Abstract
Pathogenic variants of the KCNJ13 gene are known to cause Leber congenital amaurosis (LCA16), an inherited pediatric blindness. KCNJ13 encodes the Kir7.1 subunit that acts as a tetrameric, inwardly rectifying potassium ion channel in the retinal pigment epithelium (RPE) to maintain ionic homeostasis and allow photoreceptors to encode visual information. We sought to determine whether genetic approaches might be effective in treating blindness arising from pathogenic variants in KCNJ13. We derived human induced pluripotent stem cell (hiPSC)-RPE cells from an individual carrying a homozygous c.158G>A (p.Trp53∗) pathogenic variant of KCNJ13. We performed biochemical and electrophysiology assays to confirm Kir7.1 function. We tested both small-molecule readthrough drug and gene-therapy approaches for this “disease-in-a-dish” approach. We found that the LCA16 hiPSC-RPE cells had normal morphology but did not express a functional Kir7.1 channel and were unable to demonstrate normal physiology. After readthrough drug treatment, the LCA16 hiPSC cells were hyperpolarized by 30 mV, and the Kir7.1 current was restored. Similarly, we rescued Kir7.1 channel function after lentiviral gene delivery to the hiPSC-RPE cells. In both approaches, Kir7.1 was expressed normally, and there was restoration of membrane potential and the Kir7.1 current. Loss-of-function variants of Kir7.1 are one cause of LCA. Using either readthrough therapy or gene augmentation, we rescued Kir7.1 channel function in iPSC-RPE cells derived from an affected individual. This supports the development of precision-medicine approaches for the treatment of clinical LCA16.
Keywords: ion-channels, induced pluripotent stem cells (iPSC), Leber congenital amaurosis (LCA), KCNJ13, Kir7.1, retinal pigment epithelium (RPE), gene-therapy, translational readthrough, loss-of-function, blindness, genetic disorders
Introduction
Pathogenic variants of at least 21 genes expressed in the outer retina, i.e., the photoreceptors and retinal pigment epithelium (RPE), cause a severe form of early-childhood inherited blindness known as Leber congenital amaurosis (LCA). Within the last decade, multiple autosomal-recessive pathogenic variants of KCNJ13 (MIM: 603208) (chromosomal location: 2q37.1) have been identified in individuals with the LCA phenotype (LCA16, MIM: 614186; the 16th gene shown to cause LCA).1, 2, 3 LCA16 pathogenic variants include c.158G>A (p.Trp53∗), c.359T>C (p.Ile120Thr), c.458C>T (p.Thr153Ile), c.496C>T (p.Arg166∗), and c.722T>C (p.Leu241Pro).1, 2, 4 In addition, the compound-heterozygous KCNJ13 pathogenic variants c.314 G>T (p.Ser105Ile) and c.655C>T (p.Gln219Ter) were associated with early-onset retinal dystrophy in one individual.3 An autosomal-dominant KCNJ13 pathogenic variant, c.484C>T (p.Arg162Trp), causes the early-onset blindness called snowflake vitreoretinal degeneration (SVD, MIM: 193230).5
KCNJ13 encodes the inwardly rectifying potassium channel Kir7.1, and it is expressed in several tissues.6, 7 In the retina, Kir7.1 is localized exclusively in the RPE apical processes, where it controls retinal function and health.8, 9 The major contribution of the RPE Kir7.1 channel is the compensation of K+ concentration changes by epithelial transport, as modeled by LaCour.10 Light activation of photoreceptors reduces the RPE extracellular K+ concentration from 5 mM to 2 mM. The conductance of the Kir7.1 channel increases when extracellular K+ decreases and vice versa.11 Thus, KCNJ13 loss-of-function variants directly impact K+ buffering in the tight subretinal space and thereby alter photoreceptor function. A non-functional Kir7.1 channel will cause a sustained RPE cell depolarization that leads to cell death, an effect that has been reported after Kcnj13 knockout or suppression in mice.9, 12 The role of the Kir7.1 channel in other organs remains to be elucidated, but to date, all variants show a single ophthalmologic phenotype of blindness.13, 14
This loss-of-function phenotype for KCNJ13, as with other channelopathies, is a convenient gene-therapy target compared to the dominant negative SVD variant that alters wild-type channel function (Pattnaik et al., 2010, ARVO, abstract). Technical challenges include controlling the level of expression and successfully translating, trafficking, and assembling the ion channel as a functional unit in the membrane. Likewise, nonsense codons that give rise to misincorporation of an amino acid during translational readthrough must somehow be overcome to allow for the synthesis of a full-length protein and complete functionality. To address these challenges, we adopted two precision-medicine approaches where we used induced pluripotent stem cell (iPSC)-RPE cells derived from an affected individual to model LCA16 and explore novel, fast-track approaches to variant-specific therapies.
Material and Methods
Ethical Guidance
This study was approved by the University of Wisconsin-Madison’s institutional review board in accordance with federal regulations, state laws, and local and University policies.
Differentiation of hiPSC-RPE Cells
Fibroblasts from two subjects were reprogrammed to induced pluripotent stem cells and cultured using established methods.15, 16, 17 The proband carried the p.Trp53∗ homozygous variant in KCNJ13, and a non-symptomatic family member was heterozygous for this variant. Both the wild-type human induced pluripotent stem cells (hiPSCs) and human fetal RPE (hfRPE) cells we used as controls in our experiment have been previously characterized in our laboratory.17, 18
The hiPSC lines were differentiated to RPE via protocols described earlier.16, 17, 19, 20 In brief, hiPSCs were cultured either on mouse embryonic fibroblasts (MEFs) in iPS cell medium (Dulbecco’s modified Eagle’s medium [DMEM]: F12 (1:1), 20% KnockOut Serum, 1% minimal essential medium [MEM] non-essential amino acids, 1% GlutaMAX, β-mercaptoethanol, 20 ng/mL fibroblast growth factor 2 [FGF-2]), or on Matrigel with mTeSR1 medium. Cells were lifted enzymatically and grown as embryoid bodies (EBs) in iPS medium without FGF-2, and at day 4 changed to neural induction medium (NIM; DMEM:F12; 1% N2 supplement, 1% MEM non-essential amino acids, 1% L-glutamine, 2 μg/mL heparin), or grown in mTeSR1 and gradually transitioned to NIM by day 4. There were no differences observed in RPE differentiation between these two approaches. At day 7, free-floating EBs were plated on laminin-coated culture plates so that they could continue to differentiate as adherent cultures. At day 16, the 3D neural structures were removed, and the medium was switched to retinal differentiation medium (DMEM/F12 [3:1], 2% B27 supplement (without retinoic acid), 1% Antibiotic-Antimycotic). Remaining adhered cells were allowed to continue differentiation for an additional 45 days, followed by microdissection and passaging of pigmented RPE patches so that purified monolayers of RPE could be obtained, as described earlier.20 MEFs, Matrigel, and FGF-2 were purchased from WiCell, and all other tissue culture reagents were purchased from ThermoFisher. Gbanding karyotyping was performed via chromosome analysis by WiCell in accordance with general principles developed by the International Stem Cell Banking Initiative.
