Abstract
Primary defects in lung branching morphogenesis, resulting in neonatal lethal pulmonary hypoplasias, are incompletely understood. To elucidate the pathogenetics of human lung development, we studied a unique collection of samples obtained from deceased individuals with clinically and histopathologically diagnosed interstitial neonatal lung disorders: acinar dysplasia (n = 14), congenital alveolar dysplasia (n = 2), and other lethal lung hypoplasias (n = 10). We identified rare heterozygous copy-number variant deletions or single-nucleotide variants (SNVs) involving TBX4 (n = 8 and n = 2, respectively) or FGF10 (n = 2 and n = 2, respectively) in 16/26 (61%) individuals. In addition to TBX4, the overlapping ∼2 Mb recurrent and nonrecurrent deletions at 17q23.1q23.2 identified in seven individuals with lung hypoplasia also remove a lung-specific enhancer region. Individuals with coding variants involving either TBX4 or FGF10 also harbored at least one non-coding SNV in the predicted lung-specific enhancer region, which was absent in 13 control individuals with the overlapping deletions but without any structural lung anomalies. The occurrence of rare coding variants involving TBX4 or FGF10 with the putative hypomorphic non-coding SNVs implies a complex compound inheritance of these pulmonary hypoplasias. Moreover, they support the importance of TBX4-FGF10-FGFR2 epithelial-mesenchymal signaling in human lung organogenesis and help to explain the histopathological continuum observed in these rare lethal developmental disorders of the lung.
Keywords: 17q23.1q23.2 recurrent deletion, 5p12 deletion, aplasia of lacrimal and salivary glands, lacrimoauriculodentodigital (LAAD) syndrome, lung hypoplasia, neonatal lung disease, T-box transcription factor 4, fibroblast growth factor 10
Introduction
Diffuse developmental disorders of the lung comprise a group of rare primary defects in lung branching morphogenesis and vasculogenesis, including acinar dysplasia (AcDys), congenital alveolar dysplasia (CAD), and alveolar capillary dysplasia with misalignment of pulmonary veins (ACDMPV [MIM: 265380]) (Figure 1).1, 2 Diagnosis of these disorders has been based largely on their histopathological appearance at lung biopsy or autopsy, which demonstrate a spectrum of developmental arrest in lung growth and maturation. Further characterization of these idiopathic disorders has been hampered by their rarity and pleiotropic manifestations, as well as inconsistent use of disease definition and nomenclature.3, 4
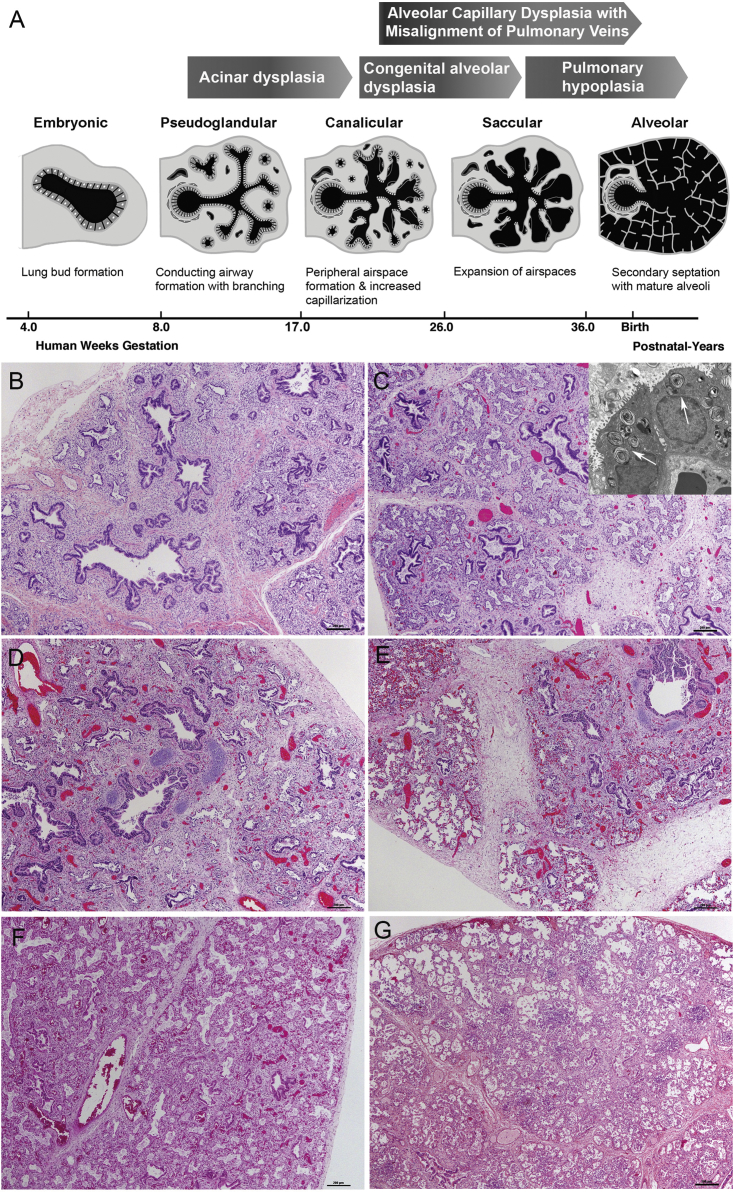
Figure 1.
Phases of Human Lung Development and Histopathological Characterization of the Lung Sections
(A) Schematic representation of phases of human lung development and stages of lung growth arrest in particular disorders (adapted from Kimura and Deutsch83).
(B–G) Histologic sections of autopsy lung. TBX4 mutations largely resemble the earlier stages of lung development when the majority of lung is composed of conducting airways (pseudoglandular stage).
(B) (P026) The distal acinar tubules are dilated and more complex with abundant intervening mesenchyme (canalicular stage).
(C) (P006) Despite the immature appearance, well-formed lamellar bodies were seen in a single case by electron microscopy: arrows denote lamellar bodies, original magnification 4,800×) and there was robust expression of surfactant related proteins (thyroid transcription factor 1, surfactant protein B, and pro surfactant protein C by immunostaining [n = 3], data not shown).
(D and E) Two case subjects (P025 depicted) showed a marked variation in histologic appearance with areas of acinar dysplasia (D) juxtaposed to more normal saccular spaces (E).
(F and G) Lungs from subjects with FGF10 mutations resemble later phases of development when distal airspaces are subdivided by secondary crests containing a double-walled capillary network (saccular stage), suggestive of congenital alveolar dysplasia in a term infant P042 (F), and mature alveoli are polygonal with thin interalveolar septa and a single capillary bed (alveolar stage). (G) (P076) More mature appearing lung architecture, but a reduced number of alveolar spaces, characteristic of pulmonary hypoplasia.
To date, only 18 subjects diagnosed with AcDys of different ethnic backgrounds have been reported; the mortality rate approaches 100% (Table S1).4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17 AcDys lungs show diffuse maldevelopment with bronchial and bronchiolar structures embedded in loose mesenchyme. Acinar structures, when present, are immature with no alveoli and limited formation of saccules.1, 4 The lungs are frequently small in size and have thickened interlobular septa. Based on these features, it has been hypothesized that AcDys reflects lung growth arrest in the pseudoglandular or early canalicular stage of lung development (Figure 1).1, 2
CAD is an even rarer condition with only a few cases reported to date.3, 16, 18, 19 Newborns with CAD are born at term and manifest with respiratory failure early in life; the mortality rate also approaches 100%. Compared to AcDys, CAD lungs contain easily identifiable distal acinar spaces, suggesting that lung growth arrest occurred at the late canalicular or early saccular stage of development (Figure 1).1 Whereas the lung weight is usually normal or even increased from congestion, the architecture is notably immature for age with simplified acini and abundant intervening mesenchyme and without well-formed alveoli. The histologic appearance is similar to the lobular maldevelopment often seen in ACDMPV, but vein misalignment and marked hypertensive changes of the pulmonary arteries are absent. Due to the spectrum of immaturity in CAD, the diagnosis cannot be made with certainty in premature infants or those with suspected pulmonary hypoplasia.
In contrast to ACDMPV caused by loss-of-function (LoF) of FOXF1 (MIM: 601089),20, 21 the molecular etiology of AcDys and CAD is largely unknown. However, we have identified a de novo heterozygous missense TBX4 (MIM: 601719, GenBank: NM_018488.3) variant c.256G>C (p.Glu86Gln) in a newborn with AcDys,6 and most recently, a de novo 4 base deletion (c.524_527del [p.Asn175Thrfs∗52]) in TBX4 and a 2.2 Mb deletion at 17q23.1q23.2 encompassing TBX4 have been reported in infants with CAD and alveolar growth abnormality, respectively.19 Moreover, a homozygous missense FGFR2 (MIM: 176943, GenBank: NM_000141.4) variant c.764G>A (p.Arg255Gln) has been described in an individual with AcDys and ectrodactyly.5 Recurrence of disease and reported consanguinity in some of the pedigrees have suggested an autosomal-recessive pattern of inheritance.4, 5
Here, we report the clinical, histopathological, and molecular findings in 26 deceased individuals with a spectrum of AcDys, CAD, and other rare lethal pulmonary hypoplasia.
Subjects and Methods
Subjects
In total, 26 individuals with AcDys spectrum (n = 14), CAD (n = 2), or other rare lung hypoplasia (n = 10) and 17 of their family members were recruited following informed consent (Table S2). Control individuals with 17q23.1q23.2 (n = 13), 5p12 (n = 3), or an intragenic TBX4 (n = 1) deletion but without developmental lung disease and healthy parents of one control individual were ascertained from the Baylor Genetics database of 25,550 reported copy-number variants (CNVs) from 19,537 subjects referred for clinical array comparative genomic hybridization (aCGH), the Database of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources (DECIPHER),22 and from our collaborators. The study protocol was approved by the Institutional Review Board for Human Subject Research at Baylor College of Medicine (BCM; H36612, H42409, H42680).
Histopathological Evaluation
Histopathological evaluation was performed using hematoxylin-eosin-stained slides from formalin-fixed paraffin-embedded (FFPE) lung obtained during autopsy and/or biopsy.
DNA and RNA Extraction
Genomic DNA was extracted from peripheral blood, saliva, skin, FFPE lung or liver, or frozen lung using Gentra Puregene Blood Kit (QIAGEN), DNeasy Blood & Tissue Kit (QIAGEN), or standard proteinase K/phenol-chloroform extraction-based protocol.23 Total RNA was extracted from frozen lung using the miRNeasy Mini Kit (QIAGEN).
