Abstract
Bone marrow-derived mesenchymal stem cells (BMSCs) have the potential to differentiate into osteoblasts or adipocytes, and the shift between osteogenic and adipogenic differentiation determines bone mass. The aim of this study was to identify whether lncRNAs are involved in the differentiation commitment of BMSCs during osteoporosis. Here, we found ORLNC1, a functionally undefined lncRNA that is highly conserved, which exhibited markedly higher expression levels in BMSCs, bone tissue, and the serum of OVX-induced osteoporotic mice than sham-operated counterparts. Notably, a similar higher abundance of lncRNA-ORLNC1 expression was also observed in the bone tissue of osteoporotic patients. The transgenic mice overexpressing lncRNA-ORLNC1 showed a substantial increase in the osteoporosis-associated bone loss and decline in the osteogenesis of BMSCs. The BMSCs pretreated with lncRNA-ORLNC1-overexpressing lentivirus vector exhibited the suppressed capacity of osteogenic differentiation and oppositely enhanced adipogenic differentiation. We then established that lncRNA-ORLNC1 acted as a competitive endogenous RNA (ceRNA) for miR-296. Moreover, miR-296 was found markedly upregulated during osteoblast differentiation, and it accelerated osteogenic differentiation by targeting Pten. Taken together, our results indicated that the lncRNA-ORLNC1-miR-296-Pten axis may be a critical regulator of the osteoporosis-related switch between osteogenesis and adipogenesis of BMSCs and might represent a plausible therapeutic target for improving osteoporotic bone loss.
Keywords: osteoporosis, BMSCs, lncRNA, miR-296, Pten, osteogenic differentiation, adipogenic differentiation, bone formation
BMSCs have the potential of multi-lineage differentiation. The imbalance between osteogenesis and adipogenesis of BMSCs is associated with osteoporosis. Yang et al. first demonstrate that lncRNA-ORLNC1 upregulation led to the inhibition of osteogenesis and enhancement of adipogenesis of BMSCs. Thus, ORLNC1 is suggested as a new target for treating osteoporosis.
Introduction
Osteoporosis is a systemic bone metabolism disease characterized by decreased bone mineral density (BMD) and deterioration of bone microarchitecture, conferring a high risk of bone fragility and fractures.1 To date, the incidence of osteoporosis has dramatically increased with each passing year.2 Studies showed that approximately one in two women and up to one in four men aged 50 and above endure osteoporosis.3 It has been an enormous public health problem all over the world.4 Bone tissue is a balanced internal microenvironment with bone formation and resorption, which is critically required for the maintenance of the bone homeostasis.5 Accumulating evidence supports the theory that osteoporosis is the result of decreased osteogenesis combined with increased adipogenesis.6 It is, therefore, clear that strengthening the function of osteoblasts to restore the normal bone homeostasis is of paramount significance for the treatment of osteoporosis.
Bone marrow-derived mesenchymal stem cells (BMSCs) are a type of multipotent stem cells, which have the capacity of self-renewal and the potential of multi-lineage differentiation.7 In certain inductive microenvironments, BMSCs can differentiate into a variety of cell types, such as osteoblasts, adipocytes, chondrocytes, neurons, myocytes, myoblasts, etc.8 Such a property makes BMSCs an attractive candidate for cell therapy and tissue engineering.9 Osteoporosis is a consequence of the imbalance between osteogenic and adipogenic differentiation of BMSCs.10 As the progenitor cells of osteoblasts and adipocytes, BMSCs are considered seed cells for the treatment of osteoporosis.11 However, the molecular mechanisms underlying the switch-like diversion between osteogenic and adipogenic differentiation in BMSCs remain largely unknown.
Long non-coding RNAs (lncRNAs) are a recently defined family of transcripts of longer than 200 nt.12, 13 Numerous studies have indicated that lncRNAs are involved in cell proliferation, differentiation, and apoptosis, as well as in the pathogenesis of various diseases, including cardiovascular conditions, neurological disorders, and cancers.14, 15 Recently, several lncRNAs were found to participate in osteogenesis, osteoarthritis, and osteosarcoma.16, 17, 18 As another class of non-coding RNAs, microRNAs (miRNAs) of 18–22 nt in length are involved in regulating gene expression by directly binding to the 3′ UTR of mRNAs.19 One of the mechanisms for the effects of lncRNAs is to act as competitive endogenous RNAs (ceRNAs) to sponge miRNAs by sequence complementarity and, thereby, reduce the functional availability of the targeted miRNAs. Such a lncRNA-miRNA interaction forms a regulatory network to control the expression of downstream target genes.20, 21 For instance, lncRNA-LINCMD1 was shown to activate the expression of muscle-specific genes (MAML1 and Mef2c) by absorbing miR-133 and miR-135, leading to the differentiation of myoblasts.22 Yet, whether lncRNAs are involved in regulating osteogenic and adipogenic potentials of BMSCs has remained unclear.
To shed light on this aspect, we conducted the present study using a mouse model of osteoporosis induced by ovariectomy (OVX) surgery. We analyzed the lncRNA expression profile to identify the dysregulated lncRNAs in osteoporotic tissues. We focused on lncRNA-ORLNC1 (osteoporosis-related lncRNA 1) that was found overexpressed in bone tissue, serum, and BMSCs of OVX mice, by characterizing the role of this lncRNA in osteoporotic differentiation and delineating the underlying cellular and molecular mechanisms.
Results
Osteoporosis Induces lncRNA-ORLNC1 Overexpression in Mice and Humans
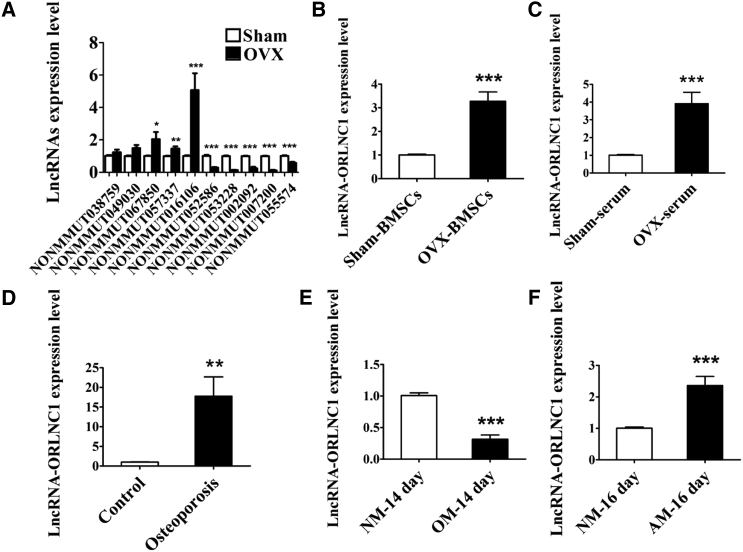
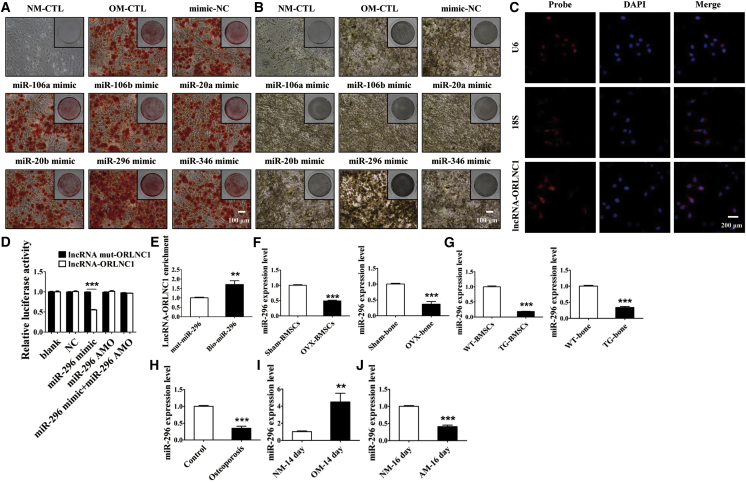
As the first step toward understanding the role of lncRNAs in osteoporosis, we conducted microarray analysis to identify the differentially expressed lncRNAs in a mouse model of osteoporosis induced by OVX (Figures S1A–S1E). This allowed us to identify a subset of lncRNAs that were differentially expressed between the osteoporotic mice and the sham-operated control counterparts (Figure S1E). Among lncRNAs with high conservative property from the microarray data, we chose the top five lncRNAs from the highest expression and the top five ones from the lowest expression in OVX-induced osteoporotic mice compared with the sham group. The differential expression of ten lncRNAs was verified by qRT-PCR (Figure 1A). Among these lncRNAs, NONCODE: NONMMUT016106 was the most significantly upregulated one, with a 2.5-fold increase in microarray analysis and a 5-fold upregulation by using qRT-PCR (Figure S1E; Figure 1A). We therefore centered our subsequent experiments on this lncRNA. For convenience, we named this lncRNA ORLNC1. Notably, upregulation of lncRNA-ORLNC1 was also consistently observed in the BMSCs and serum of osteoporotic mice by qRT-PCR (Figures 1B and 1C).
Figure 1.
lncRNA-ORLNC1 Is Associated with Osteoporosis and Differentiation of BMSCs
(A) Real-time qPCR verification of differential expression of ten lncRNAs chosen from microarray analysis in the bone tissues from sham and OVX mice. (B and C) Upregulation of lncRNA-ORLNC1 in BMSCs (B) and serum (C) from OVX mice compared with the sham group. (D) Real-Time qPCR was performed to assess the expression of lncRNA-ORLNC1 in the osteoporotic patients (n = 5). (E and F) The mRNA levels of lncRNA-ORLNC1 in BMSCs, which were induced with OM and AM during osteogenic (E) and adipogenic (F) differentiation. NM, normal growth medium; OM, osteogenic induced medium; AM, adipogenic induced medium. *p < 0.05, **p < 0.01, and ***p < 0.001.
Intriguingly, the sequences of lncRNA-ORLNC1 are conserved across species, including human and mouse. Furthermore, upregulation of lncRNA-ORLNC1 in the bone tissues from osteoporotic patients relative to healthy controls was also confirmed (Figure 1D).
To further investigate the possible role of lncRNA-ORLNC1 in osteogenesis and adipogenesis, BMSCs were induced into osteoblasts and adipocytes using osteogenic induced medium (OM) and adipogenic induced medium (AM) in a time-dependent manner (Figures S2A–S2E). Interestingly, qRT-PCR revealed that the expression level of lncRNA-ORLNC1 was notably decreased during the osteoblast mineralization phase and significantly elevated over the course of adipogenic differentiation in the BMSCs of 8-week-old C57BL/6J mice (Figures 1E and 1F).