RT-PCR and Restriction Fragment-Length Polymorphism
Total RNA was isolated from mature hiPSC-RPE cells from both the affected individual and the carrier; wild-type hiPSC-RPE cells; and hfRPE cells with the RNeasy kit according to the manufacturer’s instructions (QIAGEN). The quality and the concentration of the isolated RNA was measured with a Nanodrop (ThermoFisher), and 200 ng of RNA was used for cDNA synthesis with the Superscript III first-strand cDNA synthesis kit according to the manufacturer’s instructions (ThermoFisher). PCR was performed with MyTaqHS master mix (Bioline) in a final volume of 25 μL under the following conditions: one cycle of initial denaturation at 95° C for 5 min followed by 35 cycles of denaturation at 95° C for 15 s, annealing at 55° C for 30 s, and extension at 72° C for 30 s. A final extension step was done for 10 min at 72° C, and amplification products were visualized by electrophoresis on a 2% agarose gel containing Midori green advanced stain (Nippon Genetics Europe). For the RFLP assay, PCR was performed as described with primers specific to the full-length KCNJ13 mRNA (forward 5′- GCTTCGAATTCCGACAGCAGTAATTG-3′ and reverse 5′-ATCCGGTGGATCCTTATTCTGTCAGT-3′). The PCR products were digested with NheI restriction enzyme (ThermoFisher) and visualized via electrophoresis on a 2% agarose gel containing Midori green advanced stain (Nippon Genetics Europe).
Transmission Electron Microscopy
Monolayers of hiPSC-RPE cells on transwell inserts (Corning, cat# 3470) were fixed with a solution of 2.5% glutaraldehyde and 2.0% paraformaldehyde in 0.1 M sodium phosphate buffer (PB) (pH 7.4), for ∼1 hr at room temperature (RT). Samples were rinsed five times for 5 min each in 0.1 M PB. The rinsed cultures were post-fixed in 1% osmium tetroxide (OsO4), 1% potassium ferrocyanide in PB for 1 hr at RT. After post-fixation, samples were rinsed in PB as before, followed by three 5 min rinses in distilled water so the phosphates would be cleared. The samples were stained en bloc in uranyl acetate for 2 hr at RT and dehydrated via an ethanol series. The membrane was cut from the transwell support, placed in an aluminum weighing dish, transitioned in propylene oxide (PO), and allowed to polymerize in fresh PilyBed 812 (Polysciences). Ultrathin sections were prepared from these polymerized samples and processed before images were captured and documented with an FEI CM120 transmission electron microscope mounted with an AMT BioSprint12 (Advanced Microscopy Techniques) digital camera. All electron microscope images were subjected to morphometry analysis of the number of mitochondria and the length of apical processes. We used a 10 μm2 area of the image and manually counted the number of mitochondria by using a blinded approach. We used the ImageJ program to define the length of each apical process. All data from at least seven different images were averaged in Excel (Microsoft).
Immunocytochemistry
Transwell inserts with a monolayer of hiPSC-RPE cells from either the affected individual or the control were fixed as follows: the transwell membrane was cut out and fixed via immersion in 4% paraformaldehyde in phosphate-buffered saline for 10 min in the dark. The membrane with cells was washed with chilled PBS twice and blocked for 2 hr in blocking solution that contained 5% goat serum and 0.25% Tween-20 in 1× PBS. For confocal microscopy, the cells were incubated for 24–48 hr with primary antibodies raised against Kir7.1 (mouse monoclonal IgG, 1:250; Santa Cruz Biotechnology), and ZO-1 (rabbit polyclonal, 2.5 μg/mL; ThermoFisher) prepared in incubation solution (blocking solution diluted in 1:3 with 1× PBS). After incubation with primary antibody, the membranes were washed thrice with chilled 1× PBS and incubated with conjugated secondary antibodies (donkey anti-goat Alexa Fluor 488, donkey anti-rabbit Alexa Fluor 594, and DAPI, 1:500) in incubation solution for 1 hr in the dark. A no-primary-antibody control was included for all experiments. Immunostained samples were imaged on a Nikon C2 confocal microscope (Nikon Instruments).
Immunoblotting Protein was isolated from >60-day-old hiPSC-RPE cells on transwells with radioimmunoprecipitation assay (RIPA) lysis buffer (ThermoFisher) along with sonication, and it was processed as described previously.1 The primary antibodies used for immunoblotting were anti-Kir7.1 (mouse monoclonal, 1:1000; Santa Cruz Biotechnology), anti-Bestrophin1 (mouse monoclonal, 1:1000; Novus Biologicals), anti-RPE65 (mouse monoclonal, 1:1000; ThermoFisher), and anti-GFP (mouse monoclonal, 1:1000; NeuroMab); both anti-GAPDH (rabbit monoclonal, 1:1000) and anti-β-actin (rabbit monoclonal, 1:1000; Cell Signaling Technology) were used as loading controls. Blots were imaged in the Odyssey Imaging System.
Phagocytosis Assay
The labeled photoreceptor outer segments (POSs) were prepared as previously described and fed to transwell-grown hiPSC-RPE cells that had a transepithelial electrical resistance (TEER) of >150 Ωcm2.15, 21 The cells were fed POSs for either 4 hr or 24 hr, after which cells were washed three times with DMEM so that any POSs that had not been phagocytosed would be removed. The cells were incubated for 24 hr or 6 days, respectively, before imaging took place. The images were captured and analyzed with NIS-Elements on a Nikon C2 confocal microscope (Nikon Instruments).
hiPSC-RPE Cell Transduction
Lentivirus custom engineered to be devoid of pathogenic elements was generated by Cyagen Biosciences and used for transduction; this lentivirus carried KCNJ13, fused at the N terminus with GFP, under the control of the EF1a promoter.22 The LCA-16 hiPSC-RPE cells in monolayer culture were infected with pLV-EF1α Kir7.1- GFP at an MOI of 200. The cells were cultured for 4–5 days after infection, then used for immunocytochemistry and immunoblotting (as described above).
Electrophysiology
Standard whole-cell patch clamp on hiPSC-RPE cells, transduced hiPSC-RPE cells, or NB84-treated hiPSC-RPE cells, all of which had distinct apical processes, was performed as described elsewhere.1 Data acquisition and the holding-potential parameters were controlled with the Axopatch 200-B, Digidata 1550, and Clampex software (Axon Instruments). During patch clamping, Ringer’s solution was continuously perfused as an external solution, and all drugs were suspended through this solution.
Statistical Analysis
The statistical analysis was performed in Origin (version 9.1) with a two-tailed Student’s t test so that the significant differences could be assessed. p < 0.05 was considered statistically significant. An ANOVA and a Tukey’s post-hoc test were also used for multiple comparisons. The data are expressed as the means ± SEM.