Chromosomal Microarray Analyses
aCGH was performed using GenomeDx v5 custom designed array (GeneDx) (P006), Agilent Sureprint C3Hmn 400K array (P019), or 60K Agilent array (the ISCA v.2 design) (P003, P009, P012, P015/16, P046, and P073) (Agilent Technologies). CNVs in individuals P026, P033, P035, P038, C039, P042–045, and P048 were analyzed using customized high-resolution 180K microarrays (Agilent Technologies) with probes targeting genes involved in lung development. SNP microarray analyses of subjects P006, P009, P012, P019, P022, P025, and P026 and control individuals C051–055, C058, and C059 were performed using Affymetrix CytoScan HD array containing 750,000 genotype-able SNPs (Applied Biosystems).
PCR and Sanger Sequencing
Deletion junctions were amplified with LA Taq DNA polymerase (TaKaRa Bio) using two-step long range PCR in a final volume of 25 μL. PCR conditions included 30 cycles of 98°C for 10 s and 68°C for 60–420 s. Primers for long-range PCR were design using the Primer3 software. PCR products were treated using FastAP Thermosensitive Alkaline Phosphatase and Exonuclease I (Thermo Scientific). The amplicons were directly sequenced using BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). The sequence data were analyzed using Sequencher 5.4.6 software (Gene Codes Corporation).
Parental Origin of CNVs and SNVs
Parental origin of the identified deletions was determined using informative single-nucleotide variants (SNVs). To determine the parental origin of the TBX4 point mutations, amplicons containing the SNV of interest and the neighboring informative marker were cloned into the pGEM-T vector (Promega) and transformed into Escherichia coli strain DH5α competent cells (Invitrogen). Ten clones for each construct were used for plasmid isolation using the PureLink Quick Plasmid Miniprep Kit (Invitrogen) and Sanger sequenced.
Real-Time Quantitative PCR Analysis
RNA extracted from frozen lung obtained at autopsy from affected subject with 17q23.1q23.2 deletion (P035) was reverse-transcribed using SuperScript III First-Strand Synthesis System (Invitrogen). TBX2 (MIM: 600747) and TBX4 transcript levels were normalized to GAPDH (MIM: 138400) and ACTB (MIM: 102630). qPCR was repeated three times using TaqMan probes and TaqMan Universal PCR Master Mix (Applied Biosystems). qPCR conditions included 40 cycles of 95°C for 15 s and 60°C for 1 min. For relative quantification of the studied transcripts, the comparative CT method was used. Normal fetal lung was designated as a calibrator sample.
Exome Sequencing (ES)
Fifteen subjects with hypoplastic lungs (P003, P009, P012, P015, P025–028, P033, P042–046, and P048) were analyzed by ES. ES in individuals P003 and P025–028 was performed at BCM Human Genome Sequencing Center (BCM-HGSC) through the Baylor Hopkins Center for Mendelian Genomics (BHCMG) initiative, according to previously described protocol.24 ES in subject P033 was analyzed at GeneDx; in subjects P009, P012, P042, and P044 at Oxford Gene Technology using SureSelect XT Human all exon V5; and in individuals P015, P043, and P045–P048 at Institut du cerveau et de la moelle épinière, Hôpital de la Pitié Saléptrière using Medexome Nimblegen 47 Mb (Roche NimbleGen) followed by Illumina sequencing (Illumina). Sequence variants obtained for each individual were filtered in a stepwise manner to exclude synonymous or non-exonic SNV/indels and variants with minor allele frequency (MAF) > 1% in the Exome Variant Server, the 1000 Genomes Project, and in our internal exome database. Variants predicted as neutral by MutationTaster and PolyPhen-2 tools or variants with a negative conservation scores in PhyloP analysis were parsed and filtered out.
Whole-Genome Sequencing (WGS)
WGS was performed in 26 deceased subjects with lung disease and in 13 control subjects without any severe lung phenotype. Libraries were prepared with a TruSeq Nano DNA HT Library Prep Kit (Illumina) according to the manufacturer’s protocol, followed by sequencing on the HiSeqX platform (Illumina) at CloudHealth Genomics. The raw sequencing data were processed according to the specification of bcl2fastq package from Illumina. Short reads obtained during sequencing were processed using Trimmomatic25 to remove adapter sequences. Data were aligned and mapped to the human genome reference sequence (hg38) using the BWA 0.7.12 tool.26 Variants were called using the GATK 3.7 software.27 The genome annotations were converted to the GRCh37/hg19 human genome reference sequence.
Bioinformatic Analyses
Reference DNA sequences, coordinates of regulatory elements, transcription factor binding sites, long non-coding RNAs (lncRNAs), structural variants, conservation, and ChIP-seq data for IMR-90 and NHLF cell lines were accessed using the UCSC Genome Browser (GRCh37/hg19) and Roadmap. The eQTL variants were analyzed using the GTEx Portal. NMDEscPredictor was used to predict the effect of the premature termination codon.28 The pLI scores and MAFs were obtained from the ExAC and gnomAD (r2.0.2) databases, respectively. Protein structures were analyzed using Phyre2 bioinformatic tool and Swiss-Model. The chromatin interaction data were visualized using the 3D Genome Browser.
Variant Enrichment Analysis
To verify the enrichment of non-coding variants within and upstream to TBX4, we selected variants carried by at least two individuals with lung disease and 17q23.1q23.2 deletion (P006, P009, P012, P019, P026, P035, and P073) which were absent in 13 control individuals with the same deletion but without any structural lung abnormalities. Upon further consideration, we excluded the most common variants (MAF > 10%, gnomAD r2.0.2). To test whether there is an excess of selected variants in a given region A, we used a Monte Carlo approach. We estimated the empirical distribution of the number of variants selected in the previous step that fall into randomly selected genomic intervals of the fixed size (equal to the size of region A) sampled from the 17q23.1q23.2 deletion region. p value was calculated by dividing the number of intervals containing the same number or more variants than in the region A by the total number of sampled intervals. Haplotypes were analyzed using LDlink. Probability of distribution of SNPs rs35827636 and rs192153557 (frequencies 7.9% and 3.5%, respectively) which were observed in four (P006, P012, P022, P035) and two (P009, P035) subjects, respectively, and absent in the control individuals was calculated using a formula: [0.921ˆ13∗ 0.079ˆ4 ∗0.921ˆ5 ∗ factorial (9) / (factorial (4)∗ factorial (5)].
Results
Clinical and Histopathological Findings
A total of 26 deceased individuals from 23 unrelated families with a lethal developmental lung disorder were enrolled into the study (Table S2, Supplemental Note). Pregnancy histories were predominantly uneventful except for intrauterine growth restriction in 4/26 (15%) subjects. Lung hypoplasia was detected prenatally in 4/26 (15%) case subjects and resulted in voluntary medical termination of two pregnancies. The remaining children were born at term (>37 weeks), except six individuals born between 32 and 36 weeks. Lifespans ranged between a few minutes and 10 weeks. Recurrence in siblings was observed in four families, and consanguinity was reported in one family.
Twenty-three subjects had autopsy lung available for review by one pathologist (G.D.). Two of these subjects also had a surgical biopsy prior to demise that showed similar features to the subsequent lung histology at autopsy. In all cases in which lung weight/body weight was documented (n = 19), criteria for pulmonary hypoplasia were met.29 Evaluation of lung sections revealed a variable degree of abnormal lung development, ranging from AcDys, to CAD, to pulmonary hypoplasia. In two case subjects, the degree of abnormal lung development could not be determined due to early gestational age (Table S2).
CNV Deletions on 17q23 and 5p12
For CNV analyses, we applied aCGH and WGS. A heterozygous recurrent ∼2.2 Mb CNV deletion on 17q23.1q23.2, involving TBX2 and TBX4 and de novo heterozygous nonrecurrent ∼2.12 Mb CNV deletion on 17q23.2q23.3, also involving TBX2 and TBX4, were found in six (P006, P009, P012, P019, P026, and P073) and one (P035) affected individuals, respectively (Figure 2, Tables 1 and S3). In two siblings, P015 with CAD and P016 with AcDys spectrum, we identified a small ∼8.6 kb heterozygous intragenic frameshifting deletion, involving exons 4 and 5 of TBX4 (Figure 2B), inherited from their healthy mother. In one subject (P038), an ∼10.45 kb heterozygous CNV deletion on 17q23.2, involving a portion of intron six of BCAS3 (MIM: 607470) (Figure 2), inherited from the apparently healthy father was detected (Tables 1 and S3). This small deletion was found in two individuals in the 1000 Genomes database, suggesting that it may be a nonpathogenic polymorphism. Moreover, in two unrelated families, overlapping heterozygous deletions at 5p12 (∼2.18 Mb and ∼2.32 Mb in size) including FGF10 were identified. In both cases, deletions were inherited from a parent presenting with lacrimoauriculodentodigital (LADD) syndrome (MIM: 149730) (Figure 3, Tables 1 and S3).
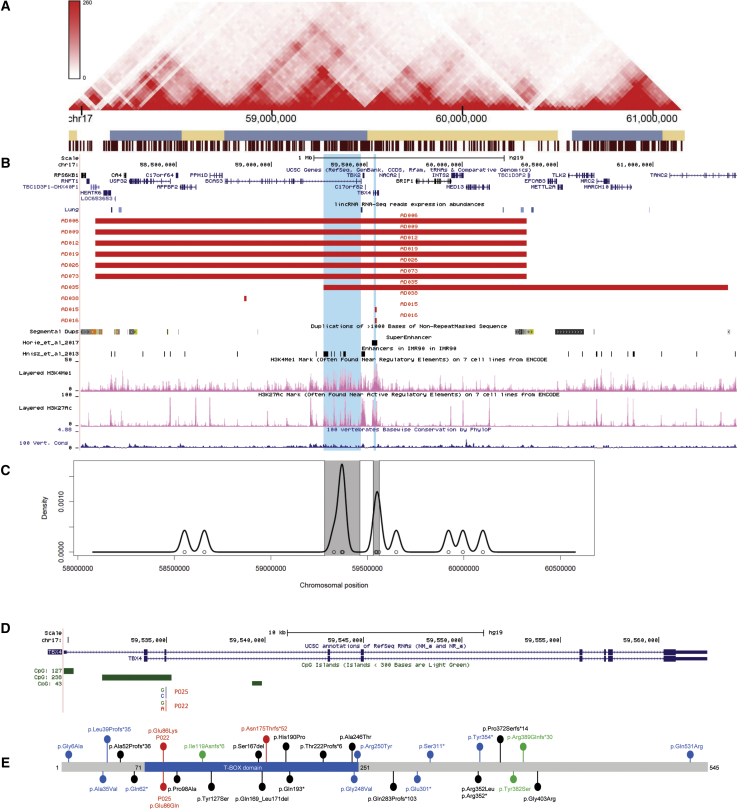
Figure 2.
