Abstract
Axl receptor tyrosine kinase is involved in the growth and metastasis and is an indicator of poor prognosis in several cancers including lung cancers. Although a mitogen-activated protein kinase (MAPK) pathway and an epithelial-to-mesenchymal transition (EMT) program are critical, molecular mechanisms underlying the Axl-driven cancer progression have not been fully elucidated. We aimed to identify molecules up-regulated by Axl kinase in lung adenocarcinomas. Through the global gene expression analysis and the functional annotation clustering, we found that AXL expression positively correlated with mRNA expressions of immune checkpoint molecules and chemokine receptors in non-small-cell lung cancers. Validation cohorts including our biobank confirmed that the AXL expression significantly correlated with expression of genes encoding programmed death-ligand1 (PD-L1) and CXC chemokine receptor 6 (CXCR6) in lung adenocarcinoma, especially in epidermal growth factor receptor (EGFR) mutation-positive adenocarcinoma. Pharmacological inhibition of Axl kinase activity decreased mRNA expressions of PD-L1 and CXCR6 in EGFR mutation-positive cell lines. Our data indicates the novel role of Axl kinase as a driver of immune checkpoint molecules and chemokine signalling pathways in the progression of lung adenocarcinomas. This study also highlights the necessity of clinical trials in order to test the efficacy of Axl kinase inhibition in the Axl-highly expressing subset of lung adenocarcinomas.
Electronic supplementary material
The online version of this article (10.1186/s12943-019-0953-y) contains supplementary material, which is available to authorized users.
Keywords: Non-small-cell lung cancer, Axl receptor tyrosine kinase, Immune checkpoint molecules, Chemokine signalling, Global gene expression array
Results and discussion
Gene signatures of NSCLC with higher AXL mRNA expression
Activation of Axl receptor tyrosine kinase has a key role in the growth and metastasis of several cancers [1]. In lung adenocarcinomas, the protein expression of Axl and its ligand, growth arrest specific-6 (Gas6), is a critical indicator for the poor prognosis [2]. Moreover acquisition of Axl leads to resistance to epidermal growth factor receptor (EGFR)-targeted therapy for lung adenocarcinomas [3]. Based on these studies, the combination therapy of a selective Axl kinase inhibitor (BGB324) and an EGFR tyrosine kinase inhibitor (Erlotinib) for patients with Stage IIIB or IV non-small cell lung cancers (NSCLC) has currently been in a phase I/II clinical trial (NCT02424617).
Recent studies reported that intracellular kinases (e.g. mitogen-activated protein kinases, MAPKs) and epithelial-to-mesenchymal transition (EMT)-initiating transcription factors are involved in the Axl-driven survival and motility of cancers [1]. In addition a recent report indicates that Axl also up-regulates the expression of an immune checkpoint molecule, programmed death-ligand1 (PD-L1, or CD274) in head and neck cancers [4]. These studies suggest that the activation of Axl controls diverse molecular pathways contributing to a microenvironment beneficial to tumor progression. However the diverse array of molecules under Axl kinase has not been fully elucidated in lung cancer.
In order to characterise molecular phenotypes of NSCLC with higher AXL expression, we sought to identify genes whose expressions significantly correlated with AXL mRNA expression in a lung cancer tissue biobank (GSE accession number, GSE42127, n = 176 subjects, Additional file 1: Materials and Methods). In this discovery cohort, we found that 935 genes positively correlated with AXL expression (rp > 0.4; Additional file 2: Table S1), whereas 137 genes were negatively correlated (rp < − 0.4; Additional file 2: Table S2). A functional annotation clustering analysis revealed that gene ontology terms, “chemokine mediated signalling pathway” and “antigen processing and presentation”, were enriched in the 935 genes positively correlating with AXL mRNA expression (Additional file 2: Table S3). We failed to detect gene ontology terms in the 137 genes negatively correlating with AXL expression.