Results
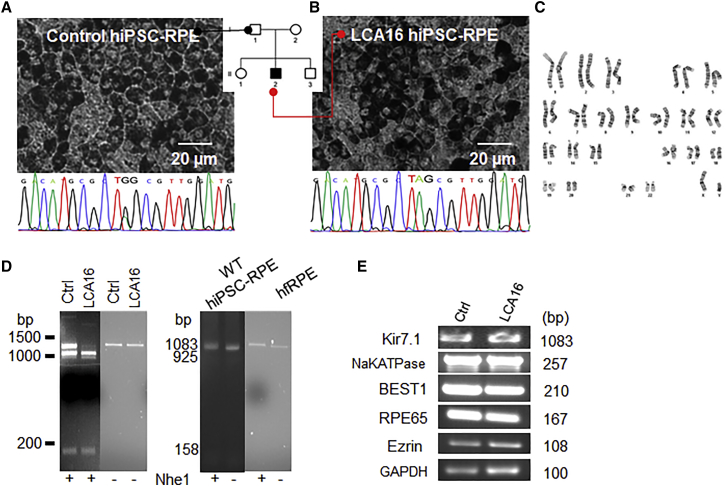
An analysis of hiPSC-RPE cells derived from an unaffected family member (herein referred to as control hiPSC-RPE cells) and an affected individual (herein referred to as LCA16 hiPSC-RPE cells) showed that these cells, had normal RPE morphology, including a cobblestone appearance and pigmentation (Figures 1A and 1B, respectively), and were identical to homozygous wild-type hiPSC cells (Figure S1A). DNA sequencing confirmed that the control hiPSC-RPE cells were from a heterozygous carrier, although the LCA16 pathogenic variant cells were apparently homozygous for the variant c.158G>A. The LCA16 hiPSC cells had a normal karyotype (Figure 1C), confirming that there was no gross chromosomal abnormality that might affect cellular behavior or response to therapy. This assay would not detect chromosomal abnormality below the level of resolution of the karyotype. The variant (c.158G>A) introduced a restriction site for Nhe1, enabling the mutant sequence to be identified in hiPSC-RPE cells specific to the affected individual, further verifying the presence of an apparently homozygous variant (Figure 1D). The control hiPSC-RPE cells were similarly analyzed and were found to be carrying an undigested 1,083 bp band and two digested bands, consistent with the genotype of the donor (Figure 1D). A single full-length transcript band of 1,083 bp was found in both wild-type, unrelated hiPSC-RPE cells, and commercially available hfRPE cells (Figure 1D, right panel). Because the presence of KCNJ13 transcript was comparable to that in both carrier and wild-type hiPSC-RPE cells (Figures S1C and S1D), we believe there was no nonsense-mediated decay in LCA16 hiPSC-RPE cells. There was no difference in the expression of RPE-specific genes between the two cell types (Figure 1E). Thus, the hiPSC-RPE cells derived from the affected individual conformed to the genotype of inherited retinal dystrophy and were a suitable source for establishing a disease-specific cellular model.1
Figure 1.
hiPSC-RPE Cells Derived from an Affected Individual Show the LCA16 Phenotype
(A and B) Bright-field images of a mature hiPSC-RPE cell monolayer derived from (A) an unaffected family member showing a heterozygous sequence, and (B) an LCA16-affected proband with the TAG sequence. A pedigree chart shows sources of hiPSCs.
(C) Normal karyotype in the affected individual’s hiPSC line showing no chromosomal abnormality.
(D) Control, LCA16, and wild-type hiPSC-RPE cells and hfRPE cells showing Nhe1 digestion product (lanes marked +Nhe1) and undigested (lanes marked −Nhe1). The full-length Kir7.1 sequence is 1,083 base pairs (bp) in length, and the digested products are 925 and 158 bp in length. Note that the PCR product for both wild-type hiPSC-RPE cells and hfRPE cells (+Nhe1 lanes) runs at a slightly higher molecular weight in the presence of Nhe1.
(E) RPE-specific gene expression in hiPSC-RPE cells.
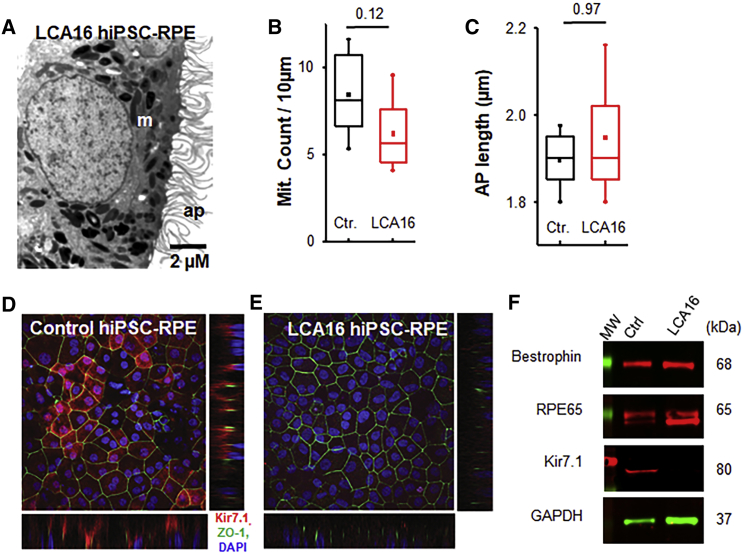
Kir7.1 channels are located within the highly specialized apical membrane processes of the RPE.8, 9 We therefore examined the intactness of apical membrane structures through electron microscope image analysis. The hiPSC-RPE cells had a polarized structure, including intact basal membrane infoldings and elongated apical processes that measured 1.49 ± 0.05 μm in length in the control hiPSC-RPE cells and 1.50 ± 0.14 μm in length in the proband’s hiPSC-RPE cells (p = 0.96, n = 7) (Figures 2A, 2C, and S2). The distribution and number of mitochondria in the two cell lines appeared normal, averaging 8.4 ± 1.0 and 6.2 ± 0.8 in the control and LCA16 proband’s hiPSC-RPE cells, respectively (p = 0.12, n = 6) (Figures 2A and 2B). Pigment granules correctly distributed apically in both cell types. Kir7.1 protein expression was detected on the apical membrane of mature control hiPSC-RPE cells, but not in LCA16 hiPSC-RPE cells (Figures 2D and 2E). Immunoblot analysis aimed at detecting the C-terminal end of the Kir7.1 protein revealed that, in the LCA16 disease model, there is a complete lack of Kir7.1, whereas other RPE proteins showed equal expression between control hiPSC-RPE and LCA16 hiPSC-RPE cells (Figure 2F). The c.158G>A (p.Trp53∗) variant is located within the second exon of the three-exon KCNJ13 sequence. Our previous work demonstrates that this variant generates a truncated protein product;1 thus, LCA16 hiPSC-RPE cells are expected to lack Kir7.1 channel function.
Figure 2.
Impaired Kir7.1 Protein Expression in LCA16 hiPSC-RPE Cells
(A) Electron micrograph of a representative LCA16 iPSC-RPE cell.
(B) Comparison of average mitochondria (Mit.) count within 10 μm2 of the cell.
(C) Evaluation of average length of RPE apical processes (AP).
