Abstract
Using exome sequencing, we have identified de novo variants in MAPK8IP3 in 13 unrelated individuals presenting with an overlapping phenotype of mild to severe intellectual disability. The de novo variants comprise six missense variants, three of which are recurrent, and three truncating variants. Brain anomalies such as perisylvian polymicrogyria, cerebral or cerebellar atrophy, and hypoplasia of the corpus callosum were consistent among individuals harboring recurrent de novo missense variants. MAPK8IP3 has been shown to be involved in the retrograde axonal-transport machinery, but many of its specific functions are yet to be elucidated. Using the CRISPR-Cas9 system to target six conserved amino acid positions in Caenorhabditis elegans, we found that two of the six investigated human alterations led to a significantly elevated density of axonal lysosomes, and five variants were associated with adverse locomotion. Reverse-engineering normalized the observed adverse effects back to wild-type levels. Combining genetic, phenotypic, and functional findings, as well as the significant enrichment of de novo variants in MAPK8IP3 within our total cohort of 27,232 individuals who underwent exome sequencing, we implicate de novo variants in MAPK8IP3 as a cause of a neurodevelopmental disorder with intellectual disability and variable brain anomalies.
Keywords: MAPK8IP3, de novo, neurodevelopmental disorder, intellectual disability, developmental delay, brain anomalies, polymicrogyria
Introduction
The prevalence of intellectual disability (ID) in the general population is estimated to be around 1% and has become the most frequent reason for referral to pediatric genetic services.1, 2 Genetically, ID is extremely heterogeneous, and de novo variants detected via trio exome sequencing are the major etiology in children of unrelated parents.3, 4 Disease-related genes of developmental delay (DD) or ID can be identified through the widespread use of next-generation-sequencing followed by collaborative efforts and the use of matchmaking platforms to prove causation through accumulating a sufficient number of individuals with overlapping phenotypes and de novo variants in the same gene, as well as through functional analyses in model organisms. After the initial identification of a de novo missense variant in MAPK8IP3 (MIM 605431) in an individual with ID, the gene became an appealing candidate for further investigation in light of the established role that MAPK8IP3 orthologs play in axonal transport in vertebrates and invertebrates.5 In addition, three de novo variants had been separately published in individuals with a neurodevelopmental disorder.4, 6, 7 Through international collaboration, we identified 13 individuals with DD and/or ID who harbor heterozygous de novo variants in MAPK8IP3. Through in silico molecular modeling and functional analyses in Caenorhabditis elegans, we provide further evidence for pathogenicity and causality, as well as primary insights into the specific role of certain domains in MAPK8IP3′s diverse functions.
Material and Methods
Study Subjects
This study was approved by the ethics committee of the University of Leipzig (approval code: 402/16-ek). Through matchmaking8 and international collaborative efforts, we identified 13 individuals who harbored heterozygous de novo variants in MAPK8IP3 and had an overlapping phenotype of DD and/or ID; these individuals were referred from different centers in Brazil, Germany, Israel, Italy, the USA, and the Netherlands. Referring physicians provided detailed clinical information via a uniform clinical questionnaire. Informed consent was obtained from all examined individuals or their legal guardians. In some cases, testing was done as part of routine clinical care and therefore institutional ethics approval was not required. If done in a research setting, the testing was approved by the ethics committees of the respective centers. All families provided informed consent for clinical testing and publication.
Identification and Evaluation of Variants
Exome sequencing via standard commercial products was performed within diagnostic or research settings in a total cohort of 27,232 affected individuals (Supplemental Material and Methods). Candidate variants were prioritized on the basis of gene and variant attributes, minor-allele frequency, effect on protein function, status determined by in silico prediction tools, phenotype, family history, and inheritance according to the local proceedings in the respective centers. Individuals 1 and 4 were identified in a cohort of 5,926 individuals with neurodevelopmental disorders (NDDs); individual 2 was identified in a cohort of 4,600 individuals with a variety of clinical indications; individuals 3, 6, 9, 10, and 12 were identified in a cohort of 14,113 individuals with NDD; individuals 5 and 8 were identified in a cohort of 393 individuals with NDD; individual 7 was identified in a cohort of 200 individuals with NDD; individual 11 was identified in a cohort of 1,400 individuals with NDD; and individual 13 was identified in a cohort of 600 individuals with NDD. De novo occurrence was confirmed in all individuals. The families of all individuals, except for individual 2, underwent trio exome sequencing. The de novo nonsense variant in individual 2 was identified by single-exome sequencing and a subsequent search for truncating variants among highly constrained genes with a probability of loss-of-function intolerance score (pLI-score) >0.95.9 The de novo occurrence was confirmed with Sanger sequencing. The Exome Aggregation Consortium (ExAC) database and the Genome Aggregation Database (gnomAD) served as control populations.9
Statistical Analysis for Enrichment of De Novo Variants in MAPK8IP3
The statistical analysis was done with R. We used a binomial test and an established framework of gene-specific mutation rates10 to test whether the occurrence of the number of de novo variants in MAPK8IP3 in our cohort was significantly greater than would be expected by chance.
Structural Modeling
Structural analysis of the variants p.Leu444Pro and p.Glu461Gly7 was performed on the basis of the known crystal structures of the leucine zipper domain of murine MAPK8IP3 (PDB: 6EJN11), which exhibits 99% sequence identity to human MAPK8IP3. We modeled the structure of the MAPK8IP3 WD40 repeat that harbors the variants p.His994Gln and p.Arg1146Cys by using the homologous domain from Ciao1 (PDB: 3FM012) as a template. For the remaining variants, modeling was not possible because of the lack of suitable template structures. Amino acid exchanges were modeled with SwissModel,13 and structure visualization was performed with RasMol.14 All sequence positions reported for the structures were adjusted to match those of the MAPK8IP3 isoform investigated in the present study.
