Abstract
Cardiolipin (Ptd2Gro) is a complex, double-charged phospholipid located in the inner mitochondrial membrane where it plays an essential role in regulating bioenergetics. Abnormalities in Ptd2Gro content or composition have been associated with mitochondrial dysfunction in a variety of disease states. Herein, we developed an adapted high-resolution data independent acquisition (DIA) MS/MSALL shotgun lipidomic method to enhance the accuracy and reproducibility of Ptd2Gro molecular species quantitation from biological samples. Utilizing the double charged molecular ions and the isotopic pattern under negative electrospray ionization mass spectrometry (ESI-MS) mode using an adapted MS/MSALL approach, we profiled more than 150 individual Ptd2Gro species, including monolysocardiolipin. The method described in this study demonstrated high reproducibility, sensitivity, and throughput with a wide dynamic range. This high-resolution MS/MSALL shotgun lipidomics approach could be extended to screening aberrations of Ptd2Gro metabolism involved in mitochondrial dysfunction in various pathological conditions and diseases.
Keywords: Cardiolipin, mass spectrometry, data independent acquisition, MS/MSALL, shotgun lipidomics
Introduction
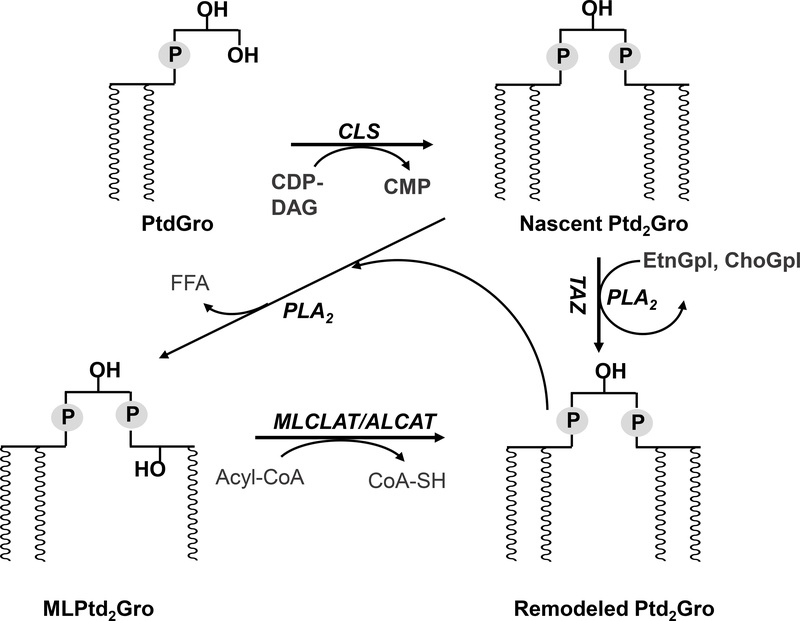
In mammalian cells, cardiolipin (Ptd2Gro) is found almost exclusively in the inner mitochondrial membrane where it plays an essential role in mitochondrial bioenergetics [1–3]. In eukaryotes, Ptd2Gro is biosynthesized from phosphatidylglycerol (PtdGro) and cytidine diphosphate-diacylglycerol (CDP-DAG) by Ptd2Gro synthase on the inner face of the inner mitochondrial membrane [1]. Nascent Ptd2Gro is remodeled by a deacylation-reacylation process, which cleaves acyl chains from Ptd2Gro, generating monolysocardiolipin (MLPtd2Gro) and, via reacylation from donor chains (Figure 1), allows for the enrichment of certain acyl chains, especially linoleic acid (18:2), on the backbone of Ptd2Gro in the majority of mammalian tissues [4]. Additional remodeling pathways of Ptd2Gro involve the acyl CoA independent transfer of 18:2 acyl chains directly from choline glycerophospholipids (ChoGpl) or ethanolamine glycerophospholipids (EtnGpl) via tafazzin (TAZ) [5].
Figure 1. Cardiolipin remodeling in mammalian cells.
ALCAT, acyl-CoA:lysocardiolipin acyltransferase; CDP-DAG, cytidine diphosphate-diacylglycerol; Ptd2Gro, cardiolipin; CLS, cardiolipin synthase; CMP, cytidine monophosphate; MLPtd2Gro, monolysocardiolipin; MLCLAT, monolysocardiolipin acyltransferase; ChoGpl, choline glycerophospholipids; EtnGpl, ethanolamine glycerophospholipids; PtdGro, phosphatidylglycerol; PLA2, phospholipase A2; TAZ, tafazzin.
The molecular structure and composition of Ptd2Gro enhances the functional activity of mitochondrial respiratory chain complexes by maintaining the balance of biophysical and charge properties of the mitochondrial inner membrane [3, 6]. In addition to its role in mitochondrial bioenergetics, Ptd2Gro also plays an important regulatory role in cytochrome C release, which triggers downstream signaling events in apoptosis [7]. Abnormalities in Ptd2Gro composition have been associated with mitochondrial dysfunction in multiple tissues in a variety of pathological conditions, including Barth syndrome [8], ischemia [9], thyroid neoplasia [10], traumatic brain injury [11], and brain tumors [12].
