Abstract
The role of long non-coding RNA (lncRNA) in idiopathic pulmonary fibrosis (IPF) is poorly understood. We found a novel lncRNA-ITPF that was upregulated in IPF. Bioinformatics and in vitro translation verified that lncITPF is an actual lncRNA, and its conservation is in evolution. Northern blot and rapid amplification of complementary DNA ends were used to analyze the full-length sequence of lncITPF. RNA fluorescence in situ hybridization and nucleocytoplasmic separation demonstrated that lncITPF was mainly located in the nucleus. RNA sequencing, chromatin immunoprecipitation (ChIP)-qPCR, CRISPR-Cas9 technology, and promoter activity analysis showed that the fibrotic function of lncITPF depends on its host gene integrin β-like 1 (ITGBL1), but they did not share the same promoter and were not co-transcribed. Luciferase activity, pathway inhibitors, and ChIP-qPCR showed that smad2/3 binds to the lncITPF promoter, and TGF-β1-smad2/3 was the upstream inducer of the fibrotic pathway. Furthermore, RNA-protein pull-down, liquid chromatography-mass spectrometry (LC-MS), and protein-RNA immunoprecipitation showed that lncITPF regulated H3 and H4 histone acetylation in the ITGBL1 promoter by targeting heterogeneous nuclear ribonucleoprotein L. Finally, sh-lncITPF was used to evaluate the therapeutic effect of lncITPF. Clinical analysis showed that lncITPF is associated with the clinicopathological features of IPF patients. Our findings provide a therapeutic target or diagnostic biomarker for IPF.
Keywords: IPF, lncRNA, lncITPF, ITGBL1, TGF-β1-smad2/3, RNA-sequencing, RNA-protein pull-down, CRISPR-Cas9 technology
Although more and more lncRNAs have been identified, their expression patterns, characteristics, and mechanism in IPF remain largely unexplored.
This study showed that lncITPF can epigenetically regulate its host gene ITGBL1 by binding to hnRNP-L, and it provides a potential therapeutic target or diagnostic biomarker for IPF.
Introduction
Idiopathic pulmonary fibrosis (IPF) is a specific form of chronic, progressive, and fibrosing interstitial pneumonia of unknown cause.1 Activated fibroblast migration, proliferation, and extracellular matrix secretion are the prominent features and key drivers of IPF development.2, 3 Thus, fibroblasts have become the focus of research interest in pulmonary fibrosis therapy. However, the underlying molecular mechanisms that mediate fibroblast migration, proliferation, and extracellular matrix secretion still remain unclear.4, 5 Recent reports indicated that IPF is related to epigenetic mechanisms involving long non-coding RNA (lncRNA) and post-translational modification of histones. Thus, researchers have begun to study the restoration of epigenetically modified genes as a new therapeutic strategy.6, 7
The lncRNA comprises more than 200 nt, and it has attracted considerable attention because of its wide range of biological regulatory functions, such as cell migration, proliferation, differentiation, apoptosis, extracellular matrix secretion, and immune response.8, 9 Emerging evidence has purported that lncRNAs are involved in the progression of various diseases, such as cancer, steatohepatitis, and myocardial and liver fibrogenesis.10, 11, 12 For example, the lncRNA Haunt regulates the Hoxa gene cluster during embryonic stem cell differentiation.13 The interaction of nuclear factor κB (NF-κB) with lncRNA is linked to breast cancer metastasis and patient prognosis.14 Although a set of human lncRNAs has been identified, their expression patterns, characteristics, and mechanism in IPF remain largely unexplored. Recently, lncRNA AP003419.16 was reported to be involved in the IPF process, but its underlying working mechanism still needs to be elucidated.15
This study is a follow-up to previous research conducted in our laboratory. Our previous study was the first to report the different lncRNA profiles in IPF.16, 17 However, two critical issues remained to be addressed. First, the clinical significance of lncRNAs for IPF patients was not evaluated. Second, the molecular mechanisms of these RNAs are poorly elucidated. In the present work, we investigated a novel lncITPF molecular mechanism, and we evaluated its potential clinical significance in IPF. We found that lncITPF expression is significantly upregulated in a transforming growth factor β (TGF-β)1-smad2/3-dependent manner. By directly binding to heterogeneous nuclear ribonucleoprotein L (hnRNP-L), lncITPF can epigenetically regulate its host gene integrin β-like 1 (Itgbl1). Clinical analysis showed that lncITPF is associated with the clinicopathological features of patients with IPF. Our findings elucidated the mechanism of the lncRNA in IPF progression, and they provide a potential therapeutic target or diagnostic biomarker for IPF.
Results
lncITPF Verification, Expression, and Conservation
Using microarray analysis, we revealed the differentially expressed profiles of lncRNAs in bleomycin-induced pulmonary fibrosis in rats.16, 17 The RNA transcript of MRAK053938 has significantly higher expression compared with other transcripts. Thus, it was selected for further study and was renamed lncITPF (lncRNA regulates its host gene Itgbl1 during pulmonary fibrogenesis). To verify the results of microarray analysis, we investigated the lncITPF expressions. Hydroxyproline (HYP), a physiological marker of fibrogenesis, was also tested in rats. lncITPF and HYP were highly expressed in lung tissues from rats treated with bleomycin for 7, 14, 21, and 28 days compared with those in the sham group (Figures S1A and S1B). Pearson correlation coefficient indicated that lncITPF was positively correlated with HYP (Figure S1C), thereby suggesting that lncITPF was associated with lung fibrosis development.
In actual cases, many predicted lncRNAs are not real lncRNAs because they can still encode proteins. To verify that lncITPF has no protein-coding potential, we first analyzed its sequence using the open reading frame (ORF) finder from the NCBI. However, we failed to predict a protein of more than 48 amino acids. We further searched the amino acid sequences for the conserved domains from NCBI, and we found that lncITPF does not contain a valid Kozak sequence (Figure S1D). In addition, we evaluated the protein-coding potential of lncITPF using the following tools: Coding Potential Calculator (http://cpc.cbi.pku.edu.cn/), Coding Potential Assignment Tool (http://lilab.research.bcm.edu/cpat/index.php), and Coding-Non-Coding Index (CNCI) tool (https://github.com/www-bioinfo-org/CNCI). The protein-coding ability scores of lncITPF were −1.23513, 0.000109432, and −0.00041, indicating that lncITPF is devoid of protein-coding potential.18, 19, 20 lncITPF translation activities were further measured in an in vitro translation system, which revealed that this gene has no translation activities (Figure 1A). These analyses strongly verified that lncITPF is an actual lncRNA.
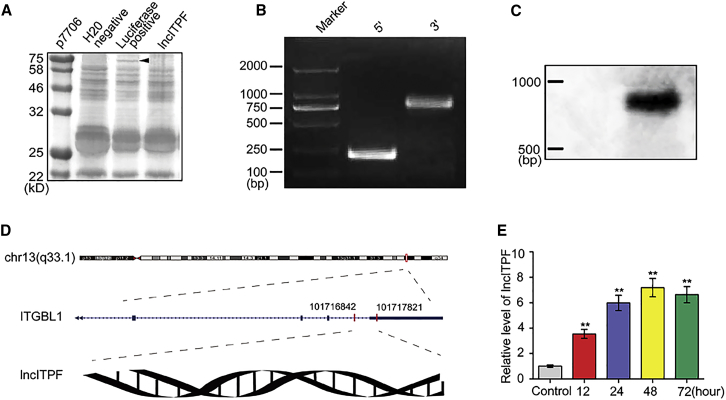
Figure 1.
lncITPF Verification, Expression, and Conservation
(A) In vitro translation assay showed that lncITPF did not exhibit translation activities. The black arrow indicates that the positive control translated a 75-kDa protein. (B) The full-length sequence of lncITPF in the human genome was analyzed via RACE. (C) The size of lncITPF was detected via northern blot and was close to the 1,000-bp length of the human genome. (D) lncITPF maps to chromosome 13 and contains introns and exons of the ITGBL1 gene in the human genome. (E) lncITPF was upregulated in MRC-5 cells treated with 5 ng/mL TGF-β1 for 12, 24, 48, and 72 hr. Each bar represents the mean ± SD; n = 6; **p < 0.01.
To evaluate the lncITPF clinical value for the future research, we first analyzed its conservation in evolution with human orthologs. lncITPF is located in chromosome 15 from 113,759,358 to 113,760,900 in the rat genome. Using a BLAST search of the lncITPF sequence against the human genome, we found a highly homologous sequence located on chromosome 13 of the human genome (Figure S1E). Then, rapid amplification of complementary DNA ends (RACE) was used to characterize the potential 5′ and 3′ ends of this RNA transcript and the full-length sequence in the human genome. lncITPF has a size of 980 bp (Figure 1B; Figure S1F), and this size was confirmed via northern blot (Figure 1C). The lncITPF transcription start and 3′ end sites were mapped in the last intron and exon of a protein-coding gene Itgbl1 in the human genome (Figure 1D). Itgbl1 is also adjacent to the lncITPF loci in rat genomes. These data indicated that Itgbl1 is the host gene of lncITPF.
