant number HICF-1009-003); see www.ddduk.org/access.html for full acknowledgment. This study used data shared through MyGene2.org. Funding for MyGene2 was provided by the National Institutes of Health/National Human Genome Research Institute and National Heart Lung and Blood Institute through the University of Washington Center for Mendelian Genomics (2UM1HG006493-05) and by Wellcome Trust, U.S. National Institutes of Health, and Howard Hughes Medical Institute through the Open Science Prize. H.C.M. has grant funding from NIH (NINDS NS069605). Sequencing at University of Washington was provided by the University of Washington Center for Mendelian Genomics (UW-CMG) and was funded by NHGRI and NHLBI grants UM1 HG006493 and U24 HG008956. The content is therefore solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Published: January 10, 2019
Footnotes
Supplemental Data include seven figures, two tables, Supplemental Note (detailed clinical history), and Supplemental Material and Methods and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.12.007.
Contributor Information
Raphael Carapito, Email: carapito@unistra.fr.
Seiamak Bahram, Email: siamak@unistra.fr.
Accession Numbers
The ZMIZ1 variants reported in this paper have been submitted to ClinVar: SUB4881332.
Web Resources
DECIPHER, https://decipher.sanger.ac.uk/
ESEfinder, http://exon.cshl.edu/ESE/
ExAC Browser, http://exac.broadinstitute.org/
GATK, http://software.broadinstitute.org/gatk/guide/article?id=1247
GeneMatcher, https://genematcher.org/
gnomAD Browser, http://gnomad.broadinstitute.org/
Matchmaker Exchange, https://www.matchmakerexchange.org/
MutationTaster, http://www.mutationtaster.org/
MyGene2, https://mygene2.org/MyGene2/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server (accessed August 2018), http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
PolyPhen-2 v.2.2.2, http://genetics.bwh.harvard.edu/pph2/
SIFT v.1.03, http://sift.bii.a-star.edu.sg/
Supplemental Data
References
- 1.Maulik P.K., Mascarenhas M.N., Mathers C.D., Dua T., Saxena S. Prevalence of intellectual disability: a meta-analysis of population-based studies. Res. Dev. Disabil. 2011;32:419–436. doi: 10.1016/j.ridd.2010.12.018. [DOI] [PubMed] [Google Scholar]
- 2.Gilissen C., Hehir-Kwa J.Y., Thung D.T., van de Vorst M., van Bon B.W., Willemsen M.H., Kwint M., Janssen I.M., Hoischen A., Schenck A. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511:344–347. doi: 10.1038/nature13394. [DOI] [PubMed] [Google Scholar]
- 3.Deciphering Developmental Disorders Study Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519:223–228. doi: 10.1038/nature14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wright C.F., Fitzgerald T.W., Jones W.D., Clayton S., McRae J.F., van Kogelenberg M., King D.A., Ambridge K., Barrett D.M., Bayzetinova T., DDD study Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet. 2015;385:1305–1314. doi: 10.1016/S0140-6736(14)61705-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Izumi K. Disorders of transcriptional regulation: An emerging category of multiple malformation syndromes. Mol. Syndromol. 2016;7:262–273. doi: 10.1159/000448747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parenti I., Teresa-Rodrigo M.E., Pozojevic J., Ruiz Gil S., Bader I., Braunholz D., Bramswig N.C., Gervasini C., Larizza L., Pfeiffer L. Mutations in chromatin regulators functionally link Cornelia de Lange syndrome and clinically overlapping phenotypes. Hum. Genet. 2017;136:307–320. doi: 10.1007/s00439-017-1758-y. [DOI] [PubMed] [Google Scholar]
- 7.Yuan B., Pehlivan D., Karaca E., Patel N., Charng W.L., Gambin T., Gonzaga-Jauregui C., Sutton V.R., Yesil G., Bozdogan S.T. Global transcriptional disturbances underlie Cornelia de Lange syndrome and related phenotypes. J. Clin. Invest. 2015;125:636–651. doi: 10.1172/JCI77435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shuai K., Liu B. Regulation of gene-activation pathways by PIAS proteins in the immune system. Nat. Rev. Immunol. 2005;5:593–605. doi: 10.1038/nri1667. [DOI] [PubMed] [Google Scholar]
