Abstract
Hypusine is formed post-translationally from lysine and is found in a single cellular protein, eukaryotic translation initiation factor-5A (eIF5A), and its homolog eIF5A2. Biosynthesis of hypusine is a two-step reaction involving the enzymes deoxyhypusine synthase (DHPS) and deoxyhypusine hydroxylase (DOHH). eIF5A is highly conserved throughout eukaryotic evolution and plays a role in mRNA translation, cellular proliferation, cellular differentiation, and inflammation. DHPS is also highly conserved and is essential for life, as Dhps-null mice are embryonic lethal. Using exome sequencing, we identified rare biallelic, recurrent, predicted likely pathogenic variants in DHPS segregating with disease in five affected individuals from four unrelated families. These individuals have similar neurodevelopmental features that include global developmental delay and seizures. Two of four affected females have short stature. All five affected individuals share a recurrent missense variant (c.518A>G [p.Asn173Ser]) in trans with a likely gene disrupting variant (c.1014+1G>A, c.912_917delTTACAT [p.Tyr305_Ile306del], or c.1A>G [p.Met1?]). cDNA studies demonstrated that the c.1014+1G>A variant causes aberrant splicing. Recombinant DHPS enzyme harboring either the p.Asn173Ser or p.Tyr305_Ile306del variant showed reduced (20%) or absent in vitro activity, respectively. We co-transfected constructs overexpressing HA-tagged DHPS (wild-type or mutant) and GFP-tagged eIF5A into HEK293T cells to determine the effect of these variants on hypusine biosynthesis and observed that the p.Tyr305_Ile306del and p.Asn173Ser variants resulted in reduced hypusination of eIF5A compared to wild-type DHPS enzyme. Our data suggest that rare biallelic variants in DHPS result in reduced enzyme activity that limits the hypusination of eIF5A and are associated with a neurodevelopmental disorder.
Keywords: exome sequencing, DHPS, hypusine, deoxyhypusine synthase, eIF5A, hypusination, neurodevelopmental disorder, polyamine pathway, inborn errors of metabolism, DOHH
Introduction
Hypusination is a post-translational process that results in the formation of the unique amino acid hypusine [N(epsilon)-(4-amino-2-hydroxybutyl)-lysine].1, 2 To date, only eukaryotic initiation factor 5A (eIF5A) and its homolog eIF5A2, which shows tissue-restricted expression, are known to contain hypusine. The hypusination of eIF5A occurs as a two-step reaction requiring the enzymes deoxyhypusine synthase (DHPS) and deoxyhypusine hydroxylase (DOHH).3 DHPS functions as a tetramer to catalyze the initial step, transferring the 4-aminobutyl moiety from the polyamine spermidine to a specific lysine residue (Lys50 in human eIF5A) of eIF5A, thereby forming the intermediate deoxyhypusine residue. Subsequently, DOHH hydroxylates this intermediate to complete the synthesis of hypusine and the activation of eIF5A.
eIF5A is conserved throughout evolution, and the high level of conservation around the hypusine residue suggests an important role for this modification. Studies using human cell lines, yeast, and zebrafish have implicated eIF5A, the hypusinated form of eIF5A (eIF5AHyp) and DHPS, in cellular proliferation and differentiation.3, 4, 5, 6, 7 eIF5AHyp has also been shown to function in mRNA translation.3, 7, 8 In particular, eIF5AHyp was found to relieve ribosome stalling during mRNA translation of sequences encoding stretches of polyprolines,4 suggesting that without eIF5AHyp, the translation of certain transcripts may be interrupted. The exact mRNA transcripts that require eIF5AHyp for efficient mRNA translation are unknown. Understanding the cell-specific complement of genes regulated by eIF5AHyp could elucidate the function of eIF5A and DHPS in various tissues. Of note, homozygous whole-body deletion of any of the three genes Eif5a, Dhps, or Dohh in mice is embryonic lethal,9, 10, 11 providing evidence that eIF5A and the enzymes responsible for its hypusination (DHPS and DOHH) are essential for embryonic development.
To date, DHPS (MIM: 600944), DOHH (MIM: 611262), and EIF5A (MIM: 600187) have not been associated with any Mendelian human genetic disorders. Herein, we report five affected individuals from four independent families who have biallelic variants in DHPS, identified by exome sequencing. Furthermore, we demonstrate the biochemical effects of two of these variants via in vitro functional studies that assay DHPS activity and the deleterious impact of these variants on the hypusination of eIF5A in cells.
Material and Methods
Recruitment and Molecular Genetic Studies
The four families in this study had clinical exome sequencing at the same laboratory using the same methods. Genomic DNA was extracted from peripheral blood of affected individuals and their parents. Exome sequencing was performed using genomic DNA from the proband and parents. The exonic regions and flanking splice junctions of the genome were captured using the SureSelect Human All Exon V4 (50 Mb), the Clinical Research Exome kit (Agilent Technologies), or the IDT xGen Exome Research Panel v.1.0. Sequencing was performed on an Illumina system with 100 bp or greater paired-end reads. Sequencing reads were aligned to human genome build GRCh37/UCSC hg19 and analyzed for sequence variants using a custom-developed analysis tool. Additional sequencing technology and variant interpretation protocol has been previously described.12
For family 1, for affected individual 1, the clinical exome analysis performed at the clinical laboratory in 2013 was non-diagnostic. In 2017, sequencing data were re-analyzed at Columbia University Medical Center. The study was approved by the Institutional Review Board of Columbia University, and written informed consent was obtained for all the individuals. The re-analysis of the exome sequencing for family 1 (two affected individuals and both parents) included mapping and aligning sequence to human genome build GRCh37/UCSC hg19 and analyzing for sequence variants using a custom-developed analysis tool as previously described.13 Specifically, the DHPS variants were prioritized based on their inheritance pattern, low allele frequencies, the absence of homozygotes in internal and publicly available population databases, and multiple bioinformatics prediction algorithms including metaSVM,14 Combined Annotation Dependent Depletion (CADD),15 SIFT, PolyPhen 2, MutationTaster, and Mutation Assessor (Table S2). The DHPS variants were added to GeneMatcher. Families 2, 3, and 4 were identified through GeneMatcher.16
Haplotype Analysis
VCF-merge was used to merge the individual VCF files of exome-sequencing data for each family. A 2.7 Mb (chr19:11221454–13988416) region around the shared DHPS variants were used in the haplotype analysis. Common SNPs with Mendelian inheritance, with an average genotype quality GQ ≥ 90, and heterozygous or homozygous alternative variants in at least 8 out of 12 family members were analyzed. Applying these filters, 88 SNPs were included in the haplotype analysis (Table S4). PHASE v2.1 trio mode was used to reconstruct the haplotypes.17
Functional Studies on the DHPS c.1014+1G>A Splice Variant
Peripheral blood samples were collected from the affected siblings, an unaffected sibling, and both parents of family 1. DNA was extracted using a QIAGEN Kit, and RNA was extracted using a PAXgene blood RNA kit (QIAGEN) according the manufacturer’s instructions. cDNA was reverse transcribed from the RNA using Transcriptor First Strand cDNA Synthesis kit (Roche) using mixed random hexamer primers and an anchored-oligo(dT)18 primer.