(D) Immunofluorescence localization of Kir7.1 (red), ZO-1 (green), and DAPI (blue) in control hiPSC-RPE cells. Both the lower and side panels reveal a polarized distribution of Kir7.1 in reference to ZO-1 and DAPI (confocal z stack images).
(E) Localization of Kir7.1 (red), ZO-1 (green), and DAPI (blue) in LCA16 hiPSC-RPE cells.
(F) Immunoblot showing the expression of RPE-cell-specific proteins in both tissue samples. Using a C-terminal-specific antibody against Kir7.1, we detected Kir7.1 protein in whole-cell lysates from the control hiPSC-RPE cells but not in those from the LCA16 hiPSC-RPE cells.
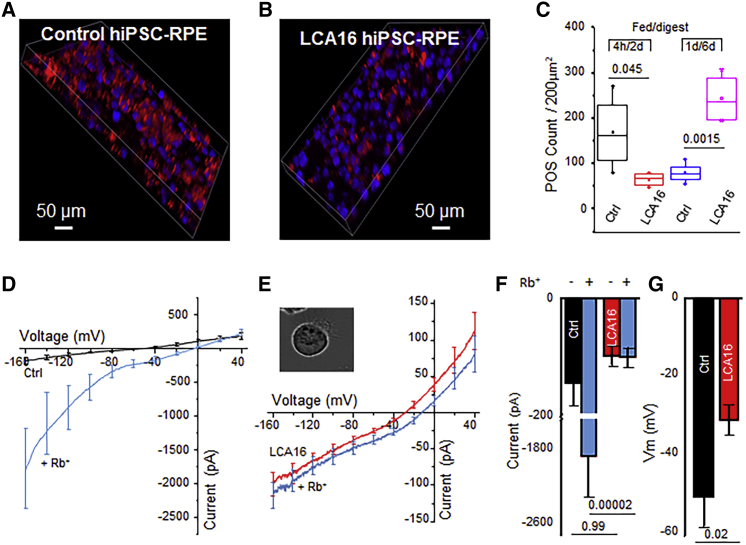
One of the key physiological functions of RPE cells is the daily phagocytosis of the POSs, a process which contributes to POS renewal. To test whether the absence of normal Kir7.1 protein alters phagocytosis, we fed control and LCA16 hiPSC-RPE cell cultures with fluorescently labeled POSs. The cells were fed labeled POSs for 4 hr, and phagosome digestion by RPE cells was allowed for an additional 24 hr. Control hiPSC-RPE cells showed a higher rate of phagosomal uptake than LCA16 hiPSC-RPE cells (169 ± 40 versus 66.5 ± 7.4, p = 0.04, n = 4) (Figures 3A–3C). In addition, when cells were fed with POSs for 1 day and then allowed to process phagosomes for 6 days, the LCA16 hiPSC-RPE cells failed to digest POSs (80.2 ± 11.1 versus 244.2 ± 27.6 counts within a 200 μm2 field, p = 0.001, n = 4) (Figure S2). This finding suggests that the fundus pigmentation observed in LCA16-affected individuals most likely results from an inability to normally phagocytose POSs, which therefore accumulate over time in the retinas of affected individuals.
Figure 3.
LCA16 hiPSC-RPE Cells Exhibit Aberrant Physiology
(A and B) Phagosome (red) localization within (A) control hiPSC-RPE cell and (B) LCA16 iPSC-RPE cell samples.
(C) Plot of the average phagosome count within a fixed 200 μm2 area in the control cells and LCA16 hiPSC-RPE cells after 4 hr of feeding and a subsequent 48 hr digestion period or after 1 day of feeding followed by 6 days of digestion.
(D) Plot of the average current-voltage (I/V) curve for Kir7.1 currents in the presence of normal external K+ (black) or high external Rb+ (light blue) in control hiPSC-RPE cells.
(E) An average I/V curve in the presence of normal K+ (red) or high Rb+ (light blue) in LCA16 hiPSC-RPE cells.
(F) Average plot of an inward current amplitude measured at −150 mV. Color representation as shown in (D) and (E).
(G) Comparison of the average membrane potential of the control (black) cells to depolarized LCA16 (red) hiPSC-RPE cells.
By using differentiated hiPSC-RPE cells, taken from an asymptomatic carrier family member, that develop apical processes and localize Kir7.1 to the apical membrane, we were able to detect a small, but measurable Kir7.1 current (−144.2 ± 51 pA) in control hiPSC-RPE cells. Normal function was confirmed by a 13-fold increase in Rb+ permeability (−1887.3 ± 589.6 pA, n = 5) (see cells in Figures 3D and 3F and compare to results for wild-type hiPSC cells in Figure S1B), which is a specific property of the Kir7.1 channel.1, 23 However, in LCA16 hiPSC-RPE cells, we did not detect any change in the current amplitude mediated by Rb+ conductance (−98.1 ± 15.7 pA and −99.6 ± 15.7 pA, n = 9) (Figures 3E and 3F. A direct comparison of both current amplitude (p = 0.0006 with Rb+) and cell membrane potential (−50.0 ± 5.1 versus −30.6 ± 3.7 mV, p = 0.0005; as shown in Figure 3G) supports our hypothesis that the cause of blindness is a nonfunctional Kir7.1 channel that depolarizes LCA16 hiPSC-RPE cells. This hypothesis is also consistent with observations in an in vivo Kir7.1-deficient mouse model or an in vitro cell line exogenously expressing the human Trp53∗ Kir7.1 channel.1, 9
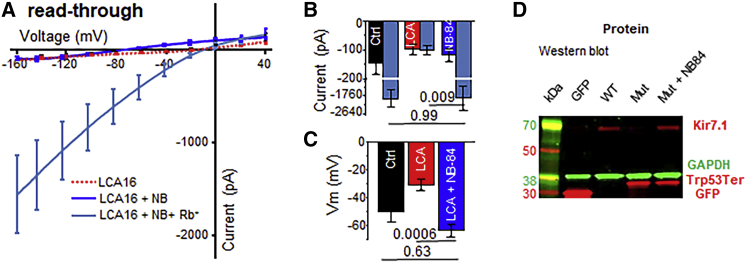
We next assessed the functional consequences of the NB84-mediated readthrough on the Kir7.1 current in LCA16 hiPSC-RPE cells. After we treated the LCA16 hiPSC-RPE cells with 500 μM NB84, we obtained a measurable current of −94.3 ± 24.0 pA; the current was enhanced 10-fold upon the introduction of the permeant ion Rb+ (−1,562.7 ± 546.7 pA, p = 0.005, n = 8; Figures 4A and 4B). A significant recovery of membrane potential from −30.6 ± 3.7 mV in the untreated cells to –56.3 ± 3.6 mV (p = 0.0001, n = 10; Figure 4C) in the NB84-treated cells further supports the potential of readthrough drug therapy for restoring channel function. In a subgroup of cells, we noticed rescue in membrane potential without any significant change in current amplitude. One possible explanation for this finding is the heterogeneity in the incorporation of the near-cognate amino acid during Kir7.1 translation (Figure S3).
Figure 4.