Schematic Representations of SNVs and CNVs Involving TBX4
(A) Topologically associating domains (TADs) detected in fetal lung fibroblasts at 17q23.1q23.2
(B) The 17q23.1q23.2 region depicting deletions identified in nine subjects with pulmonary hypoplasia (red bars) overlapping the enhancers identified in IMR-90 cell line84 or the super-enhancer in lung fibroblasts32 (black bars). Complex LCRs flanking the recurrent 17q23.1q23.2 deletions are shown.58 H3KMe1 and H3KMe3 marks in the fetal lung, conservation scores, and lncRNAs are shown below deletion track. Regions enriched in non-coding variants are highlighted in blue.
(C) Distribution of variants in the 17q23.1q23.2 deletion region showing SNV enrichment (variants with MAF < 10% shared by at least two affected subjects with 17q23.1q23.2 deletion and two affected subjects with de novo TBX4 missense variant and absent in 13 control individuals with the same deletion but without lung abnormalities).
(D) The TBX4 gene and variants identified in two subjects mapping in CpG island.
(E) The TBX4 protein showing T-box domain (blue). Missense mutations and 4 bp deletion identified in three unrelated subjects with lung hypoplasia (red). Previously reported variants identified in individuals with pulmonary hypertension (PAH), ischiocoxopodopatellar syndrome, or PAH with coexisting ischiocoxopodopatellar syndrome (black, blue, and green, respectively).6, 54, 55, 56, 57, 58, 59
Table 1.
Genetic Findings in Studied Individuals with Lung Hypoplasia
| Subject | Diagnosis | Deletion CNV Coordinates (hg19) | Repetitive Element at the Breakpoints | SNV | WGS | ES | aCGH |
|---|---|---|---|---|---|---|---|
| 17q23.1q23.2 Deletions Involving Entire TBX4 | |||||||
| P006 | AcDys | chr17:58,089,454/58,090,137–60,346,028/60,346,711 | LCR/LCR | − | x | x | x |
| P009 | AcDys | chr17:58,090,283/58,090,656–60,346,857/60,347,230 | LCR/LCR | − | x | x | x |
| P012 | AcDys | chr17:58,088,933/58,089,453–60,345,508/60,346,028 | LCR/LCR | − | x | x | x |
| P019 | NA | ∼chr17:58,167,485–60,174,066 | LCR/LCR | − | x | − | x |
| P026 | AcDys | chr17:58,088,933/58,089,453–60,345,508/60,346,028 | LCR/LCR | BCLAF1 (NM_001077440.1); c.1615G>A (p.Asp539Asn) | x | x | x |
| P073 | NA | chr17:58,086,876/58,087,936–60,343,456/60,344,516 | LCR/LCR | − | x | − | x |
| P035 | AcDys | chr17:59,272,842/59,272,846–61,392,993/61,392,997 | AluJb/- | − | x | − | x |
| TBX4 Intragenic Deletion at 17q23.2 | |||||||
| P015/P016 | CAD/AcDys Spectrum | chr17:59,542,891/59,542,894–59,551,500/59,551,503 | −/− | TBX5 (NM_000192.3); c.331G>T (p.Asp111Tyr) | x | x | x |
| TBX4 Point Mutations | |||||||
| P022 | AcDys | NA | NA | TBX4 (NM_018488.3); c.256G>A (p.Glu86Lys) | x | − | − |
| P025 | marked variation with AcDys ranging to near normal | NA | NA | TBX4 (NM_018488.3); c.256G>C (p.Glu86Gln) | x | x | − |
| 17q23 Deletions Involving BCAS3 | |||||||
| P038 | AcDys | chr17:58,857,889/58,857,898–58,868,328/58,868,337 | AluSx1/AluSx | − | x | − | x |
| 5p12 Deletions Involving FGF10 | |||||||
| P040/P041 | pulmonary hypoplasia/CAD versus pulmonary hypoplasia | chr5:43,957,152/43,957,220–46,135,141/46,135,209 | L1PA4/L1PA4 | − | x | − | − |
| P076 | pulmonary hypoplasia | chr5:42,985,023−45,244,787 | −/L1PA15 | − | x | − | − |
| FGF10 Mutations | |||||||
| P033 | AcDys | NA | NA |
FGF10 (NM_004465.1); c.526delA (p.Met176Cysfs∗5) STRA6 (NM_001142617.1); c.653T>C (p.Phe218Ser) |
x | x | x |
| P042 | CAD | NA | NA |
FGF10 (NM_004465.1); c.577C>T (p.Arg193∗) FRAS1 (NM_025074.6); c.10245G>C (p.Gln3415His) |
x | x | x |
| Other Mutations | |||||||
| P003 | marked variation with AcDys ranging to near normal | NA | NA |
BTBD7 (NM_018167.4); c.1075G>A (p.Ala359Thr) FRAS1 (NM_025074.6); c.4648C>T (p.Leu1550Phe); c.7039C>T (p.Val2347Phe) |
x | x | x |
| P027 | AcDys | NA | NA | FRAS1 (NM_025074.6); c.7451G>T (p.Thr2484Met) | x | x | − |
| P028 | AcDys | NA | NA | DSPP (NM_014208.3); c.3660_3661insATCT (p.Asp1221Ilefs∗2); c.3734_3742delGACAGCAGCA (p.Asn1248_Ser1250del) | x | x | − |
| P046 | AcDys | NA | NA | TCF21 (NM_003206.3); c.329C>T (p.Pro110Leu) | x | x | x |
Abbreviations are as follows: −, absent; aCGH, array comparative genomic hybridization; AcDys, acinar dysplasia; CAD, congenital alveolar dysplasia; CNV, copy number variant; ES, exome sequencing; LCR, low-copy repeats; SNV, single-nucleotide variant; WGS, whole-genome sequencing; LCR, low-copy repeats; NA, not applicable.
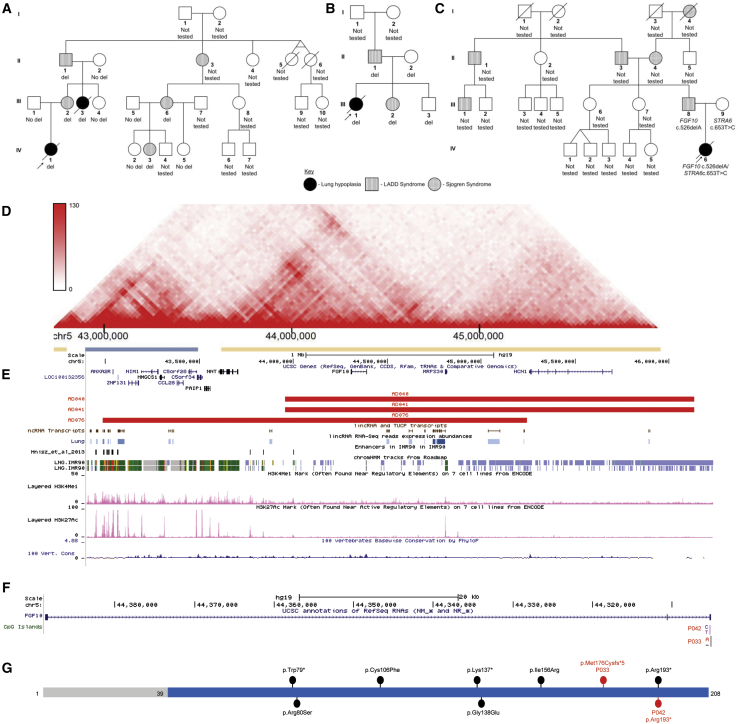
Figure 3.
Schematic Representations of SNVs and CNVs Involving FGF10
(A–C) Pedigrees of families with 5p12 CNV deletions (A) (P040/P041), (B) (P076), and SNV (C) (P033) involving FGF10 are shown.
(D) Topologically associating domains (TADs) detected in fetal lung fibroblasts in the region of 5p12 deletion.
(E) The 5p12 genomic region depicting CNV deletions identified in three individuals from two unrelated families with pulmonary hypoplasia (red bars) overlapping the enhancers identified in IMR-90 cell line.84 H3KMe1 and H3KMe3 marks in the human lung, chromatin state annotation based on ChIP-seq mapping (Roadmap) in the IMR-90 cell line, conservation scores (PhyloP) and lncRNAs are shown below deletion track.
(F) The FGF10 gene and variants identified in two subjects with lung hypoplasia.
(G) The FGF10 protein showing FGF domain (blue). Variants identified in two AcDys subjects are indicated in red. Previously reported variants identified in individuals with LADD syndrome or aplasia of lacrimal and salivary glands (ALSG) are shown in black.49, 62, 63, 64, 65
The recurrent 17q23.1q23.2 deletions flanked by large complex low-copy repeats (LCRs) were likely mediated by nonallelic homologous recombination (NAHR). Using long-range PCR with primers flanking the directly oriented paralogous subunit pairs, we narrowed the predicted NAHR junctions to an ∼15 kb subunit (core duplicon; chr17:58,083,346–58,098,450/chr17:60,339,929–60,355,017) responsible for genomic instability on chromosome 1730 (Figure 2, Tables 1 and S3).
The mutational signatures and features of the sequenced breakpoints of four nonrecurrent CNV deletions are consistent with being derived by a microhomology-mediated break induced replication (MMBIR) mechanism (Tables 1 and S3).31
Identification of SNVs in the Coding Regions of TBX4, FGF10, and Other Genes Involved in Lung Development
We further examined SNVs in the coding portions of the candidate genes involved in lung development. Analysis of TBX4 (GenBank: NM_018488.3) on 17q23.2 revealed a de novo missense variant c.256G>A (p.Glu86Lys) at a CpG site (subject P022) (Figure 2) which is predicted to invert the polarity of amino acids from negative to positive and might affect the stabilization of the hydrophobic protein core close to the active site, compromising binding ability of TBX4 (Figure S1). A de novo missense variant at the same nucleotide position (c.256G>C) but resulting in a different amino acid substitution (p.Glu86Gln) was previously reported in subject P025 (Figure 2).6
In FGF10 (GenBank: NM_004465.1) on 5p12, two variants were identified: a heterozygous nonsense variant c.577C>T (p.Arg193∗) of unknown parental origin (P042) and a heterozygous frameshift deletion c.526delA (p.Met176Cysfs∗5) (P033, IV-6 in Figure 3C) inherited from the father with LADD syndrome (Figure 3, Tables 1, S3, and S4). This paternally inherited frameshift variant is predicted to escape nonsense-mediated decay.28
In siblings P015 and P016, in addition to the intragenic frameshifting deletion in TBX4 inherited from the healthy mother (Figure 2), a rare heterozygous c.331G>T (p.Asp111Tyr) variant in TBX5 (MIM: 601620; GenBank: NM_000192.3) inherited from the healthy father was identified (Figure S1, Tables 1 and S4). In five other subjects (P003, P026, P027, P028, and P048), deleterious SNVs were identified in TCF21 (MIM: 603306), BTBD7 (MIM: 610386), DSPP (MIM: 125485), and BCLAF1 (MIM: 612588) (Tables 1 and S4). Absence of heterozygosity (AOH) analyses revealed that one subject (P048) was from a consanguineous family, confirming clinical findings (Tables S2 and S5).
