Abstract
Chimeric antigen receptor (CAR)-T cell immunotherapy is under intense preclinical and clinical investigation, and it involves a rapidly increasing portfolio of novel target antigens and CAR designs. We established a platform that enables rapid and high-throughput CAR-screening campaigns with reporter cells derived from the T cell lymphoma line Jurkat. Reporter cells were equipped with nuclear factor κB (NF-κB) and nuclear factor of activated T cells (NFAT) reporter genes that generate a duplex output of enhanced CFP (ECFP) and EGFP, respectively. As a proof of concept, we modified reporter cells with CD19-specific and ROR1-specific CARs, and we detected high-level reporter signals that allowed distinguishing functional from non-functional CAR constructs. The reporter data were highly reproducible, and the time required for completing each testing campaign was substantially shorter with reporter cells (6 days) compared to primary CAR-T cells (21 days). We challenged the reporter platform to a large-scale screening campaign on a ROR1-CAR library, and we showed that reporter cells retrieved a functional CAR variant that was present with a frequency of only 6 in 1.05 × 106. The data illustrate the potential to implement this reporter platform into the preclinical development path of novel CAR-T cell products and to inform and accelerate the selection of lead CAR candidates for clinical translation.
Keywords: NF-κB/NFAT reporter cells, chimeric antigen receptor, library screening, cancer immunotherapy, ROR1
Analysis of novel CAR designs and constructs mainly depends on laborious and time-consuming testing campaigns in primary T cells. Rydzek et al. present an NF-κB/NFAT reporter cell platform to identify lead constructs from CAR libraries, with minimum time and effort to accelerate progress in CAR research.
Introduction
Adoptive immunotherapy with gene-engineered T cells expressing a chimeric antigen receptor (CAR) is under intense preclinical and clinical investigation.1, 2 CARs are synthetic designer molecules that redirect the specificity of T cells to surface molecules on target cells. CAR specificity is determined by an extracellular antigen-binding domain that typically incorporates the variable heavy (VH) and variable light (VL) chain domains of a monoclonal antibody (mAb) specific for the intended target antigen.3 In addition, several parameters have been identified that affect CAR sensitivity and the ensuing antitumor function of T cells. These parameters include antigen-binding domain affinity, length and composition of the extracellular CAR spacer domain,4, 5, 6 and composition of the co-stimulatory domains included in the intracellular signaling module.7
At present, the evaluation of novel CAR designs and constructs is performed empirically and depends on testing campaigns in primary T cells. This approach is laborious and time consuming, and it is confounded by donor-to-donor variability in T cell subset composition, genomic insertion, and ensuing CAR surface expression.8, 9, 10 These factors contribute to a high degree of variability between individual testing campaigns and poor reproducibility between testing campaigns that are performed in different laboratories.
In pharmaceutical small molecule and antibody discovery, lead compounds are commonly identified in library-screening campaigns using standardized platforms.11, 12 Indeed, a standardized platform that is capable of identifying CAR constructs with the desired specificity and function from a large pool of candidates, thereby informing the selection of lead candidates for preclinical and clinical development, would substantially accelerate the development of novel CAR-T cell products. Ideally, a CAR-screening platform would work with an established cell line to eliminate the need for primary T cell culture, and it would provide an easy-to-measure, quantitative readout as a surrogate for primary T cell function.
Several recent studies suggest that CAR stimulation activates characteristic T cell receptor (TCR) signaling pathways in primary human T cells that are ignited by the phosphorylation of immunoreceptor tyrosine-based activation motifs (ITAMs) of the CAR CD3ζ domain. Accordingly, CAR stimulation activates Src family kinases, leads to the phosphorylation of ZAP-70, and induces downstream signaling proteins that are known to control the transcription factors nuclear factor κB (NF-κB) and nuclear factor of activated T cells (NFAT).13, 14, 15 Intriguingly, the signaling machinery and transcription factor network of primary T cells are highly conserved in the T cell lymphoma line Jurkat. This includes the transcription factor family of NF-κB, which is associated with inhibitor of NF-κB (IκB) family members that retain NF-κB in the cytosol. Upon activation, IκB is degraded, which allows NF-κB to translocate into the nucleus where it binds to its target genes.16 Further, TCR activation induces the calcium-calmodulin-dependent phosphatase calcineurin, which dephosphorylates members of the NFAT family, leading to their translocation to the nucleus. There are four calcium-regulated members (NFATc1 to NFATc4) that remain inactive in the cytosol until the nuclear localization signal is unmasked by dephosphorylation.17 Both NF-κB and NFAT transcription factors are indicators for the strength and duration of T cell activation that has been received through the TCR. For instance, NFAT induction correlates positively with TCR stimulation and is a regulator of the resulting T cell response.18, 19, 20 In addition, it has been demonstrated that NF-κB induction depends on the magnitude of the TCR signal, affecting cytokine production, T cell proliferation, and survival.21, 22, 23
Therefore, we reasoned that Jurkat cells, equipped with NF-κB- and NFAT-responsive enhanced CFP (ECFP) and EGFP reporter genes, may be useful for establishing a CAR-screening platform. We demonstrate that Jurkat NF-κB/NFAT reporter cells, modified with ROR1-specific and CD19-specific CARs, generate a rapid and specific ECFP and EGFP reporter signal. Further, we performed screening campaigns involving small (n = 3) and large (n > 1 × 106) CAR libraries, and we show that NF-κB/NFAT reporter cells are capable of rapidly identifying lead CAR constructs that confer maximum antitumor reactivity in primary T cells.
Results
NF-κB and NFAT Are Activated upon CAR Engagement in Primary T Cells
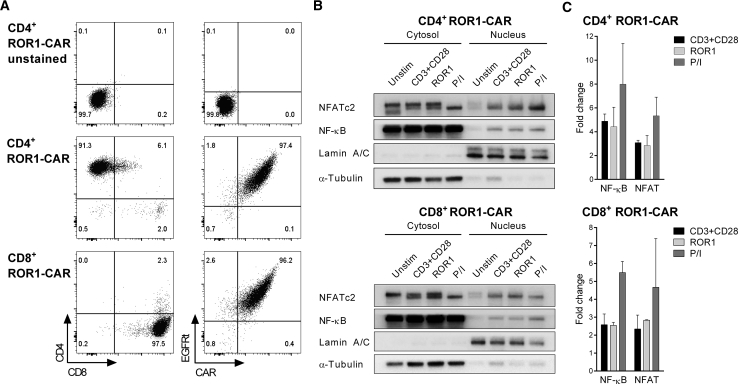
We hypothesized that CAR engagement would lead to the translocation of NF-κB and NFAT from the cytosol into the nucleus of primary T cells. Therefore, we modified CD8+ and CD4+ T cells with a ROR1-specific CAR recognizing the R12 epitope. We enriched CAR-expressing T cells using the truncated epidermal growth factor receptor (EGFRt) transduction marker, and we confirmed specific binding to soluble ROR1 protein (Figure 1A).
Figure 1.
Activation of NF-κB and NFAT in ROR1-CAR-T Cells after Antigen Stimulation
(A) Phenotype of CD4+ and CD8+ ROR1-CAR-T cells. Staining was performed with anti-EGFR mAb to detect the EGFRt transduction marker and AF647-labeled ROR1 protein to detect the CAR. (B) NF-κB and NFAT were detected by western blot in the nuclear and cytosolic fractions of ROR1-CAR-T cells. ROR1-CAR-T cells were either unstimulated or stimulated for 120 min with anti-CD3 and anti-CD28 mAbs, immobilized ROR1 protein, or PMA and ionomycin (P/I). Lamin A/C and α-tubulin served as loading controls for the nuclear and cytosolic fractions, respectively. (C) Nuclear enrichment of NF-κB and NFAT as quantified by densitometric analysis of western blots (n = 3 donors for CD4+ and n = 2 donors for CD8+ ROR1-CAR-T cells). Data were normalized to corresponding loading controls and presented as fold change ± SD relative to unstimulated T cells.
ROR1-CAR-T cells were then stimulated either with immobilized ROR1 protein or with anti-CD3 and anti-CD28 mAbs for 2 hr. At the end of the stimulation period, the T cells were lysed, and the cytosolic and nuclear fractions were obtained to track NF-κB and NFAT by western blot. The data show that both transcription factors were highly enriched in the nuclear fraction of CD4+ and CD8+ T cells after stimulation with ROR1 antigen (Figure 1B). In ROR1-CAR-T cells that had received no stimulus, only a minute fraction of NF-κB and NFAT could be detected in the nuclear fraction compared to the cytosolic fraction. Quantitative analysis revealed that the amounts of NF-κB and NFAT that could be detected in the nuclear fraction after stimulation with ROR1 protein through the CAR were similar to that obtained after stimulation with anti-CD3 and anti-CD28 mAbs and comparable in CD4+ and CD8+ T cells (Figure 1C). Concurrently, decreases in both transcription factors were observed in the cytosol by quantitative analysis (data not shown).