To identify the cell type in which lncITPF was expressed, lung tissues of rats were obtained, separated, and cultured via primary culture method. We screened out fibroblasts and verified them based on primary cell culture (Figures S2A and S2B). qRT-PCR was used to analyze lncITPF expression in primary fibroblasts and MRC-5, a fibroblast cell line. The data showed that lncITPF was upregulated in primary fibroblasts (Figure S2C) and MRC-5 treated with TGF-β1 for 12, 24, 48, and 72 hr (Figure 1E). These findings indicated that lncITPF expression increased in fibroblasts in vivo and in vitro, which coincided with the results obtained from animal models. Therefore, MRC-5 can be used as a cell model to further study lncITPF function and mechanism.
lncITPF Function in Pulmonary Fibrosis
To determine the function of lncITPF, gain- and loss-of-function analysis was carried out using an lncITPF smart silencer (lncITPF RNAi) and an lncITPF overexpression vector, which was constructed by cloning the full-length sequence of lncITPF into a pcDNA3.1 vector (recombinant plasmid [RP]). qRT-PCR revealed that lncITPF RNAi effectively interfered with lncITPF expression, whereas lncITPF overexpression increased the levels of lncITPF in vitro (Figure S3A). One of the phenomena that occurs during the pulmonary fibrosis process is the manifestation of mesenchymal characteristics, such as α-smooth muscle actin (α-SMA), collagen, and vimentin. Immunofluorescence analysis revealed that α-SMA expression was prevented by lncITPF RNAi and promoted by lncITPF overexpression (Figure 2A). Western blot results showed that the expression levels of α-SMA, collagen, and vimentin were decreased by lncITPF RNAi and increased by lncITPF overexpression (Figure 2B). Another common phenomenon that occurs during the process of pulmonary fibrosis is the high level of migration of activated fibroblasts. A real-time cellular migration analysis system was used to test the migration of activated fibroblasts. The lncITPF RNAi reduced migration, whereas lncITPF overexpression promoted it compared with the control (Figure 2C).
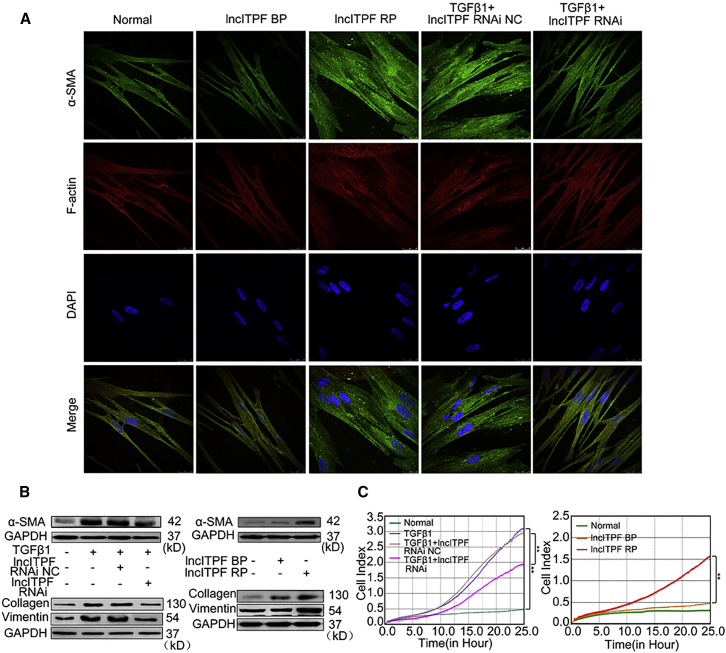
Figure 2.
lncITPF Promoted Pulmonary Fibrosis
(A) α-SMA expression was decreased by lncITPF RNAi and increased by lncITPF overexpression (lncITPF RP). Typical α-SMA staining revealed that cytoplasmic fibers partially co-localized with cell skeleton F-actin. F-actin co-localization was used to validate the α-SMA. α-SMA around the nuclear membrane was stained with FITC (green). The nuclei were counterstained with Hoechst 33258 (blue). F-actin was stained red. (B) Western blot showed that lncITPF RNAi decreased α-SMA, collagen, and vimentin expression levels, whereas lncITPF overexpression increased α-SMA, collagen, and vimentin expression levels. (C) Real-time migration analysis system showed that lncITPF RNAi decreased migration, whereas lncITPF overexpression increased migration. GAPDH served as the control. NC indicates a negative control, BP indicates blank plasmid, and RP indicates the recombinant plasmid of the overexpressed lncITPF. Each bar represents the mean ± SD; n = 6; **p < 0.01.
Regulatory Mechanism of lncITPF on Its Host Gene
Approximately 40% of lncRNAs intersect in protein-coding loci and exhibit a strong pattern of co-expression.21 Thus, lncRNAs exert their regulatory function by specifically interacting with their host genes. So we examined whether lncITPF affects its host gene Itgbl1. First, Itgbl1 expression was determined by using RNA sequencing. Itgbl1 was found to be increased in pulmonary fibrosis and decreased under lncITPF RNAi treatment (Figure 3A). The upregulation of Itgbl1 was confirmed via qRT-PCR in TGF-β1-stimulated cells at different time points (Figure 3B). The expression of Itgbl1 was tested by lncITPF knockin and knockdown. Itgbl1 was evidently downregulated or upregulated when lncITPF was knocked down or knocked in at the mRNA (Figure 3C) and protein levels (Figure 3D), respectively. These data suggested that co-expression exists between lncITPF and Itgbl1.
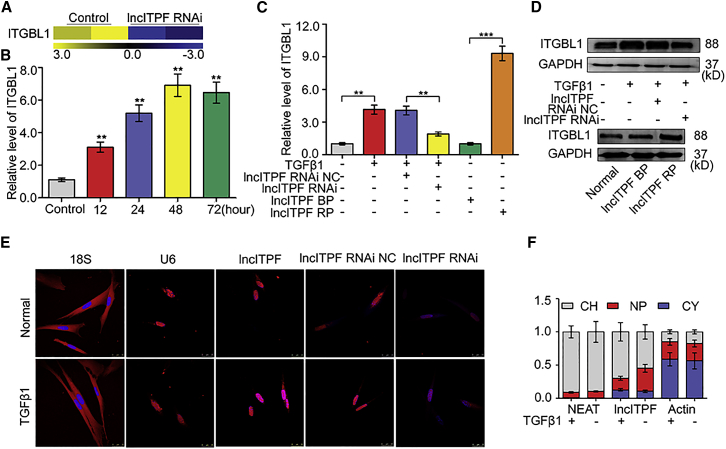
Figure 3.
Regulatory Mechanism of lncITPF in Its Host Gene
(A) Itgbl1 expression was higher in fibrotic tissue than in normal tissue on the basis of RNA sequencing. (B) Itgbl1 was upregulated in cells treated with 5 ng/mL TGF-β1 for 12, 24, 48, and 72 hr. (C) qRT-PCR analysis showed that Itgbl1 was decreased by lncITPF RNAi and increased by lncITPF overexpression. (D) Western blot showed that ITGBL1 was decreased by lncITPF RNAi and increased by lncITPF overexpression. (E) Single-molecule RNA-FISH detecting the location of lncITPF (red) in cells. U6 and 18S RNA were used as cytoplasmic and nuclear localization markers, respectively. DNA (blue) was stained with DAPI. A representative image is shown. (F) qRT-PCR analysis of RNAs purified from nucleoplasmic (red), chromatin nuclear (gray), and cytosolic (blue) compartments in cells. NC indicates a negative control, BP indicates blank plasmid, and RP indicates the recombinant plasmid of the overexpressed lncITPF. Each bar represents the mean ± SD; n = 6; **p < 0.01 and ***p < 0.001 versus the control group.
lncRNAs control their host gene expression via transcriptional or post-transcriptional mechanisms.22 The transcription process usually happens in the nucleus, whereas post-transcription happens in the cytoplasm. Thus, we examined lncITPF localization by performing RNA fluorescence in situ hybridization (FISH) (Figure 3E) and nuclear-cytosol fractionation (Figure 3F). All data showed that lncITPF was predominant in the nucleus with or without TGF-β1 treatment. In addition, more lncITPFs were present in chromatin-associated fractions compared with free nucleoplasmic fractions, especially under TGF-β1 condition. As expected, mature β-actin transcripts were predominantly localized in the cytoplasm, whereas NEAT1 (a nuclear long intergenic ncRNA [lincRNA]) was confined to the nucleus (Figure 3F). Thus, we inferred that lncITPF regulates Itgbl1 transcription.
lncRNA can regulate its host gene by affecting histone modification in the promoter regions. Hence, histone acetylation on Itgbl1 promoters was analyzed under TGF-β1, lncITPF RNAi, and lncITPF overexpression treatments via chromatin immunoprecipitation (ChIP)-qPCR. After immunoprecipitation using antibodies against acetylated H3 and H4, qPCR was used to amplify the chromatin DNAs in the ITGBL1 promoter region. The H3 and H4 histone acetylation levels across the Itgbl1 promoter region increased in both TGF-β1-stimulated and lncITPF-overexpressed cells. The lncITPF RNAi reversed the TGF-β1-induced change, which was concomitant with Itgbl1 deregulation, compared with the control (Figure 4A). However, no changes were found in the total H3 and H4 acetylation levels across the GAPDH after transfecting with lncITPF RNAi or overexpression vector (Figure 4B). These data proved that lncITPF regulates its host gene Itgbl1 by affecting H3 and H4 histone acetylation in its promoter regions.