Quantitative analysis of Ptd2Gro, particularly at the molecular species level, is largely based on electrospray ionization mass spectrometry (ESI-MS), either coupled with normal phase high performance liquid chromatography (HPLC) [13, 14], or by direct infusion, i.e. shotgun lipidomics [15]. Under negative mode ESI, cardiolipin species are easily formed as double-charged ions with a specific isotope distribution ([M-2H]2-, [M-2H]2-+0.5, and [M-2H]2-+1), which could be used for the quantification and identification of Ptd2Gro species [15]. However, under analysis with triple quadrupole (QQQ) mass spectrometers, the lower resolution mass to charge ratio (m/z) of the double charged ion and its isotopic peaks require deconvolution to provide accurate quantification [15]. Furthermore, traditional QQQ MS-based lipidomics requires pre-selection of specific ions for analysis of diverse lipids [15, 16]. Recently, the strategy of data-independent acquisition (DIA) has been developed to overcome this shortcoming by recording all the fragments in isolation windows with various m/z widths [17]. MS/MSALL is one variant of DIA, which utilizes a hybrid quadrupole high-resolution time-of-flight (qTOF) with a 1 m/z isolation window for profiling lipids in biological samples [18, 19]. Herein, we developed an adapted high-resolution MS/MSALL shotgun lipidomics method specifically for Ptd2Gro molecular species with an analysis time of 6 mins. The isolation window and collision energy were adjusted to accommodate the isotopic peak [M-2H]2-+0.5. The adapted Ptd2Gro MS/MSALL shotgun lipidomics method exhibited a lower coefficient of variation (CV) for Ptd2Gro molecular species compared with the conventional MS/MSALL method for other negative lipids.
Utilizing the high-resolution MS/MSALL shotgun lipidomics approach described in this study, the characteristic Ptd2Gro molecular species in lipid extracts of mouse heart, kidney, liver, muscle, pancreas, brown adipose tissue (BAT), white adipose tissue (WAT), and brain were quantified. The high sensitivity and wider dynamic range for quantification is valuable for studying alterations in Ptd2Gro metabolism in a variety of metabolically diverse biological samples.
Materials and Methods
Mice
All animal procedures were approved by the Institutional Animal Use and Care Committee at Joslin Diabetes Center. C57BL/6J male mice (Stock no. 000664) were purchased from The Jackson Laboratory and used at 12 weeks of age. All mice were housed in the Joslin Diabetes Center Animal Care Facility. At 25 weeks of age, mice were sacrificed and dissected. Tissues were snap frozen in liquid nitrogen.
Materials
Ptd2Gro 14:0–14:0–14:0–14:0 standard, heart Ptd2Gro, egg PtdOH, and egg PtdGro standards were purchased from Avanti Polar Lipids (Alabaster, AL). All solvents were HPLC or LC/MS grade and were purchased from Fisher Scientific (Waltham, MA) or VWR International (Radnor, PA). Mouse tissues were provided by Joslin Diabetes Center and were frozen before the extraction. All animal procedures were approved by the Institutional Animal Use and Care Committee at Joslin Diabetes Center.
Sample Preparation and Extraction
20 mg mouse tissues samples were transferred to an OMNI bead tube with 0.8 mL 10× diluted PBS and were homogenized for 2 min at 4 °C using the OMNI Cryo Bead Ruptor 24 (OMNI International, Inc., Kennesaw, GA). Protein concentration was measured using BCA assay, and 1 mg of protein was used for extraction with Ptd2Gro 14:0–14:0–14:0–14:0 added as the internal standard (IS). Lipid extraction was performed using a modified Bligh and Dyer method as previously described [19]. Extraction was automated using a customized sequence on a Hamilton Robotics STARlet system (Hamilton, Reno, NV) to meet the high-throughput needs. Lipid extracts were dried under N2 and reconstituted in chloroform/methanol (1:1, by vol). Samples were flushed with N2 and stored at −20 °C.
To study the dynamic range of Ptd2Gro under the ESI-MS, the synthetic Ptd2Gro standard, Ptd2Gro 14:0–14:0–14:0–14:0, was spiked at different serial concentrations into 1 mg protein extract from mouse heart homogenate to generate the calibration curves. For assay reproducibility, 5 aliquots of 1 mg heart homogenate were extracted with IS and analyzed using two different methods – one for negative lipids, including phosphatidylserine (PtdSer), phosphatidylglycerol (PtdGro), phosphatidylinositol (PtdIns), and phosphatidic acid (PtdOH), and the other method dedicated for the Ptd2Gro species. The coefficient of variation (CV) was calculated for comparison.
To assess carry over, aliquots of 10 nmol each for heart Ptd2Gro, egg PtdOH, and egg PtdGro extracts with the appropriate mixture of internal standards cocktail were prepared in 0.4 mL chloroform/methanol (1:1, v/v), separately and analyzed using the described cardiolipin mass spectrometry method.