Taken together, these data demonstrate that the transcription factors NF-κB and NFAT are translocated into the nucleus of CD4+ and CD8+ ROR1-CAR-T cells after CAR engagement, suggesting they can be exploited as surrogates of CAR activation in a reporter system.
Expression of NF-κB and NFAT Reporter Genes in Jurkat Cells
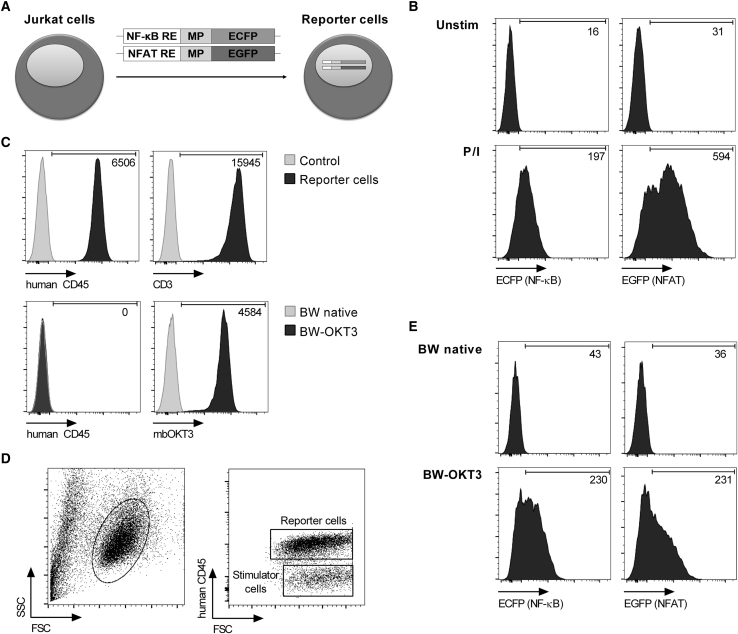
We reasoned that activation of NF-κB and NFAT after CAR engagement may be modeled in the human T cell lymphoma line Jurkat. To provide a simple and quantitative measurement of CAR activation, we constructed two reporter genes that induce expression of ECFP and EGFP under the control of NF-κB and NFAT, respectively (Figure 2A). Each of these genes contains a response element for the respective transcription factor and a minimal promoter that, by default, is inactive. We integrated both reporter genes in Jurkat cells, which from here on are termed reporter cells. As a proof of concept, we performed stimulation with phorbol 12-myristate 13-acetate (PMA) and ionomycin for 24 hr, and we detected a strong and uniform increase in ECFP and EGFP signals in reporter cells by flow cytometry. Without PMA and ionomycin stimulation, no reporter signal was detectable (Figure 2B).
Figure 2.
NF-κB and NFAT Activation in Jurkat Cells Detected with Reporter Genes
(A) Schematic representation of reporter cells with NF-κB- and NFAT-inducible ECFP and EGFP reporter genes, respectively. RE, response element; MP, minimal promoter. (B) MFI of ECFP and EGFP after stimulation of reporter cells with PMA and ionomycin (P/I). (C) Phenotype of reporter cells and BW5147 stimulator cells. The membrane-bound anti-CD3-scFv of BW-OKT3 cells was detected by its CD14 stem domain. Numeric values represent the MFI of transduced cells minus the MFI of control. (D) Co-culture of reporter cells with BW5147 stimulator cells distinguished by human CD45 detection during flow cytometry analysis. (E) MFI of ECFP and EGFP after stimulation of reporter cells with BW-OKT3 cells at a 2.5:1 ratio for 24 hr.
Next, we established a stimulator cell line that could be used in conjunction with reporter cells to express the antigen of interest in CAR-screening campaigns. We selected the mouse thymoma cell line BW5147, which expresses murine activating and inhibitory ligands that do not cross-react with human receptors on Jurkat cells, and thus it only induces negligible background signal.24 In co-culture experiments, reporter cells and BW5147 stimulator cells were distinguished by their differential expression of human CD45 (Figures 2C and 2D). To provide a positive control, we used BW5147 cells expressing a membrane-bound anti-CD3-single-chain fragment variable (scFv) derived from mAb OKT3 (from here on termed BW-OKT3 cells) to engage CD3 on reporter cells (Figure 2C). As anticipated, co-culture of reporter cells with BW-OKT3 stimulator cells resulted in high-level ECFP and EGFP reporter gene induction (Figure 2E).
In summary, these data show that NF-κB and NFAT reporter genes are active in Jurkat cells and enable quantitative readouts of cellular activation by flow cytometry.
CAR Stimulation Induces a Specific NF-κB and NFAT Signal in Reporter Cells
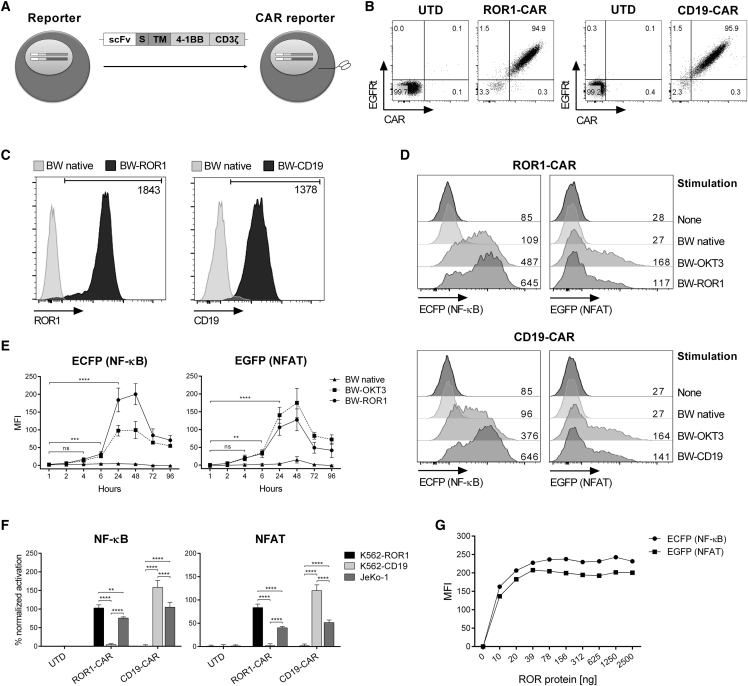
In the next set of experiments, we sought to determine the magnitude and kinetics of NF-κB and NFAT activation in reporter cells after CAR stimulation. Therefore, we modified reporter cells with either a ROR1-specific or a CD19-specific CAR (Figure 3A). We confirmed ROR1-CAR and CD19-CAR expression by staining with soluble ROR1 protein and anti-c-Myc mAb, respectively (Figure 3B).
Figure 3.
CAR-Mediated Activation of NF-κB and NFAT Reporter Genes
(A) Schematic representation of CAR-modified reporter cells. scFv, single-chain fragment variable; S, spacer; TM, transmembrane domain. (B) CAR expression of transduced reporter cells. ROR1-CAR was directly stained with AF647-labeled ROR1 protein, and CD19-CAR was directly detected by its N-terminal c-Myc tag. UTD, untransduced. (C) Target antigen expression of transduced BW5147 stimulator cells in comparison to native BW5147 cells. Numeric values represent the MFI of transduced cells minus the MFI of native cells. (D) MFI of ECFP and EGFP after stimulation of ROR1-CAR and CD19-CAR reporter cells with stimulator cells at a 2.5:1 ratio for 24 hr. (E) ECFP and EGFP expression kinetics of stimulated ROR1-CAR reporter cells. Data represent MFI ± SD. (F) NF-κB and NFAT activation of ROR1-CAR and CD19-CAR reporter cells co-cultured with human K562 cells expressing either ROR1 or CD19 or JeKo-1 cells expressing both antigens. The activation in percent ± SD was calculated by normalizing CAR stimulation to the positive control (stimulation with BW-OKT3). (G) MFI of ECFP and EGFP after a 24-hr stimulation of ROR1-CAR reporter cells with plate-bound ROR1 protein. Statistical significance for (E) (n = 4) and (F) (n = 3) was determined using two-way ANOVA with Holm-Sidak post hoc test; ns, not significant; **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Then, we performed co-culture experiments of CAR reporter cells and BW5147 stimulator cells that had been transduced to express ROR1 or CD19 (termed BW-ROR1 and BW-CD19) (Figure 3C). For both, ROR1-CAR and CD19-CAR reporter cells, we detected a specific and strong reporter signal after 24 hr of stimulation (Figure 3D). No reporter gene activation was observed after co-culture with native BW5147 cells. Analysis of reporter gene activation over time showed that the ROR1-CAR-induced ECFP and EGFP signal was first detectable after 4 hr and significantly increased after 6 hr of stimulation (Figure 3E). The reporter gene signal continued to increase at the 24-hr-analysis time point, and it reached its maximum after 48 hr. At the subsequent analysis time points of 72 and 96 hr, the reporter signal had decreased but was still higher compared to baseline signal and the signal obtained with native BW5147 cells, which were included in the assay as a reference. Of interest, the amplitude of the reporter signal after antigen-specific stimulation with BW-ROR1 cells relative to BW-OKT3 cells was much higher for NF-κB compared to NFAT, likely because the CAR construct contained a 4-1BB co-stimulatory domain (Figure 3E).