Caenorhabditis elegans Functional Analyses
De novo variants in MAPK8IP3 have so far been reported on three separate occasions in three individuals with neurodevelopmental disorders. The reported variants are c.281A>G (p.Tyr94Cys),6 c.1382A>G (p.Glu461Gly),7 and c.3436C>T (p.Arg1146Cys).4 These three variants from the literature and the variants in our cohort (the missense variants c.1331T>C [p.Leu444Pro] and c.1574G>A [p.Arg525Gln] and the truncating variant c.111C>G [p.Tyr37∗]) affect amino acid positions that are conserved up to C. elegans, enabling us to perform functional analyses (Figure 3A). In C. elegans, the MAPK8IP3 ortholog is known as UNC-16, which is encoded by the unc-16 gene. C. elegans unc-16 loss-of-function mutations are recessive and are associated with sluggish locomotion and impaired clearance of organelles from axons.15, 16, 17 In mutants lacking UNC-16, axonal transport is disrupted such that a subset of cell soma organelles, including lysosomes, early endosomes, and Golgi, aberrantly enter axons and accumulate in the synaptic region. This function of UNC-16 is conserved in mammals; as an example, mouse neurons lacking MAPK8IP3 (known as JIP3 in mammals) accumulate lysosomes in their axons.18
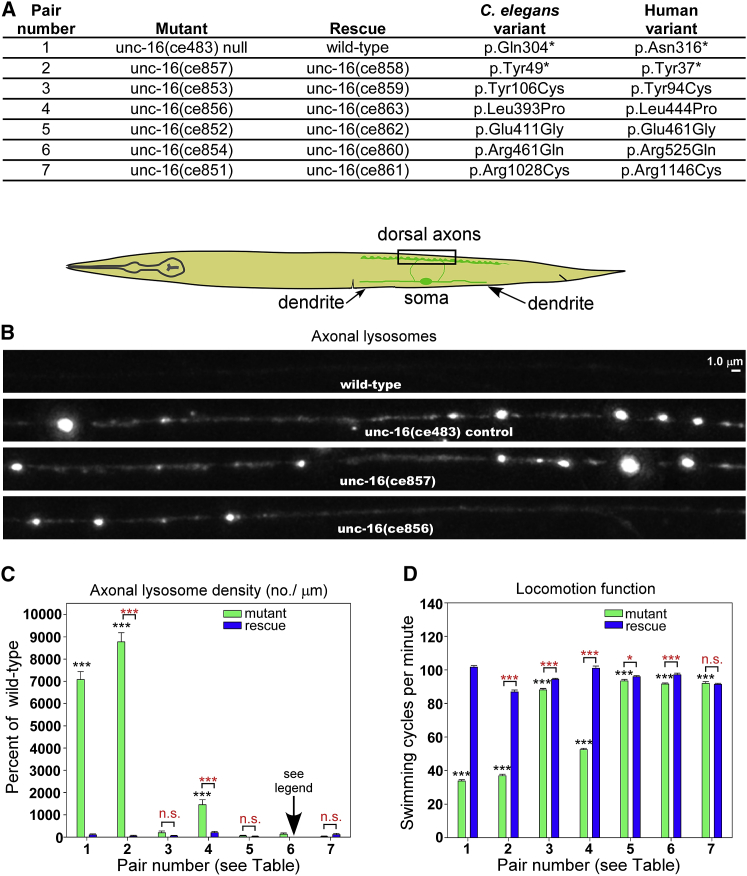
Figure 3.
Impact of Variants on Axonal Lysosome Density and Locomotion
(A) List of all C. elegans unc-16 mutations produced in the study, along with their corresponding wild-type revertants. “Pair number” refers to numbers on the X axes of the graphs. “Rescue” means that the mutation has been reverted to the wild-type amino acid sequence. The amino acid positions of each mutation on the UNC-16 protein in C. elegans and MAPK8IP3 in H. sapiens (GenBank: NM_015133.4) are indicated.
(B and C) Representative images and quantification of lysosome density in DB6 and DA6 motor neuron axons in the indicated genotypes. The rectangle in the drawing indicates the imaged region. The lysosome marker CTNS-1-RFP is expressed from the integrated transgene ceIs56. Representative images were chosen on the basis of density per μm and are identically scaled. Graph data depict lysosome number per μm of axon length exceeding a threshold intensity and are means and standard errors (SEMs) from 57–68 animals each. No rescue data were obtained for “6” because it was not significantly different from the wild-type. Black asterisks compare the marked bar to the wild-type value. Red asterisks compare the indicated two bars in a group. Unmarked bars are not significantly different from the wild-type or the other bar in the group. ∗∗∗ indicates p values that are < 0.001. n.s. = not significant.
(D) Swimming cycles per min for the indicated genotypes. Data are means and SEMs from 72 animals for each strain. Black asterisks compare the marked bar to the wild-type value. Red asterisks compare the indicated two bars in a group. ∗ and ∗∗∗ indicate p < 0.05 or < 0.001, respectively. n.s. = not significant.
To determine the extent to which these six variants disrupt the function of UNC-16, we used CRISPR genome editing to engineer the six variants onto the native C. elegans unc-16 gene. Then we assayed the six mutant strains for locomotion function and lysosome accumulation in the axons. We then compared the strains to wild-type and an unc-16 reference null mutant.15 To control for off-target effects, we again used CRISPR genome editing to reverse-engineer each mutant allele back to its wild-type sequence; this produced “rescued” alleles, which were subjected to the same assays.
To visualize individual lysosomes in neurons, we crossed mutants with a genomically integrated transgene that expresses fluorescently tagged lysosomes in a set of nine ventral cord cholinergic motor neurons. We then counted lysosomal puncta exceeding a pre-defined threshold.15, 17
Placing C. elegans in liquid over a smooth agar surface induces an escape response in which the animal alters its locomotion pattern and attempts to escape the liquid via high frequency swimming. For each strain, we counted swimming cycles in 72 animals for 3 min per animal and obtained averages. At 23° C, wild-type has ∼100 swimming cycles per min (i.e., over 20,000 cycles for the 72 animals). To assay locomotion function in the mutants, we quantified the rate of swimming cycles. We performed swimming-cycle assays by using a modification of the originally described “thrashing assay.”19 When counting swimming cycles, we focused on one side of the animal in the middle of the body and counted each time that part went through a complete cycle. We started the timer when the animal was beginning the cycle, such that the first count represents one complete cycle (for further details on C. elegans functional analyses, see Supplemental Material and Methods).