Putative Kir7.1 Loss-of-Function Rescue Through Nonsense Suppression
(A–C) (A) Average I/V relationship before (red) and after (dark blue) treatment with NB84. The Rb+ current measured is shown as a light-blue trace. Evaluation of the average inward current measured at −150 mV (B) and of the membrane potential (C) to demonstrate the effect of NB84.
(D) Immunoblot showing detection of eGFP fusion proteins, via anti-GFP antibody, in cell lysates from CHO cells transfected with empty vector (GFP), wild-type Kir7.1 coding sequence (WT), or Trp53∗ coding sequence (Mut). A partial restoration of the full-length protein product was observed in the latter after NB84 treatment (Mut + NB84).
Amounts of endogenous Kir7.1 in the NB84-treated hiPSC-RPE cells were below the detection level of immunoblots, so we transfected Chinese hamster ovary (CHO) cells with a mutant Kir7.1-eGFP fusion construct to detect formation of the full-length protein. After treating these cells with NB84, we detected full-length protein in addition to truncated protein on an immunoblot, and both membrane potential and current deficits were corrected (Figures 4D and S4), demonstrating that NB84 potentiates the specific readthrough of the recessive Trp53∗ variant.
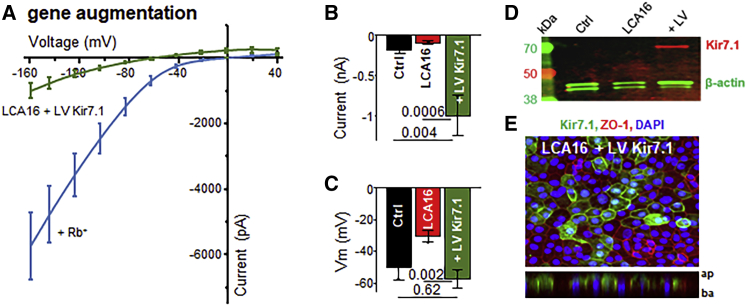
We designed a lentiviral vector with an EF1a-promoter-driven, N-terminal-GFP-fused human Kir7.1 open-reading frame;24 we subsequently used this vector to infect hiPSC-RPE monolayers. Intriguingly, wild-type Kir7.1-expressing cells produced normal Kir7.1 currents or even slightly higher amplitudes than those observed in the control cells (−921 ± 223 pA, p = 0.001, n = 8). This current was further potentiated by the introduction of Rb+ (−5,453 ± 929 pA), as expected for a normally functioning Kir7.1 channel (Figures 5A and 5B). In addition to K+ currents, the membrane potential of LCA16 iPSC-RPE cells was approaching normal values (−57.5 ± 5.4 mV, p = 0.0008) (Figure 5C). Moreover, newly expressed full-length Kir7.1 (Figure 5D) was localized to the apical membranes of affected-individual-specific hiPSC-RPE cells (Figure 5E and Video S1).
Figure 5.
hiPSC-RPE Cells Regain Kir7.1 Function after Gene Augmentation
(A–C) Plot of the average I/V curve for Kir7.1 currents measured in LCA16 hiPSC-RPE GFP-positive cells expressing a normal copy of the human Kir7.1 clone (A). Both K+ (green) and Rb+ (light blue) traces are shown. Average plots of the (B) current amplitudes measured at −150 mV and (C) membrane potential show rescue after gene augmentation.
(D) Immunoblot analysis of lysate from control hiPSC-RPE cells (Ctrl), LCA16 hiPSC-RPE cells (LCA16), or LCA16 hiPSC-RPE cells infected with lentivirus coding for the eGFP-WT Kir7.1 fusion protein (LV) via anti-GFP antibody.
(E) Cultured LCA16 hiPSC-RPE cells showing wild-type Kir7.1 (green), ZO-1 (red), and DAPI (blue) proteins. Z stack planes are shown in the lower panel.
Discussion
We have reported previously that targeted inhibition of Kir7.1 in the mouse retina (induced using either small interfering RNA (siRNA) or a pharmacological blocker) gave rise to an altered electroretinogram phenotype consistent with that observed in LCA16- affected individuals.9 In the current report, we extend our findings by describing the development of a novel LCA16 disease-in-a-dish model. We developed a robust hiPSC-RPE cell culture derived from skin biopsies from one LCA16 proband carrying an apparently homozygous nonsense variant (p.Trp53∗) in exon 2 of KCNJ13 and developed a control cell culture from an unaffected family member.15 The highly pigmented RPE cells from both the control and the affected individual appeared normal, expressed RPE markers, and were immunopositive for several RPE-specific proteins. As intended, the LCA16 hiPSC-RPE cells lacked mutation-specific Kir7.1 expression. Our rescue of Kir7.1 function through the use of single-nonsense-variant hiPSC-RPE cells is a proof of concept that can be translated to other similar loss-of-function KCNJ13 variants.
Our study determined that the major pathogenic alteration affecting the RPE cells is the lack of a functional Kir7.1 channel. We were also able to establish a genotype-phenotype (normal) correlation in an asymptomatic carrier individual. Our use of carrier (control) hiPSC-RPE cells from within the family also accounts for modifier gene functions, epigenetic changes, and environmental contributions. We have previously determined that cultured hfRPE cells produce an Rb+-sensitive Kir7.1 current similar to that recorded from wild-type and carrier hiPSC-RPE cells.18 We also found that heterozygous expression of equal amounts of wild-type and non-functional p.Trp53∗ mutant protein did not influence the cellular physiology outcome.1 Given these findings, we reasoned that carrier hiPSC-RPE cells are a suitable control for comparing test results from both gene restoration and translational readthrough.
Our hiPSC-RPE disease model is particularly valuable given that a Kcnj13 knockout in mice causes embryonic lethality, suggesting that Kcnj13 in mice or animal models has a different function that complicates the experimental study of disease pathobiology.12, 25, 26 We showed that hiPSC-RPE cells from an affected individual had normal apical processes where Kir7.1 is expressed. We thus confirmed that these cultured hiPSC-RPE cells were mature, unlike the ones generated for previous studies of ciliopathy.27 Compared to control hiPSC-RPE cells, mature LCA16 hiPSC-RPE cells did not show Kir7.1 function, their membrane potential was depolarized, and they were unable to phagocytose POSs. A defect in the phagocytosis of shed POSs by LCA16 hiPSC-RPE cells is thus secondary, and it is consistent with the slow progression toward blindness in LCA16-affected individuals; such a progression has been documented through clinical manifestations, including electroretinogram abnormalities and retinal pigmentation.1, 2, 3, 4 Previously, it was challenging to study ion channels in iPSCs because of the lack or low level of expression. However, our study demonstrates that stem cell technology is a powerful tool for overcoming those barriers and analyzing the pathophysiology of LCA16.28
Nonsense variants, such as KCNJ13 c.158G>A (p.Trp53∗), account for about 11% of inherited genetic disorders.29 Several studies have suggested the potential usefulness of readthrough therapy as an intervention that could improve vision.30, 31, 32 Designer aminoglycosides such as NB84 bind to the eukaryotic ribosomal-RNA decoding site.33 This binding promotes the transition of the 16S rRNA decoding center from a tRNA binding conformation to a productive state, resulting in an increase in the rate of readthrough at a stop codon.34 Here we show that a KCNJ13 p.Trp53∗ nonsense variant is suppressed by the incorporation of a near-cognate aminoacyl tRNA in the presence of the small-molecule readthrough designer aminoglycoside NB84.32, 35, 36 Both in vitro and in vivo toxicity studies demonstrate that NB84 is ready for translational therapy in animal models and human subjects.37 We foresee further development of a non-toxic formulation of NB84, perhaps through the use of poly-L-aspartate to increase cytoplasmic concentrations and allow for a lower dosage.38 Formulations that allow topical application to the eye could avoid risks to other organs. As expected, only a fraction of cells showed complete recovery of both membrane potential and current amplitude, and another subgroup demonstrated only partial recovery of function. Readthrough therapy also has a significant clinical advantage in the management of LCA16 diagnosed at an early stage.