Parental Origin of 17q23.1q23.2 Deletion CNVs and TBX4 SNVs
To determine whether the abnormal phenotypes of individuals with the recurrent 17q23.1q23.2 CNV deletion result from the parent-of-origin effect, we investigated their origin in an individual with lung disease (P006) and a control subject without any reported lung anomalies (C051). The analyses showed that both CNVs arose de novo on maternal chromosome 17, arguing against genomic imprinting at this locus. In agreement, de novo missense variants in TBX4 in affected subjects P022 and P025 occurred on maternal and paternal chromosome 17, respectively.
TBX2 and TBX4 Expression
To investigate the influence of the 17q23.1q23.2 CNV deletion on TBX2 and TBX4 expression, we applied quantitative PCR. Analysis of TBX2 and TBX4 mRNA extracted from the frozen lung in a subject with 17q23.1q23.2 deletion (P035) showed an 11.9-fold change lower expression of TBX2 and 7.7-fold change lower expression of TBX4, when compared to control lung (Figure S2).
Enrichment of the Non-coding Variants in the 17q23.1q23.2 Locus
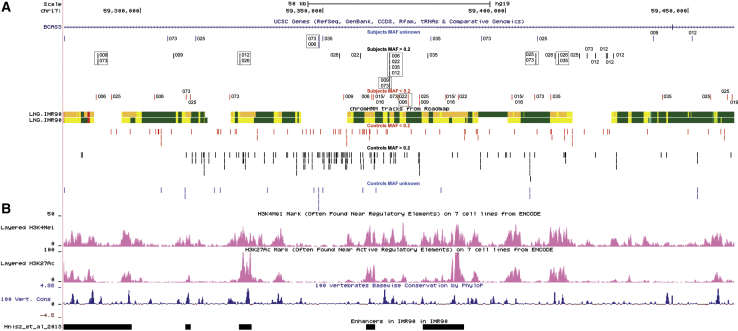
Given the phenotypic differences between subjects and control individuals carrying the TBX4 and FGF10 null alleles, we hypothesized that additional genetic modifiers are required to cause severe lung disease. To this aim, we first performed SNP microarray analyses of DNA from two subjects with the TBX4 missense variant (P022 and P025), five affected individuals with the recurrent 17q23.1q23.2 deletion (P006, P009, P012, P019, and P026), and five control subjects with the same deletion but without any structural lung anomalies (C051, C054, C055, C058, and C059). Results of these studies showed enrichment of non-coding variants mapping within and upstream to TBX4 in individuals with lung abnormalities (Table S6, Figure S3). These data were validated using WGS analyses in the larger group of affected and control individuals. Interestingly, we observed enrichment of non-coding variants mapping within (chr17:59,457,361–59,562,471, p = 0.0598) and upstream to (chr17:59,279,024–59,462,062, p = 0.0169) TBX4, shared by at least two affected individuals with the 17q23.1q23.2 CNV deletion (P006, P009, P012, P019, P026, P035, and P073) or TBX4 missense variant (P022, P025) and absent in 13 control individuals with the same deletion (C051, C052, C054, C055, C058–65, C072) (Figures 2, 4, S4, and S5, Table 2). The above regions overlap the predicted regulatory elements identified in human fetal lung fibroblasts (IMR-90) (Figures 2 and 4), including a lung-specific super-enhancer.32 In fetal lung fibroblasts, they are located in the same topologically associating domain (TAD), but in two different subdomains33 (Figure 2).
Figure 4.
Lung-Specific Enhancer Region Located Upstream to TBX2 and TBX4
(A) Chromatin state annotation based on ChIP-seq mapping (Roadmap) in the IMR-90 cell line within the chr17:59,279,024–59,462,062 genomic region. SNVs identified in subjects are presented in the top of chromatin state annotation scheme, while SNVs identified in controls are shown below this track. SNVs with gnomAD (r2.0.2) MAF ≥ 0.2 are shown in red; SNVs with MAF > 0.2 are shown in black, and SNVs with unknown MAF are shown in blue. The variants identified in more than one individual with lung disease are indicated by black dashed rectangles.
(B) H3KMe1 and H3KMe3 marks in the IMR-90 cell line and fetal lung, conservation scores (PhyloP), and the enhancers identified in IMR-90 cell line within the chr17:59,279,024–59,462,062 genomic region.84
To investigate the possibility of common SNVs contributing to the lung phenotype, we performed haplotype analyses in seven individuals with TBX4 deletion CNVs and found different-sized haplotype blocks in all of them (Figure S6). Analysis of the enriched SNVs mapping in the region upstream to TBX4 revealed two very closely located (113 bp apart) SNPs—rs35827636 and rs192153557 (population frequency 7.9% and 3.5%, respectively)—in the last intron of BCAS3. These SNVs are present in 4 (P006, P012, P022, P035) and 2 (P009, P035) subjects, respectively, and absent in 13 control individuals (Figure 4, Table 2). The probability of such distribution is 0.001115712. Analysis of the region within TBX4 revealed a block of six non-coding SNVs (Figure S7), which was observed in full (n = 5) or partially (n = 3) in subjects with coding TBX4 CNVs or SNVs (Table S7). However, these six SNVs were also found in two control subjects (C060 and C061) with the 17q23.1q23.2 CNV deletion, but without any lung abnormalities, making this haplotype unlikely to contribute to the lethal lung phenotype (Table S7).
Table 2.
Non-coding SNVs Identified in Affected Individuals with Heterozygous Coding CNVs and Point Mutations Involving TBX4 Absent in the Control Individuals with 17q23.1q23.2 Deletion
| Position [hg19] | rsa | Ref | Alt | MAFb | P006 | P009 | P012 | P015/016 | P019 | P022 | P025 | P026 | P035 | P073 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| chr17:59279120–59279120 | NA | C | CTT | NA | − | − | − | − | − | − | − | − | − | + |
| chr17:59287811–59287811 | 145662401 | G | T | 0.0039 | + | − | − | − | − | − | − | − | − | − |
| chr17:59288406–59288406 | 8070692 | T | G | 0.2246 | − | + | − | − | − | − | − | − | − | + |
| chr17:59292085–59292085 | 117188060 | C | A | 0.0063 | − | − | − | − | − | − | + | − | − | − |
| chr17:59303786–59303786 | 139983813 | G | A | 0.0051 | + | − | − | − | − | − | − | − | − | − |
| chr17:59307503–59307503 | NA | T | TACAC | NA | − | − | − | − | − | − | − | − | − | + |
| chr17:59309085–59309085 | 72832589 | T | C | 0.0810 | − | + | − | − | − | − | − | − | − | − |
| chr17:59312457–59312457 | 138660616 | G | A | 0.0106 | − | − | − | − | − | − | − | − | − | + |
| chr17:59313654–59313654 | 150043642 | T | C | 0.0003 | − | − | − | − | − | − | + | − | − | − |
| chr17:59315155–59315155 | 940861097 | A | G | N/A | − | − | − | − | − | + | − | − | − | − |
| chr17:59324435–59324435 | 753135645 | C | T | 0.0002 | − | − | − | − | − | − | − | − | − | + |
| chr17:59327165–59327165 | 35636245 | G | GA | 0.0407 | − | − | + | − | − | − | − | + | − | − |
| chr17:59348785–59348785 | NA | A | ATTTTTTTTTTTTTTT | NA | + | − | − | − | − | − | − | − | − | + |
| chr17:59349997–59349997 | NA | C | CAAAA | NA | − | − | − | − | − | − | − | − | + | − |
| chr17:59354561–59354561 | 75380888 | T | C | 0.0248 | − | − | − | − | − | − | − | + | − | − |
| chr17:59355734–59355734 | 567208829 | G | A | 0.0027 | − | + | − | − | − | − | − | − | − | − |
| chr17:59360179–59360179 | 146403465 | T | C | 0.0217 | − | − | − | − | − | + | − | − | − | − |
| chr17:59363288–59363288 | 117484839 | C | T | 0.0063 | + | − | − | − | − | − | − | − | − | − |
| chr17:59363880–59363880 | 117798644 | G | A | 0.0072 | − | − | − | + | − | − | − | − | − | − |
| chr17:59368180–59368180 | 35827636 | T | C | 0.0793 | + | − | + | − | − | + | − | − | + | − |
| chr17:59368293–59368293 | 192153557 | C | A | 0.0347 | − | + | − | − | − | − | − | − | − | + |
| chr17:59370539–59370539 | 112164816 | A | T | 0.0102 | − | − | − | − | − | − | − | − | − | + |
| chr17:59373345–59373345 | 148383088 | C | T | 0.0194 | + | − | − | − | − | + | − | − | − | − |
| chr17:59376344–59376344 | 139134582 | C | T | 0.0020 | − | − | − | − | − | − | + | − | − | − |
| chr17:59376380–59376380 | 561102192 | C | T | 0.0003 | − | + | − | − | − | − | − | − | − | − |
| chr17:59378757–59378757 | 34867966 | G | A | 0.1443 | − | − | − | − | − | − | − | − | + | − |
| chr17:59379480–59379480 | NA | T | C | NA | − | − | − | − | − | − | − | − | + | − |
| chr17:59383687–59383687 | 117259668 | G | A | 0.0107 | − | − | − | + | − | − | − | − | − | − |
| chr17:59387086–59387086 | 973627683 | G | A | 0.0001 | − | − | − | − | − | + | − | − | − | − |
| chr17:59393463–59393463 | NA | C | T | NA | − | − | − | − | − | − | − | − | − | + |
| chr17:59401781–59401781 | 117993484 | G | A | 0.0076 | − | − | − | + | − | − | − | − | − | − |
| chr17:59408027–59408027 | 113520216 | C | T | 0.0102 | − | − | − | − | − | − | − | − | − | + |
| chr17:59408341–59408341 | 3785850 | G | A | 0.1219 | − | − | − | − | − | − | + | − | − | + |
| chr17:59408765–59408765 | 190888982 | G | C | NA | − | − | − | − | − | − | + | − | − | − |
| chr17:59412341–59412341 | 117088470 | C | T | 0.0069 | − | − | − | − | − | − | − | − | + | − |
| chr17:59413482–59413482 | 7224107 | C | T | 0.1016 | − | − | − | − | − | − | − | + | − | − |
| chr17:59414473–59414473 | 566255513 | C | CAA | 0.1022 | − | − | − | − | − | − | − | + | + | − |
| chr17:59420152–59420152 | 35383405 | G | T | 0.1169 | − | − | − | − | − | − | + | − | − | − |
| chr17:59422277–59422277 | 143541906 | T | TAC | 0.0937 | − | − | − | − | − | − | − | − | − | + |
| chr17:59424604–59424604 | 143968095 | G | A | 0.0662 | − | − | + | − | − | − | − | − | − | − |
| chr17:59427643–59427643 | 75073226 | G | A | 0.1128 | − | − | + | − | − | − | − | − | − | − |
| chr17:59427829–59427829 | 116271272 | G | A | 0.1074 | − | − | + | − | − | − | − | − | − | − |
| chr17:59429503–59429503 | 79390380 | G | A | 0.0741 | − | − | + | − | − | − | − | − | − | − |
| chr17:59440490–59440490 | 918478913 | G | A | NA | − | + | − | − | − | − | − | − | − | − |
| chr17:59442994–59442994 | 116842887 | C | T | 0.0078 | − | − | − | − | − | − | − | − | + | − |
| chr17:59451090–59451090 | NA | G | GCCCCC | NA | − | − | + | − | − | − | − | − | − | − |
| chr17:59456218–59456218 | 80207525 | C | T | 0.0019 | − | − | − | − | − | − | + | − | − | − |
| chr17:59460811–59460811 | 188999860 | G | C | 0.0001 | − | − | − | − | − | − | + | − | − | − |
| chr17:59462062–59462062 | 117518238 | C | T | 0.0180 | − | − | − | − | + | − | − | − | − | − |
Abbreviations are as follows: +, present; −, absent; Alt, altered allele; MAF, minor allele frequency; NA, not applicable; Ref, reference allele.