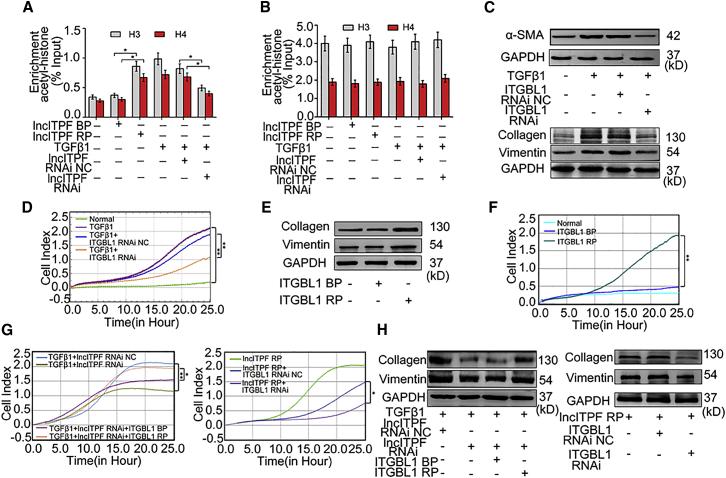
Figure 4.
Regulatory Mechanism of lncITPF in Its Host Gene
ChIP analysis of the level of H3 and H4 histone acetylation of ITGBL1 (A) and GAPDH (B) promoter regions using anti-acetyl-histone H3 and H4. Enrichment was determined relative to input controls. GAPDH was used as the control. (C) Western blot showed that ITGBL1 RNAi decreased α-SMA, vimentin, and collagen expression levels. GAPDH served as the control. (D) Real-time migration system showed that ITGBL1 RNAi decreased the migration ability of cells. Migration was monitored in the xCELLigence DP system. (E) Vimentin and collagen were increased by ITGBL1 overexpression. (F) Migration was increased by ITGBL1 overexpression. (G) Rescue experiment showed that the suppression or induction of lncITPF on migration was partly reversed by ITGBL1. (H) Rescue experiment showed that the suppression or induction of collagen and vimentin expression levels by lncITPF RNAi or lncITPF overexpression was partly reversed by ITGBL1. NC indicates a negative control, BP indicates blank plasmid, and RP indicates the recombinant plasmid of the overexpressed lncITPF or ITGBL1. Each bar represents the mean ± SD; n = 6; *p < 0.05 and **p < 0.01.
From the above findings, we hypothesized that lncITPF acquires its fibrotic function through its host gene Itgbl1. We performed gain- and loss-of-function assays by transfection with Itgbl1-overexpressing vector and specific ITGBL1-small interfering RNA (siRNA) with or without TGF-β1. The efficiency of Itgbl1-siRNA and Itgbl1 overexpression was measured through qRT-PCR (Figure S3B). The three siRNAs could all inhibit Itgbl1 expression. We used the most efficient Itgbl1-siRNA1, namely, Itgbl1 RNAi, to knock down Itgbl1 in the following studies. The knockdown of Itgbl1 (Itgbl1 RNAi) decreased the levels of α-SMA, vimentin, and collagen in TGF-β1-stimulated cells (Figure 4C). Real-time cellular migration analysis showed that Itgbl1 RNAi reduced the migration in TGF-β1-stimulated cells (Figure 4D). On the contrary, Itgbl1 knockin (Itgbl1-overexpressing vector) increased the levels of vimentin, collagen, and migration in ITGBL1-overexpressing cells (Figures 4E and 4F). To determine whether Itgbl1 is necessary for the enhanced function of lncITPF in pulmonary fibrosis, we performed rescue experiments. Cells were simultaneously transfected with lncITPF siRNA and Itgbl1-overexpressed vector under TGF-β1 treatment, and lncITPF-overexpressed vectors and Itgbl1 siRNAs. The suppression or induction of lncITPF on migration, collagen, and vimentin synthesis was partly reversed by Itgbl1 (Figures 4G and 4H). All these data indicated that the fibrotic function of lncITPF depends on its host gene Itgbl1.
Upstream Mechanism of lncITPF in Pulmonary Fibrosis
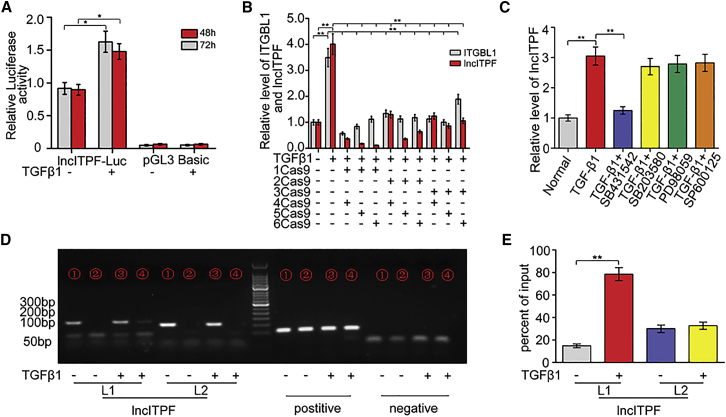
To further elucidate the signaling pathway by which lncITPF regulates pulmonary fibrosis, we evaluated the upstream and downstream mechanisms of lncITPF. Many studies have documented that the TGF-β1-smad2/3 pathway is involved in the upstream signaling pathway of fibrosis.23, 24 Hence, we detected whether the TGF-β1-smad2/3 is an upstream inducer of lncITPF. The promoter region of lncITPF was cloned into the pGL3 luciferase reporter, and the plasmids were transfected into cells with or without TGF-β1 treatment. The plasmids displayed significantly higher luciferase activity than the empty pGL3. Moreover, TGF-β1 increased the lncITPF promoter activity, thereby suggesting that TGF-β1 activated lncITPF transcription (Figure 5A). Considering that lncITPF lies within Itgbl1, we determined whether they are co-transcribed or they share the same promoter. Using the CRISPR-Cas9 technology to knock out the sequence between Itgbl1 and the lncITPF transcription start site with or without TGF-β1, we found that lncITPF had its own promoter located inside the Itgbl1 gene and that lncITPF and Itgbl1 did not share the same promoter nor were they co-transcribed (Figure 5B).
Figure 5.
Upstream Mechanism of lncITPF
(A) TGF-β1 increased the lncITPF promoter activity. Cells transfected with pGL3-lncITPF vectors displayed significantly higher luciferase activity than empty pGL3. Similarly, cells treated with TGF-β1 displayed significantly higher luciferase activity than those without. Cells were transfected with pGL3-lncITPF vectors or an empty vector with or without TGF-β1 treatment for 48 and 72 hr. Luciferase activity was normalized to renilla. (B) Result of Cas9 technology showed that lncITPF had its own promoter located inside the Itgbl1 gene, and they could not be co-transcribed. (C) Signal pathway inhibitors were used to detect the change in lncITPF expression. Only the inhibitor SB431542 of the smad2/3 pathway blocked the expression of lncITPF. (D) ChIP-qPCR analysis of the binding of smad2/3 at the lncITPF promoter region with or without TGF-β1 treatment. ChIP-purified DNA targeting of the indicated genes was analyzed by qPCR. L1 and L2 indicated the different sequences of lncITPF. (1) MRC-5 input; (2) MRC-5 chip DNA; (3) MRC-5 input treated with TGF-β1; (4) MRC-5 chip DNA treated with TGF-β1. (E) The ChIP-qPCR gel images were quantified and analyzed. Each bar represents the mean ± SD; n = 6; *p < 0.05, **p < 0.01, and ***p < 0.001.
Then, we explored which TGF-β1-activated pathway was important by using chemical inhibitors, namely, SB431542, SB203580, PD98059, and SP600125; these inhibitors are specific to smad2/3-, P38MAPK-, ERK-, and JNK-signaling pathways, respectively. Only SB431542 blocked the expression of lncITPF (Figure 5C). To further explore the possibility of smad2/3 functioning as a transcriptional regulator of the lncITPF gene, we tested for smad2/3 binding over the lncITPF locus by ChIP-qPCR. Data obtained with primers, which covered the promoter region of lncITPF, showed increased enrichment of smad2/3 by direct or indirect occupation in the presence of TGF-β1 (Figures 5D and 5E). Thus, lncITPF expression is mediated by the TGF-β1-smad2/3-signaling pathway.