Direct Infusion Quadruple Time of Flight (QTOF) MS/MSALL Mass Spectrometry
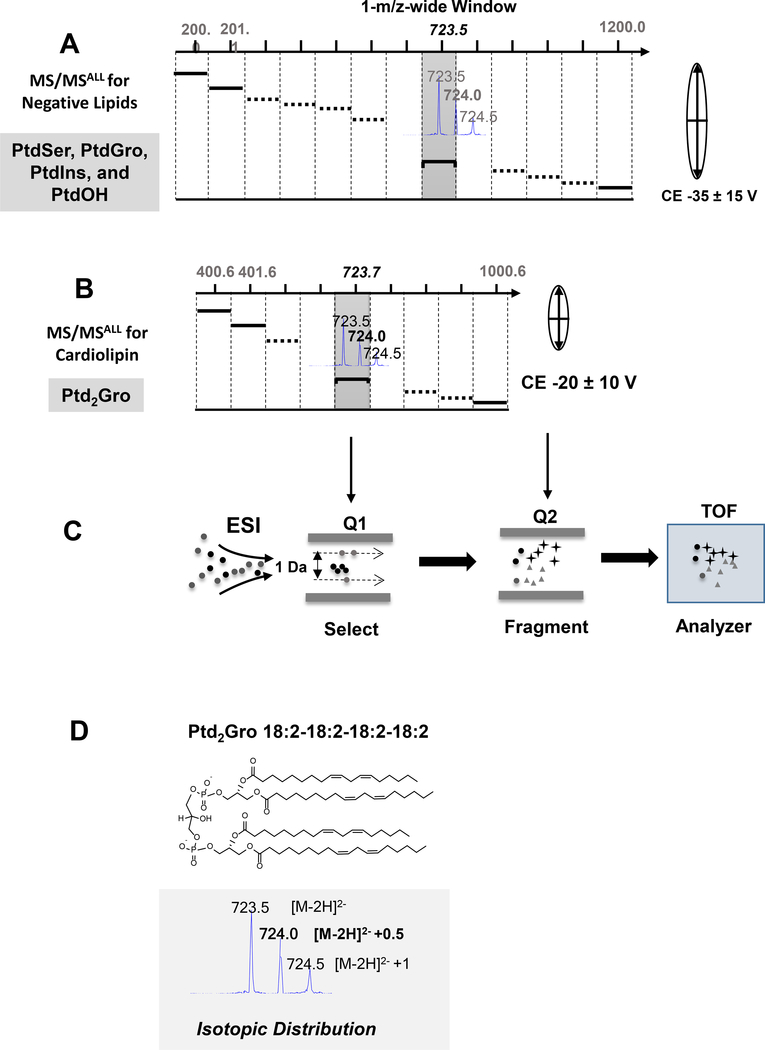
Concentrated samples were diluted 50× in isopropanol-methanol-acetonitrile-H2O (3:3:3:1, by vol) with 2 mM ammonium acetate, and 50 μL diluted lipid extract was automatically loaded and directly delivered to the ESI source using an Ekspert microLC 200 system (SCIEX, Framingham, MA) with a flow rate of 6 μL/min on a customized sample loop. The mass spectrometry experiments were carried out on a TripleTOF 5600+ (SCIEX). ESI source parameters included nebulizing gasses GS1 at 10, GS2 at 10, curtain gas at 15, ion spray voltage at −4500 V, declustering potential at 100 V, and temperature at 300 °C. The atmospheric pressure chemical ionization probe was connected to a calibrant pump which delivers mass calibration solution for MS and MS/MS. The MS/MSALL data acquisition was controlled by Analyst® software (SCIEX). For the conventional negative lipid MS/MSALL analysis [18, 19] all precursors are selected in the Q1 quadrupole at 1 Da isolation window in the MS range was 200–1200, in which the precursor ions are equally spread in one of the isolation windows (Figure 2). The collision energy for negative lipids MS/MSALL was −30 ± 15 V. For the Ptd2Gro species, the MS range was shortened to 400–1000 with the isolation window shifting to fit the double charged isotopologue Ptd2Gro species (Figure 2). In addition, the optimal collision energy for Ptd2Gro species was −20 ± 10 V. The identification of Ptd2Gro species was based on the high-resolution molecular weight, the [M-2H]2- +0.5 isotope, and the specific fatty acyl chain fragments. The quantification was performed by comparing the peak area of molecular species isotope, M+0.5, to that of the internal standard within a linear dynamic range with the isotopic corrections [15] and normalized to 1 mg protein across different tissues.
Figure 2. Data independent acquisition MS/MSALL schemes for common negative lipids and cardiolipin.
(A) MS/MSALL for common negative lipids. All precursors are selected in the Q1 at a 1-m/z wide isolation window across 200 to 1200 m/z. Each isolation window is selected to accommodate singly charged negative lipid species with a higher collision energy. (B) MS/MSALL for Ptd2Gro. The Ptd2Gro precursors are selected in Q1 at a 1-m/z wide isolation window across 400 to 1000 m/z with low collision energy and each single adjusted isolated window to fit the doubly charged Ptd2Gro species and isotopic peaks (see inset). (C) ESI-QTOF-MS/MSALL workflow. (D) The molecular structure and isotopic distribution of Ptd2Gro 18:2–18:2–18:2–18:2 under negative ESI conditions. CE, collision energy; ESI, electrospray ionization; PtdOH, phosphatidic acid; PtdGro, phosphatidylglycerol; PtdIns, phosphatidylinositol; PtdSer, phosphatidylserine; Q, quadrupole; TOF, time of flight mass spectrometer.
Results
Data independent acquisition (DIA) MS/MSALL for cardiolipin molecular species
Under negative ESI, Ptd2Gro molecular species demonstrate different ionization compared to other negatively charged phospholipids through the formation of a double charged molecular ion. This double charged molecular ion demonstrated an isotopic pattern with [M-2H]2-, [M-2H]2-+0.5, [M-2H]2-+1 peaks (Figure 2D). Using a previously assigned strategy for shotgun lipidomics [15], the diagnostic [M-2H]2-+0.5 peak was exploited for the identification and quantification of Ptd2Gro using the high-resolution DIA MS/MSALL shotgun lipidomics strategy. Applying an adapted MS/MSALL approach for Ptd2Gro compared to the conventional analysis method for negative phospholipids (Figure 2A), the Ptd2Gro precursors were selected at a shifted 1-m/z wide isolation window in Q1 in order to accommodate the double charged [M-2H]2-+0.5 peak with a narrow scan range from 400 to 1000 m/z (Figure 2B). In addition, the collision energy (CE) for Ptd2Gro was lower than the other negative phospholipids in order to preserve both double charged molecular ions and fragments. This process is illustrated in Figure 2C in which the precursor ions are isolated in a 1-m/z wide window in Q1, fragmented in Q2 using ramping CE and the molecular ions and fragment ions were all analyzed by the high-resolution TOF MS analyzer at a high scan speed.