Overexpression of lncRNA-ORLNC1 Suppresses Osteogenesis and Promotes Adipogenesis of BMSCs
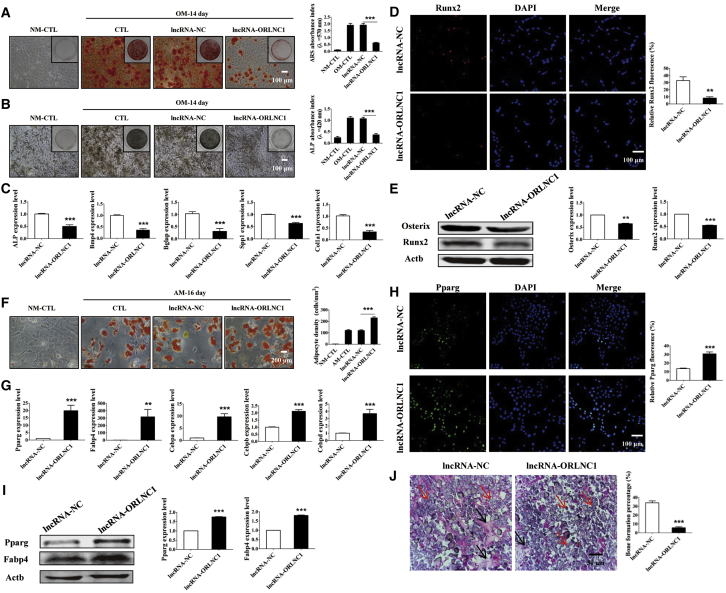
The above results suggest that the expression of lncRNA-ORLNC1 is upregulated in osteoporotic mice and patients and may be involved in regulating the balance between osteogenic and adipogenic differentiation of BMSCs. To determine the role of overexpression of lncRNA-ORLNC1 in the osteogenic differentiation of BMSCs, the cells were transfected with lncRNA-ORLNC1 overexpressing lentivirus successfully (Figure S3A). As shown in Figure 2A, alizarin red S (ARS) staining showed that transfection of overexpressed lncRNA-ORLNC1 inhibited the mineralized nodule formation in BMSCs after osteogenic differentiation of 14 days, but the negative control (NC) did not produce any appreciable effects (Figure 2A). Alkaline phosphatase (ALP) staining confirmed that the BMSCs treated with lncRNA-ORLNC1 were slightly stained after 14-day induction, indicating that the cells failed to differentiate into mature osteoblasts (Figure 2B). Quantitative analysis of ARS and ALP stainings showed that the overexpression of lncRNA-ORLNC1 suppressed osteogenesis of BMSCs (Figures 2A and 2B). Moreover, the overexpression of lncRNA-ORLNC1 reduced the mRNA levels of five important osteogenic marker genes after 14-day osteogenic differentiation, including ALP, bone morphogenetic protein 4 (Bmp4), osteocalcin (Bglap), osteopontin-like protein (Spp1), and collagen type I, alpha 1 (Col1a1), known to be crucial for osteogenesis (Figure 2C). Immunofluorescence assay further supported that lncRNA-ORLNC1 overexpression repressed the expression of runt-related transcription factor 2 (Runx2) after osteogenesis of 3 days (Figure 2D). In addition, western blot confirmed that transfection of lncRNA-ORLNC1 decreased the expression of Osterix and Runx2 after osteogenic induction of 7 days (Figure 2E).
Figure 2.
lncRNA-ORLNC1 Inhibits Osteogenesis and Stimulates Adipogenesis of BMSCs
(A) ARS staining and quantification deriving from osteogenic BMSCs with lentivirus overexpressing lncRNA-ORLNC1 or lncRNA-NC were performed to assess the osteogenic capacity after 14 days. Cells grown in the normal growth medium were shown as a reference. NM, normal growth medium; OM, osteogenic induced medium. Scale bar, 100 μm. (B) BMSCs stained with ALP staining showed calcium deposits in the presence of lncRNA-ORLNC1 overexpression compared with NC. Scale bar, 100 μm. (C) The expression levels of ALP, Bmp4, Bglap, Spp1, and Col1a1 significantly decreased after treatment with overexpression of lncRNA-ORLNC1. (D) Immunostaining of BMSCs transfected with lncRNA-ORLNC1 overexpression exhibited decreased Runx2-positive cells. Runx2, red; DAPI, blue. Scale bar, 100 μm. (E) Relative protein levels of Runx2 and Osterix after treatment with lncRNA-ORLNC1 overexpression or lncRNA-NC at day 7 of the osteogenic differentiation. (F) Increase of adipogenic differentiation by lncRNA-ORLNC1 overexpression treatment. Scale bar, 200 μm. (G) The mRNA levels of adipogenic genes. (H) Images showed Pparg expression in BMSCs treated with lncRNA-ORLNC1 overexpression compared to NC. Pparg, green; DAPI, blue. Scale bar, 100 μm. (I) Western blot was applied to ensure the protein levels of Pparg and Fabp4 in BMSCs. (J) H&E staining and quantification of bone regeneration. Relative bone growth surface area compared to total surface. The black arrows indicate the bone formation, and the red arrows indicate hydroxyapatite. Scale bar, 50 μm. **p < 0.01, and ***p < 0.001.
For adipogenic differentiation, the adipogenic potential of BMSCs was investigated by oil red O (ORO) staining after 16 days. As shown in Figure 2F, lncRNA-ORLNC1 overexpression promoted lipid droplet formation of BMSCs 16 days after adipogenic differentiation. The mRNA levels of five adipocyte markers, including peroxisome proliferator activated receptor gamma (Pparg), fatty acid-binding protein 4 (Fabp4), CCAAT/enhancer-binding protein alpha (Cebpa), CCAAT/enhancer-binding protein beta (Cebpb), and CCAAT/enhancer-binding protein delta (Cebpd), were elevated by lncRNA-ORLNC1 overexpression after adipogenic differentiation of 16 days (Figure 2G). In the confocal fluorescence images, lncRNA-ORLNC1 treatment increased Pparg-positive BMSCs 4 days after adipogenic differentiation, which was in accordance with the results of western blot analysis performed after 8-day adipogenic differentiation (Figures 2H and 2I).
To determine whether lncRNA-ORLNC1 treatment leads to the decline of bone formation, the BMSCs transfected with lncRNA-ORLNC1 overexpression or NC were implanted into the backside of nude mice. After 8 weeks, the implants were collected to examine whether lncRNA-ORLNC1 affected the bone formation. H&E staining showed that BMSC-ORLNC1 formed less bone tissue than BMSC-NC in immunodeficient mice after 8-week transplantation (Figure 2J).
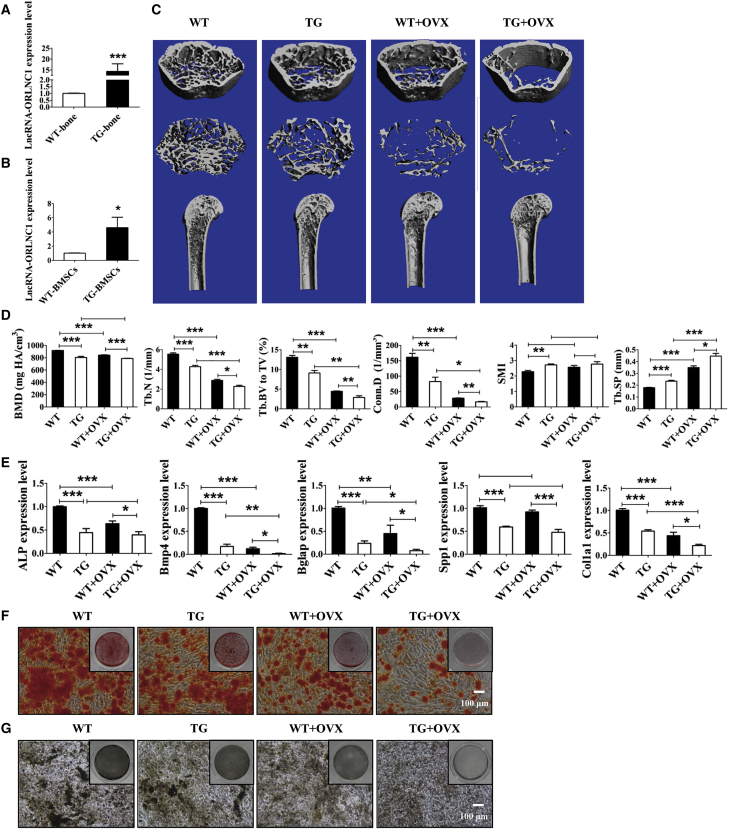
We then continued to investigate whether lncRNA-ORLNC1 is a physiologically relevant regulator of osteoporosis. To this end, we generated Osterix-derived lncRNA-ORLNC1 transgenic (TG) mice using the conventional gene-targeting technique for detailed analysis. The expression levels of lncRNA-ORLNC1 were significantly increased in the bone tissue and BMSCs of TG mice, as detected by qRT-PCR, relative to wild-type (WT) littermates (Figures 3A and 3B). Bone microarchitecture in the femora of WT mice, TG mice, WT mice with OVX, and TG mice counterparts was analyzed by micro-computed tomography (microCT) (Figures 3C and 3D). MicroCT demonstrated that the BMD, trabecular number (Tb.N), connectivity density (Conn.D), and trabecular bone volume relative to tissue volume (Tb.BV to TV) were much lower, but the trabecular separation (Tb.SP) was higher, in the femora of TG mice than in those of the age-matched WT mice (Figure 3D). Additionally, TG mice exhibited decreased bone mass, and increased structure model index (SMI), which indicates the relative amount of plates (Figure 3D). Furthermore, overexpression of lncRNA-ORLNC1 suppressed mRNA expression of the osteogenic markers in the bone tissue of TG mice compared to WT mice (Figure 3E). To confirm these observations, we isolated BMSCs from TG and WT mice and induced BMSCs into osteoblasts for 14 days. Consistent with the ALP staining, ARS staining demonstrated that BMSCs of TG mice displayed reduced potential of osteogenic differentiation (Figures 3F and 3G).
Figure 3.
Overexpression of lncRNA-ORLNC1 Results in Osteoporosis and Impairs Osteogenic Capacity in a Mouse Model of Osteoporosis
(A and B) lncRNA-ORLNC1 expression was markedly upregulated in the bone tissue (A) and BMSCs (B) from lncRNA-ORLNC1-overexpressing transgenic mice. WT, wild-type mice; TG, transgenic mice. (C and D) Representative μCT images (C) and quantitative analysis (D) of the femur from WT and TG mice indicated the bone loss of TG mice compared to WT mice. WT + OVX, wild-type mice with OVX surgery; TG + OVX, transgenic mice with OVX surgery. (E) The mRNA levels of ALP, Bmp4, Bglap, Spp1, and Col1a1 in the bone tissue of TG mice compared with WT mice. (F and G) ARS (F) and ALP (G) stainings exhibited impaired osteogenic differentiation potential of BMSCs in TG mice and TG mice with OVX surgery. Scale bar, 100 μm. *p < 0.05, **p < 0.01, and ***p < 0.001.