The c.1014+1G>A is a canonical splice donor site variant located after coding exon 8 (GenBank: NM_001930.3). Primers were designed to amplify exons 3 to 9 of the cDNA (Table S1). The PCR products were visualized by agarose gel electrophoresis, and the PCR products from the mother and one of the affected children were cloned into a TOPO vector (Invitrogen). Clones were purified by plasmid Miniprep purification (QIAGEN) and were Sanger sequenced using T3 and T7 universal primers.
Purification of DHPS Wild-Type and Variant Enzymes
Bacterial vectors encoding His-tagged DHPS wild-type and mutant enzymes were generated by site-directed mutagenesis (GenScript). Recombinant His-tagged Deoxyhypusine Synthase wild-type (DHPSWT) enzyme and the two mutant enzymes corresponding to the p.Asn173Ser (DHPSN173S) and p.Tyr305_Ile306del (DHPSY305_I306del) variants were individually expressed in E. coli BL21(DE) cells transformed with a pET15b-hDHPS vector encoding each form. The cells transformed with each vector were cultured in LB medium containing 100 μg/mL ampicillin (Sigma-Aldrich), and the enzyme expression was induced by addition of 1 mM IPTG (isopropyl-β-D-1-thiogalactopyranoside) (Sigma-Aldrich) when cell density reached 0.6 OD (optical density). After 4 h of induction, the cells were harvested by centrifugation. Cell pellets were either frozen immediately or lysed by sonication in 50 mM Tris HCl, 0.1 M NaCl buffer at pH 7.5 containing Halt Protease Inhibitor Cocktail (Thermo Scientific) and 1 mM PMSF (phenylmethylsulfonyl fluoride) (Sigma-Aldrich). The sonicated cell lysates were clarified by centrifugation in a refrigerated microfuge at 15,000 × g for 20 min at 4°C. The supernatant was separated. The insoluble pellets from total lysates contained most of the recombinant enzyme protein in the form of inclusion body. The enzymes were extracted from the insoluble pellets by resuspension in 0.2 M Tris (pH 12.5 containing 2 M urea). After removal of insoluble debris in the re-suspended mixture by centrifugation, the recombinant enzyme was obtained in a highly pure and active form (>95% purity). The solubilized enzyme was used directly or after removal of urea by equilibration with buffers (0.2 M Tris buffer; pH 12.5, pH 9.0, or pH 7.5). The enzyme exhibited higher activity upon equilibration in 0.2 M Tris solution (pH 12.5) than in 0.2 M Tris HCl buffer (pH 9.0 or pH 7.5). The level of enzyme expression and the purity were determined by protein assay (Biorad) and SDS-PAGE (sodium dodecyl sulfate polyacrylamide gel electrophoresis).
DHPS In Vitro Activity Assay
The assay was performed as described previously18 with minor modifications. eIF5A(Lys) was purified as described previously.19 In brief, the reaction mixture (25 μL) contained 0.4 mM NAD, 1 mg/mL BSA, 4 μg of eIF5A(Lys), 3 μCi of [1,8-terminal methylenes-3H] spermidine (PerkinElmer/NEN), and 1 mM DTT in 0.2 M glycine NaOH buffer (pH 9.5) with the indicated amounts of cell lysates or purified enzyme. After incubation at 37°C for 1.5 h, 0.5 mg of BSA (bovine serum albumin) carrier protein was added and the reaction was immediately stopped by the addition of 1 mL of 10% TCA (trichloroacetic acid) containing 1 mM of each of the three polyamines (Sigma-Aldrich); putrescine, spermidine, and spermine. The TCA-precipitated proteins were collected by centrifugation at 15,000 × g in a refrigerated microcentrifuge for 5 min. After removal of supernatants containing unreacted radioactive spermidine, the precipitates were re-suspended in a 10% TCA solution containing polyamines. The wash was repeated twice to remove all of the unreacted radioactive spermidine. The TCA-washed protein precipitates were then dissolved in 0.1 mL of 0.15 M NaOH, and the radioactivity incorporated into eIF5A protein was measured using a Beckman liquid scintillation counter. The enzyme activity was calculated after subtraction of the background radioactivity bound to TCA precipitates of the reaction mixture lacking any enzyme.
Construction of HA-Tagged Expression Vectors for Co-transfection Studies
To differentiate ectopic versus endogenous DHPS, expression vectors were generated that expressed N-terminal hemagglutinin (HA)-tagged forms of wild-type and the DHPS variants. First, primer sets were designed to insert the DHPSN173S and DHPSY305_I306del variants into the previously published GFP-DHPS construct20 using the Quick Change II Site-Directed Mutagenesis Kit (Stratagene) and Q5 Site-Directed Mutagenesis Kit (New England Biolabs), respectively (Table S1). Next, the HA-tag with an EcoRV restriction site was inserted into the pcDNA3.3-DHS-HA vector20 at the XbaI and PmeI restriction sites. The DHPS sequences were amplified starting with the first nucleotide downstream of the start codon through the stop codon using the wild-type and DHPS variant constructs as template. Finally, these sequences were inserted at the EcoRV site of the HA-tagged plasmid (pcDNA3.3HA-EcorV) using the In-Fusion HD cloning kit (Takara Bio). The open reading frame for each plasmid was Sanger sequenced to confirm DHPS tagging and insertion of variants. A construct expressing GFP-tagged eIF5A was also used in co-transfection studies.20
Transfection
HEK293T cells (ATCC #CRL-3216) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS (Hyclone; Fisher Scientific), 1% penicillin/streptomycin, and 2 mM L-glutamine (Fisher Scientific). Cells were grown to 60%–70% confluency and transiently transfected using Lipofectamine 3000 (Invitrogen #L3000008) with equal amounts of GFP-eIF5A (500 ng into 300,000 cells/well, 6-well dish) and HA-DHPS variant(s) (HADHPS, HAN173S, or HAY305_I306del; 500 ng) DNA. At 24 h after transfection, cells were washed with PBS and lysed on ice for western blot analysis.
For co-transfection experiments, to further evaluate the activity of the HAN173S, we reduced the amount of each DHPS construct by serial dilution from 500 ng to 16 ng (data not shown). The choice to transfect 16 ng of construct was based upon our observation that even with transfection of this amount of wild-type HADHPS construct, hypusination of eIF5A could be observed above the endogenous level. 100,000 cells/well were plated (24-well dish) and a stable amount of GFP-eIF5A (250 ng) was co-transfected with a varying amount of HADHPS or HAN173S (500 ng or 16 ng). At 24 h after transfection, cells were washed with PBS and lysed on ice for western blot analysis.