Results
Clinical Description
An overview of the main clinical symptoms of 13 individuals harboring de novo variants in MAPK8IP3 is presented in Table 1 (see Table S1 for detailed phenotypic descriptions). The onset of symptoms was noted in infancy or early childhood; all 13 individuals displayed DD and a variable degree of ID between mild (4/13; 31%), moderate (6/13; 46%), and severe (3/13; 23%). Furthermore, two individuals were diagnosed with an autism spectrum disorder (ASD). 11 individuals showed abnormalities of muscular tone (hypotonia in nine and spasticity in four; two individuals showed both). Two individuals presented with signs of ataxia, and one presented with an unstable gait. In addition, two individuals had cortical visual impairment, and three individuals had scoliosis. Growth development of individuals in our cohort was mostly normal, although short stature in two and microcephaly of −2.5, −2.5, and −2.7 standard deviations in three individuals were exceptions. One individual had recurrent generalized seizures, and three individuals had events involving one generalized seizure only. None of the individuals was reported to show signs of developmental regression, and there was no consistent finding of facial dysmorphism.
Table 1.
Summary of Clinical Features of Individuals with Causative De Novo Variants in MAPK8IP3.
| Individual | De Novo Variant | p. | Sex | Age | DD and ID (IQ) | ASD | Sz | Neurological Exam | Microcephaly | Brain Anomalies | Other |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | c.65delG | p.(Gly22Alafs∗3) | M | 14 yr | moderate (48) | + | – | MH, ataxia | – | cerebellar atrophy, white-matter hyperintensity in posterior limbs of internal capsules | scoliosis |
| 2 | c.79G>T | p.(Glu27∗) | M | 4 yr | severe | – | – | MH, ataxia | – | no | – |
| 3 | c.111C>G | p.(Tyr37∗) | M | 4 yr | moderate | – | – | MH | – | no | – |
| 4 | c.1198G>A | p.(Gly400Arg) | M | 7 yr | mild | + | – | – | – | NA | – |
| 5 | c.1331T>C | p.(Leu444Pro) | M | 10 yr | moderate (49) | – | 1× gSz | MH | – | perisylvian polymicrogyria | scoliosis |
| 6 | F | 9 yr | mild | – | – | MH | – | perisylvian polymicrogyria | – | ||
| 7 | c.1574G>A | p.(Arg525Gln) | F | 3 yr | mild | – | – | – | −2.7 SD. | no | – |
| 8 | c.1732C>T | p.(Arg578Cys) | F | 5 yr | severe | – | 1× gSz | MH, SP | −2.5 SD. | cerebral atrophy, white-matter volume loss, thin CC with hypoplasia of rostrum and splenium, cerebellar atrophy | short stature |
| 9 | F | 6 yr | moderate | – | 1× gSz | MH, SP | −2.5 SD. | cerebral atrophy, white-matter volume loss, thin CC, cerebellar hypoplasia predominant in inferior vermis | short stature | ||
| 10 | c.2982C>G | p.(His994Gln) | M | 4 yr | moderate | – | gSz | MH | – | no | – |
| 11 | c.3436C>T | p.(Arg1146Cys) | F | 11 yr | mild | – | – | SP | – | mild cerebral atrophy, white-matter volume loss, thin CC | cortical visual impairment |
| 12 | F | 4 yr | severe | – | – | SP | – | short CC | – | ||
| 13 | F | 19 yr | moderate (49) | – | – | MH, unstable gait | – | white-matter volume loss, thin CC | cortical visual impairment, scoliosis |
Variant nomenclature corresponds to GenBank: NM_015133.4. Abbreviations are as follows: ASD = autism spectrum disorder; CC =corpus callosum; DD = developmental delay; ID = intellectual disability; F = female; M = male; MH = muscular hypotonia; NA = not available; SP = spasticity; and gSz = generalized seizures. Further clinical details are provided in Table S1.
Brain Imaging
Cranial MRI was performed for all individuals, apart from individual 4. The MRIs of four individuals (individuals 2, 3, 7 and 10) were reported to be unremarkable. The cranial MRIs of all remaining individuals (individuals 1, 5, 6, 8, 9, 11, 12, and 13) were evaluated for this study and showed a variety of brain anomalies affecting both cerebral and cerebellar structures (Figure 1). Two individuals presented with bilateral perisylvian polymicrogyria with otherwise normal brain structures. One individual showed mild cerebellar atrophy and white-matter hyperintensity in the posterior limbs of the internal capsules. A very thin or short corpus callosum was noted in five individuals, three of whom also showed signs of cerebral atrophy, and four of whom presented with considerable white-matter volume loss.