We also confirmed that ROR1-CAR and CD19-CAR reporter cells were able to respond to antigen on human cancer cell lines, as exemplified by specific, high-level NF-κB and NFAT reporter signal after stimulation with JeKo-1 lymphoma cells (ROR1+ CD19+) and K562 cells that had been transduced to express ROR1 (K562-ROR1) or CD19 (K562-CD19), respectively (Figure 3F). Further, we confirmed that reporter cells were able to respond to purified antigen, as exemplified by ROR1-CAR reporter cells that we probed against titrated amounts of plate-bound ROR1 protein (Figure 3G).
Collectively, these data demonstrate that CAR reporter cells respond to their cognate antigen with a specific and readily detectable NF-κB and NFAT reporter signal. The reporter assay has a rapid turnaround time and delivers results within 6–24 hr after assay setup.
Reporter Cells Rapidly Identify the Lead CAR Construct in a Small-Scale Screening Campaign
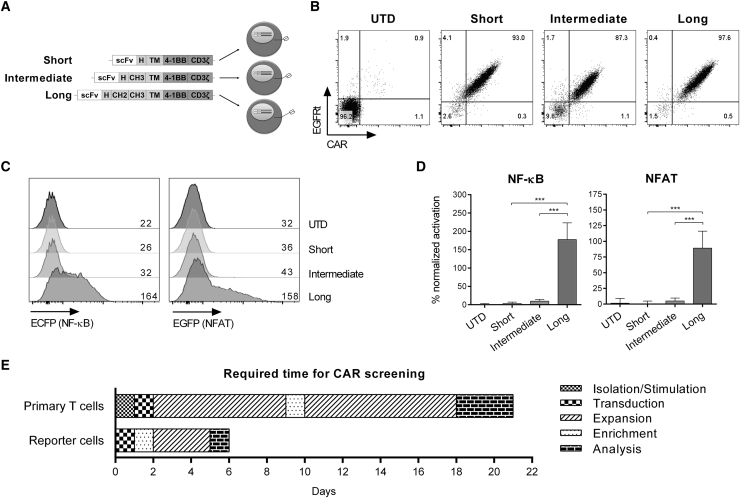
Next, we employed our CAR reporter cells in a screening campaign with the objective to identify the optimal CAR construct from a small panel of candidates. In previous work, we had demonstrated that a ROR1-specific CAR targeting the R11 epitope requires a long IgG4-Fc spacer consisting of hinge, CH2, and CH3 domains to bind membrane-bound ROR1 protein on tumor cells.5 We therefore expressed a small library of 3 R11 ROR1-CAR constructs, comprising either the long spacer or the alternative intermediate (hinge and CH3) and short (hinge only) spacer variants in reporter cells (Figure 4A). We confirmed uniform expression of each CAR construct on the cell surface using soluble ROR1 protein (Figure 4B), and then we co-cultured the CAR reporter cells with BW-ROR1 cells for 24 hr. The CAR reporter cells readily identified the functional R11 ROR1-CAR with the long spacer domain, and they distinguished it from the non-functional R11 ROR1-CAR variants with the intermediate or short spacer domain (Figure 4C). We repeated the reporter assay 3 times and obtained highly reproducible results (Figure 4D).
Figure 4.
Small-Scale Screening Campaign with a ROR1-CAR Spacer Library in Reporter Cells
(A) Schematic of the ROR1-CAR spacer library with short, intermediate, and long spacer domains derived from IgG4-Fc. scFv, single-chain fragment variable; H, hinge; CH, constant heavy domain. (B) CAR expression of transduced reporter cells. ROR1-CAR was directly stained with AF647-labeled ROR1 protein. UTD, untransduced. (C) MFI of ECFP and EGFP after stimulation of CAR reporter cells with BW-ROR1 cells at a 2.5:1 ratio for 24 hr. (D) NF-κB and NFAT activation in percent ± SD of BW-ROR1 stimulated ROR1-CAR reporter cells normalized to the positive control (stimulation with BW-OKT3). Statistical significance (n = 3) was determined using one-way ANOVA with Holm-Sidak post hoc test; ***p < 0.001. (E) Time required for a small-scale screening campaign with reporter cells or primary human T cells.
Notably, the entire screening campaign to identify the functional CAR construct from this small ROR1-CAR spacer library was completed in 6 days, which compares favorably to the amount of time (approximately 21 days) typically required in a screening campaign with primary T cells (Figure 4E). Together, these results show that small-scale library-screening campaigns can be conveniently performed on the reporter cell platform and substantially shorten the time required for distinguishing functional from non-functional CAR constructs compared to testing in primary T cells.
Reporter Cells Support Large-Scale Screening Campaigns with CAR scFv Libraries
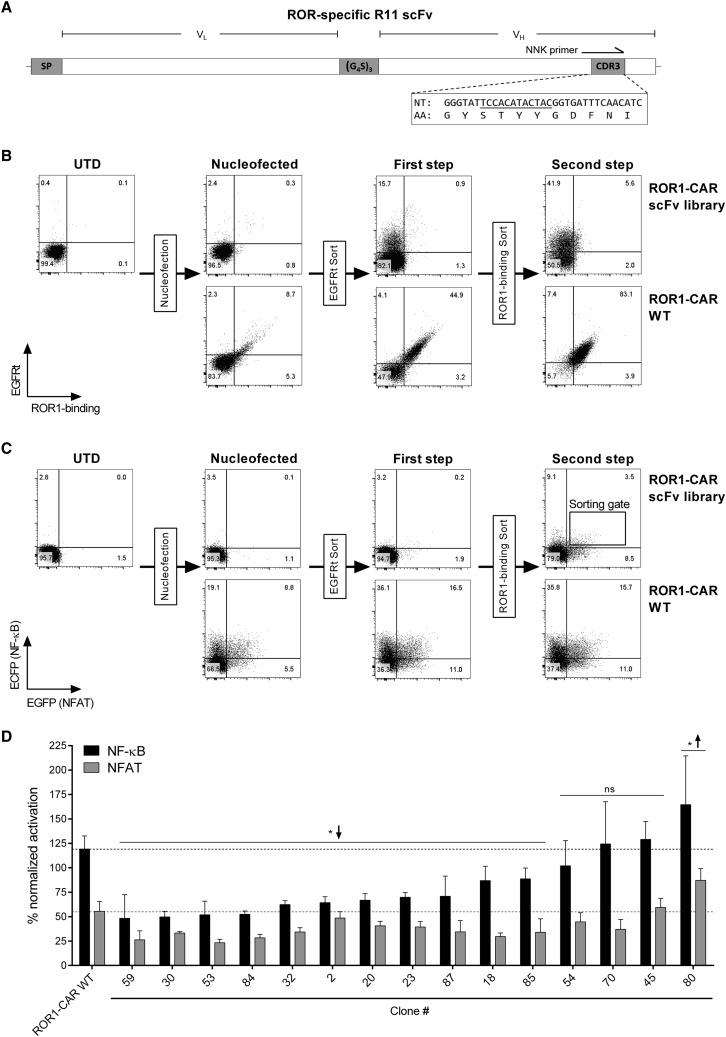
Finally, we challenged reporter cells to a large-scale screening campaign with a CAR scFv library to identify the optimal CAR construct from a large panel of candidates. We established a library from the ROR1-specific R11 scFv using PCR-based site-restricted mutagenesis with NNK primers to introduce nucleotide changes in the VH domain complementarity-determining region 3 (CDR3) (Figure 5A) that is critical for binding to ROR1, as we have recently shown.25 We estimated that our mutagenesis generated a total of 1.05 × 106 (NNK)4 nucleotide sequence variants, resulting in approximately 2 × 105 distinct amino acid sequence variants. From the 1.05 × 106 (NNK)4 nucleotide sequence variants, only six variants would still encode the wild-type (WT) amino acid sequence. Our PCR-based mutagenesis was not expected to re-generate the WT nucleotide sequence, which we, however, confirmed to be present in our library prior to initiating the screening campaign.