rs numbers based on dbSNP v.150
MAF based on the GnomAD database (r2.0.2)
Comparison of the 5p12 region in affected members of two unrelated families with overlapping FGF10 deletions (Figure 3) and in the control individuals carrying differently sized FGF10 deletions but without structural lung anomalies revealed no significant variants on the non-deleted alleles. In one family, WGS showed 21 non-coding SNVs located on the remaining allele, shared by two individuals with lung hypoplasia (P040 and P041, IV-1 and III-3 in Figure 3A, respectively) and absent in the individual with the same 5p12 CNV deletion and LADD syndrome (C039, III-2 in Figure 3A) (Table S8). In the second family with the overlapping 5p12 CNV deletion, none of these variants were found in subject P076 (III-1 in Figure 3B), her father with LADD syndrome (C074, II-1 in Figure 3B), or her sister (C077, III-2 in Figure 3B), also with LADD syndrome (Figure 3). Importantly, while subject P041 (III-3 in Figure 3A) and her sister without lung abnormalities (C039, III-2 in Figure 3A) inherited the alternative 5p12 alleles from their healthy mother, in the other family both affected and healthy children with the deletion inherited the same allele from their mother. With the exception of breast cancer,34 no lung-specific enhancer has been predicted in the 5p12 deleted region.35 Thus, in these patients we elected to study the 17q23.1q23.2 region to search for potential variants that could contribute to the abnormal lung phenotype. Notably, analysis of the predicted lung-specific enhancer region, located upstream to TBX4 in affected individuals with FGF10 SNVs or CNVs deletion, revealed the presence of rare non-coding variants that were absent in the control 17q23.1q23.2 deletion samples (Figure S8).
Analysis of the lung-specific expression quantitative trait loci (eQTLs) SNVs mapping within the deleted regions at 17q23.1q23.2 and 5p12 revealed no specific haplotype (Table S9).
Discussion
In contrast to other developmental anomalies such as congenital heart defects associated with hundreds of genes in numerous syndromic and non-syndromic disorders,36 only a few genes have been implicated as contributing to developmental lung diseases.1, 37, 38 These genes include four T-box genes (TBX2, TBX3, TBX4, and TBX5) and FGF10.39, 40, 41, 42, 43
The T-box protein family encodes transcription factors characterized by a conserved DNA-binding motif (T-box domain). TBX3 (MIM: 601621) and TBX5 on chromosome 12q24.21 as well as TBX2 and TBX4 on 17q23.2 are closely localized gene sets that are products of evolutionary gene duplications.44 While in vitro depletion of Tbx4 in murine lung organ cultures results in reduction of lung branching, simultaneous depletion of Tbx4 and Tbx5 completely inhibits formation of new lung branches.41 Similar results have been obtained in vivo, suggesting that regulation of lung branching is mediated by interactions between these T-box genes.39 Tbx2-deficient mice also have hypoplastic lungs, indicating that Tbx2 is one of the key members of the network regulating mouse lung organogenesis.40
In addition to T-box genes, mesenchyme-expressed FGF10 is required for lung branch formation.45, 46 In the developing lung, FGF10 is regulated by SHH epithelial mesenchymal signaling and is dependent on its own receptor FGFR2.45, 47 Animal studies have demonstrated that decreased expression of Tbx4 and Tbx5 in murine lungs or Tbx4 in chicken embryos suppress Fgf10 expression, indicating that Fgf10 is likely a downstream target of Tbx4.39, 48 Whereas heterozygous Fgf10 knockout leads to aplasia of lacrimal glands and hypoplasia of salivary glands in mice,49 homozygous Fgf10 knockout mice die shortly after birth due to complete disruption of pulmonary branching morphogenesis.50
With three exceptions,6, 19 variants in TBX2, TBX4, and FGF10 have not been reported in subjects with severe pulmonary hypoplasia. Recently, missense SNVs in TBX2 have been described in individuals with a syndromic cardiovascular and skeletal developmental disorder,51 whereas SNVs or CNVs involving TBX4 have been associated with pulmonary hypertension (PAH),52, 53, 54 ischiocoxopodopatellar syndrome (MIM: 147891),53, 55, 56, 57 and developmental delay with coexisting PAH,58 heart defects, and limb abnormalities52, 53, 58, 59, 60 (Table S10). The pLI score61 for TBX4 is 0.41, indicating it is more tolerant for LoF variants than TBX2 whose pLI score is 0.96. Since TBX2 and TBX4 are located in two different subdomains of the same TAD identified in fetal lung fibroblasts, and the decrease of TBX2 expression was larger than TBX4 in the subject with the 17q23.1q23.2 CNV deletion, we hypothesize that the putative hypomorphic variants in the predicted lung-specific enhancer located upstream to these two genes may affect TBX2 more than TBX4.
FGF10 is also predicted to be intolerant for LoF variants (pLI score 0.92) and heterozygous SNVs and CNVs deletions are associated with aplasia of lacrimal and salivary glands (ALSG [MIM: 180920])49, 62, 63 and LADD syndrome, indicating that, similar to murine organs, during human organogenesis, lacrimal and salivary glands are more dosage sensitive than lungs.64, 65 However, whereas children with ALSG do not show lung defects, adult ALSG-affected individuals with LoF variants in FGF10 had decreased spirometric values, indicating that FGF10 defects might manifest later in life with lung dysfunction.66 The presence of a phenotypic difference in the described affected and control individuals suggests variable phenotypic expressivity of LoF involving TBX4, TBX2, and FGF10, a phenomenon well known to other microdeletion syndromes.67, 68, 69
Several lines of evidence support our hypothesis that compound heterozygosity of coding variants involving TBX4 or FGF10 and an additional non-coding variant in trans on the other allele or a genetic modifier(s) elsewhere in the genome may be responsible for AcDys, CAD, or other rare pulmonary hypoplasias. For example, as we noted from identification of variants in TBX4, heterozygous variation of TBX4 alone is not sufficient to cause disease. Similarly, occurrence of heterozygous FGF10 SNVs and CNVs in subjects with severe lethal lung hypoplasia inherited from the parents with LADD syndrome, or the presence of the same nonsense variant in subject with CAD which was previously found in a family with ALSG,49 suggests that these lung phenotypes cannot be explained by FGF10 haploinsufficiency alone. Taken together, these data support the possibility of compound inheritance in the described lung hypoplasias.
There is growing evidence that along with coding variants, non-coding changes (de novo or inherited) within regulatory elements can be responsible for diverse disease manifestation.70, 71, 72, 73 Precedent for compound inheritance of rare variant pathogenic coding and common non-coding variants has been demonstrated for thrombocytopenia absent radius (TAR) syndrome with recurrent 1q21 deletion and congenital scoliosis with recurrent 16p11.2 deletion, both in which compound coding and in trans non-coding variant alleles at the same locus are required for phenotypic manifestation.72, 73, 74, 75, 76 In this study, we have identified a statistically significant enrichment of the non-coding variants (either common or rare) on the other allele in the subjects with AcDys, CAD, or pulmonary hypoplasia and heterozygous SNVs or CNVs involving TBX4. Many of the identified variants mapping within TBX4 overlap the putative regulatory elements, including enhancers specific for the gene expression in mouse lung77 or human lung fibroblasts, discovered in the previous Roadmap large-scale epigenomics study. On the other hand, variants located upstream to TBX4 overlap the hindlimb-specific enhancer in mice77 and the putative enhancers specific for human lung fibroblasts. Interestingly, in addition to histone marks indicative of regulatory potential, the region upstream to TBX2/TBX4 also harbors lncRNAs highly expressed in the human lung (Figures 2 and S9). Fetal lung-specific RNAs identified in the enhancer region upstream to FOXF1 at 16q24.1 have been proposed to play an important role in its regulation, and disruption of this process may result in ACDMPV.20, 21, 78 Identification of rare SNVs in the enhancer region upstream to TBX2 and TBX4 in affected subjects with FGF10 SNVs or deletion CNVs, as well as the presence of double heterozygous TBX4 and TBX5 variants in two affected siblings, suggest epistatic interactions of protein variants from the same signaling pathway. However, stochastic or environmental factors influencing the phenotypic manifestation should also be considered.
In addition to TBX4 or FGF10 variants found in more than 60% of the studied case subjects, we have also identified exonic variants in TCF21, BTBD7, DSPP, and BCLAF1 (Tables 1, S3, and S4). While all of these genes are known to play a role in lung development,79, 80, 81, 82 identified changes are predicted as deleterious using only in silico tools. Thus, we cannot conclude that those variants are sufficient for causing the phenotype.