Western Blot Analysis
Transfected cells were lysed in buffer containing 50 mM Tris (pH 8.0), 150 mM NaCl, 0.05% deoxycholate, 0.1% IGEPAL CA-630, 0.1% SDS, 0.2% sarcosyl, 10% glycerol, 1 mM DTT, 1 mM EDTA, 10 mM NaF, protease inhibitors (Roche, # 11836170001), phosphatase inhibitors (Roche, #4906845001), 2 mM MgCl2, 0.05% v/v Benzonase) and subjected to protein quantification using the DC Protein Assay Kit II (Bio-Rad, #5000112). Protein (20 μg) was separated by SDS-PAGE (4%–20% gel) and transferred to PVDF membranes. After blocking in Odyssey Blocking Buffer (LI-COR Biosciences, #927-40100) at room temperature for 1 h, the membrane was incubated with primary antibodies diluted in Odyssey Blocking Buffer at 4°C overnight. After washing with TBST buffer three times, blots were incubated with near infrared fluorescent dye-conjugated secondary antibodies at room temperature for 1 h. Following three washes with TBST buffer, imaging was performed on an Odyssey CLx Imaging System and images were analyzed using the CLx Image Studio Version 5.2 Software (LI-CORE Biosciences). The following primary antibodies were used at the dilutions indicated: mouse anti-deoxyhypusine synthase (1:2,000; Santa Cruz, #sc-365077), rabbit anti-beta tubulin (1:2,000; Cell Signaling Technology, #2146S), mouse anti-eIF5A (1:2,000; BD Biosciences, #611977), goat anti-HA (1:2,000; Novus Technologies, #NB600362), and rabbit anti-eIF5AHyp (1:2,000; antibody reported in Maier et al.20).
Results
Clinical Presentations
Clinical findings of the five affected individuals from four independent families are summarized in Table 1. Ages of individuals range between 5 and 24 years. Features common to the most of the individuals with DHPS variants are developmental delay/intellectual disability (5/5), abnormalities in muscle tone (5/5), abnormal EEG (5/5), clinical seizures (5/5), pregnancy issues (4/5), coordination and walking difficulties (4/5), mild dysmorphic facial features (4/5) (Figure 2), and behavioral issues (3/5).
Table 1.
Clinical Features Seen in the Affected Individuals with Biallelic DHPS Variants
| Individual 1 | Individual 2 | Individual 3 | Individual 4 | Individual 5 | |
|---|---|---|---|---|---|
| Age at last visit | 9 years | 5 years | 7 years | 8 years | 24 years |
| Gender | female | male | female | female | female |
| Ethnicity | mat, Irish; pat, Scottish/Swedish | mat, Irish; pat, Scottish/Swedish | mat, Irish/English; pat, French/Belgium/English | mat, Irish/English; pat, Eastern European/French | mat, Slovakian; pat, English |
| Variants | p.Asn173Ser (mat); c.1014+1G>A (pat) | p.Asn173Ser (mat); c.1014+1G>A (pat) | p.Asn173Ser (mat); p.Tyr305_Ile306del (pat) | p.Asn173Ser (mat); c.1014+1G>A (pat) | p.Met1? (mat); p.Asn173Ser (pat) |
| Prenatal | increased NT, HELLP syndrome, IUGR | increased NT | pregnancy-induced hypertension | pre-eclampsia | oligohydramnios, low blood pressure |
| Natal | prematurity (32 weeks), intraventricular hemorrhage | unremarkable | low birth weight, neonatal hypoglycemia, temperature instability | prematurity (34 weeks), 12 days stay at NICU | unremarkable |
| Anthropometric Measures | |||||
| Birth | Len: 38.1 cm (12%) Wt: 1.39 kg (21%) OFC: 30.5 cm (87%) |
Len: 53.34 cm (96%) Wt: 3.13 kg (35%) OFC: 35 cm (25%) |
Len: 47.5 cm (18%) Wt: 2.55 kg (6%) OFC: 33 cm (21%) |
Len: n/a Wt: 2.07 kg (45%) OFC: n/a |
Len: 54.61 cm (97%) Wt: 4.026 kg (91%) OFC: n/a |
| Last visit | Ht: 124 cm (2%) Wt: 24 kg (4%) OFC: 52 cm (81%) |
Ht: 113 cm (44%) Wt: 21.1 kg (60%) OFC: 53 cm (87%) |
Ht: 117 cm (13%) Wt: 20.5 kg (15%) OFC: 48 cm (<1%) |
Ht: 111 cm (1%) Wt: 19.7 kg (8%) OFC: 48 cm (<1%) |
Ht: 157.8 cm (20%) Wt: 49.6 kg (12%) OFC: n/a |
| Psychomotor Milestones | |||||
| Sitting | 9 months | 10 months | >9 months | 6 months | 12–18 months |
| Speaking | limited language until 3 years of age; currently minimally verbal | spoke at 17–18 months | first words at 15 months, can speak some longer phrases | non-verbal | possible first word (mimicking) at 3-year old; never communicated verbally |
| Walking | 20 months | 14–15 months | 25 months | 18 months | 30 months; now wheelchair bound |
| Behavioral & sleep | unremarkable | unremarkable | hand flapping, grunting, growling, pica, mouthing of objects & reduced REM sleep | autism, happy child, hand flapping, hyperacusis | hand flapping, rocking when excited & sleep problems during childhood, now sleeps >10 h a day |
| Neurologic | truncal hypotonia, appendicular hypertonia, balance & coordination problems, high threshold for pain, hemiplegia, hand apraxia | hypotonia, balance & coordination problems | appendicular hypertonia, spastic gait & postural instability, hypertonia, spasticity (more severe in lower limbs) | hypotonia (upper limbs), high threshold for pain | diagnosed with cerebral palsy, history of hypotonia, now hypertonia/stiffness in lower limbs, wheelchair bound |
| Clinical seizures | yes (at night) | yes | unclear (staring spells) | yes | yes |
| EEG | ESES | partial benign refractory seizures in sleep originating in the occipital lobe; absence seizures on EEG | focal epileptiform discharges from left temporal and occipital quadrants but no correlation with staring spells | frequent spikes independently over right and left central and right parietal regions during sleep | absence seizures, tonic-clonic seizures |
| Brain MRI | normal | not performed | normal | normal | normal |
| Dysmorphic findings | deep-set eyes, sacral dimple | deep-set eyes, sacral dimple | deep set eyes, prominent infraorbital creases, medial eyebrow flare, prominent nasal bridge, thin upper lip, mildly high palate, bilateral 5th finger clinodactyly | microcephalic, normally spaced and deep-set eyes, prominent infraorbital creases, medial eyebrow flare, mildly low set ears, smooth philtrum, thin upper lip | none |
| Dermatologic | easy bruising, skin sensitivity | easy bruising, skin sensitivity | mild eczema, occasional hives | mild erythema on hands | normal |
| Other | borderline low IgA & IgG levels; constipation; hyperopia | low IgA & IgG levels; tracheomalacia; subglottic stenosis | constipation (on PEG); cold/mottled hands | gynecomastia; cold/mottled/bluish hands | loss of bladder control |
Abbreviations: Mat, maternal; Pat, paternal; HELLP, hemolysis, elevated liver enzymes, and low platelet count occurring in pregnancy; IUGR, intrauterine growth retardation; NT, nuchal translucency; Len, length; Ht, height; Wt, weight; OFC, occipito-frontal circumference; REM, rapid eye movements; EEG, electroencephalography; ESES, electrical status epilepticus in sleep; PEG, percutaneous endoscopic gastrostomy.