Knockdown of lncRNA-ORLNC1 Increases Osteogenic Differentiation and Decreases Adipogenic Differentiation of BMSCs
We have shown that the gain of function of lncRNA-ORLNC1 suppressed osteogenesis and promoted adipogenesis of BMSCs. It was thus anticipated that the loss of function of lncRNA-ORLNC1 should produce the opposite results. Our subsequent experiments indeed supported this notion.
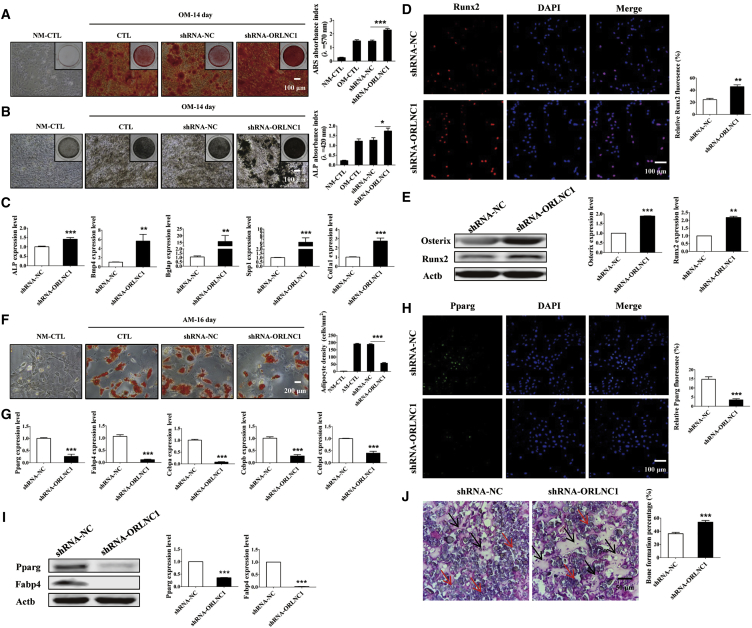
To identify the effect of knockdown of lncRNA-ORLNC1 in the osteogenic differentiation of BMSCs, the cells were transfected with shRNA-ORLNC1 (short hairpin RNA-ORLNC1 as an inhibitor of lncRNA-ORLNC1) successfully (Figure S3A). First, BMSCs treated with shRNA-ORLNC1 had strengthened osteogenic potential of BMSCs with increased mineralization, as indicated by ARS and ALP stainings (Figures 4A and 4B). Consistently, the mRNA levels of osteoblast-specific genes (ALP, Bmp4, Bglap, Spp1, and Col1a1) were increased by shRNA-ORLNC1 during subsequent osteoblast differentiation for 14 days (Figure 4C). In agreement with the changes of their mRNA levels, the expression levels of the osteoblastic genes were also elevated after transfection with shRNA-ORLNC1, as revealed by western blot and immunofluorescence techniques (Figures 4D and 4E). On the other hand, shRNA-ORLNC1 decreased the number and cell surface area of adipocytes after the 16-day adipogenic differentiation of BMSCs (Figure 4F). Meanwhile, the mRNA levels of adipogenic differentiation-related genes (Pparg, Fabp4, Cebpa, Cebpb, and Cebpd) were significantly downregulated by shRNA-ORLNC1 in BMSCs treated with AM for 16 days (Figure 4G). Moreover, BMSCs treated with shRNA-ORLNC1 exhibited fewer Pparg-positive cells than the NC group (Figure 4H). Pparg and Fabp4 protein levels were also significantly diminished by the knockdown of lncRNA-ORLNC1 after adipogenic differentiation of 8 days (Figure 4I). This result agreed with qRT-PCR analysis and immunofluorescence assay.
Figure 4.
Knockdown of lncRNA-ORLNC1 Increases Osteoblast Maturation and Decreases Adipogenesis of BMSCs
(A and B) ARS (A) and ALP (B) stainings and quantitative analysis (A and B) of matrix mineralization showed BMSCs treated with shRNA-ORLNC1 increased the osteogenic ability compared with BMSCs pretreated with NC after osteogenic induction for 14 days. Scale bar, 100 μm. (C) Real-time qPCR analysis showed the increased levels of osteogenic markers in BMSCs with treatment of shRNA-ORLNC1 compared with NC as indicated. (D) Immunofluorescence staining for osteogenic protein Runx2 in BMSCs induced into osteoblasts for 3 days after transfection with shRNA-ORLNC1, normalized to the negative control group. Runx2, red; DAPI, blue. Scale bar, 100 μm. (E) Western blot analysis showed the increased levels of osteoblast genes in BMSCs after treatment with shRNA-ORLNC1. (F) ORO staining was performed to determine the number of adipocytes in BMSCs after adipogenic differentiation of 16 days. Scale bar, 200 μm. (G) The mRNA levels of adipogenesis-related genes were attenuated in the presence of shRNA-ORLNC1, as determined by real-time qPCR. (H and I) Immunofluorescence staining (H) and western blot (I) analysis indicated that the protein expression of adipocyte-specific genes were inhibited by shRNA-ORLNC1 treatment. Pparg, green; DAPI, blue. Scale bar, 100 μm. (J) H&E-stained sections of heterotopic bone showed the increased bone formation after treatment with shRNA-ORLNC1. The black arrows indicate the bone formation, and the red arrows indicate hydroxyapatite. Scale bar, 50 μm. *p < 0.05, **p < 0.01, and ***p < 0.001.
Then, the pro-osteogenic effects of shRNA-ORLNC1 were reproduced under in vivo conditions. The BMSCs infected with shRNA-ORLNC1 were transplanted into the backside of immunodeficient mice, and H&E staining was performed. Noticeably, the results clearly showed that lncRNA-ORLNC1 knockdown rendered enhanced new bone formation relative to NC (Figure 4J).
In all cases, the BMSCs transfected with NC did not show any meaningful alterations. The efficacy of shRNA-ORLNC1 to knock down endogenous lncRNA-ORLNC1 and the failure of NC to affect lncRNA-ORLNC1 were confirmed (Figure S3A).
lncRNA-ORLNC1 Acts by Interacting with miR-296
With the above-presented data strongly suggesting the critical role of lncRNA-ORLNC1 in regulating osteogenic and adipogenic differentiation of BMSCs both in in vitro and in vivo models, we went on to elucidate the mechanisms on how lncRNA-ORLNC1 controls osteogenic and adipogenic differentiation. It is known that lncRNAs regulate gene expression by multiple and diverse mechanisms, of which ceRNA appears to be a common mode of action for many lncRNAs.
To see if such a mechanism applies to lncRNA-ORLNC1 in our models, we first performed computational analysis to search for the miRNAs that could be bound by this lncRNA. In this way, we were able to identify six miRNAs that have the potential to interact with lncRNA-ORLNC1, because they have excellent sequence complementarity to the latter with high free energy. Among six miRNAs, miR-106a, miR-106b, miR-20a, miR-20b, and miR-346 were not able to affect osteogenesis (Figures 5A and 5B). However, miR-296 possessed the ability to regulate osteogenic differentiation of BMSCs (Figures 5A and 5B). To further investigate the underlying mechanism of lncRNA-ORLNC1 regulated miR-296, we tested the distribution of lncRNA-ORLNC1 using fluorescence in situ hybridization (FISH). The results of FISH indicated that lncRNA-ORLNC1 was found to be located in both the nucleus and the cytoplasm of BMSCs (Figure 5C). We therefore conducted the following experiments to establish the relationship between lncRNA-ORLNC1 and miR-296. The expression of miR-296 in BMSCs transfected with miR-296 mimic or miR-296 anti-miRNA oligonucleotide (AMO) is shown in Figure S3B.
Figure 5.
lncRNA-ORLNC1 Directly Targets miR-296, and miR-296 Possesses the Ability to Regulate Osteogenic Differentiation of BMSCs
(A and B) The effects of miR-106a, miR-106b, miR-20a, miR-20b, miR-296-3p, and miR-346 on the osteogenic differentiation of BMSCs were detected by ARS (A) and ALP (B) stainings. Scale bar, 100 μm. (C) FISH images showing localization of lncRNA-ORLNC1 in BMSCs. 18S, probe for 18S rRNA; U6, probe for U6 snRNA. Scale bar, 200 μm. (D) Luciferase reporter assay was used to assess the relationship between lncRNA-ORLNC1 and miR-296. (E) The expression level of lncRNA-ORLNC1 was upregulated in BMSCs transfected with biotin-labeled miR-296. (F) The expression of miR-296 was significantly decreased in the BMSCs (left) and bone tissue (right) of osteoporotic mice normalized to the control group. (G) The level of miR-296 was markedly decreased in the BMSCs (left) and bone tissue (right) of osteoporotic transgenic mice compared with WT mice. (H) Quantification of miR-296 in the bone tissue of patients with osteoporosis compared with control. (I and J) The expression level of miR-296 was highly increased during osteogenic differentiation (I) and obviously decreased after adipogenic differentiation (J) of BMSCs. **p < 0.01, and ***p < 0.001.
We first demonstrated the reciprocal relationship between lncRNA-ORLNC1 and miR-296 in terms of their expression levels. We then continued to test if the observed reciprocal expression could be attributed to a direct interaction between lncRNA-ORLNC1 and miR-296. For this purpose, we constructed luciferase reporters that carried either a WT lncRNA-ORLNC1 or the mutant lncRNA-ORLNC1 that contains base replacement to disrupt the putative miR-296-binding site. Transfection of miR-296 mimic diminished the luciferase activities elicited by the WT lncRNA-ORLNC1 vector, but not by the mutant vector (Figure 5D). When the binding site of miR-296 in the lncRNA-ORLNC1 sequence was mutated, the regulation of lncRNA-ORLNC1 was abolished (Figure 5D).
Next, BMSCs were transfected with biotinylated NC or biotinylated miR-296 for 48 hr. The cells were harvested for a biotin-based pull-down assay. qRT-PCR analysis demonstrated that lncRNA-ORLNC1 expression level was augmented in BMSCs treated with biotinylated miR-296 (Figure 5E). As illustrated in Figure 5F, the expression of miR-296 was downregulated in the bone tissues and cells of OVX-induced osteoporotic mice, which was opposite to the upregulation of lncRNA-ORLNC1 in the same models. Moreover, miR-296 level was considerably lower in lncRNA-ORLNC1-overexpressing TG mice than in WT littermates (Figure 5G). Strikingly, miR-296 level was also substantially reduced in the bone tissues of osteoporotic patients compared with control subjects (Figure 5H). During the progress of osteogenesis, while the level of lncRNA-ORLNC1 was decreased, that of miR-296 was increased in BMSCs after osteogenic differentiation (Figure 5I). Besides, the expression level of miR-296 was decreased in BMSCs after adipogenic differentiation of 16 days (Figure 5J).