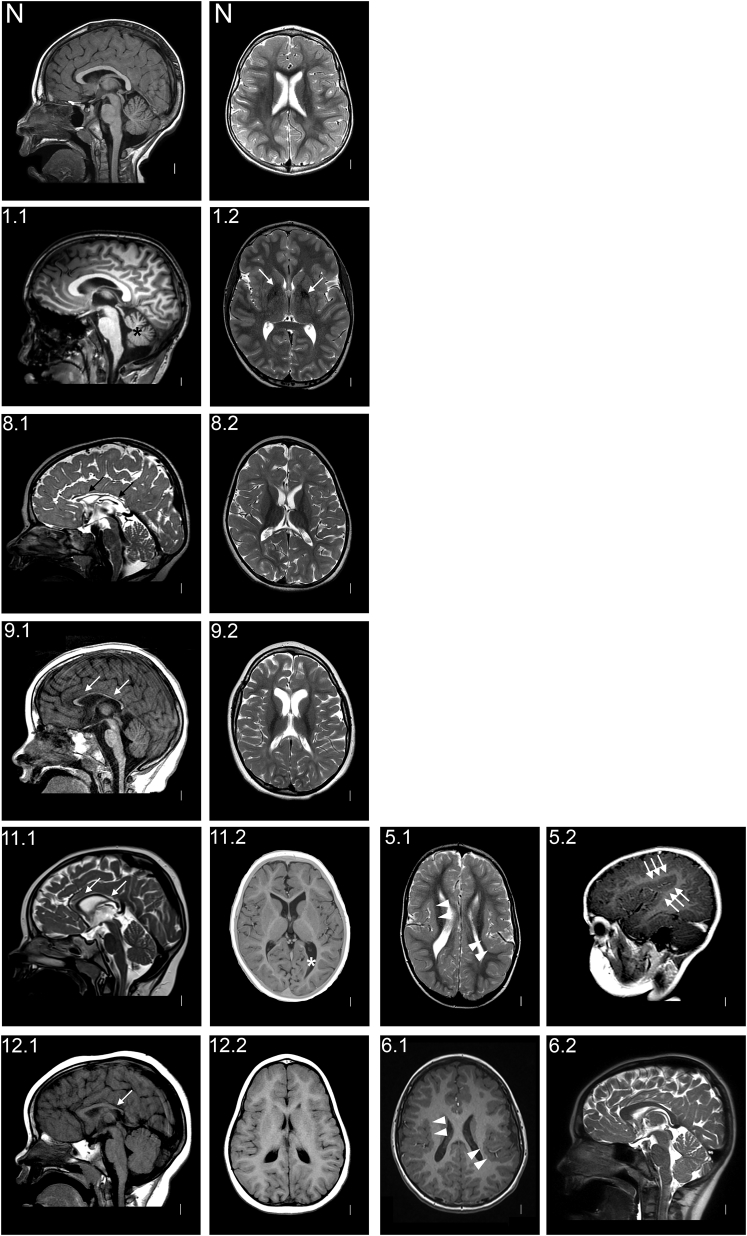
Figure 1.
MRI of Individuals with Brain Anomalies
Images of affected individuals are labeled with numbers as listed in Table 1, and there are two images for each individual. N represents a T1 sagittal midline (left) and a T2 axial (right) image of a healthy individual.
(1.1 and 1.2) T1 midline sagittal and T2 axial images taken at the age of 10 years show mild cerebellar vermis hypoplasia (black asterisk) and hyperintensity in the posterior limbs of the internal capsules (white arrows).
(8.1 and 8.2) T2 midline sagittal and axial images taken at the age of 3 years demonstrate very thin corpus callosum with hypoplasia of the rostrum and splenium (black arrows), cerebral atrophy with broad sulci, and reduced white-matter volume and cerebellar atrophy.
(9.1 and 9.2) T1 midline sagittal and T2 axial images taken at the age of 5 years show a thin corpus callosum (white arrows) and cerebellar and cerebral atrophy with white-matter volume loss.
(11.1 and 11.2) T2 midline sagittal and T1 IR axial images taken at the age of 3 years show a thin corpus callosum (white arrows) and mild cerebral atrophy with white-matter volume loss, predominantly in the occipital lobes, as well as enlarged lateral ventricles (white asterisk).
(12.1 and 12.2) T1 midline sagittal and axial images taken at the age of 26 months demonstrate short and thin corpus callosum but otherwise normal morphology.
(5.1 and 5.2) T2 axial image and a T1 sagittal image (viewed through the right sylvian fissure) taken at the age of 3 years show bilateral perisylvian polymicrogyria (arrow heads and triple arrows).
(6.1 and 6.2) T1 axial and T2 midline sagittal images taken at the age of 8 years show bilateral perisylvian polymicrogyria (white arrowheads) with otherwise unremarkable midline structures.
Causative Variants
By using exome sequencing, we identified de novo protein-affecting, single-nucleotide variants in MAPK8IP3 in 13 individuals (Figure 2A). All identified variants were absent from gnomAD. All three of the truncating variants are located in the first exon of MAPK8IP3; thus, nonsense-mediated mRNA decay is likely.20 The six missense variants, three of which are recurrent, affect amino acids that are highly conserved up to zebrafish, or even further, and all are predicted to be deleterious by multiple in-silico prediction programs (Table S2). According to data from ExAC, MAPK8IP3 is a gene with a significantly reduced number of truncating and missense variants in controls, indicating a selective constraint on both types of variants in a control population that lacks severe, early-onset phenotypes such as DD and ID (pLI score = 1.00; z score for missense variants = 4.06).9
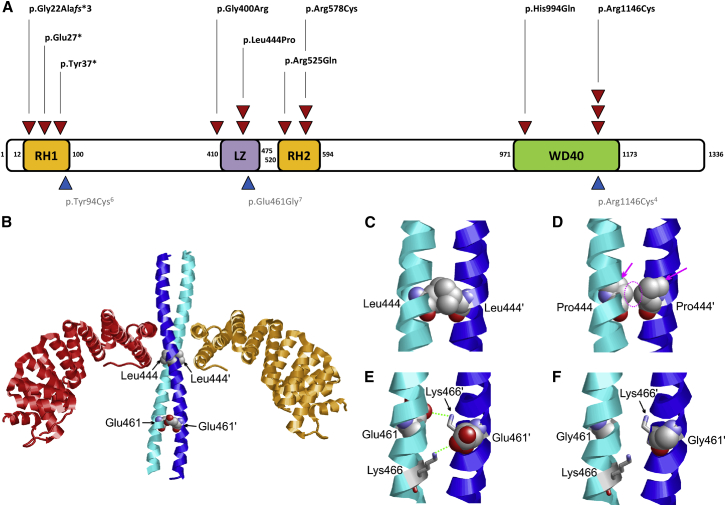
Figure 2.
De Novo Variants in MAPK8IP3 and Structural Effects in the Leucine Zipper Domain
(A) Location of de novo single-nucleotide variants in MAPK8IP3 with respect to domain structure (GenBank: NM_015133.4). Red triangles = individuals from this cohort. Blue triangles = published variants in individuals with a neurodevelopmental disorder. Abbreviations are as follows: LZ =leucine zipper; RH1 and RH2 = Rab-interacting lysosomal protein (RILP) homology 1 and 2.
(B) Crystal structure of the MAPK8IP3 leucine zipper (cyan and blue) in complex with two TPR domains of the kinesin light chain (orange and red). The sites of the sequence variants detected in the present study are shown in space-filled presentation.