Positive correlation of AXL expression with immune checkpoint molecules and chemokine receptors in lung adenocarcinomas
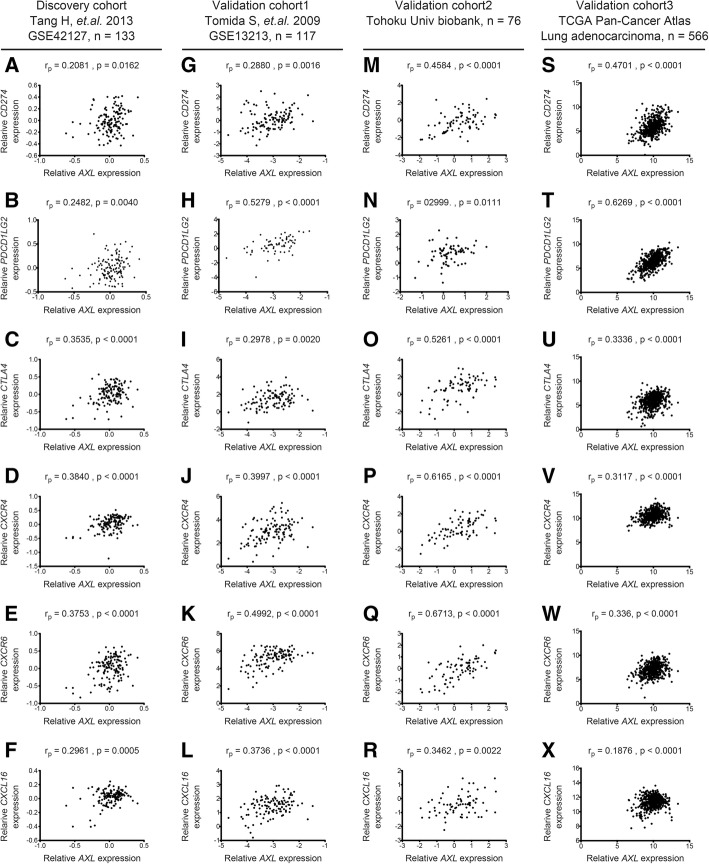
Our unbiased analysis of the discovery cohort microarray data suggests that chemokine signalling pathways and molecules associated with antigen processing and presentation (e.g., major histocompatibility complex (MHC) genes) are relevant to de novo AXL-highly expressing NSCLC. A recent report indicated that genes encoding MHC class I molecules positively correlated to PD-L1 mRNA expression in ovarian tumor cells [5]. Thus we sought to further validate the contribution of immune checkpoint molecules and chemokine signalling pathways to Axl-expressing adenocarcinomas. We have established Tohoku University Biobank (Additional file 2: Table S4). Using three independent validation cohorts of lung adenocarcinomas (Validation cohort1: GSE13213 in the Gene Expression Omnibus (GEO) of the National Center for Biotechnology Information (NCBI), n = 117; Validation cohort2: Our cohort of Tohoku University Biobank, n = 76; Validation cohort3: Lung Adenocarcinoma in The Cancer Genome Atlas (TCGA), Pan-Cancer Atlas, n = 566), we found that AXL expression significantly correlated with the expressions of genes encoding immune checkpoint molecules (CD274, PDCD1LG2 and CTLA4), chemokine receptors (CXCR4 and CXCR6) or a chemokine (CXCL16) in the discovery cohort as well as the three validation cohorts (Fig. 1). These chemokine receptors have been attributed to the invasion and metastases of cancers [6].
Fig. 1.
Genes encoding immune checkpoint molecules and chemokine/chemokine receptors were enriched in Axl-highly expressing NSCLC. Correlations of mRNA expressions between AXL and genes encoding PD-L1 (CD274; a, g, m and s), PD-L2 (PDCD1LG2; b, h, n and t), CTLA-4 (CTLA4; c, i, o and u), CXCR4 (CXCR4; d, j, p and v), CXCR6 (CXCR6; e, k, q and w) and CXCL16 (CXCL16; f, l, r and x). Microarray data from three cohorts (discovery cohort (a-f, GSE42127), validation cohort1 (g-l, GSE13213), validation cohort3 (s-x, TCGA Pan-Cancer Atlas) and qRT-PCR data from our cohort of Tohoku University Biobank (m-r) are shown. rp is a Pearson correlation coefficient. The x-y axes indicate log2 transformed expression values of genes indicated
Higher expression of genes encoding PD-L1, CXCR6 and CXCL16 in Axl-expressing adenocarcinomas with mutated EGFR.