Optimization of High-resolution MS/MSALL shotgun lipidomics for Ptd2Gro molecular species
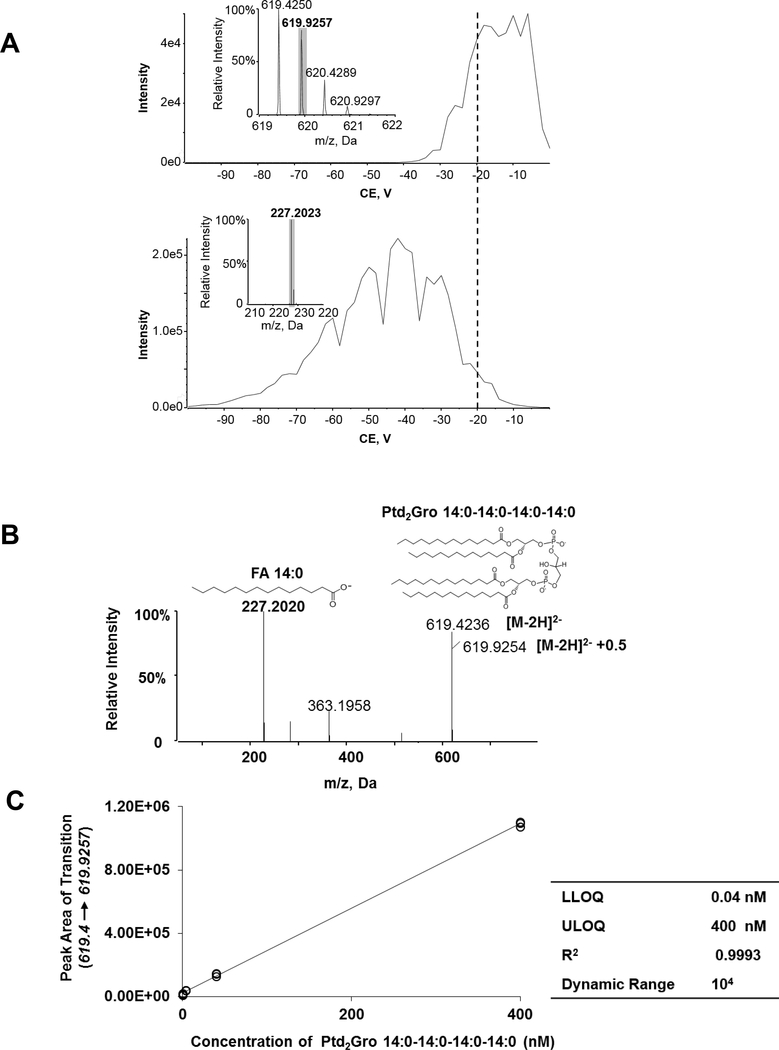
Ptd2Gro species were isolated in the 1-m/z isolation window by the Q1, and then dissociated in the collision cell (Figure 2). The collision energy (CE) is a key factor that influences the isotopic peaks and the fragments of the Ptd2Gro species and requires optimization. 1 nM Ptd2Gro 14:0–14:0–14:0–14:0 solution was infused into the mass spectrometer and the double charged molecular ions were fragmented under a ramping CE ranging from −100 to 0 V (Figure 3A). The m/z 619.9 of the isotopic peak [M-2H]2-+0.5 and the m/z 227.2 of the fatty acid fragment peak were monitored during the infusion. The optimal CE range for the [M-2H]2-+0.5 peak was between −20 and −5 V. We chose a ramping CE as −20 ± 10 V as our final CE for Ptd2Gro species to produce both stable [M-2H]2-+0.5 and the diagnostic fatty acid fragment (Figure 3B). Quantification using the isotopic peak [M-2H]2-+0.5 under lower resolution mass spectrometers was reported previously [8, 15]. To determine the linear dynamic range and sensitivity of the high-resolution DIA method for Ptd2Gro species quantification, a serial dilution of Ptd2Gro 14:0–14:0–14:0–14:0 in one aliquot of 1 mg mouse heart homogenate was analyzed and the calibration curve ranging from 0.04–400 nM was established with an R2 regression value of 0.9993 (Figure 3C).
Figure 3. Optimization of high-resolution shotgun lipidomics for cardiolipin.
(A) Plot of Intensity of the isotopic [M-2H]2- +0.5 peak (m/z 619.9257) (upper panel) and of the fatty acid fragment m/z 227.2023 (lower panel) of Ptd2Gro 14:0–14:0–14:0–14:0 against varying rolling collision energy (CE). (B) MS/MS spectra and fragments of Ptd2Gro 14:0–14:0–14:0–14:0 at the CE=−20 V. (C) The peak area of the isotopic ion [M-2H]2- +0.5 (619.4 → 619.9257) against the concentration of Ptd2Gro 14:0–14:0–14:0–14:0 yielded a linear curve (R2 = 0.9993) when spiked in mouse tissue. The insert table shows the related quantitative parameters. The spectra were acquired using the MS/MSALL for cardiolipin under the following conditions: GS1=10, GS2=10, CUR=15, Temp=300 °C, DP=100 V. CE, collision energy; Ptd2Gro, cardiolipin; CUR, curtain gas; DP, declustering potential; FA, fatty acid; GS, nitrogen gas flow; LLOQ, the lower limit of quantification; ULOQ, the upper limit of quantification.; Temp, temperature.
Specificity of High-resolution MS/MSALL shotgun lipidomics for Ptd2Gro molecular species
To determine the specificity of the adapted cardiolipin MS/MSALL method, we examined potential interference of other Ptd2Gro analysis with other phospholipids standards, heart Ptd2Gro vs egg PtdOH or egg PtdGro. We chose PtdOH and PtdGro extracts because both of phospholipids ionize in the negative mode and share a similar mass range that partially overlaps with Ptd2Gro. Analysis of egg PtdOH and egg PtdGro stocks revealed less than 3% interference for the analysis of cardiolipin, which could be associated with background (Supplemental Figure 1).