Figure 5.
Large-Scale Screening Campaign of a ROR1-CAR Library with scFv Mutations
(A) Schematic representation of the WT ROR1-specific R11 scFv with nucleotide (NT) and amino acid (AA) sequence of the VH CDR3 region. Nucleotides targeted by mutagenesis are underlined. SP, signal peptide; (G4S)3, linker. (B) EGFRt expression and ROR1-specific CAR expression of ROR1-CAR scFv library and ROR1-CAR WT reporter cells after nucleofection and subsequent EGFRt-based magnetic bead enrichment (first step) and flow cytometry-based sorting with AF647-labeled ROR1 protein (second step). (C) ECFP and EGFP reporter signal after stimulation of ROR1-CAR scFv library and ROR1-CAR WT reporter cells with BW-ROR1 cells at a 2.5:1 ratio for 24 hr. (D) NF-κB and NFAT activation in percent ± SD of BW-ROR1-stimulated ROR1-CAR scFv library clones normalized to the positive control (stimulation with BW-OKT3) and compared to the ROR1-CAR WT (NF-κB, black line; NFAT, gray line). Statistical significance (n = 3) in comparison to the ROR1-CAR WT was determined using two-way ANOVA with Holm-Sidak post hoc test. ↑, higher; ↓, lower; ns, not significant; *p < 0.05.
To generate the ROR1-CAR scFv library, we integrated the R11 scFv mutants into the CAR framework of the pT2/HB Sleeping Beauty transposon donor vector, and then we performed virus-free gene transfer into reporter cells. Nucleofection was performed into 64 × 106 reporter cells, which resulted in ∼2.7% EGFRt-positive cells, of which only a small fraction (<0.3%) was still capable of binding soluble ROR1 protein (Figure 5B). To reduce the amount of non-nucleofected reporter cells and reporter cells that encoded nucleotide variants comprising a stop codon, we performed two sequential sorting steps enriching for CAR reporter cells that expressed EGFRt (first step) and were able to bind ROR1 protein (second step). This increased the population of interest (double positive, i.e., EGFRt positive and binding to ROR1 protein) to 5.6%. As a reference, we also inserted the original ROR1-CAR with WT R11 scFv (ROR1-CAR WT) into reporter cells, and we confirmed that all EGFRt-positive cells showed specific binding to ROR1 protein (Figure 5B). Accordingly, this enrichment strategy increased the number of library reporter cells that displayed NF-κB ECFP and NFAT EGFP reporter signals after stimulation with BW-ROR1 cells (Figure 5C).
Reporter Cells Retrieve a Rare R11 scFv Mutant Capable of Binding ROR1
We then performed single-cell sorting for ROR1-CAR scFv library reporter cells that showed the highest level of ECFP and EGFP expression (sorting gate shown in Figure 5C). From this sorting campaign, a total of 100 cell clones were re-analyzed for reporter activity upon stimulation with BW-ROR1 cells, and the 25 clones with highest ECFP and EGFP signal intensity were selected. Of these, 10 clones had to be excluded from further analysis due to insufficient expansion or unspecific reactivity, which left 15 clones for detailed analysis (Figure 5D). In 11 of these 15 clones, the signal of at least one of the two reporter genes was significantly lower (* ↓) than the reference set by reporter cells expressing the ROR1-CAR WT. In 4 of these 15 clones (45, 54, 70, and 80), the reporter signal was similar (ns) or higher (* ↑) compared to the ROR1-CAR WT reference. These clones were, therefore, of particular interest.
In 3 of these 4 clones (45, 70, and 80), more than two nucleotide sequences were detected by VH CDR3 sequencing, apparently because multiple genomic transposon integrations per cell had occurred (Table 1). In each of the three clones, there was at least one nucleotide sequence that encoded a novel and unique CDR3 amino acid sequence. However, in each of the three clones, we also detected the WT scFv nucleotide sequence. Preliminary testing of CARs carrying the novel CDR3 amino acid motifs did not show specific binding to ROR1 protein and no recognition of ROR1+ stimulator cells, suggesting that the reporter signal had been generated by the ROR1-CAR with WT scFv in these three reporter cell clones. In clone 80, in which we detected the highest reporter signal, we also detected the highest level of CAR expression, as assessed by staining for EGFRt. This observation provides an explanation for the higher reporter signal compared to clones 45, 54, 70, and WT reference.
Table 1.
Sequence Analysis of Cell Clones with Highest NF-κB and NFAT Activation
| Library Clone | CDR3 Nucleotide Sequence | Amino Acid Sequence |
|---|---|---|
| 45 | 5′-TCCACATACTAC-3′ (WT) | STYY (WT) |
| 5′-GATACGTATTAG-3′ | DTY– | |
| 5′-ACGTTGAATTCG-3′ | TLNS | |
| 5′-GATCCGCCGCAT-3′ | DPPH | |
| 54 | 5′-TCGACTTATTAT-3′ (non-WT) | STYY (WT) |
| 70 | 5′-TCCACATACTAC-3′ (WT) | STYY (WT) |
| 5′-TAGCGTGCTCCT-3′ | –RAP | |
| 80 | 5′-TCCACATACTAC-3′ (WT) | STYY (WT) |
| 5′-TAGTTTACGGCT-3′ | –FTA | |
| 5′-GTGTGGGTTACG-3′ | VWVT | |
| 5′-ACGCCGCTGCCT-3′ | TPLP | |
| 5′-ATGACTGGGTAG-3′ | MTG– | |
| 5′-CAGGCTTGGATG-3′ | QAWM | |
| 5′-ATGAGTATGATT-3′ | MSMI | |
| Reference | 5′-TCCACATACTAC-3′ (WT) | STYY (WT) |
Analysis of clone 54 revealed only a single nucleotide sequence (n = 1), suggesting that only a single (or low number of) genomic insertion of the Sleeping Beauty (SB) transposon vector had occurred. This nucleotide sequence was distinct from the WT scFv nucleotide sequence; however, upon translating the genetic code, we found that this nucleotide sequence encoded the amino acid sequence STYY, i.e., the same motif contained in WT VH CDR3. As such, our reporter platform had retrieved an scFv nucleotide variant encoding a functional ROR1-CAR that we had calculated to occur with a frequency of 6 in 1.05 × 106.
In aggregate, these data demonstrate that reporter cells are able to identify functional CAR constructs in large-scale library-screening campaigns, where lead candidates are only present with very low frequency. In addition, the data suggest that the VH CDR3 region is critical for the reactivity of the R11 ROR1-CAR and may not be readily modified without compromising the ability of binding to ROR1 antigen. Collectively, the data highlight the potential utility of NF-κB/NFAT reporter cells as a screening tool in CAR development.
Discussion
Translational research in CAR-T cell immunotherapy involves a rapidly increasing spectrum of target antigens and CAR designs, as well as extensive testing in preclinical models in vitro and in vivo. To accelerate this process, reduce cost, and facilitate high-throughput analyses, we established a screening platform that is based on Jurkat cells, an immortalized CD4+ T cell lymphoma line. For several decades, Jurkat cells have been used to study TCR signaling and its underlying molecular mechanisms, because, with a few exceptions, Jurkat cells comprise the entire downstream signaling machinery of primary T cells, turning them into an intuitive tool for the analysis of T cell immune receptors.26, 27
We have recently derived a reporter cell line from Jurkat cells that reads out the activity of NF-κB and NFAT through ECFP and EGFP reporter genes, respectively. We have demonstrated that these reporter cells provide a quantitative measure for stimulation through the endogenous TCR as well as activating and inhibitory ligands.28, 29 Other investigators have used similar reporter gene-modified Jurkat cells to examine antibody-dependent cell-mediated cytotoxicity, inhibitory molecules, immune checkpoint molecules, as well as virus- and tumor-specific TCRs.30, 31, 32, 33 In the present study, we demonstrate that Jurkat NF-κB/NFAT reporter cells can be employed as an effective tool in CAR-screening campaigns. The ECFP and EGFP reporter genes utilized in our study are easy to read out, and they permit quantitative analyses of intact cells by flow cytometry, which allows rapid and automated acquisition of a high number of events from a high number of samples. The advantages of using fluorescent proteins as a reporter rather than luciferase activity is that several reporter signals can be analyzed simultaneously and the reporter signal can be traced back to a single cell. In addition, fluorescent protein reporter assays require considerably less hands-on time, which makes them more suitable for high-throughput screening than bioluminescence-based systems.