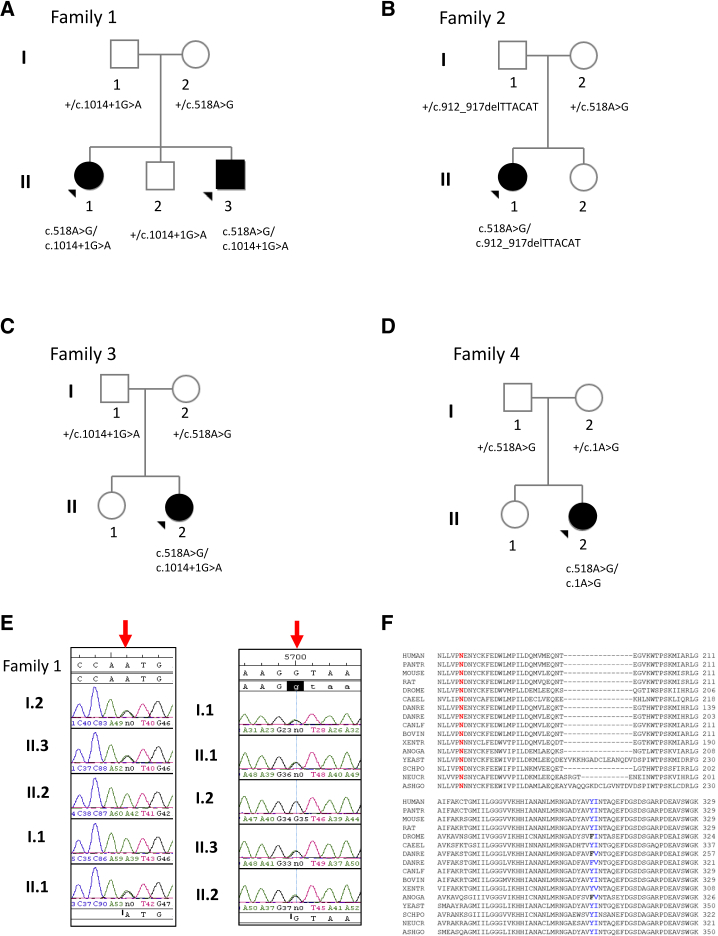
Figure 2.
Photos of Affected Individuals Showing Mild Dysmorphic Facial Features Including Deep-Set Eyes, Broad Nasal Tip, and Thin Upper Lip and a Schematic Diagram of DHPS
(A–D) Shown are individual 1 (A), individual 2 (B), and individual 3 (C and D).
(E) Localization of the detected missense variants with respect to important amino acid residues involved in the binding of spermidine (green), NAD (orange), and the critical Lys329 involved in catalysis (yellow) on the DHPS primary structure diagram.25
Pregnancy issues include HELLP syndrome, pregnancy-induced hypertension, pre-eclampsia, and oligohydramnios. Anthropometric measurements were normal at birth and there were no specific health problems in the neonatal period, but short stature was noted in two individuals at the time of last evaluation. Sitting and walking were mildly delayed but speech was the most severely affected milestones. Hypotonia was observed in two individuals while in two others hypotonia was accompanied or followed by hypertonia/stiffness in extremities, and one individual only has hypertonia/spasticity. Walking difficulties range from ataxic gait due to balance issues in two individuals and spastic gait due to hypertonia affecting the extremities in other two individuals. Absence seizures were the most frequent clinical seizure type, and tonic-clonic seizures are also reported in one individual. There were non-specific lateralized abnormal discharges present mostly during sleep in some, but there was no specific EEG finding recorded across all patients. Dysmorphic features are mild and variable and include deep-set eyes, sacral dimple, prominent infraorbital creases, medial eyebrow flare, and thin upper lip. Dermatologic issues are non-specific and range from easy bruising, mild eczema and erythema, and occasional hives to cold/mottled hands. Behavioral issues are mainly characterized by repetitive behaviors and sleep problems. High pain tolerance was noted in two individuals. Constipation was also noted in two individuals. There was a history of borderline low IgA and IgG levels in siblings, individuals 1 and 2. All of the families reported at least one parent to be of Irish and/or English ancestry. A detailed clinical history of each affected individual is given in the Supplemental Note.
Identification of DHPS Variants
All affected individuals had undergone chromosome analysis, chromosomal microarray, fragile X testing, and Prader-Willi/Angelman syndrome methylation assays, all of which were normal. Additionally, transferrin isoelectric focusing assays, intellectual disability gene sequencing panel, mitochondrial genome sequencing/deletion, and MECP2 sequencing were also normal for individuals 1 and 3.
Exome sequencing identified biallelic variants in DHPS, the gene encoding deoxyhypusine synthase. Sanger sequencing of all members of family 1 confirmed the biallelic DHPS variants in the affected siblings and showed that the unaffected sibling and both parents are carriers of a single variant (Figures 1A and 1E and Table S2). The variants seen in PLA2G4D (MIM: 612864) and PUS7 (MIM: 612864) were excluded based on their high population frequencies (Table S2). Families 2, 3, and 4 were subsequently identified through GeneMatcher. Individual 1, individual 2, and individual 4 are compound heterozygous for c.518A>G (p.Asn173Ser) and c.1014+1G>A variants; individual 3 is compound heterozygous for c.518A>G (p.Asn173Ser) and c.912_917delTTACAT (p.Tyr305_Ile306del) variants; individual 5 is compound heterozygous for c.518A>G (p.Asn173Ser) and c.1A>G (p.Met1?) variants (Tables 1 and S3; Figure 1).
Figure 1.
DHPS Variants in the Affected Families
(A–D) Pedigrees showing individuals from families 1–4 and segregation of DHPS variants.
(E) Sanger sequencing of c.518A>G (p.Asn173Ser) and c.1014+1G>A variant in members of family 1.
(F) Sequence alignment of DHPS amino acid residues Asn173 and Tyr305_Ile306 and the surrounding residues across species.
The p.Asn173Ser, p.Met1?, and c.1014+1G>A variants are present at very low frequencies in population databases with no homozygotes. The p.Tyr305_Ile306del variant is not present in any population database. The population allele frequencies and the computational prediction scores for the variants are given in Table 2. Notably, the missense and splice site variants are predominantly seen in non-Finnish European individuals in Genome Aggregation Database. The c.1014+1G>A variant is a canonical splice donor site at 3′ of coding exon 8. The p.Met1? variant alters the translation initiation codon (Figure 2E).
Table 2.
Genomic Findings of DHPS Variants
| Genomic Coordinates (hg 19) | HGVS cDNA | HGVS Protein | gnomAD Population Allele Frequencies (NFE/Total) & (Homozygote?) |
Computational Prediction Scores |
||
|---|---|---|---|---|---|---|
| CADD v1.4 | REVEL | PROVEAN | ||||
| chr19:12790510T>C | c.518A>G | p.Asn173Ser | 0.00012/0.000058 & (no) | 24.8 | 0.347 | −4.75 |
| chr19: 12786928–12786933delATGTAA | c.912_917delTTACAT | p.Tyr305_Ile306del | not present | 13.64 | n/a | −17.18 |
| chr19:12786830C>T | c.1014+1G>A | p.? | 0.00057/0.0004267 & (no) | 27.5 | n/a | n/a |
| chr19:12792580T>C | c.1A>G | p.Met1?a | 0.000067/0.00003231 & (no) | 19.78 | 0.267 | −0.53 |
Abbreviations: HGVS, human genome variation society; gnomAD, genome aggregation database; NFE, non-Finnish European; n/a, not available.
There are three heterozygous individuals in gnomAD with c.2T>C which also results in p.Met1?.