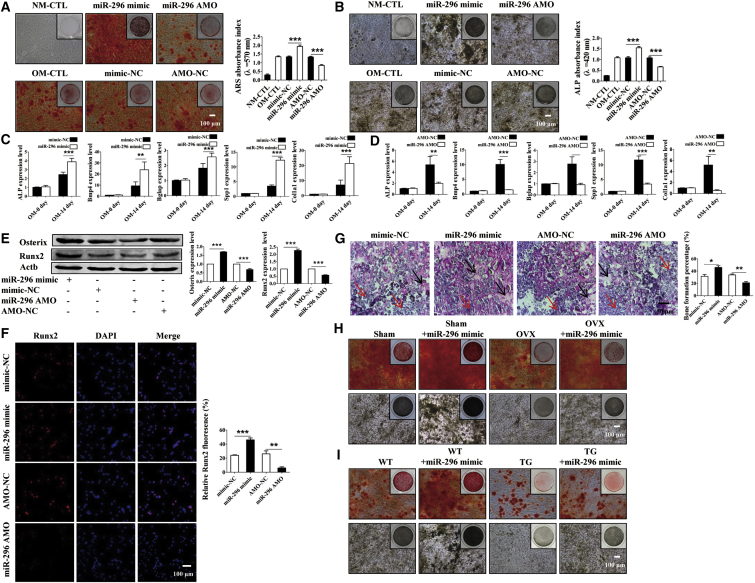
If miR-296 truly mediates the regulatory effects of lncRNA-ORLNC1 on osteogenic and adipogenic differentiations, then it is expected to elicit the opposite actions to the lncRNA. The following functional studies indeed provided the evidence in support of this notion. As shown in Figure S4A, there was no obvious effect of miR-296 in the osteogenic potential under normal condition. Also, osteogenic differentiation was promoted in the BMSCs by pretreatment with miR-296 mimic after the induction of 14 days, and this action was blunted by miR-296 AMO (Figures 6A and 6B). Consistently, the mRNA levels of master regulators of osteogenesis were upregulated by miR-296 mimic and downregulated by miR-296 AMO (Figures 6C and 6D). A similar pattern of alterations of osteoblast marker genes was observed at the protein level, as indicated by the results from western blotting and immunostaining of BMSCs (Figures 6E and 6F).
Figure 6.
miR-296 Directs toward Osteoblastic Differentiation in Immunodeficient Mice and Bone Formation from BMSCs
(A and B) ARS (A) and ALP (B) analysis revealed that BMSCs treated with miR-296 mimic potentiated osteogenesis and miR-296 AMO weakened osteoblast differentiation compared with NC after osteogenic differentiation of 14 days. Scale bar, 100 μm. (C and D) Real-time qPCR confirmed that miR-296 mimic (C) increased the mRNA levels of osteoblast genes and miR-296 AMO (D) decreased the expression relative to NC, respectively. (E) Expression of osteogenic genes in BMSCs were diminished by miR-296 AMO and increased by miR-296 mimic. (F) Fluorescent images of Runx2 revealed significantly enhanced osteogenic ability caused by miR-296 mimic while this was decreased by miR-296 AMO. Runx2, red; DAPI, blue. Scale bar, 100 μm. (G) The ratio of new bone was analyzed by H&E staining from immunodeficient mice. The black arrows indicate bone formation, and the red arrows indicate hydroxyapatite. Scale bar, 50 μm. (H and I) ARS and ALP analysis verified miR-296 can restore the osteogenic ability of BMSCs from osteoporotic mice caused by OVX surgery (H) or overexpression of lncRNA-ORLNC1 (I) compared with control mice. Scale bar, 100 μm. *p < 0.05, **p < 0.01, and ***p < 0.001.
Under in vivo conditions, the area of bone formation of the backside was markedly higher in immunodeficient mice that received miR-296 mimic compared to the same mice prior to mimic-NC treatment (Figure 6G). Furthermore, H&E staining revealed that BMSCs transfected with miR-296 AMO impaired bone formation in immunodeficient mice (Figure 6G). Exceptionally, miR-296 mimic rescued the OVX-induced impairment of the osteogenic potential of BMSCs, as uncovered by ARS and ALP staining assays (Figure 6H). Furthermore, miR-296 mimic transfection restored the decreased osteogenesis of BMSCs from lncRNA-ORLNC1-overexpressing transgenic mice (Figure 6I).
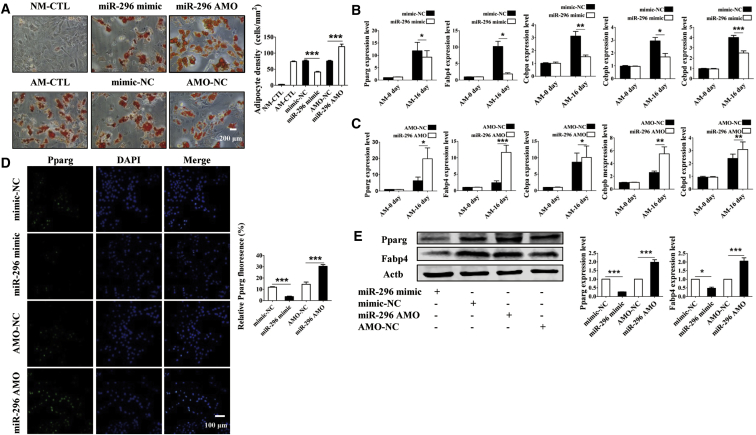
With the above data suggesting that miR-296 favors osteogenic differentiation of BMSCs, it is logical to anticipate that this miRNA should disfavor adipogenic differentiation. We therefore turned to look at the effects of miR-296 on the adipogenic potential of BMSCs. Forced expression of miR-296 in BMSCs decreased the number of adipocytes 16 days following the induction of adipogenic differentiation (Figure 7A). In contrast, miR-296 AMO treatment accelerated the adipogenic differentiation of BMSCs (Figure 7A). The mRNA levels of the selected adipogenic markers were robustly reduced by miR-296 mimic, but they were increased by miR-296 AMO (Figures 7B and 7C). Consistently, the protein levels of adipocyte-specific genes were substantially lower in the miR-296 mimic group, but they were higher in the miR-296 AMO group (Figures 7D and 7E).
Figure 7.
miR-296 Suppresses Adipogenesis of BMSCs under In Vitro Conditions
(A) ORO staining evaluated that miR-296 inhibited fat accumulation in BMSCs undergoing differentiation of 16 days. Scale bar, 200 μm. (B and C) Relative expression levels of Fabp4, Pparg, Cebpa, Cebpb, and Cebpd were regulated in BMSCs after transfection with miR-296 mimic (B) and miR-296 AMO (C). (D and E) Immunofluorescence (D) and western blot (E) analyses showed the decreased levels of adipogenic-related genes in miR-296-transfected BMSCs. Pparg, green; DAPI, blue. Scale bar, 100 μm. *p < 0.05, **p < 0.01, and ***p < 0.001.
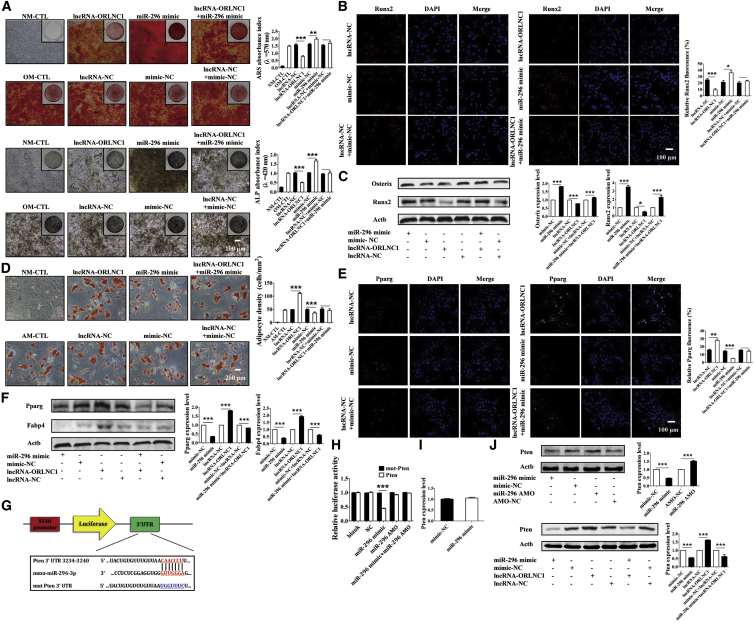
To further validate whether lncRNA-ORLNC1 could govern the osteogenic differentiation increased by miR-296, ARS and ALP stainings were performed. The results indicated that osteoblast matrix mineralization was markedly augmented in BMSCs treated with miR-296 mimic, and it was abolished by overexpression of lncRNA-ORLNC1 (Figure 8A). We next performed immunofluorescence staining and western blot, and the results confirmed that the protein levels of osteoblast-related genes were extremely increased by miR-296 mimic, which were further diminished by overexpressed lncRNA-ORLNC1 (Figures 8B and 8C). As shown in Figure 8D, lncRNA-ORLNC1 reversed the decrease in the number and area of adipocytes induced by miR-296 mimic. Meanwhile, the western blot and fluorescence staining strengthened the finding that lncRNA-ORLNC1 promoted the protein expressions of adipogenesis-related genes, which were attenuated by miR-296 mimic (Figures 8E and 8F). These data confirmed that lncRNA-ORLNC1 suppressed osteogenic differentiation and promoted adipogenic differentiation by inhibiting the role of miR-296.
Figure 8.
lncRNA-ORLNC1 Regulates Osteogenesis and Adipogenesis of BMSCs through Targeting miR-296, and Pten Is a Candidate Target Gene for miR-296
(A) ARS and ALP stainings indicated that lncRNA-ORLNC1 counteracted the increased effect of miR-296 on osteogenic differentiation of BMSCs. Scale bar, 100 μm. (B and C) Immunofluorescence staining (B) and western blot analysis (C) of relative protein expression showed increased levels of osteogenic genes in miR-296-treated BMSCs were inhibited by lncRNA-ORLNC1. Runx2, red; DAPI, blue. Scale bar, 100 μm. (D) Quantitative analysis of the number of adipocytes revealed that lncRNA-ORLNC1 increased the adipogenic differentiation, which was inhibited by miR-296. Scale bar, 200 μm. (E and F) Immunofluorescence staining (E) and western blot (F) indicated the protein levels of adipocyte-related genes in BMSCs after treatment with lncRNA-ORLNC1 and miR-296. Pparg, green; DAPI, blue. Scale bar, 100 μm. (G) The binding sites between miR-296-3p and Pten. (H) Luciferase reporter assay revealed the relationship between miR-296 and Pten. (I) The relative mRNA level of Pten was not changed after treatment with miR-296 mimic and NC. (J) The protein level of Pten was largely decreased by miR-296 mimic, while it was increased by miR-296 AMO; and, lncRNA-ORLNC1 and miR-296 mimic regulated the protein expression of Pten. *p < 0.05, **p < 0.01, and ***p < 0.001.