The histologic appearance of the described subjects’ lungs reflects a spectrum of lung maturational arrest, ranging from the morphologic pseudoglandular to saccular stages of development. Whereas variation exists both between and within individual cases, the phenotype of individuals with TBX4 variants are more severe, within the spectrum of AcDys, while the FGF10 group has more developed lungs, resembling CAD and pulmonary hypoplasia. This suggests that the dosage of TBX4 is more crucial for early phases of lung development. However, both genes have been found to be expressed in the newly formed lung buds in mice at E9.5 (equivalent to embryonic days 22–23 in humans), suggesting that both of them are required for normal lung development around the same time.39, 46 The histopathological continuum between AcDys, CAD, and pulmonary hypoplasia supports the notion that these rare disorders share a common pathway and require genetic interrogation for disease classification. However, assessment of additional case subjects will be required to assess the frequency of these variants and spectrum of pathology.
Conclusions
The observed concomitance of coding and non-coding SNVs or CNVs involving TBX4 or FGF10 loci in our subjects with lethal lung maldevelopment, including AcDys and CAD spectrum, supports the previously proposed role of a TBX4-FGF10-FGFR2 epithelial-mesenchymal signaling in lung organogenesis. Our studies also demonstrate that while heterozygous coding CNV deletions or SNVs involving FGF10 co-segregate with the LADD syndrome phenotype, and those involving TBX4 co-segregate in families with childhood-onset PAH, ischiocoxopodopatellar syndrome, or 17q23.1q23.2 deletion syndrome, these variants also can confer a significantly increased risk for lethal developmental lung disorders along a spectrum of growth arrest. However, the presence of a LoF variant per se cannot be used as a predictor of the likely phenotype in the subjects, since the additional modifier may be required for lung disease manifestation.
We provide evidence that biallelic variation at TBX4 or FGF10, as a compound inheritance model with rare coding and rare or common non-coding variant alleles, can result in a mutational burden and perturbation of the epithelial-mesenchymal signaling pathway involved in lung organogenesis, resulting in lethal lung disease. Functional characterization of non-coding regulatory variants in vitro or in animal models is necessary to gain further insight into their mechanistic role underlying human genetic disorders.
Declaration of Interests
J.R.L. has stock ownership in 23andMe and Lasergen, is a paid consultant for Regeneron Pharmaceuticals, and is a co-inventor on multiple US and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, and bacterial genomic fingerprinting. C.G.G.-J. is a full-time employee of the Regeneron Genetics Center and receives stock options as part of compensation. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from the chromosomal microarray analysis and clinical exome sequencing offered in the Baylor Genetics Laboratory.
Acknowledgments
We thank Drs. Neil Hanchard, Claire Langston, Pengfei Liu, Feng Zhang, and Jill Rosenfeld for helpful discussion and Rodger Song for technical assistance.
This work was supported by grants awarded by the US National Institutes of Health (NIH), National Heart, Lung, and Blood Institute (NHLBI) R01HL137203 to P. Stankiewicz, the US National Human Genome Research Institute (NHGRI)/NHLBI grant number UM1HG006542 to the Baylor-Hopkins Center for Mendelian Genomics (BHCMG), the National Institute of Neurological Disorders and Stroke (NINDS) R35 NS105078 to J.R.L., National Institute of General Medical Sciences of NIH Postdoctoral Training Program in Medical Genetics 5T32GM007454 to J.N.D. and A.S.F., a grant from the JPB Foundation to W.K.C., and from Polish budget funds for science in years 2016–2019, Iuventus Plus grant IP2015 019874 to T.G. This study makes use of data generated by the DECIPHER community. A full list of centers who contributed to the generation of the data is available from https://decipher.sanger.ac.uk and via email from decipher@sanger.ac.uk. Funding for the project was provided by the Wellcome Trust.
Published: January 10, 2019
Footnotes
Supplemental Data include nine figures, ten tables, and Supplemental Note (case reports) and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.12.010.
Contributor Information
Cedric Le Caignec, Email: cedric.lecaignec@chu-nantes.fr.
Paweł Stankiewicz, Email: pawels@bcm.edu.
Accession Numbers
The CNV calls presented in this paper can be accessed through the NCBI dbVar database under accession number nstd164.
Web Resources
1000 Genomes, http://www.internationalgenome.org/
3D Genome Browser, http://promoter.bx.psu.edu/hi-c/view.php
DECIPHER, https://decipher.sanger.ac.uk/
ENCODE, https://www.encodeproject.org/
ExAC Browser, http://exac.broadinstitute.org/
gnomAD Browser, http://gnomad.broadinstitute.org/
GTEx Portal, https://gtexportal.org/home/
MutationTaster, http://www.mutationtaster.org/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
NMDEscPredictor, https://nmdprediction.shinyapps.io/nmdescpredictor
OMIM, http://www.omim.org/
Phyre2, http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
Primer3, http://bioinfo.ut.ee/primer3
Roadmap, http://www.roadmapepigenomics.org/
SWISS-MODEL, http://swissmodel.expasy.org/
UCSC Genome Browser, https://genome.ucsc.edu
Supplemental Data
References
- 1.Nogee L.M. Interstitial lung disease in newborns. Semin. Fetal Neonatal Med. 2017;22:227–233. doi: 10.1016/j.siny.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Langston C., Dishop M.K. Diffuse lung disease in infancy: a proposed classification applied to 259 diagnostic biopsies. Pediatr. Dev. Pathol. 2009;12:421–437. doi: 10.2350/08-11-0559.1. [DOI] [PubMed] [Google Scholar]
- 3.Hegde S., Pomplun S., Hannam S., Greenough A. Nonfatal congenital alveolar dysplasia due to abnormalities of NO synthase isoforms. Acta Paediatr. 2007;96:1248–1250. doi: 10.1111/j.1651-2227.2007.00394.x. [DOI] [PubMed] [Google Scholar]
- 4.Chow C.W., Massie J., Ng J., Mills J., Baker M. Acinar dysplasia of the lungs: variation in the extent of involvement and clinical features. Pathology. 2013;45:38–43. doi: 10.1097/PAT.0b013e32835b3a9d. [DOI] [PubMed] [Google Scholar]
- 5.Barnett C.P., Nataren N.J., Klingler-Hoffmann M., Schwarz Q., Chong C.-E., Lee Y.K., Bruno D.L., Lipsett J., McPhee A.J., Schreiber A.W. Ectrodactyly and lethal pulmonary acinar dysplasia associated with homozygous FGFR2 mutations identified by exome sequencing. Hum. Mutat. 2016;37:955–963. doi: 10.1002/humu.23032. [DOI] [PubMed] [Google Scholar]
- 6.Szafranski P., Coban-Akdemir Z.H., Rupps R., Grazioli S., Wensley D., Jhangiani S.N., Popek E., Lee A.F., Lupski J.R., Boerkoel C.F., Stankiewicz P. Phenotypic expansion of TBX4 mutations to include acinar dysplasia of the lungs. Am. J. Med. Genet. A. 2016;170:2440–2444. doi: 10.1002/ajmg.a.37822. [DOI] [PubMed] [Google Scholar]
- 7.Rutledge J.C., Jensen P. Acinar dysplasia: a new form of pulmonary maldevelopment. Hum. Pathol. 1986;17:1290–1293. doi: 10.1016/s0046-8177(86)80576-7. [DOI] [PubMed] [Google Scholar]
- 8.Chambers H.M. Congenital acinar aplasia: an extreme form of pulmonary maldevelopment. Pathology. 1991;23:69–71. doi: 10.3109/00313029109061444. [DOI] [PubMed] [Google Scholar]
- 9.Davidson L.A., Batman P., Fagan D.G. Congenital acinar dysplasia: a rare cause of pulmonary hypoplasia. Histopathology. 1998;32:57–59. doi: 10.1046/j.1365-2559.1998.00325.x. [DOI] [PubMed] [Google Scholar]
- 10.Moerman P., Vanhole C., Devlieger H., Fryns J.P. Severe primary pulmonary hypoplasia (“acinar dysplasia”) in sibs: a genetically determined mesodermal defect? J. Med. Genet. 1998;35:964–965. doi: 10.1136/jmg.35.11.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Al-Senan K.A., Kattan A.K., Al-Dayel F.H. Congenital acinar dysplasia. Familial cause of a fatal respiratory failure in a neonate. Saudi Med. J. 2003;24:88–90. [PubMed] [Google Scholar]
- 12.Gillespie L.M., Fenton A.C., Wright C. Acinar dysplasia: a rare cause of neonatal respiratory failure. Acta Paediatr. 2004;93:712–713. doi: 10.1111/j.1651-2227.2004.tb03003.x. [DOI] [PubMed] [Google Scholar]
- 13.Stuhrmann S., Sachweh J., Bindl L., Vázquez-Jiménez J., Hermanns-Sachweh B., Seghaye M.-C. Congenital cystic adenomatoid malformation type 0-a rare cause of neonatal death. Pediatr. Crit. Care Med. 2007;8:580–581. doi: 10.1097/01.PCC.0000288803.19741.59. [DOI] [PubMed] [Google Scholar]
- 14.DeBoer E.M., Keene S., Winkler A.M., Shehata B.M. Identical twins with lethal congenital pulmonary airway malformation type 0 (acinar dysplasia): further evidence of familial tendency. Fetal Pediatr. Pathol. 2012;31:217–224. doi: 10.3109/15513815.2011.650284. [DOI] [PubMed] [Google Scholar]
- 15.Langenstroer M., Carlan S.J., Fanaian N., Attia S. Congenital acinar dysplasia: report of a case and review of literature. AJP Rep. 2013;3:9–12. doi: 10.1055/s-0032-1329126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Don M., Orsaria M., Da Dalt E., Tringali C., Sacher B. Rapidly fatal “congenital lung dysplasia”: a case report and review of the literature. Fetal Pediatr. Pathol. 2014;33:109–113. doi: 10.3109/15513815.2013.878009. [DOI] [PubMed] [Google Scholar]