Assessment of variation for the Ptd2Gro MS/MSALL method for quantifying molecular species
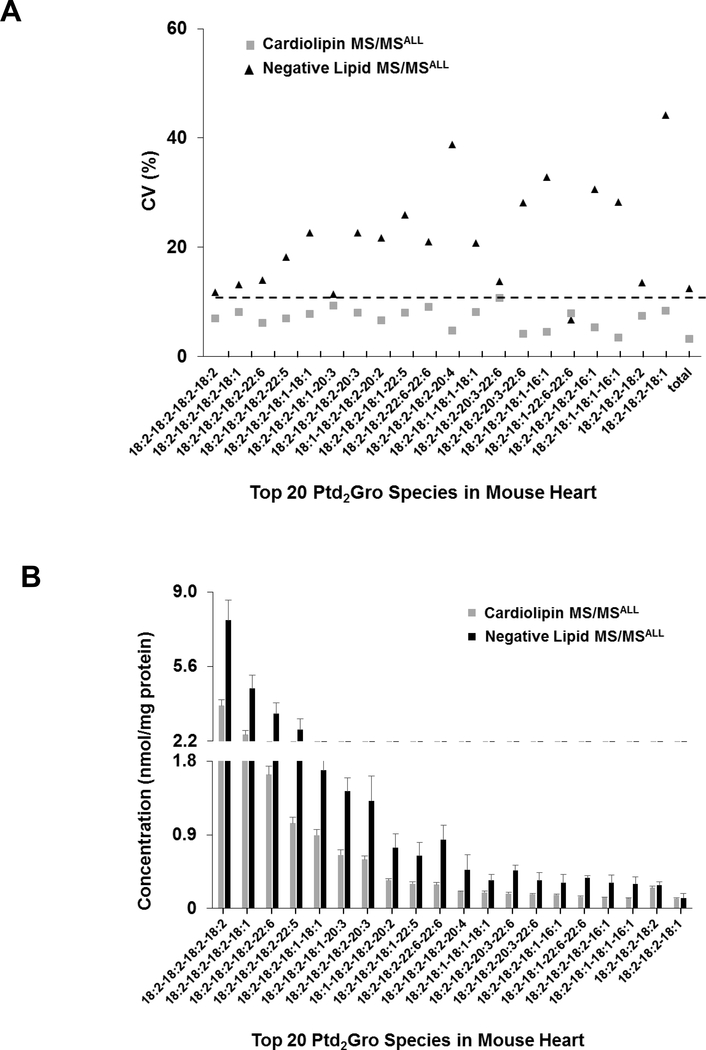
To assess the reproducibility of Ptd2Gro adapted MS/MSALL method, 5 aliquots of 1 mg mouse heart extract were analyzed using the method for the conventional negative lipid analysis as well as the adapted method for Ptd2Gro analysis (as described in the methods section). The coefficient of variation (CV) of the top 20 Ptd2Gro species in the mouse heart tissue using the novel Ptd2Gro method were less than 10%, whereas the CV of most Ptd2Gro using the method for other negative lipids was higher than 10% (Figure 4), most likely because the isotopic peaks are near the edge of the isolation window and these ions become less stable than the ions in the middle of the window. The quantitative value of cardiolipin species also demonstrated minimal variation in replicates from the adapted MS/MSALL Ptd2Gro workflow compared to negative MS/MSALL analysis (Figure 4B). The lower variation in the new adapted cardiolipin method compared to the standard negative MS/MSALL method, may in part be due to lower collision energy creating more stable ions for the 0.5 isotopologue compared to the higher CE in the negative MS/MSALL method which might be more instable creating more variability.
Figure 4. Assessment of the two different methods-MS/MSALL for negative lipids and MS/MSALL for cardiolipin.
(A) The coefficient of variation (CV) and (B) the concentration profile of the top 20 Ptd2Gro species from mouse heart tissue. Mass spectrometry conditions: GS1=10, GS2=10, CUR=15, Temp=300 °C, DP=100 V, CE=−35 ± 15 V and −20 ± 10 V for the negative lipid MS/MSALL and the Ptd2Gro MS/MSALL methods, respectively. Data are expressed as mean ± SD (n=5).
Analysis of Ptd2Gro molecular species from different mouse tissues
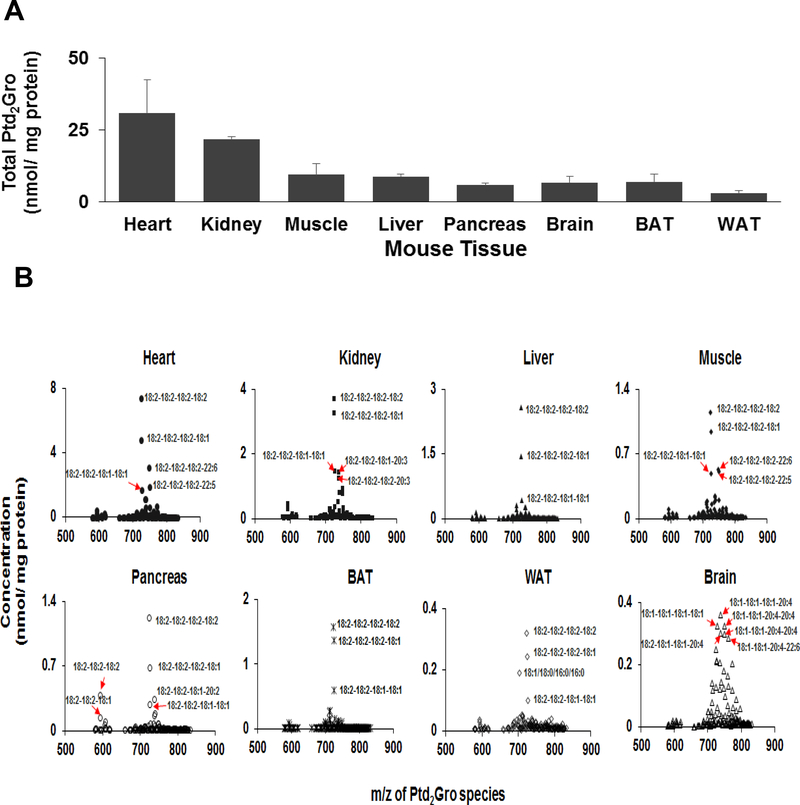
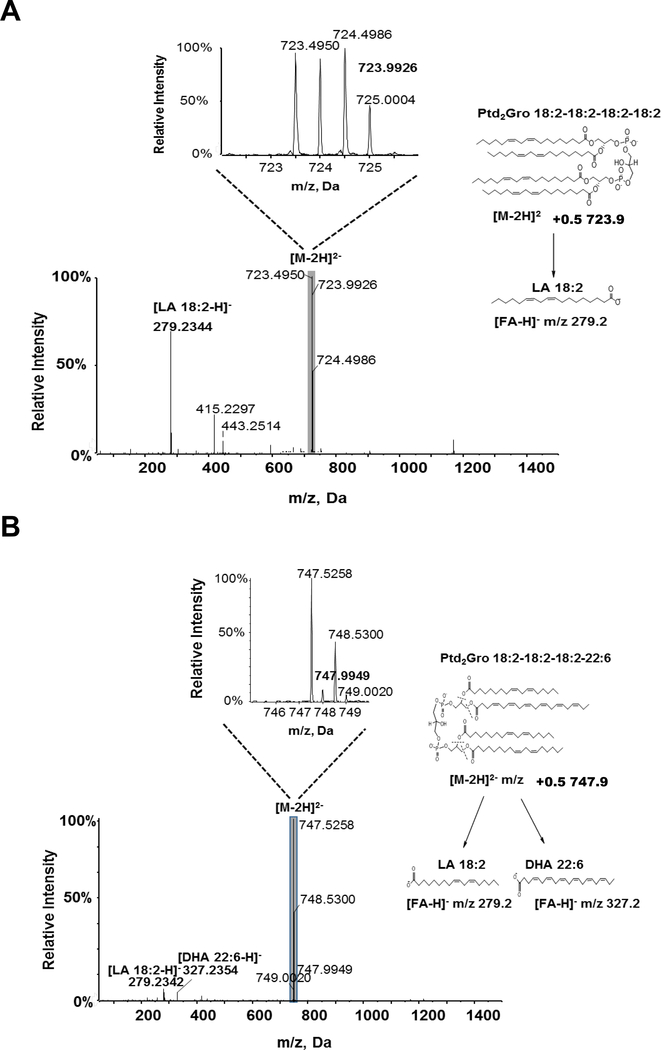
We applied the adapted MS/MSALL DIA Ptd2Gro shotgun lipidomics method to quantify the Ptd2Gro molecular species from various mouse tissues, including heart, kidney, liver, muscle, pancreas, brown adipose tissue (BAT), white adipose tissue (WAT), and brain (Figure 5). The molecular structure of Ptd2Gro species was identified by the [M-2H]2- 0.5 isotopic peaks and the MS/MS spectra, as illustrated in Figure 6 where endogenous Ptd2Gro 18:2–18:2–18:2–18:2 and Ptd2Gro 18:2–18:2–18:2–22:6 in the mouse heart were confirmed. Across the profile of Ptd2Gro molecular species, two separate clusters of MLPtd2Gro (around m/z 600) and Ptd2Gro (between m/z 650–850 ranges) were clearly present. Amongst all tissues, the heart contained the highest content of Ptd2Gro per milligram of protein, followed by kidney and liver (Figure 5A). The dominant Ptd2Gro species in all tissues (Supplemental Table 1), except brain (Supplemental Table 2), were 18:2–18:2–18:2–18:2 and 18:2–18:2–18:2–18:1 as previously reported [15] (Figure 5B). For mouse brain tissue, the major Ptd2Gro species were enriched with oleic acid (FA 18:1) and polyunsaturated fatty acids (PUFA), such as arachidonic acid (ARA) and docosahexaenoic acid (DHA).