To determine whether the EGFR mutation status is influential in the correlation of AXL expression with the immune checkpoint molecules and the chemokine/chemokine receptors, we evaluated Pearson correlation coefficients between EGFR mutation-positive and wild type lung adenocarcinomas in our cohort of Tohoku University Biobank. We found higher correlations of gene expressions between AXL and three genes (CD274, CXCR6 and CXCL16) in EGFR-mutation positive adenocarcinomas than in wild type (Fig. 2).
Fig. 2.
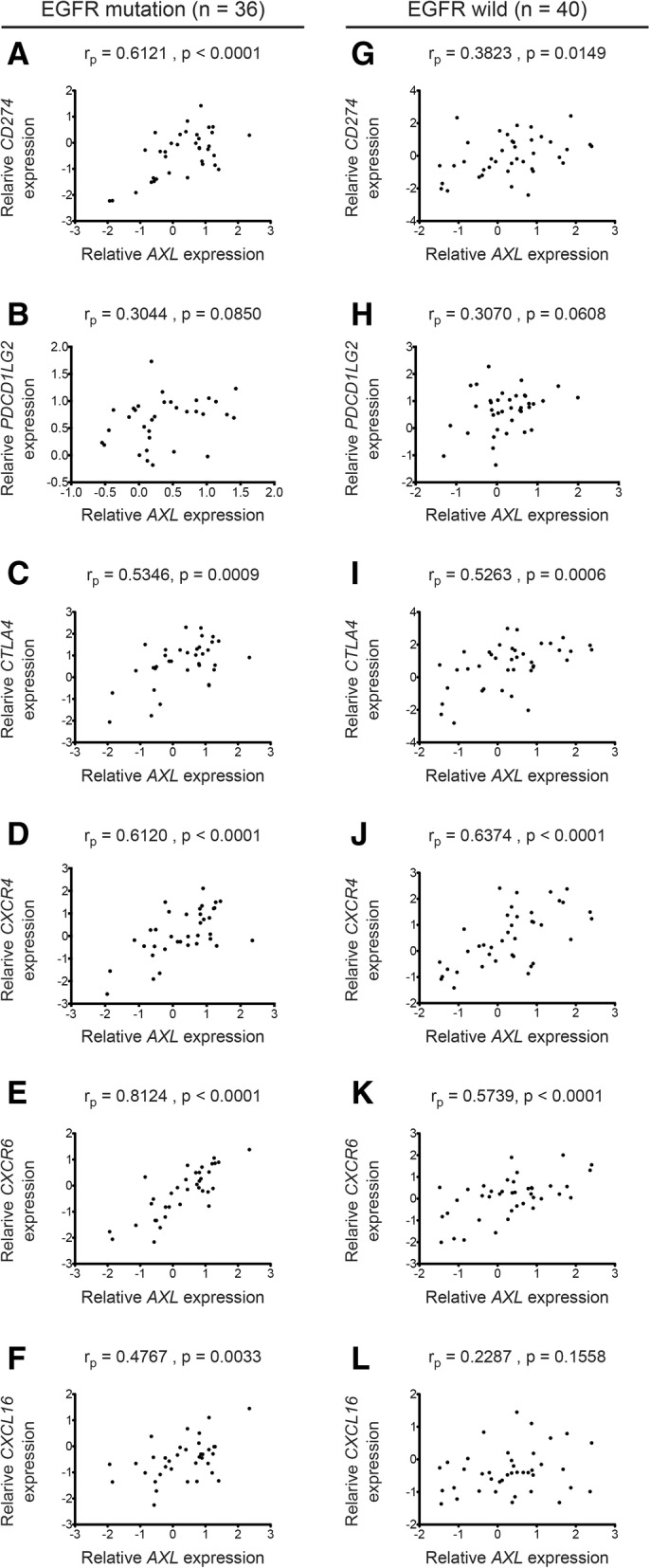
Genes encoding PD-L1 and CXCR6 correlated with AXL mRNA in EGFR-mutated lung adenocarcinomas. Correlations of mRNA expressions between AXL and genes encoding PD-L1 (CD274; a and g), PD-L2 (PDCD1LG2; b and h), CTLA-4 (CTLA4; c and i), CXCR4 (CXCR4; d and j), CXCR6 (CXCR6; e and k) and CXCL16 (CXCL16; f and l) in EGFR mutation-positive (a-f) and EGFR wild-type (l-l) lung adenocarcinomas from Tohoku University Biobank. rp is a Pearson correlation coefficient. The x-y axes indicate log2 transformed expression values of genes indicated
Pharmacological inhibition of Axl kinase activity decreases the mRNA expressions of CD274, PDCD1LG2 and CXCR6 in EGFR mutation-positive lung adenocarcinoma cell lines