Downstream Mechanism of lncITPF in Pulmonary Fibrosis
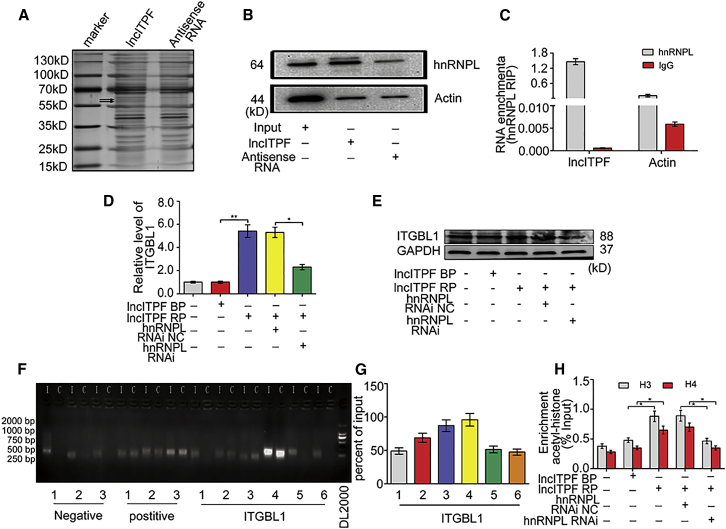
Because lncRNAs regulate their host genes by targeting RNA-binding proteins, we performed biotin RNA-protein pull-down assays by incubating in vitro-transcribed biotinylated lncITPF or its antisense RNA with extracts from TGF-β1-treated cells. lncITPF-bound complex was captured using streptavidin magnetic beads and analyzed using SDS-PAGE. The gel was stained with silver. The specifically enriched protein bands (55–70 kDa) were excised and subjected to mass spectrometry for identification (Figure 6A). This approach identified hnRNP-L as one of lncITPF’s binding targets. The ability of hnRNP-L to bind lncITPF was further confirmed by western blot (Figure 6B). We confirmed the lncITPF-hnRNP-L interaction by purifying endogenous hnRNP-L and analyzing the co-purified RNA by qRT-PCR. The RNA immunoprecipitation assays showed that lncITPF was enriched in hnRNP-L-immunoprecipitated RNAs compared with the control in TGF-β1-treated cells (Figure 6C).
Figure 6.
Downstream Mechanism of lncITPF
(A) SDS-PAGE analysis of proteins purified from in vitro binding assay using biotinylated lncITPF or antisense control RNA. The arrow indicates that the highlighted protein bands were subjected to LC-MS. (B) Protein levels of hnRNP-L in immunoprecipitates with lncITPF RNA were evaluated by western blot. (C) RNA immunoprecipitation assays followed by qRT-PCR analysis of co-purified RNAs with hnRNP-L in cross-linked cells. lncITPF RNA expression levels are presented as fold enrichment values relative to IgG immunoprecipitates. (D) Overexpression of lncITPF increased Itgbl1 mRNA expression, which was impaired in hnRNP-L knockdown. (E) Overexpression of lncITPF increased ITGBL1 protein expression, which was impaired in hnRNP-L knockdown. (F) ChIP-qPCR analysis of hnRNP-L occupancy in ITGBL1 promoter in cells. (G) The ChIP-qPCR gel images were quantified and analyzed. (H) ChIP analysis of the effect of hnRNP-L on H3 and H4 histone acetylation of ITGBL1 promoter regions. NC indicates a negative control, BP indicates blank plasmid, and RP indicates the recombinant plasmid of the overexpressed lncITPF. Each bar represents the mean ± SD; n = 6; *p < 0.05 and **p < 0.01.
Next, we knocked down hnRNP-L using three independent small hairpin RNA (shRNA) lines with distinct non-overlapping sequences (Figure S3C) to detect whether hnRNP-L is involved in the regulation of lncITPF on Itgbl1 expression. As expected, an overexpressed lncITPF-induced increase of Itgbl1 mRNA and protein levels was impaired in hnRNP-L knockdown cells compared with the control cells (Figures 6D and 6E). We further performed ChIP-qPCR using six primers to explore whether hnRNP-L directly binds to the promoter regions of ITGBL1. The hnRNP-L covered the promoter region of Itgbl1 in a direct or indirect manner (Figures 6F and 6G). Moreover, the specific hnRNP-L siRNA caused decreased histone H3 and H4 acetylation levels at the Itgbl1 promoter compared with control cells under lncITPF overexpression (Figure 6H). These results indicated that lncITPF positively regulates the H3 and H4 acetylation of the Itgbl1 gene by interacting with hnRNP-L.
lncITPF as a Potential Therapeutic Target for Pulmonary Fibrosis
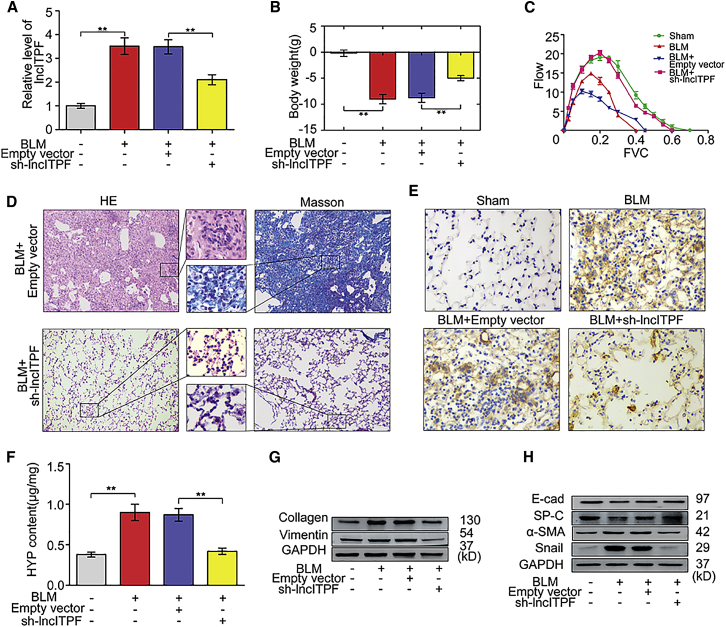
To evaluate the potential of lncITPF as a therapeutic target, we synthesized the sh-lncITPF packaged into the adenovirus vector in animal experiments. The interference efficiency of sh-lncITPF was detected via qRT-PCR (Figure 7A). Body weight monitoring revealed that bleomycin-treated mice lost substantial body mass compared with sham mice. However, sh-lncITPF effectively blocked this loss compared with the control vectors (Figure 7B). Lung function assessment demonstrated that the forced vital capacity (FVC) was improved in sh-lncITPF mice compared with mice in the bleomycin-exposed groups (Figure 7C). H&E and Masson staining showed that the bleomycin-induced distortion of lung architecture and collagen deposition was partially restored by sh-lncITPF (Figure 7D). To further investigate the antifibrotic action of sh-lncITPF in vivo, we tested the expression of mesenchymal markers α-SMA, collagen, and vimentin; epithelial cell markers E-cadherin and SP-C; and other indicators of pulmonary fibrosis, HYP and transcription repressor Snail, after sh-lncITPF treatment. sh-lncITPF decreased the expression levels of α-SMA, HYP, collagen, vimentin, and Snail and increased those of E-cadherin and SP-C in the sh-lncITPF group compared with the bleomycin group (Figures 7E–7H).
Figure 7.
Knockdown of lncITPF Suppressed the Process of Lung Fibrosis in Mice
(A) The inference of lncITPF in mice was detected via qRT-PCR. (B) Body weight monitoring showed that sh-lncITPF blocked the lost body mass compared with the control mice. (C) The FVC result showed that sh-lncITPF improved the lung function of mice compared with the control group. (D) Histological images of lung paraffin sections stained with H&E. Collagen deposition in the lung tissue was detected by Masson’s trichrome staining. Scale bars, 20 μm. (E) Representative immunohistochemical staining of α-SMA in serial lung sections. The brown color indicates positive staining. (F) HYP content was decreased by sh-lncITPF. Fibrotic markers, including mesenchymal markers and epithelial cell markers, such as collagen and vimentin (G), E-cadherin, SP-C, α-SMA, and Snail (H) in lung tissue tested via western blot. Each bar represents the mean ± SD; n = 6; **p < 0.01.
Clinical Significance of lncITPF in Patients with IPF
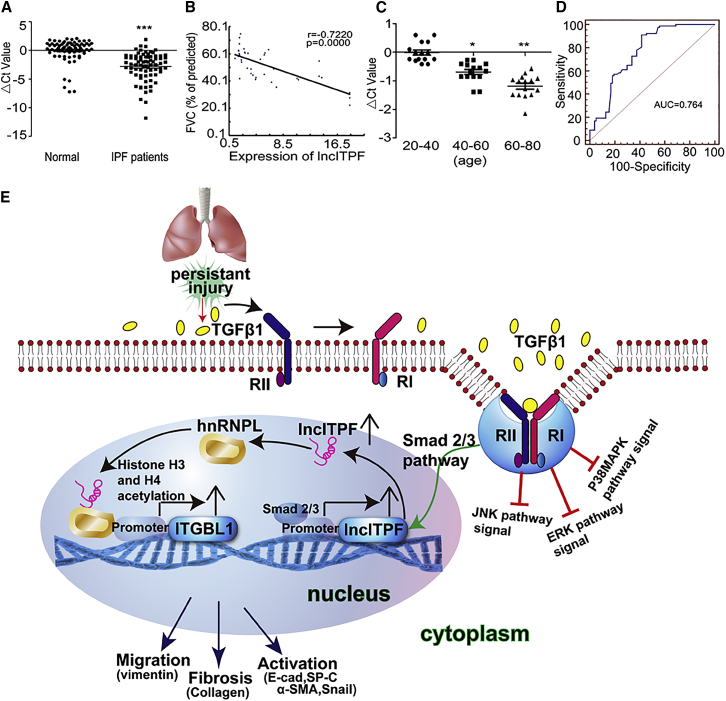
Finally, we assessed lncITPF expression levels in IPF patients. The characteristics and physiologies of patients with IPF are shown in Table 1. qRT-PCR was conducted to examine lncITPF expression levels in plasma from patients with IPF. lncITPF expression levels were significantly increased in patients with IPF compared with normal individuals (Figure 8A). Pearson correlation analysis showed that the expression levels of lncITPF were associated with FVC% predicted (Figure 8B). lncITPF expression levels increased with age (Figure 8C), probably because IPF is an age-related disease. The receiver operating characteristic (ROC) curve showed that the sensitivity and specificity values were 64.3 and 95, respectively. The area under the ROC curve was 0.804 (Figure 8D), which indicated the potential value of lncITPF as a biomarker or therapeutic target for IPF.