(C) The Leu444 sidechains of the homodimeric leucine zipper interact with each other (a prime denotes residues of the second subunit).
(D) In the p.Leu444Pro variant, these interactions are lost as a result of the shorter proline sidechain (dotted circle). In addition, the cyclic proline sidechain causes a steric clash with the backbone of adjacent helix residues (magenta arrows). Both effects are expected to cause a drastic destabilization of the helix and to impede homodimer formation.
(E) The Glu461 side chain forms weak inter-subunit electrostatic interactions with Lys466 (green dotted lines).
(F) The p.Glu461Gly variant cannot form these polar interactions because there is no charged sidechain.
Enrichment of De Novo Variants in MAPK8IP3 in 27,232 Individuals
Altogether, we performed exome sequencing in 27,232 individuals, a majority of whom had been diagnosed with a neurodevelopmental disorder. Because the estimated probability of mutation (missense, nonsense, frameshift) for MAPK8IP3 is roughly 0.629×10−4,10 the occurrence of 13 de novo variants is highly unlikely to have occurred by chance (p = 6.165×10−5). This is probably a conservative estimate; the de novo nonsense variant in individual 2 was identified through single-exome sequencing in a cohort of 4,600 individuals with a variety of clinical indications and a subsequent segregation analysis of only truncating variants in genes with a pLI > 0.95.9
In Silico Structural Modeling
A structural analysis of MAPK8IP3 indicates that the variant-affected amino acid positions Leu444 and Glu461 are located in the leucine zipper domain, which forms a homodimeric structure consisting of two α helices oriented in a parallel fashion. Leu444 is located immediately adjacent to the binding site of the KLC2 tetratricopeptide (TPR) domain (Figure 2B). A closer inspection of the zipper structure reveals that the Leu444 side chains from both subunits form contacts with each other (Figure 2C), but these contacts are absent in the p.Leu444Pro variant (Figure 2D). In addition, proline is known to act as a strong α helix breaker, and thus this exchange is expected to severely affect both the structure of the helical subunits and the homodimer formation and, consequently, the interaction with TPR-domains.
In contrast, the structural effect of the p.Glu461Gly7 variant appears to be less severe. The Glu461 side chain forms weak electrostatic interactions with Lys466; these interactions are absent in the p.Glu461Gly variant (Figures 2E and 2F). In addition, p.Glu461Gly might have a negative effect on helix stability because of enhanced backbone flexibility.
Inspection of the WD40-domain model indicates that His994 and Arg1146 are oriented toward the solvent and do not form significant interactions with other residues of the domain. This observation, together with the high sequence-conservation among orthologs, might indicate that the respective residues play a role for protein-protein interactions rather than for WD40-domain stability. WD40 domains are a large group of protein-interaction domains, which recognize protein ligands via divergent surface patches.21
Axonal Lysosomal Density in C. elegans
The MAPK8IP3 mutant corresponding to p.Tyr37∗ had an axonal lysosomal density about 88-fold higher than that of wild-type (Figures 3B and 3C). This was slightly greater than that of the reference null mutant, which corresponds to p.Asn316∗ in MAPK8IP3. The p.Tyr37∗ mutant phenotype was entirely caused by the disrupted function of UNC-16: reverse engineering the sequence back to the wild-type sequence corrected the lysosomal density to the wild-type density (Figure 3C). In addition to modeling the nonsense variant, we engineered five missense mutants, but the remaining three variants were not conserved in C. elegans and were thus not included. Of the five missense mutants, only that corresponding to p.Leu444Pro had a significantly elevated axonal lysosome density (∼15-fold higher than that of wild-type; Figures 3B and 3C). Reverse engineering the proline 444 back to leucine corrected the axonal lysosome density to a level not significantly different from that of the wild-type (Figure 3C). The remaining four variants produced non-significant results.
Locomotion Assay in C. elegans
The unc-16 reference null mutant had a swimming-cycle rate that was ∼35% of the wild-type rate (Figure 3D). Overall, five of the six investigated mutants had swimming-cycle rates that were significantly lower than their reverse-engineered counterparts. The mutant corresponding to MAPK8IP3 p.Tyr37∗ had a swimming rate similar to that of the reference null mutant (Figure 3D). The mutant corresponding to MAPK8IP3 p.Leu444Pro had a swimming rate that was ∼50% of the wild-type rate and only 55% higher than that of the reference null mutant, again indicating a strong loss of function for this mutant (Figure 3D). Three of the other mutants had swimming rates that were 93%–94% of their reverse-engineered counterparts, but this was still highly significant because of the large sample size and large number of swimming cycles counted for each strain (Figure 3D). The mutants with milder, but statistically significant, locomotion phenotypes correspond to MAPK8IP3 p.Tyr94Cys, p.Arg525Gln, and p.Glu461Gly. The remaining mutant, corresponding to p.Arg1146Cys, showed non-significant results.
Discussion
In this study, we describe in detail 13 individuals with intellectual disability and variable brain anomalies. All individuals harbor de novo truncating or missense variants in MAPK8IP3; this represents a significant statistical enrichment within our cohort in light of the fact that this gene is under constraint of such variants. To further underscore pathogenicity, we modeled adverse structural effects of variants located in the leucine zipper domain, and we functionally characterized a subset of variants in C. elegans showing increased lysosomal density and abnormal locomotion. Thus, taking all lines of evidence together, we firmly establish de novo variants in MAPK8IP3 as a cause of developmental delay and/or intellectual disability with variable brain anomalies such as perisylvian polymicrogyria, as well as cerebral and cerebellar atrophy.