- 17.Lertsburapa T., Vargas D., Lambert-Messerlian G., Tantravahi U., Gündoğan F., DeLaMonte S., Coyle M.G., De Paepe M.E. Lethal hypoplasia and developmental anomalies of the lungs in a newborn with intrauterine adrenal hemorrhage and cerebral infarcts: a proposed pulmonary disruption sequence. Pediatr. Dev. Pathol. 2014;17:374–381. doi: 10.2350/14-05-1485-CR.1. [DOI] [PubMed] [Google Scholar]
- 18.MacMahon H.E. Congenital alveolar dysplasia; a developmental anomaly involving pulmonary alveoli. Pediatrics. 1948;2:43–57. [PubMed] [Google Scholar]
- 19.Suhrie K., Pajor N.M., Ahlfeld S.K., Dawson D.B., Dufendach K.R., Kitzmiller J.A., Leino D., Lombardo R.C., Smolarek T.A., Rathbun P.A. Neonatal lung disease associated with TBX4 mutations. J. Pediatr. 2018 doi: 10.1016/j.jpeds.2018.10.018. Published online November 7, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szafranski P., Dharmadhikari A.V., Brosens E., Gurha P., Kolodziejska K.E., Zhishuo O., Dittwald P., Majewski T., Mohan K.N., Chen B. Small noncoding differentially methylated copy-number variants, including lncRNA genes, cause a lethal lung developmental disorder. Genome Res. 2013;23:23–33. doi: 10.1101/gr.141887.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szafranski P., Gambin T., Dharmadhikari A.V., Akdemir K.C., Jhangiani S.N., Schuette J., Godiwala N., Yatsenko S.A., Sebastian J., Madan-Khetarpal S. Pathogenetics of alveolar capillary dysplasia with misalignment of pulmonary veins. Hum. Genet. 2016;135:569–586. doi: 10.1007/s00439-016-1655-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Firth H.V., Richards S.M., Bevan A.P., Clayton S., Corpas M., Rajan D., Van Vooren S., Moreau Y., Pettett R.M., Carter N.P. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am. J. Hum. Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J., Fritsch E.F., Maniatis T., Laboratory C.S.H. Cold Spring Harbor Laboratory Press; New York: 1989. Molecular Cloning : A Laboratory Manual. [Google Scholar]
- 24.Lupski J.R., Gonzaga-Jauregui C., Yang Y., Bainbridge M.N., Jhangiani S., Buhay C.J., Kovar C.L., Wang M., Hawes A.C., Reid J.G. Exome sequencing resolves apparent incidental findings and reveals further complexity of SH3TC2 variant alleles causing Charcot-Marie-Tooth neuropathy. Genome Med. 2013;5:57. doi: 10.1186/gm461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coban-Akdemir Z., White J.J., Song X., Jhangiani S.N., Fatih J.M., Gambin T., Bayram Y., Chinn I.K., Karaca E., Punetha J., Baylor-Hopkins Center for Mendelian Genomics Identifying genes whose mutant transcripts cause dominant disease traits by potential gain-of-function alleles. Am. J. Hum. Genet. 2018;103:171–187. doi: 10.1016/j.ajhg.2018.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Archie J.G., Collins J.S., Lebel R.R. Quantitative standards for fetal and neonatal autopsy. Am. J. Clin. Pathol. 2006;126:256–265. doi: 10.1309/FK9D-5WBA-1UEP-T5BB. [DOI] [PubMed] [Google Scholar]
- 30.Ou Z., Jarmuz M., Sparagana S.P., Michaud J., Décarie J.-C., Yatsenko S.A., Nowakowska B., Furman P., Shaw C.A., Shaffer L.G. Evidence for involvement of TRE-2 (USP6) oncogene, low-copy repeat and acrocentric heterochromatin in two families with chromosomal translocations. Hum. Genet. 2006;120:227–237. doi: 10.1007/s00439-006-0200-7. [DOI] [PubMed] [Google Scholar]
- 31.Hastings P.J., Ira G., Lupski J.R. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horie M., Miyashita N., Mikami Y., Noguchi S., Yamauchi Y., Suzukawa M., Fukami T., Ohta K., Asano Y., Sato S. TBX4 is involved in the super-enhancer-driven transcriptional programs underlying features specific to lung fibroblasts. Am. J. Physiol. Lung Cell. Mol. Physiol. 2018;314:L177–L191. doi: 10.1152/ajplung.00193.2017. [DOI] [PubMed] [Google Scholar]
- 33.Schmitt A.D., Hu M., Jung I., Xu Z., Qiu Y., Tan C.L., Li Y., Lin S., Lin Y., Barr C.L., Ren B. A compendium of chromatin contact maps reveals spatially active regions in the human genome. Cell Rep. 2016;17:2042–2059. doi: 10.1016/j.celrep.2016.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghoussaini M., French J.D., Michailidou K., Nord S., Beesley J., Canisus S., Hillman K.M., Kaufmann S., Sivakumaran H., Moradi Marjaneh M., kConFab/AOCS Investigators. NBCS Collaborators Evidence that the 5p12 variant rs10941679 confers susceptibility to estrogen-receptor-positive breast cancer through FGF10 and MRPS30 regulation. Am. J. Hum. Genet. 2016;99:903–911. doi: 10.1016/j.ajhg.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prince L.S. FGF10 and human lung disease across the life spectrum. Front. Genet. 2018;9:517. doi: 10.3389/fgene.2018.00517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sifrim A., Hitz M.-P., Wilsdon A., Breckpot J., Turki S.H.A., Thienpont B., McRae J., Fitzgerald T.W., Singh T., Swaminathan G.J., INTERVAL Study. UK10K Consortium. Deciphering Developmental Disorders Study Distinct genetic architectures for syndromic and nonsyndromic congenital heart defects identified by exome sequencing. Nat. Genet. 2016;48:1060–1065. doi: 10.1038/ng.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morrisey E.E., Cardoso W.V., Lane R.H., Rabinovitch M., Abman S.H., Ai X., Albertine K.H., Bland R.D., Chapman H.A., Checkley W. Molecular determinants of lung development. Ann. Am. Thorac. Soc. 2013;10:S12–S16. doi: 10.1513/AnnalsATS.201207-036OT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ardini-Poleske M.E., Clark R.F., Ansong C., Carson J.P., Corley R.A., Deutsch G.H., Hagood J.S., Kaminski N., Mariani T.J., Potter S.S., LungMAP Consortium LungMAP: The Molecular Atlas of Lung Development Program. Am. J. Physiol. Lung Cell. Mol. Physiol. 2017;313:L733–L740. doi: 10.1152/ajplung.00139.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arora R., Metzger R.J., Papaioannou V.E. Multiple roles and interactions of Tbx4 and Tbx5 in development of the respiratory system. PLoS Genet. 2012;8:e1002866. doi: 10.1371/journal.pgen.1002866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lüdtke T.H., Rudat C., Wojahn I., Weiss A.-C., Kleppa M.-J., Kurz J., Farin H.F., Moon A., Christoffels V.M., Kispert A. Tbx2 and Tbx3 act downstream of Shh to maintain canonical Wnt signaling during branching morphogenesis of the murine lung. Dev. Cell. 2016;39:239–253. doi: 10.1016/j.devcel.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 41.Cebra-Thomas J.A., Bromer J., Gardner R., Lam G.K., Sheipe H., Gilbert S.F. T-box gene products are required for mesenchymal induction of epithelial branching in the embryonic mouse lung. Dev. Dyn. 2003;226:82–90. doi: 10.1002/dvdy.10208. [DOI] [PubMed] [Google Scholar]
- 42.Lüdtke T.H.-W., Farin H.F., Rudat C., Schuster-Gossler K., Petry M., Barnett P., Christoffels V.M., Kispert A. Tbx2 controls lung growth by direct repression of the cell cycle inhibitor genes Cdkn1a and Cdkn1b. PLoS Genet. 2013;9:e1003189. doi: 10.1371/journal.pgen.1003189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takahashi T., Friedmacher F., Zimmer J., Puri P. Expression of T-box transcription factors 2, 4 and 5 is decreased in the branching airway mesenchyme of nitrofen-induced hypoplastic lungs. Pediatr. Surg. Int. 2017;33:139–143. doi: 10.1007/s00383-016-4005-z. [DOI] [PubMed] [Google Scholar]
- 44.Agulnik S.I., Garvey N., Hancock S., Ruvinsky I., Chapman D.L., Agulnik I., Bollag R., Papaioannou V., Silver L.M. Evolution of mouse T-box genes by tandem duplication and cluster dispersion. Genetics. 1996;144:249–254. doi: 10.1093/genetics/144.1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abler L.L., Mansour S.L., Sun X. Conditional gene inactivation reveals roles for Fgf10 and Fgfr2 in establishing a normal pattern of epithelial branching in the mouse lung. Dev. Dyn. 2009;238:1999–2013. doi: 10.1002/dvdy.22032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bellusci S., Grindley J., Emoto H., Itoh N., Hogan B.L. Fibroblast growth factor 10 (FGF10) and branching morphogenesis in the embryonic mouse lung. Development. 1997;124:4867–4878. doi: 10.1242/dev.124.23.4867. [DOI] [PubMed] [Google Scholar]
- 47.Li C., Hu L., Xiao J., Chen H., Li J.T., Bellusci S., Delanghe S., Minoo P. Wnt5a regulates Shh and Fgf10 signaling during lung development. Dev. Biol. 2005;287:86–97. doi: 10.1016/j.ydbio.2005.08.035. [DOI] [PubMed] [Google Scholar]
- 48.Sakiyama J., Yamagishi A., Kuroiwa A. Tbx4-Fgf10 system controls lung bud formation during chicken embryonic development. Development. 2003;130:1225–1234. doi: 10.1242/dev.00345. [DOI] [PubMed] [Google Scholar]
- 49.Entesarian M., Matsson H., Klar J., Bergendal B., Olson L., Arakaki R., Hayashi Y., Ohuchi H., Falahat B., Bolstad A.I. Mutations in the gene encoding fibroblast growth factor 10 are associated with aplasia of lacrimal and salivary glands. Nat. Genet. 2005;37:125–127. doi: 10.1038/ng1507. [DOI] [PubMed] [Google Scholar]
- 50.Sekine K., Ohuchi H., Fujiwara M., Yamasaki M., Yoshizawa T., Sato T., Yagishita N., Matsui D., Koga Y., Itoh N., Kato S. Fgf10 is essential for limb and lung formation. Nat. Genet. 1999;21:138–141. doi: 10.1038/5096. [DOI] [PubMed] [Google Scholar]