Haplotype Analysis
Two of the DHPS variants were common across at least two families (chr19:12790510T>C, c.518A>G [p.Asn173Ser] and chr19:12786830C>T, c.1014+1G>A). With the exome-sequencing data and PHASE v2.1,17 we performed haplotype analysis using single-nucleotide variants. For c.518A>G, the longest shared haplotype was 853,693 bp shared by three parents of families 1, 3, and 4 (Figure S2A; Table S4). In family 2, c.518A>G (p.Asn173Ser) is on a different haplotype (Figure S2A). For the c.1014+1G>A variant, two parents share a 781,592 bp long haplotype containing the variant (Figure S2B; Table S4).
Analysis of the DHPS c.1014+1G>A Canonical Splice Site Variant
To understand the effect of the DHPS c.1014+1G>A splice donor variant (3′ to coding exon 8), we performed DHPS transcript analysis from peripheral blood samples collected from family 1. Gel electrophoresis of amplified cDNA sequences showed two PCR products in the mother, whereas three PCR products (two amplicons of the same size as in maternal sample) were present in the samples of the father, the two affected children, and the unaffected son, all of whom carry the c.1014+1G>A variant (Figures S3A–S3D). The PCR products from the maternal and affected proband’s sample (family 1-II.1) were cloned. Sequencing of multiple clones from the mother’s sample identified two DHPS inserts of 657 bp and 553 bp. These sequences corresponded to the canonical isoform (657 bp; GenBank: NM_001930.3) and a noncoding RNA (553 bp; GenBank: NR_038192.1), and approximately half of these sequenced clones had the c.518A>G (p.Asn173Ser) variant (Figures S3A–S3D). Sequencing the clones derived from affected proband’s sample (family 1-II.1) showed the two isoforms seen in the mother and an additional novel fragment of 427 bp corresponding to an isoform lacking coding exons 7 and 8, resulting in a frameshift following the 261st amino acid and premature termination after the introduction of three novel amino acids. Sequences of the two normal isoforms from the affected individual carried the p.Asn173Ser variant whereas the 427 bp fragment seen in the carriers of the c.1014+1G>A variant did not harbor it, confirming that these two variants are in trans.
In human DHPS, the enzyme-substrate intermediate formation at Lys329 is reported to be critical for catalysis (Figure 2E). Taken together, these data suggest that if the novel c.1014+1G>A variant is translated, it is predicted to result in a truncated DHPS protein (containing amino acids 1-261+3 amino acids) that would lack the active site and therefore enzyme activity.
The DHPSN173S and DHPSY305_I306del Variants Alter Enzyme Activity
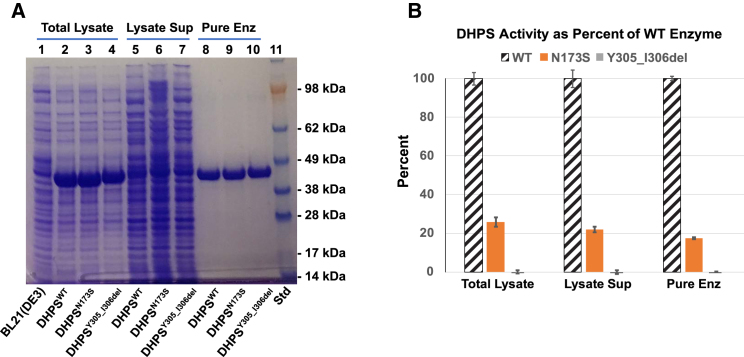
To determine the effect of the DHPSN173S and the DHPSY305_I306del variants on enzyme function, we performed an in vitro enzyme activity assay. E. coli BL21(DE3) cells were used to express wild-type, DHPSN173S, and DHPSY305_I306del recombinant enzymes. Figure 3A shows that all three enzymes were highly expressed in E. coli upon induction with IPTG (Figure 3A, total lysates). Clarification of the lysates by centrifugation resulted in a significant portion of each recombinant enzyme in the insoluble pellet (containing inclusion bodies) and only small amounts in each lysate supernatant (Figure 3A, Lysate Sup). The enzymes were therefore purified by extraction of the insoluble inclusion bodies using a 0.2 M Tris buffer (pH 12.5) containing 2 M urea. After removal of urea, highly pure enzymes were obtained (Figure 3A, Pure Enz). The mobility of the DHPSY305_I306del was slightly reduced compared with wild-type or DHPSN173S consistently in the total lysate, supernatant, and after purification (Figure 3A). The finding suggests that there are subtle differences in the unfolded, SDS-coated conformation between DHPSY305_I306del and the other two forms of the enzyme.
Figure 3.
Purification and the Activities of the Wild-Type DHPS (DHPSWT), p.Asn173Ser (DHPSN173S), and p.Try305_Ile306del (DHPSY305_I306del) Variants
The recombinant enzymes were expressed in BL21(DE3) cells and the enzyme activity was measured as described under Material and Methods.
(A) Coomassie Blue-stained patterns of proteins in total lysates (lanes 1–4), lysate supernatants (lanes 5–7), or purified enzymes (lanes 8–10) are shown. Lysate proteins of BL21(DE3) not transformed with a DHPS expression vector is shown in lane 1, for comparison with those of DHPS-overexpressing lysates.
(B) Comparison of enzyme activities. The enzyme activity assays were performed with total lysates (0.2 μg proteins in each sample), lysate supernatant (0.1 μg proteins in each sample), or purified DHPS enzymes (0.02 μg each sample) as described under Material and Methods. The activities of the mutant enzymes are expressed as percent of the wild-type enzyme in each group. The experiments were repeated three times with similar results. Data from a representative experiment with duplicate samples is shown with error bars.
The enzyme activity for the wild-type, DHPSN173S, and DHPSY305_I306del enzymes was assayed using total lysate, lysate supernatant, and purified protein. Compared with wild-type DHPS, the mutant enzymes displayed impaired biosynthesis of deoxyhypusine. Specifically, DHPSY305_I306del was consistently devoid of enzyme activity in the total lysate, the lysate supernatant, and the purified form (Figure 3B). DHPSN173S exhibited partial activity, ranging from 18% to 25% of that of wild-type DHPS enzyme when measured in total lysate, lysate supernatant, and the purified form (Figure 3B). These findings suggest that the DHPSY305_I306del has no enzyme activity in vitro, whereas the DHPSN173S has partial activity in vitro amounting to approximately 20% of wild-type.