- 9.Lee J., Beliakoff J., Sun Z. The novel PIAS-like protein hZimp10 is a transcriptional co-activator of the p53 tumor suppressor. Nucleic Acids Res. 2007;35:4523–4534. doi: 10.1093/nar/gkm476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharma M., Li X., Wang Y., Zarnegar M., Huang C.Y., Palvimo J.J., Lim B., Sun Z. hZimp10 is an androgen receptor co-activator and forms a complex with SUMO-1 at replication foci. EMBO J. 2003;22:6101–6114. doi: 10.1093/emboj/cdg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pinnell N., Yan R., Cho H.J., Keeley T., Murai M.J., Liu Y., Alarcon A.S., Qin J., Wang Q., Kuick R. The PIAS-like coactivator Zmiz1 is a direct and selective cofactor of Notch1 in T cell development and leukemia. Immunity. 2015;43:870–883. doi: 10.1016/j.immuni.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rakowski L.A., Garagiola D.D., Li C.M., Decker M., Caruso S., Jones M., Kuick R., Cierpicki T., Maillard I., Chiang M.Y. Convergence of the ZMIZ1 and NOTCH1 pathways at C-MYC in acute T lymphoblastic leukemias. Cancer Res. 2013;73:930–941. doi: 10.1158/0008-5472.CAN-12-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franklin J.L., Berechid B.E., Cutting F.B., Presente A., Chambers C.B., Foltz D.R., Ferreira A., Nye J.S. Autonomous and non-autonomous regulation of mammalian neurite development by Notch1 and Delta1. Curr. Biol. 1999;9:1448–1457. doi: 10.1016/s0960-9822(00)80114-1. [DOI] [PubMed] [Google Scholar]
- 14.Gaiano N., Nye J.S., Fishell G. Radial glial identity is promoted by Notch1 signaling in the murine forebrain. Neuron. 2000;26:395–404. doi: 10.1016/s0896-6273(00)81172-1. [DOI] [PubMed] [Google Scholar]
- 15.Angulo-Rojo C., Manning-Cela R., Aguirre A., Ortega A., López-Bayghen E. Involvement of the Notch pathway in terminal astrocytic differentiation: role of PKA. ASN Neuro. 2013;5:e00130. doi: 10.1042/AN20130023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Córdova-Fletes C., Domínguez M.G., Delint-Ramirez I., Martínez-Rodríguez H.G., Rivas-Estilla A.M., Barros-Núñez P., Ortiz-López R., Neira V.A. A de novo t(10;19)(q22.3;q13.33) leads to ZMIZ1/PRR12 reciprocal fusion transcripts in a girl with intellectual disability and neuropsychiatric alterations. Neurogenetics. 2015;16:287–298. doi: 10.1007/s10048-015-0452-2. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y., Liang Y., Cicek A.E., Li Z., Li J., Muhle R.A., Krenzer M., Mei Y., Wang Y., Knoblauch N. A statistical framework for mapping risk genes from de novo mutations in whole-genome-sequencing studies. Am. J. Hum. Genet. 2018;102:1031–1047. doi: 10.1016/j.ajhg.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodriguez-Magadán H., Merino E., Schnabel D., Ramírez L., Lomelí H. Spatial and temporal expression of Zimp7 and Zimp10 PIAS-like proteins in the developing mouse embryo. Gene Expr. Patterns. 2008;8:206–213. doi: 10.1016/j.modgep.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 19.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Firth H.V., Richards S.M., Bevan A.P., Clayton S., Corpas M., Rajan D., Van Vooren S., Moreau Y., Pettett R.M., Carter N.P. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am. J. Hum. Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Philippakis A.A., Azzariti D.R., Beltran S., Brookes A.J., Brownstein C.A., Brudno M., Brunner H.G., Buske O.J., Carey K., Doll C. The Matchmaker Exchange: a platform for rare disease gene discovery. Hum. Mutat. 2015;36:915–921. doi: 10.1002/humu.22858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwarz J.M., Cooper D.N., Schuelke M., Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat. Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 23.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cartegni L., Wang J., Zhu Z., Zhang M.Q., Krainer A.R. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bowling K.M., Thompson M.L., Amaral M.D., Finnila C.R., Hiatt S.M., Engel K.L., Cochran J.N., Brothers K.B., East K.M., Gray D.E. Genomic diagnosis for children with intellectual disability and/or developmental delay. Genome Med. 2017;9:43. doi: 10.1186/s13073-017-0433-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carapito R., Konantz M., Paillard C., Miao Z., Pichot A., Leduc M.S., Yang Y., Bergstrom K.L., Mahoney D.H., Shardy D.L. Mutations in signal recognition particle SRP54 cause syndromic neutropenia with Shwachman-Diamond-like features. J. Clin. Invest. 2017;127:4090–4103. doi: 10.1172/JCI92876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nambot S., Thevenon J., Kuentz P., Duffourd Y., Tisserant E., Bruel A.L., Mosca-Boidron A.L., Masurel-Paulet A., Lehalle D., Jean-Marçais N., Orphanomix Physicians’ Group Clinical whole-exome sequencing for the diagnosis of rare disorders with congenital anomalies and/or intellectual disability: substantial interest of prospective annual reanalysis. Genet. Med. 2018;20:645–654. doi: 10.1038/gim.2017.162. [DOI] [PubMed] [Google Scholar]