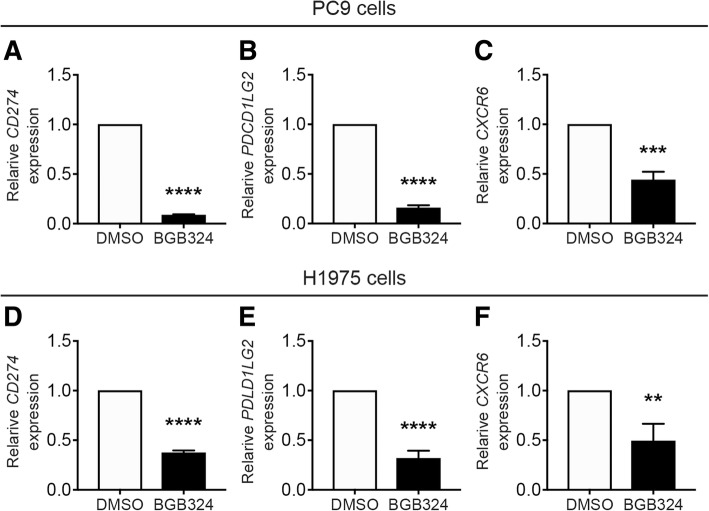
To determine whether and how Axl regulates the mRNA expressions of PD-L1, PD-L2 and CXCR6 in EGFR-mutated lung adenocarcinomas, we treated two EGFR mutation-positive lung adenocarcinoma cell lines (PC9 cells and H1975 cells) with a selective Axl kinase inhibitor BGB324 or with siRNA targeting Axl. We confirmed mRNA and protein expressions of Axl in both PC9 cells and H1975 cells (data not shown) and verified that the Axl inhibitor BGB324 decreased phosphorylation of Axl in these cell lines in a dose-dependent manner (Additional file 3: Figure S1). We found that BGB324 significantly decreased the expressions of CD274 and PDCD1LG2 and CXCR6 in both cell lines (Fig. 3). This was confirmed by siRNA-mediated Axl silencing (Additional file 3: Figure S2). We further confirmed that BGB324 reduced phosphorylation of extracellular signal-regulated kinase (ERK) and AKT in PC9 cells and H1975 cells (Additional file 3: Figure S3). A selective MEK inhibitor (U1206) or an allosteric AKT inhibitor (MK-2206) decreased mRNA expression of PD-L1 (Additional file 3: Figure S4), suggesting that ERK and AKT were at least partly involved in downstream signalling pathways of Axl [7].
Fig. 3.