Altered DHPS Enzyme Activity Impairs Hypusination of eIF5A
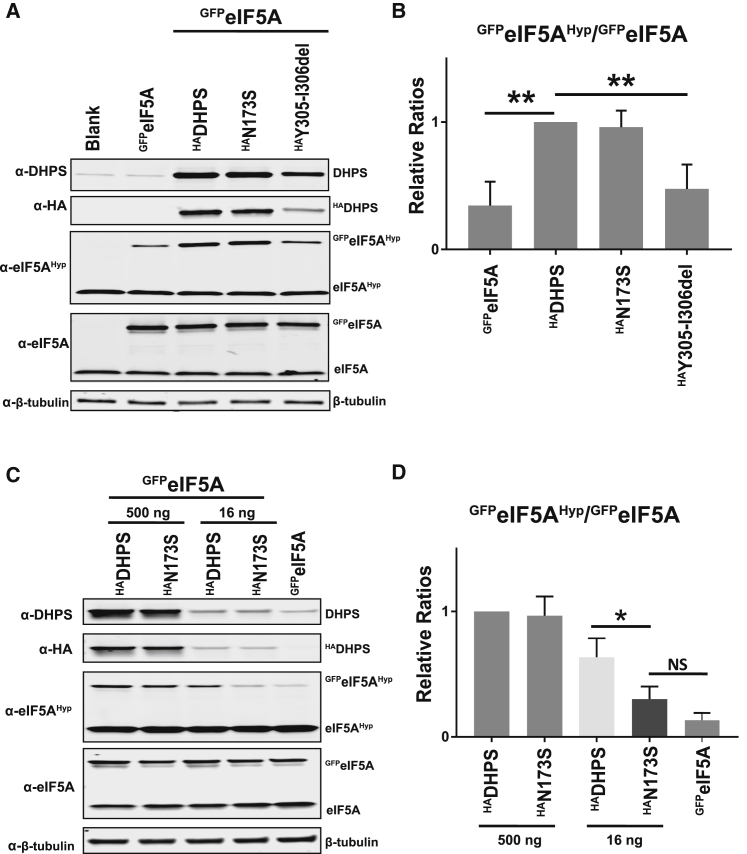
To evaluate the effect of the DHPSN173S and DHPSY305_I306del variants on enzyme function, we co-transfected constructs overexpressing HA-tagged DHPS (HADHPS, HAN173S, HAY305-I306del) along with a vector encoding a GFP-eIF5A fusion protein (GFPeIF5A) into HEK293T cells. Of note, transfection of an ectopic copy of eIF5A is required to assess DHPS enzymatic activity as most of the endogenous eIF5A is already hypusinated in mammalian cells such as HeLa cells.21 At 24 h after transfection, cells were lysed, and DHPS, GFPeIF5AHyp, GFPeIF5A, eIF5AHyp, and eIF5A were assessed by western blot (Figure 4A). As expected, a significant increase in the amount of GFPeIF5AHyp was observed with co-transfection of GFPeIF5A and HADHPS as compared to GFPeIF5A alone (Figure 4B). Conversely, a significant decrease in GFPeIF5AHyp was observed with co-transfection of GFPeIF5A and the HAY305_I306del variant (500 ng) as compared to co-transfection of GFPeIF5A and wild-type HADHPS (p < 0.01; Figures 4A and 4B). These data are consistent with the total inactivity of the DHPSY305_I306del enzyme seen in the in vitro recombinant enzyme activity assay (Figure 3B). Further, the levels of HAY305-I306del were noted to be lower than the wild-type HADHPS enzyme (Figure 4A, panel 1, lanes 3 and 5), suggesting that the DHPSY305_I306del variant might be unstable in HEK293T cells, which was confirmed using in vitro stability assays (data not shown).
Figure 4.
The DHPS p.Tyr305_Ile306del and p.Asn173Ser Variants Cause Decreased Hypusination of GFPeIF5A
(A) HEK293T cells were co-transfected with 500 ng of HA-tagged DHPS (HADHPS, HAN173S, HAY305_I306del) constructs and 500 ng of GFP-eIF5A construct (GFPeIF5A). At 24 h after transfection, cells were lysed and 20 μg protein was used for western blot analysis to assess expression levels of DHPS, HA (transfected HADHPS), eIF5A (all forms; hypusinated, nonhypusinated, and GFP-tagged), and β-tubulin (loading control). A previously published antibody20 was utilized to detect the hypusinated forms of eIF5A, which permitted the estimation of hypusinated GFPeIF5A. This figure is a representative blot.
(B) Relative levels of GFPeIF5AHyp to total GFPeIF5A after normalization to the HADHPS control. Data are means ± SEM, n = 3, ∗∗p < 0.005.
(C) HEK293T cells were co-transfected with either 500 ng or 16 ng of HA-tagged DHPS (HADHPS, HAN173S) constructs and 250 ng of GFPeIF5A construct. Western blot analysis was performed as described above. This is a representative blot.
(D) Relative protein levels of GFPeIF5AHyp to total GFPeIF5A after normalization to the HADHPS control. Data are means ± SEM, n = 3, ∗p < 0.05. The bars in these graphs represent the average signal ratio from three independent experiments. NS indicates not significant.
Co-transfection of GFPeIF5A and HAN173S constructs (500 ng) resulted in an increase of GFPeIF5AHyp compared to GFPeIF5A alone and this was comparable to the co-transfection with wild-type enzyme construct, HADHPS (Figure 4A, lane 3 versus 4). Given that the in vitro recombinant DHPS enzyme activity assay showed reduced enzyme activity for DHPSN173S (Figure 3B), we reasoned that transfection of a saturating amount of HAN173S construct leading to overproduction of the partially active mutant enzyme in cells might have masked the differences in enzyme activity. Therefore, we performed co-transfection experiments wherein the transfected amount of GFPeIF5A construct was kept constant (250 ng) but the amount of HADHPS or HAN173S was either limiting (16 ng) or abundant (500 ng). Consistent with the above data (Figures 4A and 4B), co-transfection of 500 ng of either HADHPS or HAN173S construct together with GFPeIF5A resulted in a similar increase in GFPeIF5AHyp compared with transfection of GFPeIF5A alone (Figures 4C and 4D). In contrast, co-transfection of GFPeIF5A with limiting amount (16 ng) of HAN173S construct resulted in a reduction in GFPeIF5AHyp compared to the co-transfection of the same amount (16 ng) of HADHPS wild-type construct along with GFPeIF5A (Figures 4C and 4D). Taken together, these data support that the HAN173S has partial activity in cells, which is consistent with the in vitro recombinant enzyme assay (Figure 3B).
Discussion
We identified biallelic inherited rare variants in DHPS associated with decreased DHPS enzyme activity and reduced eIF5A hypusination in five individuals from four independent families, primarily with neurodevelopmental delay and seizures. In some cases, the phenotype also includes short stature, microcephaly, abnormal tone (axial hypertonia or spasticity), and prenatal maternal hypertension.
Trio exome sequencing identified that each affected individual carries a combination of two of the four DHPS variants (Tables 1 and 2). Interestingly, the c.518A>G (p.Asn173Ser) variant is seen in trans with another variant in all affected individuals, and five individuals from the four families share this variant, which raised the question of whether the c.518A>G variant might be a founder variant (Table 1, Figure 1). Across four carrier parents, c.518A>G is located on two distinct haplotypes. The haplotype shared by three carriers (families 1, 3, and 4) is about 853 kb and therefore is unlikely to be an identity-by-descent (IBD) segment from a common ancestor within the last 20 generations but could represent an older common ancestor (Figure S2A).22 The c.518A>G variant is carried on a distinct haplotype by one carrier (family 2), and this is likely due to a recurrent mutation. The c.518A>G variant is predominantly seen in non-Finnish Europeans in gnomAD (Table 2). In two carrier parents (families 1 and 3), the c.1014+1G>A variant is present on a haplotype that is about 782 kb and similarly does not represent a recent common ancestor but could represent an older common ancestor (Figure S2B).