In previous studies, screening campaigns were performed with Jurkat cells to detect TCR-dependent cytosolic calcium flux that provides a rapid signal, but the analysis procedure is substantially more complex, is only moderately quantitative, and provides a single output.34, 35 Another approach that has been pursued in Jurkat cells is the analysis of activation markers like CD25 and CD95 or cytokines like interleukin-2 (IL-2) and tumor necrosis factor alpha (TNF-α), but their expression is often low, and their detection requires antibodies, making them inappropriate for platform-based screenings.36, 37 We therefore focused on NF-κB and NFAT, both crucial transcription factors that are strongly induced upon stimulation through the endogenous TCR in primary human T cells and in Jurkat cells.18, 23, 28 Because CARs integrate structural and functional elements of the TCR and engage similar signaling molecules upon stimulation,13, 38 we reasoned that NF-κB and NFAT would serve as indicators and surrogates of CAR-mediated activation. Indeed, several studies reported NFAT activation via inducible reporter gene systems or inducible cytokine secretion in T cells and Jurkat cells,39, 40, 41, 42 and, similarly, the induction of NF-κB signaling after CAR triggering has been described.43, 44 These observations are supported by our data demonstrating an accumulation of NF-κB and NFAT in the nucleus of primary T cells and an activation of reporter genes in Jurkat cells after CAR stimulation.
In the present study, we equipped reporter cells with CAR constructs specific for ROR1, which is expressed in several hematologic malignancies, including chronic lymphocytic leukemia and mantle cell lymphoma, as well as several prevalent epithelial cancers, including lung adenocarcinoma and triple-negative breast cancer.45 We also modified reporter cells with a CD19-specific CAR that has obtained clinical proof of concept in patients with acute lymphoblastic leukemia, non-Hodgkin lymphoma, and chronic lymphocytic leukemia.46 For each of these CAR constructs, the reporter cells generated a specific and high-level NF-κB and NFAT reporter signal. The reporter signal was detectable as early as 6 hr following CAR stimulation, and it reached its maximum between 24 and 48 hr. The CAR reporter cells provided a clear distinction between functional and non-functional CAR constructs in a rapid and highly competitive turnaround time. Our data show that reporter cells can be used to screen cell preparations where the antigen of interest is expressed on the cell surface and protein preparations where the antigen is plate bound. Therefore, it is conceivable to engage CAR reporter cells with one or multiple known or even unknown specificities in the analyses of primary tumor cell samples and tumor cell lysates and in ligandome analyses to discover novel tumor antigens.
We conducted several screening campaigns with our CAR reporter cell platform. In one example, we performed a small-scale screening involving a CAR spacer library, which was established in earlier work, showing that the extracellular spacer domain is a decisive feature in CAR design.5 Indeed, the ROR1-CAR targeting the membrane-proximal R11 epitope in the ROR1 kringle domain requires a long IgG4-Fc spacer to function. Using a panel of R11 ROR1-CARs with short, intermediate, and long spacer designs, reporter cells readily identified the functional CAR construct due to specific and high-level NF-κB and NFAT reporter signals. Compared to conventional analyses with primary T cells, the screening campaign with CAR reporter cells required less than one-third of the time for identifying the lead construct.
In a second example, we performed a large-scale screening campaign involving a CAR scFv library that was generated from the ROR1-specific R11 scFv by site-restricted mutagenesis. We have recently obtained a co-crystal structure of the R11 scFv in complex with the kringle domain of ROR1, confirming that VH CDR3 is critical for binding.25 Accordingly, introducing mutations to VH CDR3 was anticipated to cause a loss of specificity and affinity in the majority of cases. This provided the opportunity of challenging the reporter platform to retrieve the exceedingly rare library variants that comprised the WT amino acid sequence or an alternative amino acid sequence that was still capable of binding ROR1. Our screening campaign yielded four reporter cell clones that stood out through their high NF-κB and NFAT signals. In one of these clones, VH CDR3 contained a novel, unique nucleotide sequence that had been introduced by our mutagenesis but still encoded the WT amino acid sequence STYY. The occurrence of multiple genomic insertions in reporter cells after conventional lentiviral and transposon-based gene transfer is a technical challenge that complicated our library-screening campaign. Strategies of targeted gene insertion into distinct genomic loci are currently emerging,47, 48 and they will facilitate future screening campaigns with CAR libraries where each reporter cell only expresses a single library variant.
Several prior studies have rationally modified the VL and VH CDR regions to modulate the binding affinity and properties of mAbs and scFvs.49, 50 It is conceivable that such modifications would also alter the binding affinity and properties of CARs, thereby affecting the activation and function of CAR-T cells. In line with our finding that the VH CDR3 region of R11 is critical for binding ROR1,25 we have not yet identified novel scFv variants with improved functionality over the WT from our ROR1-CAR scFv library. It is conceivable and supported by the R11/ROR1 co-crystal structure that the STYY motif is indeed optimal in terms of the paratope-epitope interaction and that identifying novel scFv variants will therefore be challenging. Other stretches in the CDR or framework regions of VH of R11 (VL of R11 does not interact with the antigen) are likely more suitable for yielding non-WT amino acid sequences after randomization and reporter cell-based screening.
On the other hand, it should be noted that, due to our selection criteria (high NF-κB and NFAT signal, expansive growth, and specific activation), clones with low-affinity CAR variants were likely excluded from the clones that we included into our sequencing analysis. In ongoing experiments, we have adjusted the selection criteria and analyzed a larger number of clones to support the identification of novel CAR variants with altered (lower) affinity from this library. In the small- and large-scale screening campaigns reported in this study, the lead CAR constructs were functionally superior to the other candidates. In screening campaigns, where functional CAR constructs are contained with higher frequency and/or functional CAR variants differ more subtly, a higher number of sequential pre-enrichment and screening rounds may be required for identifying the optimal CAR construct. In particular, the identification of high-affinity CAR variants, which are anticipated to generate a stronger reporter signal compared to the WT, may be fostered by performing a higher number of iterative pre-enrichment steps.
In summary, we report on the development of a standardized CAR-screening platform that employs Jurkat NF-κB/NFAT reporter cells. The reporter cell system is attractive as a platform technology because it is independent of testing in primary T cells, exportable, and scalable to accommodate small- to large-scale CAR library screenings. We are, therefore, confident that this reporter cell platform will facilitate and accelerate the preclinical development and evaluation of novel CAR constructs.
Materials and Methods
Human Subjects
Peripheral blood from healthy donors was obtained after written informed consent to participate in research protocols that were approved by the Institutional Review Board of the University of Würzburg.
Cell Lines
Jurkat cells (clone E6.1) were transduced with retroviruses encoding NF-κB- and NFAT-inducible ECFP and EGFP reporter genes and clonally selected by limiting dilution.28 Jurkat NF-κB/NFAT reporter cells (reporter cells) were maintained in Iscove’s modified Dulbecco’s medium (IMDM) with 25 mM HEPES, 4 mM L-glutamine, 1× GlutaMAX, 10% fetal calf serum (FCS), and 100 U/mL penicillin-streptomycin. The murine cell line BW5147 (thymoma) and the human cancer cell lines K562 (erythroid leukemia) and JeKo-1 (mantle cell lymphoma) were cultured in RPMI-1640 medium with 25 mM HEPES, 10% FCS, 1× GlutaMAX, and 100 U/mL penicillin-streptomycin.
Reporter Genes
Genes encoding ECFP and EGFP were cloned downstream of minimal promoters controlled by response elements for NF-κB and NFAT, respectively. Both reporter gene cassettes were subcloned into the self-inactivating retroviral vector pSIRV.28
CAR Constructs
The VH and VL domains of the antibody FMC63 were fused via a Whitlow linker to generate a CD19-specific scFv,51 and a c-Myc tag (EQKLISEEDL) in duplicate was added at the N terminus to facilitate detection by flow cytometry. Similarly, the VH and VL domains of antibodies R12 and R11 were coupled via a (G4S)3 linker, resulting in ROR1-specific scFvs.52 Both CD19 scFv and ROR1 R12 scFv were cloned into lentiviral vector epHIV7 upstream of an IgG4-Fc hinge spacer, a CD28 transmembrane domain, and intracellular CD3ζ and 4-1BB domains.6 The ROR1-specific R11 scFv was cloned into lentiviral vector epHIV7 upstream of an IgG4-Fc short (hinge domain), intermediate (hinge + CH3 domain), or long (hinge + CH2 + CH3 domain) spacer, fused to a CD28 transmembrane domain, and intracellular CD3ζ and 4-1BB domains.5 All constructs included an EGFRt transduction marker separated from the CAR gene by a viral 2A sequence.