Pten Mediates the lncRNA-ORLNC1-miR-296 Axis as an Anti-osteogenic Factor
To gain further insight into the mechanism by which the lncRNA-ORLNC1-miR-296 axis regulates osteoblast activity, we continued to explore the key target gene for miR-296 action relevant to adipogenesis and osteogenesis of BMSCs. Using the RegRNA database to predict the potential targets of miR-296, Pten was identified as a candidate for our subsequent experiments (Figure 8G). We first constructed luciferase reporter vectors that had either a WT Pten 3′ UTR or a Pten 3′ UTR containing mutant sequences of the miR-296-binding site (Figure 8H). First, as depicted in Figure 8H, miR-296 mimic, but not its AMO, substantially inhibited the luciferase reporter activities elicited by the WT Pten 3′ UTR. As expected, the luciferase activities produced by the vector carrying the mutated Pten 3′ UTR was not affected by miR-296 (Figure 8H). The mRNA level of Pten was unaffected by miR-296 (Figure 8I). Moreover, at 14 days following osteogenic differentiation of BMSCs, the protein level of Pten was grossly lower in the miR-296 mimic group and higher in the miR-296 AMO group, compared to their respective NCs (Figure 8J). The downregulation of Pten expression in BMSCs treated with miR-296 was reversed by the overexpression of lncRNA-ORLNC1 (Figure 8J).
Discussion
The present study was designed to exploit the role of lncRNAs in osteoporotic differentiation and delineate the underlying cellular and molecular mechanisms. Our initial microarray analysis in conjunction with qRT-PCR allowed us to focus our detailed investigation on a particular lncRNA that we named ORLNC1. Expression of lncRNA-ORLNC1 was dramatically decreased during osteogenic differentiation and increased during adipogenic differentiation of BMSCs. We demonstrated that lncRNA-ORLNC1 regulated BMSC lineage commitment. Overexpression of lncRNA-ORLNC1 inhibited osteoblast differentiation and promoted adipocyte differentiation of BMSCs. Overexpression of lncRNA-ORLNC1 resulted in osteoporosis-related bone loss and low BMD in transgenic mice.
Overexpression of lncRNA-ORLNC1 might prevent BMSCs from differentiating into osteoblastic lineage and promote adipogenic differentiation of BMSCs. In addition, when the lncRNA-ORLNC1 was knocked down, BMSCs acquired the ability to differentiate into osteoblasts in vitro, and they showed decreased adipogenic differentiation. More importantly, our results confirmed that the expression of lncRNA-ORLNC1 was significantly upregulated in the bone tissue of osteoporotic patients. This discovery provided a new molecular insight into the dysregulated lncRNAs in osteoporosis, and it suggested that lncRNA-ORLNC1 resulted in a high incidence of osteoporosis. Similarly, several studies have reported that lncRNAs govern the differentiation of BMSCs.23, 24 Knockdown of lncRNA NONHSAT009968 ameliorates osteogenic differentiation in an inflammatory environment caused by Staphylococcal protein A (SpA) of human BMSCs (hBMSCs).25
Various studies have indicated that miRNAs are found to be involved in the regulation of osteogenesis through affecting key molecules that control osteogenic differentiation in BMSCs.26 miR-2861 enhanced Bmp2-induced osteoblast differentiation by targeting histone deacetylase 5 (Hdac5).27 However, there was no research on the role of miR-296 in the regulation of BMSC commitment. In this study, we first identified miR-296 as an important enhancer in osteoblast differentiation of BMSCs. BMSCs treated with miR-296 mimic exhibited increased osteogenic differentiation ability and decreased adipogenic differentiation capacity in BMSCs. Knockdown of miR-296 potently suppressed the osteogenesis and elevated adipogenesis of BMSCs. During ectopic bone formation, BMSCs transfected with miR-296 mimic generated more new bone formation, which was essential for bone homeostasis and fracture healing.
Pten is known as a negative regulator of osteogenesis, which has been reported to function as a negative regulator of osteoblastic differentiation.28, 29 miRNA bioinformatics analysis databases, including TargetScan and RegRNA, all predicted that Pten is a potential target of miR-296. Therefore, we hypothesized that miR-296 might control the differentiation of BMSCs into osteoblasts by targeting Pten. To verify this hypothesis, our data revealed that the overexpression of miR-296 suppressed Pten protein level without affecting the mRNA level. Luciferase reporter assay also confirmed that Pten was one of the targets for miR-296, and miR-296 could repress the expression of Pten by specifically binding to its 3′ UTR. Indeed, in the present study, the protein expression of Pten exhibited a negative correlation with the osteoblast differentiation process, indicating the increased osteogenic ability of BMSCs treated with miR-296 mimic, owing to the influence by inhibiting Pten.
In our study, the sequences of lncRNA-ORLNC1 are evolutionarily conserved across species, including human and mouse. The mature sequence of lncRNA-ORLNC1 is derived from two exons, and in particular the second exon is highly conserved between mouse and human. Interestingly, the binding site of miR-296 is located in the second exon of lncRNA-ORLNC1, which has highly evolutionarily conserved domains. Meanwhile, miR-296 is a highly conserved miRNA across different species. It suggests that the lncRNA-ORLNC1-miR-296 axis forms a highly conserved mechanism for governing BMSC commitment.
We investigated the roles of lncRNA-ORLNC1 in regulating the switch between osteogenic and adipogenic differentiation of BMSCs during osteoporosis. In addition, the expression level of lncRNA-ORLNC1 was increased in the bone tissue of osteoporotic patients. It implies that lncRNA-ORLNC1 is the potent biomarker for the diagnosis of osteoporosis. Osteoporosis is easy to be diagnosed by X-ray examination. But, the sensitive biomarker is always required for the early diagnosis of osteoporosis. Our results provide a new potential biomarker for patients with osteoporosis. However, the mechanisms of the serum lncRNA-ORLNC1 released from bone cells remain to be elucidated.
In addition, we found that knockdown of lncRNA-ORLNC1 could reverse the decreased osteogenic capacity of BMSCs in osteoporosis in vitro. Consistently, in vivo study showed that BMSCs treated with shRNA-ORLNC1 lentivirus formed more bone tissues than its control after transplanted into the backside of immunodeficient mice. Moreover, we proved that miR-296 leads to accelerated osteogenesis and suppressed adipogenesis, and, thereby, it participates in the formation of bone tissues. Collectively, these data also point to the importance of the lncRNA-ORLNC1-miR-296 axis in the lineage commitment of BMSCs, and they suggest silencing lncRNA-ORLNC1 or enhancing miR-296 as a new approach for promoting bone formation. However, in our study, miR-296 transgenic mice were not used to support the function of miR-296 in osteoporosis. This is a limitation of the present study.
Conclusions
In summary, we first confirmed that lncRNA-ORLNC1 was positively correlated with osteoporosis. lncRNA-ORLNC1 can not only inhibit osteogenic differentiation and potentiate adipogenic differentiation of BMSCs in vitro, but also effectively restrain the acceleration of miR-296 on osteoblast differentiation by targeting Pten. More importantly, our findings enriched the understanding of lncRNA-miRNA regulation of bone development and osteogenic differentiation. Furthermore, our study puts forward a new target for bone tissue engineering and especially the prevention and treatment of osteoporosis.
Materials and Methods
Clinical Data Collection
The bone tissues of osteoporotic and control patients used in this study were collected from 20- to 60-year-old men or women, from January 2016 to December 2017, in The First Affiliated Hospital of Harbin Medical University. Some pieces of human bone were obtained when the osteoporotic patients or controls underwent routine therapeutic surgery. In our study, the bone tissues were collected from 5 osteoporotic patients (T-score ≤ −2.5) and 5 controls (T-score ≥ 1) who experienced fractures caused by strike, high-altitude falling, or slip. The BMD analysis and additional examinations used for osteoporosis diagnosis were recorded according to the criteria of having experienced one or more fragility fractures and a T-score ≤ −2.5.
The participants were excluded from the study if they had suffered diseases that can affect bone metabolism, including diseases of the kidney, liver, or parathyroid; diabetes mellitus; hyperprolactinemia; oophorectomy; rheumatoid arthritis; ankylosing spondylitis; malabsorption syndromes; malignant tumors; hematological diseases; or previous pathological fractures. The bone tissues of the patients were treated with RNAlater (Vazyme, China) and stored in a −80°C freezer for subsequent analyses.
Animals
Female C57BL/6J mice (8 weeks, 18–20 g) were purchased from the Experimental Animal Center of Harbin Medical University. The animal-handling procedures conformed to the principle of the Care and Use of Laboratory Animals published by the NIH. The mice were housed under specific pathogen-free conditions (22°C, 12-hr light and 12-hr dark cycles, and 50%–55% humidity) with free access to food pellets and tap water. Additionally, all experimental procedures were carried out in strict accordance with the Ethic Committee of Harbin Medical University. All experiments related to the use of mice were approved by the Institutional Animal Care Committee of Harbin Medical University.
Isolation and Culture of BMSCs
BMSCs were isolated from female C57BL/6J mice (8 weeks, 18–20 g) and harvested as described in detail in our previous work.30 In brief, mice were anesthetized by intraperitoneal injection of pentobarbital sodium (Sigma-Aldrich, USA) at a dose of 40 mg/kg and sacrificed. Then the femur and tibia were isolated and soaked in 75% ethyl alcohol (Tianjin Fuyu Fine Chemical, China). After removal of the attached muscles and connective tissues from bones, bone marrow was flashed out with BMSC normal culture medium (Cyagen Biosciences, USA), containing 10% fetal bovine serum (FBS), 1% penicillin, and 1% glutamine, using a 1-mL syringe (Changzhou Kangfulai Medical, China) stabbed into the marrow cavity. Then, the flushing fluid was transferred into a clean ampoule and ultimately removed into a 25-cm2 culture flask (Corning, USA). BMSCs were cultured in a 37°C, 5% CO2 humidified incubator (Thermo, USA). The freshly isolated BMSCs were designated as passage 0 (P0). The medium was changed to remove non-adherent BMSCs. The normal culture medium was renewed every 3 days to support the growth of BMSCs. BMSCs were detached with 0.25% trypsin (Cyagen Biosciences, USA) at a confluence of 70%–80%, and they were subcultured to the next passage, which was considered as passage one (P1). BMSCs between the third and fifth passages were used in the following experiments.