Figure 5. Cardiolipin species profiles from different mouse tissues.
(A) The total Ptd2Gro and (B) the Ptd2Gro molecular species profile from heart, kidney, liver, muscle, pancreas, brown adipose tissue, white adipose tissue demonstrates the predominant Ptd2Gro species present is 18.2–18.2–18.2–18.2, whereas brain Ptd2Gro species are vastly different as previously described [12, 25, 26]. The spectra were acquired using the MS/MSALL for Ptd2Gro method under the following conditions: GS1=10, GS2=10, CUR=15, Temp=300 °C, DP=100 V, CE=−20 ± 10 V. Data are expressed as mean ± SD (n=3).
Figure 6. MS/MS spectra and fragmentation pattern for Ptd2Gro species identified from mouse heart.
(A) Species Ptd2Gro 18:2–18:2–18:2–18:2 and (B) 18:2–18:2–18:2–22:6 were identified from mouse heart. Insets: The enlarged isotopic distribution of [M-2H]2- of Ptd2Gro species. The spectra were obtained using the MS/MSALL for Ptd2Gro under the following conditions: GS1=10, GS2=10, CUR=15, Temp=300 °C, DP=100 V, CE=−20 ± 10 V. DHA, docosahexaenoic acid; LA, linoleic acid
Discussion
Ptd2Gro is a polyglycerol phospholipid composed of two phosphatidyl moieties and four fatty acyl chains, and plays an essential role in mitochondrial function [3, 20]. Disruption of Ptd2Gro content or composition in mammalian cells has been related to the initiation of mitochondrial dysfunction in various disease states [1, 2]. The complexity and low abundance of Ptd2Gro in most tissues poses a bioanalytical challenge to accurately quantify Ptd2Gro at the molecular species level. Under ESI conditions, Ptd2Gro forms as a double negative-charged ion with a distinctive isotopic distribution, the characteristics of which could be exploited for Ptd2Gro identification and quantification [15]. However, most previous studies were performed with lower resolution mass spectrometers, such as QQQ, and the isotopic peaks can overlap and requires deconvolution for accurate quantitation [15]. In this study, we applied an adapted high-resolution qTOF mass spectrometry along with the DIA MS/MSALL strategy to accurately profile the Ptd2Gro molecular species from different mouse tissues. The isotopic peaks were clearly identified and used for subsequent quantification. The MS/MSALL isolation window, the collision energy, and other parameters were optimized to provide a high-throughput, sensitive method for specifically quantifying Ptd2Gro molecular species from various biological samples.