Table 1.
Characteristics and Physiologies of IPF Patients and the Normal
| Characteristic | Normal | IPF |
|---|---|---|
| Number | 76 | 76 |
| Age (years) | 67.3 ± 8.7 | 67.7 ± 11.6 |
| Gender (male/female) | 56/24 | 54/22 |
| FVC (% of predicted) | 93.8 ± 8.2 | 67.6 ± 9.2 |
| FEV1/FVC (% of predicted) | 87.9 ± 3.4 | 85.3 ± 2.0 |
| TLC (% of predicted) | 89.1 ± 13.6 | 63.5 ± 10.8 |
| DLCO (% of predicted) | 88.7 ± 4.9 | 54.5 ± 11.6 |
| PaO2 (mmHg) | 94.2 ± 6.9 | 77.3 ± 6.5 |
| PaCO2 (mmHg) | 39.6 ± 3.6 | 36.6 ± 2.6 |
| Smoking history (%) | 48 | 52 |
Data are depicted as means ± SD. FVC, forced vital capacity; FEV1/FVC, the ratio of forced expiratory volume in the first second to forced vital capacity; TLC, total lung capacity; DLCO, diffusing capacity for carbon monoxide; Smoking history denotes subjects with >5 pack-years of cigarette smoking.
Figure 8.
Assessment of lncITPF Value in IPF Patients and Its Regulatory Mechanism
(A) Expression levels of lncITPF increased in 76 patients with IPF, as detected by qRT-PCR. Normal samples corresponding to the sex and age of patients with IPF were selected. (B) Negative correlation between lncITPF and FVC analyzed through Pearson’s correlation. (C) lncITPF expression increased with age. This population comprised a group of healthy volunteers. (D) ROC curve of lncITPF shows that the sensitivity and specificity values are 64.3 and 95, respectively, in IPF patients. (E) Regulatory mechanism of lncITPF. Each bar represents the mean ± SD; *p < 0.05, **p < 0.01, and ***p < 0.001.
Discussion
lncRNAs play versatile roles in biological processes and human disorders, thereby leading to a novel perspective.25, 26 The lncRNA-based mechanism is one of the main focus points of future research on disease treatment. Our previous studies have extended lncRNA research into the IPF field.16, 17 However, the lncRNA mechanism remains poorly understood. In the current study, we identified a novel lncITPF, demonstrated its function, and explored its regulatory mechanism, thereby indicating the potential value of lncITPF as a biomarker or as a therapeutic target for IPF.
Several lncRNAs have been discovered through high-throughput techniques, such as high-resolution microarray, RNA sequencing, and ChIP sequencing, by combining bioinformatics and epigenetic analysis and 5′ and 3′ extremity identification analyses.27, 28 To date, more than 15,000 lncRNAs have been annotated in the human genome. These lncRNAs are grouped into six categories, namely, antisense RNAs, lincRNAs, sense overlapping transcripts, intronic transcripts, processed transcripts, and 3′-overlapping ncRNAs, on the basis of their location with respect to protein-coding genes.29, 30 Among these lncRNAs, intronic lncRNAs are the major component of the ncRNA transcriptome.31 Through examination of bioinformatics data and experiments, lncITPF was defined as a novel intronic lncRNA that is located approximately 140 bp downstream of the Itgbl1 gene in the rat genome. In the human genome, it was transcribed from Itgbl1 intron (10/10) and exon (11/11).
lncRNAs exhibit the greatest diversity function and are considered as central regulators of gene expression in various biological settings. However, few lncRNAs (1%), including Xist, KCNQ10T1, AIR, HotAir, ANRIL, HOTTIP, MALAT1, TERRA, and HULC, have been characterized functionally in humans.32 In the present study, we gained critical insights into the functions of lncITPF through the gain- and loss-of-function approach mediated by siRNA or shRNA, ASO, and Cas9. lncITPF controls pulmonary fibrosis by regulating cell activation, migration, and extracellular matrix secretion. The functions of intronic and bidirectional lncRNAs are correlated with their associated coding genes.33 Derrien et al.21 reported the importance of lncRNAs in the regulation of nearby protein-coding genes. Our study shows that lncITPF positively regulated its nearby host gene Itgbl1. The TGF-β1-signaling pathway was found to be an upstream inducer of lncITPF, which subsequently activates Itgbl1 expression. This finding provided a theoretical rationale for the positive feedback of TGF-β1-lncITPF-ITGBL1 in the pulmonary fibrosis process.
Itgbl1 is a novel human gene encoding TIED, a β integrin-related protein, which contains 10 repeats of epidermal growth factor (EGF)-like domain.34 As an extracellular matrix protein, ITGBL1 exerts its role in the extracellular space to influence cell functions. Although highly homologous to the N-terminal EGF-like stalk fragment of β integrin, the ITGBL1 protein does not have a transmembrane domain and an Arg-Gly-Asp-binding domain. This information provides novel insights into the evolution and alternative functions of the stalk structure of integrin. ITGBL1 plays a key role in the bone metastasis of breast cancer.35 Tong et al.36 found that ITGBL1 is a target of miR-576-5p and promotes non-small cell lung cancer (NSCLC) cell migration and invasion through Wnt/PCP signaling. Our study defines a new role of ITGBL1 in pulmonary fibrosis, which explains well the profibrotic effect of lncITPF. We provided mechanistic insights into the way by which lncITPF controls ITGBL1 expression. We proved that lncITPF elevates the transcriptional activity of Itgbl1 by increasing H3 and H4 histone acetylation and by maintaining the activated chromatin conformation.
hnRNP-L, a member of hnRNP complexes, is an RNA-binding protein found inside and outside the nucleolus. hnRNPs play an important function in alternative splicing, polyadenylation, exportation of mRNA from genes lacking introns, internal ribosome entry site-mediated translation, pre-mRNA processing, and mRNA stability.37 Lv et al.38 found that hnRNP-L binds the vascular endothelial growth factor A (VEGFA) 3′ UTR CARE and prevents microRNA (miRNA)-induced silencing activity. Atianand et al.39 demonstrated that lincRNA-EPS controls nucleosome positioning and represses the transcription of immune response genes (IRGs) by interacting with hnRNP-L via a CANACA motif located in its 3′ end. Our studies showed that the lncITPF-hnRNP-L interaction is necessary for Itgbl1 transcriptional regulation, revealing that hnRNP-L is a protein partner for lncITPF-regulated networks and lncITPF-dependent effects.
The mutual regulation between lncRNA and its targeted protein can be correlated with other modes, such as a competing endogenous RNA.40 For example, lncRNA pro-fibrotic lncRNA (PFL) contributes to cardiac fibrosis by acting as a competing endogenous RNA of let-7d.41 Our previous study also reported that lncRNA PCF functions as a competing endogenous RNA to promote pulmonary fibrosis progression.16 In addition, lncRNA can act as a protein-coding gene and a non-coding RNA. Such dual-function regulation of gene expression networks reflects the diversity and complexity of the lncRNA mechanism during disease progression.42 Whether there are other unknown functional modes for lncITPF remains to be elucidated. Experiments will be designed to determine this in future research.
lncRNAs are critical in disease development. Thus, they have emerged as a major source of biomarkers and potential therapeutic targets. For example, SChLAP1 was identified and validated as a potential biomarker for treating the intensification of aggressive prostate cancer.43 lncRNA HNF1A-antisense 1 (HNF1A-AS1) was found to be significantly correlated with TNM stage, tumor size, and lymph node metastasis in lung adenocarcinoma.44 The mitochondrial lncRNA uc022bqs.1 (LIPCAR) is reportedly a novel cardiac remodeling biomarker that predicts future death in heart failure patients.45 The XIST unmethylated fragment in male plasma might be a diagnostic biomarker for testicular germ cell tumors.46 However, diagnostic and prognostic biomarkers of lncRNAs for IPF, which will be useful in the development of novel antifibrotic therapies, remain unavailable. This study demonstrated that lncITPF is positively correlated with the degree of fibrosis and can be detected in the blood of patients with IPF. Interestingly, lncITPF expression increased with age, which could be due to the fact that age is the strongest demographic risk factor for IPF. Hao et al.15 also reported that lncRNA AP003419.16 regulates its adjacent gene RPS6KB2, which is involved in the process of aging and IPF. These findings are beneficial in determining whether the abnormal lncITPF can be taken as a biomarker or a therapeutic target for IPF.
In conclusion, this study revealed the regulatory mechanism of lncITPF in IPF (Figure 8E). Our findings may provide a diagnostic biomarker or therapeutic target to ameliorate pathologies and improve the health of patients with IPF. Of course, future studies are required to expand the analysis of lncITPF to a larger cohort of patients with IPF and to determine with statistical confidence whether increased lncITPF expression correlates with worse patient outcome.