A selective inhibitor of Axl kinase decreased PD-L1, PD-L2 and CXCR6 mRNA expression in vitro. Relative expression values of CD274 (PD-L1; a, d), PDCD1LG2 (PD-L2; b, e) and CXCR6 (c, f) in PC9 cells (a-c) and H1975 cells (d-f) six hours after treatment with 0.1% DMSO or Axl inhibitor BGB324 (10 μM). Each value represents the fold-increase in gene expression of BGB324-treated cells compared to that of DMSO-treated cells in each experiment. Data are expressed as the mean ± SD of three independent experiments. ****p < 0.0001, **p < 0.01, *p < 0.05, vs. DMSO; two-tailed paired t-test. NS indicates not significant
Positive correlation of AXL expression with co-inhibitory and co-stimulatory molecules across multiple cancers
So far, through the unbiased transcriptome analyses and the subsequent in vitro cell culture assays, we identified a new role of Axl kinase in up-regulating PD-L1 in lung adenocarcinoma. We then sought to extend this finding to several cancers including EGFR-related cancers (glioblastoma and colon cancer) and other solid tumors (melanoma, renal cell carcinoma, gastric cancer and head and neck cancer), most of which Nivolumab have been approved for the treatment. In addition we wished to clarify a broader expression pattern of co-inhibitory and co-stimulatory molecules whose expressions correlated with Axl kinase. Using TCGA datasets, we found that the EGFR-related cancers exhibited more significant correlation of PD-L1 mRNA expression with AXL expression (Additional file 2: Table S6). Moreover co-inhibitory receptors (PDCD1 encoding PD-1 and HAVCR2 encoding Tim3) and co-stimulatory receptors (CD28 and CD137) showed the significant correlation with AXL across a variety of cancers (Additional file 2: Table S6).
In summary, our findings suggest that Axl kinase up-regulates several pathways such as immune checkpoint molecules/chemokine signalling involved in regulation of the immune microenvironment and tumor proliferation (Additional file 3: Figure S5). Considering the diverse non-genomic actions driven by Axl, monotherapies targeting immune checkpoint molecules such as PD-L1 or PD-L2/PD-1 or chemokines might not yield sufficient treatment efficacy in patients with Axl-positive NSCLC. This hypothesis is supported by a recent report demonstrating that Axl was up-regulated in melanomas with innate resistance to an anti-PD-1 therapy [8]. Given our results, it might be possible that Axl kinase inhibition could increase the sensitivity to a PD-1 targeted therapy. In addition NSCLC harbouring EGFR mutations are associated with poor efficacy of PD-1/PD-L1 inhibitors [9]. We therefore speculate that Axl kinase drives multiple molecular pathways promoting tumor progression especially in EGFR-mutated lung adenocarcinoma.
There are some limitations in the present study. First we did not observe correlation between Axl kinase and these immune-related molecules at a protein level. Second we have not tested if Axl kinase promotes immune checkpoint molecules and chemokines that function in in vivo settings. Third we cannot exclude a possibility that PD-L1-bearing cell-types other than cancer cells were involved in this bioinformatic analysis as we analyzed transcriptome data from whole tissue lysates but not at a single cell level. Further validation studies using immunohistochemistry, animal models and a single cell-RNA sequence are required.
In conclusion, Axl-highly expressing lung adenocarcinomas exhibit higher expressions of multiple genes encoding immune checkpoint molecules and chemokines/chemokine receptors. Our data therefore provide a rationale for further mechanistic studies to validate the role of Axl kinase as a driver in diversifying downstream molecules in NSCLC and, ultimately, to perform trials testing the efficacy of Axl inhibition in the Axl-highly expressing subset of NSCLC.