Lentiviral Vector Construction and Transfection
The packaged lncRNA-ORLNC1-overexpressing lentivirus vector and NC lentivirus vector were obtained from Cyagen Biosciences. In addition, to generate lncRNA-ORLNC1 knockdown cells, BMSCs were treated with lentivirus vector that contained shRNA targeting lncRNA-ORLNC1. All the lentivirus vectors were used for BMSC transfection at an MOI of 30–40. BMSCs were plated at a density of 3 × 105–5 × 105 cells/cm2 in each six-well plate (Corning, USA), according to the protocol manual. When BMSCs grew to 40%–50% confluence, transfection was performed in the presence of polybrene (Cyagen Biosciences, USA) and the appropriate volume of fresh F12-DMEM (HyClone, USA) without FBS to improve efficiency, according to the manufacturer’s instructions. After a 6-hr reaction, the medium was refreshed. Transfection efficiency of the lentivirus was verified by real-time qPCR.
miRNA Transfection
Mmu-miR-296-3p mimic, mmu-miR-296-3p AMO, and their respective NCs were synthesized and supplied by GenePharma (China). The sequences of mmu-miR-296-3p mimic were as follows: primary chain, 5′-GAGGGUUGGGUGGAGGCUCUCC-3′, and passenger chain, 5′-AGAGCCUCCACCCAACCCUCUU-3′. The sequence of mmu-miR-296-3p AMO was 5′-GGAGAGCCUCCACCCAACCCUC-3′. The sequences of mimic-NC were as follows: 5′-UUCUCCGAACGUGUCACGUTT-3′ and 5′-ACGUGAACAGUUCGGAGAATT-3′. The sequence of AMO-NC was 5′-CAGUACUUUUGUGUAGUACAA-3′.
After BMSCs had reached a confluence of 50%–60%, transfection of mmu-miR-296-3p mimic was conducted in Opti-MEM Reduced Serum Medium (Invitrogen, USA), according to the manufacturer’s instructions. The transfection mix was composed of 50 nM mmu-miR-296-3p mimic, 10 μL X-treme Reagent (Roche, Switzerland), and an optimal volume of Opti-MEM Reduced Serum Medium in each six-well plate. The mmu-miR-296-3p AMO was transfected into BMSCs at a final concentration of 100 nM in each well of six-well plates.
Osteogenic Differentiation and Adipogenic Differentiation of BMSCs
To induce osteogenic differentiation, BMSCs at a density of 2 × 104 or 1 × 105 cells/cm2 were cultured in the 24-well plates or 6-well plates. At confluence of approximately 60%–70%, BMSCs were treated with osteogenic induced medium (Cyagen Biosciences, USA), supplemented with 10% FBS, 1% glutamine, 1% penicillin-streptomycin, 0.2% ascorbic acid, 1% β-glycerophosphate, and 0.01% dexamethasone, to promote the osteogenic differentiation and accelerate the formation of mineralized matrix. The medium was refreshed every 3 days, and the cells were harvested after osteogenic differentiation.
For adipogenic differentiation, BMSCs at a density of 2 × 104 or 1 × 105 cells/cm2 were seeded in the 24-well or 6-well plates, and, when the cells reached about 80%–90%, the cells were induced into adipocytes in the presence of adipogenic induced medium (Cyagen Biosciences, USA), according to the protocol recommended by the manufacturer. Adipogenic induced medium A was composed of 175 mL culture medium, 10% FBS, 1% glutamine, 1% penicillin-streptomycin, 0.2% insulin, 0.1% IBMX, 0.1% rosiglitazone, and 0.1% dexamethasone. Adipogenic induced medium B contained 175 mL culture medium, 10% FBS, 1% penicillin-streptomycin, 1% glutamine, and 0.2% insulin. The cells were differentiated in adipogenic induced medium A for 3 days and medium B for the next day. Experiments were performed after adipogenic differentiation.
ARS Staining and Quantitative Analysis
The extracellular matrix calcification was detected by ARS staining 14 days after osteogenic differentiation of BMSCs. After fixation by 4% neutral buffered formalin (Tianjin Fuyu Fine Chemical) for 20 min, cells were stained by ARS solution for 30 min (Cyagen Biosciences, USA), followed by PBS washing for three times with gentle shaking. Images were recorded by a scanner (Nikon, Japan). For quantification of mineralization, the staining was solubilized by 100 mM cetylpyridinium chloride (Sigma-Aldrich, USA) for 1 hr and measured spectrophotometrically at 570 nm by a microplate reader (Tecan, Switzerland). ARS staining and quantification were repeated three times.
ALP Staining and Quantitative Analysis
On day 14 following osteogenic differentiation of BMSCs, OM was removed and ALP staining was performed to detect matrix mineralization depositions. Cells were washed three times with PBS (Solarbio, China) and fixed with 95% ethanol (Tianjin Fuyu Fine Chemical, China) at room temperature for 10 min. Then, BMSCs were embedded in the ALP incubation buffer containing 3% β-glycerophosphate disodium salt hydrate (Sigma-Aldrich, USA), 2% sodium pentobarbital (Peking Shangcai Fine Chemical, China), 2% calcium chloride (Tianjin Bodi Chemical, China), 2% magnesium sulfate (Tianjin Haijing Fine Chemical, China) at 37°C in the dark for 4 hr. After washes, cobalt nitrate (Tianjin Tianli Chemical, China) was added into 24-well plates. After 5 min, the cells were exposed to 1% ammonium sulfide (Tianjin Fuyu Fine Chemical, China) for 2 min and washed with PBS three times. Then, the calcium mineralization pictures were obtained by an inverted optimal microscope (Nikon, Japan). The black and gray granular positions were regarded as calcium deposits, which indicated the intensity of ALP staining. The cells were incubated with 10 mM p-nitrophenyl phosphate (Dalian Meilunbio, China) as the substrate at 37°C for 15 min. Afterward, ALP activity was quantified at 420 nm by a microplate reader.
ORO Staining and Quantification
ORO staining was performed to detect the number of mature adipocytes 16 days after adipogenic differentiation of BMSCs. After washing with PBS thoroughly, the cells were fixed in 4% paraformaldehyde (Solarbio, China) at 4°C for 20 min and then subjected to ORO staining with the working solution diluted in PBS (Cyagen Biosciences, China) at room temperature for 30 min, according to the manufacturer’s instructions. After washing with PBS three times, photographs of lipid droplets were taken by an inverted optimal microscope and camera system.
RNA Extraction and Real-Time qPCR
Total RNA was isolated from BMSCs and bone tissues using TRIzol reagent (Life Technology, USA), according to the manufacturer’s instructions as previously described.31 The RNA samples of 0.5 μg each were used to perform reverse transcription. The cDNA was generated using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, USA). The cycling profile of the cDNA Reverse Transcription (Roche, Switzerland) was as follows: 95°C for 10 min, 2 cycles at 37°C for 1 hr, 85°C for 5 min, and cooling at 4°C. The amplification reactions were assembled in 20-μL reaction volumes containing 10 μL FS Universal SYBR Green Master (Roche, Switzerland), 7 μL double-distilled water, 2 μL amplification primers (GenePharma, China), and a 1 μL cDNA in each amplification reaction using a Roche LightCycler 480 PCR machine. The conditions of the real-time PCR were as follows: denaturation at 95°C for 10 s and 40 cycles at 95°C for 10 s and 60°C for 30 s. The annealing temperature was set according to the primers used in the study. The level of the relative mRNA was normalized to the level of housekeeping gene (Gapdh or Actb) mRNA using the 2−ΔΔCT method and expressed as fold changes relative to the internal reference. The sequences of specific primers are listed in Table 1.
Table 1.
The Sequences of Primers
| Target Genes | Primer Sequences | |
|---|---|---|
| ALP | forward | 5′-ACAACCTGACTGACCCTTCG-3′ |
| reverse | 5′-TCATGATGTCCGTGGTCAAT-3′ | |
| Bmp4 | forward | 5′-TCGTTACCTCAAGGGAGTGG-3′ |
| reverse | 5′-ATGCTTGGGACTACGTTTGG-3′ | |
| Bglap | forward | 5′-TTCTGCTCACTCTGCTGACC-3′ |
| reverse | 5′-TTTGTAGGCGGTCTTCAAGC-3′ | |
| Spp1 | forward | 5′-ACACTTTCACTCCAATCGTCC-3′ |
| reverse | 5′-TGCCCTTTCCGTTGTTGTCC-3′ | |
| Col1a1 | forward | 5′-CAGCCGCTTCACCTACAGC-3′ |
| reverse | 5′-TTTTGTATTCAATCACTGTCTTGCC-3′ | |
| Pparg | forward | 5′-TCACAAGAGGTGACCCAATG-3′ |
| reverse | 5′-CCATCCTTCACAAGCATGAA-3′ | |
| Fabp4 | forward | 5′-TTCCTGTCGTCTGCGGTGATT-3′ |
| reverse | 5′-GATGCCTTTGTGGGAACCTGG-3′ | |
| Cebpa | forward | 5′-GTGTGCACGTCTATGCTAAACCA-3′ |
| reverse | 5′-GCCGTTAGTGAAGAGTCTCAGTTTG-3′ | |
| Cebpb | forward | 5′-TGGACAAGCTGAGCGACGAG-3′ |
| reverse | 5′-GAACAAGTTCCGCAGGGTGC-3′ | |
| Cebpd | forward | 5′-CACGACTCCTGCCATGTACG-3′ |
| reverse | 5′-GCCGCTTTGTGGTTGCTGTT-3′ | |
| Gapdh | forward | 5′-CATCACTGCCACCCAGAAGAC-3′ |
| reverse | 5′-CCAGTGAGCTTCCCGTTCAG-3′ | |
| U6 small nuclear RNA | forward | 5′-GCTTCGGCAGCACATATACTAAAAT-3′ |
| reverse | 5′-CGCTTCACGAATTTGCGTGTCAT-3′ | |
| RT primer | 5′-CGCTTCACGAATTTGCGTGTCAT-3′ | |
| mmu-miR-296-3p | forward | 5′-CGCCGGAGGGTTGGGTGGAG-3′ |
| reverse | 5′-CAGCCACAAAAGAGCACAAT-3′ | |
| RT primer | 5′CCTGTTGTCTCCAGCCACAAAAGAGCACAATATTTCAGGAGACAACAGGGGAGAGC-3′ | |
| mmu-lncRNA-ORLNC1 | forward | 5′-CAAAGCTGGGCCTGTGTACT-3′ |
| reverse | 5′-CTGGGAGGGCTGCAATATC-3′ | |
| hsa-lncRNA-ORLNC1 | forward | 5′-GTACTTATCATCATCCCACCAA-3′ |
| reverse | 5′-GCAGGCTTACTTCTTCATTCC-3′ | |
| hsa-Actb | forward | 5′-GGGAAATCGTGCGTGACATT-3′ |
| reverse | 5′-GGAACCGCTCATTGCCAAT-3′ | |
| mmu-Actb | forward | 5′-ACTGCCGCATCCTCTTCCT-3′ |
| reverse | 5′-TCAACGTCACACTTCATGATGGA-3′ | |
Western Blot Analysis
Western blotting was performed as previously described.32 Cells were washed three times with ice-cold PBS and lysed in radioimmunoprecipitation (RIPA) buffer (Sigma-Aldrich, USA) with 1% cocktail at 4°C for 30 min. Supernatants were collected, and the concentration of protein was quantified by a BCA protein assay kit (Beyotime Biotechnology, China). Total protein was separated by 12.5% SDS-PAGE (Shanghai EpiZyme Scientific, China) and then transferred to nitrocellulose membrane (Millipore, USA). Subsequently, the membrane was blocked with 5% non-fat milk in Tris-buffered saline (TBS)-Tween 20 (TBST) buffer for 2 hr at room temperature, and then it was incubated with anti-Runx2 (ab76956, Abcam, Britain), anti-Osterix (ab22552, Abcam, Britain), anti-Pparg (ab178860, Abcam, Britain), anti-Pten (ab32199, Abcam, Britain), or anti-Fabp4 (3544, Cell Signalling Technology, USA) at 4°C overnight. The membrane was washed with TBST buffer and then incubated with secondary antibodies at room temperature for 1 hr. The protein levels were detected using ImageJ software (NIH, USA). Anti-β-Actb (ab8226, Abcam, Britain) was used as a loading control.