Data independent acquisition (DIA) is a strategy of molecular analysis in which all ions within a selected m/z window are fragmented and analyzed [17]. The DIA methods has been widely used for proteomics [21], metabolomics [22], and lipidomics [18]. Compared with traditional data dependent acquisition (DDA) methods, DIA overcomes the disadvantages of irreproducibility and under-sampling issues of DDA [17] by covering all fragments of all molecules within the sample. MS/MSALL is a DIA method developed specifically for lipidomics [18, 19]. The MS/MSALL approach uses 1 m/z isolation window to select the molecular ions, which not only collects all the fragments from the precursor ions, but also maintains the connections between the precursors and fragments. For Ptd2Gro analysis, we adjusted the conventional method by shifting the window edges and lowering the collision energy to accommodate the [M-2H]2- +0.5 isotopic ion to achieve more stable signals and lower variance between signals.
More than 150 Ptd2Gro molecular species, including MLPtd2Gro and Ptd2Gro, were quantified from eight mouse tissues. Amongst the tissues investigated, heart provided the highest content of Ptd2Gro, with the dominant species of Ptd2Gro being 18:2–18:2–18:2–18:2. Our findings are in line with previous results analyzed by QQQ [15]. In addition to heart, liver, pancreas, kidney and muscle, we also determined Ptd2Gro molecular species in brown adipose tissue (BAT) and white adipose tissue (WAT). The Ptd2Gro content of BAT is more than twice that of WAT, indicating higher amounts of mitochondria and the high metabolic activity of BAT, which helps regulate energy expenditure by thermogenesis [23, 24]. The Ptd2Gro profile from brain tissue is quite different from other tissues and contains more oleic acid (18:1) and polyunsaturated fatty acids (PUFA) such as ARA and DHA. The Ptd2Gro species we detected from brain are consistent with previously reported studies [12, 25, 26].
In summary, we developed an adapted high-resolution DIA MS/MSALL approach to accurately quantify and profile Ptd2Gro molecular species from biological samples. The specialized chemical properties of the double charged isotopic molecular ions under the negative ESI mode were used to accurately analyze a multitude of individual Ptd2Gro molecular species, including MLPtd2Gro species. The method is reproducible and sensitive with limits of quantification as low as 40 pM. The workflow presented herein offers an efficient and accurate approach for quantitation of Ptd2Gro molecular species in biological samples and could be extended for screening abnormalities of Ptd2Gro metabolism in various disease states.
Supplementary Material
Major Ptd2Gro molecular species in heart Ptd2Gro, egg PtdOH, and egg PtdGro. An aliquot of each extract around 10 nmol each was analyzed using the internal standard cocktail and subsequently analyzed using the Ptd2Gro MS/MSALL method.
Acknowledgements
This work was supported in part by US National Institutes of Health (NIH) grants R01DK077097 (to Y.-H.T.) a research grant from the American Diabetes Foundation (ADA 7-12-BS-191, to Y.-H.T.) and by funding from the Harvard Stem Cell Institute (to Y.-H.T.). M.D.L was supported by NIH fellowships (T32DK007260 and F32DK102320). This study was also funded by BERG, LLC. The authors would like to thank K. Wilkinson for scientific writing assistance.
Footnotes
Conflict of Interest
FG, JM, EC, HR, CN, RS, NN and MK are current employees of BERG, LLC and have stock options. NN and RS are co-founders of BERG, LLC.
References
- 1.Chicco AJ, and Sparagna GC (2007) Role of cardiolipin alterations in mitochondrial dysfunction and disease. American journal of physiology Cell physiology 292 : C33–44 [DOI] [PubMed] [Google Scholar]
- 2.Claypool SM, and Koehler CM (2012) The complexity of cardiolipin in health and disease. Trends in biochemical sciences 37 : 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paradies G, Paradies V, De Benedictis V, Ruggiero FM, and Petrosillo G (2014) Functional role of cardiolipin in mitochondrial bioenergetics. Biochimica et biophysica acta 1837 : 408–417 [DOI] [PubMed] [Google Scholar]
- 4.Hoch FL (1992) Cardiolipins and biomembrane function. Biochimica et biophysica acta 1113 : 71–133 [DOI] [PubMed] [Google Scholar]
- 5.Xu Y, Kelley RI, Blanck TJJ, and Schlame M (2003) Remodeling of Cardiolipin by Phospholipid Transacylation. Journal of Biological Chemistry 278 : 51380–51385 [DOI] [PubMed] [Google Scholar]
- 6.Schlame M, Rua D, and Greenberg ML (2000) The biosynthesis and functional role of cardiolipin. Progress in lipid research 39 : 257–288 [DOI] [PubMed] [Google Scholar]
- 7.McMillin JB, and Dowhan W (2002) Cardiolipin and apoptosis. Biochimica et biophysica acta 1585 : 97–107 [DOI] [PubMed] [Google Scholar]
- 8.Kiebish MA, Yang K, Liu X, Mancuso DJ, Guan S, Zhao Z, Sims HF, Cerqua R, Cade WT, Han X, and Gross RW (2013) Dysfunctional cardiac mitochondrial bioenergetic, lipidomic, and signaling in a murine model of Barth syndrome. Journal of Lipid Research 54 : 1312–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lesnefsky EJ, Slabe TJ, Stoll MS, Minkler PE, and Hoppel CL (2001) Myocardial ischemia selectively depletes cardiolipin in rabbit heart subsarcolemmal mitochondria. American journal of physiology Heart and circulatory physiology 280 : H2770–2778 [DOI] [PubMed] [Google Scholar]