Additional files
Materials and Methods. (DOCX 37 kb)
Table S1. 935 genes positively correlating AXL mRNA expression in a NSCLC biobank (GSE accession number, GSE42127). Table S2. 137 genes negatively correlating AXL mRNA expression in a NSCLC biobank (GSE accession number, GSE42127). Table S3. Ontology terms enriched in the 935 genes that positively correlated AXL mRNA expression in a lung cancer biobank (GSE accession number, GSE42127). Table S4. Characteristics of patients (n = 76) with lung adenocarcinomas of our Tohoku University Biobank for the qPCR validation study. Table S5. Primer list. Table S6. Correlation of mRNA expression between AXL and genes encoding co-stimulatory/inhibitory molecules as shown by Pearson’s r. (XLSX 68 kb)
Figure S1. A dose-dependent decrease in phosphorylation of Axl in PC9 cells by the Axl kinase inhibitor. Figure S2. Knockdown of Axl decreases mRNA expression of PD-L1, PD-L2 and CXCR6 in vitro. Figure S3. Immunoblots indicating decreases in phosphorylation of ERK1/2 and AKT by the Axl kinase inhibitor. Figure S4. A selective MEK1/2 inhibitor or an AKT inhibitor reduces PD-L1 mRNA expression in vitro. Figure S5. Diverse downstream pathways driven by Axl receptor tyrosine kinase in non-small-cell lung cancer. (DOCX 2506 kb)
Acknowledgements
We are grateful to Dr. Akira Sakurada (Tohoku University) for obtaining human lung tissues. We thank the Biomedical Research Unit of Tohoku University Hospital for technical support. We very much appreciate Mr. Brent K. Bell for reading the manuscript and Ms. Mitsuru Takahashi and Ms. Masako Honda for their technical supports.
Funding
This work was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI (Grant Number: JP16H06641).
Availability of data and materials
The microarray datasets analysed in the current study are available in the NCBI GEO repository [GSE42127 and GSE13213] or cBioPortal for Cancer Genomics (http://www.cbioportal.org). The datasets used and/or analysed during the current study are available from the corresponding author on a reasonable request.
Abbreviations
- CXCL
CXC chemokine ligand
- CXCR
CXC chemokine receptor
- EGFR
Epidermal growth factor receptor
- EMT
Epithelial-to-mesenchymal transition
- ERK
Extracellular signal-regulated kinase
- Gas6
Growth arrest specific-6
- GEO
The Gene Expression Omnibus
- MAPKs
Mitogen-activated protein kinases
- MHC
Major histocompatibility complex
- NCBI
The National Center for Biotechnology Information
- NSCLC
Non-small-cell lung cancer
- PD-L1
Programmed death-ligand1
- PI3K
Phosphoinositide 3-kinase
- TCGA
The Cancer Genome Atlas
Authors’ contributions
YT performed experiments, analysed/interpreted data and wrote the manuscript. NF conceived this project, designed strategy, interpreted data and wrote the manuscript. EM interpreted data, contributed to writing the manuscript and the biobank. RS and FF contributed to the biobank and performed pathological diagnosis. KI, YK, KO, MY, TO and HS performed experiments. YO contributed to the biobank. AI and MI provided advice and supervised the project. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by the Ethnic Committee at Tohoku University School of Medicine.
(Number: 2017–1-972). Written informed consents were received from all patients before the surgery.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yoko Tsukita, Email: y-tsukita@rm.med.tohoku.ac.jp.
Naoya Fujino, Phone: +81-22-717-8539, Email: nfujino@med.tohoku.ac.jp.
Eisaku Miyauchi, Email: miyauchi@rm.med.tohoku.ac.jp.
Ryoko Saito, Email: saitoarlu@patholo2.med.tohoku.ac.jp.
Fumiyoshi Fujishima, Email: ffujishima@patholo2.med.tohoku.ac.jp.
Koji Itakura, Email: koji.itakura@rm.med.tohoku.ac.jp.
Yorihiko Kyogoku, Email: y.kyogoku@rm.med.tohoku.ac.jp.
Koji Okutomo, Email: kseric-rm@rm.med.tohoku.ac.jp.
Mitsuhiro Yamada, Email: yamitsu@med.tohoku.ac.jp.
Tatsuma Okazaki, Email: tmokazaki0808@gmail.com.
Hisatoshi Sugiura, Email: sugiura@rm.med.tohoku.ac.jp.
Akira Inoue, Email: akira.inoue.b2@tohoku.ac.jp.
Yoshinori Okada, Email: yoshinori.okada.a1@tohoku.ac.jp.
Masakazu Ichinose, Email: ichinose@rm.med.tohoku.ac.jp.