Immunofluorescence Staining
At 3 days after osteogenic differentiation, BMSCs were washed by PBS and fixed with 4% PFA at room temperature for 20 min. Then, BMSCs were rinsed with PBS three times and penetrated with 0.4% Triton X-100 (Biosharp, China) at room temperature for 60 min. The solution was withdrawn and cells were washed with PBS three times. An appropriate volume of available goat serum (Boster Biological Technology, China) was used to block the cells at room temperature for 30 min. After washing three times, the BMSCs were incubated with primary antibody against Runx2 (12556S, Cell Signaling Technology, USA) on a shaking table at 4°C overnight. The cells were rinsed with PBS again the next day and incubated with anti-mouse or anti-rabbit secondary antibodies diluted (1:500) in PBS at room temperature for 1 hr. To stain the nucleus, the cells were then further stained with DAPI (Solarbio, China) diluted in PBS in the dark at room temperature for 30 min. The cells were next visualized under a confocal microscope (Olympus, Japan) equipped with proper filters. The BMSCs were incubated with the primary antibody against Pparg (ab178860, Abcam, Britain) to demonstrate the adipogenic differentiation.
BMSC Transplantation Assay In Vivo and Histochemistry Analysis
To evaluate the role of lncRNA-ORLNC1 and miRNA in regulating new bone formation in mice, BMSCs were transplanted into immunodeficient mice following treatment with miRNA and lncRNA. BMSCs (1 × 105 cells per ampoule) were transfected with lncRNA-ORLNC1 lentivirus, shRNA-ORLNC1 lentivirus, miR-296 mimic, miR-296 AMO, or their respective NCs for 24 hr. Then, the cells were washed with PBS three times. Next, 2 × 106 cells were resuspended in 40 mg hydroxyapatite (Nanjing Emperor Nano Material, China) dissolved in physiological saline (Harbin Medical Pharmaceutical, China), and they were transplanted subcutaneously into the dorsal surface of 6-week-old female immunocompromised mice (Beijing Vital River Laboratory Animal Technology, China). After 8 weeks, the transplantations were harvested, and histochemistry analysis was performed as described previously.33 The transplantations were immersed in 4% PFA overnight, followed by decalcification in 10% EDTA for 15 days. Paraffin sections were deparaffinized, hydrated, and stained with H&E (Solarbio, China). Image-Pro Plus software was used to quantify the bone regeneration.
Generation of Transgenic Mice
Osterix+ osteoprogenitor-specific lncRNA-ORLNC1 conditional overexpressing TG mice were constructed by Cyagen Biosciences, using the conventional gene-targeting approach. To generate the lncRNA-ORLNC1-overexpressing transgenic mice, a vector containing Sp7 promoter and lncRNA-ORLNC1 plasmid was constructed. Micro-injection technology was used to inject DNA into mouse embryos and transplant the embryos into foster female mice. After serial breeding schemes, lncRNA-ORLNC1 transgenic mice were obtained. Genotypic analysis was determined by genomic DNA, which was isolated from mouse tails. WT littermates were used as controls.
OVX-Induced Osteoporotic Mouse Model
24 8-week-old female C57BL/6J mice were randomly divided into two groups of an equal number, and they underwent either bilateral OVX operation or sham surgery, as previously described.34 The mice were allowed to adapt to the environment for 1 week before surgery, and they were operated on under anesthetization with chloral hydrate (Sigma-Aldrich, USA). After surgery, all mice were housed under the same pathogen-free conditions for 4 weeks before harvest.
MicroCT Scanning and Analysis
MicroCT (or μCT) analysis was performed to evaluate bone volume and microstructure using a SCANCO μCT-100 instrument (SCANCO Medical, Switzerland) with a 90-kV microfocus X-ray source. Mouse femora were fixed in 4% PFA for 24 hr and fully rinsed with PBS three times. Then, the femora were soaked in 75% ethanol (Tianjin Fuyu Fine Chemical, China) and scanned at 1.25 μM resolution. The Scanco software was used to analyze three-dimensional structure images and the trabecular bone parameters of mouse femur in vitro, including BMD, bone volume relative to tissue volume (BV to TV), Tb.N, Conn. D, SMI, Tb.Sp.
Dual-Luciferase Reporter Analysis
The direct interaction between lncRNA-ORLNC1 and mmu-miR-296-3p was analyzed by luciferase reporter assay. The sequence of lncRNA-ORLNC1 is 5′-TAAAAAGCCAAAGCTGGGCCTGTGTACTCACAAGTGTGCAGCCATGGGCAGGCTTACTTCTTCATTCCTGCTACTGATTGTTCCTGCATATGTCCTGTCCCAGGTTACTCTGAAAGAGTCTGGCCCTGGGATATTGCAGCCCTCCCAGACCCTCAGTCTGACTTGTTCTTTCTCTGGGTTTTCACTGAGCACTTTTGGTATGGGTGTAGGCTGGATTCGTCAGCCTTCAGGGAAGGGTCTGGAGTGGCTGGCACACATTTGGTGGGATGATGATAAGTACTATAACCCAGCCCTGAAGAGTCGGCTCACAATCTCCAAG-3′. To generate Luc-3′ UTR reporter vector, a miR-296 3′ UTR region centering the lncRNA-ORLNC1 target sequence was subcloned into the psiCHECKTM vector (Promega, USA) downstream of the luciferase open reading frame. BMSCs were co-transfected with WT or mut lncRNA-ORLNC1 constructs and either miR-296 mimic or NC using Lipofectamine 2000 (Invitrogen, USA), according to the manufacturer’s instructions. The luminescence signal was quantified by a dual-luciferase reporter assay system (Promega, USA) using a luminometer (Promega, USA) for 48 hr after transfection. Values from the firefly luciferase assay were normalized to the renilla luciferase assay value.
Pull-down Assay with Biotinylated miRNA
BMSCs were harvested from 6-well plates at 72 hr post-transfection with biotinylated miR-296 at a concentration of 50 nM. After being rinsed with PBS three times, the cells were resuspended and incubated with 70 μL RIPA buffer (Sigma-Aldrich, USA) at 4°C for 15 min. The lysates were precleared by centrifugation for 1 min at 5,000 × g, and 50 μL samples were aliquoted for analysis. To prevent non-specific binding of RNA and protein complexes, the beads were washed with RIPA buffer three times. The lysates were incubated with M-280 streptavidin magnetic beads (Sigma-Aldrich, USA) for binding at 4°C for at least 3 hr. The RNA bound to the beads was purified using TRIzol Reagents for additional analysis.
Statistical Analysis
All experiments in this work were performed in at least triplicates. The data were expressed as mean ± SEM. The differences among multiple groups were by ANOVA followed by Student’s unpaired t test and between two groups were compared by Student’s t test only. A value of p < 0.05 was considered as statistically significant.
Study Approval
All animal care protocols and experiments were reviewed and approved by the Animal Care and Use Committee (KY2017-257) of the Laboratory Animal Research Center at The Second Affiliated Hospital of Harbin Medical University. All mice were maintained in the specific pathogen-free facility of the Laboratory Animal Research Center at Harbin Medical University. The clinical study was approved by the Ethics Committee (201516) of The First Affiliated Hospital of Harbin Medical University, and written informed consent was obtained from all participants.
Author Contributions
L.Y. and B.C. conceived the idea, wrote the manuscript, and designed and supervised the project. F.Y. and Y.L. carried out experiments, analyzed the data, and wrote the manuscript. C.F. and M.G. performed qRT-PCR and western blot analysis. T.L. and M.J. performed the BMSC isolation and culture experiments. D.W., Y.S., and Y.Y. collected human bone tissue samples. G.Y., R.G., and M.H. assisted in the animal experiments. E.I., W.M., Z.H., and L.Z. provided advice and technical assistance. Q.H. and F.D. performed analyses of the human patient samples and the mouse models. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by grants from the Natural Science Foundation of China (81501920 and 81573434), the Postdoctoral Science Foundation of China (2018M630370), the Heilongjiang Postdoctoral Fund (LBH-Z17134), and the Natural Science Foundation of Heilongjiang Province (LC2015027 and H2015056).
Footnotes
Supplemental Information includes Supplemental Materials and Methods and four figures and can be found with this article online at https://doi.org/10.1016/j.ymthe.2018.11.019.
Contributor Information
Lei Yang, Email: yangray83@vip.qq.com.
Benzhi Cai, Email: caibz@ems.hrbmu.edu.cn.
Fan Yang, Email: yflora915@163.com.