Materials and Methods
Blood Samples
Blood samples from IPF patients, diagnosed in accordance with the American Thoracic Society (ATS) and European Respiratory Society (ERS) criteria for IPF, and age-matched healthy controls were collected at the Affiliated Hospital of Binzhou Medical University (Binzhou and Yantai, China). All patients provided written informed consent, and ethical consent was granted from the Committee of Binzhou Medical University.47
Animal Model
Experiments on the animals studied were approved by the Committee on the Ethics of Animal Experiments of Binzhou Medical University. All animals were bred and maintained in a 12-hr light/dark cycle and allowed free access to food and water. Anesthetized male Sprague-Dawley (SD) rats or C57BL/6 mice aged 8 months old received saline or 5 mg/kg bleomycin (Nippon Kayaku, Tokyo, Japan) through aerosolized intratracheal delivery, as previously described.16, 17 Each group was composed of eight to ten mice. On days 7, 14, 21, and 28, rats were killed, and lung tissue sections were collected and immediately frozen in liquid nitrogen for further studies. Physiological measurements were performed on day 28 using the Pulmonary Function Test (DSI, St. Paul, MN, USA). Histopathological and biochemical analyses of tissues or cells derived from animal models were performed by investigators masked to the genotypes or treatments of the animals.
Cell Model
MRC-5 cell line was obtained from American Type Culture Collection and cultured at 37°C in an atmosphere containing 5% CO2 and in advanced minimum essential medium (MEM), supplemented with 10% fetal bovine serum (Gibco, Grand Island, NY, USA). The cells were treated with 5 ng/mL TGF-β1 (Invitrogen, Carlsbad, CA, USA).
RNAi and Overexpression
Specific siRNAs or smart silencer targeting lncITPF, ITGBL1, hnRNP-L, and their negative control was synthesized by Ribo Bio Technology (Guangzhou, China). shRNAs or control lentivectors were intratracheally delivered into C57BL/6 mice on the third day after bleomycin injury. The overexpression vectors for lncITPF and ITGBL1 were constructed by subcloning their full-length sequence into the pcDNA3.1 (+) vector (Obio Technology, Shanghai, China). BamHI and Hind III were jointly connected to expression vector pcDNA 3.1+ through a double-enzyme connection. Overexpression, siRNA, and smart silencer sequences are listed in Table S1. Cells were transfected with 50 nM siRNA, smart silencer, or plasmid using attractene transfection reagent (QIAGEN, Germany), in accordance with the manufacturer’s instructions with or without TGF-β1treatment.
RACE Experiment
5′-RACE and 3′-RACE analyses were performed with 5 μg total RNA using the GeneRacer kit (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions.
In Vitro Translation Reaction
T7-lncITPF DNA sample was prepared by PCR using the following: forward 5′- ACCGCCTAATACGACTCACTATAGGGACCCCACCATGGACTCAGTGATAGGAACAAAATGT-3′, reverse 5′-TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTACTAACAT AAAACAGTGTGCTAATCA-3′. The PCR product was purified by a gel extraction kit.
We rapidly thawed the TnT Quick Master Mix by hand-warming and placing on ice. The other components were thawed at room temperature and then stored on ice. We assembled the reaction components in a 0.5- or 1.5-mL microcentrifuge tube. After adding all of the components, we gently mixed by pipetting and centrifuging briefly to return the reaction to the bottom of the tube. The T7 luciferase control DNA and H2O were used as positive and negative controls, respectively. We incubated the reaction at 30°C for 60–90 min. After the SDS-PAGE, the gel was stained using InstantBlue Ultrafast Protein Staining Solution (Expedeon, UK) to analyze the results of translation.
Luciferase Analysis
Human genomic DNA was extracted by using the method of rapid isolation of mammalian DNA. The primer pair PF (5′-TTTCTCTATCGATAGGTACCACTGCTATCCTAATTTATT-3′) and PR (5′-GATCGCAGATCTCGAGACATTTTGTTCCTATCACT-3′), respectively containing ends that are homologous to the pGL3-basic vector linearized with Kpn1 and Xho1, were used to amplify the 5′ promoter sequence of lncITPF gene from the extracted human genomic DNA. The PCR was conducted at 94°C for 5 min followed by 35 cycles at 94°C for 45 s, 60°C for 30 s, and 72°C for 2 min. PCR-amplified fragment was about 1.9 kb that was subcloned into the SmaI/HindIII site of pGL3-basic vector using the In-Fusion HD cloning kit from Clontech. All PCR products were verified by DNA sequencing. Cells were transfected with the pGL3-lncITPF or pGL3-basic vector plus the Renilla luciferase plasmid (pRL-TK). Then, the cells were harvested after 48 and 72 hr under TGF-β1 condition for firefly-Renilla luciferase assays using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA), according to the manufacturer’s instructions. Luciferase activities were normalized to the cotransfected pRL-TK plasmid (mean ± SD).
Biotin Pull-Down and Mass Spectrometry
lncITPF transcripts were transcribed using T7 and SP6 RNA polymerase (Ambion) in vitro, then by using the RNeasy Plus Mini Kit (QIAGEN, Germany) and treated with TURBO DNase I. Purified RNAs were biotin-labeled using Biotin RNA Labeling Mix (Ambion Life). The biotinylated lncITPF and its antisense RNAs were mixed and incubated with cell lysates. Then, avidin magnetic beads were added to each binding reaction, and the mixtures were incubated at room temperature. Finally, the beads were washed, and the retrieved proteins were resolved in gradient gel electrophoresis followed by regular liquid chromatography-mass spectrometry (LC-MS) (Dionex’s UltiMate 3000, USA; Thermo Scientific Q Exactive Orbitrap, USA) analysis or detected by western blot analysis. Beijing Protein Innovation provided guidance and assistance for the specific procedures of LC-MS.
RNA-Binding Protein Immunoprecipitation
The RNA immunoprecipitation (RIP) was performed using an EZMagna RIP kit (Millipore) following the manufacturer’s protocol. Briefly cells were lysed in complete RIP lysis buffer, and the extract was incubated with magnetic beads conjugated with hnRNP-L antibodies or control immunoglobulin G (IgG) (Merck Millipore, Billerica, MA, USA) for 6 hr at 4°C. Next, the beads were washed and incubated with Proteinase K to remove the proteins. Finally, purified RNA was subjected to qRT-PCR analysis using specific primers for lncITPF.
Nuclear-Cytosol Fractionation
Cells (1 × 107) were collected by centrifugation at 120 × g for 4 min at 4°C and then added with 1 mL CEB-A Mix (BioVision, Milpitas, CA, USA) containing RSB, DTT, and protease inhibitors. The cells were Dounce treated gently with 10–12 strokes in a chilled B-type (tight) Dounce homogenizer and then mixed with ice-cold cytosol extraction buffer B (BioVision). Then, the tube was reincubated on ice for 1 min. After reincubation, the tube was centrifuged for 5 min at maximum speed in a microcentrifuge. The supernatant (cytoplasmic extract) fraction was immediately transferred to a clean pre-chilled tube. The nuclear pellet was gently resuspended into 200 μL ice-cold nuclei lysis buffer followed by a pulsed vortexing and incubation on ice for 1 min. Thereafter, samples were spun at 14,000 rpm in microfuge tubes for 10 min at 4°C. The supernatant from this spin represented the nucleoplasmic fraction, and it was immediately transferred to a clean pre-chilled tube, placed on ice, and then stored at −80°C for future use.
RNA FISH
lncITPF FISH probe was synthesized by Ribo Bio Technology (Guangzhou, China). FISH was performed with the FISH kit according to the manufacturer’s protocol (Ribo Bio Technology). Cells were fixed with 4% paraformaldehyde for 10 min at room temperature, and then permeabilized in PBS with 0.5% Triton X-100 on ice for 5 min, followed by pretreatment with pre-hybridization buffer at 37°C for 30 min. Subsequently, cells were hybridized with 20 μM using Cy3-labeled RNA of lncITPF or U6/18S FISH probe mix in a moist chamber at 37°C overnight. Cells were rinsed thrice in 4× saline sodium citrate (SSC) with 0.1% Tween-20 for 5 min at 42°C, followed by washing once for 5 min at 42°C in 2× SSC and then washed once for 5 min at 42°C in 1 ×SSC. After hybridization, cells were stained with DAPI for 10 min at room temperature. Finally, the images were observed with confocal microscope and analyzed with fluorescence microscope (Leica, Solms, Germany).
Real-Time qPCR
Total RNAs from whole-blood samples were extracted using TRIzol LS Reagent (Invitrogen, Carlsbad, CA, USA), whereas those from tissues and cells were isolated using RNAiso Plus (Takara, Dalian, China). RNA concentration and quality were measured using the NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA). First-strand cDNA was generated using the PrimeScript RT reagent kit (Takara). qRT-PCR was performed in accordance with a standard protocol by using the Rotor-Gene 3000 Real-time PCR system (Sydney, Australia). As endogenous controls in each sample, GAPDH was used for routine method assays. By convention, changes in expression were determined using the 2−ΔΔCT method for cell and animal samples and ΔCt for blood samples from IPF patients by Rotor-Gene 6 software.