MAPK8IP3 has been shown to be part of the axonal transport machinery, which is essential for the function and maintenance of neurons.5 It supplies the synapse with newly synthesized proteins and lipids while damaged or misfolded proteins are cleared. Defects in axonal transport can lead to progressive neuronal cell death22 and have initially been associated with a wide range of neurodegenerative diseases, for example hereditary spastic paraplegia, Parkinson disease, Alzheimer disease, and other forms of dementia.23 But certain components of the axonal transport machinery, such as the dynein heavy chain 1 (DYNC1H1) and BICD2 have also been implicated in childhood-onset neurodevelopmental disorders comprising ID and a spectrum of malformations of cortical development (MCD), such as lissencephaly and (bilateral perisylvian) polymicrogyria.24, 25, 26, 27 In addition, the disruption of proteins (such as CDK517 and NTRK228 [alternative name TRKB]) that directly interact with MAPK8IP3 has been shown to cause childhood-onset phenotypes of ID, lissencephaly, and cerebellar hypoplasia.29, 30, 31, 32 Taken together, these overlapping phenotypic effects of ID and variable brain anomalies due to the disruption of other parts of the axonal transport machinery provide another line of evidence for the causal role of MAPK8IP3 in the genesis of ID and variable brain anomalies.
Although our observations are based on a small cohort of 13 individuals, we observe several hints for genotype-phenotype correlations, which are especially striking with respect to brain anomalies in individuals harboring recurrent missense variants: (1) both individuals with the missense variant p.Leu444Pro presented with a very similar bilateral perisylvian polymicrogyria (PMG) with otherwise normal brain structures; (2) both individuals with the missense variant p.Arg578Cys presented with an overlapping phenotype of cerebral atrophy, considerable white-matter volume loss, and a very thin corpus callosum, as well as cerebellar atrophy or hypoplasia; and (3) two of three individuals with the missense variant p.Arg1146Cys showed white-matter volume loss and a thin corpus callosum. In comparison, the third individual with the p.Arg1146Cys variant showed a short corpus callosum only. This mostly consistent overlap of brain anomalies in individuals with recurrent de novo missense variants could suggest variant-specific functional consequences possibly related to MAPK8IP3′s domain constitution. Further studies with additional individuals harboring the same de novo variants will have to investigate whether these preliminary observations can be confirmed in a bigger cohort. Other aspects of a possible genotype-phenotype correlation include signs of an ataxic gait in two of three individuals with truncating variants; spasticity, short stature, and microcephaly in both individuals harboring the variant p.Arg578Cys; and the occurrence of spasticity and cortical visual impairment in two individuals with the p.Arg1146Cys variant. Although these phenotypic presentations recurred within our cohort, there is only very limited evidence that seizures are truly part of the phenotypic spectrum of MAPK8IP3-related neurodevelopment disorders: three individuals had only one event of a generalized seizure each, and two of these occurred during febrile episodes; and individual 10, who had recurrent generalized seizures, in fact harbors a second de novo missense variant in SLC6A1 (MIM: 137165), a gene associated with myotonic-atonic epilepsy (see Table S1). This could be a rare case of an individual in whom two monogenic disorders both cause an NDD phenotype.
PMG constitutes an MCD with a plethora of non-genetic causes, such as congenital infections and hypoxia, in contrast to a wide range of monogenic causes.33 The most common location for PMG is the perisylvian region.34 PMG is poorly understood in regard to its origin in brain development, and the term “polymicrogyria” has been used imprecisely in the literature.35 Presently, PMG in humans is not thought to arise because of disrupted migration of cortical neurons; instead it more likely represents a postmigration disorder involving the fusion of cortical laminae.35, 36 In a recent study,37 MAPK8IP3’s mediation of NTRK2 axonal anterograde transport implicated it in the regulation of radial migration, and the expression of MAPK8IP3 was shown to gradually increase from embryonic day 10 to postnatal day 10; especially elevated periods occurred at the time of migration.
De novo variants in MAPK8IP3 have so far been independently reported in three individuals with an NDD. Only one detailed phenotypic description is available: that of a ten-year-old female with a tentative clinical suspicion of Smith-Magenis-Syndrome.7 The girl showed moderate ID and muscular hypotonia fitting the spectrum of MAPK8IP3-related neurodevelopmental disorders described in our cohort. The other two reported de novo missense variants comprise (1) a variant found in the context of the Deciphering Developmental Delay study,4 c.3436C>T (p.Arg1146Cys), which we also identified in three individuals in our cohort, and (2) c.281A>G (p.Tyr94Cys) in an individual with autistic features.6 Interestingly, the latter de novo variant was identified in a male individual with an IQ > 90. Nonetheless, combined with the observation of autistic features in two individuals in our cohort, this strengthens the possibility that ASD is part of the spectrum of MAPK8IP3-related neurodevelopmental disorders. ID and ASD show a significant molecular overlap. However, it is unclear whether this overlap is due to shared molecular pathways between two distinct neurodevelopmental disorders or whether it represents two aspects of clinical presentation within a certain genetic disorder.2, 38
The truncating variants p.Tyr37∗ and p.Leu444Pro constitute the variants displaying the most drastic changes in the model and functional assays we studied, but the intellectual outcome was in fact more severe in other individuals. Four of the six missense variants had relatively mild functional consequences with respect to axonal lysosome accumulation and locomotion, and the variant p.Arg1146Cys showed only non-significant alterations despite constituting the most recurrent de novo variant (seen in four individuals overall). This is not surprising because MAPK8IP3 is a large protein and appears to have evolved at least two independent functions that are distinguished by different cargos and interactions with different motors.5 One function, assayed in this study, is the organelle clearance function, which appears to be conserved from C. elegans to mammals15, 18 and functions independently of the Kinesin-1 motor.5, 15 However, MAPK8IP3 also has a Kinesin-1-dependent adaptor function that appears to connect the Kinesin-1 motor to Kinesin-1 cargoes that are carried on or within small transport vesicles.5 So far, the only MAPK8IP3 adaptor cargo that has been clearly defined is NTRK2,28, 39 which is a receptor for brain-derived neurotrophic factor (BDNF) that has no homologs in C. elegans. Other studies have found that MAPK8IP3′s Kinesin-1-dependent function(s) are important for axonal elongation.40, 41, 42 One of these studies provided evidence that MAPK8IP3 itself was the relevant Kinesin-1 cargo for axonal elongation and found that MAPK8IP3 had a stimulatory effect at growing axon tips.40 In C. elegans there are no assays for the Kinesin--dependent functions of MAPK8IP3, and there is no evidence for axonal elongation defects in C. elegans mutants lacking MAPK8IP3. We hypothesize that the four variants with no detectable organelle-clearance defects disrupt protein domains that are not required for organelle clearance but that are required for one or more of MAPK8IP3′s functions, such as Kinesin-1-dependent axonal development.