- 29.Ploski R., Pollak A., Müller S., Franaszczyk M., Michalak E., Kosinska J., Stawinski P., Spiewak M., Seggewiss H., Bilinska Z.T. Does p.Q247X in TRIM63 cause human hypertrophic cardiomyopathy? Circ. Res. 2014;114:e2–e5. doi: 10.1161/CIRCRESAHA.114.302662. [DOI] [PubMed] [Google Scholar]
- 30.Tanaka A.J., Cho M.T., Millan F., Juusola J., Retterer K., Joshi C., Niyazov D., Garnica A., Gratz E., Deardorff M. Mutations in SPATA5 are associated with microcephaly, intellectual disability, seizures, and hearing loss. Am. J. Hum. Genet. 2015;97:457–464. doi: 10.1016/j.ajhg.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Y., Muzny D.M., Reid J.G., Bainbridge M.N., Willis A., Ward P.A., Braxton A., Beuten J., Xia F., Niu Z. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N. Engl. J. Med. 2013;369:1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dosztányi Z., Csizmok V., Tompa P., Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 34.Lieutaud P., Canard B., Longhi S. MeDor: a metaserver for predicting protein disorder. BMC Genomics. 2008;9(Suppl 2):S25. doi: 10.1186/1471-2164-9-S2-S25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hnisz D., Shrinivas K., Young R.A., Chakraborty A.K., Sharp P.A. A phase separation model for transcriptional control. Cell. 2017;169:13–23. doi: 10.1016/j.cell.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yunus A.A., Lima C.D. Structure of the Siz/PIAS SUMO E3 ligase Siz1 and determinants required for SUMO modification of PCNA. Mol. Cell. 2009;35:669–682. doi: 10.1016/j.molcel.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beliakoff J., Sun Z. Zimp7 and Zimp10, two novel PIAS-like proteins, function as androgen receptor coregulators. Nucl. Recept. Signal. 2006;4:e017. doi: 10.1621/nrs.04017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Friedman M.J., Shah A.G., Fang Z.H., Ward E.G., Warren S.T., Li S., Li X.J. Polyglutamine domain modulates the TBP-TFIIB interaction: implications for its normal function and neurodegeneration. Nat. Neurosci. 2007;10:1519–1528. doi: 10.1038/nn2011. [DOI] [PubMed] [Google Scholar]
- 39.Kovar H. Dr. Jekyll and Mr. Hyde: the two faces of the FUS/EWS/TAF15 protein family. Sarcoma. 2011;2011:837474. doi: 10.1155/2011/837474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patel A., Lee H.O., Jawerth L., Maharana S., Jahnel M., Hein M.Y., Stoynov S., Mahamid J., Saha S., Franzmann T.M. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- 41.Chong S., Dugast-Darzacq C., Liu Z., Dong P., Dailey G.M., Cattoglio C., Heckert A., Banala S., Lavis L., Darzacq X., Tjian R. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science. 2018;361:eaar2555. doi: 10.1126/science.aar2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saillour Y., Broix L., Bruel-Jungerman E., Lebrun N., Muraca G., Rucci J., Poirier K., Belvindrah R., Francis F., Chelly J. Beta tubulin isoforms are not interchangeable for rescuing impaired radial migration due to Tubb3 knockdown. Hum. Mol. Genet. 2014;23:1516–1526. doi: 10.1093/hmg/ddt538. [DOI] [PubMed] [Google Scholar]
- 43.Noctor S.C., Martínez-Cerdeño V., Ivic L., Kriegstein A.R. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat. Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- 44.Beliakoff J., Lee J., Ueno H., Aiyer A., Weissman I.L., Barsh G.S., Cardiff R.D., Sun Z. The PIAS-like protein Zimp10 is essential for embryonic viability and proper vascular development. Mol. Cell. Biol. 2008;28:282–292. doi: 10.1128/MCB.00771-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu J.I., Lessard J., Olave I.A., Qiu Z., Ghosh A., Graef I.A., Crabtree G.R. Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron. 2007;56:94–108. doi: 10.1016/j.neuron.2007.08.021. [DOI] [PubMed] [Google Scholar]
- 46.Li X., Zhu C., Tu W.H., Yang N., Qin H., Sun Z. ZMIZ1 preferably enhances the transcriptional activity of androgen receptor with short polyglutamine tract. PLoS ONE. 2011;6:e25040. doi: 10.1371/journal.pone.0025040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.