We performed functional studies to assess the effects of the variants. Specifically, cDNA sequencing studies in carriers of the c.1014+1G>A variant demonstrated a novel cDNA fragment lacking exons 7 and 8 (Figure S3C), which results in an aberrant transcript, with disruption of the C-terminal region including the critical catalytic residue Lys329 and therefore is predicted to abolish enzymatic activity. In vitro enzyme activity assays showed that p.Tyr305_Ile306 variant results in total loss of activity whereas p.Asn173Ser variant retained 20% activity compared to wild-type enzyme (Figure 3B).
Since DHPS catalyzes hypusination of eIF5A, we studied the effects of p.Asn173Ser and p.Tyr305_Ile306 variants on eIF5A hypusination levels (eIF5AHyp) using co-transfection experiments in HEK293T cells. Co-transfection of GFPeIF5A and HAY305_I306del did not result in a substantial change of GFPeIF5AHyp levels compared to GFPeIF5A alone, and GFPeIF5AHyp levels were significantly lower in comparison to the co-transfection with wild-type HADHPS enzyme construct, consistent with the inactivity observed in the in vitro assay (Figures 4A and 4B). However, there was no difference in the GFPeIF5AHyp levels after co-transfection with 500 ng of GFPeIF5A and HADHPSN173S when compared with GFPeIF5A and HADHPS (500 ng). This is probably because in this cell culture model DHPSN173S is highly overexpressed compared to the endogenous DHPS enzyme (∼10-fold, Figure 4A, top panel, lane 4 versus lane 2) and the levels are sufficient to modify all GFPeIF5A and result in GFPeIF5AHyp levels indistinguishable from the wild-type HADHPS enzyme. However, co-transfection with a reduced amount of HADHPS or HAN173S construct (16 ng) induced only a ∼2-fold increase in DHPS protein compared with the endogenous enzyme level (Figure 4C). At this expression level, we observed reduced GFPeIF5AHyp with co-transfection of HAN173S compared with HADHPS wild-type enzyme. These cell culture studies are in alignment with our in vitro recombinant enzyme activity studies, which confirm that the p.Tyr305_Ile306 variant results in loss of activity whereas the p.Asn173Ser variant has reduced activity compared to wild-type enzyme.
DHPS is a 369-amino acid containing protein that catalyzes the hypusination of eIF5A using the butylamine moiety of spermidine via NAD-dependent oxidative reaction. The amino acids Asn173, Tyr305, and Ile306 altered in the p.Asn173Ser, p.Tyr305_Ile306del variants are all localized in the highly conserved region of DHPS23 (Figure 1F). Tyr305 is located at the active site (Figure S4) and Tyr305Ala substitution caused a significant reduction in the binding of spermidine and NAD, and the enzyme activity.24 Therefore, it is not surprising that deletion of this residue and Ile306 totally abolishes the enzyme activity. Asn173 is localized in the vicinity of the active site facing a critical active site residue Trp327 (Figure S4).24, 25 The substitution of Asn173 with Ser may alter the active site topology and reduce the enzyme activity by affecting the binding of substrates or enzyme tetramer formation.
Our exome sequencing and in vitro studies demonstrated that affected individuals in all four families have a combination of two alleles, one allele resulting in a complete lack of DHPS activity (p.Tyr305_Ile306del, c.1014+1G>A, or p.Met1?) and the other allele possessing significantly reduced DHPS activity (p.Asn173Ser), which is consistent with the autosomal-recessive inheritance pattern. Unaffected carrier parents and siblings along with the constraint metrics (variant distribution) from population databases suggest that haploinsufficiency of DHPS is tolerated in humans. Furthermore, the absence of individuals with homozygous loss-of-function DHPS alleles in population databases (gnomAD, TOPMed) lends support to the notion that complete DHPS deficiency is incompatible with normal human embryonic development. These observations are consistent with the lack of a severe phenotype and embryonic lethality in mice with heterozygous and homozygous deletion of Dhps, respectively.9, 10
The role of EIF5A, DHPS, or DOHH in disease models has not been extensively investigated to date, with the exception of pharmacological inhibition of Dhps in a mouse model of type 1 diabetes, which resulted in reduced inflammation and disease incidence.26 The identification of individuals with variants resulting in reduced DHPS activity provides insight into clinical features associated with DHPS deficiency and reduced eIF5AHyp in humans. eIF5A has been previously implicated in neuronal growth and survival.27 A range of neurological impairment is primarily observed in our affected individuals, including seizures, imbalance, hypotonia/hypertonia, speech delay, which indicates that severe reduction in levels of DHPS and hypusinated eIF5A affect the normal functioning of the nervous system. Additionally, there were mild dermatological and immunological issues observed in a subset of individuals which may reflect the role of DHPS and EIF5A in other cell types and is also consistent with their expression pattern in various tissues.28 Characterization of additional individuals with DHPS deficiency will allow us to determine which clinical features are consistently observed.
eIF5A has been proposed to function as a mRNA translation elongation factor that relieves ribosome stalling at polyproline stretches in yeast.4 However, more recent studies utilizing the ribosome profiling technique in eIF5A-depleted S. cerevisiae cells have provided evidence that eIF5A promotes mRNA translation termination and facilitates mRNA translation elongation not only at polyproline stretches but also at broad ranges of ribosome stalling motifs.8 Mutations in translation elongation factors, enzymes, and ribosomal proteins involved in translational regulation are increasingly recognized as causes of human developmental and neurological disorders.29 Mutations in genes including eukaryotic elongation factor 1a2 (EEF1A2 [MIM: 602959]), eukaryotic elongation factor 2 (EEF2 [MIM:130610]), and ribosomal proteins (RPS23 and RPL10) disrupt translation elongation and are associated with neurodevelopmental phenotypes.29
The clinical features of affected individuals in our study show some inter- and intra-familial variation, but the core neurological features are shared. Phenotypic variation such as that between individuals 1 and 2 within the same family is common across Mendelian conditions and can be due to other genetic and non-genetic modifiers. Future efforts are being directed at elucidating genotype-phenotype correlation, the role and mechanism of DHPS and eIF5AHyp in brain development, function, and the downstream targets of eIF5AHyp.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We thank the families for their generous contribution. We also thank Matchmaker exchange/Genematcher for connecting us with the other families. We thank Patricia Lanzano for her technical expertise. These studies were supported by grants from the SFARI at the Simons Foundation, the JPB Foundation, and the DHPS Foundation. T.L.M. was supported by a Career Development Award from the Juvenile Diabetes Research Foundation (5-CDA-2016-194-A-N). R.G.M. was supported by National Institutes of Health grants (R01 DK60581 and P30 DK097512). M.H.P. was supported by the intramural program of the National Institute of Dental and Craniofacial Research, National Institutes of Health.
Published: January 17, 2019
Footnotes
Supplemental Data include four figures, four tables, and Supplemental Note that contains clinical descriptions and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.12.017.
Accession Numbers
The accession numbers for the DHPS variants reported in this paper are ClinVar: SCV000804205, SCV000804206, SCV000804207, and SCV000804223.