- 51.Liu N., Schoch K., Luo X., Pena L.D.M., Bhavana V.H., Kukolich M.K., Stringer S., Powis Z., Radtke K., Mroske C., Undiagnosed Diseases Network (UDN) Functional variants in TBX2 are associated with a syndromic cardiovascular and skeletal developmental disorder. Hum. Mol. Genet. 2018;27:2454–2465. doi: 10.1093/hmg/ddy146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu N., Gonzaga-Jauregui C., Welch C.L., Ma L., Qi H., King A.K., Krishnan U., Rosenzweig E.B., Ivy D.D., Austin E.D. Exome sequencing in children with pulmonary arterial hypertension demonstrates differences compared with adults. Circ Genom Precis Med. 2018;11:e001887. doi: 10.1161/CIRCGEN.117.001887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kerstjens-Frederikse W.S., Bongers E.M.H.F., Roofthooft M.T.R., Leter E.M., Douwes J.M., Van Dijk A., Vonk-Noordegraaf A., Dijk-Bos K.K., Hoefsloot L.H., Hoendermis E.S. TBX4 mutations (small patella syndrome) are associated with childhood-onset pulmonary arterial hypertension. J. Med. Genet. 2013;50:500–506. doi: 10.1136/jmedgenet-2012-101152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abou Hassan O.K., Haidar W., Nemer G., Skouri H., Haddad F., BouAkl I. Clinical and genetic characteristics of pulmonary arterial hypertension in Lebanon. BMC Med. Genet. 2018;19:89. doi: 10.1186/s12881-018-0608-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bongers E.M.H.F., Duijf P.H.G., van Beersum S.E.M., Schoots J., Van Kampen A., Burckhardt A., Hamel B.C.J., Losan F., Hoefsloot L.H., Yntema H.G. Mutations in the human TBX4 gene cause small patella syndrome. Am. J. Hum. Genet. 2004;74:1239–1248. doi: 10.1086/421331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oda T., Matsushita M., Ono Y., Kitoh H., Sakai T. A novel heterozygous mutation in the T-box Protein 4 gene in an adult case of small patella syndrome. J Orthop Case Rep. 2018;8:85–88. doi: 10.13107/jocr.2250-0685.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vanlerberghe C., Boutry N., Petit F. Genetics of patella hypoplasia/agenesis. Clin. Genet. 2018;94:43–53. doi: 10.1111/cge.13209. [DOI] [PubMed] [Google Scholar]
- 58.Ballif B.C., Theisen A., Rosenfeld J.A., Traylor R.N., Gastier-Foster J., Thrush D.L., Astbury C., Bartholomew D., McBride K.L., Pyatt R.E. Identification of a recurrent microdeletion at 17q23.1q23.2 flanked by segmental duplications associated with heart defects and limb abnormalities. Am. J. Hum. Genet. 2010;86:454–461. doi: 10.1016/j.ajhg.2010.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schönewolf-Greulich B., Ronan A., Ravn K., Baekgaard P., Lodahl M., Nielsen K., Rendtorff N.D., Tranebjaerg L., Brøndum-Nielsen K., Tümer Z. Two new cases with microdeletion of 17q23.2 suggest presence of a candidate gene for sensorineural hearing loss within this region. Am. J. Med. Genet. A. 2011;155A:2964–2969. doi: 10.1002/ajmg.a.34302. [DOI] [PubMed] [Google Scholar]
- 60.Nimmakayalu M., Major H., Sheffield V., Solomon D.H., Smith R.J., Patil S.R., Shchelochkov O.A. Microdeletion of 17q22q23.2 encompassing TBX2 and TBX4 in a patient with congenital microcephaly, thyroid duct cyst, sensorineural hearing loss, and pulmonary hypertension. Am. J. Med. Genet. A. 2011;155A:418–423. doi: 10.1002/ajmg.a.33827. [DOI] [PubMed] [Google Scholar]
- 61.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Entesarian M., Dahlqvist J., Shashi V., Stanley C.S., Falahat B., Reardon W., Dahl N. FGF10 missense mutations in aplasia of lacrimal and salivary glands (ALSG) Eur. J. Hum. Genet. 2007;15:379–382. doi: 10.1038/sj.ejhg.5201762. [DOI] [PubMed] [Google Scholar]
- 63.Seymen F., Koruyucu M., Toptanci I.R., Balsak S., Dedeoglu S., Celepkolu T., Shin T.J., Hyun H.-K., Kim Y.-J., Kim J.-W. Novel FGF10 mutation in autosomal dominant aplasia of lacrimal and salivary glands. Clin. Oral Investig. 2017;21:167–172. doi: 10.1007/s00784-016-1771-x. [DOI] [PubMed] [Google Scholar]
- 64.Rohmann E., Brunner H.G., Kayserili H., Uyguner O., Nürnberg G., Lew E.D., Dobbie A., Eswarakumar V.P., Uzumcu A., Ulubil-Emeroglu M. Mutations in different components of FGF signaling in LADD syndrome. Nat. Genet. 2006;38:414–417. doi: 10.1038/ng1757. [DOI] [PubMed] [Google Scholar]
- 65.Milunsky J.M., Zhao G., Maher T.A., Colby R., Everman D.B. LADD syndrome is caused by FGF10 mutations. Clin. Genet. 2006;69:349–354. doi: 10.1111/j.1399-0004.2006.00597.x. [DOI] [PubMed] [Google Scholar]
- 66.Klar J., Blomstrand P., Brunmark C., Badhai J., Håkansson H.F., Brange C.S., Bergendal B., Dahl N. Fibroblast growth factor 10 haploinsufficiency causes chronic obstructive pulmonary disease. J. Med. Genet. 2011;48:705–709. doi: 10.1136/jmedgenet-2011-100166. [DOI] [PubMed] [Google Scholar]
- 67.Potocki L., Shaw C.J., Stankiewicz P., Lupski J.R. Variability in clinical phenotype despite common chromosomal deletion in Smith-Magenis syndrome [del(17)(p11.2p11.2)] Genet. Med. 2003;5:430–434. doi: 10.1097/01.gim.0000095625.14160.ab. [DOI] [PubMed] [Google Scholar]
- 68.McDonald-McGinn D.M., Sullivan K.E., Marino B., Philip N., Swillen A., Vorstman J.A.S., Zackai E.H., Emanuel B.S., Vermeesch J.R., Morrow B.E. 22q11.2 deletion syndrome. Nat. Rev. Dis. Primers. 2015;1:15071. doi: 10.1038/nrdp.2015.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vergaelen E., Swillen A., Van Esch H., Claes S., Van Goethem G., Devriendt K. 3 generation pedigree with paternal transmission of the 22q11.2 deletion syndrome: Intrafamilial phenotypic variability. Eur. J. Med. Genet. 2015;58:244–248. doi: 10.1016/j.ejmg.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 70.Brandler W.M., Antaki D., Gujral M., Kleiber M.L., Whitney J., Maile M.S., Hong O., Chapman T.R., Tan S., Tandon P. Paternally inherited cis-regulatory structural variants are associated with autism. Science. 2018;360:327–331. doi: 10.1126/science.aan2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Short P.J., McRae J.F., Gallone G., Sifrim A., Won H., Geschwind D.H., Wright C.F., Firth H.V., FitzPatrick D.R., Barrett J.C., Hurles M.E. De novo mutations in regulatory elements in neurodevelopmental disorders. Nature. 2018;555:611–616. doi: 10.1038/nature25983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang F., Lupski J.R. Non-coding genetic variants in human disease. Hum. Mol. Genet. 2015;24(R1):R102–R110. doi: 10.1093/hmg/ddv259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu N., Ming X., Xiao J., Wu Z., Chen X., Shinawi M., Shen Y., Yu G., Liu J., Xie H. TBX6 null variants and a common hypomorphic allele in congenital scoliosis. N. Engl. J. Med. 2015;372:341–350. doi: 10.1056/NEJMoa1406829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Albers C.A., Paul D.S., Schulze H., Freson K., Stephens J.C., Smethurst P.A., Jolley J.D., Cvejic A., Kostadima M., Bertone P. Compound inheritance of a low-frequency regulatory SNP and a rare null mutation in exon-junction complex subunit RBM8A causes TAR syndrome. Nat. Genet. 2012;44:435–439. doi: 10.1038/ng.1083. S1–S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Klopocki E., Schulze H., Strauss G., Ott C.-E., Hall J., Trotier F., Fleischhauer S., Greenhalgh L., Newbury-Ecob R.A., Neumann L.M. Complex inheritance pattern resembling autosomal recessive inheritance involving a microdeletion in thrombocytopenia-absent radius syndrome. Am. J. Hum. Genet. 2007;80:232–240. doi: 10.1086/510919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang N., Wu N., Zhang L., Zhao Y., Liu J., Liang X., Ren X., Li W., Chen W., Dong S. TBX6 compound inheritance leads to congenital vertebral malformations in humans and mice. Hum. Mol. Genet. 2018 doi: 10.1093/hmg/ddy358. Published online October 10, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Menke D.B., Guenther C., Kingsley D.M. Dual hindlimb control elements in the Tbx4 gene and region-specific control of bone size in vertebrate limbs. Development. 2008;135:2543–2553. doi: 10.1242/dev.017384. [DOI] [PubMed] [Google Scholar]
- 78.Szafranski P., Dharmadhikari A.V., Wambach J.A., Towe C.T., White F.V., Grady R.M., Eghtesady P., Cole F.S., Deutsch G., Sen P., Stankiewicz P. Two deletions overlapping a distant FOXF1 enhancer unravel the role of lncRNA LINC01081 in etiology of alveolar capillary dysplasia with misalignment of pulmonary veins. Am. J. Med. Genet. A. 2014;164A:2013–2019. doi: 10.1002/ajmg.a.36606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Quaggin S.E., Schwartz L., Cui S., Igarashi P., Deimling J., Post M., Rossant J. The basic-helix-loop-helix protein pod1 is critically important for kidney and lung organogenesis. Development. 1999;126:5771–5783. doi: 10.1242/dev.126.24.5771. [DOI] [PubMed] [Google Scholar]
- 80.Daley W.P., Matsumoto K., Doyle A.D., Wang S., DuChez B.J., Holmbeck K., Yamada K.M. Btbd7 is essential for region-specific epithelial cell dynamics and branching morphogenesis in vivo. Development. 2017;144:2200–2211. doi: 10.1242/dev.146894. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 81.Alvares K., Kanwar Y.S., Veis A. Expression and potential role of dentin phosphophoryn (DPP) in mouse embryonic tissues involved in epithelial-mesenchymal interactions and branching morphogenesis. Dev. Dyn. 2006;235:2980–2990. doi: 10.1002/dvdy.20935. [DOI] [PubMed] [Google Scholar]
- 82.McPherson J.P., Sarras H., Lemmers B., Tamblyn L., Migon E., Matysiak-Zablocki E., Hakem A., Azami S.A., Cardoso R., Fish J. Essential role for Bclaf1 in lung development and immune system function. Cell Death Differ. 2009;16:331–339. doi: 10.1038/cdd.2008.167. [DOI] [PubMed] [Google Scholar]
- 83.Kimura J., Deutsch G.H. Key mechanisms of early lung development. Pediatr. Dev. Pathol. 2007;10:335–347. doi: 10.2350/07-06-0290.1. [DOI] [PubMed] [Google Scholar]
- 84.Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-André V., Sigova A.A., Hoke H.A., Young R.A. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.