ROR1-CAR scFv Library
The ROR1-specific R11 scFv was selected for preparation of the CAR scFv library. Using NNK-doping strategy (N = A, C, G, or T; K = G or T) for site-restricted mutagenesis of its VH CDR3 region, 12 nt encoding amino acids STYY that are involved in ROR1 kringle domain binding were mutated.25 In detail, PCR of the right arm of the R11 scFv template DNA was performed using a forward degenerated primer P1 (5′-GCG ACG TAT TTC TGT GCG AGG GGG TAT NNK NNK NNK NNK GGT GAT TTC AAC ATC TGG GGT-3′), binding in the VH CDR3 region that covers the four amino acids with NNK codons to introduce mutations, and a reverse primer P2 (5′-CCC GTT AAG GTC CTG ATG-3′), binding in the IgG1-Fc spacer domain. A second PCR was performed with forward primer P3 (5′-CCT ACT CTA GAA GCT GGG TAC CG-3′) and reverse primer P4 (5′-ATA CCC CCT CGC ACA GAA ATA CGT CGC-3′) to amplify the left arm of the scFv, including the signal peptide and the VL and the residual VH domains to obtain an amplicon with overlapping sequence (underlined in P1 and P4) to the right arm amplicon. PCR products were recovered by gel purification, and the complete scFv was fused by overlapping PCR using primers P3 and P2. Then the scFv mutants were cloned via restriction sites into the pT2/HB Sleeping Beauty transposon donor vector53 upstream of a CD28 transmembrane domain and intracellular CD3ζ and 4-1BB domains by GeneArt (Thermo Fisher Scientific, Regensburg, Germany) to generate the ROR1-CAR scFv library. Library accuracy analysis confirmed a correctness of 91%, with 74 of 81 clones containing four sense codons in the correct reading frame.
CAR Gene Transfer into Reporter Cells
Reporter cells were transduced in medium supplemented with 5 μg/mL polybrene using lentiviral vectors encoding the CAR, and expression was analyzed after 2 days. CAR-expressing reporter cells were enriched via the EGFRt transduction marker using biotinylated anti-epidermal growth factor receptor (EGFR) antibodies and anti-biotin microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany). The ROR1-CAR scFv library was transfected into reporter cells using the 4D-Nucleofector X Unit and nucleofection kit SE (Lonza, Cologne, Germany), according to the manufacturer’s instructions. In brief, 5 μg CAR encoding Sleeping Beauty transposon donor vector and 2.5 μg SB100X minicircle vector53 were added per 2 × 106 cells, and nucleofection was performed with program CL-120.
Generation of Stimulator Cells
BW-OKT3 cells were generated using a previously described vector encoding membrane-bound OKT3 that was created by fusing the anti-CD3 scFv to the human CD14 stem domain.24 BW5147 and K562 cells were modified with the full-length sequences of human ROR1 (UniProt: Q01973) or human CD19 (UniProt: P15391).
CAR-T Cell Manufacturing
CD4+ and CD8+ T cells were isolated from peripheral blood mononuclear cells (PBMCs) via magnetic cell separation and activated with anti-CD3/anti-CD28 Dynabeads (Thermo Fisher Scientific, Darmstadt, Germany). The following day, 5 μg/mL polybrene was added to T cells, and transduction with lentiviral vectors encoding the CAR was performed at an MOI of 5. Dynabeads were removed after 7 days, CAR expression was analyzed, and CAR-T cells were expanded using a rapid expansion protocol.54
Flow Cytometry
Fluorophore-labeled mAbs for CD3, CD4, CD8, CD19, ROR1, and c-Myc were purchased from Miltenyi Biotec (Bergisch Gladbach, Germany). Streptavidin-phycoerythrin (SA-PE) and fluorophore-labeled mAbs for CD14 and CD45 were purchased from BioLegend (San Diego, CA, USA). The anti-EGFR mAb Cetuximab (Bristol-Myers Squibb, New York, NY, USA) was labeled in-house with biotin or AF647. Recombinant human ROR1 protein linked to fluorophore AF647 was a kind gift of Professor Johannes Huppa (Medizinische Universität Wien, Wien, Austria). Antibody staining was performed according to the manufacturer’s instructions. Measurements were done on a FACSCanto II flow cytometer (BD Biosciences, Heidelberg, Germany). FACS was performed by the Cell Sorting core facility at the Institute for Virology and Immunobiology Würzburg using a FACSAria III (BD Biosciences, Heidelberg, Germany). Data were analyzed with FlowJo software (version 10.4, Tree Star, Ashland, OR, USA).
Reporter Cell Assay
CAR reporter cells (5 × 104) and stimulator cells (2 × 104) expressing the relevant target antigen were added to round-bottom 96-well plates in duplicates. K562 and JeKo-1 cells were pre-stained with eFluor 670 dye (Thermo Fisher Scientific, Darmstadt, Germany), according to the manufacturer’s instructions. After a 24-hr incubation at 37°C, the co-cultures were harvested, and reporter cells were distinguished from murine BW5147 stimulator cells by the detection of human CD45 and from human cell lines (K562/JeKo-1) by the detection of eFluor 670 via flow cytometry. Reporter gene activation was analyzed by measuring the geometric mean fluorescence intensity (MFI) of ECFP and EGFP.
Cell Lysis, Fractionation, and Western Blot
ROR1-CAR-T cells were added to 12-well plates pre-coated with anti-CD3 mAb (2.5 μg/mL) and soluble anti-CD28 mAb (6 μg/mL), recombinant human ROR1 protein (0.6 μg/mL), or PMA (120 ng/mL) and ionomycin (3 μg/mL), and they were incubated for 2 hr at 37°C. Cells were harvested, and cytosolic and nuclear lysates were separated by rapid, efficient, and practical (REAP) fractionation.55 Protein concentration was determined using the DC Protein Assay Kit (Bio-Rad, Munich, Germany), according to the manufacturer’s instructions, and 10–15 μg protein was added to a 10% SDS polyacrylamide gel. After separation by electrophoresis, proteins were transferred to a polyvinylidene fluoride (PVDF) membrane and blocked with 5% milk in Tris-buffered saline with Tween 20 (TBS-T). Proteins were detected using antibodies for NFATc2 (clone D43B1), NF-κB p65 (clone D14E12), Lamin A/C (clone 4C11), and α-tubulin (clone 11H10), all purchased from Cell Signaling Technology (Frankfurt am Main, Germany). Protein bands were visualized using the ChemiDoc MP gel imaging system (Bio-Rad, Munich, Germany).
Genomic DNA Analysis
Sequence analysis of the R11 VH CDR3 region was performed on genomic DNA of reporter cells obtained with a genomic DNA extraction kit (Thermo Fisher Scientific, Darmstadt, Germany), according to the manufacturer’s instructions. The scFv was amplified by PCR with primers covering the 5′ and 3′ ends using Phusion High-Fidelity DNA polymerase (New England Biolabs, Frankfurt am Main, Germany). Amplified fragments were separated by electrophoresis, and PCR amplicons were purified and transferred to TOPO-TA Cloning vector (Thermo Fisher Scientific, Darmstadt, Germany) for sequencing.
Statistical Analysis
Data are presented as mean ± SD. Statistical analysis was performed with GraphPad Prism Software using one-way or two-way ANOVA with Holm-Sidak post hoc test. Significance is indicated as follows: ns, not significant; *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Author Contributions
Conceptualization, J.R., P.S., and M.H.; Methodology, J.R., T.N., and M.H.; Investigation, J.R., T.N., and H.P.; Resources, S.J., J.L., P.S., H.P., and C.R.; Writing – Original Draft, J.R. and M.H.; Writing – Review & Editing, J.R., H.P., J.L., P.S., C.R., and M.H.; Visualization, J.R. and M.H.; Supervision, P.S., H.E., and M.H.
Conflicts of Interest
M.H. is co-inventor on a patent application (PCT/US2013/055862) related to CAR spacer design that has been filed by the Fred Hutchinson Cancer Research Center (Seattle, WA) and licensed by JUNO Therapeutics, Inc. The other authors declare no competing interests.
Acknowledgments
M.H. was supported by the Young Scholar Program of the Bavarian Academy of Sciences (Junges Kolleg, Bayerische Akademie der Wissenschaften) and is an extraordinary member of the Bavarian Academy of Sciences. This work was supported by grants from the German Cancer Aid (Deutsche Krebshilfe e.V., Max Eder Program 110313 to M.H. and CONCERT Translational Oncology Program 111975 to M.H.).