References
- 1.Graham DK, DeRyckere D, Davies KD, Earp HS. The TAM family: phosphatidylserine-sensing receptor tyrosine kinases gone awry in cancer. Nat Rev Cancer. 2014;14:769–785. doi: 10.1038/nrc3847. [DOI] [PubMed] [Google Scholar]
- 2.Ishikawa M, Sonobe M, Nakayama E, Kobayashi M, Kikuchi R, Kitamura J, et al. Higher expression of receptor tyrosine kinase Axl, and differential expression of its ligand, Gas6, predict poor survival in lung adenocarcinoma patients. Ann Surg Oncol. 2013;20(Suppl 3):S467–S476. doi: 10.1245/s10434-012-2795-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Z, Lee JC, Lin L, Olivas V, Au V, LaFramboise T, et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat Genet. 2012;44:852–860. doi: 10.1038/ng.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skinner HD, Giri U, Yang LP, Kumar M, Liu Y, Story MD, et al. Integrative analysis identifies a novel AXL-PI3 kinase-PD-L1 signaling Axis associated with radiation resistance in head and neck Cancer. Clin Cancer Res. 2017;23:2713–2722. doi: 10.1158/1078-0432.CCR-16-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aust S, Felix S, Auer K, Bachmayr-Heyda A, Kenner L, Dekan S, et al. Absence of PD-L1 on tumor cells is associated with reduced MHC I expression and PD-L1 expression increases in recurrent serous ovarian cancer. Sci Rep. 2017;7:42929. doi: 10.1038/srep42929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagarsheth N, Wicha MS, Zou W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat Rev Immunol. 2017;17:559–572. doi: 10.1038/nri.2017.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen N, Fang W, Zhan J, Hong S, Tang Y, Kang S, et al. Upregulation of PD-L1 by EGFR activation mediates the immune escape in EGFR-driven NSCLC: implication for optional immune targeted therapy for NSCLC patients with EGFR mutation. J Thorac Oncol. 2015;10:910–923. doi: 10.1097/JTO.0000000000000500. [DOI] [PubMed] [Google Scholar]
- 8.Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2016;165:35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee CK, Man J, Lord S, Links M, Gebski V, Mok T, et al. Checkpoint inhibitors in metastatic EGFR-mutated non-small cell lung Cancer-a meta-analysis. J Thorac Oncol. 2017;12:403–407. doi: 10.1016/j.jtho.2016.10.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Materials and Methods. (DOCX 37 kb)
Table S1. 935 genes positively correlating AXL mRNA expression in a NSCLC biobank (GSE accession number, GSE42127). Table S2. 137 genes negatively correlating AXL mRNA expression in a NSCLC biobank (GSE accession number, GSE42127). Table S3. Ontology terms enriched in the 935 genes that positively correlated AXL mRNA expression in a lung cancer biobank (GSE accession number, GSE42127). Table S4. Characteristics of patients (n = 76) with lung adenocarcinomas of our Tohoku University Biobank for the qPCR validation study. Table S5. Primer list. Table S6. Correlation of mRNA expression between AXL and genes encoding co-stimulatory/inhibitory molecules as shown by Pearson’s r. (XLSX 68 kb)
Figure S1. A dose-dependent decrease in phosphorylation of Axl in PC9 cells by the Axl kinase inhibitor. Figure S2. Knockdown of Axl decreases mRNA expression of PD-L1, PD-L2 and CXCR6 in vitro. Figure S3. Immunoblots indicating decreases in phosphorylation of ERK1/2 and AKT by the Axl kinase inhibitor. Figure S4. A selective MEK1/2 inhibitor or an AKT inhibitor reduces PD-L1 mRNA expression in vitro. Figure S5. Diverse downstream pathways driven by Axl receptor tyrosine kinase in non-small-cell lung cancer. (DOCX 2506 kb)
Data Availability Statement
The microarray datasets analysed in the current study are available in the NCBI GEO repository [GSE42127 and GSE13213] or cBioPortal for Cancer Genomics (http://www.cbioportal.org). The datasets used and/or analysed during the current study are available from the corresponding author on a reasonable request.