The particular variant studied here, and other recessive variants that cause blindness, are potential targets for gene therapy. This premise is nearing reality, given the recent FDA approval of a gene-therapy treatment for blindness.39, 40, 41 Using a LCA16 hiPSC-RPE model, we demonstrate that the rescue of channelopathy via lentiviral gene augmentation produced a potassium current and normalized membrane potential (Figure S5). Newly expressed protein was also localized to apical aspects of LCA16 hiPSC RPE cells.
Gene therapy is a simple concept, but the problem remains that both too little and too much expression can have deleterious effects.42, 43 To characterize the optimum dosage required, we expressed copies of both wild-type and mutant protein in a heterologous expression system. We observed that as little as 25% of wild-type protein expression of total Kir7.1 protein product was sufficient to enhance both membrane potential and the potassium current, and it rescued the mutant phenotype (Figure S6). A splice variant of Kir7.1 was reported in human RPE cells that failed to produce the anticipated small protein band.44 Because the expected truncated protein consisted of the N terminus and the first transmembrane domain, the LCA16 nonsense variant p.Trp53∗ is predicted to truncate all Kir7.1 transcript variants. Our results show that the p.Trp53∗ truncated protein product does not assemble with wild-type protein to alter Kir7.1 channel function. Detection of a truncated protein product after heterologous expression also suggests it might not undergo nonsense-mediated decay.1
In this study, we have taken advantage of lentivirus transduction to mediate long-lasting transgene expression in RPE cells in vivo.45 Also, we were unable to transduce hiPSC-RPE cells with adeno-associated virus (AAV), a difficulty reported by several other groups. This might be because of the higher efficacy of the lentiviral vector in transducing post-mitotic RPE cells, as shown for non-dividing cells.46 Our third-generation lentivirus packaging system also has a reduced risk of generating replication-competent virus. In a preclinical study carried out in non-human primates, the vector stayed in the eye after subretinal delivery of a lentiviral RPE65 gene, perhaps because of the tight blood-retina barrier. These results suggested high systemic safety for this approach.47 Retinal gene therapy via lentiviral vector is in clinical trials for neovascular age-related macular degeneration, Stargardt disease, and Usher syndrome type 1B.48 We conclude that both treatments developed in this study suggest that 25% of normal protein expression within a population of RPE cells restores membrane potential with variable recovery in current amplitude. Achieving functional rescue in vivo could thus be less challenging, and residual mutant protein product is not expected to negatively influence functional outcomes.
Advances in genetic screening and improvements in access to medical care will undoubtedly improve our understanding of the array of disorders caused by channelopathies and expand our understanding of the role that KCNJ13 plays in health and disease. Genome editing holds promise for genetic disorders, but gene correction in non-dividing cells and the introduction of unwanted genomic variants at on- and off-target sites limit its potential use. Overall, we show a promising proof of concept for readthrough and gene augmentation therapies and demonstrate the potential of using a precision-medicine approach to ameliorate pediatric blindness.
Declaration of Interests
Both P.K.S. and B.R.P. are on a patent application discussing therapeutics developed in this paper. D.M.G. has an ownership interest in Opsis Therapeutics LLC. All other authors declare no competing interests.
Acknowledgments
We sincerely thank the affected individual and family members for their willingness to participate and donate biopsy skin samples and time. We also thank an anonymous donor for support of LCA16 research through the University of Wisconsin Foundation. NB84 was kindly provided by Ellen Welch. This work was also supported by the National Institutes of Health (NIH) (EY024995 awarded to B.R.P.), University of Wisconsin (UW) Vision Core grant P30 EY01665, and the Retina Research Foundation M. D. Mathews Professorship (B.R.P.) at the UW School of Medicine and Public Health and the UW Department of Pediatrics. We also thank Laura Hogan for her editorial assistance, supported through the NIH Clinical and Translational Science Award (CTSA) program (NCRR-1UL1RR025011 and NCATS-9U54TR000021). D.M.G. is supported by the McPherson Eye Research Institute Sandra Lemke Trout Chair in Eye Research and the Retina Research Foundation Emmett Humble Distinguished Directorship. We thank Kris Saha for suggestions on this manuscript and Randall J. Massey for help with electron microscopy. An unrestricted grant from Research to Prevent Blindness to the UW Madison Department of Ophthalmology and Visual Sciences also supported this work. The views of this report are primarily of the authors and were not influenced by the funding agencies. The members of the Pattnaik Lab are also recognized for their input.
Published: January 24, 2019
Footnotes
Supplemental Data include six figures and one video and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.12.019.