Western Blot
Protein samples (30 μg) were separated by 10% SDS-PAGE and then transferred onto polyvinylidene difluoride membranes. The membranes were blocked with 5% non-fat milk in Tris-buffered saline and Tween (TBST) buffer for 2 hr at room temperature. The membranes were washed three times with TBST buffer and then incubated at 4°C overnight with the specific primary antibodies against α-SMA, Snail, E-cad, Vim, and ITGBL1 (Abcam, Hong Kong, China). Horseradish peroxidase (HRP)-labeled goat anti-rabbit IgG (1:5,000; Beijing Zhong Shan-Golden Bridge Technology, Beijing, China) was used as the secondary antibody, and the signals were detected with Western Chemiluminescent HRP Substrate (Millipore, Billerica, MA, USA). Membranes were subsequently stripped and re-probed with GAPDH antibody (1:500), which served as the loading control.
Immunofluorescence Analysis
For immunocytochemistry, cells transfected with lncITPF RNAi, lncITPF overexpression, and TGF-β1 were cultured and fixed on a 24-well plate. After incubation with rabbit anti-α-SMA antibody (Abcam, 1:200) and then with fluorescein isothiocyanate (FITC) fluorescein-labeled goat anti-rabbit IgG (Abbkine, CA, USA; 1:500), the slides were mounted by adding DAPI-Fluoromount-G (Roche Molecular Biochemicals, Basel, Switzerland). All images were collected under a fluorescence microscope (Leica, Solms, Germany).
Real-Time Cellular Migration Analysis
Myofibroblast migration was performed in 16-well CIM plates in an RTCA DPlus instrument for real-time cellular migration assay system (ACEA Biosciences). The cells (4 × 104) were seeded in the upper chamber in a basal medium containing 0.1% fetal bovine serum (FBS). The lower chamber contained complete medium with growth factors and additives. Cell migration was monitored every 5 min for a period of 24 hr. The cell index representing the amount of migrated cells was calculated with the RTCA Software from ACEA Biosciences (San Diego, CA).
ChIP
ChIP was performed using the EpiQuik Acetyl-Histone H3 ChIP Kit and EpiQuik Acetyl-Histone H4 ChIP Kit (Epigentek, NY, USA) in accordance with its manual. The chromatin was immunoprecipitated using Anti-Acetyl-Histone H3 antibodies (Epigentek) and Anti-Acetyl-Histone H4 antibodies (Epigentek). Normal mouse IgG was used as a negative control. qPCR was conducted using SYBR Green Mix (Takara Bio, Otsu, Japan). Data were normalized to an unrelated genomic region and are representative of three independent experiments.
RNA Sequencing
A total amount of 2 μg RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using NEBNext Ultra RNA Library Prep Kit for Illumina (E7530L, New England Biolabs, USA), following the manufacturer’s recommendations, and index codes were added to attribute sequences to each sample. Briefly, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. Fragmentation was carried out using divalent cations under elevated temperature in NEBNext First Strand Synthesis Reaction Buffer (5×). First-strand cDNA was synthesized using random hexamer primer and RNase H. Second-strand cDNA synthesis was subsequently performed using buffer, deoxy-ribonucleoside triphosphates (dNTPs), DNA polymerase I, and RNase H. The library fragments were purified with QiaQuick PCR kits and elution with elution buffer (EB), then terminal repair. A-tailing and adaptor addition were implemented. The aimed products were retrieved by agarose gel electrophoresis and PCR was performed, then the library was completed. RNA concentration of library was measured using Qubit RNA Assay Kit in Qubit 3.0 to preliminarily quantify and then dilute to 1 ng/μL. Insert size was assessed using the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA), and qualified insert size was accurately quantified using StepOnePlus Real-Time PCR System (Library valid concentration > 10 nM). The clustering of the index-coded samples was performed on a cBot cluster generation system using HiSeq PE Cluster Kit v4-cBot-HS (Illumina), according to the manufacturer’s instructions. After cluster generation, the libraries were sequenced on an Illumina Hiseq 4000 platform, and 150-bp paired-end reads were generated.
Statistical Evaluation
Statistical analyses were performed using SPSS version 19.0 software (IBM SPSS Statistics Company, Chicago, IL, USA). Data are presented as the mean ± SD of at least three independent experiments. Unpaired Student’s t test was used for experiments comparing two groups, whereas one-way ANOVA with Student-Newman-Keuls post hoc test was applied for experiments comparing three or more groups. The association between lncRNA-ITPF expression and clinicopathological features was analyzed by the chi-square test. Statistical significance was considered at p < 0.05.
Author Contributions
C.L., T.S.B., and J.Z. designed the experiments and wrote the initial draft. X.S., P.X., and C.M. performed the experiments in vivo and in vitro. C.S., R.L., and H.L. performed the experiments in patients with IPF.
Conflicts of Interest
The authors declare no conflicts of interest.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (31670365, 81741170, 81670064, and 31470415) and the Natural Science Foundation of Shandong Province (ZR2016HP34).
Footnotes
Supplemental Information includes three figures and one table and can be found with this article online at https://doi.org/10.1016/j.ymthe.2018.08.026.
Contributor Information
Jinjin Zhang, Email: jjinzhang@126.com.
Changjun Lv, Email: lucky_lcj@sina.com.
Supplemental Information
References
- 1.King T.E., Jr., Albera C., Bradford W.Z., Costabel U., du Bois R.M., Leff J.A., Nathan S.D., Sahn S.A., Valeyre D., Noble P.W. All-cause mortality rate in patients with idiopathic pulmonary fibrosis. Implications for the design and execution of clinical trials. Am. J. Respir. Crit. Care Med. 2014;189:825–831. doi: 10.1164/rccm.201311-1951OC. [DOI] [PubMed] [Google Scholar]
- 2.Dakhlallah D., Batte K., Wang Y., Cantemir-Stone C.Z., Yan P., Nuovo G., Mikhail A., Hitchcock C.L., Wright V.P., Nana-Sinkam S.P. Epigenetic regulation of miR-17∼92 contributes to the pathogenesis of pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2013;187:397–405. doi: 10.1164/rccm.201205-0888OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.King T.E., Jr., Pardo A., Selman M. Idiopathic pulmonary fibrosis. Lancet. 2011;378:1949–1961. doi: 10.1016/S0140-6736(11)60052-4. [DOI] [PubMed] [Google Scholar]
- 4.Staab-Weijnitz C.A., Fernandez I.E., Knüppel L., Maul J., Heinzelmann K., Juan-Guardela B.M., Hennen E., Preissler G., Winter H., Neurohr C. FK506-Binding Protein 10, a potential novel drug target for idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2015;192:455–467. doi: 10.1164/rccm.201412-2233OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y.M., Nepali K., Liou J.P. Idiopathic Pulmonary Fibrosis: Current Status, Recent Progress, and Emerging Targets. J. Med. Chem. 2017;60:527–553. doi: 10.1021/acs.jmedchem.6b00935. [DOI] [PubMed] [Google Scholar]
- 6.Ota C., Yamada M., Fujino N., Motohashi H., Tando Y., Takei Y., Suzuki T., Takahashi T., Kamata S., Makiguchi T. Histone deacetylase inhibitor restores surfactant protein-C expression in alveolar-epithelial type II cells and attenuates bleomycin-induced pulmonary fibrosis in vivo. Exp. Lung Res. 2015;41:422–434. doi: 10.3109/01902148.2015.1060275. [DOI] [PubMed] [Google Scholar]
- 7.Yang I.V., Pedersen B.S., Rabinovich E., Hennessy C.E., Davidson E.J., Murphy E., Guardela B.J., Tedrow J.R., Zhang Y., Singh M.K. Relationship of DNA methylation and gene expression in idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2014;190:1263–1272. doi: 10.1164/rccm.201408-1452OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heward J.A., Lindsay M.A. Long non-coding RNAs in the regulation of the immune response. Trends Immunol. 2014;35:408–419. doi: 10.1016/j.it.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prensner J.R., Iyer M.K., Sahu A., Asangani I.A., Cao Q., Patel L., Vergara I.A., Davicioni E., Erho N., Ghadessi M. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat. Genet. 2013;45:1392–1398. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thum T. Noncoding RNAs and myocardial fibrosis. Nat. Rev. Cardiol. 2014;11:655–663. doi: 10.1038/nrcardio.2014.125. [DOI] [PubMed] [Google Scholar]
- 11.Yang X., Song J.H., Cheng Y., Wu W., Bhagat T., Yu Y., Abraham J.M., Ibrahim S., Ravich W., Roland B.C. Long non-coding RNA HNF1A-AS1 regulates proliferation and migration in oesophageal adenocarcinoma cells. Gut. 2014;63:881–890. doi: 10.1136/gutjnl-2013-305266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Atanasovska B., Rensen S.S., van der Sijde M.R., Marsman G., Kumar V., Jonkers I., Withoff S., Shiri-Sverdlov R., Greve J.W.M., Faber K.N. A liver-specific long noncoding RNA with a role in cell viability is elevated in human nonalcoholic steatohepatitis. Hepatology. 2017;66:794–808. doi: 10.1002/hep.29034. [DOI] [PubMed] [Google Scholar]