References
- 1.Maude S.L., Frey N., Shaw P.A., Aplenc R., Barrett D.M., Bunin N.J., Chew A., Gonzalez V.E., Zheng Z., Lacey S.F. Chimeric antigen receptor T cells for sustained remissions in leukemia. N. Engl. J. Med. 2014;371:1507–1517. doi: 10.1056/NEJMoa1407222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brentjens R.J., Davila M.L., Riviere I., Park J., Wang X., Cowell L.G., Bartido S., Stefanski J., Taylor C., Olszewska M. CD19-targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia. Sci. Transl. Med. 2013;5:177ra38. doi: 10.1126/scitranslmed.3005930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abate-Daga D., Davila M.L. CAR models: next-generation CAR modifications for enhanced T-cell function. Mol. Ther. Oncolytics. 2016;3:16014. doi: 10.1038/mto.2016.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qin L., Lai Y., Zhao R., Wei X., Weng J., Lai P., Li B., Lin S., Wang S., Wu Q. Incorporation of a hinge domain improves the expansion of chimeric antigen receptor T cells. J. Hematol. Oncol. 2017;10:68. doi: 10.1186/s13045-017-0437-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hudecek M., Sommermeyer D., Kosasih P.L., Silva-Benedict A., Liu L., Rader C., Jensen M.C., Riddell S.R. The nonsignaling extracellular spacer domain of chimeric antigen receptors is decisive for in vivo antitumor activity. Cancer Immunol. Res. 2015;3:125–135. doi: 10.1158/2326-6066.CIR-14-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hudecek M., Lupo-Stanghellini M.T., Kosasih P.L., Sommermeyer D., Jensen M.C., Rader C., Riddell S.R. Receptor affinity and extracellular domain modifications affect tumor recognition by ROR1-specific chimeric antigen receptor T cells. Clin. Cancer Res. 2013;19:3153–3164. doi: 10.1158/1078-0432.CCR-13-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhong X.-S., Matsushita M., Plotkin J., Riviere I., Sadelain M. Chimeric antigen receptors combining 4-1BB and CD28 signaling domains augment PI3kinase/AKT/Bcl-XL activation and CD8+ T cell-mediated tumor eradication. Mol. Ther. 2010;18:413–420. doi: 10.1038/mt.2009.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Golubovskaya V., Wu L. Different Subsets of T Cells, Memory, Effector Functions, and CAR-T Immunotherapy. Cancers (Basel) 2016;8:36. doi: 10.3390/cancers8030036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sommermeyer D., Hudecek M., Kosasih P.L., Gogishvili T., Maloney D.G., Turtle C.J., Riddell S.R. Chimeric antigen receptor-modified T cells derived from defined CD8+ and CD4+ subsets confer superior antitumor reactivity in vivo. Leukemia. 2016;30:492–500. doi: 10.1038/leu.2015.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gomes-Silva D., Mukherjee M., Srinivasan M., Krenciute G., Dakhova O., Zheng Y., Cabral J.M.S., Rooney C.M., Orange J.S., Brenner M.K., Mamonkin M. Tonic 4-1BB Costimulation in Chimeric Antigen Receptors Impedes T Cell Survival and Is Vector-Dependent. Cell Rep. 2017;21:17–26. doi: 10.1016/j.celrep.2017.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Minter R.R., Sandercock A.M., Rust S.J. Phenotypic screening-the fast track to novel antibody discovery. Drug Discov. Today. Technol. 2017;23:83–90. doi: 10.1016/j.ddtec.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 12.Janzen W.P. Screening technologies for small molecule discovery: the state of the art. Chem. Biol. 2014;21:1162–1170. doi: 10.1016/j.chembiol.2014.07.015. [DOI] [PubMed] [Google Scholar]
- 13.Karlsson H., Svensson E., Gigg C., Jarvius M., Olsson-Strömberg U., Savoldo B., Dotti G., Loskog A. Evaluation of Intracellular Signaling Downstream Chimeric Antigen Receptors. PLoS ONE. 2015;10:e0144787. doi: 10.1371/journal.pone.0144787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guedan S., Posey A.D., Jr., Shaw C., Wing A., Da T., Patel P.R., McGettigan S.E., Casado-Medrano V., Kawalekar O.U., Uribe-Herranz M. Enhancing CAR T cell persistence through ICOS and 4-1BB costimulation. JCI Insight. 2018;3:96976. doi: 10.1172/jci.insight.96976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng W., O’Hear C.E., Alli R., Basham J.H., Abdelsamed H.A., Palmer L.E., Jones L.L., Youngblood B., Geiger T.L. PI3K orchestration of the in vivo persistence of chimeric antigen receptor-modified T cells. Leukemia. 2018;32:1157–1167. doi: 10.1038/s41375-017-0008-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Q., Verma I.M. NF-kappaB regulation in the immune system. Nat. Rev. Immunol. 2002;2:725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- 17.Macian F. NFAT proteins: key regulators of T-cell development and function. Nat. Rev. Immunol. 2005;5:472–484. doi: 10.1038/nri1632. [DOI] [PubMed] [Google Scholar]
- 18.Marangoni F., Murooka T.T., Manzo T., Kim E.Y., Carrizosa E., Elpek N.M., Mempel T.R. The transcription factor NFAT exhibits signal memory during serial T cell interactions with antigen-presenting cells. Immunity. 2013;38:237–249. doi: 10.1016/j.immuni.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brogdon J.L., Leitenberg D., Bottomly K. The potency of TCR signaling differentially regulates NFATc/p activity and early IL-4 transcription in naive CD4+ T cells. J. Immunol. 2002;168:3825–3832. doi: 10.4049/jimmunol.168.8.3825. [DOI] [PubMed] [Google Scholar]
- 20.Maguire O., Tornatore K.M., O’Loughlin K.L., Venuto R.C., Minderman H. Nuclear translocation of nuclear factor of activated T cells (NFAT) as a quantitative pharmacodynamic parameter for tacrolimus. Cytometry A. 2013;83:1096–1104. doi: 10.1002/cyto.a.22401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song J., So T., Croft M. Activation of NF-kappaB1 by OX40 contributes to antigen-driven T cell expansion and survival. J. Immunol. 2008;180:7240–7248. doi: 10.4049/jimmunol.180.11.7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kingeter L.M., Paul S., Maynard S.K., Cartwright N.G., Schaefer B.C. Cutting edge: TCR ligation triggers digital activation of NF-kappaB. J. Immunol. 2010;185:4520–4524. doi: 10.4049/jimmunol.1001051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thaker Y.R., Schneider H., Rudd C.E. TCR and CD28 activate the transcription factor NF-κB in T-cells via distinct adaptor signaling complexes. Immunol. Lett. 2015;163:113–119. doi: 10.1016/j.imlet.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leitner J., Kuschei W., Grabmeier-Pfistershammer K., Woitek R., Kriehuber E., Majdic O., Zlabinger G., Pickl W.F., Steinberger P. T cell stimulator cells, an efficient and versatile cellular system to assess the role of costimulatory ligands in the activation of human T cells. J. Immunol. Methods. 2010;362:131–141. doi: 10.1016/j.jim.2010.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qi J., Li X., Peng H., Cook E.M., Dadashian E.L., Wiestner A., Park H., Rader C. Potent and selective antitumor activity of a T cell-engaging bispecific antibody targeting a membrane-proximal epitope of ROR1. Proc. Natl. Acad. Sci. USA. 2018;115:E5467–E5476. doi: 10.1073/pnas.1719905115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gioia L., Siddique A., Head S.R., Salomon D.R., Su A.I. A genome-wide survey of mutations in the Jurkat cell line. BMC Genomics. 2018;19:334. doi: 10.1186/s12864-018-4718-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abraham R.T., Weiss A. Jurkat T cells and development of the T-cell receptor signalling paradigm. Nat. Rev. Immunol. 2004;4:301–308. doi: 10.1038/nri1330. [DOI] [PubMed] [Google Scholar]