Supplemental Data
References
- 1.Pattnaik B.R., Shahi P.K., Marino M.J., Liu X., York N., Brar S., Chiang J., Pillers D.A., Traboulsi E.I. A novel KCNJ13 nonsense mutation and loss of Kir7.1 channel function causes Leber congenital amaurosis (LCA16) Hum. Mutat. 2015;36:720–727. doi: 10.1002/humu.22807. [DOI] [PubMed] [Google Scholar]
- 2.Sergouniotis P.I., Davidson A.E., Mackay D.S., Li Z., Yang X., Plagnol V., Moore A.T., Webster A.R. Recessive mutations in KCNJ13, encoding an inwardly rectifying potassium channel subunit, cause leber congenital amaurosis. Am. J. Hum. Genet. 2011;89:183–190. doi: 10.1016/j.ajhg.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perez-Roustit S., Marquette V., Bocquet B., Kaplan J., Perrault I., Meunier I., Hamel C.P. Leber congenital amaurosis with large retinal pigment clumps caused by compound heterozygous mutations in Kcnj13. Retin. Cases Brief Rep. 2017;11:221–226. doi: 10.1097/ICB.0000000000000326. [DOI] [PubMed] [Google Scholar]
- 4.Khan A.O., Bergmann C., Neuhaus C., Bolz H.J. A distinct vitreo-retinal dystrophy with early-onset cataract from recessive KCNJ13 mutations. Ophthalmic Genet. 2015;36:79–84. doi: 10.3109/13816810.2014.985846. [DOI] [PubMed] [Google Scholar]
- 5.Hejtmancik J.F., Jiao X., Li A., Sergeev Y.V., Ding X., Sharma A.K., Chan C.C., Medina I., Edwards A.O. Mutations in KCNJ13 cause autosomal-dominant snowflake vitreoretinal degeneration. Am. J. Hum. Genet. 2008;82:174–180. doi: 10.1016/j.ajhg.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krapivinsky G., Medina I., Eng L., Krapivinsky L., Yang Y., Clapham D.E. A novel inward rectifier K+ channel with unique pore properties. Neuron. 1998;20:995–1005. doi: 10.1016/s0896-6273(00)80480-8. [DOI] [PubMed] [Google Scholar]
- 7.Derst C., Döring F., Preisig-Müller R., Daut J., Karschin A., Jeck N., Weber S., Engel H., Grzeschik K.H. Partial gene structure and assignment to chromosome 2q37 of the human inwardly rectifying K+ channel (Kir7.1) gene (KCNJ13) Genomics. 1998;54:560–563. doi: 10.1006/geno.1998.5598. [DOI] [PubMed] [Google Scholar]
- 8.Yang D., Pan A., Swaminathan A., Kumar G., Hughes B.A. Expression and localization of the inwardly rectifying potassium channel Kir7.1 in native bovine retinal pigment epithelium. Invest. Ophthalmol. Vis. Sci. 2003;44:3178–3185. doi: 10.1167/iovs.02-1189. [DOI] [PubMed] [Google Scholar]
- 9.Shahi P.K., Liu X., Aul B., Moyer A., Pattnaik A., Denton J., Pillers D.M., Pattnaik B.R. Abnormal electroretinogram after Kir7.1 channel suppression suggests role in retinal electrophysiology. Sci. Rep. 2017;7:10651. doi: 10.1038/s41598-017-11034-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.la Cour M. The retinal pigment epithelium controls the potassium activity in the subretinal space. Acta Ophthalmol. Suppl. 1985;173:9–10. doi: 10.1111/j.1755-3768.1985.tb06827.x. [DOI] [PubMed] [Google Scholar]
- 11.Kumar M., Pattnaik B.R. Focus on Kir7.1: Physiology and channelopathy. Channels (Austin) 2014;8:488–495. doi: 10.4161/19336950.2014.959809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roman D., Zhong H., Yaklichkin S., Chen R., Mardon G. Conditional loss of Kcnj13 in the retinal pigment epithelium causes photoreceptor degeneration. Exp. Eye Res. 2018;176:219–226. doi: 10.1016/j.exer.2018.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCloskey C., Rada C., Bailey E., McCavera S., van den Berg H.A., Atia J., Rand D.A., Shmygol A., Chan Y.W., Quenby S. The inwardly rectifying K+ channel KIR7.1 controls uterine excitability throughout pregnancy. EMBO Mol. Med. 2014;6:1161–1174. doi: 10.15252/emmm.201403944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghamari-Langroudi M., Digby G.J., Sebag J.A., Millhauser G.L., Palomino R., Matthews R., Gillyard T., Panaro B.L., Tough I.R., Cox H.M. G-protein-independent coupling of MC4R to Kir7.1 in hypothalamic neurons. Nature. 2015;520:94–98. doi: 10.1038/nature14051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh R., Shen W., Kuai D., Martin J.M., Guo X., Smith M.A., Perez E.T., Phillips M.J., Simonett J.M., Wallace K.A. iPS cell modeling of Best disease: Insights into the pathophysiology of an inherited macular degeneration. Hum. Mol. Genet. 2013;22:593–607. doi: 10.1093/hmg/dds469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyer J.S., Shearer R.L., Capowski E.E., Wright L.S., Wallace K.A., McMillan E.L., Zhang S.C., Gamm D.M. Modeling early retinal development with human embryonic and induced pluripotent stem cells. Proc. Natl. Acad. Sci. USA. 2009;106:16698–16703. doi: 10.1073/pnas.0905245106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phillips M.J., Wallace K.A., Dickerson S.J., Miller M.J., Verhoeven A.D., Martin J.M., Wright L.S., Shen W., Capowski E.E., Percin E.F. Blood-derived human iPS cells generate optic vesicle-like structures with the capacity to form retinal laminae and develop synapses. Invest. Ophthalmol. Vis. Sci. 2012;53:2007–2019. doi: 10.1167/iovs.11-9313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Halbach P., Pillers D.A., York N., Asuma M.P., Chiu M.A., Luo W., Tokarz S., Bird I.M., Pattnaik B.R. Oxytocin expression and function in the posterior retina: A novel signaling pathway. Invest. Ophthalmol. Vis. Sci. 2015;56:751–760. doi: 10.1167/iovs.14-15646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Capowski E.E., Simonett J.M., Clark E.M., Wright L.S., Howden S.E., Wallace K.A., Petelinsek A.M., Pinilla I., Phillips M.J., Meyer J.S. Loss of MITF expression during human embryonic stem cell differentiation disrupts retinal pigment epithelium development and optic vesicle cell proliferation. Hum. Mol. Genet. 2014;23:6332–6344. doi: 10.1093/hmg/ddu351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh R., Phillips M.J., Kuai D., Meyer J., Martin J.M., Smith M.A., Perez E.T., Shen W., Wallace K.A., Capowski E.E. Functional analysis of serially expanded human iPS cell-derived RPE cultures. Invest. Ophthalmol. Vis. Sci. 2013;54:6767–6778. doi: 10.1167/iovs.13-11943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parinot C., Rieu Q., Chatagnon J., Finnemann S.C., Nandrot E.F. Large-scale purification of porcine or bovine photoreceptor outer segments for phagocytosis assays on retinal pigment epithelial cells. J. Vis. Exp. 2014;94 doi: 10.3791/52100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yáñez-Muñoz R.J., Balaggan K.S., MacNeil A., Howe S.J., Schmidt M., Smith A.J., Buch P., MacLaren R.E., Anderson P.N., Barker S.E. Effective gene therapy with nonintegrating lentiviral vectors. Nat. Med. 2006;12:348–353. doi: 10.1038/nm1365. [DOI] [PubMed] [Google Scholar]