Web Resources
ClinVar, https://www.ncbi.nlm.nih.gov/clinvar/submitters/26957
gnomAD Browser, http://gnomad.broadinstitute.org/
OMIM, https://www.omim.org/
TOPMed browser, https://bravo.sph.umich.edu/freeze5/hg38/
Supplemental Data
References
- 1.Park M.H. The post-translational synthesis of a polyamine-derived amino acid, hypusine, in the eukaryotic translation initiation factor 5A (eIF5A) J. Biochem. 2006;139:161–169. doi: 10.1093/jb/mvj034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park M.H., Cooper H.L., Folk J.E. Identification of hypusine, an unusual amino acid, in a protein from human lymphocytes and of spermidine as its biosynthetic precursor. Proc. Natl. Acad. Sci. USA. 1981;78:2869–2873. doi: 10.1073/pnas.78.5.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park M.H., Nishimura K., Zanelli C.F., Valentini S.R. Functional significance of eIF5A and its hypusine modification in eukaryotes. Amino Acids. 2010;38:491–500. doi: 10.1007/s00726-009-0408-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gutierrez E., Shin B.S., Woolstenhulme C.J., Kim J.R., Saini P., Buskirk A.R., Dever T.E. eIF5A promotes translation of polyproline motifs. Mol. Cell. 2013;51:35–45. doi: 10.1016/j.molcel.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mastracci T.L., Robertson M.A., Mirmira R.G., Anderson R.M. Polyamine biosynthesis is critical for growth and differentiation of the pancreas. Sci. Rep. 2015;5:13269. doi: 10.1038/srep13269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park M.H., Wolff E.C., Lee Y.B., Folk J.E. Antiproliferative effects of inhibitors of deoxyhypusine synthase. Inhibition of growth of Chinese hamster ovary cells by guanyl diamines. J. Biol. Chem. 1994;269:27827–27832. [PubMed] [Google Scholar]
- 7.Saini P., Eyler D.E., Green R., Dever T.E. Hypusine-containing protein eIF5A promotes translation elongation. Nature. 2009;459:118–121. doi: 10.1038/nature08034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schuller A.P., Wu C.C., Dever T.E., Buskirk A.R., Green R. eIF5A functions globally in translation elongation and termination. Mol. Cell. 2017;66:194–205.e5. doi: 10.1016/j.molcel.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishimura K., Lee S.B., Park J.H., Park M.H. Essential role of eIF5A-1 and deoxyhypusine synthase in mouse embryonic development. Amino Acids. 2012;42:703–710. doi: 10.1007/s00726-011-0986-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Templin A.T., Maier B., Nishiki Y., Tersey S.A., Mirmira R.G. Deoxyhypusine synthase haploinsufficiency attenuates acute cytokine signaling. Cell Cycle. 2011;10:1043–1049. doi: 10.4161/cc.10.7.15206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sievert H., Pällmann N., Miller K.K., Hermans-Borgmeyer I., Venz S., Sendoel A., Preukschas M., Schweizer M., Boettcher S., Janiesch P.C. A novel mouse model for inhibition of DOHH-mediated hypusine modification reveals a crucial function in embryonic development, proliferation and oncogenic transformation. Dis. Model. Mech. 2014;7:963–976. doi: 10.1242/dmm.014449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Retterer K., Juusola J., Cho M.T., Vitazka P., Millan F., Gibellini F., Vertino-Bell A., Smaoui N., Neidich J., Monaghan K.G. Clinical application of whole-exome sequencing across clinical indications. Genet. Med. 2016;18:696–704. doi: 10.1038/gim.2015.148. [DOI] [PubMed] [Google Scholar]
- 13.Zhu N., Gonzaga-Jauregui C., Welch C.L., Ma L., Qi H., King A.K., Krishnan U., Rosenzweig E.B., Ivy D.D., Austin E.D. Exome sequencing in children with pulmonary arterial hypertension demonstrates differences compared with adults. Circ Genom Precis Med. 2018;11:e001887. doi: 10.1161/CIRCGEN.117.001887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong C., Wei P., Jian X., Gibbs R., Boerwinkle E., Wang K., Liu X. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum. Mol. Genet. 2015;24:2125–2137. doi: 10.1093/hmg/ddu733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stephens M., Smith N.J., Donnelly P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolff E.C., Lee S.B., Park M.H. Assay of deoxyhypusine synthase activity. Methods Mol. Biol. 2011;720:195–205. doi: 10.1007/978-1-61779-034-8_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park J.H., Dias C.A., Lee S.B., Valentini S.R., Sokabe M., Fraser C.S., Park M.H. Production of active recombinant eIF5A: reconstitution in E. coli of eukaryotic hypusine modification of eIF5A by its coexpression with modifying enzymes. Protein Eng. Des. Sel. 2011;24:301–309. doi: 10.1093/protein/gzq110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maier B., Ogihara T., Trace A.P., Tersey S.A., Robbins R.D., Chakrabarti S.K., Nunemaker C.S., Stull N.D., Taylor C.A., Thompson J.E. The unique hypusine modification of eIF5A promotes islet beta cell inflammation and dysfunction in mice. J. Clin. Invest. 2010;120:2156–2170. doi: 10.1172/JCI38924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klier H., Csonga R., Joäo H.C., Eckerskorn C., Auer M., Lottspeich F., Eder J. Isolation and structural characterization of different isoforms of the hypusine-containing protein eIF-5A from HeLa cells. Biochemistry. 1995;34:14693–14702. doi: 10.1021/bi00045a010. [DOI] [PubMed] [Google Scholar]
- 22.Speed D., Balding D.J. Relatedness in the post-genomic era: is it still useful? Nat. Rev. Genet. 2015;16:33–44. doi: 10.1038/nrg3821. [DOI] [PubMed] [Google Scholar]
- 23.Wolff E.C., Kang K.R., Kim Y.S., Park M.H. Posttranslational synthesis of hypusine: evolutionary progression and specificity of the hypusine modification. Amino Acids. 2007;33:341–350. doi: 10.1007/s00726-007-0525-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee C.H., Um P.Y., Park M.H. Structure-function studies of human deoxyhypusine synthase: identification of amino acid residues critical for the binding of spermidine and NAD. Biochem. J. 2001;355:841–849. doi: 10.1042/bj3550841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao D.I., Wolff E.C., Park M.H., Davies D.R. Crystal structure of the NAD complex of human deoxyhypusine synthase: an enzyme with a ball-and-chain mechanism for blocking the active site. Structure. 1998;6:23–32. doi: 10.1016/s0969-2126(98)00004-5. [DOI] [PubMed] [Google Scholar]
- 26.Colvin S.C., Maier B., Morris D.L., Tersey S.A., Mirmira R.G. Deoxyhypusine synthase promotes differentiation and proliferation of T helper type 1 (Th1) cells in autoimmune diabetes. J. Biol. Chem. 2013;288:36226–36235. doi: 10.1074/jbc.M113.473942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang Y., Higginson D.S., Hester L., Park M.H., Snyder S.H. Neuronal growth and survival mediated by eIF5A, a polyamine-modified translation initiation factor. Proc. Natl. Acad. Sci. USA. 2007;104:4194–4199. doi: 10.1073/pnas.0611609104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kruse M., Rosorius O., Krätzer F., Bevec D., Kuhnt C., Steinkasserer A., Schuler G., Hauber J. Inhibition of CD83 cell surface expression during dendritic cell maturation by interference with nuclear export of CD83 mRNA. J. Exp. Med. 2000;191:1581–1590. doi: 10.1084/jem.191.9.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kapur M., Ackerman S.L. mRNA translation gone awry: translation fidelity and neurological disease. Trends Genet. 2018;34:218–231. doi: 10.1016/j.tig.2017.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.