- 28.Jutz S., Leitner J., Schmetterer K., Doel-Perez I., Majdic O., Grabmeier-Pfistershammer K., Paster W., Huppa J.B., Steinberger P. Assessment of costimulation and coinhibition in a triple parameter T cell reporter line: Simultaneous measurement of NF-κB, NFAT and AP-1. J. Immunol. Methods. 2016;430:10–20. doi: 10.1016/j.jim.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 29.Jutz S., Hennig A., Paster W., Asrak Ö., Dijanovic D., Kellner F., Pickl W.F., Huppa J.B., Leitner J., Steinberger P. A cellular platform for the evaluation of immune checkpoint molecules. Oncotarget. 2017;8:64892–64906. doi: 10.18632/oncotarget.17615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng Z.J., Garvin D., Paguio A., Moravec R., Engel L., Fan F., Surowy T. Development of a robust reporter-based ADCC assay with frozen, thaw-and-use cells to measure Fc effector function of therapeutic antibodies. J. Immunol. Methods. 2014;414:69–81. doi: 10.1016/j.jim.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 31.Gonzales A.M., Orlando R.A. A Jurkat transcriptional reporter cell line for high-throughput analysis of the nuclear factor-kappaB signaling pathway. N. Biotechnol. 2009;26:244–250. doi: 10.1016/j.nbt.2009.06.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anmole G., Kuang X.T., Toyoda M., Martin E., Shahid A., Le A.Q., Markle T., Baraki B., Jones R.B., Ostrowski M.A. A robust and scalable TCR-based reporter cell assay to measure HIV-1 Nef-mediated T cell immune evasion. J. Immunol. Methods. 2015;426:104–113. doi: 10.1016/j.jim.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 33.Rosskopf S., Leitner J., Paster W., Morton L.T., Hagedoorn R.S., Steinberger P., Heemskerk M.H.M. A Jurkat 76 based triple parameter reporter system to evaluate TCR functions and adoptive T cell strategies. Oncotarget. 2018;9:17608–17619. doi: 10.18632/oncotarget.24807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bartelt R.R., Cruz-Orcutt N., Collins M., Houtman J.C.D. Comparison of T cell receptor-induced proximal signaling and downstream functions in immortalized and primary T cells. PLoS ONE. 2009;4:e5430. doi: 10.1371/journal.pone.0005430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Imboden J.B., Weiss A., Stobo J.D. The antigen receptor on a human T cell line initiates activation by increasing cytoplasmic free calcium. J. Immunol. 1985;134:663–665. [PubMed] [Google Scholar]
- 36.Khlusov I.A., Litvinova L.S., Shupletsova V.V., Dunets N.A., Khaziakhmatova O.G., Yurova K.A., Khlusova M.Y., Sharkeev Y.P. Morphofunctional changes of Jurkat T lymphoblasts upon short-term contact with a relief calcium phosphate surface. Cell Tissue Biol. 2017;11:59–64. [PubMed] [Google Scholar]
- 37.Shatrova A.N., Mityushova E.V., Aksenov N.A., Marakhova I.I. CD25 expression on the surface of Jurkat cells. Cell Tissue Biol. 2015;9:364–370. [Google Scholar]
- 38.Terakura S., Yamamoto T.N., Gardner R.A., Turtle C.J., Jensen M.C., Riddell S.R. Generation of CD19-chimeric antigen receptor modified CD8+ T cells derived from virus-specific central memory T cells. Blood. 2012;119:72–82. doi: 10.1182/blood-2011-07-366419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chmielewski M., Kopecky C., Hombach A.A., Abken H. IL-12 release by engineered T cells expressing chimeric antigen receptors can effectively Muster an antigen-independent macrophage response on tumor cells that have shut down tumor antigen expression. Cancer Res. 2011;71:5697–5706. doi: 10.1158/0008-5472.CAN-11-0103. [DOI] [PubMed] [Google Scholar]
- 40.Frigault M.J., Lee J., Basil M.C., Carpenito C., Motohashi S., Scholler J., Kawalekar O.U., Guedan S., McGettigan S.E., Posey A.D., Jr. Identification of chimeric antigen receptors that mediate constitutive or inducible proliferation of T cells. Cancer Immunol. Res. 2015;3:356–367. doi: 10.1158/2326-6066.CIR-14-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schroten C., Kraaij R., Veldhoven J.L.M., Berrevoets C.A., den Bakker M.A., Ma Q., Sadelain M., Bangma C.H., Willemsen R.A., Debets R. T cell activation upon exposure to patient-derived tumor tissue: a functional assay to select patients for adoptive T cell therapy. J. Immunol. Methods. 2010;359:11–20. doi: 10.1016/j.jim.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 42.Schaft N., Lankiewicz B., Gratama J.W., Bolhuis R.L.H., Debets R. Flexible and sensitive method to functionally validate tumor-specific receptors via activation of NFAT. J. Immunol. Methods. 2003;280:13–24. doi: 10.1016/s0022-1759(03)00067-x. [DOI] [PubMed] [Google Scholar]
- 43.Pulè M.A., Straathof K.C., Dotti G., Heslop H.E., Rooney C.M., Brenner M.K. A chimeric T cell antigen receptor that augments cytokine release and supports clonal expansion of primary human T cells. Mol. Ther. 2005;12:933–941. doi: 10.1016/j.ymthe.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 44.Li G., Boucher J.C., Kotani H., Park K., Zhang Y., Shrestha B., Wang X., Guan L., Beatty N., Abate-Daga D., Davila M.L. 4-1BB enhancement of CAR T function requires NF-κB and TRAFs. JCI Insight. 2018;3:121322. doi: 10.1172/jci.insight.121322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang S., Chen L., Wang-Rodriguez J., Zhang L., Cui B., Frankel W., Wu R., Kipps T.J. The onco-embryonic antigen ROR1 is expressed by a variety of human cancers. Am. J. Pathol. 2012;181:1903–1910. doi: 10.1016/j.ajpath.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kochenderfer J.N., Rosenberg S.A. Treating B-cell cancer with T cells expressing anti-CD19 chimeric antigen receptors. Nat. Rev. Clin. Oncol. 2013;10:267–276. doi: 10.1038/nrclinonc.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eyquem J., Mansilla-Soto J., Giavridis T., van der Stegen S.J.C., Hamieh M., Cunanan K.M., Odak A., Gönen M., Sadelain M. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature. 2017;543:113–117. doi: 10.1038/nature21405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Voigt K., Gogol-Döring A., Miskey C., Chen W., Cathomen T., Izsvák Z., Ivics Z. Retargeting sleeping beauty transposon insertions by engineered zinc finger DNA-binding domains. Mol. Ther. 2012;20:1852–1862. doi: 10.1038/mt.2012.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang W.P., Green K., Pinz-Sweeney S., Briones A.T., Burton D.R., Barbas C.F., 3rd CDR walking mutagenesis for the affinity maturation of a potent human anti-HIV-1 antibody into the picomolar range. J. Mol. Biol. 1995;254:392–403. doi: 10.1006/jmbi.1995.0626. [DOI] [PubMed] [Google Scholar]
- 50.Fellouse F.A., Wiesmann C., Sidhu S.S. Synthetic antibodies from a four-amino-acid code: a dominant role for tyrosine in antigen recognition. Proc. Natl. Acad. Sci. USA. 2004;101:12467–12472. doi: 10.1073/pnas.0401786101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nicholson I.C., Lenton K.A., Little D.J., Decorso T., Lee F.T., Scott A.M., Zola H., Hohmann A.W. Construction and characterisation of a functional CD19 specific single chain Fv fragment for immunotherapy of B lineage leukaemia and lymphoma. Mol. Immunol. 1997;34:1157–1165. doi: 10.1016/s0161-5890(97)00144-2. [DOI] [PubMed] [Google Scholar]
- 52.Yang J., Baskar S., Kwong K.Y., Kennedy M.G., Wiestner A., Rader C. Therapeutic potential and challenges of targeting receptor tyrosine kinase ROR1 with monoclonal antibodies in B-cell malignancies. PLoS ONE. 2011;6:e21018. doi: 10.1371/journal.pone.0021018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Monjezi R., Miskey C., Gogishvili T., Schleef M., Schmeer M., Einsele H., Ivics Z., Hudecek M. Enhanced CAR T-cell engineering using non-viral Sleeping Beauty transposition from minicircle vectors. Leukemia. 2017;31:186–194. doi: 10.1038/leu.2016.180. [DOI] [PubMed] [Google Scholar]
- 54.Riddell S.R., Greenberg P.D. The use of anti-CD3 and anti-CD28 monoclonal antibodies to clone and expand human antigen-specific T cells. J. Immunol. Methods. 1990;128:189–201. doi: 10.1016/0022-1759(90)90210-m. [DOI] [PubMed] [Google Scholar]
- 55.Suzuki K., Bose P., Leong-Quong R.Y.Y., Fujita D.J., Riabowol K. REAP: A two minute cell fractionation method. BMC Res. Notes. 2010;3:294. doi: 10.1186/1756-0500-3-294. [DOI] [PMC free article] [PubMed] [Google Scholar]