Supplemental Information
References
- 1.Forciea M.A., McLean R.M., Qaseem A. Treatment of Low Bone Density or Osteoporosis to Prevent Fractures in Men and Women. Ann. Intern. Med. 2017;167:904. doi: 10.7326/L17-0490. [DOI] [PubMed] [Google Scholar]
- 2.Wu Y., Xie L., Wang M., Xiong Q., Guo Y., Liang Y., Li J., Sheng R., Deng P., Wang Y. Mettl3-mediated m6A RNA methylation regulates the fate of bone marrow mesenchymal stem cells and osteoporosis. Nat. Commun. 2018;9:4772. doi: 10.1038/s41467-018-06898-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ensrud K.E., Crandall C.J. Osteoporosis. Ann. Intern. Med. 2018;168:306–307. doi: 10.7326/L17-0587. [DOI] [PubMed] [Google Scholar]
- 4.Lee J.W., Hoshino A., Inoue K., Saitou T., Uehara S., Kobayashi Y., Ueha S., Matsushima K., Yamaguchi A., Imai Y., Iimura T. The HIV co-receptor CCR5 regulates osteoclast function. Nat. Commun. 2017;8:2226. doi: 10.1038/s41467-017-02368-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X., Guo B., Li Q., Peng J., Yang Z., Wang A., Li D., Hou Z., Lv K., Kan G. miR-214 targets ATF4 to inhibit bone formation. Nat. Med. 2013;19:93–100. doi: 10.1038/nm.3026. [DOI] [PubMed] [Google Scholar]
- 6.Li C.J., Cheng P., Liang M.K., Chen Y.S., Lu Q., Wang J.Y., Xia Z.Y., Zhou H.D., Cao X., Xie H. MicroRNA-188 regulates age-related switch between osteoblast and adipocyte differentiation. J. Clin. Invest. 2015;125:1509–1522. doi: 10.1172/JCI77716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartolucci J., Verdugo F.J., González P.L., Larrea R.E., Abarzua E., Goset C., Rojo P., Palma I., Lamich R., Pedreros P.A. Safety and Efficacy of the Intravenous Infusion of Umbilical Cord Mesenchymal Stem Cells in Patients With Heart Failure: A Phase 1/2 Randomized Controlled Trial (RIMECARD Trial [Randomized Clinical Trial of Intravenous Infusion Umbilical Cord Mesenchymal Stem Cells on Cardiopathy]) Circ. Res. 2017;121:1192–1204. doi: 10.1161/CIRCRESAHA.117.310712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H., Liu P., Xu S., Li Y., Dekker J.D., Li B., Fan Y., Zhang Z., Hong Y., Yang G. FOXP1 controls mesenchymal stem cell commitment and senescence during skeletal aging. J. Clin. Invest. 2017;127:1241–1253. doi: 10.1172/JCI89511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo Y., Xie C., Li X., Yang J., Yu T., Zhang R., Zhang T., Saxena D., Snyder M., Wu Y., Li X. Succinate and its G-protein-coupled receptor stimulates osteoclastogenesis. Nat. Commun. 2017;8:15621. doi: 10.1038/ncomms15621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu J.Y., Aarnisalo P., Bastepe M., Sinha P., Fulzele K., Selig M.K., Chen M., Poulton I.J., Purton L.E., Sims N.A. Gsα enhances commitment of mesenchymal progenitors to the osteoblast lineage but restrains osteoblast differentiation in mice. J. Clin. Invest. 2011;121:3492–3504. doi: 10.1172/JCI46406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D’Souza S., del Prete D., Jin S., Sun Q., Huston A.J., Kostov F.E., Sammut B., Hong C.S., Anderson J.L., Patrene K.D. Gfi1 expressed in bone marrow stromal cells is a novel osteoblast suppressor in patients with multiple myeloma bone disease. Blood. 2011;118:6871–6880. doi: 10.1182/blood-2011-04-346775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y.G., Chang H.Y. LncRNA Seduction of GOT2 Goes Viral. Immunity. 2017;47:1021–1023. doi: 10.1016/j.immuni.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 13.Hosono Y., Niknafs Y.S., Prensner J.R., Iyer M.K., Dhanasekaran S.M., Mehra R., Pitchiaya S., Tien J., Escara-Wilke J., Poliakov A. Oncogenic Role of THOR, a Conserved Cancer/Testis Long Non-coding RNA. Cell. 2017;171:1559–1572.e20. doi: 10.1016/j.cell.2017.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Briggs J.A., Wolvetang E.J., Mattick J.S., Rinn J.L., Barry G. Mechanisms of Long Non-coding RNAs in Mammalian Nervous System Development, Plasticity, Disease, and Evolution. Neuron. 2015;88:861–877. doi: 10.1016/j.neuron.2015.09.045. [DOI] [PubMed] [Google Scholar]
- 15.Piccoli M.T., Gupta S.K., Viereck J., Foinquinos A., Samolovac S., Kramer F.L., Garg A., Remke J., Zimmer K., Batkai S., Thum T. Inhibition of the Cardiac Fibroblast-Enriched lncRNA Meg3 Prevents Cardiac Fibrosis and Diastolic Dysfunction. Circ. Res. 2017;121:575–583. doi: 10.1161/CIRCRESAHA.117.310624. [DOI] [PubMed] [Google Scholar]
- 16.Strong A.L., Shi Z., Strong M.J., Miller D.F., Rusch D.B., Buechlein A.M., Flemington E.K., McLachlan J.A., Nephew K.P., Burow M.E., Bunnell B.A. Effects of the endocrine-disrupting chemical DDT on self-renewal and differentiation of human mesenchymal stem cells. Environ. Health Perspect. 2015;123:42–48. doi: 10.1289/ehp.1408188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Q., Zhang X., Dai L., Hu X., Zhu J., Li L., Zhou C., Ao Y. Long noncoding RNA related to cartilage injury promotes chondrocyte extracellular matrix degradation in osteoarthritis. Arthritis Rheumatol. 2014;66:969–978. doi: 10.1002/art.38309. [DOI] [PubMed] [Google Scholar]
- 18.Liu Q., Huang J., Zhou N., Zhang Z., Zhang A., Lu Z., Wu F., Mo Y.Y. LncRNA loc285194 is a p53-regulated tumor suppressor. Nucleic Acids Res. 2013;41:4976–4987. doi: 10.1093/nar/gkt182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wanke K.A., Devanna P., Vernes S.C. Understanding Neurodevelopmental Disorders: The Promise of Regulatory Variation in the 3'UTRome. Biol. Psychiatry. 2018;83:548–557. doi: 10.1016/j.biopsych.2017.11.006. [DOI] [PubMed] [Google Scholar]
- 20.Song Y.X., Sun J.X., Zhao J.H., Yang Y.C., Shi J.X., Wu Z.H., Chen X.W., Gao P., Miao Z.F., Wang Z.N. Non-coding RNAs participate in the regulatory network of CLDN4 via ceRNA mediated miRNA evasion. Nat. Commun. 2017;8:289. doi: 10.1038/s41467-017-00304-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao L., Wu J., Wang J.Y., Chung H.K., Kalakonda S., Rao J.N., Gorospe M., Wang J.Y. Long Noncoding RNA uc.173 Promotes Renewal of the Intestinal Mucosa by Inducing Degradation of MicroRNA 195. Gastroenterology. 2018;154:599–611. doi: 10.1053/j.gastro.2017.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Legnini I., Morlando M., Mangiavacchi A., Fatica A., Bozzoni I. A feedforward regulatory loop between HuR and the long noncoding RNA linc-MD1 controls early phases of myogenesis. Mol. Cell. 2014;53:506–514. doi: 10.1016/j.molcel.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang Y., Zheng Y., Jia L., Li W. Long Noncoding RNA H19 Promotes Osteoblast Differentiation Via TGF-β1/Smad3/HDAC Signaling Pathway by Deriving miR-675. Stem Cells. 2015;33:3481–3492. doi: 10.1002/stem.2225. [DOI] [PubMed] [Google Scholar]
- 24.Zhuang W., Ge X., Yang S., Huang M., Zhuang W., Chen P., Zhang X., Fu J., Qu J., Li B. Upregulation of lncRNA MEG3 Promotes Osteogenic Differentiation of Mesenchymal Stem Cells From Multiple Myeloma Patients By Targeting BMP4 Transcription. Stem Cells. 2015;33:1985–1997. doi: 10.1002/stem.1989. [DOI] [PubMed] [Google Scholar]
- 25.Cui Y., Lu S., Tan H., Li J., Zhu M., Xu Y. Silencing of Long Non-Coding RNA NONHSAT009968 Ameliorates the Staphylococcal Protein A-Inhibited Osteogenic Differentiation in Human Bone Mesenchymal Stem Cells. Cell. Physiol. Biochem. 2016;39:1347–1359. doi: 10.1159/000447839. [DOI] [PubMed] [Google Scholar]
- 26.Frith J.E., Kusuma G.D., Carthew J., Li F., Cloonan N., Gomez G.A., Cooper-White J.J. Mechanically-sensitive miRNAs bias human mesenchymal stem cell fate via mTOR signalling. Nat. Commun. 2018;9:257. doi: 10.1038/s41467-017-02486-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H., Xie H., Liu W., Hu R., Huang B., Tan Y.F., Xu K., Sheng Z.F., Zhou H.D., Wu X.P., Luo X.H. A novel microRNA targeting HDAC5 regulates osteoblast differentiation in mice and contributes to primary osteoporosis in humans. J. Clin. Invest. 2009;119:3666–3677. doi: 10.1172/JCI39832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu X., Bruxvoort K.J., Zylstra C.R., Liu J., Cichowski R., Faugere M.C., Bouxsein M.L., Wan C., Williams B.O., Clemens T.L. Lifelong accumulation of bone in mice lacking Pten in osteoblasts. Proc. Natl. Acad. Sci. USA. 2007;104:2259–2264. doi: 10.1073/pnas.0604153104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang S., Tian Y.S., Lee Y.J., Yu F.H., Kim H.M. Mechanisms by which the inhibition of specific intracellular signaling pathways increase osteoblast proliferation on apatite surfaces. Biomaterials. 2011;32:2851–2861. doi: 10.1016/j.biomaterials.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 30.Yang F., Yang L., Li Y., Yan G., Feng C., Liu T., Gong R., Yuan Y., Wang N., Idiiatullina E. Melatonin protects bone marrow mesenchymal stem cells against iron overload-induced aberrant differentiation and senescence. J. Pineal Res. 2017;63:e12422. doi: 10.1111/jpi.12422. [DOI] [PubMed] [Google Scholar]
- 31.Nichterwitz S., Chen G., Aguila Benitez J., Yilmaz M., Storvall H., Cao M., Sandberg R., Deng Q., Hedlund E. Laser capture microscopy coupled with Smart-seq2 for precise spatial transcriptomic profiling. Nat. Commun. 2016;7:12139. doi: 10.1038/ncomms12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xian L., Wu X., Pang L., Lou M., Rosen C.J., Qiu T., Crane J., Frassica F., Zhang L., Rodriguez J.P. Matrix IGF-1 maintains bone mass by activation of mTOR in mesenchymal stem cells. Nat. Med. 2012;18:1095–1101. doi: 10.1038/nm.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cao L., Yang F., Liu G., Yu D., Li H., Fan Q., Gan Y., Tang T., Dai K. The promotion of cartilage defect repair using adenovirus mediated Sox9 gene transfer of rabbit bone marrow mesenchymal stem cells. Biomaterials. 2011;32:3910–3920. doi: 10.1016/j.biomaterials.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 34.Wang C., Wang J., Li J., Hu G., Shan S., Li Q., Zhang X. KDM5A controls bone morphogenic protein 2-induced osteogenic differentiation of bone mesenchymal stem cells during osteoporosis. Cell Death Dis. 2016;7:e2335. doi: 10.1038/cddis.2016.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.