- 13.Yin Y., Yan P., Lu J., Song G., Zhu Y., Li Z., Zhao Y., Shen B., Huang X., Zhu H. Opposing Roles for the lncRNA Haunt and Its Genomic Locus in Regulating HOXA Gene Activation during Embryonic Stem Cell Differentiation. Cell Stem Cell. 2015;16:504–516. doi: 10.1016/j.stem.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 14.Liu B., Sun L., Liu Q., Gong C., Yao Y., Lv X., Lin L., Yao H., Su F., Li D. A cytoplasmic NF-κB interacting long noncoding RNA blocks IκB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27:370–381. doi: 10.1016/j.ccell.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 15.Hao X., Du Y., Qian L., Li D., Liu X. Upregulation of long noncoding RNA AP003419.16 predicts high risk of aging-associated idiopathic pulmonary fibrosis. Mol. Med. Rep. 2017;16:8085–8091. doi: 10.3892/mmr.2017.7607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu H., Wang B., Zhang J., Zhang S., Wang Y., Zhang J., Lv C., Song X. A novel lnc-PCF promotes the proliferation of TGF-β1-activated epithelial cells by targeting miR-344a-5p to regulate map3k11 in pulmonary fibrosis. Cell Death Dis. 2017;8:e3137. doi: 10.1038/cddis.2017.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song X., Cao G., Jing L., Lin S., Wang X., Zhang J., Wang M., Liu W., Lv C. Analysing the relationship between lncRNA and protein-coding gene and the role of lncRNA as ceRNA in pulmonary fibrosis. J. Cell. Mol. Med. 2014;18:991–1003. doi: 10.1111/jcmm.12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kong L., Zhang Y., Ye Z.Q., Liu X.Q., Zhao S.Q., Wei L., Gao G. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35:W345–W349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L., Park H.J., Dasari S., Wang S., Kocher J.P., Li W. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013;41:e74. doi: 10.1093/nar/gkt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun L., Luo H., Bu D., Zhao G., Yu K., Zhang C., Liu Y., Chen R., Zhao Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013;41:e166. doi: 10.1093/nar/gkt646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iyer M.K., Niknafs Y.S., Malik R., Singhal U., Sahu A., Hosono Y., Barrette T.R., Prensner J.R., Evans J.R., Zhao S. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fang M., Wang J., Li S., Guo Y. Advanced glycation end-products accelerate the cardiac aging process through the receptor for advanced glycation end-products/transforming growth factor-β-Smad signaling pathway in cardiac fibroblasts. Geriatr. Gerontol. Int. 2016;16:522–527. doi: 10.1111/ggi.12499. [DOI] [PubMed] [Google Scholar]
- 24.Semren N., Welk V., Korfei M., Keller I.E., Fernandez I.E., Adler H., Günther A., Eickelberg O., Meiners S. Regulation of 26S Proteasome Activity in Pulmonary Fibrosis. Am. J. Respir. Crit. Care Med. 2015;192:1089–1101. doi: 10.1164/rccm.201412-2270OC. [DOI] [PubMed] [Google Scholar]
- 25.Liao Q., Liu C., Yuan X., Kang S., Miao R., Xiao H., Zhao G., Luo H., Bu D., Zhao H. Large-scale prediction of long non-coding RNA functions in a coding-non-coding gene co-expression network. Nucleic Acids Res. 2011;39:3864–3878. doi: 10.1093/nar/gkq1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang K.C., Chang H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meseure D., Drak Alsibai K., Nicolas A., Bieche I., Morillon A. Long Noncoding RNAs as New Architects in Cancer Epigenetics, Prognostic Biomarkers, and Potential Therapeutic Targets. Biomed Res. Int. 2015;2015:320214. doi: 10.1155/2015/320214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park C., Yu N., Choi I., Kim W., Lee S. lncRNAtor: a comprehensive resource for functional investigation of long non-coding RNAs. Bioinformatics. 2014;30:2480–2485. doi: 10.1093/bioinformatics/btu325. [DOI] [PubMed] [Google Scholar]
- 29.Su X., Malouf G.G., Chen Y., Zhang J., Yao H., Valero V., Weinstein J.N., Spano J.P., Meric-Bernstam F., Khayat D., Esteva F.J. Comprehensive analysis of long non-coding RNAs in human breast cancer clinical subtypes. Oncotarget. 2014;5:9864–9876. doi: 10.18632/oncotarget.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garzon R., Volinia S., Papaioannou D., Nicolet D., Kohlschmidt J., Yan P.S., Mrózek K., Bucci D., Carroll A.J., Baer M.R. Expression and prognostic impact of lncRNAs in acute myeloid leukemia. Proc. Natl. Acad. Sci. USA. 2014;111:18679–18684. doi: 10.1073/pnas.1422050112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Louro R., Smirnova A.S., Verjovski-Almeida S. Long intronic noncoding RNA transcription: expression noise or expression choice? Genomics. 2009;93:291–298. doi: 10.1016/j.ygeno.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 32.Hung C.L., Wang L.Y., Yu Y.L., Chen H.W., Srivastava S., Petrovics G., Kung H.J. A long noncoding RNA connects c-Myc to tumor metabolism. Proc. Natl. Acad. Sci. USA. 2014;111:18697–18702. doi: 10.1073/pnas.1415669112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Z., Chao T.C., Chang K.Y., Lin N., Patil V.S., Shimizu C., Head S.R., Burns J.C., Rana T.M. The long noncoding RNA THRIL regulates TNFα expression through its interaction with hnRNPL. Proc. Natl. Acad. Sci. USA. 2014;111:1002–1007. doi: 10.1073/pnas.1313768111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berg R.W., Leung E., Gough S., Morris C., Yao W.P., Wang S.X., Ni J., Krissansen G.W. Cloning and characterization of a novel beta integrin-related cDNA coding for the protein TIED (“ten beta integrin EGF-like repeat domains”) that maps to chromosome band 13q33: A divergent stand-alone integrin stalk structure. Genomics. 1999;56:169–178. doi: 10.1006/geno.1998.5707. [DOI] [PubMed] [Google Scholar]
- 35.Li X.Q., Du X., Li D.M., Kong P.Z., Sun Y., Liu P.F., Wang Q.S., Feng Y.M. ITGBL1 Is a Runx2 Transcriptional Target and Promotes Breast Cancer Bone Metastasis by Activating the TGFβ Signaling Pathway. Cancer Res. 2015;75:3302–3313. doi: 10.1158/0008-5472.CAN-15-0240. [DOI] [PubMed] [Google Scholar]
- 36.Gan X., Liu Z., Tong B., Zhou J. Epigenetic downregulated ITGBL1 promotes non-small cell lung cancer cell invasion through Wnt/PCP signaling. Tumour Biol. 2016;37:1663–1669. doi: 10.1007/s13277-015-3919-8. [DOI] [PubMed] [Google Scholar]
- 37.Bussone G., Dib H., Tamby M.C., Broussard C., Federici C., Woimant G., Camoin L., Guillevin L., Mouthon L. Identification of new autoantibody specificities directed at proteins involved in the transforming growth factor β pathway in patients with systemic sclerosis. Arthritis Res. Ther. 2011;13:R74. doi: 10.1186/ar3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lv D., Wu H., Xing R., Shu F., Lei B., Lei C., Zhou X., Wan B., Yang Y., Zhong L. HnRNP-L mediates bladder cancer progression by inhibiting apoptotic signaling and enhancing MAPK signaling pathways. Oncotarget. 2017;8:13586–13599. doi: 10.18632/oncotarget.14600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Atianand M.K., Hu W.Q., Satpathy A.T., Shen Y., Ricci E.P., Alvarez-Dominguez J.R., Bhatta A., Schattgen S.A., McGowan J.D., Blin J. A Long Noncoding RNA lincRNA-EPS Acts as a Transcriptional Brake to Restrain Inflammation. Cell. 2016;165:1672–1685. doi: 10.1016/j.cell.2016.05.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cesana M., Cacchiarelli D., Legnini I., Santini T., Sthandier O., Chinappi M., Tramontano A., Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang H., Pan Z., Zhao X., Liu L., Sun J., Su X., Xu C., Zhou Y., Zhao D., Xu B. LncRNA PFL contributes to cardiac fibrosis by acting as a competing endogenous RNA of let-7d. Theranostics. 2018;8:1180–1194. doi: 10.7150/thno.20846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruiz-Orera J., Messeguer X., Subirana J.A., Alba M.M. Long non-coding RNAs as a source of new peptides. eLife. 2014;3:e03523. doi: 10.7554/eLife.03523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prensner J.R., Zhao S., Erho N., Schipper M., Iyer M.K., Dhanasekaran S.M., Magi-Galluzzi C., Mehra R., Sahu A., Siddiqui J. RNA biomarkers associated with metastatic progression in prostate cancer: a multi-institutional high-throughput analysis of SChLAP1. Lancet Oncol. 2014;15:1469–1480. doi: 10.1016/S1470-2045(14)71113-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu Y., Liu H., Shi X., Yao Y., Yang W., Song Y. The long non-coding RNA HNF1A-AS1 regulates proliferation and metastasis in lung adenocarcinoma. Oncotarget. 2015;6:9160–9172. doi: 10.18632/oncotarget.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumarswamy R., Bauters C., Volkmann I., Maury F., Fetisch J., Holzmann A., Lemesle G., de Groote P., Pinet F., Thum T. Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ. Res. 2014;114:1569–1575. doi: 10.1161/CIRCRESAHA.114.303915. [DOI] [PubMed] [Google Scholar]
- 46.Kawakami T., Okamoto K., Ogawa O., Okada Y. XIST unmethylated DNA fragments in male-derived plasma as a tumour marker for testicular cancer. Lancet. 2004;363:40–42. doi: 10.1016/S0140-6736(03)15170-7. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Y.L. Guangxi Normal University Press; Guilin, China: 2014. The Value of Disease. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.