- 10.Zhang J, Yu W, Ryu SW, Lin J, Buentello G, Tibshirani R, Suliburk J, and Eberlin LS (2016) Cardiolipins Are Biomarkers of Mitochondria-Rich Thyroid Oncocytic Tumors. Cancer Research 76 : 6588–6597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ji J, Kline AE, Amoscato A, Samhan-Arias AK, Sparvero LJ, Tyurin VA, Tyurina YY, Fink B, Manole MD, Puccio AM, Okonkwo DO, Cheng JP, Alexander H, Clark RSB, Kochanek PM, Wipf P, Kagan VE, and Bayir H (2012) Lipidomics identifies cardiolipin oxidation as a mitochondrial target for redox therapy of brain injury. Nat Neurosci 15 : 1407–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiebish MA, Han X, Cheng H, Chuang JH, and Seyfried TN (2008) Cardiolipin and electron transport chain abnormalities in mouse brain tumor mitochondria: lipidomic evidence supporting the Warburg theory of cancer. Journal of Lipid Research 49 : 2545–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valianpour F, Wanders RJ, Barth PG, Overmars H, and van Gennip AH (2002) Quantitative and compositional study of cardiolipin in platelets by electrospray ionization mass spectrometry: application for the identification of Barth syndrome patients. Clinical chemistry 48 : 1390–1397 [PubMed] [Google Scholar]
- 14.Sparagna GC, Johnson CA, McCune SA, Moore RL, and Murphy RC (2005) Quantitation of cardiolipin molecular species in spontaneously hypertensive heart failure rats using electrospray ionization mass spectrometry. Journal of Lipid Research 46 : 1196–1204 [DOI] [PubMed] [Google Scholar]
- 15.Han X, Yang K, Yang J, Cheng H, and Gross RW (2006) Shotgun lipidomics of cardiolipin molecular species in lipid extracts of biological samples. Journal of Lipid Research 47 : 864–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han X, and Gross RW (2005) Shotgun lipidomics: electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass spectrometry reviews 24 : 367–412 [DOI] [PubMed] [Google Scholar]
- 17.Gillet LC, Navarro P, Tate S, Röst H, Selevsek N, Reiter L, Bonner R, and Aebersold R (2012) Targeted Data Extraction of the MS/MS Spectra Generated by Data-independent Acquisition: A New Concept for Consistent and Accurate Proteome Analysis. Molecular & Cellular Proteomics 11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simons B, Kauhanen D, Sylvänne T, Tarasov K, Duchoslav E, and Ekroos K (2012) Shotgun Lipidomics by Sequential Precursor Ion Fragmentation on a Hybrid Quadrupole Time-of-Flight Mass Spectrometer. Metabolites 2 : 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao F, McDaniel J, Chen EY, Rockwell HE, Drolet J, Vishnudas VK, Tolstikov V, Sarangarajan R, Narain NR, and Kiebish MA (2017) Dynamic and temporal assessment of human dried blood spot MS/MS(ALL) shotgun lipidomics analysis. Nutrition & Metabolism 14 : 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren M, Phoon CK, and Schlame M (2014) Metabolism and function of mitochondrial cardiolipin. Progress in lipid research 55 : 1–16 [DOI] [PubMed] [Google Scholar]
- 21.Distler U, Kuharev J, Navarro P, Levin Y, Schild H, and Tenzer S (2014) Drift time-specific collision energies enable deep-coverage data-independent acquisition proteomics. Nature methods 11 : 167–170 [DOI] [PubMed] [Google Scholar]
- 22.Tsugawa H, Cajka T, Kind T, Ma Y, Higgins B, Ikeda K, Kanazawa M, VanderGheynst J, Fiehn O, and Arita M (2015) MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat Meth 12 : 523–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cannon B, and Nedergaard J (2004) Brown adipose tissue: function and physiological significance. Physiological reviews 84 : 277–359 [DOI] [PubMed] [Google Scholar]
- 24.Cypess AM, Lehman S, Williams G, Tal I, Rodman D, Goldfine AB, Kuo FC, Palmer EL, Tseng Y-H, Doria A, Kolodny GM, and Kahn CR (2009) Identification and Importance of Brown Adipose Tissue in Adult Humans. New England Journal of Medicine 360 : 1509–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bayir H, Tyurin VA, Tyurina YY, Viner R, Ritov V, Amoscato AA, Zhao Q, Zhang XJ, Janesko-Feldman KL, Alexander H, Basova LV, Clark RS, Kochanek PM, and Kagan VE (2007) Selective early cardiolipin peroxidation after traumatic brain injury: an oxidative lipidomics analysis. Annals of neurology 62 : 154–169 [DOI] [PubMed] [Google Scholar]
- 26.Cheng H, Mancuso DJ, Jiang X, Guan S, Yang J, Yang K, Sun G, Gross RW, and Han X (2008) Shotgun Lipidomics Reveals the Temporally Dependent, Highly Diversified Cardiolipin Profile in the Mammalian Brain: Temporally Coordinated Postnatal Diversification of Cardiolipin Molecular Species with Neuronal Remodeling. Biochemistry 47 : 5869–5880 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Major Ptd2Gro molecular species in heart Ptd2Gro, egg PtdOH, and egg PtdGro. An aliquot of each extract around 10 nmol each was analyzed using the internal standard cocktail and subsequently analyzed using the Ptd2Gro MS/MSALL method.