- 23.Shimura M., Yuan Y., Chang J.T., Zhang S., Campochiaro P.A., Zack D.J., Hughes B.A. Expression and permeation properties of the K(+) channel Kir7.1 in the retinal pigment epithelium. J. Physiol. 2001;531:329–346. doi: 10.1111/j.1469-7793.2001.0329i.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.White M., Whittaker R., Gándara C., Stoll E.A. A guide to approaching regulatory considerations for lentiviral-mediated gene therapies. Hum. Gene Ther. Methods. 2017;28:163–176. doi: 10.1089/hgtb.2017.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhong H., Chen Y., Li Y., Chen R., Mardon G. CRISPR-engineered mosaicism rapidly reveals that loss of Kcnj13 function in mice mimics human disease phenotypes. Sci. Rep. 2015;5:8366. doi: 10.1038/srep08366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Villanueva S., Burgos J., López-Cayuqueo K.I., Lai K.M., Valenzuela D.M., Cid L.P., Sepúlveda F.V. Cleft palate, moderate lung developmental retardation and early postnatal lethality in mice deficient in the Kir7.1 inwardly rectifying K+ channel. PLoS ONE. 2015;10:e0139284. doi: 10.1371/journal.pone.0139284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.May-Simera H.L., Wan Q., Jha B.S., Hartford J., Khristov V., Dejene R., Chang J., Patnaik S., Lu Q., Banerjee P. Primary cilium-mediated retinal pigment epithelium maturation is disrupted in ciliopathy patient cells. Cell Rep. 2018;22:189–205. doi: 10.1016/j.celrep.2017.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goversen B., van der Heyden M.A.G., van Veen T.A.B., de Boer T.P. The immature electrophysiological phenotype of iPSC-CMs still hampers in vitro drug screening: Special focus on IK1. Pharmacol. Ther. 2018;183:127–136. doi: 10.1016/j.pharmthera.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 29.Mort M., Ivanov D., Cooper D.N., Chuzhanova N.A. A meta-analysis of nonsense mutations causing human genetic disease. Hum. Mutat. 2008;29:1037–1047. doi: 10.1002/humu.20763. [DOI] [PubMed] [Google Scholar]
- 30.Schwarz N., Carr A.J., Lane A., Moeller F., Chen L.L., Aguilà M., Nommiste B., Muthiah M.N., Kanuga N., Wolfrum U. Translational read-through of the RP2 Arg120stop mutation in patient iPSC-derived retinal pigment epithelium cells. Hum. Mol. Genet. 2015;24:972–986. doi: 10.1093/hmg/ddu509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goldmann T., Overlack N., Wolfrum U., Nagel-Wolfrum K. PTC124-mediated translational readthrough of a nonsense mutation causing Usher syndrome type 1C. Hum. Gene Ther. 2011;22:537–547. doi: 10.1089/hum.2010.067. [DOI] [PubMed] [Google Scholar]
- 32.Goldmann T., Overlack N., Möller F., Belakhov V., van Wyk M., Baasov T., Wolfrum U., Nagel-Wolfrum K. A comparative evaluation of NB30, NB54 and PTC124 in translational read-through efficacy for treatment of an USH1C nonsense mutation. EMBO Mol. Med. 2012;4:1186–1199. doi: 10.1002/emmm.201201438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fan-Minogue H., Bedwell D.M. Eukaryotic ribosomal RNA determinants of aminoglycoside resistance and their role in translational fidelity. RNA. 2008;14:148–157. doi: 10.1261/rna.805208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pape T., Wintermeyer W., Rodnina M.V. Conformational switch in the decoding region of 16S rRNA during aminoacyl-tRNA selection on the ribosome. Nat. Struct. Biol. 2000;7:104–107. doi: 10.1038/72364. [DOI] [PubMed] [Google Scholar]
- 35.Nudelman I., Glikin D., Smolkin B., Hainrichson M., Belakhov V., Baasov T. Repairing faulty genes by aminoglycosides: Development of new derivatives of geneticin (G418) with enhanced suppression of diseases-causing nonsense mutations. Bioorg. Med. Chem. 2010;18:3735–3746. doi: 10.1016/j.bmc.2010.03.060. [DOI] [PubMed] [Google Scholar]
- 36.Ramsden C.M., Nommiste B., R Lane A., Carr A.F., Powner M.B., J K Smart M., Chen L.L., Muthiah M.N., Webster A.R., Moore A.T. Rescue of the MERTK phagocytic defect in a human iPSC disease model using translational read-through inducing drugs. Sci. Rep. 2017;7:51. doi: 10.1038/s41598-017-00142-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shulman E., Belakhov V., Wei G., Kendall A., Meyron-Holtz E.G., Ben-Shachar D., Schacht J., Baasov T. Designer aminoglycosides that selectively inhibit cytoplasmic rather than mitochondrial ribosomes show decreased ototoxicity: A strategy for the treatment of genetic diseases. J. Biol. Chem. 2014;289:2318–2330. doi: 10.1074/jbc.M113.533588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Du M., Keeling K.M., Fan L., Liu X., Bedwell D.M. Poly-L-aspartic acid enhances and prolongs gentamicin-mediated suppression of the CFTR-G542X mutation in a cystic fibrosis mouse model. J. Biol. Chem. 2009;284:6885–6892. doi: 10.1074/jbc.M806728200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bennett J., Wellman J., Marshall K.A., McCague S., Ashtari M., DiStefano-Pappas J., Elci O.U., Chung D.C., Sun J., Wright J.F. Safety and durability of effect of contralateral-eye administration of AAV2 gene therapy in patients with childhood-onset blindness caused by RPE65 mutations: A follow-on phase 1 trial. Lancet. 2016;388:661–672. doi: 10.1016/S0140-6736(16)30371-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dalkara D., Goureau O., Marazova K., Sahel J.A. Let there be light: Gene and cell therapy for blindness. Hum. Gene Ther. 2016;27:134–147. doi: 10.1089/hum.2015.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bennett J. Taking stock of retinal gene therapy: Looking back and moving forward. Mol. Ther. 2017;25:1076–1094. doi: 10.1016/j.ymthe.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Porteus M.H., Connelly J.P., Pruett S.M. A look to future directions in gene therapy research for monogenic diseases. PLoS Genet. 2006;2:e133. doi: 10.1371/journal.pgen.0020133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guziewicz K.E., Cideciyan A.V., Beltran W.A., Komáromy A.M., Dufour V.L., Swider M., Iwabe S., Sumaroka A., Kendrick B.T., Ruthel G. BEST1 gene therapy corrects a diffuse retina-wide microdetachment modulated by light exposure. Proc. Natl. Acad. Sci. USA. 2018;115:E2839–E2848. doi: 10.1073/pnas.1720662115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang D., Swaminathan A., Zhang X., Hughes B.A. Expression of Kir7.1 and a novel Kir7.1 splice variant in native human retinal pigment epithelium. Exp. Eye Res. 2008;86:81–91. doi: 10.1016/j.exer.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kalesnykas G., Kokki E., Alasaarela L., Lesch H.P., Tuulos T., Kinnunen K., Uusitalo H., Airenne K., Yla-Herttuala S. Comparative study of adeno-associated virus, adenovirus, bacu lovirus and lentivirus vectors for gene therapy of the eyes. Curr. Gene Ther. 2017;17:235–247. doi: 10.2174/1566523217666171003170348. [DOI] [PubMed] [Google Scholar]
- 46.Naldini L., Blömer U., Gallay P., Ory D., Mulligan R., Gage F.H., Verma I.M., Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 47.Matet A., Kostic C., Bemelmans A.P., Moulin A., Rosolen S.G., Martin S., Mavilio F., Amirjanians V., Stieger K., Lorenz B. Evaluation of tolerance to lentiviral LV-RPE65 gene therapy vector after subretinal delivery in non-human primates. Transl Res. 2017;188:40–57.e4. doi: 10.1016/j.trsl.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 48.Auricchio A., Smith A.J., Ali R.R. The future looks brighter after 25 years of retinal gene therapy. Hum. Gene Ther. 2017;28:982–987. doi: 10.1089/hum.2017.164. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.