In summary, we establish de novo variants in MAPK8IP3 as a cause of ID and variable brain anomalies, further implicating defects in the axonal transport machinery in neurodevelopmental disorders.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We thank the families for their participation and support of this study. We thank Kelly Schoch for providing clinical follow-up information on one individual.
Published: January 3, 2019
Footnotes
Supplemental Data include Supplemental Material and Methods and two tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.12.008.
Web Resources
DECIPHER, https://decipher.sanger.ac.uk/
ExAC Browser, http://exac.broadinstitute.org/
GeneMatcher, https://genematcher.org/
OMIM, http://www.omim.org/
Supplemental Data
References
- 1.Maulik P.K., Mascarenhas M.N., Mathers C.D., Dua T., Saxena S. Prevalence of intellectual disability: A meta-analysis of population-based studies. Res. Dev. Disabil. 2011;32:419–436. doi: 10.1016/j.ridd.2010.12.018. [DOI] [PubMed] [Google Scholar]
- 2.Vissers L.E.L.M., Gilissen C., Veltman J.A. Genetic studies in intellectual disability and related disorders. Nat. Rev. Genet. 2016;17:9–18. doi: 10.1038/nrg3999. [DOI] [PubMed] [Google Scholar]
- 3.Vissers L.E.L.M., de Ligt J., Gilissen C., Janssen I., Steehouwer M., de Vries P., van Lier B., Arts P., Wieskamp N., del Rosario M. A de novo paradigm for mental retardation. Nat. Genet. 2010;42:1109–1112. doi: 10.1038/ng.712. [DOI] [PubMed] [Google Scholar]
- 4.Deciphering Developmental Disorders Study Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–438. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller K.G. Keeping neuronal cargoes on the right track: New insights into regulators of axonal transport. Neuroscientist. 2017;23:232–250. doi: 10.1177/1073858416648307. [DOI] [PubMed] [Google Scholar]
- 6.Iossifov I., O’Roak B.J., Sanders S.J., Ronemus M., Krumm N., Levy D., Stessman H.A., Witherspoon K.T., Vives L., Patterson K.E. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger S.I., Ciccone C., Simon K.L., Malicdan M.C., Vilboux T., Billington C., Fischer R., Introne W.J., Gropman A., Blancato J.K., NISC Comparative Sequencing Program Exome analysis of Smith-Magenis-like syndrome cohort identifies de novo likely pathogenic variants. Hum. Genet. 2017;136:409–420. doi: 10.1007/s00439-017-1767-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: A matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Samocha K.E., Robinson E.B., Sanders S.J., Stevens C., Sabo A., McGrath L.M., Kosmicki J.A., Rehnström K., Mallick S., Kirby A. A framework for the interpretation of de novo mutation in human disease. Nat. Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cockburn J.J.B., Hesketh S.J., Mulhair P., Thomsen M., O’Connell M.J., Way M. Structure; 2018. Insights into Kinesin-1 Activation from the Crystal Structure of KLC2 Bound to JIP3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu C., Min J. Structure and function of WD40 domain proteins. Protein Cell. 2011;2:202–214. doi: 10.1007/s13238-011-1018-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 14.Sayle R.A., Milner-White E.J. RASMOL: Biomolecular graphics for all. Trends Biochem. Sci. 1995;20:374. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- 15.Edwards S.L., Yu S.C., Hoover C.M., Phillips B.C., Richmond J.E., Miller K.G. An organelle gatekeeper function for Caenorhabditis elegans UNC-16 (JIP3) at the axon initial segment. Genetics. 2013;194:143–161. doi: 10.1534/genetics.112.147348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown H.M., Van Epps H.A., Goncharov A., Grant B.D., Jin Y. The JIP3 scaffold protein UNC-16 regulates RAB-5 dependent membrane trafficking at C. elegans synapses. Dev. Neurobiol. 2009;69:174–190. doi: 10.1002/dneu.20690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edwards S.L., Morrison L.M., Yorks R.M., Hoover C.M., Boominathan S., Miller K.G. UNC-16 (JIP3) acts through synapse-assembly proteins to inhibit the active transport of cell soma organelles to caenorhabditis elegans motor neuron axons. Genetics. 2015;201:117–141. doi: 10.1534/genetics.115.177345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gowrishankar S., Wu Y., Ferguson S.M. Impaired JIP3-dependent axonal lysosome transport promotes amyloid plaque pathology. J. Cell Biol. 2017;216:3291–3305. doi: 10.1083/jcb.201612148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller K.G., Alfonso A., Nguyen M., Crowell J.A., Johnson C.D., Rand J.B. A genetic selection for Caenorhabditis elegans synaptic transmission mutants. Proc. Natl. Acad. Sci. USA. 1996;93:12593–12598. doi: 10.1073/pnas.93.22.12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Popp M.W.-L., Maquat L.E. Organizing principles of mammalian nonsense-mediated mRNA decay. Annu. Rev. Genet. 2013;47:139–165. doi: 10.1146/annurev-genet-111212-133424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stirnimann C.U., Petsalaki E., Russell R.B., Müller C.W. WD40 proteins propel cellular networks. Trends Biochem. Sci. 2010;35:565–574. doi: 10.1016/j.tibs.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 22.Perlson E., Maday S., Fu M.-M., Moughamian A.J., Holzbaur E.L.F. Retrograde axonal transport: Pathways to cell death? Trends Neurosci. 2010;33:335–344. doi: 10.1016/j.tins.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Millecamps S., Julien J.-P. Axonal transport deficits and neurodegenerative diseases. Nat. Rev. Neurosci. 2013;14:161–176. doi: 10.1038/nrn3380. [DOI] [PubMed] [Google Scholar]
- 24.Schiavo G., Greensmith L., Hafezparast M., Fisher E.M.C. Cytoplasmic dynein heavy chain: The servant of many masters. Trends Neurosci. 2013;36:641–651. doi: 10.1016/j.tins.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poirier K., Lebrun N., Broix L., Tian G., Saillour Y., Boscheron C., Parrini E., Valence S., Pierre B.S., Oger M. Mutations in TUBG1, DYNC1H1, KIF5C and KIF2A cause malformations of cortical development and microcephaly. Nat. Genet. 2013;45:639–647. doi: 10.1038/ng.2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Di Donato N., Timms A.E., Aldinger K.A., Mirzaa G.M., Bennett J.T., Collins S., Olds C., Mei D., Chiari S., Carvill G. Analysis of 17 genes detects mutations in 81% of 811 patients with lissencephaly. Genet. Med. 2018;20:1354–1364. doi: 10.1038/gim.2018.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ravenscroft G., Di Donato N., Hahn G., Davis M.R., Craven P.D., Poke G., Neas K.R., Neuhann T.M., Dobyns W.B., Laing N.G. Recurrent de novo BICD2 mutation associated with arthrogryposis multiplex congenita and bilateral perisylvian polymicrogyria. Neuromuscul. Disord. 2016;26:744–748. doi: 10.1016/j.nmd.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 28.Huang S.-H., Duan S., Sun T., Wang J., Zhao L., Geng Z., Yan J., Sun H.-J., Chen Z.-Y. JIP3 mediates TrkB axonal anterograde transport and enhances BDNF signaling by directly bridging TrkB with kinesin-1. J. Neurosci. 2011;31:10602–10614. doi: 10.1523/JNEUROSCI.0436-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Magen D., Ofir A., Berger L., Goldsher D., Eran A., Katib N., Nijem Y., Vlodavsky E., Tzur S., Behar D.M. Autosomal recessive lissencephaly with cerebellar hypoplasia is associated with a loss-of-function mutation in CDK5. Hum. Genet. 2015;134:305–314. doi: 10.1007/s00439-014-1522-5. [DOI] [PubMed] [Google Scholar]
- 30.Yeo G.S.H., Connie Hung C.-C., Rochford J., Keogh J., Gray J., Sivaramakrishnan S., O’Rahilly S., Farooqi I.S. A de novo mutation affecting human TrkB associated with severe obesity and developmental delay. Nat. Neurosci. 2004;7:1187–1189. doi: 10.1038/nn1336. [DOI] [PubMed] [Google Scholar]
- 31.Miller K.A., Twigg S.R.F., McGowan S.J., Phipps J.M., Fenwick A.L., Johnson D., Wall S.A., Noons P., Rees K.E.M., Tidey E.A. Diagnostic value of exome and whole genome sequencing in craniosynostosis. J. Med. Genet. 2017;54:260–268. doi: 10.1136/jmedgenet-2016-104215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamdan F.F., Myers C.T., Cossette P., Lemay P., Spiegelman D., Laporte A.D., Nassif C., Diallo O., Monlong J., Cadieux-Dion M., Deciphering Developmental Disorders Study High rate of recurrent de novo mutations in developmental and epileptic encephalopathies. Am. J. Hum. Genet. 2017;101:664–685. doi: 10.1016/j.ajhg.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guerrini R., Parrini E. Neuronal migration disorders. Neurobiol. Dis. 2010;38:154–166. doi: 10.1016/j.nbd.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 34.Leventer R.J., Jansen A., Pilz D.T., Stoodley N., Marini C., Dubeau F., Malone J., Mitchell L.A., Mandelstam S., Scheffer I.E. Clinical and imaging heterogeneity of polymicrogyria: A study of 328 patients. Brain. 2010;133:1415–1427. doi: 10.1093/brain/awq078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barkovich A.J., Dobyns W.B., Guerrini R. Malformations of cortical development and epilepsy. Cold Spring Harb. Perspect. Med. 2015;5:a022392. doi: 10.1101/cshperspect.a022392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Judkins A.R., Martinez D., Ferreira P., Dobyns W.B., Golden J.A. Polymicrogyria includes fusion of the molecular layer and decreased neuronal populations but normal cortical laminar organization. J. Neuropathol. Exp. Neurol. 2011;70:438–443. doi: 10.1097/NEN.0b013e31821ccf1c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma H., Yu H., Li T., Zhao Y., Hou M., Chen Z., Wang Y., Sun T. JIP3 regulates neuronal radial migration by mediating TrkB axonal anterograde transport in the developing cerebral cortex. Biochem. Biophys. Res. Commun. 2017;485:790–795. doi: 10.1016/j.bbrc.2017.02.132. [DOI] [PubMed] [Google Scholar]
- 38.Moreno-De-Luca A., Myers S.M., Challman T.D., Moreno-De-Luca D., Evans D.W., Ledbetter D.H. Developmental brain dysfunction: Revival and expansion of old concepts based on new genetic evidence. Lancet Neurol. 2013;12:406–414. doi: 10.1016/S1474-4422(13)70011-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Drerup C.M., Nechiporuk A.V. JNK-interacting protein 3 mediates the retrograde transport of activated c-Jun N-terminal kinase and lysosomes. PLoS Genet. 2013;9:e1003303. doi: 10.1371/journal.pgen.1003303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun F., Zhu C., Dixit R., Cavalli V. Sunday Driver/JIP3 binds kinesin heavy chain directly and enhances its motility. EMBO J. 2011;30:3416–3429. doi: 10.1038/emboj.2011.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watt D., Dixit R., Cavalli V. JIP3 activates kinesin-1 motility to promote axon elongation. J. Biol. Chem. 2015;290:15512–15525. doi: 10.1074/jbc.M115.651885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun T., Yu N., Zhai L.-K., Li N., Zhang C., Zhou L., Huang Z., Jiang X.-Y., Shen Y., Chen Z.-Y. c-Jun NH2-terminal kinase (JNK)-interacting protein-3 (JIP3) regulates neuronal axon elongation in a kinesin- and JNK-dependent manner. J. Biol. Chem. 2013;288:14531–14543. doi: 10.1074/jbc